ABSTRACT
The HIV-1 envelope glycoprotein (Env) is a trimer of gp120/gp41 heterodimers that mediates viral entry. Env binds cellular CD4, an association which stabilizes a conformation favorable to its subsequent association with a coreceptor, typically CCR5 or CXCR4. The CD4- and coreceptor-binding sites serve as epitopes for two classes of HIV-1-neutralizing antibodies: CD4-binding site (CD4bs) and CD4-induced (CD4i) antibodies, respectively. Here we observed that, at a fixed total concentration, mixtures of the CD4i antibodies (E51 or 412d) and the CD4bs antibody VRC01 neutralized the HIV-1 isolates 89.6, ADA, SG3, and SA32 more efficiently than either antibody alone. We found that E51, and to a lesser extent 412d and 17b, promoted association of four CD4bs antibodies to the Env trimer but not to monomeric gp120. We further demonstrated that the binding of the sulfotyrosine-binding pocket by CCR5mim2-Ig was sufficient for promoting CD4bs antibody binding to Env. Interestingly, the relationship is not reciprocal: CD4bs antibodies were not as efficient as CD4-Ig at promoting E51 or 412d binding to Env trimer. Consistent with these observations, CD4-Ig, but none of the CD4bs antibodies tested, substantially increased HIV-1 infection of a CD4-negative, CCR5-positive cell line. We conclude that the ability of CD4i antibodies to promote VRC01 association with Env trimers accounts for the increase potency of VRC01 and CD4i antibody mixtures. Our data further suggest that potent CD4bs antibodies avoid inducing Env conformations that bind CD4i antibodies or CCR5.
IMPORTANCE Potent HIV-1-neutralizing antibodies can prevent viral transmission and suppress an ongoing infection. Here we show that CD4-induced (CD4i) antibodies, which recognize the conserved coreceptor-binding site of the HIV-1 envelope glycoprotein (Env), can increase the association of Env with potent broadly neutralizing antibodies that recognize the CD4-binding site (CD4bs antibodies). We further show that, unlike soluble forms of CD4, CD4bs antibodies poorly induce envelope glycoprotein conformations that efficiently bind CCR5. This study provides insight into the properties of potent CD4bs antibodies and suggests that, under some conditions, CD4i antibodies can improve their potency. These observations may be helpful to the development of vaccines designed to elicit specific antibody classes.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) uses its envelope glycoprotein (Env) to gain entry into host cells. Env is synthesized as precursor gp160 proteins which assemble as trimers before they are cleaved into gp120 and gp41 subunits (1). The gp120 subunit binds the primary HIV-1 receptor, CD4 (2), which then induces tertiary and quaternary conformational changes in Env that promote association with a coreceptor, usually CCR5 or CXCR4 (3, 4). The CD4- and coreceptor-binding sites are the two most conserved regions of gp120 (5, 6). Two classes of antibodies (Abs) with epitopes corresponding to each of these regions, have been defined: CD4-binding site (CD4bs) antibodies and CD4-induced (CD4i) antibodies. The latter are so named because CD4 binding induces a conformation that promotes their association with gp120.
The antibody b12 was the first potent CD4bs antibody described (7). However, its breadth was limited to 35% against a broad panel of HIV-1 isolates (8). Since then, broader and more potent antibodies have been identified, notably VRC01, 3BNC117, and NIH45-46, among others (8–10). These antibodies neutralize more than 90% of HIV-1 isolates assayed. The breadth and potency of NIH45-46 were increased through a G54W mutation (NIH45-46G54W), where the tryptophan targets the Phe-43 cavity of the CD4-binding site on gp120 (11). Importantly, these highly potent broadly neutralizing antibodies (bNAbs) can protect from HIV-1 challenge and reduce viral loads in infected humanized mice and rhesus macaques and in HIV-infected individuals (12–17).
Compared to CD4bs antibodies, well-characterized CD4i antibodies such as 17b are substantially less broad and potent (18–20). This inefficiency is largely a consequence of their recognition of an Env conformation that is usually inaccessible in the absence of CD4. Access to CD4-bound Env is impeded by the cellular membrane and is limited to the time frame between CD4 binding and association with coreceptor (21). Some CD4i antibodies, including E51 and 412d, mimic CCR5 by incorporating sulfotyrosines into their heavy-chain CDR3 (CDR-H3) regions (22, 23). These sulfotyrosines bind highly conserved pockets on gp120 that recognize the CCR5 amino terminus. Subsequently, the E51 CDR-H3 region was instrumental in the development of CCR5-mimetic peptides such as CCR5mim2-Ig (24, 25). The structure of gp120 in complex with 412d localizes two sulfotyrosine-binding pockets at the base of the third variable loop and in the fourth conserved domain (26). Perhaps as a consequence, E51 and 412d typically bind Env and neutralize HIV-1 more efficiently than 17b.
Because CD4 and CD4i antibodies bind the envelope glycoprotein cooperatively, we explored the relationship between the CD4i antibodies and a panel of CD4bs bNAbs. We observed that, at the same total concentrations, mixtures of E51 or 412d and the CD4bs antibody VRC01 were more potent than either antibody alone. We hypothesized that conformational changes of Env might play a role in the observed synergy. We found that CD4bs antibodies did not promote E51 and 412d binding to the Env trimer as efficiently as CD4-Ig. Consistent with this observation, CD4-Ig, but not CD4bs antibodies, could promote infection of CCR5-positive, CD4-negative cells. However, and in contrast to our observations with CD4bs antibodies, CD4i antibodies (E51, 412d, and 17b) and CCR5mim2-Ig promoted quaternary changes in Env that increased its binding to several highly potent CD4bs antibodies, including VRC01. Thus, we propose that CD4i antibodies induce favorable conformations on the HIV-1 Env trimer that can increase the potency of some CD4bs bNAbs.
MATERIALS AND METHODS
Cells and plasmids.
All cell lines were grown in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) at 37°C. TZM-bl cells were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from John C. Kappes, Xiaoyun Wu, and Tranzyme Inc (27–31). Cf2Th CCR5-positive, CD4-negative cells were provided by Hyeryun Choe. The variable heavy and light chains of E51, 412d, 17b, and CCR5mim2 were cloned into human and murine IgG1 expression vectors. The CCR5mim2-Ig, 89.6, ADA, JRFL, SA32, and ConC gp160 expression vectors have been previously described (24, 32). The 89.6, ADA, SA32, and ConC gp160-Δcytoplasmic tail expression vectors have been previously described (24). BG505 gp160 and gp160-Δcytoplasmic tail expression vectors were provided by John Moore and P. J. Klasse. pNL4-3.luc.R−.E− was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from Nathaniel Landau (33, 34). CD4-Ig (a fusion construct of CD4 domains 1 and 2 with an IgG1 Fc) has been previously described (24). Vectors expressing VRC01 heavy and light chains were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from John Mascola (10, 35). 10E8 expression vectors were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from Mark Connors (36). Purified b12 and 2G12 antibodies were obtained from Polymun Scientific. Vectors expressing NIH45-46 and 3BNC117 were provided by Michel Nussenzweig. The NIH45-46G54W heavy-chain expression vector was provided by Pamela Bjorkman.
