Abstract
GPCR-ModSim (http://open.gpcr-modsim.org) is a centralized and easy to use service dedicated to the structural modeling of G-protein Coupled Receptors (GPCRs). 3D molecular models can be generated from amino acid sequence by homology-modeling techniques, considering different receptor conformations. GPCR-ModSim includes a membrane insertion and molecular dynamics (MD) equilibration protocol, which can be used to refine the generated model or any GPCR structure uploaded to the server, including if desired non-protein elements such as orthosteric or allosteric ligands, structural waters or ions. We herein revise the main characteristics of GPCR-ModSim and present new functionalities. The templates used for homology modeling have been updated considering the latest structural data, with separate profile structural alignments built for inactive, partially-active and active groups of templates. We have also added the possibility to perform multiple-template homology modeling in a unique and flexible way. Finally, our new MD protocol considers a series of distance restraints derived from a recently identified conserved network of helical contacts, allowing for a smoother refinement of the generated models which is particularly advised when there is low homology to the available templates. GPCR- ModSim has been tested on the GPCR Dock 2013 competition with satisfactory results.
INTRODUCTION
G-protein coupled receptors (GPCRs) constitute a superfamily of seven transmembrane proteins (7TM) which are responsible for signal transduction in response to endogenous chemicals (hormones, neurotransmitters) as well as to external signals (light, odorants). It consists of more than 800 members that account for 3% of the human encoding genome, and it's further divided into 5 classes according to the most accepted GRAFS classification system (1). GPCRs are highly dynamic signaling proteins that undergo an important conformational change (activation) upon binding to an extracellular molecule (agonist), leading to intracellular signal transduction typically mediated by the G-protein. It is estimated that between 30 and 40% of marketed drugs target a GPCR (2), including many blockbusters in the pharmaceutical industry. After decades of functional characterization of GPCRs presided by molecular biology experiments, a series of paradigmatic methodologies for receptor stabilization and crystallization (3,4) has promoted an explosion of GPCR crystal structures since 2007. Although at least one crystal structure is now available for all human GPCR classes, the rhodopsin-like (class A in the GRAFS system) constitutes the best characterized class with more than 20 unique receptors captured in different conformational states: inactive (antagonist-bound), active-like (agonist-bound) and fully-active (ternary complex, including the signaling proteins G-protein, β-arrestin or intracellular mimics). As opposed to their sequence diversity, the conserved topology of 7TM helices connected by three extracellular (EL) and three intracellular loops (IL), is an important aid in 3D modeling of the vast majority of GPCRs of unknown structure. Consequently, several dedicated protocols for modeling GPCRs have been developed, the following being implemented in web servers: GPCRM (5), GPCR-SSFE (6), GOMoDo (7) and GPCR-ModSim (8) all performing homology-based modeling, while GPCR-I-TASSER (9) uses the threading paradigm. We herein present the new version of GPCR-ModSim, which emerges as the most complete of these online facilities.
First released in 2011, GPCR-ModSim, provides specifically designed solutions for homology-modeling and molecular dynamics (MD) simulations of GPCRs (8,10). It is currently being used for research and teaching purposes in several institutions worldwide (11,12) and it has been considered in reviews of the field (13,14), as well as recently linked as an external tool to the reference database in the field, the GPCRdb (15). 3D molecular models can be generated from amino acid sequence by homology-modeling techniques, considering different receptor conformations. This is done through an intuitive user interface, offering a fully automated protocol, where the philosophy is to recommend to the user the best (default) options, but always allowing (advanced) manual modification at each stage. Several visualization options and analysis tools are also included to assist users in the interpretation of their results. The different stages of the GPCR-ModSim protocol can be used as stand-alone tools or as parts of a whole process. The first stage identifies, through a multiple-sequence alignment (MSA) with the selected conformational profile of templates, the best template(s) to model the receptor (or certain regions of it) by homology modeling. This module was indeed used to select the best template and generate an initial alignment for homology modeling of the 5HT1B serotonin receptor by the wining submission of the GPCR-dock competition 2013 by the Carlsson lab (16). The homology modeling stage is based on the selected template(s), and an optional stage of loop refinement. Finally, the MD simulation stage, which uses the PyMemDyn module (10) allows the equilibration and consequent structural refinement of a GPCR structure. The homology modeling stage outlined above can be skipped, and the user can directly upload a 3D structure (PDB format) of the receptor of interest, i.e. an experimental (crystal) structure or a model generated by alternative computational methods, to be refined by our PyMemDyn module. Notably, this allows the consideration of ligands in complex (i.e. obtained by external docking) with the receptor, or even the consideration of several non-protein elements that might influence receptor stability: as allosteric modulators, structural water molecules, lipids, cholesterol molecules or ions. Such a flexible setup allowed the recent characterization of the molecular mechanism of the allosteric effect of sodium ions on the conformational equilibrium of the A2A adenosine receptor (17).
