ABSTRACT
The l-isoaspartyl protein carboxyl methyltransferase (PCM) repairs protein damage resulting from spontaneous conversion of aspartyl or asparaginyl residues to isoaspartate and increases long-term stationary-phase survival of Escherichia coli under stress. In the course of studies intended to examine PCM function in metabolically inactive cells, we identified pcm as a gene whose mutation influences the formation of ofloxacin-tolerant persisters. Specifically, a Δpcm mutant produced persisters for an extended period in stationary phase, and a ΔglpD mutation drastically increased persisters in a Δpcm background, reaching 23% of viable cells. The high-persister double mutant showed much higher competitive fitness than the pcm mutant in competition with wild type during long-term stationary phase, suggesting a link between persistence and the mitigation of unrepaired protein damage. We hypothesized that reduced metabolism in the high-persister strain might retard protein damage but observed no gross differences in metabolism relative to wild-type or single-mutant strains. However, methylglyoxal, which accumulates in glpD mutants, also increased fitness, suggesting a possible mechanism. High-level persister formation in the Δpcm ΔglpD mutant was dependent on guanosine pentaphosphate [(p)ppGpp] and polyphosphate. In contrast, persister formation in the Δpcm mutant was (p)ppGpp independent and thus may occur by a distinct pathway. We also observed an increase in conformationally unstable proteins in the high-persister strain and discuss this as a possible trigger for persistence as a response to unrepaired protein damage.
IMPORTANCE Protein damage is an important factor in the survival and function of cells and organisms. One specific form of protein damage, the formation of the abnormal amino acid isoaspartate, can be repaired by a nearly universally conserved enzyme, PCM. PCM-directed repair is associated with stress survival and longevity in bacteria, insects, worms, plants, mice, and humans, but much remains to be learned about the specific effects of protein damage and repair. This paper identifies an unexpected connection between isoaspartyl protein damage and persisters, subpopulations in bacterial cultures showing increased tolerance to antibiotics. In the absence of PCM, the persister population in Escherichia coli bacteria increased, especially if the metabolic gene glpD was also mutated. High levels of persisters in pcm glpD double mutants correlated with increased fitness of the bacteria in a competition assay, and the fitness was dependent on the signal molecule (p)ppGpp; this may represent an alternative pathway for responding to protein damage.
INTRODUCTION
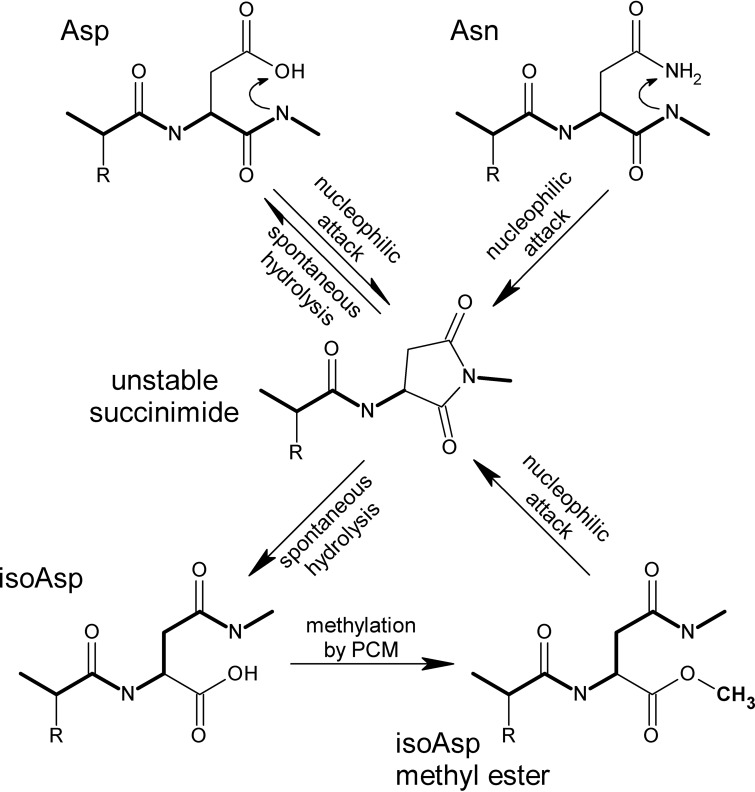
Isoaspartyl damage to proteins occurs when aspartyl or asparaginyl residues spontaneously isomerize via a succinimide intermediate (1) (Fig. 1), adversely affecting protein function (2). The damage can be repaired when the abnormal isoaspartyl (isoAsp) residue is recognized and methylated by the l-isoaspartyl protein carboxyl methyltransferase (PCM) (Fig. 1), stimulating reformation of the succinimide intermediate that can be hydrolyzed to yield normal aspartate. Repair of isoaspartyl damage has been linked to longevity in organisms ranging from bacteria to plants, insects, and mammals and also appears to be important in the prevention of autoimmune and degenerative diseases (3). PCM is found throughout the living world (spore-forming Gram-positive bacteria and budding yeast are among the few exceptions; see reference 4), and while its enzymatic activity is well studied, how PCM acts in vivo to enhance longevity and stress survival is not yet fully understood. In Escherichia coli, repair-deficient mutants exhibit impaired long-term stationary-phase survival under conditions of oxidative, osmotic, heat, or methanol stress (5).
FIG 1.
Formation and repair of isoaspartyl residues in proteins. Isoaspartyl (isoAsp) damage results from spontaneous nucleophilic attack (top) of the peptide-bond nitrogen on the side chain carbonyl group of an aspartyl (Asp) or asparaginyl (Asn) residue, resulting in an unstable succinimide intermediate (middle), which hydrolyzes spontaneously to form isoAsp or reform Asp. The formation of isoAsp “kinks” the peptide backbone (heavy line) and alters the charge (in the case of Asn) or spatial relationships. Methylation of isoAsp by PCM (bottom) stimulates reformation of the intermediate, allowing for net repair to normal Asp.
Previously, we showed that repair-proficient and repair-deficient E. coli bacteria accumulate similar levels of isoaspartyl protein damage during long-term stationary phase (6, 7) but that PCM shortens the lag phase when starving cells are resupplied with nutrients (7). We therefore hypothesized that although PCM is important for stationary-phase stress survival, it may act primarily in metabolically active subpopulations (8) during stationary phase and/or after nutrient restoration. To test this hypothesis, we proposed to use strains with larger and smaller fractions of quiescent persister cells; unexpectedly, however, we found that pcm itself is a gene whose mutation affects the persister fraction.
Persisters are subpopulations that show increased tolerance to antibiotics relative to the majority population but are not genetically antibiotic resistant (9). Persisters appear to arise stochastically during growth and are therefore present in all bacterial cultures, typically at a frequency of 0.1% or less in laboratory cultures grown in rich medium (10). Persisters may be tolerant of multiple antibiotics (potentially resulting from distinct subpopulations with different tolerances) and can be identified by viable counts after treating a culture with an antibiotic (e.g., ofloxacin) to kill nonpersister cells (11). Since many antibiotics antagonize the functions of actively growing cells (e.g., cell wall synthesis, DNA replication, and translation), reduced metabolism in the persister subpopulation is a possible explanation for antibiotic tolerance (reviewed in reference 12). The implication of inhibition of translation or other cellular processes by toxin-antitoxin (TA) systems (reviewed in reference 13) in persister formation (9) led to mechanistic support for this hypothesis, with metabolic inactivity attributed to the activation of toxins, such as HipA, RelE, or MazF. However, while metabolic inactivity appears to be a feature of most persister cells, it is not universally accepted that persister formation requires dormancy (reviewed in reference 14); in any case, persisters behave like stationary-phase cells in that they can readily resume growth when the antibiotic stress is relieved, rather than like spores or viable-but-nonculturable cells that require specific triggers to break dormancy.
Recent work on the mechanisms of persister formation suggests that the effects of TA systems are controlled by guanosine pentaphosphate [(p)ppGpp] (11, 15), placing persister formation under the control of the stringent response as a result of nutrient exhaustion (reviewed in reference 16). However, protein aggregation (17), trehalose deficiency (18), acid stress (19), biofilm formation (9), the SOS response (20), and quorum sensing (21) are among the additional factors implicated in persister formation. It therefore seems likely that persisters can arise by multiple interconnected pathways and may have selective value in response to stresses or other environmental conditions.
Here, we present evidence that a Δpcm mutant deficient in isoaspartyl protein repair maintains a prolonged peak persister fraction during stationary phase and that a Δpcm ΔglpD double mutant produces a drastically increased persister fraction. We examine the competitive fitness of repair-deficient E. coli bacteria and show for the first time that PCM provides a fitness advantage to the cell during stationary phase even in the absence of external stresses. We then show that the high-persister strain overcomes much of the competitive disadvantage of the deficiency in isoaspartyl repair and investigate the mechanisms behind this observation. This work establishes a link between PCM and the formation of persisters, a significant phenomenon both in terms of bacterial physiology and its medical importance. We then discuss the potential selective value of persister formation as a response to the stress of protein damage.
