Abstract
DNA guanine quadruplexes or G4s are non-canonical DNA secondary structures which affect genomic processes like replication, transcription and recombination. G4s are computationally identified by specific nucleotide motifs which are also called putative G4 (PG4) motifs. Despite the general relevance of these structures, there is currently no tool available that can allow batch queries and genome-wide analysis of these motifs in a user-friendly interface. QuadBase2 (quadbase.igib.res.in) presents a completely reinvented web server version of previously published QuadBase database. QuadBase2 enables users to mine PG4 motifs in up to 178 eukaryotes through the EuQuad module. This module interfaces with Ensembl Compara database, to allow users mine PG4 motifs in the orthologues of genes of interest across eukaryotes. PG4 motifs can be mined across genes and their promoter sequences in 1719 prokaryotes through ProQuad module. This module includes a feature that allows genome-wide mining of PG4 motifs and their visualization as circular histograms. TetraplexFinder, the module for mining PG4 motifs in user-provided sequences is now capable of handling up to 20 MB of data. QuadBase2 is a comprehensive PG4 motif mining tool that further expands the configurations and algorithms for mining PG4 motifs in a user-friendly way.
INTRODUCTION
Guanine quadruplexes or G4s are DNA and RNA secondary structures that are formed by breaking the canonical Watson–Crick base-pairing. Two or more planar structures, each formed by four Hoogsteen base-paired guanine nucleotides, constitute the core of G4 structures (1–4). Over the last one and a half decades, these structures have been shown to have important ramifications in genomic processes including, replication (5–8), transcription (9–11), recombination (12), nucleosome exclusion (13,14) and DNA methylation (15). These structures are also known to engage transcription factors, helicases and other nuclear proteins (8,9,16,17). Studies focusing on the role of G4s in prokaryotes have shown that these structures are prevalent and conserved in the regulatory regions of prokaryotes (18) and have important functional outcomes (19,20).
An increasing number of reports providing evidences on the in vivo existence of G4s have brought forth a renewed interest in these structures (21–23). Moreover, the presence of G4s in the promoters of several oncogenes including, but not limited to, MYC, KIT and KRAS, has led to the possibility of targeting G4s as therapeutic targets in cancer therapy, through specific G4 binding ligands like quarfloxin (24–27). G4s are largely formed by specific nucleotide motifs which are also called putative G4 (PG4) motifs. Computationally mined G4 or PG4 motifs are known to associate with promoter regions in the mammalian as well as prokaryotic genomes (18). The algorithm used in the previous version of QuadBase (28) and other studies (18,29,30), essentially looks for a pattern composed of four 2–5 mers of guanine repeats, interspersed by nucleotide sequences of varying lengths.
One of the initial strategies for mining PG4 motifs was implemented in GRS-Mapper (31). This strategy essentially involves a search for the G3-5L1-xG3-5L1-xG3-5L1-xG3-5 motif in a given nucleotide sequence, where G3-5 are guanine stems and L1-x are loops (described further in the methodology section). Similar strategies were employed in other tools to analyse the prevalence of PG4 motifs in the human genome and to identify significant over-representation of loops of specific lengths and nucleotide sequences (29,30). Based on the strategy implemented in GRS-Mapper used for mining PG4 motifs, Kikin et al. (32) developed a user-friendly web server that allows mining PG4 motifs in a single sequence at a time and requires sequence-specific information such as NCBI ID for organism-specific searches. Eddy et al. (33) developed a different strategy for mining PG4 motifs called ‘G4P Calculator’, wherein guanine (‘G’) stems of defined length and density are searched in sliding windows across the sequence. This standalone software was implemented with low-throughput in mind and is suitable only for searching a small number of sequences at a time. Later, databases of PG4 motifs were developed by Yadav et al. (28) (QuadbBase) and Kikin et al. (35) (GRSdb) for DNA and mRNA sequences respectively. More recently, to evaluate the PG4 motif conservation, Menendez et al. (34) developed a web server, QGRS-H Predictor, which finds the conserved PG4 motifs in two aligned sequences.
The previous version of QuadBase (28) provided a web interface, where users could query PG4 motifs from a database of four mammalian genomes and 146 microbial genomes. The database was divided into three modules: EuQuad, which allowed querying PG4 motifs from four eukaryotes, ProQuad, which allowed querying PG4 motifs in 146 prokaryotes and TetraplexFinder, which allowed PG4 motif search in custom sequences. In this updated version, we have radically expanded the knowledge base by allowing users to mine PG4 motifs in 178 eukaryotes and 1719 prokaryotes. Since, a database can only accommodate information for few specific PG4 motif configurations; QuadBase2 was developed as a web server which does not rely on a database of mined PG4 motifs and can process any of the large number of possible combinations of stem and loop lengths on the fly. We have organized the web server into three modules, as in the previous version, taking into consideration that user-provided, eukaryotic and prokaryotic sequences need to be handled differently and enable incorporation of different functionalities as described below. Hence, QuadBase2 fulfils the demand for a user-friendly, feature rich, interactive and a high-throughput platform for mining PG4 motifs.
METHODOLOGY OF PG4 MOTIF MINING
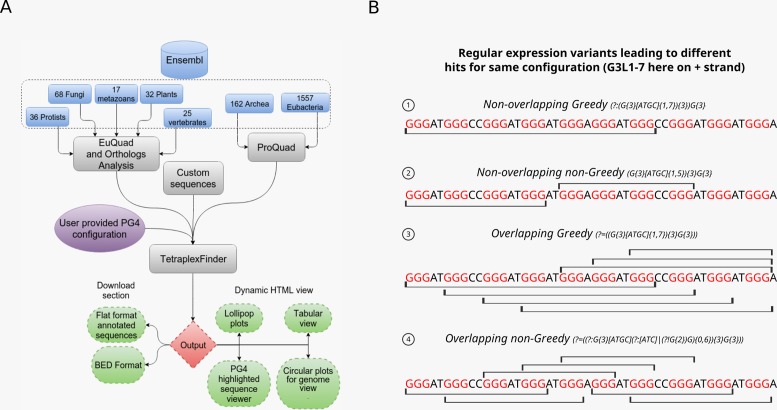
Figure 1A shows the workflow and the design of each module of the web server, which is dedicated to mining and visualization of PG4 motifs in DNA nucleotide sequences.
Figure 1.
Work flow and algorithms of PG4 motif mining (A) Schematic diagram depicting the workflow of QuadBase2 web server. Users interact with either EuQuad or ProQuad interface to select organisms and genes of interest. Information on genes and their orthologues is obtained from the current release of Ensembl. Here, the number of species from each meta-class in the current release of QuadBase2 are shown. TetraplexFinder works both as an interface and the core algorithm that performs PG4 motif mining on the server backend. The PG4 motif configurations are provided by the user in all three modules through an interactive interface. The result of PG4 motif mining can either be downloaded or visualized interactively for each gene. (B) Figure depicting the results of PG4 algorithms on an example sequence. The line over and below each sequence shows the PG4 motif that would be captured by the algorithm. The ‘greedy’ and ‘non-greedy’ algorithms detect different number and lengths of PG4 motifs. Choice of algorithm is crucial for downstream interpretation.
These motifs are of following configuration:
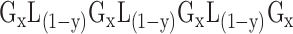 |
In the motif representation above, Gx represents contiguous ‘x’ number of guanine (G) bases, also referred to as stem length of the motif. A motif is composed of four such stems. The stem length is a constant number in in a range of two to five, however choosing a stem length of three is a common practice when looking for PG4 motifs in mammalian sequences, while stem length of two is more common for the prokaryotes. This choice, however, is based upon the fact that for a stem length of two, large number of motifs are detected in the larger genomes than the smaller ones, reflecting little biological meaning. L(1-y) represents the length of the loop which ranges from one to ‘y’, the maximum loop length. A motif is composed of three loops, each being separated by a stem. Though, in a motif search, a constant value for ‘y’ is assigned, each loop have a different length in the detected PG4 motif. Multiple evidences exist that loop length can affect the stability and the folding characteristics of G4s with a certain length, restricting them to certain types topologies (36,37). In addition, the base composition of the loops can also affect the propensity of the sequence to form the structures in vivo (38).
