Abstract
Approximately 350 million people are estimated to be persistently infected with hepatitis B virus (HBV) worldwide. HBV maintains persistent infection by employing covalently closed circular DNA (cccDNA), a template for all HBV RNAs. Chronic hepatitis B (CHB) patients are currently treated with nucleos(t)ide analogs such as lamivudine, adefovir, entecavir, and tenofovir. However, these treatments rarely cure CHB because they are unable to inhibit cccDNA transcription and inhibit only a late stage in the HBV life cycle (the reverse transcription step in the nucleocapsid). Therefore, an understanding of the factors regulating cccDNA transcription is required to stop this process. Among numerous factors, hepatocyte nuclear factors (HNFs) play the most important roles in cccDNA transcription, especially in the generation of viral genomic RNA, a template for HBV replication. Therefore, proper control of HNF function could lead to the inhibition of HBV replication. In this review, we summarize and discuss the current understanding of the roles of HNFs in the HBV life cycle and the upstream factors that regulate HNFs. This knowledge will enable the identification of new therapeutic targets to cure CHB.
Keywords: Hepatitis B virus, Hepatocyte nuclear factor, Covalently closed circular DNA, Replication
Core tip: Hepatitis B virus (HBV) infection is the leading cause of chronic liver disease and death worldwide. Persistent HBV infection is a major risk factor for chronic hepatitis and is the leading cause of liver disease, including cirrhosis and hepatocellular carcinoma. In the HBV life cycle, hepatocyte nuclear factors (HNFs) play critical roles in covalently closed circular DNA transcription. Control of HNF expression and function could regulate HBV replication. Therefore, understanding the upstream cellular factors or signals involved in the regulation of HNFs is important for controlling HBV replication.
INTRODUCTION
Chronic hepatitis B virus (HBV) infection is responsible for global public health problems and greatly increases the risk of liver diseases, including chronic hepatitis B (CHB), liver cirrhosis, and hepatocellular carcinoma (HCC). Although effective vaccination and approved nucleos(t)ide analog (NA) drugs have greatly contributed to reducing the number of newly infected individuals and the development of HBV-related liver diseases, sustained clearance of HBsAg or HBV DNA in liver tissues of CHB patients is rarely achieved using current NA drugs because of the inability of NAs to inactivate covalently closed circular DNA (cccDNA), a template for all HBV RNAs.
The HBV genome has four open reading frames (ORFs: PreC/C, P, preS1/S2/S, and X), which encode the precore and core proteins, polymerase, surface proteins, and HBx, respectively[1]. A unique feature of the HBV genome is the conversion of the 3.2-kb partially double-stranded relaxed circular DNA (rcDNA) into cccDNA in the nucleus[2,3], where five types of HBV RNAs are transcribed from the cccDNA. The production of HBV mRNAs is effectively regulated by complex interactions with various transcription factors. For example, transcription factors such as hepatocyte nuclear factors (HNFs) and CCAAT/enhancer binding protein (C/EBP) are critical for viral RNA production; these factors bind to HBV enhancers and promoters[4-7]. Interferon regulatory elements (IREs), which are present in the enhancer I (Enh I) region, regulate HBV gene expression[8]. Estrogen can suppress HBV Enh I activity by up-regulating estrogen receptor-α (ER-α), which binds to the enhancer region of HBV cccDNA and alters HNF4α binding[9]. By contrast, the androgen receptor binds to androgen-responsive elements present in HBV enhancers and thereby increases the transcription of HBV mRNAs[10]. The opposite effects of these two representative sex hormone receptors could explain the gender differences in HBV infection (males are more vulnerable than females to HBV-related HCC development). Accumulating evidence has indicated that the transcription of HBV genes is regulated by precise and ordered recruitment of chromatin modifiers and various host factors, including transcription factors, IREs, and sex hormones. Because there are no drugs that can inhibit cccDNA function, a thorough understanding of the mechanism of cccDNA transcription by cellular or viral factors will be useful for the development of drugs targeting cccDNA. This review will focus on the host factors, mainly HNFs, related directly or indirectly to the expression and regulation of HBV genes.
SHORT OVERVIEW OF THE HBV LIFE CYCLE
HBV is a prototype virus of the Hepadnaviridae family. According to the Baltimore classification, HBV is a Group VII virus, i.e., a double-stranded DNA virus that replicates through a single-stranded RNA intermediate. HBV has a complex life cycle involving reverse transcription. The genomic structure of HBV is unique and is referred to as circular partial duplex DNA, consisting of circular double-stranded DNA (dsDNA) with one strand that is only partially complete. The HBV life cycle has more complicated stages than most other viruses; these stages are explained below in a simplified form. A graphical scheme of the HBV life cycle is depicted in Figure 1. For more detail, the reader is referred to a comprehensive review of the HBV life cycle[1].
Figure 1.
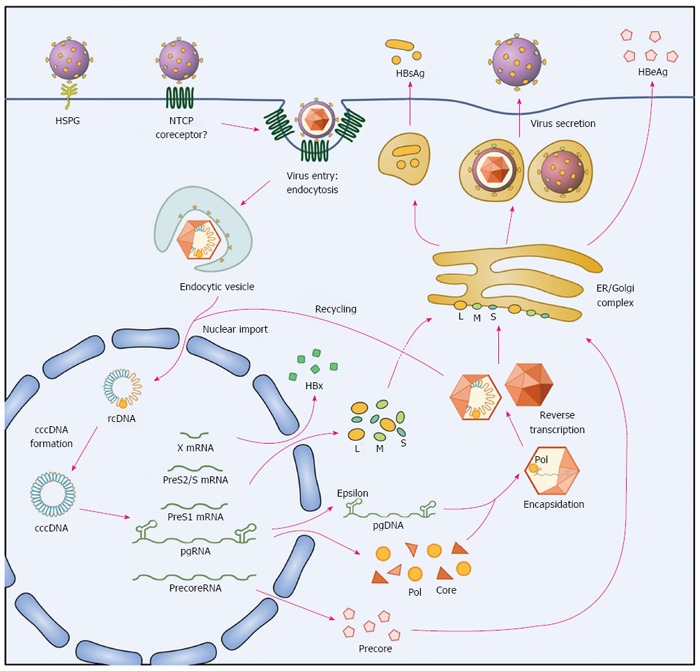
The life cycle of hepatitis B virus. Hepatitis B virus (HBV) binds to the surface receptor NTCP and enters hepatocytes, and its genome is released into the nucleus. The relaxed circular HBV genome (rcDNA) is repaired and forms covalently closed circular DNA (cccDNA), which serves as a template for the transcription of viral mRNAs (pregenomic (pg), precore, HBx, PreS1, and PreS2/S RNA). The HBV mRNAs are translated into the large (L), middle (M), and small (S) surface, precore, core, polymerase (pol), and HBx proteins. pgRNA and pol are encapsidated into the capsid, and viral DNA is reverse-transcribed. Assembled HBV virions are secreted from hepatocytes.
HBV infects hepatocytes but not hepatoma cell lines such as HepG2 or Huh7. This lack of a suitable cell infection system has hampered the study of the mechanism of virus entry (and other infection steps), and, therefore, the development of entry inhibitors. The recent identification of sodium-taurocholate cotransporting polypeptide, also known as NTCP, as a functional receptor for HBV entry has opened the door for the study of the molecular mechanisms of HBV infection[11,12]. NTCP is a transporter involved in the uptake of bile salts, and the preS1 domain of the viral large envelope protein (L-HBsAg) interacts directly with NTCP as an essential step for viral entry; therefore, poorly differentiated hepatocytes such as Huh7 or HepG2 cells, which express negligible amounts of NTCP, are not susceptible to HBV infection[13,14].
A unique feature of L-HBsAg is myristoylation at its N-terminus, which enables its association with the plasma membrane. The highly conserved motif 9-NPLGF(F/L)P-15 in the receptor-binding region of L-HBsAg is also crucial for viral infection[15,16]. Although there is limited information regarding the processes of plasma membrane fusion and endosomal migration after HBV binding, several studies have demonstrated that the virus enters cells via clathrin-mediated endocytosis[17-19].
After entering the cell, the virus undergoes uncoating and core disassembly, and its genome enters the cell nucleus[20]. Nuclear import is mediated by nuclear localization signals of capsid proteins, and nuclear entry of the encapsidated, deproteinized relaxed circular (rc) DNA uses the importin-α/importin-β receptor pathway[21]. Although the size of the genome-containing nucleocapsid of HBV is close to the functional diameter of the nuclear pore complex, the capsid was suggested not to release the viral genome before nuclear import. Unlike the capsids of other viruses, such as herpes virus, adenovirus, and influenza virus, the HBV capsid enters the nuclear basket in its intact form[22]. Only capsids with a mature genome enter the basket and consequently liberate the genome through the interaction with nucleoporin 153 (Nup153) in the nuclear basket of the nuclear pore complex[23].