(This work was presented in part at the 112th General Meeting of the American Society for Microbiology, San Francisco, CA, 16 to 19 June 2012, and the 114th General Meeting of the American Society for Microbiology, Boston, MA, 17 to 20 May 2014 [49, 50].)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All bacterial strains used in this study are derivatives of Escherichia coli K-12 strain MG1655; their genotypes and derivations are listed in Table 1. Cultures were grown in Luria-Bertani (LB) medium at 37°C under aerobic conditions for all experiments described here and maintained under the same conditions, with continued aeration for long-term stationary-phase experiments. When necessary, sterile deionized water was added to compensate for the loss of culture volume due to evaporation. The onset of stationary phase was determined by measuring the optical density at 600 nm until the change in a 30-min period was less than 5%. Streptomycin (Str), chloramphenicol (Cm), and kanamycin (Km) were added to final concentrations of 1 mg ml−1, 20 μg ml−1, and 50 μg ml−1, respectively, when needed.
TABLE 1.
Genotypes of E. coli strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| JV1120a | MG1655 rpsL150 (Strr) | Transduction of rpsL from MC1000 |
| JV1121 | JV1120 Δpcm | Recombination of a deletion plasmid construct in JV1120 |
| JV1132 | JV1121 ΔphoU755::Kmr | Transduction from JW3702-1b |
| JV1136 | JV1120 ΔglpD759::Kmr | Transduction from JW3389-1b |
| JV1154 | JV1121 ΔglpD759::Kmr | Transduction from JW3389-1b |
| JV1155 | JV1120 ΔphoU755::Kmr | Transduction from JW3702-1b |
| JV1168 | JV1121 gyrA (Nalr) | Transduction from a spontaneous Nalr variant of MM294 |
| JV1169 | JV1136 gyrA (Nalr) | Transduction from a spontaneous Nalr variant of MM294 |
| JV1170 | JV1154 gyrA (Nalr) | Transduction from a spontaneous Nalr variant of MM294 |
| JV1177 | JV1121 ΔglpD | FLP recombination to remove Kmr marker from JV1154 |
| JV1179 | JV1177 ΔhipA728::Kmr | Transduction from JW1500-2Ab |
| JV1180 | JV1177 ΔrelE785::Kmr | Transduction from JW1555-2b |
| JV1181 | JV1177 ΔmqsR720::Kmr | Transduction from JW2990-2b |
| JV1182 | JV1177 ΔcspD781::Kmr | Transduction from JW0864-1b |
| JV1189 | JV1120 ΔnadA721::Kmr | Transduction from JW0733-1b |
| JV1190 | JV1120 ΔnadA721::Kmr | Transduction from JW0733-1b |
| JV1191 | JV1120 ΔrelA782::Kmr | Transduction from JW2755-1b |
| JV1192 | JV1120 ΔphoB763::Kmr | Transduction from JW0389-1b |
| JV1193 | JV1121 ΔrelA782::Kmr | Transduction from JW2755-1b |
| JV1194 | JV1121 ΔphoB763::Kmr | Transduction from JW0389-1b |
| JV1197 | JV1177 ΔrelA782::Kmr | Transduction from JW2755-1b |
| JV1198 | JV1177 ΔphoB763::Kmr | Transduction from JW0389-1b |
| JV1199 | JV1191 ΔspoT207::cat | Transduction from CF1693c |
| JV1200 | JV1193 ΔspoT207::cat | Transduction from CF1693c |
| JV1201 | JV1197 ΔspoT207::cat | Transduction from CF1693c |
Deletion of the pcm gene was accomplished by cloning flanking sequences into the pir-dependent rpsL+ (Str-sensitive) plasmid pKAS46 (22) and selecting for streptomycin resistance in a pir null mutant host. Strains carrying other mutations were constructed by P1 transduction from mutant strains carrying resistance markers using standard methods (23). An unmarked glpD deletion was constructed from the Keio deletion collection (24) ΔglpD::Km mutant using FLP recombinase, as described previously (25).
Persister assay.
Persisters were assayed essentially as previously described (26). Cultures grown to the desired point were sampled for viable counts to determine total viable cells. To quantitate persisters, 150-μl aliquots were transferred to a 96-well plate, and ofloxacin was added to a final concentration of 5 μg ml−1. After incubation at 37°C, viable counts were again performed to determine the number of cells surviving the antibiotic treatment. Following initial time course experiments, a 24-h incubation with ofloxacin was used for all persister assays to simplify comparison. One-way analysis of variance (ANOVA) and post hoc Tukey tests were used to examine the statistical significance of observed differences between strains.
Measurement of metabolism with WST-1.
Metabolic activity was measured using the redox-sensitive tetrazolium dye WST-1, which can be reduced extracellularly to a water-soluble formazan by electrons from the electron transport chain in the presence of the electron-coupling reagent 1-methoxy-5-methyl-phenazinium methyl sulfate (27). Culture aliquots (100 μl) were mixed in a 96-well plate with 10 μl of WST-1 premixed with coupling reagent (WST-1 cell proliferation assay kit; Clontech Laboratories, Inc., Mountain View, CA). Formazan formation was monitored by absorbance at 450 nm using a microplate spectrophotometer.
Competitive survival assay and RCI.
Individual strains were grown overnight in LB broth at 37°C with aeration. Strains to be competed were diluted 1:100 in fresh LB and mixed in a 1:1 ratio (by culture volume, initially verified by viable counts) or with one strain or the other in a 10-fold excess. In each competition, one strain was marked with a nalidixic acid resistance (Nalr) marker. The mixed cultures were then grown to stationary phase at 37°C with aeration and sampled daily over 10 days for viable counts. Dilutions were plated on LB agar plates and LB agar plates containing nalidixic acid (125 μg ml−1). The number of viable cells of the Nalr strain was determined directly, and the number of viable cells of the other strain was then determined in a subtractive manner. The relative competitive index (RCI) was calculated by determining the final ratio of mutant to wild-type cells, dividing by the initial ratio (1, 0.1, or 10), and taking the log10 of the quotient (28). A one-sample t test was used to compare RCI values with the null hypothesis of equal fitness (RCI = 0).
Aggregated/aggregatable protein assay.
The amount of bulk protein that had become aggregated in vivo during growth or in stationary phase was determined by a procedure modified from that described by Gur et al. (29). Cells grown under the desired conditions were collected by centrifugation, washed in Tris-EDTA buffer containing phenylmethylsulfonyl fluoride (PMSF), and lysed by freeze-thawing, followed by centrifugation. After low-speed centrifugation to remove intact cells, the total protein concentration in the crude lysate was determined using the method of Lowry et al. (30) after trichloroacetic acid precipitation. Protein aggregates were isolated from the remaining lysate by centrifugation for 30 min at 16,000 × g, resuspended by brief sonication, and washed twice with 10% 4-nonylphenyl polyethylene glycol (Nonidet P-40 substitute; Sigma-Aldrich, St. Louis, MO) before being redissolved in 6 M guanidine hydrochloride. The protein concentration in the aggregate fraction was then determined by the Lowry method. Crude lysates were heated to 37°C for 2 h prior to aggregate isolation to quantitate proteins not initially aggregated but that could be aggregated by heating.
RESULTS
Protein repair-deficient mutants maintain a high persister frequency in stationary phase.
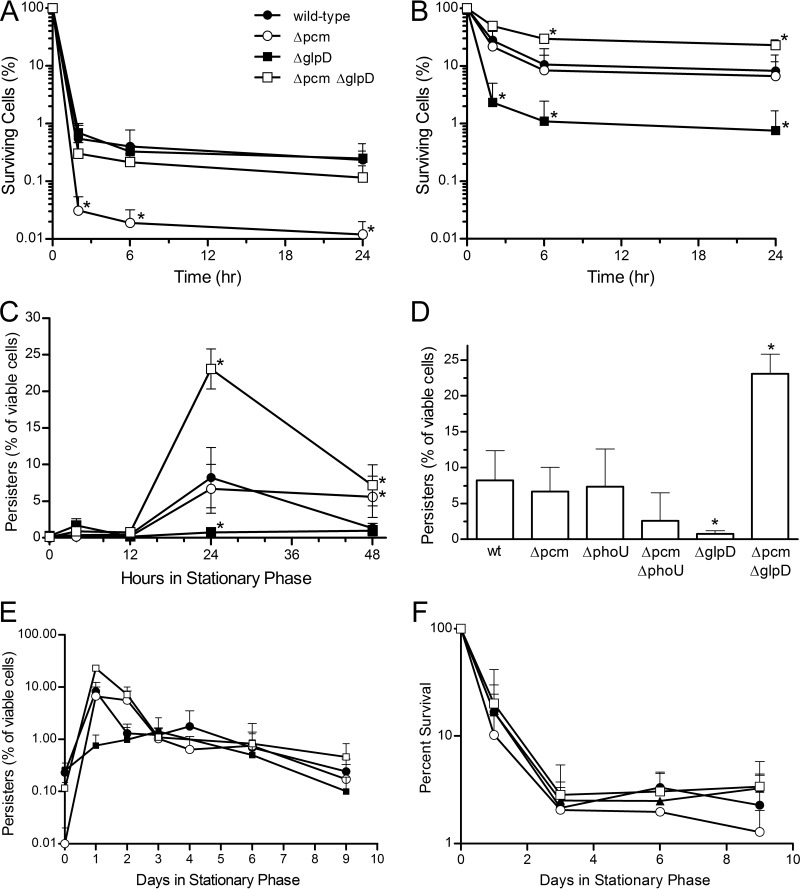
In the course of studies using persister mutants to measure repair of isoaspartyl damage to proteins by PCM under various metabolic conditions, we discovered that mutation of pcm itself altered the persister fraction in stationary-phase cells. Consequently, we sought to understand the connection between PCM and persister formation. In agreement with the report of Luidalepp et al. (31), we observed considerable variability in persister frequencies when overnight cultures were used as the starting material for persister assays, suggesting that timing is important in order to observe changes in the persister fraction. Thus, for all persister assays presented here, we used a growth curve to identify the point at which cultures entered stationary phase, defined operationally for the purposes of this experiment as the first 30-min interval after the onset of exponential growth in which the optical density at 600 nm (OD600) of the culture changed by less than 5%. As shown in Fig. 2A, ofloxacin-tolerant cells could be identified from cultures of either our wild-type strain, JV1120 (this strain differs from the sequenced E. coli K-12 strain MG1655 only by an rpsL mutation conferring streptomycin resistance, as indicated in Table 1), or an isogenic Δpcm mutant strain deficient in isoaspartyl protein repair, JV1121 (Fig. 2A, open circles), as these strains entered stationary phase. Ofloxacin had killed the majority of cells after 3 h of treatment, but further reduction in the viable cell count was slow beyond that point, showing relatively little change between 3 h and 24 h of treatment. This biphasic killing curve is characteristic of the subset of antibiotic-tolerant cells defined as persisters (9). Both strains exhibited persister frequencies in the range of 0.01 to 0.4% in early stationary phase, but we note that the repair-deficient strain showed significantly fewer persisters, suggesting that even early in stationary phase, isoaspartyl formation had altered its physiology.
