ABSTRACT
Hepadnaviruses (hepatitis B viruses [HBVs]) are the only animal viruses that replicate their DNA by reverse transcription of an RNA intermediate. Until recently, the known host range of hepadnaviruses was limited to mammals and birds. We obtained and analyzed the first amphibian HBV genome, as well as several prototype fish HBVs, which allow the first comprehensive comparative genomic analysis of hepadnaviruses from four classes of vertebrates. Bluegill hepadnavirus (BGHBV) was characterized from in-house viral metagenomic sequencing. The African cichlid hepadnavirus (ACHBV) and the Tibetan frog hepadnavirus (TFHBV) were discovered using in silico analyses of the whole-genome shotgun and transcriptome shotgun assembly databases. Residues in the hydrophobic base of the capsid (core) proteins, designated motifs I, II, and III, are highly conserved, suggesting that structural constraints for proper capsid folding are key to capsid protein evolution. Surface proteins in all vertebrate HBVs contain similar predicted membrane topologies, characterized by three transmembrane domains. Most striking was the fact that BGHBV, ACHBV, and the previously described white sucker hepadnavirus did not form a fish-specific monophyletic group in the phylogenetic analysis of all three hepadnaviral genes. Notably, BGHBV was more closely related to the mammalian hepadnaviruses, indicating that cross-species transmission events have played a major role in viral evolution. Evidence of cross-species transmission was also observed with TFHBV. Hence, these data indicate that the evolutionary history of the hepadnaviruses is more complex than previously realized and combines both virus-host codivergence over millions of years and host species jumping.
IMPORTANCE Hepadnaviruses are responsible for significant disease in humans (hepatitis B virus) and have been reported from a diverse range of vertebrates as both exogenous and endogenous viruses. We report the full-length genome of a novel hepadnavirus from a fish and the first hepadnavirus genome from an amphibian. The novel fish hepadnavirus, sampled from bluegills, was more closely related to mammalian hepadnaviruses than to other fish viruses. This phylogenetic pattern reveals that, although hepadnaviruses have likely been associated with vertebrates for hundreds of millions of years, they have also been characterized by species jumping across wide phylogenetic distances.
INTRODUCTION
The Hepadnaviridae are characterized by extremely small (3- to 3.3-kbp), partially double-stranded DNA (dsDNA) genomes. The viral particles are spherical, with a diameter of approximately 42 nm, each containing a single copy of the genome covalently linked to the viral reverse transcriptase (RT), which provides DNA polymerase activity (1–3). The hepadnaviruses are unique among animal viruses in that they replicate their DNA by reverse transcription of an RNA intermediate and comprise the only group VII animal virus (dsDNA-RT virus) of the Baltimore system, which classifies viruses according to their genome compositions and methods of replication (1, 4).
At present, the Hepadnaviridae are subdivided into two genera (5–7): the genus Orthohepadnavirus, which infects mammals, including humans, and the genus Avihepadnavirus, which infects birds (1, 8–13). Within both genera, the circular viral genomes exhibit multiple overlapping open reading frames (ORFs), comprising the polymerase, pre C/C, and pre S/S ORFs, which encode the viral polymerase (P), core (C), and surface (S) proteins, respectively. In the genus Orthohepadnavirus, a fourth ORF encodes protein X. Despite these similar genome organizations, nucleotide sequence identity between hepadnavirus genera is limited, with the exception of some highly conserved functional domains (14, 15).
Human hepatitis B virus (HuHBV) affects more than one-third of the human population, and infections have the potential to cause both severe chronic liver disease and hepatocellular carcinoma (1–3). Interestingly, chronic infection by woodchuck hepatitis B virus (WHBV) can result in similar pathological changes in that species (11, 12). Liver pathology is less commonly induced by avihepadnaviruses, although duck hepatitis B virus (DHBV) can cause liver necrosis (9). The first hepadnavirus from a bony fish, the white sucker (Catostomus commersonii), class Actinopterygii, was described in 2015, although no disease association was observed (16). To date, no exogenous reptilian or amphibian hepadnaviruses have been reported.
In addition to exogenous hepadnaviruses, a number of endogenous sequences (eHBV), in the form of endogenous viral elements (EVEs) (60), have been identified in animal genomes. Hepadnaviral EVEs have been documented in turtles, crocodiles, snakes, and birds (14, 17–19), although no mammalian, amphibian, or fish endogenous hepadnaviruses have yet been detected. The presence of EVEs has helped provide a time scale of hepadnavirus evolution, particularly as some of the endogenization events may have occurred as early as 200 million years ago (15). Hence, although there is clear evidence for some cross-species transmission (20), current data suggest that hepadnavirus evolution largely follows a pattern of virus-host codivergence that extends to at least the origin of the ray-finned fish.
To better understand the host range and evolution of the hepadnaviruses in vertebrates, particularly the extent of virus-host codivergence, we investigated new fish and amphibian (exogenous) hepadnaviral homologs that are highly divergent from the hepadnaviruses previously described in mammals and birds. They include the second fish hepadnavirus, from bluegill sunfish (Lepomis macrochirus); the first amphibian hepadnavirus from a Tibetan frog (Nanorana parkeri); and analysis of a hepadnavirus-like sequence from Lake Tanganyika African cichlid fish (Ophthalmotilapia ventralis).
MATERIALS AND METHODS
Sample collection.
The tissues used in this study were originally part of an investigation into suspected virus-induced orocutaneous neoplasms in two populations of bluegills. In total, 46 fish were examined, including 40 bluegills, five related Lepomis spp., and one largemouth bass, Micropterus salmoides (Table 1). Five bluegills from a mixed-species aquarium exhibit were submitted to the Aquatic Pathology Service at the College of Veterinary Medicine, University of Georgia, in 2009. In 2014, similar lesions were observed on bluegills by a pond owner in Waleska, GA. Between April 2014 and July 2015, 26 bluegills, 17 with lesions and 9 without, were received, along with 1 largemouth bass. An additional five bluegills, two redbreast sunfish (Lepomis auritus), and two redear sunfish (Lepomis microlophus) were received from a commercial fish hatchery in Hawkinsville, GA, in January 2015. Four bluegills and one green sunfish (Lepomis cyanellus) were received from local anglers in the Athens, GA, area in September 2015.
TABLE 1.
Molecular screening of BGHBV in bluegill, related Lepomis species, and other fish
| Source (date [mo-day-yr]) | No. | Fish | Species | Lip papillomaa | Skin lesiona | Hepadnavirus PCRb |
||
|---|---|---|---|---|---|---|---|---|
| Lip | Skin | Liver | ||||||
| Aquarium (9-9-09) | 1 | GAI-1 | L. macrochirus | + | − | − | NA | NA |
| 2 | GAI-2c | L. macrochirus | + | − | +13.4 (4) | NA | NA | |
| 3 | GAI-3 | L. macrochirus | + | − | − | NA | NA | |
| 4 | GAI-4 | L. macrochirus | + | − | − | NA | NA | |
| 5 | GAI-5 | L. macrochirus | + | − | −0.00187 (0.0009) | NA | NA | |
| Waleska, GA (4-14-14) | 6 | WA-A1 | L. macrochirus | − | + | − | NA | NA |
| 7 | WA-A2 | L. macrochirus | + | − | − | NA | NA | |
| 8 | WA-A3 | L. macrochirus | − | − | − | NA | NA | |
| 9 | WA-A4 | L. macrochirus | + | − | − | NA | NA | |
| 10 | WA-A5 | L. macrochirus | − | + | − | − | NA | |
| Waleska, GA (7-7-14) | 11 | WA-B1 | L. macrochirus | − | − | −0.0003 (0.0001) | NA | −0 (0) |
| 12 | WA-B2 | L. macrochirus | − | − | − | NA | − | |
| 13 | WA-B3 | L. macrochirus | − | − | + | NA | NA | |
| 14 | WA-B4 | L. macrochirus | − | − | NA | − | NA | |
| 15 | WA-B5 | L. macrochirus | − | − | +5.81 (1.42) | +111 (10) | +0.179 (0.0475) | |
| 16 | WA-B6 | L. macrochirus | + | − | − | NA | NA | |
| 17 | WA-B7 | L. macrochirus | + | − | − | NA | NA | |
| 18 | WA-B8 | L. macrochirus | − | − | − | NA | NA | |
| 19 | WA-B9 | L. macrochirus | + | − | + | NA | NA | |
| 20 | WA-B10 | L. macrochirus | + | − | − | NA | − | |
| 21 | WA-B11 | L. macrochirus | + | − | − | NA | NA | |
| 22 | WA-B12 | L. macrochirus | + | − | + | +146 (12.9) | +0.0076 (0.0002) | |
| 23 | WA-B13 | L. macrochirus | + | − | − | NA | NA | |
| Waleska GA (7-4-15) | 24 | WA-C1 | L. macrochirus | + | − | −0.0012 (0.001) | NA | NA |
| 25 | WA-C2 | L. macrochirus | − | − | − | NA | NA | |
| 26 | WA-C3 | L. macrochirus | + | − | − | − | − | |
| 27 | WA-C4 | L. macrochirus | + | − | − | NA | NA | |
| 28 | WA-C5 | L. macrochirus | + | − | − | NA | − | |
| 29 | WA-C6 | L. macrochirus | − | − | − | NA | NA | |
| 30 | WA-C7 | L. macrochirus | − | − | + | + | + | |
| 31 | WA-C8 | L. macrochirus | − | − | − | NA | − | |
| 32 | WA-C9 | M. salmoides | − | − | − | NA | − | |
| Hawkinsville, GA (1-16-15) | 33 | OW-1 | L. macrochirus | − | − | −0 (0) | NA | −0.012 (0.0046) |
| 34 | OW-2 | L. macrochirus | − | − | − | NA | NA | |
| 35 | OW-3 | L. macrochirus | − | − | − | − | NA | |
| 36 | OW-4 | L. macrochirus | − | − | − | NA | NA | |
| 37 | OW-5 | L. macrochirus | − | − | − | NA | NA | |
| 38 | OW-6 | L. microlophus | − | − | − | − | − | |
| 39 | OW-7 | L. microlophus | − | − | − | NA | − | |
| 40 | OW-8 | L. auritus | − | − | − | − | − | |
| 41 | OW-9 | L. auritus | − | − | − | NA | − | |
| Athens, GA (9-1-15) | 42 | SC-1 | L. macrochirus | − | − | − | NA | NA |
| 43 | SC-2 | L. macrochirus | − | − | NA | NA | − | |
| 44 | SC-3 | L. macrochirus | − | − | − | NA | NA | |
| 45 | SC-4 | L. macrochirus | − | − | − | NA | NA | |
| 46 | SC-5 | L. cyanellus | − | − | − | NA | − | |
+, lesion or nucleic acid present; −, lesion or nucleic acid not identified in the tissue.
