Abstract
The genus Torulomyces was characterised by species that typically have conidiophores consisting of solitary phialides that produce long chains of conidia connected by disjunctors. Based on the phylogenetic position of P. lagena (generic ex-neotype), the genus and its seven species were transferred to Penicillium and classified in sect. Torulomyces along with P. cryptum and P. lassenii. The aim of this study was to review the species currently classified in sect. Torulomyces using morphology and phylogenies of the ITS, BenA, CaM and RPB2 regions. Based on our results, we accept 16 species in sect. Torulomyces, including 12 new species described as P. aeris, P. austricola, P. cantabricum, P. catalonicum, P. oregonense, P. marthae-christenseniae, P. riverlandense, P. tubakianum, P. variratense, P. williamettense, P. wisconsinense and P. wollemiicola. In addition, we reclassify P. laeve and P. ovatum in sect. Exilicaulis and correct the typification of P. lagena. We provide descriptions and notes on the identification of the species.
Keywords: beta-tubulin, calmodulin, Eupenicillium, internal transcribed spacer rDNA region, low temperature Scanning Electron Microscopy (cryo-SEM), Monocillium, RNA polymerase II second largest subunit, Trichocomaceae, Wollemi pine
INTRODUCTION
Delitsch (1943) introduced the hyphomycete genus Torulomyces with T. lagena as generic type. The genus included species producing conidiophores consisting of solitary, swollen phialides borne on short, unbranched stipes (Delitsch 1943, Barron 1967, Gams 1971, Stolk & Samson 1983, Ando et al. 1998, Seifert et al. 2011). Conidia are linked together by connectives, i.e. short intercalary cylinders of cell wall material, to form very long chains (Gams 1971, Stolk & Samson 1983, Ando et al. 1998). Colonies of T. lagena and related species are brownish and their slow growth on agar media means that they are overgrown easily by other fungi and are infrequently isolated.
Stolk & Samson (1983) linked T. lagena with the sexual morph Eupenicillium limoneum, suggesting an association with Penicillium and introduced P. lagena as a new combination. Later, they proposed a sectional classification for Penicillium asexual morphs, including sect. Torulomyces for P. lagena (Stolk & Samson 1985). Pitt & Hocking (1985) argued that T. lagena did not fit the generic concept of Penicillium, at that time defined by a conidiophore with branches forming a penicillus. The transfer also was not accepted in the list of Current Names in Use for the Trichocomaceae (Pitt & Samson 1993, Pitt et al. 2000).
The taxonomy of such morphologically undifferentiated hyphomycetes is confusing, and prior to the availability of DNA-based phylogenies it was difficult to evaluate the phylogenetic significance of characters such as colony colours, phialide shape and the nature of connectives between individual conidia in chains. Apart from T. lagena, 10 similar fungi were classified as monophialidic species of Paecilomyces (Onions & Barron 1967), but following the conclusions of Gams (1971), the monograph of Samson (1974) excluded these from Paecilomyces. They were distributed by various authors into genera such as:
Monocillium S.B. Saksena (1955), sexual morphs in Niesslia (Niessliaceae, Hypocreales), including species with thick-walled, centrally swollen phialides, conidial chains lacking connectives, or often slimy rather than in chains, sometimes confused with Torulomyces (Hashmi et al. 1972);
Phialosimplex Sigler et al. (2010), sexual morphs unknown, including species with thin-walled, scarcely swollen mono- or polyphialides, and conidial chains with connectives, proposed as a syonynym of Aspergillus (Aspergillaceae, Eurotiales) by Houbraken & Samson (2011) and Samson et al. (2014), but perhaps phylogenetically distinct (Tanney, pers. comm.);
Sagenomella W. Gams (1978), sexual morphs Sagenoma (Trichocomaceae, Eurotiales), including species with thin-walled, unswollen mono- or polyphialides, and conidial chains with connectives;
Taifanglania Z.Q. Liang et al. (2009), sexual morphs unknown but Chaetomiaceae, Sordariales, including species with thin-walled, flask-shaped monophialides and conidial chains without connectives, now considered a synonym of Acrophialophora Edward (Zhang et al. 2015).
Thus, it is clear that the combination of solitary phialides with chains of ameroconidia is plesiomorphic, and its notable this combination of characters appears in several unrelated monophyletic groups, nested among species with more complex conidiophores. In particular, Phialosimplex appears to represent a reduced form of the vesiculate Aspergillus conidiophores, Taifanglania a reduced form of the verticillate Acrophialophora conidiophore, and Torulomyces a reduced form of the highly branched Penicillium conidiophore.
Following a four gene phylogenetic analysis of Penicillium, Aspergillus and related genera, Houbraken & Samson (2011) accepted Penicillium lagena as the correct classification for Delitsch’s T. lagena, and confirmed Torulomyces as a synonym of Penicillium. They transferred seven species from Torulomyces to Penicillium sect. Torulomyces, making the new combinations P. laeve, P. ovatum, P. parviverrucosum and P. porphyreum (for Monocillium humicola var. brunneum ≡ T. brunneus). Although they lack conidiophores with solitary phialides, P. cryptum and P. lassenii were included in the section. Torulomyces macrosporus was not transferred because it probably belongs in Monocillium based on the protologue of Matsushima (1987) (Ando et al. 1998). No type material was available for the second original species of Torulomyces, T. viscosus, and the original description is insufficient for determining the placement of the species (Stolk & Samson 1983, Houbraken & Samson 2011).
In this study, we re-evaluate the taxonomy of Penicillium sect. Torulomyces following the standardised methods and taxonomic approach advocated by Visagie et al. (2014b). We delineate species using phylogenies based on the nuc rDNA internal transcribed spacer region (ITS), and partial BenA (β-tubulin), CaM (calmodulin) and RPB2 (RNA polymerase II second largest subunit) gene sequences, as well as morphological characters. We describe 12 new species and provide descriptions for the six other accepted species, while also correcting the typification of P. lagena. Based on phylogenetic results, we classify P. laeve and P. ovatum in Penicillium sect. Exilicaulis. The most important morphological characters needed for species identification are summarised in table format.
MATERIALS AND METHODS
Strains
Strains used for this study (summarised in Table 1) were obtained from the CBS-KNAW Fungal Biodiversity Centre, the Netherlands (CBS) and the working collection of the Applied and Industrial Mycology department (DTO) at the same institute. Ex-type and representative cultures were also deposited in the Canadian Collection of Fungal Cultures, Ottawa (DAOMC). Reference material for P. laeve and P. ovatum was received from the Biological Resource Centre at the National Institute of Technology and Evaluation, Chiba, Japan (NBRC).
Table 1.
Strains used for the phylogenetic analysis.
| Species name | Collection accession nr. | Substrate, Locality | GenBank accession nr. |
|||
|---|---|---|---|---|---|---|
| ITS | BenA | CaM | RPB2 | |||
| P. aeris | CBS 135897T = DTO 207D4 | Unknown origin | KF303654 | KF303614 | KF303627 | KF303681 |
| P. austricola | CBS 135899 = CV 1811 = DTO 186D8 | Mite from Protea repens infructescens, Struisbaai, South Africa | KF303672 | JX091578 | JX141599 | KF303704 |
| CBS 135900T = CV 1842 = DTO 183E6 = DAOM 241066 | Mite from Protea repens infructescens, Struisbaai, South Africa | JX091466 | JX091579 | JX141600 | KF303705 | |
| CBS 135901 = CV 1902 = DTO 183F5 | Bract from Protea repens infructescens, Struisbaai, South Africa | JX091467 | JX091586 | JX141602 | KF303703 | |
| CBS 135902 = CV 1943 = DTO 183F8 = DAOMC 241067 | Mite from Protea repens infructescens, Struisbaai, South Africa | JX091468 | JX091587 | JX141603 | KF303701 | |
| CBS 135903 = CV 1954 = DTO 183F9 | Mite from Protea repens infructescens, Struisbaai, South Africa | JX091469 | JX091588 | JX141604 | KF303699 | |
| CBS 135904 = CV 1808 = DTO 183D8 = DAOMC 241065 | Mite from Protea repens infructescens, Struisbaai, South Africa | JX091465 | JX091584 | JX141598 | KF303702 | |
| CBS 136253 = CV 1968 = DTO 186G3 | Mite from Protea repens infructescens, Struisbaai, South Africa | KF303671 | JX091589 | JX141605 | KF303700 | |
| P. cantabricum | CBS 120415T = DTO 76I9 = FMR 9121 | Soil, Cantabria, Reinosa, Spain | KF303655 | KF303615 | KF303646 | KF303682 |
| P. catalonicum | CBS 110532T = DTO 78H5 | Soil, Catalonia, Montseny Natural Park, Spain | KF303650 | KF303609 | KF303644 | KF303683 |
| P. corylophilum | CBS 330.79 = IJFM 5147 | Air sample, Barcelona, Spain | GU944557 | GU944519 | GU944607 | JN406569 |
| P. cryptum | CBS 271.89T = DTO 122C9 = ATCC 60138 = IMI 296794 = NRRL 13460 | Soil from Quercus-Betula forest, New York, USA | KF303647 | KF303608 | KF303628 | JN121478 |
| P. dimorphosporum | CBS 456.70T = NRRL 5207 = ATCC 22783 = ATCC 52501 = FRR 1120 = IMI 149680 | Mangrove swamp soil, Tooraddin, Australia | AF081804 | KJ834448 | KP016783 | JN121517 |
| P. laeve | CBS 136665T = DTO 270G8 = KY 12727 = NBRC 109724 | Soil, unknown, Thailand | KF667369 | KF667365 | KF667367 | KF667371 |
| P. lagena | CBS 129212 = DTO 199C2 | Unknown, Halsey National forest, Nebraska, USA | KF303662 | KF303616 | KF303631 | KF303680 |
| CBS 129389 = DTO 202C5 | Unknown, Wisconsin, USA | KF303663 | KF303617 | KF303632 | KF303679 | |
| CBS 129620 = DTO 206D2 | Unknown, Minnesota, USA | KF303664 | KF303618 | KF303633 | KF303678 | |
| CBS 185.65T = DTO 77I8 = MUCL 8221 = JCM10149 = OAC10034 | Bog soil under Thuja plicata, Guelph, Canada | KF303665 | KF303619 | KF303634 | JN121450 | |
| P. lassenii | CBS 277.70T = DTO 95D6 = NRRL 5272 = ATCC 22054 = FRR 858 = IMI 148395 | Soil under conifers, California, USA | KF303648 | KF303607 | KF303629 | JN121481 |
| P. marthae-christenseniae | CBS 129213T = DTO 201B5 | Unknown, Wisconsin, USA | KF303651 | KF303613 | KF303645 | KF303711 |
| P. oregonensis | CBS 129775T = DTO 208A5 | Unknown, Williamette National Forest, near Blue River, Oregon, USA | KF303668 | KF303623 | KF303640 | KF303710 |
| P. ovatum | CBS 136664T = DTO 270G7 = KY 12726 | Soil under Pinus caribaea leaf litter, Kuala Lumpur, Malaysia | KF667370 | KF667366 | KF667368 | KF667372 |
| P. porphyreum | CBS 132154 = DTO 210E2 = WSF 3133 | Unknown, Wisconsin, USA | KF303661 | KF303620 | KF303635 | KF303676 |
| CBS 382.64T = DTO 78G7 = KY 12723 | Soil in community of Pinus strobus, Wisconsin, USA | KF303666 | KF303621 | KF303636 | KF303677 | |
| P. restrictum | CBS 367.48T = ATCC 11257 = FRR 1748 = IMI 040228 = NRRL 1748 | Soil, Honduras | AF033457 | KJ834486 | KP016803 | JN121506 |
| P. riverlandense | CBS 135883 = CV 1360 = DTO 183A4 = DAOMC 241061 | Bract from Protea repens infructescens, Malmesbury, South Africa | JX091458 | JX091576 | JX141594 | KF303686 |
| CBS 135884 = CV 1448 = DTO 183A9 = DAOMC 241063 | Bract from Protea repens infructescens, Malmesbury, South Africa | JX091459 | JX091582 | JX141595 | KF303684 | |
| CBS 135885 = CV 1450 = DTO 183B1 = DAOMC 241064 | Bract from Protea repens infructescens, Malmesbury, South Africa | JX091460 | JX091583 | JX141596 | KF303695 | |
| CBS 135886 = CV 1583 = DTO 186B8 | Bract from Protea repens infructescens, Malmesbury, South Africa | KF303657 | JX091577 | JX141597 | KF303689 | |
| CBS 135887 = CV 2815 = DTO 184A1 | Soil, Malmesbury, South Africa | KF303673 | JX091590 | JX141606 | KF303691 | |
| CBS 135888 = CV 2819 = DTO 184A4 | Soil, Malmesbury, South Africa | JX091462 | JX091591 | JX141607 | KF303693 | |
| CBS 135889 = CV 2822 = DTO 184A6 | Soil, Malmesbury, South Africa | JX091463 | JX091592 | JX141608 | KF303694 | |
| CBS 135890 = CV 2831 = DTO 184B4 | Soil, Malmesbury, South Africa | KF303658 | JX091593 | JX141612 | KF303696 | |
| CBS 135891 = CV 2833 = DTO 184B6 | Soil, Malmesbury, South Africa | JX091464 | JX091594 | JX141613 | KF303692 | |
| CBS 135892 = CV 2839 = DTO 184C2 | Soil, Malmesbury, South Africa | KF303659 | JX091595 | JX141615 | KF303697 | |
| CBS 135893 = CV 2849 = DTO 184C6 | Soil, Malmesbury, South Africa | KF303660 | JX091596 | JX141616 | KF303698 | |
| CBS 135894 = CV 0933 = DTO 185F9 | Soil, Malmesbury, South Africa | KF303656 | JX091575 | JX141591 | KF303690 | |
| CBS 135895 = CV 0959 = DTO 182E7 = DAOMC 241059 | Soil, Malmesbury, South Africa | JX091456 | JX091581 | JX141592 | KF303688 | |
| CBS 135896T = CV 0979 = DTO 182F6 = DAOMC 241060 | Soil, Malmesbury, South Africa | JX091457 | JX091580 | JX141593 | KF303685 | |
| DTO 183E8 = CV 1851 | Bract from Protea repens infructescens, Struisbaai, South Africa | JX091461 | JX091585 | JX141601 | KF303687 | |
| P. toxicarium | NRRL 6172 | Unknown, Strain of P. toxicarium Miyake fide Serra et al. (2008) | EF198650 | EF198620 | EF198631 | EF198499 |
| P. tubakianum | CBS 287.66T = DTO 138D9 = MUCL 8519 = IFO 8315 | Dead bark from Cyathea, New Zealand | KF303652 | KF303611 | KF303637 | KF303712 |
| P. variratense | CBS 337.97T = DTO 137C8 | Humus soil, Varirata National Park near Port Moresby, Papua New Guinea | KF303649 | KF303610 | KF303630 | KF303675 |
| P. williamettense | CBS 129774T = DTO 208A4 | Unknown, Williamette National Forest, near Blue River, Oregon, USA | KF303667 | KF303622 | KF303639 | KF303709 |
| P. wisconsinense | CBS 128279T = DTO 198H7 | Unknown, Wisconsin, USA | KF303670 | KF303624 | KF303641 | KF303706 |
| CBS 128412 = DTO 201B3 | Unknown, Wisconsin, USA | KF303674 | KF303625 | KF303642 | KF303707 | |
| CBS 128421 = DTO 198I5 | Unknown, Wisconsin, USA | KF303669 | KF303626 | KF303643 | KF303708 | |
| P. wollemiicola | CBS 137177T = DTO 297E3 | Wollemia nobilis needle debris, Australia | KJ174314 | KJ174315 | KJ174316 | KJ174313 |
DNA extraction, sequencing, and phylogenetic analysis
DNA extractions were made from colonies grown on OA for 8–10 d using the UltracleanTM Microbial DNA isolation Kit (MoBio, Solana Beach, USA). Subsequent PCR amplification and sequencing of the ITS, BenA, CaM and RPB2 gene regions, were done using methods described by Visagie et al. (2014b). Sequence contigs were assembled in DNASTAR® SeqMan v. 9.0.4. Newly generated sequences were deposited in GenBank (Table 1). Sequences were aligned in MAFFT v. 7.164 (Katoh & Standley 2013) with the L-INS-I option. For the multigene phylogeny, the datasets were concatenated in SeaView v. 4.4.1 (Gouy et al. 2010). Datasets were analysed using both Maximum Likelihood (ML) and Bayesian tree inference (BI). Trees and alignments were uploaded to TreeBASE (www.treebase.org) with submission ID 16300.