Antibody and CD4-Ig production and purification.
HEK293T cells (ATCC) seeded in T-175 flasks (Falcon) were cotransfected with 40 μg of both heavy-chain vector and light-chain vector per flask of the antibodies VRC01, NIH45-46, NIH45-46W, 3BNC117, E51, and 10E8 or with 80 μg per flask CD4-Ig vector by using a calcium phosphate transfection kit (Clontech). E51, 412d, and CCR5mim2-Ig transfections included 20 μg of TPST2 to ensure sufficient sulfation of the CDR-H3. At 12 to 16 h posttransfection, cells were washed with phosphate-buffered saline (PBS) and grown in FreeStyle 293 medium (Invitrogen). At 48 h posttransfection, medium was collected, centrifuged, and filtered with a 0.45-μm filter flask (Millipore). Antibodies were isolated with HiTrap columns (GE Healthcare) and eluted with IgG elution buffer (Thermo Scientific) into 1 M Tris-HCl buffer, pH 9.0 (G Biosciences). Buffer was exchanged with PBS and protein concentrated to 1 mg/ml with Amicon Ultura centrifugation filters (Millipore). Antibodies were stored at 4°C.
TZM-bl neutralization assays.
Pseudotyped HIV-1 was produced by coexpression of envelope glycoproteins of the indicated HIV-1 isolates with NL4-3ΔEnv, an HIV-1 expression vector lacking a functional env gene. 293T cells, grown to 50% confluence in T-175 (Falcon) flasks, were transfected with 25 μg of plasmid encoding envelope glycoprotein, 45 μg of NL4-3ΔEnv, and 5 μg each of plasmids expressing the HIV-1 tat and rev genes by the calcium phosphate technique. DMEM–10% FBS was changed at 12 h, and medium was collected at 48 h. Viral supernatants were cleared by centrifugation for 10 min at 1,500 × g, passed through a 0.45-μm syringe filter (Millipore), and stored at −80°C.
TZM-bl neutralization assays were performed as previously described (37). Briefly, HIV-1 pseudoviruses were preincubated with titrated amounts of antibody or CD4-Ig in DMEM with 10% FBS for 1 h at 37°C. TZM-bl cells were detached by trypsinization, diluted in DMEM with 10% FBS to 100,000 cells/ml, and added to the pseudovirus-inhibitor mixture. Cells were then incubated for 48 h at 37°C. Viral entry was analyzed using Britelite Plus (PerkinElmer), and luciferase was measured using a Victor X3 plate reader (PerkinElmer). Fifty percent inhibitory concentrations (IC50s) were determined with GraphPad Prism 6.0 software using sigmoidal four-parameter logistic curve analysis.
ELISA studies of monomeric HIV-1 gp120.
Enzyme-linked immunosorbent assay (ELISA) plates (Costar) were coated with 5 μg/ml HIV-1 gp120 (Immune Technology Corp.) and left overnight at 4°C. Plates were washed with PBS plus 0.05% Tween 20 (PBS-T) twice and blocked with 5% milk in PBS for 1 h at 37°C. Dilutions of murine E51, murine 412d, CD4-Ig, or CD4bs antibodies blocked with 5% milk in PBS were added to the plate and incubated for 1 h at 37°C. Samples were washed five times with PBS-T, and fixed concentrations of CD4-Ig or CD4bs antibodies (in the case of samples preincubated with murine E51 or murine 412d) or murine variants of CD4i antibodies (in the case of samples preincubated with CD4-Ig or CD4bs antibodies) were added. Plates were incubated for 1 h at 37°C. Samples were washed five times with PBS-T and labeled with a horseradish peroxidase-conjugated secondary antibody (Jackson Immuno Research) recognizing human IgG1. Plates were incubated for 1 h at 37°C and then washed 10 times with PBS-T. Tetramethylbenzidine (TMB) solution (Fisher) was added and left for 10 min at room temperature, and then the reaction was stopped with TMB stop solution (Southern Biotech). Absorbance was measured at 450 nm with a Victor X3 plate reader (PerkinElmer).
Flow cytometric analysis of cell-expressed envelope glycoprotein trimers.
HEK293T cells were transfected with plasmids expressing the indicated envelope glycoproteins lacking cytoplasmic residues 732 to 876 (HXBc2 numbering) together with a plasmid encoding the Tat protein. The transfection medium was replaced after an overnight incubation, and cells were harvested at 48 h posttransfection. Harvested cells were washed twice in flow cytometry buffer (PBS with 2% goat serum and 0.01% sodium azide). Cells were incubated with serially diluted CD4bs antibody, CD4-Ig, murine Fc variants of CD4i antibodies, or a murine Fc variant of CCR5mim2-Ig on ice for 1 h and then washed twice with flow cytometry buffer. If cells were preincubated with murine variants, cells were then incubated with 10 ng/ml of CD4bs antibody or CD4-Ig on ice for 1 h. If cells were preincubated with CD4bs antibodies or CD4-Ig, cells were then incubated with 25 ng/ml murine E51 or murine 412d on ice for 1 h. After incubation, cells were washed twice with flow cytometry buffer. Allophycocyanin (APC) or fluorescein isothiocyanate (FITC)-labeled secondary antibodies recognizing human or murine Fc (Jackson Immuno Research), respectively, were incubated with the cells for 30 min. Cells were washed twice with flow cytometry buffer and twice with PBS and then resuspended in 1% paraformaldehyde solution. Binding was analyzed with an Accuri C6 flow cytometer (BD Biosciences) and data analyzed with the C6 software (BD Biosciences).
Infection of CD4-negative cells.
HIV-1 pseudovirus expressing firefly luciferase was preincubated with titrated amounts of antibodies or CD4-Ig in DMEM with 10% FBS for 1 h at 37°C. CD4-negative Cf2Th-CCR5 cells were harvested, diluted in DMEM with 10% FBS to 100,000 cells/ml, and added to the pseudovirus-inhibitor mixture. Cells were then incubated for 48 h at 37°C. Viral entry was analyzed using Britelite Plus (PerkinElmer), and luciferase was measured with a Victor X3 plate reader (PerkinElmer).
RESULTS
Mixtures of E51 or 412d with VRC01 neutralize HIV-1 more efficiently than either antibody alone.
