ABSTRACT
We present the complete genome sequences of four members of a novel group of phages infecting Streptococcus thermophilus, designated here as the 987 group. Members of this phage group appear to have resulted from genetic exchange events, as evidenced by their “hybrid” genomic architecture, exhibiting DNA sequence relatedness to the morphogenesis modules of certain P335 group Lactococcus lactis phages and to the replication modules of S. thermophilus phages. All four identified members of the 987 phage group were shown to elicit adsorption affinity to both their cognate S. thermophilus hosts and a particular L. lactis starter strain. The receptor binding protein of one of these phages (as a representative of this novel group) was defined using an adsorption inhibition assay. The emergence of a novel phage group infecting S. thermophilus highlights the continuous need for phage monitoring and development of new phage control measures.
IMPORTANCE Phage predation of S. thermophilus is an important issue for the dairy industry, where viral contamination can lead to fermentation inefficiency or complete fermentation failure. Genome information and phage-host interaction studies of S. thermophilus phages, particularly those emerging in the marketplace, are an important part of limiting the detrimental impact of these viruses in the dairy environment.
INTRODUCTION
Streptococcus thermophilus is a globally employed dairy bacterium used in the production of a variety of cheeses and yoghurt. Having been safely consumed by humans for millennia, this bacterium is now a mainstay of the dairy industry due its favorable acidification and texturizing properties (1, 2). Despite advances in the available knowledge regarding dairy phage containment (3, 4) and S. thermophilus phage genetics and biology (5, 6), contamination of dairy production lines by S. thermophilus-infecting (bacterio)phages remains a persistent problem (for a review, see reference 7).
Classification of phages of S. thermophilus (reviewed by Mahony and van Sinderen [8]) has long been based on (i) morphology, i.e., as Siphoviridae, corresponding to group B as defined by Bradley (9), and (ii) a combination of the mode of DNA packaging (i.e., cos or pac site-containing) and major structural protein content (10). A variable genomic region thought to be (at least in part) responsible for host determination (VR2 region [11]) can also be used to categorize the majority of isolated S. thermophilus phages (12). More recently, however, a morphologically distinct and genetically divergent S. thermophilus phage named 5093, containing neither cos- or pac-defining structural elements nor a confirmed antireceptor-encoding gene, was described (13), prompting the creation of a third S. thermophilus phage group (here termed the “5093 group”). The genomic content of phage 5093 (containing several genes of nondairy streptococcal phage origin) highlights the genetic plasticity of S. thermophilus phages, thus explaining the appearance of such diverse phage lineages.
A total of 13 complete genome sequences of S. thermophilus-infecting phages have been published to date, with a large degree of conservation observed within the defined groupings. Phage groups have been defined as follows: (i) cos site-containing, with members Sfi19 and Sfi21 (lytic and temperate [14]), DT1 (lytic [15]), 7201 (lytic [16]), and Abc2 (lytic [6]); (ii) pac site-containing phages O1205 (temperate [17]), Sfi11 (lytic [18]), 2972 (lytic [19]), 858 (lytic [20]), ALQ13.2 (lytic [6]), and TP-J34 and TP-778L (temperate [5]); and (iii) the 5093 group archetype 5093 (lytic [13]).
Whole-genome sequencing of S. thermophilus-infecting phages has enabled their genome-wide, nucleotide level comparison and elucidation of their putative mechanisms of evolution. It was postulated (18) that the main modes of S. thermophilus phage evolution are represented by the rearrangement (or recombination) of discrete genomic modules, as well as by insertions, deletions, and point mutations—of which the latter is likely to function as a means to evade active clustered regularly interspaced short palindromic repeat (CRISPR) systems of their hosts (20). Consistent monitoring of phage populations in dairy plants in this manner is necessary to ensure that adequate knowledge-based rotational schemes are in place so as to avoid fermentation inconsistencies or even complete failure. This must initially include host sensitivity profiling and phage typing studies, yet may be extended to whole-phage-genome sequencing in the case of newly emerging groups and/or persistent or highly virulent phages.
Here, we present the complete genome sequences of four novel phages capable of infecting S. thermophilus ST64987, an industrial dairy starter strain. The 987 group phages were categorized as novel based on their recalcitrance to typing using a previously designed multiplex PCR protocol, their distinct morphology, and finally their genetic content, which differed from those of previously described groups of S. thermophilus phages. Comparative genomic analysis was performed for all four phages. The structural protein complement of one representative phage of this group was confirmed by mass spectrometry. The phages were further characterized by microscopic analysis and adsorption analyses, and the antireceptor of one phage was defined (as a representative) using an adsorption inhibition assay.
MATERIALS AND METHODS
Bacteriophage isolation, propagation, enumeration, and storage.
Bacterial strains were routinely grown from single colonies or reconstituted skimmed milk (RSM) stocks overnight at 30°C (Lactococcus lactis) or 42°C (S. thermophilus) in M17 broth (Oxoid, Hampshire, United Kingdom) containing 0.5% glucose (GM17; Sigma-Aldrich, St. Louis, MO, USA) for L. lactis or lactose (LM17; Sigma-Aldrich) for S. thermophilus. Phage enumeration was performed based on standard spot or plaque assay methods (21) in which LM17 broth was supplemented with 0.25% glycine (Oxoid), 10 mM CaCl2 (Oxoid), and either 10 g/liter (solid agar base) or 4 g/liter (semisolid overlay) technical agar (Merck, Darmstadt, Germany). Industrially derived whey samples from dairy plants producing fermented milk products (such as cheeses and yoghurts) were obtained (stored at −20°C) and analyzed for the presence of phages against S. thermophilus using the spot and plaque assay methods mentioned above. These samples now form part of the DSM phage collection (Delft, Netherlands). Single plaque isolates were then propagated according to the method of Moineau et al. (22) in LM17 at 42°C. The lysed culture was filtered (0.45 μm; Sarstedt, Nümbrecht, Germany) and stored at 4°C for use in subsequent assays. Single plaque isolation and propagation were performed at least twice to ensure the purity of phage preparations.
Bacteriophage purification and DNA preparation.
Individual phages were propagated in a 2-liter volume before concentration by polyethylene glycol (PEG) 8000 (Sigma-Aldrich) precipitation and purification using a discontinuous cesium chloride (CsCl; Sigma-Aldrich) block gradient as described by Sambrook et al. (23), using a Beckman 50 Ti rotor (Beckman Coulter, Brea, CA, USA). Phage DNA was prepared using a method adapted from Moineau et al. (22) and Sambrook et al. (23). Briefly, 20 μl proteinase K (20 mg/ml; Fisher Scientific, Waltham, MA, USA) was added to 500 μl of CsCl purified phage, and the mixture was heated at 56°C for 20 min. A sodium dodecyl sulfate solution (SDS; Sigma-Aldrich) was then added to a final concentration of 1.5% before heating at 65°C for 30 min. Potassium acetate was added to a final concentration of 1 M, and the mixture was placed on ice for 30 min. Centrifugation at 13,200 × g for 10 min was followed by two phenol-chloroform-isoamyl alcohol (25:24:1; Sigma-Aldrich) extractions and the addition of 0.1 volume of 3 M sodium acetate (pH 4.8; Lancaster Synthesis, Ward Hill, MA, USA) and 2.5 volumes of ice-cold 96% ethanol. Precipitated phage DNA was pelleted at 21,000 × g for 15 min and resuspended in 50 μl Tris-EDTA (TE) buffer (10 mM Tris-HCl, 1 mM EDTA [Sigma-Aldrich]; pH 7.5). Phage DNA was visualized on 1% agarose (Sigma-Aldrich) gels stained with Midori Green Advance DNA stain (Nippon Genetics Europe GmbH, Dueren, Germany) using the method of Sambrook et al. (23).
DNA sequencing and in silico analysis.
Approximately 20 μg phage DNA was extracted and verified by nanodrop (Nanodrop 2000; Thermo Scientific) quantification. Confirmatory molecular identification (ID) tests were also conducted on the DNA extract prior to shipment to the contract sequencing facility (Macrogen Inc., Seoul, South Korea). At least 100-fold sequencing coverage was obtained using pyrosequencing technology on a 454 FLX instrument. The individual sequence files generated by the 454 FLX instrument were assembled with GSassembler (454 Lifesciences, Branford, CT, USA) to generate a consensus sequence. Quality improvement of the genome sequence involved Sanger sequencing (Eurofins MWG, Ebersberg, Germany) of at least three PCR products across each entire genome to ensure correct assembly, double stranding, and the resolution of any remaining base conflicts occurring within homopolymer tracts. Genomes were annotated using a heuristic approach (Genemark) (24) and manually using the Basic Local Alignment Search Tool (NCBI) (25). Conserved protein domains (where relevant) were detected using Pfam (26), HHpred (27), and/or CDD (28). Complete genomes were visualized using Artemis (29). Phylogenetic trees were generated using the FigTree tool (http://tree.bio.ed.ac.uk/software/figtree/).
Electron microscopic analysis.
Cesium chloride phage samples were dialyzed (as described above) and subjected to further purification by ultracentrifugation (and dialysis) according to the method of Briggiler Marco et al. (30), using a Beckman VTi 65.2 rotor (Beckman Coulter). Dialysis was performed twice for 24 h and 45 min, respectively, against 2 liters of phage buffer (0.05 M Tris-HCl [pH 7.5], 0.1 M NaCl, 8 mM MgSO4) (30). Electron microscopy was performed as previously described by Casey and colleagues (31).
Structural protein identification.
