ABSTRACT
Crude glycerol, the major by-product of biodiesel production, is an attractive bioprocessing feedstock owing to its abundance, low cost, and high degree of reduction. In line with the advent of the biodiesel industry, Clostridium pasteurianum has gained prominence as a result of its unique capacity to convert waste glycerol into n-butanol, a high-energy biofuel. However, no efforts have been directed at abolishing the production of 1,3-propanediol (1,3-PDO), the chief competing product of C. pasteurianum glycerol fermentation. Here, we report rational metabolic engineering of C. pasteurianum for enhanced n-butanol production through inactivation of the gene encoding 1,3-PDO dehydrogenase (dhaT). In spite of current models of anaerobic glycerol dissimilation, culture growth and glycerol utilization were unaffected in the dhaT disruption mutant (dhaT::Ll.LtrB). Metabolite characterization of the dhaT::Ll.LtrB mutant revealed an 83% decrease in 1,3-PDO production, encompassing the lowest C. pasteurianum 1,3-PDO titer reported to date (0.58 g liter−1). With 1,3-PDO formation nearly abolished, glycerol was converted almost exclusively to n-butanol (8.6 g liter−1), yielding a high n-butanol selectivity of 0.83 g n-butanol g−1 of solvents compared to 0.51 g n-butanol g−1 of solvents for the wild-type strain. Unexpectedly, high-performance liquid chromatography (HPLC) analysis of dhaT::Ll.LtrB mutant culture supernatants identified a metabolite peak consistent with 1,2-propanediol (1,2-PDO), which was confirmed by nuclear magnetic resonance (NMR). Based on these findings, we propose a new model for glycerol dissimilation by C. pasteurianum, whereby the production of 1,3-PDO by the wild-type strain and low levels of both 1,3-PDO and 1,2-PDO by the engineered mutant balance the reducing equivalents generated during cell mass synthesis from glycerol.
IMPORTANCE Organisms from the genus Clostridium are perhaps the most notable native cellular factories, owing to their vast substrate utilization range and equally diverse variety of metabolites produced. The ability of C. pasteurianum to sustain redox balance and glycerol fermentation despite inactivation of the 1,3-PDO pathway is a testament to the exceptional metabolic flexibility exhibited by clostridia. Moreover, identification of a previously unknown 1,2-PDO-formation pathway, as detailed herein, provides a deeper understanding of fermentative glycerol utilization in clostridia and will inform future metabolic engineering endeavors involving C. pasteurianum. To our knowledge, the C. pasteurianum dhaT disruption mutant derived in this study is the only organism that produces both 1,2- and 1,3-PDOs. Most importantly, the engineered strain provides an excellent platform for highly selective production of n-butanol from waste glycerol.
INTRODUCTION
As a result of a 6-fold increase in global production between 2005 and 2012, biodiesel is presently the second most common biofuel in the world next to ethanol (1). This dramatic increase has generated an exceptional surplus of crude glycerol (80% purity), which comprises 10% (wt/wt) of final biodiesel preparations. The price of crude glycerol plummeted to roughly €0 per ton in 2009, rendering it a waste stream rather than a once-valued coproduct of biodiesel production (2, 3). Although the waste feedstock has since found use in a range of industrial processes (4), leading to a modest rebound in price, impurities arising from the transesterification reaction, namely, methanol and free fatty acids, place constraints on the utilization of crude glycerol in many applications (5). The biotechnological avenue of glycerol valorization offers a myriad of opportunities, as many microorganisms are able to tolerate moderate levels of methanol and free fatty acids (6). Impurities can also be removed or inactivated using simple cost-effective means (7, 8). In addition to its low cost and abundance, glycerol is attractive from a fermentation perspective, owing to its high degree of reductance (γD, 4.67) compared to common sugar feedstocks, such as glucose or xylose (γD, 4). Accordingly, glycerol-fermenting organisms produce a variety of highly reduced chemicals, including ethanol, 1,3-propanediol, and 2,3-butanediol.
Although many organisms in nature are capable of utilizing glycerol fermentatively as a sole source of carbon, Clostridium pasteurianum is the only organism that couples anaerobic glycerol catabolism with high-level production of n-butanol, a bulk chemical and prospective biofuel (9–11). C. pasteurianum is a strictly anaerobic endospore-forming apathogenic bacterium that can be cultivated in chemically defined medium and does not appear to be susceptible to strain degeneration processes that plague industrial exploitation of related clostridia (12). In contrast to common clostridial species, such as Clostridium acetobutylicum and Clostridium beijerinckii, C. pasteurianum has not been exploited industrially for large-scale production of solvents. Instead, biotechnological interest in C. pasteurianum has been spurred only recently in accordance with the tremendous growth experienced by the global biodiesel industry (11). Most organisms that metabolize glycerol fermentatively produce large amounts of 1,3-propanediol (1,3-PDO), the signature product of glycerol fermentation (13). Conversely, C. pasteurianum typically converts glycerol into equal amounts of 1,3-PDO and n-butanol (Fig. 1A). Given the organism's unique central metabolism, several studies have explored the glycerol-to-butanol fermentation carried out by C. pasteurianum (9–11). To enable metabolic engineering of C. pasteurianum, methodologies have recently been reported for electrotransformation (14), group-II-intron-mediated gene disruption (15), gene deletion and integration (16), antisense RNA (asRNA) gene knockdown (17), and clustered regularly interspaced short palindromic repeat (CRISPR)-associated (Cas) genome editing (18). We also sequenced the C. pasteurianum genome (19) and provided genomic analysis of the organism's central metabolism (20). C. pasteurianum genome sequences have been reported by at least five additional groups, covering the type strain (ATCC 6013/DSM 525) (16, 21–23) and two nontype strains (BC1 and NRRL B-598) (24). A method for electroporation of a unique acetone-producing aerotolerant strain was also described recently (25), demonstrating widespread interest in C. pasteurianum as a biocatalyst for crude glycerol valorization. Despite these advancements, only two targeted mutant strains possessing improved n-butanol-producing or glycerol-utilizing properties have been described. We previously downregulated the primary C. pasteurianum hydrogenase-encoding gene (hydA) using asRNA and observed superior growth characteristics, as well as enhanced titers of the reduced end products n-butanol and ethanol (17). Concurrently, Sandoval et al. (16) employed random mutagenesis and directed evolution to identify the Spo0A master transcriptional regulator as a key target for gene inactivation. Deletion of the corresponding spo0A gene yielded improved glycerol dissimilation and n-butanol yields, as well as decreased levels of 1,3-PDO. Prior to the development of the C. pasteurianum molecular genetic toolkit, however, strain engineering efforts have employed random mutagenesis and laborious screening procedures (12, 26).
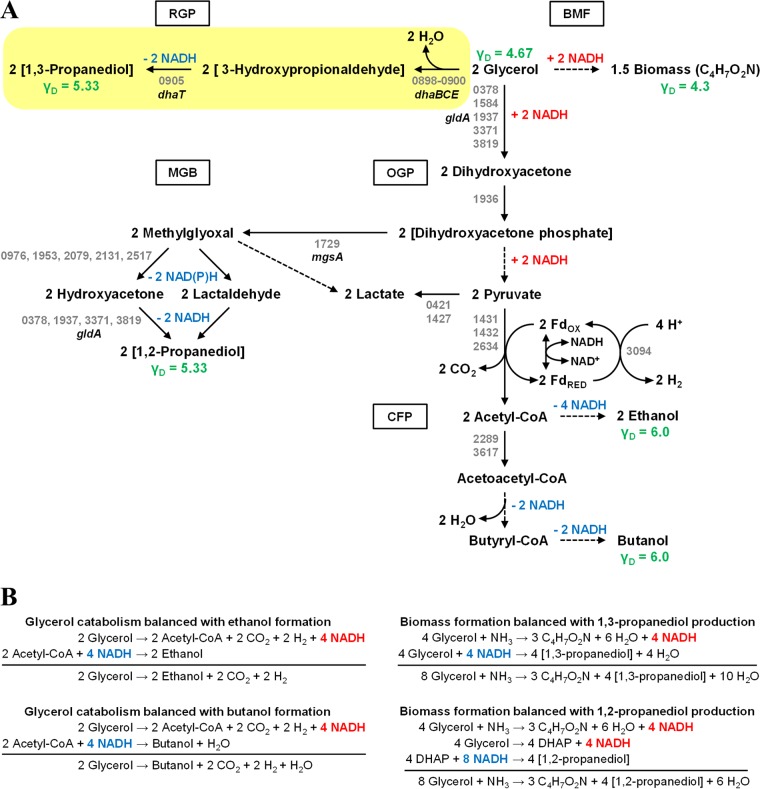
FIG 1.
Fermentative glycerol utilization in C. pasteurianum. (A) Glycerol dissimilatory pathways probed and unveiled in this study. Pathways involved in production of ethanol, n-butanol, 1,2-PDO, and 1,3-PDO are shown. Acetate and butyrate formation pathways are omitted, and the reductive 1,3-PDO pathway targeted for inactivation is highlighted. Degree of reductance (γD) of glycerol and key end products is shown in green, while NADH-forming and NADH-consuming reactions are shown in red and blue, respectively. Locus tags (CP6013 prefix is omitted) corresponding to putative genes involved in key enzymatic steps are shown in gray and are based on the C. pasteurianum genome sequence reported by Pyne et al. (19, 20). Gene abbreviations are provided for select genes discussed in this study. For enzymatic reactions in which several paralogs exist, locus tags for up to five promising candidates are provided. Putative locus tags are provided for the presumed route of 1,2-PDO formation only (via hydroxyacetone). Dashed arrows represent multiple enzymatic conversions. BMF, biomass formation; CFP, central fermentative pathways; CoA, coenzyme A; dhaBCE, glycerol dehydratase gene; dhaT, 1,3-PDO dehydrogenase gene; FDOX, oxidized ferredoxin; FDRED, reduced ferredoxin; gldA, glycerol dehydrogenase gene; MGB, methylglyoxal bypass; mgsA, methylglyoxal synthase gene; OGP, oxidative glycerol pathway; RGP, reductive glycerol pathway. (B) Fermentation balances of glycerol utilization and end product formation in C. pasteurianum. Production of ethanol and n-butanol is shown balanced with glycerol consumption (left), whereas production of 1,2- and 1,3-propanediol is shown balanced with biomass formation (right). NADH formation and consumption are shown in red and blue, respectively.
