ABSTRACT
Nuclear domain 10 (ND10) components restrict herpesviral infection, and herpesviruses antagonize this restriction by a variety of strategies, including degradation or relocalization of ND10 proteins. The rhesus monkey rhadinovirus (RRV) shares many key biological features with the closely related Kaposi's sarcoma-associated herpesvirus (KSHV; human herpesvirus 8) and readily infects cells of both human and rhesus monkey origin. We used the clustered regularly interspaced short palindromic repeat-Cas9 (CRISPR-Cas9) technique to generate knockout (ko) cells for each of the four ND10 components, PML, SP100, DAXX, and ATRX. These ko cells were analyzed with regard to permissiveness for RRV infection. In addition, we analyzed the fate of the individual ND10 components in infected cells by immunofluorescence and Western blotting. Knockout of the ND10 component DAXX markedly increased RRV infection, while knockout of PML or SP100 had a less pronounced effect. In line with these observations, RRV infection resulted in rapid degradation of SP100, followed by degradation of PML and the loss of ND10 structures, whereas the protein levels of ATRX and DAXX remained constant. Notably, inhibition of the proteasome but not inhibition of de novo gene expression prevented the loss of SP100 and PML in cells that did not support lytic replication, compatible with proteasomal degradation of these ND10 components through the action of a viral tegument protein. Expression of the RRV FGARAT homolog ORF75 was sufficient to effect the loss of SP100 and PML in transfected or transduced cells, implicating ORF75 as the viral effector protein.
IMPORTANCE Our findings highlight the antiviral role of ND10 and its individual components and further establish the viral FGARAT homologs of the gammaherpesviruses to be important viral effectors that counteract ND10-instituted intrinsic immunity. Surprisingly, even closely related viruses like KSHV and RRV evolved to use different strategies to evade ND10-mediated restriction. RRV first targets SP100 for degradation and then targets PML with a delayed kinetic, a strategy which clearly differs from that of other gammaherpesviruses. Despite efficient degradation of these two major ND10 components, RRV is still restricted by DAXX, another abundant ND10 component, as evidenced by a marked increase in RRV infection and replication upon knockout of DAXX. Taken together, our findings substantiate PML, SP100, and DAXX as key antiviral proteins, in that the first two are targeted for degradation by RRV and the last one still potently restricts replication of RRV.
INTRODUCTION
The rhesus monkey rhadinovirus (RRV) is a gamma-2-herpesvirus (rhadinovirus) naturally occurring in rhesus macaques (Macaca mulatta) and is closely related to the only human rhadinovirus, Kaposi's sarcoma-associated herpesvirus (KSHV) (1). While RRV is usually apathogenic, at least isolate 17577 of RRV has been demonstrated to induce lymphomas and mesenchymal tumors after experimental RRV infection in the context of simian immunodeficiency virus (SIV) coinfection (2). In another study, RRV and its homolog from southern pig-tailed macaques (M. nemestrina) were regularly found in abnormal cell proliferations in SIV-infected monkeys. These malignancies exhibited a high degree of similarity to those found in human immunodeficiency virus (HIV)-infected humans coinfected with KSHV (3). Therefore, RRV so far represents the closest animal and nonhuman primate model for KSHV. It shares more biological features with KSHV than the prototypic rhadinovirus herpesvirus saimiri (HVS) or the murine gammaherpesvirus 68 (MHV68). In vitro, both RRV and KSHV readily infect cells of the other host species; in vivo, on the other hand, KSHV was shown to be unable to establish latent infection in rhesus macaques (4), and there are no known cases of RRV infection in humans. While both viruses are similar with regard to genome organization, host cell tropism, and receptor usage (5) and also with regard to their associated malignancies, differences do exist. In particular, RRV more readily initiates lytic replication in cell culture in a number of both human and rhesus macaque cell types and cell lines (6). Unlike KSHV, RRV is not tightly associated with solid malignancies but is found in a broader spectrum of lymphomas, including T cell malignancies (3). Also, KSHV belongs to the RV1 rhadinovirus lineage, whereas RRV belongs to the RV2 lineage, which is absent in humans. As viruses from both lineages coexist in macaques (3, 7), they might occupy slightly different biological niches.
The nuclear domain 10 (ND10) subnuclear structure has repeatedly been observed to play a role in the infection of DNA viruses and of herpesviruses in particular. ND10 structures, also called PML bodies, contain over 160 different proteins and can differ in number, size, and composition (8). Their core scaffold is formed through large aggregates of different isoforms of the so-called PML (TRIM19) protein, probably followed in abundance by the proteins SP100 and DAXX (8, 9). While initially thought to represent a proviral structure, a robust body of research now indicates that ND10 proteins actually exert a strong antiviral effect. In many cases, the abundance of ND10 components is inversely correlated with susceptibility or permissiveness to viral infection (10), and expression of both PML and SP100 is induced by interferon (11). In line with this notion, several viruses have been found to alter the ND10 structure or composition either by dissolution of the ND10 structure itself or by relocalization and also degradation of individual ND10 components. Interestingly, the viral strategies to overcome ND10-mediated restriction are relatively diverse, from general ND10 ablation to targeting of single ND10-resident proteins, but the viruses do not pinpoint a single one of the ND10 components as a particularly preferred target. As the mechanisms of ND10-instituted immunity are still poorly understood, in particular with regard to the role of the individual components, the study of viral evasion strategies provides valuable insights into the contribution of individual ND10 components to ND10's antiviral effect. Viruses almost certainly evolve to target components that are critical for antiviral function. Therefore, the identification of cellular targets of viral counteraction strategies is one key to understanding the mechanism of ND10-instituted innate immunity.
Recently, we reported that KSHV ORF75 antagonizes ND10-instituted innate immunity through nonproteasomal degradation of ATRX and dispersion of DAXX from ND10 (12), while HVS ORF3 was shown to specifically effect proteasomal degradation of the ND10 component SP100 (13). Although the degradation mechanisms are substantially different in the two rhadinoviruses, both rhadinoviruses effect degradation through a viral homolog of the cellular enzyme FGARAT (PFAS; EC 6.3.5.3), which plays a key role in the purine synthesis pathway. We therefore analyzed whether the related RRV also attacks ND10 and targets the same or different ND10 components, whether RRV employs different or similar strategies, whether RRV is restricted by certain PML components, and whether the RRV FGARAT homolog ORF75 also functions as the viral effector protein.
MATERIALS AND METHODS
Cells and virus.
SLK cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with high glucose (Life Technologies) supplemented with 10% fetal bovine serum (FBS), 50 μg/ml gentamicin, and 10 μg/ml ciprofloxacin. Rhesus monkey primary fibroblasts were maintained in the same medium supplemented with 20% FBS for regular passaging. Infections were carried out in DMEM with 10% FBS. All cells were grown in 5% CO2 at 37°C. Yellow fluorescent protein (YFP)-tagged RRV (RRV-YFP) was generated by inserting a cytomegalovirus (CMV) immediate early (IE) promoter-driven YFP expression cassette into the RRV bacterial artificial chromosome (BAC) by two-step homologous recombination according to the protocol described by Tischer et al. (14). The cassette was inserted at nucleotide position 208 between the terminal repeats and the K1 gene locus in a noncoding region, analogous to the green fluorescent protein (GFP)-tagged RRV (RRV-GFP) construct described by Bilello et al. (15). The integrity of the L-DNA part of the genome was verified by Illumina-based next-generation sequencing. According to our analysis, our RRV-YFP BAC harbors a small deletion from positions 114350 to 114881 within the microRNA cluster of RRV that is also present in our parental BAC 17 clones. So far, we have not tested whether this deletion was already present in the original BAC 17 clone (16) or occurred during passage in Escherichia coli. RRV-YFP ORF75STOP was generated using the same mutagenesis protocol. Specifically, a recombination cassette was created from the pEPKanS template using long oligonucleotides (Ultramers; Integrated DNA Technologies [IDT], Coralville, IA) RRV75STOPsKanS_FP (TCCTCTTCTGGACTAAGGGCTGCAGCGGCCCAGCGAGGGTTCGTGCGTTAGGCCATATTTGCTAAGTCTAGCCGAGGATGACGACGATAAGTAGGG) and RRV75STOPasKanS_RP (AAAGTCCCACCCGCTTGCTTTTGCGCCGGCTAGACTTAGCAAATATGGCCTAACGCACGAACCCTCGCTGGGCCCAACCAATTAACCAATTCTGATTAG) (where the bold and underlined sequences represent the stop codon), followed by electroporation into RRV-YFP-carrying GS1783, kanamycin selection, and then a second recombination under l-arabinose-induced I-SceI expression to remove the kanamycin resistance cassette. Clones were verified by pulsed-field gel electrophoresis and restriction length fragment polymorphism analysis and by Illumina-based next-generation sequencing. The stop codon was reverted using the analogous procedure and oligonucleotides RRV75StopS-Revertant-Krp (TCCTCTTCTGGACTAAGGGCTGCAGCGGCCCAGCGAGGGTTCGTGCGTTGGGCCATATTTGCTAAGTCTAGCCGCAACCAATTAACCAATTCTGATTAG) and RRV75StopAS-Revertant-Kfp (AAAGTCCCACCCGCTTGCTTTTGCGCCGGCTAGACTTAGCAAATATGGCCCAACGCACGAACCCTCGCTGGGCCAGGATGACGACGATAAGTAGGG) (where the bold and underlined sequences represent the glutamine codon).
Infectious RRV-YFP was generated by transfection of BAC DNA into primary rhesus monkey fibroblasts using the JetPrime transfection reagent (Polyplus) per the manufacturer's instructions. In the case of the ORF75STOP and revertant mutants, an alternative protocol was used due to its higher efficiency. BAC DNA was first transfected into 293T cells using the GenJet (version II) transfection reagent (SignaGen). After 2 days, the BAC-transfected 293T cells were transferred onto a confluent rhesus monkey fibroblast monolayer and cocultivated for 2 weeks. Virus stocks were prepared by inoculating primary rhesus monkey fibroblasts at a very low multiplicity of infection (MOI; about 1 infected cell in 1,000 cells) and then letting the virus replicate until the cell monolayer was completely destroyed. The virus-containing cell supernatant was clarified by centrifugation (4,750 × g for 10 min) and then concentrated by overnight centrifugation at 4,750 × g and careful aspiration of the supernatant. The pellet was resuspended in the remaining liquid overnight. Filtration was omitted because of variable results with regard to virus retention in filter membranes. For infection experiments, the MOI was determined according to the YFP expression of the respective investigated cells after 2 days. KSHV BAC 16-GFP was prepared as described previously (12). MG132 was used at 10 μM. For the experiments whose results are shown in Fig. 8 and 10, we added 5 mM l-cysteine and 1 mM l-arginine, as we were made aware that this might mitigate the nonspecific toxicity of proteasome inhibitors (17). Cycloheximide was used at 50 μg/ml for SLK cells and human foreskin fibroblasts (HFFs) and at 100 μg/ml for rhesus monkey fibroblasts, which required higher concentrations. UV inactivation was achieved as described previously (12).
