ABSTRACT
Influenza virus infections represent a significant socioeconomic and public health burden worldwide. Although ferrets are considered by many to be ideal for modeling human responses to influenza infection and vaccination, efforts to understand the cellular immune response have been severely hampered by a paucity of standardized procedures and reagents. In this study, we developed flow cytometric and T cell enzyme-linked immunosorbent spot (ELISpot) approaches to characterize the leukocyte composition and antigen-specific T cell response within key lymphoid tissues following influenza virus infection in ferrets. Through a newly designed and implemented set of serological reagents, we used multiparameter flow cytometry to directly quantify the frequency of CD4+ and CD8+ T cells, Ig+ B cells, CD11b+ myeloid-derived cells, and major histocompatibility complex (MHC) class II-positive antigen-presenting cells (APCs) both prior to and after intranasal infection with A/California/04/09 (H1N1). We found that the leukocyte composition was altered at 10 days postinfection, with notable gains in the frequency of T cells and myeloid cells within the draining lymph node. Furthermore, these studies revealed that the antigen specificity of influenza virus-reactive CD4 and CD8 T cells was very broad, with recognition of the viral HA, NA, M1, NS1, and NP proteins, and that total reactivity to influenza virus postinfection represented approximately 0.1% of the circulating peripheral blood mononuclear cells (PBMC). Finally, we observed distinct patterns of reactivity between individual animals, suggesting heterogeneity at the MHC locus in ferrets within commercial populations, a finding of considerable interest in efforts to move the ferret model forward for influenza vaccine and challenge studies.
IMPORTANCE Ferrets are an ideal animal model to study transmission, diseases, and vaccine efficacies of respiratory viruses because of their close anatomical and physiological resemblances to humans. However, a lack of reagents has limited our understanding of the cell-mediated immune response following infection and vaccination. In this study, we used cross-reactive and ferret-specific antibodies to study the leukocyte composition and antigen-specific CD4 and CD8 T cell responses following influenza A/California/04/09 (H1N1) virus infection. These studies revealed strikingly distinct patterns of reactivity between CD4 and CD8 T cells, which were overlaid with differences in protein-specific responses between individual animals. Our results provide a first, in-depth look at the T cell repertoire in response to influenza infection and suggest that there is considerable heterogeneity at the MHC locus, which is akin to that in humans and an area of intense research interest.
INTRODUCTION
Influenza A virus infections continue to cause seasonal epidemics as well as occasional pandemics and thus remain a major cause of morbidity and mortality worldwide (1–6). While incompletely understood, it has been shown that disease severity is multifactorial and governed by distinct characteristics of the virus and host. Virulence factors include properties and/or mutations within the hemagglutinin (HA) protein, which mediates viral infectivity through regulation of receptor specificity (7, 8), transmissibility (9, 10), and susceptibility to host proteases (11, 12). Additionally, mutations within different components of the RNA polymerase complex have been demonstrated to support enhanced replication of avian viruses in mammalian cells (13–16), while others have been shown to alter pathogenicity by increasing apoptosis (17), secretion of proinflammatory cytokines (18), suppression of the innate immune response (19), and resistance to antiviral drugs (20, 21). Host factors that have been found to contribute to differences in disease severity include age (22, 23), preexisting immunity (24–26), innate and adaptive immune cell impairment (27–29), interactions with the microbial environment (30, 31), and genetic background (32, 33). Although routine vaccination has proven to be the most effective defense against drifted and shifted variants, inclusion of antigenically mismatched strains has led to poor efficacy against circulating viruses, and defining correlates of immune protection remains challenging (34–37).
Compared to other animals such as mice, outbred domestic ferrets (Mustela putorius furo) possess several important features that make them ideal for modeling human influenza virus transmission, disease, and vaccine efficacy. Ferrets are susceptible to most human isolates of influenza virus, likely due in part to similar profiles and distributions of sialic acid receptors within ferret and human respiratory tracts, and share similar anatomy at the site of viral replication, including a long upper respiratory tract. Moreover, unlike murine models of influenza infection, ferrets display human-like symptoms and pathological changes, can transmit virus via respiratory droplets, and have a relatively long life span (38–41). Collectively, these features present an excellent experimental model to study influenza immunity. Thus, information gained through the use of the ferret model has the potential to reveal key molecular and cellular signatures associated with virus evolution, mechanisms of vaccine-induced protection, and virus transmission.
Despite these considerable advantages of the ferret model, the largest obstacle to research progress has been a paucity of immunological reagents. Gene expression studies, using information gathered from the draft genome and cross-reactive probes for microarrays, have confirmed the biphasic kinetics of innate and adaptive immune responses following viral infection (42–45). While limited, serological reagents (46, 47) have been used in a number of applications, including immunohistochemical analyses to study pathological features of disease as it relates to the virus/host interface (48). Additionally, other studies have quantified antibody-secreting cells within the draining lymph node in live attenuated influenza virus vaccine (LAIV) recipients, finding that the abundance of these cells inversely correlated with virus replication during a heterologous virus challenge (49). Furthermore, in other vaccine/infection challenge experiments, the serum HA antibody titer inversely correlated with viral load, providing insight into immune correlates of protection in the ferret model (reviewed in reference 38).
Importantly, CD4+ and CD8+ T cells are also known to provide protection from influenza through multiple mechanisms, including destruction of infected cells, production of antiviral cytokines, and promoting recruitment of effectors to the site of infection (reviewed in references 50, 51, and 52). Moreover, CD4 T cells are essential for B cell production of high-affinity, neutralizing antibodies as well as generation of memory B cells and long-lived antibody-secreting cells. Previous studies using flow cytometry approaches have tracked immune cell frequency in peripheral blood over time following infection of ferrets with H1N1 and H3N2 viruses, with differences in T cell representation noted with virus strain and preexisting immunity (53). Moreover, a recent study demonstrated that when antibody responses were within the protective range, the CD3+ gamma interferon-positive (IFN-γ+) cellular response correlated best with reduced morbidity in an HA DNA prime, adenovirus boost and challenge model using A/South Carolina/1/18 (H1N1) virus (54). Finally, in a recent study using licensed vaccines, protection from infection was better correlated with the cellular response exhibited by LAIV naive animals than with the response by those receiving trivalent inactivated vaccine. The authors suggest that the cellular response may be responsible for the notable efficacy of LAIV in young children (49). While these studies and others have enhanced our understanding of the ferret cell-mediated immune response, none have directly addressed the abundance and protein specificity of influenza virus-specific CD4 and CD8 T cells in the context of primary infection or the mobilization of distinct lymphoid subsets in different tissues.
In this study, we sought to gain a deeper understanding of the ferret cellular immune response to infection with an influenza A virus. We developed a novel CD4-specific monoclonal antibody and screened commercially available antibodies for their ability to recognize key ferret-specific leukocyte antigens. Antibodies to CD4, CD8α, CD11b, major histocompatibility complex class II (MHC-II), and immunoglobulin (Ig) allowed us to detect and quantify various immune cell populations at the cellular level by flow cytometry, lending insight into the cellular composition and dynamics within lymphatic tissues pre- and postinfection. Furthermore, monoclonal antibodies to CD4 and CD8α were used for in vitro depletion experiments to perform specificity analyses of influenza virus-reactive CD8 and CD4 T cells following intranasal infection through the use of pools of overlapping peptide libraries to the viral HA, neuraminidase (NA), nucleoprotein (NP), nonstructural 1 (NS1), and matrix 1 (M1) proteins in conjunction with IFN-γ enzyme-linked immunosorbent spot (ELISpot) assays. These experiments provide a first look into the antigen-specific CD4 and CD8 T cell response, including magnitude, host variability, and potential for protein-specific preferences.
MATERIALS AND METHODS
Ethics statement.
All ferret procedures performed in this study were in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines, and animal protocols were reviewed and approved by the IACUC of the Icahn School of Medicine at Mt. Sinai (LA12-00170 and IACUC-2013-1408). All mice were maintained in a specific-pathogen-free facility at the University of Rochester Medical Center (URMC) according to institutional guidelines. All animal protocols used in this study adhere to the AAALAC International, the Animal Welfare Act, and the PHS Guide and were approved by the University of Rochester Committee on Animal Resources, Animal Welfare Assurance number A3291-01.
Animals.
Seven-month-old female ferrets were purchased from Marshall Farms (North Rose, NY) and were confirmed to be seronegative to influenza virus. A single 4-month-old male ferret purchased from Triple F Farms (Sayre, PA) was also used in a separate study; it was mock immunized and infected with A/California/4/09 (H1N1) by a natural aerosol route, and tissues were harvested at 23 days postinfection (dpi). All tissues, including lymph nodes, spleens, and whole blood, were collected and shipped overnight from Mt. Sinai (New York, NY) to URMC (Rochester, NY) at ambient temperature. For some analyses (see Fig. 7 and 8), data are related by animal identification (ear tag).
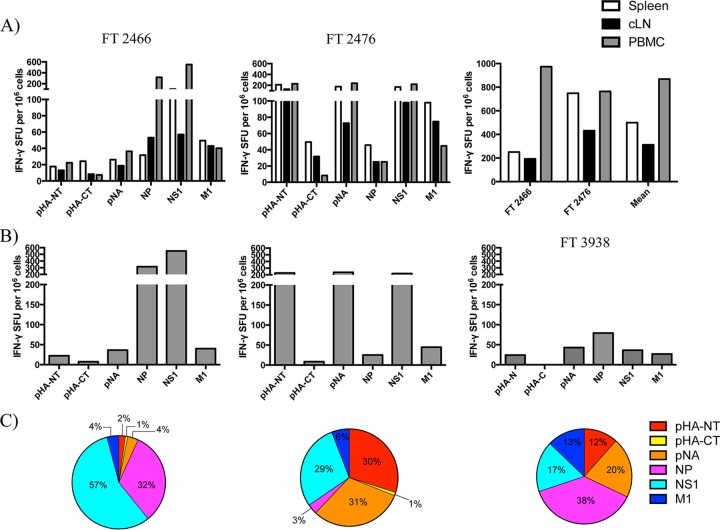
FIG 7.
