Abstract
Ageing is a complex process and a broad spectrum of physical, psychological, and social changes over time. Accompanying diseases and disabilities, which can interfere with cancer treatment and recovery, occur in old ages. MicroRNAs (miRNAs) are a set of small non-coding RNAs, which have considerable roles in post-transcriptional regulation at gene expression level. In this review, we attempted to summarize the current knowledge of miRNAs functions in ageing, with mainly focuses on malignancies and all underlying genetic, molecular and epigenetics mechanisms. The evidences indicated the complex and dynamic nature of miRNA-based linkage of ageing and cancer at genomics and epigenomics levels which might be generally crucial for understanding the mechanisms of age-related cancer and ageing. Recently in the field of cancer and ageing, scientists claimed that uric acid can be used to regulate reactive oxygen species (ROS), leading to cancer and ageing prevention; these findings highlight the role of miRNA-based inhibition of the SLC2A9 antioxidant pathway in cancer, as a novel way to kill malignant cells, while a patient is fighting with cancer.
Keywords: Ageing, Disability, Genomics, Longevity, MicroRNAs
Introduction
Ageing is a complex process and a broad spectrum of physical, psychological and social changes over time (1). It has been determined as one of the admitted risk factors for most of the human age-related diseases such as cancer, leading to approximately 100,000 people deaths around the world per day (2). Most of the disease and disability, which may interfere with cancer treatment and recovery, occur in old ages. Neoplasm is an undiscerning disease that can affect any part of the body of the human being. Roughly one third of the people are at risk to get cancer in their life (3). However, the incidence of cancer is greatly increased in an age-dependent manner. It is reported that around 60% of all cancers happen in people aged 65 years or above (2). Several molecular mechanisms have linked ageing and cancer together (4).
Micro-RNAs (miRNAs) are a set of small non-coding RNAs with considerable roles in post-transcriptional regulation at gene expression level. Recent studies revealed that miRNAs are involved in many important biological processes such as proliferation, differentiation, angiogenesis, and immune response. miRNAs are generally divided into two categories: the first category acts as cytoplasm mRNA inhibitory (e.g. miRNA-451, miRNA-31, and miRNA-150) and the second one targets nuclear gene transcription directly (e.g. miR-211) (5-7). Thus far, numerous miRNAs have been reported to be involved in different types of malignancy, such as gastric cancer, highlighting them as potential treatment targets (8,9).
In this review we attempted to summarize the current knowledge of miRNAs in ageing with mainly focus on malignancies and all underlying genetic, molecular and epigenetic mechanisms.
miRNAs and their biogenesis
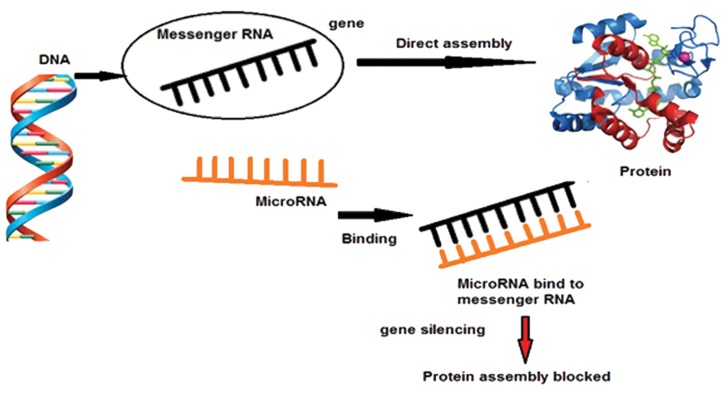
miRNAs are highly conserved RNA molecules in the cell that regulate gene expression through an interference pathway (10). RNA interference (RNAi) is the post-transcriptional silencing mechanism in eukaryotes that induces degradation of homologous mRNA through creