Abstract
Modeling loops is a critical and challenging step in protein modeling and prediction. We have developed a quick online service (http://rcd.chaconlab.org) for ab initio loop modeling combining a coarse-grained conformational search with a full-atom refinement. Our original Random Coordinate Descent (RCD) loop closure algorithm has been greatly improved to enrich the sampling distribution towards near-native conformations. These improvements include a new workflow optimization, MPI-parallelization and fast backbone angle sampling based on neighbor-dependent Ramachandran probability distributions. The server starts by efficiently searching the vast conformational space from only the loop sequence information and the environment atomic coordinates. The generated closed loop models are subsequently ranked using a fast distance-orientation dependent energy filter. Top ranked loops are refined with the Rosetta energy function to obtain accurate all-atom predictions that can be interactively inspected in an user-friendly web interface. Using standard benchmarks, the average root mean squared deviation (RMSD) is 0.8 and 1.4 Å for 8 and 12 residues loops, respectively, in the challenging modeling scenario in where the side chains of the loop environment are fully remodeled. These results are not only very competitive compared to those obtained with public state of the art methods, but also they are obtained ∼10-fold faster.
INTRODUCTION
Protein loop prediction is an essential task in protein structure modeling, structural refinement, antibody design and ion channels modeling. Accurate prediction of loops is critical because they often play key roles in molecular recognition, ligand binding, protein–protein/protein–DNA interactions and enzyme catalysis. Thus, a significant research effort has been dedicated to the development of bioinformatics tools to deal with this challenging problem. Algorithms for loop prediction have been comprehensively reviewed elsewhere (1–3). Briefly, loop structure modeling methods can be classified into template-based (database search) or ab initio (template-free) approaches and their combinations. Ab initio methods build feasible loop conformations from scratch whereas template-based methods locate a best fit from a structural library of loops extracted from the Protein Data Bank (PDB). Since the number of possible conformations grows exponentially with loop length, the template-based methods are limited to relatively short loops. In contrast, the ab initio methods overcome this problem by performing an energy-based sampling of the conformational space. Substantial progress has been recently made using ab initio or hybrid strategies. State-of-the-art methods such as KIC (4), NGK (5), HLP (6), LEAP (7), GalaxyLoop-PS2 (8) and ICMF (9) report sub-angstrom accuracy in many test cases even modeling the neighborhood of the loops. However, these approaches are computational demanding. The reported computational times for a single prediction case of 12 residue range from hundreds of hours for KIC, HLP, NGK or GalaxyLoop-PS2 to the tenths of hours for LEAP or ICMF. Logically, the computational cost also depends on the loop length. More importantly, accuracy also decreases rapidly with the length of the loop. While small and medium size loops (12 residues or less) are treatable, longer loops are significantly more challenging. In fact, the modeling of longer loops is a more complex mini folding problem.
