Abstract
Non-coding RNA transcripts such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are important genetic regulators. However, the functions of many of these transcripts are still not clearly understood. Recently, it has become apparent that there is significant crosstalk between miRNAs and lncRNAs and that this creates competition for binding between the miRNA, a lncRNA and other regulatory targets. Indeed, various competitive endogenous RNAs (ceRNAs) have already been identified where a lncRNA acts by sequestering miRNAs. This implies the down-regulation in the interaction of the miRNAs with their mRNA targets, what has been called a sponge effect. Multiple approaches exist for the prediction of miRNA targets in mRNAs. However, few methods exist for the prediction of miRNA response elements (MREs) in lncRNAs acting as ceRNAs (sponges). Here, we present spongeScan (http://spongescan.rc.ufl.edu), a graphical web tool to compute and visualize putative MREs in lncRNAs, along with different measures to assess their likely behavior as ceRNAs.
INTRODUCTION
Non-coding RNAs such as microRNAs (miRNAs) are now well established as important biological regulators. In particular, miRNAs act both to destabilize the transcripts they bind to and to block their translation. This binding event is mediated by a protein complex that recruits the mature miRNA to its target transcript and binding is established through base-pair complementarity between miRNA and a 3′UTR target transcript sequence. While many features have been associated with active miRNA binding sites, it is clear that complementarity is most important at the ‘seed’ region of the miRNA, i.e. nucleotides 2–8 of the mature miRNA (1). Complementarity between the rest of the miRNA and the target sequence is usually high, however, seed region complementarity appears to be the most critical feature of active miRNA binding sites. Once bound, miRNAs stimulate active deadenylation and decapping of the target transcript with other factors, causing the mRNA to become destabilized. Many methods have been published to detect possible miRNA target sites (e.g. TargetScan, miRanda and PicTar (2–4)), usually searching for high-complementarity, seed complementarity, conservation and other features in the 3′UTRs of mRNA sequences. More recently, it has been demonstrated that the activity of some miRNAs may be regulated through so called competitive endogenous RNAs (ceRNAs) (5). These are non-coding transcripts that harbor miRNA response elements (MREs) where miRNAs bind. If these ceRNAs possess many MREs and are expressed at high enough levels they act to sequester miRNAs reducing the number of active miRNAs that can bind mRNA regulatory targets.
Identification of ceRNAs and their target miRNAs is a challenge. Given that ceRNAs are usually non-coding and that they are likely to possess an abundance of putative binding sites in one or multiple MREs, regular miRNA target prediction tools that seek single binding site hits at 3′UTR positions are not optimized to detect ceRNAs candidates. AGO CLIP-seq as well as RNA-seq has been used to propose thousands of lncRNA–miRNA interactions (6,7) but these methods are restricted by the availability of such data for specific organisms and cell types. We sought to address these limitations by developing a novel, sequence-based, algorithm designed for the detection of MREs in non-coding transcripts which have the potential to act as ceRNAs of miRNAs that would be potentially applicable to any organism where sequence data exist. Here, we describe a new web resource—spongeScan—that provides a user-friendly interface for applying this sponge search algorithm to any set of sequences provided by the user. spongeScan also includes options to analyze gene expression data of both candidate ceRNAs and miRNAs. Important to mention is that spongeScan does not give a definitive prediction value, but rather ranks putative ceRNA–miRNAs pairs on the basis of several parameters that are indicative of sponge function (5). Our algorithm particularly identifies lncRNAs that have multiple and spread MREs. We have seen that this approach top ranks known ceRNAs acting as sponges.
ALGORITHM
spongeScan is a web resource to find highly enriched MRE binding sites in lncRNAs. Users must provide the lncRNA transcript sequences in a FASTA file. These sequences can be automatically retrieved by spongeScan from any release and species available at Ensembl (8) or be directly uploaded by the user. Additionally, an annotation GTF file is necessary. This file is used to obtain the biotype of the transcripts to filter out transcripts that are not lncRNAs, if any.