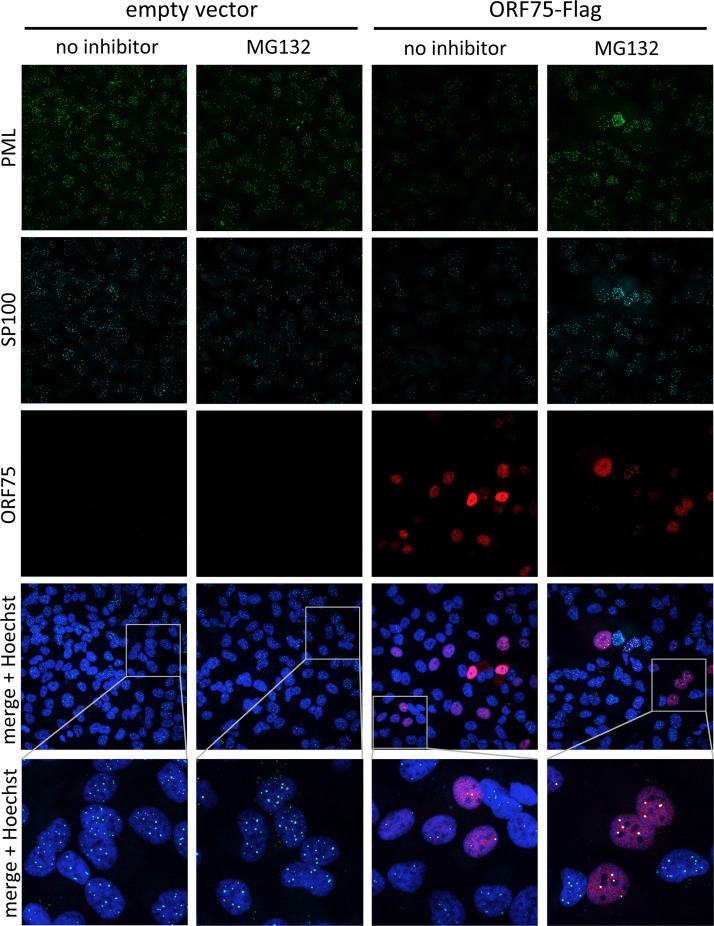
FIG 8.
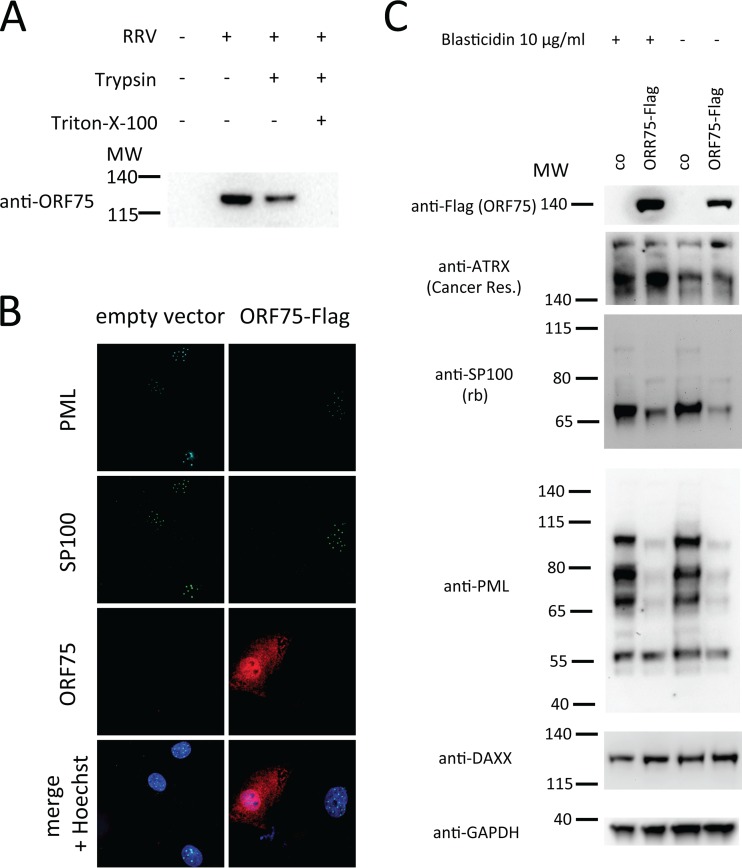
ORF75 targets SP100 and PML for proteasome-dependent degradation. SLK cells were transduced with an empty lentiviral vector or ORF75-Flag. After 3 days, the cells were either treated with MG132 or mock treated for 32 h and then subjected to immunofluorescence analysis.
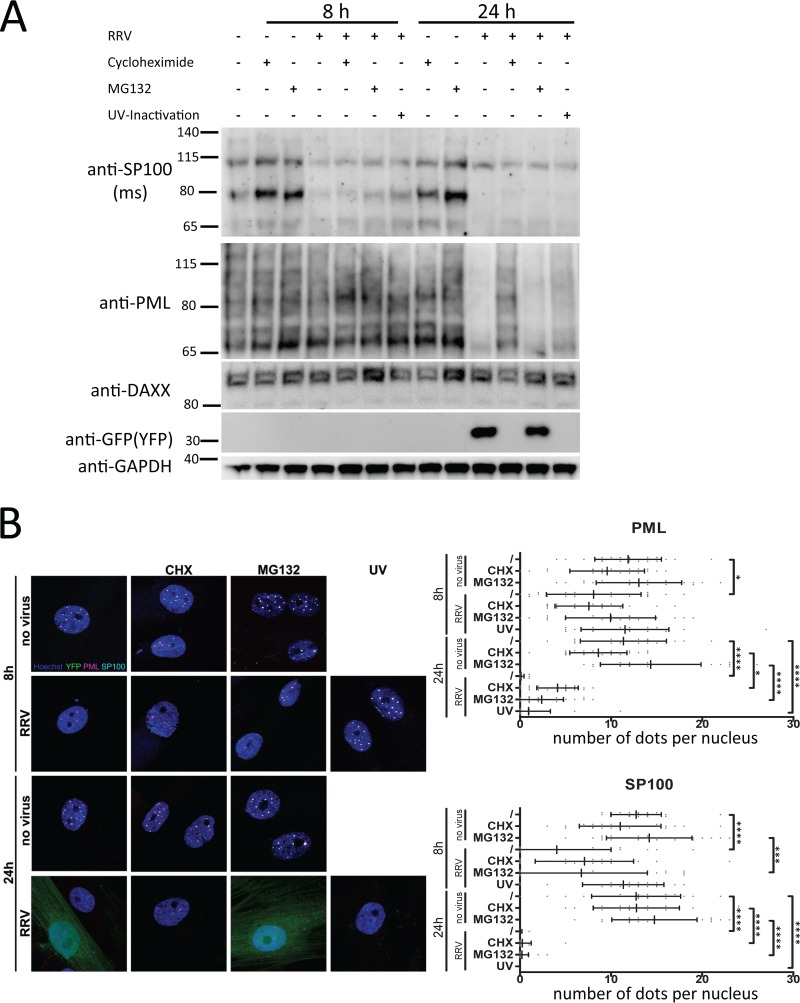
FIG 10.
Degradation of PML and SP100 in RRV-infected rhesus monkey fibroblasts. (A) Rhesus monkey fibroblasts were infected at an MOI of approximately 1 for 18 h or 24 h prior to analysis. Cycloheximide or MG132 was added to the infected cells where indicated. UV-Inactivation, inoculation with UV-inactivated RRV. The cells were harvested by trypsinization and boiled in SDS sample buffer, and the lysates were analyzed by 4 to 12% PAGE and Western blot analysis using the indicated antibodies. The numbers to the left of the gels are molecular weights (in thousands). (B) (Left) Exemplary microphotographs of rhesus monkey fibroblast nuclei after infection with RRV-YFP and immunofluorescent labeling of PML and SP100 (in merged channels, PML is pseudocolored in magenta and SP100 is pseudocolored in cyan). (Right) Quantitative analysis of SP100 and PML expression in nuclear dots in the context of RRV infection. Reductions in the number of PML/SP100 dots after virus treatment that reached significance compared with the values for the no-virus control are highlighted by asterisks (*, P ≤ 0.05; ***, P ≤ 0.001; ****, P ≤ 0.0001). Bars represent means and standard deviation.
Lentiviral expression constructs and transduction.
cDNA of RRV ORF75 was amplified using the RRV BAC as the template and inserted in pLenti CMV BLAST DEST (706–1) in frame with a C-terminal Flag epitope by Gibson Assembly. pLenti CMV BLAST DEST (706–1) was a gift from Eric Campeau (Addgene plasmid number 17451). For production of particles, one 25-cm2 flask of approximately 80% confluent 293T cells was transfected with 0.7 μg pMD2G (a vesicular stomatitis G glycoprotein expression construct), 1.8 μg psPAX2 (a Gag-Pol expression construct), and 2.5 μg pLenti CMV BLAST DEST (706–1) (the empty vector or an RRV ORF75-Flag expression construct) using the GenJet (version II) transfection reagent (SignaGen) per the manufacturer's instructions. The supernatant containing the pseudotyped lentiviral particles was harvested at 2 days posttransfection, cleared of cell debris by low-speed centrifugation, and passed through a 0.45-μm-pore-size syringe filter. Virus was used immediately or was stored short term at 4°C or long term at −80°C. For transduction, lentivirus stocks were used at a 1:2 dilution and added to the target cells overnight, and then the inoculum was replaced with fresh cell culture medium.
Clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9-mediated knockout.
The lentiviral vector pLentiCRISPRv2, a gift from Feng Zhang (Addgene plasmid number 52961) (18), was used to transduce SLK cells with Crispr-associated protein 9 (Cas9) and the respective single guide RNAs (sgRNAs). sgRNA sequences were taken from the GeCKO (version 2) library and ordered as oligonucleotides, annealed, and ligated with matching overhangs into pLentiCRISPRv2 after digestion of the vector with BsmBI. The following oligonucleotide sequences were used to be cloned as sgRNAs: AAACAGTCTTCAGGATCGTCACGAC (koATRXas), CACCGTCGTGACGATCCTGAAGACT (koATRXs), AAACCGGTAGGGGATGCGCTGCTCC (koDAXXas), CACCGGAGCAGCGCATCCCCTACCG (koDAXXs), AAACCGTAGTCGCGCTGGTACCGCC (koPMLex3as), CACCGGCGGTACCAGCGCGACTACG (koPMLex3s), AAACTGATCGTGATCTCATCACAAC (koSP100as), CACCGTTGTGATGAGATCACGATCA (koSP100s), AAACCCAAGGTGCAATCGTGCGAAC (koNTas), and CACCGTTCGCACGATTGCACCTTGG (koNTs), where NT indicates nonspecifically targeting, the suffixes s and as in the sgRNA designations represent sense and antisense, respectively, and ko represents knockout.
Flow cytometry.
Infection assays were performed as described previously (12). A minimum of 10,000 cells was analyzed per sample on an LSRII flow cytometer (BD Biosciences). Data were analyzed using FCS Express software (De Novo Software). The plots in Fig. 9 were created using the R (The R Foundation for Statistical Computing), flowCore (19), and flowViz (20) packages.