Although C. pasteurianum growth medium can be manipulated to favor 1,3-PDO as the chief fermentation end product, yielding titers up to 5 g liter−1 (27), other glycerol-utilizing organisms, particularly Clostridium butyricum (28, 29), are superior producers of 1,3-PDO in terms of both yield and titer. In this regard, the 1,3-PDO pathway becomes a prominent target for metabolic engineering to enhance n-butanol yield and selectivity. However, current models of glycerol dissimilation in clostridia dictate that the reductive 1,3-PDO pathway is essential for fermentative glycerol utilization by facilitating redox balance (9, 30). Whereas the production of n-butanol (degree of reductance [γD], 6.0) and ethanol (γD, 6.0) from glycerol represents redox-balanced pathways, the conversion of glycerol to biomass generates an excess of reducing equivalents, as cell mass (γD, 4.3) is more oxidized than glycerol (γD, 4.67) (Fig. 1B). Hence, in order to sustain glycerol fermentation, the cell must employ a means of oxidizing the surplus of reductant generated from cell mass formation, a requirement typically fulfilled by the 1,3-PDO (γD, 5.33) pathway in glycerol-fermenting species. This reductive pathway results in the net oxidation of 1 mol of NADH per 1 mol of 1,3-PDO formed. The fundamental redox role of the 1,3-PDO pathway is further evidenced from work involving metabolic engineering of C. acetobutylicum (30, 31). Although the wild-type strain is unable to utilize glycerol as a sole source of carbon, the introduction of a heterologous 1,3-PDO formation pathway provided the capacity to ferment glycerol without the use of cosubstrates. Finally, supplying C. pasteurianum with exogenous electric current was found to enhance the production of 1,3-PDO (32), thus offering additional justification for the role of this pathway in maintaining redox balance by disposing of excess reductant. Based on these reports, it is presumed that inactivation of the reductive 1,3-PDO pathway in C. pasteurianum would lead to an inability to ferment glycerol due to redox imbalance. An exception to this rationale is provided by the fermentative glycerol metabolism unveiled in Escherichia coli (33, 34), an organism lacking a 1,3-PDO pathway. In this case, production of 1,2-propanediol (1,2-PDO) (γD = 5.33), rather than 1,3-PDO (γD = 5.33), balances the excess of reducing equivalents generated from the conversion of glycerol to cell mass. The 1,2-PDO pathway branches from the ubiquitous methylglyoxal bypass and, in a manner similar to 1,3-PDO synthesis, results in the net oxidation of 1 mol of NADH per mole of 1,2-PDO formed (Fig. 1A).
In this report, we investigate inactivation of the 1,3-PDO pathway on fermentative glycerol utilization and end product distribution in C. pasteurianum. Despite current models of anaerobic glycerol utilization in clostridia, cell growth and glycerol catabolism were unaffected by disruption of the 1,3-PDO dehydrogenase gene (dhaT) in C. pasteurianum. In addition to elevated levels of n-butanol, the engineered mutant was found to exhibit, to our knowledge, the lowest 1,3-PDO titer by C. pasteurianum reported to date. Furthermore, inactivation of the 1,3-PDO pathway triggered the production of 1,2-PDO, illuminating a new model of glycerol utilization in the clostridia characterized by reciprocal 1,2- and 1,3-PDO pathways for maintenance of redox balance during fermentative glycerol dissimilation.
MATERIALS AND METHODS
Bacterial cultivation and electrotransformation.
The bacterial strains and plasmids employed in this study are listed in Table 1. C. pasteurianum ATCC 6013 was obtained from the American Type Culture Collection (ATCC) and was cultivated anaerobically at 37°C without agitation. For routine culture growth, C. pasteurianum was grown in 2× YTG medium (pH 6.3) (15) which contained (per liter): 16 g of tryptone, 10 g of yeast extract, 5 g of glucose, and 4 g of NaCl. Static flask cultivations were performed in 50 ml of standard semidefined medium (9, 35), which contained (per liter): 2 g of (NH4)2SO4, 1 g of yeast extract, 22 g of KH2PO4, 6.6 g of K2HPO4, 0.2 g of MgSO4·7H2O, 20 mg of CaCl2·2H2O, 5 mg of FeSO4·7H2O, 2 ml of trace element solution SL7, 1 mg of resazurin, and 0.5 g of cysteine-HCl. The medium pH was 6.0 to 6.1 prior to sterilization. Glycerol was autoclaved separately as a 200 g liter−1 stock solution and added to a final concentration of 40 g liter−1 in flasks containing 50 ml of medium. Prior to inoculation, biotin, p-aminobenzoic acid (PABA), and vitamin B12 were added to final concentrations of 0.02 mg liter−1, 0.02 mg liter−1, and 2 mg liter−1, respectively. Concentrations of (NH4)2SO4 (7.35 g liter−1), FeSO4·7H2O (60 mg liter−1), and yeast extract (5.08 g liter−1) were increased in the medium formulation optimized for n-butanol production (27). Cultures of recombinant C. pasteurianum contained 5 μg ml−1 thiamphenicol or 2 μg ml−1 clarithromycin. C. pasteurianum was electrotransformed according to a previous report (14). Static flask fermentations were carried out for 96 to 120 h, at which time a single endpoint sample was removed for optical density at 600 nm (OD600) measurement and metabolite analysis. Escherichia coli strains were grown aerobically at 37°C with agitation at 250 rpm in lysogeny broth (LB). E. coli strain DH5α was employed for plasmid construction and propagation, and strain ER1821 was utilized for in vivo methylation of C. pasteurianum-E. coli shuttle vectors using the gene encoding M.FnuDII methyltransferase expressed from the pFnuDIIMKn plasmid (14). Recombinant E. coli strains were selected using 100 μg ml−1 ampicillin, 30 μg ml−1 chloramphenicol, or 30 μg ml−1 kanamycin, whereby antibiotic concentrations were reduced by 50% for strains harboring two plasmids.
TABLE 1.
Strains and plasmids employed in this study
| Strains or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG Φ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK− mK+) λ− | Lab stock |
| E. coli ER1821 | F− endA1 glnV44 thi-1 relA1? e14−(mcrA) rfbD1? spoT1? Δ(mcrC-mrr)114::IS10 | Lab stock; New England Biolabs |
| Clostridium pasteurianum ATCC 6013 | Wild type | American Type Culture Collection |
| Clostridium pasteurianum dhaT::Ll.LtrB | Disruption mutant generated by inserting the Ll.LtrB intron into position 503a of the dhaT gene encoding 1,3-propanediol dehydrogenase | This study |
| Plasmids | ||
| pFnuDIIMKn | M.FnuDII methyltransferase plasmid for methylation of E. coli-C. pasteurianum shuttle vectors (Kmr; p15A ori) | 14 |
| pMTL85141ermB | E. coli-Clostridium shuttle vector (Cmr/Tmr Emr; ColE1 ori; pIM13 ori) | 14 |
| pSY6catP-ltrA | Ll.LtrB deletion derivative of pSY6catP | 15 |
| pSYltrA | Control plasmid expressing the LtrA IEP and lacking the Ll.LtrB intron | 15 |
| pSYCP | C. pasteurianum intron disruption plasmid expressing the Ll.LtrB intron and LtrA IEP from the C. pasteurianum thiolase promoter. | 15 |
| pSYCP-dhaC(391a) | Intron disruption plasmid targeting site 391a within the dhaC coding sequence | This study |
| pSYCP-dhaT(503a) | Intron disruption plasmid targeting site 503a within the dhaT coding sequence | This study |
| pSYCP-dhaB(664s) | Intron disruption plasmid targeting site 664s within the dhaB coding sequence | This study |
| pSYCP-dhaB(758a) | Intron disruption plasmid targeting site 758a within the dhaB coding sequence | This study |
| pMTLdhaT | dhaT complementation plasmid expressing the C. pasteurianum dhaT gene from its native promoter | This study |
Kmr, kanamycin resistant; Cmr, chloramphenicol resistant; Tmr, thiamphenicol resistant; Emr, erythromycin resistant; ?, unknown activity. The notation concerning mcrA indicates a loss of function.
DNA isolation and manipulation.
The oligonucleotides employed in this study were purchased from Integrated DNA Technologies (IDT; Coralville, IA) and are listed in Table 2. DNA manipulation was employed using established methods (36). Restriction endonucleases and Phusion and Taq DNA polymerases were purchased from New England BioLabs (NEB; Ipswich, MA). Commercial DNA purification and agarose gel extraction kits were obtained from Bio Basic, Inc. (Markham, Ontario, Canada). C. pasteurianum genomic DNA was isolated using a Qiagen DNeasy blood and tissue kit (Venlo, The Netherlands).
TABLE 2.
Oligonucleotides employed in this study

Underlined sequences represent relevant restriction endonuclease recognition sequences.
Plasmid pSYCP (15), expressing the Ll.LtrB group II intron and LtrA intron-encoded protein (IEP), was retargeted to 1,3-PDO pathway genes by reprogramming the Ll.LtrB intron using synthetic gBlock gene fragments or splicing by overlap extension-PCR (SOE-PCR). Synthetic gBlocks were purchased for intron retargeting to dhaC(391a) and dhaT(503a), amplified using primers pSYCP-gBlock.XhoI.S and pSYCP-gBlock.BsrGI.AS, and cloned into the XhoI and BsrGI sites of plasmid pSYCP, yielding pSYCP-dhaC(391a) and pSYCP-dhaT(503a), respectively. (Terms in parentheses represent predicted intron insertion sites, where “s” indicates sense and “a” indicates antisense.) Intron retargeting to dhaB(664s) in plasmid pSYCP-dhaB(664s) and dhaB(758a) in plasmid pSYCP-dhaB(758a) was achieved by SOE-PCR using primer sets dhaB(664s).IBS and EBS.Universal, and dhaB(664s).EBS2 and dhaB(664s).EBS1d for target site dhaB(664s), and primer sets dhaB(758a).IBS and EBS.Universal, and dhaB(758a).EBS2 and dhaB(758a).EBS1d for target site dhaB(758a). Full-length mutated PCR products were gel purified and cloned into the XhoI and BsrGI sites of plasmid pSYCP. A dhaT gene complementation plasmid, pMTLdhaT, was derived by PCR amplifying the native dhaT gene promoter and coding sequence from C. pasteurianum genomic DNA using primers dhaT.PvuI.S and dhaT.PvuI.AS. The resulting 1.5-kb PCR product was cloned into the PvuI site within plasmid pMTL85141ermB.