Inside the nucleus, rcDNA is repaired (see below) by the host repair system to form cccDNA, which is stable and difficult to remove throughout HBV infection and therefore plays an important role in viral persistence and recurrence[24]. The cccDNA acts as a transcriptional template for five types of viral RNAs: the pregenomic RNA (pgRNA) and precore RNA (both 3.5-kb), 2.4-kb and 2.1-kb HBsAg RNAs, and 0.7-kb HBx RNA. The precore RNA is the template for the production of HBeAg; the core and polymerase proteins are translated from pgRNA, which also serves as a template for reverse transcription to produce the HBV DNA.
Packaging of the pgRNA and polymerase protein into the viral capsid is initiated by a cis-acting element called epsilon, which acts as the packaging signal[25]. The epsilon, polymerase, and core proteins form the nucleocapsid. This process is referred to as encapsidation; when encapsidation is completed, polymerase begins reverse transcription of the pgRNA to generate the minus-strand DNA. The template pgRNA is simultaneously degraded by RNase H activity of polymerase before the synthesis of the plus-strand begins. After several rounds of strand transfer, formation of the circular genomic DNA is complete[26]. Genome-containing nucleocapsids interact with the envelope proteins in the ER-Golgi complex and form enveloped mature virions. The preS1, preS2, and S domains of surface proteins have different functions according to their transmembrane topology. The preS1 domain is located partially at the surface and partially on the internal side during virion maturation. The internal side of PreS1, which slightly overlaps with the PreS2 domain, interacts with core particles necessary for envelope formation[27], whereas the surface side of PreS1 interacts with a cellular receptor for virus entry. The infectious mature virions, known as Dane particles, exit the cell via the ER and Golgi apparatus, although the details of this process have not yet been completely revealed.
cccDNA AS A TEMPLATE FOR HBV TRANSCRIPTION
cccDNA plays a central role in HBV transcription and replication. cccDNA forms a minichromosome in the nucleus and becomes the source for pgRNA and other viral RNAs production, and virus replication occurs through reverse transcription of the pgRNA into rcDNA. NA drugs can inhibit reverse transcription and are currently used as a treatment for HBV[28,29]. However, these drugs are unable to affect cccDNA, which is upstream of the NA target site. To inhibit cccDNA transcription and to prevent the production of viral mRNAs, it is important to better understand how cccDNA is produced and functions.
Host factors involved in cccDNA formation
HBV rcDNA is converted to cccDNA via an intermediate form, protein-free rcDNA[30,31]. This process is referred to as repair; following plus-strand DNA synthesis by gap-filling, viral polymerase and short RNA oligomers (also called RNA primers) attached to the 5’-termini of minus-strand and plus-strand DNA should be removed[32]. Identification of the host factors involved in rcDNA to cccDNA conversion will provide potential new therapeutic targets to prevent cccDNA formation. A recent in vitro study suggested that host tyrosyl-DNA-phosphodiesterase 2 (TDP2) is involved in the removal of viral polymerase covalently linked to the 5’-end of minus-strand DNA[33]. However, knockout of the TDP2 gene does not block cccDNA formation during HBV infection of permissive hepatoma cells and does not prevent intracellular amplification of duck hepatitis B virus cccDNA[34]. Intriguingly, the knockdown of TDP2 increases the formation of HBV cccDNA[34]. Although several host enzymes, such as TDP2 and topoisomerase[35], were suggested to be involved in cccDNA biogenesis, the mechanisms of rcDNA repair and the host factors associated with cccDNA formation are largely unknown. Further studies aimed at revealing these steps are highly warranted.
Role of HBx in cccDNA function
HBx is essential for cccDNA transcription; therefore, the inhibition of HBx prevents HBV replication[36]. Accumulating evidence suggests that cccDNA transcription is epigenetically controlled. cccDNA forms a minichromosome in the nucleus and associates with histones, including H2A, H2B, H3, and H4. HBV transcription and replication is regulated by the acetylation status of cccDNA-bound histones and non-histone proteins[37]. Our understanding of the role of HBx in viral replication was considerably advanced by the finding that nuclear HBx binds to cccDNA and modifies the epigenetic regulation of cccDNA function. HBx recruits chromatin regulators such as P300 and other acetyltransferases to cccDNA and enhances viral transcription[38]. Conversely, if HBx is mutated so that it is unable to recruit acetyltransferases, cccDNA acetylation is reduced by histone deacetylases (HDACs), and the level of viral transcription and replication is reduced[38]. Moreover, occult HBV infection is demonstrated by epigenetic inactivation of cccDNA, and reactivation of cccDNA from this state is controlled by the epigenetic function of HBx[32].
Elimination or inactivation of cccDNA
Complete cure of HBV infection requires the elimination of cccDNA in liver tissues. Several studies have attempted to eliminate cccDNA by applying genome editing technologies. A zinc finger nuclease (ZFN) that was genetically designed to bind specifically to HBV cccDNA was shown to inhibit viral replication[39]. Engineered transcription activator-like effector endonucleases (TALENs) efficiently inactivated the HBV genome[40]. The RNA-guided clustered regularly interspaced short palindromic repeats (CRISPR/Cas9) system has been employed to cleave cccDNA[41]. However, genome editing technologies have numerous unresolved issues, including the in vivo delivery of genome editing molecules to HBV-infected hepatocytes, which is the key hurdle for clinical application of this technology.
Cytokines were recently shown to play an important role in the control of cccDNA. APOBEC3A and 3B, which are induced by high doses of interferon-alpha (IFN-α) or by the activation of lymphotoxin-β receptor, cause cytidine deamination in HBV cccDNA, which is subsequently degraded by a cellular endonuclease[42].
Because cccDNA is epigenetically regulated by HBx[38] and by the host antiviral factor IFN-α[43], the control of epigenetic factors that control cccDNA transcription may also be used to inactivate cccDNA. Epigenetic changes in cccDNA alter the binding of various liver-enriched transcription factors to its enhancer region. Therefore, understanding the interaction between transcription factors and enhancers will be important for cccDNA inactivation.
Structure of enhancers in HBV cccDNA and their regulatory transcription factors
Although HBV has a small genome, it has a large number of transcriptional regulatory sequences that have various roles. Genome transcription and replication can be stimulated or suppressed by the association or dissociation of numerous host factors. In particular, HNFs are the host factors that are present in hepatocytes in high concentrations, interact with viral components and play important roles in viral replication.
The HBV genome has four overlapping ORFs and contains several promoters, enhancers, a polyadenylation sequence, and an encapsidation signal. Four promoters are responsible for the transcription of the four ORFs. Two regions in the HBV genome, called Enh I and Enh II, have gene enhancer activity. Enh I is a region of approximately 300 nucleotides located between the ORFs S and X[44]. Enh II is a region of approximately 200 nucleotides, is located before the core promoter and overlaps with the core upstream regulatory sequence.
General transcription factors that bind to HBV promoters or enhancers include nuclear factor 1 (NF1), specificity protein 1 (SP1), activator protein 1 (AP1), TATA-binding protein (TBP), prospero-related homeobox protein 1 (PROX1), c-AMP-response element-binding protein (CREB), nuclear factor-kappa B (NF-κB), octamer transcription factor 1 (OCT1), and nuclear respiratory factor 1 (NRF1).
The hepatotropic nature of HBV infection is primarily mediated by hepatocyte-restricted expression of the viral receptor NTCP; however, the liver-enriched transcription control factors also play essential roles in the life cycle of HBV. Representative examples include hepatocyte nuclear factor 1α (HNF1α), hepatocyte nuclear factor 3β (HNF3β), hepatocyte nuclear factor 4α (HNF4α), hepatocyte nuclear factor 6 (HNF6), and CAAT enhancer-binding protein (C/EBP). A detailed description on these factors is provided in Table 1, and the binding sites of these factors to HBV enhancers and promoters are depicted in Figure 2.
Table 1.
