Abstract
Neural crest (NC) development is controlled precisely by a regulatory network with multiple signaling pathways and the involvement of many genes. The integration and coordination of these factors are still incompletely understood. Overexpression of Wnt3a and the BMP antagonist Chordin in animal cap cells from Xenopus blastulae induces a large number of NC specific genes. We previously suggested that Potassium Channel Tetramerization Domain containing 15 (Kctd15) regulates NC formation by affecting Wnt signaling and the activity of transcription factor AP-2. In order to advance understanding of the function of Kctd15 during NC development, we performed DNA microarray assays in explants injected with Wnt3a and Chordin, and identify genes that are affected by overexpression of Kctd15. Among many genes identified we chose Duf domain containing protein 1(ddcp1), Platelet-Derived Growth Factor Receptor a (pdgfra), Complement factor properdin (cfp), Zinc Finger SWIM-Type Containing 5 (zswim5), and complement component 3 (C3) to examine their expression by whole mount in situ hybridization. Our work points to a possible role for Kctd15 in the regulation of NC formation and other steps in embryonic development.
Keywords: Neural crest, Kctd15, DNA microarray, Xenopus, gene regulation, transcription factor AP-2
Introduction
The Kctd family of proteins has received increasing attention due to its multiple biological functions and putative roles in human disease. The family is composed of over 20 genes/proteins in different vertebrate species and can be subdivided into seven subgroups on the basis of sequence similarity (Liu et al., 2013; Skoblov et al., 2013). A number of Kctd genes have been associated with human diseases, as summarized by Liu and colleagues (Liu et al., 2013). All members of the family share a Bric-a-brack, Tram-track, Broad complex (BTB) domain usually located in the N-terminal half of the protein. BTB domains, also called POZ domains, are widely involved in protein-protein interactions (Perez-Torrado et al., 2006; Stogios et al., 2005), and thus it is not surprising that acting as adapters is perhaps the best-studied molecular function of Kctd proteins. Kctd5, Kctd6 and Kctd11 bind Cullin 3 and act as adapters mediating the interactions of E3 ubiquitin ligases with their substrates (Balasco et al., 2014; Bayon et al., 2008; Canettieri et al., 2010; Chen et al., 2009; Correale et al., 2011). While other Kctd proteins also have been reported to bind Cullin 3, this has been contradicted in recent work (Smaldone et al., 2015). Kctd8, Kctd12 and Kctd16 associate with GABA receptors, modulating their activity (Rajalu et al., 2015); these genes also show striking left/right asymmetry in their expression in the brain (Gamse et al., 2005). Other family members appear to have very different molecular functions by acting in the regulation of transcription. These include Kctd10, a factor involved in heart development through the inhibition of Tbx5a activity, apparently by direct interaction between the Tbx and Kctd components (Hu et al., 2014; Tong et al., 2014). In addition Kctd10 also interacts with proliferating cell nuclear antigen and through this interaction affects proliferation (Wang et al., 2009).
We have been interested in the functions of Kctd15 in Xenopus and zebrafish development. We found that overexpression of Kctd15 leads to very effective inhibition of the formation of the neural crest (NC), suggesting the possibility that Kctd15 regulates the size of the NC domain in development (Dutta and Dawid, 2010; Groves and LaBonne, 2014). Kctd15 and the closely similar Kctd1 have two distinct molecular functions, inhibition of canonical Wnt signaling (Dutta and Dawid, 2010; Li et al., 2014) and inhibition of the function of transcription factor AP-2 (Ding et al., 2009; Zarelli and Dawid, 2013). Both of these functions may be responsible for the suppression of NC formation by Kctd15 as Wnt signaling and AP-2 activity are essential for NC induction and differentiation (Brewer et al., 2004; de Croze et al., 2011; Ikeya et al., 1997; Knight et al., 2005; Li and Cornell, 2007; Luo et al., 2003; Luo et al., 2005; Saint-Jeannet et al., 1997; Schorle et al., 1996; Simoes-Costa and Bronner, 2015). Based primarily on the strength of the inhibitory effect we believe that Kctd15 suppression of AP-2 activity may be the dominant mechanism in its blocking of NC development. Mutations in KCTD1 in humans are responsible for scalp-ear-nipple (SEN) syndrome (Marneros et al., 2013), but it is not known whether inhibition of AP-2 or Wnt signaling plays a role in the etiology of this disease. KCTD15 is not been reported as the cause of a disease, but numerous studies show an association between this gene and obesity (Gutierrez-Aguilar et al., 2012; Leon-Mimila et al., 2013; Mei et al., 2012; Willer et al., 2009; Williams et al., 2012). It is known that AP-2α regulates the activity of C/EBPα during adipogenesis (Jiang et al., 1998), and AP-2β affects other steps of adipogenesis and insulin resistance (Ikeda et al., 2006; Meng et al., 2010; Tao et al., 2006; Zhang et al., 2014). It is tempting to speculate that the association of KCTD15 with obesity is based on its ability to inhibit AP-2 proteins, but evidence for this hypothesis in vertebrates is not available to date.
Beyond NC development and adipogenesis, the two pathways affected by Kctd15, Wnt signaling and AP-2 transcriptional regulation, have wide-ranging roles in development, physiology and disease (http://web.stanford.edu/group/nusselab/cgi-bin/wnt/) (Eckert et al., 2005; Hilger-Eversheim et al., 2000; Hoffman et al., 2007; Orso et al., 2008; Wenke and Bosserhoff, 2010). The role of Kctd15 in different tissues and cell types and the global role of Kctd15 in development have not been studied so far. We investigated Kctd15 function broadly through transcriptome analysis by DNA microarray. As test tissue we chose Xenopus laevis animal explants (animal caps) that were injected with Wnt and Chordin (Chd) mRNAs, which induce the expression of NC marker genes (Saint-Jeannet et al., 1997). Recently the transcriptome of animal caps overexpressing Pax3 and Zic1 has been analyzed (Bae et al., 2014; Plouhinec et al., 2014). This procedure is more selective for NC induction (Hong and Saint-Jeannet, 2007; Milet et al., 2013), but we chose Wnt/Chd injection to test broadly for effects of Kctd15 at the neural plate border (NPB). Among genes induced by Wnt/Chd and inhibited by Kctd15 we find many well characterized NC markers as well as several hatching gland markers. We further identify several strongly affected genes not previously studied in this context whose further analysis may contribute to an understanding of the role of Kctd15 and AP-2 in NC formation and other processes in the embryo.
Materials and methods
Embryo manipulation
In vitro fertilization and X. laevis embryo culture were performed as described previously (Wang et al., 2011). Embryos were staged according to Nieuwkoop and Faber (Nieuwkoop and Faber, 1967). Experiments have been approved by the NICHD Animal Care and Use Committee. Capped mRNA for microinjection was prepared with mMessage mMachine kit (Invitrogen) and purified with RNeasy mini kit (Qiagen).