In QuadBase2, the strategy to mine PG4 motifs, as in the previous version, is based upon a ‘regular expression’ search through the sequence. However, in this updated version, the user can either choose to run a ‘greedy’ or a ‘non-greedy’ variant of the regular expression (Figure 1B). The ‘greedy’ variant employs a look-ahead assertion in the expression which tends to maximize loop length by trying to incorporate the stems into the loop definition and hence the detected motifs may contain multiple internal PG4 motifs. The ‘non-greedy’ variant, on the other hand, tries to minimize the loop length and does not lead to a motif with internal motifs. In addition, for both the search variants, the server mines overlapping and non-overlapping PG4 motifs in a given sequence separately. Non-overlapping motifs will have non-coinciding motif coordinates in a given sequence and are more useful for statistical or bioinformatics analysis, because one genomic region is counted only once. A recent report (39), which shows that G4 structures can also be formed with bulges in the stem regions, prompted us to implement an algorithm that mines PG4 motifs with bulges of user provided length. Currently, the web server only supports bulges in a stem length of three.
WEB SERVER ARCHITECTURE AND FEATURES
QuadBase2 web server backend is written in Python programming language using Flask microframework and all visualizations are generated using Matplotlib library (40). The frontend is implemented using React.js library of Javascript. The web server interfaces with Ensembl's REST API to download gene sequences for eukaryotic genomes and to obtain names of orthologues from Compara database. This allows keeping gene models up to date with Ensembl's latest release (41,42). We have further described the individual modules of the webserver in the following sub-sections.
TetraplexFinder
TertaplexFinder provides an interface to the core module of the web server that is responsible for performing the PG4 motif search. It allows users to either upload or paste multiple FASTA formatted sequences. Currently, TetraplexFinder allows up to 20 MB data to be uploaded. The configuration box enables users to select the stem length, the minimum loop size and the maximum loop size. The User may also select one of the pre-configured stem and loop length combination, labelled as low stringency (G2L1-10 which stands for stem length of 2, minimum loop length of 1 and maximum loop length of 10), medium stringency (G3L1-10) and high stringency (G3L1-5). These are relative stringency settings provided to allow a quick motif search. Users can select either ‘greedy’ or ‘non-greedy’ algorithm for motif mining.
On submission, a progress page is displayed where users can monitor how many of the input sequences have been processed. They can bookmark this page, to access the results later. On the results page, along with a summary of PG4 motifs mined, a table is displayed where sequence-wise counts of the number of PG4 motifs found are shown for each strand. Clicking on any sequence name shows a representative figure that shows the position of the mined PG4 motif on the sequence, while another panel displays the input sequence, annotated with PG4 motifs. The user can download results for all the sequences and/or only specific sequences in a BED format file for both overlapping and non-overlapping PG4 motifs. Also, each sequence can be downloaded in a text format where non-overlapping PG4 motifs are marked in upper case characters and the rest of the sequence is in lower case. Due to its improved configuration options and ability to handle bulk queries, TetraplexFinder is an improved version of PatternFinder module present in the previous version of QuadBase. The bulk mining feature, visualization and annotated sequence download options are not currently available in any other existing PG4 motif mining server/tool.