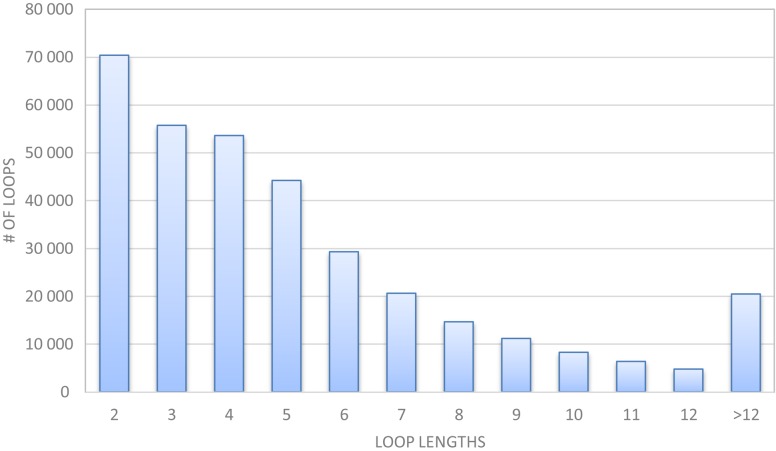
Here, we center our loop modeling efforts in treatable loop lengths up to 12 residues long to cover the majority of practical situations. The loop length distribution of high-resolution protein chains generated by the PISCES server (10) shown in Figure 1 indicates that 94% of loops have lengths smaller or equal than 12 residues. The server is based on our Random Coordinate Descent (RCD) loop closure algorithm (11) that has been demonstrated to offer an excellent balance between efficiency and sampling power. We improve the algorithm to generate more accurate native-like loops, providing an efficient way to generate a large ensemble of closed-loops for capturing the diversity of conformational space. This ensemble of decoys is scored with a new knowledge-based, coarse-grained contact potential (12) that correlates distance and orientation with pair-wise residue interaction energies. Refinement of the best representative loops employing Rosetta completes the modeling process. In a matter of few minutes from submission of the anchor residue coordinates, the top-ranked loop predictions can be interactively inspected using a user-friendly interface. In examining RCD+'s predictive ability we found comparable, if not better, results than other ab initio methods, in particular, better than the GalaxyLoop-PS2 method implemented in GalaxyLoop server, to our knowledge, the most advanced loop modeling service currently available.
Figure 1.
Distribution of loop lengths in the protein chain list generated by the PISCES server on April 13, 2016 containing 18 275 chains with 2.0A resolution, 90% sequence identity and 0.25 R-factor cutoff.
MATERIALS AND METHODS
The server is based on our original RCD loop closure algorithm (11). This ab initio algorithm solved the loop closure problem by analytically optimizing randomly selected bonds with a fast updating of loop backbone conformations based on spinor-matrices (13). The loop closure sampling was steered by a simplified Ramachandran filter that constrains the backbone φ and ψ dihedral angles, and by a simple geometric filter that prevented clashes between the loop backbone atoms and the local protein surroundings. Although this versatile tool efficiently generated large ensembles of closed loops, it has been considerably improved in both accuracy and speed to support a modeling web service. Here, we replace the original basic Ramachandran filter by neighbor-dependent probability distributions extracted using Bayesian non-parametric statistical analysis from a high-resolution data set of protein loops (14). Torsion restricted sampling was already shown to be useful in loop structure prediction for intensifying the sampling toward near-native models (5,14). To promote sampling diversification, bond lengths and angles are initially randomized from ideal values, as well as, omega torsions are normally distributed around 179.1° ± 6°. Additional improvements include geometric filters to discard clashed loops, a best workflow optimization and MPI-parallelization (detailed results of the sampling enrichment are available in the website). Once an ensemble of closed loops (typically 5000–10 000 loops for 8 and 12 residues, respectively) is efficiently generated using RCD+, it is scored using ICOSA energy function, a new pairwise coarse-grained contact potential (12) that correlates inter-residue distance and orientation using a simple icosahedral tessellation. This knowledge-based potential has accuracy and sensitivity comparable to all-atom fine-grained potentials in identifying CASP10 models and discriminating near-natives from misfolds. Moreover, ICOSA perfectly matches with RCD+ since it only needs information of the backbone atoms and it is also very efficient. The best 10% of the ensemble loops ranked by ICOSA potential are refined with PyRosetta modeling package (15) to produce accurate all-atom predictions. After side-chains are added using the standard repacking protocol, we employ five refinement cycles of side-chain repacking and gradient minimization with dfpmin_armijo_nonmonotone method and talaris2014 energy function. On each cycle we ramp the repulsive weight up and down while minimizing the loop and ultimately selecting the lowest energy loop. An equivalent minimization strategy has been already employed in MacDonald et al. (16). The refinement stage can be performed either in native (crystallographic) or modeling scenarios. In the former, only the backbone and side-chain of the loop are refined whereas in the latter the side-chains of the environment are also optimized. The environment is defined as the set of residues with any atom within 5 Å from any loop Cβ atom. It is worth to mention, in contrary to other methods, we first remove all native information of the loop as well as all surrounding sidechains within 10 Å from any of 100 pre-sampled loops. PyRosetta refinement scripts are also freely available upon request. Typical refinement times are around one minute per conformation for a 12-residues loop.