Total RNA extraction
Total RNA was extracted using the TRIzol reagent (Invitrogen) and precipitated by isopropanol. After DNase I treatment, the total RNA was purified with RNeasy purification kit (Qiagen).
Animal cap assay and RT-PCR
Animal cap assay was performed as described previously (Shi et al., 2015; Zhao et al., 2008). Briefly, animal caps were dissected from stage 9 embryos that had been injected with the indicated mRNAs, and were cultured till the sibling embryos reached stage 18. Total RNA was extracted for microarray or RT-PCR assays. cDNA was synthesized using Superscript III (Invitrogen) following the manufacturer’s manual. Primers for RT-PCR are listed in Supplementary Table S1.
Whole mount in situ hybridization
The EST clones were purchased from GE Healthcare. The preparation of digoxigenin-labeled RNA probe and whole mount in situ hybridization were performed as described previously (Shi et al., 2015; Wang et al., 2015). Embryos probed with indicated antisense RNAs were sectioned by a vibratome (Zeiss) to a thickness of 50 μm (Shi et al., 2015).
Affymetrix DNA microarray and data analysis
AC treated as described above were used to prepare RNA; three biological replicates were done for each condition. Biotinylated probe was prepared according to manufacturer’s instructions (Affymetrix). Probes were hybridized to X. laevis genome arrays 2.0 (Affymetrix). The hybridized arrays were washed by the GeneChip Fluidics station 450, and scanned with the GeneChip Scanner 3000 (Affymetrix). Gene expression profiles were analyzed by the Partek Genomics Suite software (Partek). The microarray data set was submitted to the NCBI GEO database and obtained accession number GSE72391.
Results
Identification of genes regulated by Kctd15 during NC formation
We wished to survey the range of genes that are inhibited by Kctd15 overexpression in the context of NC induction by using Affymetrix DNA microarray. Injection of RNA encoding a Wnt and a BMP inhibitor such as Chordin into Xenopus embryos followed by animal cap culture (Fig. 1A) leads to the induction of many NC and additional NPB border genes (Saint-Jeannet et al., 1997), whereas co-injection of Kctd15 inhibits induction (Dutta and Dawid, 2010). Recent analyses by other workers have focused on induction of NC genes more selectively by Pax3 and Zic1 (Bae et al., 2014; Plouhinec et al., 2014). We injected four sets of RNAs, lacZ as control (L), wnt3a+chd (WC), wnt3a+chd+kctd15 (WCK), and kctd15 (K). We tested several NC markers by PCR and found that, as predicted, wnt+chd induced, and kctd15 inhibited NC marker expression (Fig. 1B). Microarray analysis was then carried out (for raw data see GEO accession number GSE72391). Here we will deal with the two relevant comparisons: WC vs. L, representing genes activated during NPB/NC induction, although additional genes are affected; and WCK vs. WC, representing genes inhibited by Kctd15 in the context of Wnt/Chd overexpression. For further analysis we selected genes with a fold change of two or greater and a p value <0.05. Under the criteria stated above, there are 2869 Affymetrix probe sets that are induced by Wnt/Chd, while 3680 probe sets are suppressed (Supplementary Table S2). Note that Affymetrix probe sets often represent genes repetitively; nevertheless we will use “genes” to stand for “probe set” hereafter. A number of known NPB or NC genes were strongly induced by Wnt/Chd overexpression (Table 1), supporting the microarray results.
Fig. 1.
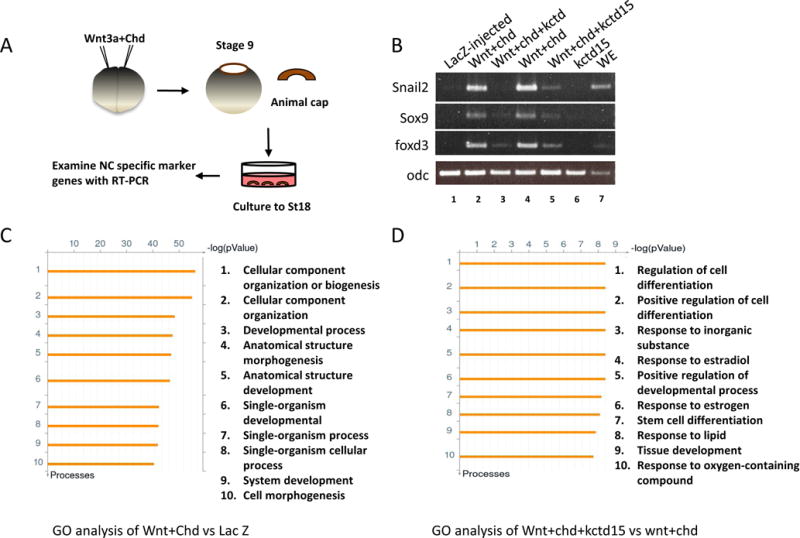
Transcriptome analysis of animal explants induced by co-injection of wnt3a and chd mRNA. (A) Schematic diagram of experimental procedures. (B) Kctd15 suppressed the expression of NC marker genes in animal cap assays. Both blastomeres of two-cell stage embryos were injected with the indicated mRNAs. RT-PCR was performed to examine expression of snai2, sox9, and foxd3. Co-overexpression of wnt3a and chd strongly induced the expression of these genes, and the induction was suppressed by addition of kctd15. Odc was the loading control. WE, whole embryos. (C,D) The top ten enriched GO terms in differentially expressed genes in the comparison of (B) induced to control (WC/L), and (C) induced/inhibited to induced (WCK/WC) animal caps.
Table 1.
Induction and repression by Kctd15 of known genes characteristic for neural border and NC.
| Gene | Unigene | WC vs L, Fold | WC vs L, p | WCK vs WC, Fold | WCK vs WC, p |
|---|---|---|---|---|---|
| foxd3a+# | Xl.525 | 144 | 1.22E-05 | −5.23 | 0.013 |
| pax3b# | Xl.45266 | 129 | 9.44E-05 | N. S. | |
| foxd3b+# | Xl.523 | 119.3 | 3.93E-05 | −5.65 | 0.018 |
|
| |||||
| zic1# | Xl.1796 | 105.3 | 4.44E-05 | N. S. | |
| pax3a# | Xl.49495 | 84.86 | 3.71E-04 | N. S. | |
| sox9a+# | Xl.1690 | 51.5 | 8.00E-05 | −14.13 | 0.0011 |
|
| |||||
| Imx1b.1+# | Xl.12464 | 38.52 | 4.39E-06 | −2.57 | 0.0013 |
| sox10# | Xl.1588 | 36.2 | 0.00359 | −12.56 | 0.02 |
| sox8+ | Xl.29789 | 34.28 | 2.34E-05 | −5.4 | 0.0011 |
|
| |||||
| snail2a+# | XL.3818 | 26.6 | 0.0009033 | −20.54 | 0.0015 |
| twist1b+# | Xl.56708 | 10.8 | 6.86E-05 | −3.21 | 0.0061 |
| tfap2e# | Xl.50785 | 4.427 | 0.000404 | −2.2 | 0.016 |
WC, Wnt+Chd, referring to RNA from animal caps injected with these mRNAs; L, LacZ; WCK, Wnt+Chd+Kctd15. N.S., change not significant.