Triplicate qPCR values are presented as femtograms (standard deviation). NA, sample not available for evaluation.
Positive bluegill lip sample used for full-genome sequencing and metagenomic analysis.
Necropsies were performed, and samples of organs and lesions were fixed in 10% neutral buffered formalin and processed routinely for histologic evaluation. Additional samples were fixed in 2% glutaraldehyde, 2% paraformaldehyde, and 0.2% picric acid in 0.1 M cacodylate-HCl buffer and processed for transmission electron microscopy (TEM). Portions of lesions were collected separately and archived in a −80°C freezer. In addition, pooled samples of liver, spleen, and kidney were collected from a subset of fish and frozen at −80°C. Select histologic sections were later used for in situ hybridization evaluation using probes designed from PCR products.
Fin clip samples from two O. ventralis cichlids were provided by a local hobbyist and archived at −80°C.
Viral metagenomic and bioinformatics analysis of next-generation sequencing (NGS) data.
In the absence of a definitive diagnosis for the skin lesions, metagenomic sequencing was performed on seven lip lesions and one nonlesioned lip, according to previously described protocols, to further investigate a potential underlying viral etiology (21–24). In brief, a tissue homogenate was centrifuged through a 0.22-μm filter to enrich viral particles by size and then treated with nucleases to deplete host nucleic acids. Nucleic acids from nuclease-resistant viral particles were extracted using the QIAquick viral-RNA column purification system, followed by sequence-independent amplification using random priming. First-strand synthesis (for both DNA and RNA) was performed with a 28-base oligonucleotide whose 3′ end consisted of eight random nucleotides (primer N1_8N, CCTTGAAGGCGGACTGTGAGNNNNNNNN), using Superscript III reverse transcriptase (Invitrogen) (21–24). A second strand was synthesized using Klenow fragment DNA polymerase (New England BioLabs). The resulting double-stranded cDNA and DNA were then PCR amplified using AmpliTaq Gold DNA polymerase and a 20-base primer (primer N1, CCTTGAAGGCGGACTGTGAG). A dual-indexed sequencing library was then prepared using the Nextera XT DNA Sample Prep kit (Illumina). After pooling, the final library was sequenced using the MiSeq sequencing system, with 250-bp paired-end sequencing reagents (Illumina MiSeq Reagents V2; 500 cycles).
A total of 11 million reads were generated and analyzed using an in-house pipeline as previously described (23). Adaptor and primer sequences were trimmed using VecScreen (25), while duplicate reads and low-sequencing-quality tails were removed using a Phred quality score of 10 as the threshold. The cleaned reads were assembled de novo using an in-house sequence assembler employing an ensemble strategy (26) that consisted of SOAPdenovo2, ABySS, meta-Velvet, and CAP3. The assembled sequence was compared with an in-house viral protein sequence database using BLASTx. Viral contigs were further inspected manually using Geneious (version R6; Biomatters).
Complete genome sequencing of the bluegill hepadnavirus (BGHBV).
To obtain the last 1% of the genome that was not covered by NGS, PCR was performed using primers BGHBV-CirF (5′-CAACGCCAACAGCATTTTTA-3′) and BGHBV-CirR (5′-TAATATCGGTCGAGACTGCG-3′), which were anchored in the polymerase and core ORFs, bridging the intergenic region. The resulting 373-bp amplicons were sequenced using Sanger methods to confirm the circularity of the genome.
Molecular screening of the fish hepadnavirus.
Tissues from 40 bluegills, three related Lepomis species, and one largemouth bass were extracted using Qiagen DNA extraction kits. Screening for BGHBV was accomplished by traditional PCR, targeting the polymerase with the primer set BGHBV-PolF (5′-TGTGGACAAAAATCCACGAA-3′) and BGHBV-PolR (5′-CGTAAAGCACCTATGGGCAT-3′), using a previously described touchdown protocol (21). Additional primers targeting the polymerase, capsid, and core proteins were also designed and verified (Table 2).
TABLE 2.
Targeted genes, primer sequences, and product sizes for BGHBV and ACHBV
| Gene | Forward |
Reverse |
Product size (bp) | ||
|---|---|---|---|---|---|
| Primer | Sequence | Primer | Sequence | ||
| Bluegill hepadnavirus | |||||
| Core | CoreF | GACCAAATTGACTCGGCTGT | CoreR | ATTTGGTCCACCAGCCATAA | 327 |
| Polyemerase and capsid | BGHBV-PolFa | TGTGGACAAAAATCCACGAA | BGHBV-PolRa | ATGCCCATAGGTGCTTTACG | 387 |
| Polyemerase and capsid | PolNestF | CACCACACTTGCCAACAAAC | PolNestR | TGCTCCCAGAACACGTACAG | 287 |
| Circle | BGHBV-CirF | CAACGCCAACAGCATTTTTA | BGHBV-CirR | CGCAGTCTCGACCGATATTA | 301 |
| Polymerase | PolQpcrFb | CCTGGCTCTGTTCGTCATACT | PolNestRb | TGCTCCCAGAACACGTACAG | 110 |
| African cichlid hepadnavirus | |||||
| Polymerase | ACHBV-PolF | TGGGCATTCAACACAAAAGA | ACHBV-PolR | GCGTGCATGACCTCTGAGTA | 302 |
| Cytochrome b | OVCytBF | TGACGCACTTGTTGACCTTC | OBCytBR | GGAGAACGTAGCCCACAAAA | 300 |
Primer used to assess the presence of BGHBV in fish surveys via endpoint PCR.
Primer designed and used for real-time PCR.
Quantitative PCR (qPCR) was used to assess the presence of viral DNA from the selected tissues, as indicated in Table 1. Primers (PolQpcrF and PolNestR) were designed from the polymerase gene to yield a 110-bp amplicon (Table 2). The primer set was used in a standard PCR with DNA extracted from bluegill GAI-2 (referred to as the positive control). The DNA was run on a 2% agarose gel, purified (Qiaquick gel extraction kit), and quantitated (NanoDrop 2000; Thermo Fisher). The DNA was adjusted to 1 ng/μl. Tenfold dilutions of this stock were made in water for qPCR standard-curve generation. Preliminary analysis indicated that the 10−1 through 10−8 dilutions (10−1 to 10−8 ng) would cover the dynamic (linear) range of the assay (R2 ≥ 0.95). qPCR was performed on a Bio-Rad IQ5 iCycler using iQ5 system software for analysis. One microliter of extracted DNA was added to each 25-μl reaction mixture containing iQ SYBR green Supermix (Bio-Rad) and 100 nmol (each) of the indicated primers. A 2-step cycling program was used as follows: initial 95°C for 3 min, followed by 35 cycles of 95°C for 10 s and 60°C for 30 s. Initial screening of all samples was performed twice, using one PCR well/sample. Final assessment of viral-DNA presence was made on samples run in triplicate.
