ABSTRACT
ZMapp, a cocktail of three monoclonal antibodies (MAbs; c2G4, c4G7, and c13C6) against the ebolavirus (EBOV) glycoprotein (GP), shows promise for combatting outbreaks of EBOV, as occurred in West Africa in 2014. Prior studies showed that Fabs from these MAbs bind a soluble EBOV GP ectodomain and that MAbs c2G4 and c4G7, but not c13C6, neutralize infections in cell cultures. Using cryo-electron tomography, we extended these findings by characterizing the structures of c2G4, c4G7, and c13C6 IgGs bound to native, full-length GP from the West African 2014 isolate embedded in filamentous viruslike particles (VLPs). As with the isolated ectodomain, c13C6 bound to the glycan cap, whereas c2G4 and c4G7 bound to the base region of membrane-bound GP. The tomographic data suggest that all three MAbs bind with high occupancy and that the base-binding antibodies can potentially bridge neighboring GP spikes. Functional studies indicated that c2G4 and c4G7, but not c13C6, competitively inhibit entry of VLPs bearing EBOV GP into the host cell cytoplasm, without blocking trafficking of VLPs to NPC1+ endolysosomes, where EBOV fuses. Moreover, c2G4 and c4G7 bind to and can block entry mediated by the primed (19-kDa) form of GP without impeding binding of the C-loop of NPC1, the endolysosomal receptor for EBOV. The most likely mode of action of c2G4 and c4G7 is therefore by inhibiting conformational changes in primed, NPC1-bound GP that initiate fusion between the viral and target membranes, similar to the action of certain broadly neutralizing antibodies against influenza hemagglutinin and HIV Env.
IMPORTANCE The recent West African outbreak of ebolavirus caused the deaths of more than 11,000 individuals. Hence, there is an urgent need to be prepared with vaccines and therapeutics for similar future disasters. ZMapp, a cocktail of three MAbs directed against the ebolavirus glycoprotein, is a promising anti-ebolavirus therapeutic. Using cryo-electron tomography, we provide structural information on how each of the MAbs in this cocktail binds to the ebolavirus glycoprotein as it is displayed—embedded in the membrane and present at high density—on filamentous viruslike particles that recapitulate the surface structure and entry functions of ebolavirus. Moreover, after confirming that two of the MAbs bind to the same region in the base of the glycoprotein, we show that they competitively block the entry function of the glycoprotein and that they can do so after the glycoprotein is proteolytically primed and bound to its intracellular receptor, Niemann-Pick C1. These findings should inform future developments of ebolavirus therapeutics.
INTRODUCTION
The world was caught off-guard when an ebolavirus emerged in West Africa in December of 2013. Ebolaviruses are hemorrhagic fever viruses that constitute a genus within the Filoviridae family. Among the five known ebolavirus species, Zaire ebolaviruses (EBOV), which include the Mayinga isolate from 1976, are the most lethal. The 2013-2014 West African outbreak was caused by EBOV-Makona, a member of this lethal EBOV species (1). At the time of the outbreak, several anti-EBOV therapeutics and vaccines were in development, but none had been approved for use in humans by the U.S. Food and Drug Administration (FDA) or an equivalent international agency. Arguably, the most promising therapeutic was ZMapp, a cocktail of three monoclonal antibodies (MAbs) directed against the glycoprotein (GP) of the Mayinga isolate of EBOV. Since this cocktail provided strong protection of nonhuman primates from ebolavirus disease, 100% survival even if given 5 days postchallenge (2), it was administered to patients stricken by EBOV-Makona on a compassionate care basis (3).
The three MAbs in the ZMapp cocktail, c13C6, c2G4 and c4G7, were sourced from two previous collections of anti-EBOV GP MAbs: MB003 and ZMAb. The immunogen for all of these MAbs was GP from EBOV-Mayinga (4, 5). c2G4 and c4G7 neutralize EBOV infections in cell culture, whereas c13C6 does not (4, 6–8). The binding sites for Fab fragments of c13C6, c2G4, and c4G7 on the soluble trimeric ectodomain of EBOV-Mayinga GP were determined by single particle electron microscopy (9), and an alanine scanning study defined residues required for binding of each MAb to EBOV-Mayinga GP (10).
Here, we provide additional insights into how the MAbs in the ZMapp cocktail bind to EBOV GP and inhibit infection. We provide the first three-dimensional (3D) images of GP from EBOV-Makona, unliganded, as well as in individual complexes with MAbs c13C6, c2G4, and c4G7. Importantly, we imaged complexes of intact c13C6, c2G4, and c4G7 IgGs with full-length Makona GP anchored in the membrane, at high density, on viruslike particles (VLPs). We next showed that c2G4 and c4G7 block entry of VLPs bearing EBOV GP (from Mayinga and Makona isolates) into the cell cytoplasm, that they compete for this function, and that they likely inhibit EBOV entry and consequent infection by blocking fusion-triggering conformational changes in proteolytic primed (19-kDa) GP bound to its intracellular receptor, Niemann-Pick C1 (NPC1).
MATERIALS AND METHODS
Cells.
HEK 293T/17 cells (University of Virginia Tissue Culture Facility, ATCC CRL-11268) and BSC-1 cells (grivet kidney; a gift from Xiaowei Zhuang, Harvard University) were propagated in high-glucose Dulbecco modified Eagle medium supplemented with 1% l-glutamine, 1% sodium pyruvate, and 1% antibiotic/antimycotic (Gibco/Life Technologies) containing 10% supplemented calf serum (HyClone) for HEK 293T/17 cells or 10% fetal bovine serum (Seradigm) for BSC-1 cells.
VLPs and MAbs.
Entry-reporter EBOV VP40-driven VLPs bearing GPs from the Mayinga and Makona isolates of EBOV were prepared as described previously (11). In brief, 293T/17 cells were transfected with cDNAs encoding EBOV GP, VP40, βlam-VP40, and mCherry-VP40. cDNA encoding Mayinga GP was described previously. Mammalian codon-optimized cDNA encoding Makona GP (GenBank accession number KM034562.1) was synthesized by DNA2.0 (Menlo Park, CA) and inserted into the vector, pWRG7077, using the NotI and BglII sites. After transfection, the cell medium was collected at 24 and 48 h and cleared of debris. VLPs in the cleared medium were then pelleted by centrifugation through a 20% sucrose cushion. After resuspension and repelleting, the VLPs were resuspended (1:100 starting volume of medium) in 10% sucrose-HM (20 mM HEPES, 20 mM morpholineethanesulfonic acid [MES], 130 mM NaCl; pH 7.4), snap-frozen, and stored at −80°C in single-use aliquots (12). Protein concentrations of final VLPs were determined by bicinchoninic acid (BCA) assay. All VLP preps were titered for entry competency on BSC-1 cells. The c13C6, c2G4, and c4G7 MAbs (intact IgGs) were prepared as described previously (2, 6).
Preparation of cryo-electron microscopy (cryo-EM) grids.
Purified Ebola VLPs were incubated on ice with or without antibodies for about 30 to 60 min at an approximate ratio of 2 μg of antibody to 1 μg of VLP. Immediately before grid preparation, 10 nm of protein A-gold was added to the sample. This mixture was then pipetted onto plasma-cleaned 200-mesh Quantifoil Multi-A carbon grids (Quantifoil). Excess buffer was blotted from the grids at room temperature and 95% humidity using a Leica EM grid plunger (Leica Microsystems). The grids were immediately plunge-frozen in liquid ethane maintained at about −180°C and stored in liquid nitrogen until use.
