Abstract
PHASTER (PHAge Search Tool – Enhanced Release) is a significant upgrade to the popular PHAST web server for the rapid identification and annotation of prophage sequences within bacterial genomes and plasmids. Although the steps in the phage identification pipeline in PHASTER remain largely the same as in the original PHAST, numerous software improvements and significant hardware enhancements have now made PHASTER faster, more efficient, more visually appealing and much more user friendly. In particular, PHASTER is now 4.3× faster than PHAST when analyzing a typical bacterial genome. More specifically, software optimizations have made the backend of PHASTER 2.7X faster than PHAST, while the addition of 80 CPUs to the PHASTER compute cluster are responsible for the remaining speed-up. PHASTER can now process a typical bacterial genome in 3 min from the raw sequence alone, or in 1.5 min when given a pre-annotated GenBank file. A number of other optimizations have also been implemented, including automated algorithms to reduce the size and redundancy of PHASTER's databases, improvements in handling multiple (metagenomic) queries and higher user traffic, along with the ability to perform automated look-ups against 14 000 previously PHAST/PHASTER annotated bacterial genomes (which can lead to complete phage annotations in seconds as opposed to minutes). PHASTER's web interface has also been entirely rewritten. A new graphical genome browser has been added, gene/genome visualization tools have been improved, and the graphical interface is now more modern, robust and user-friendly. PHASTER is available online at www.phaster.ca.
INTRODUCTION
Bacterial genomes and plasmids can contain a large fraction (>20% in some cases) of functional and non-functional bacteriophage genes (1). Indeed, these non-bacterial, prophage sequences likely account for a significant proportion of the variation within bacterial species or clades. The fact that phages and prophages allow bacteria to acquire antibiotic resistance, to adapt to new environmental niches or to become pathogenic has led to renewed interest in identifying and annotating prophage sequences in bacterial genomes. As a result, prophage finding programs and web servers have become integral to many bacterial genome annotation pipelines. One of the first web servers to be developed for phage identification and annotation was PHAST (PHAge Search Tool). First published in 2011 (2), PHAST has become one of the most popular tools for prophage identification in bacterial genomes. To date, the PHAST paper has received more than 400 citations and the PHAST website receives up to 7300 job submissions per month with up to 30% of these coming through its web API (Application Programming Interface). The popularity of the server appears to be due to a number of factors, such as its speed, accuracy, visually appealing output and the fact that it appeared just as NextGen sequencing of bacterial genomes started to ‘take off” (3).
The PHAST prophage analysis pipeline is relatively straightforward. In particular, PHAST accepts both GenBank and FASTA formatted genomic sequence data, and performs BLAST (4) searches against a custom prophage/phage database that combines protein sequences from the National Center for Biotechnology Information (NCBI) phage database and the prophage database developed by Srividhya et al. (5). Phage-like genes are then clustered into prophage regions using DBSCAN (6). Non-phage genes in these identified regions are annotated by a second BLAST search against a non-redundant bacterial protein database, and the prophage regions are assigned a completeness score based on the proportion of phage genes in the identified region. All of the annotated prophage data is then displayed on the web through a variety of hyperlinked tables and colored chromosomal maps.
PHAST was introduced at a time of increasing interest in prophage identification, which had already seen the development of several stand-alone phage search programs, namely Prophage Finder (7), Phage_Finder (8) and Prophinder (9). Since PHAST's original release, phage finding has continued to develop, and several other high quality phage identification tools have since been published. Some of the newer phage and prophage identification tools include PhiSpy (10) and VirSorter (11). PhiSpy, in addition to performing sequence similarity-based searches, analyzes several other sequence-based statistics to help identify novel phages that are not represented in existing phage databases. VirSorter is geared towards more specialized applications such as finding phages in draft genomes and Single-cell Amplified Genomes (12,13). VirSorter also handles fragmented metagenomic data as well as whole genomic data.