ML analyses were performed using MEGA v. 6.06 (Tamura et al. 2013). The most suitable substitution model for each dataset was selected using the model-test within MEGA, based on the lowest Bayesian information criterion (BIC) value. An initial tree was calculated with the Bio-Neighbour-Joining (BioNJ) option, with the subsequent Heuristic search done with Nearest-Neighbour-Interchange (NNI). A bootstrap analysis with 1 000 replicates was used for calculating node support.
For BI analyses, the most suitable substitution model for each dataset, according to the lowest Akaike information criterion (AIC) value, was selected using MrModeltest v. 2.3 (Nylander et al. 2004). Each analysis was run in MrBayes v. 3.2.1 (Ronquist & Huelsenbeck 2003), with two sets of four chains until the standard deviation of split frequencies reached 0.01. Sample frequency was set at 100 and 25 % of trees removed as burn in. ML phylograms were used for representing phylogenetic results, with both the bootstrap (bs) and posterior probability (pp) values, higher than 80 % bs and/or 0.95 pp given above thickened branch nodes.
Morphology
Standardised techniques for culturing conditions were used as described by Visagie et al. (2014b). Colony characters were recorded from strains grown on CYA (Czapek yeast autolysate agar), MEA (malt extract agar, Oxoid), YES (yeast extract sucrose agar), DG18 (dichloran 18 % glycerol agar), CYAS (CYA supplemented with 5 % NaCl), OA (oatmeal agar), SNA (Spezieller Nährstoffarmer agar) and CREA (creatine sucrose agar). All plates were incubated at 25 °C, with additional CYA plates incubated at 30 and 37 °C. Colour names and codes used for descriptions refer to the Methuen Handbook of Colour (Kornerup & Wanscher 1967).
Microscopic characters were recorded using both light-microscopy and low-temperature scanning electron microscopy (SEM). Microscopic preparations were made from 7–14-d-old cultures grown on OA, with 60 % lactic acid used as mounting fluid. Examinations were done using a Zeiss Axioskop 2 Plus light-microscope fitted with a Nikon microscope camera and pictures captured and analysed using Nikon NIS-elements D v. 4.0 software. In species descriptions, the average measurements and standard deviations are provided in brackets. Two-week-old colonies grown on OA were used for SEM observations. The technique used for SEM is similar to that described in Visagie et al. (2013). Photo plates were prepared using Adobe® Photoshop® Creative Suite v. 6. Photomicrographs were modified for aesthetic purposes using the healing brush tool, without altering areas of scientific significance.
RESULTS
Phylogeny
Phylogenies of all strains were prepared for the ITS region, and the partial BenA, CaM and RPB2 gene regions, and compared using the Genealogical Concordance Phylogenetic Species Recognition (GCPSR) concept (Taylor et al. 2000; Fig. 1). A multigene phylogeny of concatenated ITS, BenA, CaM and RPB2 sequences (Fig. 2) was prepared to evaluate the overall phylogenetic structure and the strength of support for nodes in sect. Torulomyces. The lengths of aligned datasets and substitution models selected for the analyses are provided in Table 2. Tree topologies did not differ between ML and BI analyses and ML trees are used here for presenting results, with BI pp values marked on relevant branches.
Fig. 1.
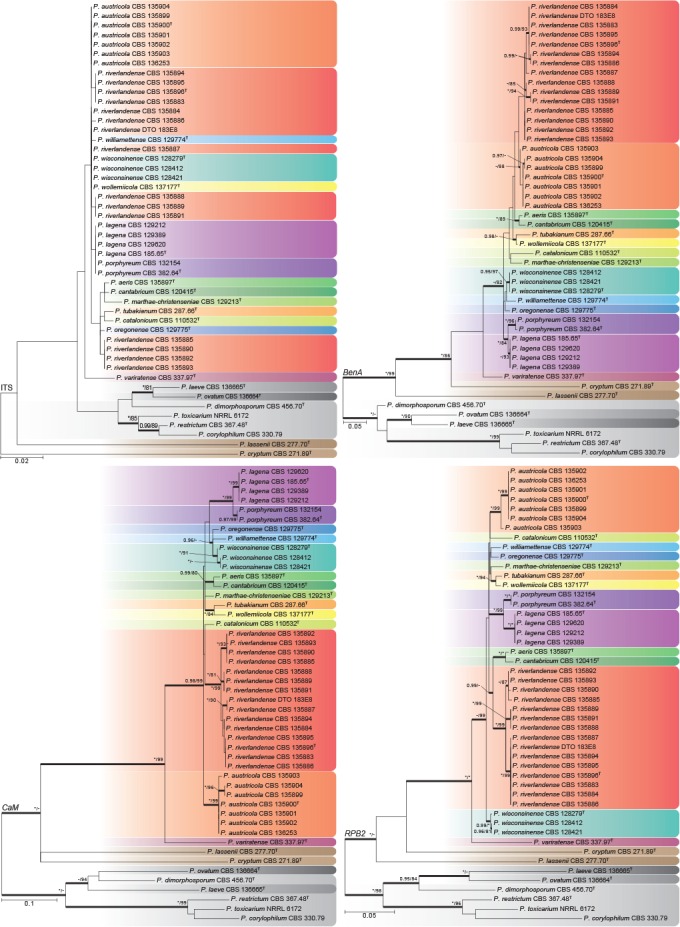
Phylogenetic trees for ITS, BenA, CaM and RPB2 datasets of Penicillium sect. Torulomyces. Branch support in nodes higher than 80 % bs and/or 0.95 pp are indicated above thickened branches (T = ex-type; * = 100 % bs or 1.00 pp; - = support lower than 80 % bs and/or 0.95 pp). Species are indicated by coloured blocks.
Fig. 2.
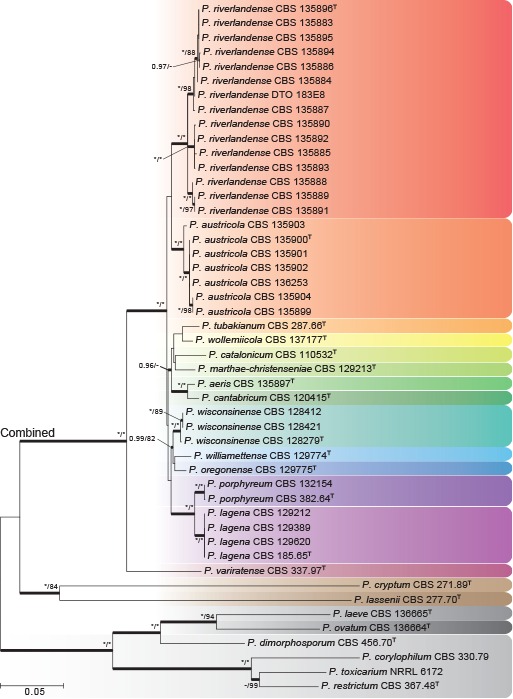
Multigene phylogeny for a combined ITS, BenA, CaM & RPB2 dataset of Penicillium sect. Torulomyces. Branch support in nodes higher than 80 % bs and/or 0.95 pp are indicated above thickened branches (T = ex-type; * = 100 % bs or 1.00 pp; - = support lower than 80 % bs and/or 0.95 pp). Species are indicated by coloured blocks.
Table 2.
Length of datasets and models used for phylogenetic analysis.
| ITS | BenA | CaM | RPB2 | Concatenated | |
|---|---|---|---|---|---|
| Length | 513 bp | 368 bp | 551 bp | 753 bp | 2185 bp |
| Model Maximum Likelihood | T92+G | K2+G | T92+G+I | TN93+G+I | TN93+G+I |
| Model Bayesian inference | GTR+G | HKY+G+I | HKY+G+I | GTR+G+I | GTR+G+I |
Abbreviations used in this table: T92 = Tamura 3-parameter; K2 = Kimura 2-parameter; TN93 = Tamura-Nei; GTR = General Time Reversible;
HKY = Hasegawa-Kishino-Yano; +G = Gamma distribution; +I = Invariant sites.
Phylogenies of BenA, CaM and RPB2 were congruent and variable enough to distinguish all species (Fig. 1b–d). The multigene phylogeny (Fig. 2) indicated the presence of 14 species in the main clade of sect. Torulomyces, with P. cryptum and P. lassenii distantly related. In the main clade, GCPSR confirmed the distinct nature of the previously described species P. lagena and P. porphyreum. A further 12 clades were recognized, and were congruent across the different phylogenies. These could be distinguished morphologically and are described as new species in the Taxonomy section below.
From the ITS phylogeny (Fig. 1a) it is apparent that the formal barcode for fungi (Schoch et al. 2012) does not work well for species identification in the section, with P. austricola, P. wisconsinense, P. wollemiicola and some strains of P. riverlandense sharing identical ITS sequences. The three alternative genes used here can distinguish between all species in the section, but we follow Visagie et al. (2014b) in proposing the use of BenA as an identification marker.
Some degree of variation in gene sequences occurred within P. austricola and P. riverlandense, the two species recovered from ecological surveys of the fynbos and Protea repens infructescences in South Africa. In the ITS tree, P. riverlandense strains had five different genotypes resolved into three main clades (red blocks in Fig. 1a). This conflicts with the other gene trees, where the same clades also occur, but are united in a monophyletic clade that is supported in the CaM (Fig. 1c) and RPB2 (Fig. 1d) phylogenies; all three clades are well-supported in the multigene phylogeny (Fig. 2). In P. austricola, there is one clade in the ITS tree, two in RPB2 and three in the CaM and BenA trees. These clades do not conflict, although one of them includes only a single strain (CBS 135903). Although it would be feasible to apply GCPSR and designate these clades within P. austricola and P. riverlandense as phylogenetic species, the lack of diagnostic phenotypic characters and their geographical and ecological coherence argues against this. Increased sampling of strains and genes should indicate whether our conservative approach is correct.
ITS sequences of P. lassenii and P. cryptum show that these species might not belong to sect. Torulomyces as defined here. They were basal to the section in the other phylogenies. Similarly, P. laeve and P. ovatum are shown to belong to sect. Exilicaulis with P. dimorphosporum as their closest relative.
Morphology
Strains used for phylogenetic analyses were grown for morphological analysis under standardised conditions, and the clades corresponded well with morphological differences. Colony growth rates on different media incubated at different temperatures were the most useful characters for identification. Conidiophores were essentially identical among the species of the section. The exceptions were P. lassenii, which produced mono- to biverticillate conidiophores, and P. cryptum, which produced mostly monoverticillate conidiophores, with a small proportion consisting of solitary phialides. Conidial shape varies from globose to ellipsoidal and is taxonomically informative. Conidial ornamentation was informative for species identification, but SEM observation is required for this. Penicillium cryptum, P. lassenii, P. laeve and P. ovatum, the species excluded from the section based on molecular results, produces smooth-walled conidia, not observed for other species in sect. Torulomyces s.str.
Morphological differences between the species described below are summarised in Table 3.
Table 3.
