ABSTRACT
Rhizobia are best known for nodulating legume roots and fixing atmospheric nitrogen for the host in exchange for photosynthates. However, the majority of the diverse strains of rhizobia do not form nodules on legumes, often because they lack key loci that are needed to induce nodulation. Nonnodulating rhizobia are robust heterotrophs that can persist in bulk soil, thrive in the rhizosphere, or colonize roots as endophytes, but their role in the legume-rhizobium mutualism remains unclear. Here, we investigated the effects of nonnodulating strains on the native Acmispon-Bradyrhizobium mutualism. To examine the effects on both host performance and symbiont fitness, we performed clonal inoculations of diverse nonnodulating Bradyrhizobium strains on Acmispon strigosus hosts and also coinoculated hosts with mixtures of sympatric nodulating and nonnodulating strains. In isolation, nonnodulating Bradyrhizobium strains did not affect plant performance. In most cases, coinoculation of nodulating and nonnodulating strains reduced host performance compared to that of hosts inoculated with only a symbiotic strain. However, coinoculation increased host performance only under one extreme experimental treatment. Nearly all estimates of nodulating strain fitness were reduced in the presence of nonnodulating strains. We discovered that nonnodulating strains were consistently capable of coinfecting legume nodules in the presence of nodulating strains but that the fitness effects of coinfection for hosts and symbionts were negligible. Our data suggest that nonnodulating strains most often attenuate the Acmispon-Bradyrhizobium mutualism and that this occurs via competitive interactions at the root-soil interface as opposed to in planta.
IMPORTANCE Rhizobia are soil bacteria best known for their capacity to form root nodules on legume plants and enhance plant growth through nitrogen fixation. Yet, most rhizobia in soil do not have this capacity, and their effects on this symbiosis are poorly understood. We investigated the effects of diverse nonnodulating rhizobia on a native legume-rhizobium symbiosis. Nonnodulating strains did not affect plant growth in isolation. However, compared to inoculations with symbiotic rhizobia, coinoculations of symbiotic and nonnodulating strains often reduced plant and symbiont fitness. Coinoculation increased host performance only under one extreme treatment. Nonnodulating strains also invaded nodule interiors in the presence of nodulating strains, but this did not affect the fitness of either partner. Our data suggest that nonnodulating strains may be important competitors at the root-soil interface and that their capacity to attenuate this symbiosis should be considered in efforts to use rhizobia as biofertilizers.
INTRODUCTION
Rhizobia are heterotrophic soil bacteria with diverse lifestyles. Some rhizobial lineages have acquired the capacity to form nodules on legume roots and fix atmospheric nitrogen for these hosts (1). Nodulating rhizobia are attracted to flavonoids released by legumes. In response, the rhizobia secrete nod factors that provoke morphological changes to the roots, enabling the bacteria to enter root cortical cells, become encased by a plant-derived membrane, differentiate into bacteroids, and fix nitrogen (2, 3). Among nodulating rhizobia, nodulation genes are typically carried on symbiosis plasmids (4, 5) or on a genomic island (the “symbiosis island”) (6–9). However, soil populations consistently include rhizobia that do not individually nodulate legume hosts (10–18), often because they lack key loci that are needed to induce nodulation (19, 20).
Rhizobial strains that nodulate host roots can have dramatic effects on legume fitness, but these nodulating symbionts must compete with other inhabitants of rhizosphere communities and their relative abundance compared to that of other microbes can vary (21). More specifically, the relative frequency of nonnodulating versus nodulating rhizobia also varies, but nonnodulating genotypes typically dominate and can encompass as much as 99% of the total rhizobial population (10–18). Nonnodulating strains can reduce the number of nodules formed by nodulating strains on legume hosts (22, 23) and can invade nodule tissues in the presence of closely related nodulating strains (24, 25). This suggests that they may be able to reduce nodulating strain fitness through competitive exclusion at the root surface. Nonnodulating strains have also been shown to promote plant growth on nonlegume hosts (26–28), but the direct effects of nonnodulating rhizobia on legume host performance remain unclear, either in isolation or when in competition with nodulating strains. Moreover, it is unknown whether nonnodulating rhizobia affect legume host performance, while in the rhizosphere or by gaining access to host resources in planta as endophytes.
Here, we investigated the effects of nonnodulating strains on native hosts and symbionts of the Acmispon-Bradyrhizobium mutualism in California. We inoculated Acmispon strigosus (formerly Lotus strigosus) hosts with sympatric Bradyrhizobium isolates to examine the effects of nonnodulating strains on both host and symbiont performance. Experimental treatments included clonal inoculations with either nonnodulating or nodulating strains, mixed inoculations of nodulating and nonnodulating strains, and inoculations with water as a control. Strain treatments were organized into 16 unique sympatric coinoculation strain pairs, with 8 pairs each sourced from independent host populations in Northern and Southern California. To investigate factors that might influence the fitness outcomes of interstrain competition and the ability of nonnodulating strains to coinfect legume nodules, coinoculation strain pairs varied in terms of genetic relatedness between competing strains and in terms of estimated abundance of each strain in the sampled populations (10, 14). We conducted separate experiments with different coinoculation ratios. One matched empirical estimates of nodulating versus nonnodulating strain abundance in the A. strigosus rhizosphere (10, 14). The other was extremely biased toward nonnodulating strains to maximize the potential for observing nodule coinfection and the effects of interstrain competition on modulating the benefits of the legume-rhizobium mutualism. Our goals in these experiments were to examine (i) the growth effects of nonnodulating strains on hosts in isolation, (ii) the effects of competing nonnodulating strains on host performance and nodulating symbiont fitness, and (iii) the genotype-specific effects of the nodulating versus nonnodulating strains in determining the fitness outcomes of interstrain competition.
MATERIALS AND METHODS
Selection of Bradyrhizobium strains and inbred Acmispon hosts.
Bradyrhizobium isolates were previously collected from the nodules and the soil-root interface of A. strigosus host plants at Bodega Marine Reserve (BMR) in Northern California and the University of California Riverside (UCR) in Southern California (10, 14). All isolates were previously tested for nodulation ability in greenhouse inoculation assays and were genotyped at multiple loci, including genes present on the chromosome (i.e., present in all Bradyrhizobium) and genes carried on the symbiosis island to confirm its presence or absence (10, 14). Strains for this study were chosen in order to examine the effects of (i) field site of origin (BMR versus UCR), since the sites varied in the relative frequencies of nodulating versus nonnodulating strains (10, 14), (ii) relatedness between competing strains (identity at chromosomal loci versus unrelated), and (iii) strain abundance of each strain tested in their sampled habitat (i.e., rare versus abundant) (10). Strains were also selected in order to vary antibiotic resistance profiles, which were used to identify coinfecting strains in vitro (29).
From each field site, 8 sympatric strains were chosen, composed of 3 nodulating and 5 nonnodulating strains, resulting in a total of 16 Bradyrhizobium strains (some strains were used in more than one coinoculation strain pair). Strains from each field site were grouped into 8 sympatric strain pairs to be experimentally coinoculated, each comprising one nodulating strain and one nonnodulating strain (Table 1). Since the primary focus was to investigate the effects of nonnodulating strains, we did not test coinoculation pairs containing only nodulating or only nonnodulating strains. The antibiotics used to differentiate nodulating and nonnodulating strains within each pair included chloramphenicol (100 μg/ml), carbenicillin (100 μg/ml), gentamicin (100 μg/ml), kanamycin (100 μg/ml), and streptomycin (100 μg/ml). Four of the strain pairs had identical genotypes at two chromosomal loci (recA and glnII) but differed in nodulation ability, modulated by the presence or absence of symbiosis island loci (10). The 12 remaining pairs consisted of diverged nodulating and nonnodulating strains that varied in the number of chromosomal single nucleotide polymorphisms (SNPs) and in local abundance (Table 1). Results of a pilot study found no evidence of horizontal gene transfer of the symbiosis loci between nodulating and nonnodulating strains; recovered nonnodulating genotypes were resequenced to confirm their identity, and PCR on these isolates consistently failed to amplify symbiosis loci (nolL and nodDA), suggesting that they did not incorporate the symbiosis island.
