Abstract
The taxonomy of Talaromyces rugulosus, T. wortmannii and closely related species, classified in Talaromyces sect. Islandici, is reviewed in this paper. The species of Talaromyces sect. Islandici have restricted growth on MEA and CYA, generally have yellow mycelia and produce rugulosin and/or skyrin. They are important in biotechnology (e.g. T. rugulosus, T. wortmannii) and in medicine (e.g. T. piceus, T. radicus). The taxonomy of sect. Islandici was resolved using a combination of morphological, extrolite and phylogenetic data, using the Genealogical Concordance Phylogenetic Species Recognition (GCPSR) concept, with special focus on the T. rugulosus and T. wortmannii species complexes. In this paper, we synonymise T. variabilis, Penicillium concavorugulosum and T. sublevisporus with T. wortmannii, and introduce four new species as T. acaricola, T. crassus, T. infraolivaceus and T. subaurantiacus. Finally, we provide a synoptic table for the identification of the 19 species classified in the section.
Keywords: multi-gene phylogeny, Penicillium rugulosum, Penicillium variabile, Talaromyces acaricola, Talaromyces crassus, Talaromyces infraolivaceus, Talaromyces subaurantiacus
INTRODUCTION
Penicillium s.l. is an important group of molds associated with a wide range of habitats, where it acts as degraders of organic material (Pitt 1980, Frisvad & Samson 2004). Penicillium subg. Biverticillium species are phylogenetically resolved in a well-supported monophyletic clade together with the teleomorphic genus Talaromyces, distinct from other Penicillium subgenera (LoBuglio et al. 1993, Houbraken & Samson 2011, Samson et al. 2011). Following the recent move to single name nomenclature in fungi, Samson et al. (2011) subsequently combined all accepted species belonging to Penicillium subg. Biverticillium into Talaromyces. Yilmaz et al. (2014), using a polyphasic approach, accepted 88 species in Talaromyces and based on a multi-gene phylogeny classified them into seven sections. One of these sections is sect. Islandici and this section is in the focus of this study.
Talaromyces sect. Islandici includes species that grow restrictedly on most media, have predominately yellow mycelia, and produce characteristic mycotoxins. Previously, Pitt (1980) introduced the Penicillium sect. Simplicium ser. Islandica for species which grow restrictedly on malt extract agar (MEA) and Czapek yeast extract agar (CYA). He included T. brunneus, T. erythromellis, T. islandicus, T. loliensis, T. piceus, T. primulinus, T. rugulosus and T. variabilis. However, a multi-gene phylogeny showed that T. erythromellis and T. primulinus are located in sect. Trachyspermi and Talaromyces, respectively (Yilmaz et al. 2014) and that the remaining species were included in sect. Islandici (Yilmaz et al. 2014).
This group of species typically produces rugulosin and/or skyrin (except for T. scorteus) (Yilmaz et al. 2014). Rugulosin is a bis-anthraquinoid pigment described by Breen et al. (1955) with a specific antibacterial activity against Staphylococcus aureus and moderate activity against the parasitic fungus-like Chromistan Pythium intermedium. Rugulosin was also indicated as a weak hepato-carcinogen (Ueno et al. 1980). A recent study showed that rugulosin extracted from T. radicus had antimicrobial activity against methicillin resistant S. aureus (Yamazaki et al. 2010a–c). Even though it has been classified as a mycotoxin, erythroskyrin was also reported to be an antitumor agent (Kenkyusho 1983). Rubroskyrin and flavoskyrin are also classified as toxins (Kawai et al. 1984, Mori et al. 1996) and are produced by some sect. Islandici species. The rugulovasines (Antipova et al. 2008) were referred to as mycotoxins but toxicity data are scarce (Cole & Cox 1981).
Talaromyces sect. Islandici includes important enzyme producers such as T. wortmannii (= T. variabilis) producing urethanase (Zhou et al. 2013) and T. rugulosus producing beta-rutinosidase and phosphatase (Reyes et al. 1999, Narikawa et al. 2000). Talaromyces wortmannii also produces high concentrations of uncharacterised bioactive natural compounds. Bara et al. (2013) showed that six compounds isolated from T. wortmannii exhibited antibacterial activity, predominantly directed against S. aureus, including (multi) drug-resistant isolates. However, other Gram-positive bacteria such as Streptococcus, Enterococcus and Bacillus were only moderately affected (Bara et al. 2013). Several compounds were isolated from T. wortmannii by Pretsch et al. (2014). A metabolite labelled as Compound C was found an effective antimicrobial against Propionibacterium acnes and had anti-inflammatory properties (Pretsch et al. 2014). It was thus suggested that this substance, or the crude extract, could represent alternative treatments for antibiotic/anti-inflammatory therapy for acne (Pretsch et al. 2014).
Some species of Talaromyces sect. Islandici may be potential opportunistic pathogens because of their ability to grow at 37 °C and higher (Yilmaz et al. 2014). Previous studies reported that T. piceus caused fungaemia (Horré et al. 2001) and rib osteomyelitis in an X-linked chronic granulomatous disease (X-CGD) (Santos et al. 2006). Talaromyces radicus caused a fatal infection in a German shepherd (de Vos et al. 2009). Corneal ulcer caused by T. rugulosus was reported by Swietliczkowa et al. (1984). Talaromyces islandicus can also grow at 37 °C, but until now has not been isolated from humans. It is more important for agriculture because it produces mycotoxins such as cyclochlorotine, islanditoxin, erythroskyrine and luteoskyrin, which are hepatotoxic agents and also carcinogenic (Uraguchi et al. 1961, 1972, Uraguchi 1962, Ueno & Ishikawa 1969, Bouhet et al. 1976, Stark et al. 1978, Pitt & Hocking 2009). This species also causes yellowing of rice in Japan (Saito et al. 1971, Sakai et al. 2005, Oh et al. 2008).
The diverse range of species in sect. Islandici species, and their importance in medicine, agriculture and biotechnology, make correct identifications crucial. The aim of this study, was thus to complete a multigene phylogenetic study of the section, and apply Genealogical Concordance Phylogenetic Species Recognition (GCPSR, Taylor et al. 2000) by adding to the internal transcribed spacer (ITS) and β-tubulin (BenA) data published in Yilmaz et al. (2014) and studying extrolites produced by the species, with a special focus on the T. wortmannii and T. rugulosus species complexes. The phylogenies resulted in the identification of four unique clades that we describe here as new species. Strains of these four new species mainly originate from a biodiversity study of Fynbos soil, Protea repens infructescences and air, in the Western Cape of South Africa (Visagie et al. 2009, 2013, 2014c, Visagie & Jacobs 2012). In addition to the multi-gene phylogenies, we compare the morphological characters and extrolite data of the new species with others in the section and provide notes to facilitate their identification.
MATERIAL AND METHODS
Isolates
Isolates used in this study were obtained from the culture collections of the CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands; the culture collection of Center for Microbial Biotechnology at Department of Systems Biology, Technical University of Denmark, Lyngby, Denmark (IBT); the Agricultural Research Service Culture Collection, Peoria, Illinois, USA (NRRL); the Canadian Department of Agriculture – Mycology Culture Collection, Ottawa, Canada (DAOM); and isolates deposited in the working collection of the Department of Applied and Industrial Mycology (DTO), housed at CBS-KNAW. Isolates are listed in Table 1.
Table 1.
Strains used for this study.
| Name | Collection no. | Substrate and origin | GenBank accession numbers |
|||
|---|---|---|---|---|---|---|
| BenA | CaM | ITS | RPB2 | |||
| T. acaricola | CBS 137367 = DTO 61-H2 | Air sample, beer producing factory, Kaulille, Belgium | KF984567 | KF984720 | KF984862 | KF984953 |
| CBS 137369 = DTO 66-H7 | Unknown, unknown | KF984568 | KF984721 | KF984860 | KF984954 | |
| CBS 137374 = DTO 77-C7 | Apple concentrate, The Netherlands | KF984569 | KF984722 | KF984861 | KF984955 | |
| CBS 137386T = DTO 183-B3 = DAOM 241025 = IBT 32387 | Mites from Protea repens infructescens, Malmesbury, South Africa | JX091610 | JX140729 | JX091476 | KF984956 | |
| CBS 137387 = DTO 183-B4 | Mites from Protea repens infructescens, Malmesbury, South Africa | JX091611 | JX140730 | JX091477 | KF984957 | |
| CBS 137388 = DTO 183-C1 = DAOM 241029 | Protea repens infructescence, Malmesbury, South Africa | JX091612 | JX140731 | JX091478 | KF984958 | |
| CBS 137390 = DTO 183-E3 = DAOM 241022 | Mites from Protea repens infructescens, Struisbaai, South Africa | JX091613 | JX140732 | JX091479 | KF984959 | |
| T. allahabadensis | CBS 137361 = DTO 55-F9 | Swab sample in vaccin producing factory, The Netherlands | KF984608 | KF984763 | KF984867 | KF985000 |
| CBS 137362 = DTO 55-G3 | Indoor air sample in vaccin producing factory, The Netherlands | KF984610 | KF984765 | KF984869 | KF985002 | |
| CBS 137373 = DTO 77-C3 | Guava pure imported to The Netherlands | KF984615 | KF984769 | KF984871 | KF985007 | |
| CBS 137397 = DTO 245-E3 | House dust, Mexico | KF984605 | KF984761 | KF984864 | KF984998 | |
| CBS 137399 = DTO 267-H6 | House dust, Thailand | KF984607 | KF984762 | KF984866 | KF984997 | |
| CBS 178.81 = DTO 247-D9 = ATCC 48474 = FRR 3579 = IMI 253805 | Type of P. zacinthae, crepis zacintha, Alicante, Spain | KF984612 | KF984767 | KF984863 | KF985004 | |
| CBS 441.89 = DTO 247-D5 | Seed groud, Denmark | KF984613 | KF984759 | KF984872 | KF985005 | |
| DTO 055-G1 | Indoor air sample in vaccin producing factory, The Netherlands | KF984609 | KF984764 | KF984868 | KF985001 | |
| DTO 067-F7 | Indoor air sample in vaccin producing factory, The Netherlands | KF984611 | KF984766 | KF984870 | KF985003 | |
| DTO 245-I6 | House dust, Mexico | KF984606 | KF984760 | KF984865 | KF984999 | |
| CBS 453.93T = DTO 93-B7 = CBS 304.63 = ATCC 15067 = NRRL 3397 = FRR 3397 = IBT 3926 = IBT 10824 | Cultivated soil, Allahabad, India | KF984614 | KF984768 | KF984873 | KF985006 | |
| T. atricola | CBS 255.31T = DTO 278-F1 = NRRL1052 = FRR 1052 = Thom 4640.439 = ATCC 52257 = IBT 4489 | Unknown, unknown | KF984566 | KF984719 | KF984859 | KF984948 |
| T. brunneus | CBS 227.60T = DTO 284-G1 = ATCC 18229 = FRR 646 = IFO 6438 = IMI 078259 = IBT 4490 | Milled rice impoted into Japan, Thailand | KJ865722 | KJ885264 | JN899365 | KM023272 |
| T. columbinus | CBS 137393 = DTO 189-A5 = IBT 13019 | Chicken feed (Unga), Nairobi, Kenya | KF984659 | KF984671 | KF984794 | KF984897 |
| NRRL 58644 | Air, Maryland, USA | KF196842 | KF196880 | KF196899 | KF196987 | |
| NRRL 62680 | Corn grits, Illinois, USA | KF196844 | KF196882 | KF196901 | KF196988 | |
| T. crassus | CBS 137379 = DTO 181-B1 | Protea repens infructescence, Stellenbosch, South Africa | JX091606 | JX140726 | JX091473 | KF984912 |
| CBS 137380 = DTO 181-B2 | Mite from Protea repens infructescence, Stellenbosch, South Africa | JX091607 | JX140725 | JX091474 | KF984913 | |
| CBS 137381T = DTO 181-C5 = DAOM 241027 = IBT 32814 | Protea repens infructescence, Stellenbosch, South Africa | JX091608 | JX140727 | JX091472 | KF984914 | |
| T. infraolivaceus | CBS 137385T = DTO 182-I2 = DAOM 241024 = IBT 32487 | Mite from Protea repens infructescence, Stellenbosch, South Africa | JX091615 | JX140734 | JX091481 | KF984949 |
| CBS 137389 = DTO 183-D2 = DAOM 241023 | Mite from Protea repens infructescence, Struisbaai, South Africa | JX091616 | JX140735 | JX091482 | KF984950 | |
| CBS 137391 = DTO 183-F1 = DAOM 241030 | Protea repens infructescence, Struisbaai, South Africa | JX091617 | JX140736 | JX091483 | KF984951 | |
| CBS 137392 = DTO 183-G4 | Mite from Protea repens infructescence, Struisbaai, South Africa | JX091618 | JX140737 | JX091484 | KF984952 | |
| T. islandicus | CBS 338.48T = DTO 107-H2 = ATTC 10127 = FRR 1036 = IMI 040042 = NRRL 1036 = IBT 14884 = IBT 4476 | Unknown, Cape Town, South Africa | KF984655 | KF984780 | KF984885 | KF985018 |
