ABSTRACT
APOBEC3 knockout and human APOBEC3A and -3G transgenic mice were tested for their ability to be infected by the herpesviruses herpes simplex virus 1 and murine herpesvirus 68 and the parvovirus minute virus of mice (MVM). Knockout, APOBEC3A and APOBEC3G transgenic, and wild-type mice were equally infected by the herpesviruses, while APOBEC3A but not mouse APOBEC3 conferred resistance to MVM. No viruses showed evidence of cytidine deamination by mouse or human APOBEC3s. These data suggest that in vitro studies implicating APOBEC3 proteins in virus resistance may not reflect their role in vivo.
IMPORTANCE It is well established that APOBEC3 proteins in different species are a critical component of the host antiretroviral defense. Whether these proteins also function to inhibit other viruses is not clear. There have been a number of in vitro studies suggesting that different APOBEC3 proteins restrict herpesviruses and parvoviruses, among others, but whether they also work in vivo has not been demonstrated. Our studies looking at the role of mouse and human APOBEC3 proteins in transgenic and knockout mouse models of viral infection suggest that these restriction factors are not broadly antiviral and demonstrate the importance of testing their activity in vivo.
INTRODUCTION
Given the frequent encounters of vertebrates with viruses, it is not surprising that there has been selection for host defense systems. In 2002, human apolipoprotein B editing complex 3G (APOBEC3G) was discovered and shown to confer intrinsic immunity to HIV-1 (1). The APOBEC3G gene belongs to a family of genes encoding DNA- and RNA-editing enzymes characterized by the presence of at least one cytidine deaminase (CDA) domain (2). The number of APOBEC3 genes varies from species to species, from 1 gene in mice to 7 genes (the APOBEC3A, -3B, -3C, -3DE, -3F, -3G, and -3H genes) in primates (2). Moreover, there are multiple allelic differences in APOBEC3 gene coding regions; notably, there are at least 2 alleles in mice that differ in both their protein-coding regions and levels of expression (3, 4). These allelic variants confer weak or strong resistance to murine retrovirus infection in vivo (3, 5–7).
Several human APOBEC3 proteins inhibit HIV-1 lacking the vif gene, which encodes a protein expressed at high levels late in infection (8–12). In vif-deficient-HIV producer cells, APOBEC3 proteins are packaged into progeny virions via interaction with the nucleocapsid (NC) protein and viral RNA. Vif prevents packaging by binding APOBEC3 proteins, targeting them for ubiquitination and degradation in the proteasome (13–17). APOBEC3 proteins inhibit infection by deaminating deoxycytidine residues on minus-strand DNA, inducing hypermutation in newly reverse-transcribed HIV-1 DNA; this leads both to degradation of viral DNA prior to integration and to G-to-A coding-strand mutations in viral genes in the integrated provirus (18). APOBEC3 proteins also inhibit replication by a number of CDA-independent mechanisms (19). APOBEC3 proteins restrict animal retroviruses (20–23) and other human retroviruses, such as human T cell leukemia virus 1 (HTLV-1), but it has also been suggested that they prevent zoonotic transmission (18, 24, 25). Indeed, several human APOBEC3 proteins have been shown to be more effective at inhibiting mouse retroviruses than the murine APOBEC3 protein, both in vitro and in vivo (26–28).
The selective pressure placed on APOBEC3 genes, which led to the expansion of the human APOBEC3 locus through gene duplication as well as to polymorphisms in the genes in many species, may also be the result of infections by other viruses in addition to retroviruses. There have been numerous reports of APOBEC3 proteins restricting members of several virus families by both deaminase-dependent and -independent mechanisms (29–31). For example, APOBEC3A but not APOBEC3G was shown to inhibit infection by the parvoviruses adeno-associated virus and minute virus of mice (MVM) in cultured cells overexpressing these proteins, in this case by deaminase-independent means (32, 33). Evidence of APOBEC3A-, 3C-, and 3G-mediated editing of herpes simplex virus 1 (HSV-1) was detected when these factors were overexpressed in cultured cells, and HSV-1 and Epstein-Barr virus (EBV) showed signs of cytidine deamination in buccal swabs or immortalized cell lines, respectively (30). Moreover, a recent study showed that several human APOBEC3s, including APOBEC3A, inhibited replication of murine gammaherpesvirus 68 (MHV68) when the viral genome was transfected into cultured cells but not when it was introduced by infection (34). However, APOBEC3 knockout (A3KO) and wild-type (WT) mice were equally resistant to infection by this virus (34).
Here, we took advantage of A3KO and human APOBEC3A- and 3G-expressing transgenic mouse lines developed by our lab several years ago to study in vivo infection by HSV-1, MHV68, and MVM and to determine if the mouse or human proteins could contribute to inhibition of zoonosis (27). These transgenic mouse strains, each of which was backcrossed onto a mouse A3KO background, express myc-tagged human APOBEC3A and -3G under the control of the β-actin promoter, and thus the transgenes are expressed in multiple tissues, including the brain, spleen, and liver, as well as sentinel cells (macrophages, dendritic cells, and lymphocytes) and fibroblasts. Importantly, the level of expression of the transgenes was similar to that seen in normal human tissue (27). The human APOBEC3 proteins were fully functional in vivo; both APOBEC3A and APOBEC3G restricted infection by the murine retroviruses mouse mammary tumor virus (MMTV) and murine leukemia virus (MLV). Interestingly, the restriction occurred by different mechanisms: APOBEC3G was packaged into virions and caused extensive deamination of the retrovirus genomes, while APOBEC3A was not packaged, did not deaminate, and restricted infection when expressed in cells that were targets of infection. While the human APOBEC3 transgenic mice showed much lower infection by the murine retroviruses, here we show that this is not the case for mouse or human herpesviruses and that only the parvovirus MVM was modestly inhibited by APOBEC3A in vivo.
