Abstract
Arthrogryposis by definition has multiple congenital contractures. All types of arthrogryposis have decreased in utero fetal movement. Because so many things are involved in normal fetal movement, there are many causes and processes that can go awry. In this era of molecular genetics, we have tried to place the known mutated genes seen in genetic forms of arthrogryposis into biological processes or cellular functions as defined by gene ontology. We hope this leads to better identification of all interacting pathways and processes involved in the development of fetal movement in order to improve diagnosis of the genetic forms of arthrogryposis, to lead to the development of molecular therapies, and to help better define the natural history of various types of arthrogryposis.
Key Words: Arthrogryposis, ClueGO, Connective tissue, Cytoscape, Enrichment analysis, Fetal movement, Gene ontology, Molecular pathways, Multiple congenital contractures, Reactome
Arthrogryposis is the term that has been used for the last century to describe individuals born with multiple congenital contractures (e.g., 2 or more areas in different body parts with limitation of movement present at birth) [Hall, 2014]. Multiple congenital contractures have been recognized at birth for hundreds of years, particularly because there is often difficulty with delivery, and the contractures are obviously present in the newborn. Often in the past, severely affected individuals did not survive. During the last century, the term arthrogryposis multiplex congenita was often used for multiple congenital contractures. However, arthrogryposis and arthrogryposis multiplex congenita are both descriptive terms or signs rather than a specific diagnosis [Hall, 2012, 2014].
What makes arthrogryposis so interesting is that anything which interferes with normal fetal movement may lead to congenital contractures. In the severest form, fetal akinesia deformation sequence, secondary deformations of multiple tissues are seen (craniofacial changes, pulmonary hypoplasia, polyhydramnios, decreased gut mobility and shortened gut, short umbilical cord, skin changes, and multiple joints with limitation of movement, including limbs, jaw, and spine) [Hall, 2009].
Nowadays, the recognition of an affected infant is possible prenatally utilizing real-time ultrasound studies; however, the presence of joint contractures is most often missed [Filges and Hall, 2013]. In arthrogryposis, delivery is often breech and difficult, leading to C-section. In spite of a C-section, fractures of the long bones occur in the perinatal period in at least 10% of affected infants [Hall et al., 2014].
Arthrogryposis is not all that infrequent occurring in about 1/3,000 pregnancies [Lowry et al., 2010]. Of these children, about 1/3 will primarily have limbs affected, 1/3 will have limbs plus other body areas affected with normal intelligence, and 1/3 will have central nervous system dysfunction (in the past, half of these would die at birth or in the first year, or have such severe involvement as to be lethal) [Hall, 2012]. Amyoplasia is the most common form of arthrogryposis occurring in about 1/10,000 live births [Hall et al., 2014]. Amyoplasia is recognized by its unique clinical features. Individuals with this form usually do surprisingly well and have normal to high intelligence, but it appears to be totally sporadic (although increased in one of monozygotic twins). Most other recognizable types of arthrogryposis have a genetic basis (i.e., single gene mutation) [Michalk et al., 2008; Hall, 2014; Hunter et al., 2015; Bayram et al., 2016].
Over the last 40 years, the heterogeneity and diversity of specific disorders has begun to be recognized and delineated. Over 400 different specific conditions with arthrogryposis have been recognized and over 320 genes implicated [Michalk et al., 2008; Hall, 2014; Hunter et al., 2015; Bayram et al., 2016]. There is a need to annotate and functionally group these genes into known pathways and biological processes. Such a grouping has potential for identification and prioritization of other candidate disease genes. Additionally, it should inform the development of better molecular diagnostic techniques and specific therapeutic options. Until now, nonspecific physical therapy to loosen contractures and realign joints, casting, and surgeries to improve joint function have been the standard therapy.
It is clear that anything which limits or interferes with fetal movement may lead to congenital contractures (limitation of joint movement) [Hall, 2012, 2014]. These include myopathic processes with structural elements, ion channels, and mechanosensing elements; neuropathic processes including central and peripheral nerves, anterior horn cells, and brain organization and function; myelin deficiency; neuromotor endplate abnormalities; connective tissue disorders; limitation of space and constraint in utero; vascular compromise (decreased blood flow to the placenta or to the embryo/fetus); teratogenic exposure, and maternal illnesses. Any of the above-mentioned processes or clinical situations may lead to decreased fetal movement. Even hypotonia of the fetus may be severe enough to decrease in utero movement sufficiently, leading to contractures at birth.
We performed an enrichment analysis (EA) to identify overrepresented functional biological groupings within the list of assembled 320 genes (table 1). EA is a common bioinformatic technique to describe common biological aspects associated with a list of genes. Gene lists are often the output of high-throughput genomics experiment or, in the case here, a listing of genes associated with a disease process. EA involves computing an enrichment statistic across a corpus of gene sets to identify over- and/or underrepresented gene sets in the gene list being interrogated. A corpus of gene sets is a collection of genes categorized together based on some biological aspect or property. The outcome of EA results in a list of statistically enriched gene sets describing the biological properties common within the given gene list. This allows for biologist interpretation of gene lists, whether it is a differential gene expression list or a list of genes associated with a disease state, such as the syndrome of arthrogryposis detailed in this review.
Table 1.
