Abstract
Chaetomium globosum, the type species of the genus, is ubiquitous, occurring on a wide variety of substrates, in air and in marine environments. This species is recognised as a cellulolytic and/or endophytic fungus. It is also known as a source of secondary metabolites with various biological activities, having great potential in the agricultural, medicinal and industrial fields. On the negative side, C. globosum has been reported as an air contaminant causing adverse health effects and as causal agent of human fungal infections. However, the taxonomic status of C. globosum is still poorly understood. The contemporary species concept for this fungus includes a broadly defined morphological diversity as well as a large number of synonymies with limited phylogenetic evidence. The aim of this study is, therefore, to resolve the phylogenetic limits of C. globosum s.str. and related species. Screening of isolates in the collections of the CBS-KNAW Fungal Biodiversity Centre (The Netherlands) and the China General Microbiological Culture Collection Centre (China) resulted in recognising 80 representative isolates of the C. globosum species complex. Thirty-six species are identified based on phylogenetic inference of six loci, supported by typical morphological characters, mainly ascospore shape. Of these, 12 species are newly described here. Additionally, C. cruentum, C. mollipilium, C. rectum, C. subterraneum and two varieties of C. globosum are synonymised under C. globosum s.str., and six species are resurrected, i.e. C. angustispirale, C. coarctatum, C. cochliodes, C. olivaceum, C. spiculipilium and C. subglobosum. Chaetomium ascotrichoides is segregated from C. madrasense and the genus name Chaetomidium is rejected. Five species, including C. globosum s.str., are typified here to stabilise their taxonomic status. A further evaluation of the six loci used in this study as potential barcodes indicated that the 28S large subunit (LSU) nrDNA and the internal transcribed spacer regions and intervening 5.8S nrRNA (ITS) gene regions were unreliable to resolve species, whereas β-tubulin (tub2) and RNA polymerase II second largest subunit (rpb2) showed the greatest promise as DNA barcodes for differentiating Chaetomium species. This study provides a starting point to establish a more robust classification system for Chaetomium and for the Chaetomiaceae.
Keywords: DNA barcode, epitypification, multi-gene phylogeny, species complex, systematics
INTRODUCTION
The genus Chaetomium was established by Kunze (Kunze & Schmidt 1817), based on C. globosum. Due to the poorly-informative original description, C. globosum has been re-defined on several occasions, and many similar species have been subsequently described, mainly based on the morphology of ascomatal hairs (Corda 1840, Fries 1849, Zopf 1881, Chivers 1915, Skolko & Groves 1953, Udagawa 1960, Ames 1963, Seth 1970). The discovery of cylindrical asci by Fuckel (1869) and ascospore germ pores by Zopf (1881), however, provided better insights into the morphological definition of the genus Chaetomium. On the other hand, the taxonomic value of ascomatal hair characteristics has been considered unreliable by several authors (Tschudy 1937, Hawksworth & Wells 1973, Dreyfuss 1976, Von Arx et al. 1984). Sörgel (1960) and Dreyfuss (1976) suggested the combined morphological traits of ascospores, asci and surface structure of the ascomatal wall for the classification of Chaetomium. Millner (1977) and Millner et al. (1977) attempted to classify Chaetomium species using features of ascospore germ pores and the growth responses of species to different temperatures. Based on a limited sampling, Dreyfuss (1976) divided the genus Chaetomium into 10 species groups. In a detailed comparative study of the C. globosum group, he noticed continuous variation in ascomatal hair morphology of C. globosum, and hence emphasised ascospore morphology for species delimitation. The monographic studies by Von Arx et al. (1984, 1986), which form the basis of contemporary classification of the genus Chaetomium, summarised the previous studies and placed emphasis on the morphology of asci, ascospores, the germ pores on ascospores, and the structure of the ascomatal wall, but paid less attention to the morphology of ascomatal hairs. Based on this classification, C. globosum was characterised by globose, ovate or obovate ostiolate ascomata; ascomatal wall of textura intricata; ascomatal hairs erect, flexuous or coiled; asci evanescent, clavate or slightly fusiform; ascospores limoniform, bilaterally-flattened, 9–12 × 8–10 × 6–8 μm (length × width × thickness) in size, with an apical germ pore. Twenty-eight species were reduced to synonymy under C. globosum, and two additional species were tentatively maintained: C. cruentum as an albino form of C. globosum, and C. spirochaete slightly deviating from C. globosum by more regularly coiled and thicker ascomatal hairs. Several species, including C. elatum and C. subaffine, were also considered as close relatives of C. globosum. The definition of C. globosum sensu Von Arx, however, was considered by subsequent researchers as being too broad (Seth et al. 1987, Asgari & Zare 2011, Doveri 2013).
Based on a three-gene phylogeny, which mainly included Iranian isolates, Asgari & Zare (2011) recognised five species groups within the genus Chaetomium. Eleven species were included in their C. globosum group, constituting C. coarctatum, C. cruentum, C. elatum, C. globosum, C. madrasense, C. megalocarpum, C. subaffine and four newly described species. The sequence data, however, only included three isolates of C. globosum sensu Von Arx and failed to clarify the species concept of C. globosum.
As the non-ostiolate counterpart genus of Chaetomium, Chaetomidium is characterised by cleistothecial ascomata bearing usually long and flexuous ascomatal hairs, and ellipsoidal to limoniform, single-celled ascospores with a single apical germ pore. This genus currently includes 12 species (Von Arx 1975, Stchigel et al. 2004, Greif & Currah 2007). Recently, a phylogenetic analysis including nine Chaetomidium species using sequence data of three gene regions revealed that the studied species were scattered throughout the Chaetomiaceae and Lasiosphaeriaceae, indicating that Chaetomidium is polyphyletic (Greif et al. 2009). As Chd. fimeti, the type species of Chaetomidium, and Chd. subfimeti, formed a strongly supported clade in all three analyses, it was suggested that Chaetomidium should be restricted to Chd. fimeti and Chd. subfimeti. However, the phylogenetic placement of Chaetomidium sensu Greif et al. (2009) was inconsistent in the three gene regions analysed. Analysis of the RNA polymerase II second largest subunit (rpb2) revealed a highly supported clade that included Chaetomidium sensu Greif et al. (2009), Chd. pilosum, C. elatum and C. globosum, forming a sister clade to the clade which included both Chaetomium and Chaetomidium species. Both the 28S large subunit (LSU) nrDNA and β-tubulin (tub2) sequence data also did not support the segregation of Chaetomidium from Chaetomium.
Despite the inconsistency and contradiction in delimitation of C. globosum, it is, undoubtedly, one of the most important Chaetomium species due to its various positive and negative impacts on humans and the environment. Chaetomium globosum sensu Von Arx is reported to be cosmopolitan, and occurs in a great variety of environments which include soil, dung, a wide variety of plant materials and other cellulose-rich substrates, as well as in air and marine environments (Ames 1963, Carter 1982, Kopytina 2005, Momesso et al. 2008, Kharwar et al. 2011, Yamada et al. 2012). This species is also well known for its cellulolytic ability, having potential use in biodegradation of waste plant material and other industrial applications (Umikalsom et al. 1998, El-Gindy et al. 2003, Ahammed et al. 2008, Prokhorov & Linnik 2011, Longoni et al. 2012, Singh et al. 2013, Sharma et al. 2014). In order to adapt to diverse environments, C. globosum is capable of producing various enzymes and secondary metabolites, displaying a wide range of biological activities. These include antifungal, antibacterial, antioxidant, anti-inflammatory and anticancer activities that are of potential use in the agricultural, medicinal and industrial fields (Udagawa et al. 1979, Sekita et al. 1981, Park et al. 2005, Ding et al. 2006, Kim & Hwang 2007, Ge et al. 2008, Momesso et al. 2008, Phonkerd et al. 2008, Kaewchai et al. 2009, Zhang et al. 2010, 2012, 2013, Kharwar et al. 2011, Yamada et al. 2012, Kumar et al. 2013, Shanthiyaa et al. 2013, Awad et al. 2014, Yan et al. 2014). As a common contaminant in indoor environments, C. globosum has been recognised as a health hazard mainly due to the production of mycotoxins, microbial volatile organic compounds and airborne fungal fragments or ascospores that, when inhaled, may contribute to the development of symptoms of rhinitis, asthma and other health problems (Gonianakis et al. 2005, Vesper et al. 2007, Apetrei et al. 2009, Polizzi et al. 2009, Ayanbimpe et al. 2010, Mason et al. 2010, Andersen et al. 2011, Miller & McMullin 2014). Chaetomium globosum has also been reported to infect humans, and is most commonly associated with onychomycosis, a disease with increasing incidence reports worldwide over recent decades (Hoppin et al. 1983, Naidu et al. 1991, Stiller et al. 1992, Yeghen et al. 1996, Lesire et al. 1999, Aspiroz et al. 2007, Latha et al. 2010, Tullio et al. 2010, Hubka et al. 2011, Hwang et al. 2012, Lagacé & Cellier 2012, Kim et al. 2013).
Clarification of the species concepts of C. globosum and allied taxa is of indispensable importance not only for taxonomy of the genus, but also to obtain a better understanding of the economical importance of the species. Therefore, the aim of the present study is to resolve the species concept of C. globosum s.str. and its relationship with allied species using phylogenetic inference based on six loci in combination with morphological features.
MATERIALS AND METHODS
Isolates
The Chaetomium isolates used in this study are housed in the collections of the CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands (CBS), and the China General Microbiological Culture Collection Centre, Institute of Microbiology, Beijing, China (CGMCC). Overall, 800 strains assigned to Chaetomium species were screened for strains belonging to the C. globosum species complex. Based on a preliminary phylogenetic analysis (data not shown) of the rpb2 and tub2 gene regions, 80 representative strains were selected for further study (Table 1).
Table 1.
Details of isolates and their sequences employed in this study. The newly generated sequences in this study are shown in bold.
| Species | Isolate codea, b | Country | Substrate / Locality | MGTc (°C) | GenBank accession numbers |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| LSU | ITS | tub2 | tef1 | rpb1 | rpb2 | |||||
| Chaetomium afropilosum | CBS 145.38 (T) | – | – | – | KT214605 | KT214574 | KT214751 | KT214713 | KT214639 | KT214675 |
| C. angustispirale | CBS 137.58 (T) | Russia | Fraxinus sp., Tellerman forest, Baleshev region | 33 | JN209862 | JN209862 | JN256141 | KF001734 | KF001779 | KF001824 |
| C. ascotrichoides | CBS 113.83 (T) | Argentina | Gossypium humitectum | 38–39 | KC109752 | KC109752 | KC109770 | KF001742 | KF001787 | KF001832 |
| CBS 110.83 (T of C. gibberosporum) | Israel | Soil | 38–39 | KC109753 | KC109753 | KC109771 | KF001743 | KF001788 | KF001833 | |
| CGMCC 3.11378 | China | Soil, Yuli county, Korla region, Xinjiang | 38–39 | JN209900 | JN209900 | JN256174 | KF001746 | KF001791 | KF001836 | |
| CGMCC 3.11392 | China | Sheep wool, Aksu region, Xinjiang | 38–39 | JN209903 | JN209903 | JN256176 | KF001744 | KF001789 | KF001834 | |
| CGMCC 3.12884 | China | Sheep dung, Aksu region, Xinjiang | 38–39 | JN209904 | JN209904 | JN256177 | KF001745 | KF001790 | KF001835 | |
| C. capillare | CBS 128489 (T) | USA | Animal hair, California | – | KT214614 | KT214583 | KT214760 | KT214724 | KT214650 | KT214686 |
| C. cervicicola | CBS 128492 (T) | USA | Neck of Homo sapiens, Texas | – | KT214592 | KT214558 | KT214735 | KT214697 | KT214623 | KT214662 |
| C. citrinum | CBS 693.82 (T) | Japan | Rice field soil, Tochigi | – | KT214617 | KT214587 | KT214764 | KT214730 | KT214656 | KT214691 |
| C. coarctatum | MUCL 18697 = CBS 162.62 (T) | Russia | Seed of Cappanula medium, St. Petersburg | 38 | JN209863 | JN209863 | JN256142 | KF001712 | KF001757 | KF001802 |
| CGMCC 3.14293 | China | Unknown animal dung, Huairou, Beijing | 37–38 | JN209923 | JN209923 | JN256193 | KF001713 | KF001758 | KF001803 | |
| CGMCC 3.14299 | China | Dead stem of unknown plant, Xiangshan Park, Beijing | 37–38 | JN209924 | JN209924 | JN256194 | KF001714 | KF001759 | KF001804 | |
| C. cochliodes | CBS 155.52 (epiT) | USA | Animal dung | 38 | KC109754 | KC109754 | KC109772 | KF001721 | KF001766 | KF001811 |
| CGMCC 3.9440 | China | Tuber of Panax notoginseng, Wenshan, Yunnan Province | 38 | JN209866 | JN209866 | JN256145 | KF001724 | KF001769 | KF001814 | |
| CGMCC 3.9471 | China | Rhizospheres of Panax notoginseng, Wenshan, Yunnan Province | 38 | JN209868 | JN209868 | JN256147 | KF001723 | KF001768 | KF001813 | |
| CGMCC 3.14296 | China | Discarded cloth, Ulanqab City, Inner Mongolia | 38 | JN209865 | JN209865 | JN256144 | KF001722 | KF001767 | KF001812 | |
| C. contagiosum | CBS 128494 (T) | USA | Cornea of Homo sapiens, North East | – | KT214589 | KT214555 | KT214732 | KT214694 | KT214620 | KT214659 |
| C. cucumericola | CBS 378.71 (T) | Turkey | –, Izmir | – | KT214610 | KT214579 | KT214756 | KT214718 | KT214644 | KT214680 |
| IRAN 1642C = CBS 126777 | Iran | Petiole of Cucumis sativus, Hashtgerd, Alborz Province | – | HM365247 | HM365247 | KT214757 | KT214719 | KT214645 | KT214681 | |
| C. elatum | CBS 910.70 (T of C. ramipilosum) | Germany | Leaves and dead stems of Ammophila arenaris, Helgoland | 35–36 | KC109757 | KC109757 | KC109775 | KF001731 | KF001776 | KF001821 |
| CBS 374.66 (T of C. virgecephalum) | USA | Decomposing leaf, Aptos, California | 35–36 | KC109758 | KC109758 | KC109776 | KF001730 | KF001775 | KF001820 | |
| C. fimeti | DSM 62108 = CBS 139034 (epiT) | Germany | Soil | – | KT214593 | KT214559 | KT214736 | KT214698 | KT214624 | KT214663 |
| CBS 153.77 | Japan | – | – | KT214594 | KT214561 | KT214738 | KT214700 | KT214626 | KT214664 | |
| CBS 168.71 | Canada | Decaying hay, Nashville, Ontario | – | FJ666358 | KT214560 | KT214737 | KT214699 | KT214625 | FJ666389 | |
| C. globosporum | CBS 108.83 (T) | India | Green leaf of Triticum aestivum | 40–41 | KC109750 | KC109750 | KC109768 | KF001735 | KF001780 | KF001825 |
| C. globosum | CBS 160.62 (neoT) | Germany | Compost | – | KT214596 | KT214565 | KT214742 | KT214704 | KT214630 | KT214666 |
| CBS 105.40 | Netherlands | Mouldy book, Amsterdam | – | KT214597 | KT214566 | KT214743 | KT214705 | KT214631 | KT214667 | |
| CBS 132.30 (T of C. subterraneum) | USA | Clay soil, Illinois | 37–38 | KC109755 | KC109755 | KC109773 | KF001702 | KF001747 | KF001792 | |
| CBS 147.60 (T of C. mollipilium) | USA | Raincoat, Jeffersonville, Indiana | 38 | JN209909 | JN209909 | JN256179 | KF001703 | KF001748 | KF001793 | |
| CBS 148.51 | USA | Stored cotton, Washington DC | 37–38 | GU563363 | GU563374 | JF772459 | KC485028 | KC485058 | KF001801 | |
| CBS 164.62 (T of C. rectum ) | Poland | –, Bydgoszcz Botanic garden | 38 | JN209920 | JN209920 | JN256190 | KF001706 | KF001751 | KF001796 | |
| CBS 371.66 (T of C. cruentum) | USA | Paper, Fort Belvoir, Virginia | 37 | JN209871 | JN209871 | JN256148 | KF001705 | KF001750 | KF001795 | |
| CGMCC 3.9994 | China | Finger nail of Homo sapiens, Beijing | 38 | JN209894 | JN209894 | JN256168 | KF001708 | KF001753 | KF001798 | |
| MUCL 39526 (T of C. globosum var. flavoviride) | Hungary | Dead stem of Juncus sp. | 37–38 | JN209875 | JN209875 | JN256152 | KF001710 | KF001755 | KF001800 | |
| MUCL 39527 (T of C. globosum var. griseum) | Hungary | Dead stem of Juncus sp. | 37–38 | JN209899 | JN209899 | JN256173 | KF001709 | KF001754 | KF001799 | |
| C. graminiforme | CBS 506.84 (T) | Canada | Acer sp., Muskoka District, Ontario | – | KT214615 | KT214584 | KT214761 | KT214725 | KT214651 | KT214687 |
| C. grande | IRAN 1064C = CBS 126780 (T) | Iran | Leaf of Triticum aestivum, Naghadeh, West Azerbaijan Province | – | HM365253 | HM365253 | HM365273 | KT214692 | KT214618 | KT214657 |
| CGMCC 3.9414 = CBS 119758 | China | Desert soil, Bayingolin, Xinjiang Autonomous Region | 38 | KC109749 | KC109749 | KC109767 | KF001736 | KF001781 | KF001826 | |
| IRAN 1208C = CBS 126781 | Iran | Straw of Triticum aestivum, Bilesavar, Ardabil Province | – | KT214588 | KT214554 | KT214731 | KT214693 | KT214619 | KT214658 | |
| C. interruptum | IRAN 1278C = CBS 126660 (T) | Iran | Seed of Triticum aestivum, Hadishahr, East Azerbaijan Province | – | HM365246 | HM365246 | KT214741 | KT214703 | KT214629 | KT214665 |
| C. madrasense | CBS 315.74 (T) | India | Rhizosphere of Pennisetum typhoides, Tamil Nadu, Madras | 38 | KC109751 | KC109751 | KC109769 | KF001741 | KF001786 | KF001831 |
| C. megalocarpum | MUCL 9589 = CBS 149.59 (epiT) | Greece | Leaf of Ficus carica | 40 | KC109744 | KC109744 | KC109762 | KF001738 | KF001783 | KF001828 |
| CBS 778.71 | India | Humus-rich soil | 40 | KC109747 | KC109747 | KC109765 | KF001737 | KF001782 | KF001827 | |
| CGMCC 3.3595 | China | Horse dung, Yinchuan Province, Ningxia City | 40 | KC109746 | KC109746 | KC109764 | KF001740 | KF001785 | KF001830 | |
| CGMCC 3.9443 | China | Soil, Shanhaiguan, Hebei Province | 40 | KC109748 | KC109748 | KC109766 | KF001739 | KF001784 | KF001829 | |
| C. novozelandicum | CBS 124555 (T) | New Zealand | Dead decaying twig, Otaki | – | KT214607 | KT214576 | KT214753 | KT214715 | KT214641 | KT214677 |
| CBS 124556 | New Zealand | Dead decaying twig, Otaki | – | KT214608 | KT214577 | KT214754 | KT214716 | KT214642 | KT214678 | |
| CBS 128484 | USA | Scalp of Homo sapiens, California | – | KT214609 | KT214578 | KT214755 | KT214717 | KT214643 | KT214679 | |
| C. nozdrenkoae | CBS 163.62 (T) | Russia | Soil, Novosibirsk region | – | KT214590 | KT214556 | KT214733 | KT214695 | KT214621 | KT214660 |
| CBS 809.68 | Germany | Greenhouse soil, Giessen | – | KT214591 | KT214557 | KT214734 | KT214696 | KT214622 | KT214661 | |
| C. olivaceum | CBS 418.80A | India | Nilgai dung, Delhi | 38 | JN209914 | JN209914 | JN256184 | KF001716 | KF001761 | KF001806 |
| CGMCC 3.9465 | China | Soil, Changchun, Jilin Province | 38 | JN209913 | JN209913 | JN256183 | KF001715 | KF001760 | KF001805 | |
| CGMCC 3.12883 | China | Camel dung, Aksu region, Xinjiang | 37 | JN209911 | JN209911 | JN256181 | KF001717 | KF001762 | KF001807 | |
| C. pilosum | CBS 335.67 (T) | Australia | Grain of Triticum aestivum, Perth, Western Australia | – | FJ666356 | KT214586 | KT214763 | KT214729 | KT214655 | FJ666387 |
| C. pseudocochliodes | CGMCC 3.9441 (T) | China | Roots of Panax notoginseng, Wenshan, Yunnan Province | 38 | JN209925 | JN209925 | JN256195 | KF001726 | KF001771 | KF001816 |
| CGMCC 3.9469 | China | Rhizosphere of Panax notoginseng, Wenshan, Yunnan Province | 37–38 | JN209926 | JN209926 | JN256196 | KF001725 | KF001770 | KF001815 | |
| C. pseudoglobosum | CBS 574.71 (T) | – | – | – | KT214604 | KT214573 | KT214750 | KT214712 | KT214638 | KT214674 |
| C. rectangulare | IRAN 1641C = CBS 126778 (T) | Iran | Leaf of Hordeum vulgare, Salmas, West Azerbaijan Province | – | HM365239 | HM365239 | HM365285 | KT214726 | KT214652 | KT214688 |
| CGMCC 3.9409 | China | Animal dung, Kanas Lake, Xinjiang | 35 | JN209873 | JN209873 | JN256150 | KF001732 | KF001777 | KF001822 | |
| IRAN 855C = CBS 126658 | Iran | Stem of Hordeum vulgare, Shabestar, East Azerbaijan Province | – | HM365240 | HM365240 | HM365286 | KT214727 | KT214653 | KT214689 | |
| C. spiculipilium | CBS 373.66 (T) | USA | Decaying vegetable debris, California | 34–35 | KC109756 | KC109756 | KC109774 | KF001719 | KF001764 | KF001809 |
| C. spirochaete | CBS 730.84 (epiT) | USA | Animal dung, Great Smokey Mountains, Tennessee | 38 | JN209921 | JN209921 | JN256191 | KF001729 | KF001774 | KF001819 |
| CBS 165.52 | – | Animal dung | – | KT214616 | KT214585 | KT214762 | KT214728 | KT214654 | KT214690 | |
| C. subaffine | CBS 637.91 (T) | USSR | Cereal | 39 | JN209929 | JN209929 | JN256199 | KF001727 | KF001772 | KF001817 |
| CGMCC 3.14297 | China | Unknown plant stem, Xingtai, Hebei Province | 39 | JN209928 | JN209928 | JN256198 | KF001728 | KF001773 | KF001818 | |
| C. subfimeti | CBS 370.66 (T, T of Chaetomidium subfimeti) | Wales | Paper and vegetable material, Cardiff | – | FJ666354 | KT214562 | KT214739 | KT214701 | KT214627 | FJ666385 |
| CBS 169.71 | USA | Soil, Kern County, California | – | FJ666357 | KT214563 | KT214740 | KT214702 | KT214628 | FJ666388 | |
| C. subglobosum | MUCL 18694 = CBS 149.60 (T) | Russian | Dead herbaceous stem, St. Petersburg | 38 | JN209930 | JN209930 | JN256200 | KF001718 | KF001763 | KF001808 |
| CBS 483.73 | Turkey | Eriobotrya japonica, Izmir | – | KT214612 | KT214581 | KT214758 | KT214722 | KT214648 | KT214684 | |
| C. telluricola | CBS 151.59 (T) | United Kingdom | Soil, Suffolk, Lakenheath Warren | – | KT214613 | KT214582 | KT214759 | KT214723 | KT214649 | KT214685 |
| C. tenue | CBS 139.38 (T) | – | – | – | KT214599 | KT214568 | KT214745 | KT214707 | KT214633 | KT214669 |
| CBS 138.38 | – | – | – | KT214600 | KT214569 | KT214746 | KT214708 | KT214634 | KT214670 | |
| CBS 140.38 | – | – | – | KT214601 | KT214570 | KT214747 | KT214709 | KT214635 | KT214671 | |
| CBS 142.38 | – | – | – | KT214602 | KT214571 | KT214748 | KT214710 | KT214636 | KT214672 | |
| CBS 143.38 | – | – | – | KT214603 | KT214572 | KT214749 | KT214711 | KT214637 | KT214673 | |
| C. umbonatum | CBS 293.83 (T) | Canada | Soil, Nova Scotia | – | KT214606 | KT214575 | KT214752 | KT214714 | KT214640 | KT214676 |
| C. undulatulum | IRAN 857C = CBS 126775 (T) | Iran | Leaf of Hordeum vulgare, Bonab, East Azerbaijan Province | – | HM365251 | HM365251 | HM365279 | KT214720 | KT214646 | KT214682 |
| IRAN 1071C = CBS 126776 | Iran | Leaf of Triticum aestivum, Miandoab, West Azerbaijan Province | – | HM365250 | HM365250 | HM365278 | KT214721 | KT214647 | KT214683 | |
| C. unguicola | CBS 128446 (T) | USA | Nail of Homo sapiens, Los Angeles | – | KT214598 | KT214567 | KT214744 | KT214706 | KT214632 | KT214668 |
| Achaetomium strumarium | CBS 333.67 (T) | India | Soil, Lucknow | – | AY681170 | AY681204 | AY681238 | KC503252 | KC503253 | KC503254 |
a CBS: CBS-KNAW Fungal Diversity Centre, Utrecht, The Netherlands; CGMCC: China General Microbiological Culture Collection Centre in the Insitute of Microbiology, Beijing, China; DSM: Deutsche Sammlung von Mikrorganismen und Zellkulturen GmbH, Braunschweig, Germany; IRAN: Iranian Research Institute of Plant Protection, Tehran, Iran; MUCL: Mycothèque de l’Université Catholique de Louvain, Belgium. Additional culture collection numbers are available where applicable under the species notes in the Taxonomy section.
b T: ex-type strain; epiT: ex-epitype strain; neoT: ex-neotype strain.
c Maximum Growth Temperature.
DNA phylogeny
Genomic DNA was extracted from mycelium harvested from cultures grown on 2 % (w/v) malt extract agar (MEA) for 7–14 d at room temperature using the E.Z.N.A.™ HP Fungal DNA Kit (Omega Bio-Tek, Norcross, GA), or the CTAB extraction method (Damm et al. 2008) with minor modification: after adding the CTAB extraction buffer, samples were subjected to three cycles of freezing in liquid nitrogen and thawing in a water bath, instead of incubating at 100 °C for 3 min. The primers used for PCR-amplification and sequencing included ITS5 & ITS4 for the internal transcribed spacer regions and intervening 5.8S nrRNA gene region (ITS; White et al. 1990); NL1 & NL4 for the D1/D2 domains of the 28S nrRNA gene region (LSU; O’Donnell 1993); T1 (O’Donnell & Cigelnik 1997) & T222 (Glass & Donaldson 1995) for the partial tub2 gene region; EF1-983F & EF1-2218R (S. Rehner, AFTOL, http://aftol.org/) for the partial translation elongation factor 1-α (tef1) gene region; gRPB1-A & fRPB1-C (Matheny et al. 2002) for partial fragments of the largest subunit of the RNA polymerase II (rpb1) gene; RPB2AM-1bf & RPB2AM-7R (Miller & Huhndorf 2005) for partial fragments of the rpb2 gene region. The PCR mixtures (12.5 μL) contained 10–20 ng of genomic DNA, 1× GoTaq® Flexi buffer (Promega, Madison, WI, USA), 1 mM MgCl2 (2.5 mM for rpb2), 40 μM dNTPs (60 μM for rpb2), 0.2 μM of each primer (0.12 μM for rpb2) and 0.5 Unit GoTaq® Flexi DNA polymerase (Promega, Madison, WI, USA). The PCR conditions for ITS, LSU, rpb1, tef1 and tub2 were the same as those described by Wang et al. (2014). The cycle conditions for amplification of the partial rpb2 gene included cycles of 94 °C/3 min (initial denaturation); 94 °C/45 s, 60 °C/45 s, 72 °C/2 min (5×); 94 °C/45 s, 58 °C/45 s, 72 °C/2 min (5×); 94 °C/45 s, 56 °C/45 s, 72 °C/2 min (35×) and 72 °C/8 min (final extension). The PCR products were purified and sequenced in both directions using the BigDye® Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems Life Technologies, Carlsbad, CA, USA) and an ABI Prism® 3730xl Genetic Analyzer (Applied Biosystems). Consensus sequences were determined using MEGA v. 6 (Tamura et al. 2013). Novel sequences generated in this study were deposited in GenBank (http://www.ncbi.nlm.nih.gov, Table 1).
In addition to the sequences generated in this study, other sequences from previous studies (Greif et al. 2009, Asgari & Zare 2011) were retrieved from GenBank. The sequence datasets were initially aligned using MAFFT v. 7 (Katoh & Standley 2013), and were manually optimised using BioEdit v. 5.0.9 (Hall 1999). Congruency of the six loci was tested using the 70 % reciprocal bootstrap criterion (Mason-Gamer & Kellog 1996) as described by Gueidan et al. (2007) and Lombard et al. (2010).
Phylogenetic analyses of individual gene alignments and the concatenated six-locus dataset were based on Bayesian inference (BI), Maximum Likelihood (ML) and Maximum Parsimony (MP) analyses. For BI, the best evolutionary model for each partition was determined using MrModeltest v. 2 (Nylander 2004) and incorporated into the analyses. A Markov Chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees using MrBayes v. 3.2.1 (Ronquist & Huelsenbeck 2003) under optimal criteria for each locus. The MCMC analysis continued until the average standard deviation of split frequencies came below 0.01 with trees saved every 1 000 generations. The first 25 % of saved trees were discarded as the ‘burn-in’ phase and posterior probabilities (PP) were determined from the remaining trees. The MP analysis was performed using PAUP v. 4.0b10 (Phylogenetic Analysis Using Parsimony; Swofford 2003). Phylogenetic relationships were estimated by heuristic searches with 1 000 random addition sequences. Tree bisection-reconnection was used, with the branch swapping option set on ‘best trees only’, with all characters weighted equally and alignment gaps treated as fifth character state. The tree length (TL), consistency index (CI), retention index (RI) and rescaled consistence index (RC) were calculated for the MP phylogenies and the bootstrap analysis (Hillis & Bull 1993) was based on 1 000 replications. The ML analysis was performed using RAxML-VI-HPC v. 7.0.3 (Stamatakis 2006) on the CIPRES Science Gateway (https://www.phylo.org) with nonparametric bootstrapping using 1 000 replicates. Trees were viewed in FigTree v. 1.1.2 (Rambaut 2009). The alignment and derived trees were deposited in TreeBASE (submission ID 17816; http://treebase.org/treebase-web/home.html).
Kimura-2-parameter values
To evaluate the efficiency of each gene region for species delimitation, individual alignments of each locus were analysed using MEGA v. 6 (Tamura et al. 2013), generating both inter- and intraspecific distance matrices using the Kimura-2-parameter model, with substitutions including transitions and transversions. Uniform rates among sites were used and gaps were completely deleted. The obtained distance values were exported in a Microsoft Excel workbook format and then sorted into frequency distribution bins (from distance 0–0.2 with intervals of 0.008 between bins). The frequency distribution mean was calculated according to the formula x = Σ(f.b)/Σ(f), in which f is the frequency and b is the bin. The distance between the mean of the inter- and intraspecific distance distributions represents the barcoding gap (Fig. 2).
Fig. 2.
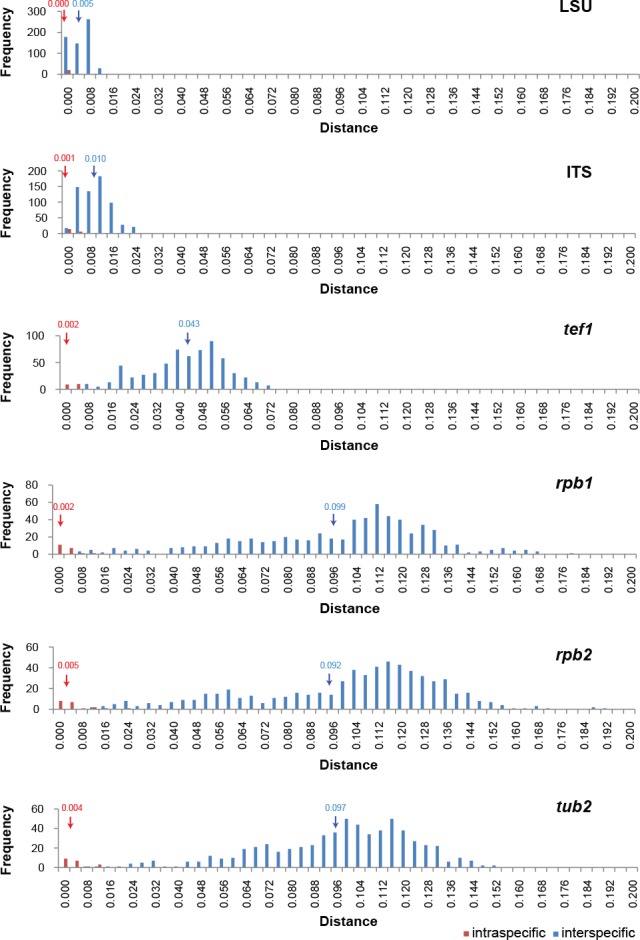
The frequency distribution graphs of the Kimura-2-parameter distances (barcoding gaps) for the six individual loci. The blue arrow indicates the average interspecific distance and the red arrow indicates the average intraspecific distance with corresponding mean values above both arrows.
Morphology
All the representative isolates were inoculated onto 3 % oatmeal (w/v) agar (OA; Crous et al. 2009), and incubated in the dark at 25 °C until the ascomata matured. Isolates that appeared to be sterile, were also inoculated onto cornmeal agar (CMA), MEA, as well as water agar (WA) and OA supplemented with sterile filter paper strips, barley leaves or elm stems. Cultures were incubated at room temperature (fluctuating from day to night), 25 °C or 28 °C in the dark or under continued UV-light in order to induce sporulation. Colonies and ascomata were observed using a Nikon SMZ 1500 dissecting-microscope and colony colours were determined using the colour charts of Rayner (1970). Microscopic features were studied using a Nikon Eclipse 80i compound microscope equipped with differential interference contrast (DIC) illumination. Shear’s mounting medium was used to observe the asci from young or newly-matured ascomata. Microscopic features of ascomata, ascomatal hairs and ascospores were determined in lactic acid with the exception of the ascospores of C. angustispirale, which were studied in water. Lactic acid mounts were gently heated to remove air bubbles and prevent ascospore shrinkage. At least 30 measurements were made for all morphologically informative features. The ascospore measurements include the extreme values given in parentheses and, in between, the 95 % confidence interval of 30 individual measurements, for the three dimensions of length, width and thickness.
Fifty-four isolates representing 17 species were compared for their maximum growth temperatures (MGT) using the methods presented in Wang et al. (2014). Taxonomic information and nomenclature for new species were deposited in MycoBank (www.MycoBank.org; Crous et al. 2004).
RESULTS
Phylogeny
The phylogenetic analyses included 80 ingroup taxa, with Achaetomium strumarium (CBS 333.67, ex-type) as outgroup. No topological conflicts were observed when the 70 % bootstrap reciprocal tree topologies of the analysed loci were compared, except for the ITS and LSU which failed to resolve most of the phylogenetic species recovered by the remaining four protein-coding gene regions. However, all six loci were combined following the argument of Cunningham (1997) that combining incongruent partitions could increase phylogenetic accuracy. The combined alignment consisted of 4 128 characters including alignment gaps. Of these, 2 671 characters were constant, 359 parsimony-uninformative and 1 098 parsimony-informative. For the Bayesian inference, a GTR+I+G model was selected for ITS, rpb1, rpb2 and tef1, and a HKY+I+G model for LSU and tub2. A total of 2 332 trees were generated during the Bayesian inference from which 582 trees were discarded as the ‘burn-in phase’ and posterior probabilities (PP) were calculated from the remaining 1 750 trees. Both the BI consensus tree and PP confirmed the tree topologies and bootstrap support (BS) values obtained with the ML and MP analyses. The MP analysis resulted in four equally most parsimonious trees (TL = 3 616; CI = 0.554; RI = 0.866; RC = 0.480). The BI consensus tree is presented here (Fig. 1) with the relevant BS values of the MP and ML analyses shown at the nodes.
Fig. 1.
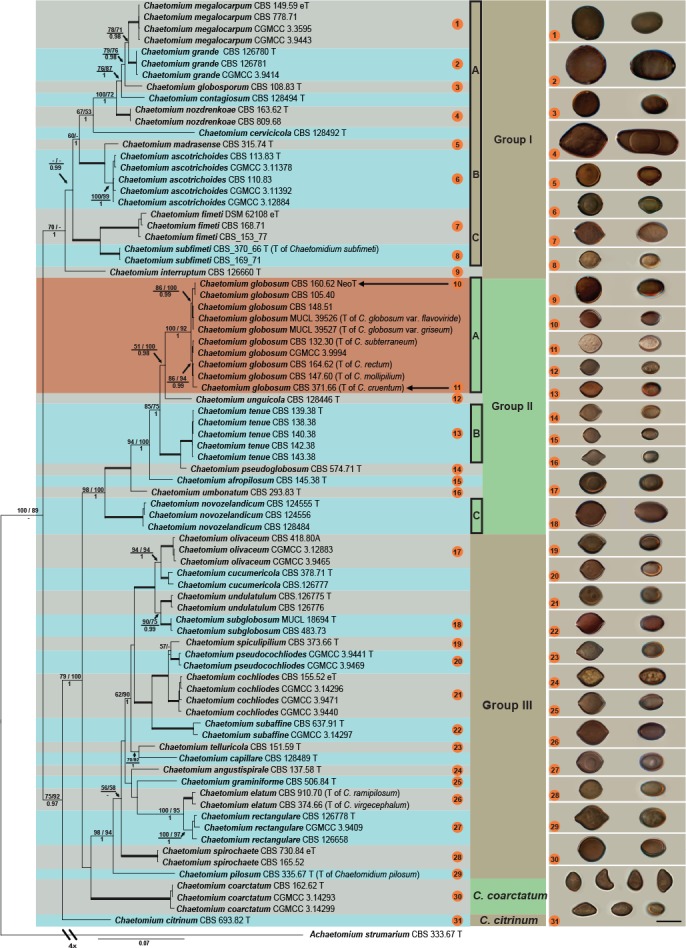
Consensus phylogram resulting from a Bayesian analysis of the concatenated rpb2, tub2, tef1, rpb1, ITS and LSU gene region alignment, with the confidence values of bootstrap (BS) proportions from the MP analysis (before the backslash), the ML analysis (after the backslash) above branches, and the posterior probabilities (PP) from the Bayesian analysis below branches. The ‘-’ indicates lacking statistical support (< 50 % for ML-BS and MP-BS analyses; < 0.90 for PP from Bayesian analyses). The branches with full statistical support (MP-BS = 100 %; ML-BS = 100 %; PP = 1.0) are highlighted with thickened branches. The tree is rooted to Achaetomium strumarium. Each species clade is discriminated with boxes in a different colour. Ascospores of all sporulating species treated in this study are illustrated at the right side of tree (scale bar = 10 μm; ascospores face view on the left and side view on the right, except for C. citrinum in the last line). The ascospores are correlated with each corresponding species using the same numbers in orange circles.
The phylogenetic tree (Fig. 1) resolved 36 well-supported clades representing possible cryptic species within the C. globosum species complex (MP-BS = 100; ML-BS = 89; PP < 0.9). The species complex was divided into two main clades, which was further divided into three groups (Fig. 1, Groups I–III). The first main clade represented Group I (MP-BS = 70; ML-BS = < 50; PP = 1.0), with C. interruptum forming a basal sister lineage to the remaining members of this group. The other taxa in Group I were further divided into three well-supported subclades (A–C). The largest of these (Group IA; MP-BS = 67; ML-BS = 53; PP = 1.0) included six well-supported lineages, two of which (CBS 128492 and CBS 128494) represent possible novel taxa. The second subclade (Group IB; MP-BS = 100; ML-BS = 100; PP = 1.0) includes C. ascotrichoides (ex-type culture CBS 113.83) forming a clade (MP-BS = 100; ML-BS = 99; PP = 1.0) separate from the ex-type culture of C. madrasense (CBS 315.74) in the same subclade. The third subclade (Group IC; MP-BS = 100; ML-BS = 100; PP = 1.0) includes Chaetomidium (Chd.) fimeti (ex-epitype culture DSM 62108), the type species of the genus, and Chd. subfimeti (ex-type culture CBS 370.66).
The second main clade (including Group II and III; MP-BS = 75; ML-BS = 92; PP = 0.97) includes C. citrinum (ex-type culture CBS 693.82), as a basal lineage to the clade. The remaining taxa (MP-BS = 79; ML-BS = 100; PP = 1.0) are divided into three monophyletic subclades. The first subclade (Group II; MP-BS = 98; ML-BS = 100; PP = 1.0) includes several single-isolate lineages (CBS 128446, CBS 574.71 and CBS 145.38, respectively) as possible novel taxa, and the ex-type culture (CBS 293.83) of C. umbonatum. Representative strains of C. globosum s.str., the type species of the genus Chaetomium, clustered together in a well-supported clade (Group IIA; MP-BS = 100; ML-BS = 92; PP = 1.0). The remaining isolates clustered in two well-supported clades (Group IIB and IIC; both with MP-BS = 100; ML-BS = 100; PP = 1.0; containing CBS 139.38 and CBS 124555, respectively), each clade representing possible novel phylogenetic species.
The second subclade (Group III; MP-BS = 98; ML-BS = 94; PP = 1.0) includes 16 well-supported phylogenetic species, from which six isolates (CBS 373.66, CBS 151.59, CBS 128489, CBS 137.58, CBS 506.84 and CBS 335.67) represent unique single-isolate lineages. Of these, three strains (CBS 151.59, CBS 506.84 and CBS 128489) are possible novel phylogenetic species. These single-isolate lineages also include the ex-type culture of Chd. pilosum (CBS 335.67), for which a new combination is required. Two clades in Group III, one represented by CBS 378.71 (MP-BS = 100; ML-BS = 100; PP = 1.0), and the other by CGMCC 3.9441 (MP-BS = 100; ML-BS = 100; PP = 1.0) are also possible novel phylogenetic species.
Kimura-2-parameter values
The individual loci showed varying degrees of overlap in their K2P distribution graphs (Fig. 2). In these datasets, tub2 showed the best barcode gap distance between the inter- and intraspecific distances, followed by rpb1, rpb2 and tef1, respectively. Of the latter three loci, rpb2 was chosen over rpb1 due to ease of amplification across the Chaetomiaceae.
Taxonomy
The phylogenetic inference resulted in the recognition of 36 species within the C. globosum species complex. Of these, 12 species are described as novel species. The genus Chaetomidium is synonymised under Chaetomium since the type species, Chd. fimeti, was shown to belong to Chaetomium based on our phylogenetic analyses. Therefore, new combinations are provided here for Chd. fimeti, Chd. pilosum and Chd. subfimeti in the genus Chaetomium. Several isolates (CBS 119758 of C. grande, CBS 126660 of C. interruptum, CBS 108.83 of C. globosporum and CBS 483.73 of C. subglobosum) only produced viable ascomata on OA supplemented with sterile elm stems. Six phylogenetic species failed to produce any viable ascomata containing ascospores under all conditions tested in our study, five of which represent novel taxa and C. undulatulum. These five novel taxa are described here based on sequence data only, following the approach of Gomes et al. (2013) for Diaporthe. Furthermore, 23 existing species are re-described based on their morphology on OA.
Chaetomium afropilosum X. Wei Wang, Crous & L. Lombard, sp. nov. — MycoBank MB812942; Fig. 3
Fig. 3.
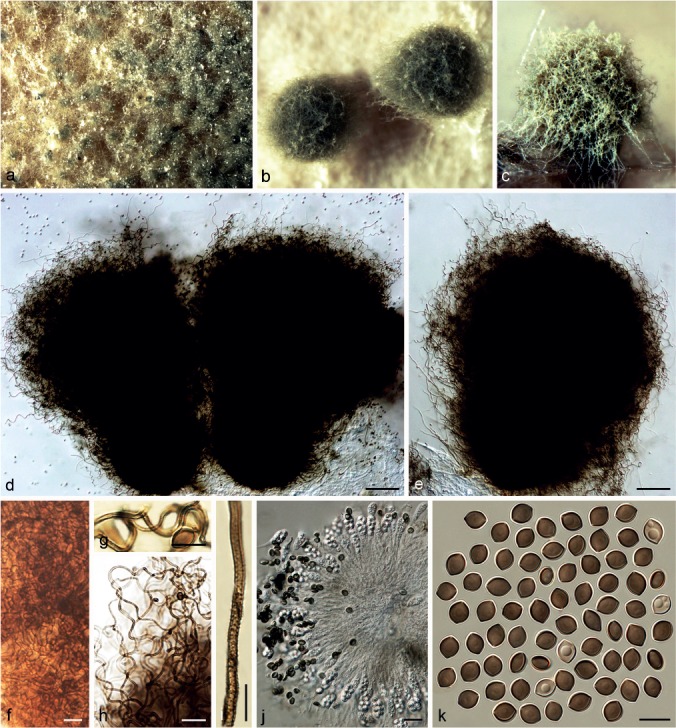
Chaetomium afropilosum (CBS 145.38, ex-type culture). a. Part of the colony on OA; b. ascomata on OA, top view; c. ascoma on OA, side view; d, e. ascomata mounted in lactic acid; f. structure of ascomatal wall in surface view; g, h. upper part of terminal ascomatal hairs; i. basal part of a terminal ascomatal hair; j. asci; k. ascospores. — Scale bars: d, e = 100 μm; h, j = 20 μm; f, g, i, k = 10 μm.
Etymology. Refers to the ‘afro’-like appearance of the ascomatal hairs.
Ascomata superficial, often covered by sparse aerial hyphae, ostiolate, pale citrine to grey-olivaceous in reflected light owing to ascomatal hairs, globose or ovate, 210–360 μm high, 180–310 μm diam. Ascomatal wall brown, composed of hypha-like or amorphous cells, textura intricata or textura epidermoidea in surface view. Terminal hairs abundant, forming a dense, nearly globose head covering the ostiole, verrucose, olivaceous brown, fading towards the tips, undulate or slightly coiled, erect or flexuous at lower part, 3–4.5 μm near the base, tapering towards the tips. Lateral hairs similar to terminal hairs, but more flexuous. Asci fasciculate, clavate or slightly fusiform, spore-bearing part 18–24 × 9–11.5 μm, stalks 15–24 μm long, with eight biseriate ascospores, evanescent. Ascospores olivaceous brown when mature, limoniform, biapiculate, bilaterally flattened, (6.5–)7–8 × (5–)5.5–6(–6.5) × 4–5 μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with sparse white aerial hyphae, sometimes forming thick white hyphae in the centre, producing apricot to orange exudates diffusing into the medium; reverse fulvous to sienna.
Material examined. UNKNOWN, substrate and collection details unknown, isolated and deposited in CBS by R.H. Tschudy in June 1938 (holotype CBS H-22192, culture ex-type CBS 145.38 = DAOM 19448).
Notes — Phylogenetic inference shows that C. afropilosum forms a unique lineage in Group II (Fig. 1), closely related to C. globosum s.str., C. unguicola, C. tenue, C. pseudoglobosum and C. umbonatum. Chaetomium afropilosum can be distinguished by its distinct ascomatal hair structure and by its smaller ascospores compared to those of C. globosum s.str. (8.5–10.5 × 7–8 × 5.5–6.5 μm), C. unguicola (7.5–9 × 6.5–7 × 4.5–5.5 μm), C. tenue (7.5–10 × 6–7 × 4.5–5.5 μm), C. pseudoglobosum (9–10 × 6.5–7.5 × 5–6 μm) and C. umbonatum (8–11 × 5.5–7 × 4–5 μm). This species has the smallest ascospores of all known species in the C. globosum species complex.
Chaetomium angustispirale Sergeeva, Not. Syst. sect. Crypt. Inst. Bot. Acad. Sci. U.S.S.R. 11: 115. 1956. — Fig. 4
Fig. 4.
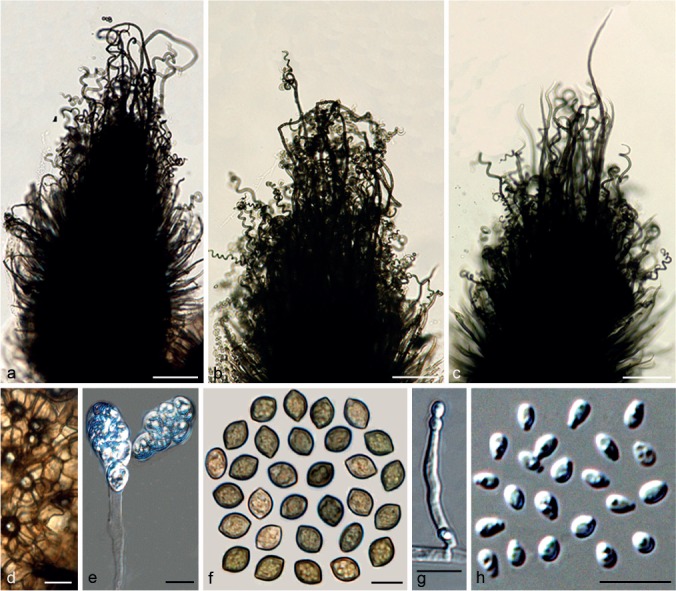
Chaetomium angustispirale (CBS 137.58, ex-type culture). a–c. Ascomata mounted in lactic acid; d. structure of ascomatal wall in surface view; e. asci; f. ascospores; g, h. asexual morph: g. conidiophore; h. conidia. — Scale bars: a–c = 100 μm; d–h = 10 μm.
Ascomata superficial, ostiolate, dark olivaceous in reflected light owing to ascomatal hairs, ellipsoid to subglobose, 270–400 μm high, 220–380 μm diam. Ascomatal wall brown, composed of irregular or angular cells, textura angularis in surface view. Terminal hairs brown, verrucose, partly long and thick, 5–7 μm diam near the base, erect, often circinate or coiled in the upper part, sometimes branched; partly short and thin, 3–5 μm diam near the base with relatively long coils in the upper part, often branched. Lateral hairs hypha-like, erect or flexuous, tapering towards the tips. Asci fasciculate, clavate or fusiform, spore-bearing part 28–35 × 11–19 μm, stalks 26–48 μm long, with eight biseriate ascospores, evanescent. Ascospores olivaceous brown when mature, limoniform, usually slightly umbonate at both ends, bilaterally flattened, (9–)9.5–11.5(–12) × (7.5–)8–9 × (5.5–)6–7 μm, with an apical germ pore. Asexual morph acremonium-like. Conidiophores discrete and simple; conidiogenous cells phialidic, hyaline. Conidia formed in basipetal succession, aseptate, smooth, hyaline, ovate or ellipsoid, usually with truncated base and rounded apex, (2.5–)3–4.5 × 2–3 μm.
Culture characteristics — Colonies on OA with greyish white to white aerial hyphae, often producing olivaceous exudates diffusing into the medium; reverse olivaceous to cinnamon.
Material examined. RUSSIA, Baleshev region, Tellerman Forest, from Fraxinus sp., 1956, K.S. Sergejeva (culture ex-type CBS 137.58 = IMI 074952 = VKM F-1942).
Notes — Chaetomium angustispirale is only known from its ex-type culture (CBS 137.58), and it was difficult to induce sporulation. Ascomata were only obtained by growing the isolate on OA supplemented with sterile elm stem pieces at the beginning of this study, and ascospores were studied using water as mounting medium. All attempts to induce sporulation again, for better morphological data, failed. Ames (1963) provided a description of C. angustispirale and noted the two types of terminal hairs as mentioned above, but did not mention its asexual morph. Von Arx et al. (1986) suggested this species to be a heterothallic relative of C. globosum, but at the same time listed it in the synonyms of C. globosum. The phylogeny suggests that this species is in Group III (Fig. 1), relatively distant from C. globosum s.str. (Group IIA).
Chaetomium ascotrichoides Calviello, Revista Mus. Argent. Cien. Nat. B. Aires, Bot. 3: 372. 1972. — Fig. 5, 6
Fig. 5.