Summary of the most important morphological characters for species identification.
| Growth rate (in mm) |
Conidia |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CYA37°C | CYA30°C | CYAS | CREA | CYA | OA | MEA | YES | SNA | DG18 | Brown soluble pigments | Acid on CREA | Conidiophore branching pattern | Ornamentation | Shape | Tubercle size (μm) | Connectives | |
| P. aeris | 4–5 | 9–10 | 6–7 | no growth | 11–12 | 9–10 | 6–7 | 10–11 | 8–9 | 11–12 | absent | absent | solitary phialide | rough | globose | 0.21–0.4 | long, lack visible rings |
| P. austricola | no growth | 10–11 | 8–10 | 3–4 | 10–13 | 9–10 | 4–8 | 10–11 | 9–11 | 9–11 | present | absent | solitary phialide | rough | globose | 0.19–0.30 | long, lack visible rings |
| P. cantabricum | 3–5 | 12–13 | 3–4 | 3–4 | 13–14 | 10–11 | 6–7 | 9–10 | 7–8 | 8–9 | absent | weak | solitary phialide | rough | globose | 0.21–0.33 | present, lack visible rings |
| P. catalonicum | no growth | no growth | 2–3 | no growth | 6–8 | 6–7 | 4–5 | 5–6 | 6–7 | 7–8 | absent | absent | solitary phialide | rough, in parallel rings | subglobose to broadly ellipsoid | 0.12–0.18 | short, visible rings present |
| P. cryptum | no growth | 6–9 | no growth | no growth | 6–7 | 5–6 | 6–7 | 5–6 | 5–6 | 1–2 | absent | absent | solitary phialide to monoverticillate | smooth | broadly ellipsoidal | n.a. | short, lack visible rings |
| P. laeve | no growth | 4–5 | no growth | no growth | 8–9 | 7–8 | micro-colonies | 8–9 | 7–8 | 5–7 | absent | absent | solitary phialide | smooth | globose | n.a. | long, lack visible rings |
| P. lagena | no growth | 8–10 | no growth to 3 | no growth | 10–13 | 8–12 | 8–10 | 11–13 | 10–13 | 11–13 | present | absent | solitary phialide | rough | globose | 0.17–0.29 | long, lack visible rings |
| P. lassenii | no growth | 4–5 | 4–5 | 3–4 | 7–9 | 5–6 | 9–10 | 9–10 | 5–6 | 10–11 | absent | absent | mono- to biverticillate | smooth | broadly ellipsoidal | n.a. | n.a. |
| P. marthaechristenseniae | no growth | 5–6 | no growth | no growth | 7–8 | 9–10 | 4–5 | 7–8 | 11–12 | 9–11 | absent | absent | solitary phialide | rough | subglobose | 0.22–0.32 | present, visible rings present |
| P. oregonense | no growth | no growth | no growth | no growth | 4–5 | 9–10 | 4–5 | 5–7 | 7–8 | 8–9 | absent | absent | solitary phialide | rough, in parallel rings | globose | 0.18–0.24 | present, lack visible rings |
| P. ovatum | no growth | 12–14 | no growth | no growth | 10–11 | 10–11 | 7–8 | 13–14 | 8–10 | 9–11 | absent | absent | solitary phialide | smooth | ellipsoidal | n.a. | present, lack visible rings |
| P. porphyreum | no growth | 11–12 | 2–3 | no growth | 10–11 | 10–11 | 9–10 | 10–12 | 11–12 | 12–13 | present | absent | solitary phialide | rough | globose | 0.16–0.23 | long, visible rings present |
| P. riverlandense | 4–7 | 9–14 | 8–9 | 3–4 | 10–14 | 10–14 | 6–8 | 10–15 | 10–12 | 10–12 | present | absent | solitary phialide | rough | globose | 0.19–0.35 | long, lack visible rings |
| P. tubakianum | no growth | 5–6 | 2–4 | no growth | 8–9 | 8–9 | 4–5 | 7–8 | 5–7 | 9–10 | absent | absent | solitary phialide | rough | globose | 0.17–0.28 | present, lack visible rings |
| P. variratense | no growth | 10–12 | 8–9 | 2–3 | 9–10 | 8–9 | 4–5 | 7–8 | 9–10 | 7–8 | absent | absent | solitary phialide | finely rough | broadly ellipsoidal | 0.08–0.14 | short, lack visible rings |
| P. williamettense | no growth | no growth | no growth | no growth | 7–8 | 5–6 | 4–5 | 7–8 | 6–7 | 7–8 | absent | absent | solitary phialide | rough | globose | 0.23–0.42 | long, lack visible rings |
| P. wisconsinense | no growth | 3–5 | no growth,sometimes 2 | no growth | 7–10 | 9–11 | 3–6 | 7–11 | 9–10 | 8–10 | absent | absent | solitary phialide | rough | globose | 0.24–0.38 | short, lack visible rings |
| P. wollemiicola | no growth | 6–7 | 4–5 | no growth | 7–8 | 9–10 | 4–5 | 8–9 | 7–9 | 9–11 | present | absent | solitary phialide | rough | globose | 0.22–0.41 | long, lack visible rings |
TAXONOMY
The following species are accepted in Penicillium sect. Torulomyces. Penicillium ovatum and P. laeve, previously classified in the section, are classified in Penicillium sect. Exilicaulis. Art. 9.3. of the Code (McNeill et al. 2012) explicitly includes published illustrations among ‘original material’, which precludes neotypification (Art. 9.7). Therefore the erroneous neotypification of Torulomyces lagena by Stolk & Samson (1983) is corrected by the designation of one of Delitsch’s (1943) original figures as lectotype and designation of the ‘neotype’ strain CBS 185.65 as the epitype. Penicillium parviverrucosum is considered a dubious species.
Penicillium aeris Visagie & Samson, sp. nov. — MycoBank MB808262; Fig. 3a–g, 4a–c
Fig. 3.
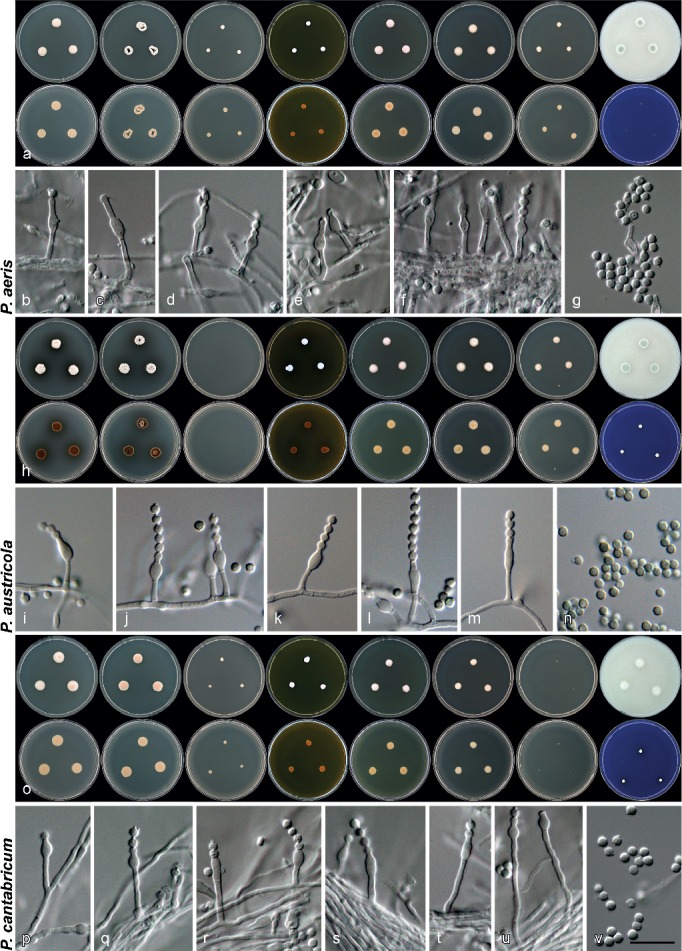
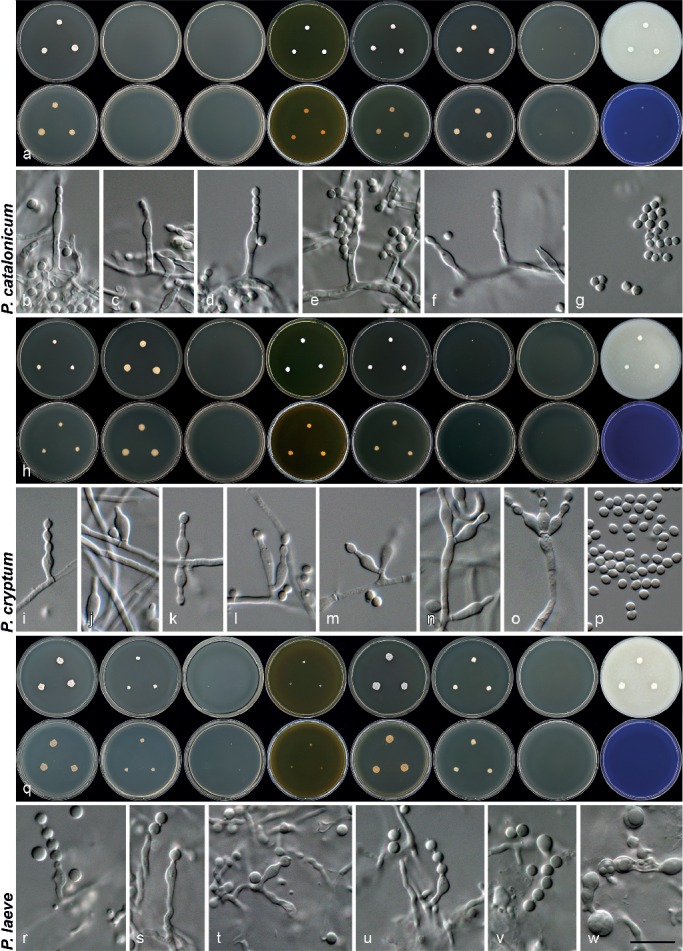
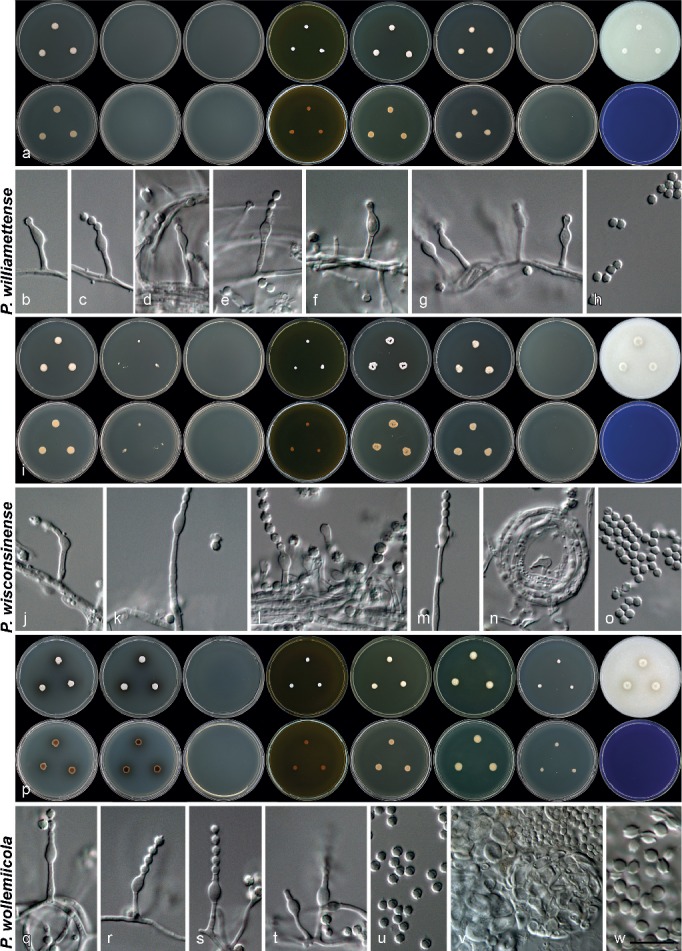
Morphology of species characterised in this study. a, h, o. Colony morphology from left to right, top row: CYA, CYA 30 °C, CYA 37 °C, MEA, YES, DG18, CYAS, OA; from left to right, bottom row: reverse colonies on CYA, CYA 30 °C, CYA 37 °C, MEA, YES, DG18, CYAS and obverse on CREA; b–g, i–n, p–v. conidiophores and conidia produced on OA. — Scale bar: v = 10 μm, applies to all microscope pictures.
Fig. 4.
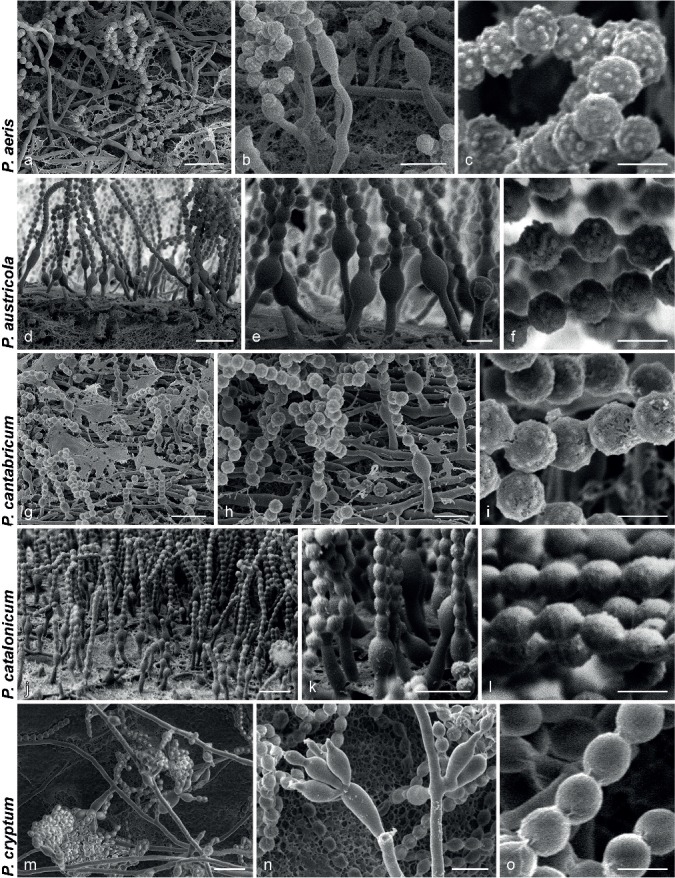
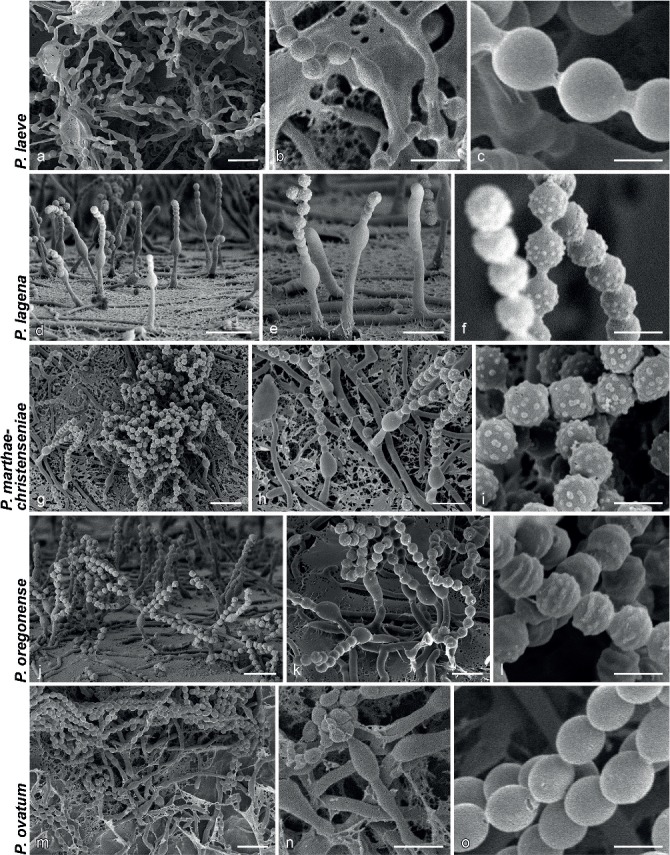
SEM micrographs showing characters of monophialidic Penicillium species. a, d, g, j, m. Overview of conidiogenous areas; b, e, h, k, n. phialides; c, f, i, l, o. conidia. — Scale bars: a, d, g, j, m = 10 μm; b, e, h, k, n = 5 μm; c, f, i, l, o = 2 μm.
