ABSTRACT
Polyurethane (PU) is widely used in many aspects of modern life because of its versatility and resistance. However, PU waste disposal generates large problems, since it is slowly degraded, there are limited recycling processes, and its destruction may generate toxic compounds. In this work, we isolated fungal strains able to grow in mineral medium with a polyester PU (PS-PU; Impranil DLN) or a polyether PU (PE-PU; Poly Lack) varnish as the only carbon source. Of the eight best Impranil-degrading strains, the six best degraders belonged to the Cladosporium cladosporioides complex, including the species C. pseudocladosporioides, C. tenuissimum, C. asperulatum, and C. montecillanum, and the two others were identified as Aspergillus fumigatus and Penicillium chrysogenum. The best Impranil degrader, C. pseudocladosporioides strain T1.PL.1, degraded up to 87% after 14 days of incubation. Fourier transform infrared (FTIR) spectroscopy analysis of Impranil degradation by this strain showed a loss of carbonyl groups (1,729 cm−1) and N—H bonds (1,540 and 1,261 cm−1), and gas chromatography-mass spectrometry (GC-MS) analysis showed a decrease in ester compounds and increase in alcohols and hexane diisocyanate, indicating the hydrolysis of ester and urethane bonds. Extracellular esterase and low urease, but not protease activities were detected at 7 and 14 days of culture in Impranil. The best eight Impranil-degrading fungi were also able to degrade solid foams of the highly recalcitrant PE-PU type to different extents, with the highest levels generating up to 65% of dry-weight losses not previously reported. Scanning electron microscopy (SEM) analysis of fungus-treated foams showed melted and thinner cell wall structures than the non-fungus-treated ones, demonstrating fungal biodegradative action on PE-PU.
IMPORTANCE Polyurethane waste disposal has become a serious problem. In this work, fungal strains able to efficiently degrade different types of polyurethanes are reported, and their biodegradative activity was studied by different experimental approaches. Varnish biodegradation analyses showed that fungi were able to break down the polymer in some of their precursors, offering the possibility that they may be recovered and used for new polyurethane synthesis. Also, the levels of degradation of solid polyether polyurethane foams reported in this work have never been observed previously. Isolation of efficient polyurethane-degrading microorganisms and delving into the mechanisms they used to degrade the polymer provide the basis for the development of biotechnological processes for polyurethane biodegradation and recycling.
INTRODUCTION
Polyurethane (PU), one of the most versatile polymers invented by humans, is synthesized from a wide variety of precursors generating almost infinite types of materials, from elastomers and varnishes to highly resistant components, such as automotive parts, foams, semirigid plastics, and textile fibers. Due to their versatility, PUs are widely used in different human activities as a substitute for wood or metal and in other innovative products. However, the advantages provided by these polymers are correlated with the problem caused by its accumulation after the end of its useful life. In Europe alone, 59 million tonnes of plastics were produced in 2014, from which 4.4 million tonnes were PU; 25.8 million tonnes of postconsumer plastic waste ended up in the waste upstream, and from that, 30.8% went to landfill sites after its useful life (1). In Mexico in 2011, more than 4 million tonnes of plastic waste was sent to landfills (2). In addition, PUs are slowly degraded, and it has been estimated that it takes hundreds of years for PU molecules to return to the environment (3, 4). Also, no clean or effective systems for their disposal exist; incineration generates toxic fume emissions, and recycling has the inconvenience that after a few rounds, the material becomes useless, ending in dump sites (5).
PUs are synthesized from polyols and isocyanates, and depending on what type of polyols is used, PUs can be of the polyester (PS) type or polyether (PE) type. Biodegradation of PUs by bacteria and fungi has been studied since the 1980s, and although different enzymatic activities have been identified (4, 6–8), the underlying mechanisms of biodegradation are still unknown. Many of the PS-PU-degrading fungi have been collected from PS-PU coupons buried in soil under laboratory conditions or in compost systems (9–14), whereas others have been found growing in PU-contaminated sand (15) or have been isolated from plants (endophytic fungi) (16).
PE-PU foams are extensively used because of their resistance and low cost of production. The foam's protective and cushioning properties make it the ideal material for packaging applications, and its inherent comfort, support, and resilience make it the clear choice for construction, automotive appliances, and apparel, health care, medical, sports, and leisure products. This material, however, is more recalcitrant to fungal attack than PS-PU (17). Although several works have documented the degradation of solid PS-PU by fungi (9–14, 17), scarce research on solid PE-PU biodegradation has been carried out. Early work reported that PE-PU foams were unaffected by Aspergillus niger and Cladosporium herbarum growth (18). Moreover, when incubated in compost piles (11, 19) or agricultural soils (19), where fungi play an important role in degradation, samples of PE-PU turned yellow and brownish but were not physically deteriorated. The only work reporting an effective microorganism, Alternaria sp. strain PURDK2, which is able to degrade ether-PU cubes as much as 27.5% after 10 weeks of exposure, was published in 2010. When incubated with substrates that contained urethane or urea bonds, this strain caused the release of some degradation products into the medium, probably by the action of urethane- and urea bond-degrading enzymes, which have been suggested as the activities that enabled the strain to degrade the ether-PU cubes (20).
Therefore, there is an urgent need to develop biotechnological processes for PU biodegradation, particularly for the highly recalcitrant PE-PU. For that, the first requirement is to isolate microorganisms with high capability to attack PU and to characterize their biological activity on the polymer. Here, we report eight novel fungal strains isolated from environmental samples, which are able to degrade Impranil (a PS-PU varnish) and Poly Lack (a PE-PU varnish) and to reduce the weight (up to 65%) of highly recalcitrant solid PE-PU foams. We also provide evidence by Fourier transform infrared (FTIR) spectroscopy, gas chromatography-mass spectrometry (GC-MS), enzymatic activity analyses, and scanning electron microscopy (SEM) of the biological action displayed by the best PU-degrading fungi on the polymer. The identification of degradation products generated by fungal action, which may be recovered to be used as the substrates for new PU synthesis, would make the development of biotechnological processes for PU biodegradation and recycling more valuable.
MATERIALS AND METHODS
Culture media.
For isolation and qualitative evaluation of PU degradation, fungi were grown in the mineral medium (MM) modified from that in reference 15: 19 mM NaH2PO4, 33.5 mM K2HPO4, 7.6 mM (NH4)2SO4, 250 μM MgSO4·7H2O, 147 μM FeCl3·6H2O, 14 μM ZnCl2·4H2O, 12 μM CoCl2·6H2O, 12 μM Na2MoO4·2H2O, 10 μM CaCl2·2H2O, 11 μM CuCl2, 12 μM MnCl2, 12 μM H3BO3, and 1.8 mM HCl; this was supplemented with different PU substrates as the only source of carbon. The reagents used for medium preparation were American Chemical Society (ACS) grade from J.T.Baker Chemicals. Agar plates were made by adding 15 g/liter agar. Potato dextrose agar (PDA) and potato dextrose broth (PDB) media were used for growth and maintenance of filamentous fungi. Since fungal growth was similar between 25 and 30°C, these temperatures were used indiscriminately for incubation.
Isolation of fungi able to grow in PU.
The different PU-related compounds used to isolate fungi able to degrade PU were as follows: Rymsapol 200/107 L-83 (Resinas y Materiales S.A. de C.V.) (diethylene-glycol polyadipate), a polyester polyol (PP) used as the substrate for PU synthesis; Impranil DLN (Bayer) (I), a PS-PU; Hydroform (PolyForm de México, S.A. de C.V.) (H), a PS-PU; and Poly Lack (Sayer Lack Mexicana, S.A. de C.V.) (PL), a PE-PU. Impranil, Hydroform, and Poly Lack are commercial PU varnishes formulated as colloidal water-based PU suspensions, whose exact compositions are unknown. Plates with solid MM and one of the PU-related compounds at 0.3% solids as carbon sources were inoculated with environmental samples collected from different sources: garden soil (T), decomposing PU foams collected at a municipal dump site (Bordo Poniente, Ciudad Nezahualcóyotl, Estado de México, México) (BP), natural airflow from laboratory indoors (A1) and outdoors (A2), and fungal colonies growing on the PU insulation of a cold room wall (A3). For the Bordo Poniente dump site samples, chloramphenicol (50 μg/ml) (Sigma-Aldrich) was added to solid medium to prevent bacterial growth. After cultivation on these media for 7 days at 25 to 30°C, all the morphologically different fungi were streaked on PDA medium for several rounds until their isolation. Conidia from each isolate were preserved by desiccation in silica gel. Fungal strains were named after their sampling source (T, BP, A1, A2, or A3) and substrate used for their isolation (PP, I, H, or PL).
