Abstract
Phasins are the major polyhydroxyalkanoate (PHA) granule-associated proteins. They promote bacterial growth and PHA synthesis and affect the number, size, and distribution of the granules. These proteins can be classified in 4 families with distinctive characteristics. Low-resolution structural studies and in silico predictions were performed in order to elucidate the structure of different phasins. Most of these proteins share some common structural features, such as a preponderant α-helix composition, the presence of disordered regions that provide flexibility to the protein, and coiled-coil interacting regions that form oligomerization domains. Due to their amphiphilic nature, these proteins play an important structural function, forming an interphase between the hydrophobic content of PHA granules and the hydrophilic cytoplasm content. Phasins have been observed to affect both PHA accumulation and utilization. Apart from their role as granule structural proteins, phasins have a remarkable variety of additional functions. Different phasins have been determined to (i) activate PHA depolymerization, (ii) increase the expression and activity of PHA synthases, (iii) participate in PHA granule segregation, and (iv) have both in vivo and in vitro chaperone activities. These properties suggest that phasins might play an active role in PHA-related stress protection and fitness enhancement. Due to their granule binding capacity and structural flexibility, several biotechnological applications have been developed using different phasins, increasing the interest in the study of these remarkable proteins.
INTRODUCTION
Polyhydroxyalkanoic acids (PHAs) are polymers synthesized by many bacteria that function as intracellular carbon and energy storage compounds. According to the length of the monomer, these polymers are classified as short (C3 to C5)- or medium (C6 to C16)-chain-length PHAs (1, 2). The best known and most common PHA is poly(3-hydroxybutyrate) (PHB), composed of C4 monomers. PHAs are accumulated as intracellular insoluble granules that are surrounded by an organized protein layer composed of several granule-associated proteins (PGAPs). Among the proteins associated with PHA granules are PHA synthases, PHA depolymerases, and a group of low-molecular-weight proteins known as phasins. The presence of structural, biosynthetic, catabolic, and regulatory proteins in the granule surface indicates that they are organized and complex subcellular structures that were designated carbonosomes (3).
The first phasin was identified in 1994 by Pieper-Fürst and Steinbüchel when they found a low-molecular-weight protein (GA14) associated with PHA granules in Rhodococcus ruber. GA14 was the predominant protein present in the granules, forming a layer at their surface. Due to these properties, the name phasin (PhaP) was proposed, analogous to the oleosins at the surface of triacylglycerol inclusions in plants (4).
Since then, phasins have been found in the surfaces of the granules of all natural PHA-producing microorganisms studied (5), constituting the most abundant and widespread granule-associated proteins. Several phasins were identified in many PHA-producing bacteria, such as Ralstonia eutropha (also known as Cupriavidus necator) (5–7), Bacillus megaterium (8), R. ruber (4), Paracoccus denitrificans (9), Pseudomonas putida (10), Azotobacter vinelandii (11), and Synechocystis sp. strain PCC 6803 (12), among many others. Phasins have also been identified in Archaea such as Haloferax mediterranei (13).
This review summarizes the main characteristics of phasins, the multiple roles associated with these proteins, and their biotechnological applications.
PHASIN PROTEIN FAMILIES
Although phasins do not constitute a highly conserved group of proteins, and early reports indicated that the degree of conservation among them was very low, several protein motifs have been defined using the great number of phasins that have already been described, and several types of phasin families have been distinguished based on sequence similarity.
Considering the Pfam database (http://pfam.xfam.org/), there are four phasin-related families, each containing a characteristic domain (Table 1). The first family (PF09361) is the largest one and includes sequences found in bacteria belonging to Alpha-, Beta-, and Gammaproteobacteria, such as the most studied phasin, PhaP1 from R. eutropha (PhaPRe). The second (PF09602) corresponds to phasins found in Bacillus species, and the third (PF09650) contains a diverse group of mostly uncharacterized proteins belonging to different Proteobacteria. The last family (PF05597) contains proteins from different Proteobacteria and includes all characterized phasins belonging to Pseudomonas that accumulate medium-chain-length PHAs (PHAmcl), such as PhaF and PhaI from P. putida.
TABLE 1.
Phasin families according to the Pfam database
| Pfam database family name | Domain | Representative taxon/taxa | Representative protein(s) |
|---|---|---|---|
| PF09361 | Phasin_2 | Ralstonia, Azotobacter | PhaP from Ralstonia eutropha |
| PF09602 | PhaP_Bmeg | Bacillus | PhaP from Bacillus megaterium |
| PF09650 | PHA_gran_rgn | Diverse Proteobacteria | Uncharacterized group of phasins |
| PF05597 | Phasin | Pseudomonas | PhaF from Pseudomonas putida |
Although most phasins described to date belong to one of these four families, there are a few that show very little similarity to the rest and contain no recognizable phasin-related domains. Among them is the first phasin described, GA14 from R. ruber, PhaP from Synechocystis sp. PCC 6803, and archaeal phasins. Analysis of PhaP from Synechocystis revealed that it is similar to thylakoid-associated proteins from different algal species and to a protein from Arthrospira platensis that contains a Phasin_2 motif (12). Many phasins have been identified in Archaea, but to date, only that of H. mediterranei has been experimentally studied. This phasin has been reported to be relatively similar to other putative archaeal phasins but not to those from bacteria (13).