DESCRIPTION OF THE WEB SERVER
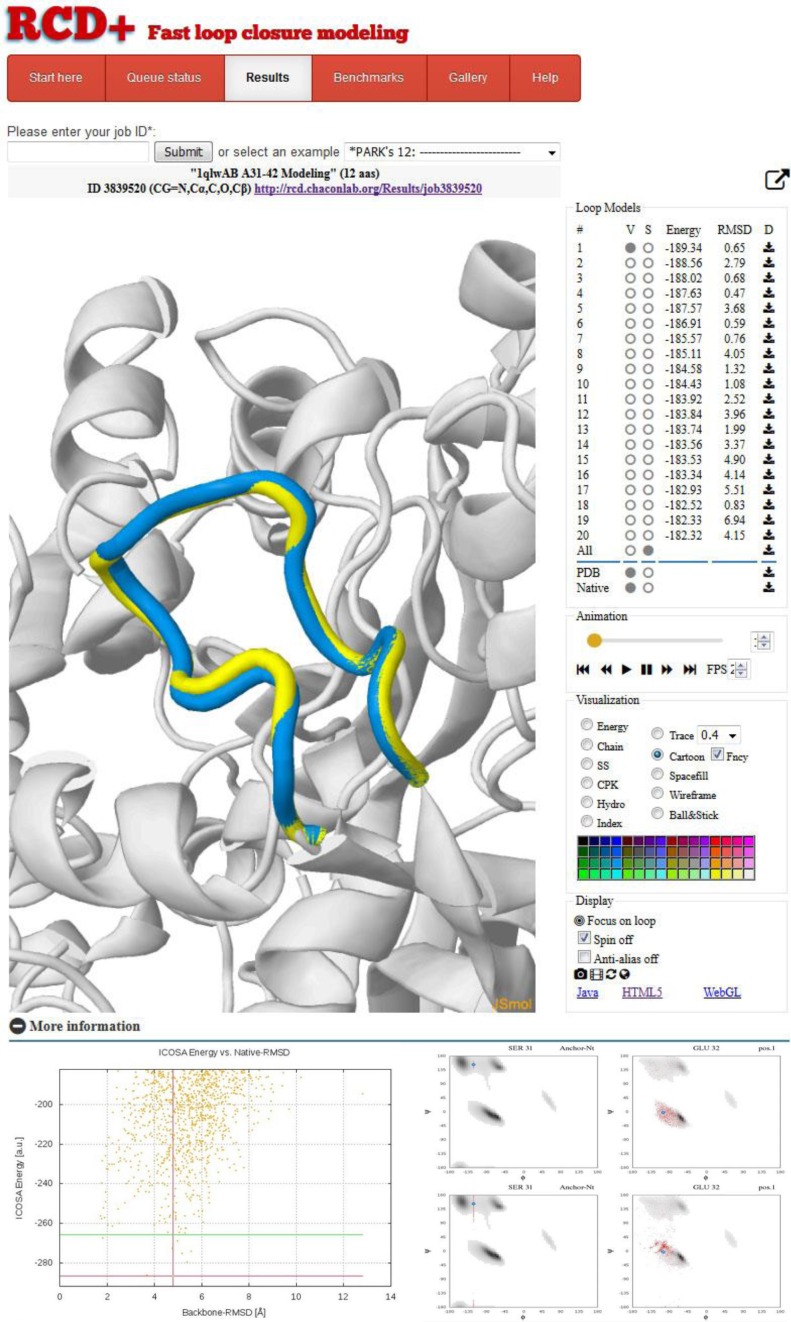
The web interface of the RCD+ server is very intuitive and responsive to all major browsers. The input is quite simple; the user must introduce the atomic coordinates of the protein, chain id, the sequence and start/end indices of the loop residues to be modeled. The atomic coordinates can be either uploaded (PDB format v3.x) or fetched directly from the PDB using the corresponding entry ID. By contrast to other algorithms, the server fully models from scratch all residues from Start to End indices (inclusive). The only requirement is the presence of the N- and C-terminal anchor residues (indices Start-1 and End+1). For example, if the user wants to model an 8-residue loop from index 270 to 277 the provided structure must include at least the N, Cα and C backbone atoms of the N-terminal (index 269) and the C-terminal (index 278) residues. Then, one of the following two prediction scenarios must be selected: (i) native, when the conformations of the side-chains of the loop environment are reliable, e.g. for predicting the missing loops in atomic structures, or (ii) modeling, when the side-chains of the loop environment could be in a different conformation and should be remodeled, e.g. if the submitted structure is an homology model. In either case, if the loop region is close to some other protein chain it should be included in the input PDB file to sterically constrain the search space. Finally, the modeled loop sequence must be typed or pasted in one letter format. Although the cis/trans isomerization is not considered in the modeling, the user can select the modeled proline isomer in the sequence by using the lower and upper case of the letter p to choose cis or trans proline, respectively. Upon submission, the prediction job will be queued in our cluster and the user will be immediately redirected to the Queue status tab to check the job status in real time. Once the jobs are completed, direct links to the results are generated in the list of finished jobs and optionally sent to the user by email. The results page (see a representative layout in Figure 2) includes a JSmol (17) visualization section in which the 20 lowest-energy models can be interactively inspected and compared in 3D through HTML5, WebGL or Java interfaces. The user can easily customize color and representation method to facilitate comparisons. Below this section, all computed files can be downloaded, including the all-atom models of the 20 best energy loops and the initial raw ensemble of energy-filtered loops. The Ramachandran plots of both the refined and the raw RCD+ energy-filtered loops can be also visualized and compared. Finally, and just for validation purposes, if the