Abstract
Psychiatric research has entered the age of ‘Big Data’. Datasets now routinely involve thousands of heterogeneous variables, including clinical, neuroimaging, genomic, proteomic, transcriptomic and other ‘omic’ measures. The analysis of these datasets is challenging, especially when the number of measurements exceeds the number of individuals, and may be further complicated by missing data for some subjects and variables that are highly correlated. Statistical learning-based models are a natural extension of classical statistical approaches but provide more effective methods to analyse very large datasets. In addition, the predictive capability of such models promises to be useful in developing decision support systems. That is, methods that can be introduced to clinical settings and guide, for example, diagnosis classification or personalized treatment. In this review, we aim to outline the potential benefits of statistical learning methods in clinical research. We first introduce the concept of Big Data in different environments. We then describe how modern statistical learning models can be used in practice on Big Datasets to extract relevant information. Finally, we discuss the strengths of using statistical learning in psychiatric studies, from both research and practical clinical points of view.
Key words: Machine learning, outcome prediction, personalized medicine, predictive modelling, statistical learning
The ‘data explosion’ in psychiatry
Once the problem of psychiatric research was that there were not enough data. Now, with the pace of technological advances that have occurred in the present century in neuroimaging, genomics, transcriptomics, proteomics and all the other ‘omics’, we are in danger of being overwhelmed by a volume of data that the human brain, aided only by ‘traditional’ statistical methods, cannot assimilate and integrate. For example, genome-wide association studies (GWAS) now typically and routinely generate millions of data points on tens of thousands of subjects. This has led to some breath-taking advances, notably the finding, based on data from 37 000 patients, that over 100 different genetic loci have a role in schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics, 2014). Similar large-scale studies are underway for other common disorders and, in the UK alone, plans are in place to sequence the entire genomes of 100 000 subjects (http://www.genomicsengland.co.uk). The standard statistical analyses of GWAS are, in principle, straightforward involving χ2 tests comparing genetic marker frequencies in cases and controls and applying a stringent correction for multiple testing. However GWAS findings tend to throw up many other problems that will not be solved by such simple analyses. For example, none of the hundred-plus genome-wide significant loci is necessary or sufficient to cause schizophrenia, so this poses a series of new questions. What combinations of loci in interplay with what environmental insults might be useful in predicting who becomes affected in at-risk groups? What combinations of loci relate to what symptom patterns, courses’ outcomes or responses to treatment? What combinations of genetic loci influence structural or functional brain-imaging characteristics? (This is a particularly thorny problem since imaging studies typically generate many more data points even than genomics.) We suggest that a set of solutions to 21st century psychiatry's information overload problems is offered by machine learning (ML) and in particular from a branch that is now often called statistical learning (SL).
A world of Big Datasets and the role of SL
Although many of us are probably unaware of it, SL is happening all around us. Social media developers, committed to retaining their users and encouraging their online activity are constantly storing information about users and their daily actions in huge datasets, and employ specific methods of analysis designed to deduce what users might ‘like’ next (e.g. new people to incorporate as ‘friends’ or pages that might be of interest). In a similar way, commercial websites such as Amazon aim to predict what product we would next like to buy by thoroughly collecting our history of shopping baskets in databases and investigating our pattern of shopping and comparing it with persons of similar shopping patterns. Another and more individual example is that many of us now use voice recognition software as an alternative to manual typing. Such software not only learns to interpret what we say into printed word but also learns our personal vocabulary, idioms and patterns of expression.
The datasets involved in such processes have three main aspects in common: they occupy vast amounts of computer memory, measured in Terabytes (trillions of bytes), they are heterogeneous containing information coming from a variety of sources, for example a combination of text messages, images and videos, and they are constantly and quickly being updated with new information. These three aspects have been proposed by some authors as the main characteristics of Big Datasets and summarized as the three Vs – volume, variety and velocity (Laney, 2001).
Large-scale datasets from clinical trials and cohort studies, electronic health records or national health registries are becoming increasingly available in biomedical research. They are becoming the focus of research studies that aim to better understand genotype–phenotype relationships, find factors that can predict disease risk, discover profiles of patients that are better responders to a treatment and discover or define disease categories. In general, these datasets meet the three Vs definition, so we can state that biomedical research has definitely entered the Big Data world.
The urgent need of methods that can help to understand such complex Big Datasets has led to a revolution in statistical sciences. Whereas statistics has focused primarily on what conclusions can be inferred from data, Big Datasets have raised other questions about what computational architectures and algorithms can be more efficient to extract maximum information from data in a computationally tractable way (Mitchell, 2006). ML (Soler Artigas et al. 2011) refers to a discipline that offers a set of tools built within the intersection of computer sciences and statistics that are capable of coping with the requirements of the Big Data world. These ‘statistical-computational’ systems improve their performance at particular tasks by experience (Mitchell, 1997, 2006; Soler Artigas et al. 2011), which is they are capable of learning from data.
Other terms commonly used in the area of ML, but showing slight conceptual differences include artificial intelligence, which encompasses natural language processing, knowledge representation and automated reasoning (Barr et al. 1981; Ripley, 1996; Russell & Norvig, 2010), deep learning, a new type of ML algorithm based on neural networks with the aim of discerning higher level features from data (LeCun et al. 2015). Other approaches include pattern recognition, a branch of ML focused on the recognition of patterns and regularities in data (Bishop, 2006) and data mining, the process of exploring data in search of consistent patterns and/or systematic relationships between variables (Hand et al. 2001).
SL is a fairly recently coined term (Hastie et al. 2009) that refers to a vast set of statistical and computational methods to understand complex data. These are based on a range of approaches, from classical concepts belonging to the first half of the 20th century such as linear regression modelling and discriminant analysis, to the latest advanced computational-based approaches including modern ML. Hence SL is a broad term that emphasizes the essential role of statistics within ML in the context of Big Data analysis.