Identification of fungi.
Microcultures of the best PU-degrading fungi were made by inoculating PDA medium cubes (0.5 by 0.5 by 0.5 cm) with conidia on the four sides. Cubes were placed on slides and cover glasses and incubated in petri dishes containing a glass triangle as support and filter paper soaked with sterile water. Microcultures were incubated for 7 days at 25°C. Water was exchanged for formaldehyde to fix the reproductive structures to the slides and incubated for 1 h. PDA medium cubes were removed from the slides and stained with lactophenol cotton blue to observe the reproductive structures under the microscope. The genera of the fungi were determined based on the morphology and pigmentation of conidia and conidiophores, according to the Saccardo system of classification (21). For species identification, molecular techniques were used. All the reagents used in DNA techniques were molecular biology grade from Sigma-Aldrich. Fungi were grown in PDB for 1 week at 30°C. DNA was extracted according to the method in reference 22, with the following modifications: 0.2 g of filtered mycelium was pulverized with liquid nitrogen, resuspended in 600 μl of extraction buffer (2% cetyltrimethylammonium bromide [CTAB], 1.4 M NaCl, 200 mM EDTA, and 100 mM Tris-HCl [pH 8]), and incubated at 65°C for 30 min. The mixture was cooled at 25 to 30°C, and 250 μl of 5 M potassium acetate was added to complete cell lysis. Cell debris was removed by centrifugation (15,000 × g) for 5 min in an Eppendorf centrifuge (model 5427C), and the supernatant was extracted twice with phenol-chloroform. DNA was precipitated overnight with absolute ethanol and resuspended in 100 μl of sterile deionized water. Genomic DNA integrity was verified by agarose gel electrophoresis, and DNA purity was determined by A260/A280 and A260/A230 ratios. An aliquot (250 ng) of genomic DNA was used as a template for PCR amplification of part of the rRNA gene cluster (approximately 1,200 bp) spanning the internal transcribed spacer 1 (ITS1), the 5.8S rRNA gene, the internal transcribed spacer 2 (ITS2), and the 28S rRNA gene region D1-D2 (23), using the ITS1 forward primer (5′-TCCGTAGGTGAACCTGCGG-3′) (24) and the D2 reverse primer (5′-TTGGTCCGTGTTTCAAGACG-3′) (25). For a precise identification of the strains belonging to the Cladosporium cladosporioides complex, partial sequences of actin (ACT) (approximately 230 bp) and translation elongation factor (TEF) 1α (approximately 240 bp) genes were amplified (26) using the primers ACT-512F/ACT-783R and EF1-728F/EF1-986R, respectively (27). PCR was performed with Phusion high-fidelity DNA polymerase (Thermo Scientific), according to the manufacturer's instructions for genomic DNA amplification (10 μM each primer, 10 mM each deoxynucleoside triphosphate [dNTP], 1× Phusion buffer, and 1 U of Phusion DNA polymerase). The PCR program was one initial denaturation cycle at 98°C for 3 min; 30 cycles of denaturation at 98°C for 10 s, annealing at 64.4°C for 30 s for reactions with ITS-D2 primers, at 63°C for 15 s for reactions with ACT primers, or at 58°C for 15 s for reactions with TEF primers, and extension at 72°C for 36 s; and a final extension cycle at 72°C for 7 min. PCR products were gel purified by using the GeneJET PCR purification kit (Thermo Scientific). Sequencing was performed at Macrogen, Inc. (Seoul, South Korea) with the same primers used for the PCR amplifications. Identification was made by comparison of the obtained sequences to the GenBank database using Basic Local Alignment Search Tool (BLAST) (28).
Impranil degradation assay.
Strains were inoculated on plates containing MM with Impranil and incubated at 25 to 30°C for 5 days. Fungal utilization of Impranil as a carbon source and hydrolysis, by clearing halo formation on Impranil plates, were qualitatively evaluated based on reference 14. For quantitative Impranil degradation assays, tubes containing 10 ml of MM with Impranil were inoculated with mycelium (80 mg [fresh weight] obtained from static cultures grown in PDB for 1 week at 30°C in darkness) from each selected fungus. The mycelial samples were washed prior to inoculation to remove any residual medium. After incubation without agitation for 14 days at 30°C in darkness, cultures were filtered through Whatman grade 41 paper and the filtrate measured turbidimetrically at λ600 nm, using distilled water as a blank. Noninoculated tubes with 10 ml of MM with Impranil were set as controls. The concentration of residual Impranil was calculated according to values obtained from a standard curve done with different Impranil concentrations. The standard curve was linear (r2 = 1) up to 1 mg/ml at λ600 nm. Three biological replicates were quantified for each fungal strain.
FTIR spectroscopy analysis of Impranil degradation.
The eight fungi with the highest capability to degrade Impranil were inoculated with 80 mg (fresh weight) of mycelium obtained from static cultures grown in PDB for 1 week at 30°C, in tubes containing 10 ml of MM with Impranil, and incubated without agitation for 14 days at 30°C in darkness. The cultures were thoroughly mixed, mycelium was eliminated by filtration through Whatman grade 41 filter paper, and 1 ml of each filtrate was analyzed by FTIR spectroscopy in a 1605 PerkinElmer spectrometer at 2,500 to 800 cm−1. Three biological replicates were analyzed for each selected fungal strain. Spectral interpretation was performed according to references 29 and 30.
Identification of Impranil degradation products.
Fungal mycelia (80 mg [fresh weight] of mycelium obtained from static cultures grown in PDB for 1 week at 30°C in darkness) were cultured in 125-ml flasks with 40 ml of MM with Impranil, without agitation at 25°C in darkness; noninoculated MM with Impranil flasks were used as controls. Analysis of inoculated and control media was carried out at 0, 7, and 14 days of incubation. For each analysis time, cultures were thoroughly mixed, and mycelium was eliminated from the media by filtering through Whatman grade 41 filter paper. The filtrates were centrifuged at 12,000 × g for 15 min in a Sorvall RC-5C Plus with rotor type SS-34. Supernatants were then filtered through 0.45-μm-pore-size Millipore membranes to eliminate conidia and residual Impranil. The filtrates were extracted three times with 10 ml of dichloromethane. The three extracts were pooled and the solvent evaporated to dryness at 25 to 30°C. The dried samples were resuspended in 50 μl of methanol-chloroform (1:1 [vol/vol]), and 1 μl was injected (injector temperature, 300°C) with a split ratio of 1:50 in an Agilent 6890N gas chromatograph with a DB-5MS column (5% phenylmethyl silicone; length, 20 m; internal diameter, 0.18 mm). The oven program was initial temperature of 40°C (held for 3 min), which increased by 20°C/min up to 300°C (held for 15 min). All the solvents used were ACS grade from J.T.Baker Chemicals. Helium was used as the carrier gas at a flow rate of 1 ml/min. Time of flight mass spectrometry (TOF-MS) analysis was performed in a Leco Pegasus 4D model apparatus. The electronic ionization energy was 70 eV, and the mass range scanned was 45 to 550 atomic mass units (amu). The delay time was 300 s. The scan rate was 20/s. Mass spectra of the compounds detected in the analyzed extracts were compared to the National Institute of Standards and Technology (NIST) Mass Spectrometry Data Center 2002 library. Compounds with mass spectrum similarity values of >850 were considered the same compound as the library hit; below this similarity value, they were considered related compounds.
Enzymatic assays.