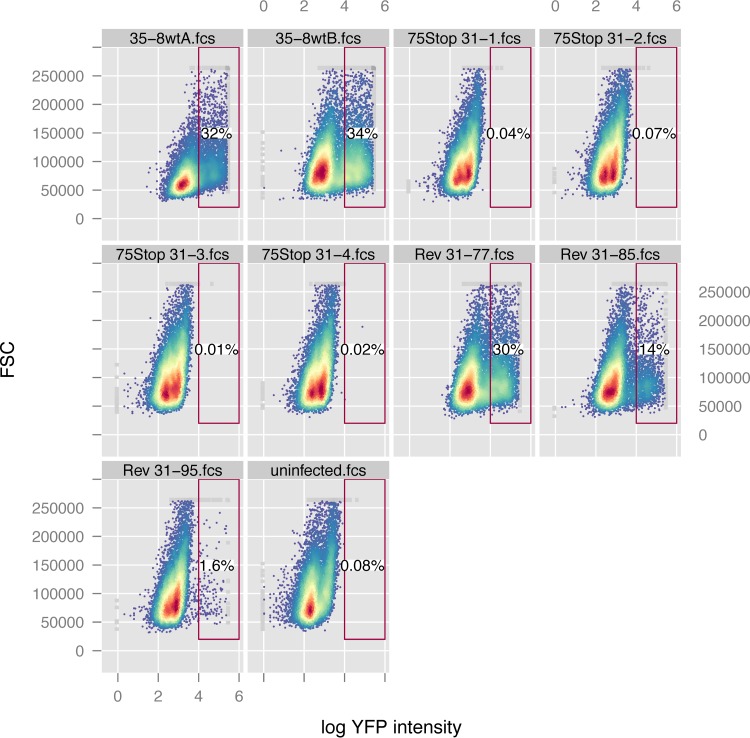
FIG 9.
RRV ORF75 is essential. Four subclones of an RRV ORF75STOP mutant (75Stop31-1, 75Stop31-2, 75Stop31-3, 75Stop31-4), three revertants of clone 31 (Rev 31-77, Rev 31-85, Rev 31-95), or two wild-type RRV-YFP clones (35-8wtA and 35-8wtB) were transfected into 293T cells, followed by coculture with rhesus monkey fibroblasts. The cell culture supernatants, including detached cells and debris, were subjected to one freeze-thaw cycle and then inoculated onto fresh rhesus monkey fibroblasts. After 1 week, the cells were harvested for flow cytometric analysis. FSC, forward scatter.
Quantitative real-time PCR-based gene expression analysis.
RNA was extracted by harvesting the cells directly in the RNAzol reagent (Sigma) per the manufacturer's instructions, followed by purification using a Direct-Zol RNA Miniprep kit with on-column DNase treatment (Zymo Research). cDNA was synthesized using a SensiFAST cDNA synthesis kit (Bioline). Real-time PCR was performed on a StepOne Plus cycler (Thermo) in 20-μl reaction mixtures using a SensiFAST probe Hi-ROX kit (Bioline) (cycling conditions were 3 min of an initial denaturation at 95°C and then 40 cycles of 95°C for 10 s and 60°C for 35 s). The following primer-probe sets were used (all from IDT, Coralville, IA): for ORF73, primers AAAGATGACTCCGTGACACC and GCGATACCCCATTCCCATAC and probe 5′-6-FAM/CCCAGAAAA/ZEN/TACCGCCCACAGAGAA-3′-IABkFQ; for ORF29, primers GCATAAAACCCAGAATCGCG and GTGCGACGTGCTTCAATAAG and probe 5′-6-FAM/CGTCTGTCC/ZEN/CCGGATGCTGT-3′-IABkFQ; for ORF50, primers TGGATACCGACGACAATCAG and CTTATCCCTGAGCGGTTCG and probe 5′-6-FAM/CTGCGTGGA/ZEN/CAACTTTTGCGGA-3′-IABkFQ; for ORF75, primers CCTTTCTGTAGAGTTCCTGCG and CTATTCACAGACCCTCACTTCG and probe 5′-6-FAM/CCCATGAGT/ZEN/ACCACTGTTCGTCCG-3′-IABkFQ; and for hypoxanthine phosphoribosyltransferase (HPRT), primers TGCTGAGGATTTGGAAAGGG and ACAGAGGGCTACAATGTGATG and probe 5′-6-FAM/TCGAGATGT/ZEN/GATGAAGGAGATGGGAGG-3′-IABkFQ/. In the probe sequences, 6-FAM represents 6-carboxyfluorescein, ZEN represents ZEN internal dark quencher, and IABkFQ represents Iowa black fluorescein quencher. Samples were analyzed in technical triplicate. Relative expression levels were calculated using the delta-delta-CT threshold cycle (CT) method after one calibration run with a serial template dilution for all primer-probe sets to verify exponential amplification.
Quantitative real-time PCR-based viral genome copy number analysis.
Cells were infected for 24 h, followed by a medium change on day 1 and day 2 postinfection. For harvest, the cells were washed twice with phosphate-buffered saline (PBS), followed by treatment with trypsin-EDTA until complete detachment of the monolayer. Trypsin activity was stopped by addition of cell culture medium with FBS, the cells were pelleted by low-speed centrifugation, and all the supernatant was removed. The cell pellets were stored at −20°C for further analysis. Total DNA was extracted using an Isolate II genomic DNA extraction kit (Bioline) per the manufacturer's instructions. The DNA concentration was measured spectrophotometrically. Real-time PCR was performed as described above, using the ORF73-specific primer-probe set, which detects genomic viral DNA, and a serial dilution of the RRV-YFP BAC as a standard.
Antibodies.
For detection of PML by Western blotting, we used mouse monoclonal antibody clone G8 (Santa Cruz Biotechnology) at 1:500 for cell lysates of human origin and 1:250 for cell lysates of rhesus monkey origin. For detection of SP100 by Western blotting, we used either anti-SP100 rabbit polyclonal antibody (Proteintech) or anti-SP100 mouse polyclonal antibody (MaxPab; Abnova) at 1:1,000 for cell lysates of human origin or anti-SP100 mouse polyclonal antibody at 1:500 for cell lysates of rhesus monkey origin. For detection of DAXX, we used rabbit polyclonal antibody to DAXX (Sigma) at 1:2,000. For detection of ATRX, we used either mouse monoclonal antibody clone D5 (Santa Cruz Biotechnology) at 1:250 or mouse monoclonal antibody to ATRX (clone ATRX-2/39f/B8) from Cancer Research at 1:1,000. Primary antibodies were diluted in NET gelatin buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris, 0.05% Triton X-100, 0.25% gelatin) with 0.05% sodium azide or in PBS supplemented with 0.05% Tween (PBS-T) and 5% dry milk powder. For immunodetection by Western blotting, horseradish peroxidase-conjugated secondary antibodies (Dako) and Alexa Fluor 647-conjugated secondary antibodies (Life Technologies) were used at 1:2,000 in PBS-T supplemented with 5% dry milk powder. For detection of PML by immunofluorescence assays, we used mouse monoclonal antibody clone PG-M3 to PML (Santa Cruz Biotechnology) at 1:1,000 for cells of human origin and anti-PML polyclonal antibody H-238 (Santa Cruz Biotechnology) at 1:500 for cells of rhesus monkey origin. For detection of SP100 by immunofluorescence, we used anti-SP100 rabbit polyclonal antibody at 1:1,000 for cells of human origin and anti-SP100 mouse polyclonal antibody at 1:500 for cells of rhesus origin. RRV ORF75 was detected by Western blotting using polyclonal antibodies to ORF75-derived peptides (GenScript). Flag epitope-tagged ORF75 was detected using rat monoclonal antibody anti-DYKDDDDK (clone L5; BioLegend) at 1:2,000 in Western blotting assays and 1:1,000 in immunofluorescence assays. Anti-mouse and anti-rabbit immunoglobulin Alexa Fluor 488- or Alexa Fluor 647-conjugated secondary antibodies (Life Technologies) were used at a 1:500 dilution. DyeLight549-conjugated secondary antibody to rat immunoglobulin (Jackson ImmunoResearch Laboratories) was used at a 1:500 dilution.
Western blot analysis.
For preparation of radioimmunoprecipitation assay (RIPA) lysates, cells were briefly trypsinized, followed by addition of complete medium to inhibit trypsin activity. The cells were then pelleted at 1,500 rpm for 5 min, washed once with PBS, repelleted, and lysed in RIPA buffer (1% Triton X-100, 0.5% sodium desoxycholate, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, 50 mM Tris, pH 8, to which aprotinin and leupeptin were freshly added) for 20 min on ice, using approximately 10 volumes of RIPA buffer per 1 volume of cells. The lysate was then clarified by centrifugation at 20,000 × g for 20 min. The protein content was assayed by a bicinchoninic acid assay (Pierce) for normalization purposes. The samples were then diluted with SDS sample buffer and separated by SDS-PAGE either on 10% Tris-glycine gels or on NuPAGE Novex 4 to 12% bis-Tris gradient gels with MOPS (morpholinepropanesulfonic acid) buffer (Life Technologies). Proteins were transferred by semidry blotting onto polyvinylidene difluoride membranes (Immobilon-FL; EMD Millipore) or, for the SP100 blot shown in Fig. 10, onto 0.2-μm-pore-size nitrocellulose membranes (GE Healthcare). The membranes were blocked for at least 30 min with PBS-T supplemented with 5% dry milk powder prior to incubation with primary antibody solution. For denaturing lysis, cell pellets were dissolved in 4× SDS sample buffer (Roth) and immediately heated to 95°C, followed by 40 cycles of sonication at room temperature (Branson 450 sonifier with a cup horn). Samples were then directly subjected to PAGE or frozen for further analysis.
Immunofluorescence.
Cells were seeded on 12-mm-diameter coverslips (Thermo) in 24-well plates. For immunofluorescence analysis, the cells were briefly washed with PBS and were fixed in 4% paraformaldehyde in PBS for 15 min. After two washes with PBS, the cells were permeabilized and nonspecific binding was blocked by incubation with IF buffer (5% FBS, 0.1% Triton X-100 in PBS) for at least 1 h. Primary antibodies in IF buffer were added for 2 h. After three washes with PBS, secondary antibodies were added in IF buffer for 2 h, followed by three washes with PBS. The coverslips were dried and then mounted using Mowiol mounting medium (Fluka) supplemented with 2 μg/ml Hoechst 33258 dye (Sigma). All images except those in Fig. 2C and 7B were recorded on a Leica TCS SP5 confocal microscope. The images in Fig. 7B were recorded on a Leica DMI 6000 B fluorescence microscope equipped with a DFC360FX digital camera. Images were processed using Fiji/ImageJ software. All pictures of one series were treated equally for each channel, except for the images in Fig. 3A, where the blue channel for the Hoechst DNA staining was adjusted and normalized automatically for each image. Live cell fluorescence microscopy for Fig. 2C was performed on a Zeiss Axio Vert.A1 FL-LED microscope.
FIG 2.
