ABSTRACT
Human immunodeficiency virus (HIV) infects and depletes CD4+ T cells, but subsets of CD4+ T cells vary in their susceptibility and permissiveness to infection. For example, HIV preferentially depletes interleukin-17 (IL-17)-producing T helper 17 (Th17) cells and T follicular helper (Tfh) cells. The preferential loss of Th17 cells during the acute phase of infection impairs the integrity of the gut mucosal barrier, which drives chronic immune activation—a key determinant of disease progression. The preferential loss of Th17 cells has been attributed to high CD4, CCR5, and CXCR4 expression. Here, we show that Th17 cells also exhibit heightened permissiveness to productive HIV infection. Primary human CD4+ T cells were sorted, activated under Th17- or Th0-polarizing conditions and infected, and then analyzed by flow cytometry. Th17-polarizing cytokines increased HIV infection, and HIV infection was disproportionately higher among Th17 cells than among IL-17− or gamma interferon-positive (IFN-γ+) cells, even upon infection with a replication-defective HIV vector with a pseudotype envelope. Further, Th17-polarized cells produced more viral capsid protein. Our data also reveal that Th17-polarized cells have diminished expression of RNase A superfamily proteins, and we report for the first time that RNase 6 inhibits HIV. Thus, our findings link Th17 polarization to increased HIV replication.
IMPORTANCE Our study compares the intracellular replicative capacities of several different HIV isolates among different T cell subsets, providing a link between the differentiation of Th17 cells and HIV replication. Th17 cells are of key importance in mucosal integrity and in the immune response to certain pathogens. Based on our findings and the work of others, we propose a model in which HIV replication is favored by the intracellular environment of two CD4+ T cell subsets that share several requirements for their differentiation: Th17 and Tfh cells. Characterizing cells that support high levels of viral replication (rather than becoming latently infected or undergoing cell death) informs the search for new therapeutics aimed at manipulating intracellular signaling pathways and/or transcriptional factors that affect HIV replication.
INTRODUCTION
Recent advances in the field of T helper cell development have shed new light on how human immunodeficiency virus (HIV) pathogenesis causes AIDS.
The rapid and preferential loss of Th17 cells—so named for their secretion of interleukin-17 (IL-17)—from the gut-associated lymphoid tissue (GALT) during acute HIV infection represents a critical aspect of HIV immunopathology (1). Recent studies link the HIV-induced preferential depletion of Th17 (and Th17-like) cells to AIDS-associated opportunistic infections, gut mucosal barrier perturbation, and chronic immune activation (2, 3).
Pathogenic and nonpathogenic primate models differ in their loss of Th17 cells, and these differences suggest a central role of Th17 cell loss in driving HIV pathogenesis. For example, in simian immunodeficiency virus (SIV)-infected macaques, the peak and set point viral loads are restricted by the initial size of the Th17 compartment (4), and a higher initial Th17/Th1 ratio at mucosal sites predicts a more rapid disease progression to AIDS (5). Further, the SIV-induced loss of the gut Th17 compartment is associated with mucosal damage and the translocation/dissemination of the enteric pathogen Salmonella enterica serovar Typhimurium (2, 6). In contrast, sooty mangabeys, which do not progress to AIDS, maintain healthy mucosal function and levels of Th17 cells following SIV infection (1, 2). HIV-induced Th17 cell depletion thus facilitates the mucosal damage and subsequent chronic immune dysregulation associated with progression to AIDS.
Th17 cells bridge innate and adaptive immune signaling at mucosal surfaces, and their preferential loss during acute HIV infection undermines mucosal immunity via multiple mechanisms. Th17 cells are enriched within mucosal tissues, especially in the GALT, which is a major site of HIV replication (1, 7). Th17 cells require several cytokines for their differentiation, including IL-1β, IL-6, and IL-23, which are expressed at high levels during HIV infection (8–16). Th17 cells, like other GALT effector/memory T cells, express high levels of HIV receptors, thus conferring their susceptibility to infection (17).
T follicular helper (Tfh) cells share many characteristics with Th17 cells, including their utilization of signal transducer and activator of transcription 3 (STAT3) and interferon-regulated factor 4 (IRF4) activity and their expression of IL-21 (18, 19). There are several notable differences between Th17 and Tfh cells: Tfh cells express their own master transcription factor, Bcl6, and the Th17-destabilizing transcription factor c-Maf (20). Tfh cells also express the chemokine receptors CXCR5 and CCR7, which promote Tfh homing to germinal centers. Although Tfh cells constitute a major site of viral production during HIV infection (21), they do not express CCR5 (22). Nonetheless, both cell types are preferentially infected during acute HIV infection, and the resulting, combined loss of IL-21-producing Th17 and Tfh cells during HIV infection stifles B cell development (23). Thus, the depletion of IL-17- and IL-21-expressing cells could represent a central mechanism by which HIV disrupts mucosal immunity during the early stages of infection and promotes opportunistic infections at mucosal sites that are associated with chronic immune activation and disease progression.
Despite effective viral suppression with combined antiretroviral therapy and the heightened in vivo availability of Th17-polarizing cytokines and antigens following HIV infection, the Th17 compartment often fails to reconstitute to preinfection levels in the gut and cervical mucosae (3, 24). The early loss of the enterocyte-proliferative and antimicrobial peptide-inducing cytokines produced by Th17 cells impairs the integrity of the gut mucosal barrier, driving microbial translocation and chronic immune activation—key determinants of disease progression to AIDS (25, 26).
The mechanisms underlying the preferential loss of Th17 cells in vivo during HIV infection remain unclear.
Prior studies examining the susceptibility of Th17 cells to HIV infection focused on entry-level factors such as HIV receptor expression. Th17 cells reportedly lack the expression of the HIV-inhibitory chemokine MIP-1β and also express high levels of the HIV-binding proteins α4β7 integrin, CD4, and CXCR4 (17). Th17 cells are enriched in CCR6 expression, and CCR6+ cells express significantly higher levels of the HIV coreceptor CCR5 than do CCR6− cells (27, 28).
However, even upon successful viral entry, CD4+ T cells vary in ability to become productively infected. Differences in the intracellular environment ultimately determine whether an HIV-infected cell will die, become latently infected, or become a factory for the production of viral progeny. Interestingly, CCR6+ and Th17-polarized cells also showed higher rates of infection when infected with replication-defective, pseudotyped HIV vectors in a vesicular stomatitis virus glycoprotein (VSV-G) envelope, suggesting that postentry mechanisms may also contribute to the preferential loss of Th17 cells (27–29). Using surface markers to identify Th17 cells rather than intracellular cytokine staining may overestimate the frequency of Th17 cells and result in the inclusion of cells that do not express IL-17 (30).
Here, we demonstrate that Th17 cells are highly permissive to HIV infection and replication and that Th17-polarizing cytokines enhance viral replication in vitro. We find that Th17-polarized cells express significantly lower levels of HIV-inhibitory RNases than Th0-polarized cells. Our findings support the concept that Th17 cells could be a major source of viral production during acute HIV infection, and we propose that common features of Th17 cells and some Tfh cells may help explain their proclivity to become major sources of viral production during acute HIV infection (31–33).
MATERIALS AND METHODS
CD4+ T cell isolation and CCR6 sorting.
CD4+ T lymphocytes were isolated from human peripheral blood mononuclear cells by negative selection using the Easy Sep CD4+ human T cell enrichment kit, according to the manufacturer's protocol (Stem Cell Technologies Inc., Vancouver, BC, Canada). To enrich for Th17 cells, CD4+ T cells were sometimes stained with an anti-CCR6 antibody (clone 11A9; BD Bioscience, San Jose, CA) and then sorted by fluorescently activated cell sorting (FACS).
Cell culture conditions.
Cells were maintained at 37°C and 5% CO2 in sterile-filtered RPMI medium supplemented with 10% human serum (Gemini Bio-Products, Sacramento, CA), penicillin-streptomycin-gentamicin, l-glutamine minimal essential medium (MEM) amino acids, and sodium pyruvate (Life Technologies, Grand Island, NY).
For phytohemagglutinin (PHA) activation, total CD4 T cells were cultured in complete medium containing PHA (2.5 μg/ml; Sigma-Aldrich, St. Louis, MO) and IL-2 (10 ng/ml; R&D Systems, Minneapolis, MN) for 3 days and then washed and cultured overnight in complete medium containing only IL-2 prior to HIV infection. After HIV infection (described below), cells were washed in phosphate-buffered saline (PBS) and then resuspended in medium containing IL-2 (10 ng/ml) or the Th17-polarizing cytokines IL-1β (10 ng/ml), IL-6 (10 ng/ml), transforming growth factor β (TGF-β) (2 ng/ml), and IL-23 (10 ng/ml; Peprotech, Rocky Hill, NJ).
CCR6+- or CCR6−-sorted cells from each donor were activated with antibodies to CD3 (plate bound; 5 μg/ml) and CD28 (soluble; 1 μg/ml; eBioscience, San Diego, CA) for 3 days in the presence of IL-1β and IL-6 (5 ng/ml each), TGF-β (1 ng/ml), and IL-23 (20 ng/ml; Peprotech, Rocky Hill, NJ). Following activation, cells were washed and then resuspended in fresh medium containing only IL-2 (2 ng/ml) and IL-23 (10 ng/ml). Cells from all polarizing conditions were maintained in IL-2/IL-23 medium after HIV infection.
Virus preparation.
HIV-1BaL stocks were prepared by infecting human macrophages in complete RPMI 1640 medium supplemented with 20% heat-inactivated fetal bovine serum (FBS; Gemini Bio-Products, Sacramento, CA). HIV-1IIIB was produced in PM1 cells (AIDS Reagent Program; catalog no. 3038) in IL-2-containing, complete RPMI 1640 medium. The transmitted-founder isolate HIV-1AD17, which was also produced in PM1 cells, was kindly provided by George Shaw (34, 35). Pseudotyped virions were generated by Fugene (Promega, Fitchburg, WI) cotransfection of 293T cells with two plasmids: one expressing an envelope-deficient HIV backbone obtained through the NIH AIDS Reagent Program (catalog no. 4692; Division of AIDS, NIAID, NIH) and the other expressing an amphotropic murine leukemia virus (AMLV) envelope. Supernatants were analyzed by p24 enzyme-linked immunosorbent assay (ELISA) and stored at −80°C.