Intron-mediated gene disruption, enrichment, and plasmid curing.
Disruption of 1,3-PDO pathway genes using targeted Ll.LtrB intron constructs was performed as previously described (15). Predicted intron insertion sites were identified and ranked using an algorithm from TargeTronics, LLC (Austin, TX). Following intron reprogramming within pSYCP, approximately 5 to 10 μg of plasmid DNA was transferred to C. pasteurianum by electrotransformation. Transformants were initially screened for intron disruption using one locus-specific and one intron-specific primer to identify colonies harboring a chromosomal intron insertion. These colonies were further screened using locus-specific primers flanking the intron insertion site to determine if colonies were mosaic or harbored a homogeneous population of intron disruptants (15). Mosaic colonies were subjected to 10 rounds of intron enrichment in selective 2× YTG medium, and serial dilutions were plated on selective 2× YTG agar plates. Enrichment colonies were then PCR screened for intron insertion using locus-specific primers flanking the predicted intron insertion site. Intron insertion was deemed unsuccessful if colonies generated a wild-type PCR product or a mixture of wild-type and intron insertion products. Curing of plasmid pSYCP-dhaT(503a) was attempted through seven transfers in nonselective 2× YTG medium, as described previously (15). Colonies were then patched onto nonselective and selective 2× YTG agar plates to assess curing.
Identification of ectopic intron insertion by arbitrary colony PCR.
Ectopic intron insertion within the dhaT::Ll.LtrB mutant strain was assessed using arbitrary colony PCR according to a previous method (37). A first round of PCR using Taq DNA polymerase was performed using one of five partially degenerate arbitrary PCR primers (ARB1 to ARB5; 5 μM final concentration) with primer LtrB.Fw (0.5 μM final concentration). Roughly 0.5 μl of these reaction mixtures was utilized as the template in a second PCR using Taq DNA polymerase and primers ARB.nested and LtrB.Fw2. Products were column purified and sequenced using primer LtrB.Fw2. DNA sequence flanking the Ll.LtrB intron region was queried against the C. pasteurianum genome using BLAST.
Analytical methods.
Growth of static flask cultures was assessed by measuring the OD600 using a Beckman Coulter DU 520 spectrophotometer (Fullerton, CA). Concentrations of glycerol, acetate, lactate, butyrate, ethanol, n-butanol, and 1,3-propanediol were determined using a Shimadzu LC-10AT HPLC (Kyoto, Japan) equipped with an RID-10A refractive index detector and an Aminex HPX-87H column from Bio-Rad Laboratories (Richmond, CA). The mobile phase consisted of 5 mM H2SO4 (pH 2.0) maintained at a flow rate of 0.6 ml min−1. Data processing was performed using Clarity Lite from DataApex (Prague, Czech Republic). The production of 1,2-PDO was validated and quantified using NMR, as previously described (38). Briefly, culture samples were prepared for analysis by adding an internal standard to bring the sample up to 10% (vol/vol) D2O and 0.5 mM DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid). The samples were scanned on a Bruker Avance 600 MHz spectrometer with triple resonance probe (TXI 600) using the first increment of a one-dimensional (1D)-nuclear Overhauser effect spectroscopy (NOESY) pulse sequence. Spectra were analyzed using Chenomx NMR Suite 8.0 (Edmonton, Alberta, Canada). Compound concentrations were calculated from the areas of ideal NMR peaks generated from the software library and superimposed on the observed spectra. 1,3-Propanediol was added to the library using the Compound Builder utility.
RESULTS
Intron-mediated disruption of the 1,3-PDO pathway genes.
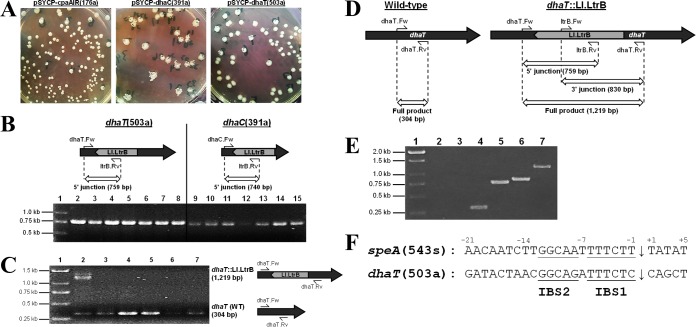
To probe the glycerol catabolic pathways of C. pasteurianum and enhance n-butanol selectivity through rational metabolic engineering, we set out to disrupt the reductive 1,3-propanediol (1,3-PDO) formation pathway in C. pasteurianum. We recently adapted the clostridial group II intron gene disruption system for use in C. pasteurianum (15), which employs plasmid pSYCP expressing the Ll.LtrB group II intron and LtrA intron-encoded protein. The conversion of glycerol to 1,3-PDO involves a two-step transformation by glycerol dehydratase (DhaBCE), encoded by dhaBCE, and 1,3-PDO dehydrogenase (DhaT), encoded by dhaT (Fig. 1A). We targeted both genes for inactivation through programming of the mobile Ll.LtrB intron within pSYCP to chromosomal target sites dhaB(664s), dhaB(758a), dhaC(391a), and dhaT(503a). Collectively, the predicted intron insertion sites possessed scores of 6.1 to 7.8 and targeted both sense (S) and antisense (AS) strands of the C. pasteurianum 1,3-PDO regulon. The corresponding plasmids pSYCP-dhaB(664s), pSYCP-dhaB(758a), pSYCP-dhaC(391a), and pSYCP-dhaT(503a) were used to transform C. pasteurianum. All four gene disruption plasmids produced poor transformation efficiencies and led to abnormal colony morphologies compared to a control intron vector targeting the C. pasteurianum cpaAIR restriction endonuclease gene (pSYCP-cpaAIR) (15). The transformants harboring pSYCP-dhaC(391a) exhibited the most irregular colony morphology, characterized by a flat nebulous shape and rough edges (Fig. 2A). Transformants of pSYCP-dhaT(503a) displayed the least-altered morphology, yet colonies were still distinct from those harboring the control plasmid pSYCP-cpaAIR. Nevertheless, seven transformant colonies were screened for intron insertion using one intron-specific primer and one locus-specific primer for each of the four dha regulon intron disruption plasmids. Several transformants (43% to 100%) corresponding to all four Ll.LtrB intron target sites produced a PCR product consistent with successful intron insertion. Notably, transformants harboring pSYCP-dhaT(503a) and pSYCP-dhaC(391a) produced the expected intron insertion product in 7/7 transformant colonies (Fig. 2B). However, subsequent PCR screening using two locus-specific primers flanking the predicted intron insertion sites produced wild-type products for all colonies screened corresponding to intron insertion sites dhaB(664s), dhaB(758a), and dhaC(391a) (data not shown). Conversely, one transformant harboring pSYCP-dhaT(503a) produced both wild-type and intron insertion PCR products using primers flanking the predicted dhaT(503a) intron insertion site (Fig. 2C). We have previously observed mosaic intron disruption colonies, composed of both wild-type and intron insertion cells, using the Ll.LtrB intron in C. pasteurianum, which we have ascribed to poor retrohoming efficiency in this host (15). In an attempt to isolate pure intron insertion mutants, we performed an intron enrichment procedure starting with one mosaic colony of each of the four pSYCP-dha disruption transformants. Following 10 successive transfers in selective 2× YTG medium, cultures were serially diluted and plated onto nonselective medium. Roughly 28 enrichment colonies from each dha-targeting transformation were subsequently screened for intron insertion using locus-specific primers flanking the intron insertion site. While target sites dhaB(664s), dhaB(758a), and dhaC(391a) produced the wild-type product in 100% of the colonies screened (data not shown), 1/24 dhaT(503a) enrichment colonies produced the expected intron insertion PCR product with no evidence of the wild-type product. This mutant colony was further validated by screening both overlapping chromosome-intron junctions (Fig. 2D and E) and Sanger DNA sequencing of the resulting products (data not shown). The resulting C. pasteurianum mutant strain was designated dhaT::Ll.LtrB.
FIG 2.
Confirmation of mosaic and homogeneous intron disruption in C. pasteurianum dhaC and dhaT genes, respectively. (A) Colony morphology of transformants harboring intron donor constructs targeting sites cpaAIR(176a), dhaC(391a), and dhaT(503a). (B) Genomic verification of dhaT(503a) and dhaC(391a) intron insertion using one locus-specific primer and one intron-specific primer. Lane 1, marker; lanes 2 to 8, pSYCP-dhaT(503a) transformants; lanes 9 to 15, pSYCP-dhaC(391a) transformants. (C) Confirmation of mosaic intron insertion at site dhaT(503a) using locus-specific primers flanking the predicted intron insertion site. Lane 1, marker; lanes 2 to 7, pSYCP-dhaT(503a) transformants. The transformant screened in lane 2 shows a mosaic intron insertion. (D) Expected PCR products of the wild-type (left) and dhaT::Ll.LtrB mutant (right) strain. Insertion of the Ll.LtrB intron into the dhaT gene leads to a 915-bp increase in size of the full-length PCR product generated using primers flanking the 503a intron insertion site (dhaT.Fw and dhaT.Rv). Both 5′- and 3′-gene-intron junction PCR products can be detected in dhaT::Ll.LtrB mutant cells using primer sets dhaT.Fw and ltrB.Rv, and ltrB.Fw and dhaT.Rv, respectively. (E) Genomic verification of a single positive dhaT::Ll.LtrB mutant colony by amplification of all three PCR products depicted in Fig. 2A (5′ junction, 3′ junction, and full product). A wild-type C. pasteurianum colony was included as a control for all three PCR primer sets. Lane 1, marker; lanes 2 to 4, wild-type colony; lanes 5 to 7, dhaT::Ll.LtrB mutant colony; lanes 2 and 5, 5′ junction; lanes 3 and 6, 3′ junction; lanes 4 and 7, full product. (F) Comparison of chromosomal intron insertion sites corresponding to speA(543s) and dhaT(503a). The predicted intron insertion site is shown with an arrow. Sequence similarities in intron binding site 1 (IBS1) and IBS2 involved in intron base pairing with target DNA are underlined.