Phage protein extraction (including methanol-chloroform extraction), SDS-PAGE visualization, and preparation of phage structural protein samples were performed as described by Casey et al. (31). Electrospray ionization-tandem mass spectrometry (ESI-MS/MS) was performed as previously described (32, 33). The coverage levels of at least two unique peptides for each structural protein or 5% of the total protein length were used as cutoff values when identifying gene products as components of the viral particle (31).
Adsorption assays.
Quantification of phage adsorption to bacterial strains was determined using a method adapted from Garvey et al. (34). A 10-ml volume of LM17 or GM17 broth was inoculated (2 to 4% depending on the strain) with the appropriate S. thermophilus strain (either ST64987 as the sample strain or ST67368 as the adsorption control strain) or L. lactis strain (LL64981 as the sample strain) from a fresh overnight culture and grown at 42°C or 30°C, respectively, until the optical density at 600 nm (OD600) reached a value between 0.5 and 0.54. A 700-μl volume of the growing culture was transferred to a microcentrifuge tube and centrifuged at 5,000 × g for 10 min to pellet the cells. The supernatant was removed, and the cells were resuspended in 700 μl of one-quarter-strength Ringer's solution (Merck). An equal volume of the appropriate phage lysate (diluted to an approximate titer of 105 to 106 PFU/ml) was added to the tube or to 700 μl one-quarter-strength Ringer's solution (Merck), which served as a negative control. The mixture was incubated at 30°C or 42°C for 12 min and centrifuged at 15,000 × g for 3 min to remove bacterial cells before 200 μl of the residual phage-containing supernatant was removed for enumeration as described above. Calculation of adsorption levels (as a percentage of the total number of phages present) was performed as follows: [(control phage titer − free phage titer in supernatant)/control phage titer] × 100.
Antireceptor purification and adsorption inhibition assays.
The protein product of ORF199871 (where ORF is open reading frame), predicted to encode the phage antireceptor (termed here the receptor binding protein or RBP9871), was purified using a previously described method (35). Briefly, the ORF199871 gene was amplified using Phusion polymerase (New England BioLabs, Ipswich, MA, USA) and employing primers that incorporate a sequence encoding an N-terminal His6 purification tag and appropriate restriction enzyme sites (namely RBP9871F, 5′-AGCAGCCCATGGCACACCATCACCATCACCATTCTTCTGGTGAACATAAGATAATTTTAAGT-3′, and RBP9871R, 5′-AGCAGCTCTAGATTAATATATACTTGGATATGA-3′) and cloned behind the nisin-inducible promoter of plasmid pNZ8048 (36). The ligation mixture was dialyzed against sterile distilled (sd) H2O for 10 min and introduced into electrocompetent L. lactis NZ9000 cells (36). Plasmid DNA was then extracted using a GeneJet Plasmid Miniprep kit (Thermo Scientific) and subjected to Sanger sequencing (as described above) to verify the integrity of the DNA sequence. For target protein induction, NZ9000 strains containing the required plasmid were grown to an OD600 of 0.2 prior to the addition of nisin (10 ng/ml) using Nisaplin (Danisco, Copenhagen, Denmark). Growth was continued for 3.5 h prior to cell lysis and sonication as per the method of Collins et al. (35), with the following modifications: the concentration of CaCl2 (Sigma-Aldrich) in the lysis buffer (10 mM Tris, 300 mM NaCl, 10 mM CaCl2, 25 mg/ml lysozyme [Sigma-Aldrich]; pH 8) was increased to 50 mM, and a further 200 μl 1 M CaCl2 was added to the lysed cells prior to sonication (Soniprep 150; MSE, London, United Kingdom) cycles. Sonicated cells were then centrifuged, and target protein purification was performed using an Ni-nitrilotriacetic acid agarose (Qiagen, Hilden, Germany) column (Bio-Rad, Hercules, CA, USA), using various concentrations of imidazole buffer (10 mM Tris-HCl, 50 mM CaCl2, 300 mM NaCl, 50 to 200 mM imidazole; pH 7.5) according to the manufacturer's instructions. Protein fractions were visualized by separation on a 12.5% SDS-PAGE gel at 160 V for 90 min. Fractions containing bands of the correct size with minimal contamination were dialyzed against 100 ml protein buffer (as above) three times for 40 min each to remove remaining imidazole. Dialyzed fractions were stored at 4°C for use in subsequent adsorption inhibition assays.
Adsorption inhibition assays were performed as described by Collins et al. (35), with the following modification: both the antireceptor incubation and phage adsorption temperatures were increased to 42°C. Adsorption to wild-type (WT) and antireceptor-incubated cells was calculated as described above. Adsorption inhibition, expressed as a percentage of phage adsorption to WT cells, was calculated as follows: [(% adsorption on WT − % adsorption on preincubated cells)/% adsorption on WT] × 100.
Accession number(s).
Whole-genome sequence data for phages 9871, 9872, 9873, and 9874 are available in the GenBank database under the following accession numbers: KU678389 (9871), KU678390 (9872), KU678391 (9873), and KU678392 (9874).
RESULTS AND DISCUSSION
Isolation of phages.
The bacterial strains and phage isolates (Table 1), which formed part of a larger industrial starter strain and phage sample collection (an exception being NZ9000), were initially subjected to a phage-host survey to determine the host ranges of isolated phages. This was followed (in the case of the phages) by further characterization and genome sequencing of a representative selection, the results of which will be published elsewhere. The phage isolates characterized as part of the current study, named 9871, 9872, 9873, and 9874 (together referred to here as “9871-4” or the 987 phage group), originated from distinct dairy fermentation samples from a range of geographical locations and time points, specifically: Portugal in 2008 (isolate 9871), Slovakia in 2008 (isolate 9872), United Kingdom in 2009 (isolate 9873), and Australia in 2010 (isolate 9874). Of 90 industrial strains tested, phages 9871-4 were found to infect just a single strain (named ST64987), with subsequent experiments revealing the ability to cause low-level infection of a second strain (ST47795) by 9872, 9873, and 9874 only (data not shown). This observed narrow host range is typical of S. thermophilus phages (12, 37). The four phages were shown to reach a high titer during standard propagations (approximately 109 PFU/ml), and DNA could readily be extracted from both crude lysate and CsCl-purified preparations. However, a standard cos or pac phage-typing PCR (38) on either lysate or DNA preparations repeatedly failed to yield a product (data not shown). For this reason, these phages were identified as phage isolates that potentially belong to a novel group, and they were therefore subjected to genome sequencing.
TABLE 1.
Bacterial strains and bacteriophages used in this study
| Bacterial strain or phage | Description | Origin |
|---|---|---|
| Bacterial strains | ||
| ST64987 | S. thermophilus host for phages 9871-4 | DSM, Netherlands |
| LL64981 | Lactococcus lactis subsp. lactis host for phage 98103 | DSM, Netherlands |
| ST67368 | S. thermophilus host for phage 3681, as adsorption control | DSM, Netherlands |
| NZ9000 | Transformation host | Kuipers et al., 1998 (36) |
| Phages | ||
| 9871 | Lytic phage of S. thermophilus ST64987 | DSM, Netherlands |
| 9872 | Lytic phage of S. thermophilus ST64987 | DSM, Netherlands |
| 9873 | Lytic phage of S. thermophilus ST64987 | DSM, Netherlands |
| 9874 | Lytic phage of S. thermophilus ST64987 | DSM, Netherlands |
| 3681 | Lytic phage of S. thermophilus ST67368, as adsorption control | DSM, Netherlands |
| 98103 | Lytic phage of L. lactis LL64981, as adsorption control | DSM, Netherlands |
Genome analysis. (i) General characteristics.
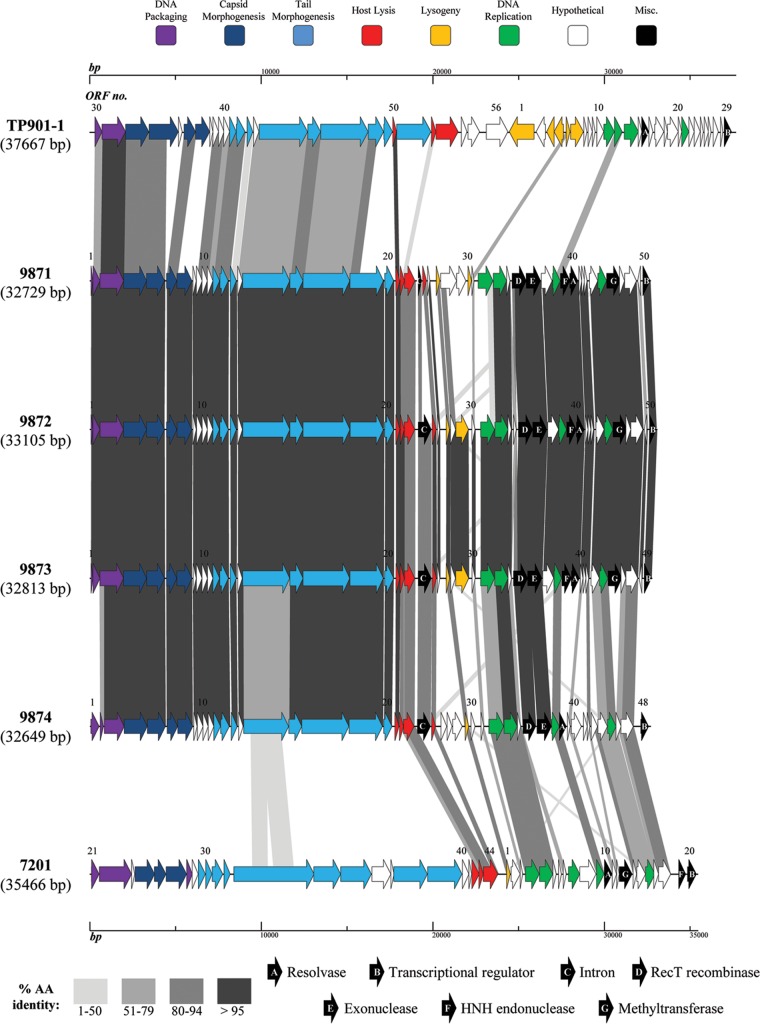
The salient genome characteristics of phages 9871-4 are outlined in Table 2, with a detailed list of top BLAST identities provided for phage 9871 (as a representative of the group, due to overall conservation of the four genome structures) in Table S1 in the supplemental material. Genome sizes ranged from 32.6 to 33.1 kilobase pairs (kbp), making these genomes the shortest thus far described for S. thermophilus phages. An initial analysis of the DNA sequences revealed a high level of nucleotide identity (greater than 90% across approximately one-third of the length of their genomes) with phage ul36 (39), and also with Tuc2009, TP901-1, and the archetype P335, which are related phages, all belonging to P335 subgroup II (40, 41). In contrast, the rightward end of each of the 9871-4 phage genomes appears to bear more similarity to S. thermophilus phage replication modules (42) (Fig. 1). Each module of this apparent “hybrid” 987 phage group is discussed below.