In addition to the appearance of new algorithms for phage searching and new kinds of user demands by phage scientists (i.e. handling metagenomic data), other important developments in the field have taken place. These include the rapid growth in the number of sequenced bacterial and phage genomes. Since 2011, the size of the NCBI phage/prophage database has grown by a factor of four while the number of sequenced bacteria has grown by nearly five fold. This has led to a massive increase in both database sizes and search times. Likewise, the ease with which bacterial genomes are now sequenced has led to a 12× growth in the number of queries being handled by PHAST (and other phage searching programs) over the past five years. In the last year alone, visits to PHAST have increased by ∼60% and the server now processes one job every ∼6 min.
These rapid developments in both bacterial genomics and the phage/prophage field led us to revisit PHAST and to actively explore methods to enhance and upgrade the PHAST server. These efforts led to the creation of PHASTER (PHAge Search Tool – Enhanced Release), a faster, better, more modern version of PHAST. To improve PHAST's processing speed, we made a number of algorithmic (software) and hardware improvements. We also expanded and enhanced the way PHAST's phage and bacterial databases are prepared, read and handled. Improvements were also made to the user interface to make the website more visually appealing and far more convenient to use. Further, by rewriting much of the PHAST codebase, we were able to make the website easier to maintain into the future. These and other improvements are described in more detail below.
WHAT'S NEW
Software upgrades and optimizations
Phage and prophage searching is a computationally intensive task with most of the time and computational resources being consumed with large scale sequence comparisons and alignments. The original version of PHAST used an older, legacy version of BLAST that was both slower and less able to handle large numbers of query sequences. By upgrading to the latest version of BLAST (known as BLAST+, version 2.3.0+ (14)) we were able to overcome a number of previous limitations and improve PHAST's overall performance. In particular, BLAST+ is specially designed to handle longer and greater numbers of sequences more efficiently and to do its alignments more quickly than earlier versions of BLAST. Although much faster alternatives to BLAST exist, they are generally less sensitive. Tests with one such alternative, RAPSearch2 (15), led to a ∼7% loss in sensitivity (data not shown). Upgrading to BLAST+ maintained prediction accuracy while achieving a 15–25% speedup to the server's BLAST search phases.
BLAST search speeds were further improved through various computing cluster optimizations. The old PHAST employed a grid scheduler but with only minimal optimizations, resulting in frequently idle CPU cores. PHASTER now prepares input to its grid scheduler such that all CPU cores are used much more efficiently, particularly in cases when the cluster is handling a single user submission and must complete it as quickly as possible. PHASTER now divides its bacterial database into several parts, allowing computing nodes to specialize in particular sub-databases. In PHASTER, each query sequence is now searched against each sub-database component, and the results are subsequently combined, with appropriate adjustments made to the BLAST expectation values (e-values) for the true total size of the database searched. Each database component can be handled by multiple cluster nodes, providing flexibility to the cluster's job scheduler, and ensuring redundancy in case of hardware failure. In addition, the set of query sequences is partitioned evenly into a small number of sub-groups, such that the sum of the sequence lengths in each sub-group is approximately the same, and each sub-set is run separately. With these optimizations, smaller individual BLAST+ jobs can be more readily distributed to available CPU cores as they become available. The optimal number of CPU cores per individual BLAST job and the number of groups into which to partition the query sequences were determined through careful parameter optimization. It is also important to note that the improved runtimes and enhanced parallelism did not compromise the accuracy of the prophage identifications (see below).
PHASTER now provides the option to search for phage regions in contigs assembled from metagenomic data. In this mode, complete and partial genes are predicted using FragGeneScan (16), and the predicted prophages are arranged by contig in the presented results. We assessed contigs of various lengths derived from a test genome (Escherichia coli O157:H7, GenBank accession NC_002655) to determine both the minimum and the optimal contig lengths for detecting prophages in metagenomic data. We found that a small number of phage regions (∼15%) could be identified in contigs as short as 2000 bp, while ∼90% of prophages could be identified in contigs >20 000 bp long. Based on these data we set a threshold minimum contig length of 2000 bp (although we obviously recommend longer contig lengths, if available). The full set of contig performance data are available on the PHASTER website.