Gene table
| Gene | Entrez Gene ID | Aliases | Functions |
|---|---|---|---|
| ABCC8 | 6833 | ABC36, HHF1, HI, HRINS, MRP8, PHHI, SUR, SUR1, SUR1delta2, TNDM2 | sarcolemma, synaptic transmission |
| ACTA1 | 58 | ACTA, ASMA, CFTD, CFTD1, CFTDM, MPFD, NEM1, NEM2, NEM3 | striated muscle thin filament, muscle filament sliding |
| ACTB | 60 | BRWS1, PS1TP5BP1 | axon guidance, regulation of body fluid levels |
| ACTG1 | 71 | ACT, ACTG, BRWS2, DFNA20, DFNA26, HEL-176 | striated muscle cell development, muscle cell development |
| ADAMTS10 | 81794 | ADAM-TS10, ADAMTS-10, WMS, WMS1 | proteinaceous extracellular matrix |
| ADAMTSL2 | 9719 | GPHYSD1 | lung development, respiratory system development |
| ADCY6 | 112 | AC6, LCCS8 | sarcolemma, regulation of neurogenesis |
| ADGRG6 | 57211 | APG1, DREG, GPR126, PS1TP2, VIGR | axon ensheathment |
| ADSL | 158 | AMPS, ASASE, ASL | carbohydrate derivative biosynthetic process |
| AIMP1 | 9255 | EMAP2, EMAPII, HLD3, SCYE1, p43 | regulation of epithelial cell proliferation, epithelial cell proliferation |
| AKT1 | 207 | AKT, CWS6, PKB, PKB-ALPHA, PRKBA, RAC, RAC-ALPHA | Schwann cell development, Schwann cell differentiation |
| ALG2 | 85365 | CDGIi, CMS14, CMSTA3, NET38, hALPG2 | mannosylation, glycoprotein biosynthetic process |
| ALG3 | 10195 | CDG1D, CDGS4, CDGS6, D16Ertd36e, NOT56L, Not56, not | mannosylation, glycoprotein biosynthetic process |
| ANTXR2 | 118429 | CMG-2, CMG2, HFS, ISH, JHF | endoplasmic reticulum part |
| AP1S2 | 8905 | DC22, MRX59, MRXS21, MRXS5, MRXSF, PGS, SIGMA1B | neuromuscular process, connective tissue development |
| APLNR | 187 | AGTRL1, APJ, APJR, HG11 | heart development, embryonic morphogenesis |
| ARX | 170302 | CT121, EIEE1, ISSX, MRX29, MRX32, MRX33, MRX36, MRX38, MRX43, MRX54, MRX76, MRX87, MRXS1, PRTS | cerebral cortex cell migration, cerebral cortex development |
| ASXL1 | 171023 | BOPS, MDS | bone development, lung development |
| ATM | 472 | AT1, ATA, ATC, ATD, ATDC, ATE, TEL1, TELO1 | neuron apoptotic process, regulation of neuron death |
| ATN1 | 1822 | B37, D12S755E, DRPLA, HRS, NOD | neuron apoptotic process, neuron death |
| ATP7A | 538 | DSMAX, MK, MNK, SMAX3 | collagen fibril organization, central nervous system neuron differentiation |
| ATRX | 546 | ATR2, JMS, MRX52, MRXHF1, RAD54, RAD54L, SFM1, SHS, XH2, XNP, ZNF-HX | limb morphogenesis, limb development |
| ATXN2 | 6311 | ASL13, ATX2, SCA2, TNRC13 | neuromuscular process, central nervous system neuron differentiation |
| ATXN3 | 4287 | AT3, ATX3, JOS, MJD, MJD1, SCA3 | actin filament-based process, synaptic transmission |
| B3GAT3 | 26229 | GLCATI, glcUAT-I | chondroitin sulfate metabolic process, proteoglycan metabolic process |
| BAG3 | 9531 | BAG-3, BIS, CAIR-1, MFM6 | spinal cord development, I band |
| BICD2 | 23299 | SMALED2, bA526D8.1 | organelle localization |
| BIN1 | 274 | AMPH2, AMPHL, SH3P9 | sarcolemma, I band |
| CANT1 | 124583 | DBQD, SCAN-1, SCAN1, SHAPY | proteoglycan metabolic process, glycoprotein biosynthetic process |
| CAPN3 | 825 | CANP3, CANPL3, LGMD2, LGMD2A, nCL-1, p94 | muscle cell cellular homeostasis, sarcolemma |
| CASK | 8573 | CAGH39, CAMGUK, CMG, FGS4, LIN2, MICPCH, MRXSNA, TNRC8 | regulation of cellular response to growth factor stimulus, synaptic membrane |
| CD24 | 100133941 | CD24A | neuromuscular synaptic transmission, neuroblast proliferation |
| CD6 | 923 | TP120 | response to wounding |
| CDK5 | 1020 | LIS7, PSSALRE | Schwann cell development, Schwann cell differentiation |
| CHAT | 1103 | CHOACTASE, CMS1A, CMS1A2, CMS6 | developmental growth, synaptic transmission |
| CHMP1A | 5119 | CHMP1, PCH8, PCOLN3, PRSM1, VPS46-1, VPS46A | organelle localization |
| CHRNA1 | 1134 | ACHRA, ACHRD, CHRNA, CMS1A, CMS1B, CMS2A, FCCMS, SCCMS | muscle cell cellular homeostasis, neuromuscular synaptic transmission |
| CHRNB1 | 1140 | ACHRB, CHRNB, CMS1D, CMS2A, CMS2C, SCCMS | neuromuscular synaptic transmission, skeletal muscle contraction |
| CHRND | 1144 | ACHRD, CMS2A, CMS3A, CMS3B, CMS3C, FCCMS, SCCMS | neuromuscular synaptic transmission, skeletal muscle contraction |
| CHRNE | 1145 | ACHRE, CMS1D, CMS1E, CMS2A, CMS4A, CMS4B, CMS4C, FCCMS, SCCMS | neuromuscular synaptic transmission, skeletal muscle contraction |
| CHRNG | 1146 | ACHRG | neuromuscular synaptic transmission, chemical synaptic transmission, postsynaptic |
| CHST14 | 113189 | ATCS, D4ST1, EDSMC1, HNK1ST | chondroitin sulfate metabolic process, proteoglycan metabolic process |
| CHST3 | 9469 | C6ST, C6ST1, HSD | chondroitin sulfate metabolic process, proteoglycan metabolic process |
| CHUK | 1147 | IKBKA, IKK-alpha, IKK1, IKKA, NFKBIKA, TCF16 | skeletal muscle contraction, striated muscle contraction |
| CNTN1 | 1272 | F3, GP135, MYPCN | positive regulation of epithelial cell proliferation, regulation of epithelial cell proliferation |
| CNTNAP1 | 8506 | CASPR, CNTNAP, NRXN4, P190 | axon ensheathment, neuromuscular process, regulation of membrane potential |
| COG7 | 91949 | CDG2E | glycoprotein biosynthetic process, glycoprotein metabolic process |
| COL11A2 | 1302 | DFNA13, DFNB53, FBCG2, HKE5, PARP, STL3 | fibrillar collagen trimer, complex of collagen trimers |
| COL1A1 | 1277 | EDSC, OI1, OI2, OI3, OI4 | fibrillar collagen trimer, complex of collagen trimers |
| COL1A2 | 1278 | OI4 | fibrillar collagen trimer, complex of collagen trimers |
| COL2A1 | 1280 | ANFH, AOM, COL11A3, SEDC, STL1 | fibrillar collagen trimer, complex of collagen trimers |
| COL3A1 | 1281 | EDS4A | fibrillar collagen trimer, complex of collagen trimers |
| COL6A1 | 1291 | OPLL | complex of collagen trimers, collagen metabolic process |
| COL6A2 | 1292 | PP3610 | collagen metabolic process, sarcolemma |