TABLE 2.
General characteristics of the genomes of phages 9871-4
| Characteristic | 9871 | 9872 | 9873 | 9874 |
|---|---|---|---|---|
| Length (bp) | 32,729 | 33,105 | 32,813 | 32,649 |
| No. of predicted ORFs | 50 | 50 | 49 | 48 |
| Coding (%) | 92.2 | 91.4 | 91.7 | 89.0 |
| GC content (%) | 37.06 | 36.84 | 36.9 | 36.62 |
FIG 1.
Comparative analysis of the genetic organization and content of phages 9871-4 with archetypes TP901-1 (L. lactis-infecting phage species P335) and 7201 (cos-containing S. thermophilus phage). Predicted ORFs (indicated by arrows) and gene products (putative function indicated by color coding) are aligned with adjacent genomes according to percent amino acid identity (indicated by shaded boxes). Gene products considered to be notable are marked in black (see color and shade keys).
(ii) Structural modules and structural protein determination.
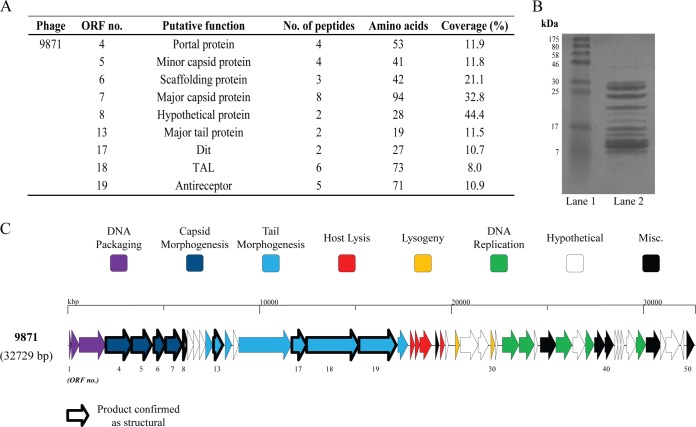
The structural gene module in the four 987 group members in each phage, spanning the region starting at the putative portal protein-encoding gene to the predicted serine acetyltransferase-encoding gene, are remarkably conserved at the deduced amino acid level (Fig. 1). For this reason, structural proteins present in purified phage particles prepared from a cell lysate were determined by mass spectrometry for phage 9871 as a representative of the group (Fig. 2), and this phage alone will be discussed further (unless otherwise indicated). The deduced products of ORF49871 to ORF89871 were all confirmed as structural proteins and are presumed to be involved in phage head morphogenesis (based on their positions in the genome, as well as amino acid identities to known phage head proteins [Fig. 2]). The proteins encoded by ORF99871, ORF109871, and ORF119871 were not detected during mass spectrometric analysis, possibly due to their low abundance in the 9871 particle. ORF99871 and ORF109871 appear to encode a so-called head-tail connector or adapter (43, 44), based on conserved phage head-tail connector domains (specifically, those present in proteins GP15 and GP16 of the well-characterized Bacillus subtilis phage SPP1) detected using CDD (ORF99871) and HHPred (ORF109871) (Fig. 1).
FIG 2.
Structural proteome analysis of phage 9871. (A) Deduced structural proteins (and corresponding ORF number) as identified by ESI-MS/MS (threshold, two unique peptides or 5% ORF coverage). No. of peptides, number of distinct polypeptide strings identified during the analysis; amino acids, the total number of amino acids identified in each protein; coverage, the number of amino acids identified expressed as a percentage of the number of amino acids in the entire protein. (B) SDS-PAGE gel (12%) showing the structural protein profile of phage 9871. Lane 1, broad-range protein ladder (New England BioLabs); lane 2, phage 9871 protein extraction. (C) ORF schematic of phage 9871 highlighting confirmed structural protein-encoding genes (bold outline).
The defined tail morphogenesis gene cluster in phage 9871 commences with ORF119871, which encodes a putative tail component (based on the presence of domains detected using CDD and Pfam). ORF129871 was not confirmed as a structural protein-encoding gene; however, a homologue of this gene product present in lactococcal phage TP901-1 has recently been annotated as the tail terminator protein (45), with apparently unchecked tail extension observed in mutant phages containing a stop codon in this gene. ORF139871 specifies the presumed major tail protein, with ORF149871 and ORF159871 encoding putative tail assembly chaperone proteins (46, 47) of ORF169871 (predicted to encode the tail tape measure protein). Indeed, ORF149871 shows evidence of a “slippery sequence” (5′-AAAAAAA-3′), a feature present in some genes involved in tail assembly that leads to an alternative frame translation (46) and production of an essential tail chaperone in bacteriophage λ (48). The product of ORF169871 (TMP9871) was not confirmed as a structural protein, suggesting that it is present in small amounts in the phage particle.
ORF179871, encoding the putative distal tail protein (the product of which was confirmed as a structural protein [Fig. 2]), is homologous to its functional equivalent in the lactococcal phage TP901-1, with the latter forming the core of the phage tail tip (49). The putative tail-associated lysin (TAL) is encoded by ORF189871 (confirmed as a structural protein [Fig. 2]) and shares significant amino acid similarity (particularly at the N terminus) with the corresponding genes in phages Tuc2009, TP901-1 (in which it was defined as the tail fiber), ul36, and P335 (39, 50–53). The location of the endopeptidase-encompassing domain (M23 family), including the catalytic His residue (residing within amino acid sequence ATGVHLHF, being the equivalent of VTGPHLHF, where the His residue is underlined, in Tuc2009 and TP901-1 [54, 55]), in this protein appears to be conserved in phage 9871, based on CDD (28) search results. Previously, it has been reported that the TAL of L. lactis phage Tuc2009 undergoes autocleavage at a specific GGSSG*GG amino acid sequence, where the asterisk (*) indicates the cleavage site (54, 55). In TAL9871, this site appears to be replaced by AASGGGG, with underlined residues indicating amino acid substitutions relative to the site in TALTuc2009.
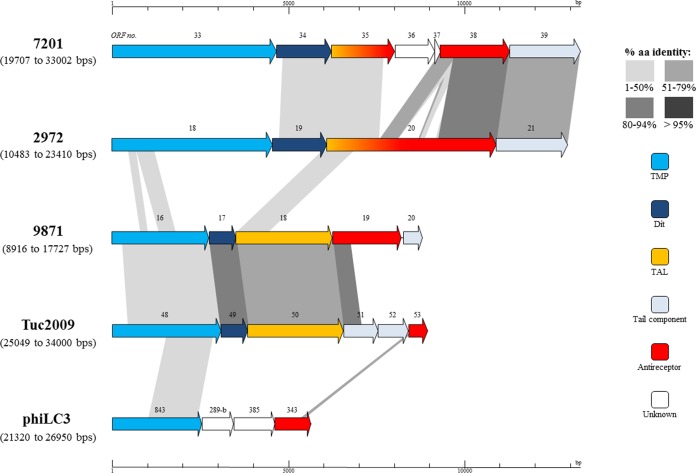
The final structural protein of phage 9871 as determined by mass spectrometry is the product of ORF199871, which encodes the putative receptor binding protein, here referred to as RBP9871. While the “tripods” (as defined by Veesler et al. [49]) of L. lactis phages that are closely related to the 987 group phages are encoded by at least two genes (e.g., Tuc2009 [35, 56] and TP901-1 [57]), the baseplate in phage 9871 appears to be encoded by a single gene (for reasons outlined below), perhaps akin to the arrangement in several P335 phages of Lactococcus lactis, including BK5-T, LC3, and BM13 and Q33, which belong to the P335 subgroups I, III, and IV, respectively (41). Several S. thermophilus phages (including DT1) apparently share this arrangement, with a single antireceptor gene containing at least one variable region, (one of) which (termed ‘VR2′) was shown to be correlated to host specificity (11). Here, the N-terminal end of RBP9871 shares a high level of amino acid identity (approximately 85%) with the N-terminal portion of the upper baseplate protein (BppU) of TP901-1, Tuc2009, P335, and ORF322 of ul36 and then appears to be extended (relative to BppU) at the C-terminal end. This composite arrangement is visualized in Fig. 3. A parallel beta helix domain at the C-terminal end of RBP9871 (identified using Pfam) is a member of clan CL0268, members of which include glycosyl hydrolases, pectate lyases, pectin esterases, and Salmonella phage P22-like tail spikes. Similarly, using a CDD search, pectate lyase domains were found to be present toward the C-terminal end of the protein and bear similarity to glycosyl hydrolase family 28, members of which hydrolyze glycosidic bonds in the heteropolysaccharide pectin (58). Taken together, these findings suggest that RBP9871 has a carbohydrate binding function, leading us to hypothesize that this protein incorporates the receptor binding activities of the BppU and BppL proteins of TP901-1, where BppL is known to be responsible for host interaction and specificity (59). Importantly, the other three members of the 987 group each encompass an ORF199871 homologue, and these homologues exhibit near-complete nucleotide identity to each other, being consistent with the extremely narrow host range of these phages.