Antigen-specific T cell responses are heterogeneous between individuals and have broad protein specificity across multiple tissue compartments. (A) Viral protein-specific IFN-γ-secreting T cell responses from ferrets (FT) 2466 and 2476 at 10 dpi with A/California/04/09 (H1N1) in the spleen, cLN, and PBMC. T cell responses were not detectable in influenza virus-naive ferret tissues (data not shown). (B) T cell responses from PBMCs of FT 2466 and 2476 as well as FT 3938 (aerosol contact infected, 23 dpi). (C) Antigen-specific T cell reactivity from the PBMCs, represented as a percentage of the total response for each individual tested, depicted as a pie graph (left to right, FT 2466, FT 2476, and FT 3938). pHA-NT, peptides spanning the HA1 domain from pandemic hemagglutinin (subtype H1); pHA-CT, peptides spanning the HA2 domain from pandemic H1; pNA, N1 subtype of neuraminidase from the pandemic H1N1 virus; NP, nucleoprotein; NS1, nonstructural protein 1; M1, matrix 1 protein.
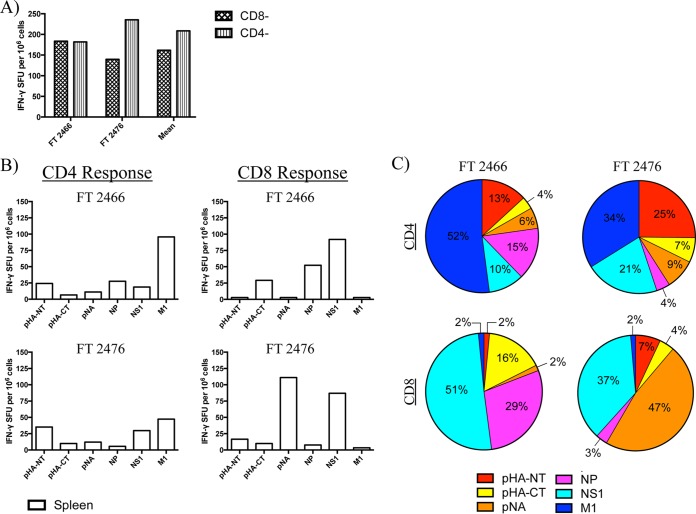
FIG 8.
Specificity of influenza virus-reactive CD4 and CD8 T cell responses are broad, display protein-specific preferences, and overlay with heterogeneity between individual animals. (A) Magnitudes of total influenza virus-specific T cell responses from individual ferrets sampled from the spleen at 10 dpi with A/California/04/09 (H1N1). CD4− and CD8− indicate depletion of that cell type for selective interrogation of CD8 and CD4 T cell responses, respectively. (B) Magnitude of antigen-specific T cell responses from individual animals. (C) Viral protein-specific T cell responses from the spleen, represented for each ferret as a percentage of the total response.
Viruses and infection.
A/California/4/09 (H1N1) was propagated in 8-day-old specific-pathogen-free embryonated hen's eggs (Charles River, North Franklin, CT, USA). Ferrets were provided water and food (high-density ferret diet [5L14]; LabDiet, St. Louis, MO) ad libitum. Ferrets were anesthetized by intraperitoneal injection of ketamine (10 mg/kg) and xylazine (2 mg/kg) and infected intranasally with 10,000 PFU of virus. Productive virus replication in infected ferrets was confirmed by measuring viral titers collected from nasal washes at 1, 3, and 5 dpi (data not shown).
Isolation of PBMC and preparation of single-cell suspensions from ferret tissues.
Blood samples were collected from ferrets in EDTA collection tubes and shipped overnight. Whole blood was diluted to a final ratio of 4 parts blood to 1 part Hanks buffered salt solution (HBSS) (Corning CellGro) and layered over Ficoll-Paque Plus (GE Healthcare). Samples were centrifuged at 800 × g for 20 min at room temperature with the brake off. The buffy coat was collected and washed, and cells were counted for use in ELISpot and flow cytometry assays. Tissues were excised, mechanically dissociated, and passed through 40-μm nylon mesh filters to produce single-cell suspensions in nutrient-supplemented Dulbecco modified Eagle medium (DMEM) (Gibco) including 1% penicillin-streptomycin–gentamicin and 10% heat-inactivated fetal bovine serum (FBS). Contaminating red blood cells (RBC) present in the single-cell suspensions were then lysed with RBC lysis buffer (Multispecies; Affymetrix eBioscience) and washed prior to shipment. The average cell recovery for each tissue was the following, represented as x/y, where x denotes naive animals and y denotes animals at 10 days postinfection: peripheral blood mononuclear cells (PBMC), 2.4 × 106/ml/2.9 × 106/ml; spleen, 4.1 × 108/1.3 × 109; cervical lymph node (cLN), 2.5 × 107/7.0 × 107; and bronchoalveolar lavage (BAL) fluid, 3.1 × 106/5.7 × 106.
Derivation of ferret anti-CD4 antibody.
Female BALB/c mice were purchased from Charles River Laboratories and used at 2 to 4 months of age. Mice were first vaccinated subcutaneously in the rear footpad with a complete Freund's adjuvant emulsion of ferret CD4 protein (25 μg/mouse), purchased from Sino Biologicals. Mice were then boosted sequentially at two 4-week intervals with 25 μg CD4 in incomplete Freund's adjuvant (IFA) in the footpad, and finally 25 μg CD4 intravenously. At 3 days postboost, the mice were sacrificed and single-cell suspensions were prepared from the spleens. Fusion was performed as previously described (55), except that the fusion partner used was SP2-0 that had been transfected with the gene encoding interleukin-6 (IL-6) (ATCC catalog number CRL-2016). After fusion, cells were plated by limiting-dilution cloning and then initially screened based on enzyme-linked immunosorbent assay (ELISA) reactivity with the ferret CD4 compared to an unrelated recombinant protein (HA). Positive cells were subcloned again by limiting-dilution cloning, and those cells arising from single cells were expanded, retested by ELISA as described above for antibody production, and cryopreserved. A single, high-producing clone was selected and expanded in DMEM with low-IgG-containing serum (Gibco Life Technologies), and the supernatant was collected for antibody purification when the culture was approximately 50% viable. The antibody was purified from the supernatant via binding to PGS Plus (Thermo Fisher), elution with McIlvaine's buffer (pH 3.0) (0.2 M Na2HPO4, 0.1 M citric acid) in 1-ml fractions, and neutralization of eluted samples with 2 M Tris (pH 8.0). The eluted antibody was dialyzed with phosphate-buffered saline (PBS) to remove all Tris and concentrated using Amicon spin concentrators to a final concentration of 1 mg/ml. Purified antibody (100 μg) was directly conjugated to Alexa Fluor 488, 555, or 647 using Molecular Probes (Life Technologies) labeling kits according to the manufacturer's instructions.
Synthetic peptides and libraries.
Seventeen-mer peptides overlapping by 11 amino acids to encompass the entire sequences of the viral proteins were obtained from the NIH Biodefense and Emerging Infections Research Repository, NIAID, NIH. They were reconstituted as 10 mM stocks using a dimethyl sulfoxide (DMSO)-water solution and dithiothreitol (DTT) for cysteine-containing peptides. The peptide arrays used included influenza virus A/California/04/09 (H1N1) HA protein (NR-15433), A/California/04/09 (H1N1) NA proteins (NR-18975), A/New York/444/2001 NS1 (NR-2612), and A/New York/348/2003 (H1N1) NP (NR-2611) and M1 (NR-2613). A pool of irrelevant peptides from Sin Nombre virus (NR-4764) was used as a negative control to account for any suppression from large pools of peptides. Peptides were pooled such that each peptide was at a final concentration of 1 μM in the assay. For the HA peptides, the pool was broken into two pools comprising the amino-terminal portion (peptides 1 to 78) and the carboxy-terminal portion (peptides 79 to 139) of the protein.
Flow cytometry.
Single-cell suspensions were transferred into a 96-well microtiter plate at a concentration of 10 to 20 million cells per ml and washed twice in fluorescence-activated cell sorting (FACS) buffer (Dulbecco's PBS [DPBS] plus 2% heat-inactivated FBS), centrifuging at 400 × g at 4°C using a Sorvall RT6000B with a HT1000B rotor. The LIVE/DEAD Fixable Aqua dead-cell stain kit (ThermoFisher) was used as a viability dye where specified according to the manufacturer's instructions. Cells were protected from light and surface stained in a final volume of 50 μl for 30 min at 4°C using antibodies shown in Table 1. Antibodies to CD4 (CL3.1.5), CD8 (OK-T8), CD11b (M1/70), MHC-II (L243), Ig (polyclonal antibody to IgA, IgM, and IgG), and Ly6c (AL-21) were directly conjugated to fluorochromes and used to identify T cells, B cells, myeloid-derived cells, and professional antigen-presenting cells (APCs). Cells were then washed two times in FACS buffer and resuspended for data acquisition. Antibodies to CD20 (71) and CD79A (HM47) were used for intracellular staining using the BD Cytofix/Cytoperm kit. For analytical staining experiments, events were collected using a 12-color BD LSR-II instrument with 488-, 633-, and 405-nm lasers and FACS Diva software. For FACS, a BD FACSAria instrument with 488-, 633-, 407-, and 522-nm lasers was used, and events were collected through a 100-μm nozzle under purity settings. All data files were analyzed using FlowJo software, version 8.8.7.
TABLE 1.