Cryo-EM.
VLPs were imaged in a Titan Krios transmission electron microscope (FEI) operated at 300 kV and equipped with a GIF Quantum energy filter (Gatan) at a slit width of 20 eV. Images were recorded on a K2 Summit camera (Gatan) at a pixel size of 2.2 Å. Tilt series were collected at ±60° in 2° increments at a magnification of ×64,000 and a defocus range of 2 to 3 μm. The total dose used per tilt series was approximately 120 e−/Å2.
Tomographic image analysis.
Fiducial-based reconstruction of tomograms using weighted back-projection techniques was performed, as previously described (13–15). Between 11 and 26 tilt series were collected for each sample (see Table 1 for a data summary). Glycoprotein spikes (1,000 to 2,500 spikes per complex) were picked automatically for subvolume averaging, as previously described (14–16), but tomograms were first binned by 8 in this study. For each structure, spikes were selected from a central region along the entire length of each VLP. For the unbound GP data set, we verified that the spikes chosen represent the global population by picking spikes from the entire surface of each VLP. This increased the number of picked subvolumes by nearly 10-fold, yet resulted in a structure extremely similar to the map shown in Fig. 1. Alignment, classification and 3D averaging of the subvolumes were performed as previously described (14, 15, 17). For the GP+c2G4 structure, subvolumes were clustered into 10 classes per iteration. With each round of refinement, classes that showed the most clearly defined features in the region of the spike were combined and used as a reference for the next round. The final map was obtained after seven iterations of refinement. All other structures were determined using the collaborative alignment method (17). Tomographic subvolumes were subjected to successive rounds of alignment and classification until the particles converged into multiple similar structural classes of approximately 100 particles each. A representative map from each data set was chosen for presentation. A class size of 100 subvolumes was chosen in order to distinguish heterogeneity within the data. For all data sets, we first verified that subvolumes displayed 3-fold symmetry in the absence of enforced symmetry. Once the inherent symmetry of the subvolumes was established, 3-fold symmetry was enforced on the averaged subvolumes.
TABLE 1.
Tomographic image analysisa
| Data set | No. |
||
|---|---|---|---|
| Tomograms | Subvolumes picked | Subvolumes contributing to final map | |
| EBOV-Makona | 11 | 1,043 | 244 |
| EBOV-Makona + c13c6 | 13 | 1,591 | 75 |
| EBOV-Makona + c2G4 | 26 | 2,470 | 1,998 |
| EBOV-Makona + c4G7 | 14 | 1,674 | 135 |
For each data set, the numbers of tomograms, subvolumes picked, and subvolumes contributing to the structure presented are shown.
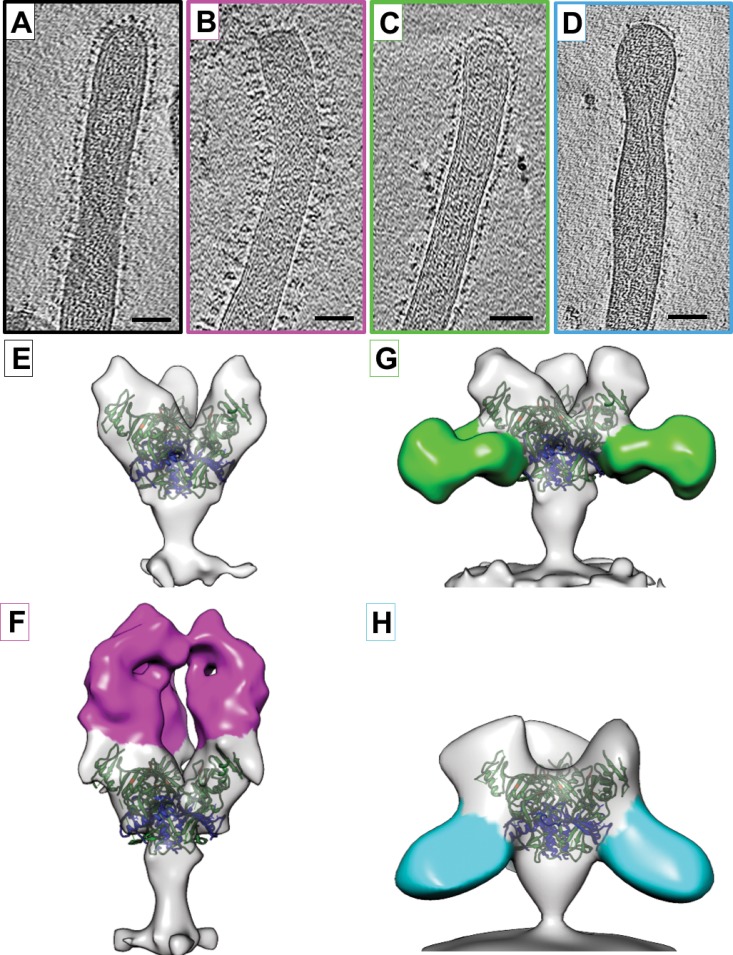
FIG 1.
Structures of EBOV-Makona GP bound to ZMapp antibodies. (A to D) Tomographic slices of EBOV-Makona VLPs are shown unbound (A) or bound to ZMapp antibodies c13C6 (B), c4G7 (C), or c2G4 (D). (E to H) Isosurface representations of density maps derived from tomographic subvolume averaging of glycoprotein spikes expressed on the VLP surface for unbound GP (E) or GP bound to c13C6 (F), c4G7 (G), or c2G4 (H). Density assigned to the antibody Fab region is colored magenta (c13C6), green (c4G7), or cyan (c2G4). All structures have been fitted with the crystal coordinates for EBOV-Mayinga GP (Protein Data Bank [PDB] accession number 3CSY), which lacks the mucin-like domain and the transmembrane domain (21). X-ray coordinates are colored with the GP1 region in green, GP2 in blue, and residues important for binding to NPC1 in orange. Scale bars, 50 nm.
Determination of X-ray coordinate fits.
All of the EM maps in this study were aligned to the unbound GP EM map. X-ray coordinates were fit manually into the EM maps so that residues known to be important in antibody binding (escape mutant residues from Davidson et al. [10]) are aligned with the location of observed antibody density in the EM maps. The same X-ray coordinate position was used in each GP map for consistency.
Measurement of GP density and spacing on the VLP surface.
The density and spacing of GP on the surface of EBOV-Makona VLPs were determined using tomographic reconstructions where clearly visible top views of trimeric GP molecules were visible. VLP surface areas were measured in six tomographic slices, and the number of GP spikes within each surface area (SA) was counted. Spike density (per 100 nm2) was determined based on the SA calculation. The distance between top views of trimeric GP molecules was determined using the same tomographic slices. The center-to-center distance was measured between each visible trimer. In total, 75 interspike distance measurements were averaged. All measurements were taken using IMOD software.
VLP entry assay.
VLP entry into the cytoplasm was monitored using entry-reporter VLPs bearing Mayinga or Makona EBOV GP as described previously (11). In brief, VLPs were pretreated for 1 h at 37°C with the indicated concentrations of the indicated MAbs and bound to the surface of BSC-1 cells by centrifugation (250 × g) for 1 h at 4°C. Cells with bound VLPs were then incubated for 3 h at 37°C (in a 5% CO2 incubator) to allow for VLP entry. The cells were loaded with the β-lactamase substrate CCF2-AM and then processed and analyzed for cytoplasmic entry (cleavage of the CCF2 dye) by flow cytometry. An amount of EBOV GP VLPs was added to attain entry in 30 to 60% of cells. Data were included from experiments in which entry values in untreated EBOV GP VLP samples ranged from 30 to 60%.