ITS Barcode. KF303654 (alternative markers: BenA = KF303614; CaM = KF303627; RPB2 = KF303681).
Etymology. Latin, meaning of the air, in reference to the type strain isolated from an air sample.
Type specimen. GERMANY, indoor air, 2012, coll. I. Ilyashenko (CBS H-21608 holotype, culture ex-type CBS 135897 = DTO 207D4).
Diagnosis — Growth present on CYA at 37 °C, no growth on CREA, conidia rough-walled, globose, 2–2.5 μm diam.
Description — Colony diam, 7 d, in mm: CYA 11–12; CYA 30 °C 9–10; CYA 37 °C 4–5; MEA 6–7; YES 10–11; OA 9–10; DG18 11–12; CYAS 6–7; SNA 8–9; CREA no growth.
Colonies at 25 °C, 7 d: CYA obverse pale orange (5A3), reverse pale orange (5A3); MEA obverse orange white (5A2), reverse yellowish brown (5D6); YES obverse greenish white (25A2), reverse brownish orange (5C5); OA obverse greyish green (26E5); DG18 obverse yellowish white (3A2), reverse yellowish white (3A2); CYAS obverse white, reverse white; SNA obverse greenish grey (26B2), reverse white to light greyish; CREA acid not produced.
Conidiophores as solitary phialides; stipes smooth-walled, 4–13.5 × 1.5–2 μm; phialides ampulliform, 3.5–6.5 × 2–3 μm (4.9 ± 0.5 × 2.4 ± 0.2); conidia rough to spiny, globose, 2–2.5 × 2–2.5 μm (2.1 ± 0.2 × 2.1 ± 0.2, n = 34), average width/length = 0.99; sclerotia not produced.
SEM observations: Conidia rough-walled with tubercles present, connectives long without visible rings, tubercles 0.21–0.4 μm diam (0.31 ± 0.05).
Penicillium austricola Visagie & K. Jacobs, sp. nov. — MycoBank MB805184; Fig. 3h–n, 4d–f
ITS Barcode. JX091466 (alternative markers: BenA = JX091579; CaM = JX141600; RPB2 = KF303705).
Etymology. Latin, meaning resident from the south, in reference to the species isolated close to the most southern tip of Africa at Struisbaai.
Type specimen. SOUTH AFRICA, Struisbaai, from mite inside Protea repens infructescence, Aug. 2009, coll. C.M. Visagie (CBS H-21605 holotype, culture ex-type CBS 135900 = CV 1842 = DTO 183E6 = DAOMC 241066).
Diagnosis — No growth on CYA at 37 °C, growth on CYA at 30 °C and CREA, colonies typically produces brown soluble pigments, conidia rough-walled, globose, 1.5–2.5 μm diam.
Description — Colony diam, 7 d, in mm: CYA 10–13; CYA 30 °C 10–11; CYA 37 °C no growth; MEA 4–8; YES 10–11; OA 9–10; DG18 9–11; CYAS 8–10; SNA 9–11; CREA 3–4.
Colonies at 25 °C, 7 d: CYA obverse greyish to dull green (25B3–26D3), reverse olive to dark brown (4E3–6F8), brown soluble pigment produced in some isolates; MEA obverse greyish green (25B3–26B3), reverse light to dark brown (6D6–F8), brown soluble pigment produced in some isolates; YES obverse orange white (5A2) when not sporulating, greyish green (25C4) when sporulating, reverse greyish yellow (4B4); OA obverse greyish green (25D5–E5); DG18 obverse greenish white to pale green (25A2–A3), reverse dull green (29D3); CYAS obverse greenish grey (25B2), reverse brown (6E6); SNA obverse dull green (29D4), reverse greenish grey (29B2); CREA acid not produced.
Conidiophores as solitary phialides; stipes smooth-walled, 4–11 × 1–1.5 μm; phialides ampulliform, 5–8 × 2–3 μm (6.1 ± 0.6 × 2.7 ± 0.2); conidia rough-walled, globose, 1.5–2.5 × 1.5–2.5 μm (2.1 ± 0.1 × 2.1 ± 0.1, n = 25), average width/length = 0.98; sclerotia not produced.
SEM observations: Conidia rough-walled with tubercles present, connectives long without visible rings, tubercles 0.19–0.30 μm diam (0.23 ± 0.03).
Additional material examined. South Africa, Struisbaai, from mite inside Protea repens infructescence, Aug. 2009, coll. C.M. Visagie, cultures CBS 135904 (CV 1808 = DTO 183D8 = DAOMC 241065), CBS 135899 (CV 1811 = DTO 186D8), CBS 135902 (CV 1943 = DTO 183F8 = DAOMC 241067), CBS 135903 (CV 1954 = DTO 183F9), CBS 136253 (CV 1968 = DTO 186G3); Struisbaai, bract from Protea repens infructescence, Aug. 2009, coll. C.M. Visagie, culture CBS 135901 (CV 1902 = DTO 183F5).
Penicillium cantabricum Visagie & Samson, sp. nov. — MycoBank MB808263; Fig. 3o–v, 4g–i
ITS Barcode. KF303655 (alternative markers: BenA = KF303615; CaM = KF303646; RPB2 = KF303682).
Etymology. Latin, named after Cantabria, from where the type was isolated.
Type specimen. SPAIN, Cantabria, Reinosa, from soil, June 1996, isol. A.M. Stchigel (CBS H-21612 holotype, culture ex-type CBS 120415 = DTO 76I9 = FMR 9121).
Diagnosis — Growth present on CYA at 37 °C, very weak acid produced within colony periphery on CREA, conidia finely rough-walled, globose, 2–2.5 μm.
Description — Colony diam, 7 d, in mm: CYA 13–14; CYA 30 °C 12–13; CYA 37 °C 3–5; MEA 6–7; YES 9–10; OA 10–11; DG18 8–9; CYAS 3–4; SNA 7–8; CREA 3–4.
Colonies at 25 °C, 7 d: CYA obverse pale orange (6A3), reverse pale orange (5A3), pinkish mycelia produced; MEA obverse orange white (5A2), reverse yellowish brown (5D6); YES obverse orange white (5A2), reverse light orange (5A4); OA obverse greyish green (25B3); DG18 obverse yellowish white (3A2), reverse yellowish white (3A2); CYAS obverse yellowish white (3A2), reverse yellowish white (3A2); SNA obverse white to light greyish, reverse white to light greyish; CREA acid very weakly produced within colony periphery.
Conidiophores as solitary phialides; stipes smooth-walled, 4.5–24 × 1.5–2 μm; phialides ampulliform, 3.5–5.5 × 2.5–3 μm (4.5 ± 0.5 × 2.6 ± 0.2); conidia finely rough-walled, globose, 2–2.5 × 2–2.5 μm (2.2 ± 0.1 × 2.2 ± 0.1, n = 41), average width/length = 0.99; sclerotia not produced.
SEM observations: Conidia rough-walled with tubercles present, connectives present without visible rings, tubercles 0.21–0.33 μm diam (0.26 ± 0.03).
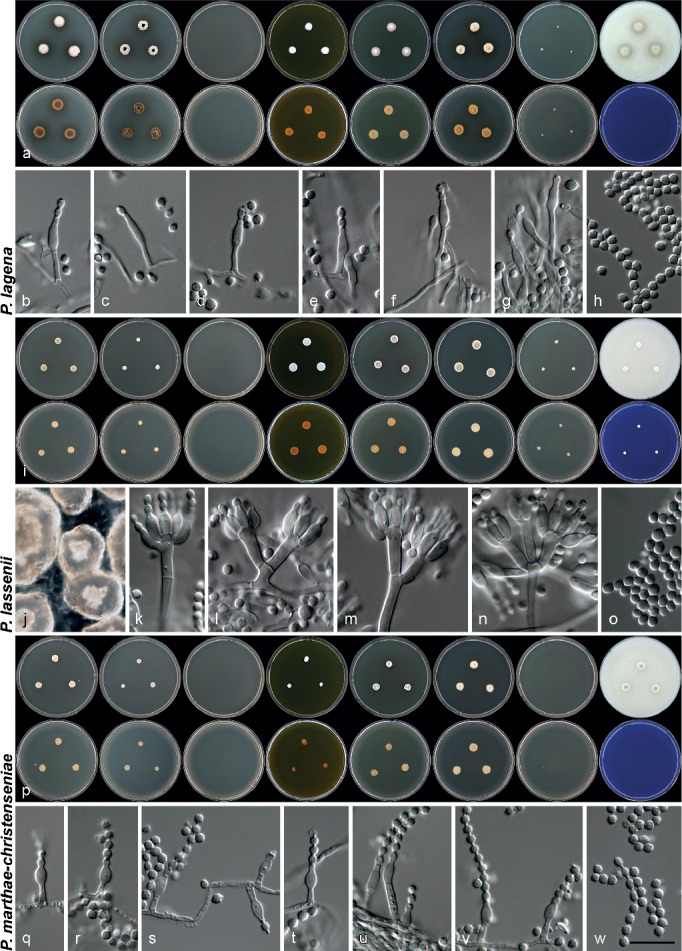
Penicillium catalonicum Visagie & Samson, sp. nov. — MycoBank MB808265; Fig. 4j–l, 5a–g
Fig. 5.
Morphology of species characterised in this study. a, h, q. Colony morphology from left to right, top row: CYA, CYA 30 °C, CYA 37 °C, MEA, YES, DG18, CYAS, OA; from left to right, bottom row: reverse colonies on CYA, CYA 30 °C, CYA 37 °C, MEA, YES, DG18, CYAS and obverse on CREA; b–g, i–p, r–w. conidiophores, conidia, asci and ascospores (in w) produced on OA. — Scale bar: w = 10 μm, applies to all microscope pictures.
ITS Barcode. KF303650 (alternative markers: BenA = KF303609; CaM = KF303644; RPB2 = KF303683).
Etymology. Latin, named after Catalonia, from where the type was isolated.
Type specimen. SPAIN, Catalonia, Montseny Natural Park, from soil, unknown date, isol. A.M. Stchigel & M. Calduch (CBS H-21610 holotype, culture ex-type CBS 110532 = DTO 78H5).
Diagnosis — No growth on CYA at 30 °C, growth on CYAS, conidia finely rough-walled, with tubercles forming rings around conidia under SEM, subglobose to broadly ellipsoidal, 2–2.5 × 2–2.5 μm.
Description — Colony diam, 7 d, in mm: CYA 6–8; CYA 30 °C no growth; CYA 37 °C no growth; MEA 4–5; YES 5–6; OA 6–7; DG18 7–8; CYAS 2–3; SNA 6–7; CREA no growth.
Colonies at 25 °C, 7 d: CYA obverse greenish white (25A2), reverse yellowish white (4A2); MEA obverse orange white (5A2), reverse yellowish brown (5D6); YES obverse turquoise white to greenish white (24A2–25A2), reverse olive brown (4D5); OA obverse greyish green (26C4); DG18 obverse yellowish white (3A2), reverse yellowish white (3A2); CYAS obverse yellowish white (3A2), reverse yellowish white (3A2); SNA obverse white to light greyish, reverse white to light greyish; CREA acid not produced.
Conidiophores as solitary phialides; stipes smooth-walled, 4.5–11 × 1.5–2 μm; phialides ampulliform, 5–7 × 2–3 μm (5.9 ± 0.6 × 2.4 ± 0.2); conidia finely rough, subglobose to broadly ellipsoidal, 2–2.5 × 2–2.5 μm (2.16 ± 0.1 × 2.11 ± 0.1, n = 41), average width/length = 0.98; sclerotia not produced.
SEM observations: Conidia more ellipsoidal than when observed under light microscope, finely rough-walled with very small tubercles present, sometimes in ring like pattern, connections short and not well defined, tubercles 0.12–0.18 μm diam (0.14 ± 0.02).
Penicillium cryptum Goch., Mycotaxon 26: 349. 1986 — MycoBank MB103648; Fig. 4m–o, 5h–p
Basionym. Eupenicillium cryptum Goch., Mycotaxon 26: 349. 1986.
ITS Barcode. KF303647 (alternative markers: BenA = KF303608; CaM = KF303628; RPB2 = JN121478).
Type specimen. USA, New York, Long Island, from soil in Quercus-Betula forest, Oct. 1983, isol. S.E. Gochenaur (NY 769 holotype, culture ex-type CBS 271.89 = DTO 122C9 = ATCC 60138 = IMI 296794 = NRRL 13460).
Diagnosis — No growth on CYA at 37 °C or CREA, growth on CYA at 30 °C, conidiophores typically monophialidic to monoverticillate, conidia smooth-walled, broadly ellipsoidal, 2–2.5 × 1.5–2 μm.
Description — Colony diam, 7 d, in mm: CYA 6–7; CYA 30 °C 6–9; CYA 37 °C no growth; MEA 6–7; YES 5–6; OA 5–6; DG18 1–2; CYAS no growth; SNA 5–6; CREA no growth.
Colonies at 25 °C, 7 d: CYA obverse greenish white (25A2), reverse pale; MEA obverse greenish white (25A2), reverse yellowish brown (5D6); YES obverse greenish white (25A2), reverse yellowish white (3A2); OA obverse greenish white (25A2); DG18 small white to pale; SNA obverse white to light greyish, reverse white to light greyish.
Conidiophores monoverticillate with at most 3 phialides, phialides often borne directly on hyphae, conidiophores sometimes as solitary phialides; stipes smooth-walled, 5–30 × 1–2.5 μm when monoverticillate, 1–11 × 1–2.5 μm when monophialidic; phialides ampulliform, 5–9 × 2.5–3.5 μm (6.8 ± 0.9 × 2.9 ± 0.2); conidia smooth-walled, broadly ellipsoidal, 2–2.5 × 1.5–2 μm (2.3 ± 0.1 × 1.9 ± 0.1, n = 35), average width/length = 0.83; sclerotia not produced.