Endpoint PCRs were performed to test the cichlids for African cichlid hepadnavirus (ACHBV). Fin biopsy specimens from two O. ventralis cichlids were extracted using spin columns as described above. Tissue DNA was screened for the presence of cichlid hepadnavirus DNA using primers (ACHBV-PolF and ACHBV-PolR) specific for the cichlid hepadnavirus polymerase sequence (Table 2). PCR for cytochrome b was used as a positive control to verify extraction and PCR methods (Primers OVCytBF and OVCytBR) (Table 2) (27).
In silico screening of public sequence data.
The core, polymerase, and surface protein sequences from BGHBV were used as queries in a BLAST analysis against the GenBank whole-genome shotgun (wgs) and transcriptome shotgun assembly (TSA) databases in March 2016 to detect hepadnavirus homologs in amphibians and fish, employing an E value of 10e−4. The resulting sequences were then reanalyzed by reverse BLAST, ORF predication, and sequence comparison and alignment, as well as bioinformatics analysis, to validate the initial assembly. Other orthohepadnavirus and avihepadnavirus proteins used as queries detected sequences identical to that from BGHBV (data not shown).
Sequence comparisons and phylogenetic analysis.
Coding sequences of representative hepadnavirus C, P, and S genes were downloaded from GenBank and combined with those of BGHBV and Tibetan frog hepadnavirus (TFHBV). To be as broad as possible, the background GenBank data set included exogenous Avihepadnavirus, Orthohepadnavirus, and white sucker hepatitis B virus (WSHBV) sequences, as well as available avian and reptilian (crocodilian) endogenous hepadnavirus (eHBV) sequences that were of sufficient length to conduct phylogenetic analyses, although sequence availability differed by gene. Although a number of snake eHBVs have been documented (14, 15), they are highly fragmentary and contain multiple stop codons and hence were of insufficient length to be included in our phylogenetic analysis, which was based on amino acid sequences (see below). A full list of the sequences utilized is provided in Table 3).
TABLE 3.
Sequences used in phylogenetic analysis
| Virus | Reference and/or GenBank accession no.a |
||
|---|---|---|---|
| C | P | S | |
| ACHBV | NA | This study; ANN02854 | NA |
| BGHBV | This study; YP_009259540 | This study; YP_009259541 | This study; YP_009259542 |
| TFHBV | This study; ANN02856 | This study; ANN02857 | This study; ANN02855 |
| WSHBV | AKT95193 | AKT95195 | AKT95194 |
| eCRHBV1 Crocodylus porosus | 15 | 15 | NA |
| eCRHBV1 Gavialis gangeticus | 15 | 15 | NA |
| eBHBV1 Melopsittacus undulatus | 15 | 15 | NA |
| eBHBV2 Melopsittacus undulatus | 15 | 15 | NA |
| eTHBV Apalone spinifera | 15 | NA | NA |
| eTHBV Pelodiscus sp. | 15 | NA | NA |
| eZHBV Neoaves | 15 | 15 | NA |
| PHBV | YP_004956862 | YP_004956864 | YP_004956865 |
| RHGBV | AAR89922 | YP_024968 | YP_024969 |
| StHBV | AJ251934 | CAC80820 | AJ251934 |
| HHBV | NP_040997 | NP_040998 | NP_040999 |
| CrHBV | NA | CAD29588 | CAD29589 |
| ShHBV | YP_024973 | YP_024974 | YP_024975 |
| DHBV | ADP55743 | 1803562C | NP_039824 |
| SGHBV | YP_031693 | AAD21995 | YP_031696 |
| TBHBV | YP_009046002 | KC790381 | NA |
| WHBV | NP_671816 | AAA19183 | AAA19182 |
| GSHBV | NP_040993 | NP_040994 | NA |
| BtHBV | YP_007678002 | YP_007677999 | YP_007678000 |
| HBHBV | YP_009045998 | KC790377 | NA |
| RBHBV | YP_009045994 | YP_009045991 | NA |
| Woolly monkey HBV | NA | AAO74855 | NA |
| HuHBV genotype A | BAD91278 | CCK33754 | Q4R1S6 |
| HuHBV genotype B | BAO96185 | BAO96176 | BAK32999 |
| HuHBV genotype C | BAU25817 | BAO96196 | BAQ95566 |
| HuHBV genotype D | CCH63726 | ABC87304 | BAJ51643 |
| HuHBV genotype E | BAD91272 | CCK33758 | CCK33757 |
| HuHBV genotype F | CCK33700 | CCK86729 | CCK33685 |
| HuHBV genotype G | BAM05705 | CCK86644 | BAD91285 |
| HuHBV genotype H | BAF49207 | BAN75948 | BAN75949 |
| Chimpanzee HBV | P12901 | P12900 | P12911 |
| Gibbon HBV | P89951 | P87744 | AAG01444 |
| Orangutan HBV | AAF33123 | AAF33121 | AAF33124 |
NA, not applicable.
Amino acid sequence alignment of the C, P, and S data sets was inferred using multiple cycles of the MUSCLE algorithm (28). Because the highly divergent nature of some sequences could compromise phylogenetic accuracy, alignment gaps and ambiguously aligned sequences were removed using the Gblocks program with relatively relaxed settings (i.e., allowing smaller final blocks and less strict flanking regions) (29). This resulted in the following final multiple-sequence alignment lengths: (i) P, 35 taxa, 272 amino acids; C, 34 taxa, 110 amino acids; S, 24 taxa, 187 amino acids. Based on these alignments, maximum-likelihood (ML) phylogenetic trees were estimated using PhyML (30), employing the LG+Γ model of amino acid substitution and 1,000 bootstrap replicates. Finally, pairwise sequence similarities were calculated using the translated amino acid sequences with the sequence demarcation tool (31) (Table 4).
TABLE 4.
Percent amino acid identities, sizes, and GenBank accession numbers of predicted and published hepadnavirus core, polymerase, and surface proteins of BGHBV, ACHBV, and TFHBV compared to hepadnaviruses partitioned by open reading frames
| Virusa | Polymerase protein |
Surface protein |
Core protein |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GenBank accession no. | Size (amino acids) | % amino acid identity |
GenBank accession no. | Size (amino acids) | % amino acid identity |
GenBank accession no. | Size (amino acids) | % amino acid identity |
|||||
| BGHBV | TFHBV | ACHBV | BGHBV | TFHBV | BGHBV | TFHBV | |||||||
| BGHBV | YP_009259541 | 781 | 35 | 30 | YP_009259542 | 328 | 34 | YP_009259540 | 181 | 37 | |||
| ACHBV | ANN02854 | 828 | 30 | 25 | |||||||||
| WSHBV | AKT95195 | 789 | 35 | 34 | 30 | AKT95194 | 346 | 39 | 31 | AKT95193 | 213 | 24 | 31 |
| TFHBV | ANN02857 | 744 | 35 | 25 | ANN02855 | 443 | 34 | ANN02856 | 266 | 37 | |||
| StHBV | CAC80820 | 790 | 36 | 36 | 27 | AJ251934 | 337 | 35 | 37 | AJ251934 | 305 | 33 | 33 |
| HHBV | NP_040998 | 788 | 35 | 35 | 23 | NP_040999 | 335 | 36 | 34 | NP_040997 | 305 | 32 | 35 |
| PHBV | YP_004956864 | 795 | 31 | 38 | 33 | YP_004956865 | 375 | 34 | 31 | YP_004956862 | 305 | 35 | 29 |
| DHBV | NP_039822 | 788 | 35 | 36 | 25 | NP_039824 | 330 | 33 | 36 | ADP55743 | 262 | 33 | 33 |
| CrHBV | CAD29588 | 785 | 35 | 37 | 25 | CAD29589 | 327 | 32 | 35 | ||||
| SGHBV | YP_031695 | 787 | 36 | 37 | 25 | YP_031696 | 329 | 36 | 36 | YP_031693 | 305 | 32 | 31 |
| WHBV | NP_671813 | 884 | 41 | 30 | 35 | NP_671814 | 431 | 37 | 33 | NP_671816 | 188 | 42 | 32 |
| BtHBV | YP_007677999 | 853 | 40 | 33 | 29 | YP_007678000 | 399 | 35 | 28 | YP_007678002 | 217 | 43 | 24 |
| GSHBV | NP_040994 | 881 | 41 | 30 | 29 | NP_040995 | 282 | 39 | 39 | NP_040993 | 217 | 44 | 35 |
| HuHBV | NP_647604 | 843 | 42 | 31 | 28 | YP_355333 | 400 | 37 | 32 | YP_355335 | 212 | 43 | 36 |
White sucker HBV (WSHBV), stork HBV (STHBV), heron HBV (HHBV), parrot HBV (PHBV), duck HBV (DHBV), crane hepatitis B virus (CHBV), snow goose HBV (SGHBV), woodchuck HBV (WHBV), bat HBV (BtHBV), ground squirrel HBV (GSHBV), and human HBV (HuHBV).