Curing of pSYCP-dhaT(503a) and identification of speA ectopic intron insertion.
Following construction and validation of the C. pasteurianum dhaT::Ll.LtrB mutant, we attempted to cure the strain of the pSYCP-dhaT(503a) intron donor plasmid. Using a previously reported procedure (15), one colony of the dhaT::Ll.LtrB mutant was transferred in nonselective 2× YTG medium three times, serially diluted, and spread onto nonselective 2× YTG agar plates. A total of 24 single colonies were then picked and streaked onto nonselective and selective 2× YTG agar to assess plasmid curing. Although this method led to a curing efficiency of 50% for plasmid pSYCP-cpaAIR(176a) (15), all 24/24 dhaT::Ll.LtrB mutant colonies produced growth on selective 2× YTG agar. In a second attempt at plasmid curing, we subcultured the dhaT::Ll.LtrB mutant using 10 successive transfers and screened 16 resultant colonies for plasmid loss. Once again, all 16 colonies screened grew on selective 2× YTG agar. We attempted a final plasmid curing experiment by cultivating the dhaT::Ll.LtrB mutant under dramatically altered growth conditions in the event that specific medium components were responsible for our inability to cure mutant cells of the pSYCP-dhaT(503a) plasmid. In place of 2× YTG, we cultivated the dhaT::Ll.LtrB mutant in a semidefined medium (9) containing 1 g liter−1 yeast extract and 40 g liter−1 sodium gluconate as a carbon source. The dhaT::Ll.LtrB mutant was transferred eight times in nonselective semidefined medium, and 100 colonies were assessed for plasmid curing. All 100 colonies screened in this manner grew on selective semidefined agar, again indicating an inability to cure plasmid pSYCP-dhaT(503a) from the dhaT::Ll.LtrB mutant.
The inability to cure intron donor plasmid from intron-disrupted cells suggests that the intron has inserted into the sense strand of an essential gene (39). Whereby antisense-oriented intron insertions produce unconditional disruptions, as transcription from the target gene promoter generates the complement sequence of the intron RNA, introns inserted into the sense orientation are transcribed into functional intron RNA (40, 41). If a source of the LtrA IEP is present, such sense-oriented introns can then be spliced from target mRNA, leading to the production of an intact and functional protein product despite insertional inactivation at the DNA level. Hence, essential genes can only be disrupted using a sense-oriented intron and if a source of the LtrA protein is supplied to mediate RNA splicing (39). Since the dhaT(503a) insertion carried by the dhaT::Ll.LtrB mutant constitutes an antisense intron insertion, we speculated that in addition to the intended dhaT insertion, the mutant also accumulated an off-target intron insertion within the sense strand of a gene that is essential to C. pasteurianum. To address this hypothesis, we used arbitrary PCR to identify potential ectopic intron insertions, since the technique is widely employed to rapidly genotype mutants generated via random transposon mutagenesis (42). We utilized five partially degenerate arbitrary PCR primers (Table 2), along with an intron-specific primer to amplify small regions of DNA sequence flanking any intron insertion that may be present within the genome of the dhaT::Ll.LtrB mutant. The resulting PCR products were sequenced, and intron-flanking sequences obtained were queried against the C. pasteurianum genome using BLAST. While only one of five arbitrary PCR primers amplified sequence flanking the validated dhaT(503a) intron insertion site, the remaining four arbitrary PCR primers generated the same off-target sequence corresponding to the arginine decarboxylase (speA) coding sequence. The Ll.LtrB intron was found to be inserted in the same position within the speA gene [speA(543s)] in all four PCR products. A comparison of the nucleotide sequences flanking the speA intron insertion site to the dhaT(503a) target site unveiled substantial sequence similarity in the intron binding site 2 (IBS2) and IBS1 regions required for base pairing between Ll.LtrB intron RNA and target DNA (Fig. 2F). Importantly, Ll.LtrB insertion occurred in the sense strand of the speA coding sequence, providing justification for our inability to cure the dhaT::Ll.LtrB mutant of plasmid pSYCP-dhaT(503a). We propose that the C. pasteurianum speA gene is essential for growth under the conditions employed in this study, whereby intron-mediated speA disruption is conditional on the production of the LtrA protein from plasmid pSYCP-dhaT(503a). The presence of plasmid pSYCP-dhaT(503a) and, therefore, LtrA protein, facilitates excision of the Ll.LtrB intron from speA at the mRNA level, generating intact functional SpeA protein (39). On the other hand, the disruption of dhaT in the dhaT::Ll.LtrB mutant is permanent and stable, as the Ll.LtrB intron is incapable of splicing from antisense-oriented DNA insertions (40, 41). Given the inability to cure cells of the intron donor plasmid, we proceeded with a characterization of solvent production using the dhaT::Ll.LtrB mutant.
Solvent production by C. pasteurianum dhaT::Ll.LtrB in standard and butanol-optimized media.
To assess the effect of dhaT disruption on the growth and product distribution of C. pasteurianum, we first performed static flask cultivations of wild-type and dhaT::Ll.LtrB mutant strains using a standard semidefined growth medium containing 40 g liter−1 glycerol and 1 g liter−1 yeast extract. Under these conditions, growth on glycerol as a sole source of carbon was unaffected by dhaT disruption in the dhaT::Ll.LtrB mutant compared to the wild-type strain, as the two cultures reached a similar final cell density (OD600, 3.0 to 3.1) and consumed roughly the same amount of glycerol (approximately 28 g liter−1). Conversely, metabolite analysis of culture supernatants revealed dramatic differences in the central fermentative metabolism between the engineered mutant and the wild-type strain (Table 3). Whereas the wild-type strain converted glycerol to nearly equal quantities of 1,3-PDO and n-butanol (4.6 and 5.5 g liter−1, respectively), the dhaT::Ll.LtrB mutant produced 83% less 1,3-PDO (0.79 g liter−1) and 29% more n-butanol (7.1 g liter−1). Accordingly, wild-type C. pasteurianum utilized 47.7% and 19.4% of glycerol for n-butanol and 1,3-PDO production, respectively, while the dhaT::Ll.LtrB mutant converted 62.8% of substrate to n-butanol and only 3.4% to 1,3-PDO. n-Butanol selectivity in the engineered mutant was 0.74 g n-butanol g−1 of total solvents (butanol, ethanol, 1,2-PDO, and 1,3-PDO), compared to 0.43 g n-butanol g−1 of total solvents in the wild-type strain (butanol, ethanol, and 1,3-PDO), corresponding to 72% improvement. Both strains generated small amounts of ethanol (1.4 to 2.7 g liter−1) and only trace amounts of acetate, lactate, and butyrate (0.07 to 0.31 g liter−1 of total acids). The dhaT::Ll.LtrB mutant was capable of growth in a chemically defined medium containing biotin and p-aminobenzoic acid (PABA) (data not shown), indicating that growth was not dependent on the presence of yeast extract.
TABLE 3.
HPLC metabolite analysis of the wild-type and dhaT::Ll.LtrB mutant strains of C. pasteurianum in standard and butanol-optimized growth media
| Strain by medium type | Cultivation time (h) | Growth (OD600) | Glycerol consumed (g liter−1) | Products formed (g liter−1 [%])a |
Carbon recovery in products (%)c | Butanol selectivity (g g−1)d | |||
|---|---|---|---|---|---|---|---|---|---|
| Butanol | 1,3-PDO | Ethanol | Acidsb | ||||||
| Standard growth medium | |||||||||
| Wild type | 112 | 3.1 | 28.5 | 5.5 ± 0.9 (47.7) | 4.6 ± 0.2 (19.4) | 2.7 ± 0.9 (18.7) | 0.07 ± 0.06 (0.4) | 86.4 | 0.43 |
| dhaT::Ll.LtrB mutant | 97 | 3.0 | 28.0 | 7.1 ± 2.0 (62.8) | 0.79 ± 0.31 (3.4) | 1.4 ± 0.9 (9.7) | 0.31 ± 0.16 (1.8) | 79.1 | 0.74 |
| dhaT::Ll.LtrB [pMTLdhaT] | 107 | 2.8 | 24.4 | 5.0 ± 0.7 (50.9) | 1.6 ± 0.2 (7.7) | 2.6 ± 0.4 (21.3) | 0.25 ± 0.07 (1.5) | 81.9 | 0.54 |
| Optimized growth medium | |||||||||
| Wild type | 102 | 4.1 | 31.8 | 6.7 ± 0.4 (52.1) | 3.6 ± 0.5 (13.7) | 2.9 ± 1.6 (18.2) | 0.72 ± 0.21 (3.6) | 87.7 | 0.51 |
| dhaT::Ll.LtrB mutant | 94 | 4.8 | 32.6 | 8.6 ± 0.1 (65.5) | 0.58 ± 0.07 (2.1) | 0.7 ± 0.1 (4.3) | 0.35 ± 0.07 (1.9) | 75.4 | 0.83 |
Product titers are reported as average ± standard deviation of the results from three biological replicates. The percent moles of glycerol consumed in the production of each metabolite is provided in parentheses.
Combined titer of acetate, butyrate, and lactate.
Calculated as the percent moles of carbon in total liquid products (including 1,2-PDO and excluding biomass and gaseous products) per mole of carbon in glycerol consumed.
Reported as grams of n-butanol produced per gram of total solvents (butanol, ethanol, 1,2-PDO, and 1,3-PDO).
In an attempt to complement DhaT deficiency in the dhaT::Ll.LtrB mutant, we transformed the engineered strain with plasmid pMTLdhaT containing the C. pasteurianum dhaT gene expressed from its native promoter. Although the DhaT rescue strain produced twice as much 1,3-PDO as the dhaT::Ll.LtrB mutant, the complementation was weak, since the wild-type strain produced almost 3-fold-higher levels of 1,3-PDO than the complemented strain (Table 3). While n-butanol production was enhanced in the dhaT::Ll.LtrB mutant compared to the wild-type strain, the n-butanol titer following dhaT complementation was similar to that in the wild-type strain, wherein roughly half of the consumed glycerol was metabolized to n-butanol. Based on these data, plasmid-borne dhaT was only partially effective in complementing inactivated dhaT in the engineered mutant.