For enzymatic analyses, 80 mg (fresh weight) of mycelium obtained from static cultures grown in PDB for 1 week at 30°C in darkness was cultured in 125-ml flasks with 40 ml of MM with Impranil, without agitation at 25°C in darkness. Cultures were harvested after 0, 7, and 14 days of incubation (3 replicates for each time) and filtered through four layers of cheesecloth. The filtrates were centrifuged at 12,000 × g for 15 min in a Beckman J2-21 centrifuge with a JA-20 rotor. Cell-free supernatants, the source for extracellular enzymatic activity, were filtered through 0.45-μm-pore-size Millipore membranes and then concentrated to 3 ml in Amicon Ultra 10K devices. Next, three dialysis rounds were sequentially carried out against 50 mM potassium phosphate (pH 7.0) for 2 h (in 1 liter of buffer), overnight (in 2 liter of buffer), and two more hours (in 1 liter of buffer). After that, samples were concentrated to 400 μl in an Amicon Ultra 10K, and protein levels were determined according to Bradford's method by using bovine serum albumin (BSA) as a standard. For 0 and 7 days of incubation, 1 μg of protein was used, whereas for 14 days of incubation, 4 μg of protein was used for the enzymatic assays. Esterase activity was determined spectrophotometrically by hydrolysis of p-nitrophenyl acetate (p-NPA) (catalog no. L00314; Alfa Aesar) at λ405 nm, as previously reported (6). The reaction mixture contained 50 mM potassium phosphate (pH 7.0), 100 μl of enzyme extract, and 5 mM p-NPA in a final volume of 1 ml. Lipase from Pseudomonas fluorescens (catalog no. 28602; Sigma-Aldrich) was used as a positive control. Protease activity was determined spectrophotometrically by casein hydrolysis at λ280 nm, as previously reported (6), except that the reaction mixture contained 100 mM potassium phosphate (pH 7.0), 50 μl of enzyme extract and 0.5% casein in a final volume of 1 ml, and it was incubated at 37°C overnight. Proteinase K (catalog no. EO0491; Thermo Scientific) was used as a positive control. Urease activity was determined spectrophotometrically by using a phenol hypochlorite assay, which measured ammonia release at λ636 nm (31). The reaction mixture contained 15 mM potassium phosphate (pH 7.0), 100 μl of enzyme extract, and 3.8 mM urea in a final volume of 1.3 ml. Jack bean urease (catalog no. U-1500; Sigma-Aldrich) was used as a positive control.
Degradation of PE-PU foams.
Two types of PE-PU foams (A and B), used for commercial production of mattress cushioning (kindly provided by Espumas Industriales Monterrey, S.A. de C.V.), were tested for fungal degradation. Both foams were synthesized from Caradol SC56-22 (Schell Chemicals) (propylene oxide/ethylene oxide-based polyether polyol) and Caradol MD30-45 (Schell Chemicals) (propylene oxide/ethylene oxide-based polyether polyol grafted with a styrene-acrylonitrile copolymer for higher strength) used at different proportions and polymerized with Mondur TD 80 grade A (toluene diisocyanate) (Bayer). Foam A contained a halogenated flame retardant, CP2 LV [Tris-(1,3-dichloroisopropyl) phosphate (TDCPP)]. Preweighed pieces of foam (8 by 30 by 15 mm) were washed, dried, autoclaved, inoculated with 80 mg (fresh weight) of 1-week-old mycelium grown in PDB in static cultures at 30°C in darkness, and incubated for 21 days at 25 to 30°C in petri dishes containing 20 ml of 50% PDB. The PDB was refreshed every 5 days. Mycelium growing around the foam pieces was carefully eliminated with sterile bistoury and forceps at every medium change. After the completion of incubation time, weight was recorded to determine biodegradation level. For that, foam pieces were treated with 0.88% (wt/vol) sodium hypochlorite for 18 h to destroy the remaining mycelium, washed out four times with 30 ml of distilled water, and vacuum dried at 25 to 30°C until constant weight. Noninoculated control foam pieces were similarly treated.
Scanning electron microscopy analysis.
Replicate samples of PU foam pieces used for quantifying biodegradation were analyzed by scanning electron microscopy (SEM). The mycelium-containing foams were fixed in 2.5% glutaraldehyde in 0.05 M phosphate buffer (pH 7.3) overnight. Samples were washed three times with 0.05 M phosphate buffer and then dehydrated in ethanol at 30%, 50%, 70%, 90%, and 100% for 5 min each. After this treatment, foams were incubated in a vacuum oven at 25 to 30°C for 72 h. For the mycelium-free foams, superficial mycelium was carefully removed with a razor blade, and internal mycelium was eliminated by submerging the foam in 50 ml of 0.88% (wt/vol) sodium hypochlorite for 18 h. The foams were then washed six times with 20 ml of distilled water by shaking at 125 rpm for 2 min each time. Vacuum filtration over a Buchner funnel (each side of the foam for 1 min) and incubation in a vacuum oven (72 h at 25 to 30°C) were used to remove the remaining water until complete dryness. Foams with and without mycelium were coated with gold and analyzed under a high vacuum without microanalysis in a Jeol JSM-5900-LV electron microscope. In each sample, 5 fields were analyzed as representative data.
Statistical analyses.
Impranil and PE-PU foam biodegradation were statistically analyzed by analysis of variance (ANOVA). Mean comparison was performed with Tukey's test for balanced experiments (P < 0.05).
Accession number(s).
Sequences were deposited in GenBank (accession numbers for partial sequences of rRNA genes, KU605792, KU605788, KU605794, KU605789, KU605793, KU605790, KU605787, and KU605791; for partial sequences of the actin gene, KU605800, KU605796, KU605799, KU605797, KU605795, and KU605798; and for partial sequences of the translation elongation factor 1α gene, KU605786, KU605782, KU605785, KU605783, KU605781, and KU605784). Table S1 in the supplemental material shows the percentage of the highest matches obtained and has the GenBank accession numbers for reference strains and the strains reported in this work.
RESULTS
Isolation of fungi.
A variety of fungi isolated from samples of diverse environments grew on mineral media containing different PU-related compounds used as carbon sources. A total of 73 morphologically different colonies were isolated, with 42 filamentous fungi and 31 yeast-like strains. PU degradation qualitative assays were performed by inoculating all the strains on agar plates with the different PU substrates. None of the yeast-like and five of the filamentous fungi were able to grow in any of the tested media; therefore, no further analysis was made with these strains. From the remaining 37 tested strains, 31 filamentous fungi were able to grow and produce clearing halos on MM with Impranil and MM with Rymsapol, indicating the capability to hydrolyze a polyester PU and a polyester polyol, respectively. From these 31 halo-forming strains, 22 were also able to grow on MM with Poly Lack, a PE-PU varnish. Since Poly Lack does not clarify when degraded, it fails as an indicator for quantitative assays, although it provides a means to select possible PE-PU-degrading microorganisms. Therefore, because Impranil clarifies when microorganisms hydrolyze it, it is a visual and reliable method for quantitative analysis (7, 16, 32, 33), and so we chose this substrate for further quantitative degradation experiments.
Biodegradation of Impranil and identification of fungi.
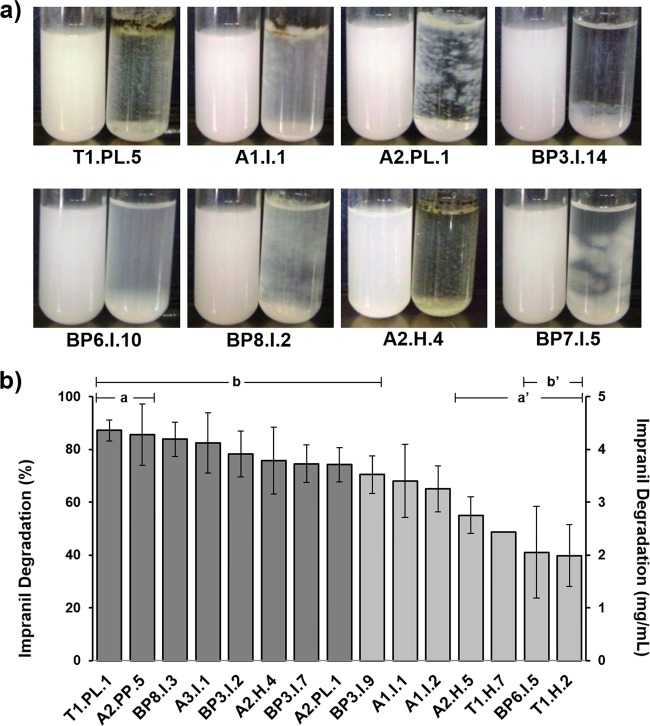
All the 31 fungi that grew and formed degradation halos on Impranil solid media were evaluated for quantitative turbidimetric analysis of Impranil degradation. The growth of representative strains in liquid MM with Impranil after 14 days of culture is depicted in Fig. 1a. All the fungal strains were able to degrade Impranil to different extents, with values that ranged from 6% to 99% (data not shown). An interesting correlation was that all nine fungi unable to grow on Poly Lack exhibited the lowest Impranil degradation values. Based on these results, a second quantitative Impranil degradation experiment with three replicates was performed, in which the 15 fungal isolates that exhibited the highest Impranil degradation levels were analyzed. This analysis showed degradation values from 40% to 87% (Fig. 1b). Isolates T1.PL.1, A2.PP.5, BP8.I.3, and A3.I.1 showed degradation levels above 80%, whereas isolates BP3.I.2, A2.H.4, BP3.I.7, and A2.PL.1 displayed degradation levels between 74% and 78%. ANOVA showed statistically significant differences on Impranil degradation between strains. Tukey's test demonstrated that only the two most Impranil-degrading strains, T1.PL.1 and A2.PP.5 (>80% degradation) (group a), showed statistically significant differences compared to the four least Impranil-degrading strains, A2.H.5, T1.H.7, BP6.I.5, and T1.H.2 (39 to 55% degradation) (group a′). When the nine most Impranil-degrading strains (>70% degradation) were considered as a group (group b), they showed statistically significant differences only compared to the two least-degrading strains (39 to 41% degradation) (group b′).
