Abstract
Sarcomas are infrequent malignant mesenchymal neoplasms characterized by notable morphological and molecular heterogeneity. At present, the diagnosis of sarcoma is based on morphology, immunohistochemistry, and clinicopathological correlation. Molecular studies in the clinical setting provide refinements to morphologic sarcoma classification, and contribute diagnostic information (frequently), prognostic stratification (rarely) and predictive information concerning specific therapies (occasionally). In this review, we summarize the major molecular mechanisms that underlie sarcoma pathogenesis, highlighting molecular alterations that provide diagnostic, prognostic or predictive information. The discussion focuses on representative mesenchymal tumor types, following a pathogenic classification combining cytogenetic / genomic information and molecular biologic features. We will address five major molecular alterations frequent in sarcoma, including 1) the presence of chimeric transcription factors, in vascular tumors; 2) abnormal kinase signaling, in gastrointestinal stromal tumor; 3) epigenetic deregulation, either by oncometabolites, as a result of metabolic enzyme mutations, or as primary events, in chondrosarcoma, chondroblastoma, and other tumor types; 4) deregulated cell survival and proliferation, due to extreme copy number alterations, in dedifferentiated liposarcoma; and 5) extreme genomic instability in conventional osteosarcoma, as a representative example of sarcomas with highly complex karyotype.
Keywords: molecular diagnostics, molecular markers, soft tissue tumor, bone, sarcoma, GIST
Overview
Sarcomas are infrequent malignant mesenchymal neoplasms characterized by notable morphological and molecular heterogeneity. The current WHO classification recognizes over 100 soft tissue and bone tumor types, over 70 of which are sarcomas,1 illustrating a nosologic complexity that reflects biologic complexity and leads to substantial challenges in diagnosis and clinical management. Sarcoma diagnosis is based on morphology, immunohistochemistry, and clinicopathological correlation. In addition, molecular studies in the clinical setting provide refinements to morphologic sarcoma classification, and contribute diagnostic information (frequently), prognostic stratification (rarely) and predictive information concerning specific therapies (only occasionally, but most excitingly). Much of the current molecular understanding of sarcomas derives from conventional karyotypic analysis, which has been extremely fruitful in this field over the last twenty-five years.2 At the cytogenetic level, a binary distinction between sarcomas with simple karyotype versus those with complex karyotype has been long recognized, and provides a simple conceptual framework of some academic value but limited clinicopathological significance.3 The molecular correlates of these cytogenetic presentations are recurrent genomic rearrangements and activating gene mutations for sarcomas with relatively simple karyotype, and multiple, diverse genomic events including gene amplifications and non-recurrent rearrangements, for those with complex karyotype. Biologically, oncogenic mechanisms are better understood for sarcomas with simple karyotype, and fall typically into two broad categories: transcriptional deregulation or deregulated signaling. This is in contrast to sarcomas with highly complex karyotypes, which typically do not harbor single “driver” genetic alterations, and rather display non-specific molecular changes that promote oncogenic traits, such as cell cycle de-regulation or genomic instability.
In this review, we summarize the major molecular mechanisms that underlie sarcoma pathogenesis, highlighting those alterations that provide diagnostic, prognostic or predictive information (the so-called “clinically-actionable” alterations). The discussion will focus on representative mesenchymal tumor types, following a pathogenic classification combining cytogenetic / genomic information and molecular biologic features, as summarized in Table 1. We will address five major molecular alterations, including the presence of chimeric transcription factors, in vascular tumors; deregulated kinase signaling, in gastrointestinal stromal tumor (GIST); epigenetic deregulation by oncometabolites, as a result of metabolic enzyme mutations in chondrosarcoma and other tumor types; deregulated cell survival and proliferation, due to extreme copy number alterations, in dedifferentiated liposarcoma (DDLPS); and extreme genomic instability in conventional osteosarcoma, as a representative example of sarcomas with highly complex karyotype. Table 2 provides a non-comprehensive list of clinically-actionable genetic alterations commonly encountered in sarcoma.
Table 1.
Molecular genetic categories of soft tissue tumors and sarcoma
|
Table 2.
Diagnostic, Prognostic and Predictive Molecular Markers in Sarcoma
| Type of alteration | Genes | Entities | Clinical value |
|---|---|---|---|
| Recurrent rearrangements | Fusion oncogenes | Sarcoma-type specific (reviewed by Mertens)4 | Diagnostic / Prognostic |
| Point mutations or small indels | KIT / PDGFRA, SDHA / B | GIST | Predictive / Diagnostic |
| CTNNB1 | Desmoid tumor | Diagnostic | |
| IDH1, IDH2 | Enchodroma / chondrosarcoma | Diagnostic | |
| SUZ12, EED | Malignant peripheral nerve sheath tumor | Diagnostic | |
| PIK3CA | Myxoid liposarcoma | Predictive | |
| KDR | Angiosarcoma | Diagnostic | |
| NRAS, KRAS, HRAS, FGFR4 | Embryonal rhabdomyosarcoma | Diagnostic / Predictive | |
| MYOD1 | Spindle cell rhabdomyosarcoma | Diagnostic | |
| MED12 | Leiomyoma (and small subset of leiomyosarcoma) | Diagnostic | |
| NF1 | MPNST and others | Diagnostic | |
| Copy number gain / Amplification | MDM2, CDK4 | WD/DDLPS | Diagnostic |
| CDK4 | Predictive | ||
| MYC | Postradiation sarcoma | Diagnostic | |
| MYOCD | Leiomyosarcoma | Diagnostic | |
| Copy number loss / Deletion | TP53 | Osteosarcoma, leiomyosarcoma, and others | Prognostic |
| SMARCB1 | Rhabdoid tumor, epithelioid sarcoma | Diagnostic | |
| SMARCA4 | SMARCA4-deficient thoracic sarcomas | Diagnostic | |
| CDKN2A | MPNST, fibrosarcomatous DFSP, advanced GIST | Predictive | |
| RB1 | Spindle cell lipoma, and others | Diagnostic | |
| High-grade sarcomas with complex karyotype | Predictive | ||
| NF1 | MPNST and others | Diagnostic / Predictive |
Sarcomas with simple genome
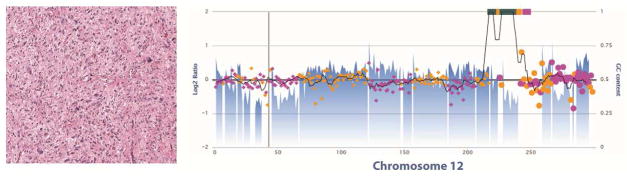
Sarcomas with simple genomic profiles usually harbor a recurrent molecular aberration, either a balanced chromosomal rearrangement or a mutation in a known oncogene or tumor suppressor gene, which is critical for tumorigenesis and is considered the main oncogenic driver. These alterations are usually present in the context of a relatively stable genome, with a low mutational load and a (near) diploid karyotype; additional point mutations or copy number alterations may occur during tumor progression, frequently following reproducible patterns, in contrast with the remarkable variability observed in genomically-complex sarcomas (Figure 1).
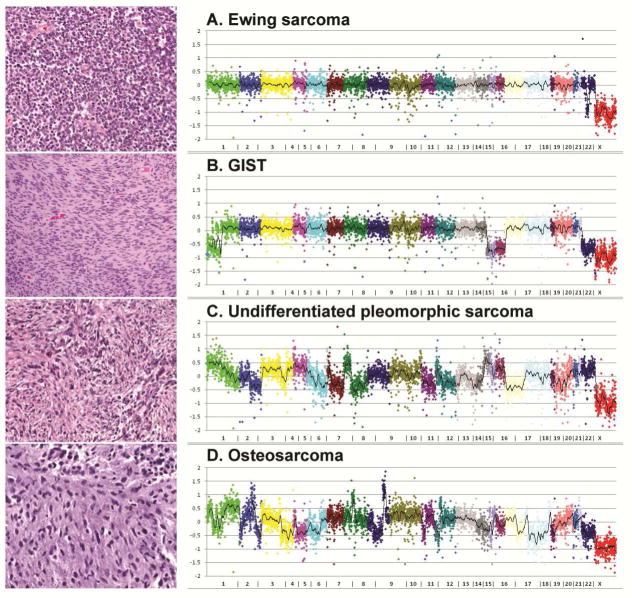
Figure 1.
Copy number profiles of sarcomas with simple genome (top) in comparison with sarcomas with complex genome (bottom), as determined by a next generation sequencing platform.118 A. Ewing sarcoma affecting a 9 year old boy. Note the simple genomic profile. This tumor harbored a EWSR1-FLI1 fusion, identified by this assay. B. High-risk, spindle cell intestinal GIST in a 60 year old male patient. The tumor harbored a KIT K642E mutation detected by the assay. Note a relatively simple genomic profile, with near-diploid karyotype and loss of chromosomes 1p, 14q, 15q and 22q, characteristic of advanced GIST. C. Undifferentiated pleomorphic sarcoma arising in the deltoid of a 55 year old man and D. Conventional osteosarcoma in the femur of a 7 year old boy: multiple chromosomal gains and losses in a non-recurrent pattern. Both these tumors showed alterations in TP53 (copy number loss and truncating mutations).