Chaetomium ascotrichoides (CBS 113.83, ex-type culture). a. Part of the colony on OA; b. ascomata on OA, top view; c. ascomata and masses of ascospores on OA, side view; d, e. ascomata mounted in lactic acid; f. structure of ascomatal wall in surface view; g. terminal ascomatal hairs around the ostiole; h. asci; i. ascospores. — Scale bars: d, e = 100 μm; f–i = 10 μm.
Fig. 6.
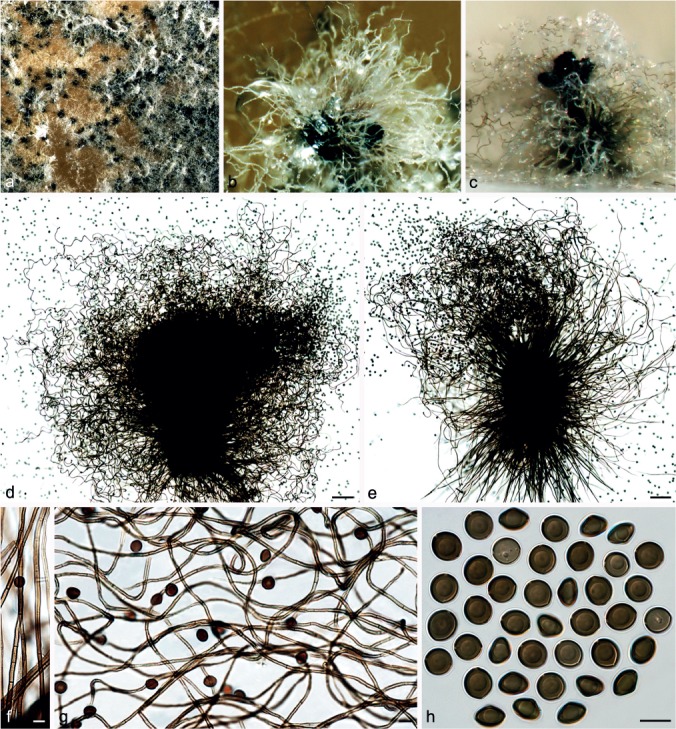
Chaetomium ascotrichoides (CBS 110.83, ex-type of C. gibberosporum). a. Part of the colony on OA; b. ascoma on OA, top view; c. ascoma and mass of ascospores on OA, side view; d, e. ascomata mounted in lactic acid; f. basal part of terminal ascomatal hairs; g. upper part of terminal ascomatal hairs; h. ascospores. — Scale bars: d, e = 100 μm; f–h = 10 μm.
= Chaetomium gibberosporum Dreyfuss ex Sedlar et al., Arch. Mikrobiol. 92: 105. 1973 (nom. inval., Art. 38).
Ascomata, superficial, ostiolate, pale olivaceous buff, or occasionally rosy buff in reflected light owing to ascomatal hairs, later becoming black due to ascospore masses on ascomata, ellipsoid, ovate or obovate, 170–290 μm high, 130–250 μm diam. Ascomatal wall brown, composed of hypha-like or amorphous cells, textura intricata or textura epidermoidea in surface view. Terminal hairs finely verrucose, relatively sparse, brown, flexuous, undulate, sometimes simply branched, 2.5–3.5 μm near the base, hairs around ostiole often relatively short, flexuous or geniculate, constricted at septa, irregularly branched in the upper part. Lateral hairs hypha-like, flexuous, tapering towards the tips. Asci fasciculate, fusiform or clavate, spore-bearing part 30–45 × 11–19 μm, stalks 18–35 μm long, with eight biseriate ascospores, evanescent. Ascospores olivaceous brown when mature, broad limoniform, slightly apiculate at both ends, bilaterally flattened, usually triangle-shaped in side view due to a lateral bulge, (8.5–)9.5–10.5(–11) × (8–)8.5–9.5(–10) × (6–)6.5–7(–7.5) μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with sparse, white aerial hyphae, and without coloured exudates; reverse uncoloured.
Materials examined. ARGENTINA, from Gossypium humitectum, Jan. 1983, B.O. Calviello (culture ex-type CBS 113.83 = IMI 182725). – CHINA, Xingjiang, Yuli County, Korla region, from soil, June 2007, F.-J. Liu, CGMCC 3.11378; Asku region, from sheep dung, June 2007, F.-J. Liu, CGMCC 3.12884; from sheep wool, June 2007, F.-J. Liu, CGMCC 3.11392. – ISRAEL, M. Dreyfuss, deposited in CBS by O. Petrini, Jan. 1983 (isotype of C. gibberosporum CBS H-6870, ex-isotype culture of C. gibberosporum CBS 110.83 = ETH 7714).
Notes — Chaetomium ascotrichoides is morphologically similar to C. madrasense, and was treated as a synonym of the latter by Von Arx et al. (1986). This species can be distinguished by flexuous or irregularly branched ascomatal hairs compared to the coiled hairs of C. madrasense and narrower ascospores in lateral view (6.5–7 μm) than those of C. madrasense (7.5–8.5 μm). Isolate CBS 110.83 was originally attributed to C. gibberosporum without description, rendering this species name invalid under the International Code of Nomenclature for algae, fungi and plants (ICN; Art. 38; McNeill et al. 2012). Although isolate CBS 110.83 has relatively numerous and undulate to slightly coiled ascomatal hairs (Fig. 6), the presence of branched ascomatal hairs and narrow ascospores indicate that it must be conspecific with CBS 113.83, the ex-type culture of C. ascotrichoides, as was shown by phylogenetic inference (Group IB, Fig. 1). Several Chinese isolates of C. ascotrichoides possess only a few ascospores with a lateral bulge and, therefore, may be confused with C. globosum or C. coarctatum. However, the ascospores of C. ascotrichoides (9.5–10.5 × 8.5–9.5 × 6.5–7 μm) are wider than those of C. globosum (8.5–10.5 × 7–8 × 5.5–6.5 μm) and narrower than those of C. coarctatum (10–11 × 9–10 × 6.5–7.5 μm).
Chaetomium capillare X. Wei Wang, Crous & L. Lombard, sp. nov. — MycoBank MB812975
Etymology. Refers to animal hair from which this fungus was first collected.
Cultures sterile. Chaetomium capillare forms a unique lineage in Group III (Fig. 1), sister to C. telluricola. This species differs from the closest phylogenetic lineage, C. telluricola, by several fixed unique single nucleotide polymorphisms (SNP) in the six loci used in this study: rpb2 positions 21(A), 60(C), 69(C), 120(G), 132(A), 147(G), 165(T), 177(T), 195(C), 198(T), 222(T), 227(T), 228(G), 240(T), 246(T), 249(C), 265(C), 273(A), 282(C), 291(C), 294(T), 300(G), 324(C), 333(A), 351(A), 373(C), 405(C), 409(A), 411(G), 420(C), 477(T), 513(T), 546(T) and 582(T); tub2 positions 9(T), 12(G), 14(A), 15(G), 16(C), 28(T), 71(C), 90(indel), 97(T), 127(indel), 228(C), 264(T), 265(T), 331(C), 337(T), 360(G), 368(A), 370(indel), 371(indel), 372(indel), 373(indel), 405(G), 561(A), 571(G), 577(A), 593(A) and 601(C); tef1 positions 33(C), 216(C), 363(C), 399(C), 411(C), 459(G), 501(T), 683(G), 846(T), 909(T); rpb1 positions 107(G), 122(indel), 160(C), 202(C), 286(C), 292(T), 319(T), 331(C), 343(G), 370(C), 388(C), 436(T), 442(T), 455(A), 505(A), 535(T), 544(T), 574(A), 592(C), 610(G), 628(C), 631(C), 676(T), 697(C), 706(T) and 709(T); ITS positions 31(C), 81(C), 89(A), 105(T), 162(A); LSU position 441(A).
Culture characteristics — Colonies on OA with white floccose aerial hyphae, and without coloured exudates; reverse uncoloured.
Material examined. USA, California, isolated from animal hair, collection date unknown, deposited in CBS by D.A. Sutton, 29 Sept. 2010 (holotype CBS H-22187, culture ex-type CBS 128489 = UTHSC 03-1339 = dH 21593).
Notes — All attempts to induce sporulation on OA failed, even with the addition of sterile elm twig pieces. Phylogenetic inference and SNP analysis indicate that this isolate belongs to Group III, and it forms a sister lineage to C. telluricola (Fig. 1), representing a novel phylogenetic species, introduced here as C. capillare.
Chaetomium cervicicola X. Wei Wang, Crous & L. Lombard, sp. nov. — MycoBank MB812976
Etymology. Refers to the neck of Homo sapiens, from which this fungus was isolated.
Cultures sterile. Chaetomium cervicicola forms a unique lineage in Group IA (Fig. 1), sister to a clade, which includes the five species, C. megalocarpum, C. grande, C. globosporum, C. contagiosum and C. nozdrenkoae. This species differs from the latter species by several unique fixed SNPs for the six loci used in this study: rpb2 positions 36(T), 42(T), 45(C), 64(C), 66(G), 69(C), 72(T), 108(C), 135(G), 138(A), 147(A), 153(T), 180(A), 184(A), 207(A), 210(C), 213(A), 222(G), 231(A), 264(T), 267(A), 285(C), 300(C), 339(A), 345(C), 349(C), 350(A), 360(T), 366(A), 367(C), 368(A), 378(T), 387(T), 429(C), 435(G), 447(C), 450(G), 456(T), 468(T), 504(A), 525(A), 537(G), 555(G), 579(G) and 582(G); tub2 positions 22(C), 29(indel), 39(indel), 40(indel), 41(indel), 66(C), 72(A), 73(A), 76(T), 79(indel), 80(indel), 81(indel), 82(indel), 94(G), 103(T), 146(G), 147(A), 152(A), 153(G), 156(G), 161(A), 164(A), 167(indel), 172(G), 173(T), 178(C), 183(T), 226(T), 233(C), 250(T), 251(C), 264(G), 269(C), 278(C), 279(A), 321(C), 322(indel), 323(indel), 324(indel), 325(indel), 326(indel), 327(indel), 336(A), 440(A), 450(T), 456(C), 465(T), 477(T), 494(C), 560(T), 563(C), 568(indel), 573(indel), 577(G), 589(T), 594(A), 595(A) and 604(A); tef1 positions 18(C), 24(T), 78(T), 129(T), 255(C), 333(T), 376(T), 387(T), 459(T), 627(C), 636(T), 675(C), 679(T), 687(G), 864(C), 918(C) and 927(G); rpb1 positions 65(A), 83(A), 85(T), 94(G), 107(A), 108(C), 125(G), 127(indel), 131(A), 137(G), 138(A), 139(G), 229(T), 234(A), 235(C), 251(T), 252(G), 253(G), 256(G), 262(T), 271(G), 272(A), 273(A), 278(G), 286(G), 288(C), 289(C), 290(G), 291(G), 292(A), 294(G), 295(G), 296(A), 297(C), 298(C), 300(T), 301(C), 303(A), 310(G), 337(T), 412(G), 472(C), 475(C), 490(C), 493(T), 535(A), 587(C), 613(T), 634(T), 670(C), 685(C), 691(G), 715(T), 718(T) and 724(C); ITS positions 105(C), 146(C), 452(C), 483(G), 489(G), 491(indel), 504(indel), 505(indel), 506(indel), 507(indel); LSU positions 403(G), 411(A), 424(T), 433(C), 477(G), 517(C), 520 (C), 521(G) and 522 (C).
Culture characteristics — Colonies on OA with white floccose aerial hyphae, and without coloured exudates; reverse uncoloured.
Material examined. USA, Texas, isolated from neck of Homo sapiens, deposited in CBS by D.A. Sutton, 29 Sept. 2010 (holotype CBS H-22188, culture ex-type CBS 128492 = UTHSC 07-3593 = dH 21625).
Notes — All attempts to induce sporulation of this isolate during this study failed, even with the addition of sterile elm twig pieces. Phylogenetic inference indicates that this species forms a basal branch in Group IA (Fig. 1), and represents a novel phylogenetic species, which is further supported by SNP analysis.
Chaetomium citrinum Udagawa & T. Muroi, Trans. Mycol. Soc. Japan 22: 15. 1981. — Fig. 7
Fig. 7.
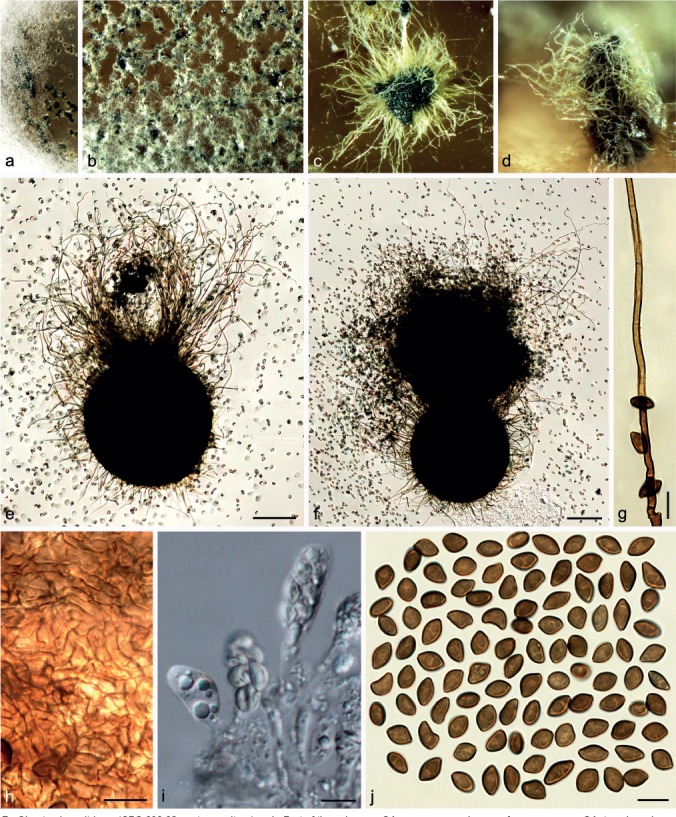
Chaetomium citrinum (CBS 693.82, ex-type culture). a, b. Part of the colony on OA; c. ascoma and mass of ascospores on OA, top view; d. ascoma and mass of ascospores on OA, side view; e, f. ascomata mounted in lactic acid; g. terminal ascomatal hair; h. structure of ascomatal wall in surface view; i. asci; j. ascospores. — Scale bars: e, f = 100 μm; g–j = 10 μm.
Ascomata covered by thick aerial hyphae or exposed, ostiolate, citrine-green to pale amber in reflected light owing to ascomatal hairs, globose, 200–380 μm diam. Ascomatal wall brown, composed of hypha-like cells, textura intricata in surface view. Terminal hairs finely punctate to verrucose, pale brown, hypha-like, flexuous or undulate, sometimes geniculate, 3–5 μm near the base. Lateral hairs similar to terminal hairs, but shorter. Asci fasciculate, clavate to fusiform, spore-bearing part 13.5–28 × 6.5–13 μm, stalks 10–40 μm long, with eight biseriate ascospores, evanescent. Ascospores pale brown when mature, irregularly fusiform, limoniform, ovate, lunate or triangular, bilaterally flattened, (7–)8–10(–12) × (4–)5–6(–7) × 4–5(–5.5) μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with profuse, floccose, white aerial hyphae often covering ascomata, producing ochreous to luteous exudates diffusing into the medium; reverse cinnamon to fulvous.
Material examined. JAPAN, Tochigi, Nasu-gun, Nishinasuno-machi, from rice-field soil, collector and collection date unknown, isolated by S. Udagawa, 23 Apr. 1978 (culture ex-type CBS 693.82 = NHL 2873).
Notes — Chaetomium citrinum is only known from its ex-type strain. It is characterised by irregular and relatively small ascospores. Von Arx et al. (1986) suggested that this species is closely related to C. globosum and allied species, especially C. madrasense. Phylogenetic analysis indicates C. citrinum to be a distinct species basal to Group III (Fig. 1).
Chaetomium coarctatum Sergeeva, Not. Syst. sect. Crypt. Inst. Bot. Acad. Sci. U.S.S.R. 14: 146. 1961. — Fig. 8
Fig. 8.
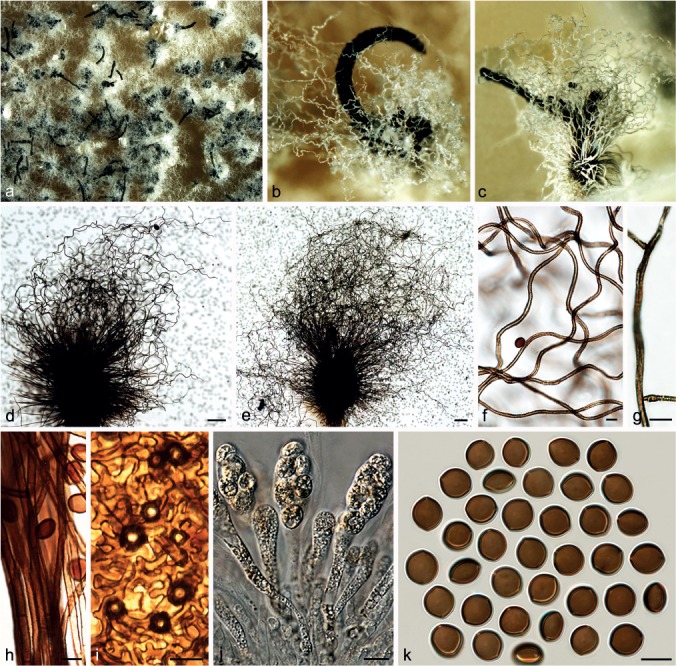
Chaetomium coarctatum (CBS 162.62, ex-type culture). a. Part of the colony on OA; b. ascoma and mass of ascospores on OA, top view; c. ascoma and mass of ascospores on OA, side view; d, e. ascomata mounted in lactic acid; f. upper part of terminal ascomatal hairs; g. branched middle part of a terminal ascomatal hair; h. basal part of terminal ascomatal hair; i. structure of ascomatal wall in surface view; j. asci; k. ascospores. — Scale bars: d, e = 100 μm; f–k = 10 μm.
Ascomata superficial, ostiolate, pale grey to olivaceous buff in reflected light owing to ascomatal hairs, obovate to subglobose, 260–420 μm high, 190–330 μm diam. Ascomatal wall brown, composed of amorphous or hypha-like cells, textura epidermoidea or textura intricata in surface view. Terminal hairs verrucose, brown, undulate or slightly coiled, sometimes branched, 3–4 μm near the base and tapering. Lateral hairs erect or flexuous, tapering towards the tips. Asci fasciculate, fusiform or clavate, spore-bearing part 28–43 × 14–20 μm, stalks 30–53 μm long, with eight biseriate ascospores, evanescent. Ascospores olivaceous brown when mature, broad limoniform to nearly globose, biapiculate, bilaterally flattened, (9.5–)10–11(–11.5) × 9–10(–10.5) × 6.5–7.5(–8) μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with sparse white aerial hyphae and pale orange to slightly dark brick exudates diffusing into the medium; reverse fulvous to sienna.
Materials examined. CHINA, Beijing, Huairou District, from animal dung, Aug. 2009, J. Li, CGMCC 3.14293; Xiangshan Park, from unknown plant stem, Aug. 2009, J. Li, CGMCC 3.14299. – RUSSIA, St. Petersburg, from seed of Campanula medium, collector and collection date unknown, isolated by K.S. Sergejeva, deposited in CBS by K.S. Sergejeva, Feb. 1962 (culture ex-type CBS 162.62 = ATCC 14530 = IMI 090491 = MUCL 18697 = VKM F-1946).
Notes — Von Arx et al. (1986) treated C. coarctatum as a synonym of C. globosum. However, C. coarctatum has broader limoniform to nearly globose and larger ascospores (10–11 × 9–10 × 6.5–7.5 μm vs 8.5–10.5 × 7–8 × 5.5–6.5 μm). Phylogenetic inference indicated that C. coarctatum has a basal position to the second main clade and is sister to Group III (Fig. 1), but its closest relatives remain unclear.
Chaetomium cochliodes Palliser, N. Amer. Fl. 3: 61. 1910. — Fig. 9
Fig. 9.