TABLE 1.
Summary of strain features and antibiotic resistancea
| Host population | Pair no. | No. of SNPs between strains | Strain | Genotype | Genotype abundance | Nodulation ability | Antibiotic resistanceb |
|---|---|---|---|---|---|---|---|
| BMR | 1 | 0 | 45 | G01_R01 | 24 | + | CHLr |
| 15 | − | CHLs | |||||
| 2 | 0 | 37 | G05_R02 | 24 | + | CHLs | |
| 80 | − | CHLr | |||||
| 3 | 83 | 49 | G03_R01 | 355 | + | GENr | |
| 1 | G17_R17 | 8 | − | GENs | |||
| 4 | 40 | 49 | G03_R01 | 355 | + | GENr | |
| 41 | G112_R09 | 1 | − | GENs | |||
| 5 | 58 | 49 | G03_R01 | 355 | + | STRs | |
| 64 | G16_R16 | 2 | − | STRr | |||
| 6 | 64 | 37 | G05_R02 | 24 | + | GENr | |
| 1 | G17_R17 | 8 | − | GENs | |||
| 7 | 17 | 37 | G05_R02 | 24 | + | GENr | |
| 41 | G112_R09 | 1 | − | GENs | |||
| 8 | 48 | 37 | G05_R02 | 24 | + | STRs | |
| 64 | G16_R16 | 2 | − | STRr | |||
| UCR | 9 | 0 | 107 | G03_R01 | 355 | + | GENr |
| 98 | − | GENs | |||||
| 10 | 0 | 131 | G11_R07 | 62 | + | CARr | |
| 110 | − | CARs | |||||
| 11 | 73 | 87 | G03_R01 | 355 | + | KANs | |
| 102 | G36_R35 | 20 | − | KANr | |||
| 12 | 108 | 87 | G03_R01 | 355 | + | GENr | |
| 109 | G42_R47 | 5 | − | GENs | |||
| 13 | 12 | 87 | G03_R01 | 355 | + | CARr | |
| 112 | G04_R21 | 1 | − | CARs | |||
| 14 | 77 | 131 | G11_R07 | 62 | + | KANs | |
| 102 | G36_R35 | 20 | − | KANr | |||
| 15 | 110 | 131 | G11_R07 | 62 | + | GENr | |
| 109 | G42_R47 | 5 | − | GENs | |||
| 16 | 12 | 131 | G11_R07 | 62 | + | GENr | |
| 112 | G04_R21 | 1 | − | GENs |
The numbers of SNPs between strains and genotype abundances were previously determined by Hollowell and colleagues (10).
CHL, chloramphenicol; GEN, gentamicin, STR, streptomycin; CAR, carbenicillin; KAN, kanamycin.
Inbred lines of A. strigosus were generated from each field site following published protocols (14), except that seedlings were transplanted into 1-gallon pots with enriched soil (UCR number 3 soil). Plants were grown for 5 months (19 November 2013 to 17 April 2014) in UCR greenhouse 11 (33.972798, −117.323548), and fruits were picked as they developed (∼1,500 seeds per plant). No supplemental lighting was used to alter day length. We chose one inbred line of hosts per site for inoculation (BMR04.02; UCR09.03). All Bradyrhizobium strains were inoculated onto sympatric hosts.
Preparation of Bradyrhizobium inocula.
Each Bradyrhizobium strain was initiated from ∼2 μl of frozen stock and streaked onto plates with modified arabinose gluconate medium (MAG) (14). A single colony of each strain was spread onto 5 MAG plates and incubated until lawns formed (29°C, ∼8 days). Bacteria were scraped from each plate and resuspended in liquid MAG, and concentrations were estimated via optical density (30). The resuspended cells were centrifuged (4,000 rpm, 20 min) to remove media and resuspended again in sterile double-distilled water (ddH2O) at 108 cells ml−1.
Inoculation experiments.
Seed preparation and planting followed previously published methods (14). Inoculated plants received a total of 5 × 108 rhizobial cells in 5 ml of ddH2O (equivalent by mass to ∼106 cells g−1 soil), which is higher than most estimates of natural rhizobial soil populations (up to 105 nodulating cells g−1 soil) (31–33), but compensates for rhizobial attrition that occurs during the stressful inoculation process (10, 14, 20, 30, 34–36).
Two separate experiments were conducted with different coinoculation ratios. The “ecological experiment” used coinoculation ratios that matched the empirical population estimates of nodulating versus nonnodulating strain abundance in A. strigosus rhizospheres (i.e., 1:3 at BMR; 1:95 at UCR) (10, 14). The “extreme experiment” used a coinoculation ratio of 1:500, nodulating to nonnodulating rhizobia, for both host population sources to maximize the potential for competition and nodule coinfection by nonnodulating rhizobia.
For each host population, axenic A. strigosus seedlings were separately arranged by size and size-matched seedlings were randomly assigned to sympatric inoculation treatments and greenhouse locations. For each coinoculation pair, bacterial treatments consisted of (i) clonal inoculation of the nodulating strain, (ii) clonal inoculation of the nonnodulating strain, (iii) coinoculation of both strains, and (iv) inoculation with water (ecological experiment: 4 treatments per pair × 16 strain pairs × 4 replicate plants per treatment combination × 2 harvest points, 256 plants per host population; extreme experiment: 4 treatments per pair × 16 strain pairs × 4 replicate plants per treatment combination, 128 plants per host population). Plants were inoculated on 3 October 2014 (BMR) and 9 October 2014 (UCR).
Harvest and coinfection analysis.
During harvest, plants were removed from the pots, and soil was separated from the roots by washing with tap water. Nodules were dissected, counted, and photographed. Roots, shoots, and nodules were separated and oven dried (60°C, >4 days) prior to weighing. We harvested all plants prior to flower formation since this is when nodule senescence in A. strigosus often begins in the greenhouse (i.e., around 9 weeks postinoculation, but this can range from 8 to 24 weeks in the field depending on rainfall) (20). Additionally, results of a pilot study suggested that coinfection varied with plant developmental stage. To maximize the potential to observe coinfection, half of the plants from each host population in the ecological experiment were harvested 4 weeks after inoculation (n = 128 per host population) and the remaining half at 8 weeks (n = 128 per host population). All plants in the extreme experiment from both host populations were harvested 6 weeks after inoculation (n = 128 per host population). Therefore, at each harvest point, 16 plants per coinoculation pair (4 plants per inoculation treatment) were harvested.
The frequencies of coinfected nodules and the relative proportion of each rhizobial strain within the nodules were estimated for each coinoculation strain pair at each harvest point. Two (of 4) plants were randomly selected for nodule culturing for each strain pair and harvest week (n = 96). For each sampled plant, 4 randomly selected nodules were chosen for bacterial culturing. We cultured bacteria from a total of 24 nodules per coinoculation strain, 8 at each harvest week, from 96 test plants (n = 384). The proportion of plants per treatment and the number of nodules per plant selected for culturing were chosen in order to complete harvests in a timely manner and to be consistent with those in our previous studies (30, 35). Nodules were surface sterilized following previously described methods, crushed, and spread onto 3 MAG plates (30). To estimate the relative proportions of the nodulating and nonnodulating strains within the nodule, 100 randomly selected colonies were replica plated onto MAG plates containing the appropriate antibiotic with plain MAG plates as controls for growth (Table 1; detection limit of nonnodulating strains of 1%) (30, 35). If fewer than 100 colonies were present, they were all tested for resistance traits.
To confirm that all plated colonies came from the internal portions of the nodules, nodule surface sterilization efficiency was confirmed experimentally. Briefly, 8 A. strigosus nodules (collected from 3 separate plants) were dissected from plant roots, and each unsterilized nodule was individually rolled over 1 MAG plate using a sterile loop. Nodules were then surfaced sterilized in undiluted bleach (6% sodium hypochlorite) for 2 min, rinsed 3 times in sterile water, and subsequently rolled over a second MAG plate to confirm the absence of surface contaminants. Nodules were then crushed using a sterile glass rod, and bacteria were plated onto a third MAG plate to confirm rhizobial viability within each surface sterilized nodule. Original surface contaminants were present on all nodules tested (i.e., growth on the first MAG plate) and effectively removed in all cases (i.e., no growth on the second MAG plate). Rhizobial viability was confirmed in 7 (of 8) nodules (i.e., growth on the third MAG plate).