MATERIALS AND METHODS
Mice.
A3KO, A3Ahigh, A3Glow, and A3Ghigh transgenic mice on an A3KO (C57BL/6N) background were previously described (20, 27). All transgenic lines were maintained by breeding with mouse A3KO mice, so the transgenes were carried in heterozygotes on the A3KO background. This allowed us to generate nontransgenic, matched controls for infection studies. Tmem173 mice (C57BL/6J background) were obtained from the Jackson Laboratory, and BALB/cN and C57BL/6N mice were obtained from Charles River; all 3 strains were bred at the University of Pennsylvania. The mice were housed according to the policies of the Institutional Animal Care and Use Committee of the University of Pennsylvania. The experiments performed with mice in this study were approved by this committee (IACUC protocol 801594). All virus infections were carried out under animal biosafety level 2 conditions.
HSV-1 infections and analysis.
Mice were intraocularly (i.o.) or intraperitoneally (i.p.) inoculated with 1 × 106 PFU of HSV-1 (McKrae strain), and then sacrificed at 5.7 days postinoculation (dpi) to harvest the tissue samples. Moribund mice (as defined by a hunched posture, dehydration, and lethargy) were sacrificed before 5.7 days, as noted below. For i.o. inoculations, mice were anesthetized with ketamine-xylazine, and then 2 to 10 μl of virus was pipetted onto the eyes. For mice inoculated by the i.o. route, DNA was isolated from the trigeminal ganglia, eyeballs, and spleen, and real-time quantitative PCR (qPCR) was performed using primers specific for the HSV-1 thymidine kinase (TK) gene (5′-GAGTTTCACGCCACCAAGAT-3′/5′-CTATGATGACACAAACCCCG-3′). HSV-1 values were normalized to mouse GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (27). For mice inoculated by the i.p. route, daily body weight and survival were ascertained, and brain tissue samples obtained at euthanasia were homogenized in 1 ml culture medium. The supernatants of homogenates were 10-fold serially diluted, and titers were determined on Vero cells. At 2 h postinfection, the inoculum was replaced with culture medium containing 2% fetal calf serum (FCS) and 5% methylcellulose. The cells were fixed with methanol and stained with crystal violet at 3 dpi. Plaques were counted, and the viral titers were calculated as PFU/ml.
MHV68 infections and analysis.
MHV68 (103 PFU; WUMS strain) was intranasally inoculated into mice. The mice were sacrificed at 7 and 16 dpi for acute- and latent-infection analysis, respectively (34). Plaque assays were conducted to measure the virus titers during acute infection in the lungs. The lungs were homogenized in 1 ml culture medium. The supernatants of homogenates were 10-fold serially diluted. The titer of each dilution was determined on NIH3T12 cells. At 2 h postinfection, the inoculum was replaced with culture medium containing 5% FCS and 2.5% methylcellulose. The cells were fixed with methanol and stained with crystal violet at 7 dpi. Plaques were counted, and the viral titers were calculated as PFU/ml. MHV68 latency was evaluated by qPCR of spleen and lung DNA, using the specific primer set for the MHV68 ORF12 gene (5′-GTCTACAACAGGATCTGCATTT-3′/5′-AAAACTCTACCGTGACTGTGAA-3′), as described by Minkah et al. (34). The values were normalized to mouse GAPDH.
MVMi infections and PCR.
Newborn C57BL/6N, BALB/cN, A3A, and A3KO mice were infected oranasally with equal amounts of the lymphotropic-specific variant of MVM, MVMi (35). Mice were sacrificed at 5, 9, or 13 days postinfection. Spleens, lungs, kidneys, and brains were isolated from the infected mice, and DNA was isolated from the organs using a DNeasy kit (Qiagen). qPCR was performed to examine the MVMi levels in the infected organs. MVMi primers used for qPCR detection were 5′-AAGGTACGATGGCGCCTC-3/5′-GTGCTCTTTGGCAGC-3′. MVMi values were normalized to GAPDH.
3DPCR and sequencing.
PCRs to amplify edited viral DNA were performed using a gradient cycler (Veriti 96-well thermal cycle; Applied Biosystems) on DNA isolated from the indicated organs. To examine deamination in HSV-1, differential DNA denaturation PCR (3DPCR) on organ DNA was conducted using GoTaq Green master mix (Promega) with a specific primer set for the HSV-1 ICP22 gene, 5′-CGACGCGGGCCCGAGCRTATRCTYYAT-3′/5′-GGAAATGGCGGACACCTTCCTGGAYAYYAT-3′, as described by Suspène and colleagues (30). The denaturation temperature of the PCR cycle was programmed to range from 89 to 95°C, and reactions were carried out for 40 cycles. For MHV68, 3DPCR was performed using Q5 Hot Start high-fidelity DNA polymerase (New England BioLabs) with a specific PCR primer set for a GC-rich region of the MHV68 genome (bp 88884 to 89613) (5′-ACGACCCTGACAACATCAAC-3′/5′-TCTTGTTCCCAGGTGGCCTTAA-3′) (34). The denaturation temperature of the PCR cycle ranged from 83 to 98°C. For MVMi, a 946-bp region of the genome was amplified with the primers 5′-CTGTCCACTCAGCTGCAAGA-3′/5′-ACTCACCCAGTTAACCCCCA-3′. A temperature gradient of 85 to 89°C was applied. To increase the likelihood of detecting deamination events, the PCR products amplified at the lowest denaturation temperature for all the viruses were purified from agarose gels using a Qiagen QIAquick gel extraction kit and were cloned into pCR-Blunt II-TOPO vector as directed by the manufacturer (Invitrogen, Inc.). DNA was isolated from the resultant colonies using the TempliPhi kit (GE Healthcare Life Sciences, Inc.). Clones were then sequenced using a BigDye Terminator v3.1 cycle sequencing kit from Applied Biosystems. The numbers of target sites for the different APOBEC3s in the regions of HSV-1, MHV68, and MVMi sequenced are shown in Table 1.