The different similarity groups in which phasins can be classified seem to reflect both the phylogenetic origin of the phasins and the kind of PHAs to which they bind. Most phasins characterized to date belong to Proteobacteria. Among them, PF09650 and PF09361 group phasins are related to short-chain PHAs, while phasins related to PHAmcl, mostly belonging to Pseudomonas species, constitute a separate group (PF05597 family). Although most Pseudomonas spp. accumulate only PHAmcl, several strains, such as Pseudomonas sp. 61-3 (14), P. extremaustralis (15), and P. pseudoalcaligenes (16), have been observed to accumulate both PHAmcl and PHB. These bacteria contain separate biosynthesis gene clusters for each polymer and different phasins that belong to different phasin families. A recent study that characterized granules containing different polymer compositions in Pseudomonas sp. 61-3 determined that phasins PhaF and PhaI, which have the Phasin domain (Table 1), were found on the surface of PHAmcl granules while PhbP, which belongs to the largest phasin family (PF09361) containing the Phasin_2 domain, was bound to PHB granules in this organism (14), suggesting that phasins have a certain degree of specificity. The ability of Pseudomonas strains to accumulate PHB could have been acquired by horizontal gene transfer (15, 16), so it is possible that genes encoding the PHB-related phasins in these bacteria could have also been acquired in this manner.
STRUCTURAL ASPECTS OF PHASINS
The structure of phasins has been analyzed in relatively few studies. Low-resolution structural studies and in silico predictions were performed in order to elucidate the structure of different phasins, such as PhaP from R. eutropha (PhaPRe) (17), PhaP from Azotobacter sp. strain FA8 (PhaPAz) (18), PhaF from P. putida (PhaFPp) (19), PhaP from Aeromonas hydrophila (PhaPAh) (20), and PhaP from Synechocystis sp. PCC 6803 (PhaPSp) (12).
Secondary-structure in silico predictions for phasins PhaP1 to PhaP4 from R. eutropha, PhaP1 from Ralstonia solanacearum, PhaP from Ralstonia metallidurans (17), PhaPAz (18), and PhaPSp (12) revealed that these phasins have a high percentage (close to 90%) of amino acids in an α-helix conformation. This was found to be a general characteristic of phasins (18–20). However, the proportion of residues in an α-helix conformation was observed to change according to the environment in experiments performed with PhaPAz and the N-terminal domain of PhaFPp, as this proportion was shown to increase in the presence of structure-stabilizing solvents, like 2,2,2-trifluoroethanol, or with the addition of sodium oleate, which creates a hydrophobic environment that mimics PHB (18, 19).
A structural feature that has been recently recognized in phasins is the presence of disordered regions, which are predicted in many of these proteins (18, 19). Intrinsically disordered proteins or disordered regions are interconverting dynamic ensembles of structures that do not fold in a single structure and are important for the regulatory functions of the proteins because of their highly flexible nature (21). Maestro et al. proposed that the multifunctional phasin PhaFPp belonged to a new family of intrinsically disordered proteins due to the unstructured regions predicted in its N-terminal domain, which was also predicted to contain a long α-helix that is partially disordered in the absence of PHA (19). Unstructured regions were also found in PhaPAz. It was estimated that around 40 to 45% of this phasin could be disordered when it is not binding any target, and this value would decrease to 23 to 30% when interacting with other molecules. The capacity of PhaPAz to change according to the environment could be attributed to the presence of disordered regions that provide flexibility to the protein (18). Disordered regions might contribute to the functional diversity of these proteins.
All phasins form oligomers, and most studies have indicated that these proteins are normally tetramers in solution (12, 18–20). However, Neumann et al. proposed that PhaPRe occurs as a trimer (17), and some phasins have been reported to form other kinds of oligomers, such as dimers and dodecamers (corresponding to PhaP5 and PhaM from R. eutropha, respectively) (22). A common structural characteristic of phasins is the presence of coiled-coil interacting regions. Bioinformatics predictions have revealed that many of them have a high coiled-coil probability (>75%), such as phasins from R. eutropha and from different species of Pseudomonas and Bacillus (19). On the other hand, other phasins have an intermediate coiled-coil probability (25% to 75%), such as PhaPAz (18) and a phasin from Azotobacter vinelandii (19). These regions have been proposed to be involved in oligomerization and/or interaction with other proteins (18, 19). Although coiled-coil regions have been found in almost all phasins described to date, the location and extension of these regions differ greatly, even among phasins belonging to the same family (18). This suggests that the interaction between monomers and with PHA granules, other phasins, and other proteins might differ for different phasins. PhaPSp is the only phasin devoid of coiled-coil regions described to date, suggesting that the mode of oligomerization of this protein, which was observed to form tetramers in solution, is different from that described for other phasins (12).
INTERACTION WITH PHA GRANULES
As phasins are proteins that bind to hydrophobic PHA granules, the presence of hydrophobic domains that could mediate interactions with the polymer has been analyzed in many of them. GA14 from R. ruber contains two hydrophobic domains in the C terminus that were proposed to be interaction domains with PHA based on experimental results (4). A later study that analyzed PhaP1 to PhaP4 from R. eutropha revealed that these phasins do not have clear hydrophobic domains in common, although they share homologous hydrophobic regions. Experimental results obtained with PhaPRe mutants revealed that the entire protein is involved in the interaction with polymer granules and that there is no particular region responsible for this interaction (17). The absence of hydrophobic domains appears to be a common characteristic of most phasins, despite their lipid granule binding function. Analysis of the primary structure of other phasins, like PhaPAz, PhaFPp, and PhaPSp, revealed that they lack clear hydrophobic domains, and their ability to bind to PHA granules was associated with the amphiphilic nature of at least one of their helices (12, 18, 23). Furthermore, PhaP from B. megaterium (PhaPBm) was observed to be an extremely hydrophilic phasin that can effectively bind to the granules in spite of the lack of hydrophobic domains (8). The hydrophobic nature of GA14, which has no characteristic phasin domains, seems to be uncommon among phasins (18). This protein has been observed to interact with granules containing both PHA and triacylglycerols in R. ruber (24), so its characteristics might be associated with the particular composition of the lipid granules accumulated by this oleaginous bacterium. However, phasins that lack clear hydrophobic domains, such as PhaPRe, have been observed to bind to triacylglycerol inclusions when expressed in R. opacus and Mycobacterium smegmatis (25). Amphipathic helices in phasins might be important not only for the interactions of these proteins with the polymer but also for interactions with other granule-associated proteins and with hydrophobic regions of misfolded proteins and inclusion bodies (IBs) (23). Pfeiffer and Jendrossek studied the interactions between known PHB granule-associated proteins from R. eutropha in vitro. According to their results, phasins not only are able to form homo-oligomers but can also associate with each other to form hetero-oligomers, and they are also able to interact with other proteins, such as the regulator PhaR and the PHB depolymerase PhaZ1 (26).