We herein present an updated version of GPCR-ModSim, including an overhaul of the software and hardware infrastructure behind the webserver, the revision of the available templates for homology modeling and two new available features. One is the possibility to create homology models using multiple templates. These templates can easily be chosen and mixed by picking desired topological regions at the users will and best judgment. The other new feature is the refinement of our MD equilibration protocol which now uses a network of pair-distance restraints, derived from a comprehensive analysis of GPCR crystal structures (18). Besides the equilibration process, the use of such type of restraints can improve MD sampling to relevant active or inactive states by an order of magnitude (19), and can easily be extended to consider particular sets of restraints for a given receptor. The overall modeling process can then be defined as a unique combination of sequence derived (homology-modeling) and biophysical refinement (MD relaxation), which is especially suitable when there is low sequence identity to available templates. GPCR-ModSim has been tested on the GPCR Dock 2013 competition with satisfactory results (16).
MATERIALS AND METHODS
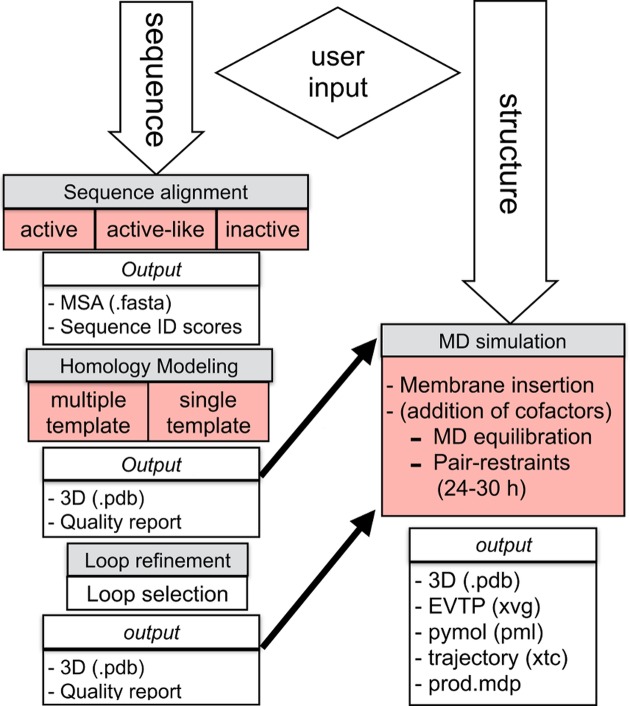
A general outline of the whole pipeline available at GPCR-ModSim can be seen in Figure 1. It takes advantage of the Python programing language, using the Django web framework, to create the input/output dialogs as well as to communicate with the calculation cluster and with the following academic-free bioinformatics software: ClustalW2 (20) (sequence alignment), Modeller (21) (homology modeling and loop refinement), MolProbity (22) (protonation and quality assessment), Gromacs (23) (MD simulations) and Structural Alignment of Multiple Proteins (STAMP) (24). It also integrates several other bioinformatics tools along the different modeling stages such as Biopython (25) (Sequence parsing), Jalview (26) (Multiple sequence alignment editor) and JSmol (27) (Molecular Viewer).
Figure 1.
Diagram illustrating the workflow of the GPCR-ModSim webserver. Each stage is depicted in gray boxes. The functionalities updated in the current version are highlighted in red.