Database preparation
As noted earlier, the rapidly growing size of both bacterial and phage/prophage sequence databases resulted in substantial increases in the runtimes for PHAST's BLAST searches. For instance, the number of bacterial prophage sequences has increased from around 45 000 as of January 1, 2011 to over 187 000 today. In addition, the bacterial sequence database, which is used in PHAST's annotation step, has grown by a factor of nearly 5X since PHAST was first released. The latest version of the bacterial sequence database consists of >16 million protein sequences from RefSeq (17). However, the bacterial sequence database has a high degree of sequence redundancy with many of the individual gene sequences being very similar to one another. This sequence redundancy adds little value to the annotation process while needlessly slowing down the BLAST searches. For PHASTER, we reduced the size of the bacterial sequence database by removing sequences with >70% sequence identity to any other sequence in the database, using CD-HIT (18). The resulting database now contains just under nine million bacterial protein sequences, or 55% of the pre-filtered database size. This database reduction led to a two-fold reduction in the search time for this phase of the PHASTER annotation pipeline.
A surprisingly high percentage of queries submitted to PHAST consist of genomes that had been previously annotated by PHAST. Rather than treating each query as a completely novel sequence and spending several minutes repeating the same annotation process that had been completed previously, we have implemented a ‘quick query check”. With this new checking routine in place, sequences or sequence files that are submitted to PHASTER are rapidly compared against a local database of >14 000 non-redundant, previously annotated (via PHAST) bacterial or plasmid genomes. In the quick query check the query sequence's nucleotide frequencies and total sequence length are computed and rapidly matched against a database of these same statistics for all cached sequences. Potential sequence matches are isolated (usually just one, if any) and then aligned against the query sequence to ensure that only exact sequence matches are used. Having identified a query that is identical to an entry in PHASTER's database, the annotations are transferred and the result is returned to the user in a few seconds. As a result, while the average de novo query to PHASTER may take 2–3 min, a significant number of queries can be returned in 2–3 s.
Hardware upgrades
There is only so much software optimization that can be performed before one reaches a point of diminishing returns. As a result, a number of hardware upgrades had to be made to the PHASTER server. In particular, we have expanded the number of CPU cores in the computing cluster available from 32 (in the original PHAST) to 112 (in PHASTER). The PHASTER cluster now has 32 Intel Xeon 5150 @ 2.66 GHz, 32 AMD Opteron 6320, and 48 AMD Opteron 6348 processing cores. To further enhance PHASTER's performance, we have removed a number of other web servers from the PHASTER cluster. This has freed up more CPU cycles to accommodate the server's heavy daytime use. All of PHASTER's gene prediction and BLAST+ search analyses are now run on this 112 core cluster. In addition, the front-end for the PHASTER website has been installed on a much quicker virtual server which is leased through the Google Compute Engine. This front-end server features 2 Intel Xeon CPUs @ 2.30GHz and a local solid-state drive. The front-end performs many of PHASTER's other computations, and now does so ∼50% faster. Moreover, because PHASTER now has a dedicated front-end server, it is able to accommodate two jobs simultaneously for the most memory-intensive portions of the pipeline. This provides faster results during periods of heavier use.