Core (capsid) protein modeling.
The structures of the capsid dimer and homohexamer were based on the published structure of the HuHBV virion (Protein Data Bank [PDB] accession no. 3J2V and 5E0I, respectively) (32, 33). Protein modeling and color manipulation were performed using PyMOL software (version 1.8.0.0) (http://www.pymol.org). No protein crystal has been resolved in any fish or amphibian HBV, but they all share conserved residues with HuHBV.
Membrane protein prediction.
The PreS/surface gene encodes three envelope proteins: L, M, and S. Since L is the largest and contains the sequence and the membrane configuration of M and S (34), we focused on that protein. Transmembrane prediction was performed on known (HuHBV and DHBV) and putative (BGHBV, WSHBV, and TFHBV) L protein sequences using hidden Markov models in TMHMM (35, 36). The results were compared to an established transmembrane model (34) to predict membrane topology. Alternative start codon positions for the envelope protein (L) were detected in TFHBV, resulting in two potential sizes (361 and 490 amino acids), so both protein sequences were analyzed.
Accession numbers.
The sequences were deposited in GenBank under accession no. KX058433 to KX058435.
RESULTS
Histopathology, electron microscopy, and in situ hybridization findings.
Histopathological examination revealed well-differentiated papillomas on the lips of 20/40 bluegills. Despite the hepatotropic nature of hepadnaviral infections and the results of metagenomic and PCR analysis described below, there was no microscopic evidence of hepatitis, and viral particles were not observed in skin or liver tissues. Similarly, in situ hybridization (ISH) attempts were unsuccessful in identifying BGHBV in PCR-positive livers, likely due to the detection limit of the ISH method.
Viral metagenomics of a divergent hepadnavirus in a bony fish.
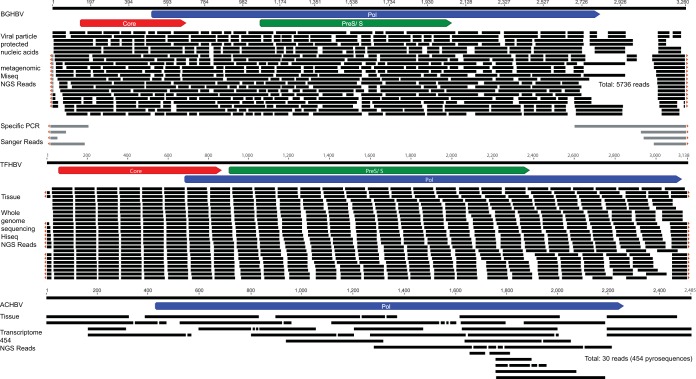
A sequence-independent metagenomic approach was attempted to identify viral sequences within the neoplastic lesions to circumvent the lack of known viral genetic information in teleosts and the paucity of cell lines for culture. Accordingly, a novel virus, denoted BGHBV (GenBank accession no. KX058433), was identified in the next-generation sequence data from one lip lesion sample (Fig. 1 and 2). Over 5,000 NGS reads covered 99% of the genome, with more than 10× coverage. The remaining sequence, as well as the circular nature of the viral genome, was confirmed by PCR and Sanger sequencing, using primers anchored in the polymerase and core ORFs that spanned the entire noncoding region (Fig. 2). The majority of the remaining NGS reads appeared to be bluegill host genome, including a number of endogenous retroviral element sequences typical of vertebrate genomes (data not shown).
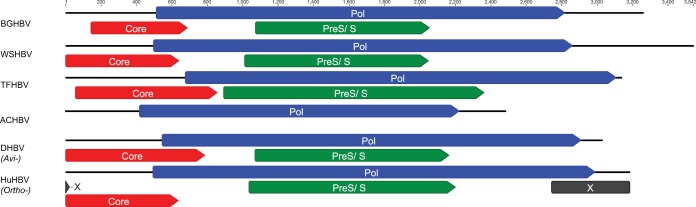
FIG 1.
Genome organization of the hepadnaviruses. Open reading frames encoding the polymerase (Pol), core, surface (PreS/S), and X proteins are indicated by colors. Circular genomes are linearized, with the exception of the partial sequence of ACHBV.
FIG 2.
Coverage map for BGHBV, TFHBV, and ACHBV. The circular genomes of BGHBV and TFHBV are linearized, and sequence coverages over 15 reads are collapsed for display purpose. The overlapping sequences confirming the circular nature of the genomes are indicated by small orange triangles.
Molecular screening of BGHBV in Lepomis species.
Forty-five Lepomis sp. fish, including 40 bluegills, and one largemouth bass were screened by endpoint and real-time PCR to investigate whether BGHBV was endogenized in the Lepomis sp. genomes. At least one tissue from all 46 fish was screened by both techniques, and 12 samples were selected for qPCR replicate analysis, which ranged from 0 fg to the highest concentration of 146 fg in the skin of one fish (Table 1). Among the four Lepomis spp. examined, BGHBV was identified by PCR in only 6/40 bluegills, and with the exception of one archived lip sample, the five positive fish came from a single pond. Fish from two additional locations were all PCR negative. Although the quantity of viral DNA in each tissue varied, BGHBV nucleic acid was identified in 3/18 grossly visible lip lesions, 3/22 nonlesioned lip samples, 3/12 pooled organ samples, and 3/7 skin samples (Table 1). These prevalence data did not indicate that BGHBV was associated with the lip papillomas, even though it was initially discovered in a diseased individual. Taken together with the circular nature of the BGHBV genome as indirect evidence against endogenization, the prevalence results indicate that BGHBV is not derived from the germ line of the bluegill.
Characterization of a prototype amphibian hepadnavirus.
In silico screening of GenBank for novel hepadnaviruses identified hepadnavirus-like sequences from the whole-genome sequence data of the Tibetan frog (37) that shared 33% protein sequence similarity with the polymerase protein of BGHBV. The initial contig (GenBank accession number JYOU01126907) was analyzed using the SOAPdenovo assembler in the original report (37), resulting in a linear sequence of 3,137 bp. Our subsequent analysis using the original 4.4B Illumina Hiseq reads extracted from GenBank confirmed the circular nature of the sequence by identifying overlapping read coverage at both sequence termini (Fig. 2), resulting in a complete genome sequence with a length of 3,138 bp (TFHBV; GenBank accession no. KX058435). The Tibetan frog data set contained 13 whole-genome-sequencing (DNA) runs, of which only a small portion (<0.003%) of the total reads were hepadnaviral (Table 5). All the runs were performed on a single muscle sample (37), so all the data sets contained TFHBV sequences. No other hepadnavirus-like sequences were identified in other amphibian whole-genome or transcriptome assembled data sets at the time of analysis.
TABLE 5.
Whole-genome sequence data of the Tibetan frog (N. parkeri) with initial contig (GenBank accession number JYOU01126907) (37)a
| Accession no. | Total no. of reads | No. of TFHBV reads | % TFHBV reads |
|---|---|---|---|
| SRX514761 | 374,982,742 | 4497 | 0.001199 |
| SRX514762 | 309,742,412 | 6200 | 0.002002 |
| SRX514763 | 313,197,132 | 7213 | 0.002303 |
| SRX514764 | 389,751,764 | 1928 | 0.000495 |
| SRX514765 | 433,440,774 | 1694 | 0.000391 |
| SRX514766 | 366,201,646 | 156 | 0.000043 |
| SRX514767 | 354,666,118 | 101 | 0.000028 |
| SRX514768 | 273,217,750 | 322 | 0.000118 |
| SRX514769 | 321,698,530 | 745 | 0.000232 |
| SRX514770 | 336,644,766 | 2128 | 0.000632 |
| SRX514771 | 337,281,318 | 1454 | 0.000431 |
| SRX514772 | 302,142,366 | 4287 | 0.001419 |
| SRX514773 | 267,159,900 | 3657 | 0.001369 |
The Tibetan frog data set contained 13 whole-genome-sequencing (DNA) runs.
Identification of hepadnavirus in cichlids.
A hepadnavirus-like sequence was also identified from the transcriptome data set of the African cichlid Ophthalmotilapia ventralis (GenBank accession number JL559376) (16, 38). Notably, this is the only hepadnaviral sequence in the entire 454 pyrosequencing transcriptome of O. ventralis, comprising the polymerase polyprotein (Fig. 1). Using the original reads, our analysis obtained a final sequence of 2,485 bp (KX058434). This is clearly a partial sequence in which a circular genome could not be obtained with the available data. To further investigate if this African cichlid hepadnavirus-like sequence (ACHBV) was endogenized into the host cellular genome, we examined cellular DNA from skin samples of two O. ventralis fish using endpoint PCR. PCRs targeting the hepadnavirus-like sequence were negative in both, while the positive-control PCR targeting the cytochrome b gene of O. ventralis was validated, indicating that ACHBV was not incorporated into the cellular genome in the samples investigated.
Genome organization of fish and amphibian hepadnaviruses.
