ABSTRACT
The four brome mosaic virus (BMV) RNAs (RNA1 to RNA4) are encapsidated in three distinct virions that have different disassembly rates in infection. The mechanism for the differential release of BMV RNAs from virions is unknown, since 180 copies of the same coat protein (CP) encapsidate each of the BMV genomic RNAs. Using mass spectrometry, we found that the BMV CP contains a complex pattern of posttranslational modifications. Treatment with phosphatase was found to not significantly affect the stability of the virions containing RNA1 but significantly impacted the stability of the virions that encapsidated BMV RNA2 and RNA3/4. Cryo-electron microscopy reconstruction revealed dramatic structural changes in the capsid and the encapsidated RNA. A phosphomimetic mutation in the flexible N-terminal arm of the CP increased BMV RNA replication and virion production. The degree of phosphorylation modulated the interaction of CP with the encapsidated RNA and the release of three of the BMV RNAs. UV cross-linking and immunoprecipitation methods coupled to high-throughput sequencing experiments showed that phosphorylation of the BMV CP can impact binding to RNAs in the virions, including sequences that contain regulatory motifs for BMV RNA gene expression and replication. Phosphatase-treated virions affected the timing of CP expression and viral RNA replication in plants. The degree of phosphorylation decreased when the plant hosts were grown at an elevated temperature. These results show that phosphorylation of the capsid modulates BMV infection.
IMPORTANCE How icosahedral viruses regulate the release of viral RNA into the host is not well understood. The selective release of viral RNA can regulate the timing of replication and gene expression. Brome mosaic virus (BMV) is an RNA virus, and its three genomic RNAs are encapsidated in separate virions. Through proteomic, structural, and biochemical analyses, this work shows that posttranslational modifications, specifically, phosphorylation, on the capsid protein regulate the capsid-RNA interaction and the stability of the virions and affect viral gene expression. Mutational analysis confirmed that changes in modification affected virion stability and the timing of viral infection. The mechanism for modification of the virion has striking parallels to the mechanism of regulation of chromatin packaging by nucleosomes.
INTRODUCTION
The timing of viral genome release into the infected host cell is critically important to the outcome of infection, as it initiates the race between viral processes and the cellular immune responses against the virus. Even small changes in the timing of viral genome release can result in decreased fitness of the virus (1, 2). The regulation of the release of the viral RNAs is poorly understood, especially for icosahedral viruses.
Brome mosaic virus (BMV) is a plant-infecting RNA virus that has served as a model system to study the regulation of RNA virus infection (3). A particularly intriguing feature of BMV is that its tripartite positive-strand RNA genome is encapsidated in three distinct virions. All three virions are required for successful infection. Upon entry into cells, RNA1 and RNA2 direct the translation of the viral replication-associated proteins, while RNA3 encodes the movement protein required for cell-to-cell spread and the coat protein (CP). The CP is translated from subgenomic RNA4. In a typical infection, RNA4 is coencapsidated with RNA3 in a 1:1 ratio (4), although the host species can influence the encapsidation of the viral RNAs (5).
The BMV CP has multiple regulatory activities during infection (6, 7). Each BMV capsid contains 180 subunits of the CP arranged in a T=3 symmetry. A CP subunit has structural features similar to those of a histone protein. Both have an intrinsically disordered N-terminal arm rich in positively charged residues followed by sequences that fold into a globular domain (7). The N-terminal arm contributes to the differential encapsidation of BMV RNA, has the ability to translocate from the internal cavity to the outside of the virion, and is preferentially cleaved during proteolysis (8, 9). Virions formed by CPs that lack the first 8 residues were found to encapsidate RNA2 well but were labile for the encapsidation of RNA1 (8).
The BMV virions can be separated by their density into two populations, one enriched for virions containing RNA1, called B1 virions, and the other enriched for virions that encapsidate RNA2 and RNA3/4, called B2.3/4 virions (9). The B1 virions release RNA1 more rapidly than the B2.3/4 virions, likely to enable the more rapid expression of the protein needed to form the viral replication factory (9, 10). The B2.3/4 virions are much more stable than the B1 virions, and the capsid appears to interact more stably with the encapsidated RNAs in B2.3/4 virions (8, 9), raising the question of whether the release of RNA2, RNA3, and RNA4 requires an additional layer of control.
The stability of histone protein binding to DNA is regulated by posttranslational modifications (PTMs) of the N-terminal tail of histone proteins (11, 12). We hypothesize that PTMs, specifically, phosphorylation, regulate the differential BMV RNA release. Herein, we show that the BMV virions contain distinct patterns of PTMs. Furthermore, phosphorylation of the BMV virions at the N-terminal arm of the B2.3/4 virions impacts their stability and the timing of viral replication. Changes in PTMs were also affected by the growth conditions of the plant hosts.
MATERIALS AND METHODS
Plant growth.
Wheat plants were grown with a 16-h light and 8-h dark cycle under 3,000-lx fluorescent lights. Growth chambers were maintained at 25°C for growth at the typical temperature and at 32°C for growth at an elevated temperature.
Virion purification.
Wheat plants were 7 days old when they were infected. Plants were infected by the rubbing leaves with the standard inoculum concentration of BMV (0.05 mg/ml) amended with carborundum. Infected leaves were harvested at 7 days postinfection. Infected leaves were homogenized in a virus buffer (VB) containing 250 mM sodium acetate and 10 mM MgCl2 (pH 5.2). Insoluble plant debris was removed by centrifugation at 5,000 × g. Impurities were removed from the lysate by phase separation with 10% (wt/vol) chloroform. The supernatant containing the virions was layered on a 10% (wt/vol) sucrose cushion, and the virus was pelleted by centrifugation for 3 h at 28,000 rpm with a Beckman SW32 rotor. The pellet was resuspended overnight in VB and applied to a 45% CsCl gradient using a Beckman TLA110 rotor. The white band containing the virus was collected and dialyzed with three changes of VB. The virions were dissolved in a VB solution with 45% CsCl and centrifuged at 50,000 rpm for 24 h. The virions formed two distinct bands that were collected and reapplied to 45% CsCl for another round of centrifugation at 50,000 rpm for 20 to 24 h. Separated bands were collected and dialyzed against VB.
Analysis of posttranslational modifications.
For intact protein analysis, a Waters Synapt G2 high-definition mass spectrometer (MS; Milford, MA) with an ion mobility cell was used. For each sample, 5 μl of a 1-mg/ml solution of separated virions was loaded into the mass spectrometer. The resulting multiply charged spectra were deconvoluted using the MaxEnt tool of the MassLynx software package, which uses a maximum-entropy approach (13).
For MS and tandem MS analyses of peptides, a Thermo Scientific Orbitrap XL MS with nano-electrospray ionization was used. Virions were digested with trypsin (Trypsin Gold; Promega) at a 1:20 (wt/wt) ratio for 3 h, and 10 picomoles was analyzed on the mass spectrometer. Data files containing both the MS and tandem MS spectra were converted to MASCOT generic format (MGF) and analyzed using the MASCOT program (14). Parent ion spectra were matched within 100 ppm, and tandem MS spectra were matched within 0.5 Da. Relative ion abundance was determined using the ProtQuant Suite from the National Center for Glycomics and Glycoproteomics Institute (15). Virions purified from plants grown at different temperatures were processed identically but were analyzed on a Thermo Scientific LTQ Velos MS, and the parent ion spectra were matched within 0.3 Da.
Phosphatase treatment of the BMV CP.
Dephosphorylation of the BMV CP was performed in NEBuffer 2 buffer (New England BioLabs), composed of 10 mM Tris-HCl (pH 7.9), 50 mM NaCl, 10 mM MgCl2, and 1 mM dithiothreitol. Virions were mixed with 10 units of calf intestinal phosphatase (CIP; New England BioLabs) at 30°C for 60 min. As a control, aliquots of the separated virions were exposed to the same conditions described above without enzyme. To stop the reaction, samples were acidified with 1 μl of 10% trifluoroacetic acid.
Phosphopeptide enrichment.
Enrichment used Pierce TiO2 phosphopeptide enrichment spin tips (Thermo Scientific) and followed the protocol provided by the manufacturer. In short, BMV virions were digested with trypsin and applied to the tips. The resin within the tip was first washed with an acetonitrile buffer, followed by washing with a lactic acid buffer. Phosphorylated peptides were eluted from the column twice. The first elution was with an ammonium hydroxide buffer, and the second elution was with a pyrrolidine buffer. Each eluted sample was split in half, with one half being acidified with trifluoroacetic acid immediately and the other half being acidified after 30 min. C18 spin columns (Thermo Scientific) were used to purify and concentrate the peptides for MS analysis. MS/MS data were obtained using an LTQ Orbitrap XL MS (Thermo Scientific) and analyzed using either Xcalibur (Thermo Scientific) or MaxQuant (16) software.