FIG 3.
Schematic representation of ORFs predicted to encode the tail proteins of S. thermophilus phage 7201 (cos containing), 2972 (pac containing), 9871 (987 group), and Tuc2009 and phiLC3 (L. lactis phage P335 group). Predicted ORFs (indicated by arrows) and gene products (putative function indicated by color coding) are aligned with adjacent genomes according to percent amino acid identity (indicated by shaded boxes).
ORF209871 (highly conserved in the 987 group phages [Fig. 1]) was not detected during mass spectrometry, and its function is currently unknown, though it appears (using a BLAST search) to be related to a family of serine acetyltransferases. A search using the CDD database confirms the presence of the serine acetyltransferase domain as well as a sugar O-acetyltransferase domain of the NeuD family, identified as a sialic acid O-acetyltransferase in group B streptococci (60). O-acetylation has been shown to be present at precise locations of the sialic acid component of the capsular polysaccharide of group B Streptococcus (61). The presumed sugar interaction of the predicted O-acetyltransferase enzyme may thus be significant in the context of the outer cell layer encountered by the phage during host adsorption, in which the product of ORF209871 may perhaps play an accessory role in host recognition, similar to that exhibited by BppA in Tuc2009 (35, 62), particularly considering its proposed position in the tail morphogenesis module of the 987 group phage genomes.
Lysis and lysogeny modules.
Approximately one-half of the currently sequenced S. thermophilus phages possess two distinct holin-encoding genes, being largely conserved, with the exception of phage 2972 (19). The 987 group phages also appear to possess two distinct holin-encoding genes, one gene product being closely related to holins found in L. lactis phages and one to those found in S. thermophilus phages (Fig. 1; see also Table S1 in the supplemental material). The lysin-encoding gene of 9871 (ORF239871) is located immediately downstream of the holin-encoding genes and appears to be interrupted by a putative group I intron, a feature previously described in other phages of S. thermophilus (63). This is indicated by the presence of a predicted endonuclease-encoding open reading frame, known to be a feature of certain group I introns (64), as well as the presence of a 14-bp consensus sequence (surrounding the predicted intron splice site) correlated with intron possession (63) in all four phages, with various levels of nucleotide identity.
The predicted lysogeny modules present in the 9871-4 phages appear in each case to have been subjected to genetic decay and therefore to be redundant, based on the small size of the region relative to that of proven lysogenic phages (5) and the absence of certain genes (most notably, in this case, an integrase-encoding gene) typically associated with these modules in genuine temperate phages (65). These regions are commonly known as lysogeny “replacement” modules, which are a feature of lytic S. thermophilus phages (14, 17).
Replication modules.
The gene products encoded by the individual replication modules present in phages 9871-4 (downstream of the lysogeny replacement modules) are largely conserved at the amino acid level (Fig. 1) and appear to belong to the “7201-like” grouping (66), which has previously been identified in phages 7201 (42), Abc2 (6), and more recently, 5093 (13). Despite this general conservation, however, the replication module of phage 9874 (more so than those of phages 9871 to 9873) is characterized by deletions, insertions, and point mutations, a common feature of this region in S. thermophilus phage genomes (18). Various genes encoding proteins of apparent non-streptococcal phage origin are positioned downstream of the replication module (detailed in the legend of Fig. 1; see also Table S1 in the supplemental material), including a RecT recombinase-encoding gene. Interestingly, these genes are often associated with exonuclease-encoding genes that together form so-called “recombination modules” (67). Indeed, this is the case for the 987 group phages, with the exonuclease-encoding gene being located immediately downstream of the recombinase-encoding genes (Fig. 1). Phages 9871, 9872, and 9873 are also predicted to encode a cytosine-5 methyltransferase (ORF469871). In general, phage-encoded methyltransferases are thought to be an antidefensive response to the DNA-targeting activity of restriction-modification systems in bacterial hosts, but potentially they also function in other viral and cellular processes (for a review, see reference 68).
The proposed “terminal” ORFs are defined here as those ORFs preceding the small subunit of the terminase in the genomes of the 987 group phages (ORF509871/9872, ORF499873, or ORF489874) (Fig. 1). These ORFs are defined as “terminal” based on homologues being present upstream of the defined cos site in several cos-containing phages of S. thermophilus such as DT1 (15), Sfi19 (14), 7201 (16), Sfi21 (14), and Abc2 (6), the cos site, in turn, being located upstream of the small subunit of the terminase. The protein products of the terminal ORFs in the 987 group phages appear to be conserved in 9871 to 9873, with that of 9874 being divergent. A Pfam search using these proteins in phages 9871 to 9873 indicates that they belong to the DUF1492 family, which was recently found to be one of several major groups of “late transcriptional regulators” (ltr) in phages of Gram-positive bacteria (69). Similarly, transcriptional regulation appears to be the primary function of the product of ORF489874, which shows approximately 50% amino acid identity with ArpU family transcriptional regulators of various streptococcal species (69).
Morphological characteristics.
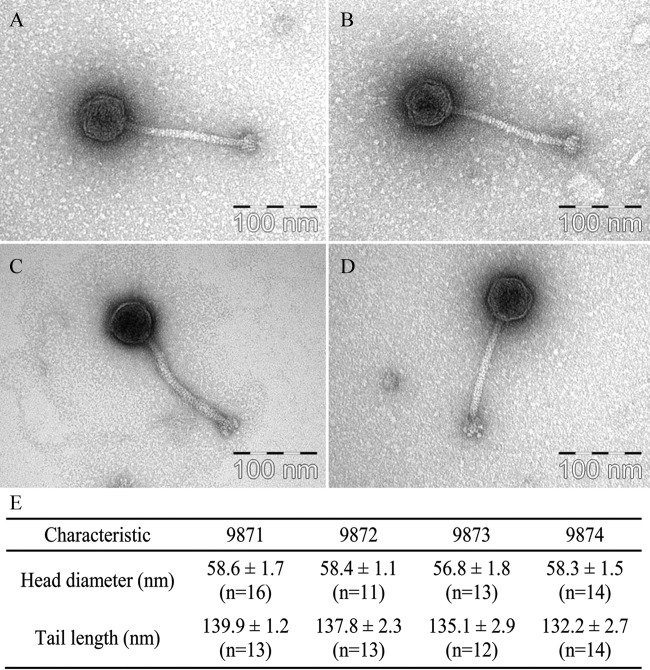
The morphology of the Siphoviridae family has been well documented (group B as defined by Bradley [9]), and siphophages infecting L. lactis and S. thermophilus exhibit the expected morphology, with icosahedral heads and noncontractile tails. Phages infecting S. thermophilus in general possess longer tails than their lactococcal counterparts (with an exception being the 949 group of lactococcal phages [40, 70]), their long tails being consistent with their long TMP-encoding genes (71). Upon electron microscopic analysis, it was found that phages 9871-4 exhibit icosahedral heads and relatively short tails (Fig. 4E shows exact dimensions). The distal tail-associated baseplate, which generally functions in the attachment of the phage to the bacterial cell (discussed above), is clearly visible (Fig. 4A to D). The presence and observed features of a baseplate are consistent with those previously observed in P335 species L. lactis phages such as Tuc2009 and TP901-1 (57, 71, 72) and indeed with the observed similarity between the tail structural gene products and those encoded by the 987 group phages. The measured head diameters are similar among each of the 987 group phages yet slightly larger than those previously reported for Tuc2009 and TP901-1; similarly, the tail lengths of the phages in the 987 group are within the same range relative to each other but slightly shorter than previously reported for those P335 phages (72–74). In keeping with this distinction between phages infecting S. thermophilus and L. lactis P335 group phages, the observed tail lengths of the 987 group phages are lower than those previously reported for S. thermophilus phages (71).
FIG 4.
Uranyl acetate-stained transmission electron micrographs of phages 9871 (A), 9872 (B), 9873 (C), and 9874 (D) and discerned head and tail measurements (E).
Adsorption and adsorption inhibition assays.
Consistent with the genetic composition of the structural module (and, in particular, the antireceptor-encoding genes) of the four 987 group phages, it appears that these phages are able to adsorb to certain L. lactis strains as well as the primary S. thermophilus host. Ten L. lactis strains that are routinely combined with ST64987 in industrial fermentations were initially tested for phage adsorption (as described above), of which one (LL64981) appeared to adsorb all four 987 group phages at a level of approximately 50% (Fig. 5). For this assay, a negative-control phage (3681, a cos-containing phage infecting industrial S. thermophilus strain ST67368) was used to illustrate the specific adsorption affinity of strains ST64987 and LL64981 for the 987 group phages. Following the adsorption assay, a DNA transduction experiment was performed using this phage-strain combination, as well as a positive-control combination, according to a previously reported method (75); however, no confirmed L. lactis transductants were obtained (data not shown). Adsorption to both strains of L. lactis and S. thermophilus suggests that a common cell surface molecule is recognized by these hybrid phages (discussed further below), complemented by the observed genetic similarity of the tail tip regions of the 987 group phages to those regions in phages infecting L. lactis. Indeed, a phage infecting L. lactis LL64981 (termed 98103) was also shown to exhibit adsorption affinity both to its host and to S. thermophilus ST64987 (Fig. 5).