Template selection and scoring algorithm
The user uploads a Fasta sequence (or the corresponding Uniprot ID) of a GPCR and the server performs a MSA against a selected group of templates. The basis for these MSAs are our updated and manually curated structural alignments of crystallized GPCRs, which are classified into three groups depending on the receptor conformation: inactive (22 unique templates), active-like (8 templates) and fully-active (3 templates). The alignment of the target sequence against a profile and the use of secondary structure penalties on the consensus TM regions, ensure initial pairwise alignment(s) of enough quality to detect the most suitable template(s). The sequence identity with each available template is provided as a matrix that divides the sequence into 15 pre-defined blocks, as defined by the secondary structure mask in the MSA, which are associated with the following structural regions: N-ter, (7)TM, (3)EL, (3)IL and C-ter. The receptor with highest TM sequence identity is proposed as the default template, a selection that can be freely changed. Moreover, the user can manually edit the MSA and produce a refined pairwise-sequence alignment to be used in the homology modeling stage. The sequence identity matrix is crucial to assign additional specific template(s) to particular structural elements if choosing the multiple-template modeling technique.
Multiple template selection
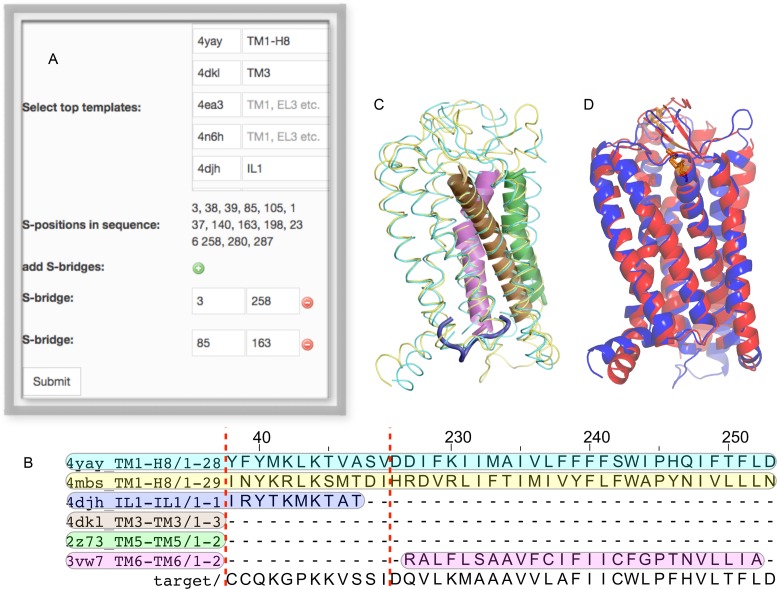
The current version of GPCR-ModSim includes a strategy for multiple-template homology modeling. Other GPCR-dedicated web servers allow this possibility, but it is not up to the user to decide the templates and/or regions to use for each template: in the GPCR-SFFE server (6), a chimeric template is created with a proposed structural template for each helical region, but with no possibility to change this selection by the user. The GPCRM server (5), on the other hand, builds a weighted average over the user-selected templates, but these are uniformly used to model the whole sequence of the target receptor (i.e. there is no possibility to use a given template for just a specified region). As opposed to these servers, GPCR-ModSim now implements a unique flexible framework that allows the user to choose several templates for each topological region. From each of the potential templates (ranked according to the overall TM sequence ID), one can select which of the defined structural regions (TM, IL, EL) should be considered for that template (if left blank, the corresponding template will not be used). The default option will select ‘all’ for the best template and ‘blank’ for the remaining templates, i.e. a single-template modeling based on top sequence ID. The user can freely play with the selections, i.e. change or remove the global template and/or add up any additional templates for each region, as depicted in Figure 2 and illustrated in the example below. The programmatic solution adopted to account for multiple-template modeling in the backend of GPCR-ModSim is to build one or more chimeric templates, both in PDB and in Fasta format, that simultaneously cover the users selection of templates and regions. This MSA with the chimeric templates maintains the rules of the original MSA alignment and this information is passed as input for the modeling stage.
Figure 2.
The multiple-template homology modeling illustrated in the example of the AT2 receptor. The template selection box (A) allows the selection of templates and topological regions. (B) The selection is reflected in the corresponding multiple-sequence alignment (MSA). In this case the fragment corresponding to IL1 is within the dashed red line and is followed by the (discontinued) region corresponding to TM6, to illustrate the different templates considered for each topological region. The full MSA is provided in the Supplementary Material. (C) Superposition of the different templates considered in this example, following the same color code as in panel B. (D) Comparison of the resulting model (blue) with the AT1 structure (red).