TABLE 1.
Numbers of APOBEC3 sequence hot spots found in the regions of HSV-1, MHV68, and MVMi sequenced
| Virus | No. of hot spots |
||
|---|---|---|---|
| Mouse APOBEC3 (TCC/GGA) | APOBEC3A (TC/GA) | APOBEC3G (CCC/GGG) | |
| HSV-1 | 39 | 106 | 83 |
| MHV68 | 27 | 112 | 11 |
| MVMi | 19 | 100 | NAa |
NA, not applicable.
Statistical analysis.
Statistical analysis was performed using GraphPad Prism software. For Fig. 1, 3, and 4, the horizontal bar denotes the geometric mean of the linear values.
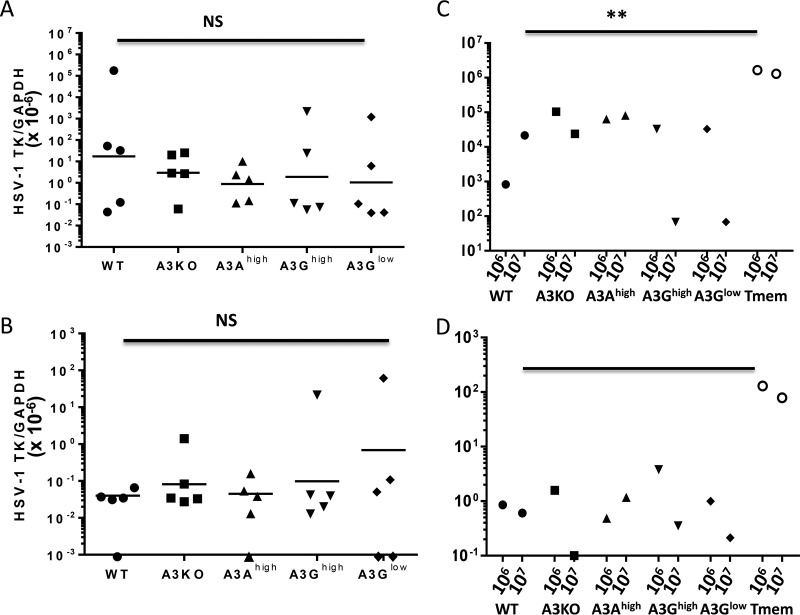
FIG 1.
APOBEC3 does not protect mice from intraocular HSV-1 infection. (A and B) HSV-1 strain McKrae (106 PFU) was intraocularly applied to mice of the indicated genotypes, and at 5.7 dpi, their eyes (A) and spleens (B) were harvested and DNA was isolated for qPCR analysis (n = 5 for all genotypes). (C and D) An additional mouse of each genotype as well as Tmem173 was inoculated via the same route with 106 or 107 PFU of HSV-1, and infection in eyes (C) and spleen (D) was analyzed. Although not included in the calculation of the geometric mean, mice with values of 0 are shown on the plot. NS, not significantly different by one-way analysis of variance (ANOVA); **, P ≤ 0.002; ****, P ≤ 0.0001 (by two-tailed unpaired t test.
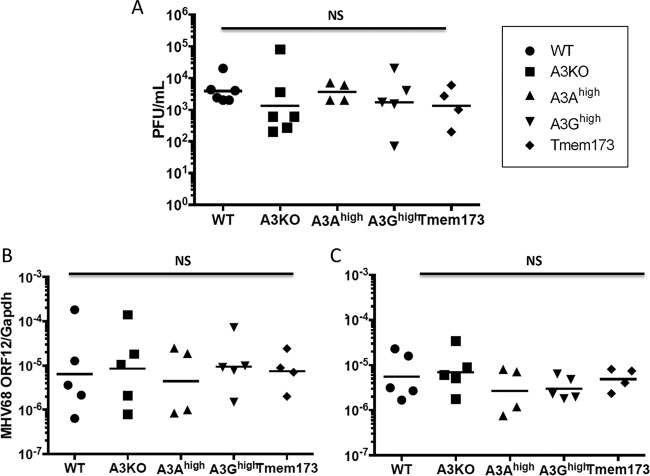
FIG 3.
APOBEC3 does not protect mice from acute or latent MHV68 infection. (A) MHV68 (103 PFU) was intranasally introduced into each genotype of mice as indicated. The mice were sacrificed, and lung tissue was collected 7 dpi. Whole lung tissues were homogenized in 1 ml culture medium, and the supernatants of homogenates were 10-fold serially diluted. The titer of each dilution was determined on NIH3T12 cells, and they were stained and fixed at 5 dpi. Plaques were counted, and the viral titers are indicated as mean PFU/ml (n = 6 for C57BL/6 and A3KO, 5 for A3Ahigh and A3Ghigh, and 4 for Tmem173). (B and C) MHV68 (103 PFU) was intranasally injected into mice of each genotype as indicated. The mice were sacrificed, and the lung (B) and spleen (C) tissues were collected 16 days after injection. DNA was isolated and subjected to PCR with primers specific to ORF12. Values are indicated as MHV68 copy numbers normalized to mouse GAPDH (n = 5, 5, 4, 5, and 4 for WT, A3KO, A3Ahigh, A3Ghigh, and Tmem, respectively). NS, not significantly different by either two-way unpaired t test or one-way ANOVA.