ROLE OF PHASINS IN PHA METABOLISM
Studies of phasin PhaPRe showed that this protein plays an important role in PHB synthesis. The first genetic analysis of the role of PhaPRe in PHB metabolism was performed by Wieczorek et al. (5), who studied the effect of phaP mutations on PHB synthesis in R. eutropha. Their study revealed that this phasin promotes PHA synthesis and affects the number and size of the granules (5), an effect that has also been observed in other bacterial producers (9, 12, 27, 28).
It has been reported that the occurrence of PhaPRe in the cells is strictly dependent on PHA biosynthesis and that cells cultivated under conditions not permissive for PHA synthesis or mutants defective in the PHA synthase structural gene do not synthesize detectable levels of PhaPRe (5, 29, 30). Intracellular levels of this phasin were observed to increase concomitantly with PHB accumulation in cells producing either low, intermediate, or high levels of the polymer (31), suggesting that in natural PHA producers the synthesis of phasins is tightly coupled with PHA synthesis. Analysis of the genetic regulation of phasin PhaPRe revealed that it is regulated by PhaR, a repressor that binds upstream of phaP and was also found to associate with PHA granules (32). Pötter et al. (33) and Pötter and Steinbüchel (34) proposed that during active PHA biosynthesis in R. eutropha, PhaR binds to PHA granules, allowing the transcription of phaP, so at the later stages of PHA accumulation, when PhaR is no longer bound to the granules, the transcription of phaP is again repressed (33, 34). In B. megaterium, PhaQ was proposed to act in a manner similar to that of PhaR from R. eutropha, as it binds to PHB granules and suffers transcriptional autoregulation (35). This mechanism is analogous to the one proposed by Prieto et al. for the regulation of the PHA biosynthesis genes in Pseudomonas oleovorans (currently known as P. putida) by phasin PhaFPp (10). PhaR was also found in H. mediterranei, but the regulation pattern of phaP by PhaR in this archaeon is slightly different from that of its bacterial counterparts, as it was shown to repress the expression of both its own gene and phaP, which are transcribed from the same operon (phaRP) (36).
Apart from its structural role as part of the PHA granule cover, many other functions related to PHA accumulation and degradation have been described for different phasins. For example, ApdA from Rhodospirillum rubrum was observed to activate PHB granules isolated from different species, including recombinant Escherichia coli, so that the granules could be hydrolyzed by soluble R. rubrum PHB depolymerase in vitro (28). This effect was demonstrated to be due to the capability of this phasin to activate the PHB depolymerase (37). In R. eutropha, the presence of PhaPRe on the granule surface was also reported to be important for PHB degradation. This study suggested that the effect of this phasin on PHB degradation might be direct, through interaction with the PHB depolymerase, or indirect, by providing access of a PHB depolymerase to the surface of the PHB granules (38).
Mms16 from Magnetospirillum gryphiswaldense was first described as a protein associated with magnetosomes, but this was later observed to be an artifact attributable to unspecific adsorption during preparation of the magnetosomes. This protein was observed to bind to PHB granules in vivo and to activate the PHB depolymerase of R. rubrum (PhaZ1) in vitro. Accordingly, the authors suggested renaming the Mms16 protein of Magnetospirillum species ApdA, the name used for the phasin from R. rubrum that activates depolymerization (39).
PhaPRe was shown to increase the activity of class II PHA synthases PhaC1 and PhaC2 from P. aeruginosa in vitro, activating PHA synthesis (40). Other phasins were also observed to affect PHA synthesis. For example, a mutant strain of Synechocystis sp. strain PCC 6803 that does not produce phasin PhaPSp showed reduced activity of the PHB synthase (12), and phasin PhaM from R. eutropha was described as the physiological activator of the PHB synthase in this microorganism (41).
The role of phasins from Aeromonas caviae (PhaPAc) and R. eutropha (PhaPRe) in the modulation of the activity of PHA synthase (PhaC) from different microorganisms was studied by in vitro polymerization assays. These experiments revealed that both phasins increased the activity of PhaC from A. caviae (PhaCAc), but they inhibited the activity of the PHB synthases from R. eutropha (PhaCRe) and Delftia acidovorans (PhaCDa) in vitro, despite the fact that the presence of PhaPAc increased in vivo polymer production in recombinant E. coli strains that express either phaCAc or phaCRe. These results led the authors to propose that there is PhaP-mediated PhaCAc activation at the molecular level (42). Another study performed using recombinant E. coli containing the PHA biosynthesis genes from R. eutropha revealed that the replacement of PhaPRe by PhaPAc affected the composition of the polymer accumulated from soybean oil, increasing the amount of 3-hydroxyhexanoate incorporated. These results suggested that the phasins affected both PHA synthase activity and its affinity for different monomers (43).
On the other hand, PhaFPp acts as a negative transcriptional regulator of PhaC in P. putida, as the disruption of phaF was observed to lead to higher expression levels of PhaC (10). Furthermore, this phasin binds to DNA nonspecifically through a histone-like domain and plays a key role in intracellular localization and equal distribution of PHA granules to daughter cells during cell division (19, 44). Last, PhaP from A. hydrophila (PhaPAh) was also observed to act as a positive regulator of the gene that encodes the PHA synthase in this organism, as the overexpression of phaP increased the expression of phaC (45).
All these results indicate that phasins have multiple and important roles in different aspects of PHA metabolism, ranging from their structural functions as components of the granule surface to regulatory roles affecting the expression and activity of different enzymes involved in PHA synthesis and depolymerization (Table 2).