FIG 1.
Growth and Impranil degradation by some of the selected fungal strains. (a) Growth of fungal strains showing different types of development in MM with Impranil. Left tube, control; right tube, fungus-inoculated medium. (b) Quantitative analysis of Impranil degradation by the 15 fungal isolates showing the highest degradation levels. All data were normalized to the negative control. The eight darker-shaded columns represent the strains that were selected for further analysis. n = 3. The error bars represent standard deviations. Statistically significant differences are indicated by a versus a′ and b versus b′.
The eight best-degrading strains were identified by classical taxonomy and by molecular biology methods. Reproductive structure morphology and sequencing of amplicons obtained by PCR from part of the rRNA genes (ITS1-D2 region) allowed the precise identification of BP3.I.7 and A2.PL.1 as Penicillium chrysogenum and Aspergillus fumigatus, respectively. In the same way, it was determined that the six best PU-degrading strains belonged to the Cladosporium cladosporioides complex, with identities between 99 and 100% (24). Further BLAST analysis of the actin and translation elongation factor 1α partial sequences from these six strains showed the highest matches with the following species: T1.PL.1 with Cladosporium pseudocladosporioides, A2.PP.5 and A3.I.1 with Cladosporium tenuissimum, BP8.I.3 and BP3.I.2 with Cladosporium asperulatum, and A2.H.4 with Cladosporium montecillanum (34).
Effect of fungal growth on Impranil degradation.
To determine the effect that fungal growth generated on Impranil, the eight selected strains were grown on MM with Impranil, and FTIR analyses were performed in cell-free filtrates obtained from 14-day-old fungal cultures. All tested fungi generated similar changes on Impranil FTIR spectra. A representative FTIR spectrum of Impranil degradation by C. pseudocladosporioides T1.PL.1 revealed a decrease in the 1,729 cm−1 signal associated with the stretching of the carbonyl group (C=O) related to both the polyester fraction and the urethane bond. Decrements in the signals of bending (1,540 cm−1) and stretching (1,261 cm−1) of the C—N—H bond in the urethane group were also observed in the fungus-treated PU (Fig. 2).
FIG 2.
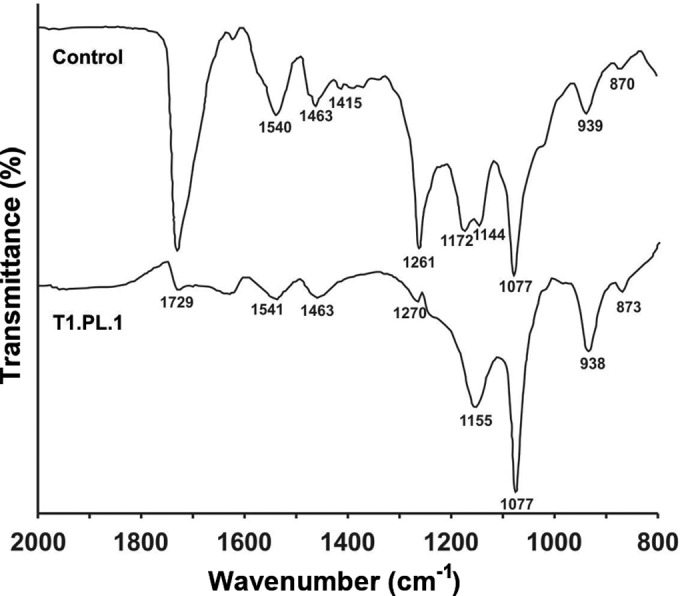
Representative FTIR spectroscopy analysis of Impranil degradation by fungi. Spectra of MM with Impranil noninoculated and inoculated with Cladosporium pseudocladosporioides strain T1.PL.1 after 14 days of incubation. All the eight best Impranil-degrading fungi presented similar results. Changes reflecting Impranil degradation are observed in the signals at 1,729 cm−1 (non-hydrogen-bonded urethane C=O stretch and ester from polyol fraction), 1,540 cm−1, and 1,261 cm−1 (urethane C—N—H bending and stretch, respectively). Signal assignments are based on references 29 and 30.
To explore the way in which fungi degrade Impranil, time course analyses of the compounds present in the media at 0, 7, and 14 days of incubation with the most Impranil-degrading strain, Cladosporium pseudocladosporioides T1.PL.1, were carried out by GC-MS. In controls, only small changes in some compounds were observed during the 14-day incubation period. However, important changes in the relative area of compounds related to PU degradation were observed in fungus-treated extracts (Table 1; see also Fig. S1 in the supplemental material). Five compounds, tentatively identified as cyclohexyl cyclohexanecarboxylate-related compound, adipic acid dicyclohexyl ester-related compound, tridecan-6-ol-related compound, tetradecan-3-yl prop-2-enoate-related compound, and cyclohexane (1-methylethyl)-related compound, disappeared in the fungus-inoculated media, most of them during the first week of incubation, whereas in the control, they remained with few changes during the 2 weeks of the experiment (Table 1 and Fig. S1, peaks 4, 6, 7, 8, and 9). Four compounds, tentatively identified as 2,2-dimethyl-1,3-propanediol, hexane-1,6-diol, hexane 1,6-diisocyanate, and adipic acid di(oct-4-yl ester)-related compound, which were present at almost constant or slightly decreasing levels in the control, increased in the cultures inoculated with the fungus (Table 1 and Fig. S1, peaks 1, 2, 3, and 5). The mass spectra of the compounds identified in these analyses are presented in Fig. S2 in the supplemental material.
TABLE 1.
Time course GC-MS analysis of Impranil degradation by the best Impranil-degrading fungal strain, Cladosporium pseudocladosporioides T1.PL1
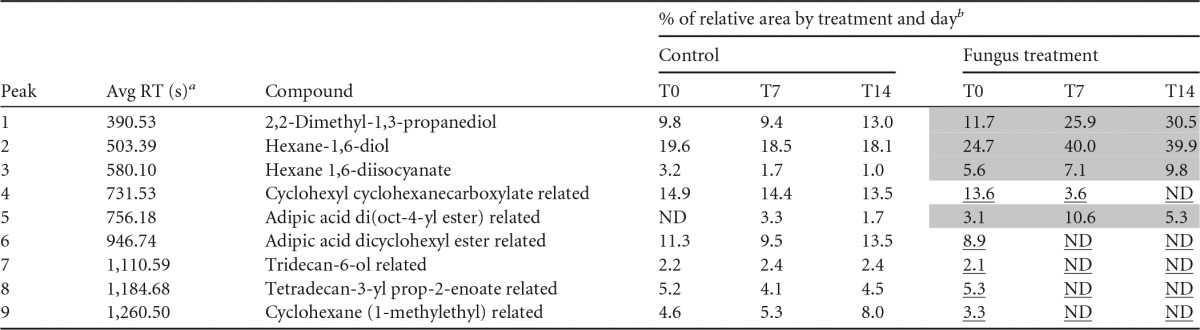
RT, retention time.
Samples from MM with Impranil culture media inoculated with C. pseudocladosporioides strain T1.PL.1 were harvested at 0 (T0), 7 (T7), and 14 (T14) days of incubation. Noninoculated samples of MM with Impranil incubated for the same times were analyzed as controls. Compounds were tentatively identified by comparison to the NIST Mass Spectrometry Data Center (2002 library). Shaded cells indicate values that increased >50% from time 0 to 14 days of incubation. Underlined values decreased to undetectable levels during the analysis. ND, nondetected.