To further limit 1,3-PDO levels and enhance n-butanol production, we investigated product distribution of the dhaT::Ll.LtrB mutant in a growth medium previously optimized for n-butanol production and low-level 1,3-PDO formation (27). Static flask fermentations were again performed using the wild-type and dhaT::Ll.LtrB mutant strains in the medium containing 40 g liter−1 glycerol. Compared to the standard growth medium, both the wild-type and engineered strain grew to a higher final cell density (OD600, 4.1 and 4.8, respectively) and consumed more glycerol (31.8 and 32.6 g liter−1, respectively) in the optimized medium (Table 3). The optimized medium proved effective in enhancing n-butanol production, as the titers were increased by 22% and 21% in the wild-type and dhaT::Ll.LtrB mutant strains, respectively, compared to the standard medium. Conversely, the use of the optimized medium led to a further 28% decrease in 1,3-PDO titer in the wild-type strain and a 34% decline in the engineered mutant. Under these conditions, the dhaT::Ll.LtrB mutant converted 65.5% of glycerol into n-butanol yet only 2.1% into 1,3-PDO, yielding final titers of 8.6 g liter−1 n-butanol and 0.58 g liter−1 1,3-PDO. n-Butanol selectivity was 0.83 g n-butanol g−1 of solvents in the engineered mutant and 0.51 g n-butanol g−1 of solvents in the wild-type strain, corresponding to a 63% increase.
Elucidation of glycerol catabolic pathways in dhaT::Ll.LtrB mutant.
During HPLC analysis of fermentation samples taken from cultures of the C. pasteurianum dhaT::Ll.LtrB mutant, we observed a peak (approximate retention time of 17.3 min) that partially overlapped the peak corresponding to 1,3-PDO (approximate retention time of 17.8 min) (Fig. 3A). This peak was observed in all dhaT::Ll.LtrB mutant samples yet was not detected in any wild-type culture supernatants, suggesting that production of the unknown metabolite is linked to disruption of dhaT. In line with this hypothesis, the area of the unknown peak was reduced in two out of three replicates of the dhaT::Ll.LtrB[dhaT] complementation strain and was not detected in the remaining replicate. In a first attempt to elucidate the identity of the unknown compound, we analyzed HPLC retention times of pure standards corresponding to key metabolites and pathway intermediates produced by C. pasteurianum (9, 10, 43), as well as compounds associated with glycerol dissimilation in other glycerol-fermenting microorganisms (2, 44, 45). Candidate metabolites included 3-hydroxypropionaldehyde, 3-hydroxypropionate, 1,2-PDO, propionate, n-propanol, acetoin, acetone, and acrolein. HPLC standards of prospective metabolites were prepared, and retention times (Table 4) were compared against chromatograms corresponding to the dhaT::Ll.LtrB mutant. Since 3-hydroxypropionaldehyde is not available commercially, a retention time of 15.6 min was assumed based on a report by Rütti et al. (46) that employed the same HPLC conditions utilized in this study. Similarly, an approximate retention time of 27.25 min for acrolein was assumed based on a report by Schaefer et al. (47), using conditions similar to those employed here. Although DhaT catalyzes the reduction of 3-hydroxypropionaldehyde to 1,3-PDO, we did not detect a buildup of this intermediate in the dhaT::Ll.LtrB mutant. We also did not identify metabolites derived from 3-hydroxypropionaldehyde, such as acrolein and 3-hydroxypropionate. Of the candidate compounds screened using HPLC, only 1,2-PDO (17.3 min) exhibited a retention time similar to that of the unknown peak (17.3 min) in dhaT::Ll.LtrB mutant culture supernatants (Fig. 3B), although a cluster of other key metabolites, namely, propionate (17.5 min), acetoin (17.8 min), and 1,3-PDO (17.8 min), were found to coelute within a very narrow retention range of 17.5 to 17.8 min. To further resolve this cluster and identify the unknown metabolite produced by the dhaT::Ll.LtrB mutant, we subjected both wild-type and mutant culture samples to NMR analysis. NMR metabolite profiling revealed eight compounds that were present at substantially higher levels (approximately 100- to 1,000-fold) than all other metabolites in both wild-type and dhaT::Ll.LtrB mutant strain supernatants. As expected, these compounds included glycerol, formate, acetate, butyrate, ethanol, n-butanol, and 1,3-PDO. Among potential candidates, only 1,2-PDO was identified in all culture supernatants derived from the dhaT::Ll.LtrB mutant and, to a lesser extent, in culture samples of the wild-type strain (Fig. 3C). Despite some overlap with other chemical species, 1,2-PDO could be quantified using resonances at both 1.13 ppm and 3.44 ppm, with a 3.9-ppm resonance used for confirmation. Using the standard growth medium, 1,2-PDO production was found to be elevated 5-fold in the dhaT::Ll.LtrB mutant (0.30 ± 0.01 g liter−1) relative to the wild-type organism (0.06 ± 0.00 g liter−1). Cultivation of the engineered mutant in medium optimized for n-butanol production led to an increased 1,2-PDO titer of 0.43 ± 0.05 g liter−1. Based on combined HPLC and NMR analyses, the identity of the unknown metabolite was unequivocally ascribed to 1,2-PDO. In other organisms, the 1,2-PDO pathway branches from the methylglyoxal bypass through two convergent pathways involving hydroxyacetone (acetol) or lactaldehyde (hydroxypropionaldehyde) (33, 48, 49) (Fig. 1A). Using pure standards to guide detection, we probed wild-type and dhaT::Ll.LtrB mutant culture supernatants for the presence of these intermediates using NMR. While the presence of lactaldehyde could not be confirmed or rejected, we observed resonances at 2.14 ppm and 4.37 ppm corresponding to the expected signals for hydroxyacetone based on spike-in experiments. In line with 1,2-PDO levels, hydroxyacetone concentration was elevated 4-fold in the engineered mutant (0.079 ± 0.05 mM) relative to the wild-type strain (0.019 ± 0.008 mM). Finally, we did not detect methylglyoxal in samples taken from either strain.
FIG 3.
HPLC and NMR confirmation of 1,2-PDO production by dhaT::Ll.LtrB mutant. (A) Detection of a unique HPLC peak in supernatants derived from the dhaT disruption mutant (dhaT::Ll.LtrB) and the dhaT-complemented mutant (dhaT::Ll.LtrB[pMTLdhaT]). The HPLC peak corresponding to the unknown metabolite is shown with black arrows and was found to partially overlap the 1,3-PDO peak. The heights of the 1,3-PDO peaks are proportionate to the 1,3-PDO concentration. (B) Comparison of the unknown metabolite (arrow) and 1,3-PDO HPLC peaks in dhaT::Ll.LtrB mutant culture supernatants with pure standards of 1,2-PDO and 1,3-PDO at concentrations of 0.5 g liter−1 and 1.0 g liter−1, respectively. The heights of the 1,3-PDO peaks are proportionate to 1,3-PDO concentration. (C) NMR resonances of 1,2-PDO used for identification and quantification. Arrows identify specific peaks attributed to 1,2-PDO. Spectra from two replicates of each strain are shown and were compared against a reference 1,2-PDO spectrum (not shown) from a standard chemical library. The chemical shift is depicted in parts per million (ppm) and is based on the change in standard resonance frequency of hydrogen (600 MHz).
TABLE 4.
HPLC retention times of relevant metabolites and pathway intermediates discussed in this study
| Metabolite | Approx retention time (min) | Reference |
|---|---|---|
| Lactate | 12.6 | This study |
| 3-Hydroxypropionate | 13.1 | This study |
| Glycerol | 13.9 | This study |
| Acetate | 15.0 | This study |
| 3-Hydroxypropionaldehydea | 15.6 | 46 |
| Unknown dhaT::Ll.LtrB mutant peak | 17.3 | This study |
| 1,2-Propanediol | 17.4 | This study |
| Propionate | 17.5 | This study |
| Acetoin | 17.8 | This study |
| 1,3-Propanediol | 17.8 | This study |
| Butyrate | 20.9 | This study |
| Acetone | 21.6 | This study |
| Ethanol | 22.2 | This study |
| Acroleinb | 27.2 | 47 |
| Propanol | 27.5 | This study |
| Butanol | 36.3 | This study |
DISCUSSION
In this report, we describe the construction and characterization of a C. pasteurianum mutant strain possessing an inactivated 1,3-PDO formation pathway. Despite the requirement for 1,3-PDO production predicted by current metabolic models of glycerol fermentation in Clostridium and related microorganisms, inactivation of 1,3-PDO dehydrogenase (dhaT) did not affect growth of the organism on glycerol as a sole source of carbon and energy. Instead, we observed dramatically reduced levels of 1,3-PDO, which was compensated by elevated n-butanol production and activation of a previously unidentified 1,2-PDO formation pathway. This work embodies the lowest 1,3-PDO titer (0.58 g liter−1) and one of the highest n-butanol selectivities (0.83 g n-butanol g−1 of solvents) reported to date for C. pasteurianum.
