Abstract
Heatmapper is a freely available web server that allows users to interactively visualize their data in the form of heat maps through an easy-to-use graphical interface. Unlike existing non-commercial heat map packages, which either lack graphical interfaces or are specialized for only one or two kinds of heat maps, Heatmapper is a versatile tool that allows users to easily create a wide variety of heat maps for many different data types and applications. More specifically, Heatmapper allows users to generate, cluster and visualize: (i) expression-based heat maps from transcriptomic, proteomic and metabolomic experiments; (ii) pairwise distance maps; (iii) correlation maps; (iv) image overlay heat maps; (v) latitude and longitude heat maps and (vi) geopolitical (choropleth) heat maps. Heatmapper offers a number of simple and intuitive customization options for facile adjustments to each heat map's appearance and plotting parameters. Heatmapper also allows users to interactively explore their numeric data values by hovering their cursor over each heat map cell, or by using a searchable/sortable data table view. Heat map data can be easily uploaded to Heatmapper in text, Excel or tab delimited formatted tables and the resulting heat map images can be easily downloaded in common formats including PNG, JPG and PDF. Heatmapper is designed to appeal to a wide range of users, including molecular biologists, structural biologists, microbiologists, epidemiologists, environmental scientists, agriculture/forestry scientists, fish and wildlife biologists, climatologists, geologists, educators and students. Heatmapper is available at http://www.heatmapper.ca.
INTRODUCTION
Thanks to rapid developments in data rendering and visualization software, heat maps have become increasingly popular routes to display information-rich data in both two and three dimensions. The concept of a heat map appears to have emerged from the widespread use of pseudo-coloured surface temperature maps (generated via infrared thermography) that became available in the 1970s and 1980s. In thermography, infrared or thermal imaging cameras are used to detect heat variations over the surface of an object. These kinds of image-based thermal heat maps actually first emerged in the 1950s (1) and have since become widely used in various fields ranging from medical imaging, weather monitoring, building inspection and materials engineering. The extension of the thermal heat map concept to assist with visualizing non-thermal data occurred in 1991 when the actual term ‘heatmaps’ was coined and later trademarked (2). Indeed, the first non-thermal or ‘data-matrix’ heat maps were used to depict real time financial market information. Since then data-matrix heat maps have been used in a wide variety of applications from molecular biology (gene and protein expression mapping), to structural biology (pairwise distance measurements), to epidemiology (disease outbreak monitoring), to environmental science (ocean salinity mapping), to climatology (mapping rainfall levels), to urban planning (traffic density mapping) and even to politics (showing regional voter preferences).
Broadly speaking heat maps fall into two classes: (i) image-based heat maps and (ii) data-matrix heat maps. Image-based heat maps display numerical information that is mapped over an image, an object or a geographic location. Examples of image-based heat maps include choropleth heat maps (3), geospatial heat maps, webpage ‘eye-scan’ heat maps and anatomical heat maps. On the other hand, data-matrix heat maps display numerical data in a pseudo-coloured tabular or matrix format. The data may be subsequently clustered using various measures of similarity or dissimilarity. Examples of data-matrix heat maps include gene/protein/metabolite expression heat maps, correlation heat maps, taxonomic heat maps and pairwise distance heat maps.
Over the past decade a number of dedicated heat mapping packages have emerged. Many are available as commercial, downloadable, stand-alone packages. Some examples include Tableau Software, Heatmap.me, Qlucore, Crazy Egg and MapBox. The vast majority of these products are quite specialized in that they are intended only for eye-scan applications (Crazy Egg, Heatmap.me), only for gene expression analysis (Qlucore) or only for choropleth mapping (MapBox). In addition to these commercial products, there is a growing number of freely available heat mapping packages. The level of detail, the type of platform and the computational requirements for these programs varies greatly depending on the type of heat map that is being generated. For instance, there is an abundance of comprehensive heat mapping software dedicated to displaying gene expression data. These include CIMminer (4), HeatmapGenerator (5), TM4 (6), GenePattern (7), MeV (8), FTreeView (9), Gitools (10) GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E/). These programs support a range of clustering options, have many customizable features and in the majority of cases need to be downloaded and run locally. Because of the heavy computational requirements, we are aware of only a couple of web-based tools that support interactive online heat mapping and clustering of expression data, namely MetaboAnalyst (11), COLOMBOS (12), GenePattern (7) and MicroScope (Khomtchouk et al. (http://dx.doi.org/10.1101/034694)).