TABLE 2.
Additional functions of phasins
| Phasin | Microorganism | Additional function(s)a | Reference(s) |
|---|---|---|---|
| PhaP1Re | Ralstonia eutropha | Increases specific activity of PhaCRe and the activity of PhaC1 and PhaC2 from Pseudomonas aeruginosa in vitro | 40 |
| Activates PHB degradation | 38 | ||
| PhaPAh | Aeromonas hydrophila | Activates transcription of phaCAh | 45 |
| PhaPSp | Synechocystis sp. PCC 6803 | Increases activity of PHB synthase in vivo | 12 |
| PhaM | Ralstonia eutropha | Physiological activator of PhaCRe | 41 |
| ApdA | Rhodospirillum rubrum | Stimulates in vitro depolymerization by PHB depolymerase of R. rubrum | 28 |
| Mms16 | Magnetospirillum gryphiswaldense | Activates PHB depolymerase from R. rubrum in vitro | 39 |
| PhbP | Azotobacter vinelandii | Increases alginate production | 11, 56, 57 |
| PhaPAc | Aeromonas caviae | Increases activity of PhaCAc but inhibits activity of PhaC from R. eutropha and Delftia acidovorans in vitro; increases the solubility of PhaCRe in vitro (chaperone-like activity) | 42 |
| PhaFPp | Pseudomonas putida | Binds to DNA through its histone-like domain in a nonspecific manner | 44 |
| Is involved in PHA granule segregation during cell division | 19 | ||
| PhaPAz | Azotobacter sp. FA8 | Protects E. coli from stress caused by PHB accumulation, heat, and paraquat; has in vitro and in vivo chaperone activity | 18 |
The abilities to bind to the PHA granule surface and to affect the number and size of the granules are considered general properties of all phasins and are not indicated in the table.
PROTECTIVE EFFECT AND CHAPERONE ACTIVITY OF PHASINS
Phasins constitute the major components of the protein layer that stabilizes PHA granules and enhances polymer synthesis (34). In natural polymer producers, phaP mutants contain one or a few large granules at the end of the accumulation phase, instead of the several small granules observed in the wild-type strains. This has been proposed to be due to the fact that naked granules exhibit a hydrophobic surface and are therefore not protected from coalescence (5, 9, 12, 27). The protective layer surrounding PHB granules might also prevent damage to cellular components due to interactions with the polymer (5).
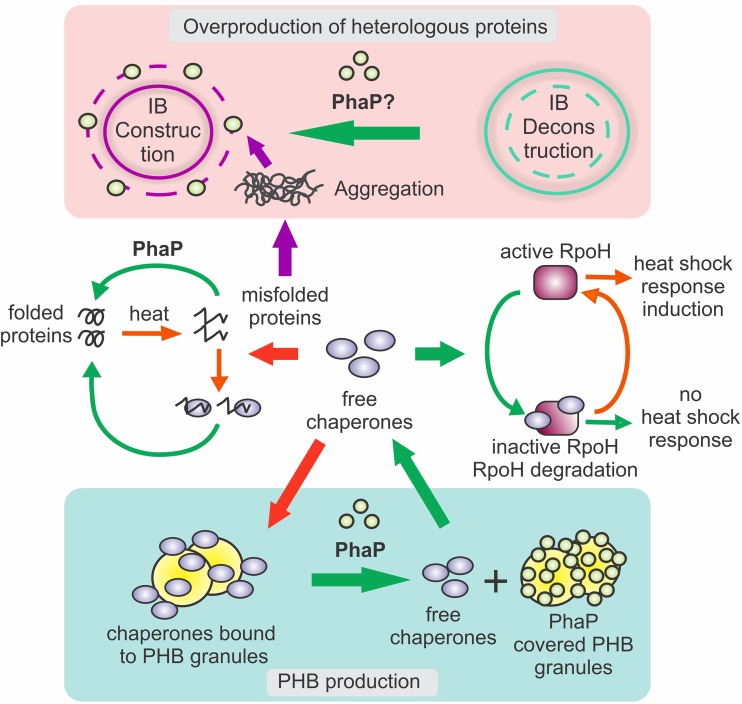
The beneficial effect of phasins on PHB production was also observed in E. coli recombinants, as the coexpression of phasins was observed to enhance growth and PHB accumulation (27, 31). PHB production was shown to cause stress in recombinant E. coli, evidenced by an increment in expression levels of chaperones, sigma factors, and other stress-related genes (46, 47). An analysis of the effect of PHB accumulation in a recombinant E. coli strain revealed that the presence of PhaPAz produces a dramatic reduction in the expression of stress-related genes, such as ibpA and dnaK, compared to the strain that does not synthesize the phasin (48). Unexpectedly, PhaPAz also had a protective effect in non-PHB-synthesizing E. coli strains under both normal and stress conditions, evidenced by a reduction in heat shock protein levels, increased growth, and higher resistance to both heat shock and superoxide stress by paraquat (48). Given that this strain does not accumulate PHB, the protective effect of the phasin could not be due to its capacity to protect cells from the consequences of polymer accumulation, and it reflected a more general protective role. The reduced heat shock protein transcription observed in strains expressing phaP, together with reduced expression of rpoH during PHB production and reduced RpoH protein levels during heat shock, suggested that PhaPAz might affect protein folding by exerting chaperone-like activity (48) (Fig. 1). PhaPAz was observed to prevent the spontaneous thermal aggregation of citrate synthase and facilitate its refolding after chemical denaturation, indicating that this phasin has in vitro chaperone activity. In addition, PhaPAz was observed to bind to inclusion bodies (IBs) formed by the heterologous protein PD, an insoluble domain of TolR from Azoarcus sp. strain CIB that aggregates when overexpressed in E. coli, and to play a role in the IB construction/deconstruction process by reducing the number and size of IBs in vivo (Fig. 1). Taken together, these results indicate that this phasin has in vivo chaperone activity (23).