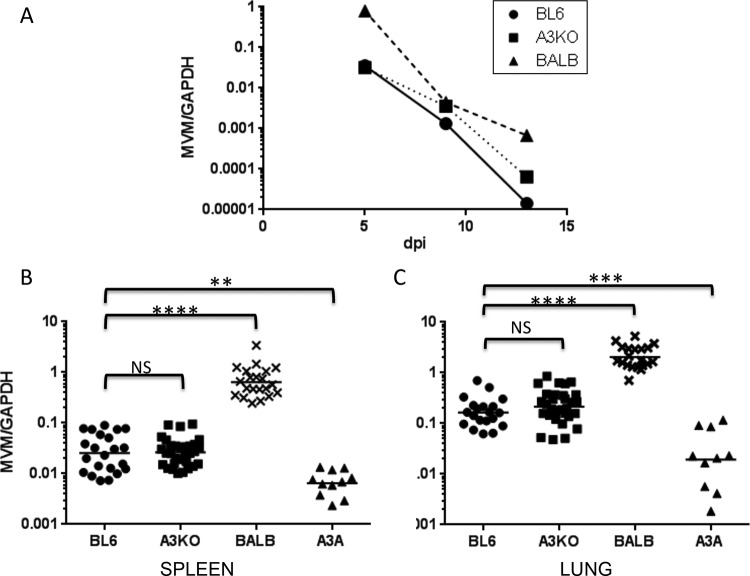
FIG 4.
APOBEC3A restricts MVMi infection in vivo. (A) MVMi genome equivalents were calculated by alkaline gel electrophoresis followed by quantitative Southern blotting. Newborn C57BL/6, BALB/c, and A3KO mice were infected oranasally with 2 × 107 genome equivalents of virus in 5 μl. Mice were sacrificed at 5, 9, or 13 dpi, and DNA was isolated from their spleens and subjected to quantitative PCR (qPCR) to determine relative levels of viral DNA, using the primers 5′-AAGGTACGATGGCGCCTC-3/5′-GTGCTCTTTGGCAGC-3′. MVMi values were normalized to GAPDH, as previously described (45). (B and C) Newborn C57BL/6, BALB/c, A3KO, and A3A mice were infected oranasally with 2 × 107 genome equivalents of virus and sacrificed at 5 dpi. DNA isolated from the spleen (B) and lungs (C) was analyzed by qPCR. Each point represents an individual mouse. ****, P < 0.0001; **, P < 0.0025; ***, P < 0.0035 (by 2-tailed unpaired t test).
RESULTS
HSV-1 infection is not affected by endogenous mouse or human APOBEC3 proteins.
Both the A3KO and human A3 transgenic mice were generated on a C57BL/6 background. Since this strain is not highly susceptible to HSV-1, we used the pathogenic McKrae strain, which is reported to more efficiently infect this mouse strain as well as to cause pathogenesis after i.o. inoculation (36). The transgenic strains used in all experiments were the A3Ghigh, A3Glow, and A3Ahigh strains described in our original publication and were expressed in a broad range of tissues (27). The A3Ghigh and A3Ahigh transgenic mice have equivalent levels of expression in mouse and human PBMCs, while the A3Glow transgenic mice express 10-fold lower levels in these cells (27). We first used i.o. inoculation (106 PFU/animal) and measured infection in the eyes and trigeminal nerve and spread to the spleen. We found no difference in infection in any of the mice (Fig. 1A and B). To confirm that the virus was indeed infectious in mice, we repeated the inoculations with both 106 and 107 PFU, this time including as a positive control STING mutant (Tmem173) mice on a C57BL/6 background, which are highly susceptible to HSV infection (37). The Tmem173 mice were more highly infected at both doses, while the other mice showed similar low levels of infection, both at the initial sites of infection (eyes) (Fig. 1C) and after spread to the spleen (Fig. 1D). Similar results for both sets of inoculations were seen in the trigeminal nerve (not shown). Interestingly, although the McKrae strain is reported to be pathogenic in C57BL/6 mice, we found that while some mice exhibited mild swelling around the forebrain after i.o. inoculation, most showed no symptoms, and with the exception of Tmem173 mice, all recovered from infection by 1 week postinfection (not shown).
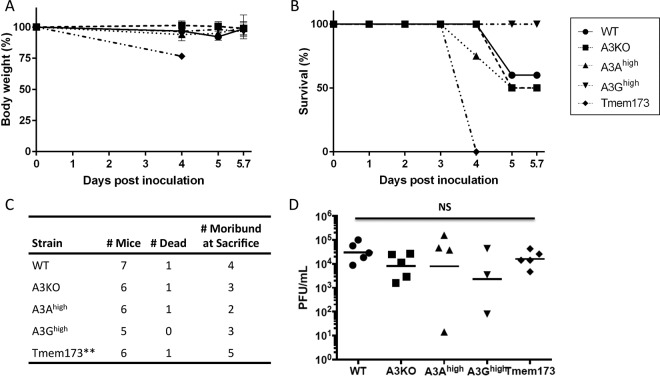
To determine if the route of infection influenced the ability of the APOBEC3 proteins to affect infection, we also inoculated the A3KO, C57BL/6, A3Ahigh, A3Ghigh, and Tmem173 mice by i.p. injection. The Tmem173 mice showed a dramatic weight loss at 4 days after i.p. infection, while all the other strains maintained their weight through day 6 (Fig. 2A). Similarly, while all the Tmem173 mutant mice rapidly succumbed to HSV-1 infection, at most 50% of the C57BL/6, A3KO, or A3Ahigh mice and none of the A3Ghigh mice succumbed at later times after infection, and there was no statistical difference in the survival curves (Fig. 2B). Moreover, approximately equal numbers of the transgenic, C57BL/6, and A3KO mice were moribund at sacrifice (Fig. 2C). In contrast to the case for mice that received i.o. inoculations, HSV-1 infection levels, as measured by viral levels in the brain, were not significantly different between the mice at day 4 (Tmem173) or 5.7 (all other strains) after i.p. inoculation (Fig. 2D), suggesting that the HSV-1–mediated mortality in Tmem173 mice was due to infection of tissues other than brain. This is in contrast to work by others demonstrating that STING KO mice on a mixed 129SvEv × C57BL/6J background sustain higher levels of brain infection after intravenous infection by HSV-1 (37).