Antibodies used for leukocyte identificationa
| Antigen | Alternative name(s) | Specificity | Clone | Isotype | Vendor |
|---|---|---|---|---|---|
| CD4 | Ly-4, L3T4, T4 | Ferret | 3.1.5 | Mouse IgG1 | Sant lab (URMC) |
| CD8α | CD8 alpha, Leu2, MAL, T8, p32 | Human | OK-T8 | Mouse IgG2a | TONBO |
| CD11b | αM integrin, Mac-1, Mo1, CR3, Ly-40, C3biR, ITGAM | Mouse/human | M1/70 | Rat IgG2b, κ | BioLegend |
| MHC-II | HLA-DR | Human | L243 | Mouse IgG2a, κ | BioLegend |
| IgA, IgG, IgM | Immunoglobulin | Ferret | Polyclonal antibody | Goat | LSBio |
| CD79ab | mb-1 | Human | HM47 | Mouse IgG1, κ | eBioscience |
| CD20b | MS4A1 | Ferret | 71 | Rabbit IgG | Sino |
| CD59 | Ly6c | Mouse | AL-21 | Rat IgM, κ | BD Pharmingen |
Single-cell suspensions derived from lymphatic tissues, including spleen and lymph nodes, were stained with each of the listed antibodies. Data were acquired using a BD LSR-II instrument and analyzed using FlowJo software, version 8.8.7.
Intracellular staining.
Reverse transcription-PCR (RT-PCR) assays.
CD4 T cells, CD8 T cells, and B cells were sorted using the BD FACSAria cell sorting system described above. A minimum of 10,000 sorted cells were pelleted, and total RNA was purified using the RNeasy Plus minikit (Qiagen) according to the manufacturer's guidelines. cDNA was synthesized from random hexamers using the SuperScript IV reverse transcriptase kit (Invitrogen) as per the manufacturer's instruction. Input cDNA was normalized between samples to 600 ng/μl, with 2 μl spiked into a 25-μl PCR mixture using Platinum Hot Start PCR 2× master mix (Invitrogen). A Bio-Rad T100 thermal cycler was used to generate double-stranded DNA (dsDNA) products using the following parameters: 94°C for 2 min (once), 94°C for 30 s (30 times), 45 to 50°C (annealing temperature 5°C below the melting temperature [Tm] for each gene-specific primer set) for 30 s (30 times), 72°C for 1 min (30 times), 72°C for 5 min (once), and then 4°C. PCR products were then loaded onto a 1% Tris-acetate-EDTA (TAE) agarose gel in the presence of loading dye containing xylene cyanol, stained with ethidium bromide, and visualized using an Alpha Innotech gel imager and AlphaEaseFC software. Primers were designed to amplify all predicted transcript variants of the target gene if more than one variant was described in NCBI's PubMed resource (see Table 3).
TABLE 3.
RT-PCR primer summary
| Gene | NCBI accession no. | Primer | Nucleotide sequence (5′→3′) | Tm (°C) | Amplicon length (bp) |
|---|---|---|---|---|---|
| CD4 | EF492055.1 | Forward | GATCCTTCCCTCTGATTATCAAG | 52.4 | 981 |
| Reverse | GGAGAAGCAGAAGACCTAAGATCC | 56.5 | |||
| CD8α | EF492056.1 | Forward | CTCCTCCAGAAGAACGAACCTG | 57.1 | 514 |
| Reverse | GTCCTCCTTGTCTGGGCATTGG | 60.2 | |||
| CD3ε | XM_004749946.2 | Forward | CATGCCCTGAGGAGGTTGAT | 57.0 | 328 |
| XM_013054918.1 | Reverse | GCGCCTTTTGCTCTCATCAC | 57.1 | ||
| CD79b | XM_004749159.2 | Forward | GGCAGAACCCACGCTTCATAG | 56.2 | 520 |
| Reverse | CGACTGACCATTTCACCTCTCC | 57.2 | |||
| CD20 | XM_004770405.2 | Forward | CTAGGCGGTCTCCTGATGATTCAC | 58.8 | 674 |
| XM_013044696.1 | Reverse | GGTGAAGGTTCCTGGTCCTG | 57.7 | 635 | |
| XM_013044697.1 | 464 | ||||
| CD14 | XM_004744791.2 | Forward | TCTGTCAGCACAAGTTCCCG | 57.4 | 457 |
| Reverse | CTCTGAAGCACCGCTAAGGT | 57.0 | |||
| CD11b | XM_013046568.1 | Forward | GTGATGTTCAGAGAGAGTGCTGG | 57.3 | 1,229 |
| Reverse | CGTCTCCATGAGAGCTGCTTCTG | 59.5 | |||
| Glyceraldehyde-3-phosphate dehydrogenase | EF392835.1 | Forward | CATGTTCCAGTATGATTCTACCCAC | 55.2 | 659 |
| Reverse | CCTGCTTCACCACCTTCTTGATGTC | 59.7 |
Ferret ELISpot assays.
ELISpot assays were performed as previously described (56, 57). Ninety-six-well filter plates (Millipore) were coated with 5 μg/ml purified anti-ferret IFN-γ (clone MTF14; MabTech) in PBS overnight and then washed and blocked with cell culture medium (DMEM; Gibco Life Technologies). Undepleted ferret lymphocytes or PBMC were plated at 300,000 cells per well with pools of peptides for 18 h at 37°C in 5% CO2. Alternatively, splenocytes (300,000 per well) that had been depleted of CD4 or CD8αhi cells using preparative flow cytometry with antibodies described above and a BD FACSAria cell sorting system were cultured with pools of peptides as described above. After incubation, cells were removed from the plates by washing. Biotinylated anti-ferret IFN-γ (clone MTF19; MabTech) was added in wash buffer with 10% FBS at 2 μg/ml and incubated at room temperature for 2 h. The plates were washed, and then streptavidin-conjugated alkaline phosphatase (Jackson Laboratories) was added, incubated for 30 min, washed, and developed using Vector Blue substrate kit III (Vector Laboratories, CA) prepared in 100 mM Tris, pH 8.2. Quantification of cytokine-secreting cells was performed with an Immunospot reader series 2A, using Immunospot software, version 5.0.9.19.
RESULTS
Flow cytometry reveals the distribution and compartmentalization of leukocytes at steady state.
Because of the interest in and value of studying the specificity, frequency, and abundance of immune cells following infection and vaccination in ferrets, our first goal was to identify and develop key antibody reagents that would allow for the study of T and B lymphocytes, myeloid-lineage cells, and antigen-presenting cells (APCs) within lymphoid organs (Table 1). Consistent with other reports (53, 58), we observed that human CD8α (OK-T8) and CD11b (M1/70) monoclonal antibodies were cross-reactive in ferrets, allowing for identification of conventional CD8 T cells and myeloid cells. Additionally, we found a human HLA-DR monoclonal antibody (L243) that was also cross-reactive, allowing the identification of both professional (dendritic cells, B cells, and macrophages) and nonprofessional APCs. Because of the importance of identifying CD4 T cells, we generated a high-affinity monoclonal ferret CD4 antibody (3.1.5) using previously described methods (55). B cells were identified using a polyclonal antibody recognizing ferret IgA, IgM, and IgG isotypes as well as CD20 and CD79a (see below).
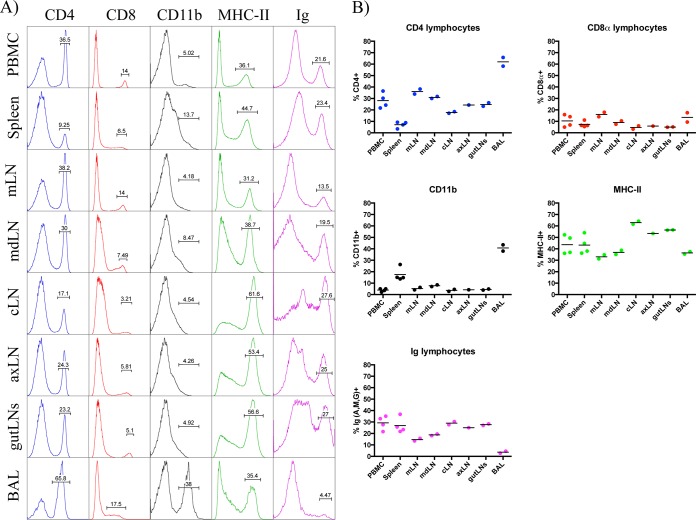
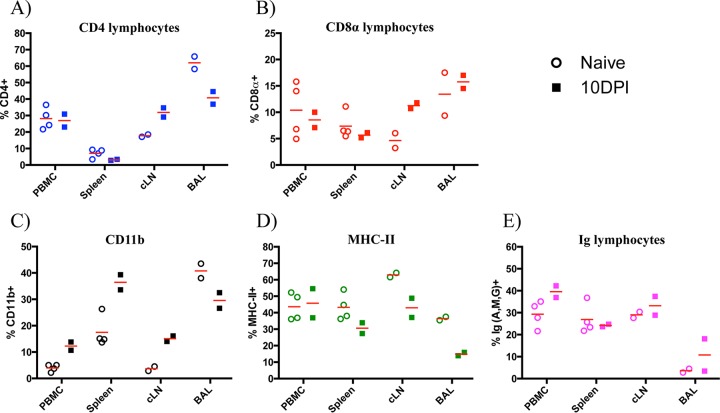
To study the contribution of the different cell types following infection in ferrets, we first sought to measure the leukocyte composition within various lymphatic tissues at steady state in animals that were previously confirmed to be seronegative to influenza virus. Shown in Fig. 1 and summarized in Table 2 are the flow cytometry staining profiles and frequencies of CD4+, CD8α+, CD11b+, MHC-II+, and Ig+ cells from peripheral blood, spleen, and bronchoalveolar lavage (BAL) fluid. We also sampled the lymph nodes that drain the upper and lower respiratory tracts and other peripheral lymph nodes. Interestingly, the frequency of cells expressing these markers varied dramatically among tissues. For example, averaged CD4 T cell frequencies ranged from approximately 7% in the spleen to 28% in the PBMC and over 60% in the BAL fluid, while CD8 T cells ranged from 5 to 16% depending on the lymph node studied and were 13% in the BAL fluid. High frequencies of MHC-II+ cells were identified in all tissues, at approximately 37% in the BAL fluid, 45% in the spleen, and 33 to 63% in lymph nodes. Using the percentages of T cells and B cells within each tissue, the relative ratios of the two populations was determined and stratified by tissue (Fig. 2). While the ratios were similar for the two cell types in the PBMC (1.3), cLN (0.8), axillary LN (axLN) (1.2), and gut LNs (1.1), we found higher T cell/B cell ratios in the mediastinal LN (mLN) (3.6), mandibular LN (mdLN) (2.1), and BAL fluid (21.0) and a lower ratio in the spleen (0.5) (Fig. 2A). Interestingly, when comparing the CD4 and CD8 T cell subsets, we found that a CD4/CD8 ratio of at least 2.0 was observed within all tissues except the spleen, which had a more balanced composition of 1.0 (Fig. 2B).