VLP trafficking to endolysosomes.
To measure the effect of MAbs c2G4 and c4G7 on EBOV VLP colocalization with NPC1, 40,000 BSC-1 cells were seeded per well of a 24-well plate containing glass coverslips that were coated with 20 μg/ml fibronectin (Sigma-Aldrich). After 18 to 24 h, the cells were pretreated with 20 μM nocodazole (ACROS organics), dimethyl sulfoxide (DMSO; mock-treated samples), or Opti-MEM1 (Gibco/Life Technologies, for MAb-treated samples) for 1 h at 37°C in a 5% CO2 incubator. Concurrently, EBOV VLPs (0.5 μg per sample) were pretreated with 50 μg/ml MAb c2G4 or 50 μg/ml MAb c4G7 as indicated for 1 h in a heat block maintained at 37°C. VLPs were bound to cells by centrifugation at 250 × g and 4°C for 1 h. The cells were gently washed once with phosphate-buffered saline (PBS) to remove unbound VLPs. The cells with bound VLPs were then incubated at 37°C in a 5% CO2 incubator for 90 min. They were then washed once with PBS, fixed in 4% paraformaldehyde (Boston Bioproducts), and stained for NPC1 as previously described (12). The coverslips were mounted on glass slides using ProLong Gold antifade reagent (Life Technologies). The samples were then imaged and analyzed as previously described (12).
Treatment of VLPs bearing EBOV GP to generate 19-kDa GP and assessment by Western blotting.
The 19-kDa GP was generated as described previously (12). In brief, VLPs bearing EBOV-Makona or EBOV-Mayinga GP were treated with 0.25 mg/ml thermolysin (VitaCyte) at 37°C for 30 min. The cleavage reaction was quenched by the addition of 500 μM phosphoramidon (Sigma-Aldrich). Cleavage of GP to 19 kDa was confirmed by Western blotting. In brief, 2 μg of VLPs were run on Mini-Protean TGX Any kD gels (Bio-Rad) and transferred to nitrocellulose membranes. After blocking (5% milk in PBS or Odyssey blocking buffer [OBB; Li-Cor]) for 1 h at room temperature, the membranes were incubated in primary antibodies overnight at 4°C on an orbital shaker. The primary antibodies were as follows (and as indicated in the figure legends): rabbit anti-GP1 (a gift from Paul Bates) diluted 1:1,000 in 5% milk-PBS, rabbit anti-GP1 F88 peptide (18) diluted 1:500 in 5% milk-PBS, or mouse H3C8 (a gift from Carolyn Wilson, FDA) diluted to 7 μg/ml in OBB plus 0.2% Tween. After one rinse with PBS, followed by three (5 min) washes with PBS (for rabbit antibodies) or PBS plus 0.1% Tween (for H3C8), the membranes were probed with Odyssey secondary (anti-mouse or anti-rabbit) antibodies diluted 1:10,000 in PBS (for rabbit primary antibodies) or OBB plus 0.2% Tween (for H3C8) for 1 h at room temperature. After rinsing and washing steps as described above, the membranes were subjected to a final quick rinse with PBS and scanned with an Odyssey infrared imaging system.
MAb binding to VLPs.
VLPs (0.5 μg/well) were bound to a high-bind enzyme-linked immunosorbent assay (ELISA) plate overnight at 4°C. The ELISA plate was then washed and blocked (3% bovine serum albumin [BSA]-PBS). After an additional wash (PBS), 50 μg/ml c2G4, c4G7, or c13C6 (PBS for control) were added, and the plate incubated for 1 h at room temperature. After another washing step in PBS, anti-human horseradish peroxidase (HRP) IgG (Thermo Scientific; 1:10,000 in PBS) was added for 1 h at room temperature, and the plate was processed and analyzed for primary antibody binding by the addition of 1-Step Ultra TMB ELISA substrate (Thermo Scientific), followed by a 2 M sulfuric acid quench. The absorbance at 450 nm (A450) was determined using a BioTek Synergy HT microplate reader. The average signals from wells containing no VLPs were subtracted, and the resulting average A450 values ± the standard deviation (SD) are shown.
Preparation of soluble NPC1 C-loop.
The soluble NPC1 C-loop was modeled after the construct described in references (19, 20). Codon-optimized DNA encoding the NPC1 C-loop, flanked by N- and C-terminal antiparallel coiled-coil domains (19) (with BglII and SalI restriction sites at the 5′ and 3′ ends, respectively), was synthesized by GenScript and cloned into the vector pDisplay, resulting in a construct with a hemagglutinin (HA) epitope at the N terminus and a myc epitope at the C terminus. Soluble C-loop protein was then produced by transfecting suspension 293F cells with this pDisplay vector. After 72 h, the protein was purified in batch using Ni-Sepharose high-performance affinity media (GE Healthcare) essentially as described previously (19).
Binding of soluble NPC1 C-loop to VLPs bearing 19-kDa GP: effect of MAbs.
Indicated VLPs were bound overnight to a high-bind ELISA plate as described above. The ELISA plate was then washed and blocked (5% milk-PBS) for 2 h at room temperature. After an additional wash (PBS), 50 μg/well c2G4, c4G7, or c13C6 (PBS for control) was added, and the plate was incubated for 1.5 h at room temperature. After further washing (PBS), 0.2 μg/ml soluble NPC1 C-loop (PBS for no NPC1 C-loop control) was added, and the plate incubated for an additional 1 h at room temperature. After washing (PBS), anti HA-HRP (Roche; 1:1,000 in PBS) was added for 1 h at room temperature. After a final wash (PBS), 1-Step Ultra TMB ELISA substrate (Thermo Scientific) was added for 30 min and quenched with 2 M sulfuric acid, and the absorbance at 450 nm was determined using a BioTek Synergy HT microplate reader. The resulting average A450 values ± the SD are shown.
Accession number(s).
Cryo-EM maps for unbound EBOV-Makona GP and EBOV-Makona GP bound to c13C6 IgG, c2G4 IgG, and c4G7 IgG have been deposited in the EMDataBank under accession numbers EMD-8225, EMD-8226, EMD-8227, and EMD-8228, respectively.
RESULTS
Cryo-EM tomographic analysis of MAbs from the ZMapp cocktail bound to EBOV-Makona GP on filamentous VLPs.
A previous single-particle EM study performed at a resolution of 21 to 24 Å located the binding sites of Fabs from the three MAbs in the ZMapp cocktail on the soluble ectodomain of GP from the Mayinga isolate of EBOV. Fab c13C6 was found to bind in the vicinity of the glycan cap, while the binding sites for Fabs c2G4 and c4G7 were localized to overlapping regions in the base of GP, near the GP1/GP2 interface (9), similar to previously characterized sites in the base regions of Mayinga GP (21) and Sudan ebolavirus GP (22) for Fabs from MAbs KZ52 and 16F6, respectively. These findings were consistent with subsequent mutagenesis data implicating residues T270 and K272 in the glycan cap for binding of c13C6 and residues C511, N550, G553, and C556 in the fusion loop of GP2 for binding of c2G4 and c4G7; c4G7 also requires D552 (10). Escape mutants to MAbs c2G4 and c4G7 mapped to residue Q508, just upstream of the fusion loop (8). We have extended these recent studies by delineating the binding sites for c13C6, c2G4, and c4G7 (intact IgGs) on EBOV-Makona GP embedded in a membrane at high density on filamentous EBOV VP40-driven VLPs.