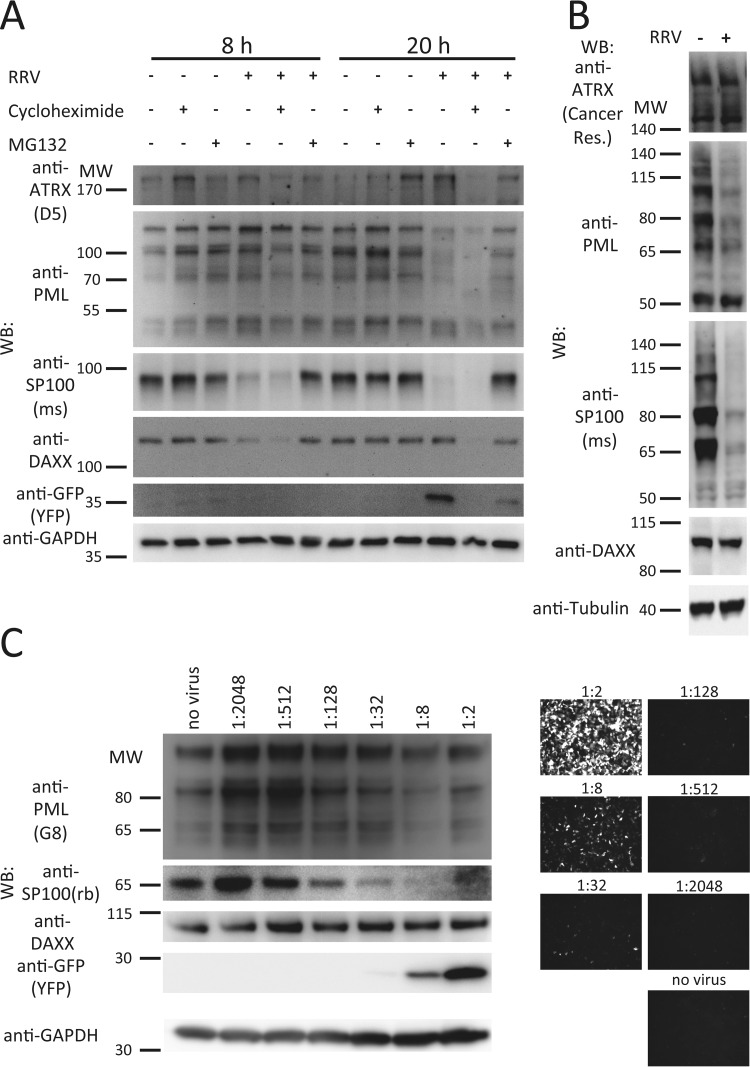
Degradation of PML and SP100 after infection with RRV. (A) SLK cells were infected with RRV at an MOI of >1 or mock infected. In addition, cycloheximide or MG132 was added to the infected cells where indicated. The cells were harvested after 8 h or 20 h, and RIPA lysates were prepared and analyzed by 10% PAGE and Western blotting (WB) for determination of protein levels of PML, SP100, DAXX, and ATRX, as well as YFP to indicate infection and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as a loading control. (B) SLK cells were infected with RRV at an MOI of >1. The cells were harvested at 24 h postinfection in SDS sample buffer, heated, and sonicated, and the samples were analyzed by 4 to 12% PAGE and Western blotting. (C) SLK cells were infected with serial dilutions of an RRV stock. The cells were harvested at 24 h postinfection and analyzed by Western blotting, as described in the legend to panel A. Microscopy images of the confluent cell layer imaged in the YFP channel are given on the right. MW, molecular weight (the numbers to the left of the gels are in thousands). ATRX was detected with clone D5 (D5) or ATRX-2/39f/B8 (Cancer Res.); SP100 was detected with polyclonal antibody raised in mouse (ms) or rabbit (rb).
FIG 7.
ORF75 exhibits characteristics of a viral tegument protein and is sufficient to induce degradation of SP100 and PML. (A) RRV ORF75 is present in the virion-containing supernatant of infected cells and is partially protected from trypsin without addition of detergent. A virus preparation from infected rhesus monkey fibroblast supernatant or cell culture medium was pelleted by centrifugation. The samples were incubated with trypsin-EDTA or trypsin-EDTA plus detergent or were mock incubated at 37°C overnight. The samples were then subjected to Western blot analysis for ORF75. (B) Primary rhesus monkey fibroblasts were transduced with a lentiviral vector encoding RRV ORF75-Flag or with an empty vector as a control at a low MOI and analyzed for expression of ORF75, PML, and SP100 at 72 h postransduction. (C) SLK cells were transduced with a lentiviral vector expressing RRV ORF75-Flag or with an empty vector as a control at a high MOI. The cells were either briefly selected with blasticidin (first and second lanes) to eliminate nontransduced cells or left untreated (third and fourth lanes). After 72 h, the cells were harvested for Western blot analysis. MW, molecular weight (the numbers to the left of the gels are in thousands).
FIG 3.
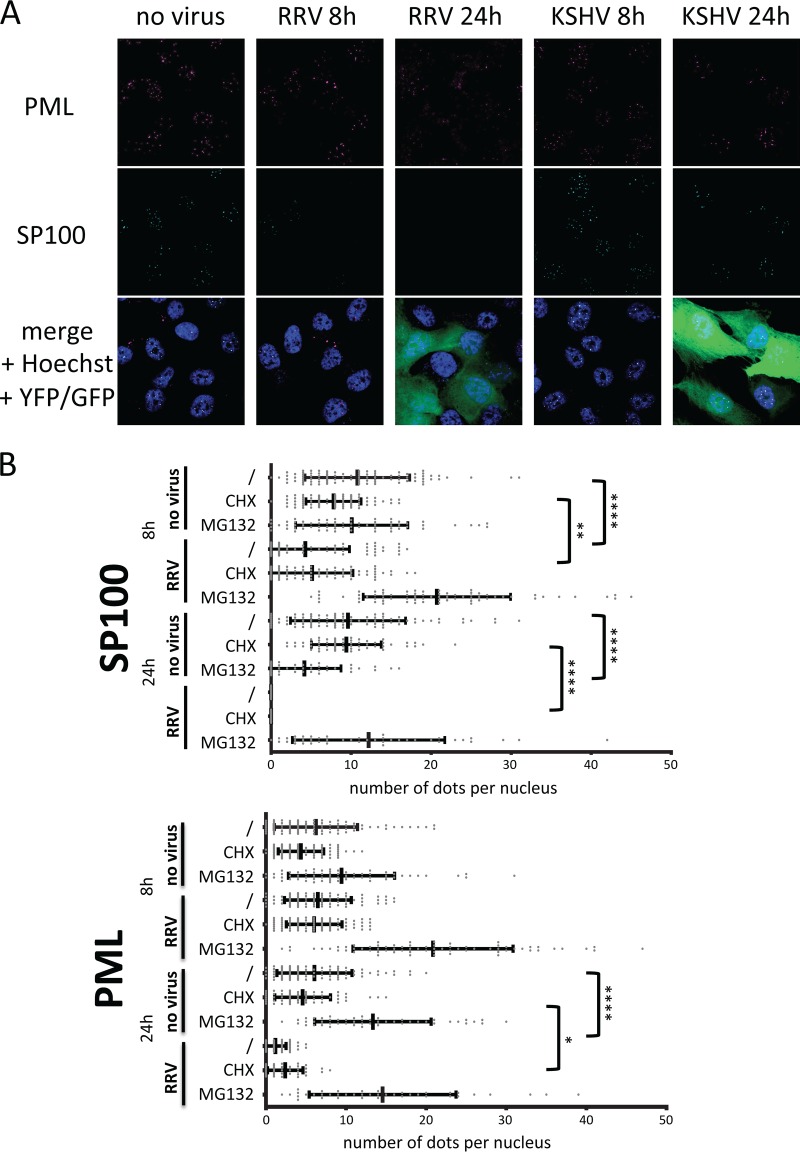
Depletion of SP100 and PML from ND10 by RRV. (A) Immunofluorescence analysis of SP100 and PML in SLK cells infected with RRV-YFP or KSHV BAC 16-GFP for 8 h or 24 h. Representative fields of view are shown. (B) Quantitative analysis of SP100 and PML expression in nuclear dots in the context of RRV infection. Reductions in the number of PML/SP100 dots after virus treatment that reached significance compared with the result for the no-virus control are highlighted by asterisks (*, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001). CHX, cycloheximide.
Image analysis and statistical analysis.
Images were processed and analyzed using the Fiji/ImageJ package. Statistical analysis was done using Microsoft Excel software (Student's t test) or GraphPad Prism (version 6) software. For the experiments whose results are shown in Fig. 3 and 6, approximately 50 to 100 cells were recorded in stacks of confocal images encompassing the whole-cell layer from top to bottom, followed by maximum projection. For the experiments whose results are shown in Fig. 10, approximately 20 to 30 cells were recorded. The number of single-point maxima determined by the use of Fiji software was calculated for each nucleus. Extreme outliers were removed by the GraphPad Prism ROUT 1% algorithm for the data in Fig. 3 and 6. The data were then analyzed using the Kruskal-Wallis test corrected (Dunn) for multiple comparisons within the respective groups, testing the respective uninfected versus infected samples for each treatment.
FIG 6.
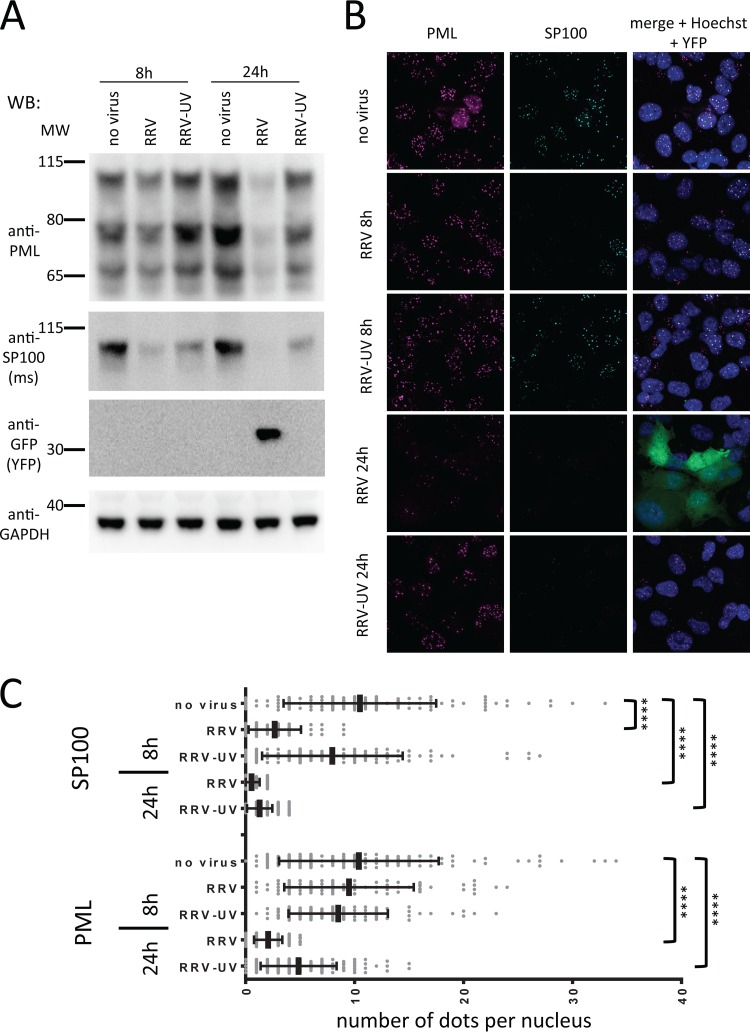
A component of the viral inoculum induces degradation of SP100 and PML. (A) SLK cells were inoculated with RRV or UV-inactivated RRV at an MOI of ∼1. The cells were harvested either at 8 h or at 24 h postinoculation and subjected to Western blot analysis with the indicated antibodies. (B) SLK cells were seeded on coverslips and inoculated as described in the legend to panel A. The cells were fixed after 8 h and 24 h and subjected to immunofluorescence analysis using the indicated antibodies. (C) Number of dots positive for SP100 or PML per nucleus. Significant differences between samples and the no-virus control are denoted by asterisks (****, P ≤ 0.0001). Bars represent means and standard deviations.