HIV infections.
Equal numbers of PHA-activated and CCR6-sorted cells were infected with HIVBaL, HIVIIIB, and HIVAD17 for 2.5 h and then washed and resuspended in medium containing IL-2 or Th17-polarizing cytokines (PHA activated) or IL-2 and IL-23 (CCR6-sorted, prepolarized cells), as indicated in Results. For replication-defective, pseudotype virus infections, 500 μl of supernatant virus (p24, 130 ng/ml) was added per million cells in 1.5 ml of the indicated medium, and cells were then analyzed by flow cytometry 4 days postinfection.
Flow cytometry staining and analysis.
Four to 6 days after infection (unless otherwise noted), cells from each condition were stimulated with phorbol myristate acetate (PMA; 50 ng/ml), ionomycin (500 ng/ml; Sigma-Aldrich), and BD GolgiPlug (BD Bioscience) for 5 h. To assess viability, cells were stained with Live/Dead Aqua (Life Technologies) according to the manufacturer's instructions, immediately prior to extracellular marker staining and fixation. For intracellular staining, cells were permeabilized according to the BD Perm/Wash protocol; then stained for p24 (Beckman Coulter; clone KC57), IL-17A, and gamma interferon (IFN-γ) (eBioscience and BioLegend, respectively); and then washed prior to analysis.
Compensation and data collection were achieved using BD CompBeads (BD Bioscience) and BD FACSDiva software version 6.0 (BD Bioscience). Data analysis and flow cytometry plots were generated using FlowJo software version 9.7.2 (FlowJo, LLC, Ashland OR). Sorting and data collection were performed at the University of Maryland—Baltimore flow cytometry core facilities at the Institute of Human Virology and at the Greenebaum Cancer Center.
RNA purification and microarray.
Total RNA was extracted from cell lysates using Qiagen Quickspin columns and DNase treatment, according to the manufacturer's protocol (Qiagen Inc., Valencia, CA). RNA quality assessment and microarray analysis were performed by the University of Maryland Greenebaum Cancer Center Translational Genomics core facility, using Affymetrix HuGene 2.0 chips (Affymetrix Inc., Santa Clara, CA). Normalized gene expression estimates were obtained with the Frozen Robust Multiarray Analysis (fRMA) method (36). A generalized linear model approach, coupled with empirical Bayes standard error shrinkage and including coefficients for data heterogeneity as derived from surrogate variable analysis (SVA) (37), was used for identifying differentially expressed genes in Th17-polarized cells relative to Th0-polarized cells. Correction for multiple testing was performed using the Benjamini-Hochberg method. The identification of pathways and biological processes differentially expressed was performed using gene set enrichment analysis (GSEA) (38) and analysis of functional annotation (AFA) as previously described (39–41).
Immunoblot protein analysis.
CCR6+ or CCR6− cells were activated for 3 days using anti-CD3/CD28 as described above, under Th0- or Th17-polarizing conditions, and then maintained in medium containing IL-2 or IL-23 for an additional 3 days. Cells were then washed in phosphate-buffered saline (PBS) and pelleted, then lysed in RIPA buffer containing an EDTA-free protease and phosphatase inhibitor cocktail (Sigma-Aldrich), flash-frozen, and stored at −80°C. Equal amounts of total protein were boiled for 10 min in the presence of 4× dithiothreitol (DTT) loading buffer and loading dye and then loaded into 1% polyacrylamide gels (Life Technologies). Protein from the gel was transferred to polyvinylidene difluoride (PVDF) membranes (Life Technologies), blocked with a 5% (wt/vol) solution of powdered milk, washed, and then incubated at 4°C overnight in the presence of anti-human RNase antibodies (Abnova Inc., Taipei, Taiwan). Bound RNase antibodies were probed with anti-mouse or anti-rabbit horseradish peroxidase (HRP)-linked secondary antibodies, depending on the source of the primary antibody. As a loading control, blots were also probed with an HRP-conjugated antibody to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (ThermoFisher Scientific Inc., Rockville, MD).
p24 ELISA.
Four days postinfection, infected cells were pelleted by centrifugation at 300 × g for 7 min, and supernatant was then collected and frozen at −80°C. Several dilutions of each supernatant were diluted into 1× lysis buffer in a 96-well plate, in triplicate. The Institute of Human Virology Core Facility (University of Maryland—Baltimore, Baltimore, MD) performed ELISA using commercially available p24 ELISA plates (PerkinElmer Inc., Boston, MA). Measured p24 values were normalized to a standard curve of known concentrations and were used only if they fell within the detectable range.
Luciferase assays.
TZM-BL cells (AIDS Reagent Program; catalog no. 8129) were seeded at 5,000 cells/well in a 96-well plate (Sigma-Aldrich) and incubated overnight at 37°C (5% CO2) in Dulbecco's modified Eagle's medium (DMEM) containing penicillin-streptomycin and l-glutamine (Life Technologies) and 10% fetal bovine serum (FBS) (Gemini Bio-Products). Thawed supernatant from sorted, polarized, infected T cells was added to the preseeded TZM-BL cells such that the final p24 concentration was 5 ng/ml. As controls, an equivalent volume of donor- and polarization-matched supernatants from uninfected cells was added. Each condition was done in triplicate, and samples were then incubated at 37°C (5% CO2) for 48 h. Finally, the TZM-BL cells were washed and lysed in Steady-Glo luciferase detection buffer (Promega), according to the manufacturer's instructions. Luciferase activity was measured in a Veritas luminometer (Promega). The p24-specific luciferase activity was evaluated by subtracting the relative light units (RLU) of the uninfected supernatant-treated cells from the RLU of the condition-matched, infected supernatants.
Statistical analyses and figure generation.
Raw experimental data were analyzed, graphed, and rendered using Microsoft Excel (Microsoft Inc., Redmond, WA), GraphPad Prism (GraphPad Inc., La Jolla, CA), and Microsoft PowerPoint software. Depending on whether we were analyzing data from different conditions or different donors, paired and unpaired Student t tests were used to measure significant differences among normally distributed data sets.
RESULTS
In vitro HIV infection of PHA-activated T cells expands rapidly upon the addition of Th17-polarizing cytokines.
Th17-polarizing cytokines IL-1β and IL-6 have been reported to increase HIV infection in vitro (42, 43). Another recent in vitro study suggested that Th17-polarizing cytokines, in the presence of antiretroviral therapy, could replenish the Th17 cells lost during HIV infection, but the authors did not show the effects of the Th17-polarizing cytokines on infection in the absence of antiretroviral treatment (44). Further, their Th17-polarizing cytokines were added in addition to IL-2. IL-2 not only increases HIV infection in vitro more than the Th1- or Th2-polarizing cytokines IL-12 and IL-4 but also inhibits IL-17 expression and promotes the plasticity of Th17 cells (45, 46). Therefore, we set out to compare the dynamics of T cell function and HIV infection in the presence of IL-2 or Th17-polarizing cytokines, hypothesizing that Th17 polarization would increase the percentage of infected cells.
First, we compared the kinetics of HIVBAL infection when IL-2 (10 ng/ml) or the Th17-polarizing cytokines IL-1β, IL-6, and IL-23 (all 10 ng/ml), and TGF-β (2 ng/ml) were added after HIV infection (Fig. 1A). Cells from two donors were analyzed 3, 5, and 7 days postinfection (Fig. 1B). Three days postinfection, 2.3% and 3.7% of total CD4+ cells were infected under IL-2 and Th17-polarizing conditions, respectively. By 5 days after infection, however, the percentage of cells expressing p24 was 4-fold higher in the presence of Th17-polarizing cytokines, compared with IL-2-treated cells (Fig. 1B). Based on our kinetics experiments, we chose day 5 after infection as a time point at which the effects of Th17-polarizing cytokines were most representative, compared with IL-2 treatment; the percentage of infected cells nearly tripled from 3.18 to 8.75 (P < 0.0002, n = 10) (Fig. 1C).
FIG 1.
Effects of Th17-polarizing cytokines on the percentage of HIV-infected, CD4 T cells. (A) Human CD4+ T cells were isolated from peripheral blood by negative selection and then mitogenically activated with phytohemagglutinin (PHA; 5 μg/ml) and interleukin-2 (IL-2; 10 ng/ml) for 3 days prior to infection. Cells were then washed, infected with the indicated isolates of HIV for 2.5 h, washed again, and then resuspended in medium containing IL-2 or the Th17-polarizing cytokines IL-1β, IL-6, and IL-23 (10 ng/ml) and TGF-β (2 ng/ml). (B and C) Time course (B) and day 5 comparison (C) of the total percentages of p24+ cells from IL-2 and from Th17-polarizing conditions. D0, D3, D5, and D7, days 0, 3, 5, and 7, respectively. (D) Representative plots of cells infected for 5 days with CCR5-tropic HIVBaL, CXCR4-tropic HIVNL4-3, or CCR5-tropic, transmitted-founder isolate HIVAD17. (E) The indicated cytokines were added after infection with HIVBaL. The IL-17 expression shown was in uninfected samples. *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001.
To determine whether our findings were specific to the CCR5-tropic HIVBaL lab isolate, we repeated our experiments using two other isolates: HIVIIIB, a CXCR4-tropic lab isolate, and HIVAD17, a CCR5-tropic clone of a clinical transmitted-founder virus (47). Our results show that treatment with Th17-polarizing cytokines increased p24 expression regardless of the HIV isolate used to infect the PHA-activated T cells (Fig. 1D).