FIG 2.
APOBEC3 does not protect mice from systemic HSV-1 infection. (A and B) HSV-1 McKrae strain (106 PFU) was intraperitoneally injected into mice of each genotype, as indicated. The mice died or were sacrificed by 5.7 dpi. Weight loss (A) and survival ratio (B) of mice were examined (n = 5 for C57BL/6 [WT]), 4 for A3KO and A3Ahigh, 3 for A3Ghigh, and 5 for Tmem173). (C) Additional mice were infected with 106 PFU of HSV-1, and the overall incidence of mortality and morbidity in the different strains was analyzed. **, all of the Tmem mutant mice were dead or moribund by 4 dpi. (D) Mice from the experiments for panels A to C were sacrificed, and brain tissue was collected at 4 to 5.7 dpi. The left hemisphere was homogenized in 1 ml culture medium, and titers in the supernatants of homogenates were determined on Vero cells. Because the Tmem173 mice were moribund by 3 dpi, they were sacrificed by day 4 (n = 6 for C57BL/6 [WT] and 5 for all the other strains). One WT and A3Ahigh mouse each and 2 A3Ghigh mice had no virus titers. NS, not significantly different by either two-tailed unpaired t test or one-way ANOVA, including the uninfected mice in the analysis.
MHV68 infection is not affected by endogenous mouse or human APOBEC3 proteins.
We next tested the mice for their susceptibility to the gammaherpesvirus MHV68; as discussed above, the human gammaherpesvirus EBV shows signs of APOBEC3-mediated cytidine deamination, and a recent report suggested that APOBEC3A but not mouse APOBEC3 was able to restrict MHV68 after DNA transfection of viral genomes but not virion infection in tissue culture cells (34). Wild-type, A3Ahigh, A3Ghigh, A3KO, and Tmem173 mice received intranasal inoculation of the virus, and at 7 dpi, virus titers in lung were measured (Fig. 3A). No differences in the infection levels were seen between any of the mice. To determine whether there was a difference in establishment of latent infection between the strains, a cohort of infected mice of each genotype was sacrificed at 16 dpi. There was no difference in infection in the lungs (Fig. 3B) or spleens (Fig. 3C) in any of the mice, including Tmem173 mutant mice. The latter finding is in accord with a recent study showing that MHV68 only weakly stimulates innate immune responses via the STING pathway, although in this study the STING mutant mice were infected at about 2-fold-higher levels at 2 days dpi (38).
Lack of cytidine deamination of herpesvirus genomes in WT or human APOBEC3 transgenic mice.
Several human APOBEC3 proteins have also been implicated in deamination of herpesvirus genomes, including APOBEC3A and -3G, without affecting the level of viral DNA (30). To determine if the viral genomes of HSV-1 and MHV68 showed evidence of editing by these enzymes in vivo, we used differential DNA denaturation PCR (3DPCR) to amplify DNA isolated from the brains of the HSV-1-infected mice and the lungs of the MHV68-infected mice of different genotypes. The fragments amplified and sequenced had a significant number of target sequences for mouse APOBEC3, APOBEC3A, and APOBEC3G (Table 1). No difference was seen in the minimum denaturation temperature needed for amplification of the viral DNA by 3DPCR (not shown). When the PCR products amplified at the lowest temperatures were sequenced, no difference in deamination of the viral genomes was detected (Table 2).
TABLE 2.
Mutation analysis of HSV-1-, MHV68-, and MVMi-infected micea
| Virus and mouse strain | No. of: |
Ratio of mutations to total nucleotides |
||||||
|---|---|---|---|---|---|---|---|---|
| Mice | Total sequences | Total nucleotides | Mutations |
G to A plus C to T | Other | |||
| Total | G to A plus C to T | Other | ||||||
| HSV-1 | ||||||||
| C57BL6 | 3 | 16 | 13,621 | 72 | 28 | 44 | 0.0021 | 0.0032 |
| A3KO | 3 | 16 | 13,632 | 52 | 30 | 22 | 0.0022 | 0.0016 |
| A3Ahigh | 3 | 17 | 14,483 | 62 | 32 | 30 | 0.0022 | 0.0021 |
| A3Ghigh | 3 | 15 | 12,775 | 64 | 32 | 32 | 0.0025 | 0.0025 |
| Tmem173 | 3 | 17 | 14,484 | 72 | 39 | 33 | 0.0027 | 0.0023 |
| MHV68 | ||||||||
| C57BL6 | 3 | 18 | 13,131 | 0 | 0 | 0 | ||
| A3KO | 3 | 16 | 11,680 | 0 | 0 | 0 | ||
| A3Ahigh | 3 | 15 | 10,945 | 4 | 1 | 3 | 9.14E−05 | 0.0003 |
| A3Ghigh | 3 | 12 | 8,761 | 1 | 0 | 1 | 0.0001 | |
| Tmem173 | 3 | 15 | 10,946 | 0 | 0 | 0 | ||
| MVMi | ||||||||
| C57BL/6 | 3 | 33 | 31,152 | 56 | 16 | 40 | 0.0005 | 0.0013 |
| A3KO | 3 | 31 | 29,264 | 59 | 12 | 47 | 0.0004 | 0.0016 |
| A3Ahigh | 3 | 29 | 27,376 | 74 | 15 | 59 | 0.0005 | 0.0022 |
DNA isolated from the brains (HSV-1), lungs (MHV68), or spleens (MVMi) of the indicated mice was subjected to differential DNA denaturing PCR. The numbers of mouse APOBEC3, APOBEC3G, and APOBEC3A target sites in each of the DNA segments analyzed are presented in Table 1.