Hepatocyte nuclear factors and other transcription factors involved in hepatitis B virus transcription
| Factor | Binding site | Effect on viral enhancers/promoters | Ref. |
| HNF1α | PreS1 | Activation | [60] |
| Enh II | Activation (interaction with hB1F) | [61] | |
| Enh II | Suppression (mutant HBV core promoter) | [50] | |
| Enh II | Activation | [62] | |
| Enh II | Activation (mutant HBV core promoter) | [49] | |
| HNF3β | Enh I | Activation (interaction with STAT3) | [48] |
| Enh I | Suppression (HepG2)/ Activation (SK-Hep1) | [68] | |
| Enh II | Suppression | [67] | |
| Enh II | Activation | [66] | |
| HNF4α | EnhII/PreS1 | Activation | [73] |
| Enh II | Activation | [130] | |
| HNF6 | PreS2 | Suppression | [78] |
| C/EBP | Enh I | Suppression | [83] |
| Enh II | Activation | [81] | |
| Enh II | Activation | [46] | |
| Enh II | Activation | [80] | |
| FXR/RXR | Enh II | Activation | [99] |
| HLF | Enh II | Activation | [6] |
| NF1 | PreS2 | Activation | [101] |
| Enh I | Suppression | [102] | |
| SP1 | Enh II | Activation | [104,105] |
| PreS1 | Activation | [106] | |
| PreS2 | Activation | [107] |
HBV: Hepatitis B virus.
Figure 2.
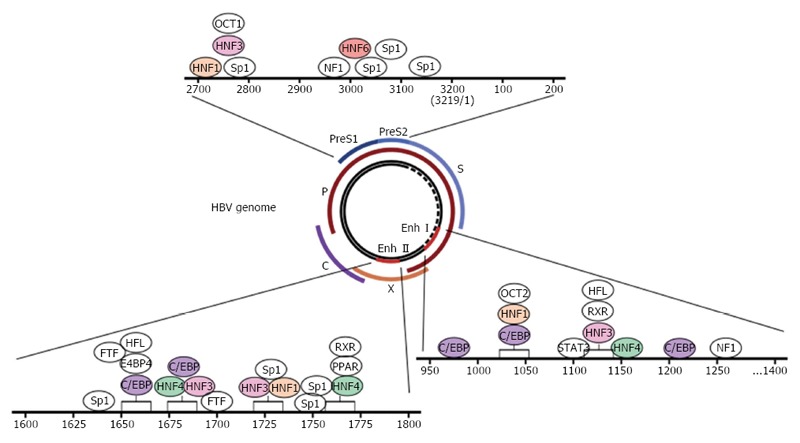
Schematic representation of transcription factor-binding regions in HBV enhancers and promoters. The inner circle represents the 3.2-kb hepatitis B virus (HBV) genome (partially double-stranded circular DNA). The outer lines indicate the four open reading frames (ORFs). There are four viral promoters located just before the ORFs and two enhancers (Enh I and Enh II). Various transcription factors can bind to the enhancers and promoters as indicated. The boxes represent competing binding regions.
REGULATION OF HBV GENE EXPRESSION BY HNFs AND miRNAs
Host factors that regulate HBV gene expression include transcription factors and microRNAs (miRNAs). Host transcription factors, especially liver-enriched factors, regulate the transcription of HBV cccDNA by binding to viral enhancers and promoters, whereas liver-enriched miRNAs regulate HBV gene expression post-transcriptionally.
HNFs are typical examples of liver-enriched transcription factors; they are highly expressed in the liver in comparison with other organs and affect viral transcription and the production of a number of liver proteins that are essential to maintain liver function and homeostasis. HNFs regulate cccDNA transcription by directly interacting with HBx, which enhances the DNA-binding activity of HNFs[4,45,46].
DNase I protection analysis revealed that several distinct ubiquitous and liver-specific cellular factors bind in concert to the HBV enhancer regions[5]. Transcription factors interact with the cognate HBV DNA sequences, and multiple transcription factors might bind competitively to the same DNA sequence region[6]. Such competitive binding can occur when transcription factors share their consensus binding motif. Several transcription factors sometimes physically interact with each other to form dimers or multimers, resulting in regulation of the transcriptional activity of target genes[47,48].
The level of HBV transcription varies with the extent of transcription factor binding to HBV DNA, which can be altered by mutations in the HBV genome. When a new mutation allows a transcription factor to bind to a new region in HBV DNA, HBV replication is stimulated, resulting in progression to fulminant hepatitis[49]. A naturally occurring double nucleotide mutation in the HBV core promoter was shown to convert a nuclear receptor binding site to an HNF1 binding site, resulting in the suppression of RNA transcription via interactions between HNF1 and the mutant HBx[50].
miRNAs play a number of essential roles as biological regulators[51-53] and are involved in various biological processes[54-56], disease progression, and pathogen infection[56-59]. miRNAs are short RNAs (approximately 20 nucleotides) that are processed by Drosha and Dicer. By binding to the untranslated or coding regions of the target transcripts, miRNAs induce mRNA degradation or inhibit translation. A large number of miRNAs are involved in the transcription and replication of HBV and can cause HBV pathogenesis. When hepatocytes are infected with HBV, the expression of cellular miRNAs increases, and they regulate HBV gene expression directly by targeting viral mRNAs, or indirectly by controlling epigenetic factors such as DNA methyl transferase (DNMT) or HDAC[44].
Understanding the relationship among transcription factors, miRNAs, regulatory sequences, and the functional consequences of their binding is very important for understanding HBV-mediated pathogenesis.
HNF1α
Although there is some controversy regarding the role of HNF1α in the regulation of HBV genes, HNF1α is known to affect HBV transcription by controlling most of the HBV regulatory elements, including preS1 promoter, core promoter, HBx promoter, and Enh II, in cccDNA. HNF1α has been reported to stimulate viral transcription by 7-fold in Huh7 cells through binding to the preS1 promoter[60]. HNF1 up-regulates Enh II activity by interacting with either the hB1F[61] or B element in Enh II[62]. In the absence of HNF1α, the concentration of HBV pgRNA is decreased, resulting in decreased genome replication[63]. The emergence of new binding sequences for HNF1 by mutation in transplant-transmitted HBV was able to lead to increased viral replication and fulminant hepatitis[49].
In contrast to these observations, HNF1α does not interact with the wild-type viral core promoter, although it binds a mutant HBV core promoter, which has an HNF1α-binding sequence, and reduces precore RNA transcription[50,64]. HNF1α has no effect on the HBx promoter or core promoter[61]. In HNF1α-null HBV transgenic mice, the levels of intracellular viral replication intermediates are increased several fold[65]. It is evident that HNF1α plays important roles in the regulation of HBV transcription, although the authentic role of HNF1α during the natural course of HBV infection remains unclear.
HNF3β
A binding motif for HNF3α and HNF3β was identified within Enh II and the TGTTTGTTT sequence was mapped as an essential motif for the specific interaction between DNA and the HNF3 protein[66]. This motif is critical for the regulation of Enh II activity by HNF3, and the introduction of mutations into this motif alters Enh II activity. Competitive binding of HNF3β and HNF4 to a region (positions 1650-1674) of HBV Enh II was also observed[66]. HNF3 cooperates with other molecules such as NF1 and STAT3. Interleukin-6 (IL-6) and epidermal growth factor (EGF) stimulate the cooperative interaction between HNF3 and STAT3, which leads to the activation of Enh I[48].
The effect of HNF3 on HBV enhancer activity differs depending on the tested system. HNF3β inhibits HBV replication in mouse NIH 3T3 fibroblasts[67] and reduces enhancer activity in HepG2 cells, whereas it increases enhancer activity in SK-Hep1 cells[68]. In mice, HNF3β inhibits HBV replication[67,69]. It appears that HNF3 inhibits HBV replication in hepatocytes in cooperation with other molecules.
HNF4α
HNF4α exists as either a homo- or heterodimer and acts as a key regulator of approximately 40% of hepatocyte genes[70]. HNF4α can also bind to various HBV enhancer regions[4,44] and is the main activator of HNF1α expression through binding to the HNF1α promoter[71]. The inhibition of HNF4α using siRNA effectively reduces HBV transcription and replication in cells and in mice[72]. When HNF4α is absent, the level of HBV pgRNA is reduced, resulting in decreased replication[64]. The overexpression of HNF4α in HepG2.1 cells increases the activity of the preS, preS2/S, and core promoters, but has no effect on the Enh I/X promoter[73]. The suppression of HBV replication by transforming growth factor-β1 (TGF-β1) can be restored by ectopic expression of HNF4α[74].
The effect of HNF4α on HBV replication appears to be clinically relevant. A retrospective study demonstrated that the HNF4α expression level was increased in CHB patients, whereas HNF3β was down-regulated[75]. The expression level of HNF4α was inversely correlated with the clinical outcomes of CHB patients[76]. Overall, HNF4α is a key transcription factor in the regulation of the HBV life cycle and in the maintenance of hepatocyte function.
HNF6
HNF6 is involved in liver homeostasis processes such as glucose metabolism, bile homeostasis, and liver cell proliferation[77]. HNF6 inhibits HBV gene expression and replication in HepG2 cells by suppressing the activity of the preS2/S promoter[78]. Interestingly, HNF6 is regulated by CYP2C12, which is expressed in a female-specific manner. Therefore, the regulation of HNF6 by CYP2C12 might explain why HBV replication is suppressed in females compared to males[79].
