ABSTRACT
Vaccinia virus (VACV) decapping enzymes and cellular exoribonuclease Xrn1 catalyze successive steps in mRNA degradation and prevent double-stranded RNA (dsRNA) accumulation, whereas the viral E3 protein can bind dsRNA. We showed that dsRNA and E3 colocalized within cytoplasmic viral factories in cells infected with a decapping enzyme mutant as well as with wild-type VACV and that they coprecipitated with antibody. An E3 deletion mutant induced protein kinase R (PKR) and eukaryotic translation initiation factor alpha (eIF2α) phosphorylation earlier and more strongly than a decapping enzyme mutant even though less dsRNA was made, leading to more profound effects on viral gene expression. Human HAP1 and A549 cells were genetically modified by clustered regularly interspaced short palindromic repeat-Cas9 (CRISPR-Cas9) to determine whether the same pathways restrict E3 and decapping mutants. The E3 mutant replicated in PKR knockout (KO) HAP1 cells in which RNase L is intrinsically inactive but only with a double knockout (DKO) of PKR and RNase L in A549 cells, indicating that both pathways decreased replication equivalently and that no additional dsRNA pathway was crucial. In contrast, replication of the decapping enzyme mutant increased significantly (though less than that of wild-type virus) in DKO A549 cells but not in DKO HAP1 cells where a smaller increase in viral protein synthesis occurred. Xrn1 KO A549 cells were viable but nonpermissive for VACV; however, wild-type and mutant viruses replicated in triple-KO cells in which RNase L and PKR were also inactivated. Since KO of PKR and RNase L was sufficient to enable VACV replication in the absence of E3 or Xrn1, the poor replication of the decapping mutant, particularly in HAP1 DKO, cells indicated additional translational defects.
IMPORTANCE Viruses have evolved ways of preventing or counteracting the cascade of antiviral responses that double-stranded RNA (dsRNA) triggers in host cells. We showed that the dsRNA produced in excess in cells infected with a vaccinia virus (VACV) decapping enzyme mutant and by wild-type virus colocalized with the viral E3 protein in cytoplasmic viral factories. Novel human cell lines defective in either or both protein kinase R and RNase L dsRNA effector pathways and/or the cellular 5′ exonuclease Xrn1 were prepared by CRISPR-Cas9 gene editing. Inactivation of both pathways was necessary and sufficient to allow full replication of the E3 mutant and reverse the defect cause by inactivation of Xrn1, whereas the decapping enzyme mutant still exhibited defects in gene expression. The study provided new insights into functions of the VACV proteins, and the well-characterized panel of CRISPR-Cas9-modified human cell lines should have broad applicability for studying innate dsRNA pathways.
INTRODUCTION
Double-stranded RNA (dsRNA) is a principal viral pathogen-associated molecular pattern that is recognized by cellular sensors, including oligoadenylate synthetase (OAS), protein kinase R (PKR), Toll-like receptors, retinoic acid-inducible gene-I (RIG-I)-like receptors, and nucleotide-binding oligomerization domain (NOD)-like receptors, resulting in activation of RNase L, phosphorylation of eukaryotic translation initiation factor alpha (eIF2α), and induction of interferon and proinflammatory responses (1–3). Many viruses produce dsRNA at some stage of their life cycles. Poxviruses are vulnerable to dsRNA pathways because of the synthesis of complementary transcripts that can anneal to form dsRNA (4, 5). Approximately 15% of the polyadenylated RNA synthesized by late times after infection with vaccinia virus (VACV), the prototype of the poxvirus family, can anneal to form long intermolecular duplexes with single-stranded RNA tails (6).
Viruses mitigate host responses to dsRNA by preventing its formation, sequestering it, degrading it, or interfering with sensing or effector pathways (2, 7). Poxviruses, including VACV, encode numerous proteins that protect against a variety of innate defenses including those triggered by dsRNA (8–10). The VACV E3 dsRNA binding protein plays an important role: mutations in the C-terminal dsRNA binding domain result in increased interferon sensitivity and a severe host range defect involving activation of PKR, RNase L, and interferon regulatory factor 3 (IRF3) (11–17). Roles of PKR and RNase L pathways were suggested by partially restoring replication of a VACV E3 deletion mutant in PKR- or RNase L-deficient mouse embryo fibroblasts (16). Knockdown (KD) of PKR substantially restored replication of E3 mutants in HeLa cells (18). Nevertheless, the mode of action of E3 and the relative roles of different dsRNA pathways in antagonizing E3 mutants are incompletely understood. Although binding of E3 to dsRNA has been demonstrated in vitro (11), the association of E3 with dsRNA in poxvirus-infected cells has not been reported. Moreover, mutations in the C-terminal region of E3 that affect dsRNA binding in vitro do not uniformly correlate with the host range function (19). In addition, the N-terminal region of E3 can interact directly with PKR (20, 21), and both the N- and C-terminal regions of E3 are required for virulence in mice (22, 23). The inactivation of another VACV protein, K3, results in enhanced interferon sensitivity and host range restriction in baby hamster kidney cells (24, 25). K3 has homology with eIF2α and competitively prevents its phosphorylation by PKR (26–28).
All poxviruses encode one or two enzymes with Nudix hydrolase motifs that can cleave the 5′ cap of mRNAs to form 5′ phosphate ends (29, 30). In VACV, these enzymes are encoded by the D9 and D10 open reading frames (ORFs), which are 25% identical in predicted amino acid sequence. Inactivation of the D9 gene has no discernible effect on VACV replication, whereas inactivation of the D10 gene reduces virus yields (31). In contrast, simultaneous mutation of both D9 and D10 causes a severe host restriction, suggesting that the two enzymes have compensatory functions (5). The defect in replication is characterized by greatly increased amounts of dsRNA and activation of RNase L and PKR, with resulting inhibition of viral protein synthesis and degradation of host mRNA and rRNA. Knockdown of the cellular 5′ exoribonuclease Xrn1 decreases VACV replication (32), resulting in defects similar to those of the D9/D10 deletion mutant (33), which suggests that decapping enzymes and Xrn1 catalyze successive steps in VACV mRNA degradation.
The present study was undertaken to compare the apparently complementary roles of E3 and D9/D10 in preventing innate immune responses and to determine whether the same host responses are important for restricting VACV replication of both mutants and cells defective in Xrn1 expression. Here, we describe results obtained using wild-type and mutant VACVs to infect human cell lines that were modified by clustered regularly interspaced short palindromic repeat-Cas9 (CRISPR-Cas9) editing of PKR, RNase L, and Xrn1 genes individually and together.
MATERIALS AND METHODS
Antibodies and chemicals.
Mouse monoclonal antibodies (MAbs) to E3 (34) and RNase L (35) were kind gifts of S. N. Isaacs (University of Pennsylvania) and R. H. Silverman (Cleveland Clinic Lerner Research Institute), respectively. Rabbit antibodies to VACV strain WR (36), A3, and D13 (37) were prepared by our laboratory. The following antibodies were purchased: phospho-(Ser396)-IRF3 (catalog number 4947), IRF3 (catalog number 11904), phospho-(Ser51)-eIF2α (catalog number 3398), and eIF2α (catalog number 5324) (all from Cell Signaling Technology); phospho (T446)-PKR (catalog number ab32036; Abcam); PKR (catalog number 700286; Life Technologies); mouse J2 MAb to dsRNA (SCICONS English and Scientific Consulting Kft); mouse anti-FLAG M2 (catalog number F1804) and actin (catalog number A2066) (both from Sigma); Xrn1 (catalog number A300-443A; Bethyl Laboratories). The 0.5- to 10-kb RNA ladder (catalog number 15623-200) was from Life Technologies. Cytosine β-d-arabinofuranoside (AraC) (catalog number C1768) was from Sigma.
Cells.
BHK-21 (ATCC CCL-10) cells were grown in minimum essential medium with Earle's balanced salts (EMEM) supplemented with 2 mM l-glutamine, 100 units of penicillin, and 100 μg of streptomycin per ml (Quality Biologicals, Inc.) and containing 10% fetal bovine serum (FBS) (Sigma-Aldrich). HAP1 control and genetically modified cell lines obtained from Horizon Discovery (Vienna) were grown in Iscove's modified Dulbecco's medium (Life Technologies) supplemented with 2 mM l-glutamine, 10% FBS, 100 units of penicillin, and 100 μg of streptomycin per ml. A549 control and modified cell lines were grown in Dulbecco's modified Eagle's medium/F-12 (DMEM/F-12) medium (Life Technologies) supplemented with 2 mM l-glutamine, 10% FBS, 100 units of penicillin, and 100 μg of streptomycin per ml. The HAP1 RNase L genetically modified cells had a 13-bp deletion in exon 2 causing a frameshift, and the PKR-modified cells had a 13-bp deletion in exon 4 causing a frameshift.
Generation of A549 CRISPR-Cas9 genetically modified cell lines.
To inactivate the RNase L, PKR, and XRN1 genes in A549 cells (ATCC CCL-185), the CRISPR-Cas9 system was adapted from a previous method (38). Targeting sequences were designed using the Web-based tool CRISPR Design (http://crispr.mit.edu/). The following target sequences were used: RNase L, CACGTCCTCCAGCGGTAGAA; PKR, ATTCAGGACCTCCACATGAT; Xrn1, GTATCCCTGTCTCAGCGAAG. The DNA sequences were synthesized (Eurofins, Luxembourg) and separately introduced into the plasmid vector pSpCas9(BB)-2A-GFP (where GFP is green fluorescent protein) from Feng Zhang (PX458, plasmid 48138; Addgene, Cambridge, MA), which drives expression of the Streptococcus pyogenes Cas9, GFP, and the chimeric guide RNA in mammalian cells. A549 cells were transfected with the recombinant plasmids using Lipofectamine 2000 (Life Technologies). Two days after transfection, single GFP-positive cells were sorted by flow cytometry to allow single-colony formation. After 14 days, total protein was prepared from individual colonies, and the absence of proteins was confirmed by Western blotting. The RNase L and PKR CRISPR-Cas9 cell line was generated by inactivating the PKR gene in the RNase L-inactivated cells. Genome sequencing indicated the addition of an A residue (shown in italics in CACGTCCTCCAGCGGTAAGAA) within the target sequence of the RNase L gene, a T residue (shown in italics in ATTCAGGACCTCCACATTGAT) within the target sequence of the PKR gene, and a deletion of two GC residues (indicated by boldface in GTATCCCTGTCTCAGCGAAG) within the target sequence of the Xrn1 gene.
Viruses.
