Abstract
Butterflies rely on color vision extensively to adapt to the natural world. Most species express a broad range of color sensitive Rhodopsins in three stochastically distributed types of ommatidia (unit eyes)1–3. The retinas of Drosophila deploy just two main types, where fate is controlled by the binary stochastic decision to express the transcription factor Spineless (Ss) in R7 photoreceptors4. We investigated how butterflies instead generate three stochastically distributed ommatidial types, resulting in a more diverse retinal mosaic that provides the basis for additional color comparisons and an expanded range of color vision. We show that the Japanese Yellow Swallowtail (Papilio xuthus, Papilionidae) and the Painted Lady (Vanessa cardui, Nymphalidae) have a second R7-like photoreceptor in each ommatidium. Independent stochastic expression of Ss in each R7-like cell results in expression of a Blue (Ss-ON) or a UV (Ss-OFF) Rhodopsin. In Papilio, these choices of Blue/Blue, Blue/UV, or UV/UV in the two R7s are coordinated with expression of additional Rhodopsins in the remaining photoreceptors, and together define the three types of ommatidia. Knocking out ss using CRISPR/Cas95,6 leads to the loss of the Blue fate in R7-like cells and transforms retinas into homogeneous fields of UV/UV-type ommatidia, with all corresponding features. Hence, the three possible outcomes of Ss expression define the three ommatidial types in butterflies. This developmental strategy allowed the deployment of an additional red-sensitive Rhodopsin in Papilio, allowing for the evolution of expanded color vision with a richer variety of receptors7,8. This surprisingly simple mechanism that makes use of two binary stochastic decisions coupled with local coordination may prove to be a general means of generating an increased diversity of developmental outcomes.
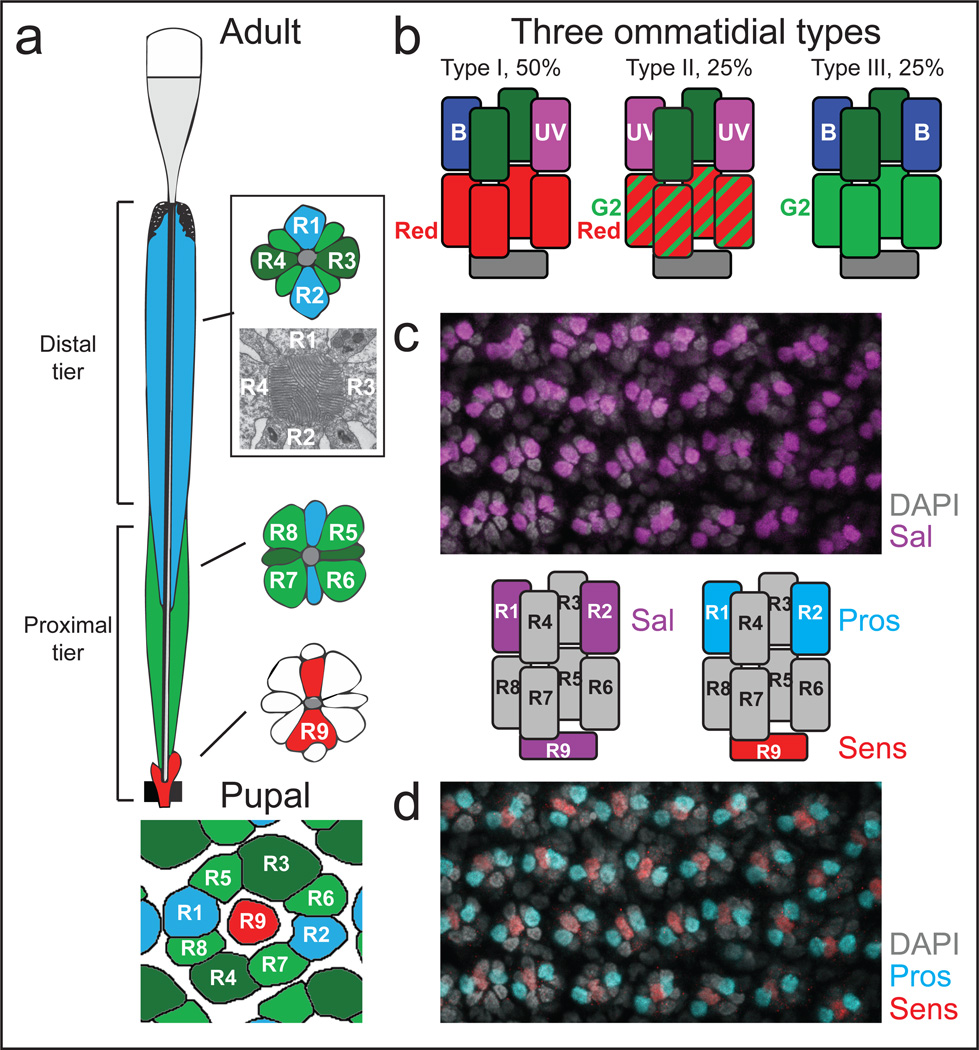
Papilio butterflies have some of the most complex retinal mosaics of any insect1,2. In Papilio xuthus (“Papilio”), three ommatidial types deploy five Rhodopsins used in color vision, motion detection, and in polarized light vision (reviewed in1, Fig.1a,b). In contrast, Vanessa cardui (“Vanessa”) exhibits a somewhat less complex pattern of expression of just three Rhodopsins2. The adult butterfly retina is tiered with four distal photoreceptors (butterfly R1–R4, bR1–4) just below the lens, four proximal photoreceptors bR5–R8 deeper in the ommatidium, and the bR9 cell below (Fig.1a)1.
Figure 1. Rhodopsin expression and ommatidial types in the Papilio retina.
a, Cell bodies of the distal tier (bR1–4) and proximal tier (bR5–8 plus bR9) photoreceptors surround a central fused rhabdom (grey; TEM shown in inset). Corresponding pupal stage photoreceptors are shown below.
b, Adult Rhodopsin expression in the three stochastically distributed ommatidial types. Combinations of UV- and Blue-sensitive Rhodopsins expressed in the distal tier are coordinated with specific long wavelength Green- and Red-sensitive Rhodopsins in the proximal tier.
c, Antibodies against Sal (magenta) in pupal eye discs label three photoreceptor nuclei per ommatidium, including the central bR9.
d, Sens (red) and Pros (cyan) expression provide evidence that there are two dR7-like cells in Papilio (bR1–2) and that bR9 is homologous to dR8: the ninth photoreceptor is a second dR7-like photoreceptor.
The three stochastically distributed ommatidial types found in butterflies are reminiscent of the simpler Drosophila eye, which contains just two stochastic types9. In Drosophila, the stochastic variations are restricted to the “inner” photoreceptors R7 and R8: in ommatidia where R7 expresses UV-sensitive Rh3, R8 expresses blue-Rh5. When UV-Rh4 is expressed in R7, R8 expresses green-Rh610,11. The remaining “outer” photoreceptors R1–6 all express the same broad spectrum Rh1 in all ommatidia and are involved in motion vision10. The stochastic decision in Drosophila is controlled by the transcription factor Spineless (Ss), whose stochastic expression in R7 directly regulates Rh44,12. This binary choice in R7 is then communicated to the underlying R8, thereby defining the two types of ommatidia11,13.