C/EBP
C/EBPα, a liver-enriched transcription factor, can form homodimers or heterodimers and plays critical roles in the regulation of hepatocyte-specific genes[45]. C/EBP binds to at least five regions of HBV promoters and enhancers (Figure 2). C/EBP promotes HBV transcription by transactivation of Enh II[80] and synergistically activates Enh II through interaction with HBx[47]. Consistent with this finding, the anti-HBV effect of interleukin 4 (IL-4) was attributed to the down-regulation of C/EBPα in Hep3B cells[81].
Low C/EBP concentrations increase the activity of the viral core promoter, whereas high concentrations suppress core promoter activity[82]. Similarly, C/EBP binds to HBV Enh I and represses HBV transcription activity[83]. Although the role of C/EBP in HBV gene regulation is controversial, C/EBP appears to act as a proviral factor activating viral enhancers and promoters.
miRNAs related to HBV gene expression
miRNAs can regulate HBV replication either indirectly, by targeting cellular proteins that are essential for HBV replication, or directly, by targeting viral RNAs. miRNA-18a prevents the expression of ER-α, which represses HBV transcription via interaction with HNF4α[84] Several miRNAs, including miRNA-1, 148a, 152, 210, and 449a, are involved in the regulation of HBV replication, mainly by targeting host epigenetic regulators such as DNMT and HDAC[85-89].
A number of miRNAs can directly target HBV transcripts. Among them, the most well-known miRNA is miRNA-122, which is abundant in hepatocytes and has received increasing attention because several studies have shown that it reduces the level of viral expression by binding to a highly conserved site in 3.5-kb pgRNA[90-92]. miRNA-199a-3p and miRNA-210 suppress HBV replication by binding to the 2.1-kb RNA and 2.4-kb RNA, respectively[93]. miRNA-125a-5p inhibits HBV translation by binding to the 2.1-kb RNA[94]. Using a 3D array system, Kohno et al. found that miRNA-1231 suppresses HBV replication by targeting the core mRNA[95]. Bioinformatics analyses have suggested several putative miRNA-binding sites on HBV RNAs, including the following: miRNA-199a-3p, 125a-5p, 210, and 345 are predicted to bind the 3.5-kb RNA; miRNA-let7, 196b, and 511, 2.4-kb RNA; miRNA-433, 2.1-kb RNA; and miRNA-205, 0.7-kb RNA[93,94,96,97].
There is a report that viral RNA can target host miRNA. Viral HBx RNA directly down-regulates the tumor suppressor miRNAs miRNA-15a and miRNA-16-1[98]. This study suggests that HBV can induce HCC development by viral RNA-mediated down-regulation of specific tumor suppressor miRNAs. The various targets of miRNAs involved in HBV replication are shown in Figure 3.
Figure 3.
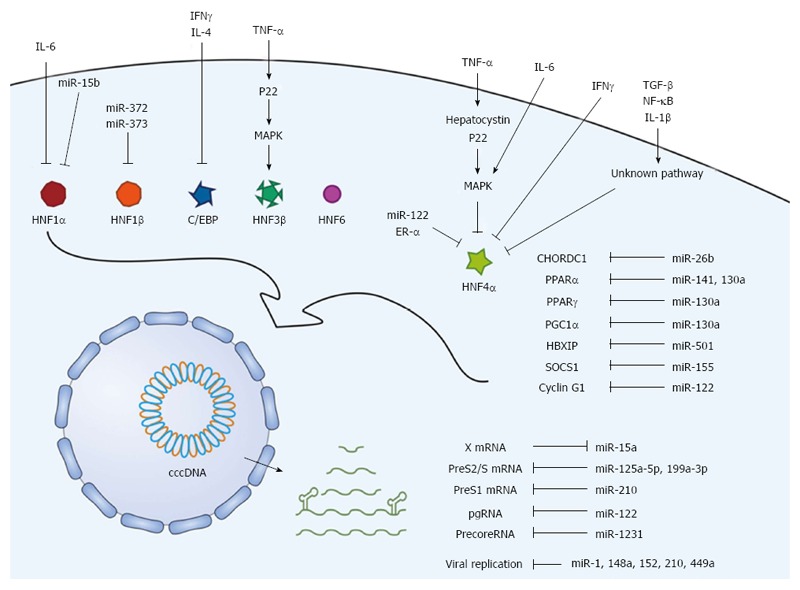
Scheme of regulatory factors associated with HBV transcription and translation. The upstream regulators, including cytokines, mediators, and signaling molecules, regulate liver-enriched transcription factors. In addition, miRNAs regulate the transcription factors and viral transcripts. These upstream factors and miRNAs exert positive or negative effects on HBV transcription and replication. HBV: Hepatitis B virus.
Others
Nuclear receptors are known to regulate the activity of HBV enhancers and promoters. Farnesoid X receptor (FXR) and retinoid X receptor (RXR) are reported to form heterodimers and increase the activity of Enh II and the core promoter[99]. Peroxisome proliferator-activated receptors (PPARs) and RXRs form heterodimers and increase the activity of the Enh I/X promoter[73,100].
Many nuclear factors have also been shown to be involved in HBV regulation. The binding of NF-1 to the S gene promoter is essential for HBV surface RNA transcription[101]; however, NF-1 binding to Enh I suppresses the activity of the HBV enhancer[102]. Nuclear factor Y (NF-Y) can activate the S promoter by binding to the CCAAT element[103]. SP1 binds to GC-rich DNA sequences on HBV enhancers and promoters and up-regulates the activity of Enh II[104,105] and the PreS1[106] and PreS2[107] promoters. HLF binds to the Enh II region and increases the transcription of pgRNA and precore RNA[6]. Testicular orphan receptor 4 (TR4) reduces core promoter activity by blocking HNF4α binding through a protein-protein interaction[108]. Cysteine- and histidine-rich domain-containing 1 (CHORDC1) binds HBV enhancers and activates gene transcription[109]. Other factors involved in HBV regulation include nuclear respiratory factor 1 (NRF1)[110], activator protein 1 (AP1)[111], TATA-binding protein (TBP)[112], CREB[113], and OCT1[45].
UPSTREAM FACTORS RELATED TO HNF REGULATION
Six families of liver-enriched transcription factors [HNF-1, HNF-3, HNF-4, HNF-6, C/EBP, and D-binding protein (DBP)] have been characterized to date. As discussed above, most of these factors, except DBP, are critically involved in HBV gene expression and replication. Therefore, understanding the upstream cellular factors or signals involved in the regulation of liver-enriched transcription factors is important to grasp the complicated cellular networks related to the HBV life cycle. For example, the activation of extracellular signal-regulated kinase (ERK) inhibits HBV replication by down-regulating HNF4α and up-regulating HNF3β[114]. A detailed description of these factors and signals is presented in Figure 3 and Table 2.
Table 2.
Upstream factors and signals involved in the regulation of hepatocyte nuclear factors and other factors related to hepatitis B virus replication
| Regulating factor | Target molecule | Effect on target molecule | Effect on HBV replication | Ref. |
| TGF-β | HNF4α | Decrease | Decrease | [74] |
| ER-α | - | Decrease | [9] | |
| NF-κB | Decrease | Decrease | [119,120] | |
| MAPK | Decrease | Decrease | [119,121] | |
| TNF-α → Hepatocystin → MAPK | Decrease | Decrease | [118] | |
| TNF-α → P22 → MAPK | Decrease | Decrease | [114] | |
| IFN-γ | Decrease | Decrease | [117] | |
| IL-6 → MAPK | Decrease | Decrease | [116] | |
| miRNA-122 | Decrease | Decrease | [124] | |
| IL-4 | C/EBP | Decrease | Decrease | [81] |
| IFN-γ | Decrease | Decrease | [117] | |
| IL-6 | HNF1α | Decrease | Decrease | [116] |
| miRNA-15b | Decrease | Increase | [122] | |
| miRNA-372, miRNA-373 | HNF1β | Decrease | Increase | [123] |
| TNF-α → P22 → MAPK | HNF3β | Increase | Decrease | [114] |
| miRNA-26b | CHORDC1 | Decrease | Decrease | [109] |
| miRNA-141 | PPARα | Decrease | Decrease | [125] |
| NF-κB → miRNA-130a | Decrease | Decrease | [128] | |
| miRNA-122 | Cyclin G1 | Decrease | Decrease | [90] |
| miRNA-501 | HBXIP | Decrease | Increase | [128] |
| miRNA-155 → JAK/STAT | SOCS1 | Decrease | Decrease | [129] |
HBV: Hepatitis B virus.