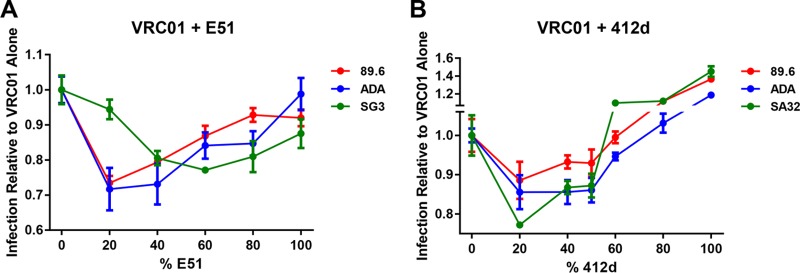
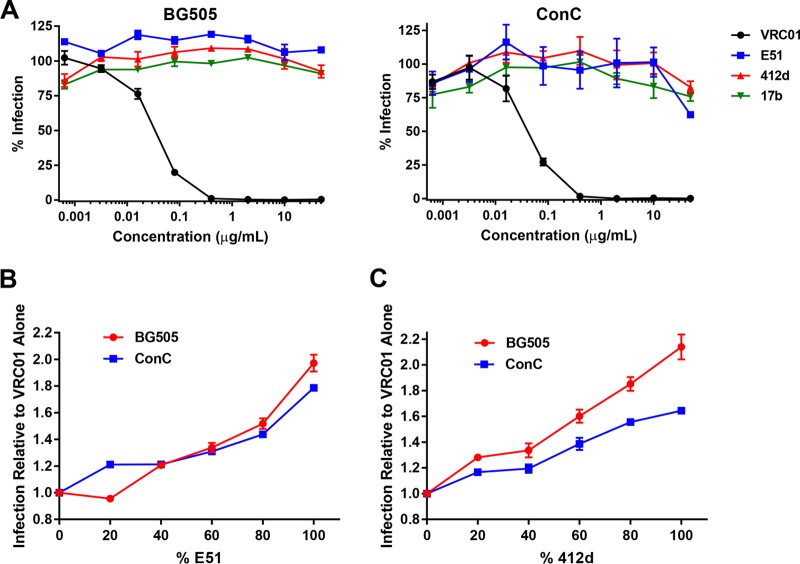
We have previously reported that mixtures of two HIV-1 entry inhibitors, CD4-Ig and CCR5mim2-Ig, which target the CD4- and coreceptor-binding sites of gp120, respectively, neutralize HIV-1 isolates better than either inhibitor individually (24). We therefore sought to determine whether, at a fixed total antibody concentration, a mixture of a CD4bs antibody and a CD4i antibody could be more potent than either antibody alone. We chose to study the CD4bs antibody VRC01 and the CD4i antibodies E51 and 412d, because they have similar potencies against a number of HIV-1 isolates. Fixing the total antibody concentration, we measured neutralization of HIV-1 using various antibody ratios. We observed that 80:20 and 60:40 ratios of VRC01 to E51 neutralized the HIV-1 isolates 89.6 and ADA more potently than either antibody alone (Fig. 1A). Ratios of 60:40 and 40:60 VRC01 to E51 were more potent against the SG3 isolate than either alone. When using 412d, ratios of 80:20 and 60:40 of VRC01 to 412d were more potent against 89.6, ADA, and the clade C isolate SA32 (Fig. 1B). However, we were unable to observe an increase in potency with mixtures of VRC01 and E51 or 412d on CD4i-resistant isolates BG505 (clade A) or consensus clade C (ConC) (Fig. 2A to C).
FIG 1.
In vitro neutralization by combinations of CD4bs antibodies with CD4i antibodies. (A) HIV-1 isolates 89.6, ADA, and SG3 were preincubated for 1 h with a fixed total concentration (0.3 μg/ml for 89.6 and ADA and 0.005 μg/ml for SG3) but various ratios of VRC01-E51 mixtures. TZM-bl cells were then added and incubated for 48 h. Infection was measured by the percent luciferase expression in the absence of inhibitor. (B) Experiment similar to that for panel A except that the indicated isolates were preincubated for 1 h with a fixed total concentration (0.3 μg/ml for 89.6 and ADA and 1.0 μg/ml for SA32) but various ratios of VRC01-412d mixtures. Error bars represent standard error of mean (SEM).
FIG 2.
In vitro neutralization by CD4i antibodies on CD4i-resistant HIV-1 isolates. (A) HIV-1 isolates BG505 and ConC were preincubated for 1 h with various concentrations of VRC01, E51, 412d, and 17b. TZM-bl cells were then added and incubated for 48 h. Infection was measured by the percent luciferase expression in the absence of inhibitor. (B) Similar to panel A except that BG505 or ConC was preincubated for 1 h with a fixed total concentration (0.033 μg/ml for BG505 or 0.045 μg/ml for ConC) but various ratios of VRC01-E51 mixtures. (C) Similar to panel A except that BG505 or ConC was preincubated for 1 h with a fixed total concentration (0.033 μg/ml for BG505 or 0.045 μg/ml for ConC) but various ratios of VRC01-412d mixtures. Error bars represent SEM.
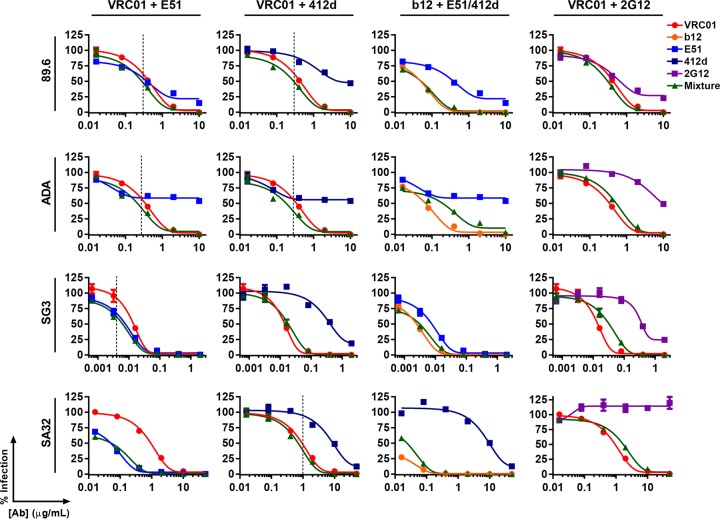
We next investigated whether combinations of VRC01 and E51 or 412d were more potent than either alone over a wider concentration range (Fig. 3). IC50 values for each antibody and mixture are summarized in Fig. 4. The E51-VRC01 mixture had a lower IC50 than either antibody alone on isolates 89.6, ADA, and SG3 (Fig. 4A), while a mixture of VRC01 and 412d had a lower IC50 on isolates 89.6, ADA, and SA32 (Fig. 4B). Similar mixtures of the CD4bs antibody b12 with E51 or 412d (Fig. 4C) or of the glycosylation-dependent antibody 2G12 with VRC01 (Fig. 4D) were assayed in parallel. These antibody mixtures did not outperform the more potent antibody at any concentration, except for the mixture of VRC01 with 2G12 on 89.6 (Fig. 4D). However, the CD4i antibodies E51 and 412d did not improve the potency of the CD4bs antibodies NIH45-46 and 3BNC117, due to the substantially greater potency of these CD4bs antibodies (not shown).