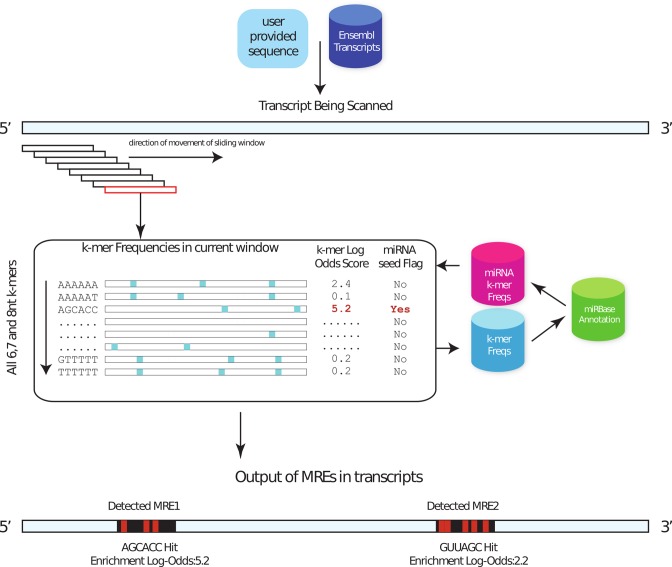
spongeScan looks for sequence complementarity between any possible k-mer of 6, 7 or 8 nucleotides and each lncRNA and identifies if any of these enriched k-mers corresponds to a known miRNAs seed sequence. To do this, the user has to indicate the species being analyzed and spongeScan will look for the corresponding miRNAs in miRBase database (9) automatically at runtime. Retrieved miRNAs are then filtered to keep the canonical seeds of 6, 7 and 8 nucleotides of only experimentally validated miRNAs (Figure 1).
Figure 1.
Flowchart showing the main strategy behind the spongeScan application. K-mers of 6, 7 and 8 nucleotides are searched for by using sliding windows of different sizes. Different k-mer frequencies are obtained for each pair k-mer – lncRNA. Highly enriched k-mers are reported and checked for correspondence with a miRNA canonical seed. Pair-wise predictions are then represented in spongeScan.
For each possible k-mer spongeScan scans for matches using sliding windows of varying sizes ranging from 50 bps to 1 kb in steps of 50 bps allowing up to one G:U wobble (Figure 1). This varying sliding window approach allows selecting the window size that returns the highest number of matches, thereby allowing for flexibility in the k-mer distribution. From k-mer frequencies, we compute a Log-Odds score (LOD, 1) to identify and report highly appearing k-mers for each lncRNA. The formula below is used to obtain the maximum number of matches for which significant pairing between a k-mer and lncRNA are found across all the sliding windows. This is compared to the maximum number of occurrences of that same k-mer in all other lncRNA sequences. This is calculated for all the possible window sizes, and reports the one with the highest LOD.
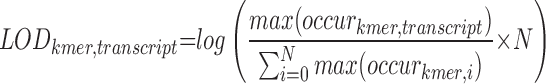 |
(1) |
lncRNA–miRNA pairs with high LOD scores are indicative of multiple MREs in the lncRNA sequence that would facility the sequestration of miRNAs by the lncRNA.
A dispersion score (2) is also calculated for every pair to evaluate the clustering of binding sites. As we are trying different window sizes, the maximum number of matches should change accordingly. For instance, if two matches of a k-mer in a window size of 50 are detected and these are approximately equally distributed, we should expect four matches to be found using a window size of 100, etc. For this reason, we build a vector containing the maximum number of occurrences normalized by the window size used and calculate the standard deviation. This value is what we called dispersion score. The lower this value is, the most equally distributed the miRNA seed matches are. This parameter allows hypothesizing on the distribution pattern on MREs along the ceRNAs. Known ceRNAs tend to have equally spaced MREs that would facilitate multiple miRNA binding (5), what implies a low dispersion score.
 |
(2) |
Finally, a complexity score (3) is calculated for all the k-mers not matching with any known miRNA canonical seed, and k-mers with low complexity scores are filtered out. The formula measures the number of single nucleotides and di-nucleotides, i.e. a k-mer containing AAAAAA would be (6 − 6) x 0.5 + 1 × 0.5 = 0.5, whereas ATGCTA would be (6 − 2) × 0.5 + 5 × 0.5 = 4.5. This score is used to filter out low complexity k-mers that may return unspecific binding.