Data analysis.
We used shoot biomass as our primary estimate of plant performance, which is the most commonly reported plant fitness component (37). We used nodule number and mass as proxies for nodulating strain fitness in our experiments. Previous work by Sachs and colleagues (20) demonstrated that both of these parameters are positively correlated with beneficial rhizobial population sizes in A. strigosus, similar to results for other systems (38–41).
The effects of inoculation treatments on host performance and nodulating strain fitness were analyzed separately for each host population at each harvest week using one-way analyses of variance (ANOVAs) (df = 3) (42). ANOVAs with significant F ratio statistics were followed by pairwise t tests (42) to test for differences among treatments. To examine the effects of nonnodulating strains on host performance, hosts receiving clonal inoculations of nonnodulating strains were compared to uninoculated controls. To test if competing nonnodulating strains altered host and symbiont fitness during symbiosis, host performance and symbiont fitness were compared between clonal inoculations of nodulating strains and coinoculation treatments. Net fitness effects were determined by combining plant data from all pairs within an inoculation treatment for each host population and harvest week (n = 128). Effects within each pair were analyzed using plants only from each respective pair for each host population and harvest week (n = 16).
Nodules and plants were scored as coinfected if ≥1 replica plated colonies were identified as a nonnodulating strain. Although several factors can influence the ability of a nonnodulating strain to coinfect legume nodules, the primary objectives in our coinfection analyses were to (i) estimate the coinfection ability of each nonnodulating strain, (ii) detect any patterns in coinfection ability, and (iii) determine if there are any host performance costs to coinfection. Thus, the coinfection ability for each nonnodulating strain was assessed using data from all harvest weeks and coinoculation ratios where each nonnodulating strain was present in the inoculum. Since half of the coinoculated plants were selected for culturing and bacteria from 4 nodules per plant were replica plated, we regard our estimates of coinfection as conservative.
Strain variations in the capacity to coinfect nodules or block coinfection were analyzed using one-way ANOVAs followed by Tukey's honestly significant difference (HSD) test among nonnodulating and nodulating strains, respectively. To test if the proportion of coinfection for chromosomally identical pairs differed from that for chromosomally diverged pairs (0, >10 SNPs) we used a generalized linear mixed model with genetic divergence (identical or diverged) as a fixed effect and coinoculation strain pair as a random effect (fit model platform in JMP 10.0). To examine if the coinfection frequency scales with the genetic distance, a correlation analysis was performed between the proportion of nodules coinfected per plant and the number of SNPs between each coinoculation strain pair.
To examine the effects of coinfection, host performance and symbiont fitness were compared using one-way ANOVAs between coinoculated plants without evidence of coinfection and coinfected plants (where nodule subculturing data were available) separately for each host population and harvest week (df = 1; n = 16).
RESULTS
Effects of nonnodulating strains on hosts in isolation.
None of the nonnodulating strains formed nodules or any detectable features on roots when inoculated in isolation (see Table S1 in the supplemental material). In no case did clonal inoculation with a nonnodulating strain affect host growth compared to that of uninoculated control plants (Table 2).
TABLE 2.
Effects of nonnodulating strains in altering host and symbiont fitnessa
| Host population and pair |
F ratio for ecological experiment, wk 8 |
F ratio for extreme experiment, wk 6 |
||||
|---|---|---|---|---|---|---|
| Shoot biomass | Nodule no. | Total nodule biomass | Shoot biomass | Nodule no. | Total nodule biomass | |
| BMR | ||||||
| Netb | 119.12***c | 185.36***c | 69.36*** | 44.73***d | 164.25***c | 90.50*** |
| 1 | 20.52*** | 32.76***c | 49.79***c | 3.67* | 15.09** | 4.40* |
| 2 | 12.67** | 13.72** | 6.46** | 3.16 | 15.87**c | 9.94**c |
| 3 | 4.60* | 16.79*** | 9.71** | 2.41 | 11.69** | 32.78*** |
| 4 | 38.16*** | 22.72*** | 31.91*** | 10.64** | 20.28*** | 11.86** |
| 5 | 25.34*** | 18.72*** | 18.23*** | 2.49 | 33.81*** | 14.10** |
| 6 | 12.78** | 15.79** | 14.16** | 6.69** | 34.16*** | 8.97** |
| 7 | 43.93*** | 43.50*** | 29.75*** | 9.13** | 27.94***c | 15.20** |
| 8 | 12.95** | 13.26** | 4.33* | 8.14**c | 19.91*** | 18.68*** |
| UCR | ||||||
| Nete | 130.82***c | 192.01***c | 140.82***c | 77.23***c | 191.68***c | 62.75***c |
| 9 | 29.34***c | 267.68***c | 32.22***c | 12.82** | 13.50** | 5.91* |
| 10 | 42.92*** | 28.26*** | 144.57*** | 8.27** | 34.76*** | 10.94** |
| 11 | 18.27***c | 18.87*** | 29.10*** | 13.67** | 95.90***c | 9.46** |
| 12 | 15.64***c | 22.29***c | 10.67**c | 8.39** | 13.79** | 12.84** |
| 13 | 17.38***c | 64.27***c | 18.78***c | 7.29** | 14.01**c | 6.17** |
| 14 | 23.98*** | 52.08***c | 33.12***c | 7.86** | 87.32***c | 12.34** |
| 15 | 6.52** | 13.36** | 10.25** | 26.54***c | 37.96***c | 56.20***c |
| 16 | 6.06** | 6.23** | 5.45* | 19.04***c | 18.37*** | 8.819** |
F ratio statistics are reported from one-way ANOVAs comparing effects of inoculation treatments (df = 3) by harvest week and net source host population (n = 128) and within coinoculation (n = 16). Asterisks indicate significant F ratio statistics (*, P < 0.05; **, P < 0.01; ***, P < 0.0001). To determine significant differences among inoculation treatments, indicated in boldface type, ANOVAs were followed by pairwise t tests comparing inoculation treatments (P < 0.05).
Refers to plants from all pairs sourced from BMR merged within treatment.
Refers to cases where the clonal nodulating treatment results are significantly higher than the coinoculation treatment results.
Refers to cases where the clonal nodulating treatment results are significantly lower than the coinoculation treatment results.
Refers to plants from all pairs sourced from UCR merged within treatment.
Effects of competing nonnodulating strains on host growth and symbiont fitness.
In the ecological experiment, coinoculation of nodulating and nonnodulating strains reduced host growth and measures of nodulating strain fitness in both host populations. For the BMR population, the coinoculated treatments reduced net host growth by ∼15%, and net nodulating strain fitness was significantly decreased in terms of total nodule number (∼18% reduction), but net total nodule biomass was not significantly affected (Fig. 1a; Table 2). Nodulating strain fitness proxies, but not host growth, varied based on strain combinations for BMR hosts (Table 2). For the UCR population, net host performance was significantly reduced by ∼28% in coinoculated treatments. Net nodulating strain fitness was also decreased in coinoculated treatments at UCR, with ∼29% and ∼32% reductions in total nodule number and total nodule biomass, respectively (Fig. 1a; Table 2). Host performance and nodulating strain fitness proxies varied based on strain combinations for UCR hosts (Table 2). The mean numbers of nodules per plant, per strain pair, and per harvest week are reported in Table S2 in the supplemental material.
FIG 1.