| COL6A3 | 1293 | DYT27 | complex of collagen trimers, collagen metabolic process |
| COL7A1 | 1294 | EBD1, EBDCT, EBR1, NDNC8 | complex of collagen trimers, collagen metabolic process |
| CRLF1 | 9244 | CISS, CISS1, CLF, CLF-1, NR6, zcytor5 | neuron apoptotic process, regulation of neuron death |
| CTSA | 5476 | GLB2, GSL, NGBE, PPCA, PPGB | lysosomal lumen, glycoprotein biosynthetic process |
| CTSL | 1514 | CATL, CTSL1, MEP | lysosomal lumen, collagen metabolic process |
| DCX | 1641 | DBCN, DC, LISX, SCLH, XLIS | cerebral cortex cell migration, hippocampus development |
| DES | 1674 | CSM1, CSM2, LGMD2R | muscle filament sliding, actin filament-based movement |
| DHCR24 | 1718 | DCE, Nbla03646, SELADIN1, seladin-1 | skin development, regulation of neuron death |
| DHCR7 | 1717 | SLOS | lung development, respiratory system development |
| DMPK | 1760 | DM, DM1, DM1PK, DMK, MDPK, MT-PK | skeletal muscle contraction, chemical synaptic transmission, postsynaptic |
| DNM2 | 1785 | CMT2M, CMTDI1, CMTDIB, DI-CMTB, DYN2, DYNII, LCCS5 | regulation of cellular response to growth factor stimulus, synaptic membrane |
| DPAGT1 | 1798 | ALG7, CDG-Ij, CDG1J, CMS13, CMSTA2, D11S366, DGPT, DPAGT, DPAGT2, G1PT, GPT, UAGT, UGAT | glycoprotein biosynthetic process, glycoprotein metabolic process |
| DPM1 | 8813 | CDGIE, MPDS | mannosylation, glycoprotein biosynthetic process |
| DST | 667 | BP240, BPA, BPAG1, CATX-15, CATX15, D6S1101, DMH, DT, EBSB2, HSAN6, MACF2 | I band, contractile fiber |
| DYM | 54808 | DMC, SMC | bone development, skeletal system development |
| DYNC1H1 | 1778 | CMT2O, DHC1, DHC1a, DNCH1, DNCL, DNECL, DYHC, Dnchc1, HL-3, SMALED1, p22 | organelle localization, glycoprotein biosynthetic process |
| DYSF | 8291 | FER1L1, LGMD2B, MMD1 | sarcolemma, muscle contraction |
| EBP | 10682 | CDPX2, CHO2, CPX, CPXD, MEND | skeletal system development, endoplasmic reticulum part |
| EGR2 | 1959 | AT591, CMT1D, CMT4E, KROX20 | Schwann cell differentiation, peripheral nervous system development |
| EMD | 2010 | EDMD, LEMD5, STA | skeletal muscle tissue development, skeletal muscle organ development |
| ERBB3 | 2065 | ErbB-3, HER3, LCCS2, MDA-BF-1, c-erbB-3, c-erbB3, erbB3-S, p180-ErbB3, p45-sErbB3, p85-sErbB3 | Schwann cell differentiation, peripheral nervous system development |
| ERCC1 | 2067 | COFS4, RAD10, UV20 | developmental growth, embryo development |
| ERCC2 | 2068 | COFS2, EM9, TFIIH, TTD, TTD1, XPD | glial cell development, spinal cord development |
| ERCC6 | 2074 | ARMD5, CKN2, COFS, COFS1, CSB, RAD26, UVSS1 | developmental growth |
| ERLIN2 | 11160 | C8orf2, Erlin-2, NET32, SPFH2, SPG18 | endoplasmic reticulum part |
| ESCO2 | 157570 | 2410004I17Rik, EFO2, RBS | animal organ development |
| EZH2 | 2146 | ENX-1, ENX1, EZH1, EZH2b, KMT6, KMT6A, WVS, WVS2 | hippocampus development, limbic system development |
| FAM20C | 56975 | DMP-4, DMP4, GEF-CK, RNS | bone development, osteoblast differentiation |
| FBN1 | 2200 | ACMICD, ECTOL1, FBN, GPHYSD2, MASS, MFS1, OCTD, SGS, SSKS, WMS, WMS2 | extracellular matrix disassembly, regulation of cellular response to growth factor stimulus |
| FBN2 | 2201 | CCA, DA9, EOMD | embryonic limb morphogenesis, extracellular matrix disassembly |
| FBN3 | 84467 | regulation of cellular response to growth factor stimulus, proteinaceous extracellular matrix | |
| FGD1 | 2245 | AAS, FGDY, MRXS16, ZFYVE3 | actin filament-based process, cellular response to growth factor stimulus |
| FGF9 | 2254 | FGF-9, GAF, HBFG-9, HBGF-9, SYNS3 | chondrocyte differentiation, regulation of stem cell proliferation, embryonic skeletal system development |
| FGFR1 | 2260 | BFGFR, CD331, CEK, FGFBR, FGFR-1, FLG, FLT-2, FLT2, HBGFR, HH2, HRTFDS, KAL2, N-SAM, OGD, bFGF-R-1 | cerebral cortex cell migration, neuroblast proliferation |
| FGFR2 | 2263 | BBDS, BEK, BFR-1, CD332, CEK3, CFD1, ECT1, JWS, K-SAM, KGFR, TK14, TK25 | prostate gland epithelium morphogenesis, neuroblast proliferation |
| FGFR3 | 2261 | ACH, CD333, CEK2, HSFGFR3EX, JTK4 | glial cell development, bone morphogenesis |
| FHL1 | 2273 | FHL-1, FHL1A, FHL1B, FLH1A, KYOT, SLIM, SLIM-1, SLIM1, SLIMMER, XMPMA | regulation of membrane potential, muscle organ development |
| FKBP10 | 60681 | BRKS1, FKBP65, OI11, OI6, PPIASE, hFKBP65 | endoplasmic reticulum part |
| FKRP | 79147 | LGMD2I, MDC1C, MDDGA5, MDDGB5, MDDGC5 | mannosylation, sarcolemma |
| FKTN | 2218 | CMD1X, FCMD, LGMD2M, MDDGA4, MDDGB4, MDDGC4 | mannosylation, muscle organ development |
| FLNA | 2316 | ABP-280, ABPX, CSBS, CVD1, FLN, FLN-A, FLN1, FMD, MNS, NHBP, OPD, OPD1, OPD2, XLVD, XMVD | protein import, actin cytoskeleton |
| FLNB | 2317 | ABP-278, ABP-280, AOI, FH1, FLN-B, FLN1L, LRS1, SCT, TABP, TAP | hippocampus development, limbic system development |
| FUCA1 | 2517 | FUCA | lysosomal lumen, glycoprotein biosynthetic process |
| GAA | 2548 | LYAG | muscle cell cellular homeostasis, skeletal muscle contraction |
| GAD1 | 2571 | CPSQ1, GAD, SCP | synaptic transmission |
| GBA | 2629 | GBA1, GCB, GLUC | lysosomal lumen, skin development |
| GBE1 | 2632 | APBD, GBE, GSD4 | carbohydrate metabolic process |
| GCK | 2645 | FGQTL3, GK, GLK, HHF3, HK4, HKIV, HXKP, LGLK, MODY2 | actin cytoskeleton, carbohydrate derivative biosynthetic process |
| GDF5 | 8200 | BDA1C, BMP-14, BMP14, CDMP1, LAP-4, LAP4, OS5, SYM1B, SYNS2 | chondrocyte differentiation, embryonic limb morphogenesis |
| GJA1 | 2697 | AVSD3, CMDR, CX43, EKVP, GJAL, HLHS1, HSS, ODDD, PPKCA | actin filament-based movement, embryonic limb morphogenesis |
| GLI3 | 2737 | ACLS, GCPS, GLI3-190, GLI3FL, PAP-A, PAPA, PAPA1, PAPB, PHS, PPDIV | cerebral cortex cell migration, neuroblast proliferation |