Other kinds of freely available data-matrix heat mapping tools also exist. For instance, LDHeatmap generates linkage disequilibrium heat maps (13) while MatrixPlot (14) generates difference distance plots or heat maps from protein coordinate data. In contrast to data-matrix heat mapping tools, software for image-based heat maps such as those with latitude and longitude data or choropleth (geopolitical) data often have very few customizable features. As a result, several can now be found as web servers. OpenHeatMap (http://www.openheatmap.com) is one example, as is the Google Heatmap Gadget (https://code.google.com/p/google-gadget-heatmap/).
Based on our survey of existing, freely accessible heat mapping tools, we found that there are very few packages that operate as web servers, essentially none that support more than one very specialized type of heat map (expression only, choropleth only, linkage disequilibrium only, etc.) and none that allow heat map customization over an uploaded image. We also found that many of the downloadable heat mapping packages were difficult to install, hard to use and not particularly user friendly – especially for non-specialists or those unfamiliar with R. Because of recent advances in web technologies and because of the lack of a ‘general’ heat mapping tool for a wide variety of data visualization applications, we decided to create an easy-to-use, online heat map web server called Heatmapper.
Heatmapper is designed to be easily used and easily understood by both specialists and non-specialists, alike. Through its intuitive graphical user interface and a large number of example files, Heatmapper allows users to quickly create a wide variety of heat maps for many different data types. These include: (i) pairwise distance maps; (ii) correlation matrices; (iii) image overlay heat maps; (iv) latitude and longitude heat maps; (v) geopolitical (choropleth) heat maps and (vi) expression-based heat maps from transcriptomic, proteomic and metabolomic experiments. Heatmapper offers a number of simple and intuitive customization options for facile adjustments to each heat map's appearance and interactive querying of each heat map's numerical data content. Heatmapper is designed to appeal to a wide range of users, including molecular biologists, structural biologists, microbiologists, epidemiologists, environmental scientists, agriculture/forestry scientists, fish and wildlife biologists, climatologists, geologists and even social/political scientists. Additional details about Heatmapper's overall design, its features and its implementation are provided below.
GENERAL DESIGN
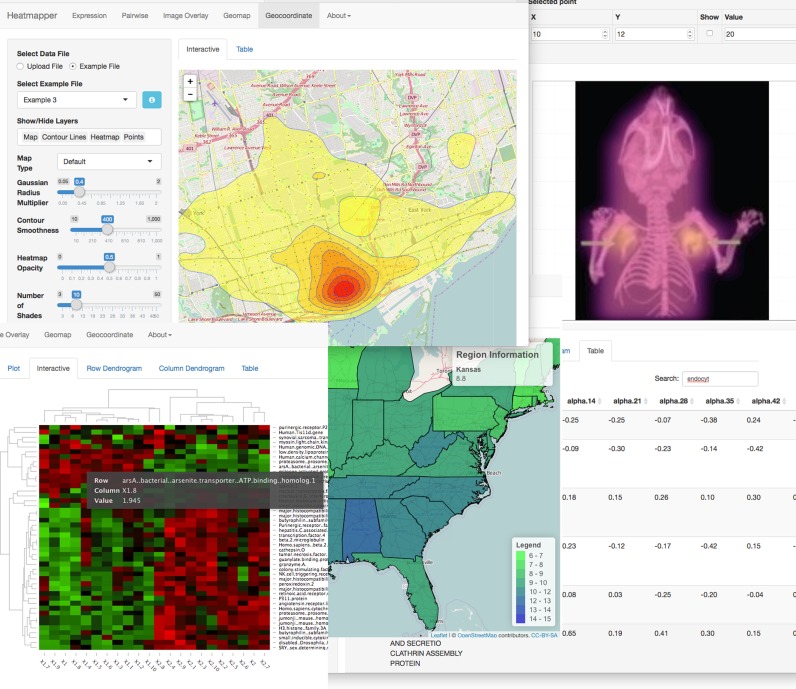
Heatmapper supports five different classes or types of heat maps. These include both image-based heat maps and data-matrix heat maps. A screen shot montage of the different kinds of heat maps generated by Heatmapper as well as some of its graphical user interface is shown in Figure 1. There are two types of data-matrix heat maps supported by Heatmapper: (i) Expression Heat Maps and (ii) Pairwise Comparison Heat Maps. Expression Maps are most useful for those doing gene, protein and/or metabolite expression analysis and appear primarily to molecular biologists, bioinformaticicans and other ‘omics’ specialists. Pairwise Comparison Maps are particularly useful for those interested in mapping geometric distances, linkage disequilibrium distances, taxonomic