Chaetomium cochliodes (CBS 155.52, ex-epitype culture). a. Part of the colony on OA; b. ascomata on OA, top view; c, d. ascomata on OA, side view; e–g. ascomata mounted in lactic acid; h. structure of ascomatal wall in surface view; i. basal part of a terminal ascomatal hair; j–l. upper parts of terminal ascomatal hairs; m. asci; n. ascospores; o. holotype sheet of C. cochliodes in New York Botanical Garden (Specimen ID 01050405); p, q. ascomatal hairs of holotype specimen. — Scale bars: e–g = 100 μm; h–l, q = 20 μm; m, n = 10 μm.
Ascomata superficial, ostiolate, greenish olivaceous in reflected light owing to ascomatal hairs, ellipsoid or subglobose, 270–450 μm high, 165–380 μm diam. Ascomatal wall brown, composed of hypha-like cells, textura intricata in surface view. Terminal hairs verrucose, dark brown, erect in the lower part, 3.5–6 μm near the base, tapering and fading towards the tips, spirally coiled in the upper part, with coils regularly tapering in diameter to appear as an elongated cone, occasionally with coiled branches. Lateral hairs brown, flexuous, undulate or coiled, tapering and fading towards the tips. Asci fasciculate, fusiform or clavate, spore-bearing part 23–32 × 13–15 μm, stalks 28–46 μm long, with eight biseriate ascospores, evanescent. Ascospores olivaceous brown when mature, limoniform, usually biapiculate at both ends, bilaterally flattened, (8–)9–10(–11) × (7–)7.5–8.5 × 5–6(–6.5) μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA without aerial hyphae, usually without coloured exudates, but occasionally producing yellowish ochreous exudates diffusing into the medium; reverse uncoloured, but grey olivaceous under ascomata.
Materials examined. CHINA, Yunnan Province, Wenshan County, from tuber of Panax notoginseng, 10 Apr. 2003, X.-Z Liu, CGMCC 3.9440; from rhizosphere of Panax notoginseng, 10 Apr. 2003, X.-Z Liu, CGMCC 3.9471; Inner Mongolia Autonomous Region, Huade County, from discarded cloth, Aug. 2009, J. Li, CGMCC 3.14296. – USA, Newfield, New Jersey, on old paper exposed to the weather, Oct. 1880 (Ellis & Everhart, North American Fungi 1541) (holotype New York Botanical Garden Specimen ID 01050405); from animal dung, isolated and deposited in CBS by L.M. Ames, Apr. 1952 (epitype designated here HMAS 244354, MBT201721, culture ex-epitype CBS 155.52).
Notes — The epitype of C. cochliodes designated here is morphologically similar to that of the holotype, particularly in morphology of the ascospores and ascomatal hairs, and originates from the same country as the type locality. Chaetomium cochliodes was once treated as a synonym of C. globosum (Von Arx et al. 1986). Here, C. cochliodes is re-introduced based on phylogenetic inference supported by morphological characters. Phylogenetic inference indicates that C. cochliodes clusters in Group III, closely related to C. pseudocochliodes and C. spiculipilium (Fig. 1). Chaetomium cochliodes can be distinguished from these species by distinctive coiled ascomatal hairs.
Chaetomium contagiosum X. Wei Wang, Crous & L. Lombard, sp. nov. — MycoBank MB812977
Etymology. Refers to the ability of this fungus to infect the cornea of Homo sapiens.
Culture sterile. Chaetomium contagiosum forms a unique lineage (Group IA, Fig. 1) closely related to C. megalocarpum, C. grande and C. globosporum and can be distinguished based on the following fixed unique SNPs: rpb2 positions 9(G), 45(A), 123(C), 233(A), 265(T), 333(T), 374(G) and 570(C); tub2 positions 12(G), 16(indel), 99(G), 277(T), 327(T), 351(T), 410(A), 472(indel), 572(G), 585(G), 594(C) and 623(C); tef1 positions 291(G), 325(A), 326(C), 332(C), 343(T), 344(C), 487(A), 633(T), 654(T), 683(C), 738(C), 747(T) and 837(C); rpb1 positions 214(A), 220(T), 234(T), 247(C), 274(G), 288(T), 324(T), 325(G), 388(C), 427(A), 455(T), 601(T), 658(G) and 721(C).
Culture characteristics — Colonies on OA with white floccose aerial hyphae, and without coloured exudates; reverse uncoloured.
Material examined. USA, North East, isolated from cornea of Homo sapiens, deposited in CBS by D.A. Sutton, 29 Sept. 2010 (holotype CBS H-22189, culture ex-type CBS 128494 = UTHSC 10-726 = dH 21640).
Notes — Phylogenetic inference and SNP analysis indicate that this species is a novel phylogenetic species in Group IA (Fig. 1). All attempts to induce sporulation on OA failed, even with the addition of sterile elm twig pieces.
Chaetomium cucumericola X. Wei Wang, Crous & L. Lombard, sp. nov. — MycoBank MB812978
Etymology. Refers to the plant host Cucumis sativus, from which this fungus was isolated.
Cultures sterile. Chaetomium cucumericola forms a unique lineage in Group III (Fig. 1), sister to C. olivaceum and is distinguished from the latter by fixed unique SNPs in four loci: rpb2 positions 48(C), 132(A), 156(C), 195(G), 203(G), 306(G), 432(A) and 507(C); tub2 positions 71(G), 217(A), 237(C), 338(G), 363(C), 378(A), 467(G), 560(indel), 570(A), 591(A) and 604(G); tef1 positions 33(T), 283(A), 347(G), 453(C) and 681(T); rpb1 positions 148(C), 169(T), 190(A), 253(A), 303(C), 307(T), 337(T), 376(T), 394(T), 397(A), 487(C), 538(C), 619(T) and 688(C).
Culture characteristics — Colonies on OA with white floccose aerial hyphae, and without coloured exudates; reverse uncoloured.
Materials examined. IRAN, Alborz Province, Hashtgerd, isolated from petiole of Cucumis sativus, 22 Oct. 2005, B. Asgari, CBS 126777 = IRAN 1642C. – TURKEY, Izmir, substrate unknown, deposited in CBS by G. Turhan, Apr. 1971 (holotype CBS H-22190, culture ex-type CBS 378.71).
Notes — Phylogenetic inference and SNP analysis indicated that both representative isolates of C. contagiosum form a lineage in Group III, sister to C. olivaceum (Fig. 1). All attempts to induce sporulation on OA failed, even with the addition of sterile elm twig pieces.
Chaetomium elatum Kunze, Deutsche Schwämme 8: 3, No. 184. 1818. — Fig. 10
Fig. 10.
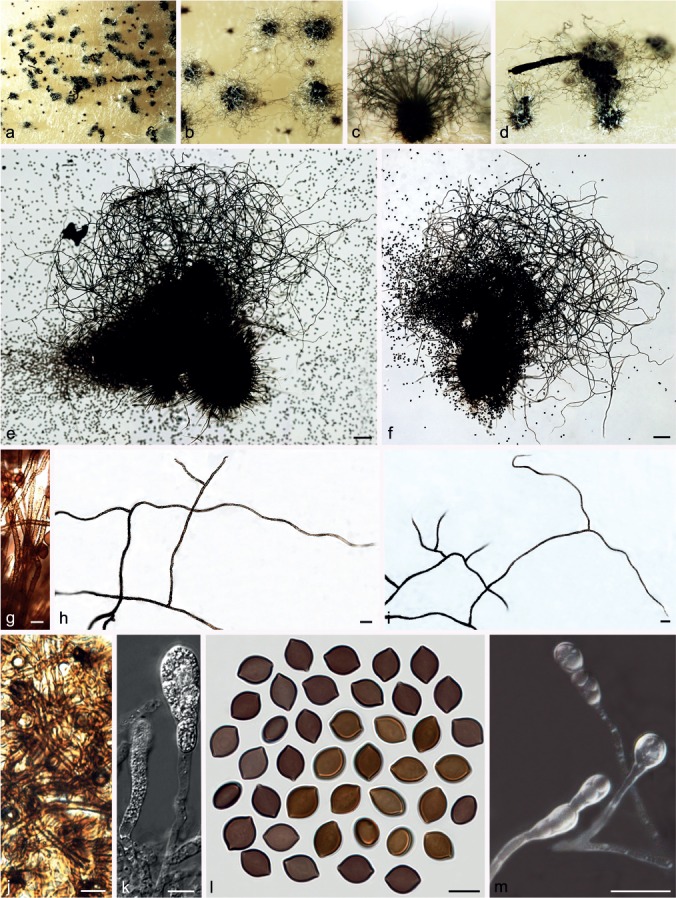
Chaetomium elatum (CBS 910.70, ex-type culture of C. ramipilosum). a. Part of the colony on OA; b. ascomata and masses of ascospores on OA, top view; c, d. ascomata and masses of ascospores on OA, side view; e, f, ascomata mounted in lactic acid; g. basal parts of terminal ascomatal hairs; h, i. upper parts of terminal ascomatal hairs; j. structure of ascomatal wall in surface view; k. asci; l. ascospores; m. asexual morph (conidiophores and conidia). — Scale bars: e, f = 100 μm; g, j–m = 10 μm; h, i = 20 μm.
= Chaetomium virgecephalum Ames, A monograph of the Chaetomiaceae: 43. 1963.
= Chaetomium ramipilosum Schaumann, Arch. Mikrobiol. 91: 98. 1973.
Ascomata superficial, ostiolate, greenish olivaceous in reflected light owing to ascomatal hairs, globose or obovate, 230–400 μm high, 175–365 μm diam. Ascomatal wall brown, composed of hypha-like or amorphous cells, textura intricata or textura epidermoidea in surface view. Terminal hairs verrucose or warty, brown, tapering and fading towards the tips, erect or flexuous in the lower part, 2.5–4.5 μm diam near the base, repeatedly and dichotomously branched at right to nearly straight angles in the upper part, with relatively flexible, flexuous or undulate terminal branches. Lateral hairs brown, flexuous, tapering towards the tips. Asci fasciculate, clavate, spore-bearing part 36–49 × 13.5–16 μm, stalks 24–55 μm long, with eight biseriate ascospores, evanescent. Ascospores brown when mature, limoniform, biapiculate or umbonate, bilaterally flattened, (11–)12–13(–14) × 9–10.5(–11) × (6–)7–8(–9) μm, with an apical germ pore. Asexual morph acremonium-like. Conidiophores formed laterally from aerial hyphae, simple, 6–18 μm long, 1.5–2.2 μm diam at the base. Conidia formed solitarily or in chains, hyaline, aseptate, smooth, globose, ellipsoidal or ovate, often with a truncated base and a rounded apex, 4.5–6.5(–7) × (3.5–)4–6 μm.
Culture characteristics — Colonies on OA with sparse aerial hyphae, and without coloured exudates; reverse uncoloured.
Materials examined. GERMANY, Helgoland, isolated from Ammophila arenaria, isolated and deposited in CBS by K. Schaumann, Nov. 1970 (culture ex-type of C. ramipilosum CBS 910.70). – USA, California, Aptos, from decomposing leaf, collection date unknown, H.K. Seth, deposited in CBS by H.K. Seth, Apr. 1966 (culture ex-isotype of C. virgecephalum CBS 374.66).
Notes — Dreyfuss (1976) restricted C. elatum to heterothallic isolates with acremonium-like asexual morphs, and classified homothallic isolates, mostly without asexual morphs, as C. virge-cephalum. Von Arx et al. (1986) reduced C. virgecephalum to synonymy with C. elatum, meaning that the species C. elatum was expanded to include both heterothallic and homothallic isolates. The phylogenetic inference in this study supports the classification of Von Arx et al. (1986). The holotype of C. elatum was originally collected in Germany, and all attempts to locate the holotype of C. elatum from B (Botanischer Garten und Botanisches Museum Berlin-Dahlem, Zentraleinrichtung der Freien Universität Berlin) were unsuccessful as a fire in 1943 destroyed parts of the ascomycete collection. Typification of this species awaits recollection from the type locality.
Chaetomium fimeti Fuckel, Enum. Fung. Nass., Ser. 1: 491. 1861. — Fig. 11
Fig. 11.
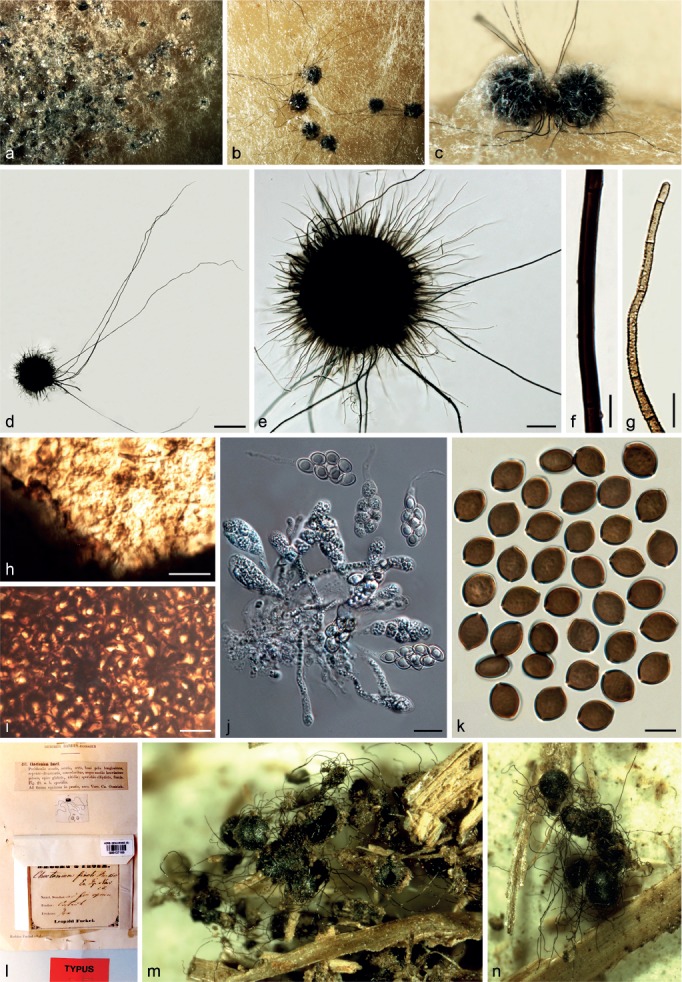
Chaetomium fimeti (CBS 153.77). a. Part of the colony on OA; b. ascomata on OA, top view; c. ascomata on OA, side view; d, e. ascomata mounted in lactic acid; f. part of terminal ascomatal hair, longer type; g. terminal ascomatal hair, shorter type; h. inner layer structure of ascomatal wall in surface view; i. external layer structure of ascomatal wall in surface view; j. asci; k. ascospores; l. holotype sheet of C. fimeti in HERB. GENAVENSE (G00127165 in Switzerland); m, n. ascomata of holotype specimen. — Scale bars: d = 500 μm; e = 100 μm; f–i, k = 10 μm; j = 20 μm.
≡ Chaetomidium fimeti (Fuckel) Sacc., Syll. Fung. 1: 39. 1882.
≡ Thielavia fimeti (Fuckel) Malloch & Cain, Mycologia 65: 1064. 1973.
Ascomata superficial, non-ostiolate, dark brown to black, with numerous short, olivaceous buff to honey ascomatal hairs, and sparse, long and black hairs in reflected light, spherical or oblate, 320–500 μm diam. Ascomata walls composed of two layers, easily separating from each other: the external wall thick, dark brown, composed of thick-walled, angular or irregular cells, textura angularis in surface view; the inner layer thin, luteous to pale brown, composed of amorphous cells, textura epidermoidea in surface view. Ascomatal hairs of two types: shorter type covering the whole ascomata, punctate to verrucose, dark brown at the lower part, fading to greyish yellow-green or pale greyish sepia at the tips, 3–4.5 μm near the base, 30–580 μm long; longer type arising from the bases of the ascomata, smooth, dark brown, 4–8.5 μm near the base, 500–4 200 μm long. Asci fasciculate, fusiform or clavate, with eight biseriate ascospores, spore-bearing part 30–50 × 14.5–19 μm, stalks 23–46 μm long, evanescent. Ascospores olivaceous brown to brown when mature, limoniform, bilaterally flattened, (11–)11.5–13.5(–16) × 9–10.5(–11) × (6–)7–8(–8.5) μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with abundant, olivaceous buff aerial hyphae, producing ochreous to pale umber exudates diffusing into the medium; reverse cinnamon.
Materials examined. CANADA, Ontario, Nashville, from decaying hay, isolated by F.R. Cain, May 1957, deposited in CBS by D.W. Malloch, Jan. 1971, CBS 168.71 = ATCC 22330 = IMI 153720 = TRTC 33005 (sterile). – GERMANY, isolated from soil, collection date unknown, Bredemeier (epitype CBS H-22198, MBT201724, culture ex-epitype DSM 62108 = CBS 139034); Oestrich, from horse dung (Hebier Fuckel 1894, holotype G00127165 from HERB. GENAVENSE (G: Conservatoire et Jardin botaniques de la Ville de Genève, Switzerland). – JAPAN, substrate unknown, collector and collection date unknown, deposited in CBS by K. Furuya, Feb. 1977, CBS 153.77 = NHL 2713 = SANK 21476.
Notes — Zopf (1881) split the genus Chaetomium into two subgenera: Euchaetomium with ostiolate ascomata and Chaetomidium with non-ostiolate ascomata. Saccardo (1882) subsequently elevated the subgenus Chaetomidium to generic level with Chaetomidium fimeti (= Chaetomium fimeti) as type species. The genus Chaetomidium was rejected by Winter (1885), Chivers (1915) and Ainsworth (1961), but was accepted by Bainier (1910). Ainsworth (1971) re-introduced this genus based on the study of Seth (1967) and following this, Malloch & Cain (1973) treated Chaetomidium as a synonym of Thielavia, which Von Arx (1975) later distinguished from Thielavia. Greif et al. (2009) revealed the polyphyly of the genus Chaetomidium using LSU, tub2 and rpb2 sequence data, and suggested that this genus be restricted to two species, the type C. fimeti and its close relative, C. subfimeti. Phylogenetic inference in this study strongly supports C. fimeti and C. subfimeti as sister lineages (Group IC, Fig 1) within the C. globosum complex, consistent with the rpb2 analysis of Greif et al. (2009). Thus, Chaetomium fimeti represents the correct species name, and the genus Chaetomidium is considered as a synonym of Chaetomium.
Chaetomium globosporum Rikhy & Mukerji, Kavaka 1: 38. 1973. — Fig. 12
Fig. 12.