Recombinant VACVs were derived from the Western Reserve (WR) strain of VACV (ATCC VR-1354). The vΔE3L virus (ΔE3L-F17R-venus) (39) was a gift of J. H. Connor (Boston University). The vD9muD10mu double mutant, with catalytic site mutations of D9 and D10, and the wild-type vD10rev (a revertant of a D10 mutant) viruses were previously described (5). The vFLAG-D9 and vD10-FLAG were made with a FLAG tag at their N and C termini, respectively (40). The vΔE3L-venus/FLAG-D9 and vΔE3L-venus/D10-FLAG viruses were constructed by homologous recombination in BHK-21 cells coinfected with 5 PFU/cell of vΔE3L-venus and 5 PFU/cell of vFLAG-D9 or vD10-FLAG. The vD10rev-E3-GFP and vD9muD10mu-E3-GFP viruses were constructed by adding the GFP ORF to the C terminus of E3 by homologous recombination with DNA generated by overlap PCR containing the GFP ORF flanked by portions of E3L and E2L. After 24 h, cells were harvested and subjected to three freeze-thaw cycles, and the lysates were diluted and used to infect new BHK-21 cell monolayers. Fluorescent plaques were picked and sequenced to identify the recombinant viruses, which were clonally purified five times. The inserted regions of the recombinant viruses were PCR amplified and sequenced to confirm their identities.
Purification of virus particles.
Recombinant viruses grown in BHK-21 cells were purified by centrifugation through a 36% sucrose cushion, followed by centrifugation through a 24% to 40% sucrose gradient, as described previously (40), and used for infections after determination of infectivity by plaque assay in BHK-21 cells.
Plaque assay and immunostaining.
VACV preparations were disrupted in a chilled water bath sonicator with two 30-s periods of vibration, followed by 10-fold serial dilutions in EMEM supplemented with 2.5% FBS. Diluted viruses were distributed to BHK-21 cell monolayers. After adsorption for 2 h, the medium was removed, and cells were covered by EMEM supplemented with 2.5% FBS and 0.5% methylcellulose. After incubation for 2 days, the cells were fixed with 1:1 methanol-acetone, washed, and incubated with anti-VACV strain WR polyclonal antibody for 2 h. Cells were washed again and incubated with peroxidase-conjugated protein A (Thermo Scientific) for 1 h, followed by the addition of the substrate dianisidine for 10 min. After plaques were visualized, the dianisidine was removed, and cells were washed.
Immunofluorescence.
Cells were seeded onto glass coverslips and mock infected or infected with 5 PFU/cell of purified virus for 13 h, after which the cells were fixed with 4% formaldehyde for 15 min and permeabilized with 0.1% Triton X-100. Samples were then blocked in 4% FBS, incubated with primary antiserum, and incubated with fluorescence-conjugated secondary antibodies (Life Technologies). Nuclei and virus factories were stained with 4′,6′-diamidino-2-phenylindole (DAPI). Coverslips were mounted on slides using ProLong Gold (Life Technologies). Images were collected on a Leica DMI6000 confocal microscope (Leica Microsystems). Images were collected and processed using Imaris, version 7.1 (Bitplane AG), and Adobe Photoshop CS3 (Adobe Systems) to adjust brightness.
Intracellular staining of dsRNA for flow cytometry.
Cells were mock infected or infected with 5 PFU/cell of virus for 13 h. The cells were fixed with intracellular (IC) fixation buffer (Life Technologies) on ice for 15 min, washed, and incubated with dsRNA J2 MAb in IC permeabilization buffer (Life Technologies) on ice for 30 min, followed by incubation with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibody (BD Biosciences) for 30 min. After being extensively washed with IC permeabilization buffer, the cells were resuspended in phosphate-buffered saline (PBS)–1% paraformaldehyde and then analyzed on a FACSCalibur (BD Biosciences). The positive cells were gated by comparison to uninfected cells, and the results are shown in histograms.
dsRNA IP.
Approximately 106 A549 cells were mock infected or infected with 5 PFU/cell of purified vD9muD10mu-E3-GFP. At 13 h after infection, cells were washed twice with cold PBS on ice, harvested by scraping, and lysed in immunoprecipitation (IP) buffer (20 mM Tris [pH 7.4], 150 mM NaCl, 2 mM EDTA, 1% Triton X-100) containing Complete EDTA-free protease inhibitor cocktail tablets (Roche) and 100 U/ml RNase inhibitor (Fermentas) on wet ice for 30 min with frequent agitation. Lysates were centrifuged for 10 min at 13,000 × g at 4°C, and the supernatant was collected. The supernatant was incubated with 2 μg of J2 MAb for 3 h at 4°C on a rotating wheel and then with 20 μl of protein G conjugated to Dynabeads magnetic beads (Thermo Fisher Scientific) for 3 h at 4°C. The beads were then washed four times with IP buffer, and proteins were eluted by boiling in reducing sample buffer.
Western blotting.
Cells were harvested, washed, and lysed in Tris-buffered saline (TBS) composed of 20 mM Tris (pH 7.4), 150 mM NaCl, and 2 mM EDTA and containing 1% Triton X-100 and Complete EDTA-free protease inhibitor cocktail tablets (Roche) on wet ice for 30 min with frequent agitation. Cell lysates were cleared by centrifugation at 13,000 × g for 10 min at 4°C; the proteins were resolved on 4 to 12% NuPAGE Bis-Tris gels (Life Technologies) and transferred to a nitrocellulose membrane with an iBlot2 system (Life Technologies). The membrane was blocked with 5% nonfat milk in TBS for 1 h, washed with TBS with 0.1% Tween 20 (TBST), and then incubated with the primary antibody in 5% nonfat milk or 5% bovine serum albumin (BSA) in TBST overnight at 4°C. The membrane was washed with TBST and incubated with the secondary antibody conjugated with horseradish peroxidase (Jackson ImmunoResearch) in TBST with 5% nonfat milk for 1 h. After the membrane was washed, the bound protein was detected with SuperSignal West Dura substrates (Thermo Scientific). For detection of phosphorylated eIF2α, PKR, and IRF3, cells were lysed with buffer containing phosphatase inhibitors as follows: 2 mM sodium orthovanadate, 2 mM sodium pyrophosphate, 50 mM glycerol-2-phosphate disodium, 50 mM sodium fluoride, and a phosphatase inhibitor mixture tablet (Roche).
Northern blotting.
Total RNA from infected cells was isolated using an RNeasy minikit (Qiagen), resolved by electrophoresis on a glyoxal gel (Life Technologies), transferred to a positively charged nylon membrane using an iBlot system (Life Technologies), and fixed by UV cross-linking. Double-stranded DNA probes to C11, F17, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) labeled with digoxigenin (DIG) were prepared with a DIG-High Prime DNA labeling and detection starter kit II (Roche) using a 400-bp PCR fragment. After hybridization with the digoxigenin-labeled probe for 20 h, the membrane was washed, blocked, incubated with alkaline phosphatase-conjugated antibody to digoxigenin (Roche), and visualized with a chemiluminescence substrate on X-ray films.
Transmission electron microscopy.
The cells were fixed with 2% glutaraldehyde and embedded in Embed 812 resin (Electron Microscopy Sciences, Hatfield, PA) as described previously (41). Specimens were viewed with a FEI Tecnai Spirit transmission electron microscope (FEI, Hillsboro, OR).
RESULTS
Colocalization of dsRNA and the E3 protein in virus factories.
The role of the VACV E3 protein during infection has been inferred from in vitro dsRNA binding studies, usually carried out with poly(I·C), as well as by activation of dsRNA pathways in cells infected with mutant viruses. E3 had previously been localized in cytoplasmic viral factories and the nucleus (42), but colocalization with dsRNA had not been demonstrated. In our previous study (5), we used a MAb specific for dsRNA of more than 40 bp (43–46) to visualize dsRNA foci by fluorescence microscopy in cells infected with VACV. Based on total fluorescence intensity, the amount of dsRNA in monkey BS-C-1 cells infected with vD9muD10mu, which has inactivating point mutations in the catalytic sites of both D9 and D10 (29, 30), was approximately 15-fold greater than that in cells infected with vD10rev (a virus generated as a revertant of a D10 mutant and used as the wild-type control) (5). Using dsRNA and E3 MAbs of different isotypes and DAPI to stain DNA, a strict coincidence of dsRNA and E3 in cytoplasmic DNA factories was visualized by confocal microscopy of human A549 cells infected with vD9muD10mu (Fig. 1A). DNA factories are closely juxtaposed to nuclei, and one was enlarged in Fig. 1A to more clearly show the site of the dsRNA. The dsRNA also colocalized with E3 in cells infected with vD10rev; however, there were fewer dsRNA foci, and some with E3 staining had barely detectable dsRNA (Fig. 1A). Only tiny dsRNA puncta, which were also associated with cytoplasmic factories, were detected in cells infected with vΔE3L (Fig. 1A).
FIG 1.
Colocalization of dsRNA and E3 protein in infected cells. (A) A549 cells on coverslips were mock infected or infected with 5 PFU per cell of vD10rev, vD9muD10mu, or vΔE3L. After 13 h, the cells were fixed, permeabilized, and stained for dsRNA with J2 MAb (green) and for E3 with E3 MAb (red), followed by isotype-specific IgG2A and IgG3 fluorescent secondary antibodies, respectively, and DAPI. The cells were imaged by confocal microscopy. Arrows point to virus factories. The factory region within the boxed area was enlarged and is shown in the row below. Scale bar, 10 μm. (B) A549 cells were infected as described for panel A, stained with J2 MAb and fluorescent secondary antibody, and analyzed by flow cytometry. (C) The experiment is the same as that described for panel A except that cells were mock infected or infected with vD10rev-E3-GFP or vD9muD10mu-E3-GFP and stained with J2 MAb to detect dsRNA (red) followed by secondary fluorescent antibodies. E3 was visualized by GFP fluorescence. The factory region in the boxed area was enlarged and is shown in the row below. (D) A549 cells were infected with vD9muD10mu-E3-GFP, and lysates were analyzed by Western blotting with antibody to GFP and RIG-I or immunoprecipitated (IP) with J2 MAb or nonspecific IgG and then analyzed by Western blotting.
Flow cytometry was used to quantitatively compare the numbers of cells with detectable dsRNA and the relative amounts of dsRNA in mock-infected A549 cells and cells infected with vD10rev, vD9muD10mu, and vΔE3L. The numbers of dsRNA-positive cells were similar in cells infected with vD9muD10mu and vΔE3L although the mean fluorescence intensity was much higher in the former (Fig. 1B). A smaller number of cells infected with vD10rev scored positive for dsRNA, but the fluorescence intensity of a minority overlapped with that of cells infected with vD9muD10mu and was higher than that of cells infected with vΔE3L (Fig. 1B). Therefore, the low levels of dsRNA in individual cells infected with vΔE3L could explain the small sizes of dsRNA puncta. However, the large number of dsRNA-positive cells infected with vΔE3L compared to that in cells infected with vD10rev was unexpected and implied that E3 might contribute to the suppression of dsRNA accumulation. We will show later that synthesis of both decapping enzymes was severely reduced in the absence of E3, possibly leading to the increased number of cells with small amounts of dsRNA.