We first sought to determine whether c13C6, c2G4, and c4G7 bind to EBOV-Makona GP on the surface of VLPs in the same locations and orientations as seen with the Fab portions of these antibodies bound to the soluble Mayinga GP ectodomain (9). Using cryo-EM tomography, we were able to visualize the Makona GP protein in a native-like state, bound to a membrane surface. GP molecules on the VLP surface were imaged in an unbound state (Fig. 1A) or in complex with intact IgG molecules, i.e., c13C6, c4G7, and c2G4 (Fig. 1B to D, respectively). GP spikes were visible on the surface of Makona VLPs in tomographic slices of each complex tested (Fig. 1A to D). Using subvolume averaging, we were able to determine the 3D structures for unbound EBOV-Makona GP (Fig. 1E), as well as the structures of GP in complex with the head-binding antibody, c13C6 (Fig. 1F), or individual base-binding antibodies, c4G7 (Fig. 1G) or c2G4 (Fig. 1H). Consistent with previous studies (9, 10), c13C6 IgG bound to the glycan cap region, perpendicular to the plane of the membrane. The dense array of c13C6 projecting perpendicularly from the VLP may therefore contribute to the Fc effector function (23), which has been proposed to elicit the protective ability of this nonneutralizing MAb (24).
c4G7 and c2G4 bind to overlapping epitopes within the base region of EBOV-Makona GP (Fig. 1G and H), as expected, at angles and orientations previously described for binding of the corresponding Fabs to the soluble Mayinga GP ectodomain (9). Visualization of density for each base-binding antibody in the expected location is consistent with our observations (described below) of functional competition between these two MAbs.
All three antibodies (c13C6, c2G4, and c4G7) are likely to bind Makona GP at high occupancy, since no unbound GP classes were resolved for any of these complexes during 3D averaging of automatically picked GP spikes. Binding of these antibodies does not appear to dramatically alter the structural conformation of GP (see the ectodomain of the GP spike in Fig. 1E to H). In structures of GP bound to c13C6, c2G4, and c4G7, the chalice-like shape of the unbound GP ectodomain is retained. Although small but significant changes to individual residues may occur upon antibody binding, our study demonstrates that, at a resolution of ∼25 Å, the structure of the ectodomain of Makona GP anchored in a membrane closely mimics that of the soluble Mayinga GP ectodomain (9, 21).
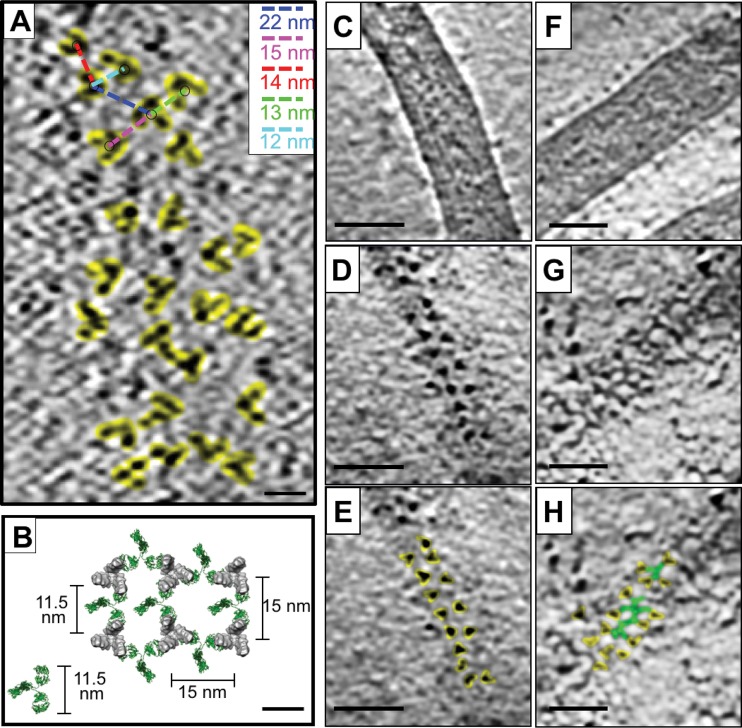
The density of GP spikes on the surface of EBOV-Makona VLPs, determined by counting only those spikes displayed in a top-view orientation, was ∼0.4 GP spikes per 100 nm2 ± 0.1 GP per 100 nm2 (SD). This corresponds to ∼1 GP spike per 250 nm2 on the VLP surface, and an average center-to-center spacing of 15 ± 4 nm (SD) between spikes (Fig. 2A). The unbound Makona GP spike structure has dimensions of ∼10 nm at the widest portion of the head domain and ∼3.5 nm at the base where c2G4 and c4G7 bind. The spacing of unbound GP on the Makona EBOV VLP surface, therefore, is such that the edge-to-edge distance between GP spikes at the c4G7 binding epitope is approximately 11.5 nm (Fig. 2A). Given that an individual IgG molecule is estimated to span ∼11.5 to 14 nm between the two Fab domains (25), we conclude that neighboring spikes can be bridged by a single antibody that targets the base region (as indicated in Fig. 2B). In fact, tomographic slices of EBOV-Makona VLPs incubated with c4G7 IgG molecules show evidence of GP bridging (Fig. 2). In the absence of antibody, top views of GP molecules on the surface of EBOV-Makona VLPs are easily distinguished in tomographic slices, and clear separation and spacing is observed between spikes (Fig. 2C to E). In contrast, after incubation with c4G7 antibody, individual GP proteins are more difficult to identify, and lines of density that appear to connect individual GP proteins are visible (Fig. 2F to H).
FIG 2.
Spacing of GP spikes on EBOV-Makona VLP surface and potential for bridging by base-specific antibodies. (A) A tomographic slice of an unbound EBOV-Makona VLP surface shows top views of trimeric GP proteins (highlighted in yellow). Center-to-center distance between spikes (indicated on representative GP spikes with colored dashed lines) is shown in the upper right corner inset. The distance corresponding to each dashed line is shown in the same color as the line. (B) A model of GP spikes on the surface of EBOV-Makona VLPs is shown using a top view of the cryo-EM structure of unbound GP. GP spikes are spaced 15 nm apart. The measured distance between the base portion of the maps is 11.5 nm, as indicated. A model IgG molecule (PDB accession number 1IGT) is shown in green. IgG molecules are placed between the GP spikes to indicate that the two Fab arms of a single IgG molecule can bridge two neighboring GP spikes. (C to E) Denoised tomographic slices through an EBOV-Makona VLP show views through the center (C) and top (D and E) of a VLP. In panel E, top views of the trimeric GP molecule are highlighted in yellow. (F to H) Denoised tomographic slices through an EBOV-Makona VLP after incubation with c4G7 IgG show views through the center (F) and top (G and H) of the VLP. In panel H, top views of the trimeric GP molecule are highlighted in yellow, while lines of density between the spikes are highlighted in green. Scale bars, 10 nm (A and B) or 50 nm (C to H).
MAbs c2G4 and c4G7 competitively block entry mediated by EBOV GP.