Genes identified in Table 2 or Table 3 of (Bae et al., 2014).
Genes identified in Table 2 or Table S1 of (Plouhinec et al., 2014).
Gene ontology (GO) analysis of the NPB/NC transcriptome revealed that GO terms of cellular component organization or biogenesis, cellular component organization, developmental process, anatomical structure morphogenesis, anatomical structure development, single-organism developmental process, single-organism process, single-organism cellular process, system development, and cell morphogenesis are highly represented (Fig. 1C).
We next surveyed the comparison of WCK vs WC, and found that 334 genes are suppressed by addition of Kctd15, while 210 genes were increased under the criteria stated above (Supplementary Table S3). Among the suppressed genes we found all genes listed in Table 1 except pax3a/b and zic1 (see Discussion). Expression of Kctd15 itself was much reduced in WCK animal caps, suggesting that Kctd15 negatively regulates its own expression. Gene ontology analysis showed that GO terms regulation of cell differentiation, positive regulation of cell differentiation, response to inorganic substance, response to estradiol, positive regulation of developmental process, response to estrogen, stem cell differentiation, response to lipid, tissue development, response to oxygen-containing compound are significantly enriched (Fig. 1D). While multiple terms are included, Kctd15 appears to affect most strongly aspects of cell differentiation.
As two recent reports examined gene expression in ACs induced to form NC by pax3 and zic1 injection (Bae et al., 2014; Plouhinec et al., 2014), we tested the overlap between the gene sets in GEO submissions associated with these references and our data, using genes changed by 2-fold or more in either direction at p<0.05. Among 203 genes increased in the data of Plouhinec et al., 154 are also increased in our data, and among 227 decreased genes, 205 in our set are likewise decreased (Supplementary Table S4). In the comparison with the work of Bae et al., of 713 increased genes 702 are also increased in our data, and among 863 decreased genes 853 are decreased in our data (Supplementary Table S5). Thus the overlap with both sets of data is high.
Developmental expression of selected genes affected by Kctd15
We next used whole mount in situ hybridization to examine the expression pattern of genes encoding Duf domain protein 1 (ddcp1), Platelet-derived growth factor receptor a (pdgfra), Complement factor properdin (cfp), Zinc finger SWIM-Type Containing 5 (zswim 5), and Complement component C3 (Table 2).
Table 2.
Microarray values for induction and inhibition of genes tested by in situ hybridization.
| Gene | Unigene | WC vs L, Fold | WC vs L, p | WCK vs WC, Fold | WCK vs WC, p |
|---|---|---|---|---|---|
| ddcp1 | Xl2.13675 | 27.0745 | 3.54E-06 | −4.09502 | 0.00135358 |
| pdgfra | Xl2.20029 | 16.6366 | 2.33E-05 | −3.5796 | 0.00230006 |
| cfp | Xl2.41202 | 8.49842 | 4.00E-06 | −2.36589 | 0.00214363 |
| zswim5 | Xl2.40416 | 3.04663 | 0.0003077 | −2.40434 | 0.00142697 |
| c3 | Xl2.44891 | 14.404 | 0.0166839 | −6.64701 | 0.0951221 |
See Table 1 for abbreviations.
Duf domain containing protein 1(ddcp1) encodes a protein that has not been characterized, and even the function of the Duf domain is unknown. Signals for ddcp1 became visible at neurula stages, forming stripes along the anterior and lateral edges of neural fold (Fig 2A,B). Two signal patches at each side with higher intensity can be observed at the future head region (arrows in Fig. 2A). Transverse sections of a stage 18 indicated that staining was localized in a region extending from ectoderm, mesoderm to the endoderm (Fig. 2B). With the progress of neural fold closure, the two signal patches seem to merge and form a short strip at the midline of the head region at tadpole and tailbud stages (Fig. 2C–E,G). Transverse sections identify an additional expression region of ddcp1 at the stomodeal-hypophyseal anlage (Fig. 2F).
Fig. 2.
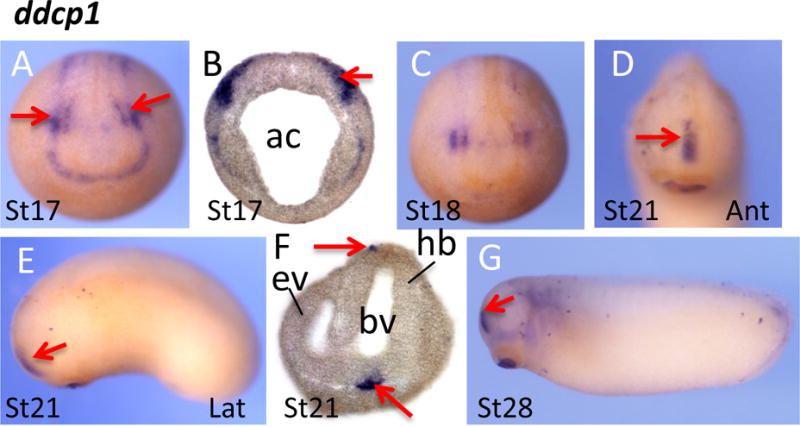
Spatial expression pattern of ddcp1 encoding a Duf domain protein. (A–C) ddcp1 was expressed along the edge of neural plate. Notably, two stronger staining patches (arrows) were detected in whole mount embryos (A) and transverse sections (B). With the closure of neural tube, the two signaling patches merged and were located at the frontal region of the head (D–G). Ant, anterior view; lat, lateral view. Ac, archerteron; ev, eye vesicle; bv, brain vesicle; hb, hindbrain.
Platelet-derived growth factor receptor (Pdgfr) is a receptor tyrosine kinase that has been identified as a pax3/zic1 target (Bae et al., 2014; Plouhinec et al., 2014). These authors also showed expression of pdgfra in the NC region at neurula stages. We found strong expression in the branchial arches in tail bud and tadpole stage embryos (Fig. 3A,B).
Fig. 3.