FIG 2.
Persister formation during stationary phase in pcm mutants. Percentages of viable cells tolerant to treatment with 5 μg ml−1 ofloxacin for the indicated times at 0 h (A) and 24 h (B) after the onset of stationary phase. The strains used are JV1120 (wild type; ●), JV1121 (Δpcm; ○), JV1136 (ΔglpD; ■), and JV1154 (Δpcm ΔglpD; □); asterisks indicate values significantly (P < 0.05) different from wild type. (C) Percentages of viable cells tolerant to 24-h treatment with ofloxacin at indicated times after cultures entered stationary phase; strains and symbols are as described for panel A. (D) Percentages of viable cells tolerant to 24-h treatment with ofloxacin 24 h after the onset of stationary phase for strains JV1120 (wild type [wt]), JV1121 (Δpcm), JV1155 (ΔphoU), JV1132 (Δpcm ΔphoU), JV1136 (ΔglpD), and JV1154 (Δpcm ΔglpD); asterisks indicate values significantly (P < 0.05) different from wild type. (E) Percentages of viable cells tolerant to 24-h treatment with ofloxacin after the indicated number of days in long-term stationary phase; strains and symbols are as described for panel A. (F) Percent survival (total viable cells) after the indicated number of days in long-term stationary phase; strains and symbols are as described for panel A. All values are averages of at least three replicates, and error bars represent one standard deviation.
For both strains, we consistently observed an increase in persister frequency as the cells aged, peaking by 24 h of incubation in stationary phase (Fig. 2B). Despite the lower persister frequency seen at earlier time points, the wild-type and Δpcm mutant strains peaked at the same level. However, whereas the peak subsided in the wild-type strain and the persister frequency was reduced to 1 to 2% of viable cells (still about 10 times higher than at the earlier time points) by 48 h (Fig. 2C; only the persister frequency following a 24-h incubation with ofloxacin is shown), the elevated persister frequency was maintained in the repair-deficient strain for an additional 24 h. For convenience in comparing these strains to those discussed below, the frequency of persisters identified after 24 h in stationary phase and following a 24-h treatment with ofloxacin is represented in Fig. 2D.
Little is known about what happens to the persister fraction as starving E. coli cells continue into long-term stationary phase, yet this is the period when the requirement of PCM for stress survival becomes evident (5). When we continued our persister assays into long-term stationary phase, we observed a persister frequency of about 1% for both strains from days 3 to 6, with a gradual decline thereafter (Fig. 2E). We observed no significant differences between the wild-type and mutant strains in this period and focused subsequently on the earlier points where there was evidence of change in the physiology of the stationary-phase cells.
Deletion of glpD increases persister frequency in a Δpcm mutant background.
As noted above, our initial intent was to manipulate the metabolic state of the cells. Because mutations in glpD and phoU affect persister frequency (26, 31, 32), we introduced deletions of these genes into our wild-type and mutant strains and assayed persisters as described above. We were surprised to find that the Δpcm mutation greatly impacted the fraction of persisters after 24 h of incubation in stationary phase in these mutant backgrounds (Fig. 2D). A phoU pcm double mutant formed about half as many persisters (P < 0.05) as its isogenic phoU mutant parent. What was most striking, however, was the effect of PCM deficiency in a glpD mutant background. Consistent with the characterization of glpD by others as a persister-promoting gene (26), we found that the deletion of glpD resulted in a significantly lower frequency of persisters after 24 h in stationary phase (Fig. 2B and D), although this strain did show a small transient increase in persisters (mean of 1.7% versus 0.37% for the wild-type strain) at the 4-h time point only (Fig. 2C). However, a Δpcm ΔglpD double mutant showed an enormous increase in persisters at 24 h (Fig. 2B to D), reaching on average nearly 25% of viable cells.
The biphasic killing curve we observed (Fig. 2B) shows that this high percentage of ofloxacin-tolerant cells were indeed persisters. In the absence of antibiotic, the fraction of surviving Δpcm ΔglpD mutant cells was not different from that of the wild-type or Δpcm mutant strain at any point over a 10-day period (Fig. 2F), suggesting that the observed persister peak represents an increase in persister formation rather than a decrease in nonpersister cells. This increase was not seen early in stationary phase (Fig. 2A), when neither the glpD nor the glpD pcm mutant was distinguishable from the wild type. Unlike that of the pcm single mutant, this persister peak was not maintained beyond the 24-h point: from 48 h through 10 days in long-term stationary phase, the double mutant had a persister profile indistinguishable from that of the other strains (Fig. 2C and E). This drastic change in persister frequency potentially represents either a cellular response to unrepaired isoAsp damage or a consequence of such damage.
High-level persister production does not result in reduced metabolic rate.
Mutants lacking PCM are sensitive to stress during long-term stationary phase but do not exhibit an overt survival defect in the absence of added stress (5) and, surprisingly, do not accumulate detectably more isoaspartate-containing protein (6, 7). Given that persisters are generally regarded to be quiescent cells (12), we hypothesized that persister formation might serve to mitigate protein damage by generating a subpopulation with reduced metabolic activity in which isoaspartyl formation would be slowed or its impact reduced until nutrients are restored and repair can occur. In a pcm mutant, increased or prolonged persister formation might limit the detrimental effect of the isoaspartyl repair deficiency.
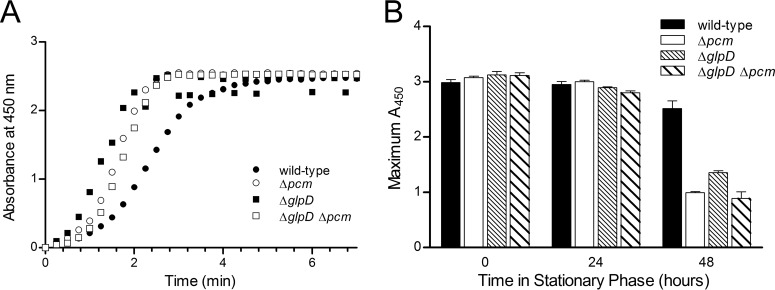
To test this idea, we probed metabolic activity in our strains with the redox-sensitive tetrazolium salt WST-1, a commonly used indicator of cellular metabolism (27); metabolic reduction of WST-1 produces formazan, which can be measured spectrophotometrically. If the large persister population that we observed were indeed composed of cells in a state of reduced metabolism, we would predict that nearly a quarter of viable Δpcm ΔglpD mutant cells should be in a metabolically inactive state by 24 h after the onset of stationary phase. Such a large effect should then be easily detectable as a decrease in the ability of the culture as a whole to reduce WST-1. We also expected the inactive fraction to be higher at 48 h in the Δpcm mutant than in the other strains. However, we observed that at 24 h after the onset of stationary phase, all four strains were capable of reducing WST-1 to nearly the same degree (Fig. 3A) and at nearly the same rate (Table 2). No correlation between the rate of WST-1 reduction and the size of the persister fraction was observed. Moreover, there was no change in metabolic capacity as measured by this method between the onset of stationary phase and the 24-h time point (Fig. 3B), during which time the persister fractions changed drastically. We did observe that the wild-type strain retained significantly (P < 0.0001) greater metabolic capacity at 48 h into stationary phase than any of the mutants (Fig. 3B), an observation that may merit further study. However, since the three mutants with their distinct persister levels behaved similarly, this effect does not appear to be connected to persister formation. By 5 days into stationary phase, all four strains had similar and low metabolic rates (data not shown). The data thus do not support the idea that persister formation limits the metabolism of a sizeable subpopulation and thereby mitigates the effects of isoaspartyl damage.