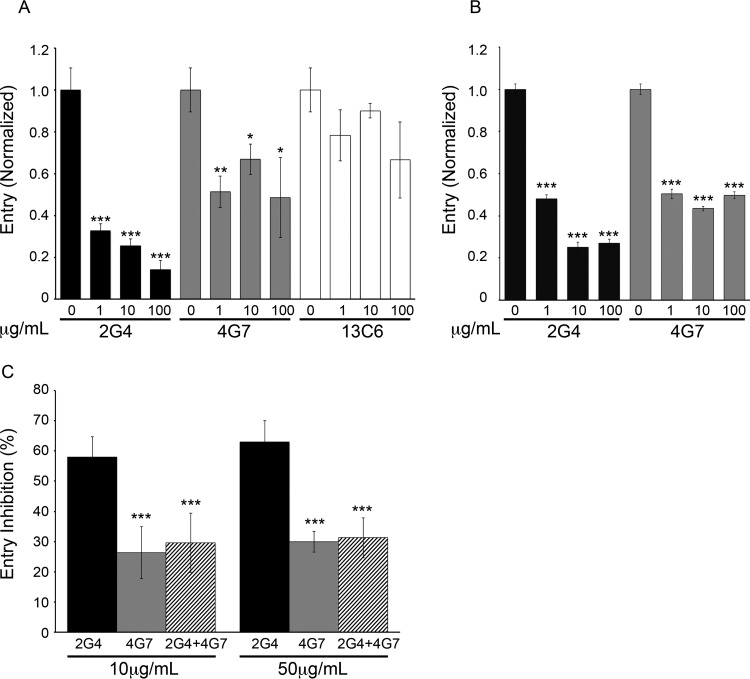
Extensive work has shown that c2G4 and c4G7 neutralize infections by EBOV and recombinant VSV expressing the GP from Mayinga EBOV (4, 6–8). Based on their neutralizing activity and binding sites (8–10), it has been proposed, but not formally shown, that these MAbs block GP-mediated entry into the cell cytoplasm. Here, using entry-reporter VLPs (11), we provide evidence that this is, indeed, the case. As seen in Fig. 3A, c2G4 and c4G7 blocked entry of VLPs bearing GP from the Mayinga isolate of EBOV. Consistent with results with viral particles (6, 8), c2G4 was more potent than c4G7. MAb c13C6 did not have a significant impact on virus entry, in line with its only weak ability to neutralize virus infection in cell cultures in the absence of complement (4, 6, 8, 24). We therefore focused on c2G4 and c4G7 in continuing studies.
FIG 3.
ZMapp MAbs c2G4 and c4G7 competitively block entry of VLPs bearing EBOV GP. VLPs bearing EBOV-Mayinga GP (A) or EBOV-Makona GP (B) were incubated with the indicated concentrations of the indicated MAb for 1 h at 37°C. VLPs were then bound to and allowed to enter BSC-1 cells, and the extent of entry determined as described in Materials and Methods. The percent entry values were normalized to mock-treated (0 μg/ml MAb) controls. Data are the averages of triplicate samples, with error bars representing the standard deviations (SD) of the mean. Mock-treated VLP entry values were 35% for (a) and (b). Similar results were seen for three and two repeat experiments for panels A and B, respectively. (C) VLPs were treated with either 10 or 50 μg/ml c2G4, c4G7, or a combination of the indicated amounts of c2G4 and c4G7 and then processed as described above. Values are the percent inhibition of entry relative to mock-treated (no MAb) controls. Data are the averages of three experiments performed in triplicate, with error bars representing the SD. The percent entry values in mock-treated samples in the three experiments were 46, 41, and 36%. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (A and B, relative to 0 μg/ml [no MAb] samples; C, relative to c2G4 alone).
In addition to their effects on entry mediated by EBOV-Mayinga GP, c2G4 and c4G7 blocked VLP entry mediated by EBOV-Makona GP (Fig. 3B). This finding is consistent with the close sequence relationship between the GP proteins from these two EBOV isolates (1, 26, 27). Previous biochemical studies showed that c2G4 and c4G7 compete for binding to EBOV-Mayinga GP, an observation consistent with their overlapping binding sites (8–10). In line with these in vitro biochemical findings, we observed that c4G7, which binds with ∼2-fold-higher affinity to GP than c2G4 (8–10), inhibited the ability of c2G4 to block VLP entry (Fig. 3C). This result indicates that c4G7 can functionally compete with c2G4 for blockade of EBOV entry and infection.
MAbs c2G4 and c4G7 block a late step of EBOV entry.
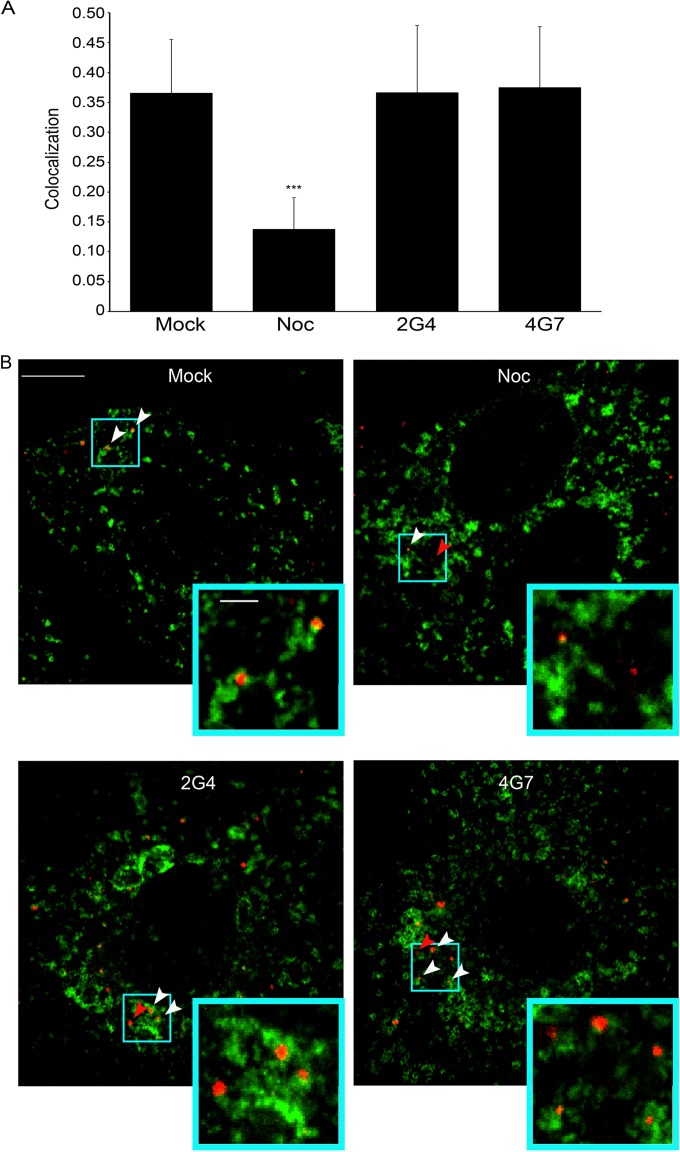
We next explored the step of EBOV entry that is blocked by c2G4 and c4G7. As seen in Fig. 4, pretreatment of EBOV GP VLPs with inhibitory concentrations of c2G4 and c4G7 did not reduce the proportion of VLPs that reach NPC1+ endolysosomes. This implies that neither MAb blocks EBOV GP VLP attachment to the cell surface, internalization from the cell surface, or trafficking to endolysosomes, the organelle where EBOV fusion occurs (12, 28, 29). Moreover, this finding indicates that c2G4 and c4G7 stay bound to GP all throughout VLP trafficking to, and proteolytic processing in, highly acidic endolysosomes found deep in the endocytic pathway.
FIG 4.