The BGHBV and the TFHBV genomes are complete circular genomes with 3,260 bp and 3,138 bp, respectively (Fig. 1). The complete circular genomes of BGHBV and TFHBV have the typical hepadnaviral organization, comprising three overlapping reading frames that encode the core, polymerase, and surface proteins (Fig. 1). Interestingly, an X protein homolog was not detected in the fish and amphibian hepadnaviruses in this study. This protein is known to be absent in the genus Avihepadnavirus. Consequently, these data confirmed that the X protein is a distinctive feature of the mammal-infecting orthohepadnaviruses (7, 39).
The hepadnavirus core gene encodes phosphoproteins that are assembled into subviral capsids. The core polyproteins are 181 amino acids (aa) (BGHBV) and 266 aa (TFHBV) in length. Signals of a core protein could not be detected in the partial genome of ACHBV (BLASTx; E value cutoff, 0.0001), suggesting the core gene may lie outside the partial sequence or is too divergent for detection using amino acid-based similarity searching. The two fish hepadnaviruses, BGHBV and the recently described WSHBV, encode some of the shortest core proteins among known hepadnaviruses (Table 4). The BGHBV core protein shares 24% amino acid identity with WSHBV, 37% amino acid identity with TFHBV, and 32 to 44% amino acid identity with avian and mammalian hepadnaviruses. Similarly, TFHBV shares 31% amino acid identity with WSHBV, 37% amino acid identity with BGHBV, and 24 to 36% amino acid identity with avian and mammalian hepadnaviruses. The C termini of BGHBV and TFHBV both contain an arginine-rich domain, a hallmark of hepadnavirus core proteins, which contains a signal for nuclear transport required for pregenome encapsidation (40, 41).
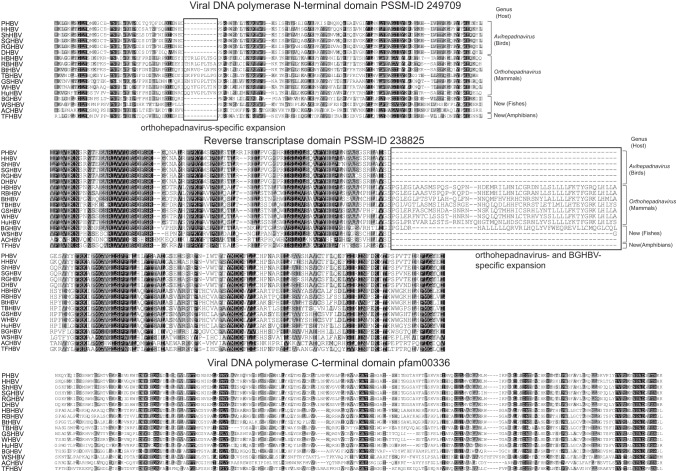
The polymerase gene encodes the viral DNA polymerase, the sole enzyme produced by hepadnaviruses. This gene, with lengths of 781 aa (BGHBV), 744 aa (TFHBV), and 828 aa (ACHBV), covers over half the hepadnavirus genome, and its open reading frame overlaps those of the core and surface proteins. The proteins from these three viruses share 23 to 42% amino acid sequence identity among themselves and other hepadnaviruses (Table 4). The newly identified amphibian and fish hepadnavirus polymerase genes contain several conserved domains homologous to known avi- and orthohepadnaviruses, including the viral DNA polymerase C (pfam00336) and N (Position-Specific Scoring Matrix [PSSM] ID 249709) termini and the reverse transcriptase long terminal repeat (LTR) (PSSM ID 238825) (Fig. 3). Mammalian orthohepadnaviruses contain an expanded reverse transcriptase domain with more than 40 additional amino acids. Strikingly, such an expansion was also observed in BGHBV, but not in other fish (WSHBV and ACHBV), amphibian (TFHBV), or avian hepadnaviruses, supporting the phylogenetic analysis that showed BGHBV shares common ancestry with the mammalian hepadnaviruses (see below). In contrast, an expansion of the viral DNA polymerase N-terminal domain was observed only in mammalian orthohepadnaviruses (Fig. 3).
FIG 3.
Conserved motifs in the polymerase proteins of mammalian, avian, amphibian, and piscine hepadnaviruses. An expanded reverse transcriptase domain is evident in mammalian orthohepadnaviruses and BGHBV. The expansion of the viral DNA polymerase N-terminal domain was observed only in mammalian orthohepadnaviruses. The degrees of sequence conservation are highlighted in grayscale. PHBV, parrot HBV; HHBV, heron HBV; ShHBV, sheldgoose HBV; SGHBV, snow goose HBV; RGHBV, Ross's goose HBV; HBHBV, horseshoe bat HBV; RBHBV, roundleaf bat HBV; BtHBV, bat HBV; TBHBV, tent-making bat HBV; GSHBV, ground squirrel HBV; WHBV, woodchuck HBV.
In known ortho- and avihepadnaviruses, the surface polyprotein gene encodes three integral transmembrane envelope glycoproteins, S, M, and L. The surface polyproteins were 328 aa and 443 aa in length for BGHBV and TFHBV, respectively, but were not detected in the partial ACHBV genome (Fig. 1). The amphibian TFHBV contains the largest PreS/S gene of all known hepadnaviruses, encoding a 490-aa protein. However, an alternative start codon was also detected that would produce a shorter, 361-aa protein. The surface proteins from BGHBV and TFHBV share 34% amino acid identity between themselves and 28 to 39% amino acid identity with other hepadnaviruses (Table 4).
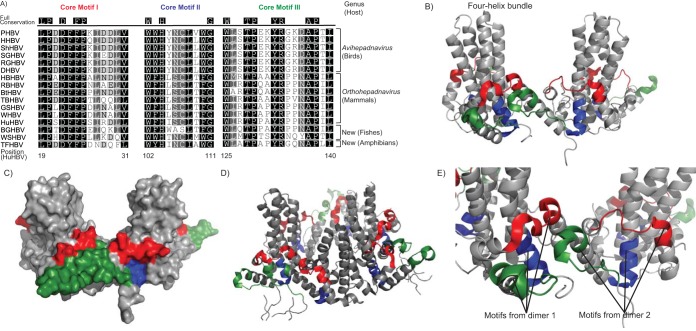
Conserved core motifs in vertebrate HBVs.
Characterization of the prototype fish and amphibian hepadnaviruses allowed us to identify family-wide conserved domains in the core protein. Besides the arginine-rich domain, several conserved motifs were identified among all avian, mammalian, fish, and amphibian hepadnaviruses. They include core motif I, LPXD(F/Y)FPXXXXX(V/L); core motif II, WXHXX(S/C)(L/I)X(W/F)G; and core motif III, WXXTPXXYRXXXAPX(I/L) (Fig. 4). Although core motif I is close to the N terminus while motifs II and III are close to the C terminus, all three motifs are in close proximity to each other when the capsid dimers are assembled in the protein model (Fig. 4B to D). In a typical HBV, two monomers associate to give a compact dimer in which the two α-helical hairpins form a four-helix bundle (Fig. 4B) (42). Residues at the antigenic sites are located near the major immunodominant tips of the four-helix bundle, and the residues that make up the four-helix bundles are not conserved among vertebrate HBVs.
FIG 4.
Conserved motifs in the core proteins of mammalian, avian, amphibian, and piscine hepadnaviruses. (A) Amino acid sequence alignment of the three conserved motifs in the core proteins. The positions in the HuHBV protein are indicated (42), and the degrees of sequence conservation are highlighted in grayscale. The accession numbers of the included HBV protein sequences are listed in Table 4. (B) Motifs I, II, and III (red, blue, and green, respectively) in the capsid protein dimer using HuHBV as a model. (C) Motif locations in the surface representation of the capsid dimer. (D and E) Homohexamer representation showing the proximity of the motifs between subunits.
In exogenous HBVs from the four classes of vertebrates, the three motifs (I, II, and III) contain a total of 15 fully conserved residues. Three additional residues, i.e., the start codon, Asp-4, and His-47 (positions according to HuHBV), are also conserved but are not included in these motifs. Visualizing the core protein using the well-established HuHBV model, all three motifs are located at the base of the capsid monomer (Fig. 4B), containing hydrophobic residues essential for folding of the capsid monomer (42). In HuHBV and other orthohepadnaviruses, the Cys-61 residues of two capsid monomers form a disulfide bond to each other at the dimer interface (43). The same Cys (C) disulfide bonds are also found in WSHBV and BGHBV. Instead of Cys, Thr (T) is found in avihepadnaviruses and His (H) is found in TFHBV in the homologous position. One possible explanation for the lack of Cys residue conservation among all vertebrate HBVs is that it is not essential for dimer or capsid formation, as evidenced by mutagenesis studies of the residue (44).