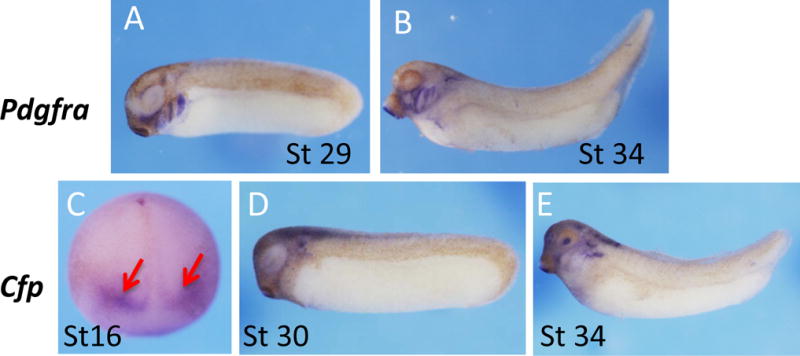
Spatial expression pattern of pdgfra and cfp. (A–B) Strong signals of pdgfra were detected in branchial arches at tail bud and tadpole stages. (C–E) Cfp signals were detected first at mid neurula stage, and appear as two stripes at the anterior neural plate indicated by red arrows (C). The signals were detected in brain, lens, and ear vesicle at tail bud and tadpole stages (D, E).
Complement factor properdin (Cfp) is a cytoplasmic glycoprotein that is implicated in regulating the alternative complement pathway of the innate immune system (Ali et al., 2014). Cfp transcripts can be detected in neurula stage embryos, presenting as two diffuse stripes extending laterally from the dorsal midline of stage16 embryos. Expression of cfp was observed in the neural tube, otic vesicle, and dorsal region of the branchial arches at tadpole stages (Fig. 3D,E).
Zinc Finger SWIM-Type Containing 5 (zswim5) encodes a protein that remains to be characterized. Whole mount in situ hybridization indicates that zswim5 was not expressed before gastrula stages (Fig. 4A). Staining was first detected in the dorsal blastopore lip at the onset of gastrulation (Fig. 4B). The positive region expanded and covered the prospective neural plate at mid gastrula stage (Fig. 4C). At the neurula stages, zswim5 was expressed at the anterior border of the neural plate, partially overlapping with the NC region. Zswim5 expression can be detected in the brain, spinal cord, and eyes at tail bud and tadpole stages (Fig. 4G,H).
Fig. 4.
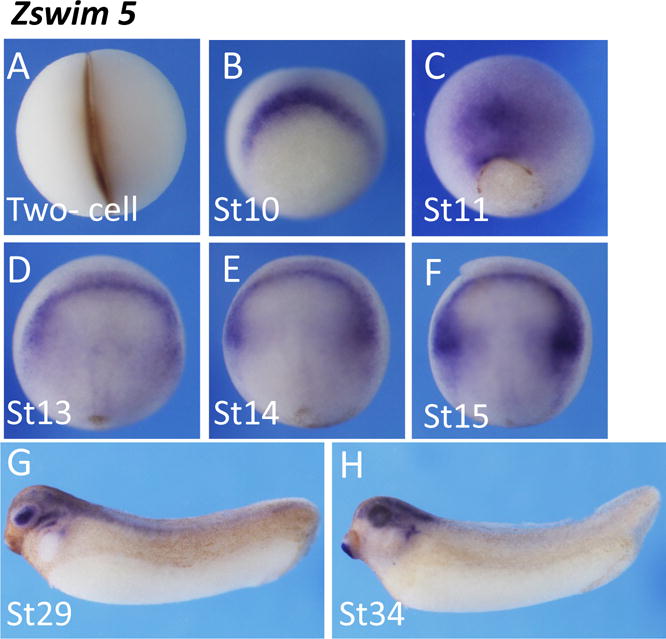
Spatial expression pattern of zswim5. Zswim5 was first detected at the dorsal blastopore lip in early gastrula (A, B), and then was preferentially expressed in the future neural plate at later gastrula stages (C). At neurula stages, zswim5 signals were restricted at the anterior neural plate, partially covering the NC region (D–F). Zswim5 was mainly expressed at anterior neural tube and lens at tail bud and tadpole stages (G, H).
Complement component 3 (C3) is one of the key components in activation of the complement system in higher vertebrates. Its activation is required for both classical and alternative complement activation pathways. C3 transcripts became visible from the early neurula stages onward (Fig. 5A,B). At stage 14, the C3 transcripts were detected at the edge of the forming neural plate. Staining for C3 was specifically observed in the NC at stage 16, and this expression was confirmed by sectioning (Fig. 5C,D). At the tail bud and tadpole stages, a strong C3 signal was observed in the trunk region, whereas the head was much less stained. Transverse sections of embryos after whole mount in situ hybridization revealed that the C3 transcripts are located in a region corresponding to the somatic and splanchnic layer of lateral plate mesoderm, and ventral mesoderm including the blood island (Fig. 5E–H). At stage 35, C3 stains the heart primordium in addition to intense endodermal expression (Fig. 5I). C3 expression in Xenopus has previously been reported by McLin and colleagues (McLin et al., 2008). While there are some differences in detail our results confirm strong NC expression at stage 16–17, and strong endodermal expression at later stages.
Fig. 5.
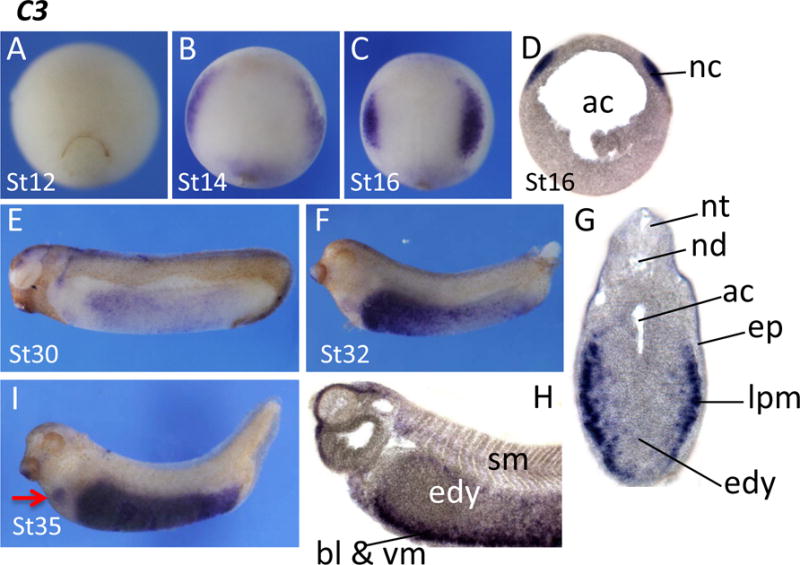
Spatial expression pattern of complement component C3 (C3). C3 signals were not detected by late gastrula stages (A). Its expression appeared at the anterior border of the neural plate, and then became restricted at the NC (B, C). NC expression was confirmed by sectioning (D). C3 expression in the NC declined quickly, and an expression domain gradually appeared at the ventral region of the trunk (E,F). Sections from stage 32 embryos indicated that C3 was expressed in the somatic and splanchnic layer of lateral plate mesoderm just internal to the epidermis. G, sagittal section; H, transversal section. Endodermal expression is prominent at later stages (I). Arrow points to an expression domain in the heart primordium. Ac, archenteron; bl, blood island; edy, endodermal yolk mass; ep, epidermis; lpm, lateral plate mesoderm; nc, neural crest; nt, neural tube; nd, notochord; sm, somite; vm, ventral mesoderm.