In response to inactivation of the 1,3-PDO pathway, the dhaT::Ll.LtrB mutant produced substantially more butanol (approximately 30%), likely a consequence of altered electron flow. As a Clostridium species, C. pasteurianum generates reductant in the form of NADH and reduced ferredoxin. During glycerol fermentation by the wild-type strain, NADH is predominantly used to drive the formation of butanol and 1,3-PDO. The fate of reduced ferredoxin is less implicit, as excess electrons can be “disposed of” through the formation of molecular hydrogen by the hydrogenase enzyme, or the reductant can be converted to NADH via ferredoxin:NAD+ oxidoreductase. It has been shown that hydrogen evolution is dramatically reduced during clostridial glycerol fermentation due to induction of the ferredoxin:NAD+ oxidoreductase enzyme, thus yielding additional NADH (50). With the 1,3-PDO and hydrogenase pathways largely inoperative, the dhaT::Ll.LtrB mutant was forced to overproduce butanol and activate the previously unidentified 1,2-PDO pathway to oxidize the excess NADH and sustain glycerol fermentation. In line with HPLC analysis, significant 1,2-PDO production (0.3 to 0.43 g liter−1) by the dhaT::Ll.LtrB mutant strain was confirmed by NMR in this study (Fig. 3). While NMR also revealed trace amounts of 1,2-PDO in culture supernatants derived from the wild-type strain (0.06 g liter−1), the levels were below the limit of detection using HPLC. Indeed, no prior studies have reported 1,2-PDO production by C. pasteurianum. Based on previous reports of fermentative glycerol utilization in E. coli (33, 34) and characterization of the dhaT::Ll.LtrB mutant in this study, we propose that the engineered mutant enlists both PDO pathways to consume reducing equivalents (i.e., NADH) generated in the synthesis of biomass during glycerol fermentation. Using a cell mass formula of C4H7O2N (9) and a dry cell weight of 0.26 g per OD600 for both C. acetobutylicum (51) and C. beijerinckii (52), the dhaT::Ll.LtrB mutant generated roughly 16.4 mM NADH through conversion of glycerol to biomass in the optimized growth medium. Thus, the formation of 16.4 mM 1,2-PDO and/or 1,3-PDO, equivalent to 1.2 g liter−1 total PDO, would be expected for the cell to restore redox balance (Fig. 1). Since the engineered strain produced 1.0 g liter−1 total PDO, it is apparent that the activity of both the 1,2- and 1,3-PDO pathways is necessary for sustaining glycerol catabolism in the engineered mutant. This outcome is corroborated by investigations into the anaerobic glycerol catabolism of E. coli (33), in which the production of only 0.04 g liter−1 1,2-PDO was sufficient for balancing reducing equivalents generated from biomass formation. Although the dhaT::Ll.LtrB mutant derived in this study consumed approximately 4-fold more glycerol (32.6 g liter−1 compared to 8.1 g liter−1) and grew to a 4-fold higher optical density (4.8 compared to 1.2, respectively) than E. coli, the dhaT::Ll.LtrB mutant produced roughly 28-fold more total PDO. Based on the above-described analyses, we propose that the activity of the dual PDO pathways provides sufficient means of balancing reducing equivalents generated in the synthesis of cell mass by the dhaT::Ll.LtrB mutant.
In the glycerol fermentation carried out by E. coli and other members of Enterobacteriaceae, 1,2-PDO production was found to be dependent on an oxidative type II glycerol dehydrogenase (glyDH-II) encoded by gldA (33). Whereas typical type I glycerol dehydrogenases oxidize glycerol to dihydroxyacetone, glyDH-II also plays an integral role in 1,2-PDO formation by reducing hydroxyacetone (acetol) to 1,2-PDO (Fig. 1A). The C. pasteurianum genome harbors five putative glycerol dehydrogenase genes (20), of which four of the corresponding proteins (loci CP6013_0378, CP6013_1937, CP6013_3371, and CP6013_3819) share 26% to 53% amino acid identities with glyDH-II from E. coli K-12 substrain MG1655 (locus b3945). At least one of these enzymes ostensibly mediates the production of 1,2-PDO in C. pasteurianum. The organism also harbors a single gene encoding methylglyoxal synthase (mgsA; locus CP6013_1729) for diversion of dihydroxyacetone phosphate from glycolysis to the methylglyoxal bypass (Fig. 1A). Methylglyoxal can then be converted to 1,2-PDO via hydroxyacetone (acetol) or lactaldehyde by sequential reduction reactions. In Clostridium sphenoides, 1,2-PDO is produced from lactaldehyde through the reduction of methylglyoxal by methylglyoxal reductase and 1,2-PDO dehydrogenase (49). In E. coli (33) and Clostridium thermosaccharolyticum (48), however, 1,2-PDO formation was found to proceed via hydroxyacetone through the action of an aldo-keto reductase, encoded by yeaE, yghZ, or yafB in E. coli (33), followed by glycerol dehydrogenase (gldA). Of these possible intermediates, we were successful in detecting only hydroxyacetone using NMR, suggesting that C. pasteurianum employs a pathway analogous to E. coli and C. thermosaccharolyticum. In this regard, C. pasteurianum harbors an assortment of genes encoding putative aldo-keto reductases (loci CP6013_0976, CP6013_1953, CP6013_2079, CP6013_2131, or CP6013_2517) for reduction of methylglyoxal to hydroxyacetone (Fig. 1A). The 1,2-PDO pathway in E. coli has been found to be induced under a very stringent set of bioprocessing conditions that includes low concentrations of potassium and phosphate, high concentrations of glycerol, acidic pH, and a low partial pressure of molecular hydrogen (33). In C. sphenoides, phosphate limitation alone is sufficient for triggering substantial production of 1,2-PDO (49), as the methylglyoxal bypass provides a nonglycolytic alternative devoid of phosphorylated intermediates. The C. pasteurianum dhaT::Ll.LtrB mutant in this study was cultivated in a medium containing very high concentrations of both potassium and phosphate (200 mM potassium phosphate buffer). While phosphate inhibits glycerol dehydrogenase (53) and methylglyoxal synthase (54, 55), both of which are key enzymes for 1,2-PDO formation, potassium enhances the toxicity of methylglyoxal (56), a highly toxic metabolite. Although glycerol utilization by the dhaT::Ll.LtrB mutant appeared to proceed unimpeded under these conditions (Table 3), it is possible that bioprocessing constraints were inadvertently placed on the mutant's capacity for 1,2-PDO formation. Because the engineered strain continued to synthesize small amounts of 1,3-PDO despite the disruption of dhaT, it is probable that the 1,2-PDO pathway alone is insufficient for maintaining redox balance under the cultivation conditions employed. Instead, a small amount of 1,3-PDO production, presumably catalyzed by alternative iron-containing dehydrogenases exhibiting low-level activity on 3-hydroxypropionaldehyde, contributed to redox balance in the engineered mutant. Indeed, the C. pasteurianum genome encodes at least 13 iron-type alcohol dehydrogenases possessing significant similarity to the canonical DhaT amino acid sequence, providing justification for 1,3-PDO production despite dhaT inactivation. Given that multiple aldehyde-alcohol dehydrogenases are also involved in formation of butyraldehyde and n-butanol in C. acetobutylicum (57) and presumably C. pasteurianum (20), it may not be possible to completely abolish 1,3-PDO formation in the dhaT::Ll.LtrB mutant without compromising the strain's capacity for high level n-butanol formation. Hence, inactivation of the upstream gene encoding glycerol dehydratase (dhaBCE) and deletion of the entire 1,3-PDO regulon represent superior strategies for developing a completely 1,3-PDO-deficient mutant of C. pasteurianum. 1,3-PDO titers could be further decreased through inactivation of the Spo0A transcriptional regulator, as recently demonstrated by Sandoval et al. (16), resulting in enhanced glycerol utilization and decreased production of 1,3-PDO (Table 5). A number of promising C. pasteurianum strains have also been generated via random mutagenesis and directed evolution, such as the hyper n-butanol-producing strain described by Malaviya et al. (12). Implementation of dhaT inactivation in these strains might result in highly productive mutants for industrial production of n-butanol from waste glycerol.
TABLE 5.
Comparison of C. pasteurianum mutant strains constructed through rational metabolic engineering and random mutagenesis
| Strain designation | Strain characteristic(s) | Products formed (g liter−1) |
Butanol yield (g of n-butanol g−1 of glycerol consumed) | Butanol selectivity (g of n-butanol g−1 of total solvents) | Reference | ||
|---|---|---|---|---|---|---|---|
| Butanol | 1,3-PDO | Total solventsa | |||||
| ATCC 6013 | Wild type | 6.7 | 3.6 | 13.2 | 0.21 | 0.51 | This study |
| dhaT::Ll.LtrB mutant | dhaT intron disruption mutant | 8.6 | 0.58 | 10.3 | 0.26 | 0.83 | This study |
| M150Bb | Evolved strain (chemical mutagenesis) | 7.1 | 1.5 | 9.1 | 0.18 | 0.78 | 16 |
| Δspo0A mutantb | spo0A deletion mutant | Comparable phenotype to evolved strain M150B | Comparable phenotype to evolved strain M150B | Comparable phenotype to evolved strain M150B | Comparable phenotype to evolved strain M150B | Comparable phenotype to evolved strain M150B | 16 |
| M150B | Evolved strain (chemical mutagenesis) | 11.7 | 0.9 | 13.1 | 0.27 | 0.89 | 16 |
| MBEL_GLY2 | Evolved strain (chemical mutagenesis) | 13.7 | 4.6 | 18.8 | 0.30 | 0.73 | 12 |
| MBEL_GLY2c | Evolved strain (chemical mutagenesis) | 17.3 | 9.5 | 27.4 | 0.30 | 0.63 | 12 |
Includes n-butanol, ethanol, and 1,3-PDO. The strains assessed in this study (wild-type ATCC 6013 and dhaT::Ll.LtrB mutant) also include 1,2-PDO.
Cultivated on crude glycerol.
Cultivated under optimized conditions.
Whereas C. pasteurianum is the only organism that combines fermentative glycerol utilization with high-level n-butanol production, C. acetobutylicum has been engineered for glycerol-to-butanol conversion through reconstitution of a full 1,3-PDO synthesis pathway from Clostridium butyricum (30, 31). Based on the model of clostridial glycerol utilization set forth in this study, wherein 1,2- and 1,3-PDO pathways exemplify complementary means of facilitating redox balance, it follows that C. acetobutylicum could be engineered for fermentative glycerol utilization through the introduction or activation of a functional 1,2-PDO formation pathway. Wild-type C. acetobutylicum harbors a methylglyoxal synthase gene (locus CA_C1604) (58), as well as multiple aldo-keto reductases (loci CA_C1958 and CA_C3378) sharing substantial amino acid identities with the corresponding proteins from E. coli. Although C. acetobutylicum also harbors a gene encoding glycerol dehydrogenase (locus CA_C1626), the corresponding protein bears little resemblance to the key glyDH-II glycerol dehydrogenase enzyme from E. coli. In principle, expression of the appropriate C. pasteurianum glycerol dehydrogenase gene in C. acetobutylicum could trigger 1,2-PDO synthesis, providing an alternative means of engineering C. acetobutylicum for fermentative glycerol utilization. In a broader sense, it would be of interest to determine how pervasive 1,2-PDO production is within the Clostridium genus and among glycerol utilizers in general. Evidence of 1,2-PDO formation by select clostridia has been reported, including C. beijerinckii, C. difficile, C. sphenoides, and C. thermosaccharolyticum (48, 49, 59), suggesting a ubiquitous role of the 1,2-PDO pathway in detoxifying methylglyoxal and surviving phosphate starvation, in addition to its function as a sink for reducing equivalents as observed in wild-type E. coli and engineered C. pasteurianum strains.