SEM observations: Conidia broadly ellipsoidal, smooth-walled, connectives present, but rather obscure.
Penicillium laeve (K. Ando & Manoch) Houbraken & Samson, Stud. Mycol. 70: 47. 2011 — MycoBank MB561960; Fig. 5q–w, 6a–c
Fig. 6.
SEM micrographs showing characters of monophialidic Penicillium species. a, d, g, j, m. Overview of conidiogenous areas; b, e, h, k, n. phialides; c, f, i, l, o. conidia. — Scale bars: a, d, g, j, m = 10 μm; b, e, h, k, n = 5 μm; c, f, i, l, o = 2 μm.
Basionym. Torulomyces laevis K. Ando & Manoch, Mycoscience 39: 317. 1998.
ITS Barcode. KF667369 (alternative markers: BenA = KF667365; CaM = KF667367; RPB2 = KF667371).
Type specimen. THAILAND, from soil, Oct. 1992, isol. K. Ando (TNS-F-238517 holotype, culture ex-type CBS 136665 = DTO 270G8 = KY 12727 = NBRC 109724).
Diagnosis — No growth on CYA at 37 °C or CREA, growth on CYA at 30 °C, conidia smooth-walled, globose, 2.5–3 μm diam. Ascomata inconspicuous, asci 3–8 μm diam, ascospores smooth-walled, globose, 1.5–2 μm diam.
Description — Colony diam, 7 d, in mm: CYA 8–9; CYA 30 °C 4–5; CYA 37 °C no growth; MEA microcolonies; YES 8–9; OA 7–8; DG18 5–7; CYAS no growth; SNA 7–8; CREA no growth.
Colonies at 25 °C, 7 d: CYA obverse white, reverse greyish yellow (3C3); MEA obverse white, reverse yellowish brown (5D6); YES obverse greenish grey (25B2), reverse brownish orange (5C4); OA obverse greyish white (1B1); DG18 obverse greyish white (1B1), reverse greyish yellow (3B3); SNA obverse white to light greyish, reverse white; CREA acid not produced.
Conidiophores as solitary phialides; stipes smooth-walled, 2–6 × 1–1.5 μm; phialides ampulliform, 4–6 × 2–3 μm (5.2 ± 0.6 × 2.4 ± 0.1); conidia smooth-walled, globose, 2.5–3 × 2.5–3 μm (2.6 ± 0.1 × 2.6 ± 0.1, n = 22), average width/length = 0.98; sexual morph produced, ascomata inconspicuous, asci 3–8 μm with smooth-walled, globose ascospores 1.5–2 × 1.5–2 μm.
SEM observations: Conidia globose, smooth-walled, long connectives present without visible rings.
Penicillium lagena (Delitsch) Stolk & Samson, Stud. Mycol. 23: 100. 1983 — MycoBank MB109162; Fig. 6d–f, 7a–h
Fig. 7.
Morphology of species characterised in this study. a, i, p. Colony morphology from left to right, top row: CYA, CYA 30 °C, CYA 37 °C, MEA, YES, DG18, CYAS, OA; from left to right, bottom row: reverse colonies on CYA, CYA 30 °C, CYA 37 °C, MEA, YES, DG18, CYAS and obverse on CREA; b–h, j–o, q–w. conidiophores, conidia and sclerotia (q) produced on OA. — Scale bar: w = 10 μm, applies to all microscope pictures.
Basionym. Torulomyces lagena Delitsch, Systematik der Schimmelpilze, Neudamm: 9. 1943.
≡ Eupenicillium limoneum Goch. & Zlattner, Stud. Mycol. 23: 100. 1983.
ITS Barcode. KF303665 (alternative markers: BenA = KF303619; CaM = KF303634; RPB2 = JN121450).
Lectotype. Fig. 233 (Delitsch, Ergebnisse der theoretischen und angewandten Mikrobiologie: Band I: Systematik der Schimmelpilze. J. Neumann, Neudamm, Germany, Tafel 30, 1943, designated here, MBT203020).
Epitype. CANADA, Guelph, from bog soil under Pacific red cedar (Thuja plicata), 1961, isol. G.L. Barron (CBS 185.65 epitype (metabolically inactive), MBT203019, culture ex-epitype CBS 185.65 = MUCL 8221 = JCM10149), designated here.
Diagnosis — No growth on CYA at 37 °C or CREA, growth on CYA at 30 °C, colonies on CYA 10–13 mm, conidia rough-walled, under SEM with small tubercles, connectives lacking visible rings, globose, 2–2.5 μm diam.
Description — Colony diam, 7 d, in mm: CYA 10–13; CYA 30 °C 8–10; CYA 37 °C no growth; MEA 8–10; YES 11–13; OA 8–12; DG18 11–13; CYAS no growth to 3; SNA 10–13; CREA no growth.
Colonies at 25 °C, 7 d: CYA obverse greenish grey (25B2), reverse dark brown (6F8), some isolates pale, brown soluble pigment produced in some isolates; MEA obverse greenish white (25A2), reverse yellowish brown (5D6), brown soluble pigment produced in some isolates; YES obverse yellowish white (3A2) when not sporulating, otherwise greyish green (25B3), reverse brownish orange (5C4) in non-sporulating cultures, otherwise olive (2D3); OA obverse greyish green (25C3–26C3), brown soluble pigment produced; DG18 obverse yellowish white (3A2) when not sporulating, otherwise (29C3), reverse brownish orange (5C5), brown soluble pigment produced; CYAS obverse white, reverse white; SNA obverse white to light greyish, reverse white to light greyish; CREA acid not produced.
Conidiophores as solitary phialides; stipes smooth-walled, 4–20 × 1–1.5 μm; phialides ampulliform, 4.5–7 × 2–2.5 μm (6.3 ± 0.7 × 2.2 ± 0.1); conidia rough-walled, globose, 2–2.5 × 2–2.5 μm (2.1 ± 0.2 × 2.1 ± 0.2, n = 27), average width/length = 0.99; sclerotia not produced.
SEM observations: Conidia rough-walled with pronounced large tubercles present, connectives long without easily visible rings, tubercles 0.17–0.29 μm diam (0.22 ± 0.04).
Penicillium lassenii Paden, Mycopathol. Mycol. Appl. 43: 266. 1971. — MycoBank MB319281; Fig. 7i–o
≡ Eupenicillium lassenii Paden, Mycopathol. Mycol. Appl. 43: 266. 1971.
ITS Barcode. KF303648 (alternative markers: BenA = KF303607; CaM = KF303629; RPB2 = JN121481).
Type specimen. USA, California, from soil under conifers, May 1969, isol. J.W. Paden (JWP 69-26 (UVIC) holotype, culture ex-type CBS 277.70 = DTO 95D6 = NRRL 5272 = ATCC 22054 = FRR 858 = IMI 148395).
Diagnosis — No growth on CYA at 37 °C, growth on CYA at 30 °C and CREA, conidiophores typically mono- to biverticillate, conidia smooth-walled, broadly ellipsoidal, 2.5–3 × 2–3 μm, produces pale to brown sclerotia.
Description — Colony diam, 7 d, in mm: CYA 7–9; CYA 30 °C 4–5; CYA 37 °C no growth; MEA 9–10; YES 9–10; OA 5–6; DG18 10–11; CYAS 4–5; SNA 5–6; CREA 3–4.
Colonies at 25 °C, 7 d: CYA obverse dull green (27D3), reverse yellowish grey (2B2); MEA obverse greyish green (27C3), reverse light brown (6D5); YES obverse dull green (27D3), reverse yellowish grey (2B2); OA obverse dull green (27D3); DG18 obverse dull green (27D3), reverse yellowish grey (2B2); CYAS obverse white, reverse white; SNA obverse white to light greyish, reverse white to light greyish; CREA acid not produced.
Conidiophores monoverticillate to sometimes biverticillate; stipes smooth-walled, 9–75 × 1.5–3.5 μm; branches 4–15 × 1.5–3.5 μm; phialides ampulliform, 6–7.5 × 2.5–3.5 μm (6.7 ± 0.6 × 2.9 ± 0.2); conidia smooth-walled, broadly ellipsoidal, 2.5–3 × 2–3 μm (2.6 ± 0.1 × 2.45 ± 0.1, n = 37), average width/length = 0.89; sclerotia pale to brownish, 105–235 × 90–170 μm.
Penicillium marthae-christenseniae Visagie & Samson, sp. nov. — MycoBank MB808267; Fig. 4g–i, 7p–w
ITS Barcode. KF303651 (alternative markers: BenA = KF303613; CaM = KF303645; RPB2 = KF303711).
Etymology. Latin, named after Martha Christensen, who isolated and deposited the ex-type strain into the CBS collection.
Type specimen. USA, Wisconsin, unknown source and date, isol. M. Christensen (CBS H-21613 holotype, culture ex-type CBS 129213 = DTO 201B5).
Diagnosis — No growth on CYA at 37 °C, CREA or CYAS, growth on CYA at 30 °C, colonies on CYA 7–8 mm and SNA 11–12 mm, conidia rough-walled, with large well-defined tubercles (average 0.26 ± 0.03 μm) under SEM, subglobose, 2–2.5 × 2–2.5 μm.
Description — Colony diam, 7 d, in mm: CYA 7–8; CYA 30 °C 5–6; CYA 37 °C no growth; MEA 4–5; YES 7–8; OA 9–10; DG18 9–11; CYAS no growth; SNA 11–12; CREA no growth.
Colonies at 25 °C, 7 d: CYA obverse white, reverse pale yellow (3B3); MEA obverse orange white (5A2), reverse yellowish brown (5D6); YES obverse greyish green (25C4), reverse yellowish grey to olive brown (4B2–E4); OA obverse dull green (27D4); DG18 obverse very light greyish green, reverse greyish yellow (4B3); SNA obverse white to light greyish, reverse white to light greyish; CREA acid not produced.
Conidiophores as solitary phialides; stipes smooth-walled, 3–10 × 1–2 μm; phialides ampulliform, 4–6.5 × 2–3 μm (5.1 ± 0.6 × 2.4 ± 0.2); conidia rough-walled, subglobose, 2–2.5 × 2–2.5 μm (2.2 ± 0.1 × 2.1 ± 0.1, n = 34), average width/length = 0.96; sclerotia not produced.
SEM observations: Conidia rough-walled with tubercles present, connectives present with visible rings, tubercles 0.22–0.32 μm diam (0.26 ± 0.03).
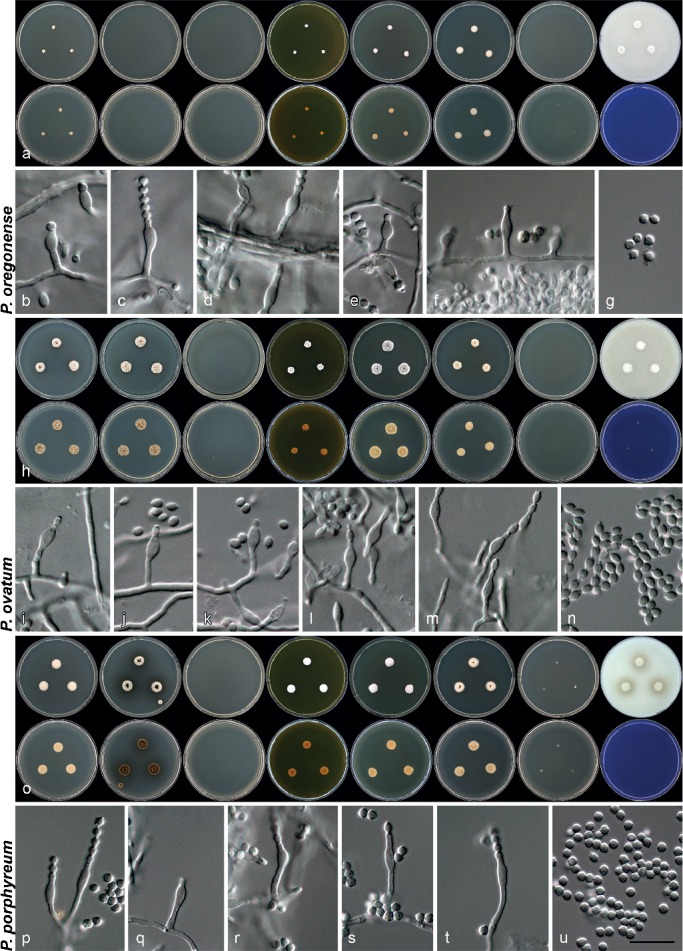
Penicillium oregonense Visagie, M. Chr. & Samson, sp. nov. — MycoBank MB808268; Fig. 6j–l, 8a–g
Fig. 8.
Morphology of species characterised in this study. a, h, o. Colony morphology from left to right, top row: CYA, CYA 30 °C, CYA 37 °C, MEA, YES, DG18, CYAS, OA; from left to right, bottom row: reverse colonies on CYA, CYA 30 °C, CYA 37 °C, MEA, YES, DG18, CYAS and obverse on CREA; b–g, i–n, p–u. conidiophores and conidia produced on OA. — Scale bar: u = 10 μm, applies to all microscope pictures.
ITS Barcode. KF303668 (alternative markers: BenA = KF303623; CMD = KF303640; RPB2 = KF303710).
Etymology. Latin, named after Oregon State, from where the type was isolated.
Type specimen. USA, Oregon, Williamette National Forest near Blue River, unknown source and date, isol. M. Christensen (CBS H-21607 holotype, culture ex-type CBS 129775 = DTO 208A5).
Diagnosis — No growth on CYA at 30 °C or CYAS, colonies on CYA at 25 °C 4–5 mm, conidia rough-walled with tubercles in rings under SEM, globose, 2–2.5 × 2–2.5 μm.
Description — Colony diam, 7 d, in mm: CYA 4–5; CYA 30 °C no growth; CYA 37 °C no growth; MEA 4–5; YES 5–7; OA 9–10; DG18 8–9; CYAS no growth; SNA 7–8; CREA no growth.
Colonies at 25 °C, 7 d: CYA obverse white, reverse pale yellow (3B3); MEA obverse orange white (5A2), reverse yellowish brown (5D6); YES obverse yellowish white (3A2), reverse yellowish white (3A2); OA obverse dull green (28E4); DG18 obverse yellowish white (3A2), reverse yellowish white (3A2); SNA obverse white to light greyish, reverse white to light greyish; CREA acid not produced.