| CBS 117284 = DTO 2-C7 | Wheat flour, The Netherlands | KF984652 | KF984777 | KF984882 | KF985015 | |
| CBS 165.81 = DTO 158-D6 = ATCC 42240 = IMI 253796 | Type of P. aurantioflammiferum, spice mixture used in sausage making industry, Spain | KF984653 | KF984778 | KF984883 | KF985016 | |
| CBS 394.50 = DTO 93-B9 | Kapok fibre, unknown | KF984656 | KF984781 | KF984886 | KF985019 | |
| DTO 158-D7 | Air contaminant, The Netherlands | KF984654 | KF984779 | KF984884 | KF985017 | |
| T. loliensis | CBS 172.91 = DTO 105-E9 | Soil, New Zealand | KF984657 | KF984782 | KF984887 | KF985020 |
| CBS 643.80T = DTO 169-F7 = ATCC 52252 = FRR 1798 = IMI 216901 = NRRL 2148 = MUCL 31325 = IBT 4546 | Rye grass (Lolium), New Zealand | KF984658 | KF984783 | KF984888 | KF985021 | |
| T. piceus | CBS 116872 = DTO 247-E1 | Production plant, The Netherlands | KF984660 | KF984678 | KF984788 | KF984903 |
| CBS 132063 = DTO 191-C5 | Straw used in horse stable, The Netherlands | KF984665 | KF984674 | KF984789 | KF984904 | |
| CBS 137363 = DTO 58-D1 | Pectin, unknown | KF984664 | KF984677 | KF984787 | KF984902 | |
| CBS 137377 = DTO 178-F3 | House dust, Cape Town, South Africa | KF984661 | KF984676 | KF984784 | KF984900 | |
| CBS 250.56 = DTO 93-G8 | Sputum from a man patient, The Netherlands | KF984666 | KF984679 | KF984790 | KF984905 | |
| CBS 354.66 = DTO 93-C6 | Unknown, United Kingdom | KF984667 | KF984672 | KF984791 | KF984907 | |
| CBS 361.48T = DTO 93-C8 = IMI 040038 = NRRL 1051 = FRR 1051 = ATCC 10519 = Thom 5633.6 = QM 7609 = IBT 4460 | Unknown, unknown | KF984668 | KF984680 | KF984792 | KF984899 | |
| CBS 435.62 = DTO 228-E5 | Sputum, The Netherlands | KF984669 | KF984681 | KF984793 | KF984906 | |
| DTO 191-C4 | Straw used in horse stable, The Netherlands | KF984662 | KF984673 | KF984785 | KF984898 | |
| DTO 191-C6 | Silage, grass, The Netherlands | KF984663 | KF984675 | KF984786 | KF984901 | |
| T. radicus | CBS 100488 = DTO 37-F6 | Wheat root, New South Wales | KF984598 | KF984772 | KF984877 | KF985012 |
| CBS 100489T = DTO 37-F7 = FRR 4718 = IBT 14379 | Root seadling, New South Wales | KF984599 | KF984773 | KF984878 | KF985013 | |
| CBS 100490 = DTO 37-F8 | Wheat root, New South Wales | KF984600 | KF984774 | KF984879 | KF985014 | |
| CBS 122887 = DTO 63-C5 | Ex infection dog, The Netherlands | KF984604 | KF984776 | KF984876 | KF985011 | |
| CBS 137382 = DTO 181-D5 | Mite from Protea repens infructescence, Stellenbosch, South Africa | KF984602 | KF984775 | KF984875 | KF985009 | |
| DTO 181-D4 | Mite from Protea repens infructescence, Stellenbosch, South Africa | KF984601 | KF984770 | KF984880 | KF985008 | |
| DTO 181-D7 | Mite from Protea repens infructescence, Stellenbosch, South Africa | KF984603 | KF984771 | KF984881 | KF985010 | |
| T. rotundus | CBS 369.48T = DTO 105-D3 = IMI 40589 = NRRL 2107 = FRR 2107 = ATCC 10493 = IBT 4829 | Wood, Panama | KJ865730 | KJ885278 | JN899353 | KM023275 |
| T. rugulosus | CBS 101423 = DTO 225-I6 | Wood, British Columbia, Vancouver, Canada | KF984597 | KF984717 | KF984856 | KF984944 |
| CBS 111.64 = DTO 93-B8 | Jute, The Netherlands | KF984573 | KF984691 | KF984857 | KF984923 | |
| CBS 137360 = DTO 14-A2 | Cardboard, Norway | KF984588 | KF984703 | KF984835 | KF984931 | |
| CBS 137366 = DTO 61-E8 | Air sample, beer producing factory, Kaulille, Belgium | KF984572 | KF984700 | KF984850 | KF984922 | |
| CBS 137369 = DTO 63-C7 | Jelly like product used in bakery, The Netherlands | KF984586 | KF984713 | KF984851 | KF984947 | |
| CBS 137372 = DTO 70-B7 | Wood of crate, United Kingdom | KF984582 | KF984697 | KF984855 | KF984943 | |
| CBS 137378 = DTO 180-A6 | House dust, Cape Town, South Africa | KF984591 | KF984704 | KF984838 | KF984933 | |
| CBS 137398 = DTO 254-A2 | Indoor air, Utrecht, The Netherlands | KF984595 | KF984695 | KF984845 | KF984937 | |
| CBS 258.37 = DTO 166-A6 = NRRL 2116 = KCTC 16068 | Unknown, unknown | KF984580 | KF984718 | KF984833 | KF984928 | |
| CBS 344.51 = DTO 93-C2 = ATCC 22352 = FRR 560 | Type of P. echinosporum, unknown, Japan | KF984574 | KF984701 | KF984858 | KF984924 | |
| CBS 371.48T = DTO 278-E8 = NRRL 1045 = FRR 1045 = IMI 040041 = ATCC 10128 = MUCL 31201 = IBT 4485 | Roating potato tubers (solanum tuberosum), USA | KF984575 | KF984702 | KF984834 | KF984925 | |
| CBS 378.48 = DTO 278-F2 = NRRL1073 = FRR 1073 = IMI 040034 = ATCC 10503 = Thom 4640.444 | Type of P. tardum & P. elongatum, decaying twigs, France | KF984579 | KF984711 | KF984832 | KF984927 | |
| DTO 066-G6 | Unknown, unknown | KF984585 | KF984714 | KF984852 | KF984940 | |
| DTO 066-G7 | Unknown, unknown | KF984584 | KF984715 | KF984853 | KF984941 | |
| DTO 070-B5 | Wood of crate, United Kingdom | KF984583 | KF984716 | KF984854 | KF984942 | |
| DTO 179-I3 | House dust, Cape Town, South Africa | KF984589 | KF984692 | KF984836 | KF984932 | |
| DTO 180-A4 | House dust, Cape Town, South Africa | KF984590 | KF984693 | KF984837 | KF984929 | |
| DTO 180-B3 | House dust, Cape Town, South Africa | KF984587 | KF984705 | KF984839 | KF984934 | |
| DTO 180-B9 | House dust, Cape Town, South Africa | KF984570 | KF984698 | KF984840 | KF984920 | |
| DTO 193-I5 = IBT 10835 = IBT 3616 | Unknown, unknown | KF984578 | KF984706 | KF984831 | KF984926 | |
| DTO 199-H3 | Material for bed for milking cows, The Netherlands | KF984576 | KF984707 | KF984841 | KF984946 | |
| DTO 244-F6 | House dust, New Zealand | KF984593 | KF984709 | KF984843 | KF984930 | |
| DTO 254-A1 | Indoor air, Utrecht, The Netherlands | KF984594 | KF984694 | KF984844 | KF984936 | |
| DTO 269-G1 | House dust, Cape Town, South Africa | KF984581 | KF984696 | KF984846 | KF984938 | |
| DTO 269-G4 | House dust, Cape Town, South Africa | KF984571 | KF984699 | KF984847 | KF984921 | |
| DTO 278-E9 = NRRL 1053 = FRR 1053 = IMI 028259 | Type of P. chrysitis, unknown, unknown | KF984577 | KF984710 | KF984848 | KF984945 | |
| DTO 61-E4 | Air sample, beer producing factory, Kaulille, Belgium | KF984596 | KF984712 | KF984849 | KF984939 | |
| T. scorteus | CBS 233.60 = DTO 278-F3 = NRRL 203 = IMI 78256 = FRR 203 = ATCC 18481 = IFO 6437 | Type of T. phialosporus, milled Californian rice, Japan | KF984562 | KF984683 | KF984895 | KF984917 |
| CBS 499.75 = DTO 247-D7 = IMI 144145 | Unknown, Nigeria | KF984563 | KF984685 | KF984894 | KF984918 | |
| CBS 500.75 = DTO 225-I5 = IMI 152168 = KCTC 16071 | Unknown, Sierra Leone | KF984564 | KF984687 | KF984896 | KF984919 | |
| DTO 270-A6 | House dust, Thailand | KF984561 | KF984686 | KF984893 | KF984915 | |
| CBS 340.34T = DTO 278-F4 = NRRL 1129 = FRR 1129 | Military equipment, Japan | KF984565 | KF984684 | KF984892 | KF984916 | |
| T. subaurantiacus | CBS 137383T = DTO 181-I2 = DAOM 241020 = IBT 32838 | Fynbos soil, Stellenbosch, South Africa | JX091609 | JX140728 | JX091475 | KF984960 |
| T. tardifaciens | CBS 250.94T = DTO 247-D6 = SUM 3017 = IBT 14986 | Paddy soil, Bhaktapur, Nepal | KF984560 | KF984682 | KF984874 | KF984908 |
| T. tratensis | CBS 113146T = DTO 140-G4 = KUFC 3383 = IBT 31982 | Soil, Trat, Thailand | KF984559 | KF984690 | KF984891 | KF984911 |
| CBS 137400 = DTO 270-F5 | House dust, Mexico | KF984557 | KF984688 | KF984889 | KF984909 | |
| CBS 137401 = DTO 278-F6 = NRRL1013 = FRR 1013 | Carbonated beverage, Washington DC, USA | KF984558 | KF984689 | KF984890 | KF984910 | |
| T. wortmannii | CBS 100258c = DTO 226-A5 | Unknown, unknown | KF984630 | KF984723 | KF984823 | KF984969 |
| CBS 116051a = DTO 92-I7 | Unknown, unknown | KF984640 | KF984736 | KF984809 | KF984995 | |
| CBS 130028c = DTO 208-I8 | Unknown, adjacent to Cinnabar Park, Medicine Bow National Forest, Wyoming, USA | KF984631 | KF984749 | KF984818 | KF984974 | |
| CBS 137364a = DTO 58-G6 | Corn kernels, imported from Brazil, The Netherlands | KF984636 | KF984732 | KF984806 | KF984980 | |
| CBS 137365a = DTO 58-H6 | Wooden crate, imported from China, The Netherlands | KF984638 | KF984734 | KF984808 | KF984993 | |
| CBS 137368c = DTO 63-C6 | Indoor air, The Netherlands | KF984617 | KF984746 | KF984815 | KF984964 | |
| CBS 137371c = DTO 67-F9 | Indoor air sample in vaccin producing factory, The Netherlands | KF984646 | KF984753 | KF984825 | KF984972 | |
| CBS 137375c = DTO 161-G6 | Bamboo, Ho Chi Minh City, Vietnam | KF984643 | KF984742 | KF984820 | KF984976 | |
| CBS 137376b = DTO 176-I7 = PF1130 | Type of T. sublevisporus, soil, Japan | KF984632 | KF984724 | KF984800 | KF984979 | |
| CBS 137384a = DTO 181-H2 = DAOM 241019 | Mite from Protea repens infructescence, Stellenbosch, South Africa | KF984622 | KF984726 | KF984802 | KF984985 | |
| CBS 137394a = DTO 189-C8 = IBT 28678 | Soil, Amazonas, Brazil | KF984624 | KF984729 | KF984795 | KF984988 | |
| CBS 137395a = DTO 189-D8 = IBT 30868 | Unknown, Brazil | KF984634 | KF984740 | KF984805 | KF984990 | |
| CBS 293.53 = DTO 108-A3 | Soil, Mozambique | KF984650 | KF984758 | KF984827 | KF984962 | |
| CBS 316.63c = DTO 93-A1 = KCTC 16056 | Polyvinyl acetate, The Netherlands | KF984620 | KF984750 | KF984819 | KF984975 | |
| CBS 319.63 = DTO 108-A4 | Unknown, unknown | KF984651 | KF984755 | KF984828 | KF984961 | |
| CBS 385.48c = DTO 92-I8 = IMI 40040 = NRRL 1048 = FRR 1048 = ATCC 10508 = IFO 6111 | Type of T. variabilis, coconut matting, Johannesburg, South Africa | KF196853 | KF196878 | KF196915 | KF196975 | |
| CBS 391.48T = DTO 278-F5 = NRRL 1017 = IMI 040047 = FRR 1017 = ATCC 10517 = IFO 7738 = Thom 4733.126.1 = IBT 4838 | Soil, Denmark | KF984648 | KF984756 | KF984829 | KF984977 | |
| CBS 553.72 = DTO 107-H4 | Soil, France | KF984649 | KF984757 | KF984830 | KF984978 | |
| CBS 777.95a = DTO 92-I6 | Rubber, Sri Lanka | KF984641 | KF984741 | KF984810 | KF984981 | |
| CBS 895.73a = DTO 92-I9 = ATCC 20201 = IFO 4683 = BCRC 31677 | Unknown, Japan | KF984626 | KF984737 | KF984811 | KF984982 | |
| CBS 896.73a = DTO 93-C7 = BCRC 31676 = IFO 9136 | Unknown, Japan | KF984642 | KF984738 | KF984799 | KF984996 | |
| CBS 898.73a = DTO 93-A2 = ATCC 20202 = IFO 6017 = BCRC 31678 | Unknown, Japan | KF984627 | KF984739 | KF984812 | KF984983 | |
| DTO 1-I3c | Wood, Amsterdam, The Netherlands | KF984616 | KF984745 | KF984814 | KF984963 | |
| DTO 127-I4c | Sauce, The Netherlands | KF984628 | KF984744 | KF984813 | KF984967 | |
| DTO 181-B7a = DAOM 241018 | Mite from Protea repens infructescence, Stellenbosch, South Africa | KF984621 | KF984725 | KF984801 | KF984984 | |
| DTO 181-I7a | Mite from Protea repens infructescence, Stellenbosch, South Africa | KF984623 | KF984727 | KF984803 | KF984986 | |
| DTO 189-C6a = IBT 27918 = NCB 1494 | Ex passito wine grape, Italy | KF984633 | KF984728 | KF984804 | KF984987 | |
| DTO 189-C9a = IBT 28726 | Soil, Amazonas, Brazil | KF984625 | KF984730 | KF984796 | KF984989 | |
| DTO 189-D9a = IBT 31242 | Wheat flour, Denmark | KF984629 | KF984743 | KF984821 | KF984968 | |
| DTO 278-E7c = NRRL 2125 = FRR 2125 | Weathering canvas, Panama | KF984635 | KF984731 | KF984797 | KF984991 | |
| DTO 55-G2c | Swab sample in vaccin producing factory, The Netherlands | KF984644 | KF984751 | KF984824 | KF984970 | |
| DTO 58-H1a | Corn kernels, imported from Brazil, The Netherlands | KF984637 | KF984733 | KF984807 | KF984992 | |
| DTO 66-H5c | Unknown, unknown | KF984639 | KF984735 | KF984798 | KF984994 | |
| DTO 67-F8c | Indoor air sample in vaccin producing factory, The Netherlands | KF984645 | KF984752 | KF984822 | KF984971 | |
| DTO 67-G1c | Indoor air sample in vaccin producing factory, The Netherlands | KF984647 | KF984754 | KF984826 | KF984973 | |
| DTO 67-G2c | Indoor air sample in vaccin producing factory, The Netherlands | KF984618 | KF984747 | KF984816 | KF984965 | |
| DTO 67-G3c | Indoor air sample in vaccin producing factory, The Netherlands | KF984619 | KF984748 | KF984817 | KF984966 | |
| T. yelensis | CBS 138209T = DTO 268-E5 | House dust, Micronesia | KJ775210 | KP119161 | KJ775717 | KP119163 |
| CBS 138210 = DTO 268-E7 | House dust, Micronesia | KJ775212 | KP119162 | KJ775719 | KP119164 | |
| Trichocoma paradoxa | CBS 788.83 | Rotting stump of cut down tree, Japan | KF984556 | KF984670 | JN899398 | JN121550 |
a Isolates previously identified as Penicillium concavorugulosum.