RESULTS
RRV infection results in degradation of SP100 and PML.
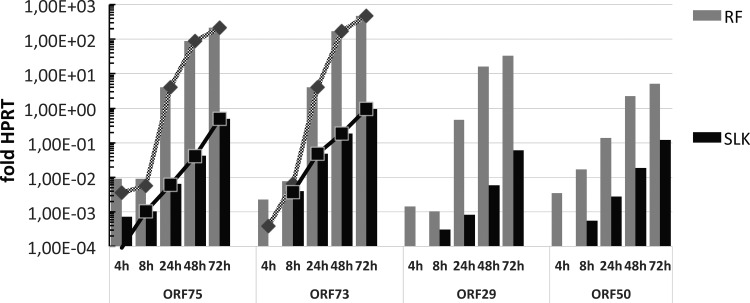
SLK cells are often used as a model system for KSHV, initially because of their hypothetical origin from a biopsy specimen from a patient with Kaposi's sarcoma. Even though all currently available SLK cells are actually identical to renal cell carcinoma (Caki-1) cells (21), they are an indispensable system for KSHV research, as they are highly permissive for entry of KSHV (22) and support the lytic replication of KSHV upon conditional expression of the lytic transactivator RTA (23). Important for our studies, SLK cells express the four major ND10 components PML, SP100, DAXX, and ATRX and form clearly distinguishable ND10 structures. High-quality antibodies that allow the detection of these proteins in cells of human origin exist. Furthermore, even though RRV infection of unaltered SLK cells mostly results in latent infection, as evidenced by a lack of a visible cytopathogenic effect (data not shown), the cells support low levels of lytic replication. As demonstrated in Fig. 1, infection of SLK cells results in detectable levels of late and structural gene expression, but the expression levels of the late genes (normalized to those of the HPRT cellular housekeeping gene) are approximately 2 to 3 log units lower than those in rhesus monkey fibroblasts. While relative ORF73 mRNA levels were comparable for rhesus monkey fibroblasts and SLK cells at the 8-h time point, relative mRNA levels of, e.g., ORF50, ORF75, and the late terminase gene (ORF29) were approximately 2 to 3 orders of magnitude lower in SLK cells than in rhesus monkey fibroblasts. The increase of late gene expression between 8 h and 48 h is approximately 3 orders of magnitude less pronounced in SLK cells than in rhesus monkey fibroblasts, and expression of the lytic switch protein ORF50 is consistently delayed and significantly lower in SLK cells than in rhesus monkey fibroblasts. We therefore chose SLK cells as our primary model for studying the interaction of RRV with ND10 during establishment of infection, to see how that compares to the interaction in related rhadinoviruses, in particular, KSHV.
FIG 1.
mRNA Expression of different RRV genes in infected rhesus monkey fibroblasts (RF) and infected SLK cells. Rhesus monkey fibroblasts and SLK cells were infected under conditions corresponding to an MOI of approximately 1. Total RNA was prepared from the cells at the indicated time points postinfection and analyzed by quantitative real-time PCR. Bars represent the level of expression relative to that of HPRT. Lines with diamonds and squares represent values corrected for residual input DNA using the respective sample not treated with reverse transcriptase (only the values for ORF75 and ORF73 were corrected, as DNase I digestion did not completely remove input viral genomic DNA in some samples; ORF29 and ORF50 were detected using primer-probe sets spanning an intron). Values represent those from technical triplicates for one biological sample.
In the first experiment, we infected SLK cells with RRV-YFP at a high MOI and analyzed the amounts of PML, SP100, DAXX, and ATRX by Western blotting (Fig. 2A). In order to assess whether changes in the expression levels of these ND10-associated proteins are dependent on viral gene synthesis or the proteasome system, infection was also carried out in the presence of an inhibitor of de novo protein synthesis, cycloheximide, and in the presence of an inhibitor of the proteasome, MG132 (12). The cells were harvested for analysis at 8 h and 20 h postinfection. While the protein levels of ATRX and DAXX remained largely constant, apart from some fluctuations in the levels of DAXX at the 8-h time point, the levels of SP100 and PML were massively altered (Fig. 2A). At 8 h as well as at 20 h postinfection, an almost complete loss of SP100 was observable. Interestingly, SP100 protein levels were not rescued by inhibition of de novo protein synthesis (Fig. 2A, lanes marked for cycloheximide treatment), hinting at a component of the viral inoculum as an effector. In contrast, degradation was prevented by inhibition of the proteasome in MG132-treated cells (Fig. 2A, lanes marked for MG132 treatment). PML levels were unchanged at 8 h postinfection but were drastically reduced by 20 h. Again, reduction of PML protein levels could not be prevented by cycloheximide but could be at least partially prevented by inhibition of the proteasome. Some posttranslational modifications, such as SUMOylation, are unstable in many buffers due to the high activity of SUMO-specific proteases, in particular, in cellular lysates, and some isoforms of nuclear proteins might not be solubilized in RIPA buffer. Therefore, we also performed Western blot analysis of RRV-infected SLK cells that were harvested in SDS sample buffer and immediately heated to 95°C to preserve posttranslational modifications, followed by sonication for maximum solubilization of proteins. Under these conditions, a slightly different band pattern was observed for PML. All except two bands that were recognized by the G8 anti-PML monoclonal antibody were strongly reduced in the RRV-infected cells (Fig. 2B); the two remaining bands were most likely nonspecific, as they also appeared in PML-knockout cells (see Fig. 4A). Three additional isoforms of SP100 could be detected after rapid denaturing lysis. Even so, all isoforms of SP100 were almost or completely absent in infected SLK cells at 1 day postinfection, regardless of their apparent molecular weight. The expression levels of ATRX and DAXX were not drastically altered across the different conditions. A very slight reduction in the levels of DAXX in infected cells was observed at the 8-h time point in Fig. 2A, but this effect was not consistently seen in different experiments; it was also not observed at later time points in SLK cells (Fig. 2A; see Fig. 4A; data not shown) and was certainly far less pronounced than the effects observed on SP100 and PML. To analyze the dose-response between virus and these effects, we infected SLK cells with a dilution series of the viral inoculum (Fig. 2C). Interestingly, we saw a clear biphasic response; at high dilutions of the virus stock, we observed an induction of the interferon-inducible proteins PML and SP100, which was then reversed with increasing amounts of input virus.
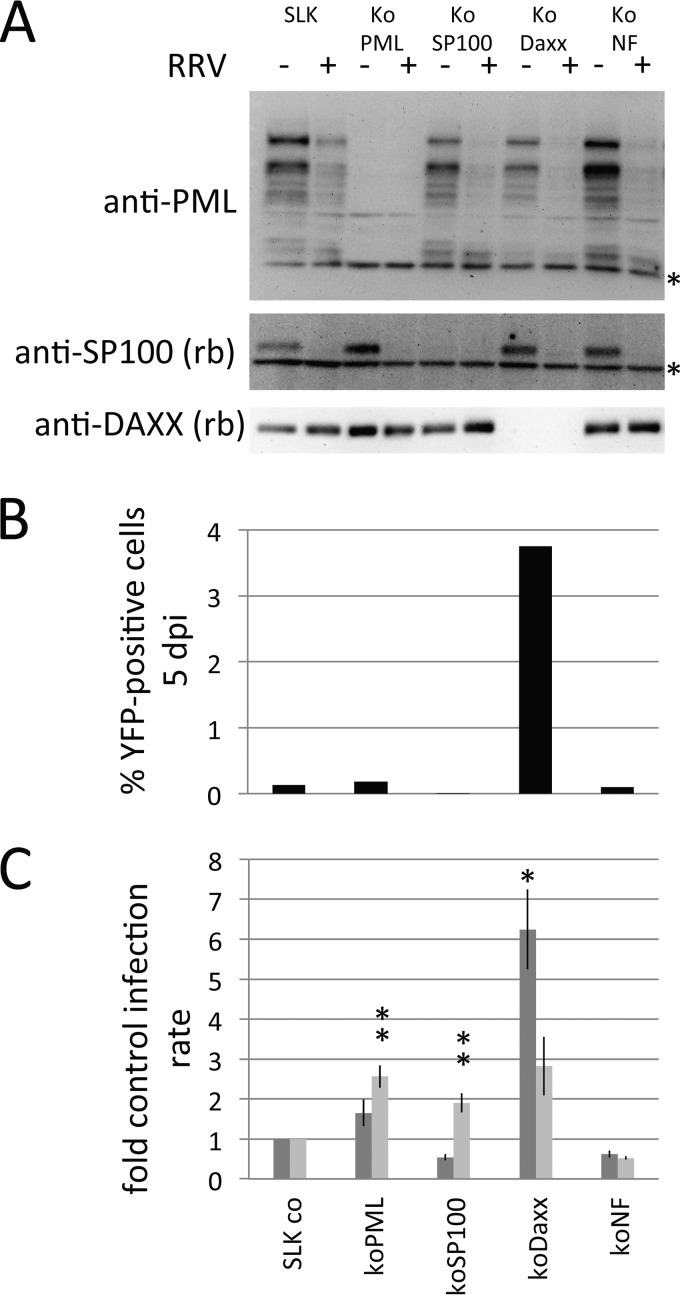
FIG 4.
Effects of PML, SP100, and DAXX knockout on RRV infection. (A) SLK cells and clonal SLK knockout cells transduced with sgRNAs to the indicated ND10 components were infected with RRV at an MOI of approximately 1 for 16 h. As an additional control, an sgRNA-Cas9-transduced clone that exhibited no knockout of the target gene ATRX was included (koNF). Asterisks, nonspecific bands. (B) The same cells used for the assay whose results are presented in panel A were infected with RRV-YFP at a very low MOI (MOI, ∼0.001). The number of YFP-positive cells was quantified by flow cytometry at 5 days postinfection (dpi). (C) Mean relative infection rate with RRV (dark gray) and KSHV (light gray) of cell clones in which the indicated proteins were knocked out. For each sgRNA, three to four clones were analyzed per experiment, and the median infection rate for the different clones was recorded and normalized to the infection rate for nontransduced SLK cells. The experiment was repeated seven times, and the mean and standard error of the mean were calculated. Significant differences relative to the infection rate in the koNF cells, as determined by one-way analysis of variance, are denoted by asterisks (*, P ≤ 0.05; **, P ≤ 0.01). Bars represent means and standard errors of the means. co, control.