| GLRA1 | 2741 | HKPX1, STHE | chemical synaptic transmission, postsynaptic, neuromuscular process |
| GLRB | 2743 | HKPX2 | neuromuscular process, synaptic membrane |
| GLUL | 2752 | GLNS, GS, PIG43, PIG59 | positive regulation of epithelial cell proliferation, regulation of epithelial cell proliferation |
| GPC3 | 2719 | DGSX, GTR2-2, MXR7, OCI-5, SDYS, SGB, SGBS, SGBS1 | body morphogenesis, chondroitin sulfate metabolic process |
| GRHL3 | 57822 | SOM, TFCP2L4, VWS2 | skin development, embryonic organ morphogenesis |
| GRN | 2896 | CLN11, GEP, GP88, PCDGF, PEPI, PGRN | neural precursor cell proliferation, positive regulation of epithelial cell proliferation |
| GUSB | 2990 | BG, MPS7 | lysosomal lumen, carbohydrate metabolic process |
| HEXA | 3073 | TSD | chondroitin sulfate metabolic process, lysosomal lumen |
| HEXB | 3074 | ENC-1AS, HEL-248, HEL-S-111 | chondroitin sulfate metabolic process, lysosomal lumen |
| HLA-DRB1 | 3123 | DRB1, DRw10, HLA-DR1B, HLA-DRB, SS1 | negative regulation of cell proliferation, response to wounding |
| HOXA13 | 3209 | HOX1, HOX1J | prostate gland epithelium morphogenesis, embryonic limb morphogenesis |
| HOXD13 | 3239 | BDE, BDSD, HOX4I, SPD | prostate gland epithelium morphogenesis, embryonic limb morphogenesis |
| HRAS | 3265 | C-BAS/HAS, C-H-RAS, C-HA-RAS1, CTLO, H-RASIDX, HAMSV, HRAS1, RASH1, p21ras | positive regulation of epithelial cell proliferation, neuron apoptotic process |
| HSPG2 | 3339 | HSPG, PLC, PRCAN, SJA, SJS, SJS1 | chondroitin sulfate metabolic process, bone morphogenesis |
| IDS | 3423 | MPS2, SIDS | chondroitin sulfate metabolic process, lysosomal lumen |
| IGF2 | 3481 | C11orf43, GRDF, IGF-II, PP9974 | digestive system development, striated muscle cell differentiation |
| IGHMBP2 | 3508 | CATF1, CMT2S, HCSA, HMN6, SMARD1, SMUBP2, ZFAND7 | spinal cord development, central nervous system neuron differentiation |
| IMPAD1 | 54928 | GPAPP, IMP 3, IMP-3, IMPA3 | chondroitin sulfate metabolic process, bone morphogenesis |
| INSR | 3643 | CD220, HHF5 | digestive system development, regulation of developmental growth |
| IRF6 | 3664 | LPS, OFC6, PIT, PPS, PPS1, VWS, VWS1 | skin development, epithelial cell proliferation |
| ISPD | 729920 | MDDGA7, MDDGC7, Nip, hCG_1745121 | mannosylation, glycoprotein biosynthetic process |
| ITGA6 | 3655 | CD49f, ITGA6B, VLA-6 | digestive tract development, digestive system development |
| ITGB4 | 3691 | CD104 | digestive tract development, digestive system development |
| KCNA1 | 3736 | AEMK, EA1, HBK1, HUK1, KV1.1, MBK1, MK1, RBK1 | neuroblast proliferation, hippocampus development |
| KCNJ11 | 3767 | BIR, HHF2, IKATP, KIR6.2, MODY13, PHHI, TNDM3 | sarcolemma, regulation of membrane potential |
| KCNK9 | 51305 | K2p9.1, KT3.2, TASK-3, TASK3 | regulation of membrane potential, synaptic transmission |
| KIAA0196 | 9897 | RTSC, SPG8 | cell development |
| KIF14 | 9928 | MKS12 | hippocampus development, limbic system development |
| KIF5C | 3800 | CDCBM2, KINN, NKHC, NKHC-2, NKHC2 | axon guidance, axonogenesis |
| KIF7 | 374654 | ACLS, AGBK, HLS2, JBTS12, UNQ340 | heart development, blood vessel development |
| KLHL40 | 131377 | KBTBD5, NEM8, SRYP, SYRP | muscle fiber development, I band |
| KLHL41 | 10324 | KBTBD10, Krp1, SARCOSIN | muscle fiber development, striated muscle contraction, striated muscle cell development |
| KLKB1 | 3818 | KLK3, PKK, PKKD, PPK | extracellular matrix disassembly, extracellular matrix organization |
| L1CAM | 3897 | CAML1, CD171, HSAS, HSAS1, MASA, MIC5, N-CAM-L1, N-CAML1, NCAM-L1, S10, SPG1 | synaptic membrane, regulation of developmental growth |
| LAMA2 | 3908 | LAMM | Schwann cell development, Schwann cell differentiation |
| LARGE | 9215 | MDC1D, MDDGA6, MDDGB6 | muscle cell cellular homeostasis, mannosylation |
| LIFR | 3977 | CD118, LIF-R, SJS2, STWS, SWS | cell-type specific apoptotic process, neuron projection morphogenesis |
| LMBR1 | 64327 | ACHP, C7orf2, DIF14, LSS, PPD2, THYP, TPT, ZRS | embryonic limb morphogenesis, limb morphogenesis |
| LMNA | 4000 | CDCD1, CDDC, CMD1A, CMT2B1, EMD2, FPL, FPLD, FPLD2, HGPS, IDC, LDP1, LFP, LGMD1B, LMN1, LMNC, LMNL1, PRO1 | striated muscle cell development, muscle cell development |
| LMX1B | 4010 | LMX1.2, NPS1 | chordate embryonic development, embryo development |
| LTBP2 | 4053 | C14orf141, GLC3D, LTBP3, MSPKA, MSTP031, WMS3 | regulation of stem cell proliferation, stem cell proliferation |
| MAGEL2 | 54551 | NDNL1, PWLS, SHFYNG, nM15 | actin filament-based process |
| MASP1 | 5648 | 3MC1, CRARF, CRARF1, MAP1, MASP, MASP3, MAp44, PRSS5, RaRF | response to wounding |
| MED12 | 9968 | ARC240, CAGH45, FGS1, HOPA, MED12S, OHDOX, OKS, OPA1, TNRC11, TRAP230 | Schwann cell development, Schwann cell differentiation |
| MEGF10 | 84466 | EMARDD | muscle cell development, skeletal muscle tissue development |
| MFN2 | 9927 | CMT2A, CMT2A2, CPRP1, HSG, MARF | organelle localization, chordate embryonic development |
| MMP2 | 4313 | CLG4, CLG4A, MMP-2, MMP-II, MONA, TBE-1 | face development, body morphogenesis |
| MNX1 | 3110 | HB9, HLXB9, HOXHB9, SCRA1 | spinal cord development, neuron migration |
| MTM1 | 4534 | CNM, MTMX, XLMTM | muscle cell cellular homeostasis, I band |
| MUSK | 4593 | CMS9, FADS | neuron apoptotic process, regulation of neuron death |
| MYBPC2 | 4606 | MYBPC, MYBPCF | muscle filament sliding, actin filament-based movement |
| MYH2 | 4620 | IBM3, MYH2A, MYHSA2, MYHas8, MYPOP, MyHC-2A, MyHC-IIa | muscle filament sliding, actin filament-based movement |
| MYH3 | 4621 | HEMHC, MYHC-EMB, MYHSE1, SMHCE | muscle filament sliding, skeletal muscle contraction |