Based on these results, and on the fact that the selection of Impranil-degrading fungi was based on their capability to form degradation halos on Impranil solid media, we hypothesized that biodegradation of the compounds present in Impranil occurred by fungal extracellular enzymatic activities. To address this possibility, culture supernatants of the most Impranil-degrading strain, C. pseudocladosporioides T1.PL.1 grown in MM with Impranil, were analyzed for enzymes whose activities have been correlated with polyurethane biodegradation: esterase, protease, and urease. Esterase activity at the initial time was almost negligible (0.057 ± 0.002 μmol p-NPA/min/mg of protein), and it increased to 84.91 ± 19.97 μmol p-NPA/min/mg of protein (1,500-fold) after 7 days of incubation and decreased 4-fold at day 14 (20.73 ± 1.41 μmol p-NPA/min/mg of protein). Protease activity was not detected at any time when analyzing aliquots of the same protein concentration as the ones used for esterase measurements. The protease-positive controls showed proper activity in the assays. Very low urease activity, at less than 3.13 ± 0.13 μm of NH3/min/μg of protein, was determined at different times during the analysis.
PE-PU foam degradation.
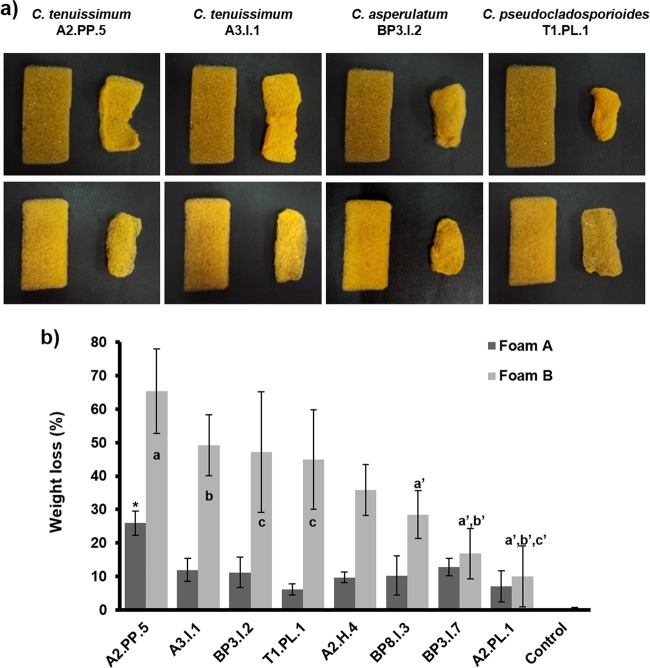
In order to explore whether the eight best Impranil-degrading strains could also be effective to degrade solid PE-PU foams, they were challenged to decompose two types of solid foams, which were similar in composition but different in density and in that foam A contained a flame retardant. Changes in the form and structure of the foam pieces were recorded, and weight losses were measured after incubation with the selected fungi. Analysis showed that the least PU-degrading strains grew only on the surface, but the best ones clearly developed their mycelia inside the foam, invading the whole piece (Fig. 3). Evidence of the capability of the best PU-degrading fungi to grow inside solid foam is shown in SEM micrographs, in which large numbers of hyphae invading the surface and the interior of foam cells can be seen (Fig. 4). After 21 days of incubation, foams inoculated with fungi changed their shape, size, and weight compared to the similarly treated but noninoculated pieces (Fig. 5a). The foam with flame retardant (foam A) was less degraded than the foam with no flame retardant (foam B), with weight losses from 6 to 25.9% and 10 to 65%, respectively (Fig. 5b). ANOVA and Tukey's test made it evident that Cladosporium strains displayed the largest foam degradation capability. Cladosporium tenuissimum A2.PP.5 was significantly the most degrading strain for both foam A (25.9%) and foam B (65.3%). C. tenuissimum A3.I.1, C. asperulatum BP3.I.2, and C. pseudocladosporioides T1.PL.1 followed in their ability to degrade foam B, at 49.3%, 47.1%, and 44.9%, respectively. Statistical analysis of foam B degradation indicates that C. tenuissimum A2.PP.5 showed significant differences with the three least-degrading strains (a′); C. tenuissimum A3.I.1 showed statistically significant differences compared to the two less-foam-degrading strains (b′), whereas C. asperulatum BP3.I.2 and C. pseudocladosporioides T1.PL.1 showed statistically significant differences only compared to the least PU-degrading strain, A. fumigatus A2.PL.1 (c′) (Fig. 5b).
FIG 3.
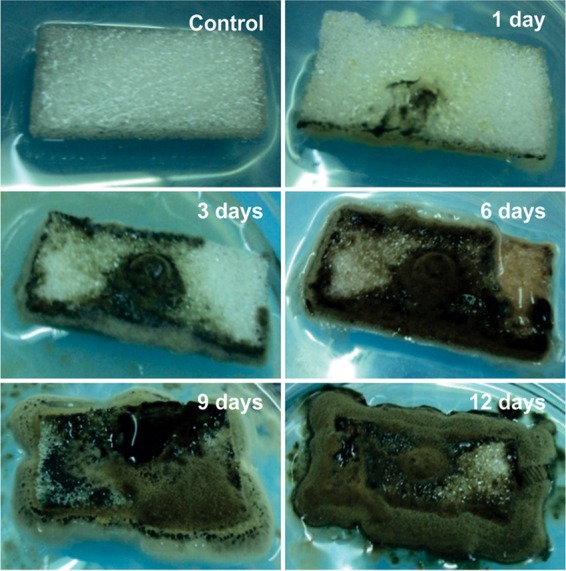
Fungal growth of Cladosporium asperulatum BP8.I.3 over and inside PE-PU foam. Representative progression of fungal growth is shown by one of the best Impranil-degrading strains, from initial colonization (1 day) on the surface of foams to extensive invasion (12 days) on the whole piece of foam. The growth on foam B (without flame retardant) is shown.
FIG 4.
Scanning electron micrographs of fungal growth inside PE-PU foam. Hyphal networks are observed in samples of foam pieces incubated for 7 days with three of the best Impranil-degrading fungi: Cladosporium tenuissimum strains A2.PP.5 and A3.I.1 and C. pseudocladosporioides strain T1.PL.1.
FIG 5.
Biodegradation of PE-PU foams by the best Impranil-degrading fungi. (a) Effects on size and form of PU foams after fungal treatment. Foam A (upper row) and foam B (lower row) were incubated with the four best Impranil-degrading fungi for 21 days. Each pair of foam pieces presents the control foam (left) compared to the fungus-treated piece (right). (b) Quantitative analysis of PE-PU foam degradation (represented as weight loss) by the best Impranil-degrading-fungi. Each value was normalized by subtracting the weight loss of noninoculated controls. n = 3. The bars represent standard deviations. For foam A degradation, an asterisk denotes a statistically significant difference. For foam B degradation, statistically significant differences are indicated by a versus a′, b versus b′, and c versus c′.