DSF.
Differential scanning fluorimetry (DSF) was performed in a real-time PCR machine (Stratagene) in 96-well plates. Each 25-μl reaction mixture contained 4 μg of separated virion and SYPRO orange at a 2.5× concentration from a stock solution (Molecular Probes). The temperature ramp ascended at a rate of 1°C/min from 25 to 95°C, and the temperature that had the fastest change in fluorescence intensity was interpreted as the apparent melting temperature of the capsid.
In vitro phosphorylation of the BMV CP.
Phosphorylation of the BMV CP by protein kinase Cα was performed in a kinase buffer containing 20 mM HEPES (pH 7.4), 100 μM CaCl2, 10 mM MgCl2, 100 μM ATP, 100 μg/μl phosphatidylserine, 0.03% Triton X-100, and purified BMV virions at a concentration of 1 μg/μl. The mixture was incubated at 30°C for 30 min. As a control, aliquots of the separated particles were exposed to the same conditions without the kinase.
Protease digestion.
Proteolysis of the separated virions was performed in a buffer optimal for proteinase K activity (50 mM Tris-HCl [pH 7.5], 75 mM NaCl, 6.25 mM EDTA, 1% SDS). Proteinase K was added to a 50-μl solution containing 25 μg CP and incubated at 37°C. Aliquots were removed at the time points indicated below and added to Laemmli sample buffer to stop the reaction. Samples were stored on ice until SDS-PAGE. The resulting gel was stained with silver, and the proteins were quantified using ImageQuant software.
Cryo-EM and 3D image reconstruction of BMV virions.
Sample preparation and cryo-electron microscopy (cryo-EM) were performed following well-established procedures as described previously (17). Briefly, 4 μl of purified BMV virions at 0.5 mg/ml was vitrified using an FEI Vitrobot apparatus. Frozen-hydrated cryo-EM specimens were transferred to a 300-kV JEOL 3200FS microscope using a Gatan 626DH cryoholder, and the grids were kept at liquid nitrogen temperature (−176°C) during the subsequent data collection. Digital micrographs were recorded under low-dose conditions (20 e−/Å2) at multiple defocus levels using a Gatan Ultrascan 4000 charge-coupled-device camera at a nominal magnification of ×80,000, which yields a pixel size of 1.5 Å at the specimen level.
The e2boxer.py program (of the EMAN2 software package) was used to semimanually extract individual virions from the digital micrographs (18). Defocus values were determined with the CTFFIND3 program (19). Images of 13,080 mock-treated B1 virions, 9,807 CIP-treated B1 virions, 32,214 mock-treated B2.3/4 virions, and 19,834 CIP-treated B2.3/4 particles were subjected to structural refinement utilizing icosahedral symmetry. The initial model for each virion type was built de novo to reduce the possible artifact of model bias and was used as the reference model to search and refine the origins and orientations in the AUTO3DEM program (20, 21). The only corrections made were for the phase-contrast during the data processing. The final reconstructions were computed from 10,464 virions for mock-treated B1, 7,846 virions for CIP-treated B1, 19,332 virions for mock-treated B2.3/4, and 13,882 virions for CIP-treated B2.3/4. At the final refinement step, the data set was randomly divided into two parts to compute two three-dimensional (3D) reconstructions, and the resolution between these two maps was estimated by Fourier shell correlation using a 0.5 threshold as the criterion. The 3D reconstructions were rendered and visualized using the RobEM (http://cryoem.ucsd.edu/programs.shtm) and Chimera (22) programs. The atomic model of BMV (Protein Data Bank [PDB] accession number 1JS9) was used to fit the maps into the cryo-EM density maps using the fit-in-map function in Chimera.
UV cross-linking and immunoprecipitation methods coupled to high-throughput sequencing (CLIP-seq).
Virions were irradiated at 400 μJ/cm2 by 254-nm UV light three times. Ultracentrifugation was used to pellet the virion. The pelleted virion was incubated in RNA fragmentation buffer (100 mM Tris-HCl [pH 7.5], 10 mM ZnCl2) at 70°C for 15 min. To quench the reaction, 20 mM EDTA was used. Anti-CP serum and protein A/G magnetic beads (Pierce) were used to immunoprecipitate the CP and CP-RNA complexes. The material captured by the beads was digested with 2 μg/ml proteinase K at 37°C for 2 h, resulting in the release of fragmented RNA. The RNA was then purified using phenol-chloroform extraction and ethanol precipitation.
To prepare the library for Illumina MiSeq sequencing, the purified RNA was treated with T4 polynucleotide kinase (New England BioLabs) to remove the 3′ phosphate and add a 5′ phosphate. The RNA was then prepared using an Illumina TruSeq small RNA preparation kit. The cDNA was amplified and purified by elution from a 1.8% agarose Tris-acetate-EDTA gel. The libraries from the CIP- and mock-treated virus samples, each of which contained a distinct barcode, were combined at equimolar ratios and sequenced using the MiSeq personal sequencer.
The MiSeq reads were cleaned by trimming the adaptor sequences and eliminating base reads with a quality score below 20 using the FASTX toolkits. Cleaned reads were aligned to the reference genome using Bowtie 2 software with default local alignment settings (23). The SAMtools program was used to obtain the total coverage of each nucleotide in the reference RNA (24). As an adjustment for the difference in sequencing depth, the actual coverage was divided by the average coverage, resulting in the normalized coverage.
RNase digestion.
RNase A was added to a 0.1-ml sample containing 0.02 mg of CP in a 20 mM Tris buffer, and the mixture was incubated at 37°C. Aliquots were removed at the times indicated below and added to phenol-chloroform to stop the reaction. The aqueous phase containing RNA was isolated following centrifugation, adjusted to 0.3 M ammonium acetate, and precipitated using ethanol. The RNA concentration was determined by spectrometry, and the RNA was treated with glyoxal, electrophoresed on a 1.2% agarose gel, and quantified using ImageQuant software.
Analysis of capsid accumulation.
Samples were harvested from wheat seedlings grown in pots 4 in. in diameter. The samples were cut into fragments, and 150 mg was homogenized with pestles that fit 1.5-ml microcentrifuge tubes. Plant debris was removed by centrifugation at 10,000 × g for 10 min. The supernatant was separated by SDS-PAGE and subjected to Western blotting with a rabbit anti-CP antibody serum.
Real-time RT-PCR analysis.
Total RNAs were extracted from 150 mg of wheat leaves using the TRIzol reagent (Life Technologies). The lysate was extracted with a 50% volume of chloroform, followed by adjustment of the mixture to 0.3 M ammonium acetate and precipitation of the RNAs with isopropanol. RNA concentrations were determined by spectrometry and confirmed by denaturing agarose gel electrophoresis.
Minus-strand BMV RNA1 [(−)RNA1] and RNA2 [(−)RNA2] were quantified using reverse transcription (RT) and real-time PCR. One microgram of total RNA was used for RT with the reverse primer for nucleotides (nt) 100 to 120 of RNA1, the reverse primer for nt 371 to 392 of RNA2, and SuperScript II reverse transcriptase, as defined by the manufacturer (Invitrogen). PCR was performed for 32 cycles with forward and reverse primers for nt 100 to 232 of RNA1 and nt 371 to 499 of RNA2.
Construction of BMV CP mutant.
BMV RNA3 expressing the mutant CP was made by PCR-mediated site-directed mutagenesis with the proper primers, which are available upon request. The sequence of the cDNA was confirmed to contain only the intended mutation. RNA1, RNA2, and mutant RNA3 in vitro transcripts were made using an AmpliCap-Max high-yield message maker kit (Cellscript). The RNA concentration was determined by spectrometry and confirmed using agarose gel electrophoresis. Week-old wheat plants were inoculated with 4 μg of each transcript in 25 μl of water per the protocol of Allison et al. (25).
Accession number(s).
The cryo-EM density maps have been deposited in EMDataBank (http://www.emdatabank.org/). The accession numbers for B1 Mock, B1 +CIP, B2.3/4 Mock, and B2.3/4 +CIP are EMD-8262, EMD-8263, EMD-8264, and EMD-8265, respectively.
RESULTS
The BMV CP contains posttranslational modifications.