Tumors with chimeric transcription factors: vascular tumors
An expanding list of mesenchymal tumors contain recurrent balanced chromosomal rearrangements, most often translocations, and most fusion genes produced by these rearrangements encode chimeric transcription factors that cause transcriptional deregulation. The best studied example is the EWSR1-FLI1 and EWSR1-ERG fusion in Ewing sarcoma. Chimeric transcription factors are thought to deregulate the expression of specific repertoires of target genes, thereby orchestrating several of the defined “hallmarks of cancer”.5 The group of sarcomas carrying a specific translocation constituted only ~15–20% of all sarcomas in 2007.6 However, using next generation sequencing approaches detecting cryptic alterations not detectable by conventional approaches, this group is rapidly expanding. Examples of the increased resolution of current methods include the detection of fusions by a paracentric inversion in solitary fibrous tumor,7 and mesenchymal chondrosarcoma,8 both of which have provided useful diagnostic immunohistochemical or molecular markers for the diagnosis of these entities. More recently, novel gene fusions have been discovered by transcriptome analysis in a series of vascular tumors, a group of lesions characteristically difficult to classify because of overlapping morphology that expand a range of biological behavior, including clinically benign, intermediate and overtly malignant tumors.
Key features of sarcomas with simple genome
Simple karyotype, low mutational rate.
Often show relatively monomorphic morphology.
Wide range of clinical behavior.
Clinically-useful diagnostic molecular markers (amenable to detection by FISH, RT-PCR, sequencing, and occasionally immunohistochemistry).
No therapies targeting chimeric transcription factors or epigenetic alterations.
Effective targeted therapies against most deregulated kinases.
Prototypical examples: Ewing sarcoma, rhabdomyosarcoma, synovial sarcoma.
Vascular tumors of bone and soft tissue constitute a heterogeneous group of tumors displaying endothelial differentiation. Tumors range from benign (hemangioma) to intermediate (various types of hemangioendothelioma) to malignant (epithelioid hemangioendothelioma and angiosarcoma). Over the past decade, with the advance of next generation sequencing techniques, the molecular background of some of these lesions has been elucidated.
Epithelioid hemangioma (previously known as angiolymphoid hyperplasia with eosinophilia or histiocytoid hemangioma) is a benign (in soft tissue) or locally aggressive (in bone) neoplasm composed of cells that have an endothelial phenotype and epithelioid morphology.1 Transcriptome sequencing of epithelioid hemangioma revealed a recurrent translocation breakpoint involving the FOS gene fused to different partners.9,10 The break was observed in exon 4 of the FOS gene and the fusion event led to the introduction of a STOP codon, truncating the FOS protein and resulting in loss of the Trans-Activation Domain.9 Atypical variants of epithelioid hemangioma were shown to harbor ZFP36-FOSB fusions.11 The distinction between epithelioid hemangioma and angiosarcoma can be challenging, and detection of FOS rearrangements may assist in the differential diagnosis.9,10,12 Treatment with curettage or marginal en bloc excision is usually sufficient for epithelioid hemangioma, while patients with angiosarcoma need more aggressive treatment.13
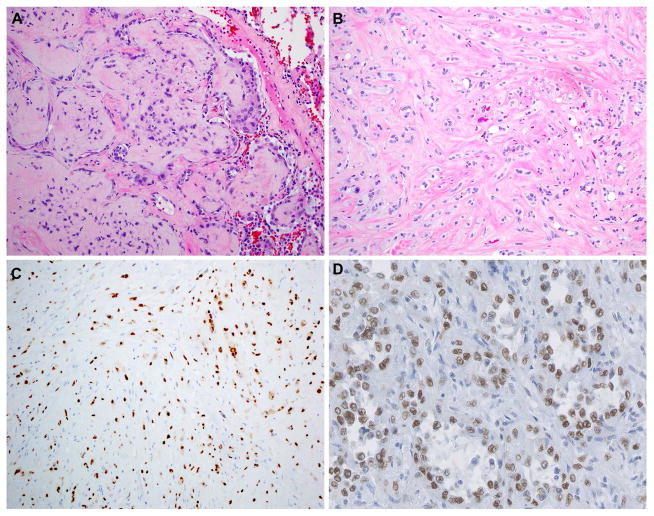
Interestingly, FOSB rearrangements are also the hallmark of pseudomyogenic haemangioendothelioma (epithelioid sarcoma-like hemangioendothelioma), which is a rare, distinctive entity frequently presenting as multiple discontiguous nodules in different tissue planes of a limb in young adult males.14,15 In epithelioid hemangioendothelioma (EHE), considered a low grade angiosarcoma, cords of epithelioid endothelial cells are seen in a distinctive myxohyaline stroma.1 EHE is characterized by the recurrent t(1;3) translocation, resulting in a WWTR1-CAMTA1 fusion gene.16,17 Interestingly, like epithelioid hemangioma, EHE also often presents as multifocal lesions, which are all monoclonal.9,16 Immunohistochemistry for CAMTA1 can be used as a surrogate marker for the translocation (Figure 2).18,19 A specific entity has been described, focally resembling EHE, with more solid architecture admixed with the formation of well-formed vascular channels, genetically characterized by YAP1-TFE3 fusions.20 These tumors have strong immunoreactivity for TFE3 (Figure 2D).20 Future studies should reveal if there is a final common pathway affected by these fusion genes that is involved in the development of these vascular tumors.
Figure 2.
Detection of molecular alterations by immunohistochemistry. Chromosomal rearrangements frequently result in overexpression of transcription factors that can be detected by immunohistochemistry. A. Pulmonary epithelioid hemangioendothelioma with WWTR1-CAMTA1 fusion, composed of small tumor nodules growing along the preexistent alveolar spaces. B. Cords and strands of endothelial epithelioid cells, with intracytoplasmic lumina, embedded in a myxohyaline stroma characteristic of epithelioid hemangioendothelioma. C. CAMTA1 expression in epithelioid hemangioendothelioma with WWTR1-CAMTA1 gene fusion. D. YAP1-TFE3-rearranged epithelioid hemangioendothelioma; TFE3 overexpression can be detected by immunohistochemistry in this unusually vasoformative variant of epithelioid hemangioendothelioma.
Unlike the epithelioid hemangiomas and the different types of hemangioendotheliomas, in angiosarcomas translocations are infrequent. Instead, angiosarcomas often show more complex genomic findings. Most radiation induced angiosarcomas show MYC gene amplification,21 which can be a helpful diagnostic tool, either using FISH or immunohistochemistry, in the differential diagnosis of atypical radiation induced vascular proliferation versus angiosarcoma.22 However, MYC amplification is not restricted to radiation induced angiosarcomas as it can also be found in a subset of primary angiosarcomas.23,24 In addition to MYC amplification, co-amplification of FLT4, or PTPRB and/or PLCG1 mutations can be found in secondary angiosarcomas.25 Angiosarcomas of the breast can present with KDR mutations (10%).26 Many of these genes involve angiogenic signaling. Recently, CIC rearrangements or mutations have been found in 9% of primary angiosarcomas, which were predominantly epithelioid with solid growth and affected younger patients, with an inferior disease-free survival.27
Tumors with deregulated kinase signaling: GIST
Deregulation of cellular signaling driving sustained proliferation is a major hallmark of cancer5 and, as such, contributes to the biology of most sarcoma types. In certain sarcoma types, however, signaling alteration due to kinase activation is the main oncogenic driver and, most likely, the initiating oncogenic event. Prominent examples amongst sarcoma include mutationally-activated receptor tyrosine kinases such as KIT in GIST, recurrent chimeric kinase fusions like ALK oncoproteins in inflammatory myofibroblastic (IMT) and ETV6-NTRK3 in infantile fibrosarcoma, or kinase receptors activated by autocrine mechanisms such as PDGFRA/B in dermatofibrosarcoma protuberans (DFSP). Advances in pharmacology have generated a collection of kinase inhibitors with variable potency, which provide tremendous clinical benefit to patients affected by this group of sarcomas. The most remarkable example of clinically effective targeted inhibition of oncogenic kinase mutations in sarcoma is inhibition of KIT/PDGFRA in GIST.