b Isolate previously identified as Talaromyces sublevisporus.
c Isolates previously identified as Talaromyces variabilis.
DNA extraction, PCR amplification and sequencing
DNA extractions were made from isolates grown for 7–14 d on MEA using the UltracleanTM Microbial DNA isolation Kit (Mo-Bio, Solana Beach, USA) and extracted DNA was stored at -20 °C. The internal transcribed spacers, including the 5.8 S rDNA (ITS), calmodulin (CaM) and RNA polymerase II (RPB2) gene regions were amplified and sequenced using previously described methods (Yilmaz et al. 2014, Visagie et al. 2014b). For BenA, primer set T10 and Bt2b (Glass & Donaldson 1995) was used with annealing temperatures of 50 and 52 °C.
Phylogeny
Sequence contigs were assembled in Seqman v. 9.0.4 (DNA-Star Inc.). The newly generated sequences were included in a dataset including sequences obtained from Peterson & Jurjević (2013) and Yilmaz et al. (2014). GenBank accession numbers for sequences used in the phylogenies are listed in Table 1. The dataset for each gene was aligned using Muscle software included within the MEGA5 software package (Tamura et al. 2011). The aligned ITS, BenA, CaM and RPB2 data were concatenated in SeaView (Gouy et al. 2010) and analysed using Maximum Likelihood (ML) and Bayesian tree Inference (BI). The model for ML was selected based on the Akaike Information Criterion (AIC) calculated in MEGA5. The analysis was initiated by calculating an initial tree using BioNJ and the subsequent Heuristic done with the Nearest-Neighbour-Interchange (NNI). Bootstrap support was calculated using 1 000 replicates. The most suitable model for BI was selected based on AIC, calculated in MrModeltest v. 2.3 (Nylander et al. 2004). The analysis was run in MrBayes v. 3.2.1 (Huelsenbeck & Ronquist 2001) with two sets of four chains (one cold, three heated), until an average deviation of split frequencies reached 0.01. The sample frequency was set at 100, with 25 % of trees removed as burn-in phase.
Morphological analysis
Macroscopic characters were studied on different media and growth conditions. Cultures were plated onto Czapek yeast extract agar (CYA), CYA supplemented with 5 % NaCl (CYAS), yeast extract sucrose agar (YES), creatine sucrose agar (CREA), dichloran 18 % glycerol agar (DG18), oatmeal agar (OA) and malt extract agar (MEA; Oxoid malt). The isolates were inoculated at three points on 90 mm Petri dishes and incubated for 7 d at 25 °C in darkness. All media were prepared as described by Visagie et al. (2014b). Additional CYA plates were incubated at 37 °C for 7 d in darkness. The isolates growing at 37 °C, were also incubated at 40 °C for 7 d in darkness. After incubation, the colony diameters on the various media were measured. The density of sporulation, obverse and reverse colony colours and the production of soluble pigments were noted. Colony colour codes refer to Kornerup & Wanscher (1967). Colonies were photographed with a Canon EOS 400D. Species were characterised microscopically by preparing slides from MEA. Lactid acid was used as mounting fluid. Specimens were examined using a Zeiss AxioSkop2 plus microscope, and the NIS-Elements D software package from Nikon was used for capturing photographs and taking measurements.
Extrolites
Extrolites were extracted from fungal strains grown on CYA and YES at 25 °C for 7 d. In some cases, extractions were made from strains also grown on MEA and OA at 25 °C for 7 d. Three agar plugs of each medium were extracted as described in Nielsen et al. (2011) and Houbraken et al. (2012). The extracts were analysed by high performance liquid chromatography with diode-array detection (HPLC-DAD) (Frisvad & Thrane 1987) for extracts made before 2011 and by UHPLC-DAD (Houbraken et al. 2012) for extracts made after 2011. The eluted compounds were identified by comparing retention time, retention index and UV spectra measured at 200–600 nm. The UV spectra were compared to a database of UV spectra (Nielsen et al. 2011), and to data from the literature.
RESULTS
Phylogeny
A multi-gene phylogeny, based on four genes, was used to infer the relationships among species in Talaromyces sect. Islandici (Fig. 1). The aligned concatenated dataset (ITS 583 bp; BenA 482 bp; CaM 541 bp; RPB2 772 bp) had a total length of 2 378 bp. The most suitable model for ML was Kimura 2-parameter (K2)+Gamma distribution (G)+evolutionarily invariable (I) and the most suitable for BI was Genereal Time Reversible (GTR)+I+G. Tree topologies for ML and BI were identical. As such, the tree obtained from ML was used to show the result with both bootstrap support (bs) and posterior probabilities (pp) indicated above branches where support was higher than 80 % (bs) and/or 0.95 (pp) (Fig. 1). The same applies to phylogenies shown in Fig. 2 and 3. Based on the phylogeny (Fig. 1), Talaromyces sect. Islandici contains 19 species, including four species that we describe here as new. Talaromyces infraolivaceus and T. acaricola are resolved in the T. rugulosus complex, while T. crassus and T. subaurantiacus are closely related to T. rotundus, T. tratensis, T. wortmannii and T. yelensis. Differences between these species are discussed in the taxonomy section. Tree topologies differed between phylogenies of different genes in the T. wortmannii clade. As such, we adopted the GCPSR concept in this clade, and this is discussed below. GCPSR was also applied for resolving the species in the T. rugulosus complex.
Fig. 1.
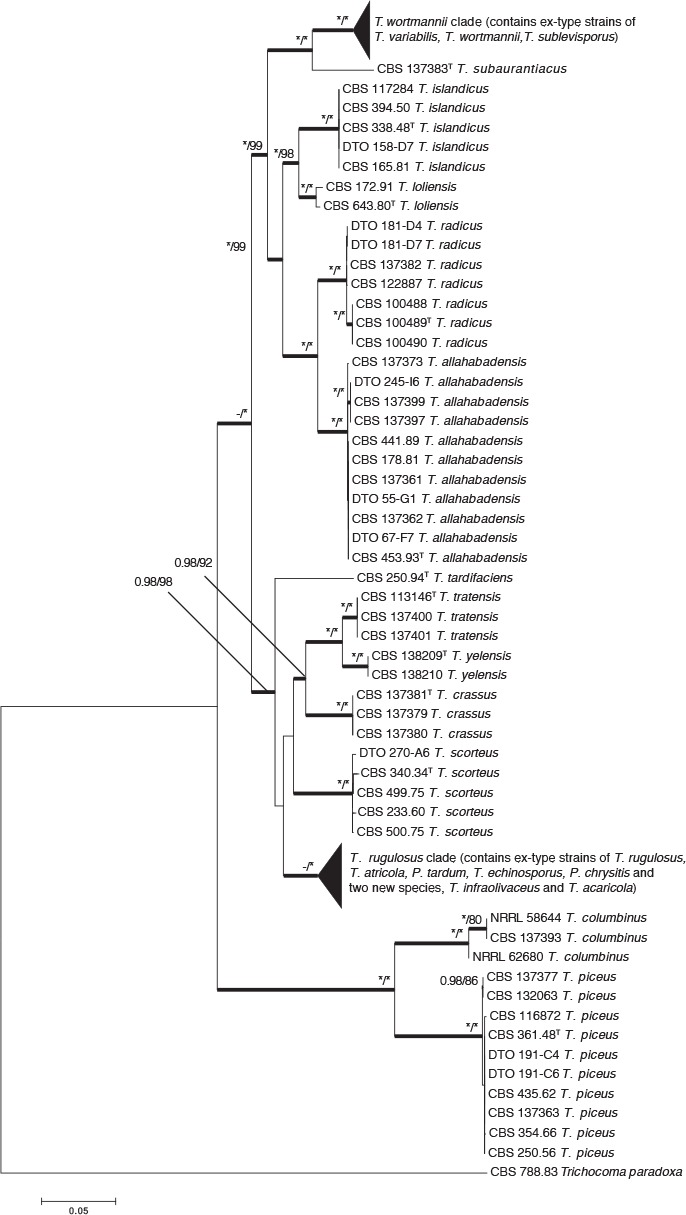
Combined phylogenetic tree comparing ITS, BenA, CaM and RPB2 of species from Talaromyces sect. Islandici. Trichocoma paradoxa was chosen as outgroup. Support in nodes is indicated above thick branches and is represented by posterior probabilities (BI analysis) of 0.95 and higher, and/or bootstrap values (ML analysis) of 80 % and higher. Full support (1.00/100 %) is indicated with an asterisk (*); support lower than 0.95/80 % is indicated with a dash (–). T = ex type.
Fig. 2.
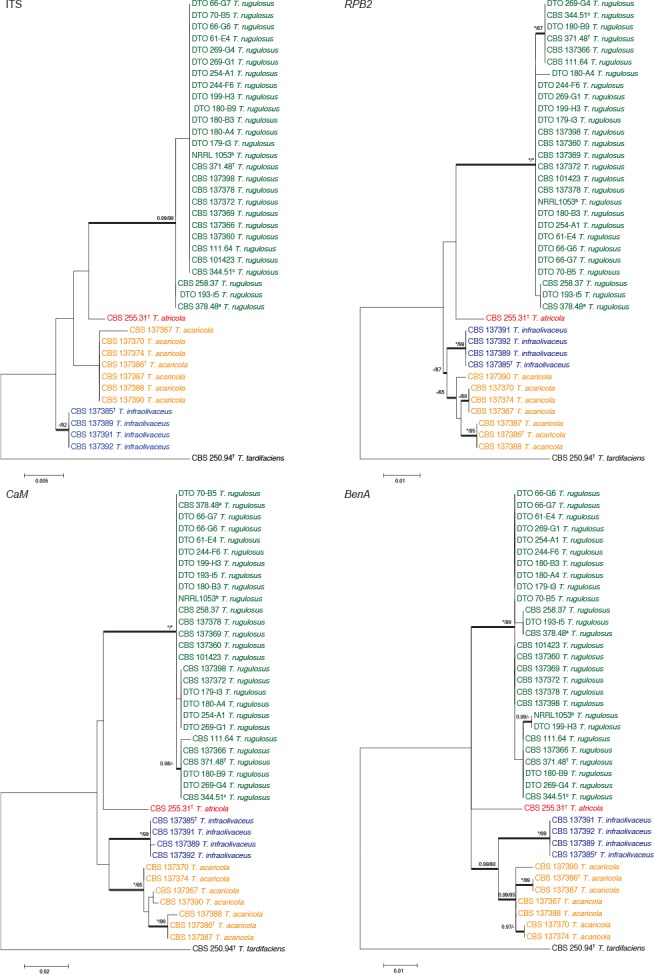
Phylogenetic trees of the ITS, BenA, CaM and RPB2 regions of strains in the T. rugulosus complex. Talaromyces tardifaciens was chosen as outgroup. Support in nodes is indicated above thick branches and is represented by posterior probabilities (BI analysis) of 0.95 and higher, and/or bootstrap values (ML analysis) of 80 % and higher. Full support (1.00/100 %) is indicated with an asterisk (*); support lower than 0.95/80 % is indicated with a dash (–). T = ex type. a = ex-type of P. elongatum and P. tardum (CBS 378.48 = NRRL 1073), b = ex-type of P. chrysitis (NRRL 1053) and c = ex-type of P. echinosporum (CBS 344.51). Colours are used to emphasise species in the clade.
Fig. 3.