EuQuad
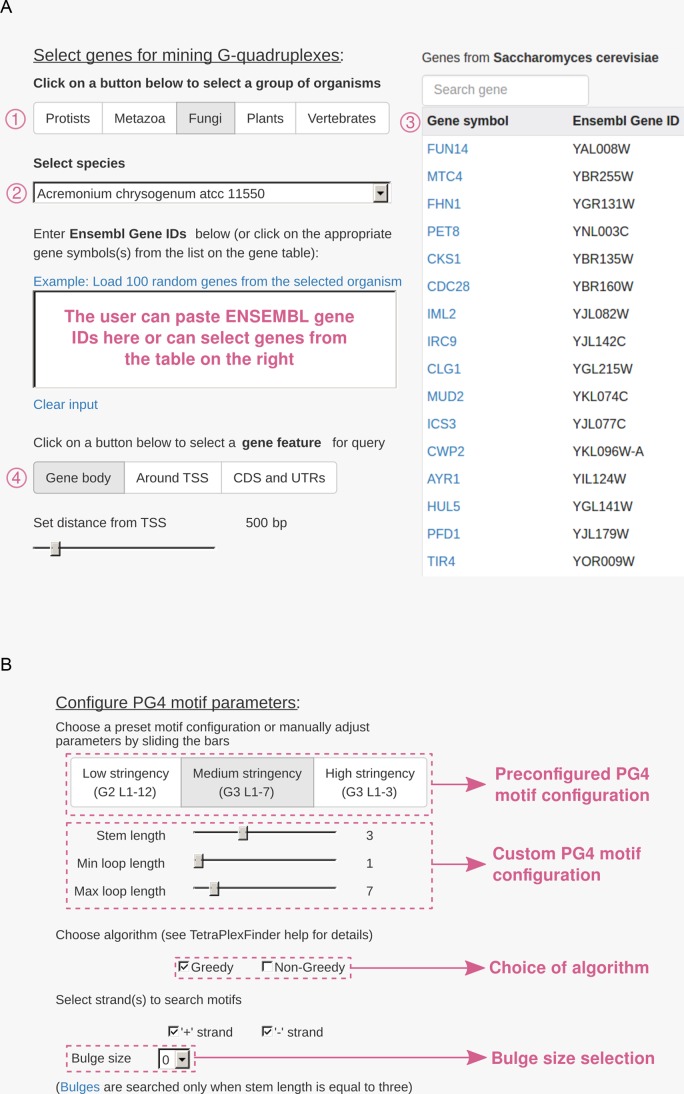
This module allows users to mine PG4 motifs in either of the 36 protists, 68 fungi, 17 metazoans, 32 plants or 25 vertebrates (Figure 1A). Once an organism group is selected, the user can choose the species which then presents a table of genes. The user can directly enter Ensembl gene IDs or can click on a gene symbol in the table which will directly update the list of genes to be mined with the corresponding Ensembl gene ID (Figure 2A). Users can choose to either mine PG4 motifs in the whole gene body or around the annotated transcription start site or just the cDNA sequence (i.e. coding sequence along with UTRs). The PG4 configuration parameters in EuQuad are the same as those present in TetraplexFinder (Figure 2B).
Figure 2.
Screenshots of example input: (A) the screenshot shows EuQuad's input interface. The user first selects the meta-class of organism (1), followed by species—strain name from dropdown menu, then user can either paste Ensembl gene ID list in the text area or can click on gene names in the provided table (3), this will automatically update the list in text area with Ensembl IDs. The user then selects the gene feature to be used for motif mining. Either gene body, flanking region of transcription start site (TSS) or the coding sequence can be chosen. In case of TSS feature, users also set the range around TSS, over which they wish to mine PG4 motifs. (B) Screenshot showing the options for PG4 configuration, users can either click on predefined configurations or build custom configuration using slider for stem, minimum loop and/or maximum loop length. Users can also choose the algorithm they want to use (‘greedy’ set by default) for PG4 mining. Moreover, when stem length is set at 3, the user can choose to include bulges in the stems which is set by selecting the length of each bulge. The user can also choose to run the search on only one particular strand.