User interface enhancements
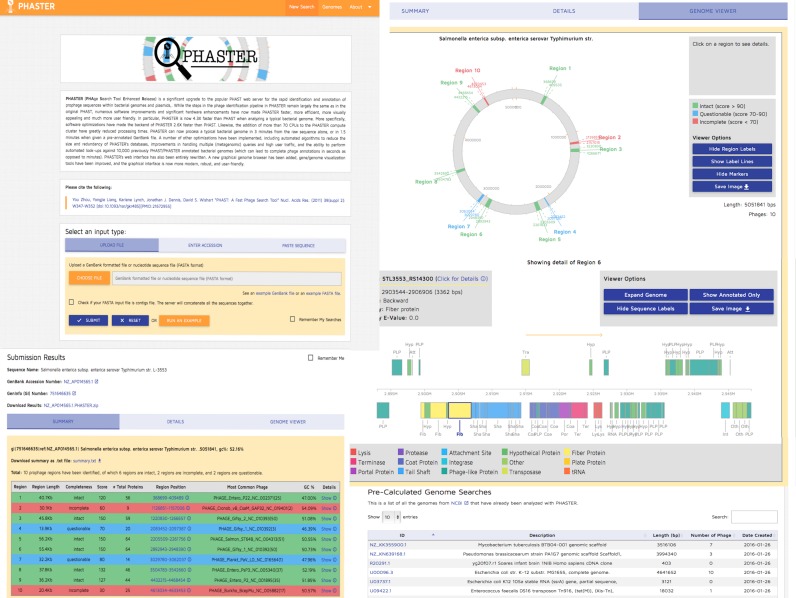
The original PHAST web interface, while very functional and utilitarian, was actually quite rudimentary and relied on a number of web technologies that are no longer current. PHASTER has a much more modern and user-friendly interface. PHAST's front-end, which was implemented using Perl and CGI, has been completely re-written for PHASTER using Ruby on Rails. PHASTER's graphical genome viewer has also been completely rewritten and modernized. The old genome viewer in PHAST provided circular and linear genome browsers implemented in Adobe Flash. Flash is a technology that is slowing being phased out by many websites and is not available on most mobile devices. As a result, PHASTER's interactive phage viewer has now been re-done using JavaScript, using AngularPlasmid (http://angularplasmid.vixis.com) for the circular genome view and D3 (http://d3js.org) for the linear genome view. As a result, PHASTER's graphical user interface is faster, more robust and better-looking (Figure 1). A more robust, threaded queuing system has also been implemented using Sidekiq (http://sidekiq.org). Another interface upgrade involved the reformatting of PHAST's vast collection of pre-computed prophage predictions for over 14 000 different bacterial genomes. PHAST presented these data in a single static table that was becoming increasingly slow and unwieldy to view or search. In PHASTER this database is now fast-loading, paginated and dynamically searchable.
Figure 1.
A screenshot montage of the upgraded PHASTER user interface.
PHASTER also allows visitors to optionally save their searches using a cookie-based storage mechanism by clicking on the appropriate check box. This will work for anyone returning to the PHASTER website using the same browser on the same computer, provided that the browser has cookies enabled. Previously submitted jobs saved in this way will be available under the new ‘My Searches” section, without any need to log in. This feature is optional, and users can still bookmark their results pages instead.
PERFORMANCE AND FEATURE EVALUATION
Comparisons of typical runtimes between PHAST and PHASTER are provided in Table 1. Note that PHAST (which is still operational) runs on 32 CPU cores while PHASTER runs on 112 CPU cores. Table 1 highlights the speed-ups obtained for each of the six different optimizations made, from the redundancy reduction of the bacterial genome database to the implementation of BLAST+ to the addition of another 80 CPU cores. In particular, reducing the bacterial database's redundancy nearly halved search times for that database, increasing the number of computing cluster cores to from 32 to 112 improves BLAST runtimes to ∼6 min for an unannotated genome, upgrading to BLAST+ further improves the total time by ∼20%, partitioning the query sequences more evenly via total amino acid count offers a ∼40% overall BLAST speed improvement, and the division of the bacterial database into separate parts to be searched separately more than halved the bacterial database search time, reducing it to just 48 seconds on average. Using a faster, dedicated front-end server, allowed processing times to be reduced from approximately 72 to 45 s. With all these enhancements, PHASTER's total runtime is now ∼210 s for a typical raw genome sequence and ∼100 s for pre-annotated GenBank input. All run times quoted in Table 1 were measured using the same test genome (E. coli O157:H7, with GenBank accession NC_002655).