submitted coordinates already contain the native loop, the modeling will completely ignore them but the results section will include the comparison of the predictions with the native loop through backbone RMSD versus energy plots to facilitate the performance evaluation of RCD+ with the ICOSA and talaris2014 energy functions. Full documentation is provided, including detailed benchmark results, a gallery of pre-computed examples and help information.
Figure 2.
Sample results page provided by the server for a bacterial hydrolase loop (PDB-ID 1qwl). In this case, for validation proposes only, the native loop (yellow) is displayed superimposed with the predicted lowest energy model in the JSmol visualization panel. On the right, the 20 top-ranked loop models are sorted by energy and can be easily selected to activate visualization and customize representation. The RMSD versus ICOSA energy plots and the Ramachandran distributions are shown in the bottom part.
PREDICTIVE PERFORMANCE
The predictive performance of our server has been tested in Table 1 using standard benchmarks of 8- and 12-residue loops (20 cases each) employed in the validation of other public state-of-the-art methods. In a native scenario, the average (or median) backbone RMSDs between the lowest-energy model and the native conformation (computed using N, Cα, C and O loop backbone atoms) are 0.6/1.0 (0.5/0.6) Å for 8/12 residues loops, respectively. These results are better than those obtained by other methods such as HLP and Galaxy-PS2 that attained 1.2/1.2 (0.6/0.6) Å and 0.9/1.6 (0.6/1.4) Å, respectively. Sub-angstrom predictions are found in the top-ranked (lowest-energy) loop for 17/16 (8/12 residues) of the cases with RCD+ whereas HLP and Galaxy-PS2 only reached 13/12 and 14/7, respectively. In the modeling scenario we employed a set of crystal structures with perturbed side-chain structures taken from Sellers et al. (6) to assess the performance in inaccurate environments. In this scenario, where side-chains of the loop and the environment are fully re-modeled from scratch, the accuracy of RCD+ is well maintained up to 0.8/1.4 (0.4/0.6) Å. The accuracy of our method with 8 residues loops is slightly lower than NGK that reaches 0.5 but with similar median (0.5–0.4). For 12 residues loops we have comparable results to HLP and NGK but with better median values (0.6 versus 0.8–0.9 Å), clearly outperforming Galaxy-PS2. In this scenario, RCD+ obtained 13 sub-angstrom predictions followed by NGK and HLP with 11. On average, the predictions of our server are better than Galaxy-PS2 server and as good as the best approaches. However, RCD+ server is able perform the predictions in 5–15 min whereas NGK and HLP require at least one order of magnitude more time.