FIG 1.
Proposed mechanism for the protective effect of phasins (based on results obtained with PhaP from Azotobacter sp. FA8 in E. coli). (Center) When misfolded proteins appear in the cell as a result of different kinds of stress, such as heat shock, production of PHB, or overproduction of heterologous proteins, chaperones bind to them, and RpoH freed from chaperones activates the heat shock response. PhaP, through its chaperone activity, exerts a protective effect that produces a reduction in the amount of misfolded proteins, diminishing the recruitment of chaperones, which are available to bind to RpoH, inactivating it. (Top) In cells that overproduce heterologous proteins, the expression of phaP reduces the number of IBs. This might occur through different mechanisms: PhaP might facilitate protein folding, preventing the aggregation of proteins in IBs; PhaP might release misfolded proteins from the IBs to the cytoplasm; or both mechanisms might occur. (Bottom) In PHB-producing cells, chaperones bind to PHB granules in the absence of phasins, reducing the pool of free chaperones, which leads to the induction of the heat shock response. On the contrary, when phaP is expressed, the presence of the phasin prevents the chaperones from binding to the granules and thus the induction of the heat shock response.
Evidence supporting the chaperone-like functions of other phasins was found while analyzing the interaction of PhaPAc and PhaPRe with different PHA polymerases. The presence of PhaPAc was observed to increase the solubility and activity of the synthase PhaCRe in in vitro assays, and this effect was attributed to a chaperone-like role of the phasin (42).
Phasin-associated phenotypes have been traditionally attributed to the capability of these proteins to form a protective layer surrounding PHB granules (5). The chaperone activity of phasins might provide an additional explanation for the positive effect of PhaP on polymer accumulation, as these proteins might help assemble the complex structure of PHB granules and achieve active PHA synthesis. Phasins might also facilitate the folding or prevent the degradation of proteins that are not involved in PHA metabolism, thus producing a general protective effect in PHA-producing cells.
As indicated in the previous section, phasins were shown to be involved in the synthesis (5) and degradation (28) of PHAs. Because of this close association, any possible effects due to the activity of phasins in natural PHA producers could be attributed to the polymer or to polymer-associated cellular components. In contrast, results obtained in a non-PHA-producing microorganism, like E. coli expressing phaPAz, have provided evidence that this phasin has a protective role that is independent from polymer metabolism (18).
PHAs are known to act as carbon and energy storage in natural producers, and the effect of PHAs in stress protection and survival has been traditionally attributed solely to the supply of energy and reducing power for stress response processes (49). However, the chaperone-like activities described for some phasins suggest that they might play an important role in stress protection, complementing the resources provided by polymer degradation with specific chaperone activities, thus participating actively in reducing the deleterious effects of different kinds of stress.
BIOTECHNOLOGICAL APPLICATIONS OF PHASINS
As indicated in the previous sections, phasins have been shown to enhance bacterial growth and polymer accumulation (31), and they have also been observed to affect polymer elongation (42) and even its composition (43). These properties highlight the importance of phasins in PHA production.
The ability of these proteins to bind to PHB granules has been exploited for a number of different applications apart from biopolymer production. For example, PhaPRe has been used to facilitate recombinant protein purification. Fusions of this protein to green fluorescent protein (GFP) and an intein, an autoclavable protein, were constructed. This fusion protein was able to bind to PHB granules. After cell lysis and differential centrifugation, this fusion protein could be obtained bound to purified PHB granules. Then, the addition of thiols activates the intein, resulting in the liberation of GFP. Similar studies have been performed with PHB-producing recombinant E. coli strains in order to produce different kinds of recombinant proteins (50, 51).
The N-terminal domain of phasin PhaFPp, denominated BioF, was used as a polypeptide tag to anchor fusion proteins to PHA granules. These granules could be easily purified by a simple centrifugation step and proteins subsequently released by treatment with mild detergents, maintaining their enzymatic activity (52). This method was proposed for the release of proteins to the environment, in particular for the liberation of the Cry1Ab toxin as an insecticide treatment (53).
Phasins were also used to develop a receptor-mediated drug delivery system using PHA nanoparticles. PhaPRe fused with different ligands was attached to PHA nanoparticles containing hydrophobic drugs, allowing the delivery of the drugs to target cells that have receptors recognized by the ligands (51). Phasins bound to PHB beads can also be easily fused to antigens for application in fluorescence-activated cell sorter (FACS) analysis (54).
Phasins were also proposed as biosurfactants because of their amphipathic nature, and PhaPAh was reported to be able to emulsify petrochemical and vegetable oils and diesel in vitro (55).
Last, the chaperone-like activity observed for PhaPAz (23), together with evidence that other phasins, such as PhaPAc (42), could have similar properties, suggests that phasins could be used to enhance the production of heterologous proteins in E. coli. These findings expand the already-wide field of potential applications for phasins.
CONCLUDING REMARKS
Phasins are proteins that were identified as major components of the PHA granule cover and found to promote growth and PHA synthesis and to affect the number, size, and distribution of the granules. Later studies revealed that they have a remarkable functional diversity, as different capabilities were described for different phasins. Apart from their PHA binding capability, phasins have been shown to interact with other proteins. Several phasins act as regulators, activating PHA depolymerization or increasing the expression and activity of PHA synthases. One phasin has been observed to exert a general protective effect in E. coli, both in the presence and in the absence of PHA, and has both in vivo and in vitro chaperone activity. Chaperone-like activities have also been proposed for other phasins in the context of PHA synthesis, and future work should help clarify whether these are common properties among phasins belonging to different groups. In natural producers, PHAs protect the cells against stress and contribute to their adaptability and survival in the environment. Although this effect has been attributed solely to the capacity of the polymer to provide carbon and energy for the different cellular processes, phasins might play an active role in reducing the deleterious effects of different kinds of stress, enhancing fitness and survival.