FIG 5.
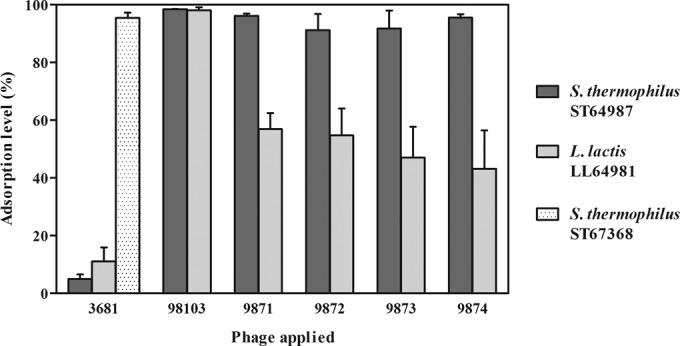
Adsorption analysis of phages 9871-4 on primary host S. thermophilus ST64987 and L. lactis LL64981 at an adsorption temperature (TA) of 42°C. Phage 3681 (a cos-containing lytic phage of S. thermophilus) is included as an adsorption-negative control for ST64987 and LL64981 and was found separately to adsorb optimally (>90%) to its primary host (S. thermophilus ST67368) at a TA of 42°C. Phage 98103 (from the P335 phage group infecting L. lactis LL64981) was also shown to exhibit adsorption affinity to both strains. Comparable adsorption data for all strains were generated at a TA of 30°C.
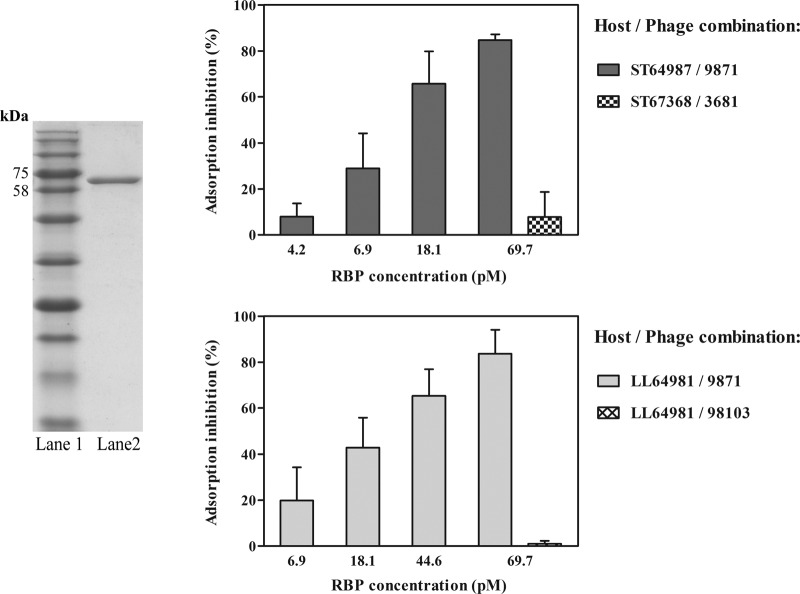
In order to further investigate the interaction between the 987 group phages and their host(s), we performed a competitive phage adsorption inhibition assay using the presumed host recognition protein RBP9871. This protein product is proposed to represent the antireceptor of this phage group based on the position of the encoding gene in all four phage genomes (Fig. 1) and its confirmed presence as a structural protein in the viral particle (Fig. 2A and C) and for reasons discussed in detail above. RBP9871 was overexpressed and purified (Fig. 6, left, lane 2) and then used in adsorption inhibition assays (as described in Materials and Methods). Figure 6 clearly shows that RBP9871, when incubated with wild-type S. thermophilus and L. lactis cells, inhibits adsorption of phage 9871 to both strains in a dose-dependent manner (Fig. 6, right panels). Maximal (an average of approximately 80%) adsorption inhibition was achieved using a concentration of 69.7 pM in both cases, a concentration comparable to that observed by Collins et al. (35) using a lactococcal phage RBP and host combination. Any observed difference in the potency of the respective RBPs may possibly be accounted for by an increased (or decreased) amount of available binding sites present on the cell surface, particularly considering the differences in host genera (for S. thermophilus ST64987) and subspecies (for L. lactis subsp. lactis LL64981).
FIG 6.
Phage 9871 adsorption inhibition analysis using various concentrations of purified RBP9871 on strains S. thermophilus ST64987 and L. lactis LL64981 by blocking assay. (Left) SDS-PAGE gel (12%) showing purified antireceptor of phage 9871. Lane 1, blue prestained protein standard, broad range (New England BioLabs); lane 2, purified 9871 antireceptor. (Right) Inhibition (%) of 9871 adsorption on ST64987 (top) and LL64981 (bottom).
These adsorption inhibition data have a number of implications. First, it may be postulated that the cell surface target used by the 987 group phages is carbohydrate in nature, considering the putative carbohydrate-binding function of the antireceptor protein (discussed above) as well as the homology of the tail tip regions to phages of L. lactis, which target cell surface carbohydrate moieties (76). These data also suggest that the phage targets expressed on the cell surface of S. thermophilus ST64987 and on L. lactis LL64981 are at least similar in nature. Considering the observed similarity between some S. thermophilus and L. lactis genes encoding exopolysaccharide (EPS) biosynthetic elements (1, 77), combined with the observed heterogeneity of the EPS clusters of S. thermophilus (1, 77, 78), this is conceivable. Furthermore, considering the genetic divergence between the antireceptors of the 987 group phages and those of previously sequenced phages such as DT1 (11) and phage 5093 (13), it is possible that alternative cell surface targets are recognized by these phages during the initial phage-host interaction.
Evolutionary aspects.
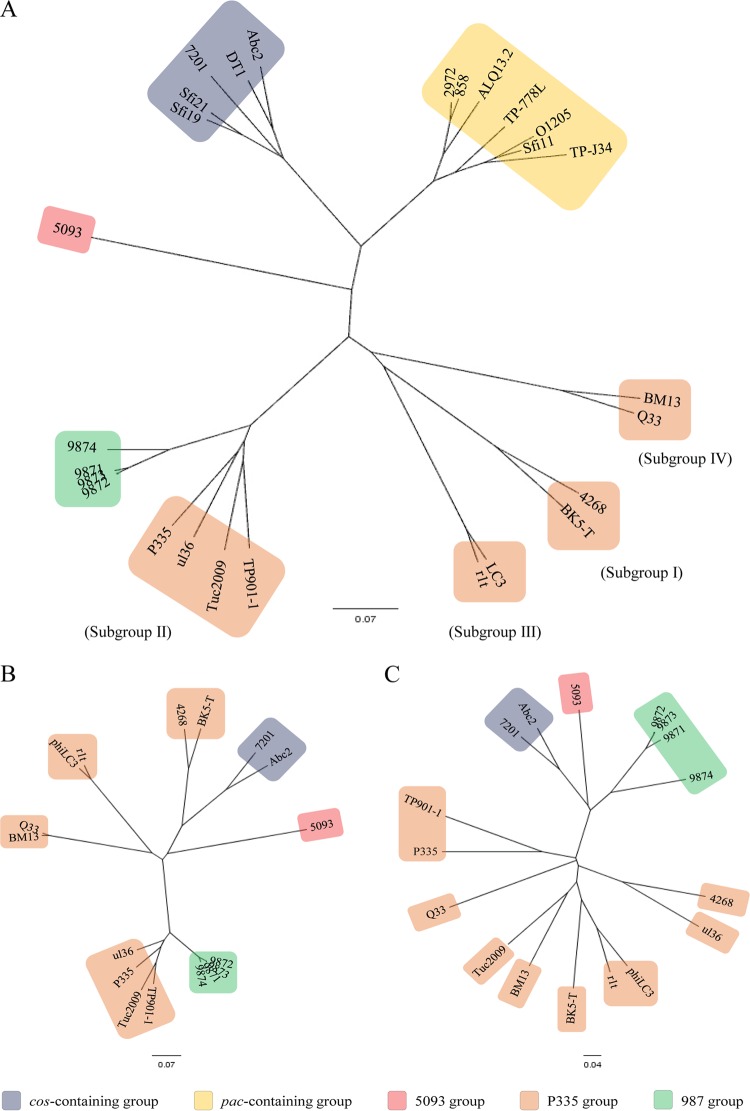
Phages 9871-4 represent a group of S. thermophilus-infecting phages that is distinct from documented cos-containing and pac-containing phage groups, as well as the more recently discovered 5093 group. While phage 5093 appears to have acquired several genes from nondairy streptococcal phages, such as those infecting Streptococcus pneumoniae, Streptococcus gordonii, and Streptococcus pyogenes (13), a genetic crossover previously observed in prophages of S. pyogenes (79), the 987 group phages appear to have been the result of a genomic recombination event between a (temperate) P335 phage of L. lactis and an unknown S. thermophilus phage. Figure 7 shows the genetic distance between the currently known fully sequenced S. thermophilus phages (numbering 13), the 10 sequenced L. lactis phages of the P335 group, and the 4 phages of 987 group. Using this (unrooted) visualization, it appears that the 987 group phages are derivatives of the P335 subgroup of which Tuc2009, TP901-1, P335, and ul36 are members and are more closely related to this group than to the other known phages that infect S. thermophilus. Considering the high level of nucleotide identity to these phages across the (generally) more-conserved structural regions, this is not surprising. Alignments of the structural modules (comprising the TerS-encoding gene to the holin-encoding gene [Fig. 7B]) and the replication modules (comprising the lysin-encoding gene to the terminal ORF [Fig. 7C]) of the 987 group with the relevant comparators, i.e., phages that also harbor group II/7201-like replication modules (42, 66), show a clear difference in clustering, indicating the diverse lineage of these respective modules in the 987 group phages.