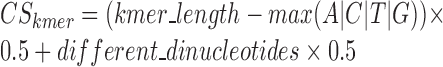 |
(3) |
The default thresholds for the Log-Odd score, dispersion and complexity scores are 1, 10 and 4, respectively. These values were obtained from the human data set, as those values at the 95% percentile of the distribution of all possible microRNA–lncRNA pairs. However, these values can be modified in the web application. For example, higher LOD score and lower dispersion score would select lncRNAs with higher number of MREs and more evenly distributed sites. Other additional and adjustable arguments are the total number of binding sites detected for a pair lncRNA:k-mer. By default, the application will be only reporting pairs where more than 20 putative binding sites have been found for a k-mer in a lncRNA sequence. In contrast to other algorithms such as DIANA-microT (6), that use PAR-CLIP data to identify putative MREs, spongeScan exclusively relies on sequence data and bases its scoring system in the number of matched sites and their distribution along the lncRNA sequence. This favors, on one hand, the detection of ceRNAs where miRNA sequestration can occur at multiple sites, and on the other hand allows application of the algorithm to any organism.
Once computations are completed, spongeScan displays identified lncRNA:k-mer pairs in a tabular format, where each row represents a different match (Figure 2). Results for k-mers of 6, 7 or 8 nucleotides are kept separately and the user can switch between them. Additionally, the results distinguish between k-mers matching known miRNAs or unknown k-mers. The results table has up to 22 different columns containing different information or statistics regarding the pairing. This table can be sorted and filtered by any of the available fields. A graphical representation of the lncRNA sequence showing the positions where the k-mer is found is also included. Matched locations can be clicked to open an integrated genome viewer (10) for closer examination of the binding sites. Additionally, if expression data have been provided, a bar plot showing the expression of the selected lncRNA and miRNA(s) will be displayed. The complete manual of the web application can be found online: http://spongescan.readthedocs.org/en/latest/Home/.
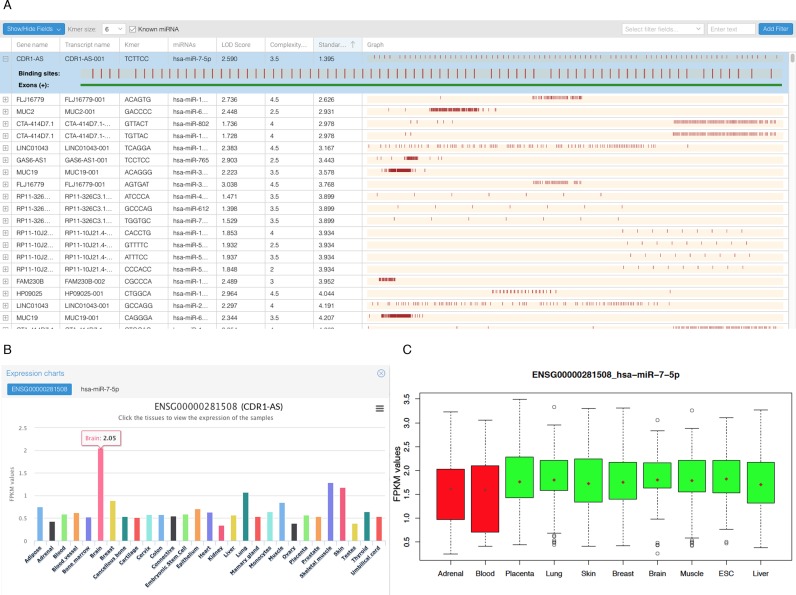
Figure 2.
spongeScan output generated for the example data set. (A) Application table showing pairwise enrichments of miRNA canonical seeds in lncRNA sequences. This view only shows a few of the total possible columns containing data and scores. (B) Expression data representation for the first pair CDR1-AS and miR-7-5p. The expression data are grouped by tissue and, when clicked, it will show the expression of all the samples in the tissue. (C) Expression levels of mRNA targets of miR-7 for different tissues as a function of the CDR1-AS expression. Red box-plots correspond to tissues where the lncRNA is not significantly expressed, whereas the green color indicates expression of the lncRNA in the tissue.