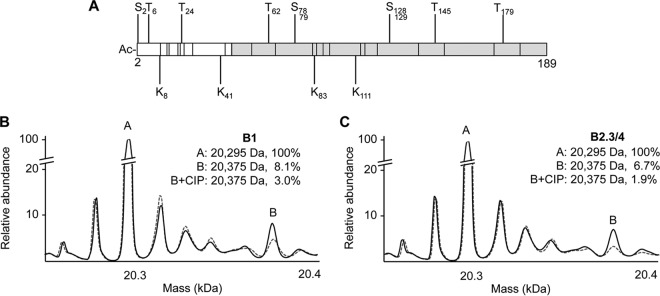
Bioinformatic analyses predict that the BMV CP can be acetylated, methylated, and phosphorylated (Fig. 1A) (26–28). MS was used to examine the presence of PTMs. The majority of the CP subunits from the B1 and B2.3/4 virions had a mass of 20,295 Da (Fig. 1B and C, peak A). This mass corresponds to a BMV CP that lacks the N-terminal methionine and an acetylated N terminus, a common modification in eukaryotic proteins (29–31). Minor amounts of CPs with lower masses, likely corresponding to CPs with single amino acid substitutions, were also observed. Additionally, approximately 25% of the CPs existed in forms with masses that indicated PTMs. The CPs from B1 and B2.3/4 virions had similar forms of the putatively modified CP, although the abundances of several forms were different between the two (Fig. 1B and C).
FIG 1.
Mass measurements of BMV CP. (A) Genetic map showing locations of BMV CP residues predicted to be phosphorylated, acetylated (Ac), and methylated. Residues predicted to be phosphorylated (S,T) are above the bar, and residues predicted to be acetylated or methylated (K) are below the bar. The location of the CP residues that form the globular region is shaded in gray. (B and C) MS was performed on the B1 (B) and B2.3/4 (C) virions to examine BMV PTMs (solid black line). Peak A corresponds to the BMV CP with the first methionine cleaved and an N-terminal acetyl group. Peak B corresponds to the putatively phosphorylated CP. CIP was used to confirm the presence of CP phosphorylation (dashed line). The abundance of the modified CP was calculated by integrating the areas encompassed by the peaks. The abundance of peak A was normalized to 100% to allow comparison to the other forms.
One form of the CP (peak B) was 80 Da heavier than the unmodified CP, a mass increase indicative of a singly phosphorylated amino acid. Peak B was 8% of the CPs from the B1 virions and 7% of those from the B2.3/4 virions (Fig. 1B and C, solid black lines), corresponding to 14 and 13 of the 180 CP subunits per virion, respectively. Treatment of the B1 and B2.3/4 virions with calf intestinal phosphatase (CIP) prior to MS analysis reduced the abundance of peak B to 3% and 1.9%, respectively (Fig. 1B and C, dashed line), confirming that the CPs were phosphorylated.
The BMV capsid contains complex PTMs.
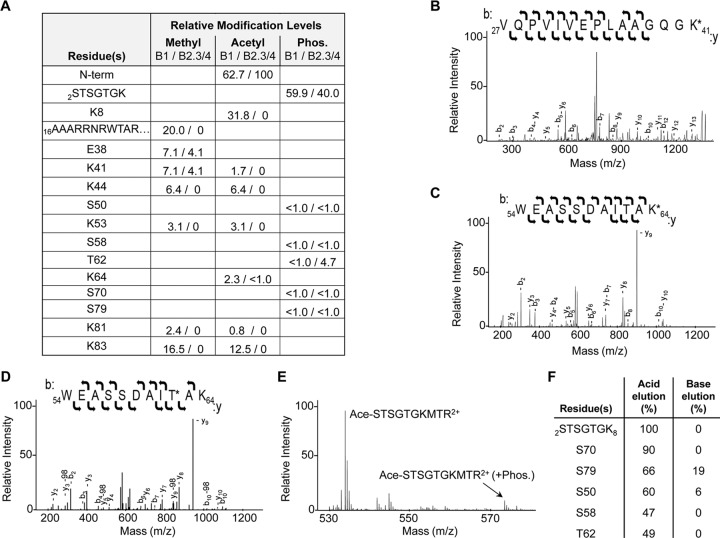
To identify the locations of PTMs within the BMV CP, the B1 and B2.3/4 virions were digested with trypsin and analyzed using tandem MS. Acetylation, methylation, and phosphorylation were observed in the BMV CP. Furthermore, the locations of the PTMs in the CPs of the B1 and the B2.3/4 virions were distinguishable (Fig. 2A). B1 virions also had a higher relative degree of methylation and acetylation than did the B2.3/4 virions. We note that the protease cleavage efficiencies and the ionization of peptides can vary; therefore, the abundances are not intended to be absolute abundances.
FIG 2.
BMV capsid modification analysis. (A) Modifications of the BMV capsid observed using MS/MS. Numbers show the relative abundance of the modified peptides compared to that of the unmodified peptide in the same sample. Phos., phosphorylation; N-term, N terminus. (B) MS/MS spectra showing methylation of K41 with the b and y ions labeled. (C) MS/MS spectra of acetylated K64 with the b and y ions labeled. (D) MS/MS spectra of phosphorylated T62 with the b and y ions, as well as those observed with neutral loss, labeled. (E) MS spectra of the partially digested N-terminal 10 residues, confirming phosphorylation of this region. Ace, acetyl. (F) Abundance of phosphorylated residues compared to that of the unmodified peptides after phosphopeptide enrichment.
The majority of the PTMs were located within residues 2 to 46 of the CP, the N-terminal arm of the BMV CP (Fig. 2A). Examples of several unambiguous modifications are shown (Fig. 2B to D). Phosphorylations were abundant in residues 2 to 8 of the CP (Fig. 2E), but the specific residue was difficult to assign due to the presence of multiple serines and threonines in this sequence (STSGTGK). The peptide contains heterogeneous phosphorylation, but the ions identified from this region were singly charged (Fig. 2A). In addition, methylation of arginines within peptides 16 to 26 could be in arginines 19, 20, and 22.
Phosphopeptides of the BMV virions were enriched using titanium oxide resin to improve assignment of the phosphorylated residues in the lower-abundance peptides (32). Half of the phosphopeptide-enriched digests was eluted and immediately acidified to preserve the phosphorylation on the peptide, while the second half was left in the basic elution buffer to hydrolyze the phosphates (32). Residues 2 to 8 were confirmed to contain abundant phosphorylated amino acids at heterogeneous positions. The peptides that had a low abundance of phosphorylation could then be more confidently assigned to residues in the globular domain of the CP (Fig. 2F).
Phosphorylation affects BMV virion stability.
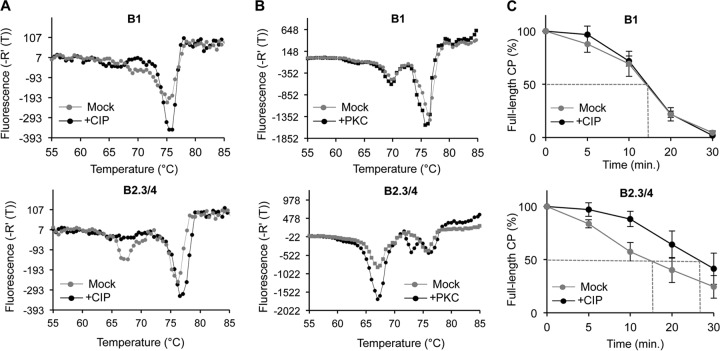
Phosphorylation can alter protein conformation and stability (33, 34). Therefore, we assessed whether phosphorylation of the CP affects the thermostability of the BMV virions using differential scanning fluorimetry (DSF) (8, 35). The denaturation profile of the B1 virions exhibited a minor transition with an apparent melting temperature (Tmapp) of 68°C and a major one of 76°C (Fig. 3A, top). CIP-treated B1 virions reduced the abundance of the minor transition and increased the abundance of the major one. The B2.3/4 virions also had a profile with two transitions. The one with a Tmapp of 66°C was more abundant than the one with a Tmapp of 76°C compared to their abundance in the B1 particles (Fig. 3A, bottom). CIP treatment of the B2.3/4 particles increased the abundance of the transition with a higher Tmapp and shifted the Tmapp to ca. 77°C (Fig. 3A, bottom). The 1°C shift was reproducible in five independent samples and was considered significant (35). These results suggest that phosphorylation decreased the stability of the B2.3/4 virions. In contrast, the stability of the B1 virions was less affected by phosphorylation.
FIG 3.
Phosphorylation of the BMV capsid affects virion stability. (A) DSF profile of B1 and B2.3/4 virions mock treated and CIP treated. -R′(T), rate of change of fluorescence. (B) Protein kinase C treatment alters the DSF profiles of BMV virions. B1 and B2.3/4 virions were mock treated or treated with protein kinase C. (C) Resistance to protease, measured by determination of the amount of intact CP, of B1 and B2.3/4 virions mock treated and CIP treated. The vertical bars show the range for 1 standard deviation.
Informatics analysis predicts that protein kinase C (PKC) can phosphorylate the BMV virion in the N-terminal arm (36). B1 virions treated with PKC showed only a minor change in their denaturation profile (Fig. 3B, top). In contrast, the denaturation profile of B2.3/4 virions treated with PKC was dramatically changed, with denaturation occurring at lower temperatures (Fig. 3B, bottom). These results confirm that phosphorylation more significantly impacts the stability of the B2.3/4 virions than that of the B1 virions.