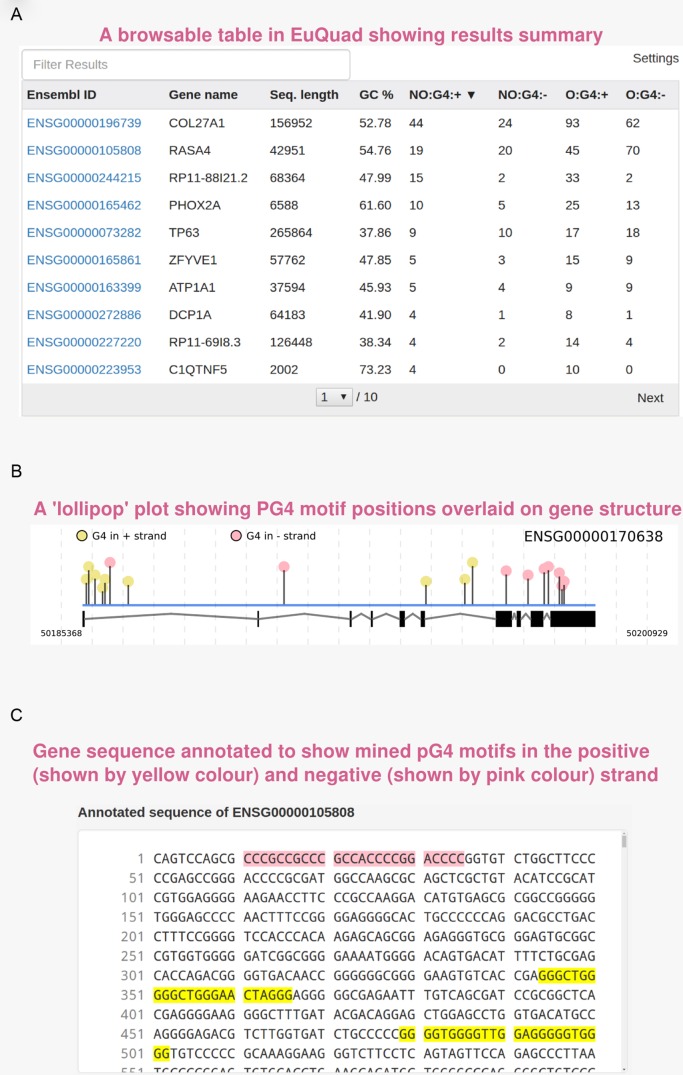
On completion of a query, a table containing the gene symbols along with the number of detected PG4 motifs, is displayed. The user can click on any gene's name in the table to view its result or perform bulk download (Figure 3A). An image (‘lollipop’ plot) representing PG4 motifs across the gene structure is displayed along with a separate box containing the annotated sequence (Figure 3B and C). In addition, orthologues of any gene can be mined by clicking on ‘Search Orthologues’ button. This reiterates the process of PG4 search among all detected orthologous genes after the web server fetches the information on orthologues from Compara database. This allows quick assessment of PG4 presence in orthologues and provides an indication towards the presence of conserved PG4 motifs.
Figure 3.
Screenshots of example output: the result page of EuQuad showing three main result elements. (A) The result table shows name of selected genes/input FASTA headers along with PG4 statistics. Each column is sortable and clicking on any row will update the ‘lollipop’ plot and sequence view of the corresponding gene. (B) ‘Lollipop’ plots in EuQuad module help visualize the relative positions of PG4 motif over the gene structure. The gene structure is shown below the motifs. The black rectangles represent exons while lines represent introns. The height of each line corresponds to relative length of the motif compared to rest motifs in the same sequence. (C) The PG4 motifs detected are annotated on the sequence such that, those on the positive strand are highlighted in yellow while those on the negative strand are highlighted in pink.