Homology modeling and loop refinement
The query-template pairwise sequence alignment (or query-templates MSA in the case of multiple-templates) generated in the template selection stage is used as a basis for homology modeling with Modeller v9.13 (21). The generated models are energetically ranked (using the DOPE-HR score) and each selected model is accompanied with stereochemical quality reports and a protonated version of the model performed with MolProbity (28). A visualization interface, based on JSmol, is implemented to check the resulting models. Alternatively, a PyMOL session is provided for a more advanced (local) structural analysis of the results. An optional stage can follow where selected loop regions are remodeled using the LoopModel (29) algorithm from Modeller. Here, an interactive menu allows the user to define each loop fragment to be considered in this optional stage. The selection and precise definition of the loop regions to be remodeled is optimally done using the visualization interfaces mentioned above.
Membrane insertion, solvation and MD equilibration with pair-distance restraints
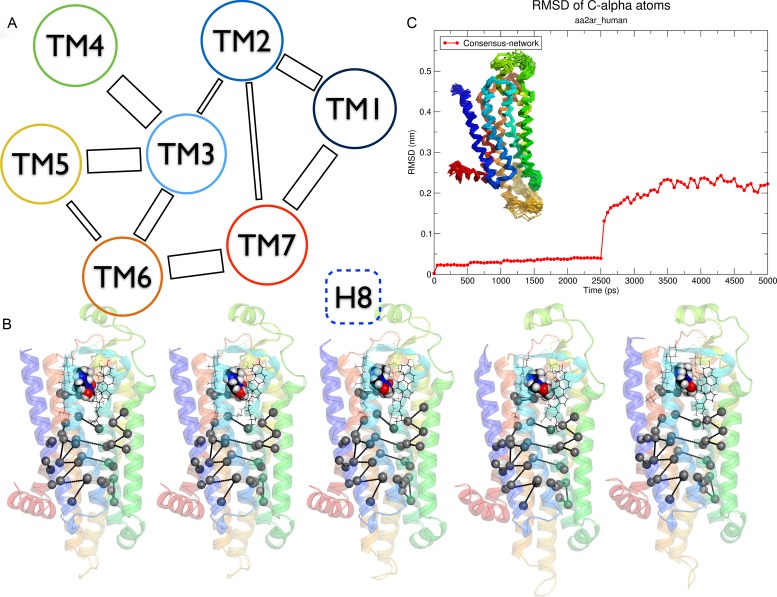
The MD protocol implemented in GPCR-ModSim (10,30) uses PyMemDyn, an in-house python module for membrane insertion and equilibration of GPCRs. There exist webservers that generate bare membranes models (31–33) without any embedded proteins, while others can embed proteins in an atomistic model of the membrane (34) but do not carry out any MD equilibration protocol, nor give the ability to upload ligands or cofactors. In GPCR-ModSim, the receptor structure is embedded in a pre-equilibrated palmitoyloleoylphosphatidylcholine (POPC) membrane model in a way that the TM bundle is parallel to the membrane vertical axis. The system is then solvated with the SPC water model, inserted into a hexagonal-prism shaped box and energy-minimized. A careful equilibration follows in the framework of periodic boundary conditions (PBC), using the OPLSAA forcefield for the protein (and ligand or cofactors if included, see below) combined with the Berger parameters for the POPC lipids. In the initial equilibration phase, which lasts 2.5 ns, positional restraints on heavy atoms are imposed and gradually released from a force constant of 1000 to 200 KJ.mol−1·nm−2 (10). As a new feature, we now include an additional 2.5 ns MD simulation where only NMR style distance restraints (35) are imposed between 24 selected pairs of C-α atoms, identified from a conserved inter-helical contact network among GPCR crystal structures (18). The restraints are applied using a piecewise style function (see Supplementary Data) where, for each pair, the target distance is defined by the average pair-distances in existing crystal structures as reported by Venkatakrishnan et al. (18), while the upper and lower limits of the function are proportional to the corresponding experimental standard deviation. The assignment is done in a fully automated way, thanks to the implementation of the common GPCR topological numbering from Ballesteros and Weinstein (36), which allow us to identify and assign the residues involved in this network in the input PDB and automatically define the list of 24 pair-distance restraints. The corresponding pair-contacts are given simply as a python list of tuples to the PyMemDyn code, and are graphically illustrated in Figure 3.
Figure 3.