FIG 3.
Metabolic capability of mutants in stationary phase. (A) Reduction of WST-1 by cells maintained 24 h in stationary phase. Normalized absorbance at 450 nm (A450) due to reduced WST-1 is shown versus time after WST-1 addition to cultures of strains JV1120 (wild type; ●), JV1121 (Δpcm; ○), JV1136 (ΔglpD; ■), or JV1154 (Δpcm ΔglpD; □). One representative experiment of three is shown. (B) Reduction of WST-1 by cells maintained in stationary phase for 0, 24, or 48 h; maximum absorbance due to reduced WST-1 is shown and was determined by the same assay used for the experiment whose results are shown in panel A for cultures of JV1120 (black bars), JV1121 (white bars), JV1136 (dense stripes), or JV1154 (wider stripes). n = 3; error bars represent one standard deviation.
TABLE 2.
Rates of reduction of WST-1 by pcm and glpD mutants
| Strain | Genotype | Production of formazan from WST-1 |
|
|---|---|---|---|
| Rate (A450 s−1) | SD | ||
| JV1120 | Wild type | 0.021 | 0.0008 |
| JV1121 | Δpcm | 0.027 | 0.0030 |
| JV1136 | ΔglpD | 0.026 | 0.0110 |
| JV1154 | Δpcm ΔglpD | 0.025 | 0.0008 |
The high-persister glpD mutation reverses the competitive defect of the Δpcm mutant.
A previous report (19) correlated persister formation with genetic alterations that decreased the environmental fitness of E. coli, leading us to hypothesize that enhanced persister formation might affect the fitness of repair-deficient pcm mutants, even in the absence of a detectable effect on metabolism. We examined the fitness of our wild-type and mutant strains using a competition assay. In other systems, similar techniques have revealed competitive advantages or disadvantages of particular genotypes in the absence of overt survival phenotypes (see, for example, reference 28).
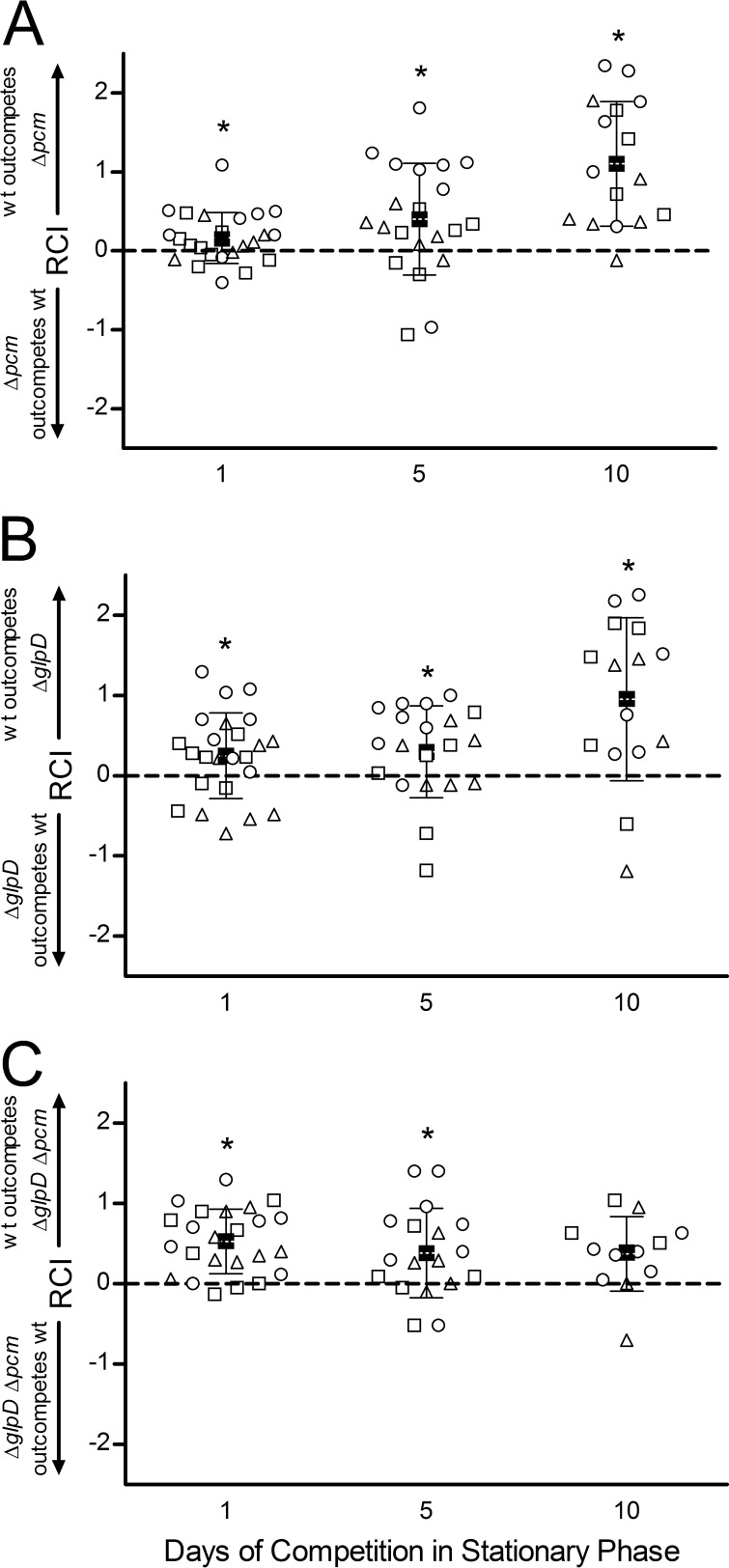
We first examined the effect of the Δpcm mutation on competitive fitness by growing mutant and wild-type cells to stationary phase, mixing the cultures at various ratios (1:10, 1:1, and 10:1) with no addition of nutrients, and then monitoring the survival of the two strains over 10 days in long-term stationary phase (Fig. 4A). In the experiments whose results are shown here, the mutant strain (Δpcm, ΔglpD, or double mutant) also carried a gyrA mutation conferring resistance to nalidixic acid (Nalr), a marker previously shown to be selectively neutral (8). To ensure that the resistance marker did not influence the results, however, reciprocal experiments were also done in which the wild-type strain was Nalr; these gave similar results (data not shown). To concisely portray the results of multiple trials with different starting ratios, Fig. 4 reports the relative competitive index (RCI; see Materials and Methods and reference 28), calculated as the log10 of the final ratio of viable wild-type cells to mutant cells divided by the initial ratio. Using this calculation, no change from the starting ratio would give an RCI of 0, whereas a positive RCI means that wild-type cells outcompeted the mutant and a negative RCI represents the opposite result, the mutant “winning” the competition. The three different starting ratios are distinguished by various symbols in Fig. 4; however, there was no significant difference among the means of the three for any strain or time point; hence, all the starting ratios have been plotted together and combined to give the overall means shown in Fig. 4.
FIG 4.
Effect of a glpD deletion on competitive fitness of a Δpcm mutant. Cultures of JV1120 (wild type) and JV1168 (Δpcm mutant) (A), JV1169 (ΔglpD mutant) (B), or JV1170 (ΔglpD Δpcm mutant) (C) were grown individually to stationary phase, mixed at 1:10 (○), 1:1 (△), or 10:1 (□) ratios and allowed to compete for 10 days. Samples were quantitated by viable counts on the indicated days and the relative competitive index (RCI) calculated as described in the text. Closed squares (■) indicate the mean RCI, with error bars representing one standard deviation in each direction. Asterisks indicate the mean RCI being different from an RCI of 0 (dashed line) with P < 0.05.