Cellular factors and signaling pathway involved in HNF regulation
Extracellular signals such as cytokines can affect HBV by dysregulating liver-enriched transcription factors. IL-4 suppresses HBV core promoter activity and inhibits pgRNA synthesis by down-regulating C/EBPα[81]. IL-6 controls HBV replication by reducing the levels of HNF4α and HNF1α[115,116]. IFN-γ also regulates the HNF4α and C/EBP levels and affects HBV replication[117]. Recently, we demonstrated that cytokine-mediated up-regulation of hepatocystin/80K-H impairs HBV replication by down-regulating HNF4α through the Ras/MAPK pathway[118] and also reported that p22-FLIP, a cleavage product of c-FLIP formed upon TNF-α stimulation, reduces the HNF4α level but increases the HNF3β level via the ERK pathway, thereby strongly impairing HBV replication[115].
NF-κB suppresses HBV replication by inhibiting HNF4α[119,120]. The activation of ERK1/2 and stress-activated protein kinase 1/c-jun NH2-terminal kinase (SAPK/JNK) also inhibits HBV replication through negative regulation of HNF4α[119,121]. ER-α suppresses HBV by physical interaction with HNF4α[9], which supports the observation that males are more vulnerable to HBV infection than females.
miRNAs involved in HNF regulation
As described above, miRNAs can directly regulate HBV by targeting viral RNAs. In this subsection, we provide a brief overview of the miRNAs that exert an indirect effect on HBV by controlling liver-enriched transcription factors that are involved in HBV replication. The control of HNF1α by miRNA-15b is reported to promote HBV replication[122], and the control of HNF1β by miRINA-372 and 373 up-regulates HBV gene expression[123]. miRNA-122 is also able to control HBV replication by inhibiting HNF4α[124].
In addition to targeting HNFs, miRNAs can also target other nuclear factors. miRNA-26b inhibits CHORDC1 expression, thereby suppressing HBV enhancer activity[109]. miRNA-141 represses HBV replication by targeting PPARα, which binds and trans-activates HBV enhancers[125]. Ectopic expression of miRNA-141 suppresses PPARα expression, decreasing viral transcription in HBV-transfected HepG2 cells. The expression of miRNA-130a is stimulated by NF-kB/p65 and inhibits HBV replication by down-regulating PGC1α and PPARγ[126]. The level of C/EBP is reduced by miR-155, the expression of which is increased by diet-induced activation of NF-κB[127].
During HBV infection, host antiviral signaling can contribute to viral clearance via the induction of miRNAs transcription. Other miRNAs that have been reported to affect HBV replication by controlling cellular proteins other than liver-enriched transcription factors include miRNA-122 targeting cyclin G1[90], miRNA-501 targeting HBXIP[128], and miRNA155 targeting SOCS1[129].
CONCLUSION
Host factors, mainly HNFs, are indispensable for the survival and maintenance of HBV in hepatocytes. These factors regulate cccDNA transcription and mediate host antiviral responses. Although our knowledge of HBV-related host factors has increased, the overall understanding of the cellular networks related to cccDNA transcription is still very limited. Numerous recent studies have revealed that miRNAs play key roles in the HBV life cycle. Systematic understanding of the complex interactions between HBV and host miRNAs is needed. Currently, there is no way to eliminate cccDNA in infected hepatocytes, which would be required for complete CHB cure. Therefore, understanding the molecular network that determines how cccDNA is activated by extrinsic and intrinsic factors will provide us a chance to inactivate or halt the cccDNA function until drugs that eliminate cccDNA are developed.
Footnotes
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: South Korea
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Conflict-of-interest statement: No conflicts of interest.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: April 5, 2016
First decision: May 12, 2016
Article in press: June 29, 2016
P- Reviewer: Cunha C, Liu LF S- Editor: Yu J L- Editor: A E- Editor: Ma S
References
- 1.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 3.Schädler S, Hildt E. HBV life cycle: entry and morphogenesis. Viruses. 2009;1:185–209. doi: 10.3390/v1020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moolla N, Kew M, Arbuthnot P. Regulatory elements of hepatitis B virus transcription. J Viral Hepat. 2002;9:323–331. doi: 10.1046/j.1365-2893.2002.00381.x. [DOI] [PubMed] [Google Scholar]
- 5.Patel NU, Jameel S, Isom H, Siddiqui A. Interactions between nuclear factors and the hepatitis B virus enhancer. J Virol. 1989;63:5293–5301. doi: 10.1128/jvi.63.12.5293-5301.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishida H, Ueda K, Ohkawa K, Kanazawa Y, Hosui A, Nakanishi F, Mita E, Kasahara A, Sasaki Y, Hori M, et al. Identification of multiple transcription factors, HLF, FTF, and E4BP4, controlling hepatitis B virus enhancer II. J Virol. 2000;74:1241–1251. doi: 10.1128/jvi.74.3.1241-1251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bar-Yishay I, Shaul Y, Shlomai A. Hepatocyte metabolic signalling pathways and regulation of hepatitis B virus expression. Liver Int. 2011;31:282–290. doi: 10.1111/j.1478-3231.2010.02423.x. [DOI] [PubMed] [Google Scholar]
- 8.Alcantara FF, Tang H, McLachlan A. Functional characterization of the interferon regulatory element in the enhancer 1 region of the hepatitis B virus genome. Nucleic Acids Res. 2002;30:2068–2075. doi: 10.1093/nar/30.9.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang SH, Yeh SH, Lin WH, Yeh KH, Yuan Q, Xia NS, Chen DS, Chen PJ. Estrogen receptor α represses transcription of HBV genes via interaction with hepatocyte nuclear factor 4α. Gastroenterology. 2012;142:989–998.e4. doi: 10.1053/j.gastro.2011.12.045. [DOI] [PubMed] [Google Scholar]
- 10.Wang SH, Yeh SH, Lin WH, Wang HY, Chen DS, Chen PJ. Identification of androgen response elements in the enhancer I of hepatitis B virus: a mechanism for sex disparity in chronic hepatitis B. Hepatology. 2009;50:1392–1402. doi: 10.1002/hep.23163. [DOI] [PubMed] [Google Scholar]
- 11.Schieck A, Schulze A, Gähler C, Müller T, Haberkorn U, Alexandrov A, Urban S, Mier W. Hepatitis B virus hepatotropism is mediated by specific receptor recognition in the liver and not restricted to susceptible hosts. Hepatology. 2013;58:43–53. doi: 10.1002/hep.26211. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Tong S. From DCPD to NTCP: the long journey towards identifying a functional hepatitis B virus receptor. Clin Mol Hepatol. 2015;21:193–199. doi: 10.3350/cmh.2015.21.3.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Fälth M, Stindt J, Königer C, Nassal M, Kubitz R, et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146:1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 14.Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JH. [Hepatitis B virus surface antigen: a multifaceted protein] Korean J Hepatol. 2004;10:248–259. [PubMed] [Google Scholar]
- 16.Le Seyec J, Chouteau P, Cannie I, Guguen-Guillouzo C, Gripon P. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J Virol. 1999;73:2052–2057. doi: 10.1128/jvi.73.3.2052-2057.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartmann-Stühler C, Prange R. Hepatitis B virus large envelope protein interacts with gamma2-adaptin, a clathrin adaptor-related protein. J Virol. 2001;75:5343–5351. doi: 10.1128/JVI.75.11.5343-5351.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang HC, Chen CC, Chang WC, Tao MH, Huang C. Entry of hepatitis B virus into immortalized human primary hepatocytes by clathrin-dependent endocytosis. J Virol. 2012;86:9443–9453. doi: 10.1128/JVI.00873-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper A, Shaul Y. Clathrin-mediated endocytosis and lysosomal cleavage of hepatitis B virus capsid-like core particles. J Biol Chem. 2006;281:16563–16569. doi: 10.1074/jbc.M601418200. [DOI] [PubMed] [Google Scholar]