To validate the microarray data, we examined the expression of zswim5 and C3 upon overexpression of kctd15 by in situ hybridization. We injected kctd15 mRNA into either both blastomeres of two-cell stage embryos or two dorsal blastomeres of four-cell stage embryos, and checked the expression of zswim5 and C3. Overexpression of kctd15 dramatically reduced zswim5 and C3 staining, consistent with our microarray data (Fig. 6; Table 2).
Fig. 6.
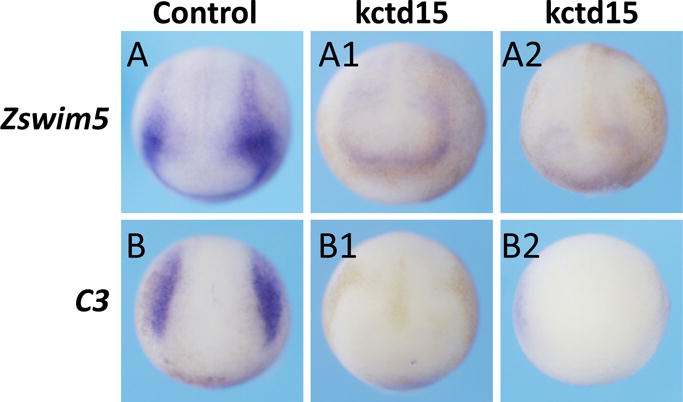
Overexpression of Kctd15 inhibits expression of zswim5 and C3. Both blastomeres of two-cell stage (A1,B1) or two dorsal blastomeres of four-cell stage embryos (A2,B2) were injected with kctd15 mRNA, and the injected embryos were collected at about stage 16. The expression zswim5 and C3 was strongly inhibited compared to those in control embryos (A,B). (A1) 72%, 13 of 18 embryos; (A2), 95%, 19 of 20 embryos; (B1), 58%, 11 of 17 embryos, and (B2) 73.7%, 14 of 19 embryos.
Discussion
Further exploration of the regulatory network that controls NC formation is essential for understanding this developmental process in the normal embryo and the pathological conditions due to disturbed NC development. Kctd15 was previously implicated in the regulation of NC formation, as overexpression of Kctd15 causes strong inhibition of NC development (Dutta and Dawid, 2010). In order to further understand the molecular events regulated by Kctd15, we performed DNA microarray analysis to identify genes that are affected by Kctd15 during NC formation.
In the system we used, Xenopus animal caps induced by inhibition of BMP and activation of Wnt signaling, NC differentiation is elicited together with other events. Yet this system allows a broad evaluation of genes that are affected during the induction process. We found activation of expression of multiple genes known to be characteristic of NC development, some of which are illustrated in Table 1. In line with previous findings, we also detected components involved in modulating Wnt (Shisa), retinoic acid (rarres1, cyp26c1, cyp26a1), and BMP (bambi) signaling, and other genes identified in earlier work. In addition, we have identified a number of novel genes that are induced under these conditions and future analysis will be needed to elucidate their possible functions in NC specification and differentiation.
During the NC specification, coordination of pax3 and zic1 gene activity is essential and sufficient to initiate this complex process (Bae et al., 2014; Garnett et al., 2012; Hong and Saint-Jeannet, 2007; Milet et al., 2013; Monsoro-Burq et al., 2005; Sato et al., 2005). Two recent papers reported the target gene profile of Pax3 and Zic1, both of which utilized inducible constructs in order to identify genes specifically involved in NC specification, EMT induction, and migration. Comparison of our results with those of (Bae et al., 2014) and (Plouhinec et al., 2014) shows a considerable degree of overlap (see Results).
The focus of our study has been to identify genes whose activation during manipulation of Wnt and BMP signaling is sensitive to Kctd15. In the subset of genes shown in Table 1, all except pax3a/b and zic1 are strongly inhibited. As Kctd15 strongly inhibits AP-2 function in addition to lesser inhibition of Wnt signaling (Dutta an d Dawid, 2010; Zarelli and Dawid, 2013), these results suggest that pax3 and zic1 expression in the neural border might be independent of AP-2. Using morpholino-mediated knock-down and enhancer studies, de Croze et al. (de Croze et al., 2011) found that expression of pax3, but not of zic1 depends on AP-2 function in this system. A possible explanation for this difference may be that different members of the AP-2 family may be involved with different Kctd15 sensitivity, or that the particular pax3 enhancer studied confers Kctd15 resistance on AP-2 molecules bound to it. In any case, it is of interest that Kctd15 can inhibit expression of multiple genes lower in the NC induction hierarchy, but does not affect two genes at its top.
Members of sox gene family, notably sox8, sox9 and sox10, are strongly activated by NC induction and are very susceptible to overexpression of kctd15. All three genes belong to the soxE group, which are known to be important to NC formation (Haldin and LaBonne, 2010). The effect of Kctd15 on soxE gene expression is likely through inhibition of AP-2 function, known to be upstream of the sox genes in NC formation (de Croze et al., 2011). In addition, Kctd15 may have a role in chondrogenesis and osteogenesis as Sox9 and Sox10 are essential in these two developmental processes (Kozhemyakina et al., 2015; Long and Ornitz, 2013). Some matrix metallopeptidases including adam33, mmp3 and mmp28a, and the genes involved in ER stress and the secretory pathway such as creb3l2 and dnajb9 are also inhibited by ectopic Kctd15 (Supplementary Table S3), suggesting again a broad spectrum of developmental processes that can be regulated by Kctd15.
We have examined the expression of some genes induced by Wnt/Chd and repressed by Kctd15, ddcp1, pdgfra, cfp, zsmim5 and C3, by in situ hybridization, and further confirmed that zswim5 and C3 are inhibited by overexpression of Kctd15. We chose these genes because their possible role in NC formation was not previously noted, or noted but not studied extensively. We find expression of these genes variously in the neural border, premigratory NC, subsequent NC derivatives, and also in unrelated tissues. These observations suggest that Kctd15 may affect the development of multiple cells and tissues in embryogenesis. The large number of genes affected by Kctd15 in our experiments further suggest that the molecular function of Kctd15 might be in the regulation of transcription rather than in other pathways as indicated for different members of the Kctd family (Balasco et al., 2014; Bayon et al., 2008; Canettieri et al., 2010; Chen et al., 2009; Correale et al., 2011). Our results provide a framework for future studies into the biological role of Kctd15.