It is likely that motif III is also important for the interactions between capsid subunits in 5-fold and 2-fold axes, as it contains the proline-rich loop (positions 128 to 136 in HuHBV) essential for such interactions (42). In particular, Tyr-132 and Pro-129 are both conserved in all the vertebrate HBVs investigated. In crystallized protein, Tyr-132 is fully buried in the capsid, which is important for proper capsid folding, while Pro-129 is important in intersubunit packaging (42).
Membrane proteins.
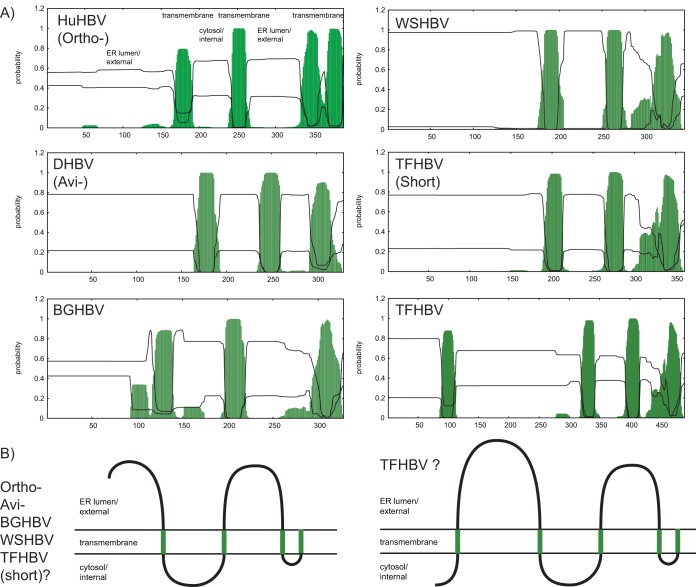
The TMHMM analysis predicted that the fish hepadnaviruses (BGHBV and WSHBV) would have membrane protein folding and topology similar to the known model for orthohepadnaviruses and avihepadnaviruses (Fig. 5) (35). The C-terminal region is hydrophobic and is most likely embedded in host membranes. Two additional hydrophobic domains were detected, forming a hairpin structure with a cytosolic loop (45). Alternative start codon positions were detected for the TFHBV PreS/S ORF, resulting in putative L envelope proteins of 490 and 361 amino acids. While analysis of the smaller L protein of TFHBV suggests that it might fold similarly to those of the other HBVs, the longer L protein was predicted to have an additional transmembrane domain near the N terminus, potentially forming an extra loop in the endoplasmic reticulum (ER) lumen and exposing the N terminus in the cytosol (Fig. 5B). The start codon usage and putative membrane folding predictions clearly need to be experimentally confirmed.
FIG 5.
Membrane protein analysis. (A) TMHMM analysis of the fish hepadnaviruses, BGHBV and WSHBV, as well as TFHBV, with known models for orthohepadnaviruses (HuHBV) and avihepadnaviruses (DHBV) (35). TMHMM probability was plotted against the length of the protein. Predicted transmembrane regions were highlighted in green. (B) Transmembrane topologies of the L protein (curved line) in the ER membrane (double horizontal lines), compared to the established model of orthohepadnavirus (45). Predicted transmembrane regions were highlighted in green. Since alternative start codon positions resulting in envelope proteins with different lengths were detected for TFHBV, the analysis was performed on both.
Evolutionary history of hepadnaviruses.
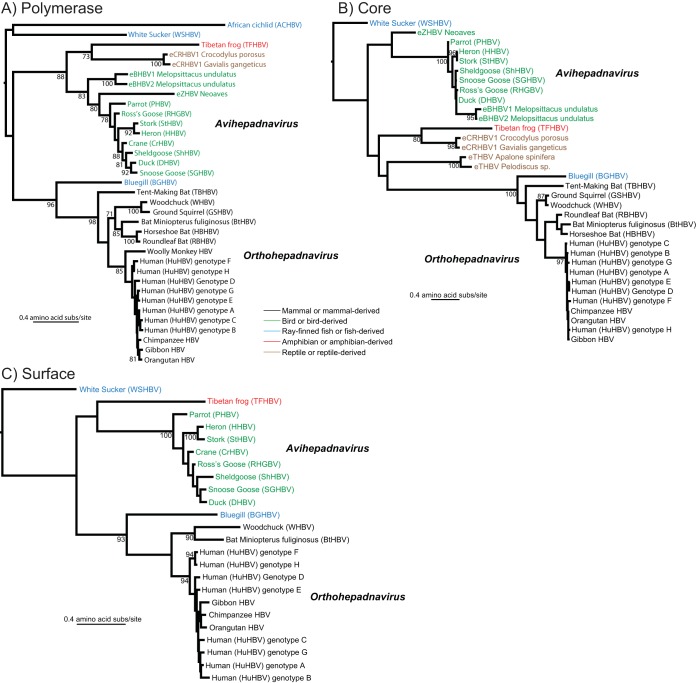
Phylogenetic analysis of ACHBV, BGHBV, and TFHBV, along with representative exogenous and endogenous hepadnaviruses from mammals, birds, reptiles, and fish (WSHBV), was performed to determine their relationships and evolutionary history. Although the (Gblocks-cleansed) sequence alignments of the polymerase, core, and surface genes are necessarily short, they are consistent in clearly showing that the three fish hepadnaviruses do not form a monophyletic group (Fig. 6). While ACHBV and WSHBV fell in divergent phylogenetic positions, both exhibiting very long branches, BGHBV is clearly more closely related to mammalian viruses of the genus Orthohepadnavirus, a relationship supported by a high level of bootstrap support (93 to 100%). Importantly, although the location of the root of these phylogenies is uncertain, no rooting position would force the fish viruses to be monophyletic. The amphibian TFHBV sequence was most closely related to the endogenous hepadnaviruses sampled from crocodilians in the P and C genes, with 73% and 80% bootstrap support, respectively. The sequences of the surface genes of these endogenous viruses were unavailable for comparison, nor could we include the snake eHBVs in the analysis due to their short sequence lengths.
FIG 6.
Maximum-likelihood phylogenetic trees of the polymerase (A), core (B), and surface (C) genes of exogenous and endogenous vertebrate hepadnaviruses. The viruses are color coded to reflect their host groups of origin. All the trees are drawn to a scale of amino acid substitutions (subs) per site and rooted on the fish (WSHBV and, where available, ACHBV) sequences, as (i) they are the most divergent and (ii) this rooting position maximizes the extent of virus-host codivergence. Bootstrap support values of >70% are shown for relevant nodes.
DISCUSSION
Relatively little is known about the host range and evolutionary history of the Hepadnaviridae. Until recently, the only described exogenous hepadnaviruses were from mammals and birds, comprising approximately 20 ortho- and avihepadnavirus genomes from humans, nonhuman primates, rodents, bats, and birds. The study describing a hepadnavirus in the white sucker fish (16) and our discovery of the second fish hepadnavirus and the first amphibian hepadnavirus are evidence that hepadnaviruses have a broader host range than previously appreciated. Indeed, the analysis of these new genomes, as well as previously described exogenous (HBV) and endogenous (eHBV) hepadnavirus (14, 15, 46) sequences, indicates that the Hepadnaviridae have been able to infect all five major groups of vertebrates, namely, mammals, birds, reptiles, amphibians, and fish (Table 6 and Fig. 6).
TABLE 6.
Current knowledge of hepadnaviral host range and viral life cycle
| Host | Virusa |
|
|---|---|---|
| Exogenous | Exclusively endogenous | |
| Mammal | Orthohepadanvirus | NA |
| Avian | Avihepadanvirus | Avian hepadnavirus EVE |
| Reptile | NA | Reptilian hepadnavirus EVE |
| Fish | WSHBV, BGHBV, and ACHBV (this study) | NA |
| Amphibian | TFHBV (this study) | NA |
Although hepadnaviral EVEs have been described in birds and reptiles (Table 6), we found no evidence that the bluegill and cichlid viruses were incorporated into the fish germ line or caused lesions. In particular, the confirmation of a circular genome and the presence of the virus in some, but not all, bluegills provides strong evidence against BGHBV being endogenous. Similarly, the absence of ACHBV in the genomes of the O. ventralis cichlids examined suggests that it is not an EVE. In addition, sequence analysis of TFHBV revealed no insertion site linking the viral genome to that of the host, and a complete circular genome was identified, again suggesting it constitutes an exogenous virus. Unfortunately, a lack of tissue specimens precluded verification of the presence or absence of the virus in additional frogs.
The genome organizations of the fish and amphibian hepadnaviruses are similar to those of orthohepadnaviruses and avihepadnaviruses, although, with the exception of the highly conserved functional domains (14, 15), the sequence identities between these virus groups are very low (Table 4). The polymerases in BGHBV, TFHBV, and ACHBV also contained conserved domains, including the viral DNA polymerase C and N termini and the reverse transcriptase LTR. Perhaps most noteworthy was the fact that the expanded reverse transcriptase domain detected in BGHBV, but not in the other fish (WSHBV and ACHBV) or amphibian (TFHBV) viruses, is concordant with the phylogenetic analysis showing that BGHBV shares common ancestry with the mammalian hepadnaviruses.