Learning from data
The methods that underlie SL learn from data, i.e. they are able to explore and retain significant structure from data that is replicable across different samples extracted from the same population. Broadly there are three categories of learning from data. The first concerns ‘supervised’ learning (Hastie et al. 2009), which typically involves building an algorithm that uses as input a dataset of candidate predictors known as features or attributes (e.g. age, cancer staging, hospital admissions) and is able to estimate a specific outcome (e.g. 6-month survival for cancer patients). Supervised learning includes classification and regression problems. In a classification problem the aim is to determine what category something belongs to, after seeing a number of examples of things from the relevant categories.
The second major category concerns ‘unsupervised’ learning (Ghahramani, 2003) when there is no predefined outcome to be predicted. The task here is deriving an algorithm able to explore data patterns and to discover structure, for example groups of patients who share similar clinical or test result profiles. The two cornerstones of unsupervised learning are clustering (Everitt et al. 2010), and dimensionality reduction (Lu et al. 2013) which includes principal components analysis and factor analysis. These methods have found important applications in medical research, particularly in psychiatric studies (Ochoa et al. 2013; Brodersen et al. 2014).
A third category, known as ‘semisupervised’ learning (Zhu & Goldberg, 2009) combines insights from supervised and unsupervised methods by exploring observations where the outcome (or label) is known only for a small amount of data (e.g. the study of the profile of patients that response positively or negatively to a drug, combined with the study of patients with unknown treatment outcome).
In the remainder of this review we will focus on supervised learning problems. Here the outcome is a variable taking either a number of levels that are often called ‘classes’ or ‘labels’ (e.g. relapsing or non-relapsing of a condition), or a quantitative value (e.g. response to treatment as measured by a rating scale). Thus when we talk about ‘labelled data’ we refer to a set of observations for which the outcome is known.
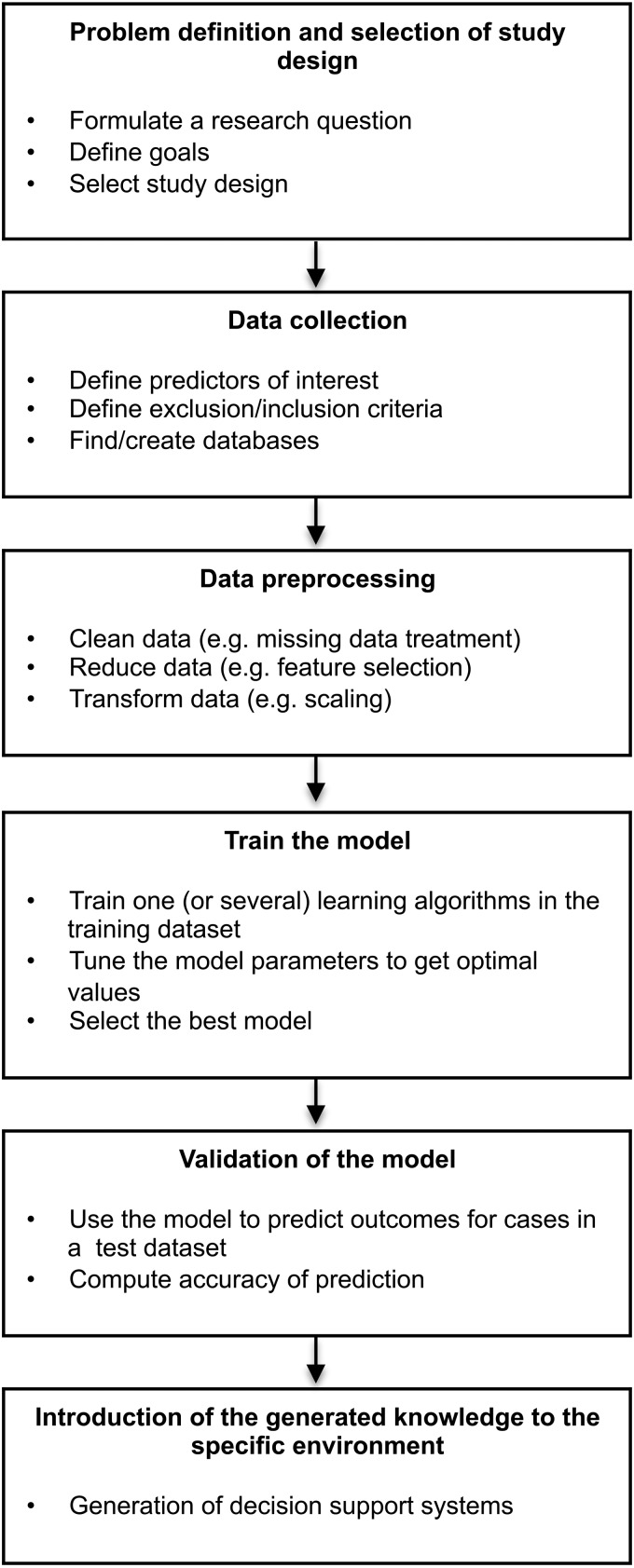
The main stages of the learning process are given below (see Fig. 1).
Fig. 1.
Main steps of the learning process.
Definition of the problem and selection of study design
The problem we aim to solve needs to be precisely defined and well understood. As with all research the starting point is critical review of the previous knowledge in the area, formulation of a research question and choice of appropriate study design (Katz, 2006). For example, a longitudinal collection of patients’ data may allow investigation of the risk of an occurrence or relapse concerning a disease over time. Designs such as the case-control that collect data of disease and healthy individuals at just one point in time, will be appropriate to test the ability of a set of factors in predicting a diagnosis.
Data collection and pre-processing
Ideally, quality data will include a well-defined selection of patients, and a rigorous collection of relevant predictors and outcomes. Before analysis, the main steps of data pre-processing include data cleaning, data reduction and data transformation.