Chaetomium globosporum (CBS 108.83, ex-type culture). a. Part of the colony on OA; b. ascomata and masses of ascospores on OA, top view; c. ascomata and masses of ascospores on OA, side view; d, e. ascomata mounted in lactic acid; f. basal parts of terminal ascomatal hairs; g. branched upper part of a terminal ascomatal hair; h. unbranched upper parts of terminal ascomatal hairs; i. structure of ascomatal wall in surface view; j. asci; k. ascospores. — Scale bars: d, e = 100 μm; f–k = 10 μm.
Ascomata superficial, ostiolate, usually covered by aerial hyphae, yellow-amber to olivaceous in reflected light owing to ascomatal hairs, soon becoming dark brown to black due to ascospore mass on ascomata, ovate, 350–510 μm high, 210–350 μm diam. Ascomatal wall brown, composed of irregular or hypha-like cells, textura epidermoidea in surface view. Terminal hairs relatively sparse, finely punctate to verrucose, brown, flexuous, occasionally branched or geniculate, 2.5–4.5 μm near the base and tapering towards the tips. Lateral hairs similar. Asci fasciculate, clavate or slightly fusiform, with eight biseriate ascospores, spore-bearing part 24–43 × 16–24 μm, stalks 11–26 μm long, evanescent. Ascospores dark brown when mature, globose to subglobose, non-apiculate, bilaterally flattened, (10–)10.5–12(–12.5) μm diam, (7–)7.5–8.5(–9) μm wide in lateral view, with one or two germ pores. Asexual morph absent.
Culture characteristics — Colonies on OA with white or pale grey aerial hyphae, producing pale ochreous exudates diffusing into the medium; reverse ochreous to fulvous.
Material examined. INDIA, isolated from green leaf of Triticum aestivum, deposited in CBS by J.N. Kapoor, Jan. 1983 (culture ex-type CBS 108.83 = ITCC 1835).
Notes — Only the ex-type strain is known for this species. Chaetomium globosporum is closely related to C. megalocarpum and C. grande (Group IA, Fig 1). This species is easily distinguished by its smaller and more regular, oblate ascospores (10.5–12 × 7.5–8.5 μm) compared to those of C. megalocarpum (13–15 × 11.5–14 × 8.5–10 μm) and C. grande (18–20.5 × 16–18 × 12–13.5 μm).
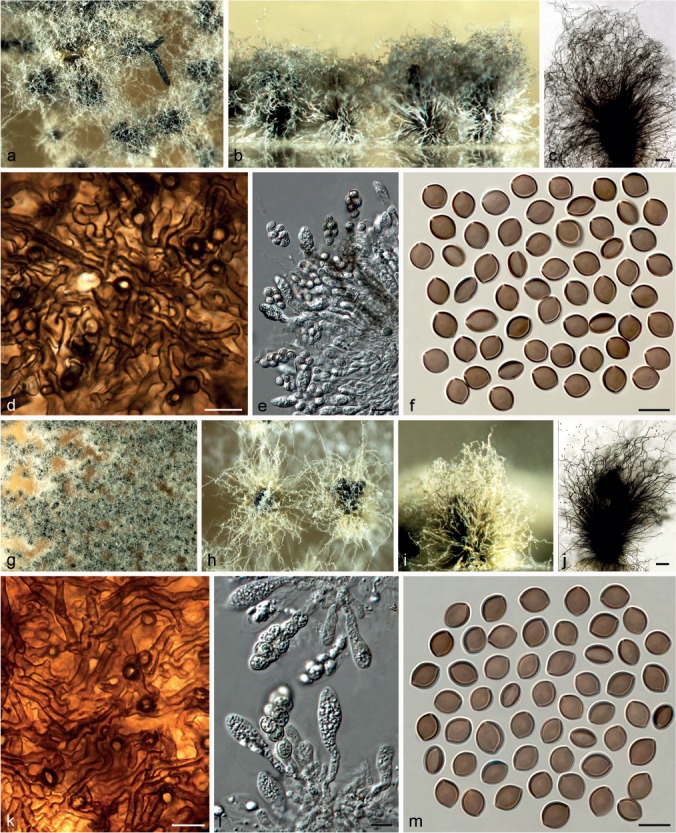
Chaetomium globosum Kunze, Mykol. Hefte 1: 16. 1817. — Fig. 13, 14, 15
Fig. 13.
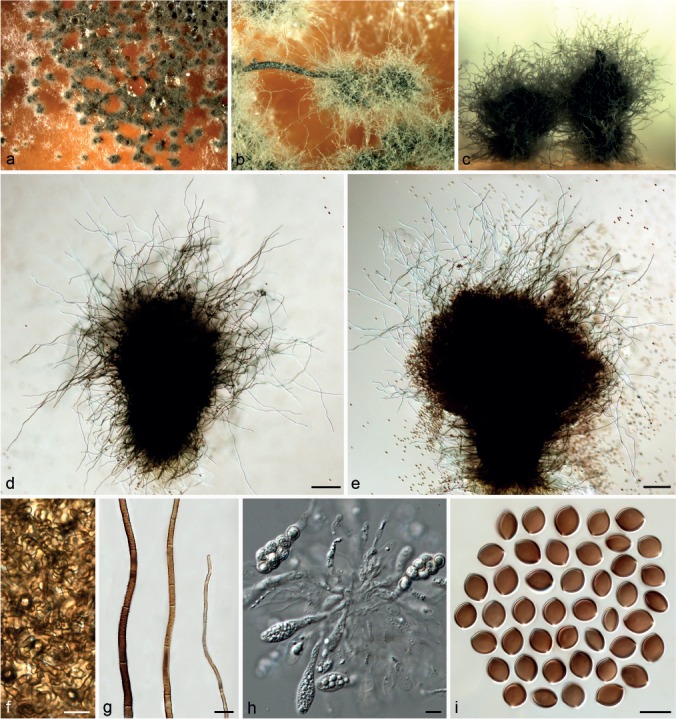
Typical morphology of Chaetomium globosum sensu stricto-1 (CBS 160.62, ex-neotype culture). a. Part of the colony on OA; b. ascomata and masses of ascospores on OA, top view; c. ascomata on OA, side view; d, e. ascomata mounted in lactic acid; f. structure of ascomatal wall in surface view; g. terminal ascomatal hairs (from left to right: lower part, middle part and upper part); h. asci; i. ascospores. — Scale bars: d, e = 100 μm; f–i = 10 μm.
Fig. 14.
Typical morphology of Chaetomium globosum sensu stricto-2. a–f. MUCL 39526 (ex-type of C. globosum var. flavoviride): a. Ascomata and masses of ascospores on OA, top view; b. ascomata on OA, side view; c. ascoma mounted in lactic acid; d. structure of ascomatal wall in surface view; e. asci; f. ascospores. – g–m. CBS 148.51 (authentic isolate of C. globosum): g. part of the colony on OA; h. ascomata and masses of ascospores on OA, top view; i. ascoma on OA, side view; j. ascoma mounted in lactic acid; k. structure of ascomatal wall in surface view; l. asci; m. ascospores. — Scale bars: c, j = 100 μm; d, f, k–m = 10 μm; e = 20 μm.
Fig. 15.

Variation of Chaetomium globosum. a–f. CBS 164.62 (ex-type of C. rectum): a. Part of the colony on OA; b. ascomata and masses of ascospores on OA, top view; c. ascomata and masses of ascospores on OA, side view; d, e. ascomata mounted in lactic acid; f. ascospores. – g–m. CBS 147.60 (ex-type of C. mollipilium): g. part of the colony on OA; h. ascomata and masses of ascospores on OA, top view; i. ascomata and masses of ascospores on OA, side view; j, k. ascomata mounted in lactic acid; l. simply-branched ascomatal hairs; m. ascospores. — Scale bars: d, e, j, k = 100 μm; l = 20 μm; f, m = 10 μm.
= Chaetomium globosum var. flavoviride E.K. Novák, Ann. Univ. Sci. Budapest. Rolando Eotvos, Sect. Biol. 8: 207. 1966.
= Chaetomium globosum var. griseum E.K. Novák, Ann. Univ. Sci. Budapest. Rolando Eotvos, Sect. Biol. 8: 207. 1966.
= Chaetomium mollipilium Ames, Mycologia 42: 642. 1950.
= Chaetomium rectum Sergeeva, Not. Syst. sect. Crypt. Inst. Bot. Acad. Sci. U.S.S.R. 14: 143. 1961.
= Chaetomium subterraneum Swift & Povah, Mycologia 21: 210. 1929.
Ascomata superficial, ostiolate, greenish olivaceous or slightly dark olivaceous buff to grey in reflected light owing to ascomatal hairs, globose, ellipsoid, ovate or obovate, 160–300 μm high, 135–250 μm diam. Ascomatal wall brown, composed of hypha-like or amorphous cells, textura intricata in surface view. Terminal hairs abundant, finely verrucose, brown, tapering and fading towards the tips, 3–5 μm diam near the base, flexuous, undulate to loosely coiled with erect or flexuous lower part, usually unbranched. Lateral hairs brown, flexuous, fading and tapering towards the tips. Asci fasciculate, fusiform or clavate, spore-bearing part 30–40 × 12–17 μm, stalks 15–25 μm long, with eight biseriate ascospores, evanescent. Ascospores olivaceous brown when mature, limoniform, usually biapiculate, bilaterally flattened, (8–)8.5–10.5(–11) × 7–8(–8.5) × 5.5–6.5(–7) μm, with an apical germ pore. Asexual state absent.
Culture characteristics — Colonies on OA without aerial hyphae or with sparse white aerial hyphae in the centre, producing luteous to orange exudates diffusing into the medium; reverse fulvous to umber, but darker under ascomata.
Materials examined. CHINA, Beijing, Peking University Third Hospital, isolated from finger nail of Homo sapiens, collection date unknown, D.-M. Li, CGMCC 3.9994. – GERMANY, from compost, isolated and deposited in CBS by A. von Klopotek, Apr. 1962, (neotype designated here: CBS H-22185, MBT201725, culture ex-neotype CBS 160.62). – HUNGARY, from dead stem of Juncus sp., 1966, E. Novak, MUCL 39526 (culture ex-type of C. globosum var. flavoviride); from dead stem of Juncus sp., 1966, E. Novak, MUCL 39527 (culture ex-type of C. globosum var. griseum). – NETHERLANDS, Bibliotheek van het Koloniaal Instituut, Amsterdam, isolated from mouldy book, collector and collection date unknown, isolated by F.H. van Beyma, CBS 105.40. – POLAND, Bydgoszcz Botanic Garden, collector and collection date unknown, isolated by K.S. Sergejeva, 1961 (culture ex-type of C. rectum CBS 164.62 = ATCC 14529 = IMI 090488 = MUCL 18692 = VKM F-1949). – USA, Illinois, isolated from clay soil at 120 cm depth, collector and collection date unknown, isolated and deposited in CBS by B.B. Kanouse, July 1930 (culture ex-type of C. subterraneum CBS 132.30); Jeffersonville, Indiana, isolated from a Japanese raincoat, collector and collection date unknown, isolated by G.W. Martin (culture ex-type of C. mollipilium CBS 147.60 = ATCC 11209 = IFO 9108 = MUCL 9596 = QM 1007 = QM 1107); Washington DC, isolated from stored cotton, isolated by H. Hunfield, 1933, CBS 148.51 = ATCC 6205 = CBS 161.52 = CEB 1218.1 = CEB 1218.2 = CECT 2701 = DSM 1962 = IFO 6347 = IHEM 3826 = IMI 045550 = MUCL 1984 = NRRL 1870 = QM 459 = UPSC 3159 = USDA 1042.4 = VTT D-81079.
Notes — Chaetomium globosum, the type species of the genus Chaetomium, was described based on an isolate collected from the stem of Dianthus carthusianorum in Leipzig, Germany. Our attempt to locate the holotype of C. globosum housed in B (Botanischer Garten und Botanisches Museum Berlin-Dahlem, Zentraleinrichtung der Freien Universität Berlin) was unsuccessful because the ascomycete collection was partly destroyed by a fire in 1943. Therefore, a dried culture, CBS H-22185 from the isolate CBS 160.62, that was collected in Germany from the same locality as the holotype, is designated here as neotype of C. globosum.
The description provided above represents the typical characteristics of C. globosum s.str., in which the morphological diversity was captured to some extent, especially in ascomatal hairs and exudate colours. For example, CBS 160.62 (the ex-neotype culture) and CBS 105.40 exhibit greenish olivaceous ascomatal hairs with flexuous to slightly undulate upper part and orange exudates diffusing into the medium (Fig. 13); while CBS 145.51 and MUCL 39526 exhibit slightly dark olivaceous buff to grey ascomatal hairs with undulate to loosely coiled upper part and luteous exudates diffusing into the medium (Fig. 14).
Ames (1950) characterised C. mollipilium by its sparse and often branched ascomatal hairs at wide angles, but did not include C. rectum in his monograph. Von Arx et al. (1986) later reduced both species to synonymy under C. globosum. However, the ex-type cultures of C. rectum (CBS 164.62; Fig. 15a–f) and C. mollipilium (CBS 147.60; Fig. 15g–m) are distinguished from one another as well as from other typical isolates of C. globosum s.str. based on the six-locus phylogeny generated in this study (Group IIA, Fig. 1). Also, the average ascospore dimensions of both CBS 164.62 (9–10.5 × 7.5–8.5 × 5.5–6 μm) and CBS 147.60 (9–10.5 × 7.5–8.5 × 5–6 μm) resemble those of C. globosum s.str. (8.5–10.5 × 7–8 × 5.5–6.5 μm). Both species are, therefore, reduced to synonyms of C. globosum s.str. However, both ex-type isolates differ from C. globosum s.str. based on their ascomatal hair morphology: sparse, erect to flexuous terminal hairs, often branching at wide or narrow angles, and also thinner (2.5–3.5 μm diam near the base) than those of typical C. globosum (3–5 μm diam near the base).
Von Arx et al. (1986) regarded C. cruentum as an albino form of C. globosum, which possesses ascospores characteristic of C. globosum s.str., but paler and slightly larger. The other morphological structures of C. cruentum also present an albinistic or degenerated morphology when compared to the typical C. globosum s.str. isolates, which make it look conspicuously different from typical C. globosum. Asgari & Zare (2011) indicated that C. cruentum and C. globosum (CBS 148.51) clustered together with high bootstrap support in a phylogenetic inference of the combined ITS, LSU and tub2 gene regions. This result was also supported in the present study (Group IIA, Fig. 1). As there is no evidence available based on the analyses of six loci to distinguish the morphological species C. cruentum from C. globosum s.str., this taxon is reduced to synonymy under C. globosum s.str. The morphological variation ascribed to ‘cruentum’, however, is described below to present its conspicuous differences from the typical morphology observed among isolates of C. globosum s.str. Future studies of the genome may reveal the genetic mechanism linked to this variation.
Chaetomium globosum morphological form ‘cruentum’ — Fig. 16
Fig. 16.

Chaetomium globosum morphological form ‘cruentum’ (CBS 371.66, ex-type of C. cruentum). a. Part of the colony on OA; b. ascomata and masses of ascospores on OA, top view; c. young ascomata on OA; d. mature ascomata and masses of ascospores on OA, side view; e. ascoma full of ascospores; f. ascoma from which ascospores have been discharged; g. structure of ascomatal wall in surface view; h. ascospores. — Scale bars: e, f = 20 μm; g, h = 10 μm.
Ascomata superficial, ostiolate, globose, ellipsoid, ovate or obovate, 210–300 μm high, 145–220 μm diam, hyaline when young, then saffron in reflected light owing to ascospore masses. Ascospore masses on the top of ascomata, rust when fresh, then slightly pale scarlet to salmon in reflected light when becoming dry. Ascomatal wall translucent, composed of amorphous or angular cells, textura epidermoidea or textura angularis in surface view. Ascomatal hairs sparse, hyaline, flexuous and delicate. Asci disappearing quickly. Ascospores pale cinnamon when mature, limoniform, usually biapiculate, bilaterally flattened, 9.5–11(–11.5) × 7.5–8.5(–9) × (6–)6.5–7 μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA without aerial hyphae, and producing yellow to yellowish amber exudates diffusing into the medium; reverse uncoloured, but ochreous or fulvous under ascomata.
Materials examined. UK, isolated from rabbit dung, collector and collection date unknown, isolated by H.K. Seth, deposited in CBS by H.K. Seth, Oct. 1968 (culture ex-type of Lophotrichus incarnatus CBS 730.68 = ATCC 18597 = IMI 135564). – UNKNOWN, substrate and collection details unknown, isolated and deposited in CBS by L.M. Ames, June 1958, CBS 145.58. – USA, Fort Belvoir, Virginia, from paper, collection date unknown, isolated by L.M. Ames, deposited in CBS by H.K. Seth, Apr. 1966 (isotype of C. cruentum CBS H-6860, culture ex-isotype CBS 371.66).
Chaetomium graminiforme X. Wei Wang, Crous & L. Lombard, sp. nov. — MycoBank MB812979; Fig. 17
Fig. 17.

Chaetomium graminiforme (CBS 506.84, ex-type culture). a. Part of the colony on OA; b. ascomata and masses of ascospores on OA, top view; c. ascomata and masses of ascospores on OA, side view; d, e. ascomata mounted in lactic acid; f. basal parts of terminal ascomatal hairs; g. upper parts of terminal ascomatal hairs; h. structure of ascomatal wall in surface view; i. asci; j. ascospores. — Scale bars: d, e = 100 μm; f–i = 20 μm; j = 10 μm.
Etymology. Refers to the grass-like ascomatal hairs formed by this fungus.
Ascomata superficial, ostiolate, luteous to amber or citrine in reflected light owing to ascomatal hairs, becoming dark due to ascospore masses on the top, ellipsoid, subglobose or ovate, 200–320 μm high, 170–260 μm diam. Ascomatal wall brown, composed of hypha-like or amorphous cells, textura epidermoidea or textura intricata. Terminal hairs sparse, olivaceous brown and fading towards the tips, punctate, erect or flexuous, sometimes simply branched, 3.5–4.5 μm diam near the base, tapering towards almost pointed tips. Lateral hairs similar. Asci fasciculate, fusiform or clavate, spore-bearing part 25.5–40 × 12.5–16 μm, stalks 14.5–29 μm long, with eight biseriate ascospores, evanescent. Ascospores olivaceous brown when mature, limoniform, bilaterally flattened, (9.5–)10–11.5(–12) × 9–10 × (5.5–)6–7 μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA usually without aerial hyphae and coloured exudates diffusing into the medium; reverse usually uncoloured, but dark grey-olivaceous under ascomata.
Material examined. CANADA, Muskoka District, Ontario, from Acer sp., 1967, D. Malloch, isolated and deposited in CBS by J.C. Krug (holotype CBS H-22193, culture ex-type CBS 506.84 = TRTC 47862).
Notes — The ascomata of C. graminiforme appears similar to the ‘rectum’-like variation of C. globosum by having sparse, erect to flexuous ascomatal hairs, but can be distinguished by larger ascospores (10–11.5 × 8–9 × 6–7 μm) compared to those of C. globosum morphological form ‘rectum’ (9–10.5 × 7.5–8.5 × 5–6 μm). Phylogenetic inference showed that C. graminiforme is distantly related to C. globosum (Group IIA, Fig 1), and clusters with C. elatum and C. rectangular. However, the relationship of C. graminiforme to C. elatum and C. rectangular is not supported (Group III, Fig. 1), and the ascomatal hair morphology of this species is also different from those of C. elatum and C. rectangular.
Chaetomium grande Asgari & Zare, Mycologia 103: 874. 2011. — Fig. 18
Fig. 18.

Chaetomium grande (CGMCC 3.9414). a. Part of the colony on OA; b. ascomata on OA, top view; c. ascoma and mass of ascospores on OA, side view; d, e. ascomata mounted in lactic acid; f. basal parts of terminal ascomatal hairs; g. upper part of a terminal ascomatal hair; h. structure of ascomatal wall in surface view; i, j. asci; k. ascospores. — Scale bars: d, e = 100 μm; f–h, j, k = 10 μm; i = 20 μm.
Description & Illustration — Based on the culture on MEA, CMA and PCA supplemented with cellulose; also see Asgari & Zare (2011).
Ascomata superficial, ostiolate, olivaceous in reflected light owing to ascomatal hairs, subglobose or ovate, 270–380 μm high, 190–310 μm diam. Ascomatal wall brown, composed of amorphous cells, textura epidermoidea in surface view. Terminal hairs finely punctate to verrucose, brown at the base, fading towards the tips, flexuous to undulate, sometimes branched, 3–4.5 μm near the base, tapering towards the tips. Lateral hairs similar. Asci fasciculate, fusiform or clavate, with eight biseriate or irregularly-arranged ascospores, spore-bearing part 51–66 × 22.5–28.5 μm, stalks 20.5–38 μm long, evanescent. Ascospores dark brown when mature, ellipsoid to subglobose, usually irregular, bilaterally flattened, (17–)18–20.5(–22.5) × (14.5–)16–18(–19) × (11–)12–13.5(–14) μm, with two apical or subapical germ pores. Asexual morph absent.
Culture characteristics — Colonies on OA with abundant white aerial hyphae, usually without exudates; reverse cinnamon to fulvous.
Materials examined. CHINA, Xinjiang Autonomous Region, Bayinguoleng, isolated from desert soil, June 2012, X.-W. Wang, CBS 119758 = CGMCC 3.9414. – IRAN, Ardabil Province, Bilesavar, isolated from straw of Triticum aestivum, 21 June 2005, B. Asgari, CBS 126781 = IRAN 1208C; West Azerbaijan province, Naghadeh, isolated from leaf of Triticum aestivum, 23 June 2005, B. Asgari (holotype IRAN 14608F, culture ex-type CBS 126780 = IRAN 1064C).
Notes — The description provided here is based on isolate CBS 119758 as the other isolates of this species, including the ex-type culture, are sterile. Chaetomium grande is closely related to C. megalocarpum and C. globosporum (Group IA, Fig 1). These three species all produce globose or subglobose ascospores without apiculate or umbonate ends, and usually with two germ pores. Chaetomium grande is easily distinguished by its much larger ascospores (18–20.5 × 16–18 × 12–13.5 μm) compared to those of C. megalocarpum (13–15 × 11.5–14 × 8.5–10 μm) and C. globosporum (10.5–12 μm diam, 7.5–8.5 μm wide in lateral view).
Chaetomium interruptum Asgari & Zare, Mycologia 103: 874. 2011. — Fig. 19
Fig. 19.