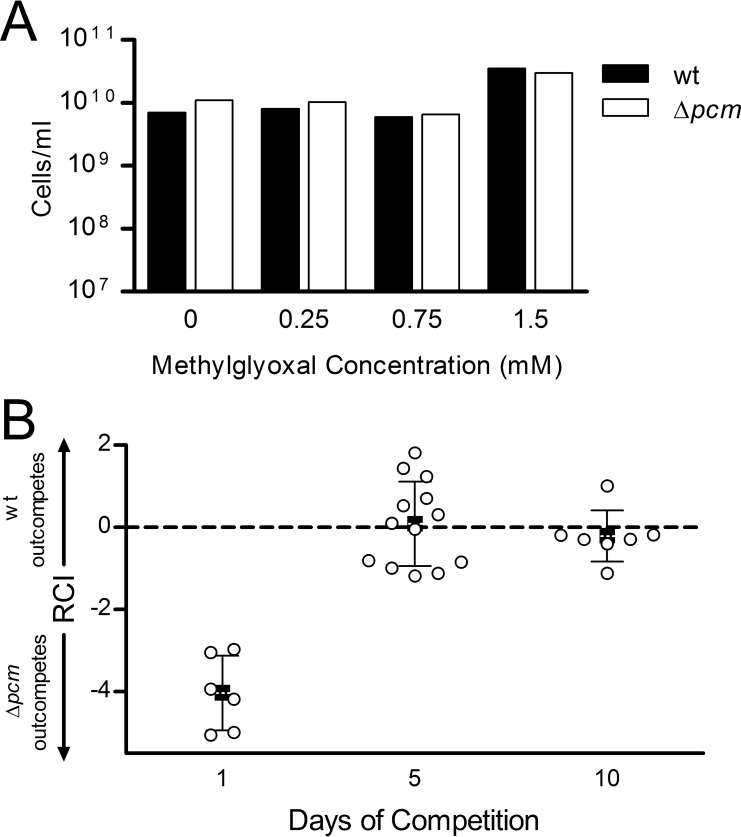
Although Δpcm mutants have no discernible survival defect when grown alone (see reference 5 and Fig. 2F), the competition assay demonstrated that this repair-deficient strain had a clear competitive disadvantage, and thus reduced fitness, relative to its wild-type parent (Fig. 4A). After 10 days in long-term stationary phase, the mean RCI (squares, Fig. 4A) was 1.1, indicating that on average, wild-type cells outcompeted the Δpcm mutant by more than 10:1. A one-sample t test supported this conclusion, showing a statistically significant difference between the experimental outcome and the null hypothesis of no survival difference between the strains (RCI = 0), with a P value of <0.0001. Furthermore, even the smaller differences observed after 1 day (wild type outcompetes Δpcm mutant by about 1.5:1) and 5 days (wild type wins by about 2.5:1) of competition were statistically significant, with P values of 0.021 and 0.017, respectively. This is the first demonstration that a deficiency in isoaspartyl protein repair is deleterious in E. coli in the absence of exogenous stress.
The results in Fig. 4A show that there is considerable variation among individual cultures in the competition assay and that the wild-type strain does not always win the competition. Long-term stationary-phase cultures achieve a stable balance between cell division and cell death (33), and many stochastic factors (e.g., reactive oxygen species and other stresses, diminished protein synthesis capacity, mutation, and selection) affect the survival of individual cells. Indeed, we always observe substantial variation among trials in our single-strain long-term survival assays (see reference 5 and Fig. 2F). However, a clear pattern emerges when the results of the competition are averaged across multiple trials and starting ratios, which we believe, given its statistical significance, is biologically relevant.
We next examined the effect of the ΔglpD mutation on competitive fitness (Fig. 4B). We found that a functional glpD allele conferred a competitive fitness advantage under these stationary-phase conditions, with the wild type outcompeting the ΔglpD mutant, on average, by 1.8:1 on day 1 (P = 0.032), 2:1 by day 5 (P = 0.031), and 9:1 by day 10 (P = 0.0026). GlpD is a glycerol 3-phosphate (G3P) dehydrogenase required for aerobic glycerol metabolism (34). One straightforward explanation for this competitive advantage would therefore be that wild-type cells are better equipped to scavenge whatever nutrients may be available in stationary phase. However, the reduced persister fraction (Fig. 2D) in this mutant might also play a role independent of metabolism.
We then turned to the ΔglpD Δpcm double mutant, which exhibits a high frequency of persisters peaking at 24 h in stationary phase. Here, two alleles that individually confer a competitive disadvantage are combined, and indeed after 1 day of stationary-phase competition with the wild-type strain, the double mutant showed less fitness than either of the two single-mutant strains, being outcompeted by the wild type by about 3.4:1 (average RCI = 0.53; Fig. 4C). Unlike the pcm and glpD mutants, however, the competitive disadvantage of this strain did not become more pronounced over time. Instead, the double mutant was more successful in competition with its wild-type parent over the long term, maintaining a relatively constant RCI over 10 days (Fig. 4C). Indeed, by 10 days in long-term stationary phase (when both single mutants had been outcompeted by the wild type by about 10:1; Fig. 4A and B), the mean RCI was statistically indistinguishable from an RCI of 0 (P = 0.062). We conclude that the introduction of a glpD mutation serves to mitigate the effect of unrepaired isoaspartyl damage on stationary-phase fitness, potentially as a consequence of high-level persister production.
Methylglyoxal production may contribute to increased fitness of ΔglpD Δpcm mutants.
Screens for genes affecting persister frequency have identified glpD both as a gene that promotes persisters (26) and as one whose mutation increases the persister fraction (35). In our hands, the deletion of glpD decreased persister formation (Fig. 2D); however, we observed a transient increase in persistence (about 4.5-fold) in this strain at 4 h after the onset of stationary phase (Fig. 2C). It is likely that both the time of incubation in stationary phase (a phenomenon also observed by others; see reference 31) and the genetic background impact the effects of glpD mutations on persisters; our results make clear that the deletion of glpD can lead to high-level persistence at least in a pcm mutant background. Inasmuch as one study that associated persistence with insertional inactivation of glpD attributed that phenomenon to an accumulation of methylglyoxal (35), we asked whether this metabolic intermediate might account for the increased fitness of our Δpcm ΔglpD mutant.
Methylglyoxal is a growth inhibitor produced from dihydroxyacetone phosphate (DHAP), the product of glycerol-3-phosphate (G3P) oxidation by GlpD as well as of glyceraldehyde-3-phosphate metabolism. In glpD mutants, the accumulation of G3P inhibits the DHAP→G3P pathway, resulting in elevated DHAP levels, leading to the production of methylglyoxal (34). Girgis et al. (35) found intracellular methylglyoxal levels of 0.7 mM in a glpD mutant and showed growth inhibition upon the addition of sublethal concentrations (1 to 2 mM) of methylglyoxal to cultures, leading them to conclude that methylglyoxal synthesis was the mechanism of increased persistence in glpD mutants. We incubated stationary-phase wild-type or Δpcm mutant cultures with methylglyoxal at concentrations up to 1.5 mM for 24 h to verify that it was not lethal and observed no reduction in survival for either strain (Fig. 5A). We then mixed wild-type E. coli with our Δpcm mutant strain in the presence of methylglyoxal and allowed them to compete in our competition assay, as described above. We found that after 5 or 10 days of competition with 1.5 mM methylglyoxal (Fig. 5B), the RCI was statistically indistinguishable from an RCI of 0 (P = 0.78 for day 5 and 0.40 for day 10), suggesting that the addition of methylglyoxal had reversed the competitive disadvantage of the repair-deficient cells. Comparable results were observed with 0.75 mM or 0.25 mM methylglyoxal (data not shown). This result supports the hypothesis that methylglyoxal production could account for the increased fitness of our pcm glpD double mutant. Surprisingly, at all the concentrations tested, we repeatably observed that the Δpcm mutant dominated the culture on day 1, outcompeting the wild type by between 103:1 and 105:1 (Fig. 5B). We discuss this phenomenon further below.
FIG 5.
Effect of methylglyoxal on competitive fitness of a Δpcm mutant. (A) Effect of methylglyoxal on survival of stationary-phase cells. Cultures of JV1120 (wild type; ■) and JV1168 (Δpcm; □) were grown to stationary phase, methylglyoxal was added to the indicated concentration, and cultures were incubated 24 h longer before measuring survival by viable counts. (B) JV1120 (wild type) and JV1168 (Δpcm mutant) were grown individually to stationary phase, mixed at 1:10, 1:1, or 10:1 ratios, and allowed to compete for 10 days in the presence of 1.5 mM methylglyoxal. Samples were quantitated by viable counts on the indicated days and the relative competitive index (RCI) calculated as described in the text. Open circles (○) represent the RCI of individual samples; closed squares (■) indicate mean RCI, with error bars representing one standard deviation in each direction.
High persister levels in glpD pcm mutants are dependent on the alarmone (p)ppGpp.
We sought to better understand the mechanisms responsible for high-level persister formation in our Δpcm ΔglpD mutant, using a genetic approach. Toxin-antitoxin systems, in which cells carry genes for growth-inhibiting toxins inhibited by the expression of a corresponding antitoxin gene (13), have been implicated in persistence (for a review, see reference 36). However, derivatives of our Δpcm ΔglpD mutant strain carrying deletions of the toxin gene hipA, relE, mqsR, or cspD all formed persisters at a high level not significantly different from the Δpcm ΔglpD mutant strain alone (data not shown). This finding is consistent with reports that multiple TA systems act together in persister formation (10).
Recent work (11, 15) implicates the alarmone (p)ppGpp and the stringent response as a master regulatory system in persister formation. Maisonneuve et al. (10, 15) found high (p)ppGpp levels in persister cells; they propose that signals, such as nutrient limitation, which increase (p)ppGpp levels lead to an accumulation of polyphosphate and subsequent activation of Lon protease to degrade the antitoxin component of type II toxin-antitoxin pairs (including hipBA, relBE, and ygiT-mqsR). Given the involvement of the stringent response in the transition to stationary phase (37), we hypothesized that our Δpcm ΔglpD mutant might be responding to stress via a (p)ppGpp signal, which might result in the activation of multiple, and perhaps redundant, TA systems (as well as possibly other pathways).