Butterflies rely on color vision for recognizing flowers, finding food, identifying mates, and in locating oviposition sites - challenging visual environments reflected by the complexity of their retinas. The increased ommatidial diversity in Papilio may have been useful for deploying a newly evolved Red Rhodopsin, which is expressed differently in each of the three ommatidial types7,14. We set out to investigate how butterflies produce more than two stochastically distributed ommatidial types.
We sequenced the genomes and adult head transcriptomes of Papilio and Vanessa. Papilio has an established history of vision research1,15, while Vanessa is a cosmopolitan species that can be maintained on artificial diet in the laboratory. Our results focus primarily on Papilio, with corresponding experiments in Vanessa described in Extended Data. We produced antibodies against factors that define photoreceptor types in Drosophila, seeking to identify homologous photoreceptors in butterflies (Fig.1)16.
The organization of butterfly photoreceptors during pupal stages resembles that of Drosophila except for the presence of nine photoreceptors instead of eight (Fig.1a). In Drosophila, the transcription factor Spalt is expressed specifically in color sensitive R7 and R8 (dR7+8) and is required for specifying these cells, which have long visual fibers (LVF) projecting to the medulla neuropil17. Outer photoreceptors dR1–6 have short visual fibers (SVF) terminating in the lamina neuropil17. A cross-reactive antibody to Drosophila Spalt labels three photoreceptor nuclei per ommatidium in pupal Papilio retinas (Fig.1c, Extended Data Fig.1c). Two of these photoreceptors become part of the distal tier of the adult ommatidium and correspond to the butterfly photoreceptors named R1 and R2 (bR1+2), while the central labeled nucleus is R9 (bR9). This expression pattern fits with previous data that suggested bR1+2 (and possibly bR9) are LVF photoreceptors18.
We identified the Papilio homolog of transcription factors Senseless (Sens), which is specifically expressed in Drosophila R819, and Prospero (Pros), which marks R7 in flies20. Antibodies against Papilio Sens specifically labelled bR9, indicating that this cell is homologous to dR8 (Fig.1c). Antibodies against Papilio Pros marked two photoreceptors in butterflies, bR1+2 (Fig.1c and Extended Data Fig.1a), which are thus homologous to two dR7 photoreceptors, as had been proposed previously based on morphology16. The presence of Sal, Pros, and Sens in developing butterfly retinas suggests that the regulatory code that defines the different photoreceptor types is deeply conserved, with the additional ninth photoreceptor of butterflies being a second dR7-type.
The three Papilio ommatidial types (Fig.1b)2,3 express different wavelength-specific Rhodopsins not only in inner, but also in outer photoreceptors, suggesting that both LVF and SVF photoreceptors are involved in color vision8,18. In all three types of Papilio ommatidia, the two distal SVF photoreceptors bR3/R4 coexpress two green Rhodopsins, Papilio xuthus G1 (PxG1) and PxG21,14. The Rhodopsins expressed in bR9, a cell that appears to play a reduced role in butterfly vision with little contribution to the rhabdom, have not been identified. In type I ommatidia (~50%), one of the dR7-like photoreceptors expresses PxUV and the other PxBlue, while the four proximal SVF photoreceptors bR5–8 express red PxR. In type II ommatidia (~25%), both R7-like photoreceptors express PxUV while bR5–8 express both PxRed and low levels of PxG221. Type III (~25%) express PxBlue in both R7-like photoreceptors, and high levels of PxG2 in bR5–8 (Fig.1b). The choices in bR5–8 SVF photoreceptors are therefore precisely coupled with the choices in the two dR7-like cells (Fig.1b). In Drosophila, a stochastic choice to express UV-Rh3 or UV-Rh4 is made in R7, and this binary choice is communicated to the underlying R8 to express Blue or Green-sensitive Rhodopsin, respectively11. All outer photoreceptors uniformly express Rh1.
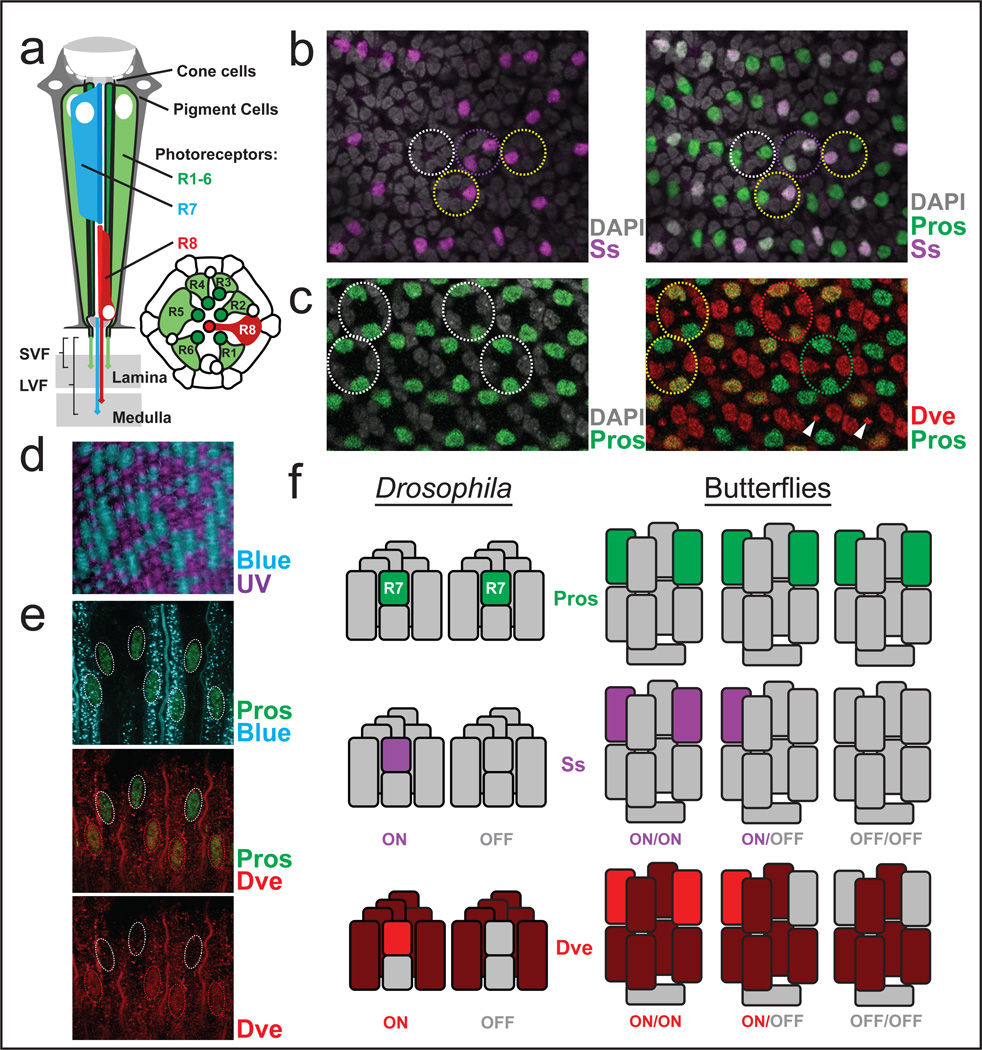
The presence of two dR7-like photoreceptors suggested a model for stochastic patterning that could produce three butterfly ommatidial types. In the Drosophila retina (Fig.2a), an independent, cell-intrinsic choice to express Ss yields two possible fates in R7, Sson or Ssoff (Fig.2f)4. Ss directly activates Rh4 as well as the transcription factor Defective proventriculus (Dve), which represses Rh322. While Dve is also expressed in all R1–6, it is on in Sson R7s and it is off in Ssoff R712. If each butterfly R7-like cell were to also make an independent choice for Ss expression, one would expect on/on, on/off, or off/off ommatidia with Dve matching Ss and Rhodopsin expression (Fig.2f).