Cleaning refers to the treatment of missing data, a common problem in psychiatric research, and this is important as inadequate missing data treatment may lead to an overestimation of prediction accuracy (Batista & Monard, 2002). Discarding individuals or variables with missing values (‘the complete-case analysis’) may bias analysis if the units with missing values differ systematically from the completely observed cases, especially if percentage of missingness is high. A preferable approach may be to estimate or ‘impute’ missing values using either classical statistics or SL. SL methods (e.g. tree-based methods; Ding et al. 2010) (Table 1) are free of assumptions and have been found to outperform classical statistical methods of imputation. For example, the methods based on SL techniques were the most suited for the imputation of missing values in a study aiming to predict cancer recurrence, and led to a significant enhancement of prognosis accuracy compared to imputation methods based on statistical procedures (Jerez et al. 2010).
Table 1.
Main properties of a set of selected statistical learning algorithms
| Machine learning algorithm | Details |
|---|---|
| General linear regression models (GLM) |
|
| Elastic net models |
|
| Naive Bayes |
|
| Classification and Regression Trees (CART) |
|
| Random forest |
|
| Support vector machines (SVM) |
|
| Artificial neural networks (ANN) |
|
All methods listed above can be used for classification (categorical outcome) and for regression (quantitative outcomes) problems. All of them can handle multiple continuous and categorical predictors.
Data reduction involves obtaining a reduced representation of the data volume that can achieve the same (or almost the same) analytical results. By creating new features as a result of the aggregation or eliminating features that are not meaningful for prediction (‘feature selection’) tasks can be made more computationally tractable. Reducing the number of features also makes models more easily interpretable. This point is critical for the success of a predictive algorithm, especially if there are thousands of features at the outset (Guyon, 2003; Witten & Tibshirani, 2010). Feature reduction can be performed as a part of pre-processing or during the modelling step using algorithms that perform an internal feature selection (elastic net regression; Zou & Hastie, 2005) or Classification and Regression Tree (CART) algorithms (Rokach & Maimon, 2008) (Table 1). The latter will usually improve reliability and increase confidence in selected features (Caruana & Niculescu-Mizil, 2006; Krstajic et al. 2014).
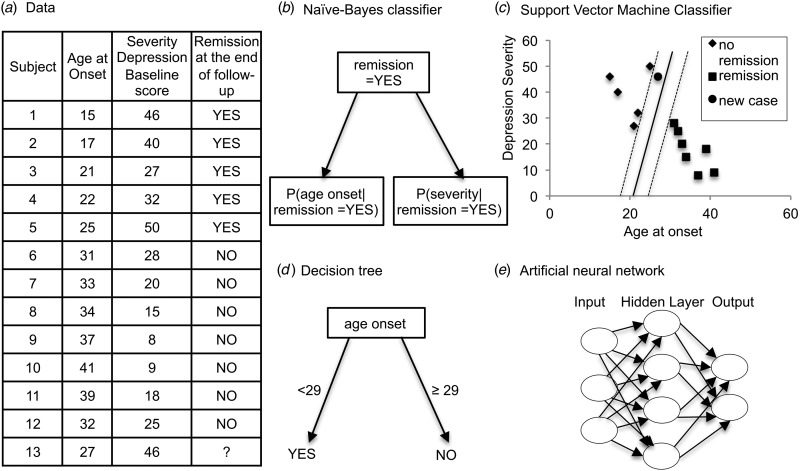
Fig. 2.
(a) Data simulated from a follow-up study of major depression patients. Age of depression onset (years) and the MADRS score at baseline ranging from 0 to 60 (0–6, normal; 7–19, mild depression; 20–34, moderate depression; >34, severe depression) are the predictor variables. The outcome is remission status at the end of the follow-up (YES or NO). (b) The Naive Bayes classifier is often represented as this type of graph. The direction of the arrows states that each class causes certain features, with a certain probability. (c) A hyper plane (a line, in dimension 2) is built at a maximal distance to every dashed line (called margin). A new case (point) will be classified as remission or non-remission depending on his relative position to the line (aka decision boundary). (d) A simple decision tree suggesting that patients with age of onset lower than 29 are more likely to reach a remission. (e) Each node represents an artificial neuron and each arrow a connection from the output of one neuron to the input of another.
Data transformation methods depend on the specific SL algorithm to be used (Kotsiantis et al. 2006). Three common data transformations are scaling, decompositions and aggregations. Many SL methods (e.g. the elastic net regression; Zou & Hastie, 2005) require all predictors to have the same scale such as between 0 and 1. Decomposition may be applied to features that represent a complex concept, as they may be more useful to a ML method when split into their constituent parts (e.g. a date can be split into day, month and year). Aggregation is appropriate when there are features that are more meaningful to the problem when combined into a single feature.
Training and validation of the model
The data used to run a learning algorithm are called training data. In supervised ML the program is told what the output should look like, for example what subjects belong to what category label. A second set of data is called the test dataset. Here the labels are again known to the researcher but in this run the program is only given the input data and the task is to correctly assign the outputs or labels. Ideally the test data and the training set should be completely independent but in practice researchers very often randomly split datasets of labelled data in two parts and arbitrarily define one part as the learning data and the other as the test set. If the algorithm is able to estimate correct labels in this new set of cases, i.e. the called prediction error is small (e.g. the number of falsely classified cases is much smaller than chance classification), the classifier may be considered to be ‘valid’ to be used in estimating outcomes for cases with unknown outcomes. As elsewhere in classification problems a variety of measures are used to assess prediction accuracy (Steyerberg et al. 2010), for example sensitivity (the proportion of correctly classified recovered cases) and specificity (the proportion of correctly not recovered cases) for binary classifications.
Wolpert & Macready (1997) consider that there is unlikely to be a single technique that will always do best for all learning problems. Hand (2006) advocated that we should base our selection on a compromise between the accuracy of the model in predicting outcomes for new cases and the interpretability of the result.
Specific ML terminologies that have been adopted by the SL community are introduced in Table 2. A more detailed set of definitions can be found in (Kohavi, 1998).