- 20.Rabe B, Vlachou A, Panté N, Helenius A, Kann M. Nuclear import of hepatitis B virus capsids and release of the viral genome. Proc Natl Acad Sci USA. 2003;100:9849–9854. doi: 10.1073/pnas.1730940100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kann M, Bischof A, Gerlich WH. In vitro model for the nuclear transport of the hepadnavirus genome. J Virol. 1997;71:1310–1316. doi: 10.1128/jvi.71.2.1310-1316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panté N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitz A, Schwarz A, Foss M, Zhou L, Rabe B, Hoellenriegel J, Stoeber M, Panté N, Kann M. Nucleoporin 153 arrests the nuclear import of hepatitis B virus capsids in the nuclear basket. PLoS Pathog. 2010;6:e1000741. doi: 10.1371/journal.ppat.1000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zoulim F. New insight on hepatitis B virus persistence from the study of intrahepatic viral cccDNA. J Hepatol. 2005;42:302–308. doi: 10.1016/j.jhep.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Pollack JR, Ganem D. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J Virol. 1993;67:3254–3263. doi: 10.1128/jvi.67.6.3254-3263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Will H, Reiser W, Weimer T, Pfaff E, Büscher M, Sprengel R, Cattaneo R, Schaller H. Replication strategy of human hepatitis B virus. J Virol. 1987;61:904–911. doi: 10.1128/jvi.61.3.904-911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponsel D, Bruss V. Mapping of amino acid side chains on the surface of hepatitis B virus capsids required for envelopment and virion formation. J Virol. 2003;77:416–422. doi: 10.1128/JVI.77.1.416-422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghany M, Liang TJ. Drug targets and molecular mechanisms of drug resistance in chronic hepatitis B. Gastroenterology. 2007;132:1574–1585. doi: 10.1053/j.gastro.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 29.Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137:1593–1608.e1-2. doi: 10.1053/j.gastro.2009.08.063. [DOI] [PubMed] [Google Scholar]
- 30.Tuttleman JS, Pourcel C, Summers J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell. 1986;47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 31.Gao W, Hu J. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J Virol. 2007;81:6164–6174. doi: 10.1128/JVI.02721-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in hepatitis B virus infection. J Hepatol. 2009;51:581–592. doi: 10.1016/j.jhep.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Königer C, Wingert I, Marsmann M, Rösler C, Beck J, Nassal M. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proc Natl Acad Sci USA. 2014;111:E4244–E4253. doi: 10.1073/pnas.1409986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui X, McAllister R, Boregowda R, Sohn JA, Cortes Ledesma F, Caldecott KW, Seeger C, Hu J. Does Tyrosyl DNA Phosphodiesterase-2 Play a Role in Hepatitis B Virus Genome Repair? PLoS One. 2015;10:e0128401. doi: 10.1371/journal.pone.0128401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang HP, Rogler CE. Topoisomerase I-mediated integration of hepadnavirus DNA in vitro. J Virol. 1991;65:2381–2392. doi: 10.1128/jvi.65.5.2381-2392.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JW, Lee SH, Park YS, Jeong SH, Kim N, Lee DH. [Inhibition of in vitro hepatitis B virus replication by lentivirus-mediated short-hairpin RNA against HBx] Korean J Hepatol. 2009;15:15–24. doi: 10.3350/kjhep.2009.15.1.15. [DOI] [PubMed] [Google Scholar]
- 37.Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, Raimondo G, Levrero M. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci USA. 2009;106:19975–19979. doi: 10.1073/pnas.0908365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weber ND, Stone D, Sedlak RH, De Silva Feelixge HS, Roychoudhury P, Schiffer JT, Aubert M, Jerome KR. AAV-mediated delivery of zinc finger nucleases targeting hepatitis B virus inhibits active replication. PLoS One. 2014;9:e97579. doi: 10.1371/journal.pone.0097579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Zhang W, Lin J, Wang F, Wu M, Chen C, Zheng Y, Peng X, Li J, Yuan Z. An efficient antiviral strategy for targeting hepatitis B virus genome using transcription activator-like effector nucleases. Mol Ther. 2014;22:303–311. doi: 10.1038/mt.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kennedy EM, Kornepati AV, Cullen BR. Targeting hepatitis B virus cccDNA using CRISPR/Cas9. Antiviral Res. 2015;123:188–192. doi: 10.1016/j.antiviral.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 42.Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221–1228. doi: 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu F, Campagna M, Qi Y, Zhao X, Guo F, Xu C, Li S, Li W, Block TM, Chang J, et al. Alpha-interferon suppresses hepadnavirus transcription by altering epigenetic modification of cccDNA minichromosomes. PLoS Pathog. 2013;9:e1003613. doi: 10.1371/journal.ppat.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quasdorff M, Protzer U. Control of hepatitis B virus at the level of transcription. J Viral Hepat. 2010;17:527–536. doi: 10.1111/j.1365-2893.2010.01315.x. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Xu Z, Zheng Y, Johnson DL, Ou JH. Regulation of hepatocyte nuclear factor 1 activity by wild-type and mutant hepatitis B virus X proteins. J Virol. 2002;76:5875–5881. doi: 10.1128/JVI.76.12.5875-5881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi BH, Park GT, Rho HM. Interaction of hepatitis B viral X protein and CCAAT/ enhancer-binding protein alpha synergistically activates the hepatitis B viral enhancer II/pregenomic promoter. J Biol Chem. 1999;274:2858–2865. doi: 10.1074/jbc.274.5.2858. [DOI] [PubMed] [Google Scholar]
- 47.Wallerman O, Motallebipour M, Enroth S, Patra K, Bysani MS, Komorowski J, Wadelius C. Molecular interactions between HNF4a, FOXA2 and GABP identified at regulatory DNA elements through ChIP-sequencing. Nucleic Acids Res. 2009;37:7498–7508. doi: 10.1093/nar/gkp823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waris G, Siddiqui A. Interaction between STAT-3 and HNF-3 leads to the activation of liver-specific hepatitis B virus enhancer 1 function. J Virol. 2002;76:2721–2729. doi: 10.1128/JVI.76.6.2721-2729.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pult I, Chouard T, Wieland S, Klemenz R, Yaniv M, Blum HE. A hepatitis B virus mutant with a new hepatocyte nuclear factor 1 binding site emerging in transplant-transmitted fulminant hepatitis B. Hepatology. 1997;25:1507–1515. doi: 10.1002/hep.510250633. [DOI] [PubMed] [Google Scholar]
- 50.Li J, Buckwold VE, Hon MW, Ou JH. Mechanism of suppression of hepatitis B virus precore RNA transcription by a frequent double mutation. J Virol. 1999;73:1239–1244. doi: 10.1128/jvi.73.2.1239-1244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 52.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 53.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 54.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab. 2007;91:209–217. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cuellar TL, McManus MT. MicroRNAs and endocrine biology. J Endocrinol. 2005;187:327–332. doi: 10.1677/joe.1.06426. [DOI] [PubMed] [Google Scholar]
- 57.Hydbring P, Badalian-Very G. Clinical applications of microRNAs. F1000Res. 2013;2:136. doi: 10.12688/f1000research.2-136.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122--a key factor and therapeutic target in liver disease. J Hepatol. 2015;62:448–457. doi: 10.1016/j.jhep.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 59.Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, Cui Q. An analysis of human microRNA and disease associations. PLoS One. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raney AK, Easton AJ, Milich DR, McLachlan A. Promoter-specific transactivation of hepatitis B virus transcription by a glutamine- and proline-rich domain of hepatocyte nuclear factor 1. J Virol. 1991;65:5774–5781. doi: 10.1128/jvi.65.11.5774-5781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cai YN, Zhou Q, Kong YY, Li M, Viollet B, Xie YH, Wang Y. LRH-1/hB1F and HNF1 synergistically up-regulate hepatitis B virus gene transcription and DNA replication. Cell Res. 2003;13:451–458. doi: 10.1038/sj.cr.7290187. [DOI] [PubMed] [Google Scholar]
- 62.Wang WX, Li M, Wu X, Wang Y, Li ZP. HNF1 is critical for the liver-specific function of HBV enhancer II. Res Virol. 1998;149:99–108. doi: 10.1016/s0923-2516(98)80085-x. [DOI] [PubMed] [Google Scholar]