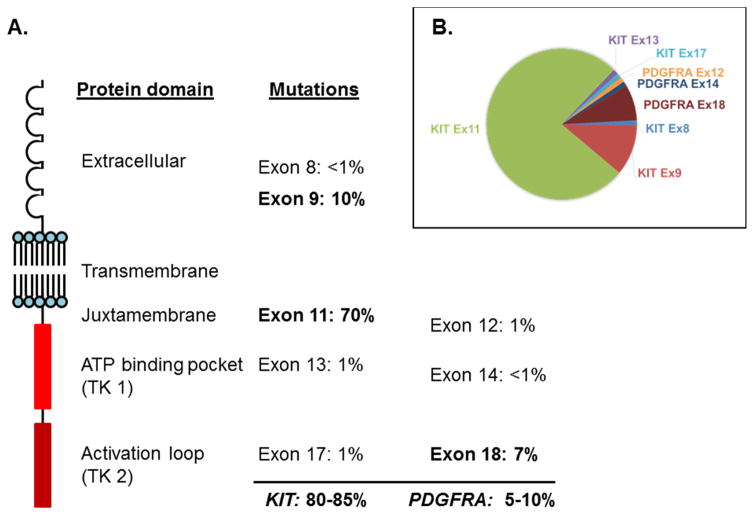
GIST is a model of oncogenic addiction: GIST cell viability and proliferation is absolutely dependent on signaling from the receptor tyrosine kinase KIT, which is constitutively active due to gain-of-function mutations in ~70–80% of cases.28 An additional ~10% of GIST are driven by analogous activating mutations in the receptor tyrosine kinase PDGFRA. PDGFRA-driven GIST shows a predilection for gastric location and an epithelioid phenotype.29–31 KIT and PDGFRA mutations, which are mutually exclusive, lead to ligand-independent activation, which in turn activates intracellular signaling pathways controlling cell differentiation, survival and proliferation.32 KIT primary mutations usually affect exon 11 (70%), exon 9 (10%), exon 13 (1%) or exon 17 (1%), whereas PDGFRA mutations affect exons 18 (5%), 12 (1%) or 14 (<1%).28 All of them are activating mutations, but the activation results from different alterations of various functional domains of the protein, which tolerate different kinds of mutational mechanisms (Figure 3): the juxtamembrane domain, encoded by exons 11 in KIT and 12 in PDGFRA, is an autoinhibitory domain that can be disrupted by in-frame deletions, in-frame insertion-deletions, or point mutations. The two domains that form the split kinase, namely the ATP-binding pocket (exons 13 to 15) and the activation loop (exons 17 and 18), are usually activated by point mutations.
Figure 3.
A. KIT mutations in untreated GISTs involve exons 11, 9, 13, and 17, encoding regions of the extracellular, juxtamembrane, ATP-binding pocket, and activation loop domains, respectively. PDGFRA mutations, found in <10% of GISTs, involve analogous domains. B. Relative frequency of the most common KIT and PDGFRA primary mutations in GIST.
The diagnostic value of KIT mutations is limited in GIST, since the diagnosis is often achievable by the detection of KIT and DOG1 expression by immunohistochemistry, in the appropriate morphological and clinical context. An infrequent exception would be rare cases of KIT-negative GIST, which may lose KIT expression during tumor progression.33,34 Regarding prognosis, some KIT/PDGFRA mutations are associated with poor clinical outcomes in untreated populations, such as in-frame deletions of codons 557–558 in exon 11.35 However, the prognostic differences between mutations are minor, in comparison to their variable sensitivity to drug inhibitors, so that the most important clinical value of mutational analysis in GIST is prediction of response to therapy.
Most mutant KIT and PDGFRA oncoproteins can be effectively inhibited by small molecule kinase inhibitors such as imatinib. The degree of inhibition, and hence the potential clinical benefit, correlates tightly with the specific mutation: in general, exon 11 KIT mutations are extremely sensitive to imatinib, while exon 9 mutations, typically a 6 nucleotide duplication affecting codons 502 and 503, are less sensitive and require a higher dose of imatinib (usually 800mg, double the standard dose).36 Approximately 80% of patients with metastatic GIST initially respond to imatinib - 50% showing partial responses, 30% with stable disease - resulting in a 3-year survival rate of 69 to 74% and a median overall survival of 5 years, compared to only 19 months in the pre-imatinib era.37–39 Primary drug resistance to imatinib results mostly from PDGFRA D842V point mutation or KIT/PDGFRA wild-type status.40 Of note, PDGFRA D842V mutation is cross-resistant to most tyrosine kinase inhibitors, with the potential exception of crenolanib that has shown activity in vitro.41 Primary resistance to imatinib due to hyperactivation of signaling effectors downstream of KIT is possible, but much less common and probably reflects the need of activation of several signaling pathways by independent mechanisms for sustained proliferation in GIST.42
Even in patients with near-complete initial response to imatinib, secondary resistance develops due to the invariable presence of residual viable GIST cells, including drug-resistant subclones which subsequently manifest as clinical progression.43 Up to 50% of GIST patients that initially respond to imatinib develop secondary resistance within 2 years of therapy. Resistance results in the majority of cases from secondary mutations that affect non-random residues in KIT, typically in either the ATP-binding pocket (exon 13–15), or the kinase activation loop (exons 17 or 18 of KIT and exon 18 of PDGFRA), which occur in cis with the primary mutation (i.e. in the same allele).44–46 These secondary KIT mutations are almost never detectable in untreated GIST, likely reflecting their negative effect in cellular fitness in the absence of pharmacologic inhibitors. In the presence of an active inhibitor, however, activating mutations in these domains are selected, and clonal expansion of tumor cells may result in the presence of different secondary mutations in different tumor cell subpopulations in a single patient. A mutation assay with high sensitivity of detection is critical in the setting of therapeutic resistance to appropriately detect heterogeneous secondary mutations, which can be missed by Sanger sequencing.
The predictive nature of KIT mutations in relation to imatinib treatment response extends to sunitinib and regorafenib –and, essentially, to every tyrosine kinase inhibitor. Sunitinib is effective against mutations in exons 13–15 of KIT (ATP-binding pocket), but is ineffective against mutations in exons 17–18 (activation loop);40 while the opposite is true for regorafenib.47,48 Therefore, the sequence of treatment for advanced GIST determined historically, first-line imatinib, followed by sunitinib and third-line regorafenib, is not surprisingly supported by the biology and the natural history of the various KIT/PDGFRA mutations in GIST. It is worth noting that sunitinib and regorafenib are less specific inhibitors than imatinib, with activity against a substantially wider range of kinases; this may explain part of their pharmacologic activity, and determines a higher incidence of significant side effects.
The three major signaling pathways activated by constitutive KIT and PDGFRA activation are the PI3K/AKT/mTOR pathway, the RAS/RAF/MAPK pathway, and the JAK/STAT pathway.32,49 The latter pathway is known to be relevant in mast cell disease harboring KIT mutations, but plays only a limited role in GIST. The PI3K/AKT/mTOR and the RAS/RAF/MAPK pathways, on the other hand, are crucial for proliferation in GIST and may be further activated and contribute to disease progression in high-risk GIST or advanced tumors.50,51 Although primary mutations in effectors downstream of KIT is not common, they can occur at the time of progression in the setting of therapeutic resistance;42,52,53 activating mutations and loss of negative regulators of these pathways can be easily detected given the low mutation rate and relatively quiet copy number variation (CNV) profile of GIST (Figure 1B).
Predictive value of molecular alterations in KIT/PDGFRA-mutant GIST
KIT exon 11 mutations are typically primary mutations, and are very sensitive to imatinib.
The most common PDGFRA mutation in GIST, D842V in exon 18, is imatinib-resistant (and cross-resistant to most tyrosine kinase inhibitors)
KIT exon 9 primary mutations, usually dup502_503, respond to higher dose of imatinib.
KIT exon 13 mutation, often K642E, is often a secondary mutation resistant to imatinib. It responds to sunitinib.
KIT activation-loop mutations, affecting exon 17, are secondary mutations cross-resistant to imatinib and sunitinib, but may respond to regorafenib.
The KIT exon 17 mutation, D816V, is resistant to virtually all inhibitors, but is extraordinarily uncommon in GIST.
Various tyrosine kinase inhibitors (sorafenib, dasatinib, nilotinib, crenolanib, ponatinib) are effective against different sets of KIT/PDGFRA mutations.