To probe further how CP phosphorylation can affect the stability of the BMV virion, we examined the sensitivity of the B1 and B2.3/4 virions to digestion by protease. The amount of the full-length CP in SDS-polyacrylamide gels was quantified to assess the rate of digestion. Approximately half of the CP in the B1 virion was digested by 14.5 min (time to 50% digestion [T50]) regardless of CIP treatment (Fig. 3C, top). However, the B2.3/4 virions became resistant to proteolysis after CIP treatment, increasing the T50 from 17.5 to 28 min (Fig. 3C, bottom). Altogether, these results suggest that phosphorylation of the B2.3/4 virions significantly impacts the stability of the virions.
Phosphorylation changes the conformation and organization of the BMV virions.
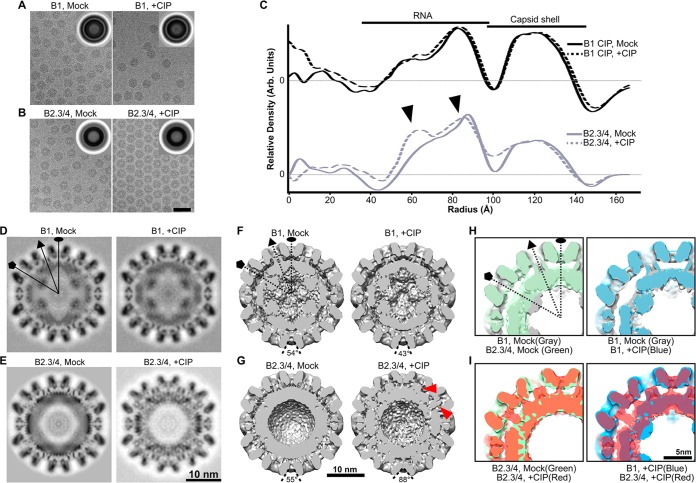
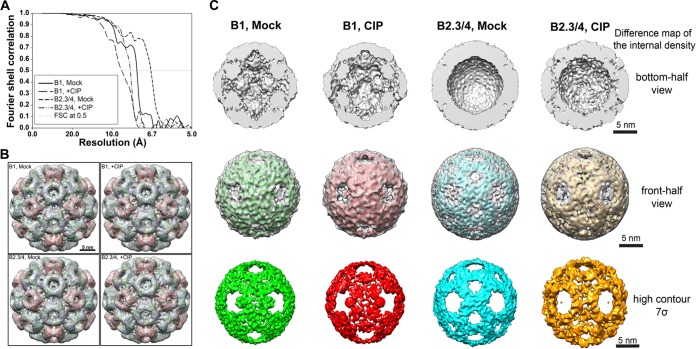
Cryo-electron microscopy (cryo-EM) was used to assess the virion structure as a function of capsid phosphorylation (Fig. 4). The morphologies of mock- and CIP-treated BMV virions were indistinguishable when they were observed in the cryo-EM micrographs, and the diameters of the virions were unchanged (Fig. 4A to C). The averaged images from over 9,800 virions showed that both the B1 and the B2.3/4 virions contained two concentric rings: a thin outer ring corresponding to the capsid shell and a thick inner ring corresponding to a combination of the capsid and the encapsidated RNA (Fig. 4A and B, insets). The one-dimensional density profile of the rotationally averaged images showed that the density of the internal shell was only marginally greater than that of the capsid shell in the B1 virions (Fig. 4C). In contrast, the density of the internal shell in B2.3/4 virions was substantially greater than that of the capsid shell (Fig. 4C). Thus, the amount of RNA and its organization are different between B1 and B2.3/4 virions.
FIG 4.
The phosphorylation state of the BMV CP can alter the conformation of the virions. (A and B) Electron micrographs of ice-embedded B1 virions (A) and B2.3/4 virions (B) mock treated and CIP treated. (Insets) Translationally aligned averaged images showing two concentric rings of the capsid and the capsid-RNA. Bar, 50 nm. (C) One-dimensional radial density profiles of the 3D reconstructions of B1 and B2.3/4 virions either mock treated or CIP treated. (D to G) Central slices (D and E) and bottom-half surface-shaded interiors (F and G) of the 3D reconstructions of mock-treated B1 virions (D and F, left), CIP-treated B1 virions (D and F, right), mock-treated B2.3/4 virions (E and G, left), and CIP-treated B2.3/4 virions (E and G, right). (H and I) Overlays of 3D maps of mock-treated B1 and B2.3/4 virions (H, left), mock- and CIP-treated B1 virions (H, right), mock- and CIP-treated B2.3/4 virions (I, left), and CIP-treated B1 and B2.3/4 virions (I, right).
Icosahedrally averaged 3D reconstructions were performed for B1 and B2.3/4 virions mock or CIP treated (Fig. 4D to G and 5A). All particles were resolved to a ca. 10-Å resolution to facilitate comparison. The 3D reconstructions of the BMV virions clearly resolved the densities corresponding to the outer capsid and the inner shell of RNA (Fig. 4D and E). The morphologies of the outer protein shells were very similar to those of previously published X-ray and cryo-EM structures (8, 37), where the capsids conformed to a T=3 surface lattice with a pentameric capsomer protrusion located at each 5-fold axis and a hexameric capsomer located at the 3-fold axis (Fig. 5B). The diameters of the mock- and CIP-treated B1 and B2.3/4 virions were virtually identical (Fig. 4C); however, the encapsidated RNAs within the B2.3/4 virions were rearranged upon CIP treatment (Fig. 4C, bottom, arrowheads).
FIG 5.
Internal density of the reconstructed BMV virions. (A) Resolution estimation of the cryo-EM 3D maps of BMV particles. The final resolutions for mock-treated B1, CIP-treated B1, mock-treated B2.3/4, and CIP-treated B2.3/4 virions are 7.6 Å, 8.9 Å, 6.7 Å, and 8.1 Å, respectively. The resolution was estimated by use of the Fourier shell correlation (FSC), determined at a 0.5 cutoff. (B) Fitting of atomic model into cryo-EM density maps of each BMV particle. (C) Difference map of the internal density. The difference map was calculated by subtracting the low-pass filtered X-ray model (using the rigid body fitting result) from each cryo-EM density map, and the substantial difference was rendered at the contour level equivalent to the original cryo-EM structure (gray) or at the high-contour level (color). (Top) Bottom-half view of internal density of each BMV particle; (middle) front-half view of the difference map shown in different contour levels (gray, low; color, high); (bottom) high front-half view of difference map rendered at the high contour level to show the dense structural features.
The previously published atomic model of BMV (PDB accession number 1JS9) was treated as a rigid body to fit into the cryo-EM density maps of B1 Mock, B1 +CIP, B2.3/4 Mock, and B2.3/4 +CIP (Fig. 5B) using the “Fit in Map” function in Chimera. The results showed that the overall capsid structures agreed strikingly well (Fig. 5B), with the exception of the CIP-treated B2.3/4 virions, where the bases of the pentameric and hexameric capsomers were tilted away from their centers (Fig. 4G). In mock- and CIP-treated B1 virions, the density of the internal shell was localized under each CP and formed a dodecahedron organization (Fig. 5C). Interestingly, at the 5-fold axis, a strong and continuous ordered density was projected from the inner surface of the pentameric capsomer and a protrusion pointed toward the center of the virion (Fig. 4F and H and 5A). To our knowledge, this structural feature is unique in B1 virions (Fig. 4F and H) and was observed for the first time in a bromovirus. However, at the current resolutions, we are unable to distinguish the entire internal density between the flexible N-terminal arm and the encapsidated RNA. Because the 3D structures of mock- and CIP-treated B1 virions are similar (Fig. 4F and H), we conclude that CIP treatment caused only a minor change in the B1 virion structure.