FIG 3.
In vitro neutralization curves of mixtures of VRC01 or b12 with E51, 412d, or 2G12. The experiment was similar to that described for Fig. 1 except that HIV-1 isolates 89.6, ADA, SG3, and SA32 was preincubated with various concentrations of indicated antibodies or mixture of antibodies. The dashed lines indicate the fixed concentration used for Fig. 1A and B. For the 89.6 and ADA neutralization curves, 50:50 mixtures of VRC01 and E51, VRC01 and 412d, b12 and E51, and VRC01 and 2G12 were used. For the SG3 neutralization curves, 25:75 mixtures of VRC01 and E51, b12 and E51, and VRC01 and 2G12 were used, while a 75:25 mixture of VRC01 and 412d was used (to accommodate for the lack of potency of 412d on SG3). For the SA32 neutralization curves, a 40:60 ratio of VRC01 and E51 was used, while 60:40 ratios of VRC01 and 412d, b12 and 412d, and VRC01 and 2G12 were used. All panels are representative of at least three independent experiments. IC50s are reported in Fig. 2. Error bars represent SEM.
FIG 4.
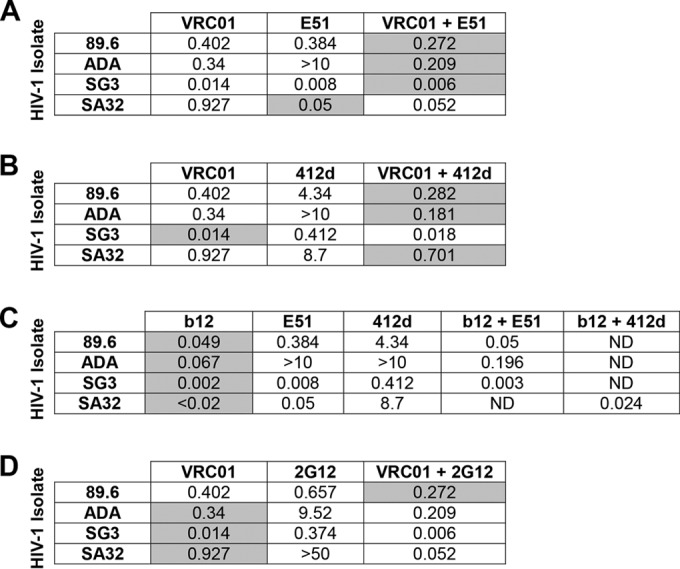
IC50s of antibodies and antibody mixtures on various HIV-1 isolates. IC50s were as determined by an experiment similar to that for Fig. 1 except that HIV-1 isolates 89.6, ADA, SG3, and SA32 were preincubated with various concentrations of the indicated antibodies or mixture of antibodies. (A) IC50s of VRC01, E51, and VRC01-E51 mixtures on the indicated isolates. (B) IC50s of VRC01, 412d, and VRC01-412d mixtures on the indicated isolates. (C) IC50s of b12, E51, and b12-E51 mixtures on the indicated isolates. (D) IC50s of VRC01, 2G12, and VRC01-2G12 mixtures on the indicated isolates. Shaded boxes indicate the most potent antibody or antibody mixture against the indicated HIV-1 isolate. Reported IC50s were determined based on the neutralization curves presented in Fig. 3.
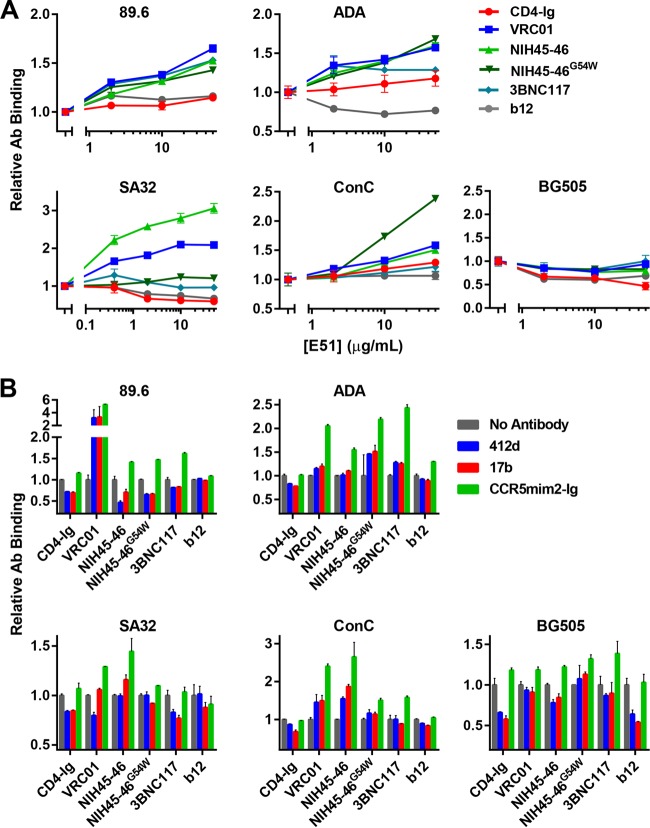
E51 and 412d do not enhance binding of CD4bs antibodies to monomeric gp120.