distances, or performing correlation mapping. These types of heat maps are most frequently used by structural biologists, taxonomists, geneticists or statisticians. Heatmapper also offers three kinds of image-based heat maps: (i) Image Overlay Heat Maps; (ii) Geomap Heat Maps and (iii) Geodensity Heat Maps. Image Overlay Maps are most useful for overlaying heat map data on custom images, such as cell, plant, animal or anatomical images, as well as satellite or aerial images. These ‘custom’ maps would be most useful to cell biologists, microbiologists, anatomists and environmental scientists. Geomap Heat Maps (or choropleth heat maps) are designed to support data mapping onto geopolitical images that depict countries, states or counties. These kinds of choropleth heat maps are particularly useful for epidemiologists, environmental scientists and social scientists. Geodensity Heat Maps are primarily used for mapping data on geospatial coordinates (latitude and longitude) corresponding to satellite images or global maps. These kinds of heat maps are often used by epidemiologists as well as ocean, weather and climate scientists.
Figure 1.
Screen shot montage of the Heatmapper web user interface.
Each of Heatmapper's five heat map types share the same general page layout, with a sidebar on left side of the page and a main viewing panel containing the heat map image (or other viewable options) to the right of it. Each sidebar is customized to the specific type of heat map being analyzed with a specific selection of widgets, such as sliders and dropdown menus, for customizing the heat map. All five heat map types offer example files, colour customization options and a variety of data export options. All the example files were obtained from papers or online data files relevant to the specific type of heat map being viewed/analyzed. Details regarding each example data file's context and provenance can be shown or hidden by clicking an ‘Information’ button. Colour customizations use a widget called jscolourR to allow different colours to be easily selected. For each heat map type a specific colour for low values and a specific colour for high values can be selected. Users can also customize the number of shades between these colour extremes to create a custom colour gradient. Heat map images can be exported in several different formats depending on the type of heat map being analyzed. Minimally, all heat maps can be exported as .png or .pdf files. Each heat map viewing panel includes a collapsible ‘Advanced Options’ menu that lists additional image manipulation features that are less commonly used. This keeps the sidebar simple and easy to navigate without limiting functionality.
The main viewing panel for each heat map type contains a number of different viewing options that can be accessed by tabs. The Plot viewing option contains a static image of the heat map. Generally this is the image that can be exported by the user. The Interactive viewing option contains an interactive image of the heat map. This means it has additional features that allow the user to directly interact with the plot, such as zooming. The Table viewing option displays the input data in the form of a table, which the user has the option to download or, in some cases, edit right in their browser window. Other viewing options that are specific to certain heat map types will be described in more detail below.