The above confocal microscopy experiments were carried out with different isotype mouse MAbs to dsRNA and E3. Although we confirmed the extremely low cross-reactivity between the isotype-specific secondary fluorescent antibodies (data not shown), we wanted to confirm the colocalization data by an alternate method involving E3 fused to GFP. Two recombinant viruses were constructed, vD10rev-E3-GFP and vD9muD10mu-E3-GFP, with GFP fused to the C terminus of E3. The ability of vD10rev-E3-GFP to replicate in human cells (data not shown) provided evidence for the function of the fusion protein. Colocalization of dsRNA and E3 in virus factories was visualized using the J2 MAb to detect dsRNA, and GFP fluorescence was used to detect E3. Again, colocalization of dsRNA and E3 occurred in cells infected with both the wild-type virus and the decapping enzyme mutant (Fig. 1C). The association of dsRNA with a virus factory is shown by the enlarged image in one row of Fig. 1C.
vD9muD10mu-E3-GFP was also used to demonstrate interaction of dsRNA with E3 by coimmunoprecipitation. The J2 MAb but not an IgG isotype control antibody was able to capture E3-GFP, which was detected by Western blotting (Fig. 1D). A small amount of RIG-I, another dsRNA binding protein, was immunoprecipitated under these conditions (Fig. 1D). Thus, our data supported the role of E3 in binding dsRNA during VACV infection and suggest a possible secondary or indirect role in preventing dsRNA accumulation.
Replicative abilities of mutant viruses in CRISPR-Cas9 RNase L and PKR KO cells.
The decapping enzyme mutant vD9muD10mu and E3 deletion mutant have similar host range defects, and both can replicate in BHK-21 cells, which were used for their propagation (5, 16). The present study was intended to compare causal relationships between cytoplasmic dsRNA sensor pathways and the host restriction of vD9muD10mu and vΔE3L by inactivation of RNase L and PKR gene expression. Our previous studies were carried out mainly with monkey BS-C-1 cells; however, human cells have advantages for studying innate immunity and for genetic modification. Since cell lines can vary in the extent to which they constitutively express proteins involved in innate immune pathways (47, 48), we chose two human cell lines, HAP1 and A549, for genome modification. HAP1 is an adherent fibroblast-like cancer cell that has been developed as a research tool because it is haploid, simplifying gene modification (49). A549 is a commonly used continuous hypotriploid alveolar epithelial carcinoma cell (50). Initial experiments verified that both human parental cell lines were permissive for wild-type virus but nonpermissive for vD9muD10mu and vΔE3L.
To evaluate the differences in innate immune responses between the HAP1 and A549 cells and their effects on VACV replication, CRISPR-Cas9 was used to genetically inactivate RNase L and PKR individually or together. Inactivation of RNase L and PKR gene expression was demonstrated by Western blotting with antibody to the encoded proteins (Fig. 2A and C) and confirmed by DNA sequencing (see Materials and Methods). Because of the absence of detectable PKR or RNase L protein, we refer to the CRISPR-Cas9 cell lines as knockouts (KOs). The parental control and genetically modified cell lines were infected with vD10rev, vD9muD10mu, and vΔE3L, and virus titers were determined at 3 h (residual input virus) and 24 h (one-step yield). In the absence of replication, the titer at 24 h decreases from the input value. As expected, vD10rev replicated well in the control parental as well as in the KO HAP1 and A549 cells (Fig. 2B and D). The vΔE3L mutant was unable to replicate in the control or RNase L HAP1 KO cells but replicated in PKR KO cells and in HAP1 cells with a double knockout (DKO) of RNase L and PKR (PKR+RNase L DKO) (Fig. 2B). In contrast, the vΔE3L mutant replicated in DKO but not in single PKR or RNase L KO A549 cells (Fig. 2D). We will show later in this report that RNase L is not activated following VACV infection of HAP1 cells, as determined by the stability of rRNA. The absence of activated RNase L explains the ability of vΔE3L to replicate in PKR as well as in DKO HAP1 cells. The inability of vΔE3L to replicate in either single PKR or single RNase L KO A549 cells indicated that activation of either pathway blocks VACV replication in the absence of E3. Furthermore, the ability of vΔE3L to replicate in PKR HAP1 and PKR+RNase L DKO A549 cells indicated that no additional dsRNA pathways severely interfered with VACV replication under one-step growth conditions.
FIG 2.
Replication of decapping enzyme mutant and E3 deletion virus in HAP1 and A549 KO cells. (A) Absence of RNase L and PKR proteins in HAP1 KO cells. Cell lysates from HAP1 control, RNase L KO, PKR KO, and DKO cells were analyzed by Western blotting using mouse polyclonal antibody to RNase L and rabbit MAb to PKR. Antibody to actin was used as a loading control. (B) One-step virus replication. HAP1 control, RNase L KO, PKR KO, and DKO cell monolayers in 12-well plates were infected in triplicate with 5 PFU/cell of purified vD10rev, vD9muD10mu, or vΔE3L and harvested at 3 and 24 h. Virus titers were determined by plaque assay in BHK-21 cells. The 3-h titers represent input virus. (C and D) Procedures were the same as those described for panels A and B except that A549 control and KO cells were used. Each bar represents the standard deviation determined from three replicate infections.
The titers of the vD9muD10mu mutant increased slightly between 3 and 24 h in the PKR KO and DKO HAP1 cells but not in the control or RNase L KO cells (Fig. 2B). However, the increases did not reach high significance (P = 0.069 and P = 0.053 for PKR KO and DKO cells, respectively). The vD9muD10mu mutant increased in titer between 3 and 24 h in the DKO A549 cells (Fig. 2D). Although this increase was significant (P = 0.005), it was much less than that of vΔE3L. The results of these and other infectivity experiments are summarized in Table 1.
TABLE 1.
Virus replication in KO cell lines
| Cell line | Virus replicationa |
||
|---|---|---|---|
| vD10Rev | vD9muD10mu | vΔE3L | |
| HAP1 | |||
| Controlb | + | − | − |
| PKR KO | + | − | + |
| RNase L KO | + | − | − |
| PKR+RNase L DKO | + | − | + |
| A549 | |||
| Control | + | − | − |
| PKR KO | + | − | − |
| RNase L KO | + | − | − |
| PKR+RNase L DKO | + | +c | + |
| Xrn1 KO | − | − | − |
| Xrn1+PKR+RNase L TKO | + | +c | + |
Cells were harvested at 24 h after infection, and virus titrations were performed by plaque assay in permissive BHK-21 cells. The presence (+) or absence (−) of replication is indicated.
Infected HAP1 parental cells had undetectable RNase L activity.
The increase in virus titer was significant but less than that of vD10rev.
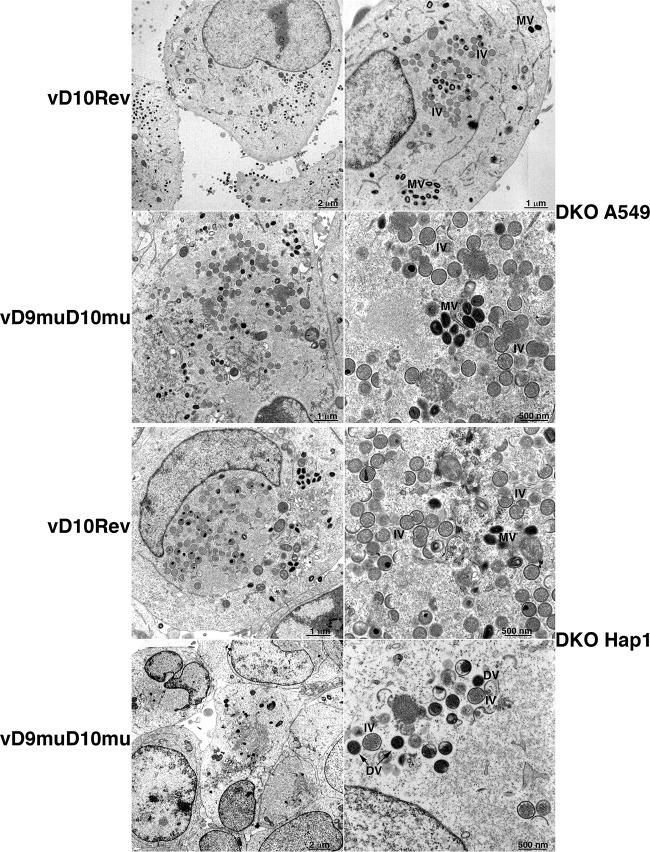
Transmission electron microscopy was performed to compare the replication defect of vD9muD10mu in DKO HAP1 and A549 cells. Large numbers of immature and mature virions were present in both DKO cell lines infected with vD10rev (Fig. 3). Immature and mature virions were also present in DKO A549 cells infected with vD9muD10mu. In contrast, mature brick-shaped virions were not seen in DKO HAP1 cells infected with vD9muD10mu; instead, low numbers of abnormal spherical, dense particles were present, consistent with the block in infectious virus formation.
FIG 3.
Transmission electron microscopy. DKO A549 (upper two rows) and HAP1 cells (lower two rows) were infected with 5 PFU per cell of vD10rev or vD9muD10mu and harvested after 13 h. The cells were fixed, cut into thin sections, and examined by transmission electron microcopy. IV, immature virion; MV, mature virion; DV, aberrant dense virion.
Further experiments were carried out to determine the basis for the restriction of replication of the decapping enzyme and E3 mutants and the roles of PKR and RNase L. For clarity, we first describe experiments with HAP1 PKR and RNase L KO cells, then with A549 PKR and RNase L KO cells, and finally with A549 Xrn1 KO cells.
Viral gene expression in HAP1 KO cells.
Time course experiments were carried out to investigate the differential replication of vD10rev, vΔE3L, and vD9muD10mu in HAP1 KO cells. Lysates, obtained at 2, 4, 8, and 12 h after infection, were analyzed by probing Western blots with broadly reactive VACV antibodies that recognize abundant postreplicative proteins (Fig. 4A) and with antibody to actin, which verified similar levels of gel loading (data not shown). In cells infected with vD10rev, major viral proteins were detected as faint bands at 4 h and increased at 8 and 12 h in control and RNase L KO cell lines, with only a slight enhancement at 4 h in the PKR and DKO cells. Faint bands were detected in the presence of cytosine β-d-arabinofuranoside (AraC), an inhibitor of DNA replication, confirming that the abundant proteins made in the absence of drug belonged to the intermediate or late class. In contrast to the results obtained with the wild-type vD10rev, viral protein synthesis in the control HAP1 cells was low or barely detectable throughout the course of infection with vD9muD10mu or vΔE3L, respectively. Viral protein synthesis was not increased in the HAP1 RNase L KO cells infected with either mutant virus, consistent with the inability of such cells to support their replication. However, in both PKR KO and DKO HAP1 cells there was a dramatic increase in wild-type viral protein levels after infection with vΔE3L and a lesser increase after infection with vD9muD10mu. The low level of viral protein synthesis in DKO HAP1 cells infected with vD9muD10mu would account for the defect in formation of mature virus particles and infectious virions.