Figure 2. Stochastic Spineless expression in flies and butterflies.
a, Schematic of an adult Drosophila ommatidium showing photoreceptor numbering and stacked dR7/R8.
b, In Papilio pupal retinas, Ss (magenta) is expressed stochastically in a subset of Pros-positive dR7-like cells (green).
c, Dve (red) is expressed stochastically in a subset of dR7-like cells (labeled by Pros in green). Dve signal also localized to rhabdomeres (arrowheads), which is also observed in Drosophila12.
d, PxB and PxUV Rhodopsins are expressed stochastically in adult Papilio dR7-like cells, leading to a mosaic of ommatidia expressing B/B, B/UV, or UV/UV, seen here in cross-section.
e, Dve expression (red) in this lateral view of adult photoreceptors can be on or off in dR7-like nuclei (Pros in green). Dve is co-expressed with Blue Rhodopsin PxBlue (cyan). The Dve signal again localized to the rhabdomeres as in Fig. 2c.
f, Left: Ss can be either on (purple) or off (grey) in Drosophila R7s (cyan). Dve is expressed in all SVF outer photoreceptors (dark red) and at lower level in R7s (light red) that express Ss.
Right: There are two dR7-like photoreceptors (green) per ommatidium in Papilio, as shown by Pros expression. Independent stochastic Ss expression in these cells could yield three outcomes. Dve follows Ss expression in each of the two dR7-like cells per ommatidium in Papilio.
Schematics shown in (a) were adapted with permission from 30.
To test this possibility, we generated antibodies against Papilio Ss and Dve as well as PxUV and PxBlue Rhodopsins. These antibodies cross-react with Vanessa proteins (see Extended Data). The Rhodopsin antibody stains showed the expected stochastic mosaic of three ommatidial types (Fig.2d). Ss expression in pupal retinas was also stochastic in the two Pros-expressing dR7-like photoreceptors, bR1 and bR2 (Fig. 2b). Ss was either on in both bR1 and bR2 (on/on), on/off, off/on, or off/off. Dve was stochastically expressed in these dR7-like cells in an identical pattern (Fig.2c, 3c), suggesting that, as in flies, it is downstream of Ss. We then attempted to determine which ommatidial types express Ss and Dve in dR7-like cells. Although Ss expression was not detected in the adult when Rhodopsins were expressed, Dve was maintained specifically in cells that express PxBlue, but not in PxUV cells (Fig.2e). Therefore, Ss, Dve, and PxBlue are expressed in the same photoreceptors. This supports a model in which stochastic expression of Ss in dR7-like cells defines the three ommatidial types: on/off (and off/on) correspond to type I, off/off to type II and on/on to type III (Fig. 2f).
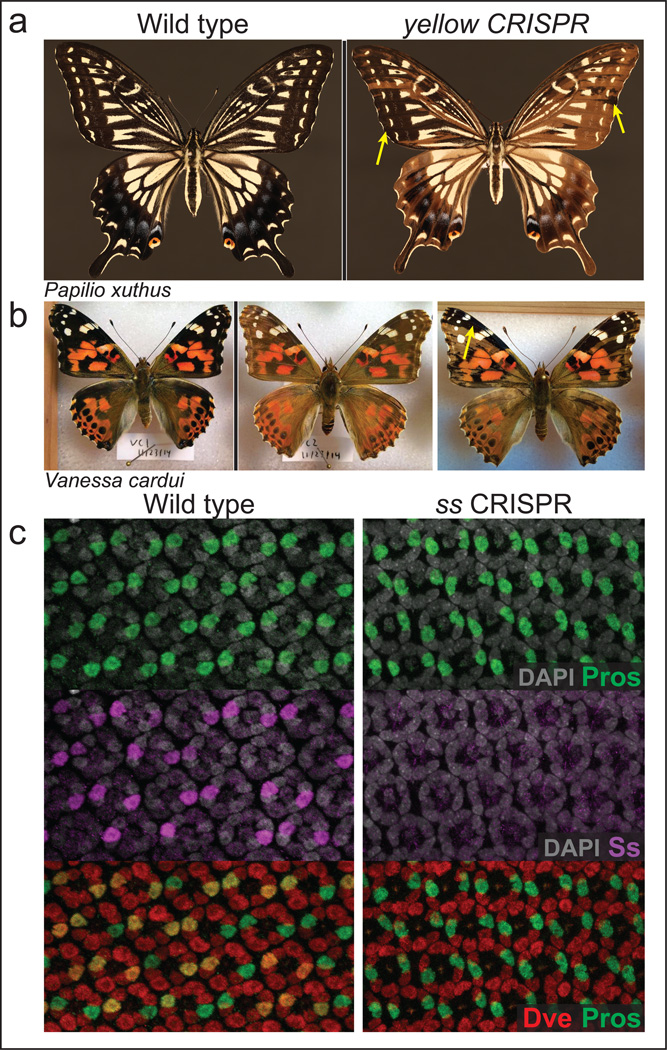
Figure 3. Targeted CRISPR/Cas9 knockout of yellow and spineless.
a, Left: wild type Papilio xuthus are yellow or black on most dorsal surfaces. Knockout of yellow (right) causes the loss of black pigmentation. Individual shown at right is a G0 mosaic showing large regions of lighter biallelic mutant tissue, with examples of darker wild-type tissue marked by arrows.
b, yellow CRISPR knockout in Vanessa cardui frequently produces fully mutant G0 animals (middle, compare to wild-type at left). Mosaics are also produced (right). Results from Vanessa are described in more detail in Extended Data.
c, Targeting spineless produces pupal retinas that lack Ss protein in large domains (middle right panel, compare to wild-type at left). Downstream expression of Dve matches Ss; compare left middle Ss (magenta) vs. bottom showing co-expression of Dve (red) in the same Pros-positive (green) nuclei. Knockout of ss (middle right) eliminates Dve expression in Pros positive cells (no coexpression in bottom right panel).
To test this model, we sought to knockout Ss expression using CRISPR/Cas95,6. First, we determined the efficiency of the system by targeting yellow, a gene involved in melanin production. Its bi-allelic knockout produced a strong wing and body yellow color phenotype (Fig.3a,b). Varying the timing of injection and concentration of guide RNAs (sgRNAs) and Cas9 protein produced a range of effects, from the loss of almost all black-pigmented regions to mixtures of mutant and non-mutant tissue (Fig.3a,b). This type of mosaic knockout is useful for testing lethal mutations like ss. We then targeted the ss locus in both Papilio and Vanessa. ss knockout animals showed defects in antenna and were missing leg bristles (Extended Data Fig.2), similar to Drosophila ss mutant phenotypes23. Sequencing revealed mutations at the target site (Extended Data Fig.2). Unexpectedly, ss also appeared to play a role in wing color patterning: colors produced via both melanin and ommochrome pathways were abolished in ss mutant wing scales in both Papilio and Vanessa (Extended Data Fig.2). Defects in the antennae and wings unfortunately meant that these animals did not eclose well and were not suitable for behavioral experiments.
In the retina, two independently tested sgRNAs targeting ss caused loss of Ss and Dve staining in dR7-like cells marked by Pros (Fig.3c and Extended Data Fig.1). Loss of Dve expression in dR7-like cells confirmed that Ss acts upstream of Dve (Fig.3c). Given the co-expression of Ss, Dve, and PxBlue (Fig. 2e), we predicted the presence of uniform UV/UV type II ommatidia in ss mutant tissues.