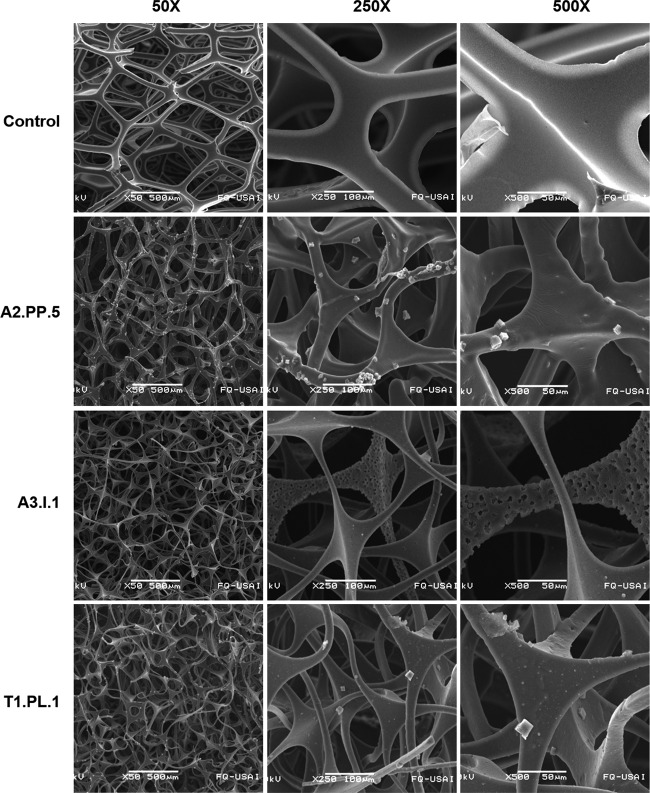
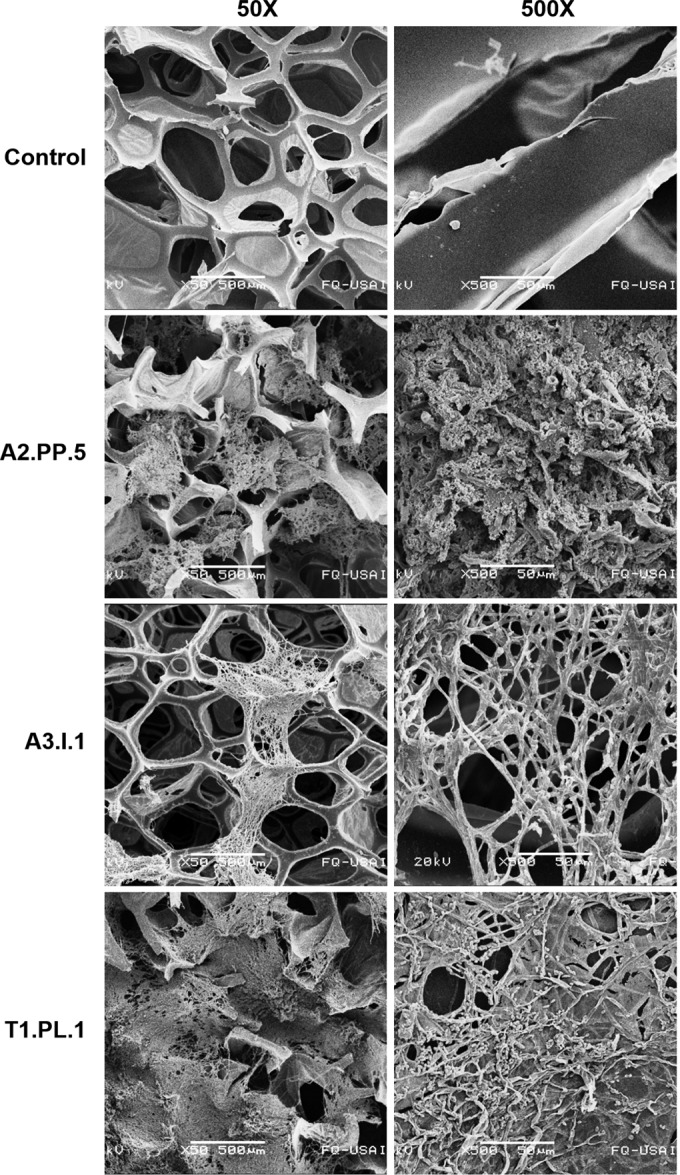
To further analyze PE-PU foam degradation, we also evaluated PU damage at a microscopic level. PU pieces of foam B incubated with the three best foam-degrading fungi (C. tenuissimum strains A2.PP.5 and A3.I.1 and C. pseudocladosporioides T1.PL.1) were treated to remove fungal mycelium, and SEM micrographs were taken. At panoramic views (50× magnification), the disruption and loss of integrity of the foam reticulated-cell structure were observed in samples incubated with the fungi (Fig. 6). At a closer view (250× magnification), thinning, bending, and relaxation of the fibers were observed in the fungus-incubated foams, in contrast to the non-fungus-treated control, in which PU cell structures remained straight and rigid. A finer detail of the fungal attack on PE-PU foam cells was observed at 500× magnification, in which pores, erosion, breaks, and bends of the reticulated-cell walls were detected in fungus-treated foams, whereas in the control, foam fibers remained smooth and rigid. Damage was more evident in PE-PU foams incubated with C. tenuissimum strain A3.I.1, in which fiber thickness diminished up to less than a half of the size of the noninoculated control (Fig. 6).
FIG 6.
Scanning electron micrographs of PE-PU foam ultrastructure after fungal growth. Foam pieces were incubated during 21 days with three of the best Impranil-degrading fungal strains: Cladosporium tenuissimum A2.PP.5 and A3.I.1 and C. pseudocladosporioides T1.PL.1; after that time, mycelia were destroyed to analyze their effect on PU structure.
DISCUSSION
The first aim of this research was to selectively isolate fungal strains able to degrade PS-PU and PE-PU, using as strategy the selection of fungi able to grow in both types of PU varnishes: Impranil DLN, a PS-PU, and Poly Lack, a PE-PU. The best filamentous fungus reached Impranil degradation levels (87%) slightly higher than those reported for some endophytic fungal strains (85%) after the same incubation time (16). However, although a diversity of fungal species isolated from different sources have been recognized to degrade PS-PU (9, 10, 12–18, 20, 35), very few have been reported to have the capability to degrade PE-PU (15, 17–20).
The six best PU-degrading strains we isolated were identified as members of the Cladosporium cladosporioides complex (26, 34). The four different identified species were collected from diverse environments: garden soil, outdoor air, a PU-insulated wall in a cold room, and a dump site. The fact that six out of the eight best PU-degrading fungal strains were isolated from different environmental sources and belong to the same genus strongly suggests that PU degradation capability might be based on the physiology, biochemistry, and genetics of the genus. Although Cladosporium herbarum (18) and Cladosporium sp. have been reported as PU degraders (35), the species described in this work and the level of PE-PU degradation they reached have not been previously reported. Even though more than 770 species have been classified as members of the genus Cladosporium (26), only the genome of Cladosporium sphaerospermum UM 843, an allergenic fungus isolated from a blood culture, has been sequenced (GenBank accession no. AIIA00000000.2, GI 666699852) (36). Genome sequencing of Cladosporium polyurethanolytic species would be of great relevance to assist in determining the mechanisms responsible for this capability and for biotechnological purposes.
Analysis of the chemical changes generated by PU-degrading fungi has been addressed only in very few papers (16, 18, 20). This type of analysis provides clues to envisage the mechanism fungi display to attack the polymer during biodegradation. The dramatic decrements in the carbonyl signal (1,729 cm−1) observed in the FTIR spectra are related to the attack of ester bonds present in the polyol fraction, as well as to the attack of urethane groups. Actually, the decrements observed in the C—N—H bond signals (1,540 and 1,261 cm−1) confirm that the urethane groups are affected (Fig. 2). In experiments performed under similar conditions, Impranil degradation by the endophytic fungus Pestalotiopsis microspora was observed as a decrease in the carbonyl signal (C=O) and, although not mentioned by the authors, the spectrum also showed decreases in the signals at 1,540 and 1,260 cm−1, which, as in our work, implies enzymatic attack on the urethane group (15). Attack on these functional groups, ester and urethane, has been suggested to be performed by bacterial hydrolytic enzymes, such as esterase, protease, and urease (4, 8). In our work, evidence of the activity of fungal enzymes on Impranil biodegradation was also detected by GC-MS analysis after the incubation of Impranil with the best PU-degrading fungus C. pseudocladosporioides T1.PL.1, an approach not explored in any other previous work, to our knowledge. Even though no quantitative conclusions can be drawn from this type of analysis, the decrements in compounds with ester bonds (Table 1, peaks 4, 6, and 8) in the fungus-treated Impranil seem to correlate with increments in compounds with alcohol groups (Table 1, peaks 1 and 2), as expected in reactions where esterases hydrolyze ester bonds. Besides, in the fungus-treated media, the increments in adipic acid di(oct-4-yl ester)-related compound (Table 1, peak 5), which can only be the result of hydrolytic reactions over aliphatic chains containing ester bonds in the polyol segment, and in hexane 1,6-diisocyanate (Table 1, peak 3) suggest that the recovery of PU precursors to be reutilized for the synthesis of new PU molecules might be feasible. It is known that the recovery of polyols from PU by chemically treating PU by glycolysis is possible (37); in addition, the recovery of polyester polyols from polyethylene terephthalate and the excess diisocyanate generated during PU synthesis to be used for the synthesis of new PUs have also been reported (38, 39). Nevertheless, in these processes, the use of chemicals and high thermal energy is required, generating significant expenses and pollution. Therefore, the use of fungal strains to degrade PU waste might be an ecological, economical, and sustainable system for recovering PU precursors for recycling. On the other hand, the aliphatic nature of tridecan-6-ol-related compound (Table 1, peak 7) and cyclohexane (1-methylethyl)-related compound (Table 1, peak 9), present in the untreated media but absent in the fungus-treated media, suggests that oxidative reactions must be involved in their degradation. Cyclohexane transformation to cyclohexanol, and subsequently to cyclohexanone by fungal peroxygenases, has been reported elsewhere (40); therefore, it may be feasible that C. pseudocladosporioides T1.PL.1 has similar enzymatic activities.