FIG 4.
Viral gene expression in HAP1 KO cells. (A to D) Western blotting. Control, RNase L KO, PKR KO, and DKO HAP1 cells were infected with 5 PFU/cell of purified vD10rev, vD9muD10mu, or vΔE3L and harvested at the indicated times (shown in hours). The lysates were analyzed by Western blotting using antibodies to VACV (A), the E3 early protein (B), the D13 intermediate protein (C), and the A3 late protein (D). Antibody to actin served as a loading control (not shown). One set of cells was infected in the presence of AraC to inhibit viral DNA replication and confirm the stage of expression of the viral proteins. (E) Northern blotting. HAP1 control, RNase L KO, PKR KO, and DKO cells were mock infected or infected with 5 PFU/cell of purified vD10rev, vD9muD10mu, or vΔE3L virus. At 13 h after infection, the cells were harvested, and the total RNAs were isolated and resolved on glyoxal gels. rRNAs were stained with ethidium bromide and detected by UV fluorescence; reverse images are shown. The RNAs were transferred to a nylon membrane, incubated with the digoxigenin-labeled probes to the viral early C11 mRNA, viral late F17 mRNA, and cellular GAPDH mRNA, detected with alkaline phosphatase-conjugated antibody to digoxigenin, and visualized with the chemiluminescence substrate and X-ray film. p.i., postinfection; Ab, antibody.
The abilities of the mutant viruses to synthesize early proteins was demonstrated by probing Western blots with antibody to the E3 protein, which was detected from 2 to 12 h after infection and in the presence of AraC (Fig. 4B). The absence of a band in cells infected with vΔE3L confirmed the KO and antibody specificity. Western blots probed with antibodies to the D13 (Fig. 4C) and A3 (Fig. 4D) proteins were used to analyze representative intermediate and late proteins, respectively. D13 and A3 were expressed similarly in all cells infected with vD10rev although there was slightly more present at the 4-h time point in the PKR and DKO cells. The D13 and A3 proteins were faint or undetectable in control and RNase L KO cells infected with vD9muD10mu or vΔE3L, respectively. Expression increased modestly in PKR KO and DKO cells infected with vD9muD10mu and more strongly upon infection with vΔE3L. Thus, the different abilities of vD9muD10mu and vΔE3L to replicate in PKR KO and DKO HAP1 cells corresponded with the levels of viral postreplicative protein synthesis, and the defect of vD9muD10mu was not due to failure to synthesize the early E3 protein.
The rRNAs and representative viral mRNAs of infected HAP1 and HAP1 KO cells were also analyzed. Activated RNase L is an endonuclease that cleaves single-stranded regions within rRNA and mRNA. However, the rRNAs remained intact in control and KO HAP1 cells, whether infected by wild-type vD10rev or mutant VACV, indicating an absence of active RNase L (Fig. 4E). Since RNase L protein was made in HAP1 cells (Fig. 2A), the lack of RNase L activity may be due to low basal levels of OAS, which was detected only after addition of exogenous interferon (data not shown). Early mRNAs are synthesized within virus cores derived from the infecting virions, whereas intermediate and late mRNAs require DNA replication and de novo viral protein synthesis. The early C11 mRNA was detected by Northern blotting of RNA from control and KO cells infected with vD10rev and both VACV mutants (Fig. 4E). The late C17 mRNA was detected in all cells infected with vD10rev and vD9muD10mu. However, KO of PKR or PKR and RNase L was required for detection of F17 mRNA in cells infected with vΔE3L. The absence of F17 mRNA in the control and RNase L KO cells infected with vΔE3L is likely due to the early and profound inhibition of viral protein synthesis, which would include transcription factors. Moreover, the dramatic increase in F17 mRNA in the PKR KO and DKO cells infected with vΔE3L paralleled the increase in viral protein synthesis. In contrast, viral late protein synthesis was less completely inhibited in A549 cells infected with vD9muD10mu, corresponding to the detection of late mRNA.
Previous studies have described greatly accelerated degradation of cellular mRNAs by VACV (51–53), which has recently been attributed to the activity of the decapping enzymes (29). The panel of mutant viruses and KO cell lines allowed us to examine this hypothesis by Northern blotting. At 13 h after infection, GAPDH mRNA (which has been reported to have a half-life of 8 h in HeLa cells [54]) was faint or undetectable in all cells infected with vD10rev, consistent with accelerated host mRNA degradation (Fig. 4E). In cells infected with vD9muD10mu, only a small amount of GAPDH RNA persisted, despite the absence of decapping enzymes. Much more GAPDH mRNA was present in control and RNase L KO cells infected with vΔE3L, in which little or no viral late mRNA or postreplicative protein synthesis was detectable. However, GAPDH mRNA was diminished when viral late mRNA and protein synthesis increased in PKR KO and DKO cells infected with vΔE3L. The decrease in GAPDH mRNA in DKO cells infected with vD9muD10mu suggested that decapping enzymes and RNase L are not the sole causes of the instability of cell mRNA and imply another mechanism dependent on synthesis of additional viral proteins.
PKR and eIF2α activation in HAP1 control and KO cells.
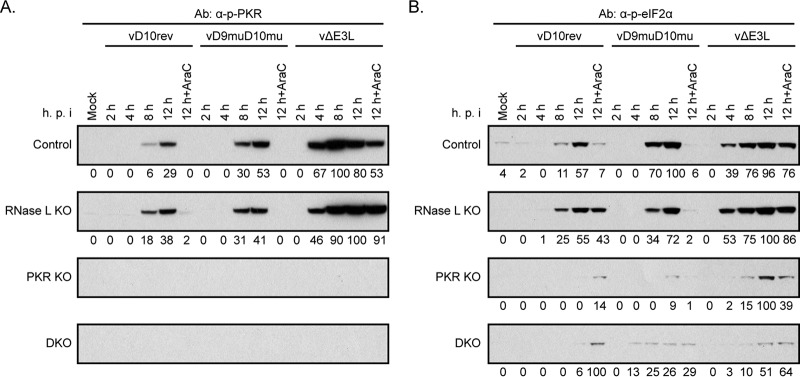
Western blotting experiments confirmed the presence of eIF2α in all cells and PKR in the control and RNase L KO cells but not in PKR KO and DKO cells (data not shown). Phosphorylated forms of PKR and eIF2α were detected with specific antibodies at 8 h, and these increased at 12 h in HAP1 control and RNase L KO cells infected with vD10rev and vD9muD10mu although the bands were somewhat stronger with the latter virus, as indicated by the quantification of their densities (Fig. 5A and B). In control and RNase L KO HAP1 cells infected with vΔE3L, phosphorylation of PKR and eIF2α was intense as early as 4 h and even in the presence of AraC (Fig. 5A and B). The very small amounts of phosphorylated eIF2α bands detected in PKR KO and DKO HAP1 cells (Fig. 5B) were presumably due to membrane-associated PKR-like endoplasmic reticulum kinase (PERK) or other kinases.
FIG 5.
Innate immune responses in HAP1 KO cells. (A) PKR. Control, RNase L KO, PKR KO, and DKO HAP1 cells were infected with 5 PFU/cell of purified vD10rev, vD9muD10mu, or vΔE3L in the absence or presence of AraC and harvested at the indicated hour after infection. The lysates were analyzed by SDS-PAGE and Western blotting using antibodies to phosphorylated PKR (α-p-PKR). Densities shown below the images were determined with ImageJ. (B) eIF2α. A Western blot prepared as described for panel A was probed with antibodies to phosphorylated eIF2α (α-p-eIF2α).
The timing and extent of PKR and eIF2α phosphorylation in HAP1 cells infected with vΔE3L and vD9muD10mu were consistent with the greater inhibition of viral late gene expression after infection with the former. However, the degree of phosphorylation of PKR and eIF2α in control and RNase L KO cells infected with vD10rev compared to that in cells infected with vD9muD10mu was surprising to us in view of the large differences in the levels of dsRNA, gene expression, and replication of the two viruses. These results indicate that relatively small amounts of dsRNA made in cells infected with vΔE3L and vD10rev can activate PKR and eIF2α and that activation at late times of infection by vD10rev does not necessarily block virus replication.
Viral gene expression in A549 CRISPR-Cas9 KO cells.
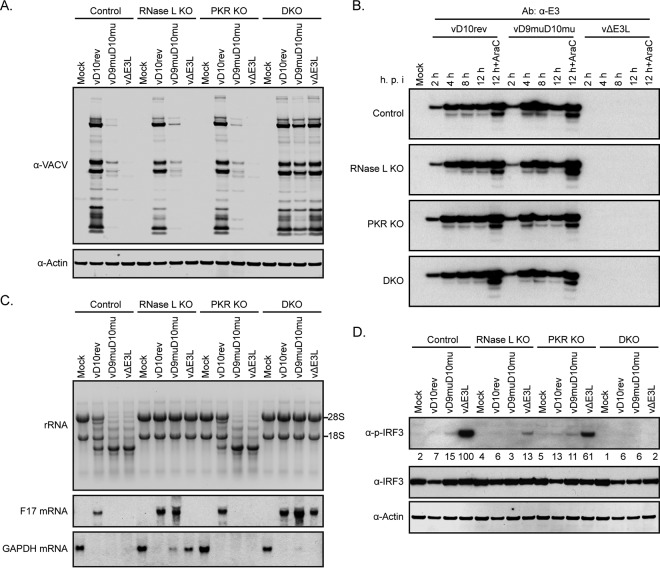
We considered that studies with A549 KO cells would allow us to compare the relative effects of RNase L and PKR on viral protein synthesis, which could not be done in HAP1 cells because of their intrinsic deficiency in RNase L activation. In addition, we sought to further investigate the basis for the improved replication of vD9muD10mu in A549 DKO cells compared to that in HAP1 DKO cells. Lysates of parental control, RNase L KO, PKR KO, and DKO A549 cells that had been infected with vD10rev, vΔE3L, or vD9muD10mu for 13 h were analyzed by Western blotting with VACV antiserum (Fig. 6A). The vD10rev blots appeared very similar regardless of cell type. In contrast, viral proteins were undetectable in control, RNase L KO, and PKR KO cells infected with vΔE3L but were remarkably restored and comparable to levels of vD10rev proteins in DKO cells. Small amounts of viral proteins were detected in control, RNase L, and PKR KO cells infected with vD9muD10mu. There was a substantial increase in viral proteins in A549 DKO cells infected with vD9muD10mu (Fig. 6A), which was greater than that in HAP1 DKO cells (Fig. 4A), although the bands were lighter than those in cells infected with vD10rev or vΔE3L. Thus, RNase L and PKR had equivalent effects on viral protein synthesis, and KO of both was required to enable full replication of vΔE3L and partial replication of vD9muD10mu.