In striking contrast, the inner shells of B2.3/4 virions showed structural features distinct from those of the B1 virions (Fig. 4G and I). The internal density in B2.3/4 virions was thicker and more amorphous than the internal density in B1 particles (Fig. 5C). Unlike the B1 particles, no density protruding from the capsid was observed under the 5-fold axis (Fig. 4G, left). Difference map analysis revealed the conformational rearrangement of the internal density within B2.3/4 virions upon CIP treatment (Fig. 5C). In the mock-treated B2.3/4 particles, the highest internal density (contoured at 7σ, 7 standard deviations above the mean density), presumably the encapsidated RNA, was located under each capsomer and connected through the 2-fold region (Fig. 5C). The internal density in CIP-treated B2.3/4 particles showed a significantly stronger density at the 3-fold axis than the mock-treated virions, even when it was rendered at a higher contour level (Fig. 5C). These high-density RNA segments seemed to move away from the 2-fold axes and formed a ring-like structure under each pentameric capsomer (Fig. 5C). Furthermore, portions of the internal density, possibly contributed by both the N-terminal arm and RNA, clearly interacted more closely with the inner capsid surface at the centers of the pentameric and hexameric capsomers (Fig. 4G, right, arrowheads, and I). In the BMV crystal structure, the N-terminal arm under the 5-fold axis in subunit A was largely disordered (37). Similar structural features were also observed in the cryo-EM density maps (38). Therefore, the density observed under the 5-fold axis suggests that the N-terminal arm in CIP-treated B2.3/4 virions gained substantial structural order. Importantly, B2.3/4 adopts a strikingly different conformation upon CIP treatment.
Phosphorylation of the BMV capsid affects capsid-RNA interaction.
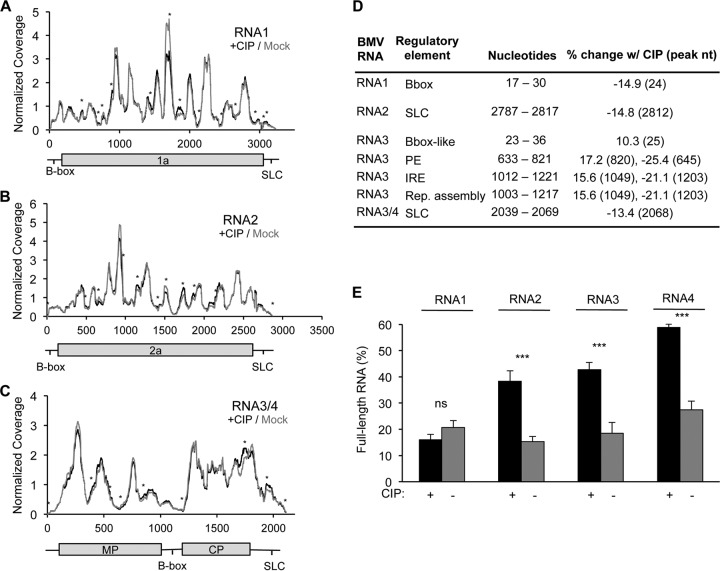
Cryo-EM analysis of the BMV virions suggests that phosphorylation affects capsid-RNA interactions. We used a modified CLIP-seq assay to examine the RNA sequences that contact the BMV capsid as a function of CIP treatment (5, 39). The nucleotides in the BMV RNA bound to the capsid were enumerated by determination of the number of sequence reads that contained the nucleotide. Phosphorylation of the capsid affected RNA contact with the capsid (Fig. 6A to C; Table 1). Interestingly, the sequences affected by phosphorylation by more than 10% included known regulatory elements in the BMV genomic RNA (Fig. 6D) (40–42). In RNA1, the B box, which regulates translation, was less bound to the CP in the CIP-treated virions (Fig. 6D). In RNA2 and RNA3/4, the core promoter for BMV minus-strand RNA synthesis (43, 44) had decreased binding to the CP in the CIP-treated virions (Fig. 6D). Several CP contacts in the regulatory intercistronic region of BMV RNA3 were also altered by CIP treatment (Fig. 6D).
FIG 6.
Phosphorylation of the BMV capsid affects the capsid-RNA interaction. (A to C) The number of RNA sequences contacting the capsid within mock-treated and CIP-treated virions identified by CLIP-seq analysis. Normalized coverage is shown. The identities of the majority of sequences that contact the capsid do not change as a function of capsid phosphorylation, but differences in intensity were observed. Peaks with an asterisk represent those with a greater than 20% change following CIP treatment. MP, movement protein; SLC, stem loop C. (D) Previously characterized BMV RNA regulatory elements that had at least a 10% change in expression following CIP treatment. The nucleotides column displays the locations of the regulatory elements. The final column shows the percent change following CIP treatment, and the nucleotide position where the change was observed is given in parentheses. PE, packaging element; IRE, intergenic replication enhancer; Rep., replication. (E) Resistance to RNase A digestion, as measured by the abundance of intact RNA derived from B1 and B2.3/4 virions before and after CIP treatment, followed by RNase treatment. Black, CIP-treated samples; gray, mock-treated samples. The range for 1 standard deviation is shown. ns, no significant difference; ***, P < 0.005.
TABLE 1.
RNA sequences that had a coverage that increased or decreased by at least 20% following CIP treatment, as observed by CLIP-seqa

In each sequence, the nucleotide with the greatest percent change is underlined and adjusted to be in the middle of the sequence, unless it is the residue at one terminus of the sequence (gray sequences). + and −, the abundance of the sequence was, respectively, increased or decreased by at least 20% relative to that for the mock-treated control.
To determine the consequences of the changes in CP-RNA interactions, virion RNA was digested with RNase A. The RNAs encapsidated in the B1 virions had similar sensitivities to RNase A digestion. However, the RNAs in the B2.3/4 virions were significantly less sensitive to RNase A after treatment with phosphatase (Fig. 6E). Altogether, these results suggest that phosphorylation of the B2.3/4 virions significantly impacts the stability of the virions and the interactions between the capsid and the encapsidated RNA.
Phosphorylation of the BMV CP affects the timing of BMV gene expression.
Changes in the stability of the virions and the CP interaction with RNA motifs suggest that phosphorylation impacts the release of the encapsidated BMV RNAs during infection. To assess the consequence of phosphorylation on BMV infection, mock- or CIP-treated BMV virions were inoculated into wheat seedlings. The amount of inoculum was kept low so that input CP could not be detected by Western blotting at 24 h postinoculation (data not shown). At 72 h, the total amount of CP accumulated from the plants on which the CIP-treated inoculum was used was reproducibly lower than that from the plants on which the mock-treated inoculum was used (Fig. 7A). The difference in the level of CP accumulation seen was reduced at 102 h postinoculation, suggesting that the BMV infection recovers from the CIP treatment during infection (Fig. 7A).
FIG 7.
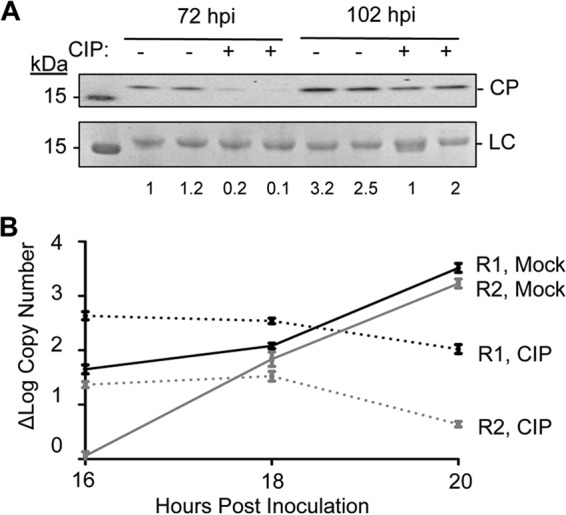
Phosphorylation affects BMV replication and virion production. (A) BMV CP accumulation in wheat seedlings inoculated for 72 and 102 h with mock- or CIP-treated BMV virions. Each sample was from a separately inoculated pot of wheat seedlings. Total wheat seedlings were harvested and processed for analysis of BMV CP accumulation by Western blotting. The CP was detected with a polyclonal antiserum that recognizes the BMV CP. BMV was undetectable at 24 and 48 h postinoculation, indicating that the signal is from CP accumulation during infection. LC, loading control. Numbers under the blots indicate quantified CP compared to LC. (B) (−)RNA1 (R1) and (−)RNA2 (R2) accumulation at 16, 18, and 20 h postinoculation in plants infected with mock-treated or CIP-treated BMV. The range for 1 standard deviation is shown at each time point.
The level of BMV RNA replication was assessed as a function of virion phosphorylation. The minus-strand BMV RNA1 and RNA2 synthesized during BMV replication were quantified using quantitative RT-PCR. RNA1 has been demonstrated to preferentially accumulate to higher levels earlier in infection than RNA2 (9). We observed this in the mock-treated virions (Fig. 7B, solid lines). The amounts of RNA1 and RNA2 then increased with time. With the CIP-treated inoculum, however, the kinetics of both minus-strand RNA1 and RNA2 accumulation were altered (Fig. 7B, dashed lines). Together these results suggest that phosphorylation affects the timing of BMV gene expression and replication.
Phosphomimetic in the N-terminal arm of the CP affected BMV infection.