FIG 1.
Leukocyte composition by tissue at steady state. (A) Representative staining profiles of CD4+, CD8αhi, MHC-II+, and Ighi cells by tissue, depicted as histogram plots. Results for peripheral blood mononuclear cells (PBMC), mediastinal LN (mLN), mandibular LN (mdLN), cervical LN (cLN), axillary LN (axLN), bronchoalveolar lavage (BAL) fluid are shown. Percentages of T and B lymphocytes were derived from a lymphocyte gate based on forward scatter (FSC) versus side scatter (SSC). (B) Frequency of positive events for each surface marker, with each symbol representing a single animal and mean values denoted by a horizontal line. Note that percent CD8α+ and Ig+ represents the fraction of cells with high mean fluorescence intensity (MFI) expression.
TABLE 2.
Summary of leukocyte composition by tissue at steady state
| Tissue | n | Frequency (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CD4 |
CD8 |
CD11b |
MHC-II |
Ig |
|||||||
| Mean | Range | Mean | Range | Mean | Range | Mean | Range | Mean | Range | ||
| PBMC | 4 | 28.2 | 21.7–36.5 | 10.4 | 4.9–15.8 | 4.0 | 2.2–5.03 | 43.7 | 36.1–52.2 | 29.3 | 21.6–35.1 |
| Spleen | 4 | 7.1 | 3.4–9.3 | 7.4 | 5.5–11.1 | 17.5 | 13.7–26.3 | 43.2 | 36.2–54.0 | 26.9 | 21.8–36.8 |
| mLN | 2 | 36.1 | 33.9–38.2 | 16.0 | 14.0–17.9 | 5.2 | 4.2–6.3 | 33.0 | 31.2–34.8 | 14.6 | 13.5–15.7 |
| mdLN | 2 | 30.9 | 30.0–31.8 | 8.9 | 7.5–10.4 | 7.8 | 7.1–8.5 | 36.9 | 35.1–38.7 | 18.8 | 18.1–19.5 |
| cLN | 2 | 17.8 | 17.1–18.5 | 4.6 | 3.2–6.0 | 3.7 | 2.8–4.5 | 62.9 | 61.6–64.2 | 29.0 | 27.6–30.4 |
| axLN | 1 | 24.3 | 5.8 | 4.3 | 53.4 | 25.0 | |||||
| Gut LNs | 2 | 24.7 | 23.2–26.2 | 5.0 | 4.9–5.1 | 4.4 | 4.0–4.9 | 56.5 | 56.3–56.6 | 27.7 | 27.0–28.4 |
| BAL fluid | 2 | 62.0 | 58.2–65.8 | 13.4 | 9.4–17.5 | 40.8 | 38.0–43.5 | 36.5 | 35.4–37.5 | 3.6 | 2.8–4.5 |
FIG 2.
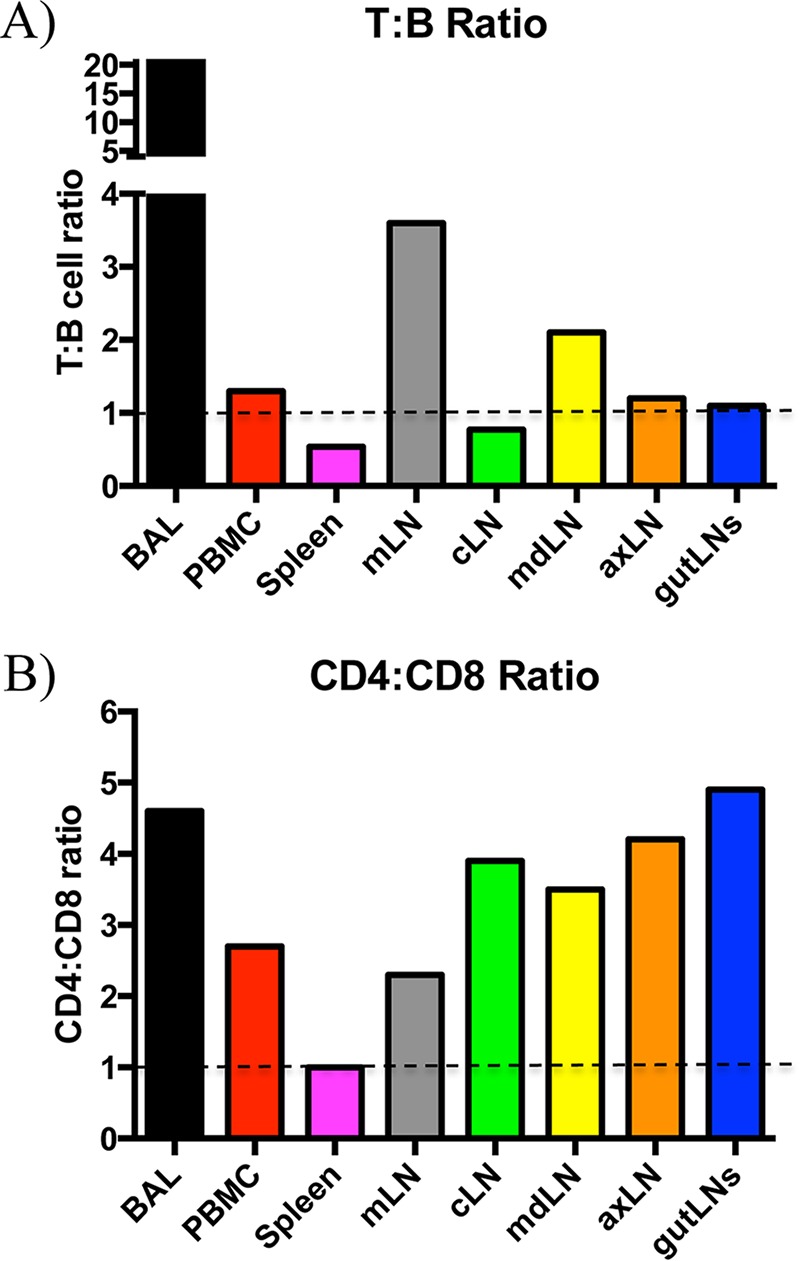
Lymphocyte frequency is distinct within multiple tissue compartments at steady state. (A) Lymphocyte composition, represented as a ratio of T cells (summed means from CD4+ and CD8αhi cells) to B cells (Ighi). (B) T cell composition, represented as a ratio of CD4+ to CD8αhi cells by tissue.
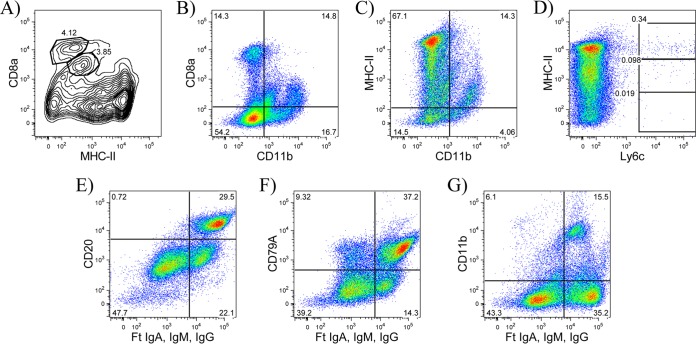
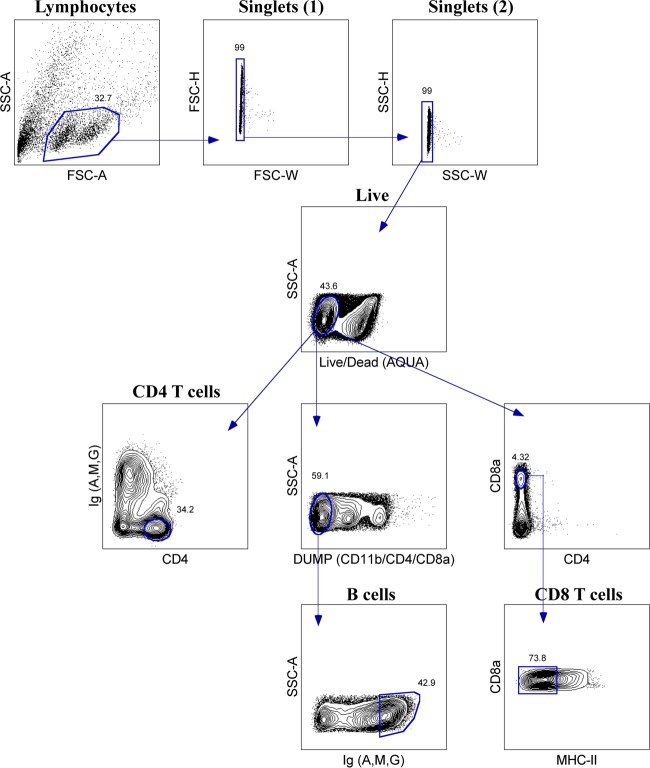
In addition to striking differences in cell frequency, we also observed heterogeneity in the patterns of cell surface expression of some markers within different tissues. Although expression of CD4+ cells appeared to be fairly uniform across tissues, we observed a wider range of expression for CD8α+ cells, particularly within the spleen and BAL fluid relative to other tissues. In mice, a subset of dendritic cells identified as CD8α+ has been well characterized (reviewed in references 59 and 60). To evaluate the possibility that some of the CD8α+ cells were APCs, we performed costaining experiments. Splenocytes stained with antibodies specific for MHC-II and CD11b revealed subpopulations of CD8αlo/int MHC-IIlo cells and CD11b+ cells, whereas conventional CD8 T cells were resolved based on high-density expression of CD8α (Fig. 3A and B). Heterogeneity in the expression of MHC-II molecules was also observed, with subsets of CD11b+ cells that were MHC-IIlo and Ly6c+ cells that were mostly MHC-IIhi, possibly identifying subsets of granulocytes and/or macrophages and dendritic cells (Fig. 3C and D). Finally, in some tissues, we observed distinct populations with intermediate and high Ig expression. Costaining with ferret CD20 and cross-reactive human CD79A monoclonal antibodies revealed that the majority of B cells were Ighi and that a subpopulation of Igint cells were CD11b+. This result suggests that some non-B cells may capture low levels of Ig through binding Fc receptors (FcRs) (Fig. 3E to G) and that B cells could be identified as Ighi cells. Currently, there is no antibody available to block ferret FcR-mediated binding. We did, however, test normal ferret serum as a way to minimize this result for ease of population delineation, but we did not observe a noticeable change in the distribution of cells detected by flow cytometry. These data collectively demonstrate that many distinct cell populations are represented within the different lymphoid tissues. These characteristics of Ighi B cells and CD8αhi T cells were used in the gating strategies to quantify B cells and CD8 T cells in these studies.