FIG 6.
Viral gene expression and innate immune response in A549 KO cells. (A) Viral protein synthesis. A549 control, RNase L KO, PKR KO, and DKO cells were mock infected or infected with 5 PFU/cell of purified vD10rev, vD9muD10mu, or vΔE3L. At 13 h after infection, the cell lysates were analyzed by Western blotting using rabbit polyclonal antibody to VACV proteins and actin. (B) E3 synthesis. Control and KO A549 cells were mock infected or infected with vD10rev, vD9muD10mu, or vΔE3L in the absence or presence of AraC. The cells were harvested at 2, 4, 8, and 12 h after infection and analyzed by Western blotting with a MAb to E3. (C) rRNA and mRNA. Total RNA was isolated from cells infected as described for panel A and resolved on glyoxal gels. The RNAs were stained with ethidium bromide and detected by UV fluorescence; reverse images are shown. RNAs were transferred from the glyoxal gel to a nylon membrane, incubated with the digoxigenin-labeled probes to the viral late F17 mRNA or cellular GAPDH mRNA, detected with alkaline phosphatase-conjugated antibody to digoxigenin, and visualized with a chemiluminescence substrate on X-ray films. (D) IRF3. Lysates from cells infected as described for panel A were analyzed by Western blotting with antibodies to phosphorylated IRF3, IRF3 protein, and actin. Densities shown below the images were determined with ImageJ.
We wanted to be certain that the effects on viral protein synthesis in A549 cells infected with vD9muD10mu were not related in part to decreased synthesis of the early E3 dsRNA binding protein. Similar amounts of E3 were detected over time by Western blotting with an E3-specific MAb in control and KO cells infected with vD10rev and vD9muD10mu (Fig. 6B). We showed earlier (Fig. 1A) that E3 colocalized with dsRNA in control A549 cells infected with vD9muD10mu. Formation of dsRNA foci also occurred in RNase L KO, PKR KO, and DKO A549 cells infected with vD9muD10mu (data not shown).
Analysis of rRNAs and viral late F17 mRNA provided further insights. Unlike the situation with HAP1 cells (Fig. 4E), cleavage of rRNA was extensive at 13 h after infection of control and PKR KO A549 cells with vD9muD10mu or vΔE3L and moderate after infection with vD10rev (Fig. 6C). The strong activation of RNase L in A549 cells infected with vΔE3L occurred despite the relatively low levels of dsRNA in individual A549 cells, as determined by flow cytometry (Fig. 7A). rRNA remained intact in the RNase L KO and DKO cells infected with wild-type and mutant VACVs, indicating that RNase L was solely responsible for degradation (Fig. 6C).
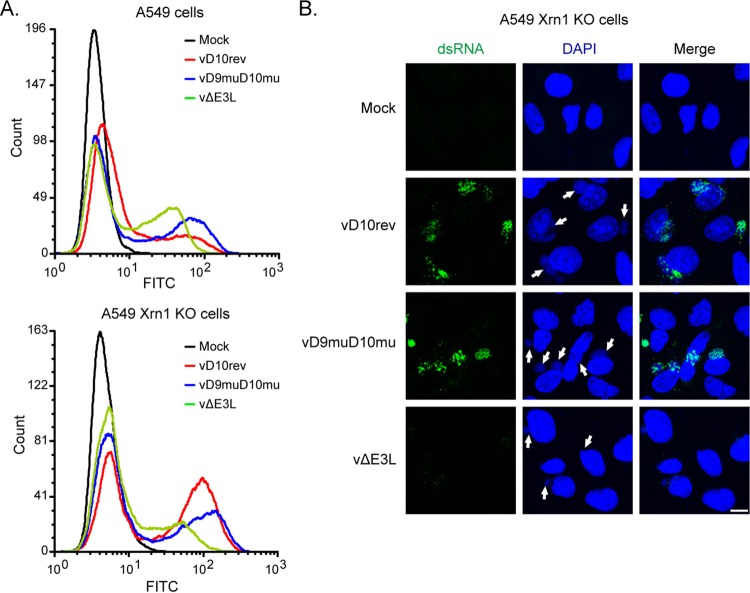
FIG 7.
Synthesis of dsRNA in Xrn1 KO cells. (A) Control and Xrn1 KO A549 cells were mock infected or infected with 5 PFU per cell of purified vD10rev, vD9muD10mu, or vΔE3L, harvested after 13 h, and stained with J2 MAb and a secondary fluorescent antibody. Fluorescent cells were analyzed by flow cytometry. (B) A549 Xrn1 KO cells were infected as described for panel A, stained with J2 MAb, secondary fluorescent antibody, and DAPI, and examined by confocal microscopy. Scale bar, 10 µm.
In contrast to rRNA, the amount of intact F17 mRNA depends on synthesis as well as degradation. Intact F17 mRNA was detected in control and KO cells infected by vD10rev, even in those displaying moderate activation of RNase L, as in shown by rRNA cleavage (Fig. 6E). For vD9muD10mu, in which rRNA cleavage was more complete in control and PKR KO cells, F17 mRNA was detected only in RNase L KO and DKO cells (Fig. 6E). For vΔE3L, F17 mRNA was detected only in the DKO cells; apparently, the drastic inhibition of viral intermediate and late protein synthesis in RNase L KO cells prevented F17 mRNA synthesis.
Analysis of GAPDH mRNA in HAP1 cells had suggested that accelerated degradation correlated with viral protein synthesis and was not entirely dependent on the VACV decapping enzymes. To confirm these impressions and determine whether activation of RNase L could contribute to instability of host mRNA, we analyzed cellular GAPDH mRNA in A549 control and KO cells. At 13 h, GAPDH RNA was undetectable in cells infected with vD10rev, regardless of whether RNase L, PKR, or both were inactivated (Fig. 6C), consistent with virus-mediated rapid turnover. In contrast, GAPDH mRNA was detected in the RNase L KO cells infected with vD9muD10mu or vΔE3L but not in PKR KO cells in which RNase L was active (Fig. 6C). GAPDH mRNA was degraded in the DKO cells infected with vD9muD10mu in which neither decapping enzymes nor RNase L was present, again suggesting a role for an additional viral protein.
We also analyzed activation of IRF3 and eIF2α in A549 KO cells. Like eIF2α, IRF3 is activated by phosphorylation. Strong IRF3 phosphorylation was induced by vΔE3L in the parental A549 cells but was reduced more in RNase L KO cells than in PKR KO cells and was almost completely abrogated in the DKO cells (Fig. 6D), in keeping with the idea that RNase L cleavage products activate this pathway (55). There was less IRF3 phosphorylation in A549 cells infected with vD10rev and vD9muD10mu (Fig. 6D), suggesting that this was prevented by E3. As in HAP1 cells, phosphorylation of PKR and eIF2α was greater in A549 cells infected with vΔE3L than in cells infected with either vD10rev or vD9muD10mu (see Fig. 8D).
FIG 8.
Replication of VACV in Xrn1 and Xrn1+PKR+RNase L TKO A549 cells. (A) Analysis of KO cells. A549 cells and A549 PKR+RNase L DKO cells were used to make Xrn1 KO and Xrn1+PKR+RNase L triple-KO (TKO) cells. Western blotting with Xrn1 and actin antibodies confirmed inactivation of the Xrn1 gene. (B) Virus replication. Control A549, DKO, Xrn1 KO, and TKO cells were infected with 5 PFU/cell of purified vD10rev, vD9muD10mu, and vΔE3L. Cells were lysed at 3 and 24 h to determine the virus input and yield, respectively, by plaque assay on BHK-21 cells. (C) Viral protein synthesis. A549 control, DKO, two independent Xrn1 KO, and TKO cells were infected with 5 PFU per cell of vD10rev, vD9muD10mu, or vΔE3L and harvested after 13 h. Lysates were analyzed by Western blotting using antibodies to VACV and actin. (D) PKR and eIF2α phosphorylation. Cells were infected as described for panel C, and Western blots were probed with antibody to phosphorylated PKR, PKR protein, phosphorylated eIF2α, and eIF2α protein. Densities shown below the images were determined with ImageJ. (E) D9 and D10 synthesis. A549 control, DKO, Xrn1 KO, and TKO cells were mock infected or infected with 5 PFU/cell of vFLAG-D9, vD10-FLAG, vFLAG-D9ΔE3L, or vD10-FLAGΔE3L. At 13 h after infection, the cells were harvested, and lysates were analyzed by Western blotting using antibodies to FLAG and E3. (F) Time course of D9 and D10 synthesis. A549 cells were mock infected or infected with vFLAG-D9 or vD10-FLAG in the absence of presence of AraC. At the indicated hours after infection, the cells were harvested, and lysates were analyzed by Western blotting with antibody to the FLAG epitope or actin.
From the experiments with A549 KO cells, we concluded that the replication defect of vΔE3L and vD9muD10mu correlated with inhibition of viral protein synthesis, which was mediated by RNase L and PKR equivalently. KO of both pathways was required for full restoration of viral protein synthesis and replication in the case of vΔE3L and for partial restoration in the case of vD9muD10mu.
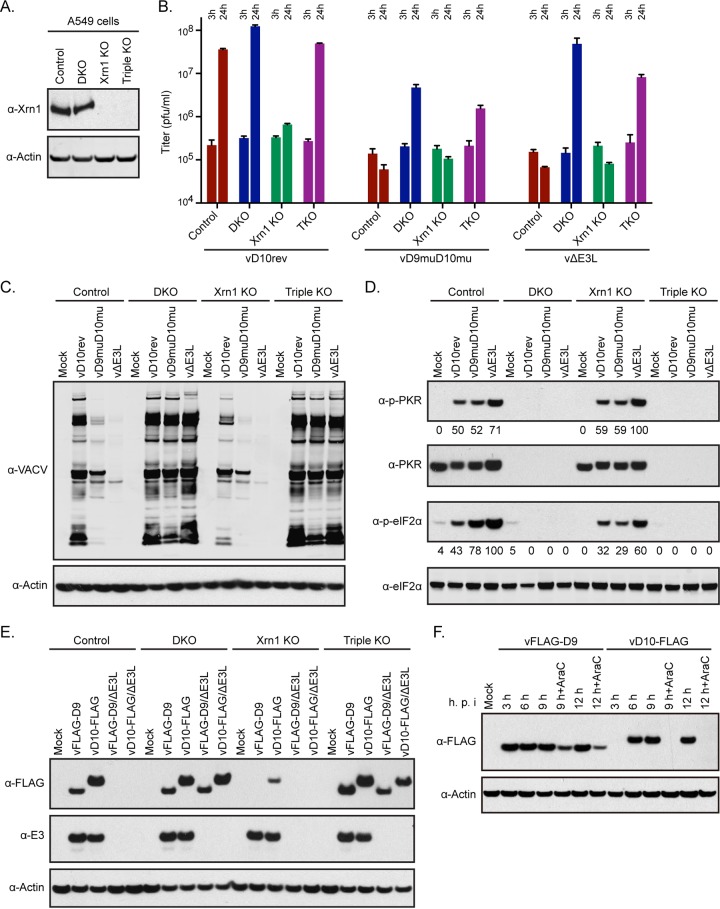
Xrn1 KO and triple knockout (TKO) of RNase L, PKR, and Xrn1 in A549 cells.