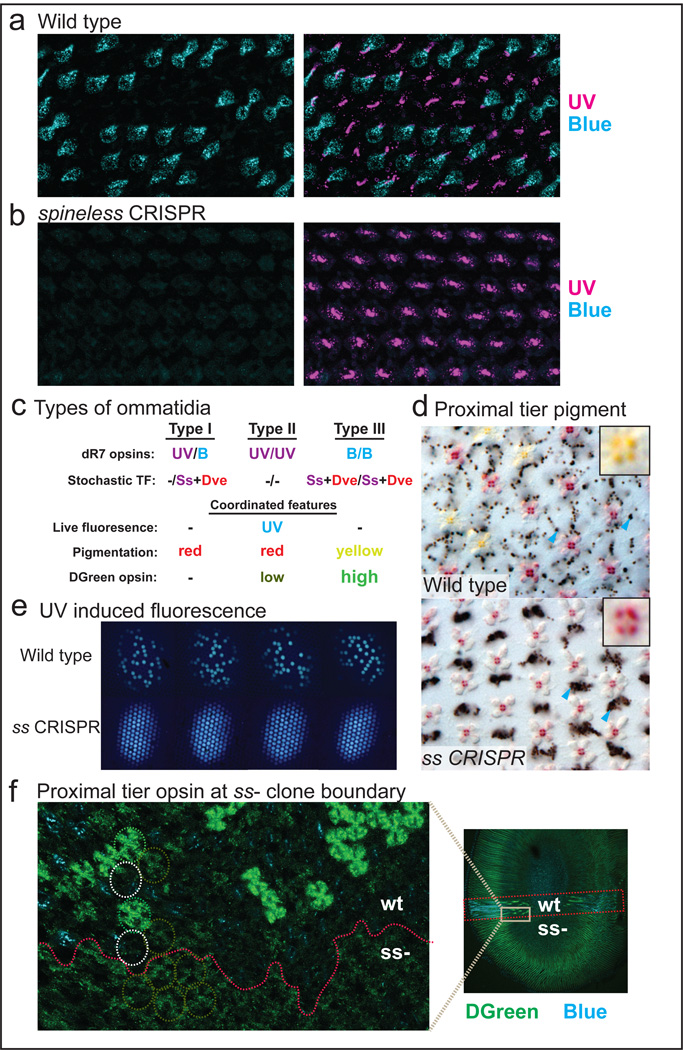
Wild type Papilio retinas showed a mosaic of PxUV and PxBlue expression in the two dR7-like photoreceptors (Fig.4a)24. In contrast, large regions in ss-knockout retinas contained only PxUV/PxUV type ommatidia with complete loss of PxBlue expression (Fig.4b). We then characterized the effect of ss knockout on other aspects of ommatidial type such as the presence of non-visual filtering pigments, UV-induced fluorescence, and the expression of PxG1, PxG2, and PxR in SVF photoreceptors.
In wild type Papilio retinas, either a red (in type I and II) or a yellow (in type III) pigment used for spectral tuning was expressed as a mosaic in the bR3–8 SVF photoreceptors (Fig.4c,d)25. In contrast, large regions in ss knockout animals contained only red pigment-expressing ommatidia, in agreement with the loss of type III ommatidia (Fig.4d).
UV light induces blueish fluorescence in type II ommatidia due to the presence of fluorescing 3-hydroxyretinol in wild type retina26 (Fig.4e). ss-knockout retinas contained large fields of ommatidia that all fluoresced blueish (Fig.4e), consistent with the loss of type I and III ommatidia.
bR5–8 photoreceptors express either PxRed (type I), PxRed and low level PxG2 (type II), or high level PxG2 (type III)21 (Fig.4c). In wild type retinas, our antibody detected PxG2 at high levels in some ommatidia, low in others, and none in the remaining ommatidia. Strikingly, ss-knockout tissue that lacked PxB expression in bR1,2 showed weak PxG2 expression in all ommatidia (Fig.4f), i.e. all type II ommatidia.
Figure 4. Outcome of spineless knockout and coordination of ommatidial fate.
a, Wild-type Papilio retinas contain a stochastic mosaic of ommatidia expressing either PxUV (magenta) or PxBlue (cyan) Rhodopsin in the dR7-like photoreceptors of the distal tier (bR1+2). Ommatidia are UV/B, UV/UV, or B/B.
b, In ss mutant tissue, all ommatidia become UV/UV and lose Blue Rhodopsin expression. c–f, Other features of ommatidial type are also coordinated, as summarized in (c).
d, Lower tier photoreceptors bR3–8 produce either a red (types I and II) or yellow pigment (type III) matching the stochastic mosaic (top). Arrowheads mark examples of dark granules of screening pigment. In ss knockout retinas only red pigment is observed, indicating loss of type III (bottom).
e, Blue fluorescence under UV light normally indicates type II fate; all ommatidia become UV fluorescing in ss knockout tissues and are therefore type II. The four panels show a series along the A/P axis across one wild type retina and one ss CRISPR knockout retina.
f, Long wavelength green-sensitive PxG2 Rhodopsin expression in the proximal tier b5–8 photoreceptors is coordinated with ommatidial type. PxG2 is highly expressed in type III, expressed at low levels in type II, and not expressed in type I. Low levels of expression in ss mutant tissue indicate type II fate. The red line indicates the approximate boundary between wild-type and ss mutant tissues; dashed circles indicate examples of the three ommatidial types in wild type (above) and mutant (below) tissues.
In summary, CRISPR knockout of ss produced ommatidia that exhibited all characterized features of type II (UV/UV) ommatidia. Therefore, Ss expression in the two Papilio dR7-like cells controls not only PxBlue vs. PxUV expression but also coordination of the stochastic decision across the entire ommatidium, effectively specifying multiple features required for visual function. While expression of Ss in Drosophila R7 instructs only R8 to express specific Rhodopsins4, Papilio dR7s influence the entire ommatidium, including the SVF outer photoreceptors.
The recruitment of two dR7-like cells that make independent binary stochastic choices allows for the production of three outcomes. Interestingly, the extra dR7-like butterfly photoreceptor is recruited in precisely the same place within the developing ommatidium where a cell known as the “mystery cell” has been described in Drosophila27. This enigmatic cell briefly begins to take on properties of a photoreceptor cell before disappearing27. It is possible that butterflies retain this extra cell during development as an additional dR7-like photoreceptor.
On average, type I is present at 50%, type II at 25%, and type III at 25% frequencies, which corresponds to a 50% probability of Ss expression in a given dR7-like photoreceptor (Extended Data Table1). Previous studies found no differences between PxBlue/PxUV and PxUV/PxBlue ommatidia (Ss on/off vs. off/on) in Papilio1,8, suggesting that the two dR7 cells are equivalent before the stochastic decision to express Ss. In Drosophila, 65% of R7 cells have Sson. Honeybees also have nine photoreceptors per ommatidium and three stochastically specified ommatidial types but have a different ratio of ommatidial types28,29. We predict that bees also have two dR7-like cells3,16,28 with a probability of 30% for Ss expression instead of 50% in butterflies (Extended Data Table 1). The mechanisms controlling stochastic Ss expression ratios are as yet unknown.