Although both genes of the 1,3-PDO pathway (dhaBCE and dhaT) were targeted for disruption in this study, we were only successful in inactivating dhaT. Nevertheless, successful intron insertion was detected in all target sites assessed (Fig. 2B), including dhaB and dhaC subunits of the glycerol dehydratase gene, yet a pure dhaBCE inactivation mutant could not be isolated. Interestingly, mosaic dhaB and dhaC intron disruption colonies exhibited a highly irregular morphology, while colonies of the dhaT::Ll.LtrB mutant also appeared altered in appearance compared to a control intron insertional mutant (Fig. 2A). These data suggest that the 1,3-PDO pathway may be active or induced under standard growth conditions in the absence of glycerol, as the disruption of this pathway appears to affect colony morphology or overall cellular fitness. In glycerol-fermenting organisms, expression of the 1,3-PDO pathway is induced by dihydroxyacetone or glycerol (60), both of which are expected to be absent from the 2× YTG medium employed for gene disruption in this study. Given our difficulties in isolating a dhaBCE gene inactivation mutant using group II intron biology, this study also highlights the overall lack of robust genetic tools available for engineering C. pasteurianum and other solventogenic clostridia (15, 61). Indeed, the dhaT intron insertional mutant in this study was found to also harbor a conditional intron insertion in the C. pasteurianum speA gene, which we propose is essential for cell viability under the culture conditions employed. Because the Ll.LtrB intron is capable of splicing from the sense-oriented speA intron insertion but not from the antisense-oriented dhaT integration (39–41), it is highly unlikely that the off-target integration contributed to the phenotypic characteristics of the engineered dhaT::Ll.LtrB mutant. Additionally, plasmid-based complementation of dhaT proved to be only partially effective, again illuminating poor performance of clostridial genetic engineering tools. While plasmid stability has not been assessed in great detail in C. pasteurianum, the pIM13 plasmid replicon used for dhaT complementation in this study has been shown to be segregationally unstable in Clostridium difficile (62) and Clostridium tyrobutyricum (63). In the report on C. tyrobutyricum, the expression of an alcohol dehydrogenase from a pIM13-based plasmid led to slow culture growth, low enzyme activity, and overall poor alcohol titers compared to other plasmid replicons assayed (63). Such plasmid instability may have contributed to weak dhaT complementation in this study. To augment the growing clostridial molecular genetic toolbox, powerful and robust genome editing technologies based on CRISPR-Cas systems have recently been reported in a number of clostridia, including C. beijerinckii (64), C. cellulolyticum (65), and C. pasteurianum (18). A methodology based on homologous recombination and mazF counterselection has also been reported recently for use in engineering C. pasteurianum (16, 66). While the intron-based strategy employed in this study predates the advent of homologous-recombination-based techniques in C. pasteurianum, the availability of more-robust genome editing methods offers new opportunities for clostridial strain engineering. Indeed, the C. pasteurianum dhaT inactivation mutant constructed in this work exemplifies an exceptional chassis for further engineering toward the realization of an industrial bioprocess based on the conversion of low-value glycerol to n-butanol as a bulk chemical and prospective biofuel.
ACKNOWLEDGMENTS
We thank Jiri Perutka of TargeTronics for helpful discussions.
D.A.C. is a founder and employee of Neemo, Inc., at which M.E.P. has also been employed. Neemo, Inc. has a financial interest in the production of biofuels using Clostridium. The remaining authors declare no competing interests.
Funding Statement
The funding by Gouvernement du Canada | Networks of Centres of Excellence of Canada (NCE) was through the BioFuelNet research network.
REFERENCES
- 1.US Energy Information Administration. 2016. International energy statistics. US Energy Information Administration, Washington, DC: http://www.eia.gov/cfapps/ipdbproject/iedindex3.cfm. [Google Scholar]
- 2.da Silva GP, Mack M, Contiero J. 2009. Glycerol: a promising and abundant carbon source for industrial microbiology. Biotechnol Adv 27:30–39. doi: 10.1016/j.biotechadv.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Yazdani SS, Gonzalez R. 2007. Anaerobic fermentation of glycerol: a path to economic viability for the biofuels industry. Curr Opin Biotechnol 18:213–219. doi: 10.1016/j.copbio.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Yang FX, Hanna MA, Sun RC. 2012. Value-added uses for crude glycerol–a byproduct of biodiesel production. Biotechnol Biofuels 5:13. doi: 10.1186/1754-6834-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson JC, He BB. 2006. Characterization of crude glycerol from biodiesel production from multiple feedstocks. Appl Eng Agric 22:261–265. doi: 10.13031/2013.20272. [DOI] [Google Scholar]
- 6.Chatzifragkou A, Papanikolaou S. 2012. Effect of impurities in biodiesel-derived waste glycerol on the performance and feasibility of biotechnological processes. Appl Microbiol Biotechnol 95:13–27. doi: 10.1007/s00253-012-4111-3. [DOI] [PubMed] [Google Scholar]
- 7.Jensen TO, Kvist T, Mikkelsen MJ, Christensen PV, Westermann P. 2012. Fermentation of crude glycerol from biodiesel production by Clostridium pasteurianum. J Ind Microbiol Biotechnol 39:709–717. doi: 10.1007/s10295-011-1077-6. [DOI] [PubMed] [Google Scholar]
- 8.Wijesekara R, Nomura N, Sato S, Matsumura M. 2008. Pre-treatment and utilization of raw glycerol from sunflower oil biodiesel for growth and 1,3-propanediol production by Clostridium butyricum. J Chem Technol Biotechnol 83:1072–1080. [Google Scholar]
- 9.Biebl H. 2001. Fermentation of glycerol by Clostridium pasteurianum–batch and continuous culture studies. J Ind Microbiol Biotechnol 27:18–26. doi: 10.1038/sj.jim.7000155. [DOI] [PubMed] [Google Scholar]
- 10.Dabrock B, Bahl H, Gottschalk G. 1992. Parameters affecting solvent production by Clostridium pasteurianum. Appl Environ Microbiol 58:1233–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taconi KA, Venkataramanan KP, Johnson DT. 2009. Growth and solvent production by Clostridium pasteurianum ATCC (R) 6013 (TM) utilizing biodiesel-derived crude glycerol as the sole carbon source. Environ Prog Sustain Energy 28:100–110. doi: 10.1002/ep.10350. [DOI] [Google Scholar]
- 12.Malaviya A, Jang YS, Lee SY. 2012. Continuous butanol production with reduced byproducts formation from glycerol by a hyper producing mutant of Clostridium pasteurianum. Appl Microbiol Biotechnol 93:1485–1494. doi: 10.1007/s00253-011-3629-0. [DOI] [PubMed] [Google Scholar]
- 13.Biebl H, Menzel K, Zeng A-P, Deckwer W-D. 1999. Microbial production of 1, 3-propanediol. Appl Microbiol Biotechnol 52:289–297. doi: 10.1007/s002530051523. [DOI] [PubMed] [Google Scholar]
- 14.Pyne ME, Moo-Young M, Chung DA, Chou CP. 2013. Development of an electrotransformation protocol for genetic manipulation of Clostridium pasteurianum. Biotechnol Biofuels 6:50. doi: 10.1186/1754-6834-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pyne ME, Moo-Young M, Chung DA, Chou CP. 2014. Expansion of the genetic toolkit for metabolic engineering of Clostridium pasteurianum: chromosomal gene disruption of the endogenous CpaAI restriction enzyme. Biotechnol Biofuels 7:163. doi: 10.1186/s13068-014-0163-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandoval NR, Venkataramanan KP, Groth TS, Papoutsakis ET. 2015. Whole-genome sequence of an evolved Clostridium pasteurianum strain reveals Spo0A deficiency responsible for increased butanol production and superior growth. Biotechnol Biofuels 8:1. doi: 10.1186/s13068-014-0179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pyne ME, Moo-Young M, Chung DA, Chou CP. 2015. Antisense-RNA-mediated gene downregulation in Clostridium pasteurianum. Fermentation 1:113–126. doi: 10.3390/fermentation1010113. [DOI] [Google Scholar]
- 18.Pyne ME, Bruder MR, Moo-Young M, Chung DA, Chou CP. 2016. Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium. Sci Rep 6:25666. doi: 10.1038/srep25666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pyne ME, Utturkar S, Brown SD, Moo-Young M, Chung DA, Chou CP. 2014. Improved draft genome sequence of Clostridium pasteurianum strain ATCC 6013 (DSM 525) using a hybrid next-generation sequencing approach. Genome Announc 2(4):e00790-14. doi: 10.1128/genomeA.00790-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pyne ME, Liu X, Moo-Young M, Chung DA, Chou CP. 2016. Genome-directed analysis of prophage excision, host defence systems, and central fermentative metabolism in Clostridium pasteurianum. Sci Rep 6:26228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poehlein A, Grosse-Honebrink A, Zhang Y, Minton NP, Daniel R. 2015. Complete genome sequence of the nitrogen-fixing and solvent-producing Clostridium pasteurianum DSM 525. Genome Announc 3(1):e01591-14. doi: 10.1128/genomeA.01591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rappert S, Song L, Sabra W, Wang W, Zeng A-P. 2013. Draft genome sequence of type strain Clostridium pasteurianum DSM 525 (ATCC 6013), a promising producer of chemicals and fuels. Genome Announc 1(1):e00232-12. doi: 10.1128/genomeA.00232-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rotta C, Poehlein A, Schwarz K, McClure P, Daniel R, Minton NP. 2015. Closed genome sequence of Clostridium pasteurianum ATCC 6013. Genome Announc 3(1):e01596-14. doi: 10.1128/genomeA.01596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolek J, Sedláø K, Provazník I, Patáková P. 2014. Draft genome sequence of Clostridium pasteurianum NRRL B-598, a potential butanol or hydrogen producer. Genome Announc 2(2):e00192-14. doi: 10.1128/genomeA.00192-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolek J, Sedlar K, Provaznik I, Patakova P. 2016. Dam and Dcm methylations prevent gene transfer into Clostridium pasteurianum NRRL B-598: development of methods for electrotransformation, conjugation, and sonoporation. Biotechnol Biofuels 9:1. doi: 10.1186/s13068-015-0423-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen TO, Kvist T, Mikkelsen MJ, Westermann P. 2012. Production of 1,3-PDO and butanol by a mutant strain of Clostridium pasteurianum with increased tolerance towards crude glycerol. AMB Express 2:44. doi: 10.1186/2191-0855-2-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moon C, Lee CH, Sang BI, Um Y. 2011. Optimization of medium compositions favoring butanol and 1,3-propanediol production from glycerol by Clostridium pasteurianum. Bioresour Technol 102:10561–10568. doi: 10.1016/j.biortech.2011.08.094. [DOI] [PubMed] [Google Scholar]