Knockdown (KD) of Xrn1 with small interfering RNA (siRNA) results in increased accumulation of dsRNA, activation of PKR and RNase L, and inhibition of viral gene expression in human fibroblasts infected with VACV (33), similar to the effects caused by inactivating the decapping enzymes (5). Burgess and Mohr (33) reported that double KD of RNase L plus Xrn1 improved viral protein synthesis compared to results with KD of Xrn1 alone, whereas a double KD with PKR plus Xrn1 had less effect, and a triple KD of RNase L, PKR, and Xrn1 was no better than KD of RNase L and Xrn1. However, in neither the double nor the triple KD was the effect on virus replication determined. Burgess and Mohr (33) also reported that viral protein synthesis was further inhibited when Xrn1 KO cells were infected with the single decapping enzyme mutant vD9mu but not with vD10mu although both mutant viruses appeared to further increase phosphorylation of PKR (31).
Since genome editing is generally more stringent than KD with siRNA, we constructed CRISPR-Cas9 Xrn1 KO A549 cells. Despite the absence of detectable Xrn1 by Western blotting as discussed and shown in the next section, the Xrn1 KO cells were viable and appeared normal. Flow cytometry was used to compare the relative amounts of dsRNA in the control and Xrn1 KO cells by staining with the J2 MAb. In the absence of VACV infection, no dsRNA was detected in either A549 or Xrn1 KO cells (Fig. 7A). There was a large increase in the number of cells that were strongly dsRNA positive in A549 Xrn1 KO cells compared to the number in the A549 control cells infected with vD10rev, whereas the differences were smaller in cells infected with vD9muD10mu and vΔE3L (Fig. 7A). Deletion of E3 actually decreased the amount of dsRNA relative to that in Xrn1 KO cells infected with vD10rev, likely due to less total viral mRNA due to rapid shutdown of gene expression, as shown below. Foci of dsRNA associated with virus factories were detected by confocal microscopy in the Xrn1 KO cells infected with vD10rev and vD9muD10mu but not in mock-infected cells or in cells infected with vΔE3L, in which less dsRNA was made (Fig. 7B).
As expected, vD10rev was unable to replicate significantly in the Xrn1 KO cells (Fig. 8B). To determine whether the deleterious effect was solely due to PKR and RNase L pathways, we used CRISPR-Cas9 to inactivate Xrn1 in our PKR+RNase L DKO A549 cells to make triple-KO (Xrn1+PKR+RNase L TKO) cells and tested the ability of wild-type and mutant viruses to replicate. The inactivation of Xrn1 in both the Xrn1 KO and TKO cells was confirmed by Western blotting (Fig. 8A). In contrast to the situation in Xrn1 KO cells, vD10rev replicated nearly as well in TKO cells as in control and DKO cells (Fig. 8B). Thus, elimination of the RNase L and PKR pathways was sufficient to counteract the adverse effects of inactivating Xrn1. Likewise, vD9muD10mu and vΔE3L were unable to replicate in Xrn1 KO cells, but their replication in TKO cells was only slightly less than that in DKO cells that expressed Xrn1 (Fig. 8B).
To further analyze the effects of Xrn1 KO on replication, viral protein synthesis was compared in control, PKR+RNase L DKO, Xrn1 KO, and TKO cells infected with vD10rev, vD9muD10mu, and vΔE3L (Fig. 8C). In Xrn1 KO cells infected with vD10rev, inhibition of viral protein synthesis was similar to that in control cells infected with vD9muD10mu, consistent with the decapping enzymes and Xrn1 each contributing to destabilization of mRNAs, resulting in increased dsRNA in the absence of either. Viral protein synthesis was more strongly inhibited in Xrn1 KO cells infected with vD9muD10mu than in similarly infected control cells, suggesting an additive effect. Consistent with our other experiments, viral protein synthesis was inhibited most in Xrn1 KO cells infected with vΔE3L. In TKO cells, viral protein synthesis was restored to levels similar to those of DKO cells in which Xrn1 was synthesized (Fig. 8C).
At 13 h, phosphorylation of PKR and eIF2α was detected in control and Xrn1 KO A549 cells infected with vD10rev, vD9muD10mu, and vΔE3L but was most intense in cells infected with vΔE3L (Fig. 8D). However, phosphorylation of eIF2α was not detected in infected DKO or TKO cells lacking PKR. Thus, the deleterious effect of Xrn1 KO on VACV replication corresponded with inhibition of viral protein synthesis and was abrogated by KO of both PKR and RNase L.
Inhibition of D9 and D10 synthesis in cells infected with vΔE3L and in Xrn1 KO cells.
We had determined that E3 was synthesized in cells infected with the vD9muD10mu mutant; however, there remained the opposite question of whether D9 and D10 were synthesized in cells infected with vΔE3L or in Xrn1 KO cells and whether a decrease in their amounts might influence the phenotypes of vΔE3L in control cells and of wild-type VACV in Xrn1 KO cells. In order to detect D9 and D10, in the absence of specific antibodies, we constructed recombinant viruses with a FLAG epitope tag on the N terminus of D9 (vFLAG-D9) and on the C terminus of D10 (vD10-FLAG) and the corresponding E3 deletion mutants (vΔE3L/FLAG-D9 and vΔE3L/D10-FLAG). vFLAG-D9 and vD10-FLAG replicated well in control, DKO, and TKO A549 cells, whereas vΔE3L/FLAG-D9 and vΔE3L/D10-FLAG replicated only in DKO and TKO cells, as determined by Western blotting with VACV antibody (data not shown). After infection with vFLAG-D9 and vD10-FLAG, the two FLAG proteins were detected in control as well as in DKO and TKO cells but only a small amount of D10-FLAG and no FLAG-D9 was detected in Xrn1 KO cells although E3 was made (Fig. 8E). After infection with vΔE3L/FLAG-D9 and vΔE3L/D10-FLAG, the two FLAG proteins were detected in the DKO and TKO cells but not in the control or Xrn1 KO cells (Fig. 8E). The small amount of D10 made in the Xrn1 KO cells infected with vD10-FLAG and the absence of D10 in cells infected with vΔE3L/D10-FLAG were expected since D10 is a late protein. However, the absence of D9 was surprising since D9, like E3, has been considered an early protein although this was based solely on analysis of the mRNAs (56, 57).
To investigate the temporal expression of D9 and D10 proteins, the FLAG-tagged D9 and D10 viruses were used in a Western blotting experiment. At 3 h, a strong D9 band was present, whereas D10 was not detected until 6 h (Fig. 8F) unless the blot was greatly over exposed (data not shown). In addition, D9 but not D10 was made in the presence of AraC. Thus, D9 is an early protein, whereas synthesis of D10 required viral DNA replication. However, the intensity of D9 was considerably less at 9 and 12 h in the presence of the DNA replication inhibitor, suggesting that D9 may be regulated by an intermediate or late promoter in addition to an early promoter, which would account in part for the strong inhibition by the E3 deletion virus and in Xrn1 KO cells. Inspection of the sequence upstream of the early D9 promoter revealed candidate intermediate or late promoters (data not shown). We concluded that failure to synthesize D9 and D10 could contribute to the phenotypes of the E3 deletion mutant in control cells and of wild-type VACV in Xrn1 KO cells.
DISCUSSION
CRISPR-Cas9 genome editing provides a new and stringent way to interrogate virus-host interactions. We focused on dsRNA, an important pathogen-associated molecular pattern, and analyzed host defense mechanisms and viral countermeasures. In this report we employed two VACV mutants, in which the E3 gene was deleted or the two capping enzymes D9 and D10 were mutated at their active sites, and six novel CRISPR-Cas9-modified human cell lines in which PKR and RNase L were inactivated separately or together and two more cell lines in which Xrn1 was inactivated alone or together with PKR and RNase L (Table 1). The major experimental findings, which will be discussed further below, were the following: (i) dsRNA colocalized with the viral E3 protein in discrete foci within virus factories in cells infected with wild-type VACV or a decapping enzyme mutant; (ii) viral late gene expression was more profoundly inhibited in cells infected with an E3 deletion mutant than in those infected with the decapping enzyme mutant, consistent with earlier time of onset and extent of PKR and eIF2α phosphorylation; (iii) in A549 cells viral late gene expression and replication of the E3 deletion mutant were restored only by inactivating both RNase L and PKR, indicating that the two pathways had comparable inhibitory effects and that no other dsRNA pathways were important; (iv) in HAP1 cells RNase L was intrinsically inactive and only KO of PKR was necessary to restore viral gene expression and replication of the E3 mutant; (v) the decapping enzyme mutant was unable to replicate significantly in RNase L and PKR DKO HAP1 cells in which viral gene expression was only slightly enhanced, suggesting additional inhibitory mechanisms; (vi) viral gene expression and replication of the decapping enzyme mutant were increased in DKO A549 cells but levels remained lower than those of wild-type virus and the E3 mutant; (vii) Xrn1 KO A549 cells were viable and resistant to VACV infection, whereas KO of PKR and RNase L in addition to Xrn1 restored VACV replication to wild-type levels; (viii) the decapping enzyme mutant expressed E3, whereas expression of decapping enzymes was markedly reduced in control cells infected with an E3 deletion mutant or in Xrn1 KO cells infected with wild-type virus.