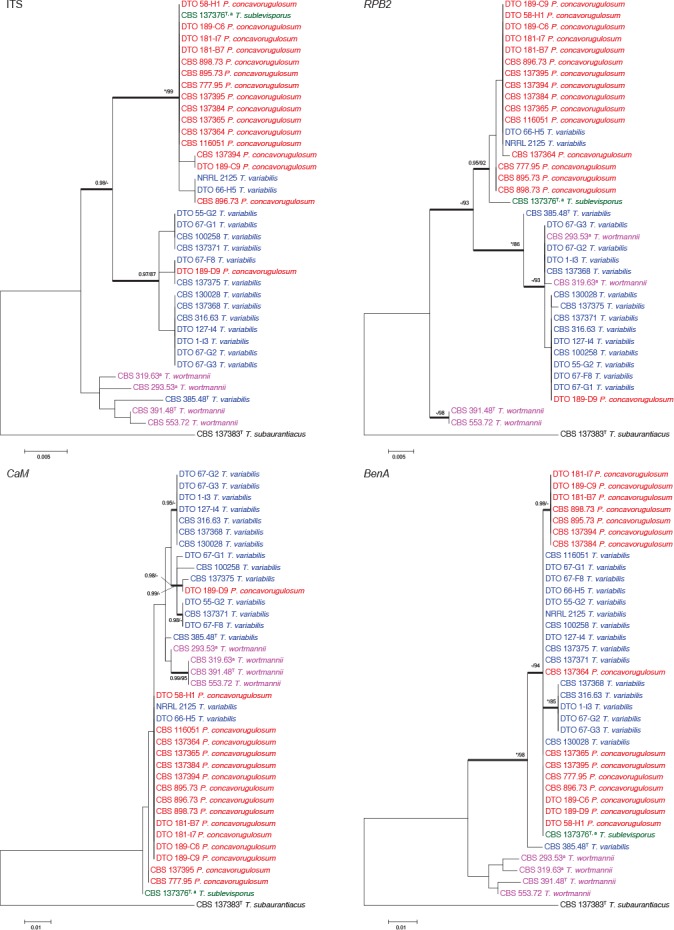
Phylogenetic trees of the ITS, BenA, CaM and RPB2 regions of strains in the T. wortmannii clade. Talaromyces subaurantiacus was chosen as outgroup. Support in nodes is indicated above thick branches and is represented by posterior probabilities (BI analysis) of 0.95 and higher, and/or bootstrap values (ML analysis) of 80 % and higher. Full support (1.00/100 %) is indicated with an asterisk (*); support lower than 0.95/80 % is indicated with a dash (–). T = ex type. Blue = isolates previously identified as T. variabilis; red = isolates previously identified as P. concavorugulosum; green = isolate of T. sublevisporus; purple = isolates previously identified as T. wortmannii and a indicates isolates which produce ascomata.
For the T. rugulosus complex, the aligned datasets were 573 (ITS), 432 (BenA), 489 (CaM) and 786 (RPB2) bp long. The most suitable models for ML were Tamura 3-parameter (T92)+G (ITS), K2 (BenA), K2+G (CaM) and K2+G (RPB2). The most suitable models for BI were Hasegawa-Kishino-Yano 1985 (HKY)+I(ITS), Symmetrical (SYM) (BenA), HKY+G (CaM) and Kimura 1980 (K80)+G (RPB2). The phylogenies (Fig. 2) show that the T. rugulosus complex contains four species. Penicillium tardum and P. chrysitis are synonyms of T. rugulosus, confirming results of Peterson & Jurjević (2013). Pitt (1980) proposed P. echinosporum as a synonym of T. rugulosus, which we confirm here. Peterson & Jurjević (2013) showed that P. rugulosum var. atricolum is a distinct phylogenetic species and introduced the new combination T. atricola, which is accepted here. We also describe two new species as T. infraolivaceus and T. acaricola. Strains of the latter two species form consistently distinct clades from all other species, which was confirmed by their unique morphological and extrolite characters.
For the T. wortmannii clade, the aligned datasets were 562 (ITS), 406 (BenA), 491 (CaM) and 766 (RPB2) bp long. The most suitable models for ML were T92+G (ITS), K2+G (BenA), K2+G (CaM) and K2+G (RPB2). The most suitable models for BI were GTR+I (ITS), K80+G (BenA), K80+G (CaM) and SYM+I (RPB2). Four previously described species are resolved in the T. wortmannii clade (Fig. 3). The four phylogenies showed different topologies between genes studied. Especially the locations of strains CBS 319.63, CBS 293.53, CBS 553.72 and CBS 391.48 varied. More noticeably, CBS 319.63 and CBS 293.53 are resolved with other T. wortmannii strains in all genes except for RPB2 which resolved them with other T. variabilis strains. Similarly, the type of T. variabilis (CBS 385.48T) is resolved within a clade of T. wortmannii for ITS. These switching positions of strains result in the only consistent branch being the one supporting the entire clade. Because this result was considered strange, DNA was extracted and strains resequenced in order to confirm the result obtained. As such, under GCPSR, strains from these four species are considered to belong to the same species. This is confirmed by our morphological studies, where conidiophores of T. sublevisporus and T. wortmannii (previously known for their teleomorphs) are identical to that of P. concavorugulosum and T. variabilis. Extrolite data also supports this. Penicillium wortmannii (1903) represents the oldest name in the clade and as a result we synonymise T. variabilis, T. sublevisporus and P. concavorugulosum with T. wortmannii.
Morphology
Species were compared morphologically, with characters distinguishing among species summarised in Table 2. The most important characters for identification include growth at 37 °C, colony texture, conidial colour, colony reverse, ascomata production and shape of ascospores. The new species identified by the phylogenetic analyses, displayed various distinct morphological features. Descriptions and distinguishing characters for each of the new species are presented below in the taxonomy section.
Table 2.
Morphological characters for the identification of Talaromyces sect. Islandici species.
| Talaromyces sp. | Colony diameter (mm) |
Reverse coloration on CYA | Texture on MEA | Conidial colour on MEA | Acid production | Conidial size (μm) | Ascomata | Shape, ornamentation and size of ascospores (μm) | Vesiculated stipes | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MEA 25 °C | YES 25 °C | CYA 25 °C | CYA 37 °C | CYA 40 °C | |||||||||
| T. acaricola | 15–20 | 13–16 | 10–15 | NG | NG | Greyish green centre fading into greyish yellow | Velvety to floccose | Dull green | A | 2.5–5.5 × 2–3 | A | A | A |
| T. allahabadensis | 20–23 | 22–23 | 20–25 | 23–25 | NG | Orange centre fading into yellow | Velvety | Dull green | P | 2.5–4.5 × 1.7–2.5 | A | A | A |
| T. atricola | 15 | 12 | 10 | NG | NG | Yellowish white | Floccose | Dull green to dark green | A | 2–5 × 2–5 | A | A | A |
| T. brunneus | 17–19 | 24–25 | 19–20 | NG | NG | Yellowish brown center fading into golden yellow | Velvety and in the center floccose | Golden brown to yellowish brown | A | 3–4(–7) × 2–4 | A | A | A |
| T. columbinus | 23–25 | 18–20 | 11–12 | 45–50 | 43 | Dark brown | Velvety and floccose | Greyish green | A | 2.5–3.5 × 3–4.5 | A | A | P |
| T. crassus | 17–20 | 15–18 | 14–16 | NG | NG | Pale yellow | Floccose | No sporulation (yellow mycelia dominant) | A to VW | 2–3 × 1.5–2.5 | A | A | A |
| T. infraolivaceus | 19–21 | 15–21 | 17–18 | NG | NG | Olive brown | Velvety and loosely funiculose in the centre | Dull green | A to VW | 2.5–4 × 1.5–3 | A | A | A |
| T. islandicus | 21–26 | 22–30 | 20–27 | 8–17 | NG | Orange to brown | Velvety and loosely funiculose | Dull green to dark green | P | 2.5–6 × 2–4.2 | A | A | A |
| T. loliensis | 13–15 | 13–15 | 10–13 | NG | NG | Deep orange centre fading into deep yellow | Loosely funiculose to floccose | Greyish green to dark green(yellow mycelia dominant) | A to VW | 3–5 × 2.4–3.5 | A | A | A |
| T. piceus | 25–27 | 15–20 | 20–27 | 30–35 | 23–27 | Orange to brown | Loosely funiculose to floccose | Greyish green | A | 2–3.8 × 2–4 | A | A | P |
| T. radicus | 15–25 | 22–25 | 15–22 | 25–30 | 3–11 | Yellowish brown | Loosely funiculose to floccose | Greyish green | A | 2–3 × 2–2.5 | A | A | A |
| T. rotundus | 15–17 | 9–10 | 9–11 | NG | NG | Greyish green circle at center fading into greenish grey | No sporulation | No sporulation (white mycelia dominant and at center yellow mycelia) | A (NG) | 3–5(–6.5) × 1.5 × 2.5 | P (2–3 weeks) | Globose, 4–5.5 × 4–5.5, spinose | A |
| T. rugulosus | 17–20 | 15–20 | 15–17 | NG | NG | Yellowish brown | Velvety | Greyish green to dark green | A to VW | 2.5–6 × 2.5–4 | A | A | A |
| T. scorteus | 10–15 | 7–16 | 8–16 | NG | NG | Olive | Velvety to floccose | Dark green | A | 3–5.5 × 2–3 | A | A | A |
| T. subauranticus | 20–21 | 17–18 | 16–18 | 7 | NG | Yellowish brown to dark brown | Floccose | Dull green | A | 2–3 × 2–2.5 | A | A | A |
| T. tardifaciens | 13–15 | 9–10 | 9–10 | NG | NG | Light orange centre fading into greyish yellow | No sporulation | No sporulation (white mycelia dominant) | A (NG) | 3–6 × 1.5–2.5 | P(3 weeks) | Broadly ovoidal, 3–3.5 × 2–3, smooth | A |
| T. tratensis | 15–20 | 12–18 | 10–12 | NG | NG | Greyish yellow to brownish orange | Floccose | No sporulation (yellow mycelia dominant) | A | 2–2.5 × 3–3.5 | P (1–2 weeks) | Ovoidal to broadly ellipsoidal 3.5–5 × 2.5–3.5 μm, thick walled slightly roughed | A |
| T. wortmannii | 15–25 | 20–30 | 18–28 | NG to 7 | NG | Reverse in various colours* | Velvety | Greyish green to dull green | A to VW | 2.5–5.8 × 1.5–3.2 | A to P (1–2 weeks) | Broadly ellipsoidal, 3.5–6 × 2.5–4 μm, thick walled, verrucose to smooth | A |
| T. yelensis | 15–16 | 20–21 | 20–22 | 14–16 | NG | Yellowish white to light yellow to brown | Floccose | No sporulation (yellow mycelia dominant) | A | 2.5–3.5 × 2.5–3 | A | A | A |
NG No Growth.
* In some isolates centre brown fading into in some isolates reddish yellow, in some isolates greyish orange to orange, in some isolates centre yellowish brown fading into in some isolates olive and in some isolates greyish yellow in some isolates with production of ascomata yellow with dark blonde dots in centre.
A Absent.
P Present.
VW Very Weak.
Strains from the T. wortmannii clade were compared morphologically (Fig. 4, 5). In our study, the only strains that produced the characteristic yellow ascomata were CBS 137376, CBS 319.63, CBS 293.53 and CBS 391.48T (Fig. 4). Ascospores of these strains are generally rough-walled. However, CBS 137376T, previously described as T. wortmannii var. sublevisporus (Yaguchi et al. 1994), produces smooth to finely roughened ascospores (Fig. 5). Yaguchi et al. (1994) mentioned that other characters of T. wortmannii var. sublevisporus and T. wortmannii were almost identical and this is clear from colony characters shown in Fig. 4. Some strains in the clade lacked ascomata and only produced conidiophores. This was typically of strains previously identified as P. concavorugulosum and T. variabilis. Abe (1956) never provided a Latin diagnosis for P. concavorugulosum species and did not make type material available. As such we synonymise this invalid name based on many strains received from other collections identified as P. concavorugulosum by their extrolite profiles. Strains of T. wortmannii characteristically produced rugulovasines, rugulosins, skyrin, wortmannilactones E, F, G and H, and mitorubrins. These exometabolites were found in several strains of isolates formerly identified as either Penicillium variabile (= T. variabilis), P. concavorugulosum or T. wortmannii.
Fig. 4.
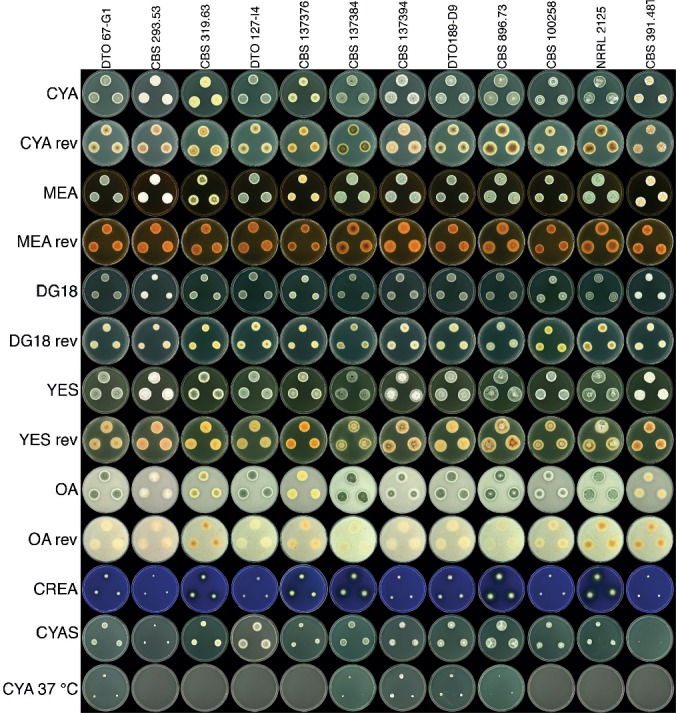
Talaromyces wortmannii colonies grown on various media at different conditions.