Pair-distance restraints connecting the TM helices. (A) Schematic representation, adapted from ref. (18). (B) 3D representation of the pair-distance restraints applied to the molecular dynamics (MD) equilibration of the Adenosine A2A receptor in complex with caffeine, on five snapshots extracted every 0.5 ns. (C) The RMSD evolution of the A2A receptor along this MD equilibration, with the structures extracted every 0.1 ns superimposed as an inset.
Implementation
The webserver is set-up using the Python based Django web framework, and Gunicorn as an HTTP request handler. It also contains a virtual environment for portability using virtualenv. For responsive design across devices, i.e. mobiles, tablets or desktops, we use the bootstrap boilerplate. The webserver connects to our local high performance computing (HPC) resources using remote procedure calls (RPC) in the local network. Our HPC resources handle homology and dynamics modeling in separate and exclusive Simple Linux Utility for Resource Management (SLURM) partitions dedicated to handle the jobs coming from the webserver, with one dedicated dual core for homology, and two nodes (32 cores) for dynamics.
GPCR-MODSIM WEBSERVER (RESULTS)
Input
For homology modeling there are two possible ways to enter a GPCR sequence. (i) copy and paste a sequence in Fasta format, or (ii) enter a a Uniprot ID directly in the sequence box. The next step is to select a profile by picking either the active, inactive or active-like group of templates. After pressing the ‘Submit’ button, the target sequence is aligned against the selected profile and the resulting MSA alignment can be visualized and edited online using Jalview, or alternatively it can be downloaded, edited locally and uploaded back to the server. The next steps allow the user to either use the template with highest TM identity score (selected by default) or select additional (and/or alternate) templates for multiple-template homology modeling. In the last case, the topological regions have to be specified for each template from a set of predefined terms, i.e. ILx, ELx, TMx, where x is the identifier of the topological region; a range can also be defined, i.e. TM1-IL3 will cover the region between these two topological elements (see Figure 2). After the homology models are completed the user can optionally follow with the loop refinement stage, by pressing the button ‘Loop’ in the corresponding model analysis window (see output section). This gives access to a menu window to input the beginning and end points for each selected loop(s) region, and the process is initiated by pressing the ‘Submit query’ button.
There are two ways to access the MD equilibration protocol (see Figure 1). In the case that the input is a homology model generated by GPCR-ModSim, there is a ‘Run dynamic’ button in the analysis window of each model. If, on the contrary, the user wants to upload an external PDB file, one can start a new project using the ‘Upload your PDB file’ button, which can be found in the ‘Model GPCR’ menu. A PDB parser will check the structure and inform the user of any format changes that need to be done, according to the Gromacs requirements. Both options give access to the MD menu, where users can optionally upload a ‘cofactor’. This is done by uploading the 3D coordinates (PDB format) of the ligand (or co-factor, i.e. structural waters, ions or lipids) of interest, together with the associated forcefield and library files (OPLSAA forcefield using Gromacs format). Currently ligand docking must be done externally, and we provide some guidelines about the format and parameter generation in the corresponding video tutorial (see below).
Output
After submitting the target sequence, the first output provided is the MSA of the target sequence with the selected group of templates. This can be visualized in the Jalview applet, and also downloaded in Fasta format (both options allow manual editing and saving of the proposed alignment). The server also provides a PDF report with detailed information of the sequence identities between the target sequence and each template, divided by topological regions. This report, which is updated everytime the alignment is manually edited, contains a bar graph of identities for the top 5 templates as well as a numerical matrix of partial identities for the whole set of templates. This is valuable information to assist on the selection of multiple templates. After submitting the homology modeling request, in a few minutes the user can see the results in two different ways: (i) A click on the corresponding frame for a generated model gives access to a window with detailed information on that model (model analysis window), which includes: an interactive view of the structure with the JSmol applet, a downloadable PDB file (before or after proton addition) and the Ramachandran plot in PDF format. (ii) A global analysis and visual inspection of the results obtained for all generated models can be performed by downloading a compressed file that contains all the structural models in PDB format together with a Pymol script, which performs structural superimposition and a suitable graphical representation. If the user chooses to refine loop regions for a given model, the same two options for model analysis are provided after loop refinement is done.
The output resulting from MD simulations comes in a downloadable bundled compressed file (MD_output.tgz). This contains a pymol script (load_gpcr.pml) to load the trajectory in pymol (used to produce Figure 3C) and two folders: (i) logs folder, with detailed Gromacs output from each equilibration step and (ii) reports folder containing plots in Xmgrace formatted xvg files for the total energy (E), volume (V), temperature (T) and pressure (P) of the system, see Figure 1. Importantly, all files needed to continue a production run using GROMACS are provided along with a README.md text file, with instructions on how to proceed in that case.