Degradation of PML and SP100 after infection with RRV, as assayed by Western blotting, was mirrored by the depletion of SP100 from ND10 domains early during infection and by dissolution of ND10 at later time points, as assayed by immunofluorescence. SLK cells were infected with RRV-YFP at an MOI of >1 and were then fixed and subjected to immunofluorescence analysis at 8 h and 24 h postinfection (Fig. 3). We also compared in parallel RRV and KSHV with respect to localization of PML and SP100 after infection (Fig. 3A). Clearly, RRV effected first the loss of SP100 and then that of PML, whereas KSHV did not alter SP100 localization at ND10 and ND10 domains remained intact. In addition to high-detail stacks of single or a few cells, we also recorded stacks of larger areas and quantified the results (Fig. 3B) to assess statistical significance. At the 8-h time point, the number of ND10 domains, as indicated by the signal for the PML protein itself, was not altered (Fig. 3). SP100, on the other hand, was almost completely lost from the ND10 domains at 8 h postinfection (Fig. 3). At 24 h postinfection, the ND10 domains themselves were practically absent, in line with the reduction in the amount of the PML protein observed by Western blotting at 20 h postinfection (Fig. 2A). Again, these effects were not reverted by treatment with cycloheximide but were reverted by inhibiting proteasomal degradation with MG132. Similar effects were observed not only in SLK cells but also in primary human foreskin fibroblasts (data not shown).
Degradation of SP100 and of PML is independent from ND10 integrity.
PML is believed to provide the major structural scaffold of ND10 domains. Therefore, the question arose whether the loss of SP100 might be a consequence of interference with PML or, more generally, whether interference with one ND10 component might cause the loss of other ND10 components. In addition, we wanted to explore whether RRV required PML to target SP100 and vice versa. To address these questions, we created knockout cells using the CRISPR-Cas9 technology. SLK cells or knockout SLK cells negative for PML, SP100, or DAXX were infected with RRV at a high MOI, and expression of the individual PML components was analyzed by Western blotting (Fig. 4A). No loss of other ND10 components was witnessed upon knockout of PML, SP100, or DAXX. Evidently, degradation of SP100 by RRV also occurred in the absence of PML (Fig. 4A, third and fourth lanes), and degradation of PML also occurred in the absence of SP100 (Fig. 4A, fifth and sixth lanes). Expression of DAXX was not appreciably altered by either knockout of SP100, knockout of PML, or infection with RRV. We also infected these cells in parallel at a very low MOI (MOI, ∼0.001 for SLK cells) and quantified the infection rate (Fig. 4B). The number of YFP-positive cells was only slightly altered by knockout of SP100 and PML and was possibly even reduced in the case of SP100, whereas it was noticeably increased in the DAXX knockout cells. This hinted at a possible restriction of RRV by DAXX.
RRV is restricted by the ND10 component DAXX.
PML, Sp100, DAXX, and ATRX have been shown to inhibit infection by several viruses. We therefore asked whether these ND10 proteins and, in particular, DAXX, which is not targeted for degradation by RRV, would constitute a barrier to RRV infection, as already suggested by our initial results in Fig. 4B. Another question was how RRV would compare in that respect to the related human rhadinovirus KSHV. We again made use of knockout cell lines created with the CRISPR-Cas9 system. Individual clones were selected, tested for the absence of protein expression, and then used in infection assays. Three or four different clones of each knockout were infected with RRV-YFP (Fig. 4C, dark gray bars) and in parallel also with KSHV-GFP (Fig. 4C, light gray bars). The median infection rate of the knockout clones normalized to the infection rate achieved with normal SLK cells was taken as a readout for each individual experiment and averaged for seven repeat experiments. Clearly, knockout of DAXX caused a pronounced, approximately 6-fold increase in the number of RRV-infected cells, as determined by measurement of the level of YFP expression, followed by PML, whose knockout still led to an approximately 2-fold increase in the number of YFP-positive cells compared to the number of parental SLK cells. Compared to the effects obtained with a nonfunctional knockout (koNF), the effects were even more pronounced. Knockout of SP100, on the other hand, had no enhancing effect on RRV infection. With regard to KSHV infection, knockout of PML and of SP100 led to a detectable increase in the level of RRV infection (approximately 3-fold and 2-fold, respectively), and knockout of DAXX also led to a comparable enhancement. The results obtained with KSHV in DAXX knockout cells were more variable and did not reach significance, which cautions against further interpretation. Nevertheless, the results obtained with KSHV are in agreement with previous results using short hairpin RNA-mediated knockdown (12).
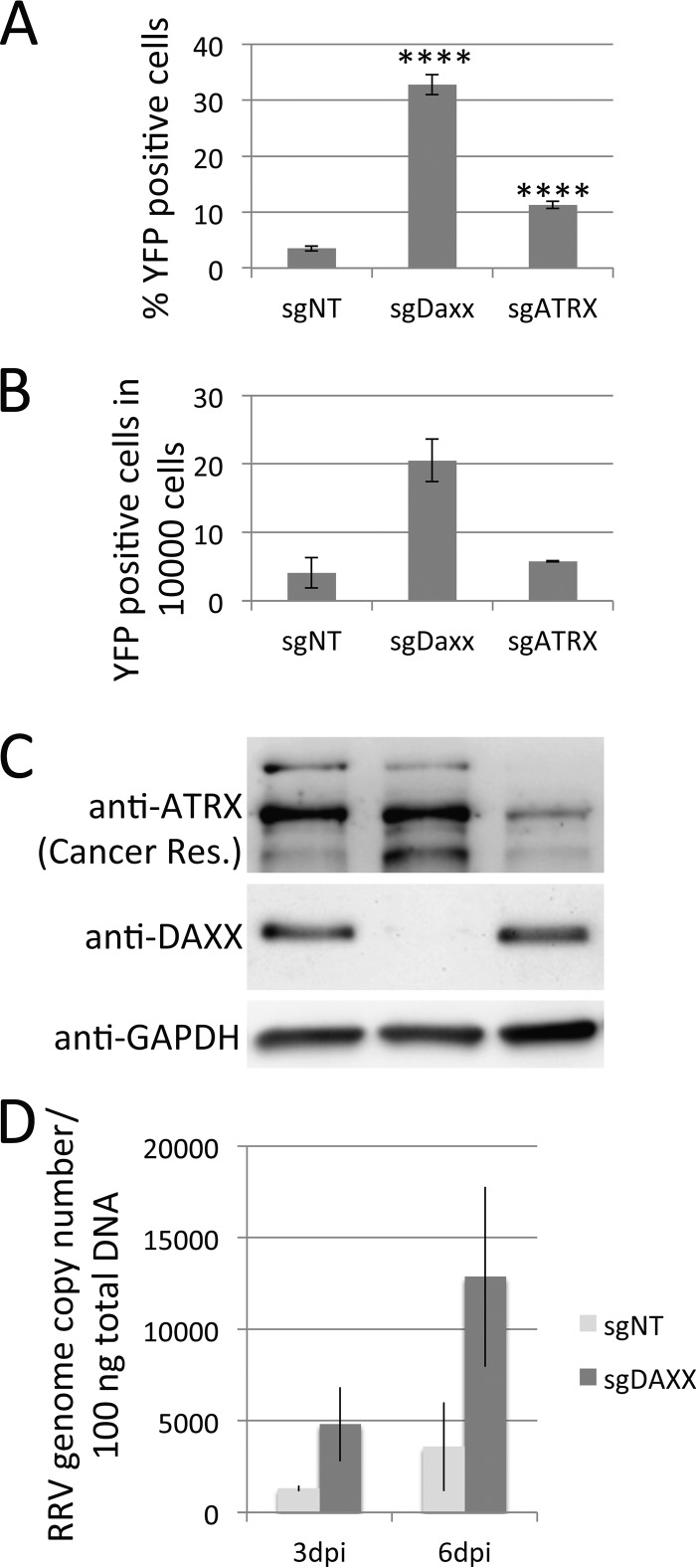
We next analyzed RRV infection not only in individual knockout clones but also in pooled cells transduced with the respective sgRNA-Cas9-expressing lentivirus at a high MOI, followed by a brief selection with puromycin to eliminate nontransduced cells (Fig. 5). Here, we also included ATRX in our analysis because, unlike with individual clones, we did not encounter problems generating a pool of transduced cells that exhibited a robust reduction in ATRX expression at the protein level (Fig. 5C). As observed before, the reduction in DAXX expression correlated with an increased permissiveness for RRV infection (Fig. 5A). While a reduction in ATRX expression also led to enhanced RRV infection, the effect was modest compared to that seen upon knockout of DAXX. In order to verify that an increased number of YFP-expressing cells really represents RRV infection and not only the differential expression of the YFP reporter gene, we made use of RRV's ability to replicate, albeit at lower levels, on SLK cells. After initial infection of the pooled knockout cells at an MOI of ∼1 and removal of the inoculum, the virus-containing supernatant was collected at days 3 to 7 postinfection and used to infect fresh SLK cells. Clearly, the knockout of DAXX led not only to increased infection of target cells in the first place but also, consequently, to the increased production of infectious progeny virus (Fig. 5B) and to an increase in viral genome copy number at days 3 and 6 postinfection (Fig. 5D), thereby validating YFP expression from RRV-YFP as a faithful reporter of viral infection, in addition to earlier reports characterizing RRV-GFP (15).
FIG 5.
RRV is restricted by DAXX. (A) RRV-YFP infection rate on pooled cells in which DAXX and ATRX were knocked out. Cells were harvested at 4 days postinfection with RRV-YFP and were analyzed for YFP expression by flow cytometry. Significant differences relative to the infection rate in the cells transduced with nonspecifically targeting sgRNA (sgNT), as determined by Student's t test, are denoted by asterisks (****, P ≤ 0.0001). Bars represent means and standard deviations. MW, molecular weight (the numbers to the left of the gels are in thousands). (B) Virus yield from protein knockout cells. Knockout SLK cells were infected with RRV-YFP at an MOI of ∼1. Cell culture supernatants were collected on days 3 to 7 postinfection and inoculated onto fresh SLK cells. The number of YFP-positive cells was assayed as described in the legend to panel A. Results are averages from two independent infections, and error bars represent the minimum and maximum. (C) Western blot analysis of knockout cell pools used for the assays whose results are presented in panels B and C. (D) Viral genome copy number analysis in DAXX and control knockout cells. Total DNA was isolated at days 3 and 6 postinfection (MOI, ∼0.1 as determined in cells transfected with nonspecifically targeting sgRNA), and the RRV genome copy number was determined by quantitative real-time PCR analysis. sgDaxx and sgATRX, sgRNA to DAXX and ATRX, respectively.
RRV ORF75 induces degradation of SP100 and of PML.
The observation that inhibition of de novo protein synthesis did not rescue SP100 and PML protein levels after infection of SLK cells and HFFs with RRV already suggested that a component of the viral inoculum effects their degradation. This notion was further corroborated by the use of UV-inactivated RRV. While it was slightly less efficient than nontreated virus, UV-inactivated RRV reduced the protein levels of SP100 and PML, as assayed by Western blotting (Fig. 6A), as well as the number of SP100 and PML dots per nucleus (Fig. 6B), as assayed by immunofluorescence. The effect on PML was slightly less pronounced than the effect on SP100. Viral FGARAT homologs are incorporated into HVS and KSHV particles (12, 13). The RRV FGARAT homolog was detected in the pelleted supernatant of infected cells and was partially resistant to trypsin treatment in the absence of detergent, compatible with a protein residing in the tegument of the virus (Fig. 7A). Therefore, we analyzed ORF75 as a candidate viral effector protein. Indeed, we found that expression of RRV ORF75 in transfected SLK cells (data not shown) and transduced rhesus monkey fibroblasts (Fig. 7B) as well as transduced SLK cells (Fig. 8) was sufficient to induce the loss of SP100 and PML, as assayed by immunofluorescence. In SLK cells transduced with a lentiviral vector expressing RRV ORF75, protein expression levels of both PML and SP100 were strongly reduced, as assayed by Western blotting (Fig. 7C). When the proteasome inhibitor MG132 was added to ORF75-expressing cells, ND10 domains were reconstituted (Fig. 8, rightmost column). Under conditions of proteasome inhibition, a pronounced colocalization of RRV ORF75 with PML and SP100 at ND10 structures could be observed (Fig. 8, rightmost column, merge + Hoechst overview and inset).