FIG 7.
Unrooted phylogenetic tree showing the genetic relatedness between the 987 group phages, cos- and pac-containing S. thermophilus-infecting phages, as well as L. lactis-infecting phages of the P335 group (a color code for the respective groupings is provided). (A) Whole-genome nucleotide comparison; (B) structural module comparison with those S. thermophilus phages also harboring a group II/7201-like replication module; (C) replication module comparison with those S. thermophilus phages also harboring a group II/7201-like replication module.
The genetic/structural similarity of these phages to those of L. lactis—with the retention of the ability to infect S. thermophilus—may be considered a form of adaptive mosaicism, a known evolutionary strategy common to phages infecting a wide range of bacteria (80, 81). Due to the close association of L. lactis and S. thermophilus both in raw milk and in the dairy processing environment, gene transfer between (the phages of) these species has been the subject of speculation, with perhaps the most striking phage example of this phenomenon being observed in the case of phage BK5-T, a temperate phage of L. lactis H2L (82), which shares significant sequence similarity with S. thermophilus phage Sfi21 (83). Further examples of this phenomenon include the genomes of phage 1358 (infecting L. lactis SMQ-388), with homology to phages infecting Listeria monocytogenes (84), phage Q54 (infecting L. lactis SMQ-562), which appears to be a hybrid of the 936 and c2 lactococcal phage species (85), and phage 1706 (infecting L. lactis SMQ-450), proposed to be derived from a number of prophages of other Firmicutes (86). The mechanisms by which such horizontal gene transfer events between phages occur have also been proposed. Moineau et al. (22) and Durmaz and Klaenhammer (87) have shown that lytic phages can evolve by acquiring segments of DNA from the host chromosome (including, potentially, remnant prophage) sometimes in response to pressure from abortive infection (Abi) phage resistance systems (88). More recently, it has been shown that transduction in L. lactis is possible using S. thermophilus phages (75), clearly demonstrating that phage-mediated horizontal DNA transfer between these two species is possible. Furthermore, considering the rapid nature of phage infection (reviewed by Quiberoni et al. [7]) and, in turn (by necessity), the acquisition of phage resistance, a common genetic lineage in phages of S. thermophilus and L. lactis phages may be reflected in the numerous phage resistance mechanisms of their hosts. Indeed, this has been shown by Sun and colleagues (89), who demonstrated that the superinfection exclusion (sie) phage resistance protein Ltp confers phage resistance to both S. thermophilus and L. lactis hosts, against their respective attacking phages (89). Such multigenus protection is indicative of coevolution of both phage and host, possibly accelerated by their continuous mutual exposure in the dairy environment.
The impact of genetic mosaicism in phages on the marketplace is illustrated by the 987 group phages above, which retain infective ability in S. thermophilus, despite having many genetic and morphological characteristics of phages of L. lactis. This is an important consideration in the dairy industry, an environment in which lactic acid bacteria are in close proximity on a regular basis and which may well present further examples of genetic mosaicism as an evolutionary strategy in dairy phages.
Conclusions.
Here we report the complete genome sequences of four novel phages infecting the dairy bacterium S. thermophilus. Comparative genomic analysis revealed a high level of nucleotide identity to the replication modules of S. thermophilus phages and the structural modules of L. lactis phages, suggesting a relatively recent horizontal gene transfer or recombination event. These genome sequences represent a significant divergence compared to the previously published 13 S. thermophilus phage genomes, being highly mosaic in nature, and are the first members of this phage group to be sequenced. The structural protein complement of one of these phages (as a representative of the group) was determined and found to be similar to previously characterized phages of L. lactis. Morphological similarity to phages of L. lactis was also observed using electron microscopic analysis, in which short tails and clawlike baseplates were observed in all four members of the group.
Adsorption studies revealed the ability of members of this group of phages to adsorb to both their native S. thermophilus hosts and an L. lactis strain with which it is routinely combined in dairy fermentations, suggesting that certain cell surface molecules are shared between the genera. This finding also hints at the event by which the hybrid genomes of these phages may have begun to be replicated, possibly being facilitated by mutually expressed cell surface proteins in combination with a favorable S. thermophilus phage coinfection or prophage-mediated evolutionary event. The phage gene product responsible for this adsorption was defined by the use of purified protein to inhibit phage adsorption to both strains, providing a more detailed analysis of the initial phage-host interaction.
Genetic mosaicism is a common trait of bacteriophages and, in the context of dairy fermentations, may represent a new challenge to phage control methods, which usually consist of traditional bacteriophage-insensitive mutant (BIM) generation and rotational schemes. In light of the rapidly increasing genetic diversity being observed in phages of S. thermophilus, continual monitoring of phage populations in dairy productions will be necessary to ensure that BIM generation methods and knowledge-based rotational systems can be used effectively to ameliorate phage spoilage of industrial fermentations.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the technical assistance of Eoghan Casey and Erik Royackers, and we thank Emiel Ver-Loren-van-Themaat for useful discussion.
We gratefully acknowledge the financial support of DSM Food Specialties. J.M. is in receipt of a Technology Innovation Development Award (TIDA) (Ref. No. 14/TIDA/2287) funded by Science Foundation Ireland (SFI). D.V.S. is supported by a Principal Investigator award (Ref. No. 13/IA/1953) through Science Foundation Ireland (SFI).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00835-16.
REFERENCES
- 1.Goh YJ, Goin C, O'Flaherty S, Altermann E, Hutkins R. 2011. Specialized adaptation of a lactic acid bacterium to the milk environment: the comparative genomics of Streptococcus thermophilus LMD-9. Microb Cell Fact 10(Suppl 1):S22. doi: 10.1186/1475-2859-10-S1-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ott A, Hugi A, Baumgartner M, Chaintreau A. 2000. Sensory investigation of yogurt flavor perception: mutual influence of volatiles and acidity. J Agric Food Chem 48:441–450. doi: 10.1021/jf990432x. [DOI] [PubMed] [Google Scholar]
- 3.Guglielmotti DM, Mercanti DJ, Reinheimer JA, Quiberoni Adel L. 2012. Review: efficiency of physical and chemical treatments on the inactivation of dairy bacteriophages. Front Microbiol 2:282. doi: 10.3389/fmicb.2011.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capra ML, Neve H, Sorati PC, Atamer Z, Hinrichs J, Heller KJ, Quiberoni A. 2013. Extreme thermal resistance of phages isolated from dairy samples: updating traditional phage detection methodologies. Int Dairy J 30:59–63. doi: 10.1016/j.idairyj.2012.11.009. [DOI] [Google Scholar]
- 5.Ali Y, Koberg S, Hessner S, Sun X, Rabe B, Back A, Neve H, Heller KJ. 2014. Temperate Streptococcus thermophilus phages expressing superinfection exclusion proteins of the Ltp type. Front Microbiol 5:98. doi: 10.3389/fmicb.2014.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guglielmotti DM, Deveau H, Binetti AG, Reinheimer JA, Moineau S, Quiberoni A. 2009. Genome analysis of two virulent Streptococcus thermophilus phages isolated in Argentina. Int J Food Microbiol 136:101–109. doi: 10.1016/j.ijfoodmicro.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Quiberoni A, Moineau S, Rousseau GM, Reinheimer J, Ackermann H-W. 2010. Streptococcus thermophilus bacteriophages. Int Dairy J 20:657–664. doi: 10.1016/j.idairyj.2010.03.012. [DOI] [Google Scholar]
- 8.Mahony J, van Sinderen D. 2014. Current taxonomy of phages infecting lactic acid bacteria. Front Microbiol 5:7. doi: 10.3389/fmicb.2014.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradley DE. 1967. Ultrastructure of bacteriophage and bacteriocins. Bacteriol Rev 31:230–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Marrec C, van Sinderen D, Walsh L, Stanley E, Vlegels E, Moineau S, Heinze P, Fitzgerald G, Fayard B. 1997. Two groups of bacteriophages infecting Streptococcus thermophilus can be distinguished on the basis of mode of packaging and genetic determinants for major structural proteins. Appl Environ Microbiol 63:3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duplessis M, Moineau S. 2001. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol Microbiol 41:325–336. doi: 10.1046/j.1365-2958.2001.02521.x. [DOI] [PubMed] [Google Scholar]