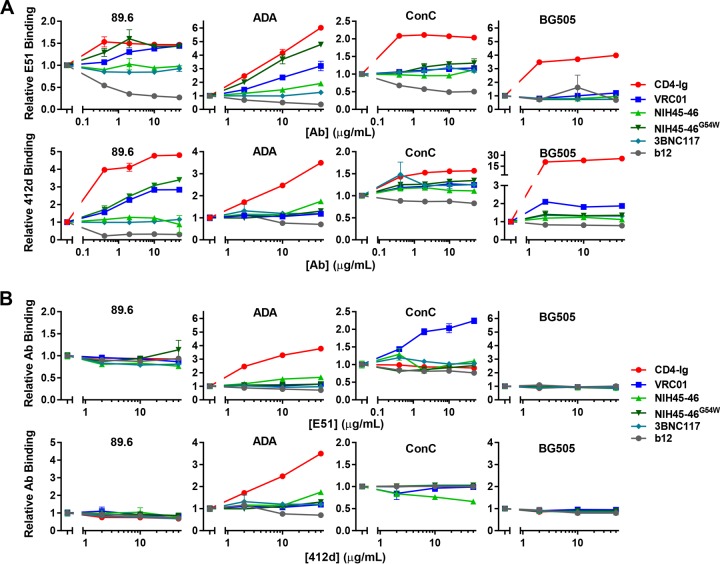
To understand the mechanism by which VRC01 combined with E51 or 412d to more potently neutralize HIV-1, we sought to determine whether either antibody could enhance association of the other to gp120 monomers or to cell-expressed Env trimers. We first examined by ELISA whether binding of E51 or 412d to immobilized gp120 was improved in the presence of increasing amounts of four CD4bs bNAbs. b12 was used as a negative control because its epitope extends into a region proximal to the E51 and 412d epitopes. To distinguish E51 and 412d from CD4bs antibodies, their Fc regions were exchanged with that of murine IgG2a. Antihuman or antimurine secondary antibodies were then used to detect CD4bs and CD4i antibody binding, respectively. As expected, CD4-Ig consistently enhanced E51 and 412d binding, while b12 inhibited binding, to all three gp120 monomers tested (Fig. 5A). The ability of the CD4bs bNAbs to promote E51 association was isolate and antibody dependent. Specifically, VRC01 and NIH45-46G54W, but not NIH45-46 or 3BNC117, enhanced binding of E51 to clade B gp120 proteins but not to a ConC gp120 (Fig. 5A). VRC01 and NIH45-46G54W also enhanced 412d binding to 89.6 but not at the levels observed with CD4-Ig. Although VRC01, NIH45-46, NIH45-46G54W, and 3BNC117 also modestly induced 412d binding to BG505 gp120 monomer, this induction was markedly less pronounced than that observed with CD4-Ig. We also assayed the ability of E51 or 412d to promote gp120 association with each of these antibodies. In general, E51 and 412d had little effect on promoting CD4bs antibody binding to gp120 monomers (Fig. 5B). However, both antibodies promoted CD4-Ig association with ADA gp120. E51 also enhanced binding of VRC01 to ConC gp120. In general, for clade B isolates, VRC01 and NIH45-46G54W modestly enhanced E51 and 412d association with gp120 monomers, but E51 and 412d had little effect on CD4bs antibody binding to these monomers.
FIG 5.
Promotion of gp120 monomeric association by CD4bs and CD4i antibodies. (A) ELISA plates were coated with 5 μg/ml of the indicated gp120 molecules. Immobilized gp120 was preincubated with serially diluted CD4-Ig or CD4bs antibodies as indicated, starting at 50 μg/ml. Wells were washed and incubated with a constant amount of E51 or 412d. Binding of E51 or 412d was measured by absorbance of an anti-horseradish peroxidase (anti-HRP) secondary antibody recognizing murine Fc at 450 nm, and data were normalized to E51 or 412d binding without preincubation of CD4-Ig or CD4bs antibody. (B) Experiment similar to that for panel A except that the indicated gp120 molecules were preincubated with serially diluted E51 or 412d starting at 50 μg/ml. Wells were washed and incubated with a constant concentration of CD4-Ig or the indicated CD4bs antibody. Data were analyzed as for panel A except that they were normalized to binding of CD4-Ig or CD4bs without E51 preincubation. Note that the scale of the y axes varies between graphs. Error bars represent SEM.
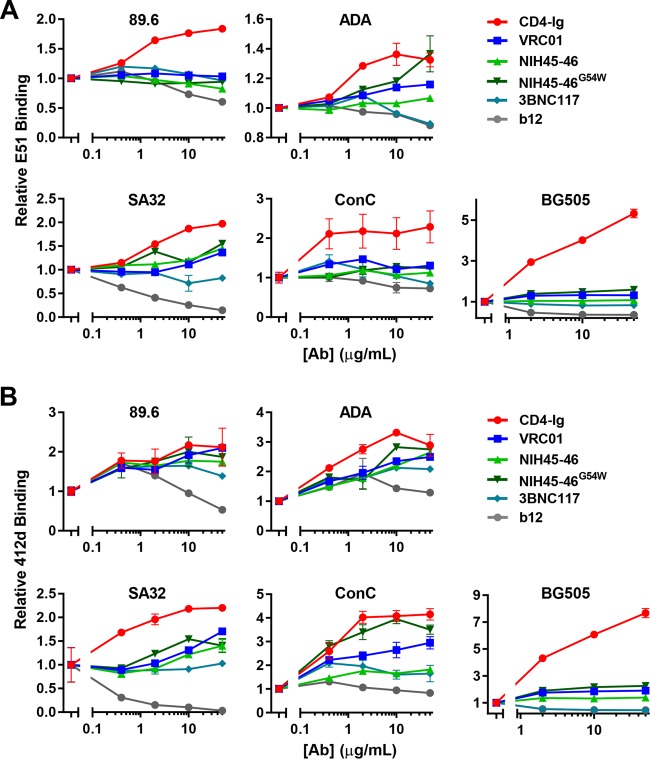
Binding of CD4i epitope by E51, 412d, 17b, and CCR5mim2-Ig enhances binding of CD4bs bNAbs to cell surface-expressed Env trimers.
We next explored whether changes in the quaternary structure of Env might be more relevant to the synergy observed in Fig. 1. To study Env trimers, we used Envs with truncations in their cytoplasmic tails. This modification greatly increases surface expression but can affect binding of some nonneutralizing antibodies, although not most CD4bs bNAbs (38). Using flow cytometry, we examined the ability of E51 and 412d to bind Env trimers expressed on the surface of HEK293T cells in the presence of increasing amounts of CD4bs antibodies prebound to Env (Fig. 6A and B). We defined induction as any increase in binding greater than 20% above baseline binding. We observed that, at various concentrations, CD4-Ig could promote binding of E51 and 412d to Env but that, in general, CD4bs bNAbs did not do so efficiently. This difference was especially clear with the BG505 Env. There were, however, two exceptions. NIH45-46G54W induced 412d binding to the ConC Env, and all CD4bs antibodies except b12 induced 412d binding to the 89.6 Env. Similar to what was observed with monomeric gp120, b12 interfered with binding of both CD4i antibodies to Env. Surprisingly, and in contrast, various concentrations of E51 enhanced binding by over 20% of all four CD4bs bNAbs to most Envs but had only very modest effects on CD4-Ig or b12 (Fig. 7A). However, E51 did not enhance binding of 3BNC117 to SA32 Env or of any CD4bs antibody to BG505 Env. We then expanded this finding by testing 412d, 17b, and CCR5mim2-Ig (Fig. 7B). Interestingly, CCR5mim2-Ig more efficiently promoted CD4bs antibody binding to all Env trimers than did 412d and 17b. VRC01 binding to 89.6 was markedly enhanced by 412d, 17b, and CCR5mim2-Ig. We also observed that all three inhibitors enhanced VRC01 and NIH45-46 binding to ConC. Based on these data, we conclude that occupation of the coreceptor-binding site facilitates association of VRC01 and other CD4bs bNAbs with the Env trimer. Taking these data together with the monomeric gp120 ELISA data (Fig. 5), we conclude that CD4i antibodies induce quaternary conformations in Env that are favorable to its association with CD4bs bNAbs. Finally, we infer that the synergy observed in Fig. 1 is largely due to the ability of CD4i antibodies to promote these quaternary conformations.
FIG 6.