- 63.Quasdorff M, Hösel M, Odenthal M, Zedler U, Bohne F, Gripon P, Dienes HP, Drebber U, Stippel D, Goeser T, et al. A concerted action of HNF4alpha and HNF1alpha links hepatitis B virus replication to hepatocyte differentiation. Cell Microbiol. 2008;10:1478–1490. doi: 10.1111/j.1462-5822.2008.01141.x. [DOI] [PubMed] [Google Scholar]
- 64.Zheng Y, Li J, Ou JH. Regulation of hepatitis B virus core promoter by transcription factors HNF1 and HNF4 and the viral X protein. J Virol. 2004;78:6908–6914. doi: 10.1128/JVI.78.13.6908-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raney AK, Eggers CM, Kline EF, Guidotti LG, Pontoglio M, Yaniv M, McLachlan A. Nuclear covalently closed circular viral genomic DNA in the liver of hepatocyte nuclear factor 1 alpha-null hepatitis B virus transgenic mice. J Virol. 2001;75:2900–2911. doi: 10.1128/JVI.75.6.2900-2911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li M, Xie Y, Wu X, Kong Y, Wang Y. HNF3 binds and activates the second enhancer, ENII, of hepatitis B virus. Virology. 1995;214:371–378. doi: 10.1006/viro.1995.0046. [DOI] [PubMed] [Google Scholar]
- 67.Tang H, McLachlan A. Mechanisms of inhibition of nuclear hormone receptor-dependent hepatitis B virus replication by hepatocyte nuclear factor 3beta. J Virol. 2002;76:8572–8581. doi: 10.1128/JVI.76.17.8572-8581.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ori A, Shaul Y. Hepatitis B virus enhancer binds and is activated by the Hepatocyte nuclear factor 3. Virology. 1995;207:98–106. doi: 10.1006/viro.1995.1055. [DOI] [PubMed] [Google Scholar]
- 69.Banks KE, Anderson AL, Tang H, Hughes DE, Costa RH, McLachlan A. Hepatocyte nuclear factor 3beta inhibits hepatitis B virus replication in vivo. J Virol. 2002;76:12974–12980. doi: 10.1128/JVI.76.24.12974-12980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuo CJ, Conley PB, Chen L, Sladek FM, Darnell JE, Crabtree GR. A transcriptional hierarchy involved in mammalian cell-type specification. Nature. 1992;355:457–461. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- 72.He F, Chen EQ, Liu L, Zhou TY, Liu C, Cheng X, Liu FJ, Tang H. Inhibition of hepatitis B Virus replication by hepatocyte nuclear factor 4-alpha specific short hairpin RNA. Liver Int. 2012;32:742–751. doi: 10.1111/j.1478-3231.2011.02748.x. [DOI] [PubMed] [Google Scholar]
- 73.Raney AK, Johnson JL, Palmer CN, McLachlan A. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J Virol. 1997;71:1058–1071. doi: 10.1128/jvi.71.2.1058-1071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hong MH, Chou YC, Wu YC, Tsai KN, Hu CP, Jeng KS, Chen ML, Chang C. Transforming growth factor-β1 suppresses hepatitis B virus replication by the reduction of hepatocyte nuclear factor-4α expression. PLoS One. 2012;7:e30360. doi: 10.1371/journal.pone.0030360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Long Y, Chen E, Liu C, Huang F, Zhou T, He F, Liu L, Liu F, Tang H. The correlation of hepatocyte nuclear factor 4 alpha and 3 beta with hepatitis B virus replication in the liver of chronic hepatitis B patients. J Viral Hepat. 2009;16:537–546. doi: 10.1111/j.1365-2893.2009.01089.x. [DOI] [PubMed] [Google Scholar]
- 76.Chen EQ, Sun H, Feng P, Gong DY, Liu C, Bai L, Yang WB, Lei XZ, Chen LY, Huang FJ, et al. Study of the expression levels of Hepatocyte nuclear factor 4 alpha and 3 beta in patients with different outcome of HBV infection. Virol J. 2012;9:23. doi: 10.1186/1743-422X-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang K, Holterman AX. Pathophysiologic role of hepatocyte nuclear factor 6. Cell Signal. 2012;24:9–16. doi: 10.1016/j.cellsig.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 78.Hao R, He J, Liu X, Gao G, Liu D, Cui L, Yu G, Yu W, Chen Y, Guo D. Inhibition of hepatitis B virus gene expression and replication by hepatocyte nuclear factor 6. J Virol. 2015;89:4345–4355. doi: 10.1128/JVI.03094-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lahuna O, Fernandez L, Karlsson H, Maiter D, Lemaigre FP, Rousseau GG, Gustafsson J, Mode A. Expression of hepatocyte nuclear factor 6 in rat liver is sex-dependent and regulated by growth hormone. Proc Natl Acad Sci USA. 1997;94:12309–12313. doi: 10.1073/pnas.94.23.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.López-Cabrera M, Letovsky J, Hu KQ, Siddiqui A. Transcriptional factor C/EBP binds to and transactivates the enhancer element II of the hepatitis B virus. Virology. 1991;183:825–829. doi: 10.1016/0042-6822(91)91019-d. [DOI] [PubMed] [Google Scholar]
- 81.Lin SJ, Shu PY, Chang C, Ng AK, Hu CP. IL-4 suppresses the expression and the replication of hepatitis B virus in the hepatocellular carcinoma cell line Hep3B. J Immunol. 2003;171:4708–4716. doi: 10.4049/jimmunol.171.9.4708. [DOI] [PubMed] [Google Scholar]
- 82.López-Cabrera M, Letovsky J, Hu KQ, Siddiqui A. Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: trans-activation and repression by CCAAT/enhancer binding protein. Proc Natl Acad Sci USA. 1990;87:5069–5073. doi: 10.1073/pnas.87.13.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pei DQ, Shih CH. Transcriptional activation and repression by cellular DNA-binding protein C/EBP. J Virol. 1990;64:1517–1522. doi: 10.1128/jvi.64.4.1517-1522.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu WH, Yeh SH, Lu CC, Yu SL, Chen HY, Lin CY, Chen DS, Chen PJ. MicroRNA-18a prevents estrogen receptor-alpha expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology. 2009;136:683–693. doi: 10.1053/j.gastro.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 85.Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490–499. doi: 10.1002/hep.23008. [DOI] [PubMed] [Google Scholar]
- 86.Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 2010;51:881–890. doi: 10.1002/hep.23381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60–70. doi: 10.1002/hep.23660. [DOI] [PubMed] [Google Scholar]
- 88.Zhang X, Zhang E, Ma Z, Pei R, Jiang M, Schlaak JF, Roggendorf M, Lu M. Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology. 2011;53:1476–1485. doi: 10.1002/hep.24195. [DOI] [PubMed] [Google Scholar]
- 89.Zhang X, Liu H, Deng W, Li B, Hou J, Lu M. 402 regulation of hbv replication by microRNA-449a through targeting CREB5 in hepatoma cells. J Hepatol. 2013;58:S165. [Google Scholar]
- 90.Wang S, Qiu L, Yan X, Jin W, Wang Y, Chen L, Wu E, Ye X, Gao GF, Wang F, et al. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G(1) -modulated P53 activity. Hepatology. 2012;55:730–741. doi: 10.1002/hep.24809. [DOI] [PubMed] [Google Scholar]
- 91.Qiu L, Fan H, Jin W, Zhao B, Wang Y, Ju Y, Chen L, Chen Y, Duan Z, Meng S. miR-122-induced down-regulation of HO-1 negatively affects miR-122-mediated suppression of HBV. Biochem Biophys Res Commun. 2010;398:771–777. doi: 10.1016/j.bbrc.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 92.Protzer U, Seyfried S, Quasdorff M, Sass G, Svorcova M, Webb D, Bohne F, Hösel M, Schirmacher P, Tiegs G. Antiviral activity and hepatoprotection by heme oxygenase-1 in hepatitis B virus infection. Gastroenterology. 2007;133:1156–1165. doi: 10.1053/j.gastro.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 93.Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 2010;88:169–175. doi: 10.1016/j.antiviral.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 94.Potenza N, Papa U, Mosca N, Zerbini F, Nobile V, Russo A. Human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen. Nucleic Acids Res. 2011;39:5157–5163. doi: 10.1093/nar/gkr067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kohno T, Tsuge M, Murakami E, Hiraga N, Abe H, Miki D, Imamura M, Ochi H, Hayes CN, Chayama K. Human microRNA hsa-miR-1231 suppresses hepatitis B virus replication by targeting core mRNA. J Viral Hepat. 2014;21:e89–e97. doi: 10.1111/jvh.12240. [DOI] [PubMed] [Google Scholar]
- 96.Liu WH, Yeh SH, Chen PJ. Role of microRNAs in hepatitis B virus replication and pathogenesis. Biochim Biophys Acta. 2011;1809:678–685. doi: 10.1016/j.bbagrm.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 97.Wu FL, Jin WB, Li JH, Guo AG. Targets for human encoded microRNAs in HBV genes. Virus Genes. 2011;42:157–161. doi: 10.1007/s11262-010-0555-7. [DOI] [PubMed] [Google Scholar]