MVMi is restricted by human APOBEC3A but not mouse APOBEC3.
While previous studies indicated that restriction of herpesviruses and papillomaviruses occurred via cytidine deamination, human APOBEC3A restriction of parvoviruses is not dependent on cytidine deamination, at least in cultured cells (32, 33). We also tested whether mouse APOBEC3 or human APOBEC3A proteins restricted MVMi infection in vivo. MVMi is a lymphotropic variant that was reported to be lethal in BALB/c but not C57BL/6 mice (35), while the prototypic MVMp which was used to study APOBEC3 restriction in cultured cells primarily infects fibroblasts and is not pathogenic in vivo (39). Interestingly, in past studies (35), susceptibility to MVMi in different mouse strains cosegregated with the weak APOBEC3 allele found in BALB/c mice, which is expressed at ∼10-fold-lower levels than that found in C57BL/6 mice; the weak BALB/c allele confers increased susceptibility to MLV and MMTV infection (3, 5–7, 40). We speculated that allelic differences in the mouse APOBEC3 gene might also play a role in MVM pathogenesis. First, we did a time course of infection of newborn C57BL/6, BALB/c, and A3KO mice after MVMi intranasal inoculation. To measure virus infection levels, we performed qPCR with DNA from different organs. There was no difference in the infection levels in A3KO versus C57BL/6 mouse spleen at all time points, indicating that mouse APOBEC3 does not restrict MVM infection (Fig. 4A); similar results were seen in lung and kidney (not shown). BALB/c mice were more highly infected than all the other mice at all dpi (Fig. 4A). This indicates that genes other than the APOBEC3 gene are responsible for the genetic susceptibility to infection in this strain. Next, we tested whether the presence of the human APOBEC3A transgene would diminish infection. Interestingly, we did find that MVMi infection was reduced by 5-fold on average in the spleens or lungs of APOBEC3Ahigh mice at 5 dpi (Fig. 4B and C); similar results were seen at 9 dpi (not shown). Surprisingly, none of the mice, including BALB/c mice, exhibited signs of pathogenesis at any time after infection up to 14 dpi, at which time all the mice had decreased infection by 2 logs or more compared to the initial infection levels (Fig. 4A); the original report for MVMi showed hemorrhaging in multiple organs of BALB/c mice by 9 dpi (35). In contrast to the previous study, all of the BALB/c mice survived infection in our study.
We looked for evidence of deamination of MVMi genomes in the APOBEC3A transgenic mice, also using 3DPCR and sequencing of the products generated by low annealing temperatures. No difference in deamination was detected (Table 2). This is in accord with studies done after infection of APOBEC3A-expressing tissue cultures with both adeno-associated virus and MVMp, demonstrating that virus restriction was independent of deaminase activity (33).
DISCUSSION
In addition to their well-established role in restricting retrovirus infection, there have been a number of reports that APOBEC3 proteins restrict other viruses, particularly DNA viruses that replicate in the nucleus. These include herpesviruses, where APOBEC3C overexpression reduced HSV-1 titers in tissue culture cells, and APOBEC3A, -3C, and 3G edited HSV-1 DNA when overexpressed, and HSV-1 and EBV in buccal swabs or immortalized cells lines, respectively, showed signs of cytidine deamination characteristic of APOBEC3 editing (30). There is also evidence that HPV is edited by APOBEC3A, -3B, and -3H, all of which are expressed in keratinocytes (41, 42). Several groups have shown that APOBEC3A, -3C, and -3H have access to the nucleus during telophase, while APOBEC3B is nuclear after mitosis (43); thus, some but not all APOBEC3 proteins can potentially affect viruses that replicate in the nucleus. Thus, it is not surprising that we found that APOBEC3G, which is found in the cytoplasm, had no significant effect on herpesvirus replication in vivo. We also found with a small cohort of mice that APOBEC3G had no impact on MVMi infection levels, although results with the low number of mice examined did not reach statistical significance (not shown). However, based on the in vitro studies and its nuclear localization, we expected that APOBEC3A might restrict infection by both herpesviruses and parvoviruses. Contrary to this expectation, only the mouse parvovirus MVMi was inhibited in vivo. While this could possibly be due to lack of transgene expression in the appropriate cell types, we think this is unlikely because both the APOBEC3A and -3G transgenes were expressed in all tissues and cell types that we have examined, including brain, lymphoid tissue, macrophages, dendritic cells, lymphocytes, and fibroblasts (27) (not shown). However, it may be that herpesviruses but not parvoviruses encode viral proteins that counteract the action of APOBEC3A in particular, since it is found in the nucleus where these viruses replicate. Since the APOBEC3 transgenes are under the transcriptional control of the β-actin regulatory region and the β-globin 3′ untranslated region and polyadenylation site, the putative viral anti-APOBEC3 proteins would have to act at the level of protein degradation, relocalization, or direct inhibition of APOBEC3 enzymatic function. This could be tested in future experiments.