The amphiphilic nature of phasins, their PHA granule binding capacity, and their structural flexibility make them suitable for diverse biotechnological applications. Among the uses that have already been described are those in protein purification and drug delivery. The increasing evidence of the multiple functions and remarkable properties of these proteins opens the way for new possible applications.
REFERENCES
- 1.Keshavarz T, Roy I. 2010. Polyhydroxyalkanoates: bioplastics with a green agenda. Curr Opin Microbiol 13:321–326. doi: 10.1016/j.mib.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Rai R, Keshavarz T, Roether JA, Boccaccini AR, Roy I. 2011. Medium chain length polyhydroxyalkanoates, promising new biomedical materials for the future. Mater Sci Eng R 72:29–47. doi: 10.1016/j.mser.2010.11.002. [DOI] [Google Scholar]
- 3.Jendrossek D. 2009. Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J Bacteriol 191:3195–3202. doi: 10.1128/JB.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pieper-Fürst U, Madkour MH, Mayer F, Steinbüchel A. 1994. Purification and characterization of a 14-kilodalton protein that is bound to the surface of polyhydroxyalkanoic acid granules in Rhodococcus ruber. J Bacteriol 176:4328–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wieczorek R, Pries A, Steinbüchel A, Mayer F. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J Bacteriol 177:2425–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pötter M, Müller H, Reinecke F, Wieczorek R, Fricke F, Bowien B, Friedrich B, Steinbüchel A. 2004. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology 150:2301–2311. doi: 10.1099/mic.0.26970-0. [DOI] [PubMed] [Google Scholar]
- 7.Pfeiffer D, Jendrossek D. 2012. Localization of poly(3-hydroxybutyrate) (PHB) granule-associated proteins during PHB granule formation and identification of two new phasins, PhaP6 and PhaP7, in Ralstonia eutropha H16. J Bacteriol 194:5909–5921. doi: 10.1128/JB.00779-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCool GJ, Cannon MC. 1999. Polyhydroxyalkanoate inclusion body-associated proteins and coding region in Bacillus megaterium. J Bacteriol 181:585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maehara A, Ueda S, Nakano H, Yamane T. 1999. Analyses of a polyhydroxyalkanoic acid granule-associated 16-kilodalton protein and its putative regulator in the pha locus of Paracoccus denitrificans. J Bacteriol 181:2914–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prieto MA, Bühler B, Jung K, Witholt B, Kessler B. 1999. PhaF, a polyhydroxyalkanoate-granule-associated protein of Pseudomonas oleovorans GPo1 involved in the regulatory expression system for pha genes. J Bacteriol 181:858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peralta-Gil M, Segura D, Guzmán J, Servı L. 2002. Expression of the Azotobacter vinelandii poly-β-hydroxybutyrate biosynthetic phbBAC operon is driven by two overlapping promoters and is dependent on the transcriptional activator PhbR. J Bacteriol 184:5672–5677. doi: 10.1128/JB.184.20.5672-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauf W, Watzer B, Roos N, Klotz A, Forchhammer K. 2015. Photoautotrophic polyhydroxybutyrate granule formation is regulated by cyanobacterial phasin PhaP in Synechocystis sp. strain PCC 6803. Appl Environ Microbiol 81:4411–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai S, Cai L, Liu H, Liu X, Han J, Zhou J, Xiang H. 2012. Identification of the haloarchaeal phasin (PhaP) that functions in polyhydroxyalkanoate accumulation and granule formation in Haloferax mediterranei. Appl Environ Microbiol 78:1946–1952. doi: 10.1128/AEM.07114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hokamura A, Fujino K, Isoda Y, Arizono K, Shiratsuchi H, Hiromi M. 2015. Characterization and identification of the proteins bound to two types of polyhydroxyalkanoate granules in Pseudomonas sp. 61-3. Biosci Biotechnol Biochem 79:1369–1377. [DOI] [PubMed] [Google Scholar]
- 15.Ayub ND, Pettinari MJ, Méndez BS, López NI. 2007. The polyhydroxyalkanoate genes of a stress resistant Antarctic Pseudomonas are situated within a genomic island. Plasmid 58:240–248. doi: 10.1016/j.plasmid.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Manso Cobos I, Ibáñez García MI, de la Peña Moreno F, Sáez Melero LP, Luque-Almagro VM, Castillo Rodríguez F, Roldán Ruiz MD, Prieto Jiménez MA, Moreno Vivián C. 2015. Pseudomonas pseudoalcaligenes CECT5344, a cyanide-degrading bacterium with by-product (polyhydroxyalkanoates) formation capacity. Microb Cell Fact 14:771–712. doi: 10.1186/s12934-015-0267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann L, Spinozzi F, Sinibaldi R, Rustichelli F, Pötter M, Steinbüchel A. 2008. Binding of the major phasin, PhaP1, from Ralstonia eutropha H16 to poly(3-hydroxybutyrate) granules. J Bacteriol 190:2911–2919. doi: 10.1128/JB.01486-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mezzina MP, Wetzler DE, Catone MV, Bucci H, Di Paola M, Pettinari MJ. 2014. A phasin with many faces: structural insights on PhaP from Azotobacter sp. FA8. PLoS One 9:e103012. doi: 10.1371/journal.pone.0103012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maestro B, Galán B, Alfonso C, Rivas G, Prieto MA, Sanz JM. 2013. A new family of intrinsically disordered proteins: structural characterization of the major phasin PhaF from Pseudomonas putida KT2440. PLoS One 8:e56904. doi: 