FIG 3.
Phenotypic characterization of immune cells by flow cytometry. (A and B) CD8α+ cells are heterogeneous, with high-MFI cells representing conventional CD8 T cells and low/intermediate-MFI cells containing subpopulations of MHC-IIlo cells and CD11b+ cells. (C and D) MHC-II+ cells are heterogeneous, with MHC-IIlo cells containing a subpopulation of CD11b+ cells and MHC-IIhi cells containing a subpopulation of Ly6c+ cells. (E and F) Ighi cells identify B cells. (G) Igint cells contain myeloid-derived cells (CD11b). Data are representative from the spleens of at least two ferrets. Ighi B cells were pregated on live lymphocytes that were SSC-Alo.
Gene expression patterns support the identity of antibody-selected leukocyte populations.
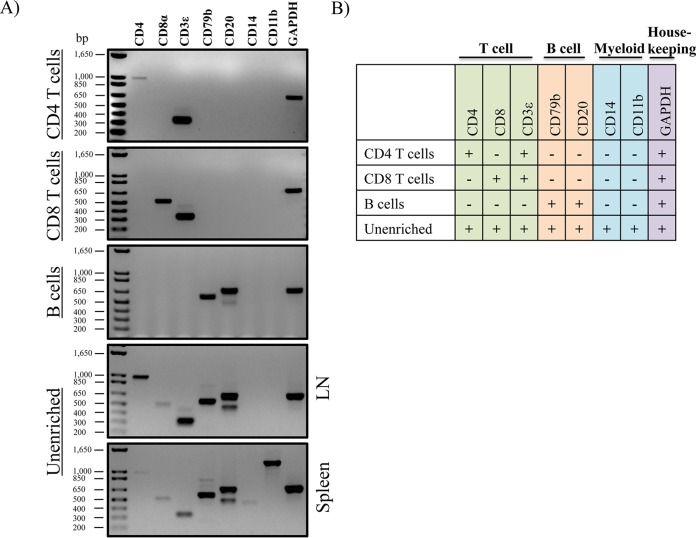
Because of the cross-reactive nature of some of the antibodies and possible cellular heterogeneity within the antibody-selected populations, we next sought to validate the specificity of the antibodies used for immunotyping (Fig. 1). To this end, we performed RT-PCR to get a qualitative assessment of the gene expression profiles of CD4 and CD8 T cells and B cells. As shown in Fig. 4, single, live lymphocyte cells were sorted to high purity into the following three populations: (i) CD4 T cells (CD4+ Ig−), (ii) CD8 T cells (CD8αhi CD4− MHC-II−), and (iii) B cells (CD11b− CD4− CD8α− Ighi). Total RNA was isolated, and contaminating genomic DNA was removed. cDNA was synthesized, and primers (Table 3) were designed to amplify a subset of genes that could distinguish T and B lymphocytes and myeloid cells. While all of the targeted genes could be detected to various expression levels between spleen and lymph node tissue in unenriched cells (including myeloid-lineage-associated genes, CD14 and CD11b), CD4 T cells were enriched in transcripts corresponding to CD4 and CD3ε, while CD8 T cell were enriched for CD8α and CD3ε, demonstrating that our monoclonal CD4 antibody and cross-reactive CD8α antibodies are antigen specific (Fig. 5). Furthermore, CD79b and CD20 were specifically expressed in sorted B cells and not in the other sorted populations, also demonstrating a high degree of specificity.
FIG 4.
FACS sorting scheme for gene expression to validate antibody specificities.
FIG 5.
Antibodies used for leukocyte identification are cell type specific. (A) Gene expression from sorted cell populations. At least 10,000 cells were sorted, total RNA was purified, and genomic DNA was removed. cDNA was synthesized, and residual RNA was removed via RNase H digestion. The cDNA concentration was normalized for each sample, and gene-specific primers were used for RT-PCR. Note that CD8 T cells and B cells were sorted from splenocytes. Due to the low frequency of CD4+ T cells in the spleen, sorting was performed using cells from lymph node tissue. (B) Summary of RT-PCR results, organized into distinct leukocyte modules.
Immune cell composition is altered by influenza virus infection.
After quantifying and validating the frequencies of distinct cell populations in naive animals, our next goal was to determine if the cellular composition in circulating blood, lymphoid tissues, and BAL fluid would be altered following an encounter with influenza virus. To this end, two ferrets were infected intranasally with 10,000 PFU of A/California/4/09 (H1N1) and sacrificed 10 days later. Figure 6 shows the mean frequency and range of responses from both ferrets within the PBMC, spleen, cLN, and BAL fluid. Compared to those in naive hosts, higher frequencies (≥2-fold) of total CD8 T cells and CD11b+ cells were found in the cLN draining the upper airway. Further, while there were decreases in total CD4 T and CD8 T cells in the spleen after infection, there was a corresponding increase in the cLN, suggesting possible priming and expansion of naive cells. Finally, at least a 2-fold increase in CD11b+ cells was observed in the peripheral blood, spleen, and cLN, reflecting cellular alterations in at least one of the following possible leukocyte populations: monocytes, macrophages, dendritic cells, natural killer cells, neutrophils, and granulocytes. Collectively, the data suggest that the host mounted a coordinated immune response involving innate and adaptive cellular participants.
FIG 6.
Leukocyte composition is altered at 10 days following influenza A virus infection. Percentages of total CD4+ (A), CD8αhi (B), CD11b+ (C), MHC-II+ (D), and Ighi (E) cells by tissue are shown. Data represent total percentages for each cell type (antigen independent); symbols represent individual animals, and horizontal lines indicate the mean frequency of positive events.
Antigen-specific CD4 and CD8 T cell responses to influenza A virus are diverse and display distinct patterns of reactivity in individual animals.
It is becoming increasingly clear that viral protein specificity may be a key determinant in the effector program of influenza virus-specific T cells (25, 61–63). However, due to a lack of ferret-specific reagents, measuring antigen-specific CD4 and CD8 T cell responses has not been possible. Therefore, unlike our understanding of influenza immunity in mice and humans, it is unknown what fraction of the T cell response in ferrets is influenza virus specific during any stage following infection. Therefore, a major goal of this work was to quantify the viral protein specificity and functionality of the elicited T cells from key sites of the adaptive response. To this end, T cells isolated from different tissues, including spleen, PBMC, and airway draining lymph nodes, were assayed for reactivity to the following viral proteins: HA (partitioned into the HA1 and HA2 domains), NA, NP, NS1, and M1. Pools of 17-mer peptides, overlapped by 11 amino acids, were selected in order to offer all potential epitopes for recall of influenza virus-specific CD4 and CD8 T cells from the repertoire. Because it is well known that the host immune response to infection is dominated by IFN-γ-secreting CD4 T helper type 1 and effector CD8 T cells (50, 54, 64), we performed IFN-γ ELISpot assays at 10 days postinfection using the same animals on which we had performed analytical staining, sampling unfractionated cells from the spleen, cLN, and PBMC.
Figure 7A shows the magnitude of the antigen-specific T cell response to each of the viral proteins described above, as well as total reactivity when responses to all specificities are summed. The assays revealed that the T cell response has broad protein specificity that is detectable across all tissues sampled, ranging from 10 to 500 spot-forming units (SFU) per million cells, depending on the individual animal, viral protein, and tissue sampled. When the responses to all protein specificities were summed (Fig. 7A, rightmost panel), it was found that as many as 0.1% of the T cells in the PBMC, 0.07% in the spleen, and 0.04% in the cLN were influenza virus specific. Although the overall abundances of influenza virus-reactive cells were comparable among the animals, the protein specificity in the individual animals was distinct. For instance, within the PBMC for ferret 2466, most of the response was focused toward NP (32%) and NS1 (57%), while T cells from ferret 2476 was focused largely toward the HA1 domain of HA (30%), NA (31%), and NS1 (29%). Heterogeneity in individual responses is further illustrated in Fig. 7A to C, which includes T cell reactivity in the PBMC from another ferret at 23 days postinfection via aerosol contact, whose antigen-specific response was more equally distributed among several viral proteins. While the magnitude of this response was lower than that at 10 dpi, as expected due to cellular contraction following viral clearance (65), it is clear that T cells specific to several viral proteins become primed and can be detected during both the effector phase and transition into memory.