Expression heat maps
The Expression Heat Map option displays unclustered (or previously clustered) data from transcriptomic (microarray or RNAseq), proteomic or metabolomic experiments in the form of a heat map. Input expression data can be uploaded in several formats including .txt, .dat, .tsv, .tab. or .xlsx. These file formats are typical of the gene/protein/metabolite expression files generated by most kinds of modern expression analysis tools. The data input must contain at least two columns and two or more (several thousand can be handled) rows with numeric data. If a column labeled ‘NAME’ exists it will be used to assign row names. Once the data file is uploaded, hierarchical clustering can also be performed and the resulting dendrogram can be viewed in Heatmapper's main viewer window. There are two additional viewing options specific to Expression Heat Maps: Row Dendrogram and Column Dendrogram. These viewing options allow users to display the row or column names and generate a dendrogram of the data when clustering is applied.
To perform the hierarchical clustering, Expression Heat Map offers users the choice of five distance measurement methods and four clustering methods. The distance measurement methods include Euclidean, Pearson, Kendall's tau, Spearman Rank correlation and Manhattan methods. The clustering methods offered are average linkage, centroid linkage, complete linkage and single linkage. Clustering can be applied to rows and/or columns. Users may also upload their own clustering files in Newick tree format, if desired. The user can further customize the heat map colours for high, low, middle and missing expression levels. Advanced options for customizing the Expression Heat Map include the ability to adjust the size of the plot, to add custom labels for each axis and to add a title.
Pairwise comparison heat maps
The Pairwise Comparison Heat Map option allows users to generate two related heat map types; (i) Pairwise distance heat maps (or matrices) and (ii) Pairwise correlation heat maps. Pairwise distance heat maps are used to compare atomic distances within molecules, between points in space or between geographic locations on a map. In structural biology pairwise distance maps have long been used to compare structures, to identify secondary structure elements and even to predict protein structure (14–16). Input coordinate data can be uploaded in several formats including .txt, .dat, .tsv, .tab., .csv, .xlsx or .pdb. Coordinates may be absolute coordinates in two dimensions (x,y), three dimensions (x,y,z) or even higher dimensions. In particular, if one provides a file with rows each with n data values, the rows will represent points in n-dimensional space. The pairwise distance program treats each row of data as a spatial coordinate, calculates distances between all pairs of coordinates and displays the matrix of distances for each pair of points as a coloured heat map. By displaying the data as a colour-coded heat map, interrelations within the data can be quickly and intuitively grasped. An added option allows structural biologists to upload a Protein Data Bank (PDB) file instead of a simple text or .csv file. When the PDB option is chosen, the 3D coordinates of the atoms are automatically parsed. When a PDB file is uploaded, users may choose to generate the resulting heat map using only C-α atoms, only backbone atoms or all atoms.
In addition to the pairwise distance heat map, we offer two other pairwise comparison options: (i) pairwise correlation heat map and (ii) alignment-free sequence comparison. Pairwise correlation heat maps permits researchers to discover variables that are positively or negatively correlated, and the use of the visualization tools in Heatmapper allows the features to be intuitively and quickly seen. When the pairwise correlation option is selected, input tables can be uploaded in several formats including .txt, .dat, .tsv, .tab and .csv. Each data column is treated as a variable that varies across a set of samples or conditions. Pearson correlation coefficients are calculated between each pair of variables (columns), resulting in a matrix of pairwise correlation coefficients that are displayed in a coloured heat map format that the user can customize to their liking. Users may quickly switch between the distance matrix and correlation options as is appropriate to their data. Another option allows users to upload a set of DNA sequences in FASTA format, from which k-mer frequencies are computed and alignment-free sequence comparison is performed. Finally, users may select to simply view their data ‘as-is.’ With this last option, no calculations are performed, allowing users to explore their own data through the aid of a heat map.
To facilitate viewing, different ‘layers’ of each heat map plot can be shown or hidden; including the legend and axis labels. One option provided for convenience allows the data in the heat map's vertical axis to be displayed in reverse order, depending on the user's desired plot arrangement. The aspect ratio of the plot can also be set. Advanced options for customizing the plot include adjusting the size of the plot, adding custom labels for each axis and adding a title.