This work provides evidence that our extensive knowledge of patterning in the Drosophila visual system allows us to understand how vision has evolved in other insects. Expression of Sal, Pros, and Sens is deeply conserved and they are reliable markers for establishing photoreceptor homology. Stochastic patterning mechanisms involving Ss and Dve are also shared. Adaptations for specific visual requirements can occur through modification of the developmental network that patterns the eye, such as the production of two dR7-like cells in butterflies. Importantly, the SVF photoreceptors must be able to determine whether none, one, or both R7-like cells have chosen to express Ss in order to choose which Rhodopsin(s) and pigments to express. This coordination is important for the function of each type of ommatidial unit. It will be interesting to determine whether the evolution of additional ommatidial types required coordinated changes in the brain for visual information processing.
Methods
Animals
Papilio xuthus were reared in the laboratory in Kanagawa, Japan using a line derived from wild-caught females. The larvae were fed on fresh citrus leaves under a light regime of 14 hours light and 10 hours dark at 28C; these conditions produce non-diapausing spring form adults. Adults eclosed approximately 10 days post-pupation.
Vanessa cardui and artificial diet for larval culture were obtained from Carolina Biological Supply. Larvae were reared individually in disposable plastic 1oz containers. Adults were maintained on sugar water and fruit slices in desktop population cages under artificial light. Egg collections were performed using stems of cut sunflowers.
Antibody generation
Proteins were produced by GenScript (Piscataway, NJ) and purified to >80% purity. Codon optimized gene synthesis was used to generate the sequences of interest for expression in bateria. GenScript performed protein injections into host animals, collected serum for testing, and performed affinity purification for each antibody (except PxDve). Antibodies were tested at a range of concentrations and fixation conditions in freshly fixed eye and embryo tissue. Embryo staining patterns provided an indication of whether staining might be specific; early transcription factor expression patterns are often highly conserved and stains showed roughly Drosophila-like patterns. Staining in the eye in subsets of the appropriate cell types was used as another indication; we show that these regulatory networks are highly conserved and the staining patterns match morphological features as well as each other (e.g. Dve correlates with Ss in R7-like photoreceptors). Further evidence of antibody specificity was provided by targeted knockout experiments showing loss of expression patterns of the target (Ss) or by changes in expression patterns of downstream targets (Dve, Rhodopsins). Rhodopsin expression patterns had been previously determined via in situ hybridization and Rhodpsin antibodies were confirmed to be mutually exclusive and in the correct patterns.
Sequences used for protein production:
PxDve: SHSPSNGSLMASNDNYAMQDRNSMKSPMQMSGSPGRYPMSIMSEDNLSNAGSDLEDDGGDLNPDDRPESPDAPL SLITTKKNNNDEEMSKQSPKPPDIIKVHDIKQELRDLTRSDQTNSPQRSPDKNSDSSHNNNNNVKEENGITDEQ DVASDDDIVQERHYRPSTPHLDRLPFPMVPNHPMFGHGIMYMSQYMTGFPGVGPVPGEGASGLNLALAGASDER RKRNRTFIDPVSEVPVLEQWFSMNTHPSHNLILKYTEELNRMPYRQKFPRLESKNVQFWFKNRRAKCKRLKMSL YEPSSPSHYSHPGHHAIA PxPros: PMHLRKAKLMFFWVRYPSSAVLKMYFPDIKFNKNNTAQLVKWFSNFREFYYIQMEKYARQAISEGLKAADDLHV AGDSELYRVLNLHYNRNNHIEVPPNFRYVVEQTLREFFRAIQGGKDTEQSWKKSIYKVISRLDDPVPEYFKSPN FLEQLE PxSs: GAYTGPYGEYPPAPALHYAPPPLDDRFLAADNLFHQYKPLPYHYTPYAPNGFLEPAPPPGYEVAPTYHRPPSRE YTYVDSTGRYMSPVTGQERRSPSVVPGPSPGGSSGSTEQDRLQQTQPSEIPRQTVLMWGAGGTAQEVPVEYSPP QVWRYHQSHYHTAEATQ VcSens: GVDHARHTPPRDDDEEELPLNLSMKNRQIWSPASVCEREQVETERESPMSRWDAHDDPDSPLELVKRCRSNPDE KRPSSTEPLRCPSAPAYHSYPPPPPPPADINISLLLKTENKNEKSFQCKQCGKCFKRSSTLSTHLLIHSDTRPY PCQYCGKRFHQKSDMKKHTYIHTGEKPHKCVVCSKAFSQSSNLITHMRKHTGYKPFSCGLCDKAFQRKVDLRRH RESQHSEVDPPSSIALSQNRYRFYGDDQTSLAPISSN PxBlue: INHPRYRAELQKRLPC PxUV: CISHPKYRQELQKRMP PxG2: CAIANLEPGMGASEA
Immunofluorescence
Antibody staining was performed as described previously for Drosophila pupal retinas31. The following antibodies concentrations were used: Guinea pig anti-Sal32,33 at 1:10,000 (kind gift of Antonia Monteiro), and the antibodies generated in this work: Rat anti-PxPros 1:100, Rabbit anti-VcSens 1:300, Rabbit anti-PxSs 1:100, Guinea pig anti-PxDve 1:400, Rabbit anti-PxBlue 1:400, Guinea pig anti-PxUV 1:100, and Rabbit anti-PxG2 1:100. Pupal retina stains were performed at 2–6 days post-pupation, with ~72 hours post-pupation providing the most optimal staging. Images were captured using standard confocal microscopy on a Leica SP5.
CRISPR/Cas9 somatic mutagenesis
We used T7 transcription-based in vitro reactions to generate single guide RNAs (sgRNAs) from target-specific gBlock DNA fragments ordered from Integrated DNA Technologies. We used a modified “F+E” sgRNA scaffold to prevent premature transcriptional termination34. These sgRNAs were injected with recombinant Cas9-NLS protein (PNA Bio).
CRISPR gRNAs
Target sequences were identified by eye and tested for uniqueness by BLASTing against the respective transcriptome and genome sequences. Regions that did not contain polymorphism at the target site were preferred. Target sequences used are as follows, with “Px” for Papilio xuthus and “Vc” for Vanessa cardui:
Px_y1_gRNA: GACAGATGCGACAGACTC Px_y2_gRNA: GGTACACTGTGGTGAGTC Px_ss2_gRNA: CAACATTTCAGGAAGAGG Px_ss3_gRNA: CACATACCAGGAATCCAG Vc_y1_gRNA: CTCTGACGAGCTCGGCTA Vc_y2_gRNA: GCCTTATCGCTTACTCCT Vc_ss1_gRNA: TAAGAGACGACGGAGGAA Vc_ss2_gRNA: TCTCGGTGCCTCTTGCTC
These targets were incorporated into the following scaffold sequence in place of the “N”s:
This sequence is composed of: M13_spacer_T7 promoter_target_scaffold_M13R. The region of target-specific sequence is underlined.
The first base pair of the target sequence was in each case changed to a “G” for efficient in vitro T7 transcription; the sequence at this 5’ position is not required for targeting. The scaffold sequence used was from 34. M13F and M13R sequences flank the fragment and are useful if additional amplification is necessary. M13F is followed by a short spacer sequence and the T7 promoter.