- 28.Biebl H, Marten S, Hippe H, Deckwer W-D. 1992. Glycerol conversion to 1,3-propanediol by newly isolated clostridia. Appl Microbiol Biotechnol 36:592–597. [Google Scholar]
- 29.González-Pajuelo M, Andrade J, Vasconcelos I. 2005. Production of 1,3-propanediol by Clostridium butyricum VPI 3266 in continuous cultures with high yield and productivity. J Ind Microbiol Biotechnol 32:391–396. doi: 10.1007/s10295-005-0012-0. [DOI] [PubMed] [Google Scholar]
- 30.González-Pajuelo M, Meynial-Salles I, Mendes F, Soucaille P, Vasconcelos I. 2006. Microbial conversion of glycerol to 1,3-propanediol: physiological comparison of a natural producer, Clostridium butyricum VPI 3266, and an engineered strain, Clostridium acetobutylicum DG1(pSPD5). Appl Environ Microbiol 72:96–101. doi: 10.1128/AEM.72.1.96-101.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.González-Pajuelo M, Meynial-Salles I, Mendes F, Andrade JC, Vasconcelos I, Soucaille P. 2005. Metabolic engineering of Clostridium acetobutylicum for the industrial production of 1,3-propanediol from glycerol. Metab Eng 7:329–336. doi: 10.1016/j.ymben.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Choi O, Kim T, Woo HM, Um Y. 2014. Electricity-driven metabolic shift through direct electron uptake by electroactive heterotroph Clostridium pasteurianum. Sci Rep 4:6961. doi: 10.1038/srep06961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez R, Murarka A, Dharmadi Y, Yazdani SS. 2008. A new model for the anaerobic fermentation of glycerol in enteric bacteria: trunk and auxiliary pathways in Escherichia coli. Metab Eng 10:234–245. doi: 10.1016/j.ymben.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Murarka A, Dharmadi Y, Yazdani SS, Gonzalez R. 2008. Fermentative utilization of glycerol by Escherichia coli and its implications for the production of fuels and chemicals. Appl Environ Microbiol 74:1124–1135. doi: 10.1128/AEM.02192-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mallette MF, Reece P, Dawes EA. 1974. Culture of Clostridium pasteurianum in defined medium and growth as a function of sulfate concentration. Appl Microbiol 28:999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning, vol 2 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 37.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 38.Sokolenko S, Aucoin MG. 2015. A correction method for systematic error in 1H-NMR time-course data validated through stochastic cell culture simulation. BMC Syst Biol 9:1. doi: 10.1186/s12918-014-0137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao J, Zhong J, Fang Y, Geisinger E, Novick RP, Lambowitz AM. 2006. Use of targetrons to disrupt essential and nonessential genes in Staphylococcus aureus reveals temperature sensitivity of Ll.LtrB group II intron splicing. RNA 12:1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frazier CL, San Filippo J, Lambowitz AM, Mills DA. 2003. Genetic manipulation of Lactococcus lactis by using targeted group II introns: generation of stable insertions without selection. Appl Environ Microbiol 69:1121–1128. doi: 10.1128/AEM.69.2.1121-1128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karberg M, Guo HT, Zhong J, Coon R, Perutka J, Lambowitz AM. 2001. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat Biotechnol 19:1162–1167. doi: 10.1038/nbt1201-1162. [DOI] [PubMed] [Google Scholar]
- 42.Knobloch JK, Nedelmann M, Kiel K, Bartscht K, Horstkotte MA, Dobinsky S, Rohde H, Mack D. 2003. Establishment of an arbitrary PCR for rapid identification of Tn917 insertion sites in Staphylococcus epidermidis: characterization of biofilm-negative and nonmucoid mutants. Appl Environ Microbiol 69:5812–5818. doi: 10.1128/AEM.69.10.5812-5818.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rainey FA, Hollen BJ, Small A. 2009. Genus I. Clostridium, p 738–830. In De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 3 Springer, New York, NY. [Google Scholar]
- 44.Forsberg CW. 1987. Production of 1,3-propanediol from glycerol by Clostridium acetobutylicum and other Clostridium species. Appl Environ Microbiol 53:639–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson DT, Taconi KA. 2007. The glycerin glut: options for the value-added conversion of crude glycerol resulting from biodiesel production. Environ Prog Sustain Energy 26:338–348. doi: 10.1002/ep.10225. [DOI] [Google Scholar]
- 46.Rütti D, Lacroix C, Jeremic T, Mathis M, Die A, Vollenweider S. 2011. Development of a reversible binding process for in situ removal of 3-hydroxypropionaldehyde during biotechnological conversion of glycerol. Biochem Eng J 55:176–184. doi: 10.1016/j.bej.2011.04.005. [DOI] [Google Scholar]
- 47.Schaefer L, Auchtung TA, Hermans KE, Whitehead D, Borhan B, Britton RA. 2010. The antimicrobial compound reuterin (3-hydroxypropionaldehyde) induces oxidative stress via interaction with thiol groups. Microbiology 156:1589–1599. doi: 10.1099/mic.0.035642-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cameron DC, Cooney CL. 1986. A novel fermentation: the production of R(—)-1,2–propanediol and acetol by Clostridium thermosaccharolyticum. Nat Biotechnol 4:651–654. doi: 10.1038/nbt0786-651. [DOI] [Google Scholar]
- 49.Tran-Din K, Gottschalk G. 1985. Formation of d(—)-1, 2-propanediol and d(—)-lactate from glucose by Clostridium sphenoides under phosphate limitation. Arch Microbiol 142:87–92. doi: 10.1007/BF00409243. [DOI] [Google Scholar]
- 50.Vasconcelos I, Girbal L, Soucaille P. 1994. Regulation of carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH on mixtures of glucose and glycerol J Bacteriol 176:1443–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu L, Zhang L, Tang W, Gu Y, Hua Q, Yang S, Jiang W, Yang C. 2012. Phosphoketolase pathway for xylose catabolism in Clostridium acetobutylicum revealed by 13C metabolic flux analysis. J Bacteriol 194:5413–5422. doi: 10.1128/JB.00713-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo T, Tang Y, Zhang QY, Du TF, Liang DF, Jiang M, Ouyang PK. 2012. Clostridium beijerinckii mutant with high inhibitor tolerance obtained by low-energy ion implantation. J Ind Microbiol Biotechnol 39:401–407. doi: 10.1007/s10295-011-1017-5. [DOI] [PubMed] [Google Scholar]
- 53.Truniger V, Boos W. 1994. Mapping and cloning of gldA, the structural gene of the Escherichia coli glycerol dehydrogenase. J Bacteriol 176:1796–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cooper R. 1984. Metabolism of methylglyoxal in microorganisms. Annu Rev Microbiol 38:49–68. doi: 10.1146/annurev.mi.38.100184.000405. [DOI] [PubMed] [Google Scholar]
- 55.Hopper D, Cooper R. 1971. The regulation of Escherichia coli methylglyoxal synthase; a new control site in glycolysis? FEBS Lett 13:213–216. doi: 10.1016/0014-5793(71)80538-0. [DOI] [PubMed] [Google Scholar]
- 56.Booth IR. 2005. Glycerol and methylglyoxal metabolism. In Brock A, Curtiss R III, Kaper JB, Neidhardt FC, Nystrom T, Rudd KE, Squires CL (ed), EcoSal-Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. [Google Scholar]
- 57.Dürre P. 2007. Biobutanol: an attractive biofuel. Biotechnol J 2:1525–1534. doi: 10.1002/biot.200700168. [DOI] [PubMed] [Google Scholar]
- 58.Huang K-X, Rudolph FB, Bennett GN. 1999. Characterization of methylglyoxal synthase from Clostridium acetobutylicum ATCC 824 and its use in the formation of 1,2-propanediol. Appl Environ Microbiol 65:3244–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liyanage H, Kashket S, Young M, Kashket ER. 2001. Clostridium beijerinckii and Clostridium difficile detoxify methylglyoxal by a novel mechanism involving glycerol dehydrogenase. Appl Environ Microbiol 67:2004–2010. doi: 10.1128/AEM.67.5.2004-2010.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forage RG, Foster MA. 1982. Glycerol fermentation in Klebsiella pneumoniae: functions of the coenzyme B12-dependent glycerol and diol dehydratases. J Bacteriol 149:413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pyne ME, Bruder M, Moo-Young M, Chung DA, Chou CP. 2014. Technical guide for genetic advancement of underdeveloped and intractable Clostridium. Biotechnol Adv 32:623–641. doi: 10.1016/j.biotechadv.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 62.Heap JT, Pennington OJ, Cartman ST, Minton NP. 2009. A modular system for Clostridium shuttle plasmids. J Microbiol Methods 78:79–85. doi: 10.1016/j.mimet.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 63.Yu MR, Du YM, Jiang WY, Chang WL, Yang ST, Tang IC. 2012. Effects of different replicons in conjugative plasmids on transformation efficiency, plasmid stability, gene expression and n-butanol biosynthesis in Clostridium tyrobutyricum. Appl Microbiol Biotechnol 93:881–889. doi: 10.1007/s00253-011-3736-y. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, Zhang Z-T, Seo S-O, Choi K, Lu T, Jin Y-S, Blaschek HP. 2015. Markerless chromosomal gene deletion in Clostridium beijerinckii using CRISPR/Cas9 system. J Biotechnol 200:1–5. doi: 10.1016/j.jbiotec.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 65.Xu T, Li Y, Shi Z, Hemme CL, Li Y, Zhu Y, Van Nostrand JD, He Z, Zhou J. 2015. Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 nickase. Appl Environ Microbiol 81:4423–4431. doi: 10.1128/AEM.00873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Al-Hinai MA, Fast AG, Papoutsakis ET. 2012. Novel system for efficient isolation of Clostridium double-crossover allelic exchange mutants enabling markerless chromosomal gene deletions and DNA integration. Appl Environ Microbiol 78:8112–8121. doi: 10.1128/AEM.02214-12. [DOI] [PMC free article] [PubMed] [Google Scholar]