Supplementary Material
Acknowledgments
This work was supported by grants from the Research Grants Council of Hong Kong N_CUHK413/12, CUHK24100414, and Lo Kwee-Seong Biomedical Research Fund (SBS-specific) to H. Z., and by the Intramural Research Program of the National Institute of Child Health and Human Development, NIH. We thank colleagues in our laboratories for helpful discussion on this project.
References
- Ali YM, Hayat A, Saeed BM, Haleem KS, Alshamrani S, Kenawy HI, Ferreira VP, Saggu G, Buchberger A, Lachmann PJ, Sim RB, Goundis D, Andrew PW, Lynch NJ, Schwaeble WJ. Low-dose recombinant properdin provides substantial protection against Streptococcus pneumoniae and Neisseria meningitidis infection. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:5301–5306. doi: 10.1073/pnas.1401011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae CJ, Park BY, Lee YH, Tobias JW, Hong CS, Saint-Jeannet JP. Identification of Pax3 and Zic1 targets in the developing neural crest. Developmental biology. 2014;386:473–483. doi: 10.1016/j.ydbio.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasco N, Pirone L, Smaldone G, Di Gaetano S, Esposito L, Pedone EM, Vitagliano L. Molecular recognition of Cullin3 by KCTDs: insights from experimental and computational investigations. Biochimica et biophysica acta. 2014;1844:1289–1298. doi: 10.1016/j.bbapap.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Bayon Y, Trinidad AG, de la Puerta ML, Del Carmen Rodriguez M, Bogetz J, Rojas A, De Pereda JM, Rahmouni S, Williams S, Matsuzawa S, Reed JC, Crespo MS, Mustelin T, Alonso A. KCTD5, a putative substrate adaptor for cullin3 ubiquitin ligases. FEBS J. 2008;275:3900–3910. doi: 10.1111/j.1742-4658.2008.06537.x. [DOI] [PubMed] [Google Scholar]
- Brewer S, Feng W, Huang J, Sullivan S, Williams T. Wnt1-Cre-mediated deletion of AP-2alpha causes multiple neural crest-related defects. Developmental biology. 2004;267:135–152. doi: 10.1016/j.ydbio.2003.10.039. [DOI] [PubMed] [Google Scholar]
- Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P, Pietrosanti L, De Smaele E, Ferretti E, Miele E, Pelloni M, De Simone G, Pedone EM, Gallinari P, Giorgi A, Steinkuhler C, Vitagliano L, Pedone C, Schinin ME, Screpanti I, Gulino A. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol. 2010;12:132–142. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, Liu L, Ding M, Peng HB, Shao F. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Molecular cell. 2009;35:841–855. doi: 10.1016/j.molcel.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Correale S, Pirone L, Di Marcotullio L, De Smaele E, Greco A, Mazza D, Moretti M, Alterio V, Vitagliano L, Di Gaetano S, Gulino A, Pedone EM. Molecular organization of the cullin E3 ligase adaptor KCTD11. Biochimie. 2011;93:715–724. doi: 10.1016/j.biochi.2010.12.014. [DOI] [PubMed] [Google Scholar]
- de Croze N, Maczkowiak F, Monsoro-Burq AH. Reiterative AP2a activity controls sequential steps in the neural crest gene regulatory network. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:155–160. doi: 10.1073/pnas.1010740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Luo C, Zhou J, Zhong Y, Hu X, Zhou F, Ren K, Gan L, He A, Zhu J, Gao X, Zhang J. The interaction of KCTD1 with transcription factor AP-2alpha inhibits its transactivation. Journal of cellular biochemistry. 2009;106:285–295. doi: 10.1002/jcb.22002. [DOI] [PubMed] [Google Scholar]
- Dutta S, Dawid IB. Kctd15 inhibits neural crest formation by attenuating Wnt/beta-catenin signaling output. Development. 2010;137:3013–3018. doi: 10.1242/dev.047548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert D, Buhl S, Weber S, Jager R, Schorle H. The AP-2 family of transcription factors. Genome Biol. 2005;6:246. doi: 10.1186/gb-2005-6-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamse JT, Kuan YS, Macurak M, Brosamle C, Thisse B, Thisse C, Halpern ME. Directional asymmetry of the zebrafish epithalamus guides dorsoventral innervation of the midbrain target. Development. 2005;132:4869–4881. doi: 10.1242/dev.02046. [DOI] [PubMed] [Google Scholar]
- Garnett AT, Square TA, Medeiros DM. BMP, Wnt and FGF signals are integrated through evolutionarily conserved enhancers to achieve robust expression of Pax3 and Zic genes at the zebrafish neural plate border. Development. 2012;139:4220–4231. doi: 10.1242/dev.081497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves AK, LaBonne C. Setting appropriate boundaries: fate, patterning and competence at the neural plate border. Developmental biology. 2014;389:2–12. doi: 10.1016/j.ydbio.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Aguilar R, Kim DH, Woods SC, Seeley RJ. Expression of new loci associated with obesity in diet-induced obese rats: from genetics to physiology. Obesity (Silver Spring) 2012;20:306–312. doi: 10.1038/oby.2011.236. [DOI] [PubMed] [Google Scholar]
- Haldin CE, LaBonne C. SoxE factors as multifunctional neural crest regulatory factors. The international journal of biochemistry & cell biology. 2010;42:441–444. doi: 10.1016/j.biocel.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilger-Eversheim K, Moser M, Schorle H, Buettner R. Regulatory roles of AP-2 transcription factors in vertebrate development, apoptosis and cell-cycle control. Gene. 2000;260:1–12. doi: 10.1016/s0378-1119(00)00454-6. [DOI] [PubMed] [Google Scholar]
- Hoffman TL, Javier AL, Campeau SA, Knight RD, Schilling TF. Tfap2 transcription factors in zebrafish neural crest development and ectodermal evolution. J Exp Zool B Mol Dev Evol. 2007;308:679–691. doi: 10.1002/jez.b.21189. [DOI] [PubMed] [Google Scholar]
- Hong CS, Saint-Jeannet JP. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Molecular biology of the cell. 2007;18:2192–2202. doi: 10.1091/mbc.E06-11-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Gan S, Xie G, Li L, Chen C, Ding X, Han M, Xiang S, Zhang J. KCTD10 is critical for heart and blood vessel development of zebrafish. Acta biochimica et biophysica Sinica. 2014;46:377–386. doi: 10.1093/abbs/gmu017. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Maegawa H, Ugi S, Tao Y, Nishio Y, Tsukada S, Maeda S, Kashiwagi A. Transcription factor activating enhancer-binding protein-2beta. A negative regulator of adiponectin gene expression. The Journal of biological chemistry. 2006;281:31245–31253. doi: 10.1074/jbc.M605132200. [DOI] [PubMed] [Google Scholar]