Preparation of injection constructs
Templates were ordered as “gBlocks” (synthesized DNA fragments) from Integrated DNA Technologies. These were PCR amplified using M13F and an internal reverse primer AAAAAAAGCACCGACTCGGTGCC. For a given sgRNA, products from four 50uL PCR reactions were pooled and purified on single Qiagen spin columns. Resulting template was used for in vitro transcription using the AmpliScribe T7-flash Transcription Kit from Epicentre/Illumina to produce sgRNAs and followed by ammonium acetate precipitation for purification. Products were resuspended in Qiagen buffer EB, quantified by spectrophotometry using a NanoDrop, and stored at −80C until injection. They were then mixed with Cas9-NLS protein from PNA Bio (catalog number CP01) at a final concentration of 250–500ng/uL of sgRNA and of Cas9 protein.
Injections
Embryo injections were performed using a Narshige micromanipulator and a 50mL syringe with pressure applied by hand. Needles were pulled using a Sutter Instrument micropipette puller and needle tips were broken by hand using forceps against a glass slide. Needles were front-loaded under the microscope with .8uL volumes of injection construct.
Embryos were collected for 0–4 hours post laying and either injected immediately for high percentage, biallelic somatic knockout or aged until 5–8 hours after eggs were laid for generation of mosaic animals. Eggs were surface sterilized using ~5 minute submersion in 8% benzalkonium chloride solution, which also helps remove the “glue” used to secure eggs upon laying. Eggs were then briefly rinsed and transferred to a glass microscope slide using a damp paint brush, and blunt forceps were used to position embryos onto thin strips of double-sided tape. After injection, embryos were placed into humid petri dishes until hatching (~4 days). Hatchlings were collected via paintbrush and transferred to leaves or artificial diet.
Yellow mutation is apparent in early larval head capsules. Wild-type head capsules in both Papilio and Vanessa are black, with mutant tissue appearing light brown or tan. In Vanessa, larval spines that are normally black in instars 3–4 become lighter in coloration in Y- tissue. Spineless mutation has no apparent effect until pupation when mutant antennae provide indication of successful ss knockout. Variability in the severity of antennal defects produced provide an early estimate of knockout efficiency. One yellow mutation was isolated and maintained as a viable line for several generations.
Mutation sequencing
DNA was isolated from tissue showing appropriate phenotypes using the Qiagen DNeasy kit. PCR products were cloned using the pGEM-T Easy Vector system and sequenced with M13R primers. Examples of the resulting sequences are presented in Extended Data Figure 2e. The primers used for PCR amplifications were:
565VcSs1MutSeqF1: ATACGAGCCAGCTTGGTACC 566VcSs1MutSeqR1: GTCTTTTGATGGTGGCTTG 567VcSs2MutSeqF1: TCAAGCCACCATCAAAAGAC 568VcSs2MutSeqR1: GACCACTTGAAAATAGCTC 569VcYMutSeqF1: CTTTAGACGCTCCCTATGAAC 570VcYMutSeqR1: ATGTTGAAGTCTCCGACAAG 595VcSs1F2: AGTTACAATTGTGCGGCTGG 596VcSs2R2: TGGTCCGTAAATAGCTGACG
Results from Vanessa cardui
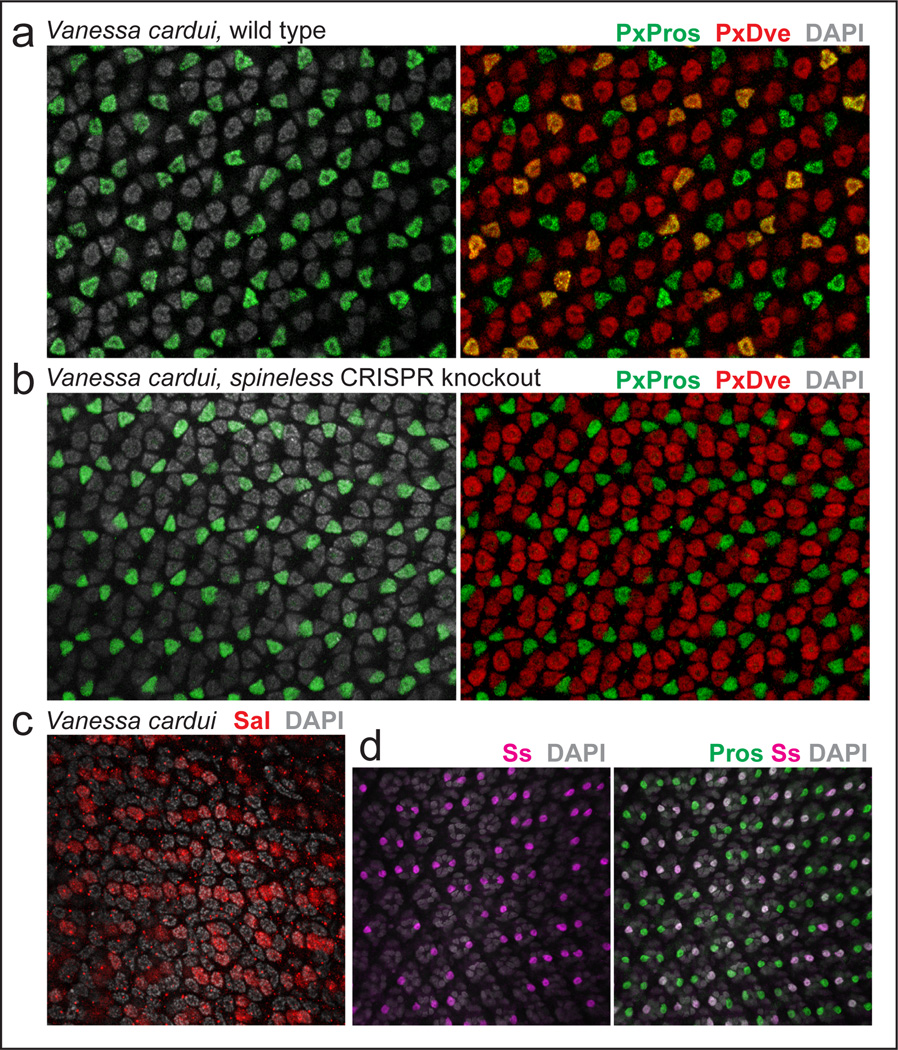
Results from Vanessa cardui are summarized in Extended Data Fig. 1. Vanessa cardui also have two photoreceptors per ommatidium labeled with Pros, three per ommatidium labeled with Sal, and a stochastic mosaic of Ss and Dve expression. Knockout of Ss via CRISPR/Cas9 produces fields of uniform Dve-negative R7s, as in Papilio xuthus. There are no known coordinated features of ommatidial type in Vanessa cardui; they express a single green Rhodopsin in all “outer”, SVF photoreceptors and have uniform pigmentation. This lack of coordinated features such as red/yellow screening pigments or differential Rhodopsin expression in the SVF, proximal tier prevented an assessment of ommatidial coordination in Vanessa. That said, Ss expression clearly causes loss of downstream Dve expression, as in Papilio, and therefore does account for known features of ommatidial type. Ss expression is presumably coordinated with the UV/UV type (of the three UV/UV, UV/B, and B/B types), as in Papilio, though this was not tested directly in Vanessa.
Adult morphology and staining
Yellow and red pigment was viewed using light microscopy of plastic sections, as described in 35. UV fluorescence was assessed by mounting live animals on a goniometer for viewing under UV illumination. Rhodopsin antibodies were initially tested on hand-dissected fragments of adult retina tissue. This approach did not provide clear images of spatial patterns and so a thick-sectioning approach was used. Corneal lenses were hand-removed from dissected retinas after an initial fixation as described in 36. Retinas were then embedded in gelatin and cut into 100uM sections, as in 36, with the overnight fixation concentration adjusted from 8% to 6%. Sectioned tissue was then stained following standard protocols31.