APOBEC3A but not APOBEC3G restricts infection by the human parvovirus adeno-associated virus, as well as by MVM (32, 33). Interestingly, we show here that APOBEC3A but not mouse APOBEC3 restricted MVM in vivo. One possibility for the lack of mouse APOBEC's antiviral activity is that the transgene is expressed in an MVM cellular target but the endogenous APOBEC3 is not; we think this unlikely because at least in lymphoid cells, we detected lower APOBEC3A transgene expression than that of the mouse APOBEC3 and yet splenic levels of MVMi infection were lower in the APOBEC3A transgenic mice (Fig. 4B). Another possibility is that the subcellular localization of the APOBEC3A (nuclear and cytoplasmic) allows it access to replicating MVM, unlike mouse APOBEC3, which is cytoplasmic (32, 44). We were surprised by the lack of MVMi lethality in any of the mouse strains tested, in contrast to previous reports (35). Moreover, the mice infected with McKrae strain HSV-1 also exhibited only modest signs of pathogenicity compared to those seen in earlier studies (36). This may be due to the purity of the virus preparations used here or perhaps because the original infections were performed with mice raised under less-stringent specific-pathogen-free criteria than are currently used.
Our previous work showed that retrovirus restriction by APOBEC3A and -3G transgenic mice provided good systems for the in vivo study of antiviral effects of these proteins. Indeed, we were able to recapitulate in our mice the ability of the HIV-1 Vif to counteract APOBEC3G (27). The results presented here suggest that only parvoviruses and not herpesviruses are likely to be targets for APOBEC3A in vivo and point to the importance of using systems that closely recapitulate infection of whole organisms to understand their biological functions. They also suggest that these and similar transgenic models might be useful for testing whether APOBEC3 proteins inhibit zoonotic viral infections.
ACKNOWLEDGMENTS
We thank Abigail Smith for invaluable advice on infecting mice with MVMi, Anthony D'Abramo for technical assistance with the MVMi, Nigel Fraser and Mark Boyer for providing us with HSV-1 and advice on carrying out the infections, and Skip Virgin and Erietta Stelekati for the MHV68.
REFERENCES
- 1.Vieira VC, Soares MA. 2013. The role of cytidine deaminases on innate immune responses against human viral infections. Biomed Res Int 2013:683095. doi: 10.1155/2013/683095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LaRue RS, Andresdottir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jonsson SR, Landau NR, Lochelt M, Malik HS, Malim MH, Munk C, O'Brien SJ, Pathak VK, Strebel K, Wain-Hobson S, Yu XF, Yuhki N, Harris RS. 2009. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol 83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okeoma CM, Petersen J, Ross SR. 2009. Expression of murine APOBEC3 alleles in different mouse strains and their effect on mouse mammary tumor virus infection. J Virol 83:3029–3038. doi: 10.1128/JVI.02536-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, Hakata Y, Takeda E, Liu Q, Iwatani Y, Kozak CA, Miyazawa M. 2012. Two genetic determinants acquired late in Mus evolution regulate the inclusion of exon 5, which alters mouse APOBEC3 translation efficiency. PLoS Pathog 8:e1002478. doi: 10.1371/journal.ppat.1002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyazawa M, Tsuji-Kawahara S, Kanari Y. 2008. Host genetic factors that control immune responses to retrovirus infections. Vaccine 26:2981–2996. doi: 10.1016/j.vaccine.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Takeda E, Tsuji-Kawahara S, Sakamoto M, Langlois MA, Neuberger MS, Rada C, Miyazawa M. 2008. Mouse APOBEC3 restricts friend leukemia virus infection and pathogenesis in vivo. J Virol 82:10998–11008. doi: 10.1128/JVI.01311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santiago ML, Montano M, Benitez R, Messer RJ, Yonemoto W, Chesebro B, Hasenkrug KJ, Greene WC. 2008. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science 321:1343–1346. doi: 10.1126/science.1161121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 9.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol 14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 10.Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J 23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liddament MT, Brown WL, Schumacher AJ, Harris RS. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol 14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 12.Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol 78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem 279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 14.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 15.Xiao Z, Ehrlich E, Yu Y, Luo K, Wang T, Tian C, Yu XF. 2006. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology 349:290–299. doi: 10.1016/j.virol.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Marin M, Rose KM, Kozak SL, Kabat D. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med 9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 17.Munk C, Jensen BE, Zielonka J, Haussinger D, Kamp C. 2012. Running loose or getting lost: how HIV-1 counters and capitalizes on APOBEC3-induced mutagenesis through its Vif protein. Viruses 4:3132–3161. doi: 10.3390/v4113132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cullen BR. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J Virol 80:1067–1076. doi: 10.1128/JVI.80.3.1067-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol 15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 20.Okeoma CM, Lovsin N, Peterlin BM, Ross SR. 2007. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature 445:927–930. doi: 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]