The ability of E3 to bind poly(I·C) was reported more than 2 decades ago, suggesting that E3 acts by sequestering dsRNA (11), yet we were unable to find a publication documenting such an association in infected cells even though specific MAbs to dsRNA (43) and E3 (42) were described at about the same time. To remedy this long delay, we first used isotype-specific dsRNA and E3 MAbs to show by confocal microscopy that all dsRNA foci colocalized with E3 in cells infected with the decapping enzyme mutant or wild-type VACV. To confirm this observation, we made a recombinant VACV that expresses an E3-GFP fusion protein so that we would not have to depend on isotype-specific MAbs and obtained the same result. In addition, E3 coprecipitated with the dsRNA MAb from infected cell lysates. In the absence of E3, confocal microscopy revealed only tiny dsRNA puncta in or near virus factories, and flow cytometry showed lower levels of dsRNA in individual cells than in wild-type VACV though the number of cells with detectable dsRNA was higher. Our demonstration that dsRNA colocalizes with E3 in infected cells lays the groundwork for assessing the significance of a recent report (19), carried out by in vitro binding of mutated E3 to poly(I·C), which concluded that dsRNA binding by E3 does not uniformly correlate with host range.
Despite the smaller amounts of dsRNA formed in cells infected with the E3 mutant than in those infected with the decapping enzyme mutant, PKR and eIF2α phosphorylations were stronger, occurred more rapidly, and led to more complete inhibition of viral gene expression. Evidently, these catastrophic events were triggered by relatively small amounts of dsRNA synthesized early due to overlapping transcription (58, 59). The rapid shutdown of viral gene expression by the E3 mutant prevented synthesis of viral intermediate and late mRNAs, which are needed to form the larger amounts of dsRNA in cells infected with wild-type virus and the decapping enzyme mutant. In HAP1 and A549 cells, the formation of dsRNA and activation of innate immune responses eventually occurred after infection with wild-type VACV. This suggests that the primary role of E3 is to delay activation of dsRNA pathways and that activation at late times does not prevent virus replication in the case of the wild-type virus. The acquisition of resistance to PKR and RNase L activation could be due to the localization of transcription and translation within viral factories (60) or to the presence of a 5′ poly(A) leader present on all intermediate and late mRNAs but on few early mRNAs (61–64), which may reduce the need for certain translation initiation factors (65–67). More dsRNA formed in cells infected with the decapping enzyme mutant than in those infected with wild-type VACV, leading to greater activation of RNase L, PKR, and eIF2α phosphorylation.
The relative contributions of PKR and RNase L pathways to the host restriction of VACV E3 and decapping enzyme mutants were investigated using CRISPR-Cas9-modified cells. Since cell lines can vary in their basal levels of innate immune defense proteins, we employed two: HAP1 and A549. We noted differences in the responses of the cell lines to VACV infection; most apparent was the absence of RNase L activation in HAP1 cells, as determined by the stability of rRNAs. Since uninfected HAP1 cells expressed RNase L, a deficiency of OAS seemed likely (47, 48). Consistent with this, OAS was detected by Western blotting only after administration of interferon (data not shown). Since HAP1 cells are defective in RNase L activation, the PKR KO was functionally a DKO. The restricted replication of the E3 mutant was overcome solely by inactivating PKR in HAP1 cells, whereas inactivation of both PKR and RNase L was necessary in A549 cells. The latter result indicated that RNase L and PKR prevented viral gene expression and replication equivalently. Furthermore, the data obtained with both the HAP1 and A549 KO cell lines indicated that only the OAS/RNase L and PKR/eIF2α dsRNA pathways significantly impaired replication of the E3 mutant. The results with the decapping enzyme mutant were quite different. This virus was unable to replicate significantly in the DKO HAP1 cell lines, and replication was suboptimal in DKO A549 cells. Transmission electron microscopy revealed only immature and abnormal-looking virus particles in DKO HAP1 cells infected with vD9muD10mu. Further analysis showed that viral protein synthesis by the decapping enzyme mutant increased more in the DKO A549 cells than in the DKO HAP1 cells, also consistent with better virus replication. Nevertheless, KO of PKR and RNase L was insufficient to completely rescue viral gene expression.
Knockdown of Xrn1 in cells infected with VACV leads to accumulation of dsRNA and activation of dsRNA pathways, suggesting that Xrn1 has a role in viral mRNA degradation (33). Here, we showed that a CRISPR-Cas9 A549 cell KO of Xrn1 was viable yet nonpermissive for VACV. In some ways, the phenotype of wild-type VACV in Xrn1 KO cells was similar to that of the decapping enzyme mutant. In both cases, excess dsRNA formed, PKR and eIF2α phosphorylation occurred to similar extents, and viral late gene expression was inhibited. However, there was a notable difference: the replication defect of wild-type VACV caused by KO of Xrn1 was overcome in a triple KO of RNase L, PKR, and Xrn1, whereas KO of RNase L and PKR in A549 cells had a lesser effect on replication of the decapping enzyme mutant.
Why inactivation of PKR and RNase L was sufficient to reverse the inhibitory effects in Xrn1 KO cells or enable replication of a VACV E3 deletion mutant but not a decapping enzyme mutant is intriguing. Assuming that degradation of viral mRNA is initiated by removal of the cap and followed by Xrn1 digestion, we would expect viral mRNA to accumulate if either step was prevented, leading to increased dsRNA, as seen in both cases. However, the RNA that accumulates at least initially would be quite different: the excess RNA would remain capped in the absence of decapping enzymes and uncapped with a 5′ monophosphate end in the absence of Xrn1. Since Northern blotting indicated that turnover of late mRNA occurred in the absence of D9/D10, we propose that capped RNA degradation products may be responsible for translational defects that persist even in the absence of PKR. Supporting such a possibility are previous studies suggesting that abortive capped transcripts severely inhibit protein synthesis by competing with mRNA for association with ribosomes or sequestering translation factors (68, 69). Thus, the role of decapping enzymes may be to regulate the stabilities of viral and cellular mRNAs, reduce the formation of dsRNA, and eliminate potentially harmful mRNA degradation products.
ACKNOWLEDGMENTS
We thank Andrea Weisberg (NIAID) and Catherine Cotter (NIAID) for electron microscopy and maintenance of cell lines, respectively. John Connor (Boston University) kindly donated the E3 deletion mutant, and Stuart Isaacs (University of Pennsylvania) and Robert Silverman (Cleveland Clinic Lerner Research Institute) generously provided antibodies.
REFERENCES
- 1.Silverman RH. 2007. Viral encounters with 2 ′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol 81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeWitte-Orr SJ, Mossman KL. 2010. dsRNA and the innate antiviral immune response. Future Virol 5:325–341. doi: 10.2217/fvl.10.18. [DOI] [Google Scholar]
- 3.Drappier M, Michiels T. 2015. Inhibition of the OAS/RNase L pathway by viruses. Curr Opin Virol 15:19–26. doi: 10.1016/j.coviro.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duesberg PH, Colby C. 1969. On the biosynthesis and structure of double-stranded RNA in vaccinia virus-infected cells. Proc Natl Acad Sci U S A 64:396–403. doi: 10.1073/pnas.64.1.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu SW, Katsafanas GC, Liu R, Wyatt LS, Moss B. 2015. Poxvirus decapping enzymes enhance virulence by preventing the accumulation of dsRNA and the induction of innate antiviral responses. Cell Host Microbe 17:320–331. doi: 10.1016/j.chom.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boone RF, Parr RP, Moss B. 1979. Intermolecular duplexes formed from polyadenylated vaccinia virus RNA. J Virol 30:365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowie AG, Unterholzner L. 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol 8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perdiguero B, Esteban M. 2009. The interferon system and vaccinia virus evasion mechanisms. J Interferon Cytokine Res 29:581–598. doi: 10.1089/jir.2009.0073. [DOI] [PubMed] [Google Scholar]
- 9.Smith GL, Benfield CT, Maluquer de Motes C, Mazzon M, Ember SW, Ferguson BJ, Sumner RP. 2013. Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J Gen Virol 94:2367–2392. doi: 10.1099/vir.0.055921-0. [DOI] [PubMed] [Google Scholar]
- 10.Haller SL, Peng C, McFadden G, Rothenburg S. 2014. Poxviruses and the evolution of host range and virulence. Infect Genet Evol 21:15–40. doi: 10.1016/j.meegid.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang HW, Watson JC, Jacobs BL. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci U S A 89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beattie E, Denzler KL, Tartaglia J, Perkus ME, Paoletti E, Jacobs BL. 1995. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J Virol 69:499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang H-W, Uribe LH, Jacobs BL. 1995. Rescue of vaccinia virus lacking the E3L gene by mutants of E3L. J Virol 69:6605–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivas C, Gil J, Melkova Z, Esteban M, Diaz-Guerra M. 1998. Vaccinia virus E3L protein is an inhibitor of the interferon (IFN)-induced 2-5A synthetase enzyme. Virology 243:406–414. doi: 10.1006/viro.1998.9072. [DOI] [PubMed] [Google Scholar]
- 15.Smith EJ, Marie I, Prakash A, Garcia-Sastre A, Levy DE. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by vaccinia virus E3L protein. J Biol Chem 276:8951–8957. doi: 10.1074/jbc.M008717200. [DOI] [PubMed] [Google Scholar]
- 16.Xiang Y, Condit RC, Vijaysri S, Jacobs B, Williams BRG, Silverman RH. 2002. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J Virol 76:5251–5259. doi: 10.1128/JVI.76.10.5251-5259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludwig H, Suezer Y, Waibler Z, Kalinke U, Schnierle BS, Sutter G. 2006. Double-stranded RNA-binding protein E3 controls translation of viral intermediate RNA, marking an essential step in the life cycle of modified vaccinia virus Ankara. J Gen Virol 87:1145–1155. doi: 10.1099/vir.0.81623-0. [DOI] [PubMed] [Google Scholar]
- 18.Zhang P, Jacobs BL, Samuel CE. 2008. Loss of protein kinase PKR expression in human HeLa cells complements the vaccinia virus E3L deletion mutant phenotype by restoration of viral protein synthesis. J Virol 82:840–848. doi: 10.1128/JVI.01891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dueck KJ, Hu YS, Chen P, Deschambault Y, Lee J, Varga J, Cao J. 2015. Mutational analysis of vaccinia virus E3 protein: the biological functions do not correlate with its biochemical capacity to bind double-stranded RNA. J Virol 89:5382–5394. doi: 10.1128/JVI.03288-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharp TV, Moonan F, Romashko A, Joshi B, Barber GN, Jagus R. 1998. The vaccinia virus E3L gene product interacts with both the regulatory and the substrate binding regions of PKR: implications for PKR autoregulation. Virology 250:302–315. doi: 10.1006/viro.1998.9365. [DOI] [PubMed] [Google Scholar]