Tumors driven by oncometabolites (via epigenetic deregulation): chondrosarcoma
Mutations in metabolic enzymes lead to deregulated cellular energetics in cancer cells and, more importantly, result in the production of metabolites that may alter tightly-regulated physiological processes such as gene expression and epigenetic regulation. Several metabolic enzymes are frequently mutated in particular tumor types. An illustrative example is the metabolic enzyme isocitrate dehydrogenase (IDH), in which somatic mutations were first described in gliomas,54 followed by other tumors55 including ~50% of chondrosarcomas.56 Heterozygous somatic mosaic mutations in IDH were later found in up to 81% of patients with multiple enchondromas (Ollier disease / Maffucci syndrome).57,58
Mutations in IDH cause epigenetic changes59–61 by the formation of a neoenzyme that catalyzes the reduction of α-ketoglutarate to D-2-hydroxyglutarate (D2HG).62–64 D2HG is considered an oncometabolite and inhibits α-ketoglutarate dependent oxygenases like TET2.65,66 This results in inhibition of DNA demethylation, causing hypermethylation. Indeed, IDH1 mutations are associated with a hypermethylated phenotype in cartilage tumors.58 D2HG also inhibits other α-ketoglutarate dependent oxygenases67,68 such as the Jumonji domain histone demethylases, thereby increasing histone methylation as well.60 These epigenetic changes are thought to affect differentiation. Indeed, when mesenchymal stem cells are treated with D2HG, or when an IDH mutation is introduced, this results in inhibition of osteogenic differentiation and stimulation of chondrogenic differentiation, explaining the development of enchondromas during bone development.69,70
Chondrosarcoma can arise secondarily within a benign enchondroma, or as a primary tumor. It is the second most frequent primary bone malignancy, predominantly affecting adults.1 The development of chondrosarcoma occurs through the acquisition of additional genetic alterations (multistep genetic progression model),71 involving amongst others the pRb pathway.72 In high grade chondrosarcomas, the IDH mutation is no longer essential for tumor growth.73,74
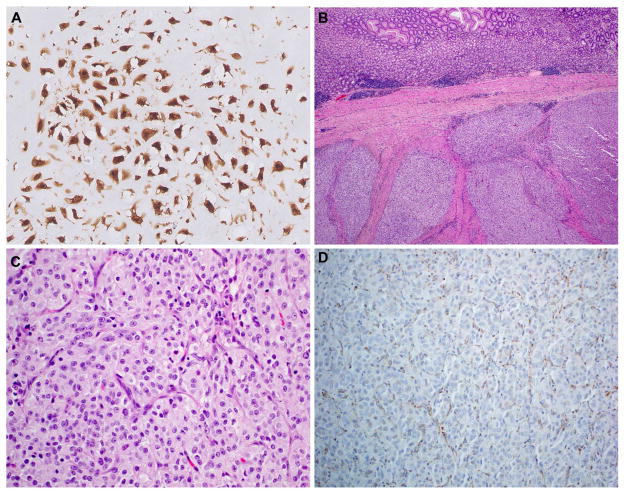
Detection of hotspot mutations in IDH1 or IDH2 can be useful in the differential diagnosis of chondrosarcoma versus chordoma or chondroblastic osteosarcoma, which can sometimes be challenging. A specific antibody against the IDH R132H mutation, widely used for the diagnosis of gliomas, permits detection of this mutation by immunohistochemistry in rare chondrosarcoma cases (Figure 4A). IDH mutations are present in 87% of Ollier- associated enchondromas, 86% of secondary central chondrosarcoma, 38–70% of primary central chondrosarcoma, ~15% of periosteal chondrosarcoma and 54% of dedifferentiated chondrosarcoma56–58,75 and are absent in peripheral chondrosarcoma, osteosarcoma and in chordoma.56,58,76,77
Figure 4.
Molecular metabolic aberrations leading to epigenetic deregulation. A. Detection of R132H mutant IDH1 in chondrosarcoma. Note that this antibody only detects the specific R132H mutation, which is, in contrast to gliomas where it is the most common mutation, infrequent in chondrosarcoma. Thus, negative immunohistochemistry does not rule out a mutation in IDH1. B. Low magnification view of SDH-deficient gastric GIST, demonstrating its characteristic multinodular growth pattern. C. Epithelioid cytomorphology and D. loss of SDHB expression in SDH-deficient gastric GIST.
Other metabolic enzymes, including succinate dehydrogenase (SDH) and fumarate hydratase (FH) are also known to be mutated in cancer and to cause defective energy metabolism as well as epigenetic deregulation in cancer. Inactivating mutations in subunits of mitochondrial complex II including the succinate dehydrogenase subunit D (SDHD), C (SDHC) and B (SDHB) genes, are found in patients with head and neck paragangliomas and pheochromocytomas.78 Also, a subset of gastrointestinal stromal tumors, lacking mutations in KIT or PDGFRA, carry mutations in one of the SDH genes79 or an SDHC epimutation80 both of which are associated with global hypermethylation.81 These gastric GIST tend to affect young patients, and are morphologically distinct, with a multinodular architecture and epithelioid cytomorphology (Figures 4B and 4C). Mutations in one of the SDH subunits destabilize the SDH complex, causing degradation and loss of SDHB. Immunohistochemistry for SDHB is therefore a surrogate marker for mutations in one of the SDH subunits (Figure 4D).82
Inactivating germline mutations of another tricarboxylic acid cycle gene, fumarate hydratase (FH), cause autosomal dominant HLRCC syndrome (hereditary leiomyomas and type 2 papillary renal cell carcinoma), including benign cutaneous and uterine leiomyomas and renal cell cancer,83 while somatic mutations are rare. The accumulation of fumarate, caused by mutations in FH, leads to aberrant succination of proteins. Positive staining for (S)-2-succinocysteine (2SC) can be used as a robust biomarker for mutations in FH.84,85 Similar to mutations in IDH, FH as well as SDH mutations affect epigenetic signaling, by inhibition of histone demethylases and the TET family of 5 hydroxymethylcytosine (5mC)-hydroxylases by accumulated fumarate and succinate, respectively.86–88 Using immunohistochemistry, loss of 5hmC and increased H3K9me3 can be shown in SDH and FH mutant tumor cells.89 In SDH mutant GIST, 5-hmC staining is also low to absent.90
Tumors driven by primary epigenetic deregulation: chondroblastoma
Epigenetic deregulation is emerging as a very prevalent oncogenic mechanism in a wide variety of tumors, beyond the effects of metabolic enzymes and oncometabolites. Molecular alterations of components of the Polycomb group, the SWI/SNF complex, and other genes involved in chromatin structure and regulation are increasingly being identified in many cancer types.91 Frequently, epigenetic deregulation is an additional feature in a cancer cell, contributing to a complex genomic environment in which several other oncogenic mechanisms are already in place (e.g. mutations in members of the PRC2 complex in malignant peripheral nerve sheath tumors).92,93 In some tumor types, however, mutations causing epigenetic deregulation seem to occur as early events, in a background of low mutational rate, and may serve as primary drivers of tumorigenesis.94 Examples include histone mutations in giant cell tumor of bone (GCTB) and chondroblastoma, SMARCB1 homozygous deletions in rhabdoid tumor,95 and SMARCA4 inactivation in SMARCA4-deficient thoracic sarcomas.96 It has recently become apparent that the main oncogenic effect of some chimeric transcription factors, specifically the SS18-SSX fusions in synovial sarcoma, is epigenetic reprograming by mechanisms such as disruption of SWI/SNF complexes;97 these two pathogenetic categories are therefore not mutually exclusive, and as our biological understanding improves, it is to be expected that other sarcomas with chimeric transcription factors may fit better in more specific pathogenetic categories.
Mutations in bone tumors affecting epigenetic signaling include histone H3.3 mutations in giant cell tumor of bone (GCTB) and chondroblastoma,98 both of which are locally aggressive bone tumors predominantly affecting young patients. In 92% of GCTBs mutations are found in the H3F3A gene, while in 95% of the chondroblastomas mutations were found in the H3F3B gene.98 Both genes encode for histone H3.3. The exact mechanism by which these histone H3.3 mutations cause the formation of these giant cell containing tumors is currently unknown. The distinction between GCTB and chondroblastoma and their distinction from other giant cell–containing lesions of bone such as aneurysmal bone cyst, telangiectatic osteosarcoma or chondromyxoid fibroma can be a challenge. Mutation analysis may be a useful diagnostic tool, in addition to the possible use of S100 or DOG1 immunohistochemistry, of which positivity would favor chondroblastoma.99,100 Moreover, in chondromyxoid fibroma, GRM1 rearrangements are found.101 When using mutation analysis for diagnosis, one should realize that the mutation is present in the mononuclear stromal cells, which constitute only a minority of the cells in the tumor. Sensitivity is therefore highly dependent on the technique used, detecting mutations in H3F3A in 69% of the giant cell tumors of bone using classical Sanger sequencing102 compared to 92% using a targeted next generation sequencing (NGS) approach.98
Sarcomas with complex genome
The majority of sarcomas show complex genomic profiles, with inconsistent, nonspecific molecular alterations. These are aggressive tumors that tend to affect older adults (with the exception of some osteosarcomas, and most radiation-associated sarcomas). Morphologically, sarcomas with complex genome are heterogeneous, usually of high histologic grade, frequently cytologically pleomorphic, and may show signs of differentiation along several mesenchymal lineages or may be undifferentiated.1 Notable examples include high grade leiomyosarcoma, pleomorphic and dedifferentiated liposarcoma, high grade myxofibrosarcoma, angiosarcoma, and undifferentiated pleomorphic sarcoma. Although the genomics of these lesions are highly variable from case to case, there is a predominance of copy number alterations over single nucleotide variants. The high chromosomal instability occurs in the context of TP53 pathway alterations, very often TP53 mutation, which is likely an early event in tumorigenesis. Additional molecular alterations include activation of the alternative lengthening of telomeres, often facilitated by loss of the chromatin remodeling factor by ATRX, and loss of multiple tumor suppressor genes. The Rb/E2F pathway is also critical for tumor development and multiple members of the pathway are frequently mutated by different mechanisms. The extreme heterogeneity and complexity explains the limited number of specific molecular markers available for these sarcomas, which at present only benefit from molecular studies in rare occasions in clinical settings. Nonetheless, the higher mutational load in these tumors potentially makes them good candidates for immunotherapy, with drugs such as immune checkpoint blockers.103
Key features of sarcomas with complex genome
Complex unbalanced karyotype. Numerous copy number changes reflecting chromosomal instability.