Image overlay heat maps
Image Overlay Heat Maps allow users to overlay quantitative data on custom images, such as cell, plant, animal or anatomical images. These types of heat maps can also be used to overlay quantitative data on satellite or aerial images. To use this heat map option users must first upload an image of interest (in .jpg, .png or .tif format). Once the image is uploaded users can either choose to upload a coordinate grid (in .txt, .dat, .tsv, .csv, .tab or .xlsx format) containing x,y values and a set of corresponding ‘temperature’ values or they can choose not to do so. Once a set of grid values is uploaded, or even if no grid is uploaded, users are free to manually edit or add temperature values using a simple user interface. The user-adjusted temperature values are immediately reflected in the updated heat map. The colours can be customized using the same methods as with the other heat maps. The heat map or ‘temperature’ information can be displayed as simple squares or as Gaussian ‘blobs’ (which are customizable). If the square grid option is chosen, each value is displayed as-is, with no neighbourhood smoothing. If the Gaussian option is chosen, each value is smoothed using a Gaussian function incorporating a weighted density of neighbourhood values. A Gaussian radius multiplier can also be set, which refers to the width used in the density estimation. In addition, the contour smoothness can be set, which adjusts the number of line segments used to draw the contour lines. Layers of the plot can be shown or hidden; including the background image, the heat map data, the heat map contour lines, the colour boundaries, the grid lines corresponding to the x and y coordinates, and the axis labels. The opacity of the overlaid ‘temperature’ data can also be adjusted. Advanced options for customizing the heat map include stretching the background image to fit the plot size, setting the number of grid points for creating a heat map without uploading a grid file and adjusting the size of the plot.
Geomap heat maps
Geomap Heat Maps allow users to map quantitative data onto geopolitical images that depict countries, states, provinces or counties. These are often called choropleth maps or choropleth heat maps. Users must upload a file (in .txt, .dat, .tsv, .tab, .csv, or .xlsx format) that contains at least two columns with region names in the first column and numeric values in the second. Note that more than one column of numeric values can be included. The second column is selected by default to generate the ‘temperature’ data for heat map. The selected column can be changed using a drop-down menu, which references different columns by the corresponding entry in the header row. This allows the user to easily switch between different data sets without needing to upload a different file. Once the data table is loaded, the user can select a map of a defined area that is saved on the server to match with their data. There are currently over 40 maps to choose from the pull down menu. Additional choropleth maps will be added based on availability and user requests.
The Geomap viewer has an additional viewing option called Region Names. This viewer contains a list of the region names (provinces, states, counties, etc.) of the selected map. This allows the user to have a better idea which names they are required to use in the data table they upload. Region Names in user uploaded files must exactly match the region names for the selected map. To handle any potential problems that this precise name matching may cause, the Table subsection has been changed slightly in order to allow the user to modify their data interactively. All Region Names specified in the uploaded file are shown in the Table viewer, with mismatched names highlighted in red. The user can then change the name manually to match one of the accepted Region Names from a drop-down menu. Once the user clicks ‘Submit’ the Region Name data is updated.
As with most other heat maps in Heatmapper, a numeric range of interest can be specified to change the heat map colour assignment. Various layers of the heat map can be shown or hidden; including the background map, the contour lines representing political boundaries and the heat map itself. The opacity of the heat map can also be set. Advanced options for customizing the plot include adding a custom legend title and adjusting the width of the contour lines.