- Ikeya M, Lee SM, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- Jiang MS, Tang QQ, McLenithan J, Geiman D, Shillinglaw W, Henzel WJ, Lane MD. Derepression of the C/EBPalpha gene during adipogenesis: identification of AP-2alpha as a repressor. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:3467–3471. doi: 10.1073/pnas.95.7.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight RD, Javidan Y, Zhang T, Nelson S, Schilling TF. AP2-dependent signals from the ectoderm regulate craniofacial development in the zebrafish embryo. Development. 2005;132:3127–3138. doi: 10.1242/dev.01879. [DOI] [PubMed] [Google Scholar]
- Kozhemyakina E, Lassar AB, Zelzer E. A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development. 2015;142:817–831. doi: 10.1242/dev.105536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Mimila P, Villamil-Ramirez H, Villalobos-Comparan M, Villarreal-Molina T, Romero-Hidalgo S, Lopez-Contreras B, Gutierrez-Vidal R, Vega-Badillo J, Jacobo-Albavera L, Posadas-Romeros C, Canizalez-Roman A, Rio-Navarro BD, Campos-Perez F, Acuna-Alonzo V, Aguilar-Salinas C, Canizales-Quinteros S. Contribution of common genetic variants to obesity and obesity-related traits in mexican children and adults. PloS one. 2013;8:e70640. doi: 10.1371/journal.pone.0070640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cornell RA. Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Developmental biology. 2007;304:338–354. doi: 10.1016/j.ydbio.2006.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chen C, Wang F, Huang W, Liang Z, Xiao Y, Wei K, Wan Z, Hu X, Xiang S, Ding X, Zhang J. KCTD1 suppresses canonical Wnt signaling pathway by enhancing beta-catenin degradation. PloS one. 2014;9:e94343. doi: 10.1371/journal.pone.0094343. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu Z, Xiang Y, Sun G. The KCTD family of proteins: structure, function, disease relevance. Cell & bioscience. 2013;3:45. doi: 10.1186/2045-3701-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, Ornitz DM. Development of the endochondral skeleton. Cold Spring Harbor perspectives in biology. 2013;5:a008334. doi: 10.1101/cshperspect.a008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Lee YH, Saint-Jeannet JP, Sargent TD. Induction of neural crest in Xenopus by transcription factor AP2alpha. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:532–537. doi: 10.1073/pnas.0237226100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Zhang Y, Khadka D, Rangarajan J, Cho KW, Sargent TD. Regulatory targets for transcription factor AP2 in Xenopus embryos. Dev Growth Differ. 2005;47:403–413. doi: 10.1111/j.1440-169X.2005.00809.x. [DOI] [PubMed] [Google Scholar]
- Marneros AG, Beck AE, Turner EH, McMillin MJ, Edwards MJ, Field M, de Macena Sobreira NL, Perez AB, Fortes JA, Lampe AK, Giovannucci Uzielli ML, Gordon CT, Plessis G, Le Merrer M, Amiel J, Reichenberger E, Shively KM, Cerrato F, Labow BI, Tabor HK, Smith JD, Shendure J, Nickerson DA, Bamshad MJ. Mutations in KCTD1 cause scalp-ear-nipple syndrome. American journal of human genetics. 2013;92:621–626. doi: 10.1016/j.ajhg.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLin VA, Hu CH, Shah R, Jamrich M. Expression of complement components coincides with early patterning and organogenesis in Xenopus laevis. The International journal of developmental biology. 2008;52:1123–1133. doi: 10.1387/ijdb.072465v. [DOI] [PubMed] [Google Scholar]
- Mei H, Chen W, Jiang F, He J, Srinivasan S, Smith EN, Schork N, Murray S, Berenson GS. Longitudinal replication studies of GWAS risk SNPs influencing body mass index over the course of childhood and adulthood. PloS one. 2012;7:e31470. doi: 10.1371/journal.pone.0031470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Kondo M, Morino K, Fuke T, Obata T, Yoshizaki T, Ugi S, Nishio Y, Maeda S, Araki E, Kashiwagi A, Maegawa H. Transcription factor AP-2beta: a negative regulator of IRS-1 gene expression. Biochem Biophys Res Commun. 2010;392:526–532. doi: 10.1016/j.bbrc.2010.01.056. [DOI] [PubMed] [Google Scholar]
- Milet C, Maczkowiak F, Roche DD, Monsoro-Burq AH. Pax3 and Zic1 drive induction and differentiation of multipotent, migratory, and functional neural crest in Xenopus embryos. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5528–5533. doi: 10.1073/pnas.1219124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsoro-Burq AH, Wang E, Harland R. Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during Xenopus neural crest induction. Developmental cell. 2005;8:167–178. doi: 10.1016/j.devcel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) Garland publishing inc; New York: 1967. [Google Scholar]
- Orso F, Penna E, Cimino D, Astanina E, Maione F, Valdembri D, Giraudo E, Serini G, Sismondi P, De Bortoli M, Taverna D. AP-2alpha and AP-2gamma regulate tumor progression via specific genetic programs. FASEB J. 2008;22:2702–2714. doi: 10.1096/fj.08-106492. [DOI] [PubMed] [Google Scholar]
- Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein-protein interaction domain. BioEssays: news and reviews in molecular, cellular and developmental biology. 2006;28:1194–1202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- Plouhinec JL, Roche DD, Pegoraro C, Figueiredo AL, Maczkowiak F, Brunet LJ, Milet C, Vert JP, Pollet N, Harland RM, Monsoro-Burq AH. Pax3 and Zic1 trigger the early neural crest gene regulatory network by the direct activation of multiple key neural crest specifiers. Developmental biology. 2014;386:461–472. doi: 10.1016/j.ydbio.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajalu M, Fritzius T, Adelfinger L, Jacquier V, Besseyrias V, Gassmann M, Bettler B. Pharmacological characterization of GABAB receptor subtypes assembled with auxiliary KCTD subunits. Neuropharmacology. 2015;88:145–154. doi: 10.1016/j.neuropharm.2014.08.020. [DOI] [PubMed] [Google Scholar]