Table 1. Detail of PHASTER's performance upgrades, and a comparison of their impact on runtimes for a 5.5 Mbp test genome (Escherichia coli O157:H7, GenBank accession NC_002655).
| Cumulative set of performance enhancements | BLAST versus viral DB runtime (s) | BLAST versus bacterial DB runtime (s) | Total runtime on GenBank annotated genome (s) | Total runtime on unannotated genome (s) |
|---|---|---|---|---|
| PHAST (baseline) current DBs, no other upgrades | 191 | 576 | 270 | 899 |
| PHASTER (upgrade 1) – filter bacterial DB | 191 | 309 | 270 | 632 |
| PHASTER (upgrade 2) – cluster upgrade | 88 | 166 | 167 | 386 |
| PHASTER (upgrade 3) – BLAST+ | 77 | 132 | 156 | 341 |
| PHASTER (upgrade 4) – partition query sequences evenly | 47 | 103 | 126 | 282 |
| PHASTER (upgrade 5) – bacterial DB, optimize parameters | 47 | 48 | 126 | 227 |
| PHASTER (upgrade 6) – faster front-end server | 47 | 48 | 100 | 205 |
Previous comparisons of PHAST to other prophage identification software have shown that PHAST's accuracy is either superior or comparable to other leading tools when analyzing complete genomic sequence data (2,11). Tests on a set of 54 bacterial genomes in 2011 showed that PHAST achieved a sensitivity of 79.4% and positive predictive value (PPV) of 86.5% on unannotated DNA sequences, and a sensitivity of 85.4% and PPV of 94.2% on GenBank annotated genomes (2). Since PHAST's original publication, larger phage databases and small refinements to PHAST's phage identification routines have led to improved accuracy. In response to user feedback, slight parameter adjustments have also been made to increase the algorithm's sensitivity so that more prophages can be identified. Tests on the same set of 54 bacterial genomes (1) used for evaluation in 2011 show that PHASTER now achieves a >5% higher sensitivity of 85.0% (versus 79.4% for PHAST) on unannotated genomes, with a slightly improved PPV of 87.3% (versus 86.5% for PHAST). On GenBank annotated genomes, PHASTER has a superior sensitivity of 86.9% (versus 85.4% for PHAST), and a PPV of 91.0% (versus 94.2% for PHAST). Because of the parameter adjustments made to increase PHASTER's sensitivity, the latest scores show a slight increase in the number of false positives. However, given that the ‘gold standard' annotations used for evaluation are now 13 years old (1), many prophages identified as ‘false positives' relative to this standard are likely to be true prophages. Indeed, manual inspection of PHASTER's predictions reveals many with highly significant hits to telltale phage genes or clusters of predicted phage genes from the same species, suggesting that 40–50% of these ‘false positive' predictions are in fact true prophages. If these corrections to the gold standard annotations were included in our calculations, PHASTER's performance would be even better. Further evaluation details are posted on the PHASTER website.
Table 2 highlights some of the main feature differences between PHAST and PHASTER. In this table we see that PHAST takes 899 s for a typical genome/phage annotation while PHASTER takes 205 s for the same genome. PHAST only uses 32 cores, while PHASTER uses 112 cores, PHAST uses BLAST while PHASTER uses BLAST+, PHAST does not handle metagenomic data while PHASTER easily handles assembled contigs from metagenomic data, PHAST lacks a ‘quick query check' look-up feature while PHASTER has quick query check capabilities, PHAST was coded in Perl and CGI while PHASTER was coded in Ruby on Rails, PHAST uses Adobe Flash for visualization while PHASTER uses more modern JavaScript tools for visualization and finally both PHAST an PHASTER achieve the same accuracy in prophage identification.
Table 2. A feature comparison between PHAST and PHASTER.