Fig. 5.
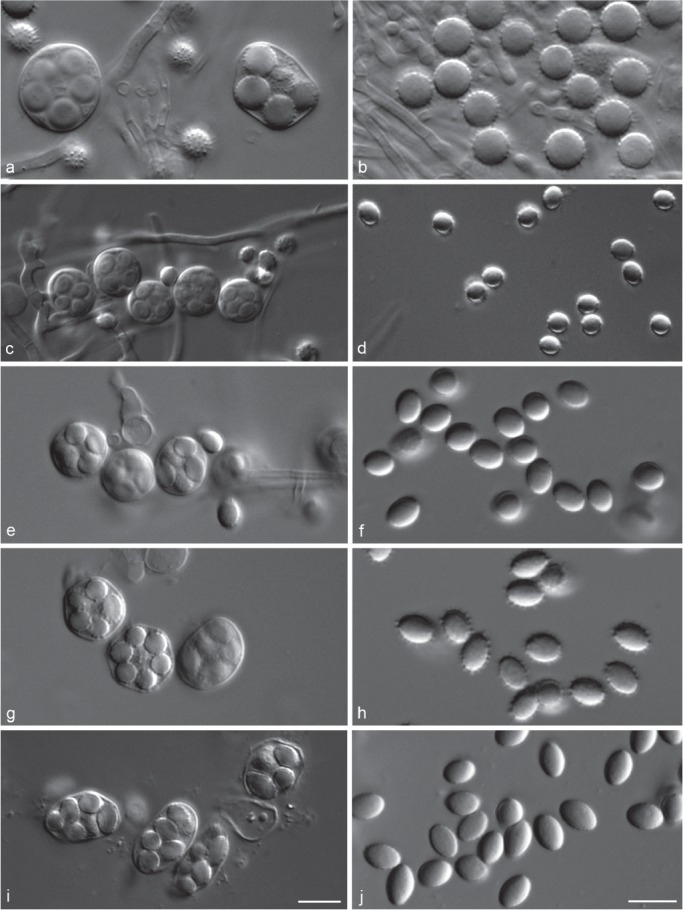
Variations of asci and ascospores produced by different species in Talaromyces sect. Islandici. a. Asci of T. rotundus (CBS 369.48T); b. ascospores of T. rotundus (CBS 369.48T); c. asci of T. tratensis (CBS 137401); d. ascospores of T. tratensis (CBS 137401); e. asci of T. tardifaciens (CBS 250.94T); f. ascospores of T. tardifaciens (CBS 250.94T); g. asci of T. wortmannii (CBS 293.53); h. ascospores of T. wortmannii (CBS 293.53); i. asci of T. wortmannii (CBS 137376 = ex-type of T. sublevisporus); j. ascospores of T. wortmannii (CBS 137376 = ex-type of T. sublevisporus).
TAXONOMY
Talaromyces acaricola Visagie, Yilmaz & K. Jacobs, sp. nov. — MycoBank MB810899, Fig. 6
Fig. 6.
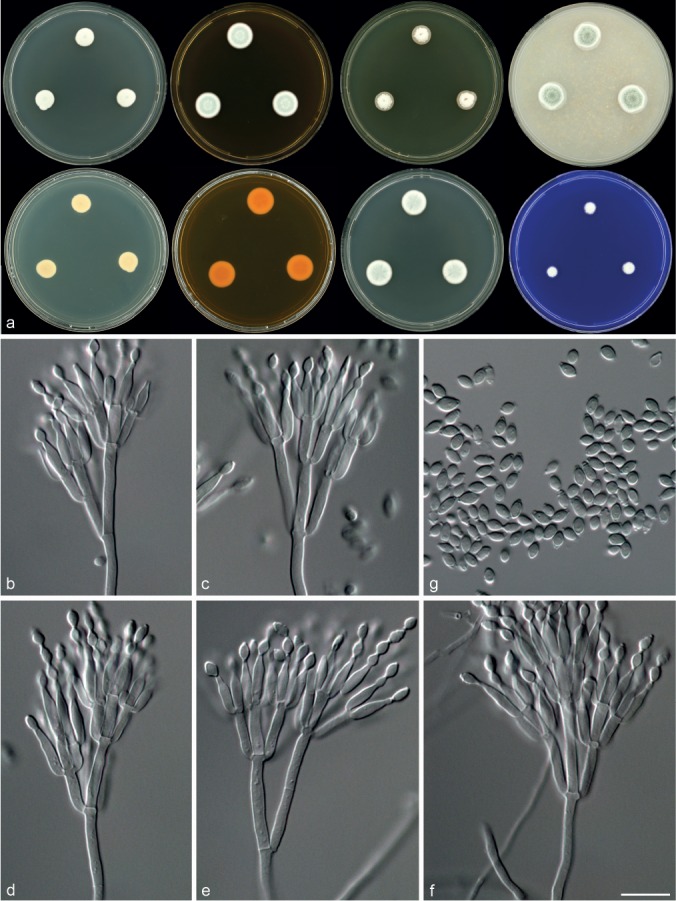
Morphological characters of Talaromyces acaricola (CBS 137386T). a. Colonies from left to right (top row) CYA, MEA, YES and OA; (bottom row) CYA reverse, MEA reverse, DG18 and CREA; b–f. conidiophores; g. conidia. — Scale bar: f = 10 μm, applies to b–g.
ITS barcode. JX091476.
Alternative markers. JX091610 (BenA), JX140729 (CaM), KF984956 (RPB2).
Etymology. Latin (acarus = mite), acaricola: meaning resident on mites, in reference strains isolated from mites inside Protea repens infructescences.
Typus. SOUTH AFRICA, Western Cape, Malmesbury, mite isolated from Protea repens infructescence, 2009, collected by C.M. Visagie (CBS-H 21632, holotype, culture ex-type CBS 137386 = DTO 183-B3 = DAOM 241025 = IBT 32387).
Diagnosis — Colonies CYA 10–15 mm, MEA 15–20 mm, acid not produced. Conidiophores biverticillate, a minor proportion with subterminal branches; phialides acerose to ampulliform; conidia rough-walled, sometimes forming ridges, ellipsoidal, 2.5–5.5 × 2–3 μm.
Colony diam, 7 d (mm) — CYA 10–15; CYA 37 °C No growth; MEA 15–20; DG18 12–17; CYAS 4–6; OA 10–20; CREA 6–10; YES 13–16.
Colony characters — CYA, 25 °C, 7 d: Colonies raised in the centre, concentrically sulcate; margins narrow (1 mm), low, entire, plane; mycelium white and yellow; texture velvety, in some isolates centre floccose; sporulation moderately dense; conidia en masse dull green to dark green (28E4–28F4); exudates absent; soluble pigment absent; reverse centre greyish green (1C4) fading into greyish yellow (1B4). MEA, 25 °C, 7 d: Colonies slightly raised in the centre, crateriform, sulcate; margins narrow (1 mm), low, entire, plane; mycelium white and yellow; texture velvety to floccose; sporulation moderately dense to dense; conidia en masse dull green (26D4–27D4); exudates absent; soluble pigment absent; reverse centre olive (1E4–1E5) fading into brownish yellow (5C7–5C8). YES 25 °C, 7 d: Colonies raised at centre, crateriform; margins narrow (1 mm), low, entire, plane; mycelium white and in some isolates yellow; texture velvety and floccose; sporulation sparse to moderately dense; conidia en masse dull green (27D4–27E4); exudates clear or yellow droplets (except CBS 137367 and CBS 137374); soluble pigment absent; reverse light yellow to greyish yellow (4A5–4B5), centre greyish green (1C4) in some isolates. DG18, 25 °C, 7 d: Colonies raised in the centre, crateriform, sulcate; margins narrow (1 mm), low, entire, plane; mycelium white and yellow; texture velvety, centre floccose in some isolates; sporulation dense; conidia en masse greyish green to dull green (25C4–25C5 to 25D4–25D5); exudates absent; soluble pigment absent; in some isolates reverse centre greyish green (1C5–1D5), in others reddish yellow to greenish yellow (4A6–4B6), fading into light yellow to greyish yellow (1A5–1B5). OA, 25 °C, 7 d: Colonies low, plane; margins narrow (1–2 mm), low, entire, plane, in some isolates with yeast like slimy margins; mycelium white and yellow; texture velvety and loosely funiculose; sporulation dense; conidia en masse greyish green to dull green (27C4–27C5 to 27D4–27D5); exudates absent; soluble pigment absent; reverse yellow to greenish yellow. CREA, 25 °C, 7 d: Acid not produced.
Micromorphology — Conidiophores biverticillate, a minor proportion with subterminal branches; stipes smooth-walled, 40–160 × 2–3 μm, branches 2–3 per stipe, 14–22 × 2–3 μm; metulae 3–5, 7.5–12 × 2–3 μm; phialides acerose to ampulliform, 3–5 per metulae, 6.5–9.5 × 2–3 μm; conidia rough-walled, sometimes forming ridges, ellipsoidal, 2.5–5.5 × 2–3 μm.
Extrolites — Talaromyces acaricola produces mitorubrins, rugulosin, skyrin, ukulactones and a polar metabolite with a chromophore similar to calbistrins.
Distinguishing characters — Talaromyces acaricola is characterised by typically floccose colonies especially on CYA and YES. The phylogenies resolve T. acaricola in the T. rugulosus complex (Fig. 2), closely related to T. rugulosus, T. atricola and T. infraolivaceus (Fig. 2). Talaromyces acaricola differs from T. rugulosus by the production of lightly coloured conidia en masse and MEA colonies that are more floccose in T. acaricola compared to the velvety colonies of T. rugulosus. It differs from T. infraolivaceus by greyish green or greyish yellow rather than dark olive reverse pigmentation and grows faster than T. atricola on most media.
Talaromyces crassus Visagie, Yilmaz & K. Jacobs, sp. nov. — MycoBank MB810900, Fig. 7
Fig. 7.
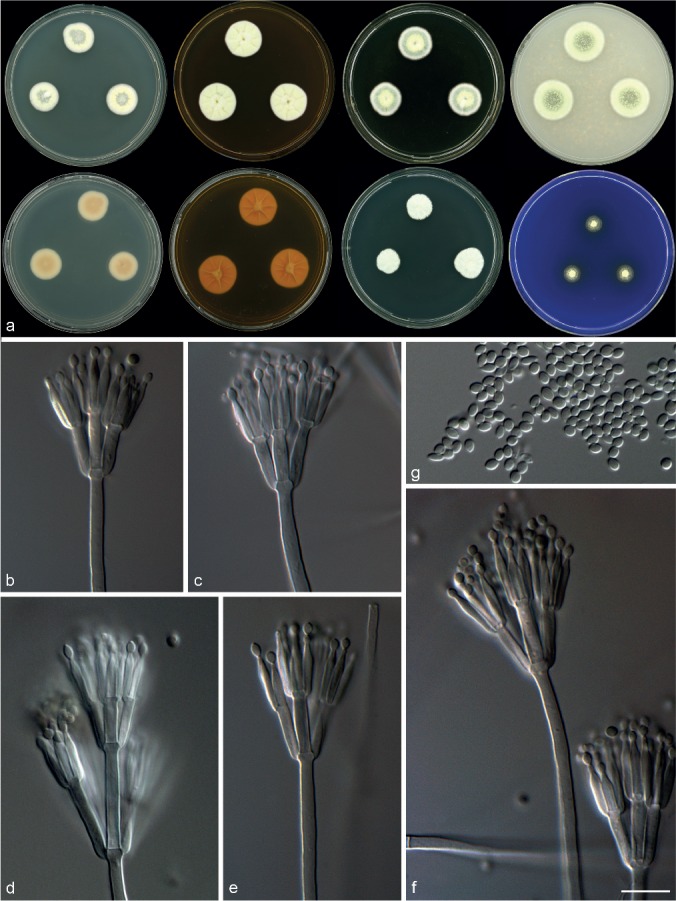
Morphological characters of Talaromyces crassus (CBS 137381). a. Colonies from left to right (top row) CYA, MEA, YES and OA; (bottom row) CYA reverse, MEA reverse, DG18 and CREA; b–f. conidiophores; g. conidia. — Scale bar: f = 10 μm, applies to b–g.
ITS barcode. JX091472.
Alternative markers. JX091608 (BenA), JX140727 (CaM), KF984914 (RPB2).
Etymology. Latin, crassus: meaning thick, in reference to the thick deep colonies produced.
Typus. SOUTH AFRICA, Western Cape, Stellenbosch, Protea repens infructescence, 2009, collected by C.M. Visagie (CBS-H 21631, holotype, culture ex-type CBS 137381 = DTO 181-C5 = DAOM 241027 = IBT 32814).
Diagnosis — Colonies on CYA 14–16 mm, MEA 17–20 mm. Acid generally not produced, some isolates weakly positive. Thick, deep, fluffy, yellow colonies on MEA. Conidiophores biverticillate, a minor proportion with subterminal branches; phialides acerose; conidia smooth-walled, ellipsoidal, 2–3 × 1.5–2.5 μm.
Colony diam, 7 d (mm) — CYA 14–16; CYA 37 °C No growth; MEA 17–20; DG18 12–16; CYAS 8–10; OA 16–20; CREA 6–10; YES 15–18.