- 21.Zielonka J, Bravo IG, Marino D, Conrad E, Perkovic M, Battenberg M, Cichutek K, Munk C. 2009. Restriction of equine infectious anemia virus by equine APOBEC3 cytidine deaminases. J Virol 83:7547–7559. doi: 10.1128/JVI.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fadel HJ, Saenz DT, Guevara R, von Messling V, Peretz M, Poeschla EM. 2012. Productive replication and evolution of HIV-1 in ferret cells. J Virol 86:2312–2322. doi: 10.1128/JVI.06035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chareza S, Slavkovic Lukic D, Liu Y, Rathe AM, Munk C, Zabogli E, Pistello M, Lochelt M. 2012. Molecular and functional interactions of cat APOBEC3 and feline foamy and immunodeficiency virus proteins: different ways to counteract host-encoded restriction. Virology 424:138–146. doi: 10.1016/j.virol.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 24.Groom HC, Yap MW, Galao RP, Neil SJ, Bishop KN. 2010. Susceptibility of xenotropic murine leukemia virus-related virus (XMRV) to retroviral restriction factors. Proc Natl Acad Sci U S A 107:5166–5171. doi: 10.1073/pnas.0913650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ooms M, Krikoni A, Kress AK, Simon V, Munk C. 2012. APOBEC3A, APOBEC3B, and APOBEC3H haplotype 2 restrict human T-lymphotropic virus type 1. J Virol 86:6097–6108. doi: 10.1128/JVI.06570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doehle BP, Schafer A, Wiegand HL, Bogerd HP, Cullen BR. 2005. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J Virol 79:8201–8207. doi: 10.1128/JVI.79.13.8201-8207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stavrou S, Crawford D, Blouch K, Browne EP, Kohli RM, Ross SR. 2014. Different modes of retrovirus restriction by human APOBEC3A and APOBEC3G in vivo. PLoS Pathog 10:e1004145. doi: 10.1371/journal.ppat.1004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rulli SJ Jr, Mirro J, Hill SA, Lloyd P, Gorelick RJ, Coffin JM, Derse D, Rein A. 2008. Interactions of murine APOBEC3 and human APOBEC3G with murine leukemia viruses. J Virol 82:6566–6575. doi: 10.1128/JVI.01357-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vartanian JP, Guetard D, Henry M, Wain-Hobson S. 2008. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science 320:230–233. doi: 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- 30.Suspene R, Aynaud MM, Koch S, Pasdeloup D, Labetoulle M, Gaertner B, Vartanian JP, Meyerhans A, Wain-Hobson S. 2011. Genetic editing of herpes simplex virus 1 and Epstein-Barr herpesvirus genomes by human APOBEC3 cytidine deaminases in culture and in vivo. J Virol 85:7594–7602. doi: 10.1128/JVI.00290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonvin M, Achermann F, Greeve I, Stroka D, Keogh A, Inderbitzin D, Candinas D, Sommer P, Wain-Hobson S, Vartanian JP, Greeve J. 2006. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 43:1364–1374. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- 32.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol 16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 33.Narvaiza I, Linfesty DC, Greener BN, Hakata Y, Pintel DJ, Logue E, Landau NR, Weitzman MD. 2009. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog 5:e1000439. doi: 10.1371/journal.ppat.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minkah N, Chavez K, Shah P, Maccarthy T, Chen H, Landau N, Krug LT. 2014. Host restriction of murine gammaherpesvirus 68 replication by human APOBEC3 cytidine deaminases but not murine APOBEC3. Virology 454-455:215–226. doi: 10.1016/j.virol.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brownstein DG, Smith AL, Jacoby RO, Johnson EA, Hansen G, Tattersall P. 1991. Pathogenesis of infection with a virulent allotropic variant of minute virus of mice and regulation by host genotype. Lab Invest 65:357–364. [PubMed] [Google Scholar]
- 36.Yao HW, Ling P, Chen SH, Tung YY, Chen SH. 2012. Factors affecting herpes simplex virus reactivation from the explanted mouse brain. Virology 433:116–123. doi: 10.1016/j.virol.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 37.Ishikawa H, Ma Z, Barber GN. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun C, Schattgen SA, Pisitkun P, Jorgensen JP, Hilterbrand AT, Wang LJ, West JA, Hansen K, Horan KA, Jakobsen MR, O'Hare P, Adler H, Sun R, Ploegh HL, Damania B, Upton JW, Fitzgerald KA, Paludan SR. 2015. Evasion of innate cytosolic DNA sensing by a gammaherpesvirus facilitates establishment of latent infection. J Immunol 194:1819–1831. doi: 10.4049/jimmunol.1402495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimsey PB, Engers HD, Hirt B, Jongeneel CV. 1986. Pathogenicity of fibroblast- and lymphocyte-specific variants of minute virus of mice. J Virol 59:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Low A, Okeoma CM, Lovsin N, de las Heras M, Taylor TH, Peterlin BM, Ross SR, Fan H. 2009. Enhanced replication and pathogenesis of Moloney murine leukemia virus in mice defective in the murine APOBEC3 gene. Virology 385:455–463. doi: 10.1016/j.virol.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henry M, Guetard D, Suspene R, Rusniok C, Wain-Hobson S, Vartanian JP. 2009. Genetic editing of HBV DNA by monodomain human APOBEC3 cytidine deaminases and the recombinant nature of APOBEC3G. PLoS One 4:e4277. doi: 10.1371/journal.pone.0004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vartanian JP, Henry M, Marchio A, Suspene R, Aynaud MM, Guetard D, Cervantes-Gonzalez M, Battiston C, Mazzaferro V, Pineau P, Dejean A, Wain-Hobson S. 2010. Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis. PLoS Pathog 6:e1000928. doi: 10.1371/journal.ppat.1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lackey L, Law EK, Brown WL, Harris RS. 2013. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle 12:762–772. doi: 10.4161/cc.23713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem 281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 45.Stavrou S, Nitta T, Kotla S, Ha D, Nagashima K, Rein AR, Fan H, Ross SR. 2013. Murine leukemia virus glycosylated Gag blocks apolipoprotein B editing complex 3 and cytosolic sensor access to the reverse transcription complex. Proc Natl Acad Sci U S A 110:9078–9083. doi: 10.1073/pnas.1217399110. [DOI] [PMC free article] [PubMed] [Google Scholar]