We then determined the relative contribution of each of the polarizing cytokines to our observed changes in IL-17 and p24 expression. Accordingly, we infected PHA-activated cells from three donors with HIVBaL and then added either IL-2 or one of the 15 possible combinations of Th17-polarizing cytokines. Our data corroborated published effects of individual cytokines on IL-17 expression and HIV infection. For example, the addition of IL-1β and IL-6 or IL-23 tended to increase the number of IL-17-producing cells, but the addition of 2 ng/ml TGF-β consistently suppressed IL-17 expression (Fig. 1E). The combination of IL-1β and IL-23 generally produced the greatest increase in the percentage of HIV-infected cells, especially in the presence of TGF-β (Fig. 1E).
Th17 cells are preferential targets for productive HIV infection.
To determine which CD4+ T cell subsets were most likely to become productively infected by HIV, we analyzed the coexpression of cytokines, T cell markers, and p24 in human T cells activated with PHA and infected with HIVBaL in the presence of IL-2 or Th17-polarizing cytokines (Fig. 2). IL-17-expressing cells were enriched among infected cells, and the coenrichment of IL-17- and p24-expressing cells was significantly higher than that observed in IFN-γ+ cells. When infected in IL-2-containing medium, IL-17+ cells constituted only 3.1% of PHA-activated T cells but accounted for 18.4% of total HIV-1BaL-infected cells (P < 0.0001). Notably, 19.8% of IL-17+ cells became productively infected, compared with only 3.2% of total T cells or 9.4% of IFN-γ+ IL-17− cells (P < 0.0001 and P < 0.003, respectively) (Fig. 2A). The coenrichment of IL-17- and p24-expressing cells was even more pronounced under Th17-polarizing conditions. Whereas 19.8% of IL-17+ cells became infected under Th0 conditions, 42.1% became infected in the presence of Th17-polarizing cytokines (P < 0.001, n = 11). Whether infected in the presence of IL-2 or of Th17-polarizing cytokines, IL-17+ cells accounted for about one-fifth of total p24+ cells. Th17-polarizing cytokines also tended to decrease the proportion of infected cells that express IFN-γ (Fig. 2C, D, and G).
FIG 2.
Comparison of total and infected IL-17- or IFN-γ-producing cells. PHA-activated CD4+ T cells were infected with HIVBaL for 5 days in IL-2 or the Th17-polarizing cytokines IL-1β, IL-6, and IL-23 (10 ng/ml) and TGF-β (2 ng/ml). (A and B) Comparison of p24 expression among total, IL-17− IFN-γ−, IL-17+, and IL-17− IFN-γ+ T cells. (C and D) Comparison of IL-17 expression among total, p24−, or p24+ T cells. (E) Fold enrichment of p24+ cells among cells that either express or lack IFN-γ or IL-17. (F) Fold enrichment of IFN-γ- or IL-17-single-positive cells among p24+ cells, relative to p24− cells. (G) Percentages of IL-17- and IFN-γ-expressing cells were measured by flow cytometry. Blue depicts the proportion of IL-17− IFN-γ− cells, red shows IL-17+ cells, and yellow shows IFN-γ+ IL-17− cells. *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001.
IFN-γ+ and IL-17+ cells were both enriched among infected cells from Th0 conditions. Nevertheless, when accounting for the proportion of total cells, IL-17+ cells were significantly more likely to become productively infected by HIV than were IFN-γ+ cells (Fig. 2A and B). In PHA-activated T cells, IL-17+ cells were infected at significantly higher rates than IFN-γ+ cells (Fig. 2E and F). IL-17+ IFN-γ− cells and IL-17+ IFN-γ+ cells had similar rates of infection (19.4% and 20.7%, respectively).
We found that Th17-polarizing cytokines rendered T cells significantly more permissive to productive HIV infection, even when washed away prior to infection. Based on this observation, we hypothesized that Th17 signaling pathways promote HIV replication intracellularly, through postentry mechanisms.
Th17 polarization renders T cells more likely to become productively infected by HIV.
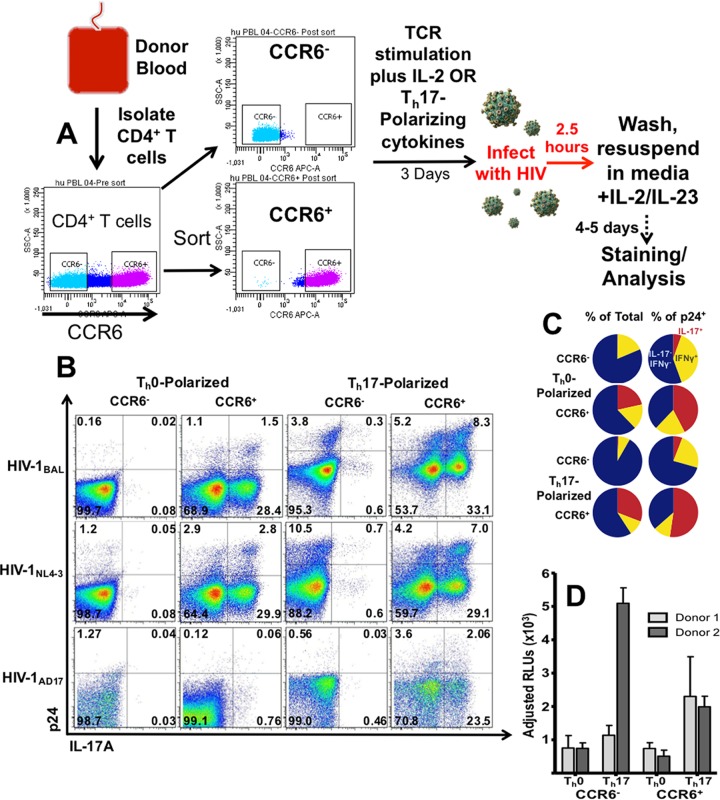
To achieve greater Th17 polarization and discern between pre- and postinfection effects of our polarizing cytokines, we polarized cells before infection as shown in Fig. 3A. Th17 cells express the chemokine receptor CCR6. Accordingly, primary human CD4+ T cells were sorted according to their CCR6 expression; activated with anti-CD3 and anti-CD28 in the presence of IL-2 or Th17-polarizing cytokines; and then washed, infected, and resuspended in medium containing only IL-2 and IL-23 (Fig. 3A).
FIG 3.
HIV infections of CCR6-sorted, prepolarized cells. (A) Cells were sorted according to their expression of CCR6 and then TCR stimulated in the presence of Th0- or Th17-polarizing cytokines (IL-2 and IL-1β, IL-6, IL-23, and TGF-β, respectively) for 3 days. Cells were then infected with the indicated isolates of HIV, washed, resuspended in medium with IL-2/IL-23, and analyzed by flow cytometry 5 days postinfection. *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001. (B) Representative flow cytometry plots depicting p24 and IL-17 expression in Th0- or Th17-polarized cells infected with the indicated isolates of HIV. (C) Percentages of IL-17- and IFN-γ-expressing cells were measured by flow cytometry. Blue depicts the proportion of IL-17− IFN-γ− cells, red shows IL-17+ cells, and yellow shows IFN-γ+ IL-17− cells. (D) Five nanograms of p24/ml from the supernatant of HIVBaL-infected T cells under the indicated conditions (or the equivalent volume from condition-matched, uninfected cells) was added to preseeded TZM-BL cells in triplicate. Luciferase activity was measured 48 h later, and relative light unit (RLU) measurements from infected supernatants were adjusted by subtracting the RLU from their corresponding uninfected supernatants.
Very few (mean = 0.43%, n = 6) of T cell receptor (TCR)-activated, CCR6− cells expressed IL-17, even when activated under Th17-polarizing conditions (0.74%). Roughly a third (30.9%) of CCR6+ cells expressed IL-17 upon activation in the presence of Th17-polarizing cytokines. A 20.5% proportion of CCR6+ cells produced IL-17 when activated in the presence of IL-2 (Fig. 3C). We observed high donor variability in the expression of IL-17 among CCR6+ cells under Th0 conditions, with some producing very few IL-17+ cells and others expressing nearly as many as in donor-matched CCR6+ cells from Th17-polarizing conditions. The percentage of IL-17-expressing cells peaked on days 3 to 5, immediately after activation, gradually decreasing to less than half of the initial peak level by day 9 (data not shown).
Representative plots in Fig. 3B that show low percentages of IL-17+ cells coexpressing p24 need to be analyzed in context. For example, in the HIVBaL-infected, CCR6−, Th0-polarized cells, the representative plot shows that only 0.02% of IL-17+ cells are p24+. However, in this example Th17 cells made up only 0.1% of total cells and yet accounted for 11% of total infected cells [0.02%/(0.02% + 0.16%)]. Furthermore, while only 0.18% of all cells were p24+, 20% of IL-17-expressing cells were infected.
An analysis combining data from all conditions from six donors reveals that the disproportionately higher rates of IL-17 and p24 coexpression were more common than that of IFN-γ and p24. Overall, IL-17+ cells had greater than a 2-fold-higher percentage of p24+ cells than total cells in 21 out of 24 samples (range, 2.1- to 14-fold). However, IFN-γ+ IL-17− cells from only 8/24 samples had greater than 2-fold increases in their percentage of p24+ cells (range, 2.2- to 6.3-fold), and 5/24 samples had fewer IFN-γ+ IL-17− cells among p24+ cells, relative to total cells.
IL-17+ cells made up only 6.2% of total T cells when all conditions were averaged but accounted for over 18% of all infected cells (P < 0.002).
IFN-γ expression was highest among cells from Th0 conditions, and levels were similar between CCR6+ and CCR6− cells (16.3% and 18.2%, respectively). Activation under Th17-polarizing conditions yielded fewer IFN-γ+ cells (7.5% for CCR6− and 10.0% for CCR6+ cells) (Fig. 3C).