Our analysis of core (capsid) and surface (membrane) proteins revealed features that unify all vertebrate hepadnaviruses, identifying conserved core protein residues or membrane protein topographies that play an important role in hepadnavirus infection and evolution. First, BGHBV and TFHBV contain an arginine-rich domain at their C termini, a hallmark of hepadnavirus core proteins (Fig. 4) (40, 41, 47). Second, 18 residues in the core proteins were fully conserved among examined vertebrate HBVs. Core motifs I, II, and III account for 15 of those conserved residues. The majority of the conserved residues are located in the hydrophobic core of the capsid, while residues at the antigenic tips, as well as the four-helix bundle, are not conserved at all. Based on protein models, the core motifs are conserved, probably because they play key roles in the formation of capsid monomers and dimers and in their intersubunit interactions (42). Structural constraints to maintain proper capsid formation seem to be a key force in hepadnaviral core capsid evolution. Since Phe-23 and Trp-102 in motifs I and II are important for interaction with replication-inhibiting drugs (32), further analysis of conserved residues as antiviral targets could be worthy of attention.
The new fish and amphibian hepadnaviruses contain three hydrophobic domains, similar to ortho- and avihepadnavirus. Therefore, it appears that all vertebrate hepadnaviruses share membrane protein topologies similar to those of orthohepadnaviruses (Fig. 5). The second half of the surface protein contains more conserved residues than the N terminus, probably due to conserved transmembrane residues.
The X gene encodes a soluble cytoplasmic X protein that is required for efficient infection by orthohepadnaviruses in vivo (48). Although it is suspected to be involved in the generation of tumors in chronic hepadnaviral infections in humans and woodchucks, its exact role in the viral replication cycle is not known (49–54). The presence or absence of an X protein represents a major genomic difference between the ortho- and avihepadnaviruses. Notably, an X protein homolog was not identified in the fish or amphibian viral genomes, providing further evidence that the X protein is a distinctive feature of orthohepadnaviruses in mammalian hosts (7, 39, 50) and that it evolved by overprinting in these taxa only (6, 7). This is consistent with the absence of a detectable X gene in lower vertebrate species, including fish, amphibians, and birds.
In addition to increasing our understanding of their genome structure and host range, the data presented here shed important new light on hepadnavirus evolution. ACHBV falls deep in all the phylogenetic trees and seemingly has a common ancestry with the white sucker virus in the polymerase gene tree (albeit with low bootstrap support), observations that are compatible with the long-term codivergence of hepadnaviruses with their vertebrate hosts over time scales spanning hundreds of millions of years. However, our data also provide compelling evidence for cross-species transmission. First, although the data are tentative, the single amphibian virus (TFHBV) is clearly most closely related to the eHBVs from crocodilians, whereas strict virus-host codivergence should place TFHBV as the sister group to viruses from reptiles, birds, and mammals. Far more dramatic, however, was the observation that BGHBV formed a strongly supported monophyletic group with the mammalian orthohepadnaviruses in all three gene trees (Fig. 6). While this is consistent with the shared presence of an expanded reverse transcriptase domain among these taxa, BGHBV differs from the orthohepadnaviruses in that it lacks both the expansion in the polymerase N-terminal domain and the X protein.
The fact that BGHBV falls as the sister group to the mammalian hepadnaviruses suggests a far more complex evolutionary history than that of strict virus-host codivergence, so that multiple species jumps need to be involved. Indeed, it is striking that the fish hepadnaviruses do not form a monophyletic group. While the precise history of these species jumps is difficult to determine, the most parsimonious scenario is that fish harbor an extensive diversity of hepadnaviruses, evident in the long branches leading to ACHBV and WSHBV, and that one of these lineages, represented by BGHBV, jumped to terrestrial vertebrates, giving rise to the mammalian orthohepadnaviruses that circulate today. If so, this would be one of the few cases in which viruses have jumped such a great taxonomic distance. Alternatively, it is possible that BGHBV represents a successful spill-back lineage from terrestrial vertebrates to fish, although this again requires a species jump across a substantial phylogenetic distance. Determining which of these, or other, evolutionary scenarios is correct will require a far wider sampling of vertebrate hepadnaviruses.
This study shows that fish carry a remarkable diversity of hepadnaviruses, one of which forms a sister group to mammalian hepadnaviruses. Although the evolution of this important group of viruses is uncertain, a clear prediction from the current study is that there are many more vertebrate hepadnaviruses to be discovered, particularly in species where there has been little active surveillance to date. For example, the observation of a hepadnavirus in a frog suggests there could be additional undiscovered hepadnaviruses with unknown significance for the health of amphibian populations. Increased viral surveillance is especially important, as amphibian populations continue to decline as a result of infectious disease and habitat loss (55–59). Finally, the increasing detection of hepadnaviruses in fish (BGHBV and WSHBV) clearly warrants additional investigations to further elucidate their host ranges and potential pathogenic effects.
ACKNOWLEDGMENTS
We thank Eric Delwart and Beatrix Kapusinszky at the University of California, San Francisco, and the Blood Systems Research Institute for assistance with sequencing.
Funding Statement
E.C.H. is funded by an NHMRC Australia Fellowship (AF30).
REFERENCES
- 1.Seeger C, Zoulim F, Mason WS. 2013. Hepadnaviruses, p 2185–2221. In Knipe DM, Howley PM (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Voyles BA. 1993. The biology of viruses. Mosby, St. Louis, MO. [Google Scholar]
- 3.Flint SJ, Enquist LW, Racaniello VR, Skalka AM, Barnum DR, de Evaluación E. 2000. Principles of virology: molecular biology, pathogenesis and control. ASM Press, Washington, DC. [Google Scholar]
- 4.Baltimore D. 1971. Expression of animal virus genomes. Bacteriol Rev 35:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orito E, Mizokami M, Ina Y, Moriyama EN, Kameshima N, Yamamoto M, Gojobori T. 1989. Host-independent evolution and a genetic classification of the hepadnavirus family based on nucleotide sequences. Proc Natl Acad Sci U S A 86:7059–7062. doi: 10.1073/pnas.86.18.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suh A, Brosius J, Schmitz J, Kriegs JO. 2013. The genome of a Mesozoic paleovirus reveals the evolution of hepatitis B viruses. Nat Commun 4:1791. doi: 10.1038/ncomms2798. [DOI] [PubMed] [Google Scholar]
- 7.van Hemert FJ, van de Klundert MA, Lukashov VV, Kootstra NA, Berkhout B, Zaaijer HL. 2011. Protein X of hepatitis B virus: origin and structure similarity with the central domain of DNA glycosylase. PLoS One 6:e23392. doi: 10.1371/journal.pone.0023392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drexler JF, Geipel A, König A, Corman VM, van Riel D, Leijten LM, Bremer CM, Rasche A, Cottontail VM, Maganga GD, Schlegel M. 2013. Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. Proc Natl Acad Sci U S A 110:16151–16156. doi: 10.1073/pnas.1308049110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saif Y. 2008. Viral diseases, p 405–448. In Diseases of poultry, vol. 12 Blackwell Publishing, Ames, IA. [Google Scholar]
- 10.Siddiqui AL, Marion PL, Robinson WS. 1981. Ground squirrel hepatitis virus DNA: molecular cloning and comparison with hepatitis B virus DNA. J Virol 38:393–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kodama KA, Ogasawara NA, Yoshikawa HI, Murakami SE. 1985. Nucleotide sequence of a cloned woodchuck hepatitis virus genome: evolutional relationship between hepadnaviruses. J Virol 56:978–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Summers J, Smolec JM, Snyder R. 1978. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc Natl Acad Sci U S A 75:4533–4537. doi: 10.1073/pnas.75.9.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prassolov A, Hohenberg H, Kalinina T, Schneider C, Cova L, Krone O, Frölich K, Will H, Sirma H. 2003. New hepatitis B virus of cranes that has an unexpected broad host range. J Virol 77:1964–1976. doi: 10.1128/JVI.77.3.1964-1976.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert C, Meik JM, Dashevsky D, Card DC, Castoe TA, Schaack S. 2014. Endogenous hepadnaviruses, bornaviruses and circoviruses in snakes. Proc Biol Sci 281:20141122. doi: 10.1098/rspb.2014.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suh A, Weber CC, Kehlmaier C, Braun EL, Green RE, Fritz U, Ray DA, Ellegren H. 2014. Early Mesozoic coexistence of amniotes and hepadnaviridae. PLoS Genet 10:e1004559. doi: 10.1371/journal.pgen.1004559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn CM, Iwanowicz LR, Cornman RS, Conway CM, Winton JR, Blazer VS. 2015. Characterization of a novel hepadnavirus in the White Sucker (Catostomus commersonii) from the Great Lakes region of the United States. J Virol 89:11801–11811. doi: 10.1128/JVI.01278-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui J, Zhao W, Huang Z, Jarvis ED, Gilbert MT, Walker PJ, Holmes EC, Zhang G. 2014. Low frequency of paleoviral infiltration across the avian phylogeny. Genome Biol 15:539. doi: 10.1186/s13059-014-0539-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert C, Feschotte C. 2010. Genomic fossils calibrate the long-term evolution of hepadnaviruses. PLoS Biol 8:e1000495. doi: 10.1371/journal.pbio.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson BH, Margolis HS. 2002. Primate hepatitis B viruses–genetic diversity, geography and evolution. Rev Med Virol 12:133–141. doi: 10.1002/rmv.348. [DOI] [PubMed] [Google Scholar]