- Saint-Jeannet JP, He X, Varmus HE, Dawid IB. Regulation of dorsal fate in the neuraxis by Wnt-1 and Wnt-3a. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13713–13718. doi: 10.1073/pnas.94.25.13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Sasai N, Sasai Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development. 2005;132:2355–2363. doi: 10.1242/dev.01823. [DOI] [PubMed] [Google Scholar]
- Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- Shi W, Xu G, Wang C, Sperber SM, Chen Y, Zhou Q, Deng Y, Zhao H. Heat shock 70-kDa protein 5 (Hspa5) is essential for pronephros formation by mediating retinoic acid signaling. The Journal of biological chemistry. 2015;290:577–589. doi: 10.1074/jbc.M114.591628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa M, Bronner ME. Establishing neural crest identity: a gene regulatory recipe. Development. 2015;142:242–257. doi: 10.1242/dev.105445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoblov M, Marakhonov A, Marakasova E, Guskova A, Chandhoke V, Birerdinc A, Baranova A. Protein partners of KCTD proteins provide insights about their functional roles in cell differentiation and vertebrate development. BioEssays: news and reviews in molecular, cellular and developmental biology. 2013;35:586–596. doi: 10.1002/bies.201300002. [DOI] [PubMed] [Google Scholar]
- Smaldone G, Pirone L, Balasco N, Di Gaetano S, Pedone EM, Vitagliano L. Cullin 3 Recognition Is Not a Universal Property among KCTD Proteins. PloS one. 2015;10:e0126808. doi: 10.1371/journal.pone.0126808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Maegawa H, Ugi S, Ikeda K, Nagai Y, Egawa K, Nakamura T, Tsukada S, Nishio Y, Maeda S, Kashiwagi A. The transcription factor AP-2beta causes cell enlargement and insulin resistance in 3T3-L1 adipocytes. Endocrinology. 2006;147:1685–1696. doi: 10.1210/en.2005-1304. [DOI] [PubMed] [Google Scholar]
- Tong X, Zu Y, Li Z, Li W, Ying L, Yang J, Wang X, He S, Liu D, Zhu Z, Chen J, Lin S, Zhang B. Kctd10 regulates heart morphogenesis by repressing the transcriptional activity of Tbx5a in zebrafish. Nature communications. 2014;5:3153. doi: 10.1038/ncomms4153. [DOI] [PubMed] [Google Scholar]
- Wang C, Kam RK, Shi W, Xia Y, Chen X, Cao Y, Sun J, Du Y, Lu G, Chen Z, Chan WY, Chan SO, Deng Y, Zhao H. The Proto-oncogene Transcription Factor Ets1 Regulates Neural Crest Development through Histone Deacetylase 1 to Mediate Output of Bone Morphogenetic Protein Signaling. The Journal of biological chemistry. 2015;290:21925–21938. doi: 10.1074/jbc.M115.644864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu Y, Chan WY, Chan SO, Grunz H, Zhao H. Characterization of three synuclein genes in Xenopus laevis. Dev Dyn. 2011;240:2028–2033. doi: 10.1002/dvdy.22693. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zheng Y, Luo F, Fan X, Chen J, Zhang C, Hui R. KCTD10 interacts with proliferating cell nuclear antigen and its down-regulation could inhibit cell proliferation. Journal of cellular biochemistry. 2009;106:409–413. doi: 10.1002/jcb.22026. [DOI] [PubMed] [Google Scholar]
- Wenke AK, Bosserhoff AK. Roles of AP-2 transcription factors in the regulation of cartilage and skeletal development. FEBS J. 2010;277:894–902. doi: 10.1111/j.1742-4658.2009.07509.x. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, Lettre G, Lim N, Lyon HN, McCarroll SA, Papadakis K, Qi L, Randall JC, Roccasecca RM, Sanna S, Scheet P, Weedon MN, Wheeler E, Zhao JH, Jacobs LC, Prokopenko I, Soranzo N, Tanaka T, Timpson NJ, Almgren P, Bennett A, Bergman RN, Bingham SA, Bonnycastle LL, Brown M, Burtt NP, Chines P, Coin L, Collins FS, Connell JM, Cooper C, Smith GD, Dennison EM, Deodhar P, Elliott P, Erdos MR, Estrada K, Evans DM, Gianniny L, Gieger C, Gillson CJ, Guiducci C, Hackett R, Hadley D, Hall AS, Havulinna AS, Hebebrand J, Hofman A, Isomaa B, Jacobs KB, Johnson T, Jousilahti P, Jovanovic Z, Khaw KT, Kraft P, Kuokkanen M, Kuusisto J, Laitinen J, Lakatta EG, Luan J, Luben RN, Mangino M, McArdle WL, Meitinger T, Mulas A, Munroe PB, Narisu N, Ness AR, Northstone K, O’Rahilly S, Purmann C, Rees MG, Ridderstrale M, Ring SM, Rivadeneira F, Ruokonen A, Sandhu MS, Saramies J, Scott LJ, Scuteri A, Silander K, Sims MA, Song K, Stephens J, Stevens S, Stringham HM, Tung YC, Valle TT, Van Duijn CM, Vimaleswaran KS, Vollenweider P, Waeber G, Wallace C, Watanabe RM, Waterworth DM, Watkins N, Witteman JC, Zeggini E, Zhai G, Zillikens MC, Altshuler D, Caulfield MJ, Chanock SJ, Farooqi IS, Ferrucci L, Guralnik JM, Hattersley AT, Hu FB, Jarvelin MR, Laakso M, Mooser V, Ong KK, Ouwehand WH, Salomaa V, Samani NJ, Spector TD, Tuomi T, Tuomilehto J, Uda M, Uitterlinden AG, Wareham NJ, Deloukas P, Frayling TM, Groop LC, Hayes RB, Hunter DJ, Mohlke KL, Peltonen L, Schlessinger D, Strachan DP, Wichmann HE, McCarthy MI, Boehnke M, Barroso I, Abecasis GR, Hirschhorn JN. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nature genetics. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MJ, Almen MS, Fredriksson R, Schioth HB. What model organisms and interactomics can reveal about the genetics of human obesity. Cell Mol Life Sci. 2012 doi: 10.1007/s00018-012-1022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarelli VE, Dawid IB. Inhibition of neural crest formation by Kctd15 involves regulation of transcription factor AP-2. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2870–2875. doi: 10.1073/pnas.1300203110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZC, Liu Y, Li SF, Guo L, Zhao Y, Qian SW, Wen B, Tang QQ, Li X. Suv39h1 mediates AP-2alpha-dependent inhibition of C/EBPalpha expression during adipogenesis. Molecular and cellular biology. 2014;34:2330–2338. doi: 10.1128/MCB.00070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Tanegashima K, Ro H, Dawid IB. Lrig3 regulates neural crest formation in Xenopus by modulating Fgf and Wnt signaling pathways. Development. 2008;135:1283–1293. doi: 10.1242/dev.015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