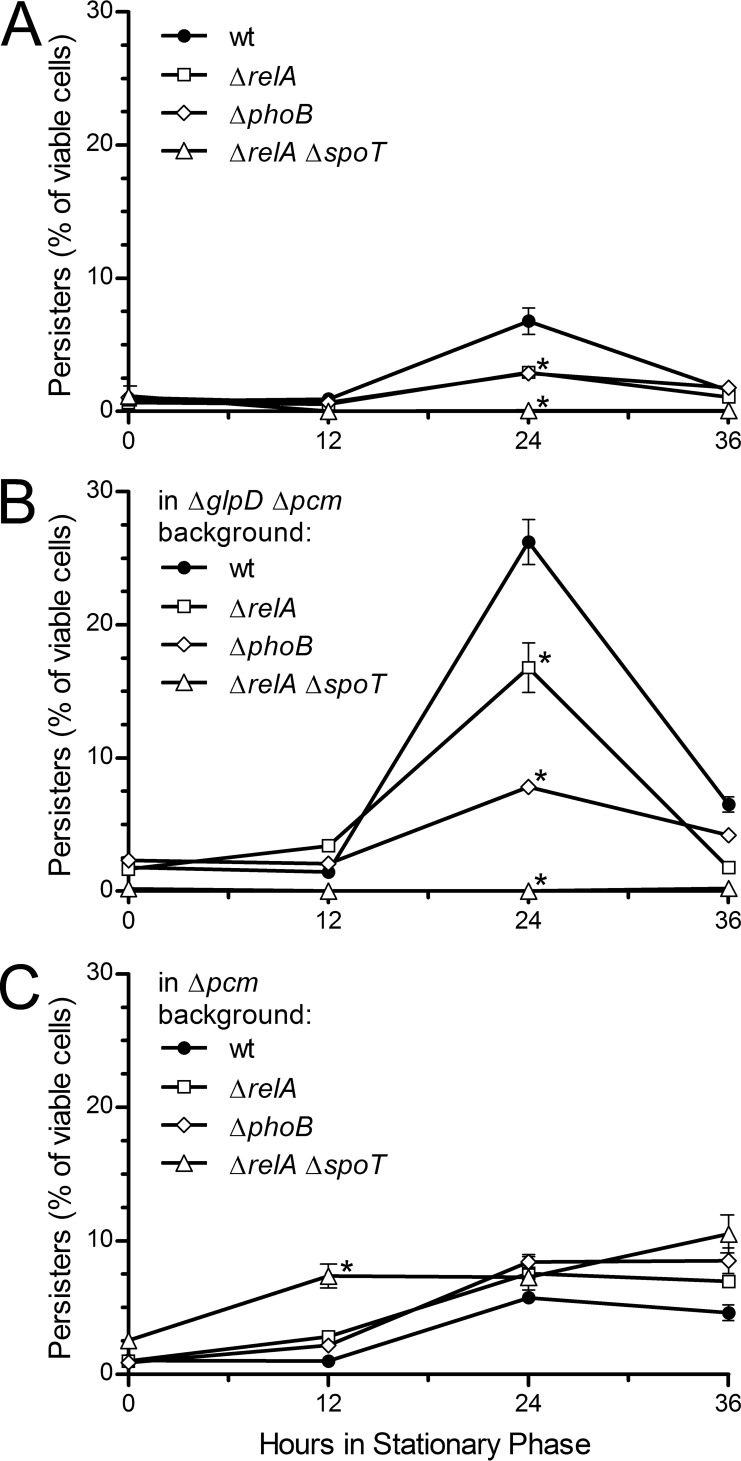
We therefore measured persister frequency at 0, 12, 24, and 36 h into stationary phase for cells carrying ΔrelA, ΔrelA ΔspoT, and ΔphoB mutations in wild-type, Δpcm mutant, and ΔglpD Δpcm mutant backgrounds. RelA is the major (p)ppGpp synthase in E. coli; SpoT is primarily a (p)ppGpp hydrolase with weak synthetic activity (16). The deletion of spoT is lethal; however, spoT relA double mutants are viable and eliminate the low level of (p)ppGpp remaining in relA mutants (38). PhoB is the major regulator of polyphosphate synthesis (39).
We found that a ΔrelA ΔspoT double mutant (Fig. 6A, triangles) eliminated the persister peak that otherwise occurred by 24 h of incubation in stationary phase in wild-type cells (Fig. 6A, circles). The ΔrelA mutant (Fig. 6A, squares) reduced this peak by about half, suggesting that this peak of persister formation is (p)ppGpp dependent and that even the low level of (p)ppGpp in a relA mutant is sufficient to initiate persister formation. The ΔphoB mutant (Fig. 6A, diamonds) likewise reduced the persister peak, implicating polyphosphate in the pathway but suggesting that persister formation under these conditions is not solely dependent on polyphosphate. We observed the same pattern for the strains carrying both the Δpcm and ΔglpD mutations (Fig. 6B); strikingly, the ability of this strain to produce a very high frequency of persisters at 24 h into stationary phase was completely eliminated by the combination of ΔspoT and ΔrelA mutations (triangles, Fig. 6B). It thus appears this high-level persister production occurs via the same (p)ppGpp-dependent mechanism that produces the stationary-phase persister peak in wild-type cells.
FIG 6.
Role of (p)ppGpp and polyphosphate in stationary-phase persister formation in isoaspartyl repair-deficient mutants. Persister frequency was determined as described at the onset of stationary phase and 12, 24, and 36 h later for ΔrelA, ΔphoB, and ΔrelA ΔspoT mutants in the following backgrounds: wild type, derivatives of JV1120 (JV1191, JV1192, and JV1199, respectively) (A); ΔglpD Δpcm mutant, derivatives of JV1177 (JV1197, JV1198, and JV1201, respectively) (B); and Δpcm mutant, derivatives of JV1121 (JV1193, JV1194, and JV1200, respectively) (C). Points are averages of the results from three trials, and the error bars represent one standard deviation. Asterisks indicate points significantly (P < 0.05) different from wild type.
The result was different when we examined persisters in a Δpcm mutant background (Fig. 6C). Here, the prolonged persister peak that we observed previously was not reduced by mutations in relA, spoT, or phoB. It therefore seems that unrepaired isoaspartyl damage leads to persister formation by a mechanism distinct from that observed in wild-type or glpD mutant cells.
Persister formation is correlated with accumulation of unstable proteins.
Whereas the (p)ppGpp-dependent persister pathway is likely activated in response to nutrient deprivation, persister formation has also been linked to other changes in the cellular environment, including in a recent report correlating persister formation and protein aggregation (17). PCM is required for maximal survival of E. coli under such potentially protein-denaturing conditions as oxidative stress (paraquat), osmotic stress, methanol, or repeated heating (5). We hypothesize that an important consequence of pcm mutations is the susceptibility of isoaspartate-containing proteins to unfolding under stress; we therefore asked whether aggregated, denatured, or destabilized proteins could trigger persister formation.
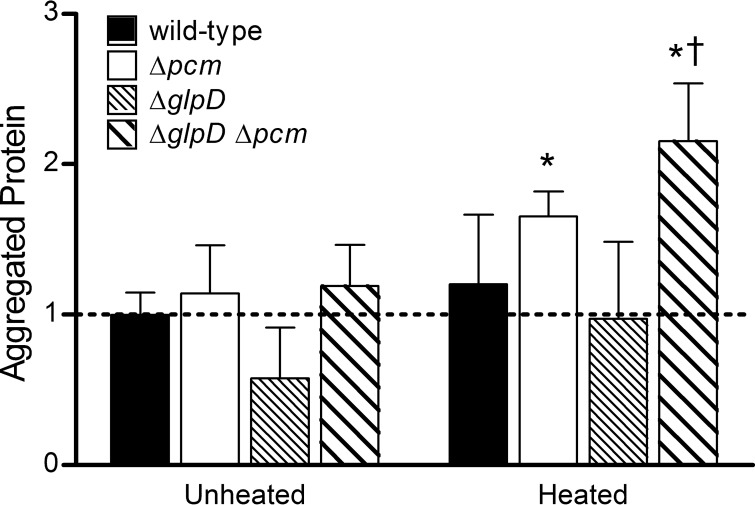
Total cytoplasmic proteins were isolated from wild-type, Δpcm mutant, ΔglpD mutant, and ΔglpD Δpcm mutant strains after 24 h of incubation in stationary phase, and aggregated proteins were recovered by differential centrifugation (see Materials and Methods). At this time point, the fraction of total protein that had aggregated during growth was similar for all strains (“unheated” bars in Fig. 7; note that all bars are normalized to the mean for the unheated wild type). We then heated aliquots of the extracted total protein to 37°C for 2 h to detect proteins that were not aggregated in vivo but were susceptible to unfolding and aggregation. This analysis revealed that the Δpcm mutant (white bars, Fig. 7) had significantly more unstable aggregatable protein than its wild-type parent and that the ΔglpD Δpcm double mutant had significantly more than either the wild-type or the Δpcm mutant (P < 0.05 for each). The smaller amount of both aggregated and heat-aggregatable protein observed in the ΔglpD single mutant (narrow stripes, Fig. 7) was repeatable but not statistically significant. It therefore appears that by 24 h into stationary phase (when persisters peak), repair-deficient strains have more conformationally unstable protein and that the amount of heat-aggregatable protein correlates with the fraction of persisters. This suggests that increased or prolonged production of persisters might be triggered by a cellular response to protein damage.