MAbs c2G4 and c4G7 do not inhibit trafficking of EBOV GP VLPs to NPC1+ endolysosomes. VLPs bearing EBOV-Mayinga GP were pretreated with 50 μg/ml c2G4 or c4G7, added to BSC1 cells, and processed for VLP trafficking and colocalization with NPC1 as described in Materials and Methods. Control cells had been preincubated with 20 μM nocodazole (Noc) or DMSO (Mock) for 1 h prior. (A) Data are the average Manders coefficients for colocalization of mCherry-VLPs (red) with NPC1 (stained green) from 27 (Mock and Noc treated) or 33 (c2G4 and c4G7 treated) microscope fields. Error bars represent the SD. ***, P < 10−15 (relative to mock samples). (B) Representative images are shown. White arrows indicate areas of VLP-NPC1 colocalization, and red arrows indicate VLPs that have not colocalized with NPC1. Scale bars, 10 μm (primary images) and 2 μm (insets). Virtually identical results were seen in two additional experiments.
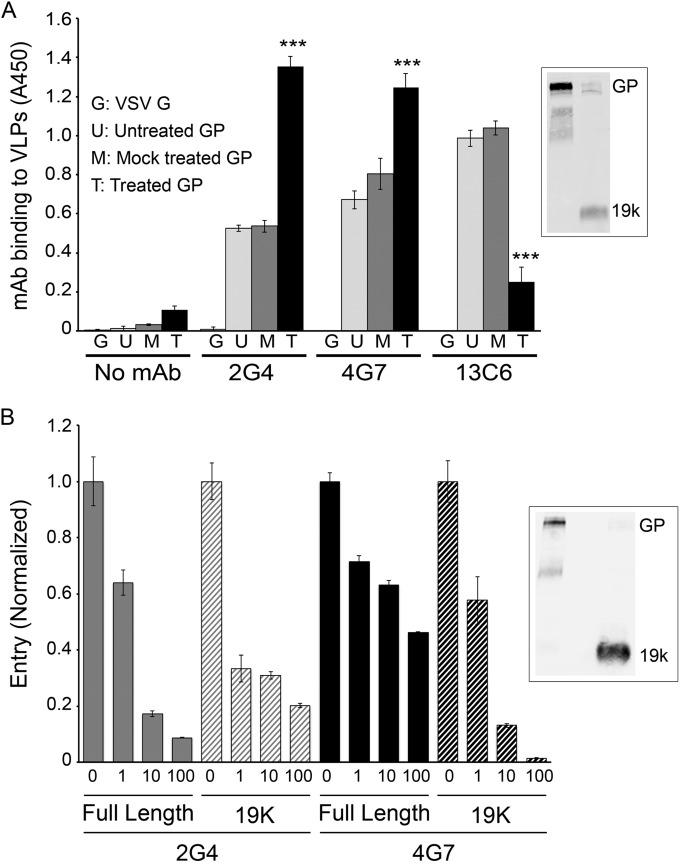
During EBOV trafficking along the endocytic pathway, the receptor binding subunit of GP (GP1) is proteolytically processed from ∼130 to ∼19 kDa (30, 31). This event, which is enacted in most cell types studied by cathepsin B and/or cathepsin L, removes the mucin domain and glycan cap from GP1 and exposes residues critical for GP binding to its intracellular receptor, NPC1 (20, 30–35). We therefore sought to determine whether c2G4 and c4G7 could bind to the 19-kDa form of GP. As seen in Fig. 5A, c2G4 and c4G7 bound to 19-kDa GP. In fact, both MAbs bound more extensively to 19-kDa GP than to full-length GP. In contrast, while c13C6 bound well to full-length GP, it showed only limited binding to VLPs with GP1 processed to 19 kDa (Fig. 5A). This is consistent with binding of c13C6 to the glycan cap (Fig. 1) (9, 10), which is removed from full-length (130 kDa) GP during proteolytic priming in the endocytic pathway. The lack of binding of MAb c13C6 to 19-kDa GP is also consistent with its inability to neutralize EBOV infections in cell cultures in the absence of complement. In line with their binding to 19-kDa GP (Fig. 5A), c2G4 and c4G7 were able to block entry mediated by 19-kDa GP (Fig. 5B).
FIG 5.
MAbs c2G4 and c4G7 bind to 19-kDa EBOV GP on VLPs and block VLP entry mediated by 19-kDa GP. (A) VLPs bearing EBOV-Makona GP were untreated, mock treated, or (thermolysin) treated and then bound (triplicate samples) to high-bind ELISA plates as described in Materials and Methods. VLPs bearing VSV-G were used as a control. After blocking and washing (see Materials and Methods), 50 μg/ml MAb c2G4, c4G7, or c13C6 (PBS for control) were added, and the plate was processed and analyzed to detect MAb binding as described in Materials and Methods. Error bars represent the SD. ***, P < 0.001 ([thermolysin]-treated GP versus mock-treated GP). For the inset, cleavage of full-length GP to 19 kDa was confirmed by Western blotting with the anti-GP1 F88 rabbit antibody as described in Materials and Methods. Lane 1, mock-treated GP; lane 2, (thermolysin)-treated GP. The band labeled GP runs at 130 kDa, indicating that it is full-length GP with its mucin-like domain intact. (b) VLPs bearing Mayinga GP were untreated or (thermolysin) treated as described in Materials and Methods. After quenching the thermolysin, the VLPs were incubated with 50 μg/ml c2G4, c4G7, or PBS (Mock), as indicated, for 1 h at 37°C. Entry was then assayed, as described in the legend to Fig. 2. The entry level of untreated VLPs with full-length GP was 60%, and that of VLPs with (thermolysin)-treated (19 kDa) GP was 35%. Error bars represent the SD. Highly similar results were seen in two additional experiments (full-length mock-treated entry levels, 45 and 40%; 19-kDa mock-treated entry levels, 20 and 25%). Cleavage of full-length GP (130 kDa) to 19 kDa was confirmed by Western blotting with MAb H3C8 as described in Materials and Methods. Lane 1, untreated; lane 2, blank; lane 3, (thermolysin) treated.
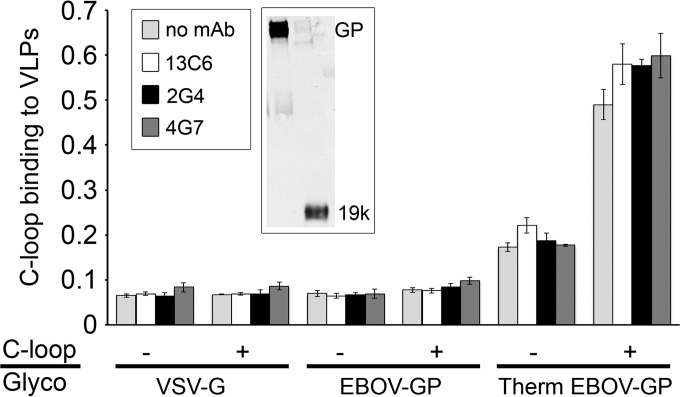
Another key event in EBOV entry is binding of the proteolytically primed (19-kDa) form of GP to NPC1 (20) in endolysosomes. Prior work has shown that the critical domain of NPC1 for this interaction is the C-loop (20), which projects into the lumen of the endolysosome. Given that the NPC1 binding site is near the distal tip of 19-kDa GP (20, 34, 36, 37), we predicted that c2G4 and c4G7, which bind to the GP base region (9, 10) (see also Fig. 1), would not interfere with NPC1 C-loop binding to 19-kDa GP. As seen in Fig. 6, this is the case: neither c2G4 nor c4G7, which bind strongly to 19-kDa GP (Fig. 5A), blocked the binding of a soluble NPC1 C-loop construct to VLPs bearing 19-kDa GP.