Higher mutational load.
Pleomorphic, high-grade morphology.
No specific molecular markers.
Frequent loss of tumor suppressor genes (most often TP53).
No effective targeted therapies.
Subsets respond to conventional chemotherapy and radiation therapy.
Prototypical examples: osteosarcoma, leiomyosarcoma, pleomorphic liposarcoma, myxofibrosarcoma, undifferentiated pleomorphic sarcoma (UPS).
Tumors with characteristic copy number alterations: DDLPS
Subsets of sarcoma cases with complex genome show somewhat reproducible pattern of chromosomal alterations. One example is well differentiated/dedifferentiated liposarcoma (WD/DDLPS), whose genome is characterized by multiple copy number changes, mostly gains and amplifications, with multiple intra and interchromosomal rearrangements. More than 90% of WD/DDLPS have characteristic neochromosomes, either linear or circular –giant markers and ring chromosomes, respectively- which are dynamic structures composed of genetic material from various distinct chromosomal regions. The composition of WD/DDLPS neochromosomes is highly variable, from case to case and during clonal evolution within an individual WD/DDLPS, but almost invariably includes a core group of genes from chromosome 12q13-15, including multiple copies of the MDM2 and CDK4 oncogenes. These represent at least two independent 12q amplicons, amongst approximately 15–20 amplicons characteristically present in WD/DDLPS cells. Some of the additional amplicons are relatively consistent, such as 1q25 or 6q21, but their extension and amplitude is highly variable. The mutational mechanisms underlying the formation of WD/DDLPS neochromosomes are the subject of intense investigations104 and can be systematized as 1) an early initiation phase, in which a single catastrophic event (chromothripsis) results in massive fragmentation, rearrangement and circularization of chromosome 12; 2) an amplification phase, in which hundreds of repetitive cycles of break-fusion-bridge allow for amplification, loss, and variable incorporation of additional chromosomal regions to the ring neochromosomes; and 3) a linearization phase, in which the neochromosomes are stabilized by capturing new chromothriptic telomeres. Understanding these events is helpful in the interpretation of copy number alterations and chromosomal rearrangements in WD/DDLPS cases.
The highly recurrent nature of MDM2 and CDK4 amplification provides a useful diagnostic marker. Detection of MDM2 amplification by FISH is currently the gold standard for diagnosis of WD/DDLPS,105 while combined immunohistochemical detection of the MDM2 and CDK4 proteins is a very useful diagnostic tool in surgical pathology routine practice.106,107 MDM2 and CDK4 amplification are readily detectable by NGS (Figure 5). These tests are particularly useful in three situations: 1) to confirm the diagnosis of WDLPS in an adipocytic lesion of minimal cytologic atypia; 2) more commonly, to establish the diagnosis of DDLPS in a relatively non-descript spindle cell or pleomorphic sarcoma in a deep somatic location, and 3) very rarely, to reclassify a high grade pleomorphic adipocytic sarcoma as DDLPS with homologous lipoblastic differentiation that could be mistaken for pleomorphic liposarcoma.108
Figure 5.
High copy number gain of MDM2 and CDK4, in two independent amplicons, in chromosomal region 12q13-15. This lesion was a dedifferentiated liposarcoma with spindle cell and pleomorphic morphologies, arising in a well-differentiated liposarcoma in the inguinal region of a 72 year old male.
Biologically, amplification of MDM2 results in inactivation of p53, while CDK4 amplification leads to cell cycle progression.109 Both alterations can be pharmacologically targeted with compounds that are at different stages of clinical development. MDM2 inhibitors restore p53 function disrupting the p53-MDM2 interaction. Several classes are being evaluated in clinical trials in DDLPS and other forms of cancer. Despite the strong biologic rationale and proven on-target activity, initial clinical experiences demonstrate that few DDLPS patients achieve disease stabilization after MDM2 inhibition, at the expense of substantial adverse effects.110 Such drug toxicities seem to be class-specific, and novel compounds are being evaluated that may overcome the limitations of current drugs. CDK4 inhibition can be achieved with compounds that typically target CDK4 and CDK6, such as palbociclib.111 In DDLPS, CDK4 inhibition provides limited benefits as single agent,112 but probably will be therapeutically effective in combination regimens, as recently demonstrated in other cancer types.113 The combination of MDM2 and CDK4 inhibition in DDLPS is an attractive concept supported by a strong biological rationale, but it may not be achievable due to the combined toxicities of these drugs.
Tumors with highly complex karyotypes: Osteosarcoma
Most pleomorphic sarcomas have complex karyotypes lacking specific genetic aberrations and recognizable chromosomal patterns. These high grade sarcomas often harbor aberrations in the Rb or p53 pathway. Conventional osteosarcoma is the most frequent primary high-grade bone tumor in humans, occurring predominantly in children and adolescents.1,114 It has a high risk of metastasis, and despite intensive treatment strategies, the chance at cure of patients with resectable osteosarcoma has remained around 60%–65% in the past 3 decades.115 At the genetic level osteosarcoma is extremely unstable with many translocations, amplifications, mutations and deletions (Figure 1D). The detection of specific driver genes and pathways is therefore extremely difficult. Recently two phenomena where described that reflect this genomic instability. One is chromothripsis,116 a cataclysmic event in which chromosomes are fragmented and subsequently aberrantly assembled. Chromothripsis was also been identified in other tumours, but is most prevalent in bone tumours.117 The other phenomenon is kataegis, reflected by a localized hypermutation area, which also occurs at a high frequency in osteosarcoma (~50%).117
KEY POINTS.
Sarcomas are characterized by notable morphological and molecular heterogeneity. Molecular studies in the clinical setting provide refinements to morphologic classification, and contribute mainly diagnostic and predictive information.
Sarcomas with simple genome can be driven by transcriptional deregulation, abnormal kinase signaling, or epigenetic reprogramming. This group of sarcomas can be identified with specific molecular markers.
Sarcomas with complex genome show multiple, non-recurrent molecular alterations. There are no molecular diagnostic markers for these tumors. Some prognostic information may be derived from loss of tumor suppressor genes. The high mutational load may make these tumors good candidates for immunotherapies dependent on neoantigens.
Acknowledgments
Financial support: Adrian Marino-Enriquez receives research support from The Sarcoma Alliance for Research through Collaboration (SARC).