- 20.Starkman SE, MacDonald DM, Lewis JC, Holmes EC, Simmonds P. 2003. Geographic and species association of hepatitis B virus genotypes in non-human primates. Virology 314:381–393. doi: 10.1016/S0042-6822(03)00430-6. [DOI] [PubMed] [Google Scholar]
- 21.Ng TF, Driscoll C, Carlos MP, Prioleau A, Schmieder R, Dwivedi B, Wong J, Cha Y, Head S, Breitbart M, Delwart E. 2013. Distinct lineage of vesiculovirus from big brown bats, United States. Emerg Infect Dis 19:1978–1980. doi: 10.3201/eid1912.121506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng TF, Kondov NO, Deng X, Van Eenennaam A, Neibergs HL, Delwart E. 2015. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J Virol 89:5340–5349. doi: 10.1128/JVI.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng TF, Marine R, Wang C, Simmonds P, Kapusinszky B, Bodhidatta L, Oderinde BS, Wommack KE, Delwart E. 2012. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J Virol 86:12161–12175. doi: 10.1128/JVI.00869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Victoria JG, Kapoor A, Dupuis K, Schnurr DP, Delwart EL. 2008. Rapid identification of known and new RNA viruses from animal tissues. PLoS Pathog 4:e1000163. doi: 10.1371/journal.ppat.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGinnis S, Madden TL. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng X, Naccache SN, Ng T, Federman S, Li L, Chiu CY, Delwart EL. 2015. An ensemble strategy that significantly improves de novo assembly of microbial genomes from metagenomic next-generation sequencing data. Nucleic Acids Res 43:e46. doi: 10.1093/nar/gkv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita M, Awata S, Yorifuji M, Ota K, Kohda M, Ochi H. 2014. Bower-building behaviour is associated with increased sperm longevity in Tanganyikan cichlids. J Evol Biol 27:2629–2643. doi: 10.1111/jeb.12522. [DOI] [PubMed] [Google Scholar]
- 28.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 30.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 31.Muhire BM, Varsani A, Martin DP. 2014. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9:e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klumpp K, Lam AM, Lukacs C, Vogel R, Ren S, Espiritu C, Baydo R, Atkins K, Abendroth J, Liao G, Efimov A. 2015. High-resolution crystal structure of a hepatitis B virus replication inhibitor bound to the viral core protein. Proc Natl Acad Sci U S A 112:15196–15201. doi: 10.1073/pnas.1513803112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu X, Jin L, Jih J, Shih C, Zhou ZH. 2013. 3.5 Å cryoEM structure of hepatitis B virus core assembled from full-length core protein. PLoS One 8:e69729. doi: 10.1371/journal.pone.0069729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruss V. 2004. Envelopment of the hepatitis B virus nucleocapsid. Virus Res 106:199–209. doi: 10.1016/j.virusres.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 35.Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 36.Möller S, Croning MD, Apweiler R. 2001. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 37.Sun YB, Xiong ZJ, Xiang XY, Liu SP, Zhou WW, Tu XL, Zhong L, Wang L, Wu DD, Zhang BL, Zhu CL. 2015. Whole-genome sequence of the Tibetan frog Nanorana parkeri and the comparative evolution of tetrapod genomes. Proc Natl Acad Sci U S A 112:E1257–E1262. doi: 10.1073/pnas.1501764112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baldo L, Santos ME, Salzburger W. 2011. Comparative transcriptomics of Eastern African cichlid fishes shows signs of positive selection and a large contribution of untranslated regions to genetic diversity. Genome Biol Evol 3:443–455. doi: 10.1093/gbe/evr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kew MC. 2011. Hepatitis B virus X protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J Gastroenterol Hepatol 26:144–152. doi: 10.1111/j.1440-1746.2010.06546.x. [DOI] [PubMed] [Google Scholar]
- 40.Yeh CT, Liaw YF, Ou JH. 1990. The arginine-rich domain of hepatitis B virus precore and core proteins contains a signal for nuclear transport. J Virol 64:6141–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nassal M. 1992. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol 66:4107–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wynne SA, Crowther RA, Leslie AG. 1999. The crystal structure of the human hepatitis B virus capsid. Mol Cell 3:771–780. doi: 10.1016/S1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 43.Yamada K, Terahara T, Kurata S, Yokomaku T, Tsuneda S, Harayama S. 2008. Retrieval of entire genes from environmental DNA by inverse PCR with pre-amplification of target genes using primers containing locked nucleic acids. Environ Microbiol 10:978–987. doi: 10.1111/j.1462-2920.2007.01518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nassal M, Rieger A, Steinau O. 1992. Topological analysis of the hepatitis B virus core particle by cysteine-cysteine cross-linking. J Mol Biol 225:1013–1025. doi: 10.1016/0022-2836(92)90101-O. [DOI] [PubMed] [Google Scholar]
- 45.Eble BE, Lingappa VR, Ganem D. 1986. Hepatitis B surface antigen: an unusual secreted protein initially synthesized as a transmembrane polypeptide. Mol Cell Biol 6:1454–1463. doi: 10.1128/MCB.6.5.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aiewsakun P, Katzourakis A. 2015. Endogenous viruses: connecting recent and ancient viral evolution. Virology 479:26–37. doi: 10.1016/j.virol.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 47.Glebe D, Urban S. 2007. Viral and cellular determinants involved in hepadnaviral entry. World J Gastroenterol 13:22. doi: 10.3748/wjg.v13.i1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zoulim F, Saputelli J, Seeger C. 1994. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol 68:2026–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feitelson MA, Lee J. 2007. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett 252:157–170. doi: 10.1016/j.canlet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Fourel G, Couturier J, Wei Y, Apiou F, Tiollais P, Buendia MA. 1994. Evidence for long-range oncogene activation by hepadnavirus insertion. EMBO J 13:2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fourel G, Trepo C, Bougueleret L, Henglein B, Ponzetto A, Tiollais P, Buendia MA. 1990. Frequent activation of N-myc genes by hepadnavirus insertion in woodchuck liver tumours. Nature 347:294–298. doi: 10.1038/347294a0. [DOI] [PubMed] [Google Scholar]
- 52.Hansen LJ, Tennant BC, Seeger CH, Ganem D. 1993. Differential activation of myc gene family members in hepatic carcinogenesis by closely related hepatitis B viruses. Mol Cell Biol 13:659–667. doi: 10.1128/MCB.13.1.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C, Mulawadi FH. 2012. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 44:765–769. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 54.Wen Y, Golubkov VS, Strongin AY, Jiang W, Reed JC. 2008. Interaction of hepatitis B viral oncoprotein with cellular target HBXIP dysregulates centrosome dynamics and mitotic spindle formation. J Biol Chem 283:2793–2803. doi: 10.1074/jbc.M708419200. [DOI] [PubMed] [Google Scholar]
- 55.Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (ed). 2005. Hepadnaviridae, p 373–384. In Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA. [Google Scholar]
- 56.Collins JP. 2013. History, novelty, and emergence of an infectious amphibian disease. Proc Natl Acad Sci U S A 110:9193–9194. doi: 10.1073/pnas.1305730110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenblum EB, James TY, Zamudio KR, Poorten TJ, Ilut D, Rodriguez D, Eastman JM, Richards-Hrdlicka K, Joneson S, Jenkinson TS, Longcore JE. 2013. Complex history of the amphibian-killing chytrid fungus revealed with genome resequencing data. Proc Natl Acad Sci U S A 110:9385–9390. doi: 10.1073/pnas.1300130110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCallum ML. 2007. Amphibian decline or extinction? Current declines dwarf background extinction rate J Herpetol 41:483–491. [Google Scholar]
- 59.Brunner JL, Storfer A, Gray MJ, Hoverman JT. 2015. Ranavirus ecology and evolution: from epidemiology to extinction, p 71–104. In Gray MJ, Chinchar VG (ed), Ranaviruses: lethal pathogens of ectothermic vertebrates. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 60.Katzourakis A, Gifford RJ. 2010. Endogenous viral elements in animal genomes. PLoS Genet 6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]