FIG 6.
MAbs c2G4 and c4G7 do not block binding of the NPC1 C-loop to primed (19-kDa) EBOV GP. VLPs bearing EBOV-Mayinga GP were mock treated or (thermolysin) treated (to 19 kDa) and then bound (triplicate samples) to a high-bind ELISA plate. After washing and blocking, MAbs c2G4, c4G7, or c13C6 (50 μg/well; PBS for the control) were added, and the plate incubated for 1.5 h at room temperature. After washing, 0.2 μg/ml soluble NPC1 C-loop (PBS for no NPC1 C-loop samples) was added, followed by incubation for 1 h at room temperature. After washing (PBS), the plate was processed to detect soluble C-loop binding as described in Materials and Methods. Data are the averages ± the SD. Cleavage of full-length GP to 19 kDa was confirmed by Western blotting with a rabbit anti-GP antibody (a gift from Paul Bates) as described in Materials and Methods. Lane 1, mock treated GP; lane 2, (thermolysin) treated GP. Band labeling is as described in the Fig. 5 legend.
DISCUSSION
The ZMapp cocktail of MAbs against the EBOV GP is a promising therapeutic for future use in patients infected with EBOV (2). In this study, we extended previous work (4, 5, 8–10) on the locations and mechanism of inhibitory activity of its three constituents, MAbs c2G4, c4G7, and c13C6.
Our structural studies (Fig. 1) are consistent with previous findings on the location and angles of binding of c13C6, c2G4, and c4G7 to the isolated GP ectodomain from the Mayinga isolate of EBOV, notably that c2G4 and c4G7 display overlapping binding sites to the base region of the EBOV-Mayinga GP ectodomain (9, 10). We extended these structural findings by showing (i) that all three MAbs bind as intact IgGs to GP from EBOV-Makona, the isolate that caused the 2014-2015 outbreak in West Africa; (ii) that all three IgGs bind, at high occupancy, to membrane-embedded GPs densely packed on a VLP; and (iii) that it is possible that the base-binding MAbs could bridge adjacent trimers (Fig. 2).
Our cell-based studies (see Fig. 3) provide the first formal evidence that c2G4 and c4G7 block entry of EBOV particles into the cell cytoplasm and, moreover, that c4G7 diminishes the ability of c2G4 to block entry. This finding is consistent with the higher (∼2-fold) affinity of c4G7 for retroviral pseudovirions bearing EBOV-Mayinga GP (10) and the ability of c4G7 to compete with c2G4 for binding to the Mayinga GP ectodomain (9). Hence, our cell-based findings, coupled with the previous binding data, raise the question of whether both c2G4 and c4G7 are required for the ZMapp cocktail to exert its protective effects (9).
Additional findings suggest that the base-binding MAbs (c2G4 and c4G7) exert their inhibitory action deep in the endocytic pathway, after binding of primed (19-kDa) GP to NPC1. First, neither base-binding MAb impedes VLP trafficking to NPC1+ endolysosomes (Fig. 4). Second, both c2G4 and c4G7 bind to, and can inhibit cytoplasmic entry of, VLPs bearing 19-kDa GP (Fig. 5) and, third, they do so without blocking binding of the NPC1 C-loop to 19-kDa GP (Fig. 6). It is interesting that c2G4 and c4G7, as well as KZ52, another neutralizing MAb (21), require C511 and C556 in the EBOV GP fusion subunit (GP2) for binding (10). These cysteine residues form the disulfide bond that clasps the ends of the fusion loop in EBOV GP (21, 38). Hence, the base-binding MAbs (c2G4 and c4G7) most likely block a structural change in 19-kDa GP bound to NPC1 that is needed to expose the fusion loop for interaction with the target membrane, an early step in the fusion cascade (39, 40). This conclusion is consistent with findings presented by Chandran and coworkers while this manuscript was in preparation showing that the ZMapp cocktail blocks the merging of EBOV GP pseudoviral and endolysosome membranes (29). Hence, the function of MAbs c2G4 and c4G7 is most likely similar to that of neutralizing antibodies that target the fusion peptide regions of influenza HA (41) and HIV Env (42).
ACKNOWLEDGMENTS
This study was supported by grants R21 AI103601 and RO1 AI114776 from the National Institutes of Health (NIH) (J.M.W.) and by the Intramural Research Program of the National Cancer Institute of the NIH (S.S.). J.A.S. was supported in part by T32AI007046.
We thank Alan Merk for assistance with microscopy, Alberto Bartesaghi and Oleg Kuybeda for assistance with image processing and helpful discussions, and Veronica Falconieri for assistance with figure preparation. We thank Ryan D'Souza and James Casanova for help with confocal microscopy analysis. This study utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
L. Zeitlin is President of Mapp Biopharmaceuticals.
REFERENCES
- 1.La Vega de M-A, Stein D, Kobinger GP. 2015. Ebolavirus evolution: past and present. PLoS Pathog 11:e1005221. doi: 10.1371/journal.ppat.1005221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, Johnson A, Morton J, Swope K, Bohorov O, Bohorova N, Goodman C, Kim D, Pauly MH, Velasco J, Pettitt J, Olinger GG, Whaley K, Xu B, Strong JE, Zeitlin L, Kobinger GP. 2014. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davey RT. 2016. Prevail II: a randomized controlled trial of ZMapp in acute Ebola virus infection, abstr 77LB. Conf Retrovir Opportunistic Infect (CROI), Boston, MA, 22 to 25 February 2016. [Google Scholar]
- 4.Wilson J, Hevey M, Bakken R, Guest S, Bray M. 2000. Epitopes involved in antibody-mediated protection from Ebola virus. Science 287:1664–1666. doi: 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- 5.Qiu X, Alimonti JB, Melito PL, Fernando L, Ströher U, Jones SM. 2011. Characterization of Zaire ebolavirus glycoprotein-specific monoclonal antibodies. Clin Immunol 141:218–227. doi: 10.1016/j.clim.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Qiu X, Fernando L, Melito PL, Audet J, Feldmann H, Kobinger G, Alimonti JB, Jones SM. 2012. Ebola GP-specific monoclonal antibodies protect mice and guinea pigs from lethal Ebola virus infection. PLoS Negl Trop Dis 6:e1575. doi: 10.1371/journal.pntd.0001575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiu X, Audet J, Wong G, Pillet S, Bello A, Cabral T, Strong JE, Plummer F, Corbett CR, Alimonti JB, Kobinger GP. 2012. Successful treatment of Ebola virus-infected cynomolgus macaques with monoclonal antibodies. Sci Transl Med 4:138ra81. doi: 10.1126/scitranslmed.3003876. [DOI] [PubMed] [Google Scholar]