Footnotes
Conflict of interest: The authors have nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fletcher CDM World Health Organization, International Agency for Research on Cancer. WHO classification of tumours of soft tissue and bone. 4. Lyon: IARC Press; 2013. p. 468. [Google Scholar]
- 2.Fletcher JA, Kozakewich HP, Hoffer FA, et al. Diagnostic relevance of clonal cytogenetic aberrations in malignant soft-tissue tumors. N Engl J Med. 1991;324(7):436–442. doi: 10.1056/NEJM199102143240702. [DOI] [PubMed] [Google Scholar]
- 3.Bovee JV, Hogendoorn PC. Molecular pathology of sarcomas: Concepts and clinical implications. Virchows Arch. 2010;456(2):193–199. doi: 10.1007/s00428-009-0828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15(6):371–381. doi: 10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7(4):233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 7.Robinson DR, Wu YM, Kalyana-Sundaram S, et al. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat Genet. 2013;45(2):180–185. doi: 10.1038/ng.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Motoi T, Khanin R, et al. Identification of a novel, recurrent HEY1-NCOA2 fusion in mesenchymal chondrosarcoma based on a genome-wide screen of exon-level expression data. Genes Chromosomes Cancer. 2012;51(2):127–139. doi: 10.1002/gcc.20937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van IJzendoorn DG, de Jong D, Romagosa C, et al. Fusion events lead to truncation of FOS in epithelioid hemangioma of bone. Genes Chromosomes Cancer. 2015;54(9):565–574. doi: 10.1002/gcc.22269. [DOI] [PubMed] [Google Scholar]
- 10.Huang SC, Zhang L, Sung YS, et al. Frequent FOS gene rearrangements in epithelioid hemangioma: A molecular study of 58 cases with morphologic reappraisal. Am J Surg Pathol. 2015;39(10):1313–1321. doi: 10.1097/PAS.0000000000000469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antonescu CR, Chen HW, Zhang L, et al. ZFP36-FOSB fusion defines a subset of epithelioid hemangioma with atypical features. Genes Chromosomes Cancer. 2014;53(11):951–959. doi: 10.1002/gcc.22206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Errani C, Zhang L, Panicek DM, Healey JH, Antonescu CR. Epithelioid hemangioma of bone and soft tissue: A reappraisal of a controversial entity. Clin Orthop Relat Res. 2012;470(5):1498–1506. doi: 10.1007/s11999-011-2070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen GP, Srivastava A, Kattapuram S, et al. Epithelioid hemangioma of bone revisited: A study of 50 cases. Am J Surg Pathol. 2009;33(2):270–277. doi: 10.1097/PAS.0b013e31817f6d51. [DOI] [PubMed] [Google Scholar]
- 14.Walther C, Tayebwa J, Lilljebjorn H, et al. A novel SERPINE1-FOSB fusion gene results in transcriptional up-regulation of FOSB in pseudomyogenic haemangioendothelioma. J Pathol. 2014;232(5):534–540. doi: 10.1002/path.4322. [DOI] [PubMed] [Google Scholar]
- 15.Trombetta D, Magnusson L, von Steyern FV, Hornick JL, Fletcher CD, Mertens F. Translocation t(7;19)(q22;q13)-a recurrent chromosome aberration in pseudomyogenic hemangioendothelioma? Cancer Genet. 2011;204(4):211–215. doi: 10.1016/j.cancergen.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Errani C, Zhang L, Sung YS, et al. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosomes Cancer. 2011;50(8):644–653. doi: 10.1002/gcc.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanas MR, Sboner A, Oliveira AM, et al. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med. 2011;3(98):98ra82. doi: 10.1126/scitranslmed.3002409. [DOI] [PubMed] [Google Scholar]
- 18.Doyle LA, Fletcher CD, Hornick JL. Nuclear expression of CAMTA1 distinguishes epithelioid hemangioendothelioma from histologic mimics. Am J Surg Pathol. 2016;40(1):94–102. doi: 10.1097/PAS.0000000000000511. [DOI] [PubMed] [Google Scholar]
- 19.Shibuya R, Matsuyama A, Shiba E, Harada H, Yabuki K, Hisaoka M. CAMTA1 is a useful immunohistochemical marker for diagnosing epithelioid haemangioendothelioma. Histopathology. 2015;67(6):827–835. doi: 10.1111/his.12713. [DOI] [PubMed] [Google Scholar]
- 20.Antonescu CR, Le Loarer F, Mosquera JM, et al. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer. 2013;52(8):775–784. doi: 10.1002/gcc.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manner J, Radlwimmer B, Hohenberger P, et al. MYC high level gene amplification is a distinctive feature of angiosarcomas after irradiation or chronic lymphedema. Am J Pathol. 2010;176(1):34–39. doi: 10.2353/ajpath.2010.090637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mentzel T, Schildhaus HU, Palmedo G, Buttner R, Kutzner H. Postradiation cutaneous angiosarcoma after treatment of breast carcinoma is characterized by MYC amplification in contrast to atypical vascular lesions after radiotherapy and control cases: Clinicopathological, immunohistochemical and molecular analysis of 66 cases. Mod Pathol. 2012;25(1):75–85. doi: 10.1038/modpathol.2011.134. [DOI] [PubMed] [Google Scholar]
- 23.Shon W, Sukov WR, Jenkins SM, Folpe AL. MYC amplification and overexpression in primary cutaneous angiosarcoma: A fluorescence in-situ hybridization and immunohistochemical study. Mod Pathol. 2014;27(4):509–515. doi: 10.1038/modpathol.2013.163. [DOI] [PubMed] [Google Scholar]
- 24.Verbeke SL, de Jong D, Bertoni F, et al. Array CGH analysis identifies two distinct subgroups of primary angiosarcoma of bone. Genes Chromosomes Cancer. 2015;54(2):72–81. doi: 10.1002/gcc.22219. [DOI] [PubMed] [Google Scholar]
- 25.Behjati S, Tarpey PS, Sheldon H, et al. Recurrent PTPRB and PLCG1 mutations in angiosarcoma. Nat Genet. 2014;46(4):376–379. doi: 10.1038/ng.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antonescu CR, Yoshida A, Guo T, et al. KDR activating mutations in human angiosarcomas are sensitive to specific kinase inhibitors. Cancer Res. 2009;69(18):7175–7179. doi: 10.1158/0008-5472.CAN-09-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang SC, Zhang L, Sung YS, et al. Recurrent CIC gene abnormalities in angiosarcomas: A molecular study of 120 cases with concurrent investigation of PLCG1, KDR, MYC, and FLT4 gene alterations. Am J Surg Pathol. 2016 doi: 10.1097/PAS.0000000000000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol. 2004;22(18):3813–3825. doi: 10.1200/JCO.2004.05.140. 0732-183; 0732-183. [DOI] [PubMed] [Google Scholar]
- 29.Heinrich MC, Corless CL, Duensing A, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299(5607):708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 30.Wardelmann E, Pauls K, Merkelbach-Bruse S, et al. Gastrointestinal stromal tumors carrying PDGFRalpha mutations occur preferentially in the stomach and exhibit an epithelioid or mixed phenotype. Verh Dtsch Ges Pathol. 2004;88:174–183. 0070-4113; 0070-4113. [PubMed] [Google Scholar]
- 31.Corless CL, Schroeder A, Griffith D, et al. PDGFRA mutations in gastrointestinal stromal tumors: Frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol. 2005;23(23):5357–5364. doi: 10.1200/JCO.2005.14.068. 0732-183; 0732-183. [DOI] [PubMed] [Google Scholar]
- 32.Duensing A, Medeiros F, McConarty B, et al. Mechanisms of oncogenic KIT signal transduction in primary gastrointestinal stromal tumors (GISTs) Oncogene. 2004;23(22):3999–4006. doi: 10.1038/sj.onc.1207525. 0950-9232; 0950-9232. [DOI] [PubMed] [Google Scholar]
- 33.Medeiros F, Corless CL, Duensing A, et al. KIT-negative gastrointestinal stromal tumors: Proof of concept and therapeutic implications. Am J Surg Pathol. 2004;28(7):889–894. doi: 10.1097/00000478-200407000-00007. 0147-5185; 0147-5185. [DOI] [PubMed] [Google Scholar]
- 34.Espinosa I, Lee CH, Kim MK, et al. A novel monoclonal antibody against DOG1 is a sensitive and specific marker for gastrointestinal stromal tumors. Am J Surg Pathol. 2008;32(2):210–218. doi: 10.1097/PAS.0b013e3181238cec. [DOI] [PubMed] [Google Scholar]
- 35.Martin J, Poveda A, Llombart-Bosch A, et al. Deletions affecting codons 557–558 of the c-KIT gene indicate a poor prognosis in patients with completely resected gastrointestinal stromal tumors: A study by the spanish group for sarcoma research (GEIS) J Clin Oncol. 2005;23(25):6190–6198. doi: 10.1200/JCO.2005.19.554. [DOI] [PubMed] [Google Scholar]
- 36.Gastrointestinal Stromal Tumor Meta-Analysis Group (MetaGIST) Comparison of two doses of imatinib for the treatment of unresectable or metastatic gastrointestinal stromal tumors: A meta-analysis of 1,640 patients. J Clin Oncol. 2010;28(7):1247–1253. doi: 10.1200/JCO.2009.24.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dematteo RP, Lewis JJ, Leung D, Mudan SS, Woodruff JM, Brennan MF. Two hundred gastrointestinal stromal tumors: Recurrence patterns and prognostic factors for survival. Ann Surg. 2000;231(1):51–58. doi: 10.1097/00000658-200001000-00008. 0003-4932; 0003-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verweij J, Casali PG, Zalcberg J, et al. Progression-free survival in gastrointestinal stromal tumours with high-dose imatinib: Randomised trial. Lancet. 2004;364(9440):1127–1134. doi: 10.1016/S0140-6736(04)17098-0. [DOI] [PubMed] [Google Scholar]