Conidiophores as solitary phialides; stipes smooth-walled, 2.5–14 × 1.5–2 μm; phialides ampulliform, 4.5–6 × 2.5–3.5 μm (5.5 ± 0.4 × 2.8 ± 0.2); conidia rough-walled, globose, 2–2.5 × 2–2.5 μm (2.15 ± 0.1 × 2.15 ± 0.1, n = 32), average width/length = 0.99; sclerotia not produced.
SEM observations: Conidia rough-walled, with tubercles forming conspicuous parallel rings, connectives moderately long and well defined without visible rings, tubercles 0.18–0.24 μm diam (0.22 ± 0.04).
Penicillium ovatum (K. Ando & Nawawi) Houbraken & Samson, Stud. Mycol. 70: 48. 2011 — MycoBank MB561961; Fig. 6m–o, 8h–n
Basionym. Torulomyces ovatus K. Ando & Nawawi, Mycoscience 39: 317. 1998.
ITS Barcode. KF667370 (alternative markers: BenA = KF667366; CaM = KF667368; RPB2 = KF667372).
Type specimen. MALAYSIA, Kuala Lumpur, from soil under Pinus caribaea, Nov. 1989, isol. K. Ando (TNS-F-238518 holotype, culture ex-type CBS 136664 = KY 12726 = DTO 270G7 = NBRC 109725).
Diagnosis — No growth on CYA at 37 °C or CREA, growth on CYA at 30 °C, conidia smooth-walled, ellipsoidal, 2–3 × 1.5–2 μm.
Description — Colony diam, 7 d, in mm: CYA 10–11; CYA 30 °C 12–14; CYA 37 °C no growth; MEA 7–8; YES 13–14; OA 10–11; DG18 9–11; CYAS no growth; SNA 8–10; CREA no growth.
Colonies at 25 °C, 7 d: CYA obverse greyish white (1B1), reverse brownish orange (5C4), colonies producing orange colours after prolonged incubation; MEA obverse greyish white (1B1), reverse yellowish brown (5D6); YES obverse greenish grey (29C2), reverse greyish yellow (4C4); OA obverse greyish white (1B1); DG18 obverse greenish grey (29C2), reverse greyish yellow (3C4); SNA obverse white to light greyish, reverse white; CREA acid not produced.
Conidiophores as solitary phialides; stipes smooth-walled, 3.5–15 × 1–2 μm; phialides ampulliform, 4.5–7 × 2–3 μm (5.7 ± 0.5 × 2.5 ± 0.25); conidia smooth-walled, ellipsoidal, 2–3 × 1.5–2 μm (2.4 ± 0.2 × 1.8 ± 0.1, n = 27), average width/length = 0.76; sclerotia not produced.
SEM observations: Conidia ellipsoidal, smooth-walled, connectives present without visible rings.
Penicillium porphyreum Houbraken & Samson, Stud. Mycol. 70: 48. 2011 — MycoBank MB561959; Fig. 8o–u, 9a–c
Fig. 9.
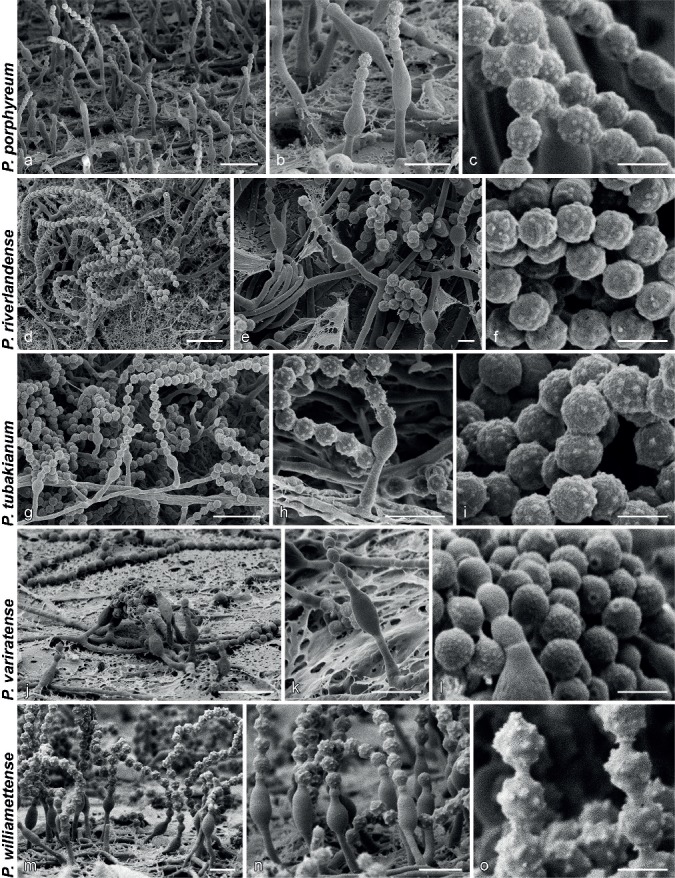
SEM micrographs showing characters of monophialidic Penicillium species. a, d, g, j, m. Overview of conidiogenous areas; b, e, h, k, n. phialides; c, f, i, l, o. conidia. — Scale bars: a, d, g, j, m = 10 μm; b, e, h, k, n = 5 μm; c, f, i, l, o = 2 μm.
≡ Monocillium humicola G.L. Barron var. brunneum M. Chr. & Backus, Mycologia 56: 498. 1964.
≡ Torulomyces brunneus (M. Chr. & Backus) K. Ando, Mycoscience 39: 314. 1998.
ITS Barcode. KF303666 (alternative markers: BenA = KF303621; CaM = KF303636; RPB2 = KF303677).
Type specimen. USA, Wisconsin, from soil in community of Pinus strobus, July 1955, isol. M. Christensen (NY barcode 00985491 holotype, culture ex-type CBS 382.64 = ATCC 15571 = DTO 78G7 = KY 12723 = WSF 9-C).
Diagnosis — No growth on CYA at 37 °C or CREA, growth on CYA at 30 °C, colonies on CYA 10–11 mm, conidia rough-walled with small tubercles (average 0.19 ± 0.02 μm), connectives have two clearly visible rings.
Description — Colony diam, 7 d, in mm: CYA 10–11; CYA 30 °C 11–12; CYA 37 °C no growth; MEA 9–10; YES 10–12; OA 10–11; DG18 12–13; CYAS 2–3; SNA 11–12; CREA no growth.
Colonies at 25 °C, 7 d: CYA obverse yellowish to orange white (4A2–5A2), reverse orange white (5A2), brown soluble pigment typical; MEA obverse yellowish to orange white (4A2–5A2), reverse light brown (6D7), brown soluble pigment typical; YES obverse turquoise white to greenish white (24A2–25A2), reverse brownish orange (5C4); OA obverse greyish green (25C3–26C3), brownish olive soluble pigment produced; DG18 obverse yellowish grey (3B2), reverse greyish orange (5C4); CYAS obverse yellowish white (3A2), reverse yellowish white (3A2); SNA obverse white to light greyish, reverse white to light greyish; CREA acid not produced.
Conidiophores as solitary phialides; stipes smooth-walled, 5–17 × 1–1.5 μm; phialides ampulliform, 4.5–7.5 × 2–2.5 μm (6 ± 0.8 × 2.3 ± 0.2); conidia rough-walled, globose, 2–2.5 × 2–2.5 μm (2.16 ± 0.1 × 2.16 ± 0.1, n = 43), average width/length = 0.99; sclerotia not produced.
SEM observations: Conidia rough-walled with pronounced large to moderately sized tubercles present, connectives long, consistently having two visible rings, tubercles 0.16–0.23 μm diam (0.19 ± 0.02).
Penicillium riverlandense Visagie & K. Jacobs, sp. nov. — MycoBank MB808269; Fig. 9d–f, 10a–g
Fig. 10.
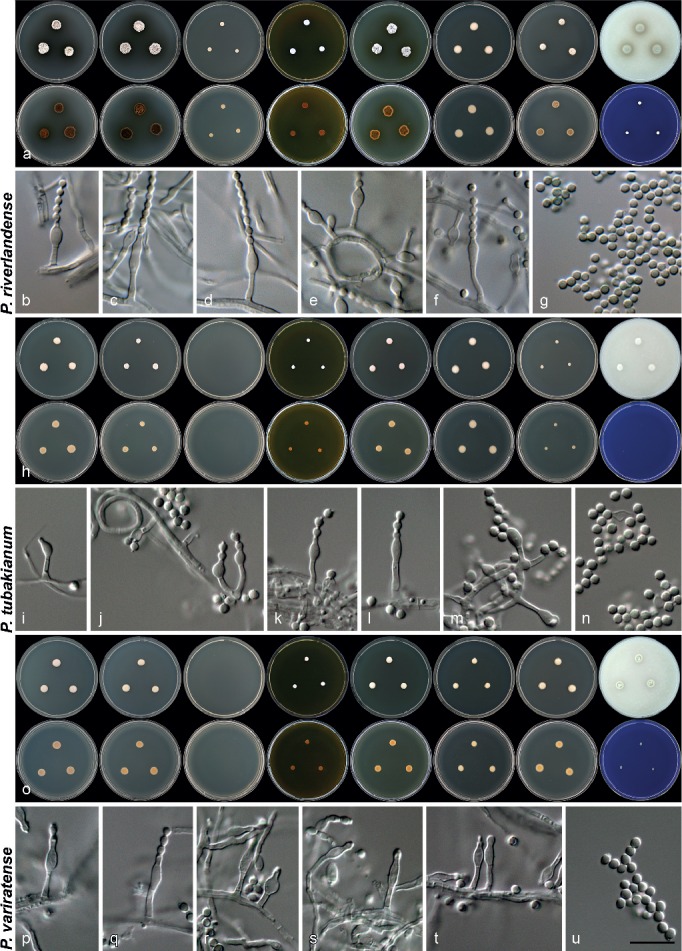
Morphology of species characterised in this study. a, h, o. Colony morphology from left to right, top row: CYA, CYA 30 °C, CYA 37 °C, MEA, YES, DG18, CYAS, OA; from left to right, bottom row: reverse colonies on CYA, CYA 30 °C, CYA 37 °C, MEA, YES, DG18, CYAS and obverse on CREA; b–g, i–n, p–u. conidiophores and conidia produced on OA. — Scale bar: u = 10 μm, applies to all microscope pictures.
ITS Barcode. JX091457 (alternative markers: BenA = JX091580; CaM = JX141593; RPB2 = KF303685).
Etymology. Latin, named after Riverlands Nature Reserve, in reference to the location of the type origin.
Type specimen. SOUTH AFRICA, Malmesbury, Riverlands Nature Reserve, from sandveld Fynbos soil, July 2009, isol. C.M. Visagie (CBS H-21606 holotype, culture ex-type CBS 135896 = CV 0979 = DTO 182F6 = DAOMC 241060).
Diagnosis — Growth observed on CYA at 37 °C, colonies producing brown soluble pigment, conidia rough-walled, globose, mostly smaller than 2 μm diam.
Description — Colony diam, 7 d, in mm: CYA 10–14; CYA 30 °C 9–14; CYA 37 °C 4–7; MEA 6–8; YES 10–15; OA 10–14; DG18 10–12; CYAS 8–9; SNA 10–12; CREA 3–4.
Colonies at 25 °C, 7 d: CYA obverse greyish green (25B2), reverse olive to dark brown (5E5–6F8), brown soluble pigment produced in all isolates; MEA obverse light to greyish green (25A5–C5), reverse dark brown (6F8), brown soluble pigment produced in all isolates; YES obverse greyish green (25B4–26B4), sporulation sometimes absent, then pale to white colonies, brown soluble pigment produced, reverse brownish orange to brown (5C5–5F8); OA obverse dull green (25D4), brownish olive soluble pigment produced; DG18 obverse white to pale yellow (3A3), reverse yellowish white to dull yellow (3A2–B4); CYAS obverse greenish grey (25B2), reverse brown (6E6); SNA obverse dull green (29D4), reverse greenish grey (29B2); CREA acid not produced.
Conidiophores as solitary phialides; stipes smooth-walled, 3.5–10 × 1–2 μm; phialides ampulliform, 4–8 × 2–3 μm (5.5 ± 0.9 × 2.6 ± 0.2); conidia rough-walled, globose, 1.5–2 × 1.5–2 μm (1.8 ± 0.1 × 1.8 ± 0.1, n = 35), average width/length = 0.98; sclerotia not produced.
SEM observations: Conidia rough-walled with tubercles present, connectives long without visible rings, tubercles 0.19–0.35 μm diam (0.27 ± 0.04).
Additional material examined. South Africa, Malmesbury, Riverlands Nature Reserve, sandveld Fynbos soil, July 2009, isol. C.M. Visagie, cultures CBS 135894 = CV 0933 = DTO 185F9, CBS 135895 = CV 0959 = DTO 182E7 = DAOMC 241059, CBS 135887 = CV 2815 = DTO 184A1, CBS 135888 = CV 2819 = DTO 184A4, CBS 135889 = CV 2822 = DTO 184A6, CBS 135890 = CV 2831 = DTO 184B4, CBS 135891 = CV 2833 = DTO 184B6, CBS 135892 = CV 2839 = DTO 184C2, CBS 135893 = CV 2849 = DTO 184C6; Riverlands Nature Reserve, bract from Protea repens infructescence, July 2009, isol. C.M. Visagie, cultures CBS 135883 = CV 1360 = DTO 183A4 = DAOMC 241061, CBS 135884 = CV 1448 = DTO 183A9 = DAOMC 241063, CBS 135885 = CV 1450 = DTO 183B1 = DAOMC 241064, CBS 135886 = CV 1583 = DTO 186B8; Struisbaai, bract from Protea repens infructescence, Aug. 2009, coll. C.M. Visagie, culture DTO 183E8 = CV 1851.
Penicillium tubakianum Visagie & Samson, sp. nov. — MycoBank MB808270; Fig. 9g–i, 10h–n
ITS Barcode. KF303652 (alternative markers: BenA = KF303611; CaM = KF303637; RPB2 = KF303712).
Etymology. Latin, name after K. Tubaki, who isolated the ex-type strain of the species.
Type specimen. NEW ZEALAND, from dead bark of Cyathea, Dec. 1963, isol. K. Tubaki (CBS H-21604 holotype, culture ex-type CBS 287.66 = DTO 138D9 = MUCL 8519 = IFO 8315).