Because the spleen is high in cellularity and has been shown to contain influenza virus-reactive T cells, we were able to selectively deplete CD4+ or CD8αhi cells for use in the ELISpot assays to separately interrogate CD8 or CD4 T cell-specific reactivity, respectively. These studies reveal striking patterns of reactivity for both T lymphocyte subsets as well as between individual ferrets. Following infection, there were differences in the magnitude and hierarchy of protein-specific responses in the animals. For ease of comparison among animals and T cell subsets, the responses to all epitopes, indicated in Fig. 8A, were summed, and then the proportions of the response dedicated to individual viral proteins were represented as a “pie chart,” where the size of the slice reflects the fractional response to individual proteins (Fig. 8C). For example, in ferret 2466, over 50% of the CD4 T cells were reactive to M1, while only 10% of the response was dedicated to NS1. In contrast, ferret 2476 had a broader distribution of reactivity among the different viral proteins tested, with reactivity distributed among epitopes within M1, NS1, and the amino-terminal half of HA (Fig. 8B and C). With respect to the CD8 response, we detected a slightly elevated range of total influenza virus reactivity (0.08 to 0.1%) in the spleen, with a high percentage (44% ± 7%) of CD8 T cell reactivity directed toward NS1 (Fig. 8B and C). Of note and similar to the observations for the CD4 response, distinct patterns of reactivity were present in individual animals. For example, although a large percentage of the CD8 response was NS1 specific in both animals, the T cell reactivity of ferret 2476 was dominated by NA-specific cells (47%), compared to just 2% reactivity exhibited by ferret 2466 (Fig. 8C). Altogether, the heterogeneity observed from the total and fractionated CD4 and CD8 T cell reactivity data sampled from multiple tissues postinfection suggests that there are likely to be many options for MHC-restricted peptide epitope display to surveying T cells in ferrets. These results also suggest that there may be considerable MHC heterogeneity in commercially available ferrets, leading to distinct patterns of CD4 and CD8 specificity among individuals. In this manner, ferrets may be more similar to humans and outbred mice than to typical inbred strains of mice that are homozygous for a given set of MHC class I and class II genes.
DISCUSSION
In this study, our goals were to develop reliable procedures and reagents to identify and quantify key cell types responsible for orchestrating adaptive immune responses and antigen-specific T cell responses following infection of ferrets with influenza virus. We used a combination of cross-reactive and ferret-specific antibodies to identify key immune cell populations, including CD4 and CD8 T cells, myeloid-lineage cells (CD11b+), MHC-II+ APCs, and B cells. Similar to what has been observed in a recent study in humans (66), we found that leukocyte subset frequency was intrinsic to the tissue site and similar across different animals and became modified in the composition of cellular components following influenza infection. When the ratios of T cells to B cells and CD4 to CD8 T cells were compared, the compositions were generally comparable for what has been described in humans and what we found in ferrets. For instance, in both humans and ferrets, the T cell/B cell ratio was lowest in the spleen and highest in the lung/BAL fluid. Furthermore, we observed a general trend of a higher CD4/CD8 ratios across all tissues except the spleen. Collectively, these results suggest that although the ferret leukocyte composition across tissues is distinct, there are some general similarities to that in humans. Moreover, these experiments reveal the potential for comprehensive tracking of distinct immune cell subsets to study their spatiotemporal regulation following encounters with influenza viruses and vaccines.
Analyses of the cell surface profiles for CD8α+, MHC-II+, and Ig+ cells suggested that the populations of cells detected by our antibodies may consist of distinct subsets. For example, we observed a range in surface expression of CD8α in cells recovered from spleen, BAL fluid, and lymph nodes. In mice and sheep, subsets of dendritic cells have been described to express CD8α (67, 68). In mice, these dendritic cells represent a key subset involved in priming T cells and particularly for cross-presentation of CD8 T cells after influenza infection and vaccination (69). Using a costaining approach, we also observed differences in expression patterns for MHC-II and Ig in some lymph nodes and BAL fluid following infection, which may reflect differences in the maturation status of B cells, since activated B cells upregulate MHC-II and plasma cells express lower levels of the Ig receptor in response to antigen stimulation (70, 71).
In addition to CD4 and CD8 T cell frequency and expression patterns, our studies revealed interesting patterns of T cell functionality and specificity. In both mice and humans, it is well documented that the CD4 T cell repertoire to influenza virus antigens is broad (72–75). While there have been multiple influenza virus specificities detected in CD8 T cells in humans (76), the breadth of CD8 T cell reactivity in mice is less well understood. CD8 reactivity in mice is generally thought to be narrower than that of their CD4 counterparts. This could be attributed to the fact that the majority of studies have utilized C57BL/6 mice, where most of the response to influenza virus is specific to two peptides, derived from NP and polymerase acidic protein (PA) (77, 78). These studies also suggest that the outbred ferrets are likely to differ in the expression of MHC proteins. The isotypic and allelic variation of MHC class I and class II proteins is currently unknown; however, the finding that the human-specific antibodies L243 and L203, reactive to the HLA-DR isotype of class II (79), detected all ferret lymphoid tissues studied so far (this report and data not shown) suggests that ferrets will express an HLA-DR/I-E homologue. Whether they also express a class II homologue to HLA-DQ/I-A or HLA-DP is not known at this time.
The information gained through these studies that dissected the abundance and specificity of antigen-specific CD8 and CD4 T cell responses after infection is important for several reasons. Our laboratory (80) and others (81) have shown that there is a deterministic linkage in specificities for T and B lymphocytes in response to enveloped viruses. In a mouse model of influenza infection, we discovered that the magnitude and kinetics of the HA-specific B cell response were accelerated when the host was armed with memory HA-specific CD4 T cells but were not helped in the presence of NP-specific CD4 cells and vice versa (80). Also, in tracking expansion of human CD4 T cells after vaccination, we determined that the HA-specific response, rather than to those specific for M1 and NP, correlated best with the neutralizing antibody response (56). This conclusion was reinforced with a recent study of human H5N1 vaccine responses performed by our group, which showed that prepandemic priming of human subjects selectively enhanced representation of HA-specific CD4 T cells, a phenotype that correlated with robust neutralizing antibody responses (82). These findings are important when considering correlates of immune protection, since the HA-specific neutralizing antibodies are thought to be the most dominant form of protection, due to their ability to achieve sterilizing immunity. However, HA-specific CD4 helper T cells to novel epitopes could be outcompeted or suppressed by the expansion of memory cells to other protein specificities such as NP that are more highly conserved and routinely encountered (83). Finally, although less fully explored, it is also possible that CD4 T cells of other specificities partition with other distinct effector programs following infection. For example, in a recent human challenge model with H3N2 or H1N1 viruses, it was noted that that preexisting CD4 T cells specific for matrix or NP correlated best with lower virus shedding and disease. These cells also possess cytotoxic activity (25), which is also thought to be a key effector activity of CD4 T cells (50, 84, 85). The reagents and methods defined here will allow us to better probe links between specificity and function in ferrets.
In summary, this study provides an in-depth analysis of the CD4, CD8, and APC composition in ferrets before and after influenza infection and has quantified the CD4 and CD8 T cell viral antigen specificity postinfection. These studies have revealed a robust adaptive T cell response and changes in distribution of key lymphoid cells postinfection. These studies have also suggested that commercially available outbred ferrets are likely to have considerable MHC heterogeneity and that individual ferrets are able to recruit CD4 and CD8 T cells of broad antigen specificity. Altogether, the progress reported here lends itself to several important avenues of investigation that can be immediately addressed, including T lymphocyte responses (e.g., phenotype and functionality) following infection with highly pathogenic avian viruses with pandemic potential (H5N1 and H7N9) as well as different formulations of influenza vaccines, which may differ by multiple factors, including replication competency, antigen composition/context (e.g., valency), and adjuvant. These studies will be critical to our understanding of how preexisting immunity and its augmentation/regulation impact protection. Furthermore, having the ability to monitor T cells in ferrets will enable studies investigating their role in controlling virus transmission, a critical area of vaccine efficacy evaluation in challenge experiments (86, 87).
ACKNOWLEDGMENTS
We are grateful for technical assistance with cell sorting experiments provided by Matt Cochran at the University of Rochester flow cytometry core facility. We also acknowledge the NIAID CEIRS Team Ferret group for support for these experiments.