| MYH7B | 57644 | MHC14, MYH14 | skeletal muscle contraction, actin filament-based movement |
| MYH8 | 4626 | DA7, MyHC-peri, MyHC-pn, gtMHC-F | muscle filament sliding, skeletal muscle contraction |
| MYO18B | 84700 | muscle fiber development, I band | |
| MYO9A | 4649 | actin cytoskeleton | |
| MYOT | 9499 | LGMD1, LGMD1A, MFM3, TTID, TTOD | sarcolemma, I band |
| NALCN | 259232 | CLIFAHDD, CanIon, IHPRF, INNFD, VGCNL1, bA430M15.1 | regulation of membrane potential |
| NEB | 4703 | NEB177D, NEM2 | striated muscle thin filament, muscle filament sliding |
| NEFH | 4744 | NFH | hippocampus development, peripheral nervous system development |
| NEU1 | 4758 | NANH, NEU, SIAL1 | lysosomal lumen, glycoprotein biosynthetic process |
| NF1 | 4763 | NFNS, VRNF, WSS | Schwann cell development, Schwann cell differentiation |
| NOG | 9241 | SYM1, SYNS1 | prostate gland epithelium morphogenesis, face development |
| OCRL | 4952 | INPP5F, LOCR, NPHL2, OCRL-1, OCRL1 | muscle system process, chordate embryonic development |
| OFD1 | 8481 | 71-7A, CXorf5, JBTS10, RP23, SGBS2 | cell projection morphogenesis, cell part morphogenesis |
| ORC4 | 5000 | ORC4L, ORC4P | actin cytoskeleton |
| ORC6 | 23594 | ORC6L | actin cytoskeleton |
| PAFAH1B1 | 5048 | LIS1, LIS2, MDCR, MDS, PAFAH | cerebral cortex cell migration, neuroblast proliferation |
| PANK2 | 80025 | C20orf48, HARP, HSS, NBIA1, PKAN | regulation of membrane potential, carbohydrate derivative biosynthetic process |
| PAX3 | 5077 | CDHS, HUP2, WS1, WS3 | spinal cord development, central nervous system neuron differentiation |
| PEX1 | 5189 | PBD1A, PBD1B, ZWS, ZWS1 | protein targeting to peroxisome, intracellular protein transmembrane import |
| PEX10 | 5192 | NALD, PBD6A, PBD6B, RNF69 | integral component of peroxisomal membrane, protein targeting to peroxisome |
| PEX12 | 5193 | PAF-3, PBD3A | integral component of peroxisomal membrane, protein targeting to peroxisome |
| PEX13 | 5194 | NALD, PBD11A, PBD11B, ZWS | integral component of peroxisomal membrane, protein targeting to peroxisome |
| PEX14 | 5195 | NAPP2, PBD13A, Pex14p, dJ734G22.2 | protein targeting to peroxisome, intracellular protein transmembrane import |
| PEX2 | 5828 | PAF1, PBD5A, PBD5B, PMP3, PMP35, PXMP3, RNF72, ZWS3 | integral component of peroxisomal membrane, protein targeting to peroxisome |
| PEX26 | 55670 | PBD7A, PBD7B, PEX26M1T, Pex26pM1T | integral component of peroxisomal membrane, protein targeting to peroxisome |
| PEX3 | 8504 | PBD10A, TRG18 | integral component of peroxisomal membrane, protein targeting to peroxisome |
| PEX5 | 5830 | PBD2A, PBD2B, PTS1-BP, PTS1R, PXR1 | protein targeting to peroxisome, intracellular protein transmembrane import |
| PEX6 | 5190 | PAF-2, PAF2, PBD4A, PDB4B, PXAAA1 | protein targeting to peroxisome, intracellular protein transmembrane import |
| PEX7 | 5191 | PBD9B, PTS2R, RCDP1, RD | protein targeting to peroxisome, intracellular protein transmembrane import |
| PFKM | 5213 | ATP-PFK, GSD7, PFK-1, PFK1, PFKA, PFKX, PPP1R122 | muscle cell cellular homeostasis, carbohydrate metabolic process |
| PIEZO2 | 63895 | C18orf30, C18orf58, DA3, DA5, FAM38B, FAM38B2, HsT748, HsT771, MWKS | regulation of membrane potential |
| PIGT | 51604 | CGI-06, MCAHS3, NDAP, PNH2 | neuron apoptotic process, neuron death |
| PIP5K1C | 23396 | LCCS3, PIP5K-GAMMA, PIP5K1-gamma, PIP5Kgamma | organelle localization, axon guidance |
| PITX1 | 5307 | BFT, CCF, LBNBG, POTX, PTX1 | embryonic limb morphogenesis, limb morphogenesis |
| PLEKHG5 | 57449 | CMTRIC, DSMA4, GEF720, Syx, Tech | chemotaxis, cellular response to growth factor stimulus |
| PLOD1 | 5351 | EDS6, LH, LH1, LLH, PLOD | extracellular matrix organization, endoplasmic reticulum part |
| PLOD2 | 5352 | BRKS2, LH2, TLH | extracellular matrix organization, endoplasmic reticulum part |
| PLOD3 | 8985 | LH3 | collagen fibril organization, lung development |
| PLP1 | 5354 | GPM6C, HLD1, MMPL, PLP, PLP/DM20, PMD, SPG2 | glial cell development, axon ensheathment |
| PMM2 | 5373 | CDG1, CDG1a, CDGS, PMI, PMI1, PMM 2 | glycoprotein biosynthetic process, glycoprotein metabolic process |
| PMP22 | 5376 | CMT1A, CMT1E, DSS, GAS-3, HMSNIA, HNPP, Sp110 | peripheral nervous system development, axon ensheathment |
| POMGNT1 | 55624 | GNTI.2, GnT I.2, LGMD2O, MEB, MGAT1.2, gnT-I.2 | glycoprotein biosynthetic process, glycoprotein metabolic process |
| POMGNT2 | 84892 | AGO61, C3orf39, GTDC2, MDDGA8 | mannosylation, neuron migration |
| POMT1 | 10585 | LGMD2K, MDDGA1, MDDGB1, MDDGC1, RT | mannosylation, extracellular matrix organization |
| POMT2 | 29954 | LGMD2N, MDDGA2, MDDGB2, MDDGC2 | mannosylation, glycoprotein biosynthetic process |
| POR | 5447 | CPR, CYPOR, P450R | bone morphogenesis, chondrocyte differentiation |
| PRG4 | 10216 | CACP, HAPO, JCAP, MSF, SZP | stem cell proliferation, animal organ development |
| PRKAR1A | 5573 | ACRDYS1, ADOHR, CAR, CNC, CNC1, PKR1, PPNAD1, PRKAR1, TSE1 | striated muscle cell development, muscle cell development |
| PRX | 57716 | CMT4F | axon ensheathment |
| PSD3 | 23362 | EFA6D, EFA6R, HCA67 | synaptic membrane |
| PTDSS1 | 9791 | LMHD, PSS1, PSSA | carbohydrate derivative biosynthetic process, endoplasmic reticulum part |
| PTH1R | 5745 | PFE, PTHR, PTHR1 | chondrocyte differentiation, cartilage development |
| RAB18 | 22931 | RAB18LI1, WARBM3 | brain development, head development |
| RAB3GAP1 | 22930 | P130, RAB3GAP, RAB3GAP130, WARBM1 | face development, body morphogenesis |
| RAPSN | 5913 | CMS11, CMS4C, FADS, RAPSYN, RNF205 | neuromuscular synaptic transmission, neuron apoptotic process |
| RBM10 | 8241 | DXS8237E, GPATC9, GPATCH9, S1-1, TARPS, ZRANB5 | cell-type specific apoptotic process, negative regulation of cell proliferation |