Chaetomium interruptum (CBS 126000, ex-type culture). a. Part of the colony on OA; b. ascomata on OA, top view; c. ascoma on OA, side view; d. ascoma mounted in lactic acid; e. basal parts of terminal ascomatal hairs; f. upper parts of terminal ascomatal hairs; g. structure of ascomatal wall in surface view; h. asci; i. ascospores. — Scale bars: d = 100 μm; e, f, h = 20 μm; g, i = 10 μm.
Description & Illustration –– Based on the culture on MEA and CMA or PCA supplemented with cellulose; also see Asgari & Zare (2011).
Ascomata superficial, ostiolate, often covered by aerial hyphae, olivaceous or pale umber in reflected light owing to ascomatal hairs, ovate or ellipsoid, 230–360 μm high, 170–240 μm diam. Ascomatal wall brown, composed of amorphous cells, textura epidermoidea in surface view. Terminal hairs smooth or finely verrucose, brown, flexuous, undulate, sometimes simply branched, 3–4.5 μm diam near the base, tapering towards the tips. Lateral hairs similar. Asci fasciculate, clavate or fusiform, with eight biseriate or irregularly arranged ascospores, spore-bearing part 30–41 × 14–28 μm, stalks 15–29 μm long, evanescent. Ascospores dark brown when mature, globose to subglobose, non-apiculate, bilaterally flattened, (10–)11–12 μm diam, (7.5–)8–9 μm wide from lateral view, with one or two germ pores. Asexual morph absent.
Culture characteristics — Colonies on OA with white, sparse to floccose aerial hyphae, producing cinnamon to fulvous exudates diffusing into the medium; reverse olivaceous.
Material examined. IRAN, East Azerbaijan province, Hadishahr, isolated from seed of Triticum aestivum, 24 June 2005, B. Asgari (holotype IRAN 14607F, culture ex-type CBS 126660 = IRAN 1278C).
Notes — Chaetomium interruptum is morphologically similar to C. globosporum. Asgari & Zare (2011) indicated that the ascospores of C. interruptum only have one indistinct, apical or slightly subapical germ pore. Our observations showed that the ascospores of C. interruptum frequently have two germ pores, which are conspicuous and often subapical or lateral. The smaller ascomata (230–360 × 170–240 μm) with abundant, smooth and undulate ascomatal hairs distinguish C. interruptum from C. globosporum, which produce larger ascomata (350–510 × 210–350 μm) with sparse, verrucose and flexuous ascomatal hairs. Phylogenetic inference also showed that C. interruptum takes a basal position to Group I (Fig. 1), and is distant from C. globosporum.
Chaetomium madrasense Natarajan, Proc. Indian Acad. Sci., B. 74: 255. 1971. — Fig. 21
Fig. 21.

Chaetomium madrasense (CBS 315.74, ex-type culture). a. Part of the colony on OA; b. ascomata and masses of ascospores on OA, top view; c. ascoma on OA, side view; d, e. ascomata mounted in lactic acid; f. structure of ascomatal wall in surface view; g. basal parts of terminal ascomatal hairs; h. upper parts of terminal ascomatal hairs; i. asci; j. ascospores. — Scale bars: d, e = 100 μm; f–j = 10 μm.
Ascomata superficial, ostiolate, olivaceous buff or rosy buff, occasionally salmon in reflected light owing to ascomatal hairs, ellipsoid, ovate or obovate, 130–300 μm high, 140–260 μm diam. Ascomatal wall brown, composed of amorphous or hypha-like cells, textura epidermoidea or textura intricata in surface view. Terminal hairs relatively abundant, brown, finely verrucose, coiled or undulate, occasionally with simple branches, 2.5–4.5 μm near the base. Lateral hairs similar. Asci fasciculate, fusiform or clavate, spore-bearing part 28–38 × 13–20 μm, stalks 16–30 μm long, with eight biseriate ascospores, evanescent. Ascospores olivaceous brown when mature, broad limoniform, often slightly apiculate at both ends, bilaterally flattened, triangle-shaped in lateral view due to a conspicuous lateral bulge, 10–11(–11.5) × (8–)9–10 × 7.5–8.5(–9) μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with sparse, white aerial hyphae, usually without exudates; reverse uncoloured, but usually black under ascomata.
Material examined. INDIA, Madras, Tamil Nadu, from rhizosphere of Pennisetum typhoides, collection date unknown, K. Natarajan, isolated by K. Natarajan, 1966 (isotype CBS H-6877, culture ex-isotype CBS 315.74).
Notes — Von Arx et al. (1986) reduced C. ascotrichoides and C. gibberosporum to synonymy under C. madrasense, both having ascospores with a lateral bulge. Phylogenetic inference in this study distinguished C. madrasense from C. ascotrichoides (and C. gibberosporum that was shown to be conspecific with C. ascotrichoides; Group IB, Fig 1). Chaetomium madrasense is, therefore, restricted here to the ex-type strain. This species can be distinguished by its coiled ascomatal hairs and more protruding lateral bulges of ascospores that appear wider than those of C. ascotrichoides (7.5–8.5 μm vs 6.5–7 μm).
Chaetomium megalocarpum Bainier, Bull. Soc. Mycol. France 25: 202. 1910. — Fig. 22
Fig. 22.
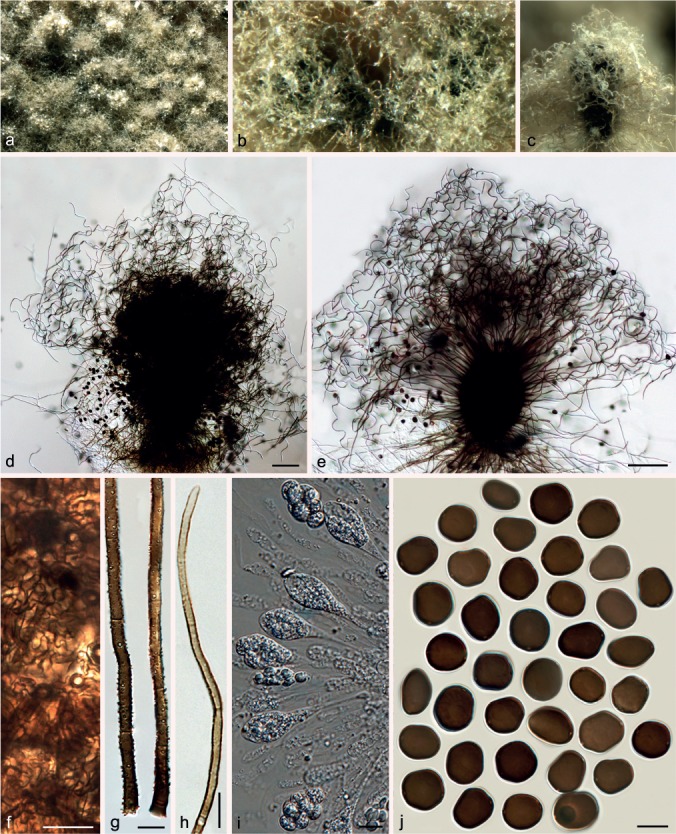
Chaetomium megalocarpum (CBS 149.59, ex-epitype culture). a. Part of the colony on OA; b. ascomata on OA, top view; c. ascoma and mass of ascospores on OA, side view; d, e. ascomata mounted in lactic acid; f. structure of ascomatal wall in surface view; g. basal parts of terminal ascomatal hairs; h. upper part of a terminal ascomatal hair; i. asci; j. ascospores. — Scale bars: d, e = 100 μm; f–h = 20 μm; i, j = 10 μm.
Ascomata superficial, ostiolate, honey to fawn in reflected light owing to ascomatal hairs, subglobose or ovate, 200–260 μm high, 148–220 μm diam. Ascomatal wall brown, composed of hypha-like or amorphous cells, textura epidermoidea or textura intricata in surface view. Terminal hairs punctate or finely verrucose, dark brown at the base, fading towards the tips, flexuous to undulate, sometimes branched, 3–5 μm near the base, tapering towards the tips. Lateral hairs similar. Asci fasciculate, fusiform or clavate, with eight biseriate or irregularly arranged ascospores, spore-bearing part 30.5–50.5 × 15–24 μm, stalks 14–30.5 μm long, evanescent. Ascospores dark brown when mature, ellipsoid to subglobose, usually irregular, bilaterally flattened, (12–)13–15(–17) × (10–)11.5–14 × (7.5–)8.5–10(–10.5) μm, with two apical, subapical or lateral germ pores. Asexual morph absent.
Culture characteristics — Colonies on OA lacking aerial hyphae, producing pale orange exudates diffusing into the medium; reverse fulvous to black under ascomata.
Materials examined. CHINA, Yinchuan Province, Ningxia City, isolated from horse dung, other collection information unknown, culture CGMCC 3.3595; Shanhaiguan, isolated from soil, other collection information unknown, culture CGMCC 3.9443. – FRANCE, lectotype of C. megalocarpum, designated here (MBT201727; Bull. Soc. Mycol. France 25: PL XVI, f. 1–4, 1910, drawn by G. Bainier based on the ex-type strain isolated from rotten paper, reproduced here as Fig. 20 after excluding the illustration of C. indicum (f. 5–14)). – GREECE, near the border to Yugoslavia, isolated from leaf of Ficus carica, collector and collection date unknown, isolated by G. Sörgel, 22 Nov. 1958 (epitype designated here CBS H-22186, MBT201728, culture ex-epitype CBS 149.59 = IMI 075851 = MUCL 9589). – INDIA, Yusmarg, Drug Tolan, isolated from humus-rich soil, collection date unknown, E. Müller, CBS 778.71 = ETH 1924.
Fig. 20.
Chaetomium megalocarpum. Illustrated by Bainier (1910, Bull. Soc. Mycol. Fr. 25: PL XVI, f. 1–4), selected as a lectotype in this study.
Notes — Chaetomium megalocarpum is differentiated from the closest species, C. grande, by possessing smaller ascospores (13–15 × 11.5–14 × 8.5–10 μm vs 18–20.5 × 16–18 × 12–13.5 μm). Phylogenetic inference showed that C. megalocarpum and C. grande form sister lineages in Group IA (Fig. 1), and are closely related to C. globosporum which produces the smallest, globose ascospores (10.5–12 μm diam, 7.5–8.5 μm wide in lateral view).
Chaetomium novozelandicum X. Wei Wang, Crous & L. Lombard, sp. nov. — MycoBank MB812980
Etymology. Refers to the country New Zealand, where this fungus was first collected.
Cultures sterile. Chaetomium novozelandicum forms a unique lineage (Group IIC, Fig. 1), basal to the C. globosum clade. This species differs by fixed unique SNPs in five loci: rpb2 positions 3(C), 9(C), 12(C), 24(A), 39(C), 51(A), 60(T), 69(T), 99(C), 124(T), 138(A), 177(G), 186(C), 220(A), 300(C), 306(A), 312(A), 372(G), 376(T), 393(T), 420(C), 450(A), 525(T), 570(T), 573(C), 579(G), 582(G) and 597(T); tub2 positions 12(C), 28(G), 97(T), 102(indel), 109(A), 142(indel), 143(indel), 144(indel), 168(C), 235(G), 236(G), 278(C), 319(T), 322(indel), 343(T), 368(A), 375(T), 378(A), 387(C), 447(indel), 459(C), 509(T), 570(G), 579(G), 656(T) and 707(T); tef1 positions 262(A), 284(T), 396(C), 465(C), 519(T), 683(C), 744(C), 762(T), 816(C) and 870(C); rpb1 positions 44(T), 59(C), 110(indel), 111(indel), 117(G), 163(C), 166(A), 175(G), 211(C), 256(C), 268(C), 272(A), 316(C), 364(T), 418(T), 427(G), 455(A), 457(C), 463(T), 487(C), 523(C), 535(T), 556(T), 580(G), 592(A), 613(C), 676(C), 685(T), 721(G) and 724(C); ITS positions 142(C) and 452 (C).
Culture characteristics — Colonies on OA with white, floccose aerial hyphae, without coloured exudates; reverse uncoloured.
Materials examined. NEW ZEALAND, town of Otaki on west coast, isolated from dead unidentified, decaying twig in a compost pile, collection date unknown, D.P. Mahoney (holotype AEB 1071, isotype CBS H-22191, culture ex-isotype CBS 124555); same collection details, CBS 124556. – USA, California, isolated from scalp of Homo sapiens, deposited in CBS by D.A. Sutton, 29 Sept. 2010, CBS 128484 = UTHSC 08-1518 = dH 21631.
Notes — Both phylogenetic inference and SNP analysis indicate that C. novozelandicum represents a novel phylogenetic species basal to Group II (Group IIC, Fig. 1). All attempts to induce sporulation on OA failed, even with the addition of sterile elm twig pieces.
Chaetomium nozdrenkoae Sergeeva, Not. Syst. sect. Crypt. Inst. Bot. Acad. Sci. U.S.S.R. 14: 140. 1961. — Fig. 23
Fig. 23.
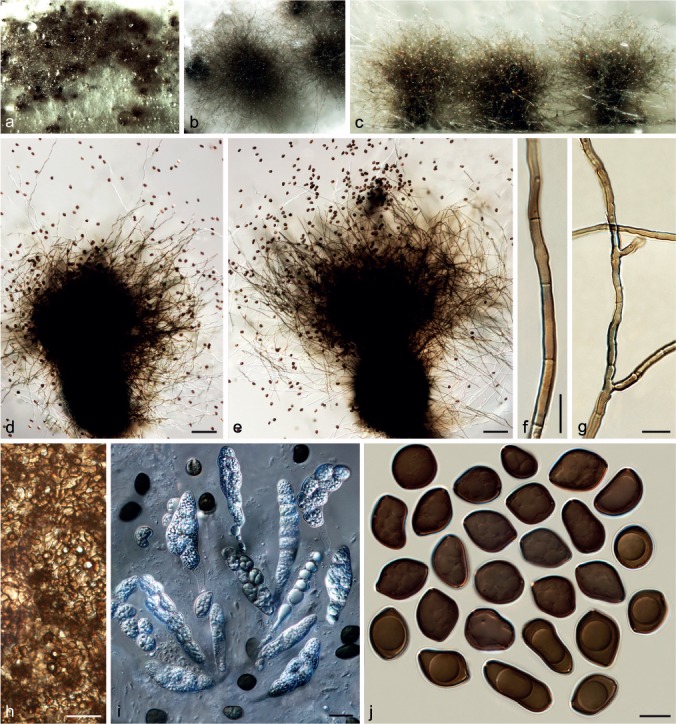
Chaetomium nozdrenkoae (CBS 163.62, ex-epitype culture). a. Part of the colony on OA; b. ascomata on OA , top view; c. ascomata on OA, side view; d, e. ascomata mounted in lactic acid; f. terminal ascomatal hair; g. branched terminal ascomatal hair; h. structure of ascomatal wall in surface view; i. asci; j. ascospores. — Scale bars: d, e = 100 μm; f, g, j = 10 μm; h, i = 20 μm.
Ascomata superficial or covered by aerial hyphae, ostiolate, umber or olivaceous to dark brick in reflected light owing to ascomatal hairs, subglobose to obovate, 280–520 μm high, 230–405 μm diam. Ascomatal wall brown, composed of amorphous cells, textura epidermoidea in surface view. Terminal hairs abundant, smooth, olivaceous brown, paler at the apices, hypha-like, flexuous, often branched, sometimes geniculate, 3–4.5 μm diam near the base. Lateral hairs similar. Asci fasciculate, fusiform or elongate clavate, with eight biseriate or irregularly-arranged ascospores, occasionally with eight ascospores uniseriately arranged in a nearly cylindrical ascus, spore-bearing part 53–93 × 13–24 μm, stalks 15–36 μm long, evanescent. Ascospores olivaceous brown when mature, irregularly limoniform to fusiform or ovate, bilaterally flattened, (12.5–)15–22(–26) × (11–)11.5–15(–17) × (9–)10–11.5(–12.5) μm, usually with two, three or occasionally four apical, subapical or lateral germ pores. Asexual morph absent.
Culture characteristics — Colonies on OA with abundant floccose, white to pale grey aerial hyphae, usually without exudates diffusing into medium; reverse uncoloured.
Materials examined. GERMANY, Giessen, isolated from greenhouse soil, collector and collection date unknown, isolated by D. Bredemeier, 1967, CBS 809.68 = IMI 180408. – RUSSIA, Novosibirsk region, isolated from virgin soil, collector and collection date unknown, isolated by K.S. Sergejeva, 1961 (culture ex-type CBS 163.62 = ATCC 14528 = IMI 090490 = IMI 090490ii = MUCL 18703 = VKM F-1953).
Notes — Chaetomium nozdrenkoae forms a unique lineage in Group IA (Fig. 1), sister to a clade including three species: C. grande, C. megalocarpum and C. globosporum. However, the latter species are distinguished from C. nozdrenkoae by having more regular, mostly globose to subglobose ascospores. All these taxa differ in their ascospores dimensions.
Chaetomium olivaceum Cooke & Ellis, Grevillea 6: 96. 1878. — Fig. 24
Fig. 24.
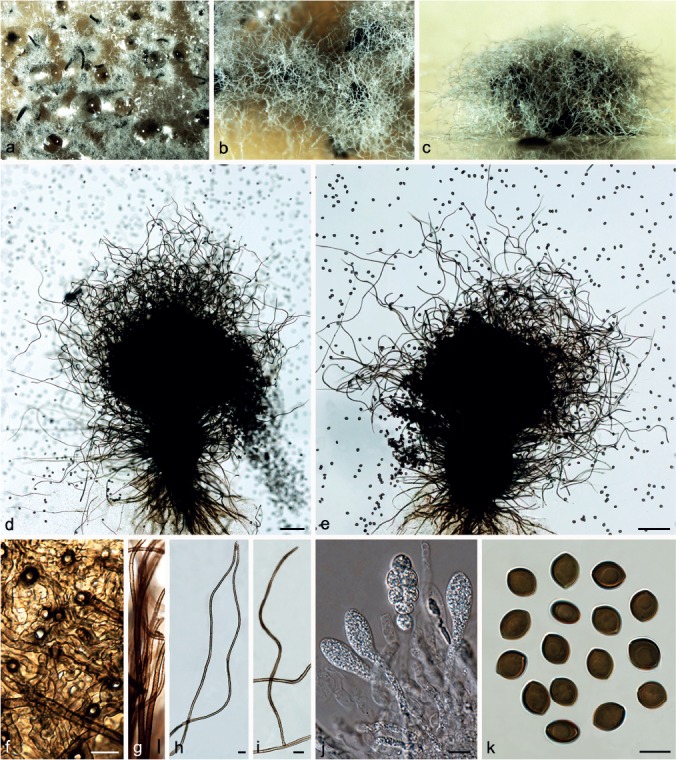
Chaetomium olivaceum (CBS 418.80A). a. Part of the colony on OA; b. ascomata on OA, top view; c. ascomata on OA, side view; d, e. ascomata mounted in lactic acid; f. structure of ascomatal wall in surface view; g. basal parts of terminal ascomatal hairs; h, i. upper parts of terminal ascomatal hairs; j. asci; k. ascospores. — Scale bars: d, e = 100 μm; f–k = 10 μm.
Ascomata superficial, ostiolate, pale olivaceous buff in reflected light owing to ascomatal hairs, subglobose to obovate, 260–440 μm high, 200–360 μm diam. Ascomatal wall brown, composed of hypha-like or amorphous cells, textura intricata or textura epidermoidea in surface view. Terminal hairs abundant, finely verrucose, brown, paler towards the apices, undulate or flexuous, occasionally branched, 2.5–4.5 μm near the base, tapering towards the tips. Lateral hairs similar. Asci fasciculate, clavate or slightly fusiform, with eight biseriate ascospores, spore-bearing part 34–41 × 13–20 μm, stalks 26–45 μm long, evanescent. Ascospores olivaceous brown when mature, limoniform to broad limoniform, usually biapiculate, bilaterally flattened, (10–)11–12(–12.5) × 8–9 × 6–7 μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with sparse white aerial hyphae, producing pale fawn exudates diffusing into the medium; reverse olivaceous, but black under ascomata.
Materials examined. CHINA, Aksu region, Xinjiang, from camel dung, Mar. 2009, F.-J. Liu, CGMCC 3.12883; Jilin Province, Changchun, from soil, Aug. 2002, X.-W. Wang, CGMCC 3.9465. – INDIA, Delhi, from nilgai (Boselaphus tragocamelus) dung, 28 Apr. 1977, K.G. Mukerji, isolated by J.A. von Arx, CBS 418.80A.
Notes — Chaetomium olivaceum was reduced to synonymy under C. globosum by Von Arx et al. (1986). This species can be distinguished by larger ascospores (11–12 × 9–10 × 6–7 μm) than those of C. globosum (8.5–10.5 × 7–8 × 5.5–6.5 μm). Phylogenetic inference indicated that C. olivaceum is in Group III (Fig. 1), closely related to C. cucumericola (sterile species), C. undulatulum and C. subglobosum. Chaetomium undulatulum (Asgari & Zare 2011) can be distinguished from C. olivaceum by smaller ascomata (230–280 μm high, 185–250 μm diam), longer ascospores (12–13.5 × 8–10 × 6–7.5 μm) and more undulate ascomatal hairs. Chaetomium subglobosum is also distinct from C. olivaceum in having larger ascospores (12–13.5 × 10.5–12 × 7–8.5 μm). The holotype of C. olivaceum was originally collected in Newfield (New Jersey, USA). No ex-type culture or isolate from the type locality is presently available. Therefore, typification of this species awaits recollection from the type locality.
Chaetomium pilosum (C. Booth & Shipton) X. Wei Wang & Crous, comb. nov. — MycoBank MB812981; Fig. 25
Fig. 25.
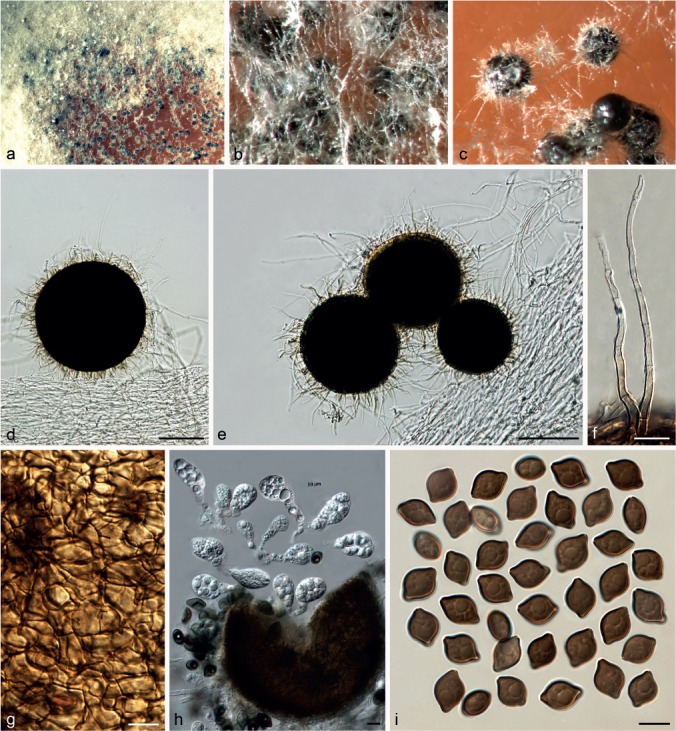
Chaetomium pilosum (CBS 335.62, ex-epitype culture). a. Part of the colony on OA; b. ascomata covered by hyphae on OA, top view; c. exposed ascomata on OA, top view; d, e. ascomata mounted in lactic acid; f. ascomatal hairs; g. structure of ascomatal wall in surface view; h. asci; i. ascospores. — Scale bars: d, e = 100 μm; f–i = 10 μm.
Basionym. Thielavia pilosa C. Booth & Shipton, Trans. Brit. Mycol. Soc. 49: 665. 1966.
≡ Chaetomidium pilosum (C. Booth & Shipton) Arx, Stud. Mycol. 8: 16. 1975.
Ascomata superficial, or covered by aerial hyphae, non-ostiolate, black in reflected light due to the dark ascomatal wall, spherical or oblate, pilose, 120–265 μm diam. Ascomatal wall brown, composed of angular cells, textura angularis in surface view. Ascomatal hairs covering the whole ascoma, hypha-like, smooth or finely verrucose, pale ochreous at the base, fading to hyaline in the upper part, 2.5–4 μm near the base, less than 120 μm long. Asci fasciculate, clavate to obovate, with eight biseriate or irregularly arranged ascospores, spore-bearing part 22–38 × 12.5–18 μm, stalks 10–24 μm long, evanescent. Ascospores olivaceous brown to brown when mature, limoniform, umbonate at both ends, bilaterally flattened, (11–)12–14.5(–16) × 9–10(–11) × (6–)7–8 μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with white to pale grey aerial hyphae, usually producing apricot to orange exudates diffusing into the medium; reverse ochreous to apricot.
Material examined. AUSTRALIA, Western Australia, Perth, isolated from grain of Triticum aestivum, collector and collection date unknown, isolated by W.A. Shipton, 1965 (isotype of Thielavia pilosa CBS H-6838, culture ex-isotype of Thielavia pilosa CBS 335.67 = IMI 113231 = VKM F-1851).
Notes — Only the ex-isotype strain is available for this species. Chaetomium pilosum forms a unique lineage basal to Group III, distant from two other species in the C. globosum complex, which have non-ostiolate ascomata, C. fimeti and C. subfimeti (Group I, Fig. 1). This species is easily distinguished by its non-ostiolate ascomata covered with hyaline hairs and distinctly umbonate ascospores.
Chaetomium pseudocochliodes X. Wei Wang, X.Z. Liu & Crous, sp. nov. — MycoBank MB812982; Fig. 26
Fig. 26.