Colony characters — CYA 25 °C, 7 d: Colonies low, plane; margins narrow (1 mm), low, entire, plane; mycelium white and predominately pale yellow; sporulation moderately dense to dense, especially in the centre; texture velvety to funiculose, conidiophores borne from aerial hyphae especially in the centre; conidia en masse dull green (25D4); exudates absent; soluble pigment absent; reverse pale yellow (4A3), in some isolates the centre greyish orange (5B3–5B4). MEA, 25 °C, 7 d: Colonies slightly raised in the centre, slightly concentrically sulcate and crateriform; margins narrow (1 mm), low, entire, plane; mycelium white and predominately yellow; sporulation none to sparse (very difficult to determine the conidia colour); texture floccose; exudates absent; soluble pigment absent; reverse brownish yellow (5C7–5C8). YES, 25 °C, 7 d: Colonies slightly raised in the centre, slightly crateriform and very slightly sulcate; margins narrow (1–2 mm), low, entire, plane; mycelium white and yellow; sporulation sparse to moderately dense; texture floccose; conidia en masse dull green (25D4–26D4); exudates small and clear droplets; soluble pigment absent; reverse butter yellow (4A5). DG18, 25 °C, 7 d: Colonies slightly raised in the centre, slightly sulcate; margins narrow (1 mm), low, entire, plane; mycelium white; sporulation none, not enough to determine colour; texture floccose; exudates absent; soluble pigment absent; reverse yellowish white and in some isolates greenish grey (1A2 and sometimes 1B2). OA, 25 °C, 7 d: Colonies low, plane; margins narrow (1 mm), low, entire, plane; mycelium white and predominately yellow; sporulation moderately dense to dense, especially in the centre; conidia en masse dull green (29E3–29E4); texture floccose and funiculose, conidiophores borne from aerial hyphae especially in the centre; exudates small and clear droplets; soluble pigment absent; reverse very pale light yellow, in some isolates centre dark green. CREA, 25 °C, 7 d: Acid production generally absent, in some isolates very weak acid production (CBS 137380).
Micromorphology — Conidiophores biverticillate, a minor proportion with subterminal branches; stipes smooth-walled, 130–390 × 2.5–3.5 μm; branches 2–3 per stipe, 13–17 × 2.5–3.5 μm, metulae 3–6, 9.5–14 × 2.5–3 μm; phialides acerose, number per metulae 3–6, 8.5–11.5 × 1.5–2.5 μm; conidia smooth-walled, ellipsoidal, 2–3 × 1.5–2.5 μm.
Extrolites — The ex-type isolate CBS 137381T, CBS 137379 and CBS 137380 only produced mitorubrins.
Distinguishing characters — Talaromyces crassus has restricted growth on most media, similar to other sect. Islandici species. Colonies are characteristically deep and consist of light yellowish mycelia that produce a fluffy texture. It cannot grow at 37 °C. Based on the multi-gene phylogeny, T. crassus is closely related to T. tratensis and the recently described T. yelensis (Visagie et al. 2014a) (Fig. 1). Morphologically all three species produce deep, fluffy yellow colonies. However, T. tratensis produces ascomata and spiny ascospores, which are absent in T. crassus and T. yelensis. Talaromyces yelensis is able to grow at 37 °C (14–16 mm), distinguishing it from T. crassus.
Talaromyces infraolivaceus Visagie, Yilmaz & K. Jacobs, sp. nov. — MycoBank MB810901, Fig. 8
Fig. 8.
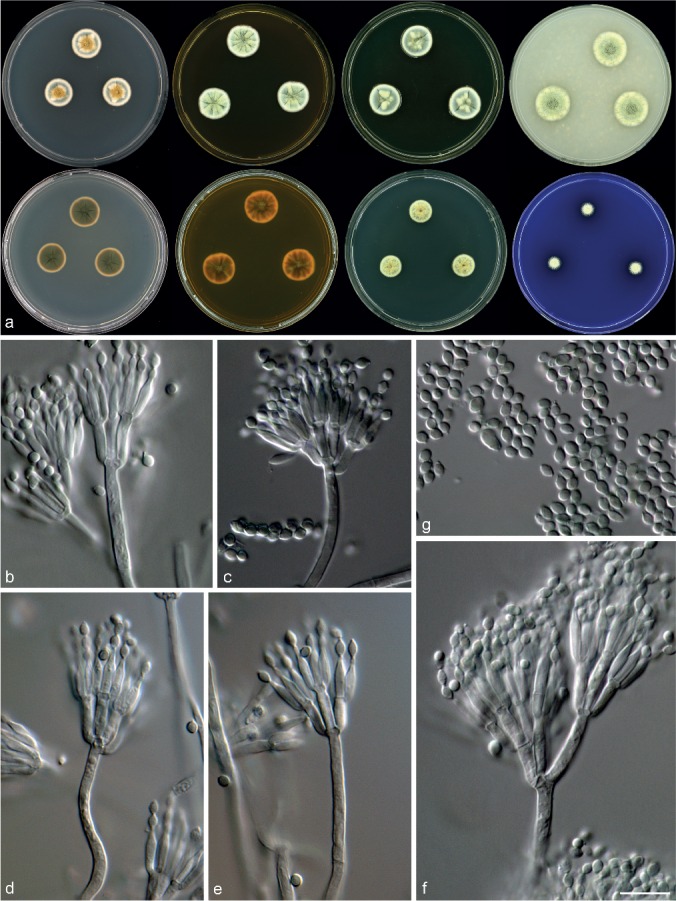
Morphological characters of Talaromyces infraolivaceus (CBS 137385T). a. Colonies from left to right (top row) CYA, MEA, YES and OA; (bottom row) CYA reverse, MEA reverse, DG18 and CREA; b–f. conidiophores; g. conidia. — Scale bar: f = 10 μm, applies to b–g.
ITS barcode. JX091481.
Alternative markers. JX091615 (BenA), JX140734 (CaM), KF984949 (RPB2).
Etymology. Latin, infraolivaceus: meaning below olive, in reference to the olive reverse of colonies.
Typus. SOUTH AFRICA, Western Cape, Malmesbury, mite isolated from Protea repens infructescence, 2009, collected by C.M. Visagie (CBS-H 21633, holotype, culture ex-type CBS 137385 = DTO 182-I2 = DAOM 241024 = IBT 32487).
Diagnosis — Colonies CYA 17–18 mm, MEA 19–21 mm. Acid generally not produced, some isolates weakly positive. Consistent production of deep olive reverses on most media. Conidiophores biverticillate, a minor proportion with subterminal branches and sometimes terverticillate; phialides acerose to ampulliform; conidia rough-walled, sometimes in ridges, ellipsoidal, 2.5–4 × 1.5–3 μm.
Colony diam, 7 d (mm) — CYA 17–18; CYA 37 °C No growth; MEA 19–21; DG18 12–13; CYAS 8–10; OA 18–20; CREA 5–8; YES 15–21.
Colony characters — CYA 25 °C, 7 d: Colonies slightly raised at centre, crateriform, radially sulcate; margins narrow (1 mm), low, entire, plane; mycelium white and very pale yellow; sporulation dense; texture velvety and loosely funiculose at centre, conidiophores borne from aerial hyphae especially in the centre; conidia en masse dull green (26D4–26E4–27E4); exudates absent; soluble pigment absent; reverse centre olive brown (4F5) fading into golden brown to light brown (5D7). MEA 25 °C, 7 d: Colonies slightly raised at centre, sulcate and in some isolates crateriform; margins narrow (1 mm), low, entire, plane; mycelium white and very pale yellow; sporulation moderately dense to dense; texture velvety and loosely funiculose, conidiophores borne from aerial hyphae especially in the centre; conidia en masse dull green (26D4–26E4–27E4); exudates absent; soluble pigment absent; reverse centre olive brown (4F5) fading into golden brown to light brown (5D7). YES 25 °C, 7 d: Colonies raised at centre, crateriform, sulcate; margins narrow (1–2 mm), low, entire, plane; mycelium white and very pale yellow; sporulation dense; texture velvety; conidia en masse dull green (26D4–26E4–27E4); exudates absent; soluble pigment absent; reverse centre olive brown (4F3–4F4 to 4D5–4E5). DG18, 25 °C, 7 d: Colonies raised in the centre, in some isolates crateriform, sulcate; margins narrow (1 mm), low, entire, plane; mycelium white and very pale yellow; texture loosely funiculose, conidiophores borne from aerial hyphae especially at centre; sporulation sparse to dense (CBS 137392, CBS 137389); conidia en masse dull green (26E4–27E4); exudates absent (except CBS 137391); soluble pigment absent; reverse centre olive brown (4F5) fading into golden brown to light brown (5D7). OA 25 °C, 7 d: Colonies low, plane; margins narrow (1–2 mm), low, entire, plane; mycelium white and very pale yellow; sporulation dense; texture velvety and loosely funiculose; conidia en masse dull green (26D4–26E4–27E4); exudates absent and in some isolates small clear droplets; soluble pigment absent; reverse brownish olive green fading into brownish yellow. CREA, 25 °C, 7 d: Acid generally not produced, some isolates weakly positive (CBS 137385T and CBS 137389).
Micromorphology — Conidiophores biverticillate, a minor proportion terverticillate and with subterminal branches; stipes smooth-walled, 12–100 × 2–3 μm, branches 2–3 per stipe when present, 11–15 × 2–3 μm; metulae 4–6, 7.5–11.5 × 2–3 μm; phialides acerose to ampulliform, 3–4 per metulae, 7–10 × 1.5–2.5 μm; conidia rough-walled, sometimes in ridges, ellipsoidal, 2.5–4 × 1.5–3 μm.
Extrolites — Isolates in this species produce mitorubrins, viomellein, vioxanthin and xanthomegnin. This is the first report of production of the xanthomegnin in Talaromyces. Xanthomegnins have formerly been found in Penicillium spp., Aspergillus spp., Trichophyton spp. and similar genera. In addition, CBS 137389 and CBS 137385T produce a compound suggesting a polar calbistrin.
Distinguishing characters — Talaromyces infraolivaceus is characterised by a consistent production of deep olive reverse on most media. Based on the phylogenies, T. infraolivaceus is resolved in the T. rugulosus complex (Fig. 1), closely related to T. rugulosus, T. atricola and T. acaricola (Fig. 2). Talaromyces infraolivaceus differs from T. rugulosus by the production of lightly coloured conidia en masse and MEA colonies that are more floccose. The most distinct feature, however, is the dark olive reverse on most media. This dark reverse was not observed in any other species from this clade.
Talaromyces subaurantiacus Visagie, Yilmaz & K. Jacobs, sp. nov. — MycoBank MB810902, Fig. 9
Fig. 9.
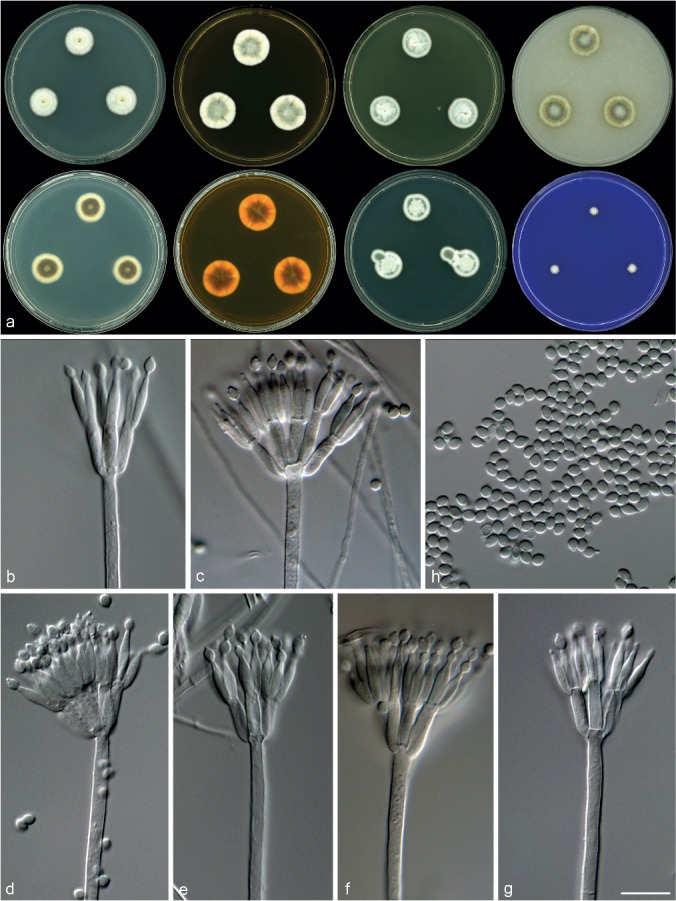
Morphological characters of Talaromyces subaurantiacus (CBS 137383T) a. Colonies from left to right (top row) CYA, MEA, YES and OA; (bottom row) CYA reverse, MEA reverse, DG18 and CREA; b–g. conidiophores; h. conidia. — Scale bar: g = 10 μm, applies to b–h.
ITS barcode. JX091475.
Alternative markers. JX091609 (BenA), JX140728 (CaM), KF984960 (RPB2).
Etymology. Latin, subaurantiacus: named in reference to the light orange mycelium produced by this species.
Typus. SOUTH AFRICA, Western Cape, Stellenbosch, Fynbos soil, 2009, collected by C.M. Visagie (CBS-H 21630, holotype, culture ex-type CBS 137383 = DTO 181-I2 = DAOM 241020 = IBT 32383).
Diagnosis — Colonies CYA 16–18 mm, MEA 20–21 mm, CYA at 37 °C 7 mm. Acid not produced. Colonies produce orange mycelia on MEA and CYA. Conidiophores biverticillate; phialides acerose; conidia finely rough-walled, ellipsoidal, 2–3 × 2–2.5 μm.
Colony diam, 7 d (mm) — CYA 16–18; CYA 37 °C 7; MEA 20–21; DG18 15–17; CYAS 9–12; OA 17–18; CREA 3–4; YES 17–18.