- 12.Binetti AG, Del Rio B, Martin MC, Alvarez MA. 2005. Detection and characterization of Streptococcus thermophilus bacteriophages by use of the antireceptor gene sequence. Appl Environ Microbiol 71:6096–6103. doi: 10.1128/AEM.71.10.6096-6103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mills S, Griffin C, O'Sullivan O, Coffey A, McAuliffe O, Meijer W, Serrano L, Ross R. 2011. A new phage on the ‘Mozzarella’ block: bacteriophage 5093 shares a low level of homology with other Streptococcus thermophilus phages. Int Dairy J 21:963–969. doi: 10.1016/j.idairyj.2011.06.003. [DOI] [Google Scholar]
- 14.Lucchini S, Desiere F, Brussow H. 1999. The genetic relationship between virulent and temperate Streptococcus thermophilus bacteriophages: whole genome comparison of cos-site phages Sfi19 and Sfi21. Virology 260:232–243. doi: 10.1006/viro.1999.9814. [DOI] [PubMed] [Google Scholar]
- 15.Tremblay DM, Moineau S. 1999. Complete genomic sequence of the lytic bacteriophage DT1 of Streptococcus thermophilus. Virology 255:63–76. doi: 10.1006/viro.1998.9525. [DOI] [PubMed] [Google Scholar]
- 16.Proux C, van Sinderen D, Suarez J, Garcia P, Ladero V, Fitzgerald GF, Desiere F, Brussow H. 2002. The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria. J Bacteriol 184:6026–6036. doi: 10.1128/JB.184.21.6026-6036.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stanley E, Fitzgerald GF, Le Marrec C, Fayard B, van Sinderen D. 1997. Sequence analysis and characterization of phi O1205, a temperate bacteriophage infecting Streptococcus thermophilus CNRZ1205. Microbiology 143(Part 11):3417–3429. [DOI] [PubMed] [Google Scholar]
- 18.Lucchini S, Desiere F, Brussow H. 1999. Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution theory. J Virol 73:8647–8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levesque C, Duplessis M, Labonte J, Labrie S, Fremaux C, Tremblay D, Moineau S. 2005. Genomic organization and molecular analysis of virulent bacteriophage 2972 infecting an exopolysaccharide-producing Streptococcus thermophilus strain. Appl Environ Microbiol 71:4057–4068. doi: 10.1128/AEM.71.7.4057-4068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lillehaug D. 1997. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J Appl Microbiol 83:85–90. doi: 10.1046/j.1365-2672.1997.00193.x. [DOI] [PubMed] [Google Scholar]
- 22.Moineau S, Pandian S, Klaenhammer TR. 1994. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl Environ Microbiol 60:1832–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 24.Besemer J, Borodovsky M. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res 27:3911–3920. doi: 10.1093/nar/27.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Sonnhammer EL, Eddy SR, Durbin R. 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405–420. doi:. [DOI] [PubMed] [Google Scholar]
- 27.Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M-A, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 30.Briggiler Marco M, Garneau JE, Tremblay D, Quiberoni A, Moineau S. 2012. Characterization of two virulent phages of Lactobacillus plantarum. Appl Environ Microbiol 78:8719–8734. doi: 10.1128/AEM.02565-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casey E, Mahony J, O'Connell-Motherway M, Bottacini F, Cornelissen A, Neve H, Heller KJ, Noben JP, Dal Bello F, van Sinderen D. 2014. Molecular characterization of three Lactobacillus delbrueckii subsp. bulgaricus phages. Appl Environ Microbiol 80:5623–5635. doi: 10.1128/AEM.01268-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ceyssens PJ, Mesyanzhinov V, Sykilinda N, Briers Y, Roucourt B, Lavigne R, Robben J, Domashin A, Miroshnikov K, Volckaert G, Hertveldt K. 2008. The genome and structural proteome of YuA, a new Pseudomonas aeruginosa phage resembling M6. J Bacteriol 190:1429–1435. doi: 10.1128/JB.01441-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanheel A, Daniels R, Plaisance S, Baeten K, Hendriks JJ, Leprince P, Dumont D, Robben J, Brone B, Stinissen P, Noben JP, Hellings N. 2012. Identification of protein networks involved in the disease course of experimental autoimmune encephalomyelitis, an animal model of multiple sclerosis. PLoS One 7:e35544. doi: 10.1371/journal.pone.0035544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garvey P, Hill C, Fitzgerald GF. 1996. The lactococcal plasmid pNP40 encodes a third bacteriophage resistance mechanism, one which affects phage DNA penetration. Appl Environ Microbiol 62:676–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins B, Bebeacua C, Mahony J, Blangy S, Douillard FP, Veesler D, Cambillau C, van Sinderen D. 2013. Structure and functional analysis of the host recognition device of lactococcal phage Tuc2009. J Virol 87:8429–8440. doi: 10.1128/JVI.00907-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuipers OP, de Ruyter PG, Kleerebezem M, de Vos WM. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol 64:15–21. doi: 10.1016/S0168-1656(98)00100-X. [DOI] [Google Scholar]
- 37.Zinno P, Janzen T, Bennedsen M, Ercolini D, Mauriello G. 2010. Characterization of Streptococcus thermophilus lytic bacteriophages from mozzarella cheese plants. Int J Food Microbiol 138:137–144. doi: 10.1016/j.ijfoodmicro.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 38.Quiberoni A, Tremblay D, Ackermann HW, Moineau S, Reinheimer JA. 2006. Diversity of Streptococcus thermophilus phages in a large-production cheese factory in Argentina. J Dairy Sci 89:3791–3799. doi: 10.3168/jds.S0022-0302(06)72420-1. [DOI] [PubMed] [Google Scholar]
- 39.Labrie S, Moineau S. 2002. Complete genomic sequence of bacteriophage ul36: demonstration of phage heterogeneity within the P335 quasi-species of lactococcal phages. Virology 296:308–320. doi: 10.1006/viro.2002.1401. [DOI] [PubMed] [Google Scholar]
- 40.Samson JE, Moineau S. 2010. Characterization of Lactococcus lactis phage 949 and comparison with other lactococcal phages. Appl Environ Microbiol 76:6843–6852. doi: 10.1128/AEM.00796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahony J, Martel B, Tremblay DM, Neve H, Heller KJ, Moineau S, van Sinderen D. 2013. Identification of a new P335 subgroup through molecular analysis of lactococcal phages Q33 and BM13. Appl Environ Microbiol 79:4401–4409. doi: 10.1128/AEM.00832-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stanley E, Walsh L, van der Zwet A, Fitzgerald GF, van Sinderen D. 2000. Identification of four loci isolated from two Streptococcus thermophilus phage genomes responsible for mediating bacteriophage resistance. FEMS Microbiol Lett 182:271–277. doi: 10.1111/j.1574-6968.2000.tb08907.x. [DOI] [PubMed] [Google Scholar]
- 43.Lurz R, Orlova EV, Gunther D, Dube P, Droge A, Weise F, van Heel M, Tavares P. 2001. Structural organisation of the head-to-tail interface of a bacterial virus. J Mol Biol 310:1027–1037. doi: 10.1006/jmbi.2001.4800. [DOI] [PubMed] [Google Scholar]
- 44.Bebeacua C, Lai L, Vegge CS, Brondsted L, van Heel M, Veesler D, Cambillau C. 2013. Visualizing a complete Siphoviridae member by single-particle electron microscopy: the structure of lactococcal phage TP901-1. J Virol 87:1061–1068. doi: 10.1128/JVI.02836-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stockdale SR, Collins B, Spinelli S, Douillard FP, Mahony J, Cambillau C, van Sinderen D. 2015. Structure and assembly of TP901-1 virion unveiled by mutagenesis. PLoS One 10:e0131676. doi: 10.1371/journal.pone.0131676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu J, Hendrix RW, Duda RL. 2004. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol Cell 16:11–21. doi: 10.1016/j.molcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Siponen M, Sciara G, Villion M, Spinelli S, Lichiere J, Cambillau C, Moineau S, Campanacci V. 2009. Crystal structure of ORF12 from Lactococcus lactis phage p2 identifies a tape measure protein chaperone. J Bacteriol 191:728–734. doi: 10.1128/JB.01363-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu J, Hendrix RW, Duda RL. 2013. A balanced ratio of proteins from gene G and frameshift-extended gene GT is required for phage lambda tail assembly. J Mol Biol 425:3476–3487. doi: 10.1016/j.jmb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veesler D, Spinelli S, Mahony J, Lichiere J, Blangy S, Bricogne G, Legrand P, Ortiz-Lombardia M, Campanacci V, van Sinderen D, Cambillau C. 2012. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc Natl Acad Sci U S A 109:8954–8958. doi: 10.1073/pnas.1200966109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seegers JF, Mc Grath S, O'Connell-Motherway M, Arendt EK, van de Guchte M, Creaven M, Fitzgerald GF, van Sinderen D. 2004. Molecular and transcriptional analysis of the temperate lactococcal bacteriophage Tuc2009. Virology 329:40–52. doi: 10.1016/j.virol.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 51.Brondsted L, Ostergaard S, Pedersen M, Hammer K, Vogensen FK. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93–109. doi: 10.1006/viro.2001.0871. [DOI] [PubMed] [Google Scholar]
- 52.Labrie SJ, Josephsen J, Neve H, Vogensen FK, Moineau S. 2008. Morphology, genome sequence, and structural proteome of type phage P335 from Lactococcus lactis. Appl Environ Microbiol 74:4636–4644. doi: 10.1128/AEM.00118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bebeacua C, Bron P, Lai L, Vegge CS, Brondsted L, Spinelli S, Campanacci V, Veesler D, van Heel M, Cambillau C. 2010. Structure and molecular assignment of lactococcal phage TP901-1 baseplate. J Biol Chem 285:39079–39086. doi: 10.1074/jbc.M110.175646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kenny JG, McGrath S, Fitzgerald GF, van Sinderen D. 2004. Bacteriophage Tuc2009 encodes a tail-associated cell wall-degrading activity. J Bacteriol 186:3480–3491. doi: 10.1128/JB.186.11.3480-3491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stockdale SR, Mahony J, Courtin P, Chapot-Chartier MP, van Pijkeren JP, Britton RA, Neve H, Heller KJ, Aideh B, Vogensen FK, van Sinderen D. 2013. The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate forms of their tail fiber into their virions for infection specialization. J Biol Chem 288:5581–5590. doi: 10.1074/jbc.M112.444901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sciara G, Blangy S, Siponen M, Mc Grath S, van Sinderen D, Tegoni M, Cambillau C, Campanacci V. 2008. A topological model of the baseplate of lactococcal phage Tuc2009. J Biol Chem 283:2716–2723. doi: 10.1074/jbc.M707533200. [DOI] [PubMed] [Google Scholar]