Upon establishing our culture system and analyzing T cells from the sorted, polarized conditions, we wanted to compare HIVBaL, HIVIIIB, and HIVAD17 infections among various subpopulations. We chose 4 to 5 days postinfection to maximize the percentage of live, polarized cells present during infection and to ensure the detectability of p24+ cells by flow cytometry.
Remarkably, IL-17+ cells from HIVBaL CCR6+, Th17-polarizing conditions made up more than half of all infected cells (Fig. 3B and C) Further, the effects of Th17-polarizing cytokines on HIV infection occurred among both CCR6+ and CCR6− cells and with all HIV isolates used (Fig. 3B). Among HIVBaL-infected cells, Th17 polarization increased the percentage of p24+ cells by 8.9- and 5.8-fold in CCR6− and CCR6+ cells, respectively (P < 0.02). CCR6+ cells had 4.0-fold (P < 0.07)- and 2.6-fold (P < 0.04)-higher percentages of total infected cells in Th0- and Th17-polarized cells, respectively (Fig. 3B and C). Sorted, polarized cells from four donors were infected with HIVIIIB, and another three were infected with transmitted-founder HIVAD17 and analyzed. Th17 polarization increased the percentage of HIV-infected cells among all three HIV isolates (Fig. 3B).
Similar to PHA-activated cells, Th17 cells from T cells that were sorted based on their expression of CCR6 and activated with TCR stimulation were preferential targets for productive HIV infection. Both IL-17+ and CCR6+ cells exhibited a higher percentage of HIV-infected cells relative to total cells, regardless of the viral isolate used. The increasing percentage of IL-17+ cells among p24+ cells resulting from Th17 polarization was proportional to that seen in total T cells. However, Th17 polarization resulted in decreased percentages of IFN-γ+ cells among infected cells (Fig. 3B and C).
To address the possibility that viruses produced under different conditions may differ in infectivity, we added a standardized p24 concentration (5 ng/ml) from supernatants of sorted, polarized, HIVBaL-infected cells to TZM-BL cells. TZM-BL cells express luciferase, dependent on HIV long terminal repeat (LTR) promoter-driven transcriptional activity (Fig. 3D). HIV-dependent luciferase activity was measured in triplicate samples of supernatants. To correct for luciferase activity that could be due to cellular effects resulting from differences in the volume or polarizing treatments of supernatant added, we subtracted the luciferase activity of uninfected control supernatants from that of their corresponding infected samples. Two of the donors showed increased luciferase activity (mean, 4.7-fold-higher relative light units) in samples treated with supernatants from Th17-polarized cells, relative to Th0-polarized cells (Fig. 3D).
Th17 cells are highly permissive to productive HIV infection by a pseudotyped HIV vector, HIVAMLV.
To control for the contribution of entry-level (susceptibility) differences to the HIV infection of Th17 cells, we infected PHA-activated or Th0/Th17-polarized cells with an amphotropic murine leukemia virus (AMLV)-pseudotyped HIV. HIVAMLV lacks the Env gene, thus preventing completion of the viral replication cycle. However, HIVAMLV is complemented with an AMLV envelope, which utilizes the widely expressed PiT2 receptor for cell entry, and can therefore infect many mammalian cell types. Despite using an HIV vector that enters cells independently of HIV receptors, the nonspreading infection still produced a disproportionately high percentage of productively infected IL-17+ cells (Fig. 4).
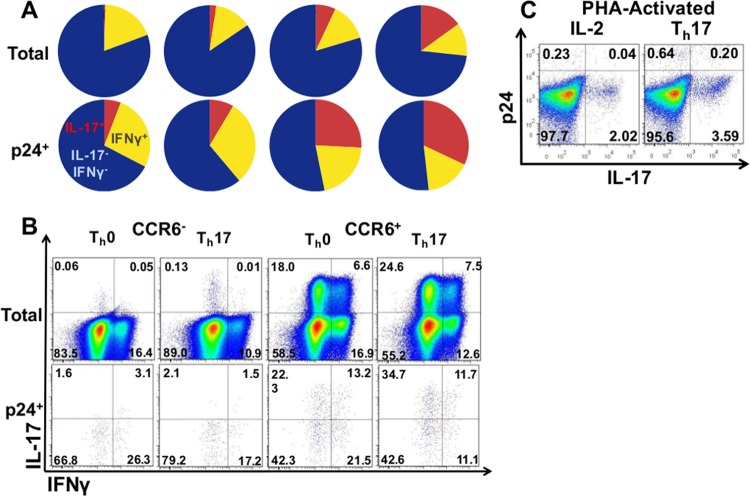
FIG 4.
IL-17 and IFN-γ expression among CCR6-sorted, prepolarized cells infected with a pseudotype HIV vector, HIVAMLV. (A) Pie charts of average percentages of IL-17- and IFN-γ-expressing cells among CCR6-sorted cells that were Th0 or Th17 polarized and then infected with an Env-deficient HIV vector in an amphotropic murine leukemia virus (AMLV) envelope for 4 days. Total and p24+-gated cells from 5 donors are shown. (B) Flow cytometry plots from the donor whose cells became most Th17 polarized. (C) Representative plots of IL-17 and p24 expression in phytohemagglutinin (PHA)-activated CD4 T cells infected for 4 days with HIVAMLV in the presence of IL-2 or Th17-polarizing cytokines.
IL-17+ cells were consistently more likely to become productively infected by HIVAMLV than IL-17− or IFN-γ+ cells, regardless of polarizing conditions. On average, Th17 cells were overrepresented among p24+ cells by 2.9-fold (n = 5, Student's t test, P < 0.001; median, 2.7-fold, and range, 1.5- to 22.1-fold increases in frequency of IL-17+ cells among p24+ cells, relative to total T cells). IFN-γ+ IL-17− cells were also more likely to become productively infected by HIVAMLV but to a lesser degree (Fig. 4A and B) (1.6-fold enrichment in p24+ cells, P < 0.03; median, 1.4-fold; range, −0.3- to 5.5-fold). When added postinfection to PHA-activated T cells, Th17-polarizing cytokines increased the percentage of productively infected cells by 262% (Fig. 4C, n = 2).
HIV replicates more efficiently in IL-17-expressing cells.
Our hypothesis that Th17 cells are more permissive to HIV replication predicts that IL-17-expressing cells will produce more virus than IL-17− cells. In our flow cytometry experiments, we noticed that infected Th17 cells tended to have a higher fluorescence intensity of p24. Therefore, we compared the geometric mean intensities of p24 from HIVBaL-infected Th17 cells and IL-17− cells.
Indeed, upon analysis of the p24 fluorescence intensity among IL-17+ cells under PHA-activating conditions, we found a significant increase in cell-associated p24 compared with IL-17− cells (Fig. 5A) (P < 0.04) or IFN-γ+ cells (data not shown; P < 0.03).
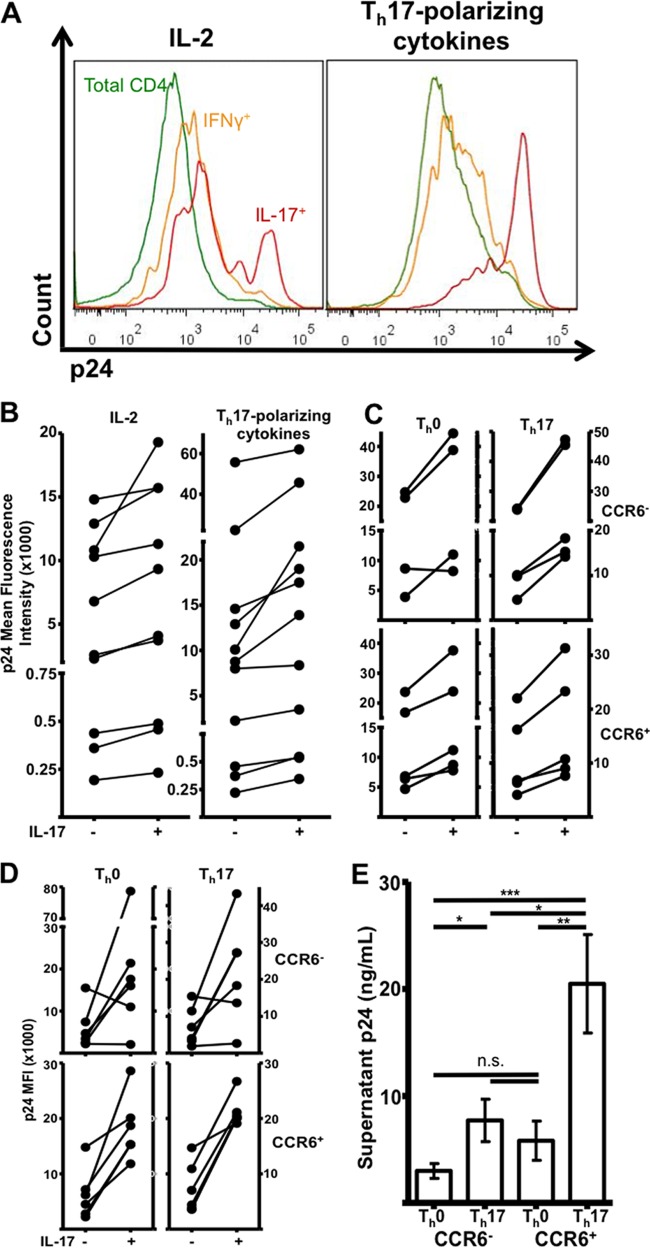
FIG 5.
p24 mean fluorescence intensities (MFIs) among infected IL-17+ and IL-17− cells and supernatant p24 from sorted, polarized cells infected with HIVBaL. (A and B) Representative histograms (A) and donor-matched plots (B) of p24 geometric MFIs among IL-17+ and IL-17−, PHA-activated cells that were infected with HIVBaL for 5 days in the presence of IL-2 or Th17-polarizing cytokines. (C and D) p24 MFI plots from sorted, polarized cells that were infected with HIVBaL (C) or HIVAMLV (D). (E) Supernatant levels of p24 5 days postinfection, as measured by enzyme-linked immunosorbent assay (ELISA; n = 5). *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001. n.s., not significant.