Table 2.
Glossary of statistical/machine learning terms used in this paper
| Term | Definition |
|---|---|
| Feature/attribute/predictor | A numerical (e.g. subset of real values) or categorical (i.e. a finite number of discrete values) value used as input to a learning algorithm |
| Outcome/response/label | A numerical or categorical value to predict from features |
| Labelled data | A set of features and labels for an observation |
| Training set | A collection of data used to train a learning algorithm |
| Test set | A collection of labelled data |
| Supervised learning | Techniques used to learn the relationship between independent attributes and a designated dependent attribute (the label) |
| Unsupervised learning | Learning techniques that group observations without a pre-specified dependent attribute. Clustering algorithms are usually unsupervised |
| Model | A structure and corresponding interpretation that summarizes or partially summarizes a set of data, for description or prediction. Most learning algorithms generate models that can then be used in a decision-making process |
| Accuracy | The rate of correct predictions made by the model over a dataset. Accuracy is usually estimated by using an independent test set that was not used at any time during the learning process. More complex accuracy estimation techniques, such as cross-validation and the bootstrap, are commonly used, especially with datasets containing a small number of observations |
| High-dimensional problem | Problems in which the number of features p is much larger than the number of observations N, often written p > N. Such problems have become of increasing importance, especially in genomics and other areas of computational biology |
| Overfitting | A modelling error that occurs when the model is too closely fit to a limited set of data points. As data being studied often has some degree of error or random noise, an overfitted model is poor in predicting new cases |
| Multicollinearity | Correlation between features, i.e. the situation where if the value of a feature change, values for the rest of features also change at some degree. When there is multicollinearity between variables in a regression model, its coefficients can become poorly determined and exhibit high variance |
| K-fold cross-validation | A method for estimating the accuracy (or error) of a learning algorithm by dividing the data into K mutually exclusive subsets (the ‘folds’) of approximately equal size. K models are trained and tested. Each time a model is trained on the data set minus a fold and tested on that fold. The accuracy estimate is the average accuracy for the K folds |
Table 1 summarizes seven popular SL algorithms. More detailed information about specific learning algorithms can be found elsewhere (Mitchell, 1997, 2006; Vapnik, 1998; Scholkopf et al. 2003; Malley et al. 2011).
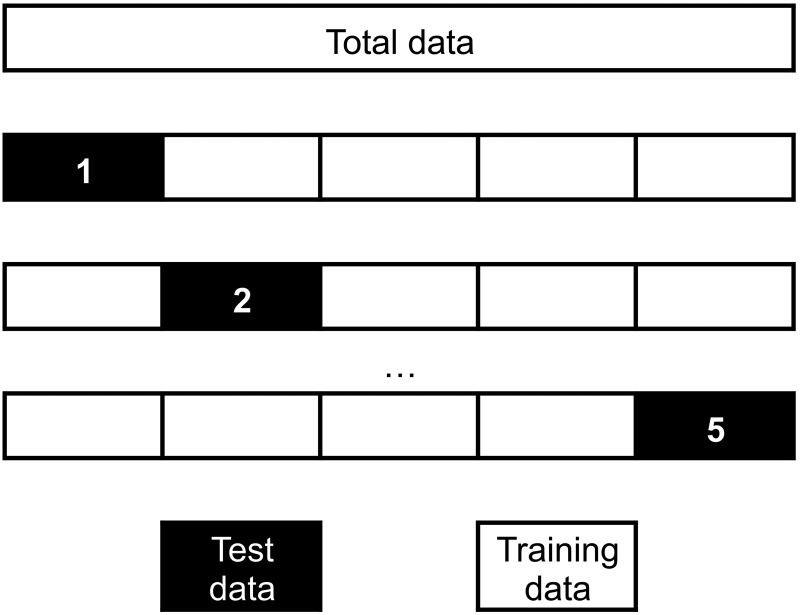
A common scheme to train different classifiers and select one based on ability to predict outcomes is the K-fold cross-validation (CV). This is a procedure where the original training sample is randomly divided in K subsamples, K-1 samples are used as a new training set and one is left out as an occasional ‘test’ set in K iterations (Fig. 3). The prediction error is then computed across test samples. Minimizing the prediction error from the CV loop is used to select the best algorithm and the best predictive model produced by the same algorithm. CV provides a nearly unbiased prediction error on new observations from the same population (Kohavi, 1995).
Fig. 3.
Example of a 5-fold cross-validation. Data are randomly split in 5-fold of equal size. At every step, one fold is selected as test dataset and the remaining four are used as training data. This procedure is repeated five times, selecting in every step a different fold as test data.
Introducing a generated predictive knowledge to a practical setting
A nice example is provided by the work of Perlis and colleagues (Perlis, 2013) who ran a prospective investigation to identify clinical predictors of antidepressant treatment resistance. The authors selected 15 easy-to-obtain features for patients with known response and adopted a SL approach. Based on the best model obtained, the team developed a web-based clinical decision support system that given the values for the 15 variables for a particular patient suffering from major depression could aid in predicting the risk of being resistant to an antidepressant treatment.
How statistical learning renders Big Data problems tractable in psychiatric research?
Dealing with heterogeneous sources of information
Data from different sources (e.g. large longitudinal clinical trials or cohort studies, electronic health records, national health registries) has a greater potential for establishing novel useful ways of categorizing patients’ groups (patient stratification) and for revealing unknown disease correlations compared to learning from each source independently (Shi et al. 2012). Specific SL algorithms have demonstrated impressive empirical performance on a wide variety of classification tasks involving heterogeneous Big Datasets (e.g. decision-tree approaches; Breiman, 1984), regularized regression models (Zou & Hastie, 2005), as well as support vector machines (Lewis et al. 2006) (Table 1). Integrating such data is a challenge that may include the problem that data are stored in many different formats. However, the handling of Big Data from a variety of sources is becoming ever more feasible and affordable, with many institutions employing their own local clusters of computers (banks of many micro-computers hooked up in parallel and providing huge computational power). ‘Cloud’ computing is another increasingly available option. This refers to using the Internet to access the vast computational resources that are offered commercially by companies such as Amazon, Google and Microsoft.