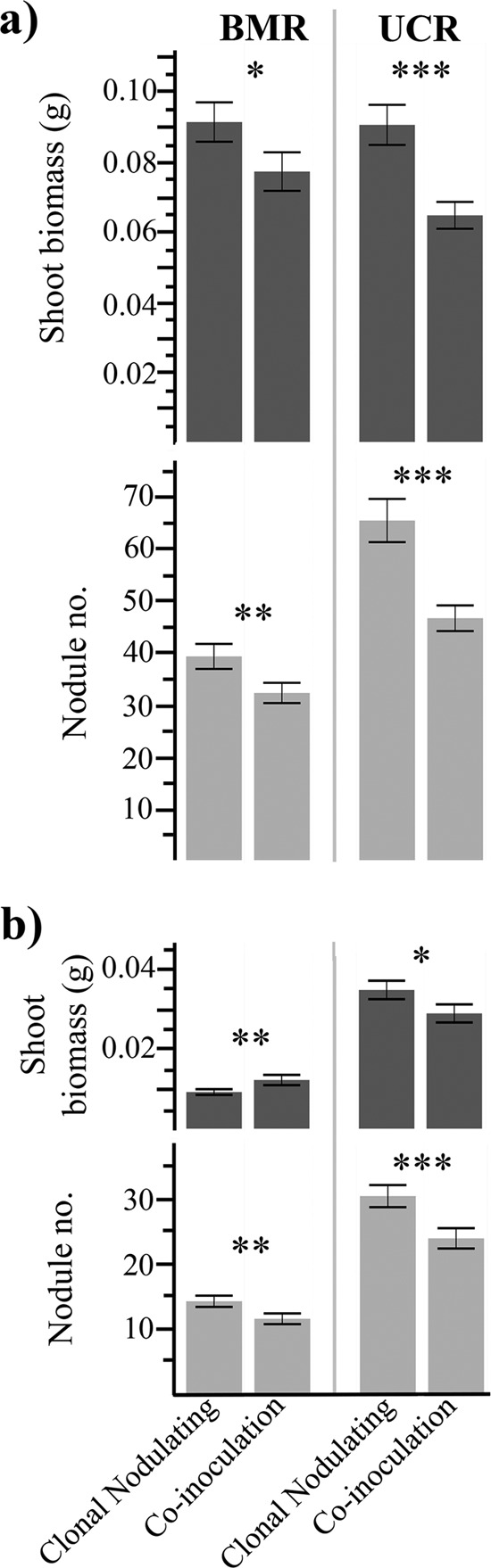
Coinoculation alters host growth and symbiont fitness. Shoot biomass and total nodule number data were merged within treatment from all pairs from the same host population. (a) Shoot biomass and the total nodule number from each host population in the ecological ratio experiment 8 weeks postinoculation. (b) Shoot biomass and the total nodule number from each host population in the extreme ratio experiment 6 weeks postinoculation. Asterisks denote significant differences between net clonal inoculations of nodulating strains and net coinoculations with nonnodulating strains within each host population (one-way ANOVAs: ***, P < 0.001; **, P < 0.01; *, P < 0.05). Error bars represent 1 standard error.
Extreme coinoculation ratios of nodulating to nonnodulating strains resulted in various effects for each host population. Net BMR host growth was significantly increased by ∼24% under extreme coinoculation conditions compared to that of clonally inoculated plants, yet the net total nodule number was reduced by ∼19% (Fig. 1b; Table 2). Similar to inoculation with an ecologically relevant ratio, net total nodule biomass was not significantly different among treatments (Table 2). All metrics of net host growth and nodulating strain fitness were significantly decreased in the UCR hosts (Fig. 1b; Table 2). Coinoculation with an extreme ratio of nonnodulating strains resulted in an ∼17% reduction in net host growth, an ∼21% reduction in net total number of nodules, and an ∼23% reduction in net total nodule biomass (Fig. 1b; Table 2). Host growth and nodulating strain fitness proxies varied based on strain combinations for both host populations under an extreme coinoculation ratio (Table 2).
Effects of nonnodulating strain coinfection on host and symbiont fitness.
Data from 296 nodules from which nodule occupancy was successfully estimated by subculturing were analyzed (Table 1; see also Table S1 in supplemental material). The coinfection ability and estimated within-nodule population proportion of each nonnodulating strain were determined using data from all harvest weeks and coinoculation ratios. All nonnodulating strains were able to colonize the nodule tissue of at least one A. strigosus nodule, and evidence of coinfection was uncovered in all coinoculation pair combinations tested, except for pairs 6, 8, and 15 (Fig. 2 and 3a). Nonnodulating strains exhibited variations in both their ability to coinfect A. strigosus nodules and their within-nodule population estimates (Fig. 3a and b). However, the capacity to coinfect nodules was not correlated with strain genotype abundance (R2 = 0.0015, P = 0.7081).
FIG 2.
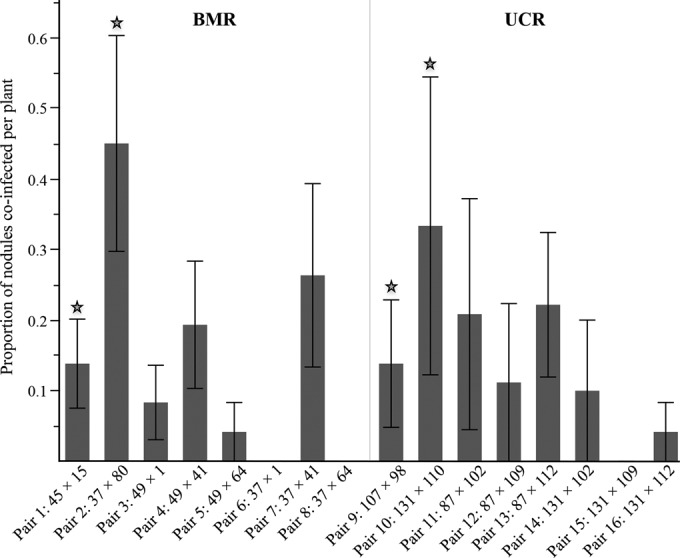
Proportion of coinfected nodules by strain combination. Coinfection proportions for each strain pair were averaged across all harvest weeks and inoculation ratios within each source host population. Stars represent coinoculation pairs where strains are genetically identical at the chromosomal level. Error bars represent 1 standard error.
FIG 3.
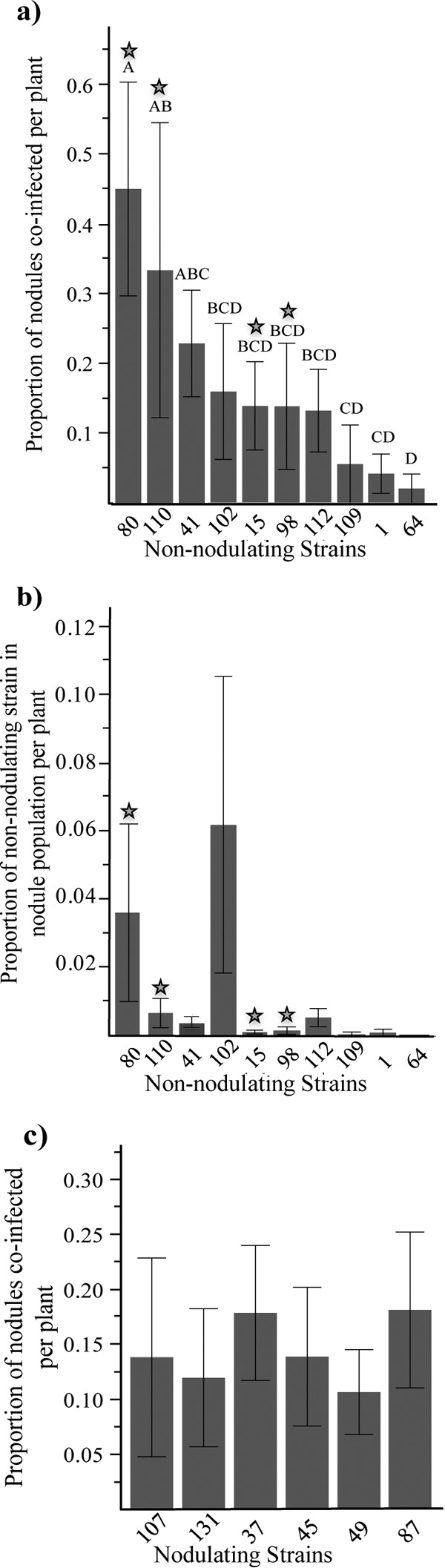
Proportion of coinfected nodules by individual strain and within-nodule proportion estimates. Coinfection proportions were averaged for each strain using data from all strain combinations. Stars represent nonnodulating strains that are genetically identical to coinoculated nodulating strains at the chromosomal level. (a) Proportion of coinfected nodules for each nonnodulating strain. Letters are significant differences among nonnodulating strains (Tukey's HSD test, P < 0.05). (b) Mean proportion of each nonnodulating strain within the total nodule population per plant. (c) Proportion of coinfected nodules for each nodulating strain. Error bars represent 1 standard error.