Prepolarized cells exhibited the same trend, regardless of CCR6 expression or polarizing conditions (Fig. 5B). Infections with HIVIIIB and transmitted-founder HIVAD17 produced similar results (Fig. 3B and data not shown).
Moreover, HIVAMLV-infected Th17 cells had higher p24 mean fluorescence intensities (MFIs) (Fig. 5D). IL-17+ CD4 T cells, independently of their CCR6 expression or polarizing conditions, tended to have a higher p24 MFI than p24+ IL-17− cells. This was especially true for CCR6+-sorted cells, which had 2.9-fold (P < 0.01)- and 3.1-fold (P < 0.008)-higher p24 MFIs than IL-17− cells. Th17 polarization generally increased overall p24 MFI, but this trend did not reach statistical significance, perhaps due to a lower n (P < 0.09). PHA-activated T cells exhibited a similar trend. The addition of Th17-polarizing cytokines during infection also resulted in heightened p24 MFI among IL-17+ cells and tended to increase overall p24 fluorescence intensity, regardless of IL-17 expression (Fig. 4C and 5A [P < 0.02] and data not shown).
To determine whether Th17-polarized cells also produced more secreted virus, we infected equal numbers of sorted, polarized cells and then subjected the supernatants to p24 ELISA and viral titer analyses. Consistent with our data suggesting that HIV replicates more efficiently in Th17 cells, the supernatant p24 level was highest from Th17-polarized cells. Among the five donors tested, we detected an average 6.9-fold increase (P < 0.0008) in supernatant p24 from infected, Th17-polarized, CCR6+ cells. This increase was relative to donor-matched, Th0-polarized CCR6− cells, which were relatively devoid of Th17 cells (mean percents IL-17+ were 30.9 and 0.4, respectively). Supernatant p24 levels from CCR6+ Th17-polarized cells were also 2.7 (P < 0.02)- and 3.5 (P < 0.007)-fold higher than those from donor-matched Th17-polarized, CCR6− cells and Th0-polarized CCR6+ cells, respectively (Fig. 5E).
Thus, Th17-polarized cells inherently produce larger amounts of secreted, infectious virus than do Th0-polarized cells.
Th17-polarized cells have diminished expression of RNase A genes and proteins.
Next, we attempted to identify changes in gene expression among Th17-polarized cells that could potentially promote viral replication intracellularly. To confirm that our sorting and polarization conditions produced meaningful changes in gene expression, we performed a microarray analysis of RNA extracted from CCR6−, Th0-polarized cells and donor-matched CCR6+, Th17-polarized cells (Fig. 6A).
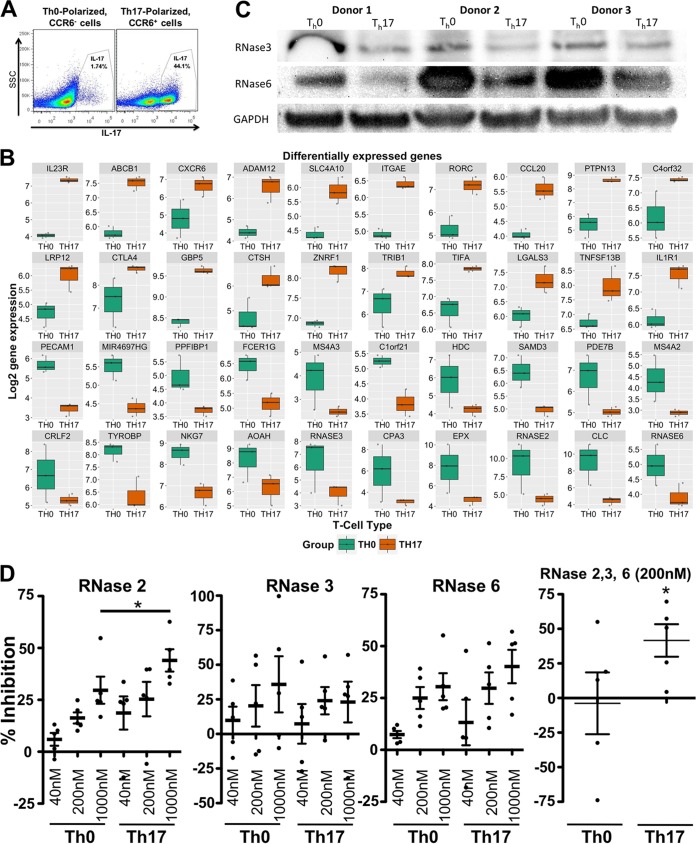
FIG 6.
Comparison of RNase expression and HIV inhibition in Th17-polarized cells with those in Th0-polarized cells. (A) Representative flow cytometry plots depicting IL-17 expression among Th0-polarized, CCR6− cells and Th17-polarized, CCR6+ cells. SSC, side scatter. (B) Box plot showing differential gene expression between Th0- and Th17-polarized cells. Total RNA from Th0- or Th17-polarized cells was analyzed by using microarrays. Relative levels of gene expression in Th17-polarized cells and Th0-polarized cells are shown. (C) Thirty micrograms of total protein from cell lysates of CCR6−, Th0-polarized and CCR6+, Th17-polarized CD4 T cells from three donors was denatured and separated according to size by SDS-PAGE, transferred to a PVDF membrane, and then probed for the indicated RNase expression with anti-RNase antibodies. The loading control was glyceraldehyde 3-phosphate dehydrogenase (GAPDH). (D) Th0- and Th17-polarized CD4 T cells were infected with HIVIIIB, washed, and then resuspended in medium containing the indicated RNases. Percent inhibition was calculated from the percentage of p24+ cells, as measured by flow cytometry.
After correction for multiple testing, our differential gene expression analysis shows that genes associated with the Th17 phenotype, such as IL-23 receptor, retinoic acid receptor (RAR)-related orphan receptor C (RORγc), and MIP-3α/CCL20, are significantly upregulated in the Th17-polarized population. Gene set enrichment analyses (GSEAs) revealed an immunologic signature in the Th17-polarized cells consistent with genes upregulated in memory CD4 T cells compared to naive cells (false-discovery rate [FDR] q value of <0.001 [48]) with a consistent pattern for downregulated genes. GSEA reactome analyses showed a signature comparable to those of reported reactomes for HIV infection (FDR = 0.023), of host interaction of HIV factors (FDR = 0.0031), and of vif-mediated degradation of APOBEC3G (FDR = 0.024), indicating that the Th17 cells are enriched in factors that favor HIV replication. Analyses of signaling pathways showed that Th17-polarized cells upregulated genes that are myc targets (FDR = 0.006), by tumor necrosis factor alpha (TNF-α) signaling via NF-κB (FDR = 0.02).
Among the genes that were most significantly downregulated in the Th17-polarized cells, we noticed two members of the RNase A family known to have antiviral activity, i.e., RNase 2 (Fig. 6B, log fold change [logFC] = −4.928, adjusted P value = 0.003) and RNase 3 (logFC = −2.533, adjusted P value = 0.02). A third member of the same family which had not previously been tested against HIV, RNase 6, had a relatively weaker downregulation and false-discovery rate (Fig. 6B) (logFC = −1.081, adjusted P value = 0.12). Known restriction factors, such as APOBEC3G and SamHD1, were not differentially expressed in Th17-polarized cells from the three donors tested (data not shown).
Based on these findings, we harvested protein from CCR6-sorted, polarized cells and compared their levels of expression of RNases 2, 3, and 6 by immunoblotting. While RNase 2 was not in detected in activated T cells with the antibody that we used, RNases 3 and 6 were expressed at higher levels in the Th0-polarized cells than in donor-matched cells from Th17-polarizing conditions. The steady-state RNase 3 and RNase 6 protein expression levels were 57% and 41%, respectively, lower in Th17-polarized cells (n = 5, P < 0.05) (Fig. 6C). Thus, our microarray data and confirmatory immunoblotting assays revealed a sharp decrease in the expression of RNase 3 and RNase 6 upon Th17 polarization.
We next activated CD4 T cells from 5 donors in the presence of IL-2 or Th17-polarizing cytokines, infected cells with HIVIIIB, washed them, resuspended them in medium, and then added recombinant human RNases (Fig. 6D). We measured p24 expression by flow cytometry, comparing RNase-treated cells with untreated controls.
Recombinant RNases 2 and 6 significantly inhibit HIV infection at 200 nM and 1 μM, respectively (Fig. 6D) (P < 0.05 at all concentrations). At the doses that we tested, RNase 3 did not significantly inhibit HIV infection in either Th0- or Th17-polarized cells. Since RNase 3 had been previously tested and found to be antiviral, we postulate that the dose of the protein that we used was not sufficient to inhibit HIV under our assay conditions. Nonetheless, the inhibition of HIV by RNases 2 and 6 was most pronounced in Th17-polarized cells. When we tested RNase 2, the heightened inhibition in Th17-polarized cells reached statistical significance (P < 0.05) compared to Th0-polarized cells. A similar trend was observed in RNase 6-treated cells. To our knowledge, these are the first data showing HIV inhibition by human RNase 6. When we tested all three RNases at 200 nM each, significant HIV inhibition was observed only in Th17-polarized cells.
DISCUSSION
Here, we show that human peripheral blood CD4+ T cells that express IL-17 are consistently more likely to become productively infected by HIV than IL-17− or IFN-γ+ cells regardless of activation method, polarizing conditions, or viral tropism (Fig. 1 to 4). Further, our data indicate that Th17 cells not only are more susceptible to HIV entry but also mediate enhanced viral replication and production compared with IL-17− CD4+ T cells.