10.1371/journal.pone.0056904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao M, Li Z, Zheng W, Lou Z, Chen GQ. 2006. Crystallization and initial X-ray analysis of polyhydroxyalkanoate granule-associated protein from Aeromonas hydrophila. Acta Crystallogr Sect F Struct Biol Cryst Commun 62:814–819. doi: 10.1107/S1744309106025000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z, Dunker AK. 2007. DisProt: the Database of Disordered Proteins. Nucleic Acids Res 35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfeiffer D, Jendrossek D. 2013. Development of a transferable bimolecular fluorescence complementation system for the investigation of interactions between poly(3-hydroxybutyrate) granule-associated proteins in gram-negative bacteria. Appl Environ Microbiol 79:2989–2999. doi: 10.1128/AEM.03965-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mezzina MP, Wetzler DE, de Almeida A, Dinjaski N, Prieto MA, Pettinari MJ. 2015. A phasin with extra talents: a polyhydroxyalkanoate granule-associated protein has chaperone activity. Environ Microbiol 17:1765–1776. doi: 10.1111/1462-2920.12636. [DOI] [PubMed] [Google Scholar]
- 24.Kalscheuer R, Wältermann M, Manuel H, Alexander A. 2001. Preparative isolation of lipid inclusions from Rhodococcus opacus and Rhodococcus ruber and identification of granule-associated proteins. Arch Microbiol 177:20–28. doi: 10.1007/s00203-001-0355-5. [DOI] [PubMed] [Google Scholar]
- 25.Hanisch J, Waltermann M, Robenek H, Steinbüchel A. 2016. The Ralstonia eutropha H16 phasin PhaP1 is targeted to intracellular triacylglycerol inclusions in Rhodococcus opacus PD630 and Mycobacterium smegmatis mc2155, and provides an anchor to target other proteins. Microbiology 152:3271–3280. doi: 10.1099/mic.0.28969-0. [DOI] [PubMed] [Google Scholar]
- 26.Pfeiffer D, Jendrossek D. 2011. Interaction between poly(3-hydroxybutyrate) granule-associated proteins as revealed by two-hybrid analysis and identification of a new phasin in Ralstonia eutropha H16. Microbiology 157:2795–2807. doi: 10.1099/mic.0.051508-0. [DOI] [PubMed] [Google Scholar]
- 27.de Almeida A, Nikel PI, Giordano AM, Pettinari MJ. 2007. Effects of granule-associated protein PhaP on glycerol-dependent growth and polymer production in poly(3-hydroxybutyrate)-producing Escherichia coli. Appl Environ Microbiol 73:7912–7916. doi: 10.1128/AEM.01900-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Handrick R, Reinhardt S, Schultheiss D, Reichart T, Jendrossek V, Jendrossek D. 2004. Unraveling the function of the Rhodospirillum rubrum activator of polyhydroxybutyrate (PHB) degradation: the activator is a PHB-granule-bound protein (phasin). J Bacteriol 186:2466–2475. doi: 10.1128/JB.186.8.2466-2475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinbuchel A, Aerts K, Babel W, Follner C, Liebergesell M, Madkour MH, Mayer F, Pieper-Furst U, Pries A, Valentin HE. 1995. Considerations on the structure and biochemistry of bacterial polyhydroxyalkanoic acid inclusions. Can J Microbiol 41(Suppl 1):S94–S105. [DOI] [PubMed] [Google Scholar]
- 30.York GM, Junker BH, Stubbe JOA, Sinskey AJ. 2001. Accumulation of the PhaP phasin of Ralstonia eutropha is dependent on production of polyhydroxybutyrate in cells. J Bacteriol 183:4217–4226. doi: 10.1128/JB.183.14.4217-4226.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.York GM, Stubbe J, Sinskey AJ. 2001. New insight into the role of the PhaP phasin of Ralstonia eutropha in promoting synthesis of polyhydroxybutyrate. J Bacteriol 183:2394–2397. doi: 10.1128/JB.183.7.2394-2397.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.York GM, Stubbe J, Sinskey AJ. 2002. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J Bacteriol 184:59–66. doi: 10.1128/JB.184.1.59-66.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pötter M, Madkour MH, Mayer F, Steinbüchel A. 2002. Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology 148:2413–2426. doi: 10.1099/00221287-148-8-2413. [DOI] [PubMed] [Google Scholar]
- 34.Pötter M, Steinbüchel A. 2005. Poly(3-hydroxybutyrate) granule-associated proteins: impacts on poly(3-hydroxybutyrate) synthesis and degradation. Biomacromolecules 6:552–560. doi: 10.1021/bm049401n. [DOI] [PubMed] [Google Scholar]
- 35.Lee T-R, Lin J-S, Wang S-S, Shaw G-C. 2004. PhaQ, a new class of poly-β-hydroxybutyrate (PHB)-responsive repressor, regulates phaQ and phaP (phasin) expression in Bacillus megaterium through interaction with PHB. J Bacteriol 186:3015–3021. doi: 10.1128/JB.186.10.3015-3021.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai S, Cai L, Zhao D, Liu G, Han J, Zhou J, Xiang H. 2015. A novel DNA-binding protein, PhaR, plays a central role in the regulation of polyhydroxyalkanoate accumulation and granule formation in the haloarchaeon Haloferax mediterranei. Appl Environ Microbiol 81:373–385. doi: 10.1128/AEM.02878-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Handrick R, Technow U, Reichart T, Reinhardt S, Sander T, Jendrossek D. 2004. The activator of the Rhodospirillum rubrum PHB depolymerase is a polypeptide that is extremely resistant to high temperature (121°C) and other physical or chemical stresses. FEMS Microbiol Lett 230:265–274. doi: 10.1016/S0378-1097(03)00919-4. [DOI] [PubMed] [Google Scholar]
- 38.Kuchta K, Chi L, Fuchs H, Pötter M, Steinbüchel A. 2007. Studies on the influence of phasins on accumulation and degradation of PHB and nanostructure of PHB granules in Ralstonia eutropha H16. Biomacromolecules 8:657–662. doi: 10.1021/bm060912e. [DOI] [PubMed] [Google Scholar]