REFERENCES
- 1.Simonsen L, Fukuda K, Schonberger LB, Cox NJ. 2000. The impact of influenza epidemics on hospitalizations. J Infect Dis 181:831–837. doi: 10.1086/315320. [DOI] [PubMed] [Google Scholar]
- 2.Horimoto T, Kawaoka Y. 2005. Influenza: lessons from past pandemics, warnings from current incidents. Nat Rev Microbiol 3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 3.Thompson WW, Comanor L, Shay DK. 2006. Epidemiology of seasonal influenza: use of surveillance data and statistical models to estimate the burden of disease. J Infect Dis 194(Suppl 2):S82–S91. doi: 10.1086/507558. [DOI] [PubMed] [Google Scholar]
- 4.Neumann G, Noda T, Kawaoka Y. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morens DM, Taubenberger JK, Fauci AS. 2013. Pandemic influenza viruses—hoping for the road not taken. N Engl J Med 368:2345–2348. doi: 10.1056/NEJMp1307009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uyeki TM, Cox NJ. 2013. Global concerns regarding novel influenza A (H7N9) virus infections. N Engl J Med 368:1862–1864. doi: 10.1056/NEJMp1304661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Childs RA, Matrosovich T, Wharton S, Palma AS, Chai W, Daniels R, Gregory V, Uhlendorff J, Kiso M, Klenk HD, Hay A, Feizi T, Matrosovich M. 2010. Altered receptor specificity and cell tropism of D222G hemagglutinin mutants isolated from fatal cases of pandemic A(H1N1) 2009 influenza virus. J Virol 84:12069–12074. doi: 10.1128/JVI.01639-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mak GC, Au KW, Tai LS, Chuang KC, Cheng KC, Shiu TC, Lim W. 2010. Association of D222G substitution in haemagglutinin of 2009 pandemic influenza A (H1N1) with severe disease. Euro Surveill 15(14):pii=19534 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19534. [PubMed] [Google Scholar]
- 9.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klenk HD, Garten W. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol 2:39–43. doi: 10.1016/0966-842X(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 12.Horimoto T, Nakayama K, Smeekens SP, Kawaoka Y. 1994. Proprotein-processing endoproteases PC6 and furin both activate hemagglutinin of virulent avian influenza viruses. J Virol 68:6074–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Wit E, Kawaoka Y, de Jong MD, Fouchier RA. 2008. Pathogenicity of highly pathogenic avian influenza virus in mammals. Vaccine 26(Suppl 4):D54–D58. doi: 10.1016/j.vaccine.2008.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tscherne DM, Garcia-Sastre A. 2011. Virulence determinants of pandemic influenza viruses. J Clin Invest 121:6–13. doi: 10.1172/JCI44947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subbarao EK, Kawaoka Y, Murphy BR. 1993. Rescue of an influenza A virus wild-type PB2 gene and a mutant derivative bearing a site-specific temperature-sensitive and attenuating mutation. J Virol 67:7223–7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bussey KA, Bousse TL, Desmet EA, Kim B, Takimoto T. 2010. PB2 residue 271 plays a key role in enhanced polymerase activity of influenza A viruses in mammalian host cells. J Virol 84:4395–4406. doi: 10.1128/JVI.02642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zamarin D, Garcia-Sastre A, Xiao X, Wang R, Palese P. 2005. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog 1:e4. doi: 10.1371/journal.ppat.0010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. 2007. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog 3:1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 20.Bloom JD, Gong LI, Baltimore D. 2010. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328:1272–1275. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ilyushina NA, Govorkova EA, Webster RG. 2005. Detection of amantadine-resistant variants among avian influenza viruses isolated in North America and Asia. Virology 341:102–106. doi: 10.1016/j.virol.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Nakaya HI, Hagan T, Duraisingham SS, Lee EK, Kwissa M, Rouphael N, Frasca D, Gersten M, Mehta AK, Gaujoux R, Li GM, Gupta S, Ahmed R, Mulligan MJ, Shen-Orr S, Blomberg BB, Subramaniam S, Pulendran B. 2015. Systems analysis of immunity to influenza vaccination across multiple years and in diverse populations reveals shared molecular signatures. Immunity 43:1186–1198. doi: 10.1016/j.immuni.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haq K, McElhaney JE. 2014. Immunosenescence: influenza vaccination and the elderly. Curr Opin Immunol 29:38–42. doi: 10.1016/j.coi.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Adachi Y, Onodera T, Yamada Y, Daio R, Tsuiji M, Inoue T, Kobayashi K, Kurosaki T, Ato M, Takahashi Y. 2015. Distinct germinal center selection at local sites shapes memory B cell response to viral escape. J Exp Med 212:1709–1723. doi: 10.1084/jem.20142284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilkinson TM, Li CK, Chui CS, Huang AK, Perkins M, Liebner JC, Lambkin-Williams R, Gilbert A, Oxford J, Nicholas B, Staples KJ, Dong T, Douek DC, McMichael AJ, Xu XN. 2012. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med 18:274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 26.Hillaire ML, van Trierum SE, Kreijtz JH, Bodewes R, Geelhoed-Mieras MM, Nieuwkoop NJ, Fouchier RA, Kuiken T, Osterhaus AD, Rimmelzwaan GF. 2011. Cross-protective immunity against influenza pH1N1 2009 viruses induced by seasonal influenza A (H3N2) virus is mediated by virus-specific T-cells. J Gen Virol 92:2339–2349. doi: 10.1099/vir.0.033076-0. [DOI] [PubMed] [Google Scholar]
- 27.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU, Garcia-Sastre A. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S, Min JY, Krug RM, Sen GC. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349:13–21. doi: 10.1016/j.virol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Belz GT, Wodarz D, Diaz G, Nowak MA, Doherty PC. 2002. Compromised influenza virus-specific CD8+-T-cell memory in CD4+-T-cell-deficient mice. J Virol 76:12388–12393. doi: 10.1128/JVI.76.23.12388-12393.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, Hakimpour P, Gill KP, Nakaya HI, Yarovinsky F, Sartor RB, Gewirtz AT, Pulendran B. 2014. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samuelson DR, Welsh DA, Shellito JE. 2015. Regulation of lung immunity and host defense by the intestinal microbiota. Front Microbiol 6:1085. doi: 10.3389/fmicb.2015.01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gazit R, Gruda R, Elboim M, Arnon TI, Katz G, Achdout H, Hanna J, Qimron U, Landau G, Greenbaum E, Zakay-Rones Z, Porgador A, Mandelboim O. 2006. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat Immunol 7:517–523. doi: 10.1038/ni1322. [DOI] [PubMed] [Google Scholar]
- 33.Srivastava B, Blazejewska P, Hessmann M, Bruder D, Geffers R, Mauel S, Gruber AD, Schughart K. 2009. Host genetic background strongly influences the response to influenza a virus infections. PLoS One 4:e4857. doi: 10.1371/journal.pone.0004857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krammer F, Cox RJ. 2013. The emergence of H7N9 viruses: a chance to redefine correlates of protection for influenza virus vaccines. Expert Rev Vaccines 12:1369–1372. doi: 10.1586/14760584.2013.850036. [DOI] [PubMed] [Google Scholar]
- 35.Ohmit SE, Petrie JG, Cross RT, Johnson E, Monto AS. 2011. Influenza hemagglutination-inhibition antibody titer as a correlate of vaccine-induced protection. J Infect Dis 204:1879–1885. doi: 10.1093/infdis/jir661. [DOI] [PubMed] [Google Scholar]
- 36.Subbarao K, Joseph T. 2007. Scientific barriers to developing vaccines against avian influenza viruses. Nat Rev Immunol 7:267–278. doi: 10.1038/nri2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie H, Wan XF, Ye Z, Plant EP, Zhao Y, Xu Y, Li X, Finch C, Zhao N, Kawano T, Zoueva O, Chiang MJ, Jing X, Lin Z, Zhang A, Zhu Y. 2015. H3N2 mismatch of 2014-15 Northern Hemisphere influenza vaccines and head-to-head comparison between human and ferret antisera derived antigenic maps. Sci Rep 5:15279. doi: 10.1038/srep15279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margine I, Krammer F. 2014. Animal models for influenza viruses: implications for universal vaccine development. Pathogens 3:845–874. doi: 10.3390/pathogens3040845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Donnell CD, Subbarao K. 2011. The contribution of animal models to the understanding of the host range and virulence of influenza A viruses. Microbes Infect 13:502–515. doi: 10.1016/j.micinf.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thangavel RR, Bouvier NM. 2014. Animal models for influenza virus pathogenesis, transmission, and immunology. J Immunol Methods 410:60–79. doi: 10.1016/j.jim.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnard DL. 2009. Animal models for the study of influenza pathogenesis and therapy. Antiviral Res 82:A110–122. doi: 10.1016/j.antiviral.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng X, Alfoldi J, Gori K, Eisfeld AJ, Tyler SR, Tisoncik-Go J, Brawand D, Law GL, Skunca N, Hatta M, Gasper DJ, Kelly SM, Chang J, Thomas MJ, Johnson J, Berlin AM, Lara M, Russell P, Swofford R, Turner-Maier J, Young S, Hourlier T, Aken B, Searle S, Sun X, Yi Y, Suresh M, Tumpey TM, Siepel A, Wisely SM, Dessimoz C, Kawaoka Y, Birren BW, Lindblad-Toh K, Di Palma F, Engelhardt JF, Palermo RE, Katze MG. 2014. The draft genome sequence of the ferret (Mustela putorius furo) facilitates study of human respiratory disease. Nat Biotechnol 32:1250–1255. doi: 10.1038/nbt.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cameron CM, Cameron MJ, Bermejo-Martin JF, Ran L, Xu L, Turner PV, Ran R, Danesh A, Fang Y, Chan PK, Mytle N, Sullivan TJ, Collins TL, Johnson MG, Medina JC, Rowe T, Kelvin DJ. 2008. Gene expression analysis of host innate immune responses during Lethal H5N1 infection in ferrets. J Virol 82:11308–11317. doi: 10.1128/JVI.00691-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carolan LA, Rockman S, Borg K, Guarnaccia T, Reading P, Mosse J, Kelso A, Barr I, Laurie KL. 2015. Characterization of the localized immune response in the respiratory tract of ferrets following infection with influenza A and B viruses. J Virol 90:2838–2848. doi: 10.1128/JVI.02797-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rowe T, Leon AJ, Crevar CJ, Carter DM, Xu L, Ran L, Fang Y, Cameron CM, Cameron MJ, Banner D, Ng DC, Ran R, Weirback HK, Wiley CA, Kelvin DJ, Ross TM. 2010. Modeling host responses in ferrets during A/California/07/2009 influenza infection. Virology 401:257–265. doi: 10.1016/j.virol.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martel CJ, Aasted B. 2009. Characterization of antibodies against ferret immunoglobulins, cytokines and CD markers. Vet Immunol Immunopathol 132:109–115. doi: 10.1016/j.vetimm.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 47.Saalmuller A, Lunney JK, Daubenberger C, Davis W, Fischer U, Gobel TW, Griebel P, Hollemweguer E, Lasco T, Meister R, Schuberth HJ, Sestak K, Sopp P, Steinbach F, Xiao-Wei W, Aasted B. 2005. Summary of the animal homologue section of HLDA8. Cell Immunol 236:51–58. doi: 10.1016/j.cellimm.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 48.Vidana B, Majo N, Perez M, Montoya M, Martorell J, Martinez J. 2014. Immune system cells in healthy ferrets: an immunohistochemical study. Vet Pathol 51:775–786. doi: 10.1177/0300985813502815. [DOI] [PubMed] [Google Scholar]