Extended Data
Extended Data Figure 1. Additional expression data and CRISPR/Cas9 spineless knock out results for Vanessa cardui.
a, Antibodies to Papilio Pros and Dve (PxPros and PxDve) cross-react in Vanessa cardui. As in Papilio, Pros labels two R7-like photoreceptors per ommatidium (green) while Dve labels bR3–8, the SVF photoreceptors equivalent to the “outer” photoreceptors of Drosophila (red), along with a stochastic subset of Pros-expressing R7-like PRs (yellow co-expression).
b, Coexpression of Dve in Pros-positive R7-like photoreceptors is lost in ss CRISPR/Cas9 knockout tissue; compare to yellow co-expression in wild-type in (a).
c, Sal antibodies label three photoreceptors per ommatidium in Vanessa (red).
d, As in Papilio, Ss antibodies (magenta) label a stochastic subset of R7-like Pros expressing photoreceptors (green).
Extended Data Figure 2. Pleiotropic effects of spineless mutation and sequencing of CRISPR/Cas9 generated mutations.
a, b, spineless mutation affects antennal development in Papilio (a) and Vanessa (b), as shown previously for Drosophila. This effect is first visible at pupal stages, where missing/shortened antennae are absent in their normal channels (red arrow) and shortened structures protrude in their place (blue arrow). Strongly affected individuals have little to no remaining antennae in adult stages (at right).
c, spineless mutation affects bristle development in Vanessa, as shown previously for Drosophila, where some bristles are missing or reduced. At left, the regular comb of bristles on the tibia is interrupted in mosaic ss mutant adults, or almost completely missing (right).
d, Mutation of ss produced an unexpected effect on wing color pattern. The wing of an animal injected with guideRNAs targeting both ss and yellow during the late blastoderm stage (7–9 hours after egg lay) is shown at left. A brown-colored yellow mutant patch of tissue is visible (yellow arrowhead), as shown in for yellow mutation in Figure 3, but lighter patches of wing scales lacking both melanin (black) and ommachromes (oranges) are also visible, example marked with white arrow. Similar clones were observed when only ss-gRNAs were injected (middle, right). This effect was observed for two independent gRNAs targeting Ss.
e, Cloning and sequencing of target regions from mutant tissues reveals a mixture of CRISPR/Cas9 generated mutations. Unmodified cDNA sequences are shown in the top line for comparison.
Extended Data Table 1. Specific probabilities of Ss expression yield specific ratios of ommatidial type.
If Spineless has a specific, single probability of being expressed and if the two R7-like cells make this decision independently then a specific ratio of the three ommatidial types will be produced. These ratios are shown for example probabilities. If on/off and off/on (type Ia and type Ib) are equivalent these types can be combined, shown below. The ratio observed for Papilio supports the idea that the two R7-like cells are independent and, on average, have a 50% probability of Spineless expression. Interestingly a different ratio that matches this model has previously been observed for the honeybee Apis mellifera (see Discussion).
- Ss+; Ss+ p1*p2
- Ss+; Ss− p1*(1-p2)
- Ss−; Ss+ (1-p1)*p2
- SS−; Ss− (1-p1)*(1-p2)
- p*p
- p*(1-p)
- p*(1-p)
- (1-p)*(1-p)
| Probability | Type II | Type Ia | Type Ib | Type III |
|---|---|---|---|---|
| 0.1 | 0.01 | 0.09 | 0.09 | 0.81 |
| 0.2 | 0.04 | 0.16 | 0.16 | 0.64 |
| 0.3 | 0.09 | 0.21 | 0.21 | 0.49 |
| 0.4 | 0.16 | 0.24 | 0.24 | 0.36 |
| 0.5 | 0.25 | 0.25 | 0.25 | 0.25 |
| 0.6 | 0.36 | 0.24 | 0.24 | 0.16 |
| 0.7 | 0.49 | 0.21 | 0.21 | 0.09 |
| 0.8 | 0.64 | 0.16 | 0.16 | 0.04 |
| 0.9 | 0.81 | 0.09 | 0.09 | 0.01 |
| With on/off and off/on combined: | |||
|---|---|---|---|
| Probability | Type II | Type I | Type III |
| 0.1 | 0.01 | 0.18 | 0.81 |
| 0.2 | 0.04 | 0.32 | 0.64 |
| 0.3 | 0.09 | 0.42 | 0.49 |
| 0.4 | 0.16 | 0.48 | 0.36 |
| 0.5 | 0.25 | 0.5 | 0.25 observed for Papilio |
| 0.6 | 0.36 | 0.48 | 0.16 |
| 0.7 | 0.49 | 0.42 | 0.09 observed for Apis |
| 0.8 | 0.64 | 0.32 | 0.04 |
| 0.9 | 0.81 | 0.18 | 0.01 |
Acknowledgments
We thank members of the Desplan and Arikawa labs for discussion, and especially Mathias Wernet and Jens Rister for suggestions. We thank Markus Friedrich for clarifying insect eye homologies, Alberto Stolfi for discussing CRISPR/Cas9 protocols, Christine Merlin for discussing butterfly injection technique, and Jacques Bothma for discussion. We thank Antonia Monteiro for providing anti-Sal and Kevin Shi at Genscript for help with antibody design. This work was supported by NIH grant EY13010 and the Center for Genomics and Systems Biology of NYU Abu-Dhabi to C.D., and the JSPS Kakenhi grants # 26251036 and # 20167232 to K.A. M.P. was supported by an NIH Ruth L. Kirschstein NRSA, a JSPS Short Term Fellowship award, and the Revson Biomedical Research Foundation Postdoctoral Fellowship.
Footnotes
Contributions
M.P. and C.D. jointly conceived the project with input from K.A. M.P., C.D., and K.A. designed experiments. M.P., M.K., and L.H. performed experiments. G.S. was responsible for genomic data analysis and de novo assembly. C.D. and K.A. supervised the project. M.P., K.A., and C.D. wrote the paper. All authors discussed the results and manuscript.