| RELN | 5649 | ETL7, LIS2, PRO1598, RL | cerebral cortex cell migration, hippocampus development |
| RET | 5979 | CDHF12, CDHR16, HSCR1, MEN2A, MEN2B, MTC1, PTC, RET-ELE1, RET51 | digestive tract development, digestive system development |
| RIPK4 | 54101 | ANKK2, ANKRD3, DIK, NKRD3, PKK, PPS2, RIP4 | morphogenesis of an epithelium, tissue morphogenesis |
| RMRP | 6023 | CHH, NME1, RMRPR, RRP2 | hippocampus development, limbic system development |
| RNASEH2A | 10535 | AGS4, JUNB, RNASEHI, RNHIA, RNHL | osteoblast differentiation, ossification |
| RNASEH2B | 79621 | AGS2, DLEU8 | chordate embryonic development, embryo development |
| RYR1 | 6261 | CCO, MHS, MHS1, PPP1R137, RYDR, RYR, RYR-1, SKRR | muscle fiber development, sarcolemma |
| SCN4A | 6329 | CMS16, HOKPP2, HYKPP, HYPP, NAC1A, Na(V)1.4, Nav1.4, SkM1 | muscle contraction, regulation of membrane potential |
| SEPN1 | 57190 | CFTD, MDRS1, RSMD1, RSS, SELN | muscle fiber development, striated muscle cell development |
| SETX | 23064 | ALS4, AOA2, SCAR1, bA479K20.2 | regulation of neurogenesis, regulation of nervous system development |
| SGCG | 6445 | 35DAG, A4, DAGA4, DMDA, DMDA1, LGMD2C, MAM, SCARMD2, SCG3, gamma-SG | sarcolemma, muscle cell development |
| SHOX | 6473 | GCFX, PHOG, SHOXY, SS | skeletal system development |
| SKI | 6497 | SGS, SKV | Schwann cell development, Schwann cell differentiation |
| SLC12A6 | 9990 | ACCPN, KCC3, KCC3A, KCC3B | blood vessel development, synaptic transmission |
| SLC26A2 | 1836 | D5S1708, DTD, DTDST, EDM4, MST153, MSTP157 | regulation of membrane potential, ossification |
| SLC39A13 | 91252 | LZT-Hs9 | connective tissue development, tissue development |
| SLC3A1 | 6519 | ATR1, CSNU1, D2H, NBAT, RBAT | carbohydrate metabolic process |
| SLC9A6 | 10479 | MRSA, NHE6 | regulation of cellular response to growth factor stimulus, developmental growth |
| SMN1 | 6606 | BCD541, GEMIN1, SMA, SMA1, SMA2, SMA3, SMA4, SMA@, SMN, SMNT, T-BCD541, TDRD16A | I band, contractile fiber |
| SOD1 | 6647 | ALS, ALS1, HEL-S-44, IPOA, SOD, hSod1, homodimer | muscle cell cellular homeostasis, Schwann cell development |
| SOX10 | 6663 | DOM, PCWH, WS2E, WS4, WS4C | neuroblast proliferation, peripheral nervous system development |
| SOX9 | 6662 | CMD1, CMPD1, SRA1, SRXX2, SRXY10 | prostate gland epithelium morphogenesis, bone morphogenesis |
| SPG20 | 23111 | SPARTIN, TAHCCP1 | neuromuscular process, connective tissue development |
| SRD5A3 | 79644 | CDG1P, CDG1Q, KRIZI, SRD5A2L, SRD5A2L1 | glycoprotein biosynthetic process, glycoprotein metabolic process |
| STAC3 | 246329 | NAM | neuromuscular synaptic transmission, skeletal muscle contraction |
| SULF1 | 23213 | SULF-1 | prostate gland epithelium morphogenesis, proteoglycan metabolic process |
| SYNE1 | 23345 | 8B, ARCA1, C6orf98, CPG2, EDMD4, MYNE1, Nesp1, SCAR8, dJ45H2.2 | contractile fiber, synaptic membrane |
| TARP | 445347 | CD3G, TCRG, TCRGC1, TCRGC2, TCRGV | cell-type specific apoptotic process |
| TBX15 | 6913 | TBX14 | embryonic skeletal system development, skeletal system morphogenesis |
| TBX5 | 6910 | HOS | embryonic limb morphogenesis, limb morphogenesis |
| TGFB3 | 7043 | ARVD, ARVD1, RNHF, TGF-beta3 | face development, body morphogenesis |
| TNNI2 | 7136 | AMCD2B, DA2B, FSSV, fsTnI | striated muscle thin filament, muscle filament sliding |
| TNNT1 | 7138 | ANM, NEM5, STNT, TNT, TNTS | striated muscle thin filament, muscle filament sliding |
| TNNT3 | 7140 | TNTF | striated muscle thin filament, muscle filament sliding |
| TPM2 | 7169 | AMCD1, DA1, DA2B, HEL-S-273, NEM4, TMSB | striated muscle thin filament, muscle filament sliding |
| TPM3 | 7170 | CAPM1, CFTD, HEL-189, HEL-S-82p, NEM1, OK/SW-cl.5, TM-5, TM3, TM30, TM30nm, TM5, TPMsk3, TRK, hscp30 | striated muscle thin filament, muscle filament sliding |
| TREX1 | 11277 | AGS1, CRV, DRN3, HERNS | endoplasmic reticulum part |
| TSC1 | 7248 | LAM, TSC | hippocampus development, limbic system development |
| TSC2 | 7249 | LAM, PPP1R160, TSC4 | protein import, morphogenesis of an epithelium |
| TWIST2 | 117581 | AMS, BBRSAY, DERMO1, FFDD3, SETLSS, bHLHa39 | face development, body morphogenesis |
| TYMP | 1890 | ECGF, ECGF1, MEDPS1, MNGIE, MTDPS1, PDECGF, TP, hPD-ECGF | blood vessel development, chemotaxis |
| UBA1 | 7317 | A1S9, A1S9T, A1ST, AMCX1, CFAP124, GXP1, POC20, SMAX2, UBA1A, UBE1, UBE1X | endoplasmic reticulum part |
| UBE3A | 7337 | ANCR, AS, E6-AP, EPVE6AP, HPVE6A | developmental growth, brain development |
| UPK3A | 7380 | UP3A, UPIII, UPIIIA, UPK3 | endoplasmic reticulum part, cellular component morphogenesis |
| UTRN | 7402 | DMDL, DRP, DRP1 | sarcolemma, synaptic membrane |
| VPS33B | 26276 | bone development, organelle localization | |
| WNT5A | 7474 | hWNT5A | prostate gland epithelium morphogenesis, face development |
| WNT7A | 7476 | chemical synaptic transmission, postsynaptic, chondrocyte differentiation | |
| ZBTB42 | 100128927 | LCCS6, ZNF925 | muscle organ development, muscle structure development |
| ZC4H2 | 55906 | HCA127, KIAA1166, WRWF, WWS | spinal cord development, central nervous system neuron differentiation |
| ZIC3 | 7547 | HTX, HTX1, VACTERLX, ZNF203 | digestive tract development, digestive system development |
| ZMPSTE24 | 10269 | FACE-1, FACE1, HGPS, PRO1, STE24, Ste24p | endoplasmic reticulum part |
| ZNF335 | 63925 | MCPH10, NIF-1, NIF1, NIF2 | neuroblast proliferation, regulation of stem cell proliferation |
This table lists all the genes associated with arthrogryposis used in the ClueGO enrichment analysis. Entrez Gene ID: Entrez Gene unique ID; Aliases: additional gene names; Functions: GO terms that are associated with the specific gene.