The IMAGEN study (Whelan et al. 2014) is a good example where researchers integrated data from very heterogeneous domains and applied a SL approach of analysis. Domains included brain structure and function, individual personality and cognitive differences, environmental factors, life experiences, and candidate genes. They applied elastic net regularized regression (Zou & Hastie, 2005) to generate models to predict current and future adolescent alcohol misuse based on such holistic characterization. This ‘regularized’ approach automatically dropped out features that were not contributing to the class predictions. Thus the final model incorporated a subset of the most relevant variables for prediction selected from all of the explored families of predictors. The favoured models pointed to life experiences, neurobiological differences and personality as important antecedents of binge drinking, suggesting possible targets for prevention. The authors reported specific predictors in their models along with their regression coefficient as a standard and interpretable measure of strength between each predictor and the outcome. The approach correctly predicted alcohol misuse for individuals not in the original dataset, emphasizing the model's capability to generalize to novel data.
The search for meaningful predictors of a psychiatric outcome in high-dimensional datasets
Big Datasets in psychiatry research can be ‘Big’ regarding volume and number of features but involving a relative smaller sample size. For example, even though GWAS typically now contain tens of thousands of subjects, there may be many millions of data points. Increasingly large-scale case-control studies also include gene expression, genome sequencing and epigenetics, proteomics or metabolomics inflating the data to research subject ratio even more. This is often called the high-dimensional data problem, or the ‘p > N’ problem (where p is the number of features and N the number of observations). Such data are commonly represented in a matrix, with more columns than rows. The classical approach of comparing thousands of single association tests and then ranking features by their statistical significance is not an optimal solution. The first concern is that multiple testing increases the risk of spurious findings due to chance. The application of stringent methods to correct this can lead to the detection of strong contributors to outcome at the expense of overlooking smaller contributors. This poses a problem in complex traits and disorders that, by their nature are multifactorial. Another related weakness is that independent analysis variable by variable does not permit inferences about combinations of variables. Generalized linear regression models (Hosmer et al. 2013) are problematic in estimating the effect of such combinations. This kind of model is in danger of explaining mainly noise instead of the relationships between variables (and so models are poor in generalizing to new datasets). This problem is known as overfitting (Table 2). A second problem for generalized linear regression is correlation between features, i.e. the situation where if one feature changes, so do one or more other features, an effect known as multicollinearity (Table 2). An example is genetic variation. Due to the fact that most of our genetic information is inherited in ‘blocks’ from our parents, the information at different positions of our genome is expected to be highly correlated within families. Blocks, albeit smaller ones, also occur within genetically homogenous populations. Multicollinearity can seriously distort the interpretation of a model, making it less accurate by introducing bias within the coefficients of the model (Maddala & Lahiri, 2009) and increasing uncertainty, as reflected in inflated standard errors (Glantz & Slinker, 2000; Miles & Shevlin, 2001).
Supervised SL models offer a means to overcome these problems and to maximize the predictive power, hence providing exciting opportunities for individualized risk prediction based on personal profiles (Ashley et al. 2010; Manolio, 2013). SL models such as the multivariate adaptive regression splines (MARS) procedure (Friedman, 1991), the CART (Breiman, 1984), elastic net regularized regression (Tibshirani, 1994; Zou & Hastie, 2005; Friedman et al. 2010) and support vector machines (Cortes & Vapnik, 1995) (Table 1) perform especially well in the high-dimensional scenario and in the presence of correlation between predictors (Libbrecht & Noble, 2015). They also allow to efficient identification of informative patterns of interactions between clinical and biomarker variables, which are known to play an important role in the development and treatment of many complex diseases (Lehner, 2007; Ashworth et al. 2011), but are often missed by single association tests (Cordell, 2009).
Models in practice: the case of stratified and personalized medicine
In recent years stratified and personalized medicine became of interest in mental health research which utilizes molecular biomarkers (Kapur et al. 2012), demographic and clinical information, including patients’ health records, to identify subgroups of patients who are likely to respond similarly to treatment using SL methods. Major depressive disorder is a prime example of a common disorder where there are many available drugs but where there is no straightforward way of deciding which treatment is likely to work for a given individual (Simon & Perlis, 2010). The Genome-based Therapeutic Drugs for Depression (GENDEP Investigators et al. 2013) project is a study aiming to test clinical and genetic data as predictors of treatment response to two antidepressant drugs (Uher et al. 2009, 2010). The need for prediction at individual level involving hundreds of thousands of variables prompted the use of SL methods (Iniesta et al. 2016). The challenge was the integration of clinical with biological markers and deriving optimal models with minimal risk of overfitting. Demographic, clinical and genetic predictors were combined in a model to predict the change in severity symptoms after a 12-week period in a sample of patients randomly treated with one of the two drugs. A linear regularized elastic net model (Zou & Hastie, 2005) looked for the best combination of variables in predicting symptoms course. Interestingly, the feature selection approach of elastic net allowed building drug-specific models that were able to predict treatment outcome with accuracy above a clinical significance threshold. The results suggested a potential for individualized indications for antidepressant drugs. The benefits of using the elastic net were several: first, the elastic net provided an efficient internal method of search and selection of predictors from a large set of variables available. Second, the iterative CV procedure used allowed the selection of predictors based on their ability in predicting outcome for unseen cases, which was the aim of this research. Third, this flexible approach reported distinct and specific models to each outcome and drug sample. Fourth, the elastic net allowed estimation of the combined predictive ability of a high number of variables while preventing the models from overfitting.