Accession codes
Sequence Read Archive
SRP071804
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Arikawa K. Spectral organization of the eye of a butterfly, Papilio. J. Comp. Physiol. A. Neuroethol. Sens. Neural. Behav. Physiol. 2003;189:791–800. doi: 10.1007/s00359-003-0454-7. [DOI] [PubMed] [Google Scholar]
- 2.Briscoe AD. Reconstructing the ancestral butterfly eye: focus on the opsins. J. Exp. Biol. 2008;211:1805–1813. doi: 10.1242/jeb.013045. [DOI] [PubMed] [Google Scholar]
- 3.Wernet MF, Perry MW, Desplan C. The evolutionary diversity of insect retinal mosaics: common design principles and emerging molecular logic. Trends Genet. 2015;31:316–328. doi: 10.1016/j.tig.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wernet MF, et al. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 2006;440:174–180. doi: 10.1038/nature04615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briscoe AD. Six opsins from the butterfly Papilio glaucus: molecular phylogenetic evidence for paralogous origins of red-sensitive visual pigments in insects. J. Mol. Evol. 2000;51:110–121. doi: 10.1007/s002390010071. [DOI] [PubMed] [Google Scholar]
- 8.Koshitaka H, Kinoshita M, Vorobyev M, Arikawa K. Tetrachromacy in a butterfly that has eight varieties of spectral receptors. Proc. Biol. Sci. 2008;275:947–954. doi: 10.1098/rspb.2007.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franceschini N, Kirschfeld K, Minke B. Fluorescence of photoreceptor cells observed in vivo. Science. 1981;213:1264–1267. doi: 10.1126/science.7268434. [DOI] [PubMed] [Google Scholar]
- 10.Rister J, Desplan C. The retinal mosaics of opsin expression in invertebrates and vertebrates. Dev. Neurobiol. 2011;71:1212–1226. doi: 10.1002/dneu.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou WH, et al. Patterning of the R7 and R8 photoreceptor cells of Drosophila: evidence for induced and default cell-fate specification. Development. 1999;126:607–616. doi: 10.1242/dev.126.4.607. [DOI] [PubMed] [Google Scholar]
- 12.Johnston RJ, et al. Interlocked feedforward loops control cell-type-specific Rhodopsin expression in the Drosophila eye. Cell. 2011;145:956–968. doi: 10.1016/j.cell.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papatsenko D, Sheng G, Desplan C. A new rhodopsin in R8 photoreceptors of Drosophila: evidence for coordinate expression with Rh3 in R7 cells. Development. 1997;124:1665–1673. doi: 10.1242/dev.124.9.1665. [DOI] [PubMed] [Google Scholar]
- 14.Kitamoto J, Sakamoto K, Ozaki K, Mishina Y, Arikawa K. Two visual pigments in a single photoreceptor cell: identification and histological localization of three mRNAs encoding visual pigment opsins in the retina of the butterfly Papilio xuthus. J. Exp. Biol. 1998;201:1255–1261. doi: 10.1242/jeb.201.9.1255. [DOI] [PubMed] [Google Scholar]
- 15.Kinoshita M, Arikawa K. Color and polarization vision in foraging Papilio. J. Comp. Physiol. A. Neuroethol. Sens. Neural. Behav. Physiol. 2014;200:513–526. doi: 10.1007/s00359-014-0903-5. [DOI] [PubMed] [Google Scholar]
- 16.Friedrich M, Wood EJ, Wu M. Developmental evolution of the insect retina: insights from standardized numbering of homologous photoreceptors. J. Exp. Zool. B. Mol. Dev. Evol. 2011;316:484–499. doi: 10.1002/jez.b.21424. [DOI] [PubMed] [Google Scholar]
- 17.Mollereau B, et al. Two-step process for photoreceptor formation in Drosophila. Nature. 2001;412:911–913. doi: 10.1038/35091076. [DOI] [PubMed] [Google Scholar]
- 18.Takemura S-Y, Kinoshita M, Arikawa K. Photoreceptor projection reveals heterogeneity of lamina cartridges in the visual system of the Japanese yellow swallowtail butterfly, Papilio xuthus. J. Comp. Neurol. 2005;483:341–350. doi: 10.1002/cne.20446. [DOI] [PubMed] [Google Scholar]
- 19.Xie B, Charlton-Perkins M, McDonald E, Gebelein B, Cook T. Senseless functions as a molecular switch for color photoreceptor differentiation in Drosophila. Development. 2007;134:4243–4253. doi: 10.1242/dev.012781. [DOI] [PubMed] [Google Scholar]
- 20.Cook T, Pichaud F, Sonneville R, Papatsenko D, Desplan C. Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila. Dev Cell. 2003;4:853–864. doi: 10.1016/s1534-5807(03)00156-4. [DOI] [PubMed] [Google Scholar]
- 21.Arikawa K, Mizuno S, Kinoshita M, Stavenga DG. Coexpression of two visual pigments in a photoreceptor causes an abnormally broad spectral sensitivity in the eye of the butterfly Papilio xuthus. J. Neurosci. 2003;23:4527–4532. doi: 10.1523/JNEUROSCI.23-11-04527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thanawala SU, et al. Regional modulation of a stochastically expressed factor determines photoreceptor subtypes in the Drosophila retina. Dev. Cell. 2013;25:93–105. doi: 10.1016/j.devcel.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burgess EA, Duncan I. Direct control of antennal identity by the spineless-aristapedia gene of Drosophila. Mol. Gen. Genet. 1990;221:347–357. doi: 10.1007/BF00259398. [DOI] [PubMed] [Google Scholar]
- 24.Kitamoto J, Ozaki K, Arikawa K. Ultraviolet and violet receptors express identical mRNA encoding an ultraviolet-absorbing opsin: identification and histological localization of two mRNAs encoding short-wavelength-absorbing opsins in the retina of the butterfly Papilio xuthus. J. Exp. Biol. 2000;203:2887–2894. doi: 10.1242/jeb.203.19.2887. [DOI] [PubMed] [Google Scholar]
- 25.Arikawa K, Stavenga D. Random array of colour filters in the eyes of butterflies. J. Exp. Biol. 1997;200:2501–2506. doi: 10.1242/jeb.200.19.2501. [DOI] [PubMed] [Google Scholar]
- 26.Arikawa K, et al. An ultraviolet absorbing pigment causes a narrow-band violet receptor and a single-peaked green receptor in the eye of the butterfly Papilio. Vision Res. 1999;39:1–8. doi: 10.1016/s0042-6989(98)00070-4. [DOI] [PubMed] [Google Scholar]
- 27.Tomlinson A, Bowtell DDL, Hafen E, Rubin GM. Localization of the sevenless protein, a putative receptor for positional information, in the eye imaginal disc of Drosophila. Cell. 1987;51:143–150. doi: 10.1016/0092-8674(87)90019-5. [DOI] [PubMed] [Google Scholar]
- 28.Ready DF. A multifaceted approach to neural development. Trends Neurosci. 1989;12:102–110. doi: 10.1016/0166-2236(89)90166-5. [DOI] [PubMed] [Google Scholar]
- 29.Wakakuwa M, Kurasawa M, Giurfa M, Arikawa K. Spectral heterogeneity of honeybee ommatidia. Naturwissenschaften. 2005;92:464–467. doi: 10.1007/s00114-005-0018-5. [DOI] [PubMed] [Google Scholar]
- 30.Rister J, Desplan C, Vasiliauskas D. Establishing and maintaining gene expression patterns: insights from sensory receptor patterning. Development. 2013;140:493–503. doi: 10.1242/dev.079095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 31.Hsiao HY, et al. Dissection and immunohistochemistry of larval, pupal and adult Drosophila retinas. J Vis Exp. 2012:e4347. doi: 10.3791/4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoehr AM, Walker JF, Monteiro A. Spalt expression and the development of melanic color patterns in pierid butterflies. Evodevo. 2013;4:6. doi: 10.1186/2041-9139-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Celis JF, Barrio R, Kafatos FC. Regulation of the spalt/spalt-related gene complex and its function during sensory organ development in the Drosophila thorax. Development. 1999;126:2653–2662. doi: 10.1242/dev.126.12.2653. [DOI] [PubMed] [Google Scholar]
- 34.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arikawa K, Scholten DGW, Kinoshita M, Stavenga DG. Tuning of Photoreceptor Spectral Sensitivities by Red and Yellow Pigments in the Butterfly Papilio xuthus. Zoolog. Sci. 1999;16:17–24. [Google Scholar]
- 36.Kinoshita M, Shimohigasshi M, Tominaga Y, Arikawa K, Homberg U. Topographically distinct visual and olfactory inputs to the mushroom body in the Swallowtail butterfly, Papilio xuthus. J. Comp. Neurol. 2015;523:162–182. doi: 10.1002/cne.23674. [DOI] [PubMed] [Google Scholar]