Promotion of CD4i antibody association with trimeric HIV-1 Env by CD4-Ig and CD4bs antibodies. (A) HEK293T cells were transfected to express the indicated envelope glycoproteins that lack a portion of the cytoplasmic domain. Cells were preincubated with various concentrations of CD4-Ig and the indicated antibodies at 50 μg/ml and serially diluted by 5-fold to 0.4 μg/ml. Cells were washed and then incubated with 0.25 μg/ml E51. Binding was analyzed by flow cytometry. Mean fluorescence intensity (MFI) data are normalized to value of E51 binding without CD4-Ig or CD4bs antibodies prebound. (B) Experiment similar to that for panel A except that cells were incubated with 0.25 μg/ml of 412d after preincubation with CD4bs antibodies. Note that the scale of the y axes varies between graphs. Error bars represent SEM.
FIG 7.
Promotion of CD4bs antibody association with trimeric Env by CD4i antibodies and CCR5mim2-Ig. (A) Experiment similar to that for Fig. 6 except that cells were preincubated with E51 at serially diluted concentrations starting at 50 μg/ml. Cells were washed and then incubated with 0.01 μg/ml of the indicated CD4bs antibody or CD4-Ig. Analysis was performed as for Fig. 4 except that MFI data were normalized to the value of each antibody or CD4-Ig binding without E51 preincubation. (B) Experiment similar to that for panel A except that cells were preincubated with 50 μg/ml of 412d, 17b, or CCR5mim2-Ig. Note that the scale of the y axes varies between graphs. Error bars represent SEM.
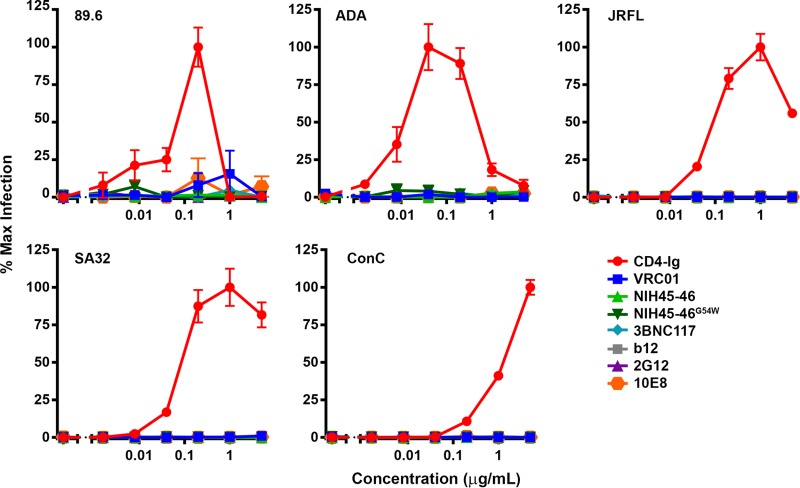
Unlike CD4-Ig, CD4bs bNAbs do not promote HIV-1 infection of CD4-negative cells.
As shown in Fig. 6 and 7, CD4-Ig has different properties than CD4bs bNAbs. It promotes E51 and 412d association to Env trimers, but its own association with Env is largely unaffected by the presence of E51, 412d, 17b, or CCR5mim2-Ig. In contrast, CD4bs bNAbs do not significantly improve CD4i antibody binding to Env, but CD4i antibodies do promote association of CD4bs bNAbs. To determine if these differences had functional consequences, we infected CCR5-positive, CD4-negative Cf2Th cells with five HIV-1 isolates in the presence of CD4-Ig, CD4bs bNAbs, b12, 10E8 (an MPER antibody), or 2G12 (Fig. 8). As expected, CD4-Ig substantially enhanced infection of these cells, with maximum enhancement peaking at 0.1 to 1 μg/ml. In contrast, little or no enhancement of infection was observed with CD4bs bNAbs antibodies or with the neutralizing antibodies b12, 2G12, and 10E8. Thus, the inefficiency with which CD4bs bNAbs promote CD4i association is consistent with their corresponding inability to promote infection of CCR5-positive, CD4-negative cells. We conclude that, unlike CD4-Ig, these antibodies do not induce Env conformations that promote binding of Env to CCR5.
FIG 8.
CD4-Ig, but not CD4bs antibodies, promotes HIV-1 entry into Cf2Th CCR5+ CD4− cells. Luciferase-expressing HIV-1 pseudotyped with the indicated Env was preincubated with the indicated antibodies. Dog thymus Cf2Th cells expressing CCR5, but not CD4, were added to the virus-antibody mixture and incubated for 48 h. Infection was measured by the percent luciferase expression in the absence of inhibitor. Error bars represent SEM.
DISCUSSION
Here we observed a modest synergy between the CD4bs bNAb VRC01 and either of two CD4i antibodies, E51 and 412d. To understand the underlying mechanism of this synergy, we studied how these CD4i antibodies interact with several VRC01-like bNAbs, as well as the CD4bs antibody b12. We observed that, as expected, CD4-Ig induced a conformation in monomeric gp120 preferred by E51 or 412d, but in general VRC01-like bNAbs did so less efficiently. Differences between CD4-Ig and VRC01-like CD4bs bNAbs were more pronounced with cell-expressed trimeric Env. CD4-Ig robustly enhanced E51 and 412d binding to Env trimers, but VRC01-like antibodies were much less efficient in promoting CD4i antibody binding (Fig. 6). Consistent with this observation, CD4-Ig, but not CD4bs antibodies enhanced HIV-1 infection of a CD4-negative, CCR5-positive cell line.