ProQuad
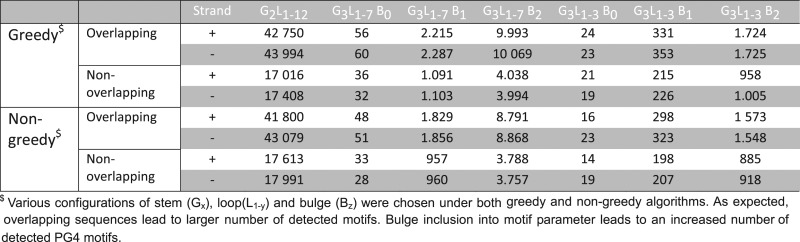
This module allows PG4 motif mining in 1719 prokaryotic species, composed of 4142 strains of Eubacteria (1557 unique species) and Archaea (162 unique species). The module allows mining PG4 motifs in the selected gene and the promoter sequences. Additionally, the user can choose to perform a whole genome query. The result of the gene-wise query is similar to that of TetraplexFinder and EuQuad, however, complete genome query allows the user to access and visualize coordinates of PG4 motifs across the genome as circular histograms. The user can visualize the strand-wise distribution of PG4 motifs vis-a-vis the GC content along the prokaryotic chromosome and can download the result in a BED format file for their downstream analysis. Table 1 provides illustrative figures for the number of PG4 motifs detected in the Escherichia coli genome as overlapping or non-overlapping motifs on using each of the two algorithms. These numbers indicate the stringency of each selected motif and help users set the motif configuration in their searches.
Table 1. Number of PG4 motifs detected in complete Escherichia coli genome by using genome-wide search feature of ProQuad.
|
CONCLUSIONS AND FUTURE DIRECTIONS
QuadBase2 is the first web server that allows multiplexed PG4 motif mining in large number of nucleotide sequences and also enables profiling complete prokaryotic genomes. The web server is linked with Ensembl's genomic datasets that allows virtually any organism's latest gene models or genomic assembly to be mined for the presence of PG4 motifs. The database also allows access to search in orthologues of eukaryotic genes through Ensembl's Compara. This web server provides intuitive graphical and sequence level representations of mined PG4 motifs and the results can be accessed later or be saved in convenient formats. The web server is designed, keeping in mind the requirements of both, the experimental and computational biologists, who want deep insights in to PG4 motif occurrences in their gene sets or genome of interest.
The methodology of motif mining allows flexible customization of stem length, loop length and inclusion of bulges. Since certain configurations will allow thermodynamically more favourable structures than others, users are advised to set the configuration carefully. QuadBase2 currently doesn't include any strategy to normalize the GC bias and skewness of the sequence and score PG4 motif based on local GC content. A recently developed method by Bedrat et al. (43) addresses such concerns and we hope to incorporate a similar strategy into QuadBase in the future.
Even with abundance of information on molecular structure of G4s derived through nuclear magnetic resonance and circular dichroism-based characterization of PG4 motifs, there is still no effective mathematical model that can accurately predict the folding propensity of PG4 motifs. In addition, a single PG4 motif can have the ability to fold into different G4 conformations (44). Future work on QuadBase2 would include a robust mathematical model that predicts stability and conformation of PG4 motifs in vitro or in vivo based upon thermodynamic properties of the sequences and machine learning systems, based on UV melting and circular dichroism data.
AVAILABILITY
The web server is freely available at quadbase.igib.res.in.
Acknowledgments
We acknowledge Mr Raghu Nandanan MV and Mr Anil Kumar from IT team at CSIR-IGIB for their assistance in deployment of the server. We also, thank members of Chowdhury lab, especially Pankaj Narang and Payal Arora, for their help, support and useful suggestions in the development of the web server. SC is senior research fellow of Wellcome Trust/DBT India Alliance.
FUNDING
CSIR Project BSC0123 ( GENCODE-A). Wellcome Trust/DBT India Alliance Research Fellowship (to S.C.); CSIR SPM Junior Research Fellowship (to P.D.). Funding for open access charge: Wellcome Trust/DBT India Alliance [500127/Z/09/Z].