Table 1. Loop-prediction performance of RCD+ and other state-of-the-art methods.
| Nativea | Modeling | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HLPb | Galaxy | RCD+ | HLP | Galaxy | Rosetta | RCD+ | ||||||
| Lengthc | Std. | PS2 | 1d | 5 | 20 | SS | PS2 | NGK | 1 | 5 | 20 | |
| Median | 0.6 | 0.6 | 0.5 | 0.3 | 0.3 | 0.9 | 1.1 | 0.4 | 0.5 | 0.4 | 0.4 | |
| 8 | Average | 1.2 | 0.9 | 0.6 | 0.5 | 0.4 | 1.3 | 1.3 | 0.5 | 0.8 | 0.7 | 0.6 |
| Sigma | 1.5 | 0.7 | 0.3 | 0.3 | 0.2 | 1.5 | 1.0 | 0.3 | 1.0 | 1.0 | 0.8 | |
| #e | 13 | 14 | 17 | 18 | 19 | 11 | 9 | 17 | 15 | 18 | 18 | |
| Median | 0.6 | 1.4 | 0.6 | 0.4 | 0.4 | 0.9 | 1.6 | 0.8 | 0.6 | 0.6 | 0.5 | |
| 12 | Average | 1.2 | 1.6 | 1.0 | 0.9 | 0.7 | 1.4 | 2.1 | 1.7 | 1.4 | 0.8 | 0.8 |
| Sigma | 1.2 | 1.3 | 1.7 | 1.0 | 1.0 | 1.4 | 1.7 | 1.8 | 1.6 | 0.9 | 0.9 | |
| #e | 12 | 7 | 16 | 16 | 17 | 11 | 4 | 11 | 13 | 17 | 17 | |
aSampling scenarios: (i) Native, the side-chains of the loop environment are kept, or (ii) Modeling, include the refinement of the loop environment side-chains.
bHLP, HLP-SS, Galaxy-PS2 and Rosetta-NGK Root Mean Squared Deviations (RMSDs) were taken from Supplementary Tables S1, S2, S4 and S5 of (8) and calculated considering the main-chain atoms N, Cα, C and O.
cNumber of residues of the loop.
dRMSD of the lowest Rosetta-energy loop predicted with RCD+ together with the best RMSD of the 5 and 20 loops of lowest Rosetta-energies.
eNumber of sub-angstrom cases.