Deletion of ORF75 results in a replication-deficient virus.
In order to further analyze the contribution of ORF75 to RRV infection, we generated a functional ORF75 knockout mutant by inserting a stop codon near the amino terminus of the ORF75 open reading frame. Upon coculture of BAC-transfected 293T cells with rhesus monkey fibroblasts, we observed the very sporadic transmission of YFP fluorescence to cells, which, on the basis of their morphology, were tentatively identified to be rhesus monkey fibroblasts. Within the fibroblasts, no further spread was observable, and no free virus could be recovered after transfer of the supernatant to fresh rhesus monkey fibroblasts. In summary, transfection of four different ORF75 knockout clones did not result in the recovery of free virus, whereas transfection of wild-type virus or of three revertants from one of the knockout clones resulted in the recovery of infectious virus, as evidenced by YFP reporter gene expression (Fig. 9). We therefore conclude that ORF75 of RRV is essential for virus replication, at least within our rhesus monkey fibroblast culture system.
Degradation of PML and SP100 by RRV in rhesus monkey fibroblasts.
RRV infection of human SLK cells leads to a predominantly latent infection in most cells, as documented by an absence of a cytopathogenic effect and a very low yield of progeny virus. In contrast, primary rhesus monkey fibroblasts are fully permissive for lytic replication of RRV, with practically every single infected cell entering the lytic cycle, as evidenced by the complete disintegration of the cell layer and high virus yields. We therefore also analyzed the fate of SP100 and PML upon RRV infection in these cells. While the general pattern was similar in these cells, with SP100 and PML being degraded within 8 h and remaining absent at 24 h postinfection, as analyzed by Western blotting (Fig. 10A) and immunofluorescence (Fig. 10B), several notable differences between SLK cells and primary rhesus monkey fibroblasts became apparent. Protein levels of SP100 were barely reconstituted by inhibition of the proteasome at the 8-h time point but not at the 24-h time point postinfection, as assayed by Western blotting and immunofluorescence (Fig. 10B), which contrasts with our results with SLK cells (Fig. 2A and 3B) and human foreskin fibroblasts (data not shown). Similar to our results in SLK cells, inhibition of de novo gene synthesis with cycloheximide did not prevent the loss of SP100 at 8 h and 24 h postinfection, as assayed by Western blotting (Fig. 10A). According to the results of immunofluorescence analysis, cycloheximide marginally stabilized SP100 levels in ND10 at 8 h postinfection (Fig. 10B) but did not prevent its complete ablation at 24 h postinfection. In contrast to our findings with other cell types (Fig. 3B and data not shown), MG132 did not prevent degradation of PML in rhesus monkey fibroblasts by 24 h postinfection, as assayed by Western blotting (Fig. 10A) and by immunofluorescence (Fig. 10B). Western blot analysis revealed partial preservation of PML protein levels by cycloheximide at the 24-h time point (Fig. 10A), which was also mirrored by the observation that at least some PML bodies were still detectable by immunofluorescence analysis in infected cells under cycloheximide treatment (Fig. 10B). UV-inactivated virus induced degradation of PML and SP100 by 24 h (Fig. 10A and B), similar to our results with SLK cells. It should be noted that detection of both SP100 and PML in lysates of rhesus monkey fibroblasts by Western blotting using antibodies to human SP100 was complicated by a higher background and lower specificity than detection in lysates of cells of human origin. Even so, at least one protein band with a molecular weight of approximately 75,000 recognized by our SP100 antibody was clearly absent in RRV-infected cells, and so were several bands recognized by the PML antibody. We also infected subconfluent rhesus monkey fibroblasts with an amount of virus that led to 100% of the cells expressing the YFP reporter gene within 24 h, indicating a very high, saturating MOI (data not shown). Consistent with our observations presented in Fig. 10, cycloheximide did not prevent the degradation of SP100 by 24 h. However, as already observed by Western blot analysis at an MOI of ∼1 (Fig. 10A), cycloheximide was, surprisingly, able to prevent degradation of PML also under conditions of a very high MOI (not shown) in rhesus monkey fibroblasts, possibly implicating de novo gene synthesis in the disruption process in rhesus cells and some sort of stabilization of PML under these conditions.
DISCUSSION
Taken together, our results demonstrate that RRV efficiently degrades both SP100 and PML through the action of its viral FGARAT (vFGARAT) homolog ORF75, with SP100 being degraded with a considerably faster kinetic. Conversely, we found that knockout of the genes for these proteins increases infection by RRV only slightly for PML or not at all for SP100, consistent with the notion that RRV escapes the antiviral effector functions of PML and SP100 through targeting them for proteasomal degradation. The other two ND10 components that were analyzed in this study, DAXX and ATRX, were not found to be targets of degradation by RRV. It has to be noted that we observed some fluctuation in the expression levels of DAXX, as assayed by Western blotting, after infection with RRV, but these minor differences were not consistently observed. Although DAXX and ATRX lose their association with ND10 as RRV degrades PML, the major structural scaffold of the ND10 structure (9), they still seem to exert a negative effect on RRV infection. This concept is corroborated by the fact that knockout of DAXX and also, to a lesser degree, knockout of ATRX increased infection with RRV. For DAXX knockout, this increase in the number of infected cells was also mirrored by an increase in RRV genome copy number (Fig. 5D) and in progeny virus after replication (Fig. 5B). Knockout of DAXX had the most pronounced effect on RRV infection at a low MOI compared to the effect of knockout of PML, SP100, or ATRX, resulting in an up to 10-fold increase in infection (Fig. 4B and C and 5A). As we observed these effects very early, before a second round of infection could lead to detectable reporter gene expression, we conclude that the DAXX-mediated restriction occurs predominantly initially, during establishment of infection. This notion is also supported by the fact that the increase in infectious progeny virus or RRV genome copy number upon knockout of DAXX is of a similar order of magnitude as the increase in infectious progeny virus or RRV genome copy number in the first place.
We found that the RRV FGARAT homolog ORF75 is sufficient to induce degradation of SP100 and PML when expressed in cells by recombinant means (Fig. 7B and C and 8). The known viral homologs of this protein are part of the virion of HVS (13), KSHV (12), and Epstein-Barr virus (EBV) (24), and we clearly detected ORF75 in the lysate of pelleted RRV-containing cell supernatant (Fig. 7A). A substantial fraction of RRV ORF75 was protected from proteolysis by trypsin in the absence of detergent, compatible with the characteristics of a tegument protein (Fig. 7A). Interestingly, a sizable fraction of ORF75 was sensitive to trypsin without detergent, hinting at a possible release also of nonenveloped ORF75 from infected cells. When expressed by means of lentiviral transduction, localization of RRV ORF75 did not seem to be particularly concentrated at ND10 bodies, albeit this was most likely attributable to the fact that ND10 bodies were relatively rapidly dissolved (Fig. 8, third column). Upon inhibition of the proteasome, RRV ORF75 prominently colocalized with PML and SP100 at ND10 (Fig. 8, rightmost column). Thus, when RRV ORF75 was expressed on its own, it targeted the ND10 structure and induced the degradation of the main ND10 components, PML and SP100. Our findings further illustrate conservation of the biological function between different viral FGARAT homologs, albeit in a broader sense. While the exact cellular targets and the mechanisms of action differ between the vFGARATs of HVS, KSHV, EBV, and MHV68, they all target components of the ND10 structure and induce their degradation, relocalization, or both.
Compared to the other rhadinoviruses analyzed so far, RRV employs a slightly different strategy to counteract ND10. RRV and KSHV are biologically very similar but are different enough, especially on the molecular level, to allow meaningful comparisons that can identify important evolutionarily conserved concepts and differences. Unlike the vFGARAT of KSHV, RRV ORF75 does not affect the protein levels of ATRX, at least under our experimental conditions. Like the vFGARAT ORF75c of MHV68, RRV ORF75 induces degradation of PML. Similar to ORF3 of HVS, one of two pirated HVS FGARAT genes, RRV ORF75 induces degradation of SP100. Mechanistically, RRV ORF75 acts more similarly to the vFGARATs of HVS and of MHV68 than to the vFGARAT of KSHV, as the former two viruses also induce proteasomal degradation of their ND10-associated targets, whereas KSHV relies on a so far elusive nonproteasomal mechanism. These differences are remarkable, as, with regard to the primary sequence, RRV ORF75 is closer to the FGARAT homolog of KSHV than to that of HVS, a T cell-transforming virus. Furthermore, RRV's lifestyle is clearly more similar to KSHV's than to HVS's, and for both RRV (Fig. 9) and KSHV (12), their respective FGARAT gene homologs are essential for replication.
Our findings unequivocally demonstrate the ability of the RRV FGARAT homolog ORF75 to induce degradation of PML and SP100. Nevertheless, the interplay between RRV and ND10 is more complex. This is evidenced by our findings in rhesus monkey fibroblasts, which support efficient lytic replication of RRV. UV-inactivated RRV was slightly less efficient than untreated virus at inducing degradation of SP100 and PML in SLK cells (Fig. 6) and rhesus monkey fibroblasts (Fig. 10), hinting at the possibility that additional viral gene products that are not present in the virion particle target these ND10 components. In infected rhesus monkey fibroblasts, expression of PML and SP100 could be at most partially restored by MG132 at the 8-h time point postinfection and only marginally or not at all at the 24-h time point (Fig. 10). This suggests the existence of an additional, proteasome-independent mechanism for degradation of SP100 and PML. These observations indicate that in cells that support efficient lytic replication, in addition to virion-associated proteins, de novo-synthesized gene products, perhaps from copackaged viral mRNA delivered with the viral particle (25) or from very early transcripts, can contribute to the degradation of SP100 and, in particular, of PML. An attractive hypothesis would be that early gene products of RRV contribute to degradation of SP100 and PML and induce their nonproteasomal degradation, in addition to the effects of the tegument protein ORF75. Alternatively, the confounding effects of interferon induction of PML and SP100 and uncharted functions of most of the eight different RRV-encoded viral interferon regulatory factors (vIRFs) could be of relevance here. Having different and redundant effectors targeting ND10 components would be similar to KSHV's strategy: the KSHV lytic switch protein RTA induces degradation of PML (26), while vIRF-3 (27) also targets PML for degradation and KSHV ORF75 (12) targets ATRX. Similarly, MHV68 encodes at least one other PML-modifying protein, ORF61 (28), despite MHV68's ability to potently induce degradation of PML through its FGARAT homolog, ORF75c. These redundant strategies might be needed, as a single effector protein may not be sufficient to ablate certain ND10 components in all cell types and under all conditions. It should be noted that effects exerted by the tegument seem to extend also to cells that do not become productively infected. In our experiments, nearly all cells treated with a viral inoculum corresponding to an MOI of ∼1 (on the basis of cell expression of YFP), which would correspond to ∼63% of infected cells, lost SP100; thus, the other third of cells do not receive a viral genome capable of mediating the YFP reporter gene or, presumably, viral gene expression or only noninfectious particles still containing a set of viral tegument proteins. This is not surprising, as all herpesviruses are known to have a high excess of noninfectious particles, characterized by particle-to-PFU ratios of 1:100 to 1:1,000 or even higher. In addition, our results demonstrate that a sizable fraction of the ORF75 found in our viral inoculum is sensitive to trypsin and indicate that ORF75 is also present as a free protein in crude virus preparations and the supernatants of infected cells (Fig. 7A). While it was not analyzed in this study, it is more likely than not that ORF75 is also present in defective viral particles, exosomes, or other vesicular fractions that are released from infected cells.