When searching C. pseudocladosporioides T1.PL.1 cultures for extracellular enzymatic activities proposed to participate in Impranil biodegradation, high esterase and low urease but not protease activities were detected. The high increment in esterase activity observed at 7 days of culture, and the fact that it still remains active after 14 days, suggest that it might be responsible for the attack on ester and urethane groups in the PU, as observed by FTIR and GC-MS analysis. Several fungi displaying esterase activity suggested to be responsible for PS-PU biodegradation were previously reported (12, 35). Although urease activity has been suggested to participate in PU biodegradation (15, 20), it seems not to have a relevant role in Impranil biodegradation.
The eight best Impranil-degrading fungi were also able to grow on the PE-PU varnish Poly Lack as the sole carbon source and to degrade solid PE-PU foams. C. tenuissimum A2.PP.5 was the most degrading strain of solid PE-PU foams, showing levels of degradation of 65.3% not previously reported for solid PE-PUs. Foam A was much less degraded than foam B (Fig. 5), probably due to the presence of the flame retardant TDCPP in foam A. TDCPP has been implicated as a cause of toxicity in vertebrates, and it has been reported to affect the growth and reproduction of Tetrahymena thermophila by downregulating genes expressing ribosomal proteins, accompanied by decreased ribosome quantity and enlarged size, in rough endoplasmic reticulum and cytoplasm (41). Even though foam A presents this toxicity, C. tenuissimum A2.PP.5 was able to degrade it to some extent (25.9%), suggesting that this strain might have some resistance to the toxic effects of TDCPP. PE-PU has been consistently reported to be highly recalcitrant to biodegradation (11, 13, 14, 17–19). In our work, weight losses from 35 to 65% were reached when PE-PU foams were incubated in 50% PDB for 3 weeks with the six different Cladosporium strains. These values are much larger than the 27.5% reported for the degradation of PE-PU foams incubated in LB-50% glucose for 10 weeks with the Alternaria sp. strain PURDK2 (20). Cultures of Cladosporium strains in MM with PE-PU foams failed to grow (data not shown), indicating that an easily available carbon source is needed to sustain fungal growth and trigger PU biodegradation capability. On the other hand, our results suggest that the extent of hyphal penetration into the PU foam structure seems to be correlated with the biodegradation of solid foam, since mycelia of the Cladosporium strains, the most degrading species, extensively invaded the tested material (Fig. 3 and 4), whereas A. fumigatus A2.PL.1 and P. chrysogenum BP3.I.7, the least degrading strains, produced a limited amount of mycelia with scarce foam penetration (data not shown). Moreover, the enzymes secreted by fungal hyphae must actively participate in biodegradation. Indirect evidence of these activities is observed in the microscopic structure of the PE-PU foams after fungal treatment; they not only looked to be broken by the physical action of mycelial growth, but they also looked thinner, rougher, and more porous than the non-fungus-treated foams (Fig. 6). Although Impranil (PS-PU) and foams (PE-PU) have different formulations, urethane and ester groups are common in all PUs; therefore, the extracellular esterase activities detected in the MM with Impranil cultures must also participate in the biodegradation of PE-PUs. However, other types of enzymes, such as amidases, oxidases, or peroxidases, must also be involved in the degradation of PE-PUs. Currently, efforts are being developed in our laboratory in order to identify the enzymatic activities involved in the biochemical processes these fungi display to biodegrade PU.
In conclusion, in this work, we report the isolation of several environmental filamentous fungi with the capability to degrade PS-PU and PE-PU varnishes, as well as PE-PU solid foams, with the highest degradation values reported so far, to our knowledge. Several species belonging to the Cladosporium cladosporioides complex were the best PU degraders. FTIR spectroscopy and GC-MS analysis showed that ester and urethane groups were attacked through the activity of fungal enzymes. Considerable esterase and low urease but not protease activities were detected during Impranil fungal degradation. Our findings represent one step forward toward the development of new biotechnological processes for more efficient PU degradation.
Supplementary Material
ACKNOWLEDGMENTS
J.Á.-B. acknowledges the Facultad de Química, UNAM Programa 121, Formación Básica en Investigación. J.Á.-B. and R.G.-H. acknowledge CONACYT for graduate studies scholarships. SEM and GC-MS analyses were carried out at Unidad de Servicios de Apoyo a la Investigación (USAI) at Facultad de Química, UNAM, by Iván Puente Lee and Georgina Duarte-Lisci. We thank Gabriel García Batarse, CEO of Espumas Industriales Monterrey, S.A. de C.V., for providing the PE-PU foams; Javier Cruz Gómez, who provided the polyester polyol; Néstor López Castillo for technical support for the FTIR analysis; and Hermilo Leal Lara for technical support on fungal culture.
This study was funded by UNAM-DGAPA-PAPIIT grants IN222811 and IN217114, Facultad de Química-PAIP grant 5000-9117, and CONACYT grant 252001. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Statement
This work, including the efforts of academic members of the Departamento de Bioquímica, Facultad de Química, UNAM, was funded by CONACYT (252001). This work, including the efforts of Herminia Loza-Tavera, was funded by Facultad de Química, UNAM (5000-9117). This work, including the efforts of Herminia Loza-Tavera, was funded by UNAM-DGAPA-PAPIIT (IN222811). This work, including the efforts of Herminia Loza-Tavera, was funded by UNAM-DGAPA-PAPIIT (IN217114). The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01344-16.
REFERENCES
- 1.Plastics Europe. 2015. Plastics—the facts 2015. Plastics Europe, Frankfurt, Germany: http://www.plasticseurope.es/Document/plastics-the-facts-2015.aspx?FolID=2. [Google Scholar]
- 2.Instituto Nacional de Estadística y Geografía. 2012. Anuario estadístico de los Estados Unidos Mexicanos 2011. Instituto Nacional de Estadística y Geografía, Aguascalientes, Mexico. [Google Scholar]
- 3.Gautam R, Bassi AS, Yanful EK. 2007. A review of biodegradation of synthetic plastic and foams. Appl Biochem Biotechnol 141:85–108. doi: 10.1007/s12010-007-9212-6. [DOI] [PubMed] [Google Scholar]
- 4.Mahajan N, Gupta P. 2015. New insights into the microbial degradation of polyurethanes. RSC Adv 5:41839–41854. doi: 10.1039/C5RA04589D. [DOI] [Google Scholar]
- 5.Ignatyev IA, Thielemans W, Vander Beke B. 2014. Recycling of polymers: a review. ChemSusChem 7:1579–1593. doi: 10.1002/cssc.201300898. [DOI] [PubMed] [Google Scholar]
- 6.Oceguera-Cervantes A, Carrillo-García A, López N, Bolaños-Nuñez S, Cruz-Gómez MJ, Wacher C, Loza-Tavera H. 2007. Characterization of the polyurethanolytic activity of two Alicycliphilus sp. strains able to degrade polyurethane and N-methylpyrrolidone. Appl Environ Microbiol 73:6214–6223. doi: 10.1128/AEM.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howard GT. 2012. Polyurethane biodegradation, p 189–211. In Singh SN. (ed), Microbial degradation of xenobiotics. Springer-Verlag, Heidelberg, Germany. [Google Scholar]
- 8.Loredo-Treviño A, Gutiérrez-Sánchez G, Rodríguez-Herrera R, Aguilar CN. 2012. Microbial enzymes involved in polyurethane biodegradation: a review. J Polym Environ 20:258–265. doi: 10.1007/s10924-011-0390-5. [DOI] [Google Scholar]