EXAMPLES OF USE
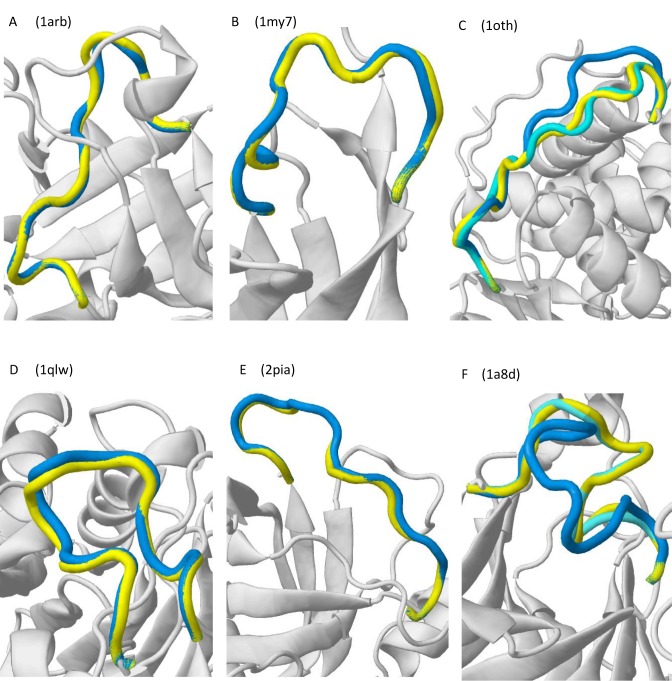
Several 12-residue test cases have been selected from those 20-case benchmark sets employed in the validation to directly illustrate usage and performance of our server (additional examples are available on the Gallery tab of the website). When a native scenario is considered, RCD+ is able to obtain sub-angstrom predictions in 16 of the 20 cases (Supplementary Table S2) in the first solution. For example, in Figure 3, the lowest energy models predicted by RCD+ (blue) of two representative cases (1arb and 1my7) are illustrated (panels A and B) together with the corresponding native conformation of the loop (yellow) and its environment (gray). Moreover, in one (1oth) of the 4 cases with RMSD significantly above 1.0 Å, a sub-angstrom structure can be found within the first 20 best predictions (panel C). In 1cnv and 1cs6 cases, other methods also fail to obtain a sub-angstrom model, indicating that these are difficult cases. In a modeling scenario, we find 13 sub-angstrom predictions in the top-ranked loops (see 1qlw and 2pia in panels D and E), but we improve up to 17 when considering the best 5 predictions. In this more challenging scenario, we are still able to recover sub-angstrom models from the top-scoring solutions sampled in two failed cases 1oth and 1oyc. Also, in the 1a8d case the best solution (5.2 Å) is dramatically improved up to 1.1 Å just by the second top-ranked model (panel F). In the 1m3s case all methods but ours fail, presumably because only we considered the loop neighboring oligomers. It is worth noting that in the remaining 1cs6 and 1cnv failed cases none tested methods obtained good solutions. Interestingly, the RCD performance can be improved in some of these cases by running several independent predictions. Thus, improving the current method to combine several independent runs will probably lead to accuracy improvements in future server versions.
Figure 3.
Illustrative cases of the server performance with benchmark test cases. In all the cases, the first ranked model (lowest energy) is depicted in blue, the native loop in yellow and the protein environment in gray. Alternative solutions found in the 12nd best (1oth) and 2nd best (1a8d) top-ranked predictions are colored in cyan.
TECHNICAL DETAILS
The web server is implemented as a combination of several PHP, python and JavaScript modules running in a dedicated Linux system with two Intel® Xeon® E5-2650 processors running at 2.00 GHz (16 cores) and equipped with 128 GB RAM. For optimal web server usage, a queue system (grid engine) is included for job management and scheduling. The calculations are performed in a modest Linux cluster with 10 nodes of 8 GB of RAM of dual Intel® Xeon® E5410 2.33 GHz processors. A typical 12-residues loop prediction costs around ∼10 CPU-hours in the cluster (equivalent to ∼6 h in a modern E5-2650 processor). Modeling results are visualized in 3D with JSmol (17).