Nonnodulating strains that were paired with genetically identical nodulating strains (strains 15, 80, 98, and 110) coinfected significantly more nodules per plant than nonnodulating strains in genetically diverged coinoculation pairs (generalized linear mixed model: F1,96 = 6.21, P = 0.0258 and SNP correlation analysis R2 = −0.0719, P = 0.0086, n = 96) (Fig. 2 and 3a). However, we did not find any significant differences in terms of estimated within-nodule populations (Fig. 3b). None of the nodulating strains were able to prevent coinfection, and the proportion of coinfected nodules per plant was not significantly different among nodulating strains (Fig. 3c).
No significant effects of coinfection were found on host or nodulating strain fitness in either host population in the ecological and extreme ratio experiments (Table 3). Coinfection was reduced over time in the ecological experiment in both host populations (harvested at 4 and 8 weeks), although this trend was not significant for BMR host plants (Fig. 4).
TABLE 3.
Effects of coinfection on host and symbiont fitnessa
| Experiment and host population | Harvest wk |
F ratio |
|||
|---|---|---|---|---|---|
| Shoot biomass | Root biomass | Nodule no. | Total nodule biomass | ||
| Ecological | |||||
| BMRb | 4 | 0.001 | 0.182 | 0.187 | 0.568 |
| 8 | 0.005 | 0.090 | 0.404 | 1.341 | |
| UCRc | 4 | 0.020 | 0.066 | 0.021 | 0.845 |
| 8 | 0.223 | 0.039 | 2.505 | 0.302 | |
| Extreme | |||||
| BMRb | 6 | 4.090 | 0.656 | 0.024 | 1.280 |
| UCRc | 6 | 0.192 | 0.385 | 0.301 | 1.553 |
F ratio statistics are reported from one-way ANOVAs comparing effects of coinfection (df = 1) by harvest week and net source host population (n = 16).
Refers to plants from all pairs sourced from BMR merged within treatment.
Refers to plants from all pairs sourced from UCR merged within treatment.
FIG 4.
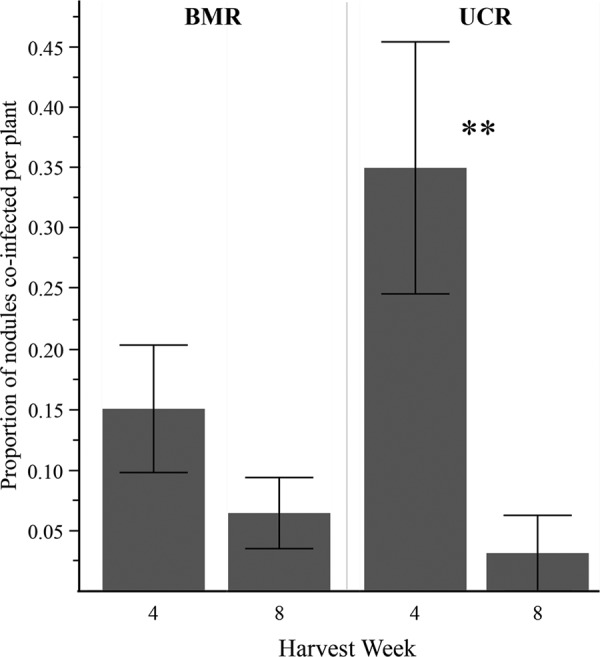
Proportion of coinfection in the ecological ratio experiment at 4 and 8 weeks postinoculation for each host population. The proportion of nodules coinfected per plant was calculated by averaging all coinoculated treatment plants from all pairs in the same host population (one-way ANOVA: **, P < 0.01). Error bars represent 1 standard error.
DISCUSSION
Rhizobia are increasingly being understood to have multifarious lifestyles, including root-nodule symbiosis, colonization of plant roots in the rhizosphere or as root endophytes, and independent growth in the soil or other habitats (24–28, 32). Yet, these lifestyles can be transient and are only partially dependent on the presence or absence of symbiosis loci, which have been a major focus of research. Nodulating strains with the canonical nodulation loci, for instance, are not capable of forming nodules on all hosts (e.g., host specificity) (14, 43, 44). Furthermore, nonnodulating strains (lacking key nodulation genes) can coinfect legume nodules, expropriating a symbiotic role (24, 25). Regardless of what factors determine lifestyle, strains that do not form nodules comprise the majority of rhizobial populations that have been sampled (10–18). Our data here suggest that nonnodulating rhizobia are not passive players in the host rhizosphere. Instead, nonnodulating rhizobia can have considerable effects on the legume-rhizobium symbiosis, most often by reducing plant performance and attenuating nodulating symbiont fitness.
Our results from clonal inoculations corroborate the results from previous work, which found no effect of nonnodulating strains on legume host growth in isolation (14, 25, 45). However, under parameters that model relative abundances of nonnodulating strains within the rhizosphere, our data set revealed substantial costs to host growth and symbiont fitness in both host populations examined (except for total nodule biomass at BMR) (Fig. 1a; Table 2). Our data are consistent with the results of previous reports of a reduction in nodulating strain fitness, measured by numbers of nodules formed by nodulating strains (22, 23), but reveal that this does not always result in a significant decrease in all rhizobial fitness estimates (i.e., total nodule biomass). The higher ratio of nonnodulating strains in ecologically relevant UCR coinocula might explain this difference and further suggests a competitive role for nonnodulating strains at the root-soil interface, but we are unable to disentangle the effects of coinoculation ratios from any differences due to host genotype.
The host growth responses to extreme ratios of nonnodulating strains differed between host populations. Coinoculation resulted in a significant increase in net BMR host growth (although this trend was only significant for strain pair 8) (Table 2), yet reductions in nodule numbers and effects on total biomass were similar compared to ecologically relevant ratios (Fig. 1a and b; Table 2). Legumes have finely tuned mechanisms to regulate nodule numbers (46), but the number of nodules formed in any interaction is nonetheless a product of the host and rhizobium genotypes (38, 47). This suggests that host control over nodule numbers is incomplete (34). Just as ineffective rhizobia form many nodules on hosts without benefiting the host (30), effective rhizobium strains might often produce more nodules on a host than is optimal for host growth. Thus, the reduction of nodule numbers by nonnodulating strains might actually increase host growth if the nodulation strains present are prolific nodule producers. However, this hypothesis cannot be explicitly tested with our data since there was no significant variation in nodule numbers for BMR populations. All estimates of host performance and nodulating strain fitness were reduced under extreme coinoculation conditions at UCR (Fig. 1b; Table 2), although they were not as pronounced as those for coinoculations with ecologically relevant ratios. Differences in host growth among treatments are more distinct as plants approach flowering (closer to the harvest at 8 weeks in the ecological experiment); thus, the magnitudes in the reductions in host performance and nodulating strain fitness might have been more comparable if the extreme experiment was harvested at a later date.