Diagnosis — No growth on CYA at 37 °C, CREA, growth on CYA at 30 °C, colonies on CYA 8–9 mm, CYAS 2–4 mm and SNA 5–7 mm, conidia rough-walled, globose, 2–2.5 μm diam, sclerotia not produced.
Description — Colony diam, 7 d, in mm: CYA 8–9; CYA 30 °C 5–6; CYA 37 °C no growth; MEA 4–5; YES 7–8; OA 8–9; DG18 9–10; CYAS 2–4; SNA 5–7; CREA no growth.
Colonies at 25 °C, 7 d: CYA obverse yellowish to orange white (4A2–5A2), reverse orange white (5A2); MEA obverse orange white (5A2), reverse yellowish brown (5D6); YES obverse pale orange (5A3), reverse pale orange (5A3); OA obverse greyish green (26C4); DG18 obverse yellowish white (3A2), reverse yellowish white (3A2); CYAS obverse yellowish white (3A2), reverse yellowish white (3A2); SNA obverse white to light greyish, reverse white to light greyish; CREA acid not produced.
Conidiophores as solitary phialides; stipes smooth-walled, 2.5–10 × 1–1.5 μm; phialides ampulliform, 3.5–6.5 × 2–3 μm (4.6 ± 0.2 × 2.5 ± 0.2); conidia rough-walled, globose, 2–2.5 × 2–2.5 μm (2.1 ± 0.1 × 2.1 ± 0.1, n = 34), average width/length = 0.99; sclerotia not produced.
SEM observations: Conidia rough-walled with tubercles present, connectives present without visible rings, tubercles 0.17–0.28 μm diam (0.23 ± 0.03).
Penicillium variratense Visagie & Samson, sp. nov. — MycoBank MB808271; Fig. 9j–l, 10o–u
ITS Barcode. KF303649 (alternative markers: BenA = KF303610; CaM = KF303630; RPB2 = KF303675).
Etymology. Latin, named after Varirata National Park in Papua New Guinea, from where the type was isolated.
Type specimen. PAPUA NEW GUINEA, Varirata National Park near Port Moresby, from humus soil, Dec. 1996, isol. A. Aptroot & A. van Iperen (CBS H-21611 holotype, culture ex-type CBS 337.97 = DTO 137C8).
Diagnosis — No growth on CYA at 37 °C, growth on CYA at 30 °C and CREA, conidia smooth-walled by light microscope, finely roughened under SEM, broadly ellipsoidal, 2–3 × 2–2.5 μm.
Description — Colony diam, 7 d, in mm: CYA 9–10; CYA 30 °C 10–12; CYA 37 °C no growth; MEA 4–5; YES 7–8; OA 8–9; DG18 7–8; CYAS 8–9; SNA 9–10; CREA 2–3.
Colonies at 25 °C, 7 d: CYA obverse greyish green (25B4), reverse greyish yellow to greyish orange (4B4–5B4); MEA obverse orange white (5A2), reverse yellowish brown (5D6); YES obverse greenish white (27A2), reverse brownish orange (5C5); OA obverse dark green (27F6); DG18 obverse pale yellow (1A3), reverse light yellow (3A4); CYAS obverse greyish green (25B4), reverse greyish yellow (3B5); SNA obverse white to light greyish, reverse white to light greyish; CREA acid not produced.
Conidiophores as solitary phialides; stipes smooth-walled, 3.5–11.5 × 1–2 μm; phialides ampulliform, 4.5–7 × 2–3.5 μm (5.7 ± 0.6 × 2.6 ± 0.2); conidia smooth-walled, broadly ellipsoidal, 2–3 × 2–2.5 μm (2.5 ± 0.2 × 2.3 ± 0.1, n = 30), average width/length = 0.91; sclerotia not produced.
SEM observations: Conidia very finely rough-walled, with very small tubercles present, connectives short, not well defined, tubercles 0.08–0.14 μm diam (0.11 ± 0.02).
Penicillium williamettense Visagie, M. Chr. & Samson, sp. nov. — MycoBank MB808272; Fig. 9m, n, 11a–h
Fig. 11.
Morphology of species characterised in this study. a, i, p. Colony morphology from left to right, top row: CYA, CYA 30 °C, CYA 37 °C, MEA, YES, DG18, CYAS, OA; from left to right, bottom row: reverse colonies on CYA, CYA 30 °C, CYA 37 °C, MEA, YES, DG18, CYAS and obverse on CREA; b–h, j–o, q–w. conidiophores, conidia, ascocarps (in v) and asci (in w) produced on OA. — Scale bar: w = 10 μm, applies to all microscope pictures, except v = 20 μm.
ITS Barcode. KF303667 (alternative markers: BenA = KF303622; CaM = KF303639; RPB2 = KF303709).
Etymology. Latin, named after Williamette National Forest in Oregon, from where the type was isolated.
Type specimen. USA, Oregon, Williamette National Forest near Blue River, unknown source and date, isol. M. Christensen (CBS H-21609 holotype, culture ex-type CBS 129774 = DTO 208A4).
Diagnosis — No growth on CYA at 30 °C or CYAS, colonies on CYA at 25 °C 7–8 mm, conidia rough-walled with big tubercles, visible with SEM, globose, 2–2.5 μm diam.
Description — Colony diam, 7 d, in mm: CYA 7–8; CYA 30 °C no growth; CYA 37 °C no growth; MEA 4–5; YES 7–8; OA 5–6; DG18 7–8; CYAS no growth; SNA 6–7; CREA no growth.
Colonies at 25 °C, 7 d: CYA obverse white, reverse pale yellow (3B3); MEA obverse orange white (5A2), reverse yellowish brown (5D6); YES obverse yellowish white (3A2), reverse yellowish white (3A2); OA obverse dull green (28E4); DG18 obverse yellowish white (3A2), reverse yellowish white (3A2); SNA obverse white to light greyish, reverse white to light greyish; CREA acid not produced.
Conidiophores as solitary phialides; stipes smooth-walled, 5–16 × 1–1.5 μm; phialides ampulliform, 4–6.5 × 2–3 μm (5.4 ± 0.5 × 2.6 ± 0.2); conidia rough-walled, globose, 2–2.5 × 2–2.5 μm (2.2 ± 0.2 × 2.2 ± 0.2, n = 25), average width/length = 0.99; sclerotia not produced.
SEM observations: Conidia rough-walled with very big tubercles present, connectives long with rings barely visible, tubercles 0.23–0.42 μm diam (0.33 ± 0.04).
Penicillium wisconsinense Visagie, M. Chr. & Samson, sp. nov. — MycoBank MB808273; Fig. 11i–o, 12a–c
Fig. 12.
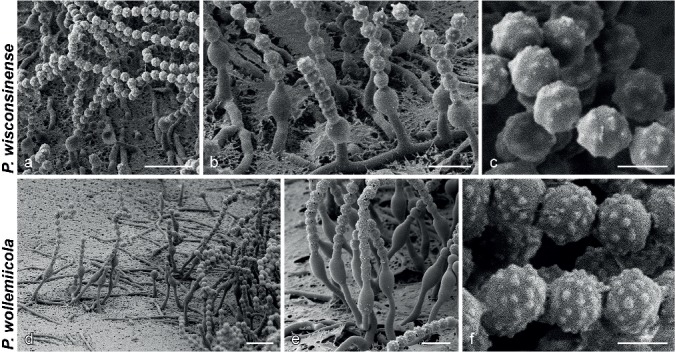
SEM micrographs showing characters of monophialidic Penicillium species. a, d. Overview of conidiogenous areas; b, e. phialides; c, f. conidia. — Scale bars: a, d = 10 μm; b, e = 5 μm; c, f = 2 μm.
ITS Barcode. KF303670 (alternative markers: BenA = KF303624; CaM = KF303641; RPB2 = KF303706).
Etymology. Latin, named after Wisconsin, from where the type was isolated.
Type specimen. USA, Wisconsin, unknown source and date, isol. M. Christensen (CBS H-21614 holotype, culture ex-type CBS 128279 = DTO 198H7 = WSF 3132).
Diagnosis — No growth on CYA at 37 °C or CREA, growth on CYA at 30 °C, colonies on CYA 7–10 mm, CYAS 0–2 mm, SNA 9–10 mm, conidia rough-walled with very big tubercles under SEM, globose, 2–2.5 × 2–2.5 μm.
Description — Colony diam, 7 d, in mm: CYA 7–10; CYA 30 °C 3–5; CYA 37 °C no growth; MEA 3–6; YES 7–11; OA 9–11; DG18 8–10; CYAS no growth, sometimes 2; SNA 9–10; CREA no growth.
Colonies at 25 °C, 7 d: CYA obverse yellowish to orange white (4A2–5A2), reverse orange white (5A2), sometimes greyish brown (5D3); MEA obverse orange white (5A2), reverse yellowish brown (5D6); YES obverse greyish turquoise to greyish green (24B3–25B3), reverse olive to olive brown (3D5–4D5); OA obverse dull green (28E4); DG18 obverse greyish turquoise to greyish green (24B3–25B3), reverse greyish green (28C3), some greyish yellow (4C4); CYAS obverse pale when growth, reverse pale when growth; SNA obverse dull green (29D4), reverse greenish grey (29B2); CREA acid not produced.
Conidiophores as solitary phialides; stipes smooth-walled, 5–45 × 1–2 μm; phialides ampulliform, 4.5–7.5 × 2.5–3 μm (5.7 ± 0.6 × 2.6 ± 0.1); conidia rough-walled, globose, 2–2.5 × 2–2.5 μm (2.3 ± 0.2 × 2.3 ± 0.2, n = 40), average width/length = 0.98; sclerotia not produced.
SEM observations: Conidia rough-walled with very large tubercles present, connectives rather short with rings not easily visible, tubercles 0.24–0.38 μm diam (0.31 ± 0.04).
Penicillium wollemiicola Visagie, Summerell, Shenoy & Seifert, sp. nov. — MycoBank MB808274; Fig. 11p–v, 12d–f
ITS Barcode. KJ174314 (alternative markers: BenA = KJ174315; CaM = KJ174316; RPB2 = KJ174313).
Etymology. Latin, named after the genus of the tree, Wollemia, in reference to the species isolated from its pine needles.
Type specimen. AUSTRALIA, from Wollemia nobilis needle debris, 12 Dec. 2007, coll. B. Summerell, isol. S. Shenoy (DAOM 675862 holotype, duplicate dried specimen deposited as CBS H-21615, ex-type CBS 137177 = DTO 297E3).
Diagnosis — No growth on CYA at 37 °C, CREA, growth on CYA at 30 °C, colonies on CYA 7–8 mm, CYAS 4–5 mm and SNA 7–9 mm, distinctly rough to spiny, globose, 2–2.5 μm diam. Ascomata pale coloured, asci not seen, ascospores lenticular, smooth-walled with two widely spaced ridges, 2.5–3.5 × 2.5–3.
Description — Colony diam, 7 d, in mm: CYA 7–8; CYA 30 °C 6–7; CYA 37 °C no growth; MEA 4–5; YES 8–9; OA 9–10; DG18 9–11; CYAS 4–5; SNA 7–9; CREA no growth.
Colonies at 25 °C, 7 d: CYA obverse white, reverse brown (5E5–5F5), inconspicuous brown soluble pigment; MEA obverse white, reverse yellowish brown (5D6); YES obverse orange white (5A2), reverse light orange (5A5); OA obverse greyish green (26C4), brown soluble pigment produced; DG18 obverse orange white (5A2), reverse yellowish white (3A2); CYAS obverse orange white (5A2), reverse yellowish white (3A2); SNA obverse dull green (26D3), reverse greenish grey (26B2); CREA acid not produced.
Conidiophores as solitary phialides; stipes smooth-walled, 4–13 × 1–1.5 μm; phialides ampulliform, 4.5–6.5 × 2.5–3 μm (5.6 ± 0.4 × 2.8 ± 0.3); conidia distinctly rough to spiny, globose, 2–2.5 × 2–2.5 μm (2.3 ± 0.1 × 2.3 ± 0.1, n = 25), average width/length = 0.99; sclerotia abundant, colourless, 30–94 × 24–90 μm; cleistothecia observed on old dry plate, similarly sized to sclerotia, wall pseudoparenchymatous, single layer, asci not observed, ascospores lenticular with two widely separated ridges, smooth-walled, 2.5–3.5 × 2.5–3 μm.
SEM observations: Conidia rough-walled with tubercles present, connectives relatively long without visible rings, tubercles 0.22–0.41 μm diam, average 0.31 ± 0.05.
DUBIOUS SPECIES
Penicillium parviverrucosum (K. Ando & Pitt) Houbraken & Samson, Stud. Mycol. 70: 48. 2011 — MycoBank MB561962
Basionym. Torulomyces parviverrucosus K. Ando & Pitt, Mycoscience 39: 317. 1998.
Type specimen. AUSTRALIA, Sydney, Gap Park, from soil, Oct. 1989, isol. K. Ando (TNS-F-0.5203 holotype, culture ex-type KY12720).
Notes — Because of commercial restrictions placed on the use of the ex-type strain, material was unavailable for examination. Morphologically, the species was distinguished from close relatives by its verrucose conidia that have small tubercles under SEM (Ando et al. 1998). Because of the overlapping characters of conidia observed during SEM examinations with other species in this section, it is clear that DNA sequence data is necessary to determine the precise taxonomic placement of P. parviverrucosum. Presently, we consider it a dubious species and should be reconsidered when material is made available.
DISCUSSION
Penicillium sect. Torulomyces was introduced by Houbraken & Samson (2011) for P. lagena (CBS 185.65) and the phylogenetically related P. lassenii and P. cryptum, with the consequence that the genus Torulomyces was confirmed as a synonym of Penicillium based on multigene phylogenetic evidence. This group of species all display restricted growth on media and the majority produce conidiophores consisting of solitary phialides. The aim of our study was to re-evaluate the taxonomy of species currently classified in the section in the context of the discovery of 12 new species, described above in the taxonomy section. This considerably extends the species sampling for this section, allowing a stringent evaluation of species concepts and the significance of inconspicuous morphological characters, and some consideration of ecological and biogeographic data in the resulting taxonomic structure. Our multigene phylogenies combined with morphological data recognize 16 species in the section. As noted above, identical ITS sequences are shared by several species; BenA was proposed as a secondary marker for Penicillium by Visagie et al. (2014b) and using BenA works well for identification of species in this section.