Geodensity heat maps
Geodensity Heat Maps are primarily used for mapping data on geospatial coordinates (latitude and longitude). These kinds of heat maps are typically generated using satellite images, as well as geological, oceanographic or atlas-derived maps. To use this heat mapping option users must upload a file (in .txt, .dat, .tsv, .tab, .csv or .xlsx format) that contains at least two columns with valid latitude and longitude coordinates. An optional third column containing values can be included for calculating a weighted density estimation instead of basing the estimation on the density of points. At least one value must be strictly > 0 if the value column is used. The resulting heat map is typically a multi-coloured ‘blob’ map expressing different colour densities. Density estimation calculations can be performed based on point densities or weighted by values specified for each coordinate. As with other heat maps, different plot layers can be shown or hidden; including the background map, the heat map itself, the points as defined in the input file, and the contour lines. Selecting one of four styles from a drop-down menu also allows users to set the map background style. Just as with the Image Overlay Heat Map described earlier, the Gaussian radius multiplier and contour smoothness can be set to suit different user preferences. Advanced options for customizing Geodensity heat maps include the ability to adjust the width of the contour lines, the point width and the point opacity.
COMPARATIVE FEATURE EVALUATION
Tables 1 and 2 compare different features and capabilities in Heatmapper to a number of existing (freeware) heat mapping packages. Table 1 compares the features and capabilities found in Heatmapper to a wide variety of popular heat mapping tools with different functionalities. As seen in this table, Heatmapper offers considerably more features and capabilities than any other tool. Table 2 compares Heatmapper to expression-only heat mapping packages. Some of the tools mentioned in this table include Heatmap Builder (17), Maple Tree (http://mapletree.sourceforge.net), Cluster 3.0 (18), Microarray Explorer or MAExplorer (19), MeV (8) and FTreeView (9). Again, the same trend is seen. While Heatmapper is quite unique in terms of its broad capabilities, interactivity and web accessibility, it tends to be a bit slower than most stand-alone programs. Furthermore, some of the advanced data processing or data visualization options found in the more specialized heat mapping programs are not found in Heatmapper.
Table 1. Comparison of Heatmapper to other freely available heat map tools.
| Feature | Heatmapper | OpenHeat Map | CIMminer | Heatmap Generator | Gitools | Gene Pattern |
|---|---|---|---|---|---|---|
| Expression, clustered heat maps | Yes | No | Yes | Yes | Yes | Yes |
| Distance matrix heat maps | Yes | No | No | No | No | No |
| Correlation matrix heat maps | Yes | No | No | No | Yes | No |
| Image overlay heat maps | Yes | No | No | No | No | No |
| Choropleth heat maps | Yes | Yes | No | No | No | No |
| longitude/latitude heat maps | Yes | Yes | No | No | No | No |
| Graphical user interface | Yes | Yes | Minimal | Yes | Yes | Yes |
| Web interface | Yes | Yes | Yes | No | No | Difficult |
| Installation requirements | None required | None required | None required | Installation required, unreliable | Installation required | None required |
| Initial heat map generation | Instantly | Submission form to receive email | Submission form to receive email | Instantly | Instantly | In several seconds |
| Interactive data editing | Yes | Yes | No | No | Yes | No |
Table 2. Comparison of Heatmapper to existing expression/microarray heat mapping tools.