Legume nodules can harbor multiple lineages of bacteria (48–56), yet few studies have considered the capacity of rhizobial strains lacking nodulation loci to invade and persist within nodule tissue (but see references 24 and 25). To our knowledge, ours is the first study to explore the potential for coinfection using native combinations of strains on sympatric hosts. All of the nonnodulating strains that we tested were able to coinfect nodules (Fig. 3a and b), and this was true in nearly every strain combination tested (Fig. 2). This suggests that coinfection with nonnodulating strains is likely to be at least as common as coinfection with nonrhizobial bacteria. These data lend support to past reports of rhizobia that were isolated from legume nodules but were subsequently found to be unable to form nodules in inoculation tests (57, 58). Our data also imply that the coinfection ability can vary, depending on both rhizobial and plant factors. First, the estimated natural abundance of nonnodulating strains did not appear to impact coinfection ability, but genetic relatedness between strains did have a significant effect. The proportion of coinfected nodules was increased in strain pairs that were more closely related (0 SNPs) (Table 1), compared to strain pairs that were more distantly related (>10 SNPs) (Table 1). One possible explanation for this result is that the similarity of critical signaling molecules during root colonization is more important than abundance for nonnodulating strains (e.g., exopolysaccharides [EPS]) (25). Recently Zgadzaj and colleagues (25) found symbiotic rhizobia with compatible EPS had an advantage over coinfecting endophytes with different EPS molecules. In Bradyrhizobium, EPS genes are chromosomally carried (not within the symbiosis island) (8). Hence, it is possible that chromosomally identical (i.e., 0 SNPs) coinoculation pairs make similar, if not identical EPS, explaining the higher coinfection rates than for strain pairs that are genetically unrelated (i.e., >10 SNPs). Second, evidence of coinfection decreased over time since inoculation (Fig. 4). Previous work has shown that legume hosts can actively sanction ineffective rhizobial strains (nonfixing), reducing nodule growth rate and within-nodule rhizobial population sizes (30, 35, 40, 59). Since the presence of nonnodulating strains within legume nodules did not increase host growth in this study (Table 3), we can consider infections by nonnodulating strains to be similar to ineffective infections. Sanction mechanisms could be one reason for the observed decline of coinfection over time, but we are unable to discern if the host is controlling nonnodulating strain population sizes via sanctions or if nonnodulating strains are poorly adapted to survival and proliferation within the nodule environment. Last, while coinfection is prevalent, the lack of any measurable effects on host growth suggests that it might not play a critical role in terms of host fitness (Table 3).
Theoretical and empirical work on the legume-rhizobium symbiosis has generally assumed that legume fitness is predominately regulated by which rhizobial strains successfully nodulate host roots (20, 30, 35, 40, 59–62). Investigations have sought to uncover the mechanisms of competition among nodulating strains of various levels of symbiotic effectiveness (i.e., nitrogen fixation capacity) and to understand how the outcomes of this competition affect host fitness. Although effective nodulating strains can be competitive for nodulation (37, 63, 64), competitive ability may not be correlated with beneficial quality (41, 61, 65–67). Researchers attempting to apply highly effective rhizobial strains to improve legume crop commonly find that these strains nodulate hosts at low rates and that the inoculant strains get outcompeted by less efficient symbionts (60–62, 68). Despite the prevalence and dominance of nonnodulating strains (10–18), such studies have neglected the impact of endemic nonnodulating strains as potential negative competitors on the mutualism. Our work illustrates that endemic nonnodulating rhizobial strains often coinfect legume nodules and, more importantly, may play an active role in modulating the legume-rhizobium mutualism. Our results also show that nonnodulating rhizobia lack effects on host growth in isolation and during nodule coinfection, suggesting that the key fitness effects of nonnodulating strains are mediated by interstrain competition at the root-soil interface before nodulation occurs. Further research is necessary to understand the specific mechanisms of interstrain competition within the microbiota of the rhizosphere, but the overall competitive effects of nonnodulating rhizobial strains and other nonnodulating rhizosphere microbes should be promptly considered both in bioinoculant development and in research.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the help of Jonathan Lyu and Eunice Adinata in creating inbred plant lines. We thank the U.C. Natural Reserve System and Bodega Marine Reserve.
We declare no conflicts of interest.
This research was supported by NSF awards 0816663 and 1150278 to J.L.S. and the University of California Riverside's Graduate Research Mentorship Fellowship and Department of Biology Newell Award to K.A.G.-C.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01116-16.
REFERENCES
- 1.Sawada H, Kuykendall LD, Young JM. 2003. Changing concepts in the systematics of bacterial nitrogen-fixing legume symbionts. J Gen Appl Microbiol 49:155–179. doi: 10.2323/jgam.49.155. [DOI] [PubMed] [Google Scholar]
- 2.Lodwig EM, Hosie AH, Bourdès A, Findlay K, Allaway D, Karunakaran R, Downie J, Poole PS. 2003. Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis. Nature 422:722–726. doi: 10.1038/nature01527. [DOI] [PubMed] [Google Scholar]
- 3.Sprent JI, Sutherland J, De Faria S, Dilworth M, Corby H, Becking J, Materon L, Drozd J. 1987. Some aspects of the biology of nitrogen-fixing organisms. Philos Trans R Soc Lond B Biol Sci 317:111–129. doi: 10.1098/rstb.1987.0051. [DOI] [Google Scholar]
- 4.Galibert F, Finan TM, Long SR, Pühler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 5.Young JPW, Crossman LC, Johnston AW, Thomson NR, Ghazoui ZF, Hull KH, Wexler M, Curson AR, Todd JD, Poole PS. 2006. The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol 7:R34. doi: 10.1186/gb-2006-7-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Göttfert M, Röthlisberger S, Kündig C, Beck C, Marty R, Hennecke H. 2001. Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J Bacteriol 183:1405–1412. doi: 10.1128/JB.183.4.1405-1412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res 7:331–338. doi: 10.1093/dnares/7.6.331. [DOI] [PubMed] [Google Scholar]
- 8.Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res 9:189–197. doi: 10.1093/dnares/9.6.189. [DOI] [PubMed] [Google Scholar]
- 9.Lee K-B, De Backer P, Aono T, Liu C-T, Suzuki S, Suzuki T, Kaneko T, Yamada M, Tabata S, Kupfer DM. 2008. The genome of the versatile nitrogen fixer Azorhizobium caulinodans ORS571. BMC Genomics 9:271. doi: 10.1186/1471-2164-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollowell AC, Regus JU, Gano KA, Bantay R, Centeno D, Pham J, Lyu JY, Moore D, Bernardo A, Lopez G, Patil A, Patel S, Lii Y, Sachs JL. 2016. Epidemic spread of symbiotic and non-symbiotic Bradyrhizobium genotypes across California. Microb Ecol 71:700–710. doi: 10.1007/s00248-015-0685-5. [DOI] [PubMed] [Google Scholar]