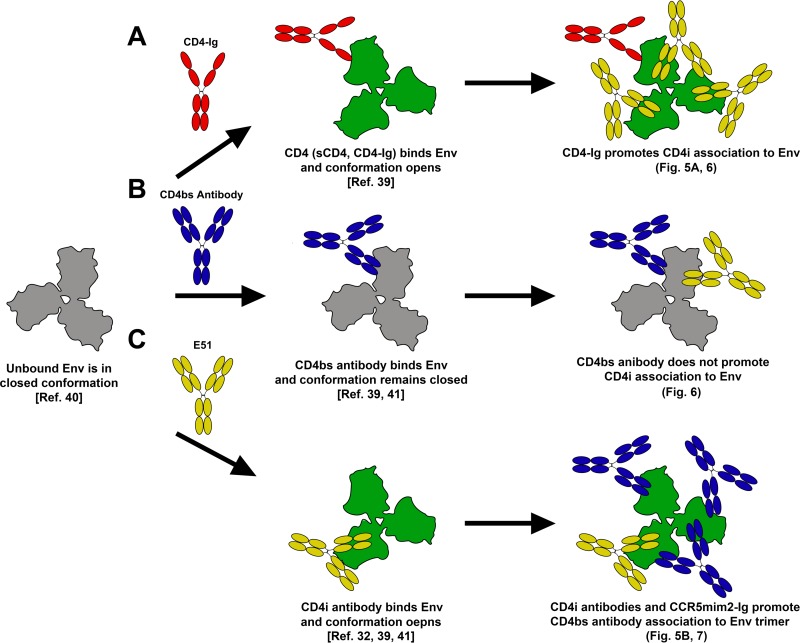
However, the most striking difference between CD4-Ig and CD4bs bNAbs was their relative abilities to bind trimeric Env in the presence or absence of CD4i antibodies or CCR5mim2-Ig. CD4i antibodies and CCR5mim2-Ig had almost no effect on the ability of CD4-Ig to bind trimers. In contrast, E51 and CCR5mim2-Ig markedly enhanced Env binding of VRC01, NIH45-46, NIH45-46G54W, and to a lesser extent 3BNC117 (Fig. 7C). This effect was less consistent with gp120 monomers. We therefore infer that occupation of the coreceptor-binding site, by either CD4i antibodies or CCR5-mimetic peptides, can promote or stabilize quaternary conformational changes in Env that facilitate access of CD4bs bNAbs to Env. These quaternary changes do not affect CD4-Ig binding and, indeed, are likely induced by CD4-Ig binding itself. These functional observations are consistent with cryo-electron microscopy studies. It has been previously shown that soluble CD4, 17b, and m36 (a CD4i antibody), but not VRC01, induced an open conformation in Env (39, 40). The open conformation induced by CD4 is preferred by CD4i antibodies for both the gp120 monomer and the Env trimer (Fig. 9A). However, CD4bs antibodies in general maintain a more closed conformation of Env, and they do not facilitate binding of CD4i antibodies or the coreceptor of trimeric Env, but in some cases CD4bs antibodies can promote CD4i binding to monomeric gp120 (Fig. 9B). Nonetheless, the current study makes clear that most potent CD4bs bNAbs preferentially bind the open conformation induced by CD4i antibodies or CCR5mim2-Ig (Fig. 9C). Consistent with this, Bartesaghi et al. have shown that in the open conformation of Env, the variable loops of Env are shifted in a way that further exposes the Env's CD4-binding site, as well as, presumably, the coreceptor-binding site (41). Similarly, Chen et al. have demonstrated that VRC01 binding to a gp120 stabilized core was enhanced when this core was fused to m36.4 (42).
FIG 9.
Model of Env induction by E51 and CD4bs antibodies. Shown is the model for induction that combines our data with the findings of Kwong et al. (32), Tran et al. (39), and Bartesaghi et al. (41). The unbound trimeric Env starts in a closed conformation (left). (A) When CD4 (or soluble CD4 or CD4-Ig) binds Env, the conformation is opened. This conformation promotes CD4i binding, like E51 or 17b, on both monomeric gp120 and trimeric Env. (B) In contrast, the binding of a CD4bs antibody (VRC01, NIH45-46, etc.) keeps the conformation in a closed state. Though this is the case, these antibodies have the ability to moderately promote CD4i binding on monomeric gp120, as seen in Fig. 5A and described by Scheid et al. (9). (C) Like for CD4, when E51 (or other coreceptor inhibitors such as 17b, m36, CCR5mim1-Ig, etc.) binds Env, the Env conformation opens. This conformation induces CD4bs antibody binding on trimeric Env but not monomeric gp120, as seen in Fig. 7 and 5B, respectively.
This study also highlights other differences between CD4-Ig and VRC01-class bNAbs. As mentioned, CD4-Ig more efficiently promotes CD4i antibody binding to monomeric gp120 and trimeric Env. This difference may be a necessary property of potent CD4bs antibodies for two possible reasons. First, an antibody selected for potency would not be expected to promote infection at low concentrations or when cellular CD4 was limiting. Second, there is likely to be an energy and/or an entropy barrier limiting access to the open conformation of Env. If an antibody can avoid the free energy penalty of inducing this conformation, it can bind with greater affinity (43). In contrast, cellular CD4 and therefore CD4-Ig must necessarily pay this penalty to facilitate coreceptor binding. CD4i antibodies, which bind gp120 and Env similarly to CCR5, must also pay this penalty, perhaps in part explaining the lower potency of these antibodies than of CD4bs bNAbs. Of course, when Env is bound to CD4, this energetic cost is already paid, the coreceptor epitope is exposed, and CD4i antibodies bind efficiently.
We show here that the synergy between VRC01 and E51 or 412d is likely the result of the ability of the CD4i antibody to induce the open state and facilitate access to the CD4-binding site. Highly potent VRC01-like bNAbs are rare. Most CD4bs antibodies in the sera of infected individuals neutralize Env less efficiently, in part because they cannot access their epitopes on the closed Env trimer and in part because they pay an energetic price for inducing the CD4-bound state of Env (43). Thus, the presence of relatively potent CD4i antibodies in sera might complement both limitations of these less potent CD4bs antibodies. Interestingly, CD4i antibodies are present in infected individuals, and some can be broad enough to neutralize HIV-2 isolates in the presence of soluble CD4 (sCD4) (44). These antibodies have been suggested to correlate with enhanced control of simian-human immunodeficiency virus (SHIV) SF162P3 in vaccinated macaques (45). Our study highlights the possibility that CD4i antibodies, by promoting exposure of the CD4-binding site, might enhance the generation of more potent CD4bs antibodies or increase the potency of less efficient CD4bs antibodies.
ACKNOWLEDGMENTS
We thank Meredith Gardner for comments and edits of the manuscript. We thank Audrey Richard for technical support with the flow cytometer.
Funding Statement
M.R.G. is a recipient of the National Institutes of Health Loan Repayment Program award.