| Heatmap Builder | Maple Tree | Functional Treeview (FTree-View) | Cluster 3.0 | Micro-Array Explorer (MAEx-plorer) | MultiEx-periment Viewer (MeV) | Metabo-Analyst | Heat-mapper | |
|---|---|---|---|---|---|---|---|---|
| Online version available | No | No | Yes (authori-zation required) | No | No | No | Yes | Yes |
| Able to display data as heatmap | Yes | Yes | Yes | No | No | Yes | Yes | Yes |
| Able to perform clustering | No | No | No | Yes | Yes | Yes | Yes | Yes |
| Major System Restrictions | Windows XP and later only | Requires Java | Requires Java | None | None | Requires Java | None | None |
| Supported Input File Types | txt (tab separated) | cdt | cdt, pcl | txt (tab separated) | mae | mev, tav, gpr, txt (tab separated) | csv, txt (tab separated) | txt, tab, FASTA, pdb, dat, tsv, csv, xlsx, Newick tree |
| Functional Annotation | No | No | Yes | No | Yes | No | Yes | No |
| Multi-variate Analysis | No | No | No | No | Yes | Yes | Yes | No |
IMPLEMENTATION
Heatmapper was built using Shiny (version 0.12.2), a framework for R (version 3.2.0). Tooltips are displayed using the shinyBS package. The custom colour selection widgets were generated using the jscolourR package. The spinner displayed on the screen when a page is loading is generated by the spin package. All of Heatmapper's heat map plots are generated using the d3heatmap, ggplot2 (20) and gplot packages. The non-geographic heat maps (such as the Expression Heat Maps, Image Overlay Heat Maps and Pairwise Comparison Heat Maps) generated by ggplot2 or gplots can be viewed using the Plot viewer option, while the Geomap and Geodensity heat maps generated by d3heatmap can be viewed using the Interactive viewer option. Dendrogram plots are generated using the ggdendro package. Colour schemes are generated using the RColorBrewer package. For geographic heat maps, such as Geomap and Geodensity, the plots are generated using the leaflet package. The jpeg, png, tiff and xlsx packages are used for importing data in different formats. The raster and reshape2 (21) packages are used to manipulate the imported data. Density estimation calculations are performed using the MASS (22), ggtern and grid packages.
Heatmapper is hosted on a dedicated server with 2 Intel Xeon E5-2630L v2 CPUs @ 2.40GHz and a solid-state drive, running Shiny Server. The Heatmapper website is compatible with all modern web browsers, including Google Chrome, Mozilla Firefox, Internet Explorer and Safari. Heatmapper can also be downloaded and run locally on any computer with R installed. Installation instructions and source code can be found at https://github.com/sbabicki/heatmapper.
CONCLUSION AND FUTURE PLANS
Heat maps have become ubiquitous in many fields of science – from molecular biology, to structural biology, to epidemiology, to climatology. They are colourful, information rich vehicles that are ideal for visualizing complex data. They are also particularly useful for quickly detecting patterns and trends. Despite their widespread appeal and their apparent ubiquity, it is still quite challenging to generate heat maps. In most cases, specialized software must be found, bought, downloaded and installed. Often unexpected platform and operating system incompatibilities must be solved or corrected. Furthermore, different heat maps require different packages, each of which has their own (often steep) learning curve and their own unique installation issues. To address these challenges we developed Heatmapper. By consolidating many common heat mapping functions into a single, freely accessible web server we believe we have helped address the accessibility and compatibility issues that still plague the heat mapping world. By developing a common, simplified, graphical interface for all of Heatmapper's plotting functions, we believe we have also helped address some of the learnability and usability issues associated with many heat mapping packages. In addition to these improvements, Heatmapper also offers some novel heat mapping techniques not found in existing tools. For example, the Image Overlay option appears to be a unique heat mapping function which could offer heat mapping options to many other fields of science. Likewise, the high level of customization available through Heatmapper makes it ideal for handling unstructured or unconventional data. In this regard we believe Heatmapper should make heat mapping much easier, much faster and much more appealing to a much wider community of scientists, educators and students. Certainly the current version of Heatmapper is not the final word on web-enabled heat mapping. In future releases of Heatmapper we expect to be adding more functionality with regard to upstream and downstream data processing. Continued efforts will also be made to optimize Heatmapper's speed, file size capacity and rendering capabilities.
Acknowledgments
The authors wish to thank the Canadian Institutes of Health Research (CIHR) and Genome Alberta, a division of Genome Canada, for financial support.
FUNDING
Canadian Institutes of Health Research (CIHR); Genome Alberta, a division of Genome Canada. Funding for open access charge: Canadian Institutes of Health Research (CIHR).
Conflict of interest statement. None declared.