- 39.Schultheiss D, Handrick R, Jendrossek D, Hanzlik M, Schüler D. 2005. The presumptive magnetosome protein Mms16 is a poly(3-hydroxybutyrate) granule-bound protein (phasin) in Magnetospirillum gryphiswaldense. J Bacteriol 187:2416–2425. doi: 10.1128/JB.187.7.2416-2425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi Q, Steinbüchel A, Rehm BHA. 2000. In vitro synthesis of poly(3-hydroxydecanoate): purification and enzymatic characterization of type II polyhydroxyalkanoate synthases PhaC1 and PhaC2 from Pseudomonas aeruginosa. Appl Microbiol Biotechnol 54:37–43. doi: 10.1007/s002530000357. [DOI] [PubMed] [Google Scholar]
- 41.Pfeiffer D, Jendrossek D. 2014. PhaM is the physiological activator of poly(3-hydroxybutyrate) (PHB) synthase (PhaC1) in Ralstonia eutropha. Appl Environ Microbiol 80:555–563. doi: 10.1128/AEM.02935-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ushimaru K, Motoda Y, Numata K, Tsuge T. 2014. Phasin proteins activate Aeromonas caviae polyhydroxyalkanoate (PHA) synthase but not Ralstonia eutropha PHA synthase. Appl Environ Microbiol 80:2867–2873. doi: 10.1128/AEM.04179-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawashima Y, Orita I, Nakamura S, Fukui T. 2015. Compositional regulation of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) by replacement of granule-associated protein in Ralstonia eutropha. Microb Cell Fact 14:1–12. doi: 10.1186/s12934-014-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galán B, Dinjaski N, Maestro B, de Eugenio LI, Escapa IF, Sanz JM, García JL, Prieto MA. 2011. Nucleoid-associated PhaF phasin drives intracellular location and segregation of polyhydroxyalkanoate granules in Pseudomonas putida KT2442. Mol Microbiol 79:402–418. doi: 10.1111/j.1365-2958.2010.07450.x. [DOI] [PubMed] [Google Scholar]
- 45.Tian S-J, Lai W-J, Zheng Z, Wang H-X, Chen G-Q. 2005. Effect of over-expression of phasin gene from Aeromonas hydrophila on biosynthesis of copolyesters of 3-hydroxybutyrate and 3-hydroxyhexanoate. FEMS Microbiol Lett 244:19–25. doi: 10.1016/j.femsle.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 46.Han M, Yoon SS, Lee SY. 2001. Proteome analysis of metabolically engineered Escherichia coli producing poly(3-hydroxybutyrate). J Bacteriol 183:301–308. doi: 10.1128/JB.183.1.301-308.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han M, Park SJ, Lee JW, Min B, Lee SY, Kim S, Yoo JS. 2006. Analysis of poly(3-hydroxybutyrate) granule-associated proteome in recombinant Escherichia coli. J Microbiol Biotechnol 16:901–910. [Google Scholar]
- 48.de Almeida A, Catone MV, Rhodius VA, Gross CA, Pettinari MJ. 2011. Unexpected stress-reducing effect of PhaP, a poly(3-hydroxybutyrate) granule-associated protein, in Escherichia coli. Appl Environ Microbiol 77:6622–6629. doi: 10.1128/AEM.05469-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kadouri D, Jurkevitch E, Okon Y, Castro-Sowinski S. 2005. Ecological and agricultural significance of bacterial polyhydroxyalkanoates. Crit Rev Microbiol 31:55–67. doi: 10.1080/10408410590899228. [DOI] [PubMed] [Google Scholar]
- 50.Banki MR, Gerngross TU, Wood DW. 2005. Novel and economical purification of recombinant proteins: intein-mediated protein purification using in vivo polyhydroxybutyrate (PHB) matrix association. Protein Sci 14:1387–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao Y-C, Zhan X-Y, Zhang J, Zou X-H, Wang Z-H, Xiong Y-C, Chen J, Chen G-Q. 2008. A specific drug targeting system based on polyhydroxyalkanoate granule binding protein PhaP fused with targeted cell ligands. Biomaterials 29:4823–4830. doi: 10.1016/j.biomaterials.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 52.Moldes C, García P, García L, Prieto MA. 2004. In vivo immobilization of fusion proteins on bioplastics by the novel tag BioF. Appl Environ Microbiol 70:3205–3212. doi: 10.1128/AEM.70.6.3205-3212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moldes C, Farinós GP, de Eugenio LI, García P, García JL, Ortego F, Hernández-Crespo P, Castañera P, Prieto MA. 2006. New tool for spreading proteins to the environment: Cry1Ab toxin immobilized to bioplastics. Appl Microbiol Biotechnol 72:88–93. doi: 10.1007/s00253-005-0257-6. [DOI] [PubMed] [Google Scholar]
- 54.Bäckström BT, Brockelbank JA, Rehm BHA. 2007. Recombinant Escherichia coli produces tailor-made biopolyester granules for applications in fluorescence activated cell sorting: functional display of the mouse interleukin-2 and myelin oligodendrocyte glycoprotein. BMC Biotechnol 7:3. doi: 10.1186/1472-6750-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei D-X, Chen C-B, Fang G, Li S-Y, Chen G-Q. 2011. Application of polyhydroxyalkanoate binding protein PhaP as a bio-surfactant. Appl Microbiol Biotechnol 91:1037–1047. doi: 10.1007/s00253-011-3258-7. [DOI] [PubMed] [Google Scholar]
- 56.Segura D, Espín G. 2004. Inactivation of pycA, encoding pyruvate carboxylase activity, increases poly-beta-hydroxybutyrate accumulation in Azotobacter vinelandii on solid medium. Appl Microbiol Biotechnol 65:414–418. [DOI] [PubMed] [Google Scholar]
- 57.Segura D, Guzmán J, Espín G. 2003. Azotobacter vinelandii mutants that overproduce poly-beta-hydroxybutyrate or alginate. Appl Microbiol Biotechnol 63:159–163. [DOI] [PubMed] [Google Scholar]