Performance
The performance of the different modeling protocols for GPCRs has been evaluated in the several editions of the Critical Assessment of GPCR Structure Modeling and Docking (GPCR-Dock) competitions since 2009. Here, in a similar fashion as in the Critical Assessment os Protein Structure Prediction (CASP) contest, researchers were asked to submit computer-generated models of a query GPCR-ligand complex, prior to the release of the corresponding crystal structure. The different modeling procedures are evaluated for their ability to recreate the structure of the target receptor as well as their prediction of the native contacts with the co-crystallized ligand. In our recent participation in the GPCR-Dock2013 competition, we tested several combinations of modeling, docking and MD refinement protocols using GPCR-ModSim (16). Our best model ranked #26 out of a total of 175 submissions, which is a good result overall and the best for a fully automated web server in the competition. It should be noted that the choice of the (external) docking technique selected affects the accuracy of some of the parameters evaluated in this competition (Ligand RMSD -Root Mean Square Deviation-, % correct receptor-ligand contacts). Thus, more specific parameters referred solely to the protein modeling are the pocket prediction accuracy, which in our case (77%) was as good as that registered for the wining model, or the overall TM backbone RMSD, which was also comparable to the top prediction (1.9 versus 1.8 Å). For a complete report on the results of the GPCR dock 2013 competition see (http://ablab.ucsd.edu/GPCRDock2013/5HT1B.html), and additional information analyzing the performance of our submission is illustrated in the Supplementary Data.
Example
An example that illustrates the capacity of both single template and multiple-template homology modeling can be extracted from our recent research on modeling the human angiotensin II receptor type 2 (AT2) (37). The native Uniprot sequence for this receptor (P50052) was downloaded and the long N-terminus (1-32) and C-terminus (336-360) were removed because of the lack of templates for these regions among the crystallized GPCRs. The edited sequence was uploaded to GPCR-ModSim and aligned against the set of inactive (default) templates. An initial set of 10 homology models was produced with the default template, the AT1 receptor (PDB code 4YAY, sequence overall/TM identity is 31/42%, respectively). One of the potential limitations of single-template homology modeling is that the structure of the resulting model can be too similar to the template selected. This problem can be counterbalanced with the choice of additional templates. Based on the provided matrix of partial similarities, we generated a second model using a number of additional templates per topological region as indicated in Figure 2 (a complete MSA with selected templates for each region is provided in the Supplementary Material). The resulting model is an adaptation to that recently reported by us, which could be used to explain the selectivity of AT2-specific ligands as compared to the crystallized AT1 receptor (37).
To illustrate the power of the MD refinement section, we selected the structurally well-characterized A2A adenosine receptor. Here, the PDB structure with the antagonist caffeine can be downloaded from the PDB (PDB code 3RFM). After some minimal editing of the file, we retained the coordinates of the protein, and the resulting file is uploaded to the server. The coordinates of the co-crystallized ligand caffeine were extracted to a separate PDB file, and used to generate the corresponding forcefield parameters in the Gromacs format (parameters are included in the Tutorial section of GPCR-ModSim). An overview of the results from the MD protocol is depicted in Figure 3, and a sample video can be seen in our YouTube channel (see below). A comparison of the results obtained with the new MD refinement and the previous equilibration protocol is provided in the Supplementary Data for four different GPCRs. It can be appreciated that the distance restraints allow the backbone structure to relax from the initial modeled configuration, while reaching equilibrium within the equilibration time.
Documentation
A detailed tutorial for the homology modeling protocol, the MD simulation (including co-factors) and some illustrative videos available at our YouTube channel, is accessible through our website (http://open.gpcr-modsim.org/tutorials/).