- 98.Wang Y, Jiang L, Ji X, Yang B, Zhang Y, Fu XD. Hepatitis B viral RNA directly mediates down-regulation of the tumor suppressor microRNA miR-15a/miR-16-1 in hepatocytes. J Biol Chem. 2013;288:18484–18493. doi: 10.1074/jbc.M113.458158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ramière C, Scholtès C, Diaz O, Icard V, Perrin-Cocon L, Trabaud MA, Lotteau V, André P. Transactivation of the hepatitis B virus core promoter by the nuclear receptor FXRalpha. J Virol. 2008;82:10832–10840. doi: 10.1128/JVI.00883-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garcia AD, Ostapchuk P, Hearing P. Functional interaction of nuclear factors EF-C, HNF-4, and RXR alpha with hepatitis B virus enhancer I. J Virol. 1993;67:3940–3950. doi: 10.1128/jvi.67.7.3940-3950.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shaul Y, Ben-Levy R, De-Medina T. High affinity binding site for nuclear factor I next to the hepatitis B virus S gene promoter. EMBO J. 1986;5:1967–1971. doi: 10.1002/j.1460-2075.1986.tb04451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spandau DF, Lee CH. Repression of the hepatitis B virus enhancer by a cellular factor. J Gen Virol. 1992;73(Pt 1):131–137. doi: 10.1099/0022-1317-73-1-131. [DOI] [PubMed] [Google Scholar]
- 103.Lu CC, Yen TS. Activation of the hepatitis B virus S promoter by transcription factor NF-Y via a CCAAT element. Virology. 1996;225:387–394. doi: 10.1006/viro.1996.0613. [DOI] [PubMed] [Google Scholar]
- 104.Zhang P, Raney AK, McLachlan A. Characterization of functional Sp1 transcription factor binding sites in the hepatitis B virus nucleocapsid promoter. J Virol. 1993;67:1472–1481. doi: 10.1128/jvi.67.3.1472-1481.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li J, Ou JH. Differential regulation of hepatitis B virus gene expression by the Sp1 transcription factor. J Virol. 2001;75:8400–8406. doi: 10.1128/JVI.75.18.8400-8406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Raney AK, McLachlan A. Characterization of the hepatitis B virus large surface antigen promoter Sp1 binding site. Virology. 1995;208:399–404. doi: 10.1006/viro.1995.1167. [DOI] [PubMed] [Google Scholar]
- 107.Raney AK, Le HB, McLachlan A. Regulation of transcription from the hepatitis B virus major surface antigen promoter by the Sp1 transcription factor. J Virol. 1992;66:6912–6921. doi: 10.1128/jvi.66.12.6912-6921.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin WJ, Li J, Lee YF, Yeh SD, Altuwaijri S, Ou JH, Chang C. Suppression of hepatitis B virus core promoter by the nuclear orphan receptor TR4. J Biol Chem. 2003;278:9353–9360. doi: 10.1074/jbc.M205944200. [DOI] [PubMed] [Google Scholar]
- 109.Zhao F, Xu G, Zhou Y, Wang L, Xie J, Ren S, Liu S, Zhu Y. MicroRNA-26b inhibits hepatitis B virus transcription and replication by targeting the host factor CHORDC1 protein. J Biol Chem. 2014;289:35029–35041. doi: 10.1074/jbc.M114.589978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tokusumi Y, Zhou S, Takada S. Nuclear respiratory factor 1 plays an essential role in transcriptional initiation from the hepatitis B virus x gene promoter. J Virol. 2004;78:10856–10864. doi: 10.1128/JVI.78.20.10856-10864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Choi BH, Park CJ, Rho HM. Insulin activates the hepatitis B virus X gene through the activating protein-1 binding site in HepG2 cells. DNA Cell Biol. 1998;17:951–956. doi: 10.1089/dna.1998.17.951. [DOI] [PubMed] [Google Scholar]
- 112.Bogomolski-Yahalom V, Klein A, Greenblat I, Haviv Y, Tur-Kaspa R. The TATA-less promoter of hepatitis B virus S gene contains a TBP binding site and an active initiator. Virus Res. 1997;49:1–7. doi: 10.1016/s0168-1702(96)01429-3. [DOI] [PubMed] [Google Scholar]
- 113.Kim BK, Lim SO, Park YG. Requirement of the cyclic adenosine monophosphate response element-binding protein for hepatitis B virus replication. Hepatology. 2008;48:361–373. doi: 10.1002/hep.22359. [DOI] [PubMed] [Google Scholar]
- 114.Park YK, Park ES, Kim DH, Ahn SH, Park SH, Lee AR, Park S, Kang HS, Lee JH, Kim JM, et al. Cleaved c-FLIP mediates the antiviral effect of TNF-α against hepatitis B virus by dysregulating hepatocyte nuclear factors. J Hepatol. 2016;64:268–277. doi: 10.1016/j.jhep.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 115.Xiang WQ, Feng WF, Ke W, Sun Z, Chen Z, Liu W. Hepatitis B virus X protein stimulates IL-6 expression in hepatocytes via a MyD88-dependent pathway. J Hepatol. 2011;54:26–33. doi: 10.1016/j.jhep.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 116.Hösel M, Quasdorff M, Wiegmann K, Webb D, Zedler U, Broxtermann M, Tedjokusumo R, Esser K, Arzberger S, Kirschning CJ, et al. Not interferon, but interleukin-6 controls early gene expression in hepatitis B virus infection. Hepatology. 2009;50:1773–1782. doi: 10.1002/hep.23226. [DOI] [PubMed] [Google Scholar]
- 117.Pasquetto V, Wieland SF, Uprichard SL, Tripodi M, Chisari FV. Cytokine-sensitive replication of hepatitis B virus in immortalized mouse hepatocyte cultures. J Virol. 2002;76:5646–5653. doi: 10.1128/JVI.76.11.5646-5653.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shin GC, Ahn SH, Choi HS, Kim J, Park ES, Kim DH, Kim KH. Hepatocystin contributes to interferon-mediated antiviral response to hepatitis B virus by regulating hepatocyte nuclear factor 4α. Biochim Biophys Acta. 2014;1842:1648–1657. doi: 10.1016/j.bbadis.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 119.Lin YC, Hsu EC, Ting LP. Repression of hepatitis B viral gene expression by transcription factor nuclear factor-kappaB. Cell Microbiol. 2009;11:645–660. doi: 10.1111/j.1462-5822.2008.01280.x. [DOI] [PubMed] [Google Scholar]
- 120.Nikolaidou-Neokosmidou V, Zannis VI, Kardassis D. Inhibition of hepatocyte nuclear factor 4 transcriptional activity by the nuclear factor kappaB pathway. Biochem J. 2006;398:439–450. doi: 10.1042/BJ20060169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zheng Y, Li J, Johnson DL, Ou JH. Regulation of hepatitis B virus replication by the ras-mitogen-activated protein kinase signaling pathway. J Virol. 2003;77:7707–7712. doi: 10.1128/JVI.77.14.7707-7712.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dai X, Zhang W, Zhang H, Sun S, Yu H, Guo Y, Kou Z, Zhao G, Du L, Jiang S, et al. Modulation of HBV replication by microRNA-15b through targeting hepatocyte nuclear factor 1α. Nucleic Acids Res. 2014;42:6578–6590. doi: 10.1093/nar/gku260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Guo H, Liu H, Mitchelson K, Rao H, Luo M, Xie L, Sun Y, Zhang L, Lu Y, Liu R, et al. MicroRNAs-372/373 promote the expression of hepatitis B virus through the targeting of nuclear factor I/B. Hepatology. 2011;54:808–819. doi: 10.1002/hep.24441. [DOI] [PubMed] [Google Scholar]
- 124.Wu Q, Liu HO, Liu YD, Liu WS, Pan D, Zhang WJ, Yang L, Fu Q, Xu JJ, Gu JX. Decreased expression of hepatocyte nuclear factor 4α (Hnf4α)/microRNA-122 (miR-122) axis in hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J Biol Chem. 2015;290:1170–1185. doi: 10.1074/jbc.M114.601203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hu W, Wang X, Ding X, Li Y, Zhang X, Xie P, Yang J, Wang S. MicroRNA-141 represses HBV replication by targeting PPARA. PLoS One. 2012;7:e34165. doi: 10.1371/journal.pone.0034165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huang JY, Chou SF, Lee JW, Chen HL, Chen CM, Tao MH, Shih C. MicroRNA-130a can inhibit hepatitis B virus replication via targeting PGC1α and PPARγ. RNA. 2015;21:385–400. doi: 10.1261/rna.048744.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K, Jacob ST. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jin J, Tang S, Xia L, Du R, Xie H, Song J, Fan R, Bi Q, Chen Z, Yang G, et al. MicroRNA-501 promotes HBV replication by targeting HBXIP. Biochem Biophys Res Commun. 2013;430:1228–1233. doi: 10.1016/j.bbrc.2012.12.071. [DOI] [PubMed] [Google Scholar]
- 129.Su C, Hou Z, Zhang C, Tian Z, Zhang J. Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol J. 2011;8:354. doi: 10.1186/1743-422X-8-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tang H, McLachlan A. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc Natl Acad Sci USA. 2001;98:1841–1846. doi: 10.1073/pnas.041479698. [DOI] [PMC free article] [PubMed] [Google Scholar]