The hoped for impact of this type of research is the introduction of a predictive model (last box in Fig. 1) as a clinical decision support system. For a model to be useful in the practical scenario there is a list of challenges we need to overcome. First, the model should have been externally validated in a test dataset. Very often the validation of models built in sample of patients with very specific characteristics (e.g. those coming from randomized clinical trials) is difficult because it is hard to find another similar sample that can work as a ‘test’ dataset. Second, as a consequence, such models tend to poorly generalize to other populations. For example, if a model was built for a homogeneous ethnical population of white Caucasian patients and ethnicity has an effect on outcome, there is no guarantee that such model will predict well for an individual of different ethnicity. Thus some authors argue matching treatments to individuals is a too ambitious aim, as given any model, there can always be a relevant-to-outcome patient characteristic that was not included nor validated. However, it is not all bad news; several studies in cancer were able to find almost perfect biomarkers for treatment selection, specifically for chemotherapy treatment and some progress towards stratified medicine is appearing feasible in psychiatry (Perlis, 2013; Iniesta et al. 2016). A third challenge is the generation of easy-to-use tools in the clinical setting. Ideally models should involve a reasonable number of easy-to-obtain variables and be implemented through tools that allow a quick introduction of patients’ data and a simple and clear display of model outputs.
We can conclude that Big Data are becoming a major challenge for statistical analysts in mental health research and a paradigm shift in methods is needed. Statistical learning provides a set of tools that can successfully help in the understanding of such complex datasets. Such methods can be useful as an alternative or in addition to ‘classical’ statistical inference methods based solely on hypothesis testing which has been criticized by many statisticians for many years (Breiman, 2001a; Nuzzo, 2014). Big Data analysis and the derivation of predictive SL models for stratified medicine in psychiatry is an emerging and hot area, and such tools have the potential to facilitate a better targeting of interventions and diagnosis of patients.
Acknowledgements
We gratefully thank Robert Tibshirani, Stanford University, for critically reading this manuscript and providing substantial comments that greatly improved the work. We also thank Professor Cathryn Lewis, King's College London, for her constant support during the work.
This work has been funded by the European Commission Framework 6 grant, EC Contract LSHB-CT-2003-503428 and an Innovative Medicine Initiative Joint Undertaking (IMI-JU) grant no. 115008 of which resources are composed of European Union and the European Federation of Pharmaceutical Industries and Associations (EFPIA) in-kind contribution and financial contribution from the European Union's Seventh Framework Programme (FP7/2007-2013).
Funding was also received from the European Community's FP7 Marie Curie Industry-Academia Partnership and Pathways, grant agreement no. 286213 (PsychDPC). Open Access for this article was funded by King's College London.
Declaration of Interest
None.
References
- Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA, Pushkarev D, Neff NF, Hudgins L, Gong L, Hodges LM, Berlin DS, Thorn CF, Sangkuhl K, Hebert JM, Woon M, Sagreiya H, Whaley R, Knowles JW, Chou MF, Thakuria JV, Rosenbaum AM, Zaranek AW, Church GM, Greely HT, Quake SR, Altman RB (2010). Clinical assessment incorporating a personal genome. Lancet 375, 1525–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth A, Lord CJ, Reis-Filho JS (2011). Genetic interactions in cancer progression and treatment. Cell 145, 30–38. [DOI] [PubMed] [Google Scholar]
- Barr A, Feigenbaum EA, Cohen PR (1981). The Handbook of Artificial Intelligence. William Kaufmann: Stanford. [Google Scholar]
- Batista G, Monard MC (2002). A study of K-nearest neighbour as an imputation method. Hybrid Intelligent Systems 87, 251–260. [Google Scholar]
- Bishop CM (2006). Pattern Recognition and Machine Learning. Springer: New York. [Google Scholar]
- Breiman L (1984). Classification and Regression Trees. Wadsworth: Belmont. [Google Scholar]
- Breiman L (2001a). Statistical modeling: the two cultures. Statistical Science 16, 199–215. [Google Scholar]
- Breiman L (2001b). Random Forests. Machine Learning 45, 5–32. [Google Scholar]
- Brodersen KH, Deserno L, Schlagenhauf F, Lin Z, Penny WD, Buhmann JM, Stephan KE (2014). Dissecting psychiatric spectrum disorders by generative embedding. NeuroImage: Clinical 4, 98–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruana R, Niculescu-Mizil A (2006). An empirical comparison of supervised learning algorithms. In 23rd International Conference on Machine Learning (ICML2006), Pittsburgh, PA, pp. 161–168.