EXAMPLE DATA SET
spongeScan contains an example data set consisting of precomputed results for the MRE search algorithm in human lncRNAs together with gene expression information for these and miRNAs across several tissues obtained by metanalysis of publicly available RNA-seq data. The human MRE search was done using the Homo sapiens ncRNA fasta file from Ensembl release 82 and the corresponding GTF annotation file. Algorithm parameters were set to k-mer complexity scores > 4, LOD > 1, standard deviation < 30, minimum number of predicted = 2 and allowing for one G:U wobble.
To obtain gene expression values for ncRNAs and miRNAs, 206 human RNA-seq data sets were downloaded from SRA and ENCODE (11) corresponding to several healthy tissues and cell lines, while 12 miRNA-seq data sets were found for the same tissues. RNA-seq data were analyzed with standard procedures (12), using Tophat (13) as mapper and htseq-count (14) as quantification tool. Quantification was obtained for a total of 13,047 lncRNAs and 1,548 microRNAs, and values were uploaded into spongeScan.
When ranking results by LOD the known sponge CDR1-AS acting on mir-7 is one of the top 50 hits and the absolute top hit when ranked by dispersion score (Figure 2A). Visualization of gene expression data reveals that CDR1-AS is preferentially expressed in brain tissue (Figure 2B) as previously described (5). To further evaluate the potential sequestration effect, we analyzed the potential effect of predicted lncRNAs with multiple MREs in sequestering to explain the down-regulatory effect of bound miRNAs over their target genes, as previously described (5). We obtained target genes for miRNAs in predicted pairs from TargetScan and compared their expression levels in tissues with or without the expression of the putative sponge, using a paired t-test. Once more, this analysis indicated that in all tissue comparisons (100%) tissues expression of mir-7 targets was upregulated when CDR1-AS was expressed (Figure 2C). Unfortunately not enough matching tissue data were available for similar analyses in other putative sponges.
Finally, we compared our results with the list of lncRNA–miRNA interactions available at lncBase (6). lncBase provides a prediction score for human lncRNA–miRNA interactions based on different evidence sources. We have observed that validated lncRNA–microRNA pairs in this database usually have scores from 0.4 to 1.0. We searched the top 100 results on our human example data and found that most (81%) of our predictions were in the lncBase, having an average score of 0.84 in this database (Supplementary Table), what supports our prediction results with an independent resource.
CONCLUSIONS
We describe spongeScan, a novel web application and algorithm able to identify putative miRNA binding patterns across lncRNA sequences. The algorithm is based on sequence complementarity and allows users to fix parameters to allow flexible search. The possibility of adding expression data to the prediction representation in the web tool, greatly facilitates downstream functional analysis. spongeScan differs from other lncRNA–miRNA interactions prediction sites that utilize CLIP-seq data (6,7) in allowing massive searchers on user provided data and in being available for any organism with sequence information. To our knowledge this is the first web resource that provides a universal searchable engine for the identification of putative lncRNAs with multiple MREs. Overall, we believe spongeScan will be extremely useful for the discovery of crosstalk between lncRNAs and miRNAs.
Supplementary Material
Acknowledgments
The authors acknowledge Stijn van Dongen, Dimitris Vitsios and Mat Davis (EMBL-EBI), Cristina Marti (CIPF, Valencia) and Oleksandr Moskalenko (UF, Florida) for useful support and discussions, and the ENCODE Consortium and the ENCODE production laboratory(s) generating the particular data set(s).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
FP7 STATegra project [agreement number 36000]; MINECO, co-funded with European Regional Development Funds (ERDF) [BIO2012-40244]; European Molecular Biology Laboratory and the European Union and ERC Seventh Framework Programme (FP7/2007-2013) [ERC-2012-StG-310325]. Funding for open access charge: University of Florida funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4 doi: 10.7554/eLife.05005. doi:10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 5.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 6.Paraskevopoulou M.D., Vlachos I.S., Karagkouni D., Georgakilas G., Kanellos I., Vergoulis T., Zagganas K., Tsanakas P., Floros E., Dalamagas T., et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44:D231–D238. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham F., Amode M.R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medina I., Salavert F., Sanchez R., de Maria A., Alonso R., Escobar P., Bleda M., Dopazo J. Genome Maps, a new generation genome browser. Nucleic Acids Res. 2013;41:W41–W46. doi: 10.1093/nar/gkt530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szcześniak M.W., Gaffney D.J., Elo L.L., Zhang X., et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anders S., Pyl P.T., Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.