- 9.Barratt SR, Ennos AR, Greenhalgh M, Robson GD, Handley PS. 2003. Fungi are the predominant micro-organisms responsible for degradation of soil-buried polyester polyurethane over a range of soil water holding capacities. J Appl Microbiol 95:78–85. doi: 10.1046/j.1365-2672.2003.01961.x. [DOI] [PubMed] [Google Scholar]
- 10.Cosgrove L, McGeechan PL, Robson GD, Handley PS. 2007. Fungal communities associated with degradation of polyester polyurethane in soil. Appl Environ Microbiol 73:5817–5824. doi: 10.1128/AEM.01083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krasowska K, Janik H, Gradys A, Rutkowska M. 2012. Degradation of polyurethanes in compost under natural conditions. J Appl Polym Sci 125:4252–4260. doi: 10.1002/app.36597. [DOI] [Google Scholar]
- 12.Mathur G, Prasad R. 2012. Degradation of polyurethane by Aspergillus flavus (ITCC 6051) isolated from soil. Appl Biochem Biotechnol 167:1595–1602. doi: 10.1007/s12010-012-9572-4. [DOI] [PubMed] [Google Scholar]
- 13.Zafar U, Houlden A, Robson GD. 2013. Fungal communities associated with the biodegradation of polyester polyurethane buried under compost at different temperatures. Appl Environ Microbiol 79:7313–7324. doi: 10.1128/AEM.02536-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zafar U, Nzeram P, Langarica-Fuentes A, Houlden A, Heyworth A, Saiani A, Robson GD. 2014. Biodegradation of polyester polyurethane during commercial composting and analysis of associated fungal communities. Bioresour Technol 158:374–377. doi: 10.1016/j.biortech.2014.02.077. [DOI] [PubMed] [Google Scholar]
- 15.Loredo-Treviño A, Garcia G, Velasco-Téllez A, Rodríguez-Herrera R, Aguilar CN. 2011. Polyurethane as substrate for fungal strains. Adv Biosci Biotechnol 2:52–58. doi: 10.4236/abb.2011.22009. [DOI] [Google Scholar]
- 16.Russell JR, Huang J, Anand P, Kucera K, Sandoval AG, Dantzler KW, Hickman D, Jee J, Kimovec FM, Koppstein D, Marks DH, Mittermiller PA, Nunez SJ, Santiago M, Townes MA, Vishnevetsky M, Williams NE, Vargas MP, Boulanger LA, Bascom-Slack C, Strobel SA. 2011. Biodegradation of polyester polyurethane by endophytic fungi. Appl Environ Microbiol 77:6076–6084. doi: 10.1128/AEM.00521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darby RT, Kaplan AM. 1968. Fungal susceptibility of polyurethanes. Appl Microbiol 16:900–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filip Z. 1979. Polyurethane as the sole nutrient source for Aspergillus niger and Cladosporium herbarum. Eur J Appl Microbiol 7:277–280. doi: 10.1007/BF00498022. [DOI] [Google Scholar]
- 19.Martens R, Domsch KH. 1981. Microbial degradation of polyurethane foams and isocyanate based polyureas in different media. Water Air Soil Poll 15:503–509. doi: 10.1007/BF00279430. [DOI] [Google Scholar]
- 20.Matsumiya Y, Murata N, Tanabe E, Kubota K, Kubo M. 2010. Isolation and characterization of an ether-type polyurethane-degrading micro-organism and analysis of degradation mechanism by Alternaria sp. J Appl Microbiol 108:1946–1953. [DOI] [PubMed] [Google Scholar]
- 21.Barnett HL, Hunter BB. 1998. Illustrated genera of imperfect fungi, 4th ed American Phytopathological Society Press, St. Paul, MN. [Google Scholar]
- 22.Doyle JJT, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus 15:12–13. [Google Scholar]
- 23.Hinrikson HP, Hurst SF, Lott TJ, Warnock DW, Morrison CJ. 2005. Assessment of ribosomal large-subunit D1-D2, internal transcribed spacer 1, and internal transcribed spacer 2 regions as targets for molecular identification of medically important Aspergillus species. J Clin Microbiol 43:2092–2103. doi: 10.1128/JCM.43.5.2092-2103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Shinsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications, Academic Press, New York, NY. [Google Scholar]
- 25.Jerome CA, Lynn DH. 1996. Identifying and distinguishing sibling species in the Tetrahymena pyriformis complex (Ciliophora, Oligohymenophorea) using PCR/RFLP analysis of nuclear ribosomal DNA. J Eukaryot Microbiol 43:492–497. doi: 10.1111/j.1550-7408.1996.tb04509.x. [DOI] [PubMed] [Google Scholar]
- 26.Bensch K, Groenewald JZ, Dijksterhuis J, Starink-Willemse M, Andersen B, Summerell BA, Shin HD, Dugan FM, Schroers HJ, Braun U, Crous PW. 2010. Species and ecological diversity within the Cladosporium cladosporioides complex (Davidiellaceae, Capnodiales). Stud Mycol 67:1–94. doi: 10.3114/sim.2010.67.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556. doi: 10.2307/3761358. [DOI] [Google Scholar]
- 28.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.McCarthy SJ, Meijs GF, Mitchell N, Gunatillake PA, Heath G, Brandwood A, Schindhelm K. 1997. In-vivo degradation of polyurethanes: transmission-FTIR microscopic characterization of polyurethanes sectioned by cryomicrotomy. Biomaterials 18:1387–1409. doi: 10.1016/S0142-9612(97)00083-5. [DOI] [PubMed] [Google Scholar]
- 30.Pergal MV, Džunuzović JV, Poręba R, Micić D, Stefanov P, Pezo L, Špírková M. 2013. Surface and thermomechanical characterization of polyurethane networks based on poly(dimethylsiloxane) and hyperbranched polyester. Express Polym Lett 7:806–820. doi: 10.3144/expresspolymlett.2013.78. [DOI] [Google Scholar]
- 31.Witte CP, Medina-Escobar N. 2001. In-gel detection of urease with nitroblue tetrazolium and quantification of the enzyme from different crop plants using the indophenols reaction. Anal Biochem 290:102–107. doi: 10.1006/abio.2000.4933. [DOI] [PubMed] [Google Scholar]
- 32.Nair S, Kumar P. 2007. Molecular characterization of a lipase-producing Bacillus pumilus strain (NMSN-1d) utilizing colloidal water-dispersible polyurethane. World J Microb Biotechnol 23:1441–1449. doi: 10.1007/s11274-007-9388-5. [DOI] [Google Scholar]
- 33.Rowe L, Howard GT. 2002. Growth of Bacillus subtilis on polyurethane and the purification and characterization of a polyurethanase-lipase enzyme. Int Biodeter Biodegr 50:33–40. doi: 10.1016/S0964-8305(02)00047-1. [DOI] [Google Scholar]
- 34.Bensch K, Groenewald JZ, Braun U, Dijksterhuis J, de Jesús Yáñez-Morales M, Crous PW. 2015. Common but different: the expanding realm of Cladosporium. Stud Mycol 82:23–74. doi: 10.1016/j.simyco.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crabbe JR, Campbell JR, Thompson L, Walz SL, Schultz WW. 1994. Biodegradation of a colloidal ester-based polyurethane by soil fungi. Int Biodeter Biodegr 33:103–113. doi: 10.1016/0964-8305(94)90030-2. [DOI] [Google Scholar]
- 36.Ng KP, Yew SM, Chan CL, Soo-Hoo TS, Na SL, Hassan H, Ngeow YF, Hoh CC, Lee KW, Yeeb WY. 2012. Sequencing of Cladosporium sphaerospermum, a dematiaceous fungus isolated from blood culture. Eukaryot Cell 11:705–706. doi: 10.1128/EC.00081-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braslaw J, Gerlock JL. 1984. Polyurethane waste recycling. 2. Polyol recovery and purification. Ind Eng Chem Process Des Dev 23:557–561. doi: 10.1021/i200026a025. [DOI] [Google Scholar]
- 38.Rossi P, Kosior E, Iovenitti P, Massod S, Sbarski I. 2003. Polyester polyols for polyurethane foams from recycled PET. Prog Rubber Plast Recyc Technol 19:51–59. [Google Scholar]
- 39.Yong H, Zhang X, Zhang X, Huang H, Chang J, Chen H. 2013. A recycling model of excess toluene diisocyanate isomers in the preparation of polyurethane prepolimer. J Appl Polym Sci 127:2176–2183. doi: 10.1002/app.37779. [DOI] [Google Scholar]
- 40.Peter S, Karich A, Ullrich R, Gröbe G, Scheibner K, Hofrichter M. 2014. Enzymatic one-pot conversion of cyclohexane into cyclohexanone: comparison of four fungal peroxygenases. J Mol Catal B Enzym 103:47–51. doi: 10.1016/j.molcatb.2013.09.016. [DOI] [Google Scholar]
- 41.Li J, Giesy JP, Yu L, Li G, Liu C. 2015. Effects of Tris(1,3-dichloro-2-propyl) phosphate (TDCPP) in Tetrahymena thermophila: targeting the ribosome. Sci Rep 5:10562. doi: 10.1038/srep10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.