Colony characters — CYA 25 °C, 7 d: Colonies raised at centre, crateriform; margins very narrow (1 mm), low, entire, plane; mycelium white and pale yellow to light orange in the centre; texture floccose; sporulation sparse; conidia en masse dull green (26D4–26E4 to 27D4–27E4); exudates absent; soluble pigment absent; reverse yellowish brown to dark brown (5F5–6F5). MEA 25 °C, 7 d: Colonies low, sulcate; margins very narrow (1 mm), low, entire, plane; mycelium white and pale light orange in the centre; texture floccose; sporulation moderately dense; conidia en masse dull green (26D4–26E4 to 27D4–27E4); exudates absent; soluble pigment absent; reverse brown (6E5–6E6). YES 25 °C, 7 d: Colonies raised at centre, crateriform; margins very narrow (1 mm), low, entire, plane; mycelium white; texture floccose; sporulation moderately dense; conidia en masse dull green (26D4–26E4 to 27D4–27E4); exudates absent; soluble pigment absent; reverse yellowish brown (5E4) in the centre fading into greyish yellow (4B4). DG18 25 °C, 7 d: Colonies, slightly raised at centre, slightly sulcate; margins very narrow (1 mm), low, entire, plane; mycelium white; texture velvety and in the centre floccose; sporulation moderately dense; conidia en masse greyish green to dull green (27E4–27E5); exudates absent; soluble pigment absent; reverse greyish orange to brownish orange (6B4–6C4) in the centre, fading into light yellow to greyish yellow (3A5–3B5). OA 25 °C, 7 d: Colonies low, plane; margins very narrow (1 mm), low, entire, plane; mycelium white and yellow; texture velvety and in the centre floccose, conidiophores borne from aerial hyphae; sporulation moderately dense; conidia en masse dark green (25F5); exudates absent; soluble pigment absent; reverse bright orange yellow. CREA, 25 °C, 7 d: Acid not produced.
Micromorphology — Conidiophores biverticillate; stipes smooth-walled, 50–285 × 2.5–3.5 μm; metulae 3–6, 9–13 × 2–3.5 μm; phialides acerose, 3–6 per metulae, 8.5–11 × 2–3 μm; conidia finely rough-walled, ellipsoidal, 2–3 × 2–2.5 μm.
Extrolites — Talaromyces subaurantiacus produces rugulovasine and an azaphilone extrolite related to sclerotiorin.
Distinguishing characters — Talaromyces subaurantiacus grows restrictedly on agar media, especially on CYA. Based on the multi-gene phylogeny, T. subaurantiacus is closely related to T. wortmannii (Fig. 1). However, orange mycelia, floccose texture on MEA and more appressed conidiophores distinguish the new species from T. wortmannii.
DISCUSSION
In this study we revised the taxonomy of Talaromyces sect. Islandici, a group easily recognised by its slow or restricted growth and conspicuous yellow aerial mycelium, using morphology, phylogeny (under GCPSR) and extrolite data. Based on our GCPSR results, sect. Islandici includes 19 species, including the four new species. These include T. acaricola with its typically floccose colonies on CYA and YES, T. crassus producing the typical deep, fluffy colonies with abundance of yellowish mycelia but unable to grow at 37 °C, T. infraolivaceus with a unique deep olive reverse on most media, and T. subaurantiacus with its generally restricted colonies, especially on CYA. The distinguishing characters for all accepted species are listed in Table 2.
Most species of sect. Islandici produce the mycotoxins rugulosin and/or skyrin, the only exceptions being T. subaurantiacus, T. scorteus and T. infraolivaceus, which seem to have lost the ability to produce these bisanthraquinones during their evolution. However, T. infraolivaceus has acquired/retained the ability to produce xanthomegnin, viomellein and vioxanthin, which are absent in all other Talaromyces species. Rugulosin/skyrin are only produced by species in this section and not in other Talaromyces species (Frisvad et al. 1990), except for T. rubicundus (Reenen-Hoekstra et al. 1990). The azaphilones known as mitorubrins are produced by nearly all species of Talaromyces but are not produced by T. subaurantiacus, T. rotundus and T. columbinus.
Generally speaking, species able to grow at body temperature (37 °C) can be considered a risk as opportunistic pathogens. Talaromyces allahabadensis, T. columbinus, T. islandicus, T. piceus, T. radicus, T. subaurantiacus and T. yelensis are able to grow at 37 °C and some strains of T. wortmannii and its synonyms T. variabilis, T. sublevisporus and P. concavorugulosum. Talaromyces piceus, T. columbinus and T. radicus are able to grow at 40 °C (Table 2). Some opportunistic pathogen cases for T. piceus and T. radicus have been previously reported (Horré et al. 2001, Santos et al. 2006, de Vos et al. 2009).
Previous studies showed a close relationship between T. wortmannii, T. variabilis, T. sublevisporus and P. concavorugulosum (Frisvad et al. 1990, LoBuglio et al. 1993, Hocking et al. 1998, Samson et al. 2011, Yilmaz et al. 2014). Hocking et al. (1998) revealed a very high similarity between T. variabilis and T. wortmannii by using a RAPD-PCR (random amplification of polymorphic DNA). Frisvad et al. (1990) and Yilmaz et al. (2014) showed that these species have many metabolites in common. Peterson & Jurjević (2013), using an RPB2 phylogeny of only ex-type strains, considered these four species distinct, but hinted that additional analyses of more isolates and more loci were required to establish a robust phylogeny for this complex. We provide this phylogeny here applying GCPSR and reveal that P. concavorugulosum, T. sublevisporus and T. variabilis should be considered synonyms of T. wortmannii (Fig. 3). In Fig. 3, it is clear that ascosporic and non-ascosporic strains are mixed within the different clades. Furthermore, Raper & Thom (1949) reported that T. variabilis (= P. variabile) strains such as NRRL 2125 were received as non-ascosporic cultures, but after numerous transfers, yellow ascomata developed in colonies after three to four weeks. According to the International Code of Nomenclature for algae, fungi and plants (ICN), after 2011, priority is given to the oldest name irrespective of whether the species was described as an anamorph or teleomorph (McNeil et al. 2012). In this case P. wortmannii, described by Klöcker (1903), is the oldest name in the clade. Talaromyces wortmannii was later introduced for the sexual state of P. wortmannii (Benjamin 1955). As such, T. wortmannii (Klöcker) C.R. Benj. (≡ Penicillium wortmannii Klöcker, ≡ Penicillium kloeckeri Pitt, = Talaromyces sublevisporus (Yaguchi & Udagawa) Samson, Yilmaz & Frisvad ≡ Talaromyces wortmannii var. sublevisporus Yaguchi & Udagawa, = Talaromyces variabilis (Sopp) Samson et al. ≡ Penicillium variabile Sopp = Penicillium concavorugulosum S. Abe (nom. inval., Art. 36)) is considered the correct name for this clade.
Pitt (1980) considered P. rugulosum var. atricolum, P. scorteum, P. concavorugulosum and P. phialosporum to be synonyms of T. rugulosus. However, Peterson & Jurjević (2013) showed that P. scorteum and T. phialosporus are the same species, with P. scorteum an older name, and hence the name T. scorteus was introduced. Peterson & Jurjević (2013) showed that P. rugulosum var. atricolum is not a synonym of T. rugulosus and introduced the new combination T. atricola. Pitt (1980) also synonymised P. echinosporum (CBS 344.51T), P. elongatum (CBS 378.48T), P. tardum (NRRL 1073T) and P. chrysitis (NRRL 1053T) with T. rugulosus and our phylogenetic results confirm their synonymy with T. rugulosus (Fig. 2). Described species and associated taxonomic conclusions of different authors are summarised in Table 3. Two of the new species, T. infraolivaceus and T. acaricola, are consistently resolved in distinct clades correlating with morphological characters discussed in the taxonomy section.
Table 3.
Overview of taxonomic treatments on Talaromyces sect. Islandici.
| Original names | Raper & Thom (1949) | Pitt (1980) | Samson et al. (2011) | Peterson & Jurjević (2013) | Current study |
|---|---|---|---|---|---|
| T. acaricola (current study) | – | – | – | – | T. acaricola |
| P. allahabadense (Mehrotra & Kumar 1962) | – | P. pinophilum | T. allahabadensis | T. allahabadensis | T. allahabadensis |
| P. rugulosum var. atricolum (Thom 1930) | P. tardum | P. rugulosum | T. rugulosus | T. atricola | T. atricola |
| P. brunneum (Udagawa 1959) | – | P. brunneum | T. brunneus | not studied | T. brunneus |
| T. columbinus (Peterson & Jurjević 2013) | – | – | – | T. columbinus | T. columbinus |
| T. crassus (current study) | – | – | – | – | T. crassus |
| T. infaolivaceus (current study) | – | – | – | – | T. infaolivaceus |
| P. islandicum (Sopp 1912) | P. islandicum | P. islandicum | T. islandicus | T. islandicus | T. islandicus |
| P. loliense (Pitt 1980) | – | P. loliense | T. loliensis | T. loliensis | T. loliensis |
| P. piceum (Raper & Fennell 1948) | – | P. piceum | T. piceus | T. piceus | T. piceus |
| P. radicum (Hocking et al. 1998) | – | – | T. radicus | T. radicus | T. radicus |
| P. rotundum (Raper & Fennell 1948) | P. rotundum | T. rotundus | T. rotundus | T. rotundus | T. rotundus |
| P. rugulosum (Thom 1910) | P. rugulosum | P. rugulosum | T. rugulosus | T. rugulosus | T. rugulosus |
| P. echinosporum (Nehira 1933) | not studied | P. rugulosum | T. echinosporus | not studied | T. rugulosus |
| P. tardum (Thom 1930) | P. tardum | P. rugulosum | T. rugulosus | T. rugulosus | T. rugulosus |
| P. elongatum (Bainier 1907) | P. tardum | P. rugulosum | T. rugulosus | not studied | T. rugulosus |
| P. chrysitis (Biourge 1923) | P. rugulosum | P. rugulosum | T. rugulosus | T. rugulosus | T. rugulosus |
| P. scorteum (Takedo et al. 1934) | P. tardum | P. rugulosum | T. rugulosus | T. scorteus | T. scorteus |
| P. phialosporum (Udagawa 1959) | – | P. rugulosum | T. phialosporus | T. scorteus | T. scorteus |
| T. subaurantiacus (current study) | – | – | – | – | T. subaurantiacus |
| T. tardifaciens (Udagawa 1993) | – | – | T. tardifaciens | not studied | T. tardifaciens |
| T. tratensis (Manoch et al. 2013) | – | – | – | not studied | T. tratensis |
| T. wortmannii var. sublevisporus (Yaguchi et al. 1994) | – | – | T. sublevisporus | not studied | T. wortmannii |
| P. concavorugulosum (Abe 1956, nom. Inval., art. 36) | – | P. rugulosum | P. concavorugulosum* | P. concavorugulosum | T. wortmannii |
| P. variabile (Sopp 1912) | P. variabile | P. variabile | T. variabilis | T. variabilis | T. wortmannii |
| P. wortmannii (Klöcker 1903) | P. wortmannii | T. wortmannii | T. wortmannii | T. wortmannii | T. wortmannii |
| T. yelensis (Visagie et al. 2014a) | – | – | – | – | T. yelensis |
* Samson et al. (2011) listed P. concavorugulosum in the ‘Taxa which need further taxonomic study’ list.
– species which were described later than the study.
Talaromyces columbinus was described by Peterson & Jurjević (2013). They isolated their strains from air samples and corn grits from the USA. One of our strains was isolated from chicken feed from Nairobi, Kenya. Our results confirm Peterson & Jurjević’s (2013) findings that the isolate IMI 392509, isolated by Santos et al. (2006) from a human and identified as T. piceus, is in fact T. columbinus. Also, Peterson & Jurjević (2013) considered CBS 102383, which was isolated from a case of fungemia and previously identified as T. piceus, as an isolate of T. columbinus. Both T. piceus and T. columbinus are able to grow at 40 °C and have vesiculate stipes. However, T. columbinus grows faster than T. piceus at 37 and 40 °C. In addition, colonies of T. columbinus have dark brown reverses and soluble pigments on YES, whereas T. piceus has orange to brownish orange reverses and lacks soluble pigments on YES.
Peterson & Jurjević (2013) mentioned problems with the amplification of BenA paralogues when using primer pairs Bt2a & Bt2b or BT2f & T22. In our study, a similar result was observed, with gel-electrophoresis revealing one band with primers Bt2a & Bt2b, but subsequent sequences with mixed electropherograms. As a result, we recommend primer set T10 & Bt2b (Glass & Donaldson 1995), at annealing temperatures of 50 or 52 °C, for the amplification and sequencing of BenA in this group of species.
Acknowledgments
We would like to acknowledge the South African Biosystematics Initiative (SABI) of the National Research Foundation (NRF) for funding provided. The Western Cape Nature Conservation Board issued permits for collecting soil and Protea repens infructescences in the fynbos. We thank the various people who have deposited strains used in this study in the various collections. We are grateful for Latin assistance provided by Uwe Braun. We would like to thank Keith A. Seifert for his valuable input in the manuscript.