Chaetomium pseudocochliodes (CGMCC 3.9441, ex-type culture). a. Part of the colony on OA; b. ascoma and mass of ascospores on OA, top view; c, d. ascomata on OA, side view; e–g. ascomata mounted in lactic acid; h. structure of ascomatal wall in surface view; i. basal parts of terminal ascomatal hairs of type I; j, k. upper parts of terminal ascomatal hairs of type I; l. terminal ascomatal hairs of type II; m. asci; n. ascospores. — Scale bars: e–g = 100 μm; h, i, k, m, n = 10 μm; j, l = 20 μm.
Etymology. Refers to the morphological similarity to C. cochliodes.
Ascomata superficial, ostiolate, citrine green to citrine in reflected light owing to ascomatal hairs, ellipsoid, ovate or subglobose, 270–425 μm high, 190–370 μm diam. Ascomatal wall brown, composed of hypha-like cells, textura intricata in surface view. Terminal hairs brown, tapering, partly type I: dark, verrucose, thick and erect in the lower part, 4–6 μm near the base, tapering and fading towards the tips, circinate (often on young ascomata) or spirally coiled in the upper part, with coils often tapering or in irregular form, sometimes with a short flexuous hypha-like extension at the tip, occasionally branched; partly type II: pale brown, finely verrucose, thinner, 3–4 μm near the base, flexuous. Lateral hairs hypha-like, flexuous, tapering towards tips. Asci fasciculate, elongated clavate, spore-bearing part 24–37 × 7–14 μm, stalks 17–42 μm long, with eight biseriate ascospores, evanescent. Ascospores olivaceous brown or brown when mature, limoniform, biapiculate, slightly umbonate at both ends, bilaterally flattened, (9–)9.5–11(–11.5) × (7–)7.5–8.5(–9) × 5.5–6.5(–7) μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with sparse white aerial hyphae or in the centre of colonies with thick, felt-like hyphae, producing luteous to rust exudates diffusing into the medium; reverse fulvous to sienna, black under ascomata.
Materials examined. CHINA, Yunnan Province, Wenshan County, from fibrous root of Panax notoginseng, 10 Apr. 2003, X.-Z Liu (holotype HMAS 244435, isotype CBS H-22197, culture ex-type CGMCC 3.9441); from the rhizosphere of P. notoginseng, 13 Apr. 2003, X.-Z Liu, CGMCC 3.9469.
Notes — Phylogenetic inference indicated that C. pseudocochliodes belongs to Group III, closely related to C. cochliodes and C. spiculipilium (Fig. 1), which is further confirmed by morphological characters. All three species produce regularly coiled ascomatal hairs and ascospores with similar dimensions. This species can be distinguished from C. cochliodes and C. spiculipilium by its more irregular and diverse ascomatal hairs as well as ascospores that usually have more protruding ends.
Chaetomium pseudoglobosum X. Wei Wang, Crous & L. Lombard, sp. nov. — MycoBank MB812983; Fig. 27
Fig. 27.
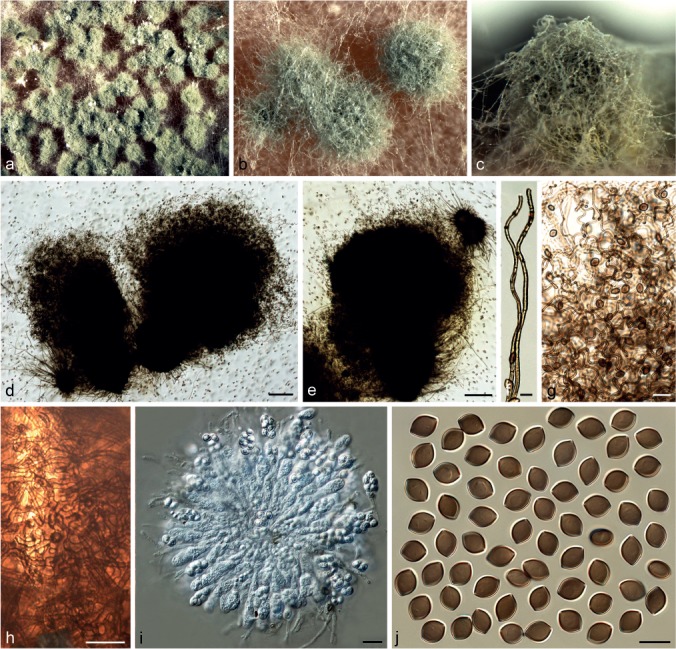
Chaetomium pseudoglobosum (CBS 574.71, ex-type culture). a. Part of the colony on OA; b. ascomata on OA, top view; c. ascoma on OA, side view; d, e. ascomata mounted in lactic acid; f. basal parts of terminal ascomatal hairs; g. upper parts of terminal ascomatal hairs; h. structure of ascomatal wall in surface view; i. asci; j. ascospores. — Scale bars: d, e = 100 μm; f, j = 10 μm; g–i = 20 μm.
Etymology. Refers to the striking resemblance to C. globosum.
Ascomata superficial or covered by sparse aerial hairs, ostiolate, olivaceous buff to greenish olivaceous in reflected light owing to ascomatal hairs, ovate to subglobose, 210–330 μm high, 165–315 μm diam. Ascomatal wall brown, composed of hypha-like cells, textura intricata in surface view. Terminal hairs abundant, forming a dense and nearly globose head over the ostiole, verrucose, olivaceous brown, fading towards the tips, loosely coiled, erect or flexuous at the lower part, 2.5–3.5 μm near the base, tapering towards the tips. Lateral hairs hypha-like, flexuous or slightly undulate, tapering and fading towards the tips. Asci fasciculate, clavate or fusiform, spore-bearing part 23–32 × 10–14 μm, stalks 17–36 μm long, with eight biseriate ascospores, evanescent. Ascospores olivaceous brown when mature, limoniform, bilaterally flattened, 9–10(–10.5) × (6–)6.5–7.5(–8) × 5–6(–6.5) μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with sparse white aerial hypha, producing pale apricot to pale orange exudates diffusing into the medium; reverse usually uncoloured, but fulvous to umber under ascomata.
Material examined. UNKNOWN, substrate and collection details unknown, deposited in CBS by J.E. Wright, Sept. 1971(holotype CBS H-10083, culture ex-type CBS 574.71).
Notes — Phylogenetic inference in this study showed that C. pseudoglobosum is in Group II, closely related to C. tenue (Group IIB, Fig 1). The latter species produces smaller ascospores (9–10 × 6.5–7.5 × 5–6 μm vs 8.5–9.5 × 6–7 × 5–5.5 μm) and less dense ascomatal hair structures. Chaetomium pseudoglobosum forms dense ascomatal hair structures covering the ascomatal ostioles, which resemble those of C. afropilosum. However, C. afropilosum produces smaller ascospores (7–8 × 5.5–6 × 4–5 μm).
Chaetomium rectangulare Asgari & Zare, Mycologia 103: 872. 2011. — Fig. 28
Fig. 28.

Chaetomium rectangulare (CBS 126658). a. Part of the colony on OA; b. ascoma on OA, top view; c. ascoma on OA, side view; d–f. ascomata mounted in lactic acid; g. basal parts of terminal ascomatal hairs; h, i. upper parts of terminal ascomatal hairs; j. structure of ascomatal wall in surface view; k. asci; l. ascospores; m. asexual morph (conidiophore and conidia). — Scale bars: d–f = 100 μm; g–i = 20 μm; j–m = 10 μm.
Ascomata superficial, ostiolate, firmly attached to the medium by well-developed rhizoids, olivaceous grey in reflected light owing to ascomatal hairs, globose to subglobose, 300–450 μm high, 215–380 μm diam. Ascomatal wall brown, composed of hypha-like or amorphous cells, textura intricata or textura epidermoidea in surface view. Terminal hairs verrucose, dark brown, erect in the lower part, 4.5–7 μm diam near the base, regularly and dichotomously branched at right to nearly straight angles in the upper part, with relatively erect and rigid spear-shaped branches, fading and tapering towards the tips. Lateral hairs brown, seta-like or sometimes terminally branched, tapering and fading towards the tips. Asci fasciculate, clavate, spore-bearing part 26–43 × 11–16 μm, stalks 23–37 μm long, with eight biseriate ascospores, evanescent. Ascospores olivaceous brown when mature, limoniform, biapiculate, bilaterally flattened, (9–)10–11(–12) × 7–9 × 6–7.5(–8) μm, with an apical germ pore. Asexual morph acremonium-like. Conidiophores formed laterally from aerial hyphae, simple, 13–29 μm long, 2–4.5 μm diam at the base. Conidia formed in chains, hyaline, aseptate, smooth, ovate or ellipsoidal, often with a truncated base and a rounded apex, (3–)3.5–5(–6) × 2–3 μm.
Culture characteristics — Colonies on OA without aerial hyphae, producing pale luteous to orange exudates diffusing into the medium; reverse cinnamon, but black under ascomata.
Materials examined. CHINA, Xinjiang Autonomous Region, Kanas Lake, from animal dung, June 2003, X.-W. Wang, CGMCC 3.9409. – IRAN, West Azerbaijan Province, Salmas, from leaf of Hordeum vulgare, 22 June 2005, B. Asgari (holotype IRAN 14606F, culture ex-type CBS 126778 = IRAN 1641C); East Azerbaijan Province, Shabestar, from stem of Hordeum vulgare, 22 May 2005, B. Asgari, CBS 126658 = IRAN 855C. – UK, from decaying hard wood, collection details unknown, CGMCC 3.5617.
Notes — The ex-type culture (CBS 126778) of C. rectangulare is sterile, and therefore the description here is based on CBS 126658. Chaetomium rectangulare is morphologically and phylogenetically close to C. elatum (Group III, Fig. 1). They are both morphologically distinguished in the C. globosum species complex by having dichotomously branched ascomatal hairs. Asgari & Zare (2011) compared both species, and distinguished C. elatum from C. rectangulare by having flexuous, irregularly branched and narrower ascomatal hairs, wider asci and larger ascospores. Our observations confirmed that C. rectangulare produces smaller asci and ascospores than those of C. elatum. However, the ascomatal hairs of both species branch at right to nearly straight angles in the upper part. Chaetomium rectangulare can also be distinguished from C. elatum by thicker, darker and rigid terminal ascomatal hairs, and well-developed rhizoids.
Chaetomium spiculipilium Ames, A Monograph of the Chaetomiaceae: 37. 1963. — Fig. 29
Fig. 29.

Chaetomium spiculipilium (CBS 373.66, ex-type culture). a. Part of the colony on OA; b. ascomata on OA, top view; c, d. ascomata on OA, side view; e–g. ascomata mounted in lactic acid; h. structure of ascomatal wall in surface view; i. basal parts of terminal ascomatal hairs; j, k. upper parts of terminal ascomatal hairs; l. ascus; m. ascospores. — Scale bars: e–g = 100 μm; h, k–m = 10 μm; i = 20 μm; j = 50 μm.
Ascomata superficial, ostiolate, citrine-green to greenish olivaceous in reflected light owing to ascomatal hairs, ellipsoid, ovate or subglobose, 370–480 μm high, 300–385 μm diam. Ascomatal wall brown, composed of amorphous or hypha-like cells, textura epidermoidea or textura intricata in surface view. Terminal hairs verrucose, dark brown, rigid, erect in the lower part, 5–8 μm diam near the base, tapering and fading towards the tips, coiled in the upper part; coils regular, sometimes slightly tapering, with a conspicuous, rigid seta-like extension at the tip, often with coiled or seta-like branches. Lateral hairs hypha-like, flexuous, fading and tapering towards the tips. Asci fasciculate, clavate, spore-bearing part 21–42 × 13–16.5 μm, stalks 27–43 μm long, with eight biseriate ascospores, evanescent. Ascospores olivaceous brown when mature, limoniform, usually biapiculate, occasionally umbonate at one or both ends, bilaterally flattened, (9–)10–13(–15) × (7–)7.5–9(–10) × 5.5–6.5(–7) μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with thick, white aerial hyphae only in the centre, producing luteous to orange or brick to vinaceous exudates diffusing into the medium; reverse fulvous to sienna.
Material examined. USA, California, Aptos, from decaying vegetable debris, collection date unknown, H.K. Seth, isolated by L.M. Ames (isotype CBS H-6893, culture ex-isotype CBS 373.66).
Notes — Chaetomium spiculipilium is closely related to C. cochliodes, C. pseudocochliodes and C. spirochaete by having regularly coiled ascomatal hairs. Phylogenetic inference also showed that C. spiculipilium, belonging to Group III, is closely related to C. cochliodes and C. pseudocochliodes (Fig. 1). Chaetomium spiculipilium can be distinguished by having thicker and more rigid ascomatal hairs (5–8 μm diam near the base) with a conspicuous seta-like extension at the tip, compared to those of C. cochliodes (3.5–6 μm near the base), C. pseudocochliodes (4–6 μm near the base for regularly coiled hairs) and C. spirochaete (3–4.5 μm near the base). The ascospores of C. spiculipilium (10–13 × 7.5–9 × 5.5–6.5 μm) are also slightly larger than those of C. cochliodes (9–10 × 7.5–8.5 × 5–6 μm) and C. pseudocochliodes (9.5–11 × 7.5–8.5 × 5.5–6.5 μm).
Chaetomium spirochaete Palliser, N. Amer. Fl. 3: 61. 1910. — Fig. 30
Fig. 30.

Chaetomium spirochaete (CBS 370.84, ex-epitype culture). a. Part of the colony on OA; b, c. ascomata on OA, top view; d. ascoma on OA, side view; e–g. ascomata mounted in lactic acid; h. structure of ascomatal wall in surface view; i. basal parts of terminal ascomatal hairs; j, k. upper parts of terminal ascomatal hairs; l. asci; m. ascospores; n. holotype sheet of C. spirochaete in New York Botanical Garden (Specimen ID 01050443); o, p. ascomatal hairs from holotype specimen. — Scale bars: e–g = 100 μm; h, l, m = 10 μm; i–k, p = 20 μm.
Ascomata superficial, ostiolate, honey to pale hazel in reflected light owing to ascomatal hairs, ellipsoid, ovate or elongate ovate, 135–230 μm high, 118–205 μm diam. Ascomatal wall brown, composed of amorphous or hypha-like cells, textura epidermoidea or textura intricata in surface view. Terminal hairs verrucose, brown, 3–4.5 μm near the base, equally diametered from the base to the tip, erect in the lower part, coiled in the upper part with coils equal in diameter, sometimes with coiled branches. Lateral hairs pale brown, flexuous, tapering towards the tips. Asci fasciculate, clavate, spore-bearing part 26–43.5 × 13.5–16 μm, stalks 18–32 μm long, with eight biseriate ascospores, evanescent. Ascospores olivaceous brown when mature, limoniform, bilaterally flattened, sometimes inequilateral, (9–)10–11(–12) × 7.5–9(–9.5) × (5.5–)6–7 μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with sparse white aerial hyphae and producing yellowish ochreous exudates diffusing into the medium; reverse uncoloured, but dark olivaceous under ascomata.
Materials examined. UNKNOWN, collection details unknown, from animal dung, isolated and deposited in CBS by L.M. Ames, Apr. 1952, CBS 165.52. – USA, Iowa, from cotton root, June 1890, L.H. Pammet (holotype New York Botanical Garden Specimen ID01050443); Tennessee, Great Smokey Mountains, unknown collection details, isolated by L.M. Ames, deposited in CBS by J.C. Krug, Nov. 1984 (epitype designated here HMAS 244438, MBT201732, culture ex-epitype CBS 730.84 = IMI 287303 = QM 6702).
Notes — The epitype of C. spirochaete, designated here, is morphologically similar to the holotype, particularly in morphology of ascospores and ascomatal hairs, and originates from the same locality as the type. Chaetomium spirochaete was synonymised under C. spirale (Chivers 1915, Ames 1963) and later re-introduced by Dreyfuss (1976). Von Arx et al. (1986) followed this treatment and rejected the species C. spirale because the type had been lost and the species could not be recognised from the original description. Chaetomium spirochaete was considered a relative of C. globosum and differed from C. globosum in having regularly coiled, relatively dark and thick (5–6 μm) ascomatal hairs (Von Arx et al. 1986). Our observations, however, showed that the ascomatal hairs of C. spirochaete (3–4.5 μm near the base) are not thicker than those of C. globosum s.str. (3–5 μm diam near the base). Chaetomium spirochaete can be distinguished from C. globosum by regularly coiled ascomatal hairs, and slightly larger ascospores. In addition, the phylogenetic inference places C. spirochaete in Group III, distant from C. globosum (Group IIA, Fig 1). Chaetomium spirochaete has regularly coiled ascomatal hairs, resembling those of C. cochliodes, C. pseudocochliodes and C. spiculipilium. Among them, C. spiculipilium has slightly larger ascospores, while the ascospores of the three remaining species are similar. However, C. spirochaete can be distinguished by the uniform diameter in both the ascomatal hairs themselves and the coils formed by the ascomatal hairs. Furthermore, the ascomatal hairs (3–4.5 μm near the base) of C. spirochaete are thinner than those of C. cochliodes (3.5–6 μm near the base), C. pseudocochliodes (4–6 μm near the base for regularly coiled hairs) and C. spiculipilium (5–8 μm diam near the base).
Chaetomium subaffine Sergeeva, Not. Syst. sect. Crypt. Inst. Bot. Acad. Sci. U.S.S.R. 14: 148. 1961. — Fig. 31
Fig. 31.

Chaetomium subaffine (CBS 637.91, ex-type culture). a. Part of the colony on OA; b. ascomata entangled by hyphae on OA, top view; c, d. ascomata and masses of ascospores on OA, side view; e, f. ascomata mounted in lactic acid; g. basal parts of terminal ascomatal hairs; h. upper part of a terminal ascomatal hair; i. structure of ascomatal wall in surface view; j. asci; k. ascospores; l. asexual morph (conidiophore and conidia). — Scale bars: e, f = 100 μm; g–l = 10 μm.
Ascomata usually covered by thick aerial hyphae, ostiolate, olivaceous or umber to dark-brick in reflected light owing to ascomatal hairs, obovate or ovate, 220–410 μm high, 180–340 μm diam. Ascomatal wall brown, composed of hypha-like cells, textura intricata in surface view. Terminal hairs verrucose, brown, erect to flexuous or slightly undulate, usually unbranched, 3.5–5 μm near the base, tapering towards the tips. Lateral hairs similar. Asci fasciculate, clavate or slightly fusiform, with eight biseriate ascospores, spore-bearing part 29–46 × 12–18 μm, stalks 32–58 μm long, evanescent. Ascospores brown when mature, limoniform, usually biapiculate, bilaterally flattened, (10.5–)11.5–13.5(–14) × 8.5–10(–10.5) × (6–)6.5–7.5(–8) μm, with an apical germ pore. Asexual morph acremonium-like. Conidiophores discrete, simple; conidiogenous cells phialidic, hyaline. Conidia formed in basipetal succession, aseptate, smooth, hyaline, ovate or spherical, usually attenuated into a narrowly truncate base, (3–)3.5–5 × 2–3 μm.
Culture characteristics — Colonies on OA with abundant, floccose white aerial hyphae without coloured exudates; reverse uncoloured.
Materials examined. CHINA, Beijing, from animal dung, Aug. 2009, J. Li, CGMCC 3.14293; Hebei Province, Xingtai, from unknown plant stem, Aug. 2009, J. Li, CGMCC 3.14297. – NETHERLANDS, Zwolle, from finger skin of Homo sapiens, collector and collection date unknown, deposited in CBS by Bact. Lab. Zwolle, Jan. 1976, CBS 111.76. – USSR, from cereal, collector and collection date unknown, deposited in CBS by K.S. Sergejeva, Nov. 1991 (culture ex-type CBS 637.91 = ATCC 14531 = IMI 90489).
Notes — Von Arx et al. (1986) maintained C. subaffine as a separate species as the ascospores (11–15 × 8–11 × 7–8.5 μm) are larger than those of C. globosum (9–12 × 8–10 × 6–8 μm), and suggested that this species is related to C. elatum. Phylogenetic inference indicates that C. subaffine is closely related to C. cochliodes, C. pseudocochliodes and C. spiculipilium (Group III, Fig. 1). However, C. subaffine can be distinguished by having abundant white mycelia covering the ascomata, mostly straight to flexuous ascomatal hairs, and having an asexual morph. The ascospores of C. subaffine are also larger than those of C. cochliodes (9–10 × 7.5–8.5 × 5–6 μm), C. pseudocochliodes (9.5–11 × 7.5–8.5 × 5.5–6.5 μm) and C. spiculipilium (10–13 × 7.5–9 × 5.5–6.5 μm).
Chaetomium subfimeti (Seth) X. Wei Wang & Crous, comb. nov. — MycoBank MB812984; Fig. 32
Fig. 32.
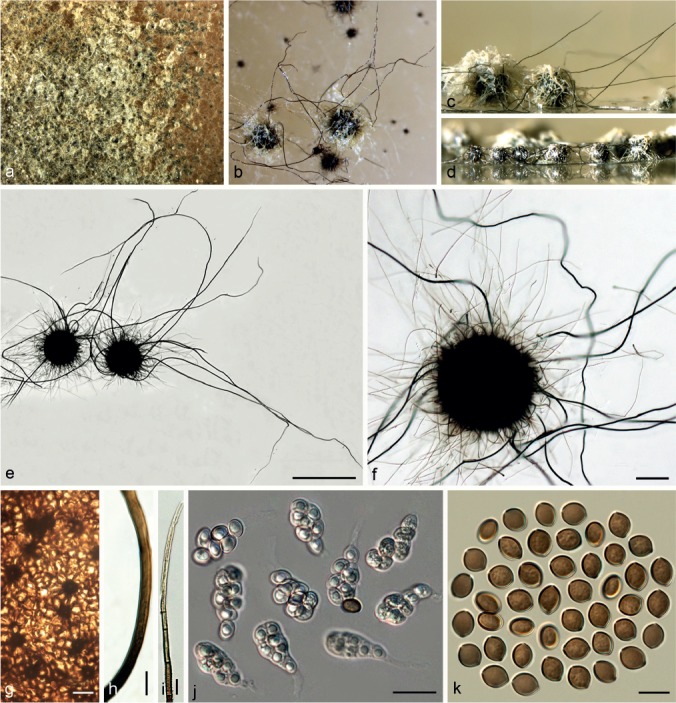
Chaetomium subfimeti (CBS 370.66, ex-type culture). a. Part of the colony on OA; b. ascomata on OA, top view; c, d. ascomata on OA, side view; e, f. ascomata mounted in lactic acid; g. structure of ascomatal wall in surface view; h. basal part of a terminal ascomatal hair; i. upper part of a terminal ascomatal hair; j. asci; k. ascospores. — Scale bars: e = 500 μm; f = 100 μm; g–i, k = 10 μm; j = 20 μm.
Basionym. Chaetomidium subfimeti Seth, Trans. Brit. Mycol. Soc. 50: 46. 1967.
≡ Thielavia subfimeti (Seth) Malloch & Cain, Mycologia 65: 1070. 1973.
Ascomata superficial or covered by thick aerial hyphae, non-ostiolate, fawn to black with numerous short, pale citrine ascomatal hairs, and sparse, long and black hairs in reflected light, spherical or oblate, 170–360 μm diam. Ascomatal wall brown, composed of thick-walled, angular or irregular cells, textura angularis in surface view. Ascomatal hairs of two types: shorter type covering the whole ascomata, less than 500 μm long, hypha-like, verrucose, dark brown at the lower part, fading to pale luteous-coloured at the apex, 2–3.5 μm near the base; longer type arising from the base of the ascomata, 200–3500 μm long, smooth, erect, flexuous or slightly undulate, dark brown, 3.5–5.5 μm near the base. Asci fasciculate, clavate or slightly fusiform, with eight biseriate ascospores, spore-bearing part 15–31.5 × 7.5–14 μm, stalks 7–18 μm long, evanescent. Ascospores olivaceous brown to brown when mature, limoniform, bilaterally flattened, (8–)8.5–9.5(–10) × (6.5–)7–7.5(–8) × 5.5–6(–6.5) μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA usually with thick, floccose to felt-like, white or pale honey aerial mycelia, sometimes covering the ascomata, producing yellowish ochreous exudates diffusing into the medium; reverse cinnamon to fulvous.
Materials examined. UK, Wales, Cardiff, isolated from paper and vegetable material, collection date unknown, isolated by H.K. Seth, 25 Dec. 1963 (isotype of Chaetomidium subfimeti CBS H-6839, culture ex-isotype of Chaetomidium subfimeti CBS 370.66 = ATCC 18209 = IMI 116692 = LCP 82.3317). – USA, California, Kern County, isolated from soil, collector and collection date unknown, isolated by G.F. Orr, CBS 169.71 = ATCC 22277 = IMI 153721.
Notes — Chaetomium subfimeti formed a sister lineage to C. fimeti (Group IC, Fig 1) as was supported by our morphological observations. Chaetomium subfimeti can be distinguished by producing smaller ascomata (170–360 μm diam vs 320–500 μm diam) and ascospores (8.5–9.5 × 7–7.5 × 5.5–6 μm vs 11.5–13.5 × 9–10.5 × 7–8 μm) than those of C. fimeti.
Chaetomium subglobosum Sergeeva, Not. Syst. sect. Crypt. Inst. Bot. Acad. Sci. U.S.S.R. 13: 172. 1960. — Fig. 33
Fig. 33.