- 11.Jarvis B, Ward L, Slade E. 1989. Expression by soil bacteria of nodulation genes from Rhizobium leguminosarum biovar trifolii. Appl Environ Microbiol 55:1426–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laguerre G, Bardin M, Amarger N. 1993. Isolation from soil of symbiotic and nonsymbiotic Rhizobium leguminosarum by DNA hybridization. Can J Microbiol 39:1142–1149. doi: 10.1139/m93-172. [DOI] [Google Scholar]
- 13.Pongsilp N, Teaumroong N, Nuntagij A, Boonkerd N, Sadowsky MJ. 2002. Genetic structure of indigenous non-nodulating and nodulating populations of Bradyrhizobium in soils from Thailand. Symbiosis 33:39–58. [Google Scholar]
- 14.Sachs JL, Kembel SW, Lau AH, Simms EL. 2009. In situ phylogenetic structure and diversity of Wild Bradyrhizobium communities. Appl Environ Microbiol 75:4727–4735. doi: 10.1128/AEM.00667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segovia L, Pinero D, Palacios R, Martinez-Romero E. 1991. Genetic structure of a soil population of nonsymbiotic Rhizobium leguminosarum. Appl Environ Microbiol 57:426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan JT, Eardly BD, Van Berkum P, Ronson CW. 1996. Four unnamed species of nonsymbiotic rhizobia isolated from the rhizosphere of Lotus corniculatus. Appl Environ Microbiol 62:2818–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sullivan JT, Patrick HN, Lowther WL, Scott DB, Ronson CW. 1995. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc Natl Acad Sci U S A 92:8985–8989. doi: 10.1073/pnas.92.19.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.VanInsberghe D, Maas KR, Cardenas E, Strachan CR, Hallam SJ, Mohn WW. 2015. Non-symbiotic Bradyrhizobium ecotypes dominate North American forest soils. ISME J 9:2435–2441. doi: 10.1038/ismej.2015.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okubo T, Tsukui T, Maita H, Okamoto S, Oshima K, Fujisawa T, Saito A, Futamata H, Hattori R, Shimomura Y, Haruta S, Morimoto S, Wang Y, Sakai Y, Hattori M, Aizawa S-i, Nagashima KVP, Masuda S, Hattori T, Yamashita A, Bao Z, Hayatsu M, Kajiya-Kanegae H, Yoshinaga I, Sakamoto K, Toyota K, Nakao M, Kohara M, Anda M, Niwa R, Jung-Hwan P, Sameshima-Saito R, Tokuda S-i, Yamamoto S, Yamamoto S, Yokoyama T, Akutsu T, Nakamura Y, Nakahira-Yanaka Y, Takada Hoshino Y, Hirakawa H, Mitsui H, Terasawa K, Itakura M, Sato S, Ikeda-Ohtsubo W, Sakakura N, Kaminuma E, Minamisawa K. 2012. Complete genome sequence of Bradyrhizobium sp. S23321: insights into symbiosis evolution in soil oligotrophs. Microbes Environ 27:306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sachs JL, Ehinger MO, Simms EL. 2010. Origins of cheating and loss of symbiosis in wild Bradyrhizobium. J Evol Biol 23:1075–1089. doi: 10.1111/j.1420-9101.2010.01980.x. [DOI] [PubMed] [Google Scholar]
- 21.Miethling R, Wieland G, Backhaus H, Tebbe C. 2000. Variation of microbial rhizosphere communities in response to crop species, soil origin, and inoculation with Sinorhizobium meliloti L33. Microb Ecol 40:43–56. doi: 10.1007/s002480000021. [DOI] [PubMed] [Google Scholar]
- 22.Singh I, Ahmad M. 1991. Competitive interaction between non-nodulating and nodulating strains for nodulation of cowpea (Vigna unguiculata). FEMS Microbiol Lett 81:157–160. doi: 10.1111/j.1574-6968.1991.tb04739.x. [DOI] [Google Scholar]
- 23.Winarno R, Lie T. 1979. Competition between Rhizobium strains in nodule formation: interaction between nodulating and non-nodulating strains. Plant Soil 51:135–142. doi: 10.1007/BF02205933. [DOI] [Google Scholar]
- 24.Pandya M, Kumar GN, Rajkumar S. 2013. Invasion of rhizobial infection thread by non-rhizobia for colonization of Vigna radiata root nodules. FEMS Microbiol Lett 348:58–65. doi: 10.1111/1574-6968.12245. [DOI] [PubMed] [Google Scholar]
- 25.Zgadzaj R, James EK, Kelly S, Kawaharada Y, de Jonge N, Jensen DB, Madsen LH, Radutoiu S. 2015. A legume genetic framework controls infection of nodules by symbiotic and endophytic bacteria. PLoS Genet 11:e1005280. doi: 10.1371/journal.pgen.1005280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaintreuil C, Giraud E, Prin Y, Lorquin J, Bâ A, Gillis M, de Lajudie P, Dreyfus B. 2000. Photosynthetic bradyrhizobia are natural endophytes of the African wild rice Oryza breviligulata. Appl Environ Microbiol 66:5437–5447. doi: 10.1128/AEM.66.12.5437-5447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yanni YG, Rizk R, Corich V, Squartini A, Ninke K, Philip-Hollingsworth S, Orgambide G, De Bruijn F, Stoltzfus J, Buckley D. 1997. Natural endophytic association between Rhizobium leguminosarum bv. trifolii and rice roots and assessment of its potential to promote rice growth. Plant Soil 194:99–114. doi: 10.1023/A:1004269902246. [DOI] [Google Scholar]
- 28.Yanni YG, Rizk RY, El-Fattah FKA, Squartini A, Corich V, Giacomini A, de Bruijn F, Rademaker J, Maya-Flores J, Ostrom P. 2001. The beneficial plant growth-promoting association of Rhizobium leguminosarum bv. trifolii with rice roots. Funct Plant Biol 28:845–870. doi: 10.1071/PP01069. [DOI] [Google Scholar]
- 29.Hollowell AC, Gano KA, Lopez G, Shahin K, Regus JU, Gleason N, Graeter S, Pahua V, Sachs JL. 2015. Native California soils are selective reservoirs for multidrug-resistant bacteria. Environ Microbiol Rep 7:442–449. doi: 10.1111/1758-2229.12269. [DOI] [PubMed] [Google Scholar]
- 30.Sachs JL, Russell JE, Lii YE, Black KC, Lopez G, Patil AS. 2010. Host control over infection and proliferation of a cheater symbiont. J Evol Biol 23:1919–1927. doi: 10.1111/j.1420-9101.2010.02056.x. [DOI] [PubMed] [Google Scholar]
- 31.Abaidoo R, Keyser H, Singleton P, Dashiell K, Sanginga N. 2007. Population size, distribution, and symbiotic characteristics of indigenous Bradyrhizobium spp. that nodulate TGx soybean genotypes in Africa. Appl Soil Ecol 35:57–67. doi: 10.1016/j.apsoil.2006.05.006. [DOI] [Google Scholar]
- 32.Denison RF, Kiers ET. 2004. Lifestyle alternatives for rhizobia: mutualism, parasitism, and forgoing symbiosis. FEMS Microbiol Lett 237:187–193. doi: 10.1111/j.1574-6968.2004.tb09695.x. [DOI] [PubMed] [Google Scholar]
- 33.Hirsch PR. 1996. Population dynamics of indigenous and genetically modified rhizobia in the field. New Phytol 133:159–171. doi: 10.1111/j.1469-8137.1996.tb04351.x. [DOI] [Google Scholar]
- 34.Regus J, Gano K, Hollowell A, Sofish V, Sachs J. 2015. Lotus hosts delimit the mutualism-parasitism continuum of Bradyrhizobium. J Evol Biol 28:447–456. doi: 10.1111/jeb.12579. [DOI] [PubMed] [Google Scholar]
- 35.Regus JU, Gano KA, Hollowell AC, Sachs JL. 2014. Efficiency of partner choice and sanctions in Lotus is not altered by nitrogen fertilization. Proc R Soc B 281:20132587. doi: 10.1098/rspb.2013.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sachs JL, Russell JE, Hollowell AC. 2011. Evolutionary instability of symbiotic function in Bradyrhizobium japonicum. PLoS One 6:e26370. doi: 10.1371/journal.pone.0026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friesen ML. 2012. Widespread fitness alignment in the legume-rhizobium symbiosis. New Phytol 194:1096–1111. doi: 10.1111/j.1469-8137.2012.04099.x. [DOI] [PubMed] [Google Scholar]