- 49.Cheng X, Zengel JR, Suguitan AL Jr, Xu Q, Wang W, Lin J, Jin H. 2013. Evaluation of the humoral and cellular immune responses elicited by the live attenuated and inactivated influenza vaccines and their roles in heterologous protection in ferrets. J Infect Dis 208:594–602. doi: 10.1093/infdis/jit207. [DOI] [PubMed] [Google Scholar]
- 50.Zens KD, Farber DL. 2015. Memory CD4 T cells in influenza. Curr Top Microbiol Immunol 386:399–421. doi: 10.1007/82_2014_401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sant AJ, McMichael A. 2012. Revealing the role of CD4(+) T cells in viral immunity. J Exp Med 209:1391–1395. doi: 10.1084/jem.20121517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doherty PC, Topham DJ, Tripp RA, Cardin RD, Brooks JW, Stevenson PG. 1997. Effector CD4+ and CD8+ T-cell mechanisms in the control of respiratory virus infections. Immunol Rev 159:105–117. doi: 10.1111/j.1600-065X.1997.tb01010.x. [DOI] [PubMed] [Google Scholar]
- 53.Music N, Reber AJ, Lipatov AS, Kamal RP, Blanchfield K, Wilson JR, Donis RO, Katz JM, York IA. 2014. Influenza vaccination accelerates recovery of ferrets from lymphopenia. PLoS One 9:e100926. doi: 10.1371/journal.pone.0100926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pillet S, Kobasa D, Meunier I, Gray M, Laddy D, Weiner DB, von Messling V, Kobinger GP. 2011. Cellular immune response in the presence of protective antibody levels correlates with protection against 1918 influenza in ferrets. Vaccine 29:6793–6801. doi: 10.1016/j.vaccine.2010.12.059. [DOI] [PubMed] [Google Scholar]
- 55.Caton AJ. 1990. A single pre-B cell can give rise to antigen-specific B cells that utilize distinct immunoglobulin gene rearrangements. J Exp Med 172:815–825. doi: 10.1084/jem.172.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nayak JL, Fitzgerald TF, Richards KA, Yang H, Treanor JJ, Sant AJ. 2013. CD4+ T-cell expansion predicts neutralizing antibody responses to monovalent, inactivated 2009 pandemic influenza A(H1N1) virus subtype H1N1 vaccine. J Infect Dis 207:297–305. doi: 10.1093/infdis/jis684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richards KA, Topham D, Chaves FA, Sant AJ. 2010. CD4 T cells generated from encounter with seasonal influenza viruses and vaccines have broad protein specificity and can directly recognize naturally generated epitopes derived from the live pandemic H1N1 virus. J Immunol 185:4998–5002. doi: 10.4049/jimmunol.1001395. [DOI] [PubMed] [Google Scholar]
- 58.Rutigliano JA, Doherty PC, Franks J, Morris MY, Reynolds C, Thomas PG. 2008. Screening monoclonal antibodies for cross-reactivity in the ferret model of influenza infection. J Immunol Methods 336:71–77. doi: 10.1016/j.jim.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shortman K, Naik SH. 2007. Steady-state and inflammatory dendritic-cell development. Nat Rev Immunol 7:19–30. doi: 10.1038/nri1996. [DOI] [PubMed] [Google Scholar]
- 60.Randolph GJ, Ochando J, Partida-Sanchez S. 2008. Migration of dendritic cell subsets and their precursors. Annu Rev Immunol 26:293–316. doi: 10.1146/annurev.immunol.26.021607.090254. [DOI] [PubMed] [Google Scholar]
- 61.DiPiazza A, Richards KA, Knowlden ZA, Nayak JL, Sant AJ. 2016. The role of CD4 T cell memory in generating protective immunity to novel and potentially pandemic strains of influenza. Front Immunol 7:10. doi: 10.3389/fimmu.2016.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leddon SA, Richards KA, Treanor JJ, Sant AJ. 2015. Abundance and specificity of influenza reactive circulating memory follicular helper and non-follicular helper CD4 T cells in healthy adults. Immunology 146:157–162. doi: 10.1111/imm.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Savic M, Dembinski JL, Kim Y, Tunheim G, Cox RJ, Oftung F, Peters B, Mjaaland S. 2016. Epitope specific T-cell responses against influenza A in a healthy population. Immunology 147:165–177. doi: 10.1111/imm.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kreijtz JH, Fouchier RA, Rimmelzwaan GF. 2011. Immune responses to influenza virus infection. Virus Res 162:19–30. doi: 10.1016/j.virusres.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 65.Masopust D, Kaech SM, Wherry EJ, Ahmed R. 2004. The role of programming in memory T-cell development. Curr Opin Immunol 16:217–225. doi: 10.1016/j.coi.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 66.Sathaliyawala T, Kubota M, Yudanin N, Turner D, Camp P, Thome JJ, Bickham KL, Lerner H, Goldstein M, Sykes M, Kato T, Farber DL. 2013. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity 38:187–197. doi: 10.1016/j.immuni.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shortman K. 2000. Burnet oration: dendritic cells: multiple subtypes, multiple origins, multiple functions. Immunol Cell Biol 78:161–165. doi: 10.1046/j.1440-1711.2000.00901.x. [DOI] [PubMed] [Google Scholar]
- 68.Beauchamp NM, Yammani RD, Alexander-Miller MA. 2012. CD8 marks a subpopulation of lung-derived dendritic cells with differential responsiveness to viral infection and toll-like receptor stimulation. J Virol 86:10640–10650. doi: 10.1128/JVI.01413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Helft J, Manicassamy B, Guermonprez P, Hashimoto D, Silvin A, Agudo J, Brown BD, Schmolke M, Miller JC, Leboeuf M, Murphy KM, Garcia-Sastre A, Merad M. 2012. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J Clin Invest 122:4037–4047. doi: 10.1172/JCI60659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pelletier N, McHeyzer-Williams LJ, Wong KA, Urich E, Fazilleau N, McHeyzer-Williams MG. 2010. Plasma cells negatively regulate the follicular helper T cell program. Nat Immunol 11:1110–1118. doi: 10.1038/ni.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Minguet S, Dopfer EP, Schamel WW. 2010. Low-valency, but not monovalent, antigens trigger the B-cell antigen receptor (BCR). Int Immunol 22:205–212. doi: 10.1093/intimm/dxp129. [DOI] [PubMed] [Google Scholar]
- 72.Crowe SR, Miller SC, Brown DM, Adams PS, Dutton RW, Harmsen AG, Lund FE, Randall TD, Swain SL, Woodland DL. 2006. Uneven distribution of MHC class II epitopes within the influenza virus. Vaccine 24:457–467. doi: 10.1016/j.vaccine.2005.07.096. [DOI] [PubMed] [Google Scholar]
- 73.Richards KA, Chaves FA, Krafcik FR, Topham DJ, Lazarski CA, Sant AJ. 2007. Direct ex vivo analyses of HLA-DR1 transgenic mice reveal an exceptionally broad pattern of immunodominance in the primary HLA-DR1-restricted CD4 T-cell response to influenza virus hemagglutinin. J Virol 81:7608–7619. doi: 10.1128/JVI.02834-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Assarsson E, Bui HH, Sidney J, Zhang Q, Glenn J, Oseroff C, Mbawuike IN, Alexander J, Newman MJ, Grey H, Sette A. 2008. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J Virol 82:12241–12251. doi: 10.1128/JVI.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richards KA, Chaves FA, Sant AJ. 2011. The memory phase of the CD4 T-cell response to influenza virus infection maintains its diverse antigen specificity. Immunology 133:246–256. doi: 10.1111/j.1365-2567.2011.03435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jameson J, Cruz J, Ennis FA. 1998. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. J Virol 72:8682–8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Belz GT, Xie W, Altman JD, Doherty PC. 2000. A previously unrecognized H-2D(b)-restricted peptide prominent in the primary influenza A virus-specific CD8+ T-cell response is much less apparent following secondary challenge. J Virol 74:3486–3493. doi: 10.1128/JVI.74.8.3486-3493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Belz GT, Xie W, Doherty PC. 2001. Diversity of epitope and cytokine profiles for primary and secondary influenza a virus-specific CD8+ T cell responses. J Immunol 166:4627–4633. doi: 10.4049/jimmunol.166.7.4627. [DOI] [PubMed] [Google Scholar]
- 79.Lampson LA, Levy R. 1980. Two populations of Ia-like molecules on a human B cell line. J Immunol 125:293–299. [PubMed] [Google Scholar]
- 80.Alam S, Knowlden ZA, Sangster MY, Sant AJ. 2014. CD4 T cell help is limiting and selective during the primary B cell response to influenza virus infection. J Virol 88:314–324. doi: 10.1128/JVI.02077-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sette A, Moutaftsi M, Moyron-Quiroz J, McCausland MM, Davies DH, Johnston RJ, Peters B, Rafii-El-Idrissi Benhnia M, Hoffmann J, Su HP, Singh K, Garboczi DN, Head S, Grey H, Felgner PL, Crotty S. 2008. Selective CD4+ T cell help for antibody responses to a large viral pathogen: deterministic linkage of specificities. Immunity 28:847–858. doi: 10.1016/j.immuni.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nayak JL, Richards KA, Yang H, Treanor JJ, Sant AJ. 2015. Effect of influenza A(H5N1) vaccine prepandemic priming on CD4+ T-cell responses. J Infect Dis 211:1408–1417. doi: 10.1093/infdis/jiu616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nayak JL, Alam S, Sant AJ. 2013. Cutting edge: Heterosubtypic influenza infection antagonizes elicitation of immunological reactivity to hemagglutinin. J Immunol 191:1001–1005. doi: 10.4049/jimmunol.1203520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brown DM, Lee S, Garcia-Hernandez Mde L, Swain SL. 2012. Multifunctional CD4 cells expressing gamma interferon and perforin mediate protection against lethal influenza virus infection. J Virol 86:6792–6803. doi: 10.1128/JVI.07172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Strutt TM, McKinstry KK, Marshall NB, Vong AM, Dutton RW, Swain SL. 2013. Multipronged CD4(+) T-cell effector and memory responses cooperate to provide potent immunity against respiratory virus. Immunol Rev 255:149–164. doi: 10.1111/imr.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wiley JA, Hogan RJ, Woodland DL, Harmsen AG. 2001. Antigen-specific CD8(+) T cells persist in the upper respiratory tract following influenza virus infection. J Immunol 167:3293–3299. doi: 10.4049/jimmunol.167.6.3293. [DOI] [PubMed] [Google Scholar]
- 87.Baker SF, Guo H, Albrecht RA, Garcia-Sastre A, Topham DJ, Martinez-Sobrido L. 2013. Protection against lethal influenza with a viral mimic. J Virol 87:8591–8605. doi: 10.1128/JVI.01081-13. [DOI] [PMC free article] [PubMed] [Google Scholar]