- Cordell HJ (2009). Detecting gene-gene interactions that underlie human diseases. Nature Reviews Genetics 10, 392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Vapnik VN (1995). Support-vector networks In Machine Learning (ed. Saitta L.), vol. 20, pp. 273–297. Kluwer Academic Publishers: Boston. [Google Scholar]
- Ding Y, Simonoff JS, Eklan C (2010). An investigation of missing data methods for classification trees applied to binary response data. Journal of Machine Learning Research 11, 131–170. [Google Scholar]
- Everitt BS, Landau S, Leese M, Stahl D (2010). Cluster Analysis. Wiley: UK. [Google Scholar]
- Friedman J, Hastie T, Tibshirani R (2010). Regularization paths for generalized linear models via coordinate descent. Journal of Statistical Software 33, 1–22. [PMC free article] [PubMed] [Google Scholar]
- Friedman JH (1991). Multivariate adaptive regression splines. Annals of Statistics 19, 1–67. [Google Scholar]
- GENDEP Investigators, MARS Investigators, STAR*D Investigators (2013). Common genetic variation and antidepressant efficacy in major depressive disorder: a meta-analysis of three genome-wide pharmacogenetic studies. American Journal of Psychiatry 170, 207–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghahramani Z (2003). Unsupervised learning. Advanced Lectures on Machine Learning 3176, 72–112. [Google Scholar]
- Glantz SA, Slinker BK (2000). Primer of Applied Regression and Analysis of Variance. McGraw-Hill: New York. [Google Scholar]
- Guyon I (2003). An introduction to variable and feature selection. Journal of Machine Learning Research 3, 1157–1182. [Google Scholar]
- Hand DJ (2006). Classifier technology and the illusion of progress. Statistical Science 21, 1–15.17906740 [Google Scholar]
- Hand DJ, Mannila H, Smyth P (2001). Principles of Data Mining. MIT Press: Cambridge. [Google Scholar]
- Hastie T, Tibshirani R, Friedman JH (2009). The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Springer: New York. [Google Scholar]
- Hosmer DW, Lemeshow S, Sturdivant RX (2013). Applied Logistic Regression. Wiley: Hoboken. [Google Scholar]
- Iniesta R, Malki K, Maier W, Rietschel M, Mors O, Hauser J, Henigsberg N, Dernovsek MZ, Souery D, Stahl D, Dobson R, Aitchison KJ, Farmer A, Lewis CM, McGuffin P, Uher R (2016). Combining clinical variables to optimize prediction of antidepressant treatment outcomes. Journal of Psychiatric Research 78, 94–102. [DOI] [PubMed] [Google Scholar]
- Jerez JM, Molina I, Garcia-Laencina PJ, Alba E, Ribelles N, Martin M, Franco L (2010). Missing data imputation using statistical and machine learning methods in a real breast cancer problem. Artificial Intelligence in Medicine 50, 105–119. [DOI] [PubMed] [Google Scholar]
- Kapur S, Phillips AG, Insel TR (2012). Why has it taken so long for biological psychiatry to develop clinical tests and what to do about it? Molecular Psychiatry 17, 1174–1183. [DOI] [PubMed] [Google Scholar]
- Katz MH (2006). Study Design and Statistical Analysis: a Practical Guide for Clinicians. Cambridge University Press: Cambridge. [Google Scholar]
- Kohavi R (1995). A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the 14th International Joint Conference on Artificial Intelligence, Montreal, Quebec, vol. 2, pp. 1137–1143.
- Kohavi R (1998). Glossary of terms. Machine Learning – Special Issue on Applications of Machine Learning and the Knowledge Discovery Process 30, 271–274. [Google Scholar]
- Kotsiantis S, Kanellopoulos D, Pintelas P (2006). Data preprocessing for supervised learning. International Journal of Computer Science 1, 111–117. [Google Scholar]
- Krstajic D, Buturovic LJ, Leahy DE, Thomas S (2014). Cross-validation pitfalls when selecting and assessing regression and classification models. Journal of Cheminformatics 6, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney D (2001). 3D data management: controlling data volume, velocity and variety. In Gartner (http://blogs.gartner.com/doug-laney/files/2012/01/ad949-3D-Data-Management-Controlling-Data-Volume-Velocity-and-Variety.pdf). Accessed 26 April 2016.
- LeCun Y, Bengio Y, Hinton G (2015). Deep learning. Nature 521, 436–469. [DOI] [PubMed] [Google Scholar]
- Lehner B (2007). Modelling genotype-phenotype relationships and human disease with genetic interaction networks. Jornal of Experimental Biology 210, 1559–1564. [DOI] [PubMed] [Google Scholar]
- Lewis DP, Jebara T, Noble WS (2006). Support vector machine learning from heterogeneous data: an empirical analysis using protein sequence and structure. Bioinformatics 22, 2753–2812. [DOI] [PubMed] [Google Scholar]
- Libbrecht MW, Noble WS (2015). Machine learning applications in genetics and genomics. Nature Reviews Genetics 16, 321–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Plataniotis KN, Venetsanopoulos A (2013). Multilinear Subspace Learning: Dimensionality Reduction of Multidimensional Data. Chapman and Hall: Florida. [Google Scholar]
- Maddala GS, Lahiri K (2009). Introduction to Econometrics. Wiley: New York. [Google Scholar]
- Malley JD, Malley KG, Pajevic S (2011). Statistical Learning for Biomedical Data. Cambridge University Press: New York. [Google Scholar]
- Manolio TA (2013). Bringing genome-wide association findings into clinical use. Nature Reviews Genetics 14, 549–606. [DOI] [PubMed] [Google Scholar]
- Miles J, Shevlin M (2001). Applying Regression & Correlation: a Guide for Students and Researchers. SAGE: London. [Google Scholar]
- Mitchell T (1997). Machine Learning. McGraw-Hill: New York. [Google Scholar]
- Mitchell T (2006). The discipline of machine learning. In CMU-ML-06-108 (http://www-cgi.cs.cmu.edu/~tom/). Accessed 31st March 2016.
- Nuzzo R (2014). Scientific method: statistical errors. Nature 506, 150–151. [DOI] [PubMed] [Google Scholar]
- Ochoa S, Huerta-Ramos E, Barajas A, Iniesta R, Dolz M, Baños I, Sánchez B, Carlson J, Foix A, Pelaez T, Coromina M, Pardo M, GENIPE group, Usall J (2013). Cognitive profiles of three clusters of patients with a first-episode psychosis. Schizophrenia Research 150, 151–157. [DOI] [PubMed] [Google Scholar]
- Perlis RH (2013). A clinical risk stratification tool for predicting treatment resistance in major depressive disorder. Biological Psychiatry 74, 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripley BD (1996). Pattern Recognition and Neural Networks. Cambridge University Press: Cambridge. [Google Scholar]
- Rokach L, Maimon OZ (2008). Data Mining with Decision Trees: Theory and Applications. World Scientific: New Jersey. [Google Scholar]
- Russell SJ, Norvig P (2010). Artificial Intelligence: a Modern Approach. Pearson: Boston. [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholkopf B, Guyon I, Weston J (2003). Statistical learning and kernel methods in bioinformatics. Artificial Intelligence and Heuristic Methods in Bioinformatics 183, 1–21. [Google Scholar]
- Shi X, Paiement JF, Grangier D, Yu PS (2012). Learning from heterogeneous sources via gradient boosting consensus. In International Conference on Data Mining, Anaheim, CA, pp. 1–12.