Conflict of interest statement. None declared.
REFERENCES
- 1.Gellert M., Lipsett M.N., Davies D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U.S.A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Majumdar A., Patel D.J. Identifying hydrogen bond alignments in multistranded DNA architectures by NMR. Acc. Chem. Res. 2002;35:1–11. doi: 10.1021/ar010097+. [DOI] [PubMed] [Google Scholar]
- 3.Phan A.T., Mergny J.-L. Human telomeric DNA: G-quadruplex, i-motif and Watson-Crick double helix. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mirkin S.M. Discovery of alternative DNA structures: a heroic decade (1979-1989) Front. Biosci. 2008;13:1064–1071. doi: 10.2741/2744. [DOI] [PubMed] [Google Scholar]
- 5.Usdin K., Woodford K.J. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 1995;23:4202–4209. doi: 10.1093/nar/23.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Besnard E., Babled A., Lapasset L., Milhavet O., Parrinello H., Dantec C., Marin J.-M., Lemaitre J.-M. Unraveling cell type–specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 2012;19:837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 7.Schiavone D., Jozwiakowski S.K., Romanello M., Guilbaud G., Guilliam T.a., Bailey L.J., Sale J.E., Doherty A.J. PrimPol is required for replicative tolerance of G Quadruplexes in vertebrate cells. Mol. Cell. 2016;61:161–169. doi: 10.1016/j.molcel.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paeschke K., Capra J.A., Zakian V.A. DNA replication through G-Quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA Helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thakur R.K., Kumar P., Halder K., Verma A., Kar A., Parent J.L., Basundra R., Kumar A., Chowdhury S. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009;37:172–183. doi: 10.1093/nar/gkn919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paramasivam M., Membrino A., Cogoi S., Fukuda H., Nakagama H., Xodo L.E. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: implications for transcription. Nucleic Acids Res. 2009;37:2841–2853. doi: 10.1093/nar/gkp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mani P., Yadav V.K., Das S.K., Chowdhury S. Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS One. 2009;4:e4399. doi: 10.1371/journal.pone.0004399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halder K., Halder R., Chowdhury S. Genome-wide analysis predicts DNA structural motifs as nucleosome exclusion signals. Mol. Biosyst. 2009;5:1703–1712. doi: 10.1039/b905132e. [DOI] [PubMed] [Google Scholar]
- 14.Wong H.M., Huppert J.L. Stable G-quadruplexes are found outside nucleosome-bound regions. Mol. Biosyst. 2009;5:1713–1719. doi: 10.1039/b905848f. [DOI] [PubMed] [Google Scholar]
- 15.Halder R., Halder K., Sharma P., Garg G., Sengupta S., Chowdhury S. Guanine quadruplex DNA structure restricts methylation of CpG dinucleotides genome-wide. Mol. Biosyst. 2010;6:2439–2447. doi: 10.1039/c0mb00009d. [DOI] [PubMed] [Google Scholar]
- 16.Gray L.T., Vallur A.C., Eddy J., Maizels N. G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat. Chem. Biol. 2014;10:313–318. doi: 10.1038/nchembio.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cogoi S., Paramasivam M., Membrino A., Yokoyama K.K., Xodo L.E. The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J. Biol. Chem. 2010;285:22003–22016. doi: 10.1074/jbc.M110.101923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rawal P., Bhadra V., Kummarasetti R., Ravindran J., Kumar N., Halder K., Sharma R., Mukerji M., Das S.K., Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: Role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cahoon L.a, Seifert H.S. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beaume N., Pathak R., Yadav V.K., Kota S., Misra H.S., Gautam H.K., Chowdhury S. Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: radioresistance of D. radiodurans involves G4 DNA-mediated regulation. Nucleic Acids Res. 2013;41:76–89. doi: 10.1093/nar/gks1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson A., Wu Y., Huang Y.C., Chavez E.a., Platt J., Johnson F.B., Brosh R.M., Sen D., Lansdorp P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaffitzel C., Postberg J., Paeschke K., Lipps H.J. Probing telomeric G-quadruplex DNA structures in cells with in vitro generated single-chain antibody fragments. Methods Mol. Biol. 2010;608:159–181. doi: 10.1007/978-1-59745-363-9_11. [DOI] [PubMed] [Google Scholar]