FIG 7.
Aggregated and unstable protein in repair and persister mutants. Insoluble protein aggregates were quantitated as the percentage of total protein for strains JV1120 (wild type; solid bars), JV1121 (Δpcm mutant; open bars), JV1136 (ΔglpD mutant; dense stripes), and JV1154 (ΔglpD Δpcm mutant; wider stripes) incubated for 24 h in stationary phase (“unheated”). To quantitate protein that was not aggregated in vivo but could be aggregated by heat (“heated”), total protein extracts were heated for 2 h at 37°C, and then insoluble aggregates were determined as described above. Bars represent the averages of the results from 4 trials, normalized to the average aggregated protein from the unheated wild-type strain. Error bars represent one standard deviation; averages significantly different (P < 0.05) from unheated wild type (*) or heated Δpcm mutant (†) are indicated.
DISCUSSION
The spontaneous formation of isoaspartate from aspartyl and asparaginyl residues (Fig. 1) is a major form of damage to proteins whose biochemistry and potential to affect enzyme activity are well understood, as is the ability of the l-isoaspartyl protein carboxyl methyltransferase to effect net repair of isoAsp to normal aspartate (reviewed in reference 3). However, despite the significant phenomena associated with PCM deficiency, including reduced longevity and stress resistance, as well as diseases ranging from autoimmunity to cancer in multicellular eukaryotes, many details of PCM's roles in the life of a cell remain to be elucidated. In this paper, we have identified a link between protein repair and persisters and provide evidence that unrepaired isoaspartyl damage may trigger increased or prolonged persister formation, potentially a cellular response that can mitigate the harmful effects of protein damage.
We observed a peak in the fraction of persister cells within aerobically grown cultures in rich medium at 24 h after the onset of stationary phase. We found that the peak was extended by a day in a pcm mutant and greatly increased in a pcm glpD double mutant (Fig. 2). In long-term competition studies (Fig. 4), we demonstrated that a pcm mutant had a clear fitness defect relative to wild-type E. coli, the first evidence that deficiency in isoaspartyl repair is detrimental to unstressed cells. We further observed that the addition of a ΔglpD mutation largely reversed the fitness defect of the Δpcm mutant strain, raising the intriguing possibility that protein damage might be one trigger leading to persister formation, which in turn may serve to protect the cell from the consequences of damage. In support of this hypothesis, we found a correlation between persister formation and the presence of conformationally unstable protein in the cell (Fig. 7). At present, we cannot distinguish between the effects of persister formation and the results of other physiological changes which might be occurring in parallel with persister formation. We further recognize that persister formation might be a consequence of protein damage (e.g., via the inactivation of an isoAsp-susceptible inhibitor of a persister pathway) and not part of a response; further study will be required to sort out the cellular phenomena involved and distinguish among these possibilities. Nevertheless, the hypothesis that persister formation might mitigate the negative effects of macromolecular damage is a provocative one, particularly in light of reports linking persister formation with various stress responses (see, for example, references 15, 17, 40, and 41).
Others have previously speculated that persister formation may represent a stress response capable of enhancing bacterial survival (17). Some corroboration for this idea is found in the report of Hong et al. (19) that reducing fitness by mutating stress-response pathways increased persister formation. Furthermore, stress response genes, including those associated with the maintenance of protein conformation (e.g., clpB, ibpA and ibpB, htpX, and htrA), are upregulated in the persister fraction (42). However, other authors suggest a stochastic mechanism of persister formation resulting in a “bet-hedging” strategy (43), or even that persisters might result from senescence (44). One study suggested that the high selective cost of persistence results in pressure to keep persister frequencies low (45). Given the ubiquity of persisters in bacterial cultures and the fact that they are always a subpopulation, it is not trivial to distinguish between stochastic activation of mechanisms that reduce metabolic activity (which seems sufficient to explain antibiotic tolerance) and an active stress response pathway. Identifying the mechanism(s) that allow (i) Δpcm mutant bacteria to maintain persisters, (ii) ΔglpD Δpcm mutant cells to drastically increase persisters, and (iii) persisters to confer a fitness advantage may help us to understand whether, and if so, how, persister formation can serve as a response to stress or mitigate the effects of protein damage.
We have begun to probe these mechanisms in this paper. Our genetic experiments implicate (p)ppGpp, polyphosphate, and the stringent response in persister formation in the ΔglpD Δpcm double mutants (Fig. 6B). This result supports the idea of persister formation as a stress response and is consistent with a recently proposed (p)ppGpp- and polyphosphate-dependent pathway of persister formation (10, 15). Additionally, the stringent response to nutrient depletion is a key component of the transition to stationary phase (37). We note, however, that persister formation in our system is only partially dependent on phoB, the major regulator of polyphosphate synthesis, suggesting that polyphosphate accumulation and the resultant Lon-mediated degradation of antitoxins (10) may not be the only (p)ppGpp-regulated pathway to persister formation. Furthermore, our PCM-deficient mutant remained capable of forming and maintaining stationary-phase persisters in a ΔrelA, ΔrelA ΔspoT, or ΔphoB mutant background (Fig. 6C). We conclude that an additional (p)ppGpp-independent pathway of persister formation is activated directly or indirectly in repair-deficient cells. It is surprising that this peak was present in Δpcm ΔrelA ΔspoT mutant cells but absent when a ΔglpD mutation was added. We do not presently understand what interactions or hierarchy among these pathways might account for this result.
Consistent with this complexity, the idea that there are multiple pathways of persister formation, not all of which necessarily involve dormancy, seems to be emerging from the recent literature (reviewed in reference 36). Our finding that even a culture with nearly 25% persisters had the same level of gross metabolic activity as the wild type (Fig. 3) supports this view and refutes the straightforward explanation that increasing the number of dormant cells could slow protein damage in the overall population or reduce the opportunity for damaged enzymes and other proteins to affect cellular processes. Taken together, our evidence suggests that at least two pathways of persister formation are operating in our system, neither of which appears to involve significant metabolic inactivity.
Of course, our persisters could be undergoing inhibition of cellular processes on a more subtle level. Indeed, we found that sublethal concentrations of methylglyoxal, a metabolic inhibitor that accumulates in glpD mutants (35), had a positive effect on the fitness of a Δpcm mutant similar to that of the glpD mutation (Fig. 5). While the best-described roles of methylglyoxal as a metabolic poison do not correspond well with our metabolic measurements or with the reduced persister frequency observed in our ΔglpD mutant, methylglyoxal has also been described as a protein-glycating agent (46), and it is interesting to speculate that the deleterious effects of glycation might be balanced either by reduced susceptibility to other protein damage or by the induction of protein maintenance and repair machinery. Possibly, a more complex effect of this nature could explain the transient but substantial increase in fitness that we observed for our Δpcm mutant in the presence of methylglyoxal.
We have thus far identified the apparent protective value of a large persister fraction only in the context of a ΔglpD Δpcm double mutant. We have also observed (Fig. 4) that the Δpcm single mutant with its prolonged-persister phenotype and the ΔglpD single mutant with its low-persister phenotype have the same effect on fitness, a seeming contradiction. We suggest, however, that the distinct pathway leading to the prolonged persister peak (Fig. 1A) identified in the Δpcm mutant could mitigate the effects of protein damage, which otherwise would render the Δpcm mutant even less fit. With increasing understanding of the physiological heterogeneity (47, 48) within bacterial cultures and an understanding of long-term stationary phase as the rise of a succession of increasingly fit genetic variants (the growth advantage in stationary-phase [GASP] phenotype; see reference 33), it is also interesting to speculate that a protective combination of mutations might arise naturally under stationary-phase conditions.
In future work, we will seek to identify the pathways of persister formation active in our system and to further examine the connections among protein damage, glpD mutations, (p)ppGpp, and fitness. One interesting avenue of further investigation would be the possible involvement of oxidative stress, one condition under which PCM becomes essential to the long-term survival of E. coli (5). The suppression of metabolic reactions that produce reactive oxygen species has been suggested as an explanation for the ability of persisters to tolerate antibiotics (40) and might suggest a direct mechanism by which persisters could mitigate the effects of protein damage.
ACKNOWLEDGMENTS
Research at an undergraduate institution is almost inevitably a cooperative effort involving multiple students whose time as lab members is frequently too short to see a body of work through to publication. While we were the primary contributors, we gratefully acknowledge the valuable contributions of other North Central College students, including, most notably, Rebecca Chung, who constructed strain JV1136 and measured the long-term survival of persister mutants, Adam Kleinman, who quantitated aggregated protein in persister mutants, and David Cabrerizo Granados, who obtained preliminary data on persister frequencies in TA mutants. We thank Mike Cashell and the E. coli Genetic Stock Center for kindly providing strains used in this work.
This work was supported by NIH grant no. 1R15AG032101 from the National Institute on Aging through the NIH AREA grant program, for which we are very grateful. We also express our gratitude to North Central College's Faculty Professional Development Committee for financial support through its summer grant program and thank the Council on Undergraduate Research for funding student travel to present some of this work at ASM conferences.