Popular gene set libraries used for EA include manually curated gene sets representing canonical signaling pathways, such as Reactome [Croft et al., 2014], and structured gene sets based on the Gene Ontology (GO) resource [Gene Ontology Consortium, 2015]. The GO is a resource, in the form of a structured ontology, which describes and categorizes gene product functions in distinct categories and the relationships between them. The GO functional categories are classified in 3 general categories: biological process, molecular function, and cellular component. The biological process category contains individual GO terms that describe processes associated with molecular events and pathways representing multiprotein-dependent functions. The molecular function category, in contrast, describes basic gene functions at the molecular level. Lastly, the cellular component category describes the location, environment, or part of the cell, so that the gene product can be located.
The EA for this review was performed using the software tool ClueGO [Bindea et al., 2009]. ClueGO calculates enrichment scores for selected gene sets against a user-provided gene list. Our analysis was performed using the biological process and cellular component categories of the gene ontology. The biological process category was selected because it captures functional descriptions that provide a better biological interpretation based on multicomponent signaling and functional groupings. The cellular component category was selected as it provides details on not only intracellular locations, but also higher ordered structures such as the ‘synaptic membrane’. The other main benefit of performing EA with ClueGO is that it groups similar GO terms and provides a network-based view of the enriched GO terms. This is important in that it aides interpretation of results by grouping related GO terms, based on shared gene members, presenting the results as a network. Since GO has a hierarchical ontological-based structure, GO terms often have overlapping gene members. When results of enrichment are presented in tabular form, this could inhibit the interpretation and summary of the results. The network visualization groups similar GO terms as nodes in the network with edges representing a measure of shared gene membership (kappa score).
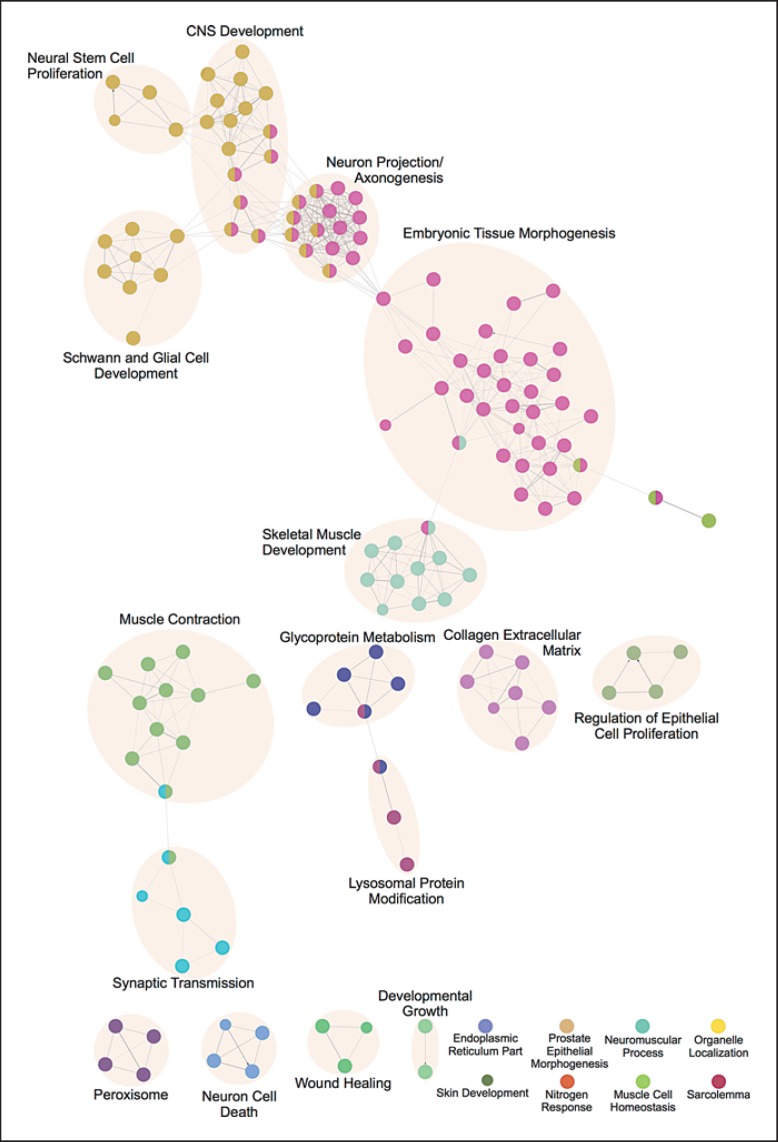
The ClueGo analysis identified 145 enriched GO terms with a corrected p < 0.001 (online suppl. table 1; see www.karger.com/doi/10.1159/000446617 for all online suppl. material). The automatic grouping of terms, performed by ClueGO, assigned them to 22 groups based on overlapping gene membership measured by the kappa score statistic (online suppl. table 2). The number of GO terms in the groups varied from a maximum of 57 to 8 groups with just 1 term each and 9 groups with shared GO terms. The network representation is displayed in figure 1. The network is composed of nodes representing enriched GO terms connected by kappa scores that are a measure of gene overlap between terms. The node color represents the membership in one of the ClueGO determined clusters. While ClueGo attempts to determine the most representative GO term to name the grouped terms, we have provided our own annotation grouping for greater clarity in interpretation and meaning (online suppl. table 3). This grouping is indicated by the shading overlaid on the network with an annotated group labeled with a summary title of the underlying GO terms in the group.