- 8.Audet J, Wong G, Wang H, Lu G, Gao GF, Kobinger G, Qiu X. 2014. Molecular characterization of the monoclonal antibodies composing ZMAb: a protective cocktail against Ebola virus. Sci Rep 4:6881. doi: 10.1038/srep06881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murin CD, Fusco ML, Bornholdt ZA, Qiu X, Olinger GG, Zeitlin L, Kobinger GP, Ward AB, Saphire EO. 2014. Structures of protective antibodies reveal sites of vulnerability on Ebola virus. Proc Natl Acad Sci U S A 111:17182–17187. doi: 10.1073/pnas.1414164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davidson E, Bryan C, Fong RH, Barnes T, Pfaff JM, Mabila M, Rucker JB, Doranz BJ. 2015. Mechanism of binding to Ebola virus glycoprotein by the ZMapp, ZMAb, and MB-003 cocktail antibodies. J Virol 89:10982–10992. doi: 10.1128/JVI.01490-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shoemaker CJ, Schornberg KL, Delos SE, Scully C, Pajouhesh H, Olinger GG, Johansen LM, White JM. 2013. Multiple cationic amphiphiles induce a Niemann-pick C phenotype and inhibit Ebola virus entry and infection. PLoS One 8:e56265. doi: 10.1371/journal.pone.0056265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mingo RM, Simmons JA, Shoemaker CJ, Nelson EA, Schornberg KL, D'Souza RS, Casanova JE, White JM. 2015. Ebola virus and severe acute respiratory syndrome coronavirus display late cell entry kinetics: evidence that transport to NPC1+ endolysosomes is a rate-defining step. J Virol 89:2931–2943. doi: 10.1128/JVI.03398-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 14.Tran EEH, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JLS, Subramaniam S. 2012. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog 8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, la Cruz de MJ, Bess VJW, Nandwani R, Hoxie JA, Lifson JD, Milne JLS, Subramaniam S. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog 6:e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran EEH, Simmons JA, Bartesaghi A, Shoemaker CJ, Nelson E, White JM, Subramaniam S. 2014. Spatial localization of the Ebola virus glycoprotein mucin-like domain determined by cryo-electron tomography. J Virol 88:10958–10962. doi: 10.1128/JVI.00870-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuybeda O, Frank GA, Bartesaghi A, Borgnia M, Subramaniam S, Sapiro G. 2013. A collaborative framework for 3D alignment and classification of heterogeneous subvolumes in cryo-electron tomography. J Struct Biol 181:116–127. doi: 10.1016/j.jsb.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ou W, Delisle J, Konduru K, Bradfute S, Radoshitzky SR, Retterer C, Kota K, Bavari S, Kuhn JH, Jahrling PB, Kaplan G, Wilson CA. 2011. Development and characterization of rabbit and mouse antibodies against ebolavirus envelope glycoproteins. J Virol Methods 174:99–109. doi: 10.1016/j.jviromet.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deffieu MS, Pfeffer SR. 2011. Niemann-Pick type C 1 function requires lumenal domain residues that mediate cholesterol-dependent NPC2 binding. Proc Natl Acad Sci U S A 108:18932–18936. doi: 10.1073/pnas.1110439108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller EH, Obernosterer G, Raaben M, Herbert AS, Deffieu MS, Krishnan A, Ndungo E, Sandesara RG, Carette JE, Kuehne AI, Ruthel G, Pfeffer SR, Dye JM, Whelan SP, Brummelkamp TR, Chandran K. 2012. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J 31:1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. 2008. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dias JM, Kuehne AI, Abelson DM, Bale S, Wong AC, Halfmann P, Muhammad MA, Fusco ML, Zak SE, Kang E, Kawaoka Y, Chandran K, Dye JM, Saphire EO. 2011. A shared structural solution for neutralizing ebolaviruses. Nat Struct Mol Biol 18:1424–1427. doi: 10.1038/nsmb.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bournazos S, DiLillo DJ, Ravetch JV. 2015. The role of Fc-FcγR interactions in IgG-mediated microbial neutralization. J Exp Med 212:1361–1369. doi: 10.1084/jem.20151267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olinger GG, Pettitt J, Kim D, Working C, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, Pauly M, Whaley KJ, Lear CM, Biggins JE, Scully C, Hensley L, Zeitlin L. 2012. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc Natl Acad Sci U S A 109:18030–18035. doi: 10.1073/pnas.1213709109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang C-C, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J Virol 77:10557–10565. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gire SK, Goba A, Andersen KG, Sealfon RSG, Park DJ, Kanneh L, Jalloh S, Momoh M, Fullah M, Dudas G, Wohl S, Moses LM, Yozwiak NL, Winnicki S, Matranga CB, Malboeuf CM, Qu J, Gladden AD, Schaffner SF, Yang X, Jiang PP, Nekoui M, Colubri A, Coomber MR, Fonnie M, Moigboi A, Gbakie M, Kamara FK, Tucker V, Konuwa E, Saffa S, Sellu J, Jalloh AA, Kovoma A, Koninga J, Mustapha I, Kargbo K, Foday M, Yillah M, Kanneh F, Robert W, Massally JLB, Chapman SB, Bochicchio J, Murphy C, Nusbaum C, Young S, Birren BW, Grant DS, Scheiffelin JS, Lander ES, Happi C, Gevao SM, Gnirke A, Rambaut A, Garry RF, Khan SH, Sabeti PC. 2014. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345:1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoenen T, Safronetz D, Groseth A, Wollenberg KR, Koita OA, Diarra B, Fall IS, Haidara FC, Diallo F, Sanogo M, Sarro YS, Kone A, Togo ACG, Traore A, Kodio M, Dosseh A, Rosenke K, de Wit E, Feldmann F, Ebihara H, Munster VJ, Zoon KC, Feldmann H, Sow S. 2015. Virology mutation rate and genotype variation of Ebola virus from Mali case sequences. Science 348:117–119. doi: 10.1126/science.aaa5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simmons JA, D'Souza RS, Ruas M, Galione A, Casanova JE, White JM. 2015. Ebolavirus glycoprotein directs fusion through NPC1+ endolysosomes. J Virol 90:605–610. doi: 10.1128/JVI.01828-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spence JS, Krause TB, Mittler E, Jangra RK, Chandran K. 2016. Direct visualization of Ebola virus fusion triggering in the endocytic pathway. Chembiochem 7:e01857-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol 80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Côté M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, Cunningham J. 2011. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dube D, Brecher MB, Delos SE, Rose SC, Park EW, Schornberg KL, Kuhn JH, White JM. 2009. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): sequence and residues critical for host cell binding. J Virol 83:2883–2891. doi: 10.1128/JVI.01956-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hood CL, Abraham J, Boyington JC, Leung K, Kwong PD, Nabel GJ. 2010. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: implications for viral entry and immunogenicity. J Virol 84:2972–2982. doi: 10.1128/JVI.02151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Shi Y, Song J, Qi J, Lu G, Yan J, Gao GF. 2016. Ebola viral glycoprotein bound to its endosomal receptor Niemann-Pick C1. Cell 164:258–268. doi: 10.1016/j.cell.2015.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bornholdt ZA, Ndungo E, Fusco ML, Bale S, Flyak AI, Crowe JE, Chandran K, Saphire EO. 2016. Host-primed Ebola virus GP exposes a hydrophobic NPC1 receptor-binding pocket, revealing a target for broadly neutralizing antibodies. mBio 7:e02154-15. doi: 10.1128/mBio.02154-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gregory SM, Harada E, Liang B, Delos SE, White JM, Tamm LK. 2011. Structure and function of the complete internal fusion loop from ebolavirus glycoprotein 2. Proc Natl Acad Sci U S A 108:11211–11216. doi: 10.1073/pnas.1104760108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White JM, Delos SE, Brecher M, Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol 43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White JM, Schornberg KL. 2012. A new player in the puzzle of filovirus entry. Nat Rev Microbiol 10:317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khanna M, Sharma S, Kumar B, Rajput R. 2014. Protective immunity based on the conserved hemagglutinin stalk domain and its prospects for universal influenza vaccine development. Biomed Res Int 2014:546274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montero M, van Houten NE, Wang X, Scott JK. 2008. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol Mol Biol Rev 72:54–84. doi: 10.1128/MMBR.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]