CONCLUSIONS
Since protein loop modeling is critical for understanding molecular mechanisms in molecular recognition, signal transduction or enzymatic reaction, it is essential having an online tool that facilitates such a challenging task. By merging a very efficient ab initio exhaustive sampling with a full-atom state-of-the-art refinement, our new web service consistently reaches sub-angstrom accuracy in 80–90% of the cases within the top 5 predictions for 8–12 residues loops. The average backbone RMSDs between the lowest-energy model and the native conformation is 0.6 Å or 0.8 Å depending if the side chains of native environment are considered or fully remodeled, respectively. The accuracy is still well maintained up to 1.0 and 1.4 Å for 12 residues loops benchmark. Our server, RCD+, is the fastest alternative to generate accurate loop predictions that can be easily explored and selected for further applications.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy and Competitiveness (BFU2013-44306P to P.C. and BFU2014-51823-R to A.J.C); National Science Foundation [1066471 to Y.L.]. Funding for open access charge: Spanish Ministry of Economy and Competitiveness (BFU2013-44306P to P.C. and BFU2014-51823-R to A.J.C); National Science Foundation [1066471 to Y.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Soto C.S., Fasnacht M., Zhu J., Forrest L., Honig B. Loop modeling: Sampling, filtering, and scoring. Proteins. 2008;70:834–843. doi: 10.1002/prot.21612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shehu A., Kavraki L.E. Modeling structures and motions of loops in protein molecules. Entropy. 2012;14:252–290. [Google Scholar]
- 3.Li Y. Conformational sampling in template-free protein loop structure modeling: An overview. Comput. Struct. Biotechnol. J. 2013;5:e201302003. doi: 10.5936/csbj.201302003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandell D.J., Coutsias E.A., Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat. Methods. 2009;6:551–552. doi: 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stein A., Kortemme T. Improvements to robotics-inspired conformational sampling in rosetta. PLoS One. 2013;8:e63090. doi: 10.1371/journal.pone.0063090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sellers B.D., Zhu K., Zhao S., Friesner R.A., Jacobson M.P. Toward better refinement of comparative models: Predicting loops in inexact environments. Proteins. 2008;72:959–971. doi: 10.1002/prot.21990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang S., Zhang C., Zhou Y. LEAP: Highly accurate prediction of protein loop conformations by integrating coarse-grained sampling and optimized energy scores with all-atom refinement of backbone and side chains. J. Comput. Chem. 2014;35:335–341. doi: 10.1002/jcc.23509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park H., Lee G.R., Heo L., Seok C. Protein loop modeling using a new hybrid energy function and its application to modeling in inaccurate structural environments. PLoS One. 2014;9:e113811. doi: 10.1371/journal.pone.0113811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnautova Y.A., Abagyan R.A., Totrov M. Development of a new physics-based internal coordinate mechanics force field (ICMFF) and its application to protein loop modeling. Proteins. 2011;79:477–498. doi: 10.1002/prot.22896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G., Dunbrack R.L. PISCES: recent improvements to a PDB sequence culling server. Nucleic Acids Res. 2005;33:W94–W98. doi: 10.1093/nar/gki402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chys P., Chacón P. Random coordinate descent with spinor-matrices and geometric filters for efficient loop closure. J. Chem. Theory Comput. 2013;9:1821–1829. doi: 10.1021/ct300977f. [DOI] [PubMed] [Google Scholar]
- 12.Elhefnawy W., Chen L., Han Y., Li Y. ICOSA: A distance-dependent, orientation-specific coarse-grained contact potential for protein structure modeling. J. Mol. Biol. 2015;427:2562–2576. doi: 10.1016/j.jmb.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 13.Chys P., Chacón P. Spinor product computations for protein conformations. J. Comput. Chem. 2012;33:1717–1729. doi: 10.1002/jcc.23002. [DOI] [PubMed] [Google Scholar]
- 14.Ting D., Wang G., Shapovalov M., Mitra R., Jordan M.I., Dunbrack R.L., Jr Neighbor-dependent Ramachandran probability distributions of amino acids developed from a hierarchical dirichlet process model. PLoS Comput. Biol. 2010;6:e1000763. doi: 10.1371/journal.pcbi.1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaudhury S., Lyskov S., Gray J.J. PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics. 2010;26:689–691. doi: 10.1093/bioinformatics/btq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacDonald J.T., Kelley L.A., Freemont P.S. Validating a coarse-grained potential energy function through protein loop modelling. PLoS One. 2013;8:e65770. doi: 10.1371/journal.pone.0065770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanson R.M., Prilusky J., Renjian Z., Nakane T., Sussman J.L. JSmol and the next-generation web-based representation of 3D molecular structure as applied to proteopedia. Isr. J. Chem. 2013;53:207–216. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.