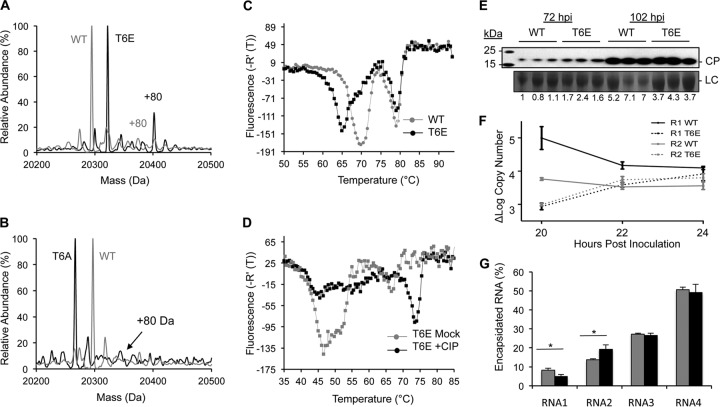
The N-terminal 8 residues of the BMV CP contain several serine and threonine residues that could be phosphorylated (Fig. 2A). The threonine at the 6th residue (T6) of the BMV CP was predicted to be a preferred residue for phosphorylation. We mutated T6 to glutamate to generate a CP with a phosphomimetic at this position. Wheat seedlings were inoculated with wild-type (WT) RNA1 and RNA2 transcripts, along with RNA3 containing the T6E substitution. CP from virions purified from infected plants contained the expected substitution (Fig. 8A). Interestingly, the CP with T6E also had an unusually high level of phosphorylation compared to WT BMV. This increase in CP phosphorylation observed in the CP with T6E was not observed in the CP with an alanine substitution at T6, T6A (Fig. 8B). The purified T6E virions were more readily dissociated in the thermodenaturation assay than the WT virions (Fig. 8C), and CIP treatment of CP with T6E, which should remove the additional phosphates in the CP, increased the stability of the virion with CP with T6E (Fig. 8D), consistent with phosphorylation of the CP reducing the stability of the virion.
FIG 8.
A phosphomimetic in the N-terminal arm of the BMV CP affects BMV infection and RNA encapsidation. (A) MS spectra of the T6E mutant. Higher levels of phosphorylation were observed for CP with the T6E mutation than WT BMV CP, as indicated by the +80 above the peak. (B) MS spectra of CP with the T6A mutation. Significantly higher levels of phosphorylation were not observed in the CP T6A mutant than WT BMV CP, as indicated by the +80 above the peak. (C) Virions with the T6E mutation dissociated more easily than the WT virions, consistent with the result that phosphorylation decreases virion stability. (D) DSF profile of virions with the T6E mutation mock treated or CIP treated. The high levels of phosphorylation significantly decrease the stability of the virions. (E) Abundance of BMV CP in wheat seedlings inoculated for 72 and 102 h with virions with the T6E mutation or WT BMV virions. Each sample was obtained from a separately inoculated pot of wheat seedlings. CP accumulation was detected by Western blotting. (F) (−)RNA1 and (−)RNA2 accumulation at 20, 22, and 24 h in plants infected with WT or T6E mutant BMV virions. The range for 1 standard deviation is shown at each time point. (G) Percentage of encapsidated RNA in the WT (gray bars) and T6E mutant (black bars) virions. The range for 1 standard deviation is shown. *, P < 0.05.
Wheat seedlings were inoculated with the WT or T6E mutant virion to assess the effects on the timing of viral infection. At 72 h, the total CP accumulation from the plants inoculated with the T6E mutant was reproducibly 2-fold higher than that from the plants treated with WT virions (Fig. 8E). The difference in CP accumulation seen was reduced at 102 h postinoculation. Notably, despite the increased CP abundance in the T6E mutant virions at early time points, a decrease in minus-strand RNA production was observed at early time points after inoculation (Fig. 8F, dashed lines). Higher levels of CP expression can suppress BMV RNA replication (45). Indeed, constitutive expression of the CP is the basis for several transgenic plants that are resistant to viral infection (46). Finally, we examined if the T6E mutation affected RNA encapsidation. Interestingly, the T6E mutant virions encapsidated less RNA1 and more RNA2 compared with amounts encapsidated by the WT virions. These results confirm that dysregulated phosphorylation of the BMV capsid can negatively affect the timing of gene expression and replication and affects the ratios of encapsidated RNA1 and RNA2.
Environmental conditions affect BMV virion posttranslational modification and infection.
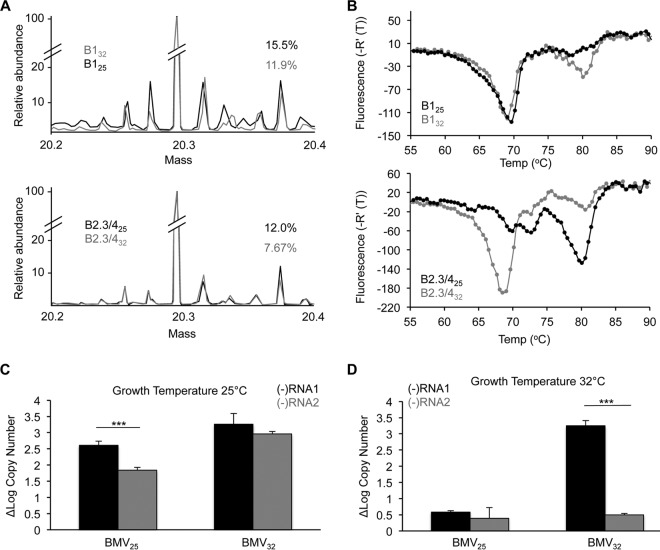
Host kinases and phosphatases are responsible for the degree of phosphorylation of the BMV capsids, as the BMV proteins are not known to have kinase or phosphatase activities. Given that the level of CP phosphorylation impacts the timing and the outcome of infection, it is highly likely that cellular kinases and/or phosphatases significantly impact the success of viral infection. Indeed, the host species was shown to affect virion accumulation and the ratios of the encapsidated RNAs (5, 47). The growth temperature of plants affects kinase activity (48–50). Therefore, we compared the PTMs on BMV virions purified from wheat grown at 25°C to those on virions purified from wheat grown at 32°C. Virions produced under the two conditions are denoted with a subscript indicating the temperature of plant growth. The BMV virions were separated using density gradients to enrich for the B1 or the B2.3/4 virions. Liquid chromatography-MS analysis revealed that the additional masses that match putative PTMs were altered in abundance as a function of the plant growth temperature (Fig. 9A). Furthermore, both B125 and B2.3/425 virions reproducibly exhibited higher levels of the phosphorylated CP than B132 and B2.3/432 virions (Fig. 9A). The higher temperature for plant growth thus decreased the phosphorylation of the BMV virions.
FIG 9.
Environmental effects on BMV CP phosphorylation and infection. (A) MS analysis of the B1 and B2.3/4 virions from plants grown at 25°C and 32°C. The abundance of phosphorylated CP decreased in the virions grown at 32°C. (B) DSF profile of the B1 and B2.3/4 virions grown at 25°C and 32°C. B1 particles grown at 32°C had a population with a Tmapp of over 80°C, while the majority of the B2.3/4 particles grown at 32°C had a Tmapp of 69°C. (C) (−)RNA1 and (−)RNA2 at 20 h postinoculation in plants grown at 25°C produced from virions at 25°C (left) and 32°C (right). (D) (−)RNA1 and (−)RNA2 at 20 h postinoculation in plants grown at 32°C produced from virions at 25°C (right) and 32°C (left) (D). The range for 1 standard deviation is shown. ***, P < 0.005.
The stabilities of the virions purified from plants grown at the two temperatures were examined using DSF (Fig. 9B). Both the B125 and the B132 virions exhibited two denaturation transitions that varied slightly (Fig. 9B, top). In contrast, the B2.3/4 virions produced at the two temperatures had dramatically different thermodenaturation profiles (Fig. 9B, bottom). CIP treatment of the B2.3/432 virions resulted in a denaturation profile similar to that of B2.3/425 (data not shown).
We examined whether BMV RNA replication by virions produced from plants grown at the two temperatures was affected. Wheat seedlings grown at 25°C or 32°C were inoculated with either BMV25 or BMV32. Plants grown at 25°C that were inoculated with BMV25 had significantly higher levels of (−)RNA1 than (−)RNA2 (Fig. 9C, left). Plants grown at 25°C and inoculated with BMV32 had levels of the two RNAs that were more comparable (Fig. 9C, right). Plants grown at 32°C and infected with BMV25 showed low levels of both (−)RNA1 and (−)RNA2 (Fig. 9D, left). Interestingly, when plants grown at 32°C were infected with BMV32, high levels of (−)RNA1 but much reduced levels of (−)RNA2 were produced (Fig. 9D, right). Together, these results suggest that the changes in PTMs due to the host growth temperature affected BMV replication. These results indicate that the epigenetic effects of host enzymes that impact PTMs could significantly alter the level of viral infection.