Chaetomium subglobosum (CBS 483.73). a. Part of the colony on OA; b. ascomata on OA, top view; c. ascoma on OA, side view; d. ascoma mounted in lactic acid; e. basal parts of terminal ascomatal hairs; f. upper part of a terminal ascomatal hair; g. branched middle parts of terminal ascomatal hairs; h. structure of ascomatal wall in surface view; i. asci; j. ascospores. — Scale bars: d = 100 μm; e–h, j = 10 μm; i = 20 μm.
Ascomata superficial or covered by aerial hyphae, ostiolate, greenish olivaceous or grey-olivaceous in reflected light owing to ascomatal hairs, subglobose to oblong, 300–450 μm high, 265–355 μm diam, firmly attached to the medium by well-developed and densely-combined rhizoids forming compact structures at the base. Ascomatal wall brown, composed of hypha-like or amorphous cells, textura intricata or textura epidermoidea in surface view. Terminal hairs abundant, finely punctate to verrucose, brown, fading towards the tips, flexuous, sometimes branched, 3–5.5 μm near the base, tapering towards the tips. Lateral hairs similar. Asci fasciculate, clavate or slightly fusiform, with eight biseriate or irregularly-arranged ascospores, spore-bearing part 32.5–45 × 13.5–18 μm, stalks 25.5–36.5 μm long, evanescent. Ascospores olivaceous brown when mature, limoniform to broad limoniform, usually biapiculate, bilaterally flattened, (11–)12–13.5(–14) × (10–)10.5–12(–13.5) × 7–8.5(–9) μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with abundant white aerial hyphae, without coloured exudates; reverse uncoloured.
Materials examined. RUSSIA, St. Petersburg, from dead herbaceous stem, in moist chamber, K.S. Sergejeva, collection date unknown, isolated and deposited in CBS by K.S. Sergejeva, June 1960 (culture ex-type CBS 149.60 = ATCC 14533 = IMI 081770 = MUCL 18694 = VKM F-1951). – TURKEY, Izmir, from Eriobotrya japonica, collection date unknown, isolated and deposited in CBS by E. Onogur, May 1973, CBS 483.73.
Notes — The description provided here is based on the isolate CBS 483.73 since the ex-type culture (CBS 149.60) is sterile. Von Arx et al. (1986) reduced C. subglobosum to synonymy under C. globosum. However, we consider C. subglobosum as a separate species based on morphological and molecular evidence. Chaetomium subglobosum can be distinguished from C. globosum s.str. by producing larger ascomata (300–450 × 265–355 μm vs 160–300 × 135–250 μm) and ascospores (12–13.5 × 10.5–12 × 7–8.5 μm vs 8.5–10.5 × 7–8 × 5.5–6.5 μm). The phylogenetic inference also showed that C. subglobosum is placed in Group III (Fig. 1), distant from C. globosum s.str. (Group IIA, Fig. 1).
Chaetomium telluricola X. Wei Wang, Crous & L. Lombard, sp. nov. — MycoBank MB812985; Fig. 34
Fig. 34.
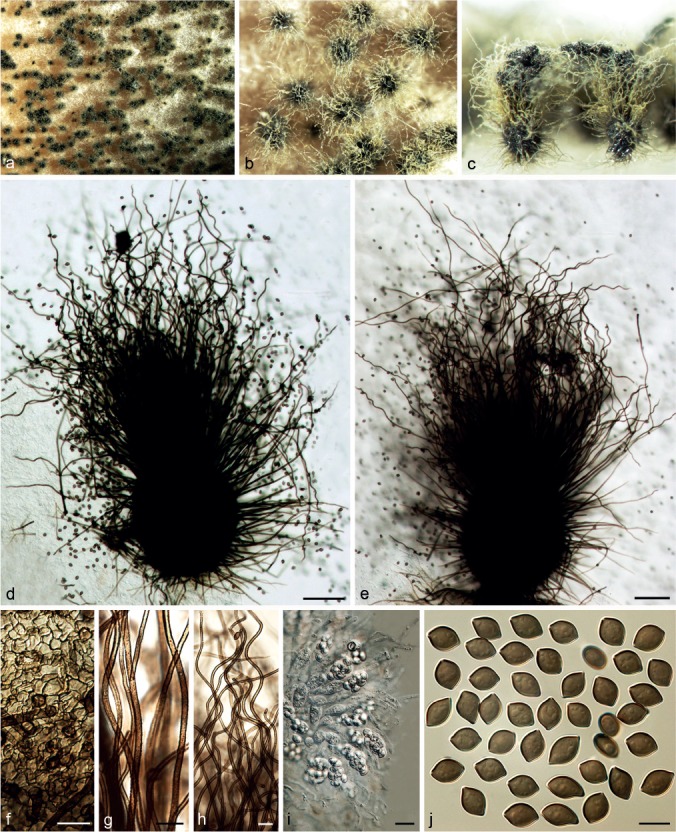
Chaetomium telluricola (CBS 151.59, ex-type culture). a. Part of the colony on OA; b. ascomata on OA, top view; c. ascomata and masses of ascospores on OA, side view; d, e. ascomata mounted in lactic acid; f. structure of ascomatal wall in surface view; g. basal parts of terminal ascomatal hairs; h. upper parts of terminal ascomatal hairs; i. asci; j. ascospores. — Scale bars: d, e = 100 μm; f, g, i = 20 μm; h, j = 10 μm.
Etymology. Refers to soil, the substrate from which this fungus was isolated.
Ascomata superficial, ostiolate, amber to citrine in reflected light owing to ascomatal hairs, globose or ovate, 140–350 μm high, 140–300 μm diam. Ascomatal wall brown, composed of angular or amorphous cells, arranged in a petal form around the bases of hairs, textura angularis or textura epidermoidea in surface view. Terminal hairs relatively sparse, verrucose, olivaceous brown, fading towards the tips, slightly tapering, erect or flexuous at the lower part, undulate at the upper part, 3–5 μm near the base. Lateral hairs hypha-like, erect or flexuous, tapering towards the tips. Asci fasciculate, clavate or slightly fusiform, spore-bearing part 24–38 × 11.5–16.5 μm, stalks 21–37 μm long, with eight biseriate ascospores, evanescent. Ascospores olivaceous brown when mature, elongate limoniform to broadly fusiform, or slightly irregular, bilaterally flattened, (9–)10–13(–15) × (6–)7.5–8(–8.5) × 5–6 μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA usually without aerial hyphae and coloured exudates; reverse uncoloured, but greenish olivaceous under ascomata.
Material examined. UK, Suffolk, Lakenheath Warren, isolated from soil, collection date unknown, J.H. Warcup, deposited in CBS by IMI, Apr. 1959 (holotype CBS H-676, culture ex-type CBS 151.59 = IMI 032543).
Notes — Chaetomium telluricola is morphologically distinct in the C. globosum species complex having elongate limoniform to broadly fusiform ascospores. Phylogenetic inference showed that C. telluricola is closely related to C. capillare in Group III (Fig. 1).
Chaetomium tenue X. Wei Wang, Crous & L. Lombard, sp. nov. — MycoBank MB812986; Fig. 35
Fig. 35.
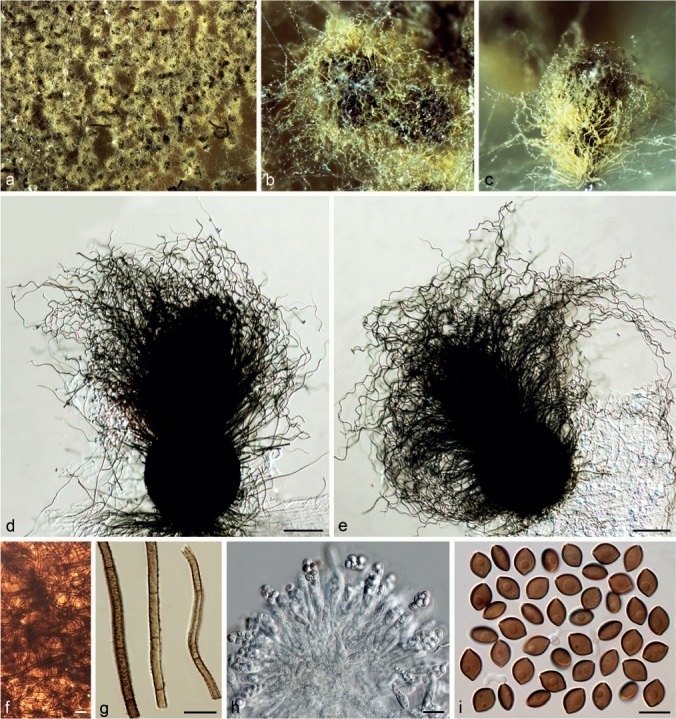
Chaetomium tenue (CBS 139.38, ex-type culture). a. Part of the colony on OA; b. ascomata on OA, top view; c. ascoma on OA, side view; d, e. ascomata mounted in lactic acid; f. structure of ascomatal wall in surface view; g. terminal ascomatal hairs (from left to right: lower part, middle part and upper part); h. asci; i. ascospores. — Scale bars: d, e = 100 μm; f, g, i = 10 μm; h = 20 μm.
Etymology. Refers to the relatively narrow ascospores formed by this fungus.
Ascomata superficial or covered by sparse aerial hyphae, ostiolate, olivaceous buff or greenish olivaceous, to pale amber or citrine-green in reflected light owing to ascomatal hairs, globose to subglobose, 165–330 μm high, 150–300 μm diam. Ascomatal wall dark brown, composed of hypha-like cells, textura intricata in surface view. Terminal hairs verrucose, olivaceous brown, fading towards the tips, undulate with erect or flexuous lower part, 3–4.5 μm diam near the base, tapering towards the tips. Lateral hairs flexuous or similar. Asci fasciculate, clavate or slightly fusiform, spore-bearing part 23–33 × 10–14 μm, stalks 16–36 μm long, with eight biseriate ascospores, evanescent. Ascospores brown when mature, elongate limoniform to broadly fusiform, biapiculate, bilaterally flattened, (7.5–)8.5–9.5(–10.5) × 6–7(–7.5) × (4.5–)5–5.5 μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with sparse to thick white aerial hyphae, usually without coloured exudates; reverse uncoloured.
Materials examined. UNKNOWN, no collection information, deposited in CBS by A.L. McAulay, Aug. 1938 (holotype CBS H-22195, culture ex-type CBS 139.38); other cultures with identical information, CBS 138.38, CBS 140.38, CBS 142.38, CBS 143.38.
Notes — Phylogenetic inference in this study showed that C. tenue is closely related to C. pseudoglobosum (Group IIB, Fig. 1). However, it is differentiated by having less dense, undulate ascomatal hairs and elongate limoniform to broadly fusiform ascospores, slightly narrower (8.5–9.5 × 6–7 × 5–5.5 μm) than those of C. pseudoglobosum (9–10 × 6.5–7.5 × 5–6 μm).
Chaetomium umbonatum D. Brewer, Proc. Trans. Nova Scotium Inst. Soc. 27: 59. 1974. — Fig. 36
Fig. 36.
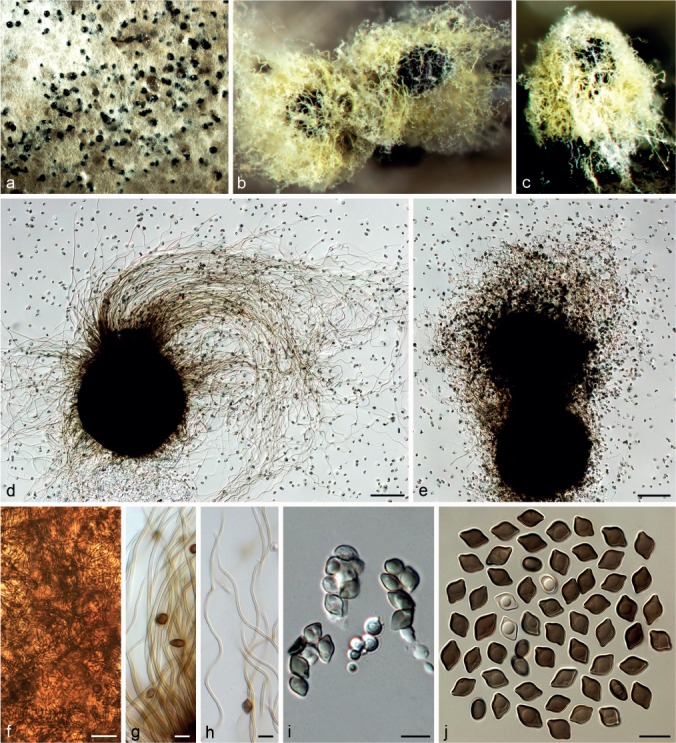
Chaetomium umbonatum (CBS 293.83, ex-type culture). a. Part of the colony on OA; b. ascomata and masses of ascospores on OA, top view; c. ascoma on OA, side view; d, e. ascomata mounted in lactic acid; f. structure of ascomatal wall in surface view; g. basal parts of terminal ascomatal hairs; h. upper parts of terminal ascomatal hairs; i. asci; j. ascospores. — Scale bars: d, e = 100 μm; f = 20 μm; g–j = 10 μm.
Ascomata superficial or covered by aerial hyphae, ostiolate, sulphur-yellow to ochreous in reflected light owing to ascomatal hairs, globose or slightly ovate, 260–360 μm high, 210–320 μm diam. Ascomatal wall brown, composed of hypha-like or amorphous cells, textura intricata or textura epidermoidea. Terminal hairs hypha-like or undulate with flexuous lower part, smooth, flexible, fulvous to pale brown at the bases, fading towards the tips, 1.5–3 μm near the base, slightly tapering towards the rounded tips. Lateral hairs similar. Asci fasciculate, clavate or slightly fusiform, with eight biseriate or irregularly-arranged ascospores, spore-bearing part 22–27 × 8–12.5 μm, stalks 13–18 μm long, evanescent. Ascospores olivaceous brown when mature, elongate limoniform, biconical, prominently umbonate at both ends, bilaterally flattened, (7.5–)8–11(–12) × (5–)5.5–7(–7.5) × (3.5–)4–5(–5.5) μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with white to pale grey aerial hyphae, usually not producing coloured exudates; reverse uncoloured.
Material examined. CANADA, Nova Scotia, isolated from soil, collection date unknown, D. Brewer (isotype CBS H-6904, culture ex-isotype CBS 293.83 = ATCC 28768 = IMI 138895).
Notes — Chaetomium umbonatum is easily recognised by its ascospores. Von Arx et al. (1986) suggested that this species is related to C. globosum, which is confirmed by the phylogenetic inference in this study. Chaetomium umbonatum is closely related to C. afropilosum in Group II (Fig. 1), which has smaller (7–8 × 5.5–6 × 4–5 μm) and biapiculate ascospores. Chaetomium umbonatum resembles C. pilosum in ascospore shape and pale ascomatal hairs, but the latter is characterised by non-ostiolate ascomata and larger ascospores (12–14.5 × 9–10 × 7–8 μm). Chaetomium pilosum is also phylogenetically distant from C. umbonatum (basal in Group III, Fig. 1).
Chaetomium undulatulum Asgari & Zare, Mycologia 103: 870. 2011
Description & Illustration — See Asgari & Zare (2011).
Materials examined. IRAN, East Azerbaijan Province, Bonab, isolated from leaf of Hordeum vulgare, 22 May 2005, B. Asgari (holotype IRAN 14605 F, culture ex-type CBS 126775 = IRAN 857C); West Azerbaijan Province, Miandoab, isolated from leaf of Triticum aestivum, 23 June 2005, B. Asgari, CBS 126776 = IRAN 1071C.
Notes — The isolates of C. undulatulum deposited in CBS are sterile. Phylogenetic inference in the present study indicated that C. undulatulum is closely related to C. subglobosum (Group III, Fig. 1). Chaetomium undulatulum can be distinguished from C. subglobosum by smaller ascomata (230–280 × 185–250 μm vs 300–450 × 265–355 μm) and narrower ascospores (12–13.5 × 8–10 × 6–7.5 μm vs 12–13.5 × 10.5–12 × 7–8.5 μm).
Chaetomium unguicola X. Wei Wang, Crous & L. Lombard, sp. nov. — MycoBank MB812987; Fig. 37
Fig. 37.
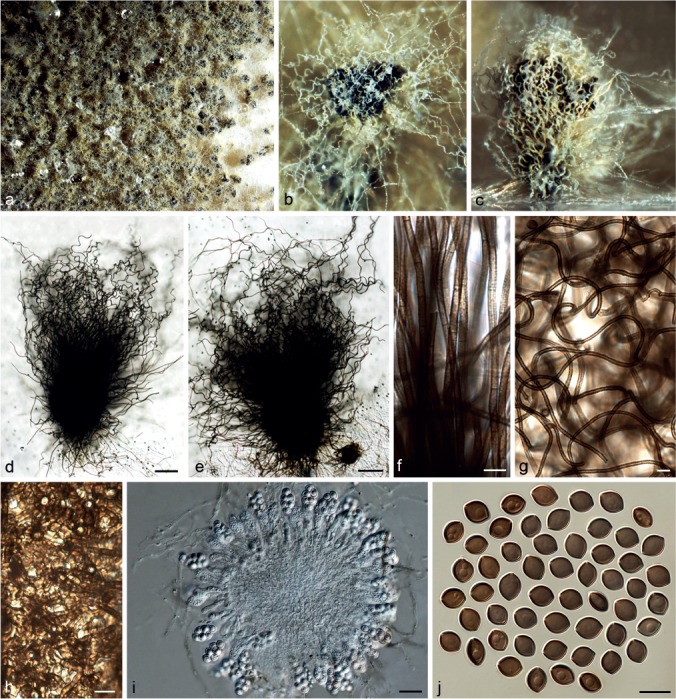
Chaetomium unguicola (CBS 128446, ex-type culture). a. Part of the colony on OA; b. ascoma and mass of ascospores on OA, top view; c. ascoma and mass of ascospores on OA, side view; d, e. ascomata mounted in lactic acid; f. basal parts of terminal ascomatal hairs; g. upper parts of terminal ascomatal hairs; h. structure of ascomatal wall in surface view; i. asci; j. ascospores. — Scale bars: d, e = 100 μm; f–h, j = 10 μm; i = 20 μm.
Etymology. Refers to a nail of Homo sapiens, the substrate from which this fungus was isolated.
Ascomata superficial or sometimes covered by sparse aerial hyphae, ostiolate, amber to citrine-green in reflected light owing to ascomatal hairs, ovate or obovate to subglobose, 170–280 μm high, 150–260 μm diam. Ascomatal wall brown, composed of amorphous or hypha-like cells, textura epidermoidea or textura intricata in surface view. Terminal hairs finely verrucose, dark olivaceous, fading towards the tips, undulate to loosely coiled with erect or flexuous lower part, 3–4.5 μm near the base, tapering towards the tips. Lateral hairs flexuous to undulate, tapering towards the tips. Asci fasciculate, fusiform or clavate, with eight biseriate or irregularly arranged ascospores, spore-bearing part 15.5–24.5 × 10–14.5 μm, stalks 11–24 μm long, evanescent. Ascospores olivaceous brown when mature, limoniform, bilaterally flattened, (7–)7.5–9 × (6–)6.5–7(–7.5) × 4.5–5.5 μm, with an apical germ pore. Asexual morph absent.
Culture characteristics — Colonies on OA with white aerial hyphae, usually not producing coloured exudates; reverse uncoloured.
Material examined. USA, Los Angeles, isolated from a nail of Homo sapiens, deposited in CBS by D.A. Sutton, 29 Sept. 2010 (holotype CBS H-22196, culture ex-type CBS 128446 = UTHSC 07-2213 = dH 21624).
Notes — Chaetomium unguicola forms a sister lineage to C. globosum (Group IIA, Fig. 1). This species is also morphologically close to C. globosum in ascomata and ascomatal hair morphology. However, C. unguicola can be distinguished by its smaller ascospores (7.5–9 × 6.5–7 × 4.5–5.5 μm vs 8.5–10.5 × 7–8 × 5.5–6.5 μm).
KEY TO SPECIES OF THE CHAETOMIUM GLOBOSUM COMPLEX
1. Parts of ascospores with more than one germ pore . . . 2
1. Ascospores with only one germ pore . . . . . . . . . 6
2. Ascospores irregularly limoniform to fusiform in front view, 12.5–26 × 11–17 × 9–12.5 μm . . C. nozdrenkoae
2. Ascospores globose to subglobose in front view . . . . 3
3. Ascospores shorter than 12.5 μm, with one or two germ pores . . . . . . . . . . . . . 4
3. Ascospores longer than 12 μm, with two or more germ pores . . . . . . . . . . . . . . . . . . . .5
4. Ascomata 350–510 × 210–350 μm, ascomatal hairs verrucose and flexuous . . . . . . . . . . . . . . . C. globosporum
4. Ascomata 230–360 × 170–240 μm, ascomatal hairs nearly smooth and undulate . . . . . . . . . C. interruptum
5. Ascospores 12–17 × 10–14 × 7.5–10.5 μm . . . . . . . . . . . . . . . . . C. megalocarpum
5. Ascospores 17–22.5 × 14.5–19 × 11–14 μm . C. grande
6. Ascospores irregular fusiform, limoniform, ovate, lunate or triangular in front view, 7–12 × 4–7 × 4–5.5 μm . . . . . . . . . . . . . . . . . . . . C. citrinum
6. Ascospores typically limoniform to broad fusiform in front view . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 7
7. Ascomata non-ostiolate . . . . . . . . . . . . . 8
7. Ascomata ostiolate . . . . . . . . . . . . . . . . . . . . 10
8. Ascomata with only short, hypha-like hairs; ascospores umbonate at both ends, 11–16 × 9–11 × 6–8 μm . . . . . . . . . . . . . . . C. pilosum
8. Ascomata with longer and shorter types of hair; ascospores biapiculate . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 9
9. Ascomata 320–500 μm diam; ascospores 11–16 × 9–11 × 6–8.5 μm . . . . . . . . . . . . . . . . C. fimeti
9. Ascomata 170–360 μm diam; ascospores 8–10 × 6.5–8 × 5.5–6.5 μm . . . . . . . C. subfimeti
10. Parts of ascospores with a lateral bulge . . . . . . . 11
10. Ascospores without a lateral bulge . . . . . . . . . 12
11. Ascospores 10–11.5 × 8–10 × 7.5–9 μm; terminal ascomatal hairs coiled . . . . C. madrasense
11. Ascospores 8.5–11 × 8–10 × 6–7.5 μm; terminal ascomatal hairs flexuous or irregularly branched . C. ascotrichoides
12. Terminal ascomatal hairs repeatedly dichotomously branched; usually with acremonium-like asexual morphs . . . . 13
12. Terminal ascomatal hairs not repeatedly dichotomously branched . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 14
13. Terminal hairs 4.5–7 μm diam near the base; ascospores 9–12 × 7–9 × 6–8 μm . . . . . . . C. rectangulare
13. Terminal hairs not more than 4.5 μm diam near the base; 11–14 × 9–11 × 6–9 μm . . . . . . . . . . C. elatum
14. Possessing regularly coiled terminal ascomatal hairs . 15
14. Terminal ascomatal hairs erect, flexuous, undulate to only slightly or loosely coiled . . . . . . . . . . . . . 19
15. With acremonium-like asexual morphs; parts of terminal hairs longer, 5–7 μm diam at base, parts of terminal hairs shorter, 3–5 μm diam near the base; ascospores 9–12 × 7.5–9 × 5.5–7 μm . . . . . . . . . C. angustispirale
15. Asexual morph absent . . . . . . . . . . . 16
16. Terminal ascomatal hairs nearly isodiametric from base towards tip, 3–4.5 μm diam, coiled in the upper part with coils equal in diameter; ascospores 9–12 × 7.5–9.5 × 5.5–7 μm . . . . . . . . . . . . . . C. spirochaete
16. Terminal ascomatal hairs tapering towards the tips . . 17
17. Terminal ascomatal hairs 5–8 μm diam near the base, with a rigid seta-like extension occurring at the tips of coiled hairs; ascospores 9–15 × 7–10 × 5.5–7 μm . . . C. spiculipilium
17. Terminal ascomatal hairs less than 6 μm diam near the base, without a rigid seta-like tip extension; ascospores 8–11.5 × 7–9 × 5–7 μm . . . . . . . . . . . . . 18
18. Ascospores biapiculate; the hairs terminal 3.5–6 μm near the bases and the coiled upper part appears as an elongate cone with coils tapering in diameter . . . . . C. cochliodes
18. Ascospores usually umbonate; parts of the terminal hairs 4–6 μm near the base, with circinate or coiled upper part; parts 3–4 μm near the base, flexuous . . . . . . . . . . . . . . . . . . . . . . . . . . . . . C. pseudocochliodes
19. Terminal ascomatal hairs less than 3 μm diam near the base, smooth; ascospores prominently umbonate, 7.5–12 × 5–7.5 × 3.5–5.5 μm . . . C. umbonatum
19. Terminal ascomatal hairs more than 3 μm diam near the base, verrucose; ascospores usually biapiculate . . . . 20
20. Ascospores broad limoniform to nearly globose . . . . . . 21
20. Ascospores not as above . . . . . . . . . . . . . 22
21. Terminal ascomatal hairs flexuous; rhizoidal structure thriving and dense; ascospores 11–14 × 10–13.5 × 7– 9 μm . . . . . . . . . . . . . . . . . C. subglobosum
21. Terminal ascomatal hairs undulate to slightly coiled; rhizoids sparser; ascospores 9.5–11.5 × 9–10.5 × 6.5 – 8 μm . . . . . . . . . . . . . . . . . C. coarctatum
22. Terminal hairs forming a dense, nearly globose head covering over the ascomatal ostiole . . . . . . . . . . . 23
22. Terminal hairs not as above . . . . . . . . . 24
23. Ascospores 6.5–8 × 5–6.5 × 4–5 μm . . . C. afropilosum
23. Ascospores 9–10.5 × 6–8 × 5–6.5 μm . . . . . . . . . . . . . . . C. pseudoglobosum
24. Ascospores elongate limoniform to broadly fusiform . 25
24. Ascospores typically limoniform . . . . . . . . . . . . . . . . . 26
25. Ascospores 9–15 × 6–8.5 × 5–6 μm . . . . C. telluricola
25. Ascospores 7.5–10.5 × 6–7.5 × 4.5–5.5 μm . . . C. tenue
26. Ascospores shorter than 11.5 μm and narrower than 8.5 μm in front view . . . . . . . . . . . . . . 27
26. Ascospores up to 12.5 μm or longer and up to 9.5 μm or wider in front view . . . . . . . . . . . . 30
27. Ascospores 7–9 × 6–7.5 × 4.5–5.5 μm . . . . C. unguicola
27. Ascospores 8–11.5 × 7–8.5 × 5–7 μm . . . . . . . . . . . . . . . . . (C. globosum s.str.) 28
28. Ascomatal wall translucent; ascomatal hairs hypha-like, sparse and hyaline; ascospore mass rust to salmon in reflected light . . . . . ‘cruentum’ morphological form of C. globosum s.str.
28. Ascomatal wall and ascomatal hairs not as above; ascospore mass black in reflected light . . . . . . . . . . . . . . . 29
29. Terminal hairs erect to flexuous, 2.5–3.5 μm diam near base . . . . . . . . . ‘rectum’-like C. globosum s.str.
29. Terminal hairs flexuous, undulate to slightly coiled, 3–5 μm diam near base . . C. globosum s.str.
30. Aerial hyphae abundant and often cover ascomata; asexual morph acremonium-like; ascomatal hairs erect to flexuous; ascospores 10.5–14 × 8.5–10.5 × 6–8 μm . C. subaffine
30. Aerial hyphae sparse or lacking; asexual morph absent . . . . . . . . . . . . .31
31. Ascospores 12–13.5 × 8–10 × 6–7.5; ascomatal hairs flexuous, undulate to loosely coiled . . . .C. undulatulum
31. Ascospores shorter than 12.5 μm . . . .32
32. Terminal hairs relatively sparse, luteous to amber or citrine in reflected light, erect or flexuous; ascospores 9.5–12 × 9–10 × 5.5–7 μm . . . . . . . . . . C. graminiforme
32. Terminal hairs abundant, slightly pale olivaceous buff in reflected light, undulate or flexuous; ascospores 10–12.5 × 8–9 × 6–7 μm . . . . . . . . . . . . C. olivaceum
DISCUSSION
The ever-increasing realisation of the importance of C. globosum and its close relatives requires the clarification of their species concepts. The broad species concept of C. globosum sensu Von Arx has resulted in extensive arguments (Seth et al. 1987, Asgari & Zare 2011, Doveri 2013). The inconsistency in species delimitation for C. globosum sensu Von Arx has constantly limited our understanding of its metabolism, function and importance. Based on phylogenetic inference of the ITS, LSU and tub2 gene regions, Asgari & Zare (2011) proposed a C. globosum species group similar to that of Dreyfuss (1976), which grouped into three clades: the C. elatum, C. globosum and C. megalocarpum clades. Eleven species were included in their C. globosum species group, although only three isolates of the broad C. globosum sensu Von Arx, i.e. CBS 162.62, CBS 371.66 and CBS 148.51, were treated. These data indicated some phylogenetic relationships between C. globosum and other Chaetomium species, but failed to resolve the boundaries of C. globosum sensu Von Arx and allied species. Based on phylogenetic inference of the LSU, tub2 and rpb2 gene regions, Greif et al. (2009) re-evaluated the genus Chaetomidium and indicated that this genus is polyphyletic. For all three loci analysed in that study, eight of the nine examined species were interspersed among species of Chaetomium, Farrowia and Thielavia within the Chaetomiaceae, whereas Chd. triangulare fell outside the Chaetomiaceae. Greif et al. (2009) failed to resolve the phylogenetic placement of most of the Chaetomidium species due to the limited sampling in the family. However, the rpb2 phylogenetic inference in that study clearly showed that Chd. fimeti, Chd. subfimeti, Chd. pilosum, C. elatum and C. globosum formed a strongly supported clade. Their molecular evidence apparently disagreed with their own suggestion to restrict Chaetomidium to its type species, Chd. fimeti, and Chd. subfimeti.
After a preliminary screening of the isolates preserved at CBS, and the isolates collected from diverse substrates in China, using partial rpb2 and tub2 gene sequences, 80 isolates representing the morphological diversity of C. globosum and related species were selected as representatives for further study. This revealed a much more expanded C. globosum complex than that of Dreyfuss (1976) and Asgari & Zare (2011). Thirty-six species were recognised in this complex, which grouped into two main clades representing three Groups. Chaetomidium fimeti, Chd. subfimeti and Chd. pilosum were shown to belong to the C. globosum species complex and cluster in two different groups. Chaetomidium triangulare clustered outside the Chaetomiaceae, while the other available Chaetomidium species were interspersed throughout the Chaetomiaceae (data not shown). In addition, many studies have indicated that fungi with cleistothecial ascomata represent a heterogeneous assemblage that evolved independently on different occasions from diverse ascomycetes (Berbee & Taylor 1992, Suh & Blackwell 1999, Stchigel & Guarro 2007).
The first main clade (Group I, Fig 1) resolved here corresponded to the C. megalocarpum clade of Asgari & Zare (2011). This Group includes several species characterised by distinct morphological features. The four species that sporulated in culture from Group IA (C. globosporum, C. grande, C. megalocarpum and C. nozdrenkoae) produce ascospores with more than one germ pore but vary in size and shape. Chaetomium madrasense and C. ascotrichoides (Group IB) are distinguished by broad limoniform ascospores with a lateral bulge. Group IC, which includes C. fimeti and C. subfimeti, is characterised by non-ostiolate ascomata possessing typical limoniform ascospores. Chaetomium interruptum, distinguished by its globose ascospores with one or two germ pores, forms a basal lineage to Group I. Except for those of C. fimeti and C. subfimeti, ascomatal hairs of species in Group I appeared typical ‘globosum-like’: flexuous, undulate to loosely coiled.
Chaetomium citrinum, a distinct species characterised by irregular ascospores, forms a basal lineage of the second main clade (Group II & III, Fig 1) in the C. globosum species complex. Group II corresponds to the C. globosum clade of Asgari & Zare (2011), which is characterised by relatively small and typical limoniform ascospores and flexuous to undulate or slightly coiled terminal ascomatal hairs. Chaetomium coarctatum can be distinguished by relatively large and broad limoniform to nearly globose and biapiculate ascospores and forms a basal lineage to Group III. Group III corresponds to the C. elatum clade of Asgari & Zare (2011) and includes 16 species, which are characterised by larger ascospores than Group II. These species exhibit a diverse morphology of terminal ascomatal hairs ranging from flexuous or undulate (C. graminiforme, C. olivaceum, C. subaffine, C. subglobosum, C. telluricola and C. undulatulum) or regularly coiled (C. cochliodes, C. cryptocochliodes, C. spiculipilium and C. spirochaete) to repeatedly dichotomously branched (C. elatum and C. rectangulare). Chaetomium pilosum, a species previously placed in the genus Chaetomidium, is characterised by non-ostiolate ascomata, and also forms part of Group III.
The C. globosum complex is shown to be monophyletic and includes a high diversity of morphological characters in the Chaetomiaceae: ascomata are ostiolate or non-ostiolate; the morphology of the ascomatal hairs embraces nearly all types in the family, ranging from hypha-like, flexuous, undulate, coiled to simply or dichotomously branched, with verrucose to smooth surface and pale to dark in colour; ascospores can be limoniform or globose to strongly irregular with one or two (occasionally three or even four) apical, subapical or lateral germ pores. The ascospores of all species in this group are bilaterally flattened. The acremonium-like asexual morph is only known for four species (C. angustispirale, C. elatum, C. rectangulare and C. subaffine) in this complex. We can, however, define this complex with the following morphological features: ascomata globose, ellipsoid to ovate or obovate, ostiolate or non-ostiolate; ascomatal wall, with a few exceptions (C. angustispirale, C. fimeti and C. subfimeti), composed of textura intricata or textura epidermoidea in surface view; asci clavate or fusiform with eight biseriate (or irregularly arranged) ascospores and evanescent; ascospores limoniform, globose to irregular, bilaterally flattened and longer than 7 μm in length; asexual morphs, if present, acremonium-like.