- 24.Yuan L., Tian T., Chen Y., Yan S., Xing X., Zhang Z., Zhai Q., Xu L., Wang S., Weng X., et al. Existence of G-quadruplex structures in promoter region of oncogenes confirmed by G-quadruplex DNA cross-linking strategy. Sci. Rep. 2013;3:1811. doi: 10.1038/srep01811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel D.J., Phan A.T., Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oganesian L., Bryan T.M. Physiological relevance of telomeric G-quadruplex formation: a potential drug target. Bioessays. 2007;29:155–165. doi: 10.1002/bies.20523. [DOI] [PubMed] [Google Scholar]
- 27.Balasubramanian S., Hurley L.H., Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yadav V.K., Abraham J.K., Mani P., Kulshrestha R., Chowdhury S. QuadBase: genome-wide database of G4 DNA–occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2008;36:D381–D385. doi: 10.1093/nar/gkm781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Todd A.K., Johnston M., Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huppert J.L., Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D'Antonio L., Bagga P. Proceedings. 2004 IEEE Computational Systems Bioinformatics Conference, 2004. CSB 2004. IEEE; Computational methods for predicting intramolecular g-quadruplexes in nucleotide sequences; pp. 561–562. [Google Scholar]
- 32.Kikin O., D'Antonio L., Bagga P.S. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eddy J., Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menendez C., Frees S., Bagga P.S. QGRS-H Predictor: a web server for predicting homologous quadruplex forming G-rich sequence motifs in nucleotide sequences. Nucleic Acids Res. 2012;40:W96–W103. doi: 10.1093/nar/gks422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kikin O., Zappala Z., D'Antonio L., Bagga P.S. GRSDB2 and GRS_UTRdb: databases of quadruplex forming G-rich sequences in pre-mRNAs and mRNAs. Nucleic Acids Res. 2008;36:D141–D148. doi: 10.1093/nar/gkm982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hazel P., Huppert J., Balasubramanian S., Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 37.Bochman M.L., Paeschke K., Zakian V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piazza A., Adrian M., Samazan F., Heddi B., Hamon F., Serero A., Lopes J., Teulade-Fichou M.-P., Phan A.T., Nicolas A. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 2015;34:e201490702. doi: 10.15252/embj.201490702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukundan V.T., Phan A.T. Bulges in G-quadruplexes: broadening the definition of G-quadruplex-forming sequences. J. Am. Chem. Soc. 2013;135:5017–5028. doi: 10.1021/ja310251r. [DOI] [PubMed] [Google Scholar]
- 40.Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]
- 41.Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., et al. Ensembl 2015. Nucleic Acids Res. 2014;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yates A., Beal K., Keenan S., McLaren W., Pignatelli M., Ritchie G.R.S., Ruffier M., Taylor K., Vullo A., Flicek P. The Ensembl REST API: Ensembl Data for any language. Bioinformatics. 2015;31:143–145. doi: 10.1093/bioinformatics/btu613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bedrat A., Lacroix L., Mergny J.-L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016;44:1746–1759. doi: 10.1093/nar/gkw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Risitano A., Fox K.R. Inosine substitutions demonstrate that intramolecular DNA quadruplexes adopt different conformations in the presence of sodium and potassium. Bioorg. Med. Chem. Lett. 2005;15:2047–2050. doi: 10.1016/j.bmcl.2005.02.050. [DOI] [PubMed] [Google Scholar]