| Feature | PHAST (as of January 2011) | PHASTER |
|---|---|---|
| Viral sequence database | ∼45 000 sequences | ∼187 000 sequences |
| Bacterial sequence database | ∼4 million sequences | ∼9 million sequences, streamlined through CD-HIT filtering |
| Computing cluster | 32 CPU cores | 112 CPU cores |
| BLAST | Legacy version 2.2.16 | BLAST+ version 2.3.0+ |
| Cluster use optimization | Rudimentary | Smart partitioning of query sequences and target bacterial DB; optimized execution parameters |
| Front-end server | Shared, single CPU | 50% faster, dedicated |
| Front-end website | Perl and CGI | Ruby on Rails |
| Genome viewer | Adobe Flash | JavaScript, AngularPlasmid and D3 |
| Queuing system | Flat file | Uses Sidekiq for threading submissions |
| Recall previous user submissions | Bookmark page | ‘My Searches’ feature or bookmark |
| Pre-computed genome results for quick query searching | 0 | >14 000 |
| Retrieve previously annotated genome results | GenBank accession or GI number only | GenBank accession, GI number, or full sequence |
| Metagenomic data handling | NA | Supported for contigs >2000 bp |
CONCLUSIONS
To handle the rapidly expanding databases sizes and increasing popularity of PHAST we decided to undertake a near complete back-end re-write and a near complete front-end overhaul of the PHAST web server. This work led to the creation of a new, enhanced release of PHAST called PHASTER. In many respects, PHASTER is faster, better, easier to use and more robust than PHAST. These performance enhancements were achieved through code optimization, improved database preparation, and hardware upgrades. We also made PHASTER's web interface more convenient, user-friendly, and substantially more robust in terms of its ability to handle higher user traffic loads. Overall, PHASTER is 4–5× faster than PHAST and, in some cases, it can be up to 400 times faster (if a previously annotated genome is submitted). PHASTER is now capable of handling assembled contig sets from metagenomic data and it offers number of useful interface enhancements for viewing its output, testing its performance, and exploring previously annotated genomes. These improvements should not only make PHASTER more appealing for users, they will also make the server far easier to maintain and sustain into the future.
FUNDING
Canadian Institutes of Health Research (CIHR); Genome Alberta, a division of Genome Canada. Funding for open access charge: CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 2.Zhou Y., Liang Y., Lynch K.H., Dennis J.J., Wishart D.S. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuster S.C. Next-generation sequencing transforms today's biology. Nat. Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Srividhya K.V., Rao G.V., Raghavenderan L., Mehta P., Prilusky J., Manicka S., Sussman J.L., Krishnaswamy S. Database and comparative identification of prophages. In: Huang D-S, Li K, Irwin GW, editors. Intelligent Control and Automation, Lecture Notes in Control and Information Sciences. Vol. 344. Berlin: Springer; 2006. pp. 863–868. [Google Scholar]
- 6.Ester M., Kriegel H., Sander J., Xu X. KDD-1996 Proceedings. Menlo Park: AAAI Press; 1996. A density-based algorithm for discovering clusters in large spatial databases with noise; pp. 226–231. [Google Scholar]
- 7.Bose M., Barber R.D. Prophage Finder: a prophage loci prediction tool for prokaryotic genome sequences. In Silico Biol. (Gedrukt) 2006;6:223–227. [PubMed] [Google Scholar]
- 8.Fouts D.E. Phage_Finder: Automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006;34:5839–5851. doi: 10.1093/nar/gkl732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lima-Mendez G., Helden J.V., Toussaint A., Leplae R. Prophinder: a computational tool for prophage prediction in prokaryotic genomes. Bioinformatics. 2008;24:863–865. doi: 10.1093/bioinformatics/btn043. [DOI] [PubMed] [Google Scholar]
- 10.Akhter S., Aziz R.K., Edwards R.A. PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012;40:e126. doi: 10.1093/nar/gks406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roux S., Enault F., Hurwitz B.L., Sullivan M.B. VirSorter: mining viral signal from microbial genomic data. PeerJ. 2015;3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamke J., Sczyrba A., Ivanova N., Schwientek P., Rinke C., Mavromatis K., Woyke T., Hentschel U. Single-cell genomics reveals complex carbohydrate degradation patterns in poribacterial symbionts of marine sponges. ISME J. 2013;7:2287–2300. doi: 10.1038/ismej.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kashtan N., Roggensack S.E., Rodrigue S., Thompson J.W., Biller S.J., Coe A., Ding H., Marttinen P., Malmstrom R.R., Stocker R., et al. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science. 2014;344:416–420. doi: 10.1126/science.1248575. [DOI] [PubMed] [Google Scholar]
- 14.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y., Haixu T., Yuzhen Y. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics. 2012;28:125–126. doi: 10.1093/bioinformatics/btr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rho M., Tang H., Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010;38:e191. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M., et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]