- Simon GE, Perlis RH (2010). Personalized medicine for depression: can we match patients with treatments? American Journal of Psychiatry 167, 1445–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler Artigas M, Loth DW, Wain LV, Gharib SA, Obeidat M, Tang W, Zhai G, Zhao JH, Smith AV, Huffman JE, Albrecht E, Jackson CM, Evans DM, Cadby G, Fornage M, Manichaikul A, Lopez LM, Johnson T, Aldrich MC, Aspelund T, Barroso I, Campbell H, Cassano PA, Couper DJ, Eiriksdottir G, Franceschini N, Garcia M, Gieger C, Gislason GK, Grkovic I, Hammond CJ, Hancock DB, Harris TB, Ramasamy A, Heckbert SR, Heliovaara M, Homuth G, Hysi PG, James AL, Jankovic S, Joubert BR, Karrasch S, Klopp N, Koch B, Kritchevsky SB, Launer LJ, Liu Y, Loehr LR, Lohman K, Loos RJ, Lumley T, Al Balushi KA, Ang WQ, Barr RG, Beilby J, Blakey JD, Boban M, Boraska V, Brisman J, Britton JR, Brusselle GG, Cooper C, Curjuric I, Dahgam S, Deary IJ, Ebrahim S, Eijgelsheim M, Francks C, Gaysina D, Granell R, Gu X, Hankinson JL, Hardy R, Harris SE, Henderson J, Henry A, Hingorani AD, Hofman A, Holt PG, Hui J, Hunter ML, Imboden M, Jameson KA, Kerr SM, Kolcic I, Kronenberg F, Liu JZ, Marchini J, McKeever T, Morris AD, Olin AC, Porteous DJ, Postma DS, Rich SS, Ring SM, Rivadeneira F, Rochat T, Sayer AA, Sayers I, Sly PD, Smith GD, Sood A, Starr JM, Uitterlinden AG, Vonk JM, Wannamethee SG, Whincup PH, Wijmenga C, Williams OD, Wong A, Mangino M, Marciante KD, McArdle WL, Meibohm B, Morrison AC, North KE, Omenaas E, Palmer LJ, Pietilainen KH, Pin I, Pola Sbreve Ek O, Pouta A, Psaty BM, Hartikainen AL, Rantanen T, Ripatti S, Rotter JI, Rudan I, Rudnicka AR, Schulz H, Shin SY, Spector TD, Surakka I, Vitart V, Volzke H, Wareham NJ, Warrington NM, Wichmann HE, Wild SH, Wilk JB, Wjst M, Wright AF, Zgaga L, Zemunik T, Pennell CE, Nyberg F, Kuh D, Holloway JW, Boezen HM, Lawlor DA, Morris RW, Probst-Hensch N, International Lung Cancer Consortium; GIANT consortium, Kaprio J, Wilson JF, Hayward C, Kahonen M, Heinrich J, Musk AW, Jarvis DL, Glaser S, Jarvelin MR, Ch Stricker BH, Elliott P, O'Connor GT, Strachan DP, London SJ, Hall IP, Gudnason V, Tobin MD (2011). Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nature Genetics 43, 1082–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, Pencina MJ, Kattan MW (2010). Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 21, 128–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R (1994). Regression shrinkage and selection via the lasso. Journal of the Royal Statistics Society, Series B 58, 267–288. [Google Scholar]
- Uher R, Maier W, Hauser J, Marusic A, Schmael C, Mors O, Henigsberg N, Souery D, Placentino A, Rietschel M, Zobel A, Dmitrzak-Weglarz M, Petrovic A, Jorgensen L, Kalember P, Giovannini C, Barreto M, Elkin A, Landau S, Farmer A, Aitchison KJ, McGuffin P (2009). Differential efficacy of escitalopram and nortriptyline on dimensional measures of depression. British Journal of Psychiatry 194, 252–260. [DOI] [PubMed] [Google Scholar]
- Uher R, Perroud N, Ng MY, Hauser J, Henigsberg N, Maier W, Mors O, Placentino A, Rietschel M, Souery D, Zagar T, Czerski PM, Jerman B, Larsen ER, Schulze TG, Zobel A, Cohen-Woods S, Pirlo K, Butler AW, Muglia P, Barnes MR, Lathrop M, Farmer A, Breen G, Aitchison KJ, Craig I, Lewis CM, McGuffin P (2010). Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. American Journal of Psychiatry 167, 555–618. [DOI] [PubMed] [Google Scholar]
- Vapnik VN (1998). Statistical Learning Theory. Wiley: New York. [Google Scholar]
- Whelan R, Watts R, Orr CA, Althoff RR, Artiges E, Banaschewski T, Barker GJ, Bokde AL, Buchel C, Carvalho FM, Conrod PJ, Flor H, Fauth-Buhler M, Frouin V, Gallinat J, Gan G, Gowland P, Heinz A, Ittermann B, Lawrence C, Mann K, Martinot JL, Nees F, Ortiz N, Paillere-Martinot ML, Paus T, Pausova Z, Rietschel M, Robbins TW, Smolka MN, Strohle A, Schumann G, Garavan H, IMAGEN Consortium (2014). Neuropsychosocial profiles of current and future adolescent alcohol misusers. Nature 512, 185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten DM, Tibshirani R (2010). A framework for feature selection in clustering. Journal of the American Statistical Association 105, 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert D, Macready W (1997). No free lunch theorems for optimization. IEEE Transactions on evolutionary computation 1, 67–82. [Google Scholar]
- Zou H, Hastie T (2005). Regularization and variable selection via the elastic net. Journal of the Royal Statistics Society, Series B 67, 301–320. [Google Scholar]
- Zhu X, Goldberg AB (2009). Introduction to semi-supervised learning. Synthesis Lectures on Artificial Intelligence and Machine Learning 3, 1–130. [Google Scholar]