REFERENCES
- Abe S. 1956. Studies on the classification of the Penicillia. Journal of General and Applied Microbiology Tokyo 2: 1–344. [Google Scholar]
- Antipova TV, Zhelifonova VP, Kochkina GA, et al. 2008. Growth and biosynthesis of rugulovasines in Penicillium variabile Sopp 1912. Microbiology 77: 446–450. [PubMed] [Google Scholar]
- Bainier G. 1907. Mycothèque de l’École de Pharmacie, IX–XI. Bulletin de la Société Mycologique de France 23: 9–27. [Google Scholar]
- Bara R, Zerfass I, Aly AH, et al. 2013. Atropisomeric dihydroanthracenones as inhibitors of multiresistant Staphylococcus aureus. Journal of Medical Chemistry 56: 3257–3272. [DOI] [PubMed] [Google Scholar]
- Benjamin CR. 1955. Ascocarps of Aspergillus and Penicillium. Mycologia 47: 669–687. [Google Scholar]
- Biourge P. 1923. Les moisissures de groupe Penicillium Link. Cellule 33: 7–331. [Google Scholar]
- Bouhet J-C, Van Chuong PP, Toma F, et al. 1976. Isolation and characterization of luteoskyrin and rugulosin, two hepatotoxic anthraquinonoids from Penicillium islandicum Sopp and Penicillium rugulosum Thom. Journal of Agricultural and Food Chemistry 24: 964–972. [DOI] [PubMed] [Google Scholar]
- Breen J, Dacre JC, Raistrick H, et al. 1955. Studies in biochemistry of micro-organisms 95. Rugulosin, a crystalline colouring matter of Penicillium rugulosum Thom. Biochemical Journal 60: 618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RJ, Cox RH. 1981. Handbook of toxic fungal metabolites. Academic Press, New York. [Google Scholar]
- Frisvad JC, Filtenborg O, Samson RA, et al. 1990. Chemotaxonomy of the genus Talaromyces. Antonie van Leeuwenhoek 57: 179–189. [DOI] [PubMed] [Google Scholar]
- Frisvad JC, Samson RA. 2004. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Studies in Mycology 49: 1–174. [Google Scholar]
- Frisvad JC, Thrane U. 1987. Standardized high-performance liquid chromatography of 182 mycotoxins and other fungal metabolites based on alkylphenone retention indices and UV-VIS spectra (diode-array detection). Journal of Chromatography 404: 195–214. [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView Version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Hocking AD, Whitelaw M, Harden TJ. 1998. Penicillium radicum sp. nov. from rhizosphere of Australian wheat. Mycological Research 102: 801–806. [Google Scholar]
- Horré R, Gilges S, Breig P, et al. 2001. Case report. Fungaemia due to Penicillium piceum, a member of the Penicillium marneffei complex. Mycoses 44: 502–504. [DOI] [PubMed] [Google Scholar]
- Houbraken J, Samson RA. 2011. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology 70: 1–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Spierenburg H, Frisvad JC. 2012. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antonie van Leeuwenhoek 101: 403–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics Applications Note 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Kawai K, Kato T, Mori H, et al. 1984. A comparative study on cytotoxicities and biochemical properties of anthraquinone mycotoxins emodin and skyrin from Penicillium islandicum Sopp. Toxicology Letters 20: 155–160. [DOI] [PubMed] [Google Scholar]
- Kenkyusho K. 1983. Antitumor agents – comprising pyrano compound obtained by culturing a Penicillium islandicum Sopp. JP 5804392-A and JP 85026372-B. Patent. Derwent Primary Accession nr. 1983-38413K.
- Klöcker A. 1903. Sur la classification du genre Penicillium et description d’une espèce nouvelle formant des asques. Comptes Rendus des Travaux du Laboratoire Carlsberg: serie Physiologique 6: 92–102. [Google Scholar]
- Kornerup A, Wanscher JH. 1967. Methuen handbook of colour. 2nd edn Sankt Jørgen Tryk, Copenhagen, Denmark. [Google Scholar]
- LoBuglio KF, Pitt JI, Taylor JW. 1993. Phylogenetic analysis of two ribosomal DNA regions indicates multiple independent losses of a sexual Talaromyces state among asexual Penicillium species in subgenus Biverticillium. Mycologia 85: 592–604. [Google Scholar]
- Manoch L, Dethoup T, Yilmaz N, et al. 2013. Two new Talaromyces species from soil in Thailand. Mycoscience 54: 335–342. [Google Scholar]
- McNeil J, BArrie FF, Buck WR, et al. (eds). 2012. International Code of Nomenclature for algae, fungi and plants (Melbourne Code). Koeltz Scientific Books, Königstein. [Regnum vegetabile no. 154]. [Google Scholar]
- Mehrotra BS, Kumar DA. 1962. A new species of Penicillium from India. Canadian Journal of Botany 40: 1399–1400. [Google Scholar]
- Mori S, Sugihara Y, Kitagawa A, et al. 1996. The respiration-impairing effect of rubroskyrin, a toxic metabolite of Penicillium islandicum, on isolated mitochondria. Mycotoxin Research 12: 91–98. [DOI] [PubMed] [Google Scholar]
- Narikawa T, Shinoyama H, Fujii T. 2000. A beta-rutinosidase from Penicillium rugulosum IFO 7242 that is a peculiar flavonoid glycosidase. Bioscience, Biotechnology and Biochemistry 64: 1317–1319. [DOI] [PubMed] [Google Scholar]
- Nehira T. 1933. On the genus Penicillium in Japan. Journal of Fermentation Technology Osaka 11: 849–866. [Google Scholar]
- Nielsen KF, Månsson M, Rank C, et al. 2011. Dereplication of microbial natural products by LC-DAD-TOFMS. Journal of Natural Products 74: 2338–2348. [DOI] [PubMed] [Google Scholar]
- Nylander AJJ, Ronquist F, Huelsenbeck JP, et al. 2004. Bayesian phylogenetic analysis of combined data. Systematic Biology 53: 47–67. [DOI] [PubMed] [Google Scholar]
- Oh JY, Kim EN, Ryoo MI, et al. 2008. Morphological and molecular identification of Penicillium islandicum isolate KU101 from stored rice. Plant Pathology Journal 24: 469–473. [Google Scholar]
- Peterson SW, Jurjević Z. 2013. Talaromyces columbinus sp. nov., and genealogical concordance analysis in Talaromyces clade 2a. PLoS ONE 8: e78084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt JI. 1980. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. Academic Press Inc., London, England. [Google Scholar]
- Pitt JI, Hocking AD. 2009. Fungi and food spoilage, 3rd ed. Springer, Dordrecht/Heidelberg. [Google Scholar]
- Pretsch A, Nagl M, Schwendinger K, et al. 2014. Antimicrobial and anti-inflammatory activities of endophytic fungi Talaromyces wortmannii extracts against acne-inducing bacteria. PLoS ONE 9: e97929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper KB, Fennell DI. 1948. New species of Penicillium. Mycologia 40: 507–546. [PubMed] [Google Scholar]
- Raper KB, Thom C. 1949. Manual of the Penicillia. Williams & Wilkins, Baltimore, USA. [Google Scholar]
- Reenen-Hoekstra ES van, Frisvad JC, Samson RA, et al. 1990. The Penicillium funiculosum complex – well defined species and problematic taxa. In: Samson RA, Pitt JI. (eds), Modern concepts in Penicillum and Aspergillus classification: 173–191. Plenum, New York. [Google Scholar]
- Reyes I, Bernier L, Simard RR, et al. 1999. Characteristics of phosphate solubilization by an isolate of a tropical Penicillium rugulosum and two UV-induced mutants. FEMS Microbiology Ecology 28: 291–295. [Google Scholar]
- Saito M, Enomoto M, Tatsuno T. 1971. Yellowed rice toxins. Luteoskyrin and related compounds, chlorine-containing compounds, and citrinin. In: Ciegler A, Kadis S, Ajl SJ. (ed), Microbial toxins: 299–308. Academic Press Inc., New York. [Google Scholar]
- Sakai A, Tanaka H, Konishi Y, et al. 2005. Mycological examination of domestic unpolished rice and mycotoxin production by isolated Penicillium islandicum. Journal of the Food Hygienic Society of Japan 46: 205–212. [DOI] [PubMed] [Google Scholar]
- Samson RA, Yilmaz N, Houbraken J, et al. 2011. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Studies in Mycology 70: 159–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos PE, Piontelli E, Shea YR, et al. 2006. Penicillium piceum infection: diagnosis and successful treatment in chronic granulomatous disease. Medical Mycology 44: 749–753. [DOI] [PubMed] [Google Scholar]
- Sopp OJ. 1912. Monographie der Pilzgruppe Penicillium mit besonderer Berücksichtigung der in Norwegen gefundenen Arten. Skrifter udgivne af Videnskabs-Selskabet i Christiania. Mathematisk-Naturvidenskabelig Klasse 11: 1–208. [Google Scholar]
- Stark AA, Townsend JM, Wogan GN, et al. 1978. Mutagenicity and antibacterial activity of mycotoxins produced by Penicillium islandicum Sopp and Penicillium rugulosum. Journal of Environmental Pathology and Toxicology 2: 313–324. [PubMed] [Google Scholar]
- Swietliczkowa I, Szusterowska-Martinowa E, Braciak W. 1984. Clinical evaluation of 1% cloteimazol ointment in the treatment of corneal mycoses. Klinika Oczna 86: 221–223. [PubMed] [Google Scholar]
- Takedo Y, Suematsu S, Nakazawa R. 1934. The moulds on military instruments II. Journal of the Agricultural Chemical Society of Japan 10: 95–121. [Google Scholar]
- Tamura K, Peterson D, Peterson N, et al. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, et al. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32. [DOI] [PubMed] [Google Scholar]
- Thom C. 1910. Cultural studies of species of Penicillium. The Bureau of Animal Industry, US Department of Agriculture, Washington, Government Printing Office. [Google Scholar]
- Thom C. 1930. The Penicillia. Baltimore, The Williams & Wilkins Company. [Google Scholar]
- Udagawa S. 1959. Taxonomic studies of fungi on stored rice grain. III. Penicillium group (penicillia and related genera) 2. Journal of Agricultural Science Tokyo Nogyo Daigaku 5: 5–21. [Google Scholar]
- Udagawa S. 1993. Three new species of Talaromyces from Nepal. Mycotaxon 48: 141–156. [Google Scholar]
- Ueno Y, Ishikawa I. 1969. Production of luteoskyrin, a hepatotoxic pigment, by Penicillium islandicum Sopp. Applied Microbiology 18: 406–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y, Sato N, Ito T, et al. 1980. Chronic toxicity and hepato-carcinogenicity of (+) rugulosin, an anthraquinoid mycotoxin from Penicillium species: preliminary surveys in mice. The Journal of Toxicological Sciences 5: 295–302. [DOI] [PubMed] [Google Scholar]
- Uraguchi K. 1962. Malignant hepatoma and so-called carcinogens, with special reference to the toxicity of luteoskyrin in a small dose. Folia Pharmacologica Japonica 58: 19. [Google Scholar]
- Uraguchi K, Miyake M, Shikata T, et al. 1961. Isolation of 2 toxic agents, luteoskyrin and chlorione-containing peptide, from metabolites of Penicillium islandicum Sopp, with properties thereof. Japanese Journal of Experimental Medicine 31: 19–46. [PubMed] [Google Scholar]
- Uraguchi K, Saito M, Noguchi Y, et al. 1972. Chronic toxicity and carcinogenicity in mice of the purified mycotoxins, luteoskyrin and cyclochlorotine. Food and Cosmetics Toxicology 10: 193–207. [DOI] [PubMed] [Google Scholar]
- Visagie CM, Hirooka Y, Tanney JB, et al. 2014a. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Studies in Mycology 78: 63–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Frisvad JC, et al. 2014b. Identification and nomenclature of the genus Penicillium. Studies in Mycology 78: 343–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Rodriques C, et al. 2013. Five new Penicillium species in section Sclerotiora: a tribute to the Dutch Royal family. Persoonia 31: 42–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Jacobs K. 2012. Three new additions to the genus Talaromyces isolated from Atlantis sandveld fynbos soils. Persoonia 28: 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Roets F, Jacobs K. 2009. A new species of Penicillium, P. ramulosum sp. nov., from the natural environment. Mycologia 101: 888–895. [DOI] [PubMed] [Google Scholar]
- Visagie CM, Seifert KA, Houbraken J, et al. 2014c. Diversity of Penicillium section Citrina within the fynbos biome of South Africa, including a new species from a Protea repens infructescence. Mycologia 106: 537–552. [DOI] [PubMed] [Google Scholar]
- Vos JP de, Garderen E van, Hensen H, et al. 2009. Disseminated Penicillium radicum infection in a dog, clinically resembling multicentric malignant lymphoma. Vlaams Diergeneeskundig Tijdschrift 78: 183–188. [Google Scholar]
- Yaguchi T, Someya A, Miyadoh S, et al. 1994. A new variety of Talaromyces wortmannii and some observation on Talaromyces assiutensis. Mycosience 35: 63–68. [Google Scholar]
- Yamazaki H, Koyama N, Omura S, et al. 2010a. New rugulosins, Anti-MRSA antibiotics, produced by Penicillium radicum FKI-3765-2. Organic Letters 12: 1572–1575. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Omura S, Tomoda H. 2010b. Xanthoradone C, a new potentiator of imipenem activity against methicillin-resistant Staphylococcus aureus, produced by Penicillium radicum FKI-3765-2. Journal of Antibiotics 63: 329–330. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Omura S, Tomoda H. 2010c. 6’-Hydroxy-3’-methoxy-mitorubrin, a new potentiator of antifungal miconazole activity, produced by Penicillium radicum FKI-3765-2. Chemical and Pharmaceutical Bulletin 58: 829–832. [DOI] [PubMed] [Google Scholar]
- Yilmaz N, Visagie CM, Houbraken J, et al. 2014. Polyphasic taxonomy of the genus Talaromyces. Studies in Mycology 78: 175–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ND, Gu XL, Tian YP. 2013. Isolation and characterization of urethanase from Penicillium variabile and its application to reduce ethyl carbamate contamination in Chinese rice wine. Applied Biochemistry and Biotechnology 170: 718–728. [DOI] [PubMed] [Google Scholar]