- 39.Blanke CD, Demetri GD, von MM, et al. Long-term results from a randomized phase II trial of standard- versus higher-dose imatinib mesylate for patients with unresectable or metastatic gastrointestinal stromal tumors expressing KIT. J Clin Oncol. 2008;26(4):620–625. doi: 10.1200/JCO.2007.13.4403. 1527-7755; 0732-183. [DOI] [PubMed] [Google Scholar]
- 40.Heinrich MC, Maki RG, Corless CL, et al. Primary and secondary kinase genotypes correlate with the biological and clinical activity of sunitinib in imatinib-resistant gastrointestinal stromal tumor. J Clin Oncol. 2008;26(33):5352–5359. doi: 10.1200/JCO.2007.15.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heinrich MC, Griffith D, McKinley A, et al. Crenolanib inhibits the drug-resistant PDGFRA D842V mutation associated with imatinib-resistant gastrointestinal stromal tumors. Clin Cancer Res. 2012;18(16):4375–4384. doi: 10.1158/1078-0432.CCR-12-0625. [DOI] [PubMed] [Google Scholar]
- 42.Serrano C, Wang Y, Marino-Enriquez A, et al. KRAS and KIT gatekeeper mutations confer polyclonal primary imatinib resistance in GI stromal tumors: Relevance of concomitant phosphatidylinositol 3-kinase/AKT dysregulation. J Clin Oncol. 2014 doi: 10.1200/JCO.2013.48.7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinrich MC, Corless CL, Blanke CD, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol. 2006;24(29):4764–4774. doi: 10.1200/JCO.2006.06.2265. [DOI] [PubMed] [Google Scholar]
- 44.Fletcher JA, Corless C, Dimitrijevic S. Mechanisms of resistance to imatinib mesylate (IM) in advanced gastrointestinal stromal tumors (GIST) Proc ASCO. 2003;22:815. [Google Scholar]
- 45.Debiec-Rychter M, Cools J, Dumez H, et al. Mechanisms of resistance to imatinib mesylate in gastrointestinal stromal tumors and activity of the PKC412 inhibitor against imatinib-resistant mutants. Gastroenterology. 2005;128(2):270–279. doi: 10.1053/j.gastro.2004.11.020. 0016-5085; 0016-5085. [DOI] [PubMed] [Google Scholar]
- 46.Liegl B, Kepten I, Le C, et al. Heterogeneity of kinase inhibitor resistance mechanisms in GIST. J Pathol. 2008;216(1):64–74. doi: 10.1002/path. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.George S, Wang Q, Heinrich MC, et al. Efficacy and safety of regorafenib in patients with metastatic and/or unresectable GI stromal tumor after failure of imatinib and sunitinib: A multicenter phase II trial. J Clin Oncol. 2012;30(19):2401–2407. doi: 10.1200/JCO.2011.39.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demetri GD, Reichardt P, Kang YK, et al. Efficacy and safety of regorafenib for advanced gastrointestinal stromal tumours after failure of imatinib and sunitinib (GRID): An international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381(9863):295–302. doi: 10.1016/S0140-6736(12)61857-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bauer S, Duensing A, Demetri GD, Fletcher JA. KIT oncogenic signaling mechanisms in imatinib-resistant gastrointestinal stromal tumor: PI3-kinase/AKT is a crucial survival pathway. Oncogene. 2007;26(54):7560–7568. doi: 10.1038/sj.onc.1210558. [DOI] [PubMed] [Google Scholar]
- 50.Floris G, Wozniak A, Sciot R, et al. A potent combination of the novel PI3K inhibitor, GDC-0941, with imatinib in gastrointestinal stromal tumor xenografts: Long-lasting responses after treatment withdrawal. Clin Cancer Res. 2013;19(3):620–630. doi: 10.1158/1078-0432.CCR-12-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel S. Exploring novel therapeutic targets in GIST: Focus on the PI3K/akt/mTOR pathway. Curr Oncol Rep. 2013;15(4):386–395. doi: 10.1007/s11912-013-0316-6. 1534-6269; 1523-3790. [DOI] [PubMed] [Google Scholar]
- 52.Yang J, Ikezoe T, Nishioka C, et al. Long-term exposure of gastrointestinal stromal tumor cells to sunitinib induces epigenetic silencing of the PTEN gene. Int J Cancer. 2012;130(4):959–966. doi: 10.1002/ijc.26095. 1097-0215; 0020-7136. [DOI] [PubMed] [Google Scholar]
- 53.Quattrone A, Wozniak A, Dewaele B, et al. Frequent mono-allelic loss associated with deficient PTEN expression in imatinib-resistant gastrointestinal stromal tumors. Mod Pathol. 2014 doi: 10.1038/modpathol.2014.53. (1530-0285; 0893-3952) [DOI] [PubMed] [Google Scholar]
- 54.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaap FG, French PJ, Bovee JV. Mutations in the isocitrate dehydrogenase genes IDH1 and IDH2 in tumors. Adv Anat Pathol. 2013;20(1):32–38. doi: 10.1097/PAP.0b013e31827b654d. [DOI] [PubMed] [Google Scholar]
- 56.Amary MF, Bacsi K, Maggiani F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224(3):334–343. doi: 10.1002/path.2913. 1096-9896; 0022-3417. [DOI] [PubMed] [Google Scholar]
- 57.Amary MF, Damato S, Halai D, et al. Ollier disease and maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet. 2011;43(12):1262–1265. doi: 10.1038/ng.994. [DOI] [PubMed] [Google Scholar]
- 58.Pansuriya TC, van Eijk R, d’Adamo P, et al. Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in ollier disease and maffucci syndrome. Nat Genet. 2011;43(12):1256–1261. doi: 10.1038/ng.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sasaki M, Knobbe CB, Munger JC, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488(7413):656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gross S, Cairns RA, Minden MD, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207(2):339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465(7300):966. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McDonough MA, Loenarz C, Chowdhury R, Clifton IJ, Schofield CJ. Structural studies on human 2-oxoglutarate dependent oxygenases. Curr Opin Struct Biol. 2010;20(6):659–672. doi: 10.1016/j.sbi.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 68.Chowdhury R, Yeoh KK, Tian YM, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suijker J, Baelde HJ, Roelofs H, Cleton-Jansen AM, Bovee JV. The oncometabolite D-2-hydroxyglutarate induced by mutant IDH1 or -2 blocks osteoblast differentiation in vitro and in vivo. Oncotarget. 2015;6(17):14832–14842. doi: 10.18632/oncotarget.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jin Y, Elalaf H, Watanabe M, et al. Mutant IDH1 dysregulates the differentiation of mesenchymal stem cells in association with gene-specific histone modifications to cartilage- and bone-related genes. PLoS One. 2015;10(7):e0131998. doi: 10.1371/journal.pone.0131998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bovee JV, Hogendoorn PC, Wunder JS, Alman BA. Cartilage tumours and bone development: Molecular pathology and possible therapeutic targets. Nat Rev Cancer. 2010;10(7):481–488. doi: 10.1038/nrc2869. [DOI] [PubMed] [Google Scholar]
- 72.Schrage YM, Lam S, Jochemsen AG, et al. Central chondrosarcoma progression is associated with pRb pathway alterations: CDK4 down-regulation and p16 overexpression inhibit cell growth in vitro. J Cell Mol Med. 2009;13(9A):2843–2852. doi: 10.1111/j.1582-4934.2008.00406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suijker J, Oosting J, Koornneef A, et al. Inhibition of mutant IDH1 decreases D-2-HG levels without affecting tumorigenic properties of chondrosarcoma cell lines. Oncotarget. 2015;6(14):12505–12519. doi: 10.18632/oncotarget.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li L, Paz AC, Wilky BA, et al. Treatment with a small molecule mutant IDH1 inhibitor suppresses tumorigenic activity and decreases production of the oncometabolite 2-hydroxyglutarate in human chondrosarcoma cells. PLoS One. 2015;10(9):e0133813. doi: 10.1371/journal.pone.0133813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cleven AH, Zwartkruis E, Hogendoorn PC, Kroon HM, Briaire-de Bruijn I, Bovee JV. Periosteal chondrosarcoma: A histopathological and molecular analysis of a rare chondrosarcoma subtype. Histopathology. 2015;67(4):483–490. doi: 10.1111/his.12666. [DOI] [PubMed] [Google Scholar]
- 76.Damato S, Alorjani M, Bonar F, et al. IDH1 mutations are not found in cartilaginous tumours other than central and periosteal chondrosarcomas and enchondromas. Histopathology. 2012;60(2):363–365. doi: 10.1111/j.1365-2559.2011.04010.x. [DOI] [PubMed] [Google Scholar]
- 77.Arai M, Nobusawa S, Ikota H, Takemura S, Nakazato Y. Frequent IDH1/2 mutations in intracranial chondrosarcoma: A possible diagnostic clue for its differentiation from chordoma. Brain Tumor Pathol. 2012;29(4):201–206. doi: 10.1007/s10014-012-0085-1. [DOI] [PubMed] [Google Scholar]
- 78.Eng C, Kiuru M, Fernandez MJ, Aaltonen LA. A role for mitochondrial enzymes in inherited neoplasia and beyond. Nat Rev Cancer. 2003;3(3):193–202. doi: 10.1038/nrc1013. [DOI] [PubMed] [Google Scholar]