- 21.Romano PR, Zhang F, Tan SL, Garcia-Barrio MT, Katze MG, Dever TE, Hinnebusch AG. 1998. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol Cell Biol 18:7304–7316. doi: 10.1128/MCB.18.12.7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brandt T, Heck MC, Vijaysri S, Jentarra GM, Cameron JM, Jacobs BL. 2005. The N-terminal domain of the vaccinia virus E3L-protein is required for neurovirulence, but not induction of a protective immune response. Virology 333:263–270. doi: 10.1016/j.virol.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Brandt TA, Jacobs BL. 2001. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J Virol 75:850–856. doi: 10.1128/JVI.75.2.850-856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beattie E, Tartaglia J, Paoletti E. 1991. Vaccinia virus encoded eIF-2a homolog abrogates the antiviral effect of interferon. Virology 183:419–422. doi: 10.1016/0042-6822(91)90158-8. [DOI] [PubMed] [Google Scholar]
- 25.Langland JO, Jacobs BL. 2002. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 299:133–141. doi: 10.1006/viro.2002.1479. [DOI] [PubMed] [Google Scholar]
- 26.Davies MV, Elroy-Stein O, Jagus R, Moss B, Kaufman RJ. 1992. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J Virol 66:1943–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies MV, Chang H-W, Jacobs BL, Kaufman RJ. 1993. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J Virol 67:1688–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carroll K, Elroy-Stein O, Moss B, Jagus R. 1993. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2 alpha-specific protein kinase. J Biol Chem 268:12837–12842. [PubMed] [Google Scholar]
- 29.Parrish S, Resch W, Moss B. 2007. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proc Natl Acad Sci U S A 104:2139–2144. doi: 10.1073/pnas.0611685104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parrish S, Moss B. 2007. Characterization of a second vaccinia virus mRNA-decapping enzyme conserved in poxviruses. J Virol 81:12973–12978. doi: 10.1128/JVI.01668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parrish S, Moss B. 2006. Characterization of a vaccinia virus mutant with a deletion of the D10R gene encoding a putative negative regulator of gene expression. J Virol 80:553–561. doi: 10.1128/JVI.80.2.553-561.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sivan G, Martin SE, Myers TG, Buehler E, Szymczyk KH, Ormanoglu P, Moss B. 2013. Human genome-wide RNAi screen reveals a role for nuclear pore proteins in poxvirus morphogenesis. Proc Natl Acad Sci U S A 110:3519–3524. doi: 10.1073/pnas.1300708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgess HM, Mohr I. 2015. Cellular 5 ′-3 ′ mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe 17:332–344. doi: 10.1016/j.chom.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weaver JR, Shamim M, Alexander E, Davies DH, Felgner PL, Isaacs SN. 2007. The identification and characterization of a monoclonal antibody to the vaccinia virus E3 protein. Virus Res 130:269–274. doi: 10.1016/j.virusres.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong B, Silverman RH. 1995. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J Biol Chem 270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 36.Davies DH, Wyatt LS, Newman FK, Earl PL, Chun S, Hernandez JE, Molina DM, Hirst S, Moss B, Frey SE, Felgner PL. 2008. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus Ankara is comparable to that of Dryvax. J Virol 82:652–663. doi: 10.1128/JVI.01706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sodeik B, Griffiths G, Ericsson M, Moss B, Doms RW. 1994. Assembly of vaccinia virus: effects of rifampin on the intracellular distribution of viral protein p65. J Virol 68:1103–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simpson-Holley M, Kedersha N, Dower K, Rubins KH, Anderson P, Hensley LE, Connor JH. 2011. Formation of antiviral cytoplasmic granules during orthopoxvirus infection. J Virol 85:1581–1593. doi: 10.1128/JVI.02247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu SW, Wyatt LS, Orandle MS, Minai M, Moss B. 2014. The D10 decapping enzyme of vaccinia virus contributes to decay of cellular and viral mRNAs and to virulence in mice. J Virol 88:202–211. doi: 10.1128/JVI.02426-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maruri-Avidal L, Weisberg AS, Moss B. 2011. Vaccinia virus L2 protein associates with the endoplasmic reticulum near the growing edge of crescent precursors of immature virions and stabilizes a subset of viral membrane proteins. J Virol 85:12431–12441. doi: 10.1128/JVI.05573-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuwen H, Cox JH, Yewdell JW, Bennink JR, Moss B. 1993. Nuclear localization of a double-stranded RNA-binding protein encoded by the vaccinia virus E3l gene. Virology 195:732–744. doi: 10.1006/viro.1993.1424. [DOI] [PubMed] [Google Scholar]
- 43.Schonborn J, Oberstrass J, Breyel E, Tittgen J, Schumacher J, Lukacs N. 1991. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res 19:2993–3000. doi: 10.1093/nar/19.11.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bonin M, Oberstrass J, Lukacs N, Ewert K, Oesterschulze E, Kassing R, Nellen W. 2000. Determination of preferential binding sites for anti-dsRNA antibodies on double-stranded RNA by scanning force microscopy. RNA 6:563–570. doi: 10.1017/S1355838200992318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Targett-Adams P, Boulant S, McLauchlan J. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J Virol 82:2182–2195. doi: 10.1128/JVI.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marshall EE, Bierle CJ, Brune W, Geballe AP. 2009. Essential role for either TRS1 or IRS1 in human cytomegalovirus replication. J Virol 83:4112–41120. doi: 10.1128/JVI.02489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stark GR, Dower WJ, Schimke RT, Brown RE, Kerr IM. 1979. 2-5A synthetase: assay, distribution and variation with growth or hormone status. Nature 278:471–473. doi: 10.1038/278471a0. [DOI] [PubMed] [Google Scholar]
- 48.Zhao L, Birdwell LD, Wu A, Elliott R, Rose KM, Phillips JM, Li Y, Grinspan J, Silverman RH, Weiss SR. 2013. Cell-type-specific activation of the oligoadenylate synthetase-RNase L pathway by a murine coronavirus. J Virol 87:8408–8418. doi: 10.1128/JVI.00769-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Essletzbichler P, Konopka T, Santoro F, Chen D, Gapp BV, Kralovics R, Brummelkamp TR, Nijman SM, Burckstummer T. 2014. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res 24:2059–2065. doi: 10.1101/gr.177220.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lieber M, Smith B, Szaki A, Nelson-Rees W, Todaro G. 1976. A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int J Cancer 17:62–70. doi: 10.1002/ijc.2910170110. [DOI] [PubMed] [Google Scholar]
- 51.Brum LM, Lopez MC, Varela JC, Baker HV, Moyer RW. 2003. Microarray analysis of A549 cells infected with rabbitpox virus (RPV): a comparison of wild-type RPV and RPV deleted for the host range gene, SPI-1. Virology 315:322–334. doi: 10.1016/S0042-6822(03)00532-4. [DOI] [PubMed] [Google Scholar]
- 52.Guerra S, Lopez-Fernandez LA, Pascual-Montano A, Munoz M, Harshman K, Esteban M. 2003. Cellular gene expression survey of vaccinia virus infection of human HeLa cells. J Virol 77:6493–6506. doi: 10.1128/JVI.77.11.6493-6506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Z, Bruno DP, Martens CA, Porcella SF, Moss B. 2010. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc Natl Acad Sci U S A 107:11513–11518. doi: 10.1073/pnas.1006594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dani C, Piechaczyk M, Audigier Y, El Sabouty S, Cathala G, Marty L, Fort P, Blanchard JM, Jeanteur P. 1984. Characterization of the transcription products of glyceraldehyde 3-phosphate-dehydrogenase gene in HeLa cells. Eur J Biochem 145:299–304. doi: 10.1111/j.1432-1033.1984.tb08552.x. [DOI] [PubMed] [Google Scholar]
- 55.Malathi K, Dong B, Gale M Jr, Silverman RH. 2007. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee-Chen GJ, Bourgeois N, Davidson K, Condit RC, Niles EG. 1988. Structure of the transcription initiation and termination sequences of seven early genes in the vaccinia virus HindIII D fragment. Virology 163:64–79. doi: 10.1016/0042-6822(88)90234-6. [DOI] [PubMed] [Google Scholar]
- 57.Lee-Chen GJ, Niles EG. 1988. Transcription and translation mapping of the 13 genes in the vaccinia virus HindIII D fragment. Virology 163:52–63. doi: 10.1016/0042-6822(88)90233-4. [DOI] [PubMed] [Google Scholar]
- 58.Yang Z, Bruno DP, Martens CA, Porcella SF, Moss B. 2011. Genome-wide analysis of the 5′ and 3′ ends of vaccinia virus early mRNAs delineates regulatory sequences of annotated and anomalous transcripts. J Virol 85:5897–5909. doi: 10.1128/JVI.00428-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Willis KL, Langland JO, Shisler JL. 2011. Viral double-stranded RNAs from vaccinia virus early or intermediate gene transcripts possess PKR activating function, resulting in NF-κB activation, when the K1 protein is absent or mutated. J Biol Chem 286:7765–7778. doi: 10.1074/jbc.M110.194704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katsafanas GC, Moss B. 2007. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2:221–228. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwer B, Visca P, Vos JC, Stunnenberg HG. 1987. Discontinuous transcription or RNA processing of vaccinia virus late messengers results in a 5′ poly(A) leader. Cell 50:163–169. doi: 10.1016/0092-8674(87)90212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bertholet C, Van Meir E, ten Heggeler-Bordier B, Wittek R. 1987. Vaccinia virus produces late mRNAs by discontinuous synthesis. Cell 50:153–162. doi: 10.1016/0092-8674(87)90211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ink BS, Pickup DJ. 1990. Vaccinia virus directs the synthesis of early mRNAs containing 5′ poly(A) sequences. Proc Natl Acad Sci U S A 87:1536–1540. doi: 10.1073/pnas.87.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahn B-Y, Moss B. 1989. Capped poly(A) leader of variable lengths at the 5′ ends of vaccinia virus late mRNAs. J Virol 63:226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mulder J, Robertson MEM, Seamons RA, Belsham GJ. 1998. Vaccinia virus protein synthesis has a low requirement for the intact translation initiation factor eIF4F, the cap-binding complex, within infected cells. J Virol 72:8813–8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gilbert WV, Zhou K, Butler TK, Doudna JA. 2007. Cap-independent translation is required for starvation-induced differentiation in yeast. Science 317:1224–1227. doi: 10.1126/science.1144467. [DOI] [PubMed] [Google Scholar]
- 67.Xia XH, MacKay V, Yao XQ, Wu JH, Miura F, Ito T, Morris DR. 2011. Translation initiation: a regulatory role for poly(A) tracts in front of the AUG codon in Saccharomyces cerevisiae. Genetics 189:469–478. doi: 10.1534/genetics.111.132068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rosemond-Hornbeak H, Moss B. 1975. Inhibition of host protein synthesis by vaccinia virus: fate of cell mRNA and synthesis of small poly(A)-rich polyribonucleotides in the presence of actinomycin D. J Virol 16:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gershowitz A, Moss B. 1979. Abortive transcription products of vaccinia virus are guanylylated, methylated and polyadenylylated. J Virol 31:849–853. [DOI] [PMC free article] [PubMed] [Google Scholar]