Characteristics of ascomatal hairs were underrated by Von Arx et al. (1986) in recognition of C. globosum and close relatives. Species with erect, flexuous to undulate or even slightly (loosely and irregularly) coiled hairs are the most predominant feature in the C. globosum species complex. Among them, the occurrence of simply branched hairs together with flexuous to undulate hairs is very common in many species (C. globosum, C. graminiforme, C. grande, C. interruptum, C. megalocarpum, C. nozdrenkoae and C. subglobosum). The variation of ascomatal hairs could be used to differentiate species to an extent, such as the dichotomously branched ascomatal hairs of C. elatum and C. rectangulare and regularly (spirally) coiled ascomatal hairs of C. cochliodes, C. spiculipilium and C. spirochaete. The detailed features of the ascomatal hairs, which include diameter, appearance of the coiled portions, smooth or with surface ornamentation (verrucose, punctate or spinulose) also help to discriminate species.
There are several other lineages within the genus Chaetomium which possess limoniform and bilaterally flattened ascospores, but these taxa all produce ascomata with walls composed of well-defined textura angularis, which include C. bostrychodes, C. seminudum, C. sphaerale and C. subspirale and the close relatives of each species. Details on ascomata, ascospore sizes or asexual morph will help to further distinguish these lineages from the C. globosum species complex (Hawksworth 1975, Von Arx et al. 1986, Untereiner et al. 2001, Wang & Zheng 2005).
Untereiner et al. (2001) used the D1/D2 regions of LSU to investigate the relationships of the genera Chaetomium and Farrowia, representing the first study using DNA sequence data for the Chaetomiaceae. Greif et al. (2009) later used the LSU, tub1 and rpb2 gene regions to re-evaluate the genus Chaetomidium. Asgari & Zare (2011) used the ITS, partial LSU, and tub2 gene regions to identify new Chaetomium species from Iran. Wang et al. (2014) provided a phylogenetic re-assessment of the C. indicum species complex based on DNA sequences of the ITS, partial LSU, tub2, tef1 and rpb1 gene regions. A useful DNA barcode should have a clearly defined barcode gap between inter- and intraspecific variation and a small to non-existing overlap between the inter- and intraspecific frequency distance distributions to ensure the adequate species resolution or a high probability of correct identification (Schoch et al. 2012). In the present study, similar tree topologies were produced for the four protein-coding gene regions, suggesting that the efficiency of each locus as possible secondary DNA barcode(s) for the C. globosum species complex could be evaluated. Using the Kimura-2-parameter analysis, the D1/D2 region of LSU showed the lowest species-level resolution. The ITS region, the standard DNA barcode marker for the Kingdom Fungi (Schoch et al. 2012), was also shown to be unreliable for species identification in the C. globosum species complex. Although all four protein-coding genes regions used in this study provided sufficient resolution for species delimitation in the C. globosum species complex, the tub2 gene region provided the best species resolution, closely followed by the rpb2, which also amplified easier across the family than rpb1. Therefore, a two-marker system including the tub2 and rpb2 genes is suggested here as secondary DNA barcode for the C. globosum species complex.
Chaetomium globosum is known as one of the causal agents of human onychomycosis (Naidu et al. 1991, Stiller et al. 1992, Aspiroz et al. 2007, Latha et al. 2010, Tullio et al. 2010, Hubka et al. 2011, Hwang et al. 2012, Lagacé & Cellier 2012, Kim et al. 2013) and skin infection of other animals (Sugiyama et al. 2008). However, whether this species and close relatives can cause systemic and deep infections remains controversial (Hoppin et al. 1983, Abbott et al. 1995, Yeghen et al. 1996, Lesire et al. 1999, Barron et al. 2003, Paterson et al. 2005, De Hoog et al. 2013). A single isolate from a clinical case of fatal brain abscess was originally identified as C. globosum (Anandi et al. 1989). Abbott et al. (1995) later re-classified this isolate as C. atrobrunneum based on morphology and ability to grow at 42 °C and suggested that infections by C. globosum are confined to cooler areas of the human body due to restricted growth at 37 °C. Growth response of a fungal species at 37 °C is used as an indicator of its potential for internal infection of humans (Abbott et al. 1995, Barron et al. 2003). In another study, Yeghen et al. (1996) reported that C. globosum caused fatal pneumonia in a patient with acute myeloid leukemia. Paterson et al. (2005) supported this diagnosis, using Southern hybridization and 18S rRNA (SSU) gene sequences. However, their data can only verify that the infection was not caused by an Aspergillus species. In this study we determined MGT for all isolates of the 17 species selected in the C. globosum species complex. Only isolates of C. globosporum, C. megalocarpum and C. subaffine can grow at 37–38 °C, whereas the growth of the other species, including C. globosum s.str., is restricted at 37–38 °C. More research, however, is required to clarify the adaptation of C. globosum and allied species to human bodies.
The present study provides both molecular and morphological knowledge for each species presently known in the C. globosum species complex, highlighting the importance of correct identification for especially medical cases. This study provides a phylogenetic backbone and framework for future studies of the genus Chaetomium. Further studies are presently underway to ascertain a definite position of the C. globosum species complex in the genus, using a wider sampling of relevant taxa.
Acknowledgments
We are very grateful to New York Botanical Garden Herbarium and Herbarium Genavense (Switzerland) for the loan of the holotypes, and also to Dr Cornelia Dilger-Endrulat (Herbarium Tubingense, Germany) for providing information about herbaria. We acknowledge the CBS-KNAW Fungal Collection for providing cultures, and the Alfred Sloan Foundation Programme on the Microbiology of the Built Environment for partially supporting the senior author. This work was jointly supported by the National Natural Science Foundation of China (Project No. 30570007) and the Ministry of Science and Technology of P.R. China (No. 2006FY120100).
REFERENCES
- Abbott SP, Sigler L, McAleer R, et al. 1995. Fatal cerebral mycoses caused by the ascomycete Chaetomium strumarium. Journal of Clinical Microbiology 33: 2692–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahammed SK, Aggarwal R, Kapoor HC. 2008. Production, partial purification and characterization of extracellar xylanase from Chaetomium globosum. Journal of Plant Biochemistry & Biotechnology 17: 95–98. [Google Scholar]
- Ainsworth GC. 1961. Ainsworth & Bisby’s Dictionary of the Fungi, 5th ed. CAB International, Wallingford. [Google Scholar]
- Ainsworth GC. 1971. Ainsworth & Bisby’s Dictionary of the Fungi, 6th ed. CAB International, Wallingford. [Google Scholar]
- Ames LM. 1950. New species of cellulose destroying fungi II. Mycologia 42: 642–645. [Google Scholar]
- Ames LM. 1963. A monograph of the Chaetomiaceae. U.S. Army Research and Development, Series 2. [Google Scholar]
- Anandi V, John TJ, Walter A, et al. 1989. Cerebral phaeohyphomycosis caused by Chaetomium globosum in a renal transplant recipient. Journal of Clinical Microbiology 27: 2226–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen B, Frisvad JC, Søndergaard I, et al. 2011. Associations between fungal species and water-damaged building materials. Applied and Environmental Microbiology 77: 4180–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetrei IC, Draganesc GE, Popescu IT, et al. 2009. Possible cause of allergy for the librarians: books manipulation and ventilation as sources of fungus spores spreading. Aerobiologia 25: 159–166. [Google Scholar]
- Asgari B, Zare R. 2011. The genus Chaetomium in Iran, a phylogenetic study including six new species. Mycologia 103: 863–882. [DOI] [PubMed] [Google Scholar]
- Aspiroz C, Gene J, Rezusta A, et al. 2007. First Spanish case of onychomycosis caused by Chaetomium globosum. Medical Mycology 45: 279–282. [DOI] [PubMed] [Google Scholar]
- Awad NE, Kassem HA, Hamed MA, et al. 2014. Bioassays guided isolation of compounds from Chaetomium globosum. Journal de Mycologie Médicale 24: e35–e42. [DOI] [PubMed] [Google Scholar]
- Ayanbimpe GM, Wapwera SD, Kuchin D. 2010. Indoor air mycoflora of residential dwellings in Jos metropolis. African Health Sciences 10: 172–176. [PMC free article] [PubMed] [Google Scholar]
- Bainier G. 1910. Monographie des Chaetomidium et des Chaetomium. Bulletin de la Société Mycologique de France 25: 191–237. [Google Scholar]
- Barron MA, Sutton DA, Veve R, et al. 2003. Invasive mycotic infections caused by Chaetomium perlucidum, a new agent of cerebral phaeohyphomycosis. Journal of Clinical Microbiology 41: 5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbee ML, Taylor JW. 1992. Convergence in ascospore discharge mechanism among Pyrenomycete fungi based on 18S ribosomal RNA gene sequence. Molecular Phylogenetics and Evolution 1: 59–71. [DOI] [PubMed] [Google Scholar]
- Carter A. 1982. A taxonomic study of the ascomycete genus Chaetomium Kunze. PhD thesis, University of Toronto, Canada. [Google Scholar]
- Chivers AH. 1915. A monograph of the genera Chaetomium and Ascotricha. Memoirs of the Torrey Botanical Club 14: 155–240. [Google Scholar]
- Corda ACJ. 1840. Icones fungorum hucusque cognitorum 4: 1–53. [Google Scholar]
- Crous PW, Gams W, Stalpers JA, et al. 2004. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ, et al. 2009. Fungal Biodiversity. CBS Laboratory Manual Series 1. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands. [Google Scholar]
- Cunningham CW. 1997. Can three incongruency tests predict when data should be combined? Molecular Biology and Evolution 14: 733–740. [DOI] [PubMed] [Google Scholar]
- Damm U, Mostert L, Crous PW, et al. 2008. Novel Phaeoacremonium species associated with necrotic wood of Prunus trees. Persoonia 20: 87–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoog GS, Ahmed SA, Najafzadeh MJ, et al. 2013. Phylogenetic findings suggest possible new habitat and routes of infection of human eumyctoma. PLoS Neglected Tropical Diseases 7: e2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G, Song YC, Chen JR, et al. 2006. Chaetoglobosin U, a cytochalasan alkaloid from endophytic Chaetomium globosum IFB-E019. Journal of Natural Products 69: 302–304. [DOI] [PubMed] [Google Scholar]
- Doveri F. 2013. An additional update on the genus Chaetomium with descriptions of two coprophilous species, new to Italy. Mycosphere 4: 820–846. [Google Scholar]
- Dreyfuss M. 1976. Taxonomische Untersuchungen innerhalb der Gattung Chaetomium. Sydowia 28: 50–133. [Google Scholar]
- El-Gindy AA, Saad RR, Fawzi E. 2003. Purification and some properties of exo-1,4-beta-glucanase from Chaetomium olivaceum. Acta Microbiologica Polonica 52: 35–44. [PubMed] [Google Scholar]
- Fries E. 1849. Summa vegetabilium scandinaviae. Stockholm & Leipzig. [Google Scholar]
- Fuckel I. 1869. Symbolae mycologicae. Wiesbaden. [Google Scholar]
- Ge HM, Zhang WY, Ding G, et al. 2008. Chaetoglobins A and B, two unusual alkaloids from endophytic Chaetomium globosum culture. Chemical Communications 45: 5978–5980. [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes RR, Glienke C, Videira SIR, et al. 2013. Diaporthe: a genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31: 1–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonianakis M, Neonakis I, Darivianaki E, et al. 2005. Airborne ascomycotina on the island of Crete: Seasonal patterns based on an 8-year volumetric survey. Aerobiologia 21: 69–74. [Google Scholar]
- Greif MD, Currah RS. 2007. Development and dehiscence of the cephalothecoid peridium in Aporothielavia leptoderma shows it belongs in Chaetomidium. Mycological Research 111: 70–77. [DOI] [PubMed] [Google Scholar]
- Greif MD, Stchigel AM, Huhndorf SM. 2009. A re-evaluation of genus Chaetomidium based on molecular and morphological characters. Mycologia 101: 554–564. [DOI] [PubMed] [Google Scholar]
- Gueidan C, Roux C, Lutzoni F. 2007. Using multigene phylogeny analysis to assess generic delineation and character evolution in Verrucariaceae (Verrucariales, Ascomycota). Mycological Research 111: 1145–1168. [DOI] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hawksworth DL. 1975. Farrowia, a new genus in the Chaetomiaceae. Persoonia 8: 167–185. [Google Scholar]
- Hawksworth DL, Wells H. 1973. Ornamentation on the terminal hairs in Chaetomium Kunze ex Fr. and some allied genera. Mycological Papers 134: 1–24. [Google Scholar]
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method of assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. [Google Scholar]
- Hoppin EC, McCoy EL, Rinaldi MG. 1983. Opportunistic mycotic infection caused by Chaetomium in a patient with acute leukemia. Cancer 52: 555–556. [DOI] [PubMed] [Google Scholar]
- Hubka V, Mencl K, Skorepova M, et al. 2011. Phaeohyphomycosis and onychomycosis due to Chaetomium spp., including the first report of Chaetomium brasiliense infection. Medical Mycology 49: 724–733. [DOI] [PubMed] [Google Scholar]
- Hwang SM, Suh MK, Ha GY. 2012. Onychomycosis due to nondermatophytic molds. Annals of Dermatology 24: 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewchai S, Soytong K, Hyde KD. 2009. Mycofungicides and fungal biofertilizers. Fungal Diversity 38: 25–50. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharwar RN, Mishra A, Gond SK, et al. 2011. Anticancer compounds derived from fungal endophytes: their importance and future challenges. Natural Product Reports 28: 1208–1228. [DOI] [PubMed] [Google Scholar]
- Kim BS, Hwang BK. 2007. Microbial fungicides in the control of plant diseases. Journal of Phytopathology 155: 641–653. [Google Scholar]
- Kim DM, Lee MH, Suh MK, et al. 2013. Onychomycosis caused by Chaetomium globosum. Annals of Dermatology 25: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopytina NI. 2005. Distribution of the fungi from the genus Chaetomium (Ascomycota) in north-western part of the Black Sea. Mikologiya I Fitopatologiya 39: 12–18. [Google Scholar]
- Kumar S, Kaushik N, Proksch P. 2013. Identification of antifungal principle in the solvent extract of an endophytic fungus Chaetomium globosum from Withania somnifera. SpringerPlus 2: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Schmidt JK. 1817. Mykologische Hefte 1. Leipzig, Germany. [Google Scholar]
- Lagacé J, Cellier E. 2012. A case report of a mixed Chaetomium globosum/Trichophyton mentagrophytes onychomycosis. Medical Mycology Case Reports 1: 76–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latha R, Sasikala R, Muruganandam N, et al. 2010. Onychomycosis due to ascomycete Chaetomium globosum: A case report. Indian Journal of Pathology and Microbiology 53: 566–567. [DOI] [PubMed] [Google Scholar]
- Lesire V, Hazouard E, Dequin PF, et al. 1999. Possible role of Chaetomium globosum in infection after autologous bone marrow transplantation. Intensive Care Medicine 25: 124–125. [DOI] [PubMed] [Google Scholar]
- Lombard L, Crous PW, Wingfield BD, et al. 2010. Phylogeny and systematics of the genus Calonectria. Studies in Mycology 66: 31–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longoni P, Rodolfi M, Pantaleoni L, et al. 2012. Functional analysis of the degradation of cellulosic substrates by a Chaetomium globosum endophytic isolate. Applied and Environmental Microbiology 78: 3693–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malloch D, Cain RF. 1973. The genus Thielavia. Mycologia 65: 1055–1077. [Google Scholar]
- Mason S, Cortes D, Horner WE. 2010. Detection of gaseous effluents and by-products of fungal growth that affect environments (RP-1243). HVAC & R Research 16: 109–121. [Google Scholar]
- Mason-Gamer R, Kellogg E. 1996. Testing for phylogenetic conflict among molecular datasets in the tribe Tiriceae (Graminae). Systematic Biology 45: 524–545. [Google Scholar]
- Matheny PB, Liu YJ, Ammirati JF, et al. 2002. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). American Journal of Botany 89: 688–698. [DOI] [PubMed] [Google Scholar]
- McNeill J, Barrie FR, Buck WR, et al. 2012. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code) adopted by the Eighteenth International Botanical Congress Melbourne, Australia, July 2011. Koeltz Scientific Books, Koenigstein. [Google Scholar]
- Miller AN, Huhndorf SM. 2005. Multi-gene phylogenies indicate ascomal wall morphology is a better predictor of phylogenetic relationships than ascospore morphology in the Sordariales (Ascomycota, Fungi). Molecular Phylogenetics and Evolution 35: 60–75. [DOI] [PubMed] [Google Scholar]
- Miller JD, McMullin DR. 2014. Fungal secondary metabolites as harmful indoor air contaminants: 10 years on. Applied Microbiology and Biotechnology 98: 9953–9966. [DOI] [PubMed] [Google Scholar]
- Millner PD. 1977. Radial growth responses to temperature by 58 Chaetomium species, and some taxonomic relationships. Mycologia 69: 492–502. [Google Scholar]
- Millner PD, Motta JJ, Lentz PL. 1977. Ascospores, germ pores, ultrastructure, and thermophilism of Chaetomium. Mycologia 69: 720–733. [Google Scholar]
- Momesso LD, Kawano CY, Ribeiro PH, et al. 2008. Chaetoglobosins produced by Chaetomium globosum, endophytic fungus found in association with Viguiera robusta Gardn (Asteraceae). Química Nova 31: 1680–1685. [Google Scholar]
- Naidu J, Singh SM, Pouranik M. 1991. Onychomycosis caused by Chaetomium globosum Kunze. Mycopathologia 113: 31–34. [DOI] [PubMed] [Google Scholar]
- Nylander JAA. 2004. MrModeltest v. 2. Programme distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- O’Donnell K. 1993. Fusarium and its near relatives. In: Reynolds R, Taylor JW. (eds), The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics: 225–233. CAB International, Wallingford. [Google Scholar]
- O’Donnell K, Cigelnik E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. [DOI] [PubMed] [Google Scholar]
- Park J-H, Choi GJ, Jang KS, et al. 2005. Antifungal activity against plant pathogenic fungi of chaetoviridins isolated from Chaetomium globosum. FEMS Microbiology Letters 252: 309–313. [DOI] [PubMed] [Google Scholar]
- Paterson PJ, Seaton S, Yeghen T, et al. 2005. Molecular confirmation of invasive infection caused by Chaetomium globosum. Journal of Clinical Pathology 58: 334. [PMC free article] [PubMed] [Google Scholar]
- Phonkerd N, Kanokmedhakul S, Kanokmedhakul K, et al. 2008. Bis-spiro-azaphilones and azaphilones from the fungi Chaetomium cochliodes VTh01 and C. cochliodes CTh05. Tetrahedron 64: 9636–9645. [Google Scholar]
- Polizzi V, Delmulle B, Adams A, et al. 2009. JEM Spotlight: Fungi, mycotoxins and microbial volatile organic compounds in mouldy interiors from water-damaged buildings. Journal of Environmental Monitoring 11: 1849–1858. [DOI] [PubMed] [Google Scholar]
- Prokhorov VP, Linnik MA. 2011. Morphological, cultural, and biodestructive peculiarities of Chaetomium species. Moscow University Biological Sciences Bulletin 66: 95–101. [Google Scholar]
- Rambaut A. 2009. FigTree v. 1.3.1. Computer program and documentation distributed by the author at http://tree.bio.ed.ac.uk/software/. [Google Scholar]
- Rayner RW. 1970. A mycological colour chart. CMI and British Mycological Society; Kew, Surrey, England. [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Saccardo PA. 1882. Sylloge Pyrenomycetum. Sylloge Fungorum 1: 1–768. [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, et al. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the USA 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekita S, Yoshihira K, Natori S, et al. 1981. Mycotoxin production by Chaetomium spp. and related fungi. Canadian Journal of Microbiology 27: 766–772. [DOI] [PubMed] [Google Scholar]
- Seth HK. 1967. Chaetomidium subfimeti sp. nov. from Wales. Transactions of the British Mycological Society 50: 45–47. [Google Scholar]
- Seth HK. 1970. A monograph of the genus Chaetomium. Beihefte zur Nova Hedwigia 37: 1–133. [Google Scholar]
- Seth HK, Chen QT, Chen YD. 1987. A view on the retention of Chaetomium globosum Kze. and C. olivaceum Cooke & Ellis as separate species. Acta Mycologica Sinica 6: 82–85. [Google Scholar]
- Shanthiyaa V, Saravanakumar D, Rajendran L, et al. 2013. Use of Chaetomium globosum for biocontrol of potato late blight disease. Crop Protection 52: 33–38. [Google Scholar]
- Sharma S, Aggarwal R, Yadav A, et al. 2014. Protein mapping of Chaetomium globosum, a potential biological control agent through proteomics approach. Journal of Plant Biochemistry and Biotechnology 23: 284–292. [Google Scholar]
- Singh RK, Tiwari MK, Kim D, et al. 2013. Molecular cloning and characterization of a GH11 endoxylanase from Chaetomium globosum, and its use in enzymatic pretreatment of biomass. Applied Microbiology and Biotechnology 97: 7205–7214. [DOI] [PubMed] [Google Scholar]
- Skolko AJ, Groves JW. 1953. Notes on seed-borne fungi VII. Chaetomium. Canadian Journal of Botany 31: 779–809. [Google Scholar]
- Sörgel G. 1960. Zum problem der trennung von arten bei pilzen, dargestellt am beispiel der ascomycetengattung Chaetomium. Archives of Microbiology 36: 51–66. [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Stchigel AM, Guarro J. 2007. A reassessment of cleistothecia as a taxonomic character. Mycological Research 111: 1100–1115. [DOI] [PubMed] [Google Scholar]
- Stchigel AM, Guarro J, Jato V, et al. 2004. Two new species of Chaetomidium (Sordariales). Studies in Mycology 50: 215–220. [Google Scholar]
- Stiller MJ, Rosenthal S, Summerbell RC, et al. 1992. Onychomycosis of the toenails caused by Chaetomium globosum. Journal of the American Academy of Dermatology 26: 775–776. [DOI] [PubMed] [Google Scholar]
- Sugiyama K, Sano A, Murakami M, et al. 2008. Three isolations of Chaetomium globosum from erythematous epilation of canine skin. Medical Mycology 46: 505–510. [DOI] [PubMed] [Google Scholar]
- Suh S-O, Blackwell M. 1999. Molecular phylogeny of the cleistothecial fungi placed in Cephalothecaceae and Pseudeurotiaceae. Mycologia 91: 836–848. [Google Scholar]
- Swofford DL. 2003. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Massachusetts, USA. [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschudy RH. 1937. Experimental morphology of some species of Chaetomium I. Use of cultural reactions in determining species characteristics. American Journal of Botany 24: 472–480. [Google Scholar]
- Tullio V, Banche G, Allizond V, et al. 2010. Non-dermatophyte moulds as skin and nail foot mycosis agents: Phoma herbarum, Chaetomium globosum and Microascus cinereus. Fungal Biology 114: 345–349. [DOI] [PubMed] [Google Scholar]
- Udagawa S. 1960. A taxonomic study on the Japanese species of Chaetomium. The Journal of General and Applied Microbiology 6: 223–251. [Google Scholar]
- Udagawa S, Muroi T, Kurata H, et al. 1979. The production of chaetoglobosins, sterigmatocystin, O-methylsterigmatocystin, and chaetocin by Chaetomium spp. and related fungi. Canadian Journal of Microbiology 25: 170–177. [DOI] [PubMed] [Google Scholar]
- Umikalsom MS, Ariff AB, Hassan MA, et al. 1998. Kinetics of cellulose production by Chaetomium globosum at different levels of dissolved oxygen tension using oil palm empty fruit bunch fibre as substrate. World Journal of Microbiology and Biotechnology 14: 491–498. [Google Scholar]
- Untereiner WA, Débois V, Naveau FA. 2001. Molecular systematics of the ascomycete genus Farrowia (Chaetomiaceae). Canadian Journal of Botany 79: 321–333. [Google Scholar]
- Vesper S, McKinstry C, Ashley P, et al. 2007. Quantitative PCR analysis of molds in the dust from homes of asthmatic children in North Carolina. Journal of Environmental Monitoring 9: 826–830. [DOI] [PubMed] [Google Scholar]
- Von Arx JA. 1975. On Thielavia and some similar genera of ascomycetes. Studies in Mycology 8: 1–32. [Google Scholar]
- Von Arx JA, Dreyfuss M, Müller E. 1984. A re-evaluation of Chaetomium and Chaetomiaceae. Persoonia 12: 169–179. [Google Scholar]
- Von Arx JA, Guarro J, Figueras MJ. 1986. The Ascomycete genus Chaetomium. Beihefte zur Nova Hedwigia 84: 1–162. [Google Scholar]
- Wang XW, Wang XL, Liu FJ, et al. 2014. Phylogenetic assessment of Chaetomium indicum and allied species, with the introduction of three new species and epitypification of C. funicola and C. indicum. Mycological Progress 13: 719–732. [Google Scholar]
- Wang XW, Zheng RY. 2005. Chaetomium ampulliellum sp. nov. (Chaetomiaceae, Ascomycota) and similar species from China. Nova Hedwigia 81: 247–255. [Google Scholar]
- White TJ, Bruns T, Lee S, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR protocols: a guide to methods and applications: 315–322. New York, Academic Press, Inc. [Google Scholar]
- Winter G. 1885. Ascomyceten: Gymnoasceen und Pyrenomyceten. Rabenhorst’s Kryptogamen-Flora von Deutschland, Oesterreich und der Schweiz 1, 2. [Google Scholar]
- Yamada T, Jinno M, Kikuchi T, et al. 2012. Three new azaphilones produced by a marine fish-derived Chaetomium globosum. The Journal of Antibiotics 65: 413–417. [DOI] [PubMed] [Google Scholar]
- Yan W, Ge HM, Wang G, et al. 2014. Pictet-Spengler reaction-based biosynthetic machinery in fungi. Proceedings of the National Academy of Sciences of the United States of America 111: 18138–18143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeghen YT, Fenelon L, Campbell CK, et al. 1996. Chaetomium pneumonia in patient with acute myeloid leukaemia. Journal of Clinical Pathology 49: 184–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GZ, Wang FT, Qin JC, et al. 2013. Efficacy assessment of antifungal metabolites from Chaetomium globosum No. 05, a new biocontrol agent, against Setosphaeria turcica. Biological Control 64: 90–98. [Google Scholar]
- Zhang J, Ge HM, Jiao RH, et al. 2010. Cytotoxic chaetoglobosins from the endophyte Chaetomium globosum. Planta Medica 76: 1910–1914. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Li HQ, Zong SC, et al. 2012. Chemical and bioactive diversities of the genus Chaetomium secondary metabolites. Mini Reviews in Medical Chemistry 12: 127–148. [DOI] [PubMed] [Google Scholar]
- Zopf W. 1881. Zur Entwicklungsgeschichte der Ascomyceten: Chaetomium. Nova Acta der Kaiserlich Leopoldinisch-Carolinisch Deutschen Akademie der Naturforscher 42: 199–292. [Google Scholar]