- 79.Janeway KA, Kim SY, Lodish M, et al. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proc Natl Acad Sci U S A. 2011;108(1):314–318. doi: 10.1073/pnas.1009199108. 1091-6490; 0027-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Killian JK, Miettinen M, Walker RL, et al. Recurrent epimutation of SDHC in gastrointestinal stromal tumors. Sci Transl Med. 2014;6(268):268ra177. doi: 10.1126/scitranslmed.3009961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Killian JK, Kim SY, Miettinen M, et al. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer Discov. 2013;3(6):648–657. doi: 10.1158/2159-8290.CD-13-0092. 2159-8290; 2159-8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kirmani S, Young WF. Hereditary paraganglioma-pheochromocytoma syndromes. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews(R) Seattle (WA): University of Washington, Seattle; 1993. NBK1548 [bookaccession] [PubMed] [Google Scholar]
- 83.Tomlinson IP, Alam NA, Rowan AJ, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30(4):406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 84.Bardella C, El-Bahrawy M, Frizzell N, et al. Aberrant succination of proteins in fumarate hydratase-deficient mice and HLRCC patients is a robust biomarker of mutation status. J Pathol. 2011;225(1):4–11. doi: 10.1002/path.2932. [DOI] [PubMed] [Google Scholar]
- 85.Ternette N, Yang M, Laroyia M, et al. Inhibition of mitochondrial aconitase by succination in fumarate hydratase deficiency. Cell Rep. 2013;3(3):689–700. doi: 10.1016/j.celrep.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cervera AM, Bayley JP, Devilee P, McCreath KJ. Inhibition of succinate dehydrogenase dysregulates histone modification in mammalian cells. Mol Cancer. 2009;8 doi: 10.1186/1476-4598-8-89. 89-4598-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smith EH, Janknecht R, Maher LJ., 3rd Succinate inhibition of alpha-ketoglutarate-dependent enzymes in a yeast model of paraganglioma. Hum Mol Genet. 2007;16(24):3136–3148. doi: 10.1093/hmg/ddm275. [DOI] [PubMed] [Google Scholar]
- 88.Xiao M, Yang H, Xu W, et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26(12):1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hoekstra AS, de Graaff MA, Briaire-de Bruijn IH, et al. Inactivation of SDH and FH cause loss of 5hmC and increased H3K9me3 in paraganglioma/pheochromocytoma and smooth muscle tumors. Oncotarget. 2015;6(36):38777–38788. doi: 10.18632/oncotarget.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mason EF, Hornick JL. Succinate dehydrogenase deficiency is associated with decreased 5-hydroxymethylcytosine production in gastrointestinal stromal tumors: Implications for mechanisms of tumorigenesis. Mod Pathol. 2013 doi: 10.1038/modpathol.2013.86. (1530-0285; 0893-3952) [DOI] [PubMed] [Google Scholar]
- 91.Kadoch C, Hargreaves DC, Hodges C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45(6):592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee W, Teckie S, Wiesner T, et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46(11):1227–1232. doi: 10.1038/ng.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.De Raedt T, Beert E, Pasmant E, et al. PRC2 loss amplifies ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature. 2014;514(7521):247–251. doi: 10.1038/nature13561. [DOI] [PubMed] [Google Scholar]
- 94.Wilson BG, Wang X, Shen X, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18(4):316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee RS, Stewart C, Carter SL, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122(8):2983–2988. doi: 10.1172/JCI64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Le Loarer F, Watson S, Pierron G, et al. SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nat Genet. 2015;47(10):1200–1205. doi: 10.1038/ng.3399. [DOI] [PubMed] [Google Scholar]
- 97.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153(1):71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Behjati S, Tarpey PS, Presneau N, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45(12):1479–1482. doi: 10.1038/ng.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cleven AH, Briaire-de Bruijn I, Szuhai K, Bovee JV. DOG1 expression in giant cell containing bone tumours. Histopathology. 2015 doi: 10.1111/his.12873. [DOI] [PubMed] [Google Scholar]
- 100.Akpalo H, Lange C, Zustin J. Discovered on gastrointestinal stromal tumour 1 (DOG1): A useful immunohistochemical marker for diagnosing chondroblastoma. Histopathology. 2012;60(7):1099–1106. doi: 10.1111/j.1365-2559.2011.04152.x. [DOI] [PubMed] [Google Scholar]
- 101.Nord KH, Lilljebjorn H, Vezzi F, et al. GRM1 is upregulated through gene fusion and promoter swapping in chondromyxoid fibroma. Nat Genet. 2014;46(5):474–477. doi: 10.1038/ng.2927. [DOI] [PubMed] [Google Scholar]
- 102.Cleven AH, Hocker S, Briaire-de Bruijn I, Szuhai K, Cleton-Jansen AM, Bovee JV. Mutation analysis of H3F3A and H3F3B as a diagnostic tool for giant cell tumor of bone and chondroblastoma. Am J Surg Pathol. 2015;39(11):1576–1583. doi: 10.1097/PAS.0000000000000512. [DOI] [PubMed] [Google Scholar]
- 103.Lim J, Poulin NM, Nielsen TO. New strategies in sarcoma: Linking genomic and immunotherapy approaches to molecular subtype. Clin Cancer Res. 2015;21(21):4753–4759. doi: 10.1158/1078-0432.CCR-15-0831. [DOI] [PubMed] [Google Scholar]
- 104.Garsed DW, Marshall OJ, Corbin VD, et al. The architecture and evolution of cancer neochromosomes. Cancer Cell. 2014;26(5):653–667. doi: 10.1016/j.ccell.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 105.Sirvent N, Coindre JM, Maire G, et al. Detection of MDM2-CDK4 amplification by fluorescence in situ hybridization in 200 paraffin-embedded tumor samples: Utility in diagnosing adipocytic lesions and comparison with immunohistochemistry and real-time PCR. Am J Surg Pathol. 2007;31(10):1476–1489. doi: 10.1097/PAS.0b013e3180581fff. [DOI] [PubMed] [Google Scholar]
- 106.Binh MB, Sastre-Garau X, Guillou L, et al. MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well-differentiated and dedifferentiated liposarcoma subtypes: A comparative analysis of 559 soft tissue neoplasms with genetic data. Am J Surg Pathol. 2005;29(10):1340–1347. doi: 10.1097/01.pas.0000170343.09562.39. [DOI] [PubMed] [Google Scholar]
- 107.Binh MB, Garau XS, Guillou L, Aurias A, Coindre JM. Reproducibility of MDM2 and CDK4 staining in soft tissue tumors. Am J Clin Pathol. 2006;125(5):693–697. doi: 10.1309/VMBP-67QU-NN6Q-3J0E. [DOI] [PubMed] [Google Scholar]
- 108.Marino-Enriquez A, Fletcher CD, Dal Cin P, Hornick JL. Dedifferentiated liposarcoma with “homologous” lipoblastic (pleomorphic liposarcoma-like) differentiation: Clinicopathologic and molecular analysis of a series suggesting revised diagnostic criteria. Am J Surg Pathol. 2010;34(8):1122–1131. doi: 10.1097/PAS.0b013e3181e5dc49. [DOI] [PubMed] [Google Scholar]
- 109.Conyers R, Young S, Thomas DM. Liposarcoma: Molecular genetics and therapeutics. Sarcoma. 2011;2011:483154. doi: 10.1155/2011/483154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ray-Coquard I, Blay JY, Italiano A, et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: An exploratory proof-of-mechanism study. Lancet Oncol. 2012;13(11):1133–1140. doi: 10.1016/S1470-2045(12)70474-6. [DOI] [PubMed] [Google Scholar]
- 111.Zhang YX, Sicinska E, Czaplinski JT, et al. Antiproliferative effects of CDK4/6 inhibition in CDK4-amplified human liposarcoma in vitro and in vivo. Mol Cancer Ther. 2014;13(9):2184–2193. doi: 10.1158/1535-7163.MCT-14-0387. [DOI] [PubMed] [Google Scholar]
- 112.Dickson MA, Tap WD, Keohan ML, et al. Phase II trial of the CDK4 inhibitor PD0332991 in patients with advanced CDK4-amplified well-differentiated or dedifferentiated liposarcoma. J Clin Oncol. 2013;31(16):2024–2028. doi: 10.1200/JCO.2012.46.5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Turner NC, Ro J, Andre F, et al. Palbociclib in hormone-receptor-positive advanced breast cancer. N Engl J Med. 2015;373(3):209–219. doi: 10.1056/NEJMoa1505270. [DOI] [PubMed] [Google Scholar]
- 114.Savage SA, Mirabello L. Using epidemiology and genomics to understand osteosarcoma etiology. Sarcoma. 2011;2011:548151. doi: 10.1155/2011/548151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Anninga JK, Gelderblom H, Fiocco M, et al. Chemotherapeutic adjuvant treatment for osteosarcoma: Where do we stand? Eur J Cancer. 2011;47(16):2431–2445. doi: 10.1016/j.ejca.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 116.Stephens PJ, Greenman CD, Fu B, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144(1):27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen X, Bahrami A, Pappo A, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014;7(1):104–112. doi: 10.1016/j.celrep.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.MacConaill LE, Garcia E, Shivdasani P, et al. Prospective enterprise-level molecular genotyping of a cohort of cancer patients. J Mol Diagn. 2014;16(6):660–672. doi: 10.1016/j.jmoldx.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]