CONCLUSION
GPCR-ModSim is a webserver specifically designed for automated modeling and MD simulation of GPCRs. It is the only online resource that provides both an adapted homology modeling protocol for this superfamily of receptors and a molecular dynamics workflow for atomistic simulations of a GPCR in a lipidic membrane. The last module, i.e. the PyMemDyn module, can be used not only for the refinement of in situ generated models but also accepts as input external structures uploaded to the server. The capacity of GPCR-ModSim to generate high quality models of GPCRs was evaluated in the last edition of the GPCR-dock 2013 competition: GPCR-ModSim was the best fully automated method and ranked within the top quartile of all submissions, nevertheless requiring the minimum human intervention from all methods submitted. In this manuscript, we describe an updated version of GRCR-ModSim that includes three main developments: (i) The target receptor can be modeled in any of the three conformational states structurally characterized (inactive, partially-active, fully active), thanks to a careful update and structural alignment of available templates; (ii) A novel multiple-template-based homology modeling protocol, which in contrast to existing GPCR modeling servers gives the user the freedom to choose how to compose chimeric templates; (iii) MD refinement now allows the receptor structure to undergo a final relaxation phase where a GPCR-conserved network of interhelical contacts is maintained through a series of pair-distance restraints, while the rest of the receptor is completely free. The overall modeling process will thus benefit from a combination of sequence derived (homology-modeling) and biophysical information (MD relaxation) to generate more accurate models for receptors with low sequence identity to available templates.
Supplementary Material
Acknowledgments
We are grateful to all members of the GLISTEN Cost action (CM1207) for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Carl Trygger Fundation [CTS 13:163, CTS KF15: 10]; Swedish strategic research programme eSSENCE. Funding for open access charge: Carl Trygger Fundation [CTS 13:163, CTS KF15: 10].
Conflict of interest statement. None declared.
REFERENCES
- 1.Fredriksson R., Lagerström M.C., Lundin L.-G., Schiöth H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 2.Overington J.P., Al-Lazikani B., Hopkins A.L. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 3.Rosenbaum D.M., Cherezov V., Hanson M.A., Rasmussen S.G., Thian F.S., Kobilka T.S., Choi H.-J., Yao X.-J., Weis W.I., Stevens R.C., et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 4.Warne T., Serrano-Vega M.J., Baker J.G., Moukhametzianov R., Edwards P.C., Henderson R., Leslie A.G., Tate C.G., Schertler G.F. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Latek D., Pasznik P., Carlomagno T., Filipek S. Towards improved quality of GPCR models by usage of multiple templates and profile-profile comparison. PloS One. 2013;8:e56742. doi: 10.1371/journal.pone.0056742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Worth C.L., Kreuchwig A., Kleinau G., Krause G. GPCR-SSFE: a comprehensive database of G-protein-coupled receptor template predictions and homology models. BMC Bioinformatics. 2011;12:185–195. doi: 10.1186/1471-2105-12-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandal M., Duy T.P., Cona M., Zung H., Carloni P., Musiani F., Giorgetti A. GOMoDo: a GPCRs online modeling and docking webserver. PLoS One. 2013;8:e74092. doi: 10.1371/journal.pone.0074092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez D., Bello X., Gutierrez-de-Teran H. Molecular modelling of G protein-coupled receptors through the web. Mol. Inform. 2012;31:334–341. doi: 10.1002/minf.201100162. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J., Yang J., Jang R., Zhang Y. GPCR-I-TASSER: a hybrid approach to G protein-coupled receptor structure modeling and the application to the human genome. Structure. 2015;23:1538–1549. doi: 10.1016/j.str.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutierrez-de-Teran H., Bello X., Rodriguez D. Characterization of the dynamic events of GPCRs by automated computational simulations. Biochem. Soc. Trans. 2013;41:205–212. doi: 10.1042/BST20120287. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez D., Ranganathan A., Carlsson J. Strategies for improved modeling of GPCR-drug complexes: blind predictions of serotonin receptors bound to ergotamine. J. Chem. Inf. Model. 2014;54:2004–2021. doi: 10.1021/ci5002235. [DOI] [PubMed] [Google Scholar]