REFERENCES
- 1.Lawson R. Implications of surface temperatures in the diagnosis of breast cancer. Can. Med. Assoc. J. 1956;75:309–310. [PMC free article] [PubMed] [Google Scholar]
- 2.United States Patent and Trademark Office, registration #75263259. 1993-09-01
- 3.Robinson A.H., Morrison J.L., Muehrke P.C., Kimmerling A.J., Guptill S.C. Elements of Cartography. 6th edn. NY: Wiley; 1995. [Google Scholar]
- 4.Weinstein J.N., Myers T., Buolamwini J., Raghavan K., van Osdol W., Licht J., Viswanadhan V.N., Kohn K.W., Rubinstein L.V., Koutsoukos A.D. Predictive statistics and artificial intelligence in the U.S. National Cancer Institute's Drug Discovery Program for Cancer and AIDS. Stem Cells. 1994;12:13–22. doi: 10.1002/stem.5530120106. [DOI] [PubMed] [Google Scholar]
- 5.Khomtchouk B.B., Van Booven D.J., Wahlestedt C. HeatmapGenerator: High performance RNAseq and microarray visualization software suite to examine differential gene expression levels using an R and C++ hybrid computational pipeline. Source Code Biol. Med. 2014;9:30. doi: 10.1186/s13029-014-0030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 7.Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 8.Chu V.T., Gottardo R., Raftery A.E., Bumgarner R.E., Yeung K.Y. MeV+R: using MeV as a graphical user interface for Bioconductor applications in microarray analysis. Genome Biol. 2008;9:R118. doi: 10.1186/gb-2008-9-7-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saldanha A.J. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 10.Perez-Llamas C., Lopez-Bigas N. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS One. 2011;6:e19541. doi: 10.1371/journal.pone.0019541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia J., Sinelnikov I.V., Han B., Wishart D.S. MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moretto M., Sonego P., Dierckxsens N., Brilli M., Bianco L., Ledezma-Tejeida D., Gama-Castro S., Galardini M., Romualdi C., Laukens K., et al. COLOMBOS v3.0: leveraging gene expression compendia for cross-species analyses. Nucleic Acids Res. 2016;44:D620–D623. doi: 10.1093/nar/gkv1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin J.H., Blay S., McNeney B., Graham J. LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Software. 2006;16:1–9. [Google Scholar]
- 14.Gorodkin J., Staerfeldt H.H., Lund O., Brunak S. MatrixPlot: visualizing sequence constraints. Bioinformatics. 1999;15:769–770. doi: 10.1093/bioinformatics/15.9.769. [DOI] [PubMed] [Google Scholar]
- 15.Richards F.M., Kundrot C.E. Identification of structural motifs from protein coordinate data: secondary structure and first-level supersecondary structure. Proteins. 1988;3:71–84. doi: 10.1002/prot.340030202. [DOI] [PubMed] [Google Scholar]
- 16.Kloczkowski A., Jernigan R.L., Wu Z., Song G., Yang L., Kolinski A., Pokarowski P. Distance matrix-based approach to protein structure prediction. J. Struct. Funct. Genomics. 2009;10:67–81. doi: 10.1007/s10969-009-9062-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King J.Y., Ferrara R., Tabibiazar R., Spin J.M., Chen M.M., Kuchinsky A., Vailaya A., Kincaid R., Tsalenko A., Deng D.X.-F., et al. Pathway analysis of coronary atherosclerosis. Physiol. Genomics. 2005;23:103–118. doi: 10.1152/physiolgenomics.00101.2005. [DOI] [PubMed] [Google Scholar]
- 18.de Hoon M.J.L., Imoto S., Nolan J., Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 19.Lemkin P.F., Thornwall G.C., Walton K.D., Hennighausen L. The microarray explorer tool for data mining of cDNA microarrays: application for the mammary gland. Nucleic Acids Res. 2000;28:4452–4459. doi: 10.1093/nar/28.22.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wickham H. ggplot2: Elegant Graphics for Data Analysis. NY: Springer; 2009. [Google Scholar]
- 21.Wickham H. Reshaping Data with the reshape Package. J. Stat. Software. 2007;21:1–20. [Google Scholar]
- 22.Venables W. N., Ripley B. D. Modern Applied Statistics with S. 4th edn. NY: Springer; 2002. [Google Scholar]