DISCUSSION
BMV encapsidates its three genomic RNAs in separate particles that have distinct physiochemical properties that impact the timing of BMV gene expression and replication (9). The particles can be physically separated by their density into two populations, one highly enriched for RNA1 (B1) and another enriched for RNA2.3/4 (B2.3/4). The two populations of virions contain numerous discernible properties (9). The B1 particles release their RNA more easily than do the B2.3/4 particles. The mechanism regulating the release of the RNAs encapsidated by the B2.3/4 particles is not known. In this work, we demonstrate that the CPs from the BMV capsid contain a large array of PTMs, especially in the highly flexible N-terminal arm that is known to bind RNA within the virion. The PTMs include phosphorylation sites that differ between the B1 and B2.3/4 virions (Fig. 1 and 2). The phosphorylation state of CPs within the virions regulates the position of the N-terminal arm of the CP and its interaction with the encapsidated RNAs. The consequences of the different PTMs impact the stability of the BMV virions, the release of the viral RNAs, and the timing of BMV gene expression and replication. Phosphorylation also affected CP binding to several regulatory RNA elements in the virions (Fig. 6D), which could alter the interaction with cellular and viral enzymes that are involved with the synthesis of BMV proteins and RNAs.
Phosphorylation of the N-terminal arm of the BMV CP could affect the localization of the N-terminal arm inside and outside the virion. In the hypophosphorylated state, the positively charged N-terminal arms of the CPs are more stably associated with the encapsidated RNAs. With phosphorylation, electrostatic repulsion would decrease the interaction energy of the N-terminal arms, supporting their display on the outside of the virions, and this finding is supported by preferential cleavage of the N-terminal arm during proteolysis (9). The exposed arms could then recruit additional cellular PTM enzymes. This proposed model explains both the differences in the densities of the B1 and B2.3/4 virions and also the increased amount of PTMs in the phosphomimetic T6E mutant virion (Fig. 8). Localization of the N-terminal arm could impact the degree of phosphorylation in the CP, perhaps affecting the number of CP subunits that are phosphorylated. Importantly, this change could regulate the timing of viral gene expression and RNA synthesis. We have evidence that the CP could interact differently with the regulatory RNA motifs within the BMV genome (Fig. 6D). The BMV CP has been shown to directly contact these RNA motifs to differentially regulate BMV RNA synthesis and protein synthesis (40–42). However, we cannot rule out the possibility of indirect effects of modified CP interacting with cellular enzymes that also impact BMV gene expression. Recently, it has also been determined that for icosahedral RNA viruses, the CP can bind to a series of low-affinity RNA-binding sites to determine the encapsidation of the viral RNA (51, 52). The selective binding of the CP to the RNA motifs has the potential to regulate the orderly release of the RNA and possibly present the viral RNA in a conformation that can influence viral gene expression. The PTM of the CP likely contributes to the regulatory events by influencing CP binding of the RNAs.
We note that in vitro reassembly of the virions to form virus-like particles does not recreate the combination of modifications found in the native BMV virions; thus, a large number of requirements for capsid-RNA interactions would be missed. The combinations of PTMs likely contribute to our previous observation that the BMV CP-RNA interaction does not strictly obey rules for charge complementarity (8).
Many other viruses have capsid proteins with positively charged and highly flexible sequences that translocate from within the virion to the external surface of the virion (7, 53). It is possible that these sequences are acted on by host proteins to regulate the disassembly of the virions and enable viral gene expression and replication. Phosphorylation has been shown to affect CP-RNA interactions in potyviruses (54, 55). The Beet black scorch virus CP was found to be phosphorylated by the protein kinase A, and mutations to prevent phosphorylation affect accumulation of the viral RNAs in plants due to decreased virion stability (56). For rubella virus, phosphorylation of the capsid has consequences on viral replication, gene expression, and RNA encapsidation (57, 58). In hepatitis B virus, modifications of the RNA-binding domain that mimic phosphorylation alter the RNA conformation (17).
For BMV, it is particularly interesting that phosphorylation had more dramatic effects in reducing the stability of the B2.3/4 particles and altered the interaction between the capsid and the RNA (Fig. 3 to 8). Phosphorylation of the B2.3/4 virions could promote the timely release of RNA2, RNA3, and RNA4 from the virions. How subsets of virions recruit host enzymes to modify the capsid remains to be determined, given that the virions have the same CP that encapsidates the RNAs. It is tempting to speculate that the BMV RNAs may promote the specific recognition of subsets of virions.
The regulation of BMV virions by PTMs has striking similarities to the regulation of the chromatin structure by nucleosomes (11, 12). The BMV CP and histones both possess a basic, flexible N-terminal arm that interacts with nucleic acid. PTM of the arms can regulate the interaction with nucleic acids to impact gene expression. Since the BMV capsids have the N-terminal arms directed inward, they can be considered inverted nucleosomes that both protect and compact the viral genome as well as respond to environmental conditions to regulate the release of the nucleic acid.
A large proportion of the genomes of eukaryotic cells is dedicated to regulating changes in the phosphorylation state of proteins, with human and Arabidopsis cells having approximately 500 and 1,000 kinase genes, respectively (59, 60). The effects of PTMs on BMV infection that we observed and our observation that the plant growth temperature can affect PTMs, virion thermostability, and viral replication rates (Fig. 9) suggest that kinases and phosphatases have an active role in modulating the success of viral infection at the level of virion disassembly. Virion modifications by cellular proteins could contribute to the host range and the outcome of viral infection.
ACKNOWLEDGMENTS
We thank the IU Proteomics Facility and Alexander Jacobs for help with mass spectrometric analysis. We thank the IU Center for Genomics and Bioinformatics for next-generation DNA sequencing. We thank Laura Kao for editing the manuscript and members of the Kao lab for helpful discussions. H.S.H. acknowledges support from the Floyd Plant and Fungal Biology Fellowship.
Funding Statement
No external funding was used in this research.
REFERENCES
- 1.Domingo E, Escarmis C, Sevilla N, Moya A, Elena SF, Quer J, Novella IS, Holland JJ. 1996. Basic concepts in RNA virus evolution. FASEB J 10:859–864. [DOI] [PubMed] [Google Scholar]
- 2.Drake JW, Holland JJ. 1999. Mutation rates among RNA viruses. Proc Natl Acad Sci U S A 96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kao CC, Sivakumaran K. 2000. Brome mosaic virus, good for an RNA virologist's basic needs. Mol Plant Pathol 1:91–97. doi: 10.1046/j.1364-3703.2000.00017.x. [DOI] [PubMed] [Google Scholar]
- 4.Rao AL. 2006. Genome packaging by spherical plant RNA viruses. Annu Rev Phytopathol 44:61–87. doi: 10.1146/annurev.phyto.44.070505.143334. [DOI] [PubMed] [Google Scholar]
- 5.Ni P, Vaughan RC, Tragesser B, Hoover H, Kao CC. 2014. The plant host can affect the encapsidation of brome mosaic virus (BMV) RNA: BMV virions are surprisingly heterogeneous. J Mol Biol 426:1061–1076. doi: 10.1016/j.jmb.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yi G, Vaughan RC, Yarbrough I, Dharmaiah S, Kao CC. 2009. RNA binding by the brome mosaic virus capsid protein and the regulation of viral RNA accumulation. J Mol Biol 391:314–326. doi: 10.1016/j.jmb.2009.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ni P, Kao CC. 2013. Non-encapsidation activities of the capsid proteins of positive-strand RNA viruses. Virology 446:123–132. doi: 10.1016/j.virol.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni P, Wang Z, Ma X, Das NC, Sokol P, Chiu W, Dragnea B, Hagan M, Kao CC. 2012. An examination of the electrostatic interactions between the N-terminal tail of the brome mosaic virus coat protein and encapsidated RNAs. J Mol Biol 419:284–300. doi: 10.1016/j.jmb.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaughan R, Tragesser B, Ni P, Ma X, Dragnea B, Kao CC. 2014. The tripartite virions of the brome mosaic virus have distinct physical properties that affect the timing of the infection process. J Virol 88:6483–6491. doi: 10.1128/JVI.00377-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, Ahlquist P. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell 9:505–514. doi: 10.1016/S1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 11.Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 12.Tropberger P, Schneider R. 2013. Scratching the (lateral) surface of chromatin regulation by histone modifications. Nat Struct Mol Biol 20:657–661. doi: 10.1038/nsmb.2581. [DOI] [PubMed] [Google Scholar]