REFERENCES
- 1.Visick JE, Clarke S. 1995. Repair, refold, recycle: how bacteria can deal with spontaneous and environmental damage to proteins. Mol Microbiol 16:835–845. doi: 10.1111/j.1365-2958.1995.tb02311.x. [DOI] [PubMed] [Google Scholar]
- 2.Clarke S. 1992. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu Rev Biochem 61:355–386. doi: 10.1146/annurev.bi.61.070192.002035. [DOI] [PubMed] [Google Scholar]
- 3.Clarke S. 2003. Aging as war between chemical and biochemical processes: protein methylation and the recognition of age-damaged proteins for repair. Ageing Res Rev 2:263–285. doi: 10.1016/S1568-1637(03)00011-4. [DOI] [PubMed] [Google Scholar]
- 4.Patananan AN, Capri J, Whitelegge JP, Clarke SG. 2014. Non-repair pathways for minimizing protein isoaspartyl damage in the yeast Saccharomyces cerevisiae. J Biol Chem 289:16936–16953. doi: 10.1074/jbc.M114.564385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visick JE, Cai H, Clarke S. 1998. The l-isoaspartyl protein repair methyltransferase enhances survival of aging Escherichia coli subjected to secondary environmental stresses. J Bacteriol 180:2623–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Visick JE, Ichikawa JK, Clarke S. 1998. Mutations in the Escherichia coli surE gene increase isoaspartyl accumulation in a strain lacking the pcm repair methyltransferase but suppress stress-survival phenotypes. FEMS Microbiol Lett 167:19–25. doi: 10.1111/j.1574-6968.1998.tb13202.x. [DOI] [PubMed] [Google Scholar]
- 7.Hicks WM, Kotlajich MV, Visick JE. 2005. Recovery from long-term stationary phase and stress survival in Escherichia coli require the l-isoaspartyl protein carboxyl methyltransferase at alkaline pH. Microbiol 151:2151–2158. doi: 10.1099/mic.0.27835-0. [DOI] [PubMed] [Google Scholar]
- 8.Finkel SE, Kolter R. 1999. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci U S A 96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 10.Maisonneuve E, Gerdes K. 2014. Molecular mechanisms underlying bacterial persisters. Cell 157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 11.Amato SM, Orman MA, Brynildsen MP. 2013. Metabolic control of persister formation in Escherichia coli. Mol Cell 50:475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Amato SM, Fazen CH, Henry TC, Mok WW, Orman MA, Sandvik EL, Volzing KG, Brynildsen MP. 2014. The role of metabolism in bacterial persistence. Front Microbiol 5:70. doi: 10.3389/fmicb.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi Y, Inouye M. 2011. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat Rev Microbiol 9:779–790. doi: 10.1038/nrmicro2651. [DOI] [PubMed] [Google Scholar]
- 14.Wood TK, Knabel SJ, Kwan BW. 2013. Bacterial persister cell formation and dormancy. Appl Environ Microbiol 79:7116–7121. doi: 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 16.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leszczynska D, Matuszewska E, Kuczynska-Wisnik D, Furmanek-Blaszk B, Laskowska E. 2013. The formation of persister cells in stationary-phase cultures of Escherichia coli is associated with the aggregation of endogenous proteins. PLoS One 8:e54737. doi: 10.1371/journal.pone.0054737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuczynska-Wisnik D, Stojowska K, Matuszewska E, Leszczynska D, Algara MM, Augustynowicz M, Laskowska E. 2015. Lack of intracellular trehalose affects formation of Escherichia coli persister cells. Microbiol 161:786–796. doi: 10.1099/mic.0.000012. [DOI] [PubMed] [Google Scholar]
- 19.Hong SH, Wang X, O'Connor HF, Benedik MJ, Wood TK. 2012. Bacterial persistence increases as environmental fitness decreases. Microb Biotechnol 5:509–522. doi: 10.1111/j.1751-7915.2011.00327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dörr T, Lewis K, Vulic M. 2009. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet 5:e1000760. doi: 10.1371/journal.pgen.1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leung V, Lévesque CM. 2012. A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J Bacteriol 194:2265–2274. doi: 10.1128/JB.06707-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skorupski K, Taylor RK. 1996. Positive selection vectors for allelic exchange. Gene 169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 23.Silhavy TJ, Berman ML, Enquist LW. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 24.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spoering AL, Vulic M, Lewis K. 2006. GlpD and PlsB participate in persister cell formation in Escherichia coli. J Bacteriol 188:5136–5144. doi: 10.1128/JB.00369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berridge MV, Herst PM, Tan AS. 2005. Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnol Annu Rev 11:127–152. doi: 10.1016/S1387-2656(05)11004-7. [DOI] [PubMed] [Google Scholar]
- 28.Morris AR, Darnell CL, Visick KL. 2011. Inactivation of a novel response regulator is necessary for biofilm formation and host colonization by Vibrio fischeri. Mol Microbiol 82:114–130. doi: 10.1111/j.1365-2958.2011.07800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gur E, Biran D, Shechter N, Genevaux P, Georgopoulos C, Ron EZ. 2004. The Escherichia coli DjlA and CbpA proteins can substitute for DnaJ in DnaK-mediated protein disaggregation. J Bacteriol 186:7236–7242. doi: 10.1128/JB.186.21.7236-7242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275. [PubMed] [Google Scholar]
- 31.Luidalepp H, Jõers A, Kaldalu N, Tenson T. 2011. Age of inoculum strongly influences persister frequency and can mask the effects of mutations implicated in altered persistence. J Bacteriol 193:3598–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fung DK, Chan EW, Chin ML, Chan RC. 2010. Delineation of a bacterial starvation stress response network which can mediate antibiotic tolerance development. Antimicrob Agents Chemother 54:1082–1093. doi: 10.1128/AAC.01218-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finkel SE. 2006. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol 4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- 34.Lin EC. 1976. Glycerol dissimilation and its regulation in bacteria. Annu Rev Microbiol 30:535–578. doi: 10.1146/annurev.mi.30.100176.002535. [DOI] [PubMed] [Google Scholar]
- 35.Girgis HS, Harris K, Tavazoie S. 2012. Large mutational target size for rapid emergence of bacterial persistence. Proc Natl Acad Sci U S A 109:12740–12745. doi: 10.1073/pnas.1205124109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helaine S, Kugelberg E. 2014. Bacterial persisters: formation, eradication, and experimental systems. Trends Microbiol 22:417–424. doi: 10.1016/j.tim.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 38.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem 266:5980–5990. [PubMed] [Google Scholar]
- 39.Rao NN, Liu S, Kornberg A. 1998. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J Bacteriol 180:2186–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim JS, Heo P, Yang TJ, Lee KS, Jin YS, Kim SK, Shin D, Kweon DH. 2011. Bacterial persisters tolerate antibiotics by not producing hydroxyl radicals. Biochem Biophys Res Commun 413:105–110. doi: 10.1016/j.bbrc.2011.08.063. [DOI] [PubMed] [Google Scholar]
- 41.Wu Y, Vulic M, Keren I, Lewis K. 2012. Role of oxidative stress in persister tolerance. Antimicrob Agents Chemother 56:4922–4926. doi: 10.1128/AAC.00921-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol 186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen NR, Lobritz MA, Collins JJ. 2013. Microbial persistence and the road to drug resistance. Cell Host Microbe 13:632–642. doi: 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klapper I, Gilbert P, Ayati BP, Dockery J, Stewart PS. 2007. Senescence can explain microbial persistence. Microbiology 153:3623–3630. doi: 10.1099/mic.0.2007/006734-0. [DOI] [PubMed] [Google Scholar]
- 45.Stepanyan K, Wenseleers T, Duéñez-Guzmán EA, Muratori F, Van den Bergh B, Verstraeten N, De Meester L, Verstrepen KJ, Fauvart M, Michiels J. 2015. Fitness trade-offs explain low levels of persister cells in the opportunistic pathogen Pseudomonas aeruginosa. Mol Ecol 24:1572–1583. doi: 10.1111/mec.13127. [DOI] [PubMed] [Google Scholar]
- 46.Pepper ED, Farrell MJ, Nord G, Finkel SE. 2010. Antiglycation effects of carnosine and other compounds on the long-term survival of Escherichia coli. Appl Environ Microbiol 76:7925–7930. doi: 10.1128/AEM.01369-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saint-Ruf C, Garfa-Traoré M, Collin V, Cordier C, Franceschi C, Matic I. 2014. Massive diversification in aging colonies of Escherichia coli. J Bacteriol 196:3059–3073. doi: 10.1128/JB.01421-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shapiro JA. 1998. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol 52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- 49.Ahn J, Visick JE. 2012. Escherichia coli deficient in an isoaspartyl protein repair enzyme increase the persister fraction by a phoU-dependent pathway, poster 2449. Abstr 112th Gen Meet Am Soc Microbiol. American Society for Microbiology, Washington, DC. [Google Scholar]
- 50.VandenBerg KE, Visick JE. 2014. High persister frequency increases competitive fitness in protein repair-deficient Escherichia coli during long-term stationary phase, poster 2190. Abstr 114th Gen Meet Am Soc Microbiol. American Society for Microbiology, Washington, DC. [Google Scholar]