- 57.Vegge CS, Brondsted L, Neve H, Mc Grath S, van Sinderen D, Vogensen FK. 2005. Structural characterization and assembly of the distal tail structure of the temperate lactococcal bacteriophage TP901-1. J Bacteriol 187:4187–4197. doi: 10.1128/JB.187.12.4187-4197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Markovic O, Janecek S. 2001. Pectin degrading glycoside hydrolases of family 28: sequence-structural features, specificities and evolution. Protein Eng 14:615–631. doi: 10.1093/protein/14.9.615. [DOI] [PubMed] [Google Scholar]
- 59.Vegge CS, Vogensen FK, Mc Grath S, Neve H, van Sinderen D, Brondsted L. 2006. Identification of the lower baseplate protein as the antireceptor of the temperate lactococcal bacteriophages TP901-1 and Tuc2009. J Bacteriol 188:55–63. doi: 10.1128/JB.188.1.55-63.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lewis AL, Hensler ME, Varki A, Nizet V. 2006. The group B streptococcal sialic acid O-acetyltransferase is encoded by neuD, a conserved component of bacterial sialic acid biosynthetic gene clusters. J Biol Chem 281:11186–11192. doi: 10.1074/jbc.M513772200. [DOI] [PubMed] [Google Scholar]
- 61.Lewis AL, Nizet V, Varki A. 2004. Discovery and characterization of sialic acid O-acetylation in group B Streptococcus. Proc Natl Acad Sci U S A 101:11123–11128. doi: 10.1073/pnas.0403010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Legrand P, Collins B, Blangy S, Murphy J, Spinelli S, Gutierrez C, Richet N, Kellenberger C, Desmyter A, Mahony J, van Sinderen D, Cambillau C. 2016. The atomic structure of the phage Tuc2009 baseplate tripod suggests that host recognition involves two different carbohydrate binding modules. mBio 7(1):e01781–15. doi: 10.1128/mBio.01781-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Foley S, Bruttin A, Brussow H. 2000. Widespread distribution of a group I intron and its three deletion derivatives in the lysin gene of Streptococcus thermophilus bacteriophages. J Virol 74:611–618. doi: 10.1128/JVI.74.2.611-618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shub DA, Goodrich-Blair H, Eddy SR. 1994. Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem Sci 19:402–404. doi: 10.1016/0968-0004(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 65.Lucchini S, Desiere F, Brussow H. 1999. Similarly organized lysogeny modules in temperate Siphoviridae from low GC content gram-positive bacteria. Virology 263:427–435. doi: 10.1006/viro.1999.9959. [DOI] [PubMed] [Google Scholar]
- 66.Brussow H, Desiere F. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol Microbiol 39:213–222. doi: 10.1046/j.1365-2958.2001.02228.x. [DOI] [PubMed] [Google Scholar]
- 67.Datta S, Costantino N, Zhou X, Court DL. 2008. Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc Natl Acad Sci U S A 105:1626–1631. doi: 10.1073/pnas.0709089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murphy J, Mahony J, Ainsworth S, Nauta A, van Sinderen D. 2013. Bacteriophage orphan DNA methyltransferases: insights from their bacterial origin, function, and occurrence. Appl Environ Microbiol 79:7547–7555. doi: 10.1128/AEM.02229-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quiles-Puchalt N, Tormo-Mas MA, Campoy S, Toledo-Arana A, Monedero V, Lasa I, Novick RP, Christie GE, Penades JR. 2013. A super-family of transcriptional activators regulates bacteriophage packaging and lysis in Gram-positive bacteria. Nucleic Acids Res 41:7260–7275. doi: 10.1093/nar/gkt508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mahony J, Randazzo W, Neve H, Settanni L, van Sinderen D. 2015. Lactococcal 949 group phages recognize a carbohydrate receptor on the host cell surface. Appl Environ Microbiol 81:3299–3305. doi: 10.1128/AEM.00143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pedersen M, Ostergaard S, Bresciani J, Vogensen FK. 2000. Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901-1 involved in tail length determination and baseplate assembly. Virology 276:315–328. doi: 10.1006/viro.2000.0497. [DOI] [PubMed] [Google Scholar]
- 72.Mc Grath S, Neve H, Seegers JF, Eijlander R, Vegge CS, Brondsted L, Heller KJ, Fitzgerald GF, Vogensen FK, van Sinderen D. 2006. Anatomy of a lactococcal phage tail. J Bacteriol 188:3972–3982. doi: 10.1128/JB.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arendt EK, Daly C, Fitzgerald GF, van de Guchte M. 1994. Molecular characterization of lactococcal bacteriophage Tuc2009 and identification and analysis of genes encoding lysin, a putative holin, and two structural proteins. Appl Environ Microbiol 60:1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johnsen MG, Neve H, Vogensen FK, Hammer K. 1995. Virion positions and relationships of lactococcal temperate bacteriophage TP901-1 proteins. Virology 212:595–606. doi: 10.1006/viro.1995.1517. [DOI] [PubMed] [Google Scholar]
- 75.Ammann A, Neve H, Geis A, Heller KJ. 2008. Plasmid transfer via transduction from Streptococcus thermophilus to Lactococcus lactis. J Bacteriol 190:3083–3087. doi: 10.1128/JB.01448-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ainsworth S, Sadovskaya I, Vinogradov E, Courtin P, Guerardel Y, Mahony J, Grard T, Cambillau C, Chapot-Chartier MP, van Sinderen D. 2014. Differences in lactococcal cell wall polysaccharide structure are major determining factors in bacteriophage sensitivity. mBio 5(3):e00880–14. doi: 10.1128/mBio.00880-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bourgoin F, Pluvinet A, Gintz B, Decaris B, Guedon G. 1999. Are horizontal transfers involved in the evolution of the Streptococcus thermophilus exopolysaccharide synthesis loci? Gene 233:151–161. doi: 10.1016/S0378-1119(99)00144-4. [DOI] [PubMed] [Google Scholar]
- 78.Pluvinet A, Charron-Bourgoin F, Morel C, Decaris B. 2004. Polymorphism of eps loci in Streptococcus thermophilus: sequence replacement by putative horizontal transfer in S. thermophilus IP6757. Int Dairy J 14:627–634. doi: 10.1016/j.idairyj.2003.12.009. [DOI] [Google Scholar]
- 79.Desiere F, McShan WM, van Sinderen D, Ferretti JJ, Brussow H. 2001. Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic streptococci: evolutionary implications for prophage-host interactions. Virology 288:325–341. doi: 10.1006/viro.2001.1085. [DOI] [PubMed] [Google Scholar]
- 80.Hatfull GF, Hendrix RW. 2011. Bacteriophages and their genomes. Curr Opin Virol 1:298–303. doi: 10.1016/j.coviro.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Casjens SR, Thuman-Commike PA. 2011. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology 411:393–415. doi: 10.1016/j.virol.2010.12.046. [DOI] [PubMed] [Google Scholar]
- 82.Boyce JD, Davidson BE, Hillier AJ. 1995. Sequence analysis of the Lactococcus lactis temperate bacteriophage BK5-T and demonstration that the phage DNA has cohesive ends. Appl Environ Microbiol 61:4089–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Desiere F, Mahanivong C, Hillier AJ, Chandry PS, Davidson BE, Brussow H. 2001. Comparative genomics of lactococcal phages: insight from the complete genome sequence of Lactococcus lactis phage BK5-T. Virology 283:240–252. doi: 10.1006/viro.2001.0857. [DOI] [PubMed] [Google Scholar]
- 84.Dupuis ME, Moineau S. 2010. Genome organization and characterization of the virulent lactococcal phage 1358 and its similarities to Listeria phages. Appl Environ Microbiol 76:1623–1632. doi: 10.1128/AEM.02173-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fortier LC, Bransi A, Moineau S. 2006. Genome sequence and global gene expression of Q54, a new phage species linking the 936 and c2 phage species of Lactococcus lactis. J Bacteriol 188:6101–6114. doi: 10.1128/JB.00581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Garneau JE, Tremblay DM, Moineau S. 2008. Characterization of 1706, a virulent phage from Lactococcus lactis with similarities to prophages from other Firmicutes. Virology 373:298–309. doi: 10.1016/j.virol.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 87.Durmaz E, Klaenhammer TR. 2000. Genetic analysis of chromosomal regions of Lactococcus lactis acquired by recombinant lytic phages. Appl Environ Microbiol 66:895–903. doi: 10.1128/AEM.66.3.895-903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Labrie SJ, Moineau S. 2007. Abortive infection mechanisms and prophage sequences significantly influence the genetic makeup of emerging lytic lactococcal phages. J Bacteriol 189:1482–1487. doi: 10.1128/JB.01111-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sun X, Gohler A, Heller KJ, Neve H. 2006. The ltp gene of temperate Streptococcus thermophilus phage TP-J34 confers superinfection exclusion to Streptococcus thermophilus and Lactococcus lactis. Virology 350:146–157. doi: 10.1016/j.virol.2006.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.