- 13.Ferroge A, Seddon M, Skilling J, Ordsmith N. 1992. The application of ‘MaxEnt’ to high resolution mass spectrometry. Rapid Commun Mass Spectrom 6:765–770. doi: 10.1002/rcm.1290061211. [DOI] [Google Scholar]
- 14.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567. doi:. [DOI] [PubMed] [Google Scholar]
- 15.Mann B, Madera M, Sheng Q, Tang H, Mechref Y, Novotny MV. 2008. ProteinQuant Suite: a bundle of automated software tools for label-free quantitative proteomics. Rapid Commun Mass Spectrom 22:3823–3834. doi: 10.1002/rcm.3781. [DOI] [PubMed] [Google Scholar]
- 16.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 17.Wang JC, Dhason MS, Zlotnick A. 2012. Structural organization of pregenomic RNA and the carboxy-terminal domain of the capsid protein of hepatitis B virus. PLoS Pathog 8:e1002919. doi: 10.1371/journal.ppat.1002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Mindell JA, Grigorieff N. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142:334–347. doi: 10.1016/S1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 20.Yan X, Dryden KA, Tang J, Baker TS. 2007. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol 157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan X, Sinkovits RS, Baker TS. 2007. AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J Struct Biol 157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 23.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allison RF, Janda M, Ahlquist P. 1988. Infection in vitro transcripts from cowpea chlorotic mottle virus cDNA clones and exchange of individual RNA components with brome mosaic virus. J Virol 62:3581–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suo SB, Qiu JD, Shi SP, Sun XY, Huang SY, Chen X, Liang RP. 2012. Position-specific analysis and prediction for protein lysine acetylation based on multiple features. PLoS One 7:e49108. doi: 10.1371/journal.pone.0049108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shien DM, Lee TY, Chang WC, Hsu JB, Horng JT, Hsu PC, Wang TY, Huang HD. 2009. Incorporating structural characteristics for identification of protein methylation sites. J Comput Chem 30:1532–1543. doi: 10.1002/jcc.21232. [DOI] [PubMed] [Google Scholar]
- 28.Blom N, Gammeltoft S, Brunak S. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 29.Frottin F, Martinez A, Peynot P, Mitra S, Holz RC, Giglione C, Meinnel T. 2006. The proteomics of N-terminal methionine cleavage. Mol Cell Proteomics 5:2336–2349. doi: 10.1074/mcp.M600225-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Giglione C, Boularot A, Meinnel T. 2004. Protein N-terminal methionine excision. Cell Mol Life Sci 61:1455–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polevoda B, Sherman F. 2000. Nalpha-terminal acetylation of eukaryotic proteins. J Biol Chem 275:36479–36482. doi: 10.1074/jbc.R000023200. [DOI] [PubMed] [Google Scholar]
- 32.Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. 2006. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat Protoc 1:1929–1935. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- 33.Groban ES, Narayanan A, Jacobson MP. 2006. Conformational changes in protein loops and helices induced by post-translational phosphorylation. PLoS Comput Biol 2:e32. doi: 10.1371/journal.pcbi.0020032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xin F, Radivojac P. 2012. Post-translational modifications induce significant yet not extreme changes to protein structure. Bioinformatics 28:2905–2913. doi: 10.1093/bioinformatics/bts541. [DOI] [PubMed] [Google Scholar]
- 35.Niesen FH, Berglund H, Vedadi M. 2007. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 36.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. 2004. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 37.Lucas RW, Larson SB, McPherson A. 2002. The crystallographic structure of brome mosaic virus. J Mol Biol 317:95–108. doi: 10.1006/jmbi.2001.5389. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Hryc CF, Bammes B, Afonine PV, Jakana J, Chen DH, Liu X, Baker ML, Kao CC, Ludtke SJ, Schmid MF, Adams PD, Chiu W. 2014. An atomic model of brome mosaic virus using direct electron detection and real-space optimization. Nat Commun 5:4808. doi: 10.1038/ncomms5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konig J, Zarnack K, Luscombe NM, Ule J. 2011. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet 13:77–83. doi: 10.1038/ni.2154. [DOI] [PubMed] [Google Scholar]
- 40.Choi SK, Hema M, Gopinath K, Santos J, Kao CC. 2004. Replicase binding sites on plus- and minus-strand BMV RNAs and identification of their roles in RNA replication in plant cells. J Virol 78:13420–13429. doi: 10.1128/JVI.78.24.13420-13429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu J, Gopinath K, Murali A, Yi GH, Hayward SD, Zhu H, Kao CC. 2007. RNA-binding proteins that inhibit RNA virus infection. Proc Natl Acad Sci U S A 104:3129–3134. doi: 10.1073/pnas.0611617104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi G, Gopinath K, Kao CC. 2007. Selective repression of translation by the brome mosaic virus 1a replication protein. J Virol 81:1601–1609. doi: 10.1128/JVI.01991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chapman MR, Kao CC. 1999. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J Mol Biol 286:709–720. doi: 10.1006/jmbi.1998.2503. [DOI] [PubMed] [Google Scholar]
- 44.Kim CH, Kao CC, Tinoco I. 2000. RNA motifs that determine specificity between a viral replicase and its promoter. Nat Struct Biol 7:415–423. doi: 10.1038/75202. [DOI] [PubMed] [Google Scholar]
- 45.Yi G, Kao CC. 2008. cis- and trans-Acting functions of brome mosaic virus protein 1a in genomic RNA1 replication. J Virol 82:3045–3053. doi: 10.1128/JVI.02390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hackland AF, Rybicki EP, Thomson JA. 1994. Coat protein-mediated resistance in transgenic plants. Arch Virol 139:1–22. doi: 10.1007/BF01309451. [DOI] [PubMed] [Google Scholar]
- 47.Kitayama M, Hoover H, Middleton S, Kao CC. 2015. Brome mosaic virus infection of rice results in decreased accumulation of RNA1. Mol Plant Microbe Interact 28:626–632. doi: 10.1094/MPMI-12-14-0389-R. [DOI] [PubMed] [Google Scholar]
- 48.Gong D, Gong Z, Guo Y, Chen X, Zhu JK. 2002. Biochemical and functional characterization of PKS11, a novel Arabidopsis protein kinase. J Biol Chem 277:28340–28350. doi: 10.1074/jbc.M107719200. [DOI] [PubMed] [Google Scholar]
- 49.Gong D, Gong Z, Zhu JK. 2002. Expression, activation, and biochemical properties of a novel Arabidopsis protein kinase. Plant Physiol 129:225–234. doi: 10.1104/pp.010776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z, Meng P, Zhang X, Ren S, Yang S. 2011. BON1 interacts with the protein kinases BIR1 and BAK1 in modulation of temperature-dependent plant growth and cell death in Arabidopsis. Plant J 67:1081–1093. doi: 10.1111/j.1365-313X.2011.04659.x. [DOI] [PubMed] [Google Scholar]
- 51.Rolfsson Ó, Middleton S, Manfield IW, White SJ, Fan B, Vaughan R, Ranson NA, Dykeman E, Twarock R, Ford J, Kao CC, Stockley PG. 2016. Direct evidence for packaging signal-mediated assembly of bacteriophage MS2. J Mol Biol 29:431–448. doi: 10.1016/j.jmb.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prevelige PE., Jr 2016. Follow the Yellow Brick Road: a paradigm shift in virus assembly. J Mol Biol 29:416–418. doi: 10.1016/j.jmb.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 53.Speir JA, Bothner B, Qu C, Willits DA, Young MJ, Johnson JE. 2006. Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics. J Virol 80:3582–3591. doi: 10.1128/JVI.80.7.3582-3591.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ivanov KI, Puustinen P, Merits A, Saarma M, Mäkinen K. 2001. Phosphorylation down-regulates the RNA binding function of the coat protein of potato virus A. J Biol Chem 276:13530–13540. [DOI] [PubMed] [Google Scholar]
- 55.Ivanov KI, Puustinen P, Gabrenaite R, Vihinen H, Ronnstrand L, Valmu L, Kalkkinen N, Mäkinen K. 2003. Phosphorylation of the potyvirus capsid protein by protein kinase CK2 and its relevance for virus infection. Plant Cell 1:2124–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao X, Wang X, Dong K, Zhang Y, Hu Y, Zhang X, Chen Y, Wang X, Han C, Yu J, Li D. 2015. Phosphorylation of the beet back scorch virus coat protein by PKA is required for assembly and stability of viral particles. Sci Rep 5:11585. doi: 10.1038/srep11585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Law L, Everitt J, Beatch M. 2003. Phosphorylation of the rubella virus capsid regulates its RNA binding activity and virus replication. J Virol 77:1764–1771. doi: 10.1128/JVI.77.3.1764-1771.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Law LJ, Ilkow CS, Tzeng WP, Rawluk M, Stuart DT, Frey TK, Hobman TC. 2006. Analyses of phosphorylation events in the rubella virus capsid protein: role in early replication events. J Virol 80:6917–6925. doi: 10.1128/JVI.01152-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 60.Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]