Among the four ND10 proteins analyzed in our study, SP100 emerges as a very prominent target for the rhadinoviruses and for herpesviruses in general. It is targeted for degradation by the gamma-2-herpesviruses HVS (13) and RRV, as demonstrated in this study. In addition, it is also targeted by the alphaherpesvirus herpes simplex virus (29) and by the betaherpesvirus human CMV (30). Even though KSHV does not degrade SP100, KSHV effects its relocalization from ND10 upon activation of the lytic cycle (12). It is tempting to speculate that SP100 might possess the ability to generally restrict DNA viruses and therefore became a prime target of viral counteraction strategies. It has also been described to inhibit infection by papillomaviruses (31). Adenovirus 5 reportedly relocalizes SP100 isoforms B, C, and HMG away from ND10, while the dominant isoform, isoform A, is recruited to adenoviral replication centers (32). RRV acts considerably less subtly and, according to our results, simply degrades all isoforms of SP100 that were detectable in the cells used in our study and does so independently of an intact ND10 structure, as shown in the absence of PML. Taken together, our observations and those of others suggest a very broad and potent antiviral effect of SP100 that necessitates viral counteraction, which warrants more detailed analysis of SP100's mechanism of action.
An obvious question is why inactivation of any one or two of the major ND10 components seems to be sufficient for individual herpesviruses to meet their replicative needs. A hypothetical explanation is that targeting too many of these proteins might induce detrimental effects that would offset the benefits of increased viral replication and that the viruses need to strike a balance here. In addition, all herpesviruses rely on lifelong persistence in the host organism and prefer to maintain the latent state in most cells. Completely unchecked replication may therefore not be favorable for this particular viral lifestyle, as it would likely result in overshooting of the immune responses or even death of the host.
ACKNOWLEDGMENTS
We thank Ronald C. Desrosiers, Myriam Scherer, Anna Reichel, and Thomas Stamminger for reagents and helpful discussions. We thank Sarah-Christin Schlagowski for technical assistance.
This work was supported by grants to A.E. by the Deutsche Forschungsgemeinschaft (DFG; SFB796 project B1), Wilhelm Sander Stfitung 2013.105.1, and BaCaTec. A.S.H. was supported by the DFG (Ha 6013/1-1 Rückkehrstipendium). A.S.H. and A.K.G. were supported by the IZKF Erlangen (project J44) and through intramural funding of the German Primate Center.
Funding Statement
This work, including the efforts of Armin Ensser, was funded by Bavaria California Technology Center (BaCaTeC; A8), by Deutsche Forschungsgemeinschaft (DFG; SFB796 project B1), and by Wilhelm Sander-Stiftung (2013.105.1). This work, including the efforts of Alexander Siegfried Hahn, was funded by IZKF Erlangen (J44), by Deutsche Forschungsgemeinschaft (DFG) (Ha 6013/1-1), and by Deutsches Primatenzentrum (DPZ). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Desrosiers RC, Sasseville VG, Czajak SC, Zhang X, Mansfield KG, Kaur A, Johnson RP, Lackner AA, Jung JU. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J Virol 71:9764–9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orzechowska BU, Powers MF, Sprague J, Li H, Yen B, Searles RP, Axthelm MK, Wong SW. 2008. Rhesus macaque rhadinovirus-associated non-Hodgkin lymphoma: animal model for KSHV-associated malignancies. Blood 112:4227–4234. doi: 10.1182/blood-2008-04-151498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruce AG, Bielefeldt-Ohmann H, Barcy S, Bakke AM, Lewis P, Tsai C-C, Murnane RD, Rose TM. 2012. Macaque homologs of EBV and KSHV show uniquely different associations with simian AIDS-related lymphomas. PLoS Pathog 8:e1002962. doi: 10.1371/journal.ppat.1002962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renne R, Dittmer D, Kedes D, Schmidt K, Desrosiers RC, Luciw PA, Ganem D. 2004. Experimental transmission of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) to SIV-positive and SIV-negative rhesus macaques. J Med Primatol 33:1–9. doi: 10.1046/j.1600-0684.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 5.Hahn AS, Desrosiers RC. 2013. Rhesus monkey rhadinovirus uses Eph family receptors for entry into B cells and endothelial cells but not fibroblasts. PLoS Pathog 9:e1003360. doi: 10.1371/journal.ppat.1003360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeMaster LK, Rose TM. 2014. A critical Sp1 element in the rhesus rhadinovirus (RRV) Rta promoter confers high-level activity that correlates with cellular permissivity for viral replication. Virology 448:196–209. doi: 10.1016/j.virol.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruce AG, Thouless ME, Haines AS, Pallen MJ, Grundhoff A, Rose TM. 2015. Complete genome sequence of pig-tailed macaque rhadinovirus 2 and its evolutionary relationship with rhesus macaque rhadinovirus and human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J Virol 89:3888–3909. doi: 10.1128/JVI.03597-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Negorev D, Maul GG. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234–7242. doi: 10.1038/sj.onc.1204764. [DOI] [PubMed] [Google Scholar]
- 9.Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF, Maul GG. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J Cell Biol 147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tavalai N, Stamminger T. 2009. Interplay between herpesvirus infection and host defense by PML nuclear bodies. Viruses 1:1240–1264. doi: 10.3390/v1031240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Regad T, Chelbi-Alix MK. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274–7286. doi: 10.1038/sj.onc.1204854. [DOI] [PubMed] [Google Scholar]
- 12.Full F, Jungnickl D, Reuter N, Bogner E, Brulois K, Scholz B, Stürzl M, Myoung J, Jung JU, Stamminger T, Ensser A. 2014. Kaposi's sarcoma associated herpesvirus tegument protein ORF75 is essential for viral lytic replication and plays a critical role in the antagonization of ND10-instituted intrinsic immunity. PLoS Pathog 10:e1003863. doi: 10.1371/journal.ppat.1003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Full F, Reuter N, Zielke K, Stamminger T, Ensser A. 2012. Herpesvirus saimiri antagonizes nuclear domain 10-instituted intrinsic immunity via an ORF3-mediated selective degradation of cellular protein Sp100. J Virol 86:3541–3553. doi: 10.1128/JVI.06992-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 15.Bilello JP, Morgan JS, Damania B, Lang SM, Desrosiers RC. 2006. A genetic system for rhesus monkey rhadinovirus: use of recombinant virus to quantitate antibody-mediated neutralization. J Virol 80:1549–1562. doi: 10.1128/JVI.80.3.1549-1562.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou F, Li Q, Wong SW, Gao S-J. 2010. Autoexcision of bacterial artificial chromosome facilitated by terminal repeat-mediated homologous recombination: a novel approach for generating traceless genetic mutants of herpesviruses. J Virol 84:2871–2880. doi: 10.1128/JVI.01734-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suraweera A, Münch C, Hanssum A, Bertolotti A. 2012. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell 48:242–253. doi: 10.1016/j.molcel.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahne F, LeMeur N, Brinkman RR, Ellis B, Haaland P, Sarkar D, Spidlen J, Strain E, Gentleman R. 2009. flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinformatics 10:106. doi: 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis B, Gentleman R, Hahne F, Le Meur N, Sarkar DD, Jiang M. 2016. flowViz: visualization for flow cytometry. R package, version 1362. The R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 21.Stürzl M, Gaus D, Dirks WG, Ganem D, Jochmann R. 2013. Kaposi's sarcoma-derived cell line SLK is not of endothelial origin, but is a contaminant from a known renal carcinoma cell line. Int J Cancer 132:1954–1958. doi: 10.1002/ijc.27849. [DOI] [PubMed] [Google Scholar]
- 22.Hahn AS, Kaufmann JK, Wies E, Naschberger E, Panteleev-Ivlev J, Schmidt K, Holzer A, Schmidt M, Chen J, Konig S, Ensser A, Myoung J, Brockmeyer NH, Sturzl M, Fleckenstein B, Neipel F. 2012. The ephrin receptor tyrosine kinase A2 is a cellular receptor for Kaposi's sarcoma-associated herpesvirus. Nat Med 18:961–966. doi: 10.1038/nm.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myoung J, Ganem D. 2011. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: maintenance of tight latency with efficient reactivation upon induction. J Virol Methods 174:12–21. doi: 10.1016/j.jviromet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, Illanes D, Sarracino D, Kieff E. 2004. Proteins of purified Epstein-Barr virus. Proc Natl Acad Sci U S A 101:16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purushothaman P, Thakker S, Verma SC. 2015. Transcriptome analysis of Kaposi's sarcoma-associated herpesvirus during de novo primary infection of human B and endothelial cells. J Virol 89:3093–3111. doi: 10.1128/JVI.02507-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izumiya Y, Kobayashi K, Kim KY, Pochampalli M, Izumiya C, Shevchenko B, Wang D-H, Huerta SB, Martinez A, Campbell M, Kung H-J. 2013. Kaposi's sarcoma-associated herpesvirus K-Rta exhibits SUMO-targeting ubiquitin ligase (STUbL) like activity and is essential for viral reactivation. PLoS Pathog 9:e1003506. doi: 10.1371/journal.ppat.1003506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcos-Villar L, Lopitz-Otsoa F, Gallego P, Muñoz-Fontela C, González-Santamaría J, Campagna M, Shou-Jiang G, Rodriguez MS, Rivas C. 2009. Kaposi's sarcoma-associated herpesvirus protein LANA2 disrupts PML oncogenic domains and inhibits PML-mediated transcriptional repression of the survivin gene. J Virol 83:8849–8858. doi: 10.1128/JVI.00339-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sewatanon J, Ling PD. 2014. Murine gammaherpesvirus 68 encodes a second PML-modifying protein. J Virol 88:3591–3597. doi: 10.1128/JVI.03081-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chelbi-Alix MK, de Thé H. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 30.Kim Y-E, Lee J-H, Kim ET, Shin HJ, Gu SY, Seol HS, Ling PD, Lee CH, Ahn J-H. 2011. Human cytomegalovirus infection causes degradation of Sp100 proteins that suppress viral gene expression. J Virol 85:11928–11937. doi: 10.1128/JVI.00758-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stepp WH, Meyers JM, McBride AA. 2013. Sp100 provides intrinsic immunity against human papillomavirus infection. mBio 4:e00845-13. doi: 10.1128/mBio.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berscheminski J, Wimmer P, Brun J, Ip WH, Groitl P, Horlacher T, Jaffray E, Hay RT, Dobner T, Schreiner S. 2014. Sp100 isoform-specific regulation of human adenovirus 5 gene expression. J Virol 88:6076–6092. doi: 10.1128/JVI.00469-14. [DOI] [PMC free article] [PubMed] [Google Scholar]