REFERENCES
- 1.Checkley MA, Luttge BG, Freed EO. 2011. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol 410:582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalgleish AG, Beverley PCL, Clapham PR, Crawford DH, Greaves MF, Weiss RA. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 3.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. 1996. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148. doi: 10.1016/S0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 4.Feng Y, Broder CC, Kennedy PE, Berger EA. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 5.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 7.Burton D, Pyati J, Koduri R, Sharp S, Thornton G, Parren P, Sawyer L, Hendry R, Dunlop N, Nara P, et al. . 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 8.Zhou T, Georgiev I, Wu X, Yang Z-Y, Dai K, Finzi A, Do Kwon Y, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TYK, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diskin R, Scheid JF, Marcovecchio PM, West AP, Klein F, Gao H, Gnanapragasam PNP, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ. 2011. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, Stephenson KE, Chang H-W, Shekhar K, Gupta S, Nkolola JP, Seaman MS, Smith KM, Borducchi EN, Cabral C, Smith JY, Blackmore S, Sanisetty S, Perry JR, Beck M, Lewis MG, Rinaldi W, Chakraborty AK, Poignard P, Nussenzweig MC, Burton DR. 2013. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature 503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burton DR, Hessell AJ, Keele BF, Klasse PJ, Ketas TA, Moldt B, Dunlop DC, Poignard P, Doyle LA, Cavacini L, Veazey RS, Moore JP. 2011. Limited or no protection by weakly or nonneutralizing antibodies against vaginal SHIV challenge of macaques compared with a strongly neutralizing antibody. Proc Natl Acad Sci U S A 108:11181–11186. doi: 10.1073/pnas.1103012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horwitz JA, Halper-Stromberg A, Mouquet H, Gitlin AD, Tretiakova A, Eisenreich TR, Malbec M, Gravemann S, Billerbeck E, Dorner M, Büning H, Schwartz O, Knops E, Kaiser R, Seaman MS, Wilson JM, Rice CM, Ploss A, Bjorkman PJ, Klein F, Nussenzweig MC. 2013. HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc Natl Acad Sci U S A 110:16538–16543. doi: 10.1073/pnas.1315295110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, Plishka R, Buckler-White A, Seaman M, Piatak M, Lifson JD, Dimitrov D, Nussenzweig MC, Martin MA. 2013. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature 503:277–280. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caskey M, Klein F, Lorenzi JCC, Seaman MS, West AP Jr, Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, Horwitz JA, Shimeliovich I, Ben-Avraham S, Witmer-Pack M, Platten M, Lehmann C, Burke LA, Hawthorne T, Gorelick RJ, Walker BD, Keler T, Gulick RM, Fatkenheuer G, Schlesinger SJ, Nussenzweig MC. 2015. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, Enama ME, Plummer S, Holman L, Hendel CS, Gordon I, Casazza J, Conan-Cibotti M, Migueles SA, Tressler R, Bailer RT, McDermott A, Narpala S, O'Dell S, Wolf G, Lifson JD, Freemire BA, Gorelick RJ, Pandey JP, Mohan S, Chomont N, Fromentin R, Chun T-W, Fauci AS, Schwartz RM, Koup RA, Douek DC, Hu Z, Capparelli E, Graham BS, Mascola JR, Ledgerwood JE. 2015. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci Transl Med 7:319ra206–319ra206. doi: 10.1126/scitranslmed.aad5752. [DOI] [PubMed] [Google Scholar]
- 18.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67:3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiang SH, Doka N, Choudhary RK, Sodroski J, Robinson JE. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res Hum Retroviruses 18:1207–1217. doi: 10.1089/08892220260387959. [DOI] [PubMed] [Google Scholar]
- 20.Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. 2004. HIV vaccine design and the neutralizing antibody problem. Nat Immunol 5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 21.Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang C-C, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol 77:10557–10565. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choe H, Li W, Wright PL, Vasilieva N, Venturi M, Huang C-C, Grundner C, Dorfman T, Zwick MB, Wang L, Rosenberg ES, Kwong PD, Burton DR, Robinson JE, Sodroski JG, Farzan M. 2003. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell 114:161–170. doi: 10.1016/S0092-8674(03)00508-7. [DOI] [PubMed] [Google Scholar]
- 23.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, Choe H. 1999. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 Entry. Cell 96:667–676. doi: 10.1016/S0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 24.Chiang JJ, Gardner MR, Quinlan BD, Dorfman T, Choe H, Farzan M. 2012. Enhanced recognition and neutralization of HIV-1 by antibody-derived CCR5-mimetic peptide variants. J Virol 86:12417–12421. doi: 10.1128/JVI.00967-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorfman T, Moore MJ, Guth AC, Choe H, Farzan M. 2006. A tyrosine-sulfated peptide derived from the heavy-chain CDR3 region of an HIV-1-neutralizing antibody binds gp120 and inhibits HIV-1 infection. J Biol Chem 281:28529–28535. doi: 10.1074/jbc.M602732200. [DOI] [PubMed] [Google Scholar]
- 26.Huang C-c, Lam SN, Acharya P, Tang M, Xiang S-H, Hussan SS-u, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. 2007. Structures of the CCR5 N Terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol 74:8358–8367. doi: 10.1128/JVI.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeuchi Y, McClure MO, Pizzato M. 2008. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J Virol 82:12585–12588. doi: 10.1128/JVI.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. 2009. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J Virol 83:8289–8292. doi: 10.1128/JVI.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwong JA, Dorfman T, Quinlan BD, Chiang JJ, Ahmed AA, Choe H, Farzan M. 2011. A tyrosine-sulfated CCR5-mimetic peptide promotes conformational transitions in the HIV-1 envelope glycoprotein. J Virol 85:7563–7571. doi: 10.1128/JVI.00630-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 34.He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol 69:6705–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barouch DH, Yang Z-y, Kong W-p, Korioth-Schmitz B, Sumida SM, Truitt DM, Kishko MG, Arthur JC, Miura A, Mascola JR, Letvin NL, Nabel GJ. 2005. A human T-cell leukemia virus type 1 regulatory element enhances the immunogenicity of human immunodeficiency virus type 1 DNA vaccines in mice and nonhuman primates. J Virol 79:8828–8834. doi: 10.1128/JVI.79.14.8828-8834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. 2012. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J, Kovacs JM, Peng H, Rits-Volloch S, Lu J, Park D, Zablowsky E, Seaman MS, Chen B. 2015. Effect of the cytoplasmic domain on antigenic characteristics of HIV-1 envelope glycoprotein. Science 349:191–195. doi: 10.1126/science.aaa9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran EEH, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JLS, Subramaniam S. 2012. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog 8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyerson JR, Tran EEH, Kuybeda O, Chen W, Dimitrov DS, Gorlani A, Verrips T, Lifson JD, Subramaniam S. 2013. Molecular structures of trimeric HIV-1 Env in complex with small antibody derivatives. Proc Natl Acad Sci U S A 110:513–518. doi: 10.1073/pnas.1214810110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartesaghi A, Merk A, Borgnia MJ, Milne JLS, Subramaniam S. 2013. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol 20:1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen W, Feng Y, Wang Y, Zhu Z, Dimitrov DS. 2012. Fusion proteins of HIV-1 envelope glycoprotein gp120 with CD4-induced antibodies showed enhanced binding to CD4 and CD4 binding site antibodies. Biochem Biophys Res Commun 425:931–937. doi: 10.1016/j.bbrc.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haim H, Salas I, McGee K, Eichelberger N, Winter E, Pacheco B, Sodroski J. 2013. Modeling virus- and antibody-specific factors to predict human immunodeficiency virus neutralization efficiency. Cell Host Microbe 14:547–558. doi: 10.1016/j.chom.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 201:1407–1419. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeVico A, Fouts T, Lewis GK, Gallo RC, Godfrey K, Charurat M, Harris I, Galmin L, Pal R. 2007. Antibodies to CD4-induced sites in HIV gp120 correlate with the control of SHIV challenge in macaques vaccinated with subunit immunogens. Proc Natl Acad Sci U S A 104:17477–17482. doi: 10.1073/pnas.0707399104. [DOI] [PMC free article] [PubMed] [Google Scholar]