- 38.Heath KD, Tiffin P. 2007. Context dependence in the coevolution of plant and rhizobial mutualists. Proc R Soc B 274:1905–1912. doi: 10.1098/rspb.2007.0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heath KD, Tiffin P. 2009. Stabilizing mechanisms in a legume-rhizobium mutualism. Evolution 63:652–662. doi: 10.1111/j.1558-5646.2008.00582.x. [DOI] [PubMed] [Google Scholar]
- 40.Kiers ET, Rousseau RA, West SA, Denison RF. 2003. Host sanctions and the legume-rhizobium mutualism. Nature 425:78–81. doi: 10.1038/nature01931. [DOI] [PubMed] [Google Scholar]
- 41.Simms EL, Taylor DL, Povich J, Shefferson RP, Sachs J, Urbina M, Tausczik Y. 2006. An empirical test of partner choice mechanisms in a wild legume-rhizobium interaction. Proc R Soc B 273:77–81. doi: 10.1098/rspb.2005.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.SAS Institute, Inc. 2012. Using JMP 10. SAS Institute, Inc., Cary, NC. [Google Scholar]
- 43.Mpepereki S, Wollum IIA, Makonese F. 1996. Diversity in symbiotic specificity of cowpea rhizobia indigenous to Zimbabwean soils. Plant Soil 186:167–171. doi: 10.1007/BF00035071. [DOI] [Google Scholar]
- 44.Turk D, Keyser HH. 1992. Rhizobia that nodulate tree legumes: specificity of the host for nodulation and effectiveness. Can J Microbiol 38:451–460. doi: 10.1139/m92-076. [DOI] [Google Scholar]
- 45.Li L, Sinkko H, Montonen L, Wei G, Lindström K, Räsänen LA. 2012. Biogeography of symbiotic and other endophytic bacteria isolated from medicinal Glycyrrhiza species in China. FEMS Microbiol Ecol 79:46–68. doi: 10.1111/j.1574-6941.2011.01198.x. [DOI] [PubMed] [Google Scholar]
- 46.Ferguson BJ, Indrasumunar A, Hayashi S, Lin MH, Lin YH, Reid DE, Gresshoff PM. 2010. Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol 52:61–76. doi: 10.1111/j.1744-7909.2010.00899.x. [DOI] [PubMed] [Google Scholar]
- 47.Porter SS, Simms EL. 2014. Selection for cheating across disparate environments in the legume-rhizobium mutualism. Ecol Lett 17:1121–1129. doi: 10.1111/ele.12318. [DOI] [PubMed] [Google Scholar]
- 48.Bai Y, Zhou X, Smith DL. 2003. Enhanced soybean plant growth resulting from coinoculation of strains with Bradyrhizobium japonicum. Crop Sci 43:1774–1781. doi: 10.2135/cropsci2003.1774. [DOI] [Google Scholar]
- 49.de Lajudie P, Willems A, Nick G, Mohamed SH, Torck U, Coopman R, Filali-Maltouf A, Kersters K, Dreyfus B, Lindström K. 1999. Agrobacterium bv. 1 strains isolated from nodules of tropical legumes. Syst Appl Microbiol 22:119–132. doi: 10.1016/S0723-2020(99)80035-6. [DOI] [Google Scholar]
- 50.Mhamdi R, Mrabet M, Laguerre G, Tiwari R, Aouani ME. 2005. Colonization of Phaseolus vulgaris nodules by Agrobacterium-like strains. Can J Microbiol 51:105–111. doi: 10.1139/w04-120. [DOI] [PubMed] [Google Scholar]
- 51.Mrabet M, Mnasri B, Romdhane SB, Laguerre G, Aouani ME, Mhamdi R. 2006. Agrobacterium strains isolated from root nodules of common bean specifically reduce nodulation by Rhizobium gallicum. FEMS Microbiol Ecol 56:304–309. doi: 10.1111/j.1574-6941.2006.00069.x. [DOI] [PubMed] [Google Scholar]
- 52.Muresu R, Polone E, Sulas L, Baldan B, Tondello A, Delogu G, Cappuccinelli P, Alberghini S, Benhizia Y, Benhizia H, Benguedouar A, Mori B, Calamassi R, Dazzo FB, Squartini A. 2008. Coexistence of predominantly nonculturable rhizobia with diverse, endophytic bacterial taxa within nodules of wild legumes. FEMS Microbiol Ecol 63:383–400. doi: 10.1111/j.1574-6941.2007.00424.x. [DOI] [PubMed] [Google Scholar]
- 53.Philipson M, Blair ID. 1957. Bacteria in clover root tissue. Can J Microbiol 3:125–129. doi: 10.1139/m57-016. [DOI] [Google Scholar]
- 54.Sturz A, Christie B, Matheson B, Nowak J. 1997. Biodiversity of endophytic bacteria which colonize red clover nodules, roots, stems and foliage and their influence on host growth. Biol Fert Soils 25:13–19. doi: 10.1007/s003740050273. [DOI] [Google Scholar]
- 55.Tokala RK, Strap JL, Jung CM, Crawford DL, Salove MH, Deobald LA, Bailey JF, Morra M. 2002. Novel plant-microbe rhizosphere interaction involving Streptomyces lydicus WYEC108 and the pea plant (Pisum sativum). Appl Environ Microbiol 68:2161–2171. doi: 10.1128/AEM.68.5.2161-2171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zakhia F, Jeder H, Willems A, Gillis M, Dreyfus B, De Lajudie P. 2006. Diverse bacteria associated with root nodules of spontaneous legumes in Tunisia and first report for nifH-like gene within the genera Microbacterium and Starkeya. Microb Ecol 51:375–393. doi: 10.1007/s00248-006-9025-0. [DOI] [PubMed] [Google Scholar]
- 57.Rangin C, Brunel B, Cleyet-Marel J-C, Perrineau M-M, Béna G. 2008. Effects of Medicago truncatula genetic diversity, rhizobial competition, and strain effectiveness on the diversity of a natural sinorhizobium species community. Appl Environ Microbiol 74:5653–5661. doi: 10.1128/AEM.01107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu LJ, Wang HQ, Wang ET, Chen WX, Tian CF. 2011. Genetic diversity of nodulating and non-nodulating rhizobia associated with wild soybean (Glycine soja Sieb. & Zucc.) in different ecoregions of China. FEMS Microbiol Ecol 76:439–450. doi: 10.1111/j.1574-6941.2011.01064.x. [DOI] [PubMed] [Google Scholar]
- 59.Denison RF. 2000. Legume sanctions and the evolution of symbiotic cooperation by rhizobia. Am Nat 156:567–576. doi: 10.1086/316994. [DOI] [PubMed] [Google Scholar]
- 60.Schumpp O, Deakin WJ. 2010. How inefficient rhizobia prolong their existence within nodules. Trends Plant Sci 15:189–195. doi: 10.1016/j.tplants.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 61.Triplett EW, Sadowsky MJ. 1992. Genetics of competition for nodulation of legumes. Annu Rev Microbiol 46:399–422. doi: 10.1146/annurev.mi.46.100192.002151. [DOI] [PubMed] [Google Scholar]
- 62.Yates RJ, Howieson JG, Reeve WG, O'Hara GW. 2011. A re-appraisal of the biology and terminology describing rhizobial strain success in nodule occupancy of legumes in agriculture. Plant Soil 348:255–267. doi: 10.1007/s11104-011-0971-z. [DOI] [Google Scholar]
- 63.Yates R, Howieson J, Real D, Reeve W, Vivas-Marfisi A, O'Hara G. 2005. Evidence of selection for effective nodulation in the Trifolium spp. symbiosis with Rhizobium leguminosarum biovar trifolii. Anim Prod Sci 45:189–198. doi: 10.1071/EA03168. [DOI] [Google Scholar]
- 64.Yates R, Howieson J, Reeve W, Brau L, Speijers J, Nandasena K, Real D, Sezmis E, O'Hara G. 2008. Host-strain mediated selection for an effective nitrogen-fixing symbiosis between Trifolium spp. and Rhizobium leguminosarum biovar trifolii. Soil Biol Biochem 40:822–833. doi: 10.1016/j.soilbio.2007.11.001. [DOI] [Google Scholar]
- 65.Bloem FJ, Law JI. 2001. Determination of competitive abilities of Bradyrhizobium japonicum strains in soils from soybean production regions in South Africa. Biol Fert Soils 33:181–189. doi: 10.1007/s003740000303. [DOI] [Google Scholar]
- 66.Hafeez F, Hameed S, Ahmad T, Malik K. 2001. Competition between effective and less effective strains of Bradyrhizobium spp. for nodulation on Vigna radiata. Biol Fert Soils 33:382–386. doi: 10.1007/s003740000337. [DOI] [Google Scholar]
- 67.Vásquez-Arroyo J, Sessitsch A, Martínez E, Peña-Cabriales JJ. 1998. Nitrogen fixation and nodule occupancy by native strains of Rhizobium on different cultivars of common bean (Phaseolus vulgaris L.). Plant Soil 204:147–154. doi: 10.1023/A:1004399531966. [DOI] [Google Scholar]
- 68.Den Herder G, Parniske M. 2009. The unbearable naivety of legumes in symbiosis. Curr Opin Plant Biol 12:491–499. doi: 10.1016/j.pbi.2009.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