Fig. 1.
GO enrichment network. The nodes in the network represent a specific GO term. The edges connecting the nodes are based on the kappa statistic that measures the overlap of shared genes between terms. The node colors correspond to the ClueGO-determined GO term clusters. The shadings represent author-annotated groupings with a summary title.
The purpose of this paper is to highlight the genes in which mutations have been identified to be associated with arthrogryposis in order to emphasize the importance of defining the other genes in their developmental pathways. This is in order to (1) develop a logical diagnostic approach and (2) to begin to think about specific therapies for specific disorders. For instance, in the disorders of neuromuscular endplate related to fetal endplate receptor, they seem to respond to increased neurotransmitters (a readily available drug used to myasthenia gravis), which then seem to allow the normal adult endplate to be able to function [Michalk et al., 2008].
Perhaps the most puzzling aspect of arthrogryposis is why extra connective tissue/fibrosis is deposited around the immobile joint(s) in the fetus. Is the process related to immobilizing that occurs with a sprain or fracture, where pain leads to an individual immobilizing the surrounding joints which then develop contractures? This process would be magnified as the fetus grows. Or is there another unique developmental process of fibrosis in young individuals? Is the process similar to tendon and ligament formation? Are connective tissue stem cells overstimulated or more susceptible in the fetus? Is this excess of connective tissue an unusual scar of some type? Is one of the connective tissue growth factors a potential therapy for arthrogryposis contractures of the future?
In this molecular era, syndromes of congenital anomalies give insight into normal developmental processes and their secondary and tertiary effects. In the case of arthrogryposis, so many of the features are secondary deformations related to fetal non-movement [Hall, 2009]. Nevertheless, all of the features which are part of the natural history of the specific disorder are important for families to know about in order to plan effectively.
The specific gene mutation in a specific disorder is acting against the rest of the individual's genome, epigenetics, and environmental history. In the course of development, the embryo fetus goes through many physiological developmental stages. The vulnerabilities, timing of insults, involved polymorphisms along a pathway, and gene action also provide insights into the human normal and abnormal developmental processes.
The work up of affected individuals [Hall, 2012, 2014] as well as the known genes are covered elsewhere; the associated syndromes are found in OMIM (http://www.omim.org/) [Hall, 2012, 2014; Hunter et al., 2015; Bayram et al., 2016].
Table 1 outlines gene ontology categories and begins to suggest prime candidates for recognizing critical pathways involved in normal fetal movement. Interestingly, many candidate genes show up in several ontology categories. This may relate to different domains of the genes, to alternative splicing, or to the ‘recycling’ of pathways for different functions.
It is hoped that this exercise is useful to those reflecting on the many mechanisms and structures involved in the development of movement, and fetal movement in particular. The listing of all genes recognized to be involved in arthrogryposis at this time is obviously an incomplete list. Some genes are involved in several disorders which were clinically thought to be distinct (perhaps related to the specific nucleotide replacement or perhaps related to various modifiers). Once a group of families with a specific mutation has their whole genome analyzed, the variation in clinical phenotype can be elucidated and important modifiers identified.
Many specific single gene disorders have intra- and interfamilial variability as to how severe the contractures are at birth, what positioning they take, or whether contractures are even present. For instance, several forms of dominant distal arthrogryposis have completely unaffected carrier individuals [Kimber et al., 2012].
It is also hoped that this listing will point to other genes involved in ontological processes that may also result in arthrogryposis and be part of the pathways leading to normal fetal movement - thereby providing better diagnostic precision among the present quagmire of interpretation of the whole genome and even exome sequencing. Ultimately, specific therapies may involve alternative pathways and enhance the affected pathway.
Disclosure Statement
The authors declare no conflicts of interest.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
References
- 1.Bayram Y, Karaca E, Coban Akdemir Z, Yilmaz EO, Tayfun GA, et al. Molecular etiology of arthrogryposis in multiple families of mostly Turkish origin. J Clin Invest. 2016;126:762–778. doi: 10.1172/JCI84457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42(Database issue):D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filges I, Hall JG. Failure to identify antenatal multiple congenital contractures and fetal akinesia - proposal of guidelines to improve diagnosis. Prenat Diagn. 2013;33:61–74. doi: 10.1002/pd.4011. [DOI] [PubMed] [Google Scholar]
- 5.Gene Ontology Consortium Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall JG. Pena Shokeir phenotype (fetal akinesia deformation sequence) revisited. Birth Defects Res A Clin Mol Teratol. 2009;85:677–694. doi: 10.1002/bdra.20611. [DOI] [PubMed] [Google Scholar]
- 7.Hall JG. Arthrogryposes (multiple congenital contractures) In: Rimoin DL, Pyeritz RE, Korf BR, editors. Emery and Rimoin's Principle and Practice of Medical Genetics. ed 6. New York: Churchill Livingstone; 2012. [Google Scholar]
- 8.Hall JG. Arthrogryposis (multiple congenital contractures): diagnostic approach to etiology, classification, genetics, and general principles. Eur J Med Genet. 2014;57:464–472. doi: 10.1016/j.ejmg.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Hall JG, Aldinger KA, Tanaka KI. Amyoplasia revisited. Am J Med Genet A. 2014;164A:700–730. doi: 10.1002/ajmg.a.36395. [DOI] [PubMed] [Google Scholar]
- 10.Hunter JM, Kiefer J, Balak CD, Jooma S, Ahearn ME, et al. Review of X-linked syndromes with arthrogryposis or early contractures - aid to diagnosis and pathway identification. Am J Med Genet A. 2015;167A:931–973. doi: 10.1002/ajmg.a.36934. [DOI] [PubMed] [Google Scholar]
- 11.Kimber E, Tajsharghi H, Kroksmark AK, Oldfors A, Tulinius M. Distal arthrogryposis: clinical and genetic findings. Acta Paediatr. 2012;101:877–887. doi: 10.1111/j.1651-2227.2012.02708.x. [DOI] [PubMed] [Google Scholar]
- 12.Lowry RB, Sibbald B, Bedard T, Hall JG. Prevalence of multiple congenital contractures including arthrogryposis multiplex congenita in Alberta, Canada and a strategy for classification and coding. Birth Defects Res A Clin Mol Teratol. 2010;88:1057–1061. doi: 10.1002/bdra.20738. [DOI] [PubMed] [Google Scholar]
- 13.Michalk A, Stricker S, Becker J, Rupps R, Pantzar T, et al. Acetylcholine receptor pathway mutations explain various fetal akinesia deformation sequence disorders. Am J Hum Genet. 2008;82:464–476. doi: 10.1016/j.ajhg.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data