Species of sect. Torulomyces are morphologically challenging to identify. Despite this, their ability or inability to grow on certain media or at higher temperatures makes identification into smaller phenotypic groups possible. Further, within small groups of phylogenetically related species, phenotypic or morphological characters were discerned to distinguish most species pairs. For example, P. aeris and P. cantabricum are consistently resolved as close relatives in our phylogenies, but P. aeris has rougher conidia than P. cantabricum (Fig. 4c, i) and does not grow on CREA. Penicillium tubakianum and P. wollemiicola are close relatives, but differ in growth rates and conidia tubercle sizes (Fig. 9i, 12f). Our multigene phylogeny places P. marthae-christenseniae, which grows on CYA 30 °C, closest to P. catalonicum, which does not. Conidial shape also differs between the two species. Penicillium williamettense and P. oregonense are close relatives, but they have different growth rates and P. oregonense has conidia that are roughened in parallel rings (Fig. 6l, 9o). When one considers P. austricola and P. riverlandense, clearly distinct species under GCPSR, the only morphological difference is that P. riverlandense produces smaller colonies at 37 °C. As the most frequently sampled species, the phylogenetic distance and coherence of each species in the single gene phylogenies support the idea that the observed ecological differences between them, discussed below, indicate they are different species rather than different populations of one species.
SEM examination of conidial ornamentations and connectives also reveals differences that distinguish species. Similar observations were made by Ando et al. (1998), who reported that P. laeve and P. ovatum have smooth conidia, but they were able to distinguish between P. lagena and P. porphyreum based on the latter’s smaller conidial tubercles. In addition to size differences in tubercles, various aspects of conidial ornamentation are informative for some species. For example, P. variratense had very finely roughened conidia compared to the rather large ornamentations in P. williamettense. In P. catalonicum and P. oregonense conidial ornamentations formed parallel rings. Connectives were also sometimes useful, with those of P. porphyreum having two distinct, clearly visible rings. Although the combination of SEM and growth characters under different conditions allows phenotype based identification, routine species identification by these means is unrealistic, because SEM is less accessible and more time consuming than routine molecular methods. Although we incorporated these observations for species delineation in this study, we suggest that DNA sequences should be used for reliable identification.
Nine of the new species are based on single isolates. This is not ideal and reflects the difficulty inherent in isolating such slow growing fungi. Although the two species from the heavily sampled fynbos soils, P. austricola and P. riverlandense, are represented by many strains, only one culture of P. wollemiicola was isolated from Wollemi pine litter among more than 700 strains isolated via dilution to extinction, a method designed to recover slow growing fungi. Future isolations should result in the recovery of more strains that can be used to better document species of sect. Torulomyces. In most cases, the genetic distances between our species, including those represented by single strains, corresponds with that observed among phylogenetic species in other groups of Penicillium. Although strict application of a phylogenetic species concept requires more than one representative of each taxon, our newly proposed species otherwise conform with GCPSR. Unfortunately, the number of singletons in the phylogenies make bootstrap values and posterior probabilities uninformative to some degree.
Determining the distribution of this group of species from the literature is hampered because until recently, most slow growing, brown, monophialidic strains were considered to represent one species, P. lagena, which was considered widespread but rarely isolated (Domsch et al. 1980). Many of the species newly described in this paper were deposited into culture collections as that species. The literature mostly reports P. lagena, and the three other previously known species, from soil surveys (Christensen & Backus 1962, Barron 1967, Gochenaur 1978, Domsch et al. 1980, Ando et al. 1998, Renker et al. 2005), although P. lagena has also been reported from other sources like dead bark and cornmeal (Domsch et al. 1980). In this study, P. riverlandense and P. austricola were frequently isolated from Protea repens infructescences and the mites living in them. Our single strain of P. wollemiicola was isolated from needle debris of Wollemia nobilis, the so-called ‘dinosaur tree’ or ‘living fossil’ that was discovered in the Blue Mountains of Australia in 1994 (Jones et al. 1995). Penicillium austricola and P. riverlandense were isolated from the fynbos biome situated in the Western Cape of South Africa. One of the fynbos sampling sites in the Riverlands Nature Reserve near Malmesbury was surveyed over a period of four years and P. riverlandense was consistently recovered from these samples. In addition, P. austricola was only found at a fynbos sampling site at Struisbaai. Our success at isolating these two species makes us optimistic that the development of appropriate sampling or selective isolation techniques will eventually lead to increased sampling of other species, and better understanding of their ecologies.
Culture independent, environmental DNA sequencing might allow enhanced detection of the distribution of this group. Because ITS, the most frequently used marker in such studies, is uninformative at the species level in this group, presently we can only consider data at the sectional level. A BLAST search with the ITS sequence of T. lagena (CBS 185.65; KF303665) on GenBank results in only 14 hits that can be assigned to the section. These hits originate from soil (Czech Republic, one unknown location), plant material (Lithuania, Ecuador), marine sediments (China), plant root tissue (Australia, South Africa), a lichen (Letharia vulpine, USA) and house dust (Finland). Most of these hits are from cultured isolates, with only two hits (dust from Finland; soil from unreported country) from culture-independent detection techniques. This seems to support that these species are widespread but occur in very low numbers as reported in Domsch et al. (1980). The diversity of identified habitats from these studies suggests an even greater diversity of trophic modes for these species than we have so far explored. Whether the use of a gene with higher species resolution, such as the BenA, CaM or RPB2 used here, will allow more sensitive detection remains to be seen.
In our study, only two of the species, P. laeve and P. wollemiicola produced ascospores in some culture transfers. The ascospores of P. laeve are smooth-walled and globose, while those of P. wollemiicola are lenticular and have two broadly spaced ridges. The latter are similar to those described for P. lagena (Stolk & Samson 1983, as Eupen. limoneum), but we did not observe ascospores in that species. Penicillium lasseni was originally described in the sexual genus Eupenicillium, but we observed only sclerotia in our strains. The paucity of ascomata observed in our study suggests that either the ability to produce the sexual morph is easily lost in culture if the species are homothallic, or that the species are heterothallic. Thus, it is difficult to confidently interpret sexual characters for identifying species, although the lack of ridges on ascospores of P. laeve, and the difference in shape from the other two species, presently seems diagnostic.
Our phylogenies reveal that P. laeve and P. ovatum, previously included in sect. Torulomyces by Houbraken & Samson (2011), should be classified in sect. Exilicaulis with P. dimorphosporum as their closest relative. The placement of P. lassenii and P. cryptum in sect. Torulomyces remains questionable, because our phylogenies resolve them as distant relatives to the well-supported clade that contains the majority of species of sect. Torulomyces (Fig. 1, 2). A BLAST search of the P. lassenii ITS sequence results in the closest match of 93 % to P. sublateritium and P. cyaneum, classified in sect. Ramigena. For P. cryptum, a BLAST search results in a 93 % match to P. donkii and P. boreae, classified in sect. Stolkia. The closest GenBank match for RPB2 of P. lassenii and P. cryptum, respectively results in a 84 % similar match to P. ornatum and 83 % similar to P. lagena. Houbraken & Samson (2011) used RPB1, RPB2, Tsr1 and Cct8 for their sectional reclassification of Penicillium, which placed both species in a clade with P. lagena. However, the branches for these three species in the clade were very long and perhaps provided deceptive support for recognising them as separate and distinct sections. In a similar situation, Visagie et al. (2014a) recently described P. alfredii, but were unable to classify it in any of the 25 sections proposed by Houbraken & Samson (2011). Although it seemed that P. alfredii represented a new section, they opted to leave it unclassified because of the uncertain phylogenetic position of P. cryptum and P. lassenii. In their RPB2 phylogeny, P. cryptum made sect. Torulomyces polyphyletic with sect. Fracta. The fact that these two species are inconsistently placed with seemingly every gene analysed, hints that they may represent two distinct sections that are presently undersampled and thus unrecognizable by cladistic approaches. Conidiophore branching patterns further support the idea that they do not belong in sect. Torulomyces, but without isolating additional strains, closely related species and/or analysing more alternative gene sequences, the placement of P. cryptum, P. lassenii and P. alfredii will remain problematic. As a result, for the time being, we believe it best to leave P. cryptum and P. lassenii in sect. Torulomyces as proposed by Houbraken & Samson (2011).
Acknowledgments
We acknowledge the South African Biosystematics Initiative (SABI) of the National Research Foundation (NRF) and the Alfred P. Sloan Foundation for financial support. We thank Uwe Braun for providing his assistance on nomenclatural issues. We are also grateful to Martha Christensen who provided many of the strains described here as new species, Brett Summerell for collecting Wollemi pine litter and Damodar Shenoy for isolating P. wollemiicola from that substrate, and Katsuhiko Ando who provided the ex-type strains of P. ovatum and P. laeve.
REFERENCES
- Ando K, Nawawi A, Manoch L, et al. 1998. Three new species and a new combination in the genus Torulomyces from soil. Mycoscience 39: 313–318. [Google Scholar]
- Barron GL. 1967. Torulomyces and Monocillium. Mycologia 59: 716–718. [PubMed] [Google Scholar]
- Christensen M, Backus MP. 1962. A new Penicillium from coniferous forest soils. Mycologia 54: 573–577. [Google Scholar]
- Delitsch H. 1943. Ergebnisse der theoretischen und angewandten Mikrobiologie: Band I: Systematik der Schimmelpilze. Neumann, Neudamm, Germany. [Google Scholar]
- Domsch KH, Gams W, Anderson T-H. 1980. Compendium of soil fungi. IHW-Verlag, Eching. [Google Scholar]
- Gams W. 1971. Cephalosporium-artige Schimmelpilze (Hyphomycetes). Gustav Fischer Verlag, Stuttgart. [Google Scholar]
- Gams W. 1978. Connected and disconnected chains of phialoconidia and Sagenomella gen. nov. segregated from Acremonium. Persoonia 10: 97–112. [Google Scholar]
- Gochenaur SE. 1978. Fungi of a Long Island oak-birch forest I. Community Organization and Seasonal Occurrence of the Opportunistic Decomposers of the A Horizon. Mycologia 70: 975–994. [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Molecular Biology and Evolution 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Hashmi MH, Kendrick B, Morgan-Jones G. 1972. Conidium ontogeny in hyphomycetes. The genera Torulomyces Delitsch and Monocillium Saksena. Canadian Journal of Botany 50: 1461–1463. [Google Scholar]
- Houbraken J, Samson RA. 2011. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology 70: 1–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WG, Hill KD, Allen JM. 1995. Wollemia nobilis, a new living Australian genus and species in the Araucariaceae. Telopea 6: 173–176. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. 1967. Methuen Handbook of Colour. Methuen & Co Ltd, Copenhagen. [Google Scholar]
- Liang ZQ, Han YF, Chu HL, et al. 2009. Studies on the genus Paecilomyces in China V. Taifanglania gen. nov. for some monophialidic species. Fungal Diversity 34: 69–77. [Google Scholar]
- Matsushima T. 1987. MMatsushima mycological memoirs no. 5: 1–100. [Google Scholar]
- McNeill J, Barrie FR, Buck WR, et al. 2012. International Code of Nomenclature for algae, fungi and plants (Melbourne Code). Koeltz Scientific Books; Konigstein. (Regnum vegetabile no. 154). [Google Scholar]
- Nylander AJJ, Ronquist F, Huelsenbeck JP, et al. 2004. Bayesian phylogenetic analysis of combined data. Systematic Biology 53: 47–67. [DOI] [PubMed] [Google Scholar]
- Onions AHS, Barron GL. 1967. Monophialidic species of Paecilomyces. Mycological Papers 107: 1–25. [Google Scholar]
- Pitt JI, Hocking AD. 1985. Interfaces among genera related to Aspergillus and Penicillium. Mycologia 77: 810–824. [Google Scholar]
- Pitt JI, Samson RA. 1993. Names in current use in the family Trichocomaceae. In: Greuter W. (ed), Names in current use in the family Trichocomaceae, Cladoniaceae, Pinaceae, and Lemnaceae: 13–57. Koeltz Scientific Books, Germany. [Google Scholar]
- Pitt JI, Samson RA, Frisvad JC. 2000. List of accepted species and their synonyms in the family Trichocomaceae. In: Samson RA, Pitt JI. (eds), Integration of modern taxonomic methods for Penicillium and Aspergillus classification: 9–79. Harwood Academic Publishers, Amsterdam, The Netherlands. [Google Scholar]
- Renker C, Otto P, Schneider K, et al. 2005. Oribatid mites as potential vectors for soil microfungi: study of mite-associated fungal species. Microbial Ecology 50: 518–528. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Saksena SB. 1955. A new fungus, Monocillium indicum gen. et sp. nov., from soil. Indian Phytopathology 8: 9–12. [Google Scholar]
- Samson RA. 1974. A monograph of Paecilomyces and allied hyphomycetes. Studies in Mycology 6: 1–119. [Google Scholar]
- Samson RA, Visagie CM, Houbraken J, et al. 2014. Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology 78: 141–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, et al. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert KA, Morgan-Jones G, Gams W, et al. 2011. The genera of Hyphomycetes. CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands. [Google Scholar]
- Sigler L, Sutton DA, Gibas CFC, et al. 2010. Phialosimplex, a new anamorphic genus associated with infections in dogs and having phylogenetic affinity to the Trichocomaceae. Medical Mycology 48: 335–345. [DOI] [PubMed] [Google Scholar]
- Stolk AC, Samson RA. 1983. The Ascomycete genus Eupenicillium and related Penicillium anamorphs. Studies in Mycology 23: 1–149. [Google Scholar]
- Stolk AC, Samson RA. 1985. A new taxonomic scheme for Penicillium anamorphs. In: Samson RA, Pitt JI. (eds), Advances in Penicillium and Aspergillus systematics: 163–192. Plenum Press, New York, USA. [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, et al. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32. [DOI] [PubMed] [Google Scholar]
- Visagie CM, Hirooka Y, Tanney JB, et al. 2014a. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Studies in Mycology 78: 63–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Frisvad JC, et al. 2014b. Identification and nomenclature of the genus Penicillium. Studies in Mycology 78: 343–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Rodriques C, et al. 2013. Five new Penicillium species in section Sclerotiora: a tribute to the Dutch Royal family. Persoonia 31: 42–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu F, Wu W, et al. 2015. A phylogenetic assessment and taxonomic revision of the thermotolerant hyphomycete genera Acrophialophora and Taifanglania. Mycologia 107: 768–779. [DOI] [PubMed] [Google Scholar]