- 12.Pruitt M.M., Lamm M.H., Coffman C.R. Molecular dynamics simulations on the Tre1 G protein-coupled receptor: exploring the role of the arginine of the NRY motif in Tre1 structure. BMC Struct. Biol. 2013;13:1–14. doi: 10.1186/1472-6807-13-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavasotto C.N., Palomba D. Expanding the horizons of G protein-coupled receptor structure-based ligand discovery and optimization using homology models. Chem. Commun. 2015;51:13576–13594. doi: 10.1039/c5cc05050b. [DOI] [PubMed] [Google Scholar]
- 14.Kowalsman N., Niv M.Y. G Protein-Coupled Receptors-Modeling and Simulation. Nederlands: Springer; 2014. GPCR and Company: databases and servers for GPCRs and interacting partners; pp. 185–204. [DOI] [PubMed] [Google Scholar]
- 15.Isberg V., Mordalski S., Munk C., Rataj K., Harpsoe K., Hauser A.S., Vroling B., Bojarski A.J., Vriend G., Gloriam D.E. GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res. 2016;44:D356–D364. doi: 10.1093/nar/gkv1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kufareva I., Katritch V., Stevens R.C., Abagyan R.others. Advances in GPCR modeling evaluated by the GPCR dock 2013 assessment: meeting new challenges. Structure. 2014;22:1120–1139. doi: 10.1016/j.str.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutierrez-de-Teran H., Massink A., Rodriguez D., Liu W., Han G.W., Joseph J.S., Katritch I., Heitman L.H., Xia L., IJzerman A.P., et al. The role of a sodium ion binding site in the allosteric modulation of the A2A adenosine G protein-coupled receptor. Structure. 2013;21:2175–2185. doi: 10.1016/j.str.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venkatakrishnan A.J., Deupi X., Lebon G., Tate C.G., Schertler G.F., Babu M.M. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 19.Raval A., Piana S., Eastwood M.P., Shaw D.E. Assessment of the utility of contact-based restraints in accelerating the prediction of protein structure using molecular dynamics simulations. Protein Sci. 2016;25:19–29. doi: 10.1002/pro.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 21.Webb B., Sali A. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics. 2014;47:5–6. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- 22.Lovell S.C., Davis I.W., Arendall W.B., de Bakker P.I., Word J.M., Prisant M.G., Richardson J.S., Richardson D.C. Structure validation by C alpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 23.Pronk S., Pall S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M.R., Smith J.C., Kasson P.M., van der Spoel D., et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell R.B., Barton G.J. Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins. 1992;14:309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- 25.Cock P.J., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2 a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanson R.M., Prilusky J., Renjian Z., Nakane T., Sussman J.L. JSmol and the next-generation web-based representation of 3D molecular structure as applied to proteopedia. Isr. J. Chem. 2013;53:207–216. [Google Scholar]
- 28.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fiser A., Do R.K.G., Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez D., Pineiro A., Gutierrez-de-Teran H. Molecular dynamics simulations reveal insights into key structural elements of adenosine receptors. Biochemistry. 2011;50:4194–4208. doi: 10.1021/bi200100t. [DOI] [PubMed] [Google Scholar]
- 31.Knight C.J., Hub J.S. MemGen: a general web server for the setup of lipid membrane simulation systems. Bioinformatics. 2015;31:2897–2899. doi: 10.1093/bioinformatics/btv292. [DOI] [PubMed] [Google Scholar]
- 32.Ghahremanpour M.M., Arab S.S., Aghazadeh S.B., Zhang J., van der Spoel D. MemBuilder: a web-based graphical interface to build heterogeneously mixed membrane bilayers for the GROMACS biomolecular simulation program. Bioinformatics. 2014;30:439–441. doi: 10.1093/bioinformatics/btt680. [DOI] [PubMed] [Google Scholar]
- 33.Stansfeld P.J., Goose J.E., Caffrey M., Carpenter E.P., Parker J.L., Newstead S., Sansom M.S. MemProtMD: automated insertion of membrane protein structures into explicit lipid membranes. Structure. 2015;23:1350–1361. doi: 10.1016/j.str.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu E.L., Cheng X., Jo S., Rui H., Song K.C., Dávila-Contreras E.M., Qi Y., Lee J., Monje-Galvan V., Venable R.M., et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tropp J. Dipolar relaxation and nuclear Overhauser effects in nonrigid molecules: the effect of fluctuating internuclear distances. J. Chem. Phys. 1980;72:6035–6043. [Google Scholar]
- 36.Ballesteros J.A., Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Recept. Mol. Biol. 1995;25:366–428. [Google Scholar]
- 37.Sallander J., Wallinder C., Hallberg A., Aqvist J., Gutierrez-de-Teran H. Structural determinants of subtype selectivity and functional activity of angiotensin II receptors. Bioorg. Med. Chem. Lett. 2016;26:1355–1359. doi: 10.1016/j.bmcl.2015.10.084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.