In their original work establishing the preferential loss of Th17 cells in HIV-infected patients, Brenchley et al. noted that peripheral blood Th17 cells were not preferentially infected compared to Th1 cells, as measured by the copy number of integrated provirus in stimulated CD27+ memory T cells (1). This seems to contradict the heightened susceptibility and permissiveness that we observed, but it is likely that cells with similar rates of successful viral integration could vary in their production of viral progeny (49). Consistent with this explanation, infected Th17-polarized cells yielded higher levels of intracellular and supernatant p24 than did Th0-polarized cells (Fig. 5D). Our use of peripheral rather than GALT CD4+ T cells may account for differences in infectivity rates via changes in receptor expression and/or cellular activation. It is also possible that unintegrated viral DNA might contribute to p24 expression (50).
The design of our Th17 enrichment strategy was based on another study (17), in which both IL-2 and IL-23 were added to the medium after polarization for all conditions, in order to maintain both Th17 and non-Th17 cells and ensure an appropriate comparison during the subsequent infection.
Th17 depletion occurs early during acute HIV infection, and the mechanisms of the loss remain unclear, possibly including direct cytocidal effects, pyroptosis, and apoptosis (51–53). The loss of these cells compromises mucosal barriers and undermines the capacity of our immune system to defend against AIDS-associated ailments (4–7, 26, 54). The Th17 cytokines IL-17 and IL-22 synergistically induce the expression of antimicrobial peptides (55). Our lab has shown that the antimicrobial peptide beta-defensin 2, which is robustly expressed in the oral mucosa of healthy individuals, is markedly decreased in HIV-infected subjects (56), possibly related to the loss of Th17 cells. Notably, combined antiretroviral therapy initiated early during acute HIV infection prevents the loss of Th17 cell numbers and polyfunctionality and reverses systemic and gut immune activation (3).
Our data suggest that Th17 polarization could promote infection by two mechanisms. First, factors expressed in Th17 cells (including transcription factors) might directly promote HIV replication. In accordance, a sequence analysis of several HIV-1 LTRs suggested the presence of putative RORγt consensus sequence binding sites (data not shown). If functional, these binding sites raise the possibility that the expression of proviral HIV may be directly modulated by RORγt activity. Additionally, STAT3 has been shown to promote HIV replication in human neonatal cord-blood mononuclear cells, and short hairpin RNA inhibition of STAT3 diminished HIV gene expression (57). Relevantly, the HIV-1 accessory protein Transactivator of Transcription (Tat) activates STAT3 in infected antigen-presenting cells, thereby driving the expression of Th17-polarizing cytokines (58). HIV-pulsed dendritic cells also activate STAT3 in T cells in vitro (59). GP120 and Nef activate STAT3 as well. Thus, HIV directly activates the Th17-related transcription factor STAT3 and supports the production of Th17-polarizing cytokines.
Second, Th17 polarization might inhibit the expression of necessary antiviral effectors (60). Our studies revealed that members of the RNase A superfamily were among the most strongly downregulated genes in Th17-polarized cells; RNase 3 and RNase 6 protein expression was also diminished (Fig. 6C). Although the regulation of the expression of these RNases is poorly understood in T cells, members of the GATA family of transcription factors promote their expression in eosinophils (61). GATA3, a key transcription factor for the differentiation of Th2 cells, is cross-inhibited by other differentiation pathways (62). Th17 cells and other subsets may inhibit GATA transcription factors to stabilize non-Th2 differentiation pathways, thereby inhibiting the expression of RNases.
The novel observation that RNase 6 inhibits HIV but is expressed at lower levels in Th17-polarized cells than in Th0-polarized cells provides new mechanistic insight into the unique vulnerability of Th17 cells to HIV infection. Further studies are needed to characterize the HIV-inhibitory activity of RNase 6 and to measure function in vivo.
Our findings suggest that RNases may constitute an important defense against the HIV infection of CD4 T cells (as depicted in Fig. 7). Members of the RNase A superfamily directly inhibit a broad range of pathogens, can be secreted, and also modulate our immune system. Of the eight human RNase proteins that have been functionally characterized, four had already been shown to have HIV-inhibitory activity (63). The secretion of RNase 4 and RNase 5/angiogenin by T cells was recently identified as a mechanism of HIV inhibition (64). Recombinant human RNase 1, RNase 2, and RNase 5 each inhibit the HIV infection of primary human T cells in vitro, whether added before or after infection. The mechanisms of viral inhibition remain unclear, but antibodies to either RNase blocked HIV-inhibitory activity (65).
FIG 7.
Potential mechanisms and effects of Th17 permissiveness during acute HIV infection. Heightened permissiveness to HIV in response to Th17- or Tfh-polarizing cytokines may promote viral replication by direct transcriptional modulation of proviral and/or antiviral response genes. The subsequent loss of Th17 cells could be attributable to programmed cell death, conversion to Tfh cells, or the cytopathic effects of high virus production. The loss of Th17 cells then promotes the disruption of mucosal immunity. APC, antigen-presenting cell.
One caveat of our study was its dependence on peripheral blood as the source of T cells. The GALT and vaginal mucosae represent unique anatomical compartments in which local antigens, cytokines, chemokines, and localization factors such as integrins differ from those present in blood. Whether or not GALT Th17 cells also demonstrate such proneness to productive HIV infection relative to other subsets remains unclear. However, our findings are consistent with an in vivo SIV transmission study showing that vaginal mucosal Th17 cells, which made up less than one-fifth of total CD4+ T cells, accounted for over four-fifths of productively infected cells (66).
A second caveat is that our CCR6-sorted cells were cultured with both IL-2 and IL-23 after TCR stimulation and polarization. We found that IL-23 was necessary to preserve the Th17 subset during infection, but IL-23 may selectively activate Th17 cells relative to other subsets in our culture system. Nonetheless, Th17 cells from our PHA/IL-2-activated cells (which were never treated with IL-23) showed disproportionately higher rates of infection than IFN-γ+ cells and also heightened p24 MFI (Fig. 1, 2, 4, and 5). Furthermore, IL-2 is an inhibitor of Th17 function, and we are comparing the infection of IL-17+ cells with that of IFN-γ+ cells, which are also highly activated.
If IL-23 contributes to increased p24 production within Th17 cells, the IL-23R-mediated activation of STAT3, which is activated in both Th17 and Tfh cells, should be further investigated for its potential role in promoting productive HIV infection (67). Such a finding would strengthen our argument that Th17 and Tfh differentiation (and unique activation pathways) are specifically linked to increased HIV permissiveness.
Our results provide deeper insight into the mechanisms underlying the preferential targeting of Th17 cells during HIV infection, compared with other CD4+ T cell subsets. In our model, given their relative abundance at sites of HIV sexual transmission, Th17 cells constitute a major source of viral production during acute infection (Fig. 7). Our data suggest that the transcriptional and translational environment of Th17 cells may directly promote the expression of HIV proteins. It is also possible that infected Th17 cells directly become Tfh cells (32, 33), which would then be capable of migration to secondary lymphoid tissues (Fig. 7), where they are capable of replication at high levels, despite their low expression of CCR5 (68, 69).
Our findings may aid in therapeutic intervention strategies aimed at preventing viral production during acute HIV infection or reducing the size of the viral reservoir in people living with HIV.
ACKNOWLEDGMENTS
For their helpful technical advice and cell sorting services, we thank Yutaka Tagaya of the Institute of Human Virology Flow Cytometry Core facility and Ferenc Livak at the Greenebaum Cancer Center Flow Cytometry Core. We are grateful to Matthew Frieman, Wuyuan Lu, Acshah Keegan, and Terez Shea-Donohue for their suggestions that helped guide our experimental approach. We also thank the AIDS Reagent Program for supplying cell lines and viral isolates.
This work was supported by the National Institute of Neurological Disorders and Stroke (award R01NS066842; principal investigator, A.G.-D.). Aaron Christensen-Quick was a trainee under institutional training grant 1T32AI095190-01A1 from the National Institute of Allergy and Infectious Diseases. Mark Lafferty was a trainee under NIAID institutional training grant T32AI007540.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, the National Institute of Neurological Disorders and Stroke, or the National Institutes of Health.
A.C.-Q. performed the majority of experiments and wrote the manuscript. M.L. and L.S. performed experiments and provided helpful suggestions and manuscript edits. L.M. analyzed microarray data, provided expert advice for its interpretation, and edited the manuscript. A.D. provided HIVAD17 stocks, designed research, and edited the manuscript. A.G.-D. designed the research, analyzed data, and edited the manuscript.
The authors have no conflicts of interest to declare.
REFERENCES
- 1.Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, Scheinberg P, Price DA, Hage CA, Kholi LM, Khoruts A, Frank I, Else J, Schacker T, Silvestri G, Douek DC. 2008. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood 112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raffatellu M, Santos RL, Verhoeven DE, George MD, Wilson RP, Winter SE, Godinez I, Sankaran S, Paixao TA, Gordon MA, Kolls JK, Dandekar S, Baumler AJ. 2008. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med 14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuetz A, Deleage C, Sereti I, Rerknimitr R, Phanuphak N, Phuang-Ngern Y, Estes JD, Sandler NG, Sukhumvittaya S, Marovich M, Jongrakthaitae S, Akapirat S, Fletscher JL, Kroon E, Dewar R, Trichavaroj R, Chomchey N, Douek DC, O'Connell RJ, Ngauy V, Robb ML, Phanuphak P, Michael NL, Excler JL, Kim JH, de Souza MS, Ananworanich J, RV254/SEARCH 010 and RV304/SEARCH 013 Study Groups. 2014. Initiation of ART during early acute HIV infection preserves mucosal Th17 function and reverses HIV-related immune activation. PLoS Pathog 10:e1004543. doi: 10.1371/journal.ppat.1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartigan-O'Connor DJ, Abel K, Van Rompay KK, Kanwar B, McCune JM. 2012. SIV replication in the infected rhesus macaque is limited by the size of the preexisting TH17 cell compartment. Sci Transl Med 4:136ra169. doi: 10.1126/scitranslmed.3003941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cecchinato V, Trindade CJ, Laurence A, Heraud JM, Brenchley JM, Ferrari MG, Zaffiri L, Tryniszewska E, Tsai WP, Vaccari M, Parks RW, Venzon D, Douek DC, O'Shea JJ, Franchini G. 2008. Altered balance between Th17 and Th1 cells at mucosal sites predicts AIDS progression in simian immunodeficiency virus-infected macaques. Mucosal Immunol 1:279–288. doi: 10.1038/mi.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klatt NR, Estes JD, Sun X, Ortiz AM, Barber JS, Harris LD, Cervasi B, Yokomizo LK, Pan L, Vinton CL, Tabb B, Canary LA, Dang Q, Hirsch VM, Alter G, Belkaid Y, Lifson JD, Silvestri G, Milner JD, Paiardini M, Haddad EK, Brenchley JM. 2012. Loss of mucosal CD103+ DCs and IL-17+ and IL-22+ lymphocytes is associated with mucosal damage in SIV infection. Mucosal Immunol 5:646–657. doi: 10.1038/mi.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med 200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thea DM, Porat R, Nagimbi K, Baangi M, St Louis ME, Kaplan G, Dinarello CA, Keusch GT. 1996. Plasma cytokines, cytokine antagonists, and disease progression in African women infected with HIV-1. Ann Intern Med 124:757–762. doi: 10.7326/0003-4819-124-8-199604150-00009. [DOI] [PubMed] [Google Scholar]
- 9.Ilyin SE, Plata-Salaman CR. 1997. HIV-1 envelope glycoprotein 120 regulates brain IL-1beta system and TNF-alpha mRNAs in vivo. Brain Res Bull 44:67–73. doi: 10.1016/S0361-9230(97)00091-9. [DOI] [PubMed] [Google Scholar]
- 10.Ketlinskii SA, Kalinina NM, Tsvetkova SN, Rakhmanova AG, Maslov VN, Khaldeeva NA. 1992. Tumor necrosis factor-alpha and interleukin-1beta in the blood plasma of patients with HIV infection. Vestn Ross Akad Med Nauk (9-10):36–41. (In Russian.) [PubMed] [Google Scholar]
- 11.Creemers PC, O'Shaughnessy M, Boyko WJ. 1986. Increased interleukin-2 receptor expression after mitogen stimulation on CD4- and CD8-positive lymphocytes and decreased interleukin-2 production in HTLV-III antibody-positive symptomatic individuals. Immunology 59:627–629. [PMC free article] [PubMed] [Google Scholar]
- 12.Hirao LA, Grishina I, Bourry O, Hu WK, Somrit M, Sankaran-Walters S, Gaulke CA, Fenton AN, Li JA, Crawford RW, Chuang F, Tarara R, Marco ML, Baumler AJ, Cheng H, Dandekar S. 2014. Early mucosal sensing of SIV infection by Paneth cells induces IL-1beta production and initiates gut epithelial disruption. PLoS Pathog 10:e1004311. doi: 10.1371/journal.ppat.1004311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alfano M, Crotti A, Vicenzi E, Poli G. 2008. New players in cytokine control of HIV infection. Curr HIV/AIDS Rep 5:27–32. doi: 10.1007/s11904-008-0005-5. [DOI] [PubMed] [Google Scholar]
- 14.Kedzierska K, Crowe SM. 2001. Cytokines and HIV-1: interactions and clinical implications. Antivir Chem Chemother 12:133–150. doi: 10.1177/095632020101200301. [DOI] [PubMed] [Google Scholar]
- 15.Louis S, Dutertre CA, Vimeux L, Fery L, Henno L, Diocou S, Kahi S, Deveau C, Meyer L, Goujard C, Hosmalin A. 2010. IL-23 and IL-12p70 production by monocytes and dendritic cells in primary HIV-1 infection. J Leukoc Biol 87:645–653. doi: 10.1189/jlb.1009684. [DOI] [PubMed] [Google Scholar]
- 16.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. 2010. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol 10:11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvarez Y, Tuen M, Shen G, Nawaz F, Arthos J, Wolff MJ, Poles MA, Hioe CE. 2013. Preferential HIV infection of CCR6+ Th17 cells is associated with higher levels of virus receptor expression and lack of CCR5 ligands. J Virol 87:10843–10854. doi: 10.1128/JVI.01838-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirota K, Ahlfors H, Duarte JH, Stockinger B. 2012. Regulation and function of innate and adaptive interleukin-17-producing cells. EMBO Rep 13:113–120. doi: 10.1038/embor.2011.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim HP, Oh J, Tunyaplin C, Carotta S, Donovan CE, Goldman ML, Tailor P, Ozato K, Levy DE, Nutt SL, Calame K, Leonard WJ. 2009. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity 31:941–952. doi: 10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, Crotty S. 2012. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J Immunol 188:3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. 2013. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, Weatherall C, Bailey M, Alcantara S, De Rose R, Estaquier J, Wilson K, Suzuki K, Corbeil J, Cooper DA, Kent SJ, Kelleher AD, Zaunders J. 2013. Simian immunodeficiency virus infects follicular helper CD4 T cells in lymphoid tissues during pathogenic infection of pigtail macaques. J Virol 87:3760–3773. doi: 10.1128/JVI.02497-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boswell KL, Paris R, Boritz E, Ambrozak D, Yamamoto T, Darko S, Wloka K, Wheatley A, Narpala S, McDermott A, Roederer M, Haubrich R, Connors M, Ake J, Douek DC, Kim J, Petrovas C, Koup RA. 2014. Loss of circulating CD4 T cells with B cell helper function during chronic HIV infection. PLoS Pathog 10:e1003853. doi: 10.1371/journal.ppat.1003853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKinnon LR, Nyanga B, Chege D, Izulla P, Kimani M, Huibner S, Gelmon L, Block KE, Cicala C, Anzala AO, Arthos J, Kimani J, Kaul R. 2011. Characterization of a human cervical CD4+ T cell subset coexpressing multiple markers of HIV susceptibility. J Immunol 187:6032–6042. doi: 10.4049/jimmunol.1101836. [DOI] [PubMed] [Google Scholar]
- 25.Mayuzumi H, Inagaki-Ohara K, Uyttenhove C, Okamoto Y, Matsuzaki G. 2010. Interleukin-17A is required to suppress invasion of Salmonella enterica serovar Typhimurium to enteric mucosa. Immunology 131:377–385. doi: 10.1111/j.1365-2567.2010.03310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pallikkuth S, Micci L, Ende ZS, Iriele RI, Cervasi B, Lawson B, McGary CS, Rogers KA, Else JG, Silvestri G, Easley K, Estes JD, Villinger F, Pahwa S, Paiardini M. 2013. Maintenance of intestinal Th17 cells and reduced microbial translocation in SIV-infected rhesus macaques treated with interleukin (IL)-21. PLoS Pathog 9:e1003471. doi: 10.1371/journal.ppat.1003471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El Hed A, Khaitan A, Kozhaya L, Manel N, Daskalakis D, Borkowsky W, Valentine F, Littman DR, Unutmaz D. 2010. Susceptibility of human Th17 cells to human immunodeficiency virus and their perturbation during infection. J Infect Dis 201:843–854. doi: 10.1086/651021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monteiro P, Gosselin A, Wacleche VS, El-Far M, Said EA, Kared H, Grandvaux N, Boulassel MR, Routy JP, Ancuta P. 2011. Memory CCR6+CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin beta7. J Immunol 186:4618–4630. doi: 10.4049/jimmunol.1004151. [DOI] [PubMed] [Google Scholar]
- 29.Gosselin A, Monteiro P, Chomont N, Diaz-Griffero F, Said EA, Fonseca S, Wacleche V, El-Far M, Boulassel MR, Routy JP, Sekaly RP, Ancuta P. 2010. Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection. J Immunol 184:1604–1616. doi: 10.4049/jimmunol.0903058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunay GA, Toth I, Eberhard JM, Degen O, Tolosa E, van Lunzen J, Hauber J, Schulze Zur Wiesch J. 2015. Parallel assessment of Th17 cell frequencies by surface marker coexpression versus ex vivo IL-17 production in HIV-1 infection. Cytometry B Clin Cytom doi: 10.1002/cyto.b.21352. [DOI] [PubMed] [Google Scholar]
- 31.Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, Foucat E, Dullaers M, Oh S, Sabzghabaei N, Lavecchio EM, Punaro M, Pascual V, Banchereau J, Ueno H. 2011. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity 34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirota K, Turner JE, Villa M, Duarte JH, Demengeot J, Steinmetz OM, Stockinger B. 2013. Plasticity of Th17 cells in Peyer's patches is responsible for the induction of T cell-dependent IgA responses. Nat Immunol 14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu KT, Kanno Y, Cannons JL, Handon R, Bible P, Elkahloun AG, Anderson SM, Wei L, Sun H, O'Shea JJ, Schwartzberg PL. 2011. Functional and epigenetic studies reveal multistep differentiation and plasticity of in vitro-generated and in vivo-derived follicular T helper cells. Immunity 35:622–632. doi: 10.1016/j.immuni.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Bar KJ, Wang S, Decker JM, Chen Y, Sun C, Salazar-Gonzalez JF, Salazar MG, Learn GH, Morgan CJ, Schumacher JE, Hraber P, Giorgi EE, Bhattacharya T, Korber BT, Perelson AS, Eron JJ, Cohen MS, Hicks CB, Haynes BF, Markowitz M, Keele BF, Hahn BH, Shaw GM. 2010. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog 6:e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ray K, Mengistu M, Yu L, Lewis GK, Lakowicz JR, DeVico AL. 2014. Antigenic properties of the HIV envelope on virions in solution. J Virol 88:1795–1808. doi: 10.1128/JVI.03048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCall MN, Bolstad BM, Irizarry RA. 2010. Frozen robust multiarray analysis (fRMA). Biostatistics 11:242–253. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leek JT, Storey JD. 2007. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 3:1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kortenhorst MS, Wissing MD, Rodriguez R, Kachhap SK, Jans JJ, Van der Groep P, Verheul HM, Gupta A, Aiyetan PO, van der Wall E, Carducci MA, Van Diest PJ, Marchionni L. 2013. Analysis of the genomic response of human prostate cancer cells to histone deacetylase inhibitors. Epigenetics 8:907–920. doi: 10.4161/epi.25574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross AE, Marchionni L, Phillips TM, Miller RM, Hurley PJ, Simons BW, Salmasi AH, Schaeffer AJ, Gearhart JP, Schaeffer EM. 2011. Molecular effects of genistein on male urethral development. J Urol 185:1894–1898. doi: 10.1016/j.juro.2010.12.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross AE, Marchionni L, Vuica-Ross M, Cheadle C, Fan J, Berman DM, Schaeffer EM. 2011. Gene expression pathways of high grade localized prostate cancer. Prostate 71:1568–1577. doi: 10.1002/pros.21373. [DOI] [PubMed] [Google Scholar]
- 42.Poli G, Bressler P, Kinter A, Duh E, Timmer WC, Rabson A, Justement JS, Stanley S, Fauci AS. 1990. Interleukin 6 induces human immunodeficiency virus expression in infected monocytic cells alone and in synergy with tumor necrosis factor alpha by transcriptional and post-transcriptional mechanisms. J Exp Med 172:151–158. doi: 10.1084/jem.172.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Harthi L, Roebuck KA, Kessler H, Landay A. 1997. Inhibition of cytokine-driven human immunodeficiency virus type 1 replication by protease inhibitor. J Infect Dis 176:1175–1179. doi: 10.1086/514110. [DOI] [PubMed] [Google Scholar]
- 44.Alvarez Y, Tuen M, Nàdas A, Hioe CE. 2012. In vitro restoration of Th17 response during HIV infection with an antiretroviral drug and Th17 differentiation cytokines. AIDS Res Hum Retroviruses 28:823–834. doi: 10.1089/aid.2011.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rani A, Greenlaw R, Runglall M, Jurcevic S, John S. 2014. FRA2 is a STAT5 target gene regulated by IL-2 in human CD4 T cells. PLoS One 9:e90370. doi: 10.1371/journal.pone.0090370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foli A, Saville MW, Baseler MW, Yarchoan R. 1995. Effects of the Th1 and Th2 stimulatory cytokines interleukin-12 and interleukin-4 on human immunodeficiency virus replication. Blood 85:2114–2123. [PubMed] [Google Scholar]
- 47.Gonzalez MW, DeVico AL, Lewis GK, Spouge JL. 2015. Conserved molecular signatures in gp120 are associated with the genetic bottleneck during simian immunodeficiency virus (SIV), SIV-human immunodeficiency virus (SHIV), and HIV type 1 (HIV-1) transmission. J Virol 89:3619–3629. doi: 10.1128/JVI.03235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abbas AR, Wolslegel K, Seshasayee D, Modrusan Z, Clark HF. 2009. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One 4:e6098. doi: 10.1371/journal.pone.0006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee GQ, Lichterfeld M. 2016. Diversity of HIV-1 reservoirs in CD4+ T-cell subpopulations. Curr Opin HIV AIDS doi: 10.1097/COH.0000000000000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jurriaans S, de Ronde A, Dekker J, Cornelissen M, Goudsmit J. 1995. Increased number of single-LTR HIV-1 DNA junctions correlates with HIV-1 antigen expression and CD4+ cell decline in vivo. J Med Virol 45:91–98. doi: 10.1002/jmv.1890450117. [DOI] [PubMed] [Google Scholar]
- 51.Cooper A, Garcia M, Petrovas C, Yamamoto T, Koup RA, Nabel GJ. 2013. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature 498:376–379. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- 52.Doitsh G, Galloway NL, Geng X, Yang Z, Monroe KM, Zepeda O, Hunt PW, Hatano H, Sowinski S, Munoz-Arias I, Greene WC. 2014. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Somasundaran M, Robinson HL. 1988. Unexpectedly high levels of HIV-1 RNA and protein synthesis in a cytocidal infection. Science 242:1554–1557. doi: 10.1126/science.3201245. [DOI] [PubMed] [Google Scholar]
- 54.Ryan ES, Micci L, Fromentin R, Paganini S, McGary CS, Easley K, Chomont N, Paiardini M. 2016. Loss of function of intestinal IL-17 and IL-22 producing cells contributes to inflammation and viral persistence in SIV-infected rhesus macaques. PLoS Pathog 12:e1005412. doi: 10.1371/journal.ppat.1005412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA. 2006. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med 203:2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun L, Finnegan CM, Kish-Catalone T, Blumenthal R, Garzino-Demo P, La Terra Maggiore GM, Berrone S, Kleinman C, Wu Z, Abdelwahab S, Lu W, Garzino-Demo A. 2005. Human beta-defensins suppress human immunodeficiency virus infection: potential role in mucosal protection. J Virol 79:14318–14329. doi: 10.1128/JVI.79.22.14318-14329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sundaravaradan V, Mehta R, Harris DT, Zack JA, Ahmad N. 2010. Differential expression and interaction of host factors augment HIV-1 gene expression in neonatal mononuclear cells. Virology 400:32–43. doi: 10.1016/j.virol.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 58.Zeng Y, Zhang X, Huang Z, Cheng L, Yao S, Qin D, Chen X, Tang Q, Lv Z, Zhang L, Lu C. 2007. Intracellular Tat of human immunodeficiency virus type 1 activates lytic cycle replication of Kaposi's sarcoma-associated herpesvirus: role of JAK/STAT signaling. J Virol 81:2401–2417. doi: 10.1128/JVI.02024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Che KF, Shankar EM, Muthu S, Zandi S, Sigvardsson M, Hinkula J, Messmer D, Larsson M. 2012. p38 mitogen-activated protein kinase/signal transducer and activator of transcription-3 pathway signaling regulates expression of inhibitory molecules in T cells activated by HIV-1-exposed dendritic cells. Mol Med 18:1169–1182. doi: 10.2119/molmed.2012.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cleret-Buhot A, Zhang Y, Planas D, Goulet JP, Monteiro P, Gosselin A, Wacleche VS, Tremblay CL, Jenabian MA, Routy JP, El-Far M, Chomont N, Haddad EK, Sekaly RP, Ancuta P. 2015. Identification of novel HIV-1 dependency factors in primary CCR4(+)CCR6(+)Th17 cells via a genome-wide transcriptional approach. Retrovirology 12:102. doi: 10.1186/s12977-015-0226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qiu Z, Dyer KD, Xie Z, Radinger M, Rosenberg HF. 2009. GATA transcription factors regulate the expression of the human eosinophil-derived neurotoxin (RNase 2) gene. J Biol Chem 284:13099–13109. doi: 10.1074/jbc.M807307200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vahedi G, Poholek AC, Hand TW, Laurence A, Kanno Y, O'Shea JJ, Hirahara K. 2013. Helper T-cell identity and evolution of differential transcriptomes and epigenomes. Immunol Rev 252:24–40. doi: 10.1111/imr.12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rugeles MT, Trubey CM, Bedoya VI, Pinto LA, Oppenheim JJ, Rybak SM, Shearer GM. 2003. Ribonuclease is partly responsible for the HIV-1 inhibitory effect activated by HLA alloantigen recognition. AIDS 17:481–486. doi: 10.1097/00002030-200303070-00002. [DOI] [PubMed] [Google Scholar]
- 64.Cocchi F, DeVico AL, Lu W, Popovic M, Latinovic O, Sajadi MM, Redfield RR, Lafferty MK, Galli M, Garzino-Demo A, Gallo RC. 2012. Soluble factors from T cells inhibiting X4 strains of HIV are a mixture of beta chemokines and RNases. Proc Natl Acad Sci U S A 109:5411–5416. doi: 10.1073/pnas.1202240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bedoya VI, Boasso A, Hardy AW, Rybak S, Shearer GM, Rugeles MT. 2006. Ribonucleases in HIV type 1 inhibition: effect of recombinant RNases on infection of primary T cells and immune activation-induced RNase gene and protein expression. AIDS Res Hum Retroviruses 22:897–907. doi: 10.1089/aid.2006.22.897. [DOI] [PubMed] [Google Scholar]
- 66.Stieh DJ, Matias E, Xu H, Fought AJ, Blanchard JL, Marx PA, Veazey RS, Hope TJ. 2016. Th17 cells are preferentially infected very early after vaginal transmission of SIV in macaques. Cell Host Microbe 19:529–540. doi: 10.1016/j.chom.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmitt N, Liu Y, Bentebibel SE, Munagala I, Bourdery L, Venuprasad K, Banchereau J, Ueno H. 2014. The cytokine TGF-beta co-opts signaling via STAT3-STAT4 to promote the differentiation of human TFH cells. Nat Immunol 15:856–865. doi: 10.1038/ni.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu H, Wang X, Malam N, Aye PP, Alvarez X, Lackner AA, Veazey RS. 2016. Persistent simian immunodeficiency virus infection drives differentiation, aberrant accumulation, and latent infection of germinal center follicular T helper cells. J Virol 90:1578–1587. doi: 10.1128/JVI.02471-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Allam A, Majji S, Peachman K, Jagodzinski L, Kim J, Ratto-Kim S, Wijayalath W, Merbah M, Kim JH, Michael NL, Alving CR, Casares S, Rao M. 2015. TFH cells accumulate in mucosal tissues of humanized-DRAG mice and are highly permissive to HIV-1. Sci Rep 5:10443. doi: 10.1038/srep10443. [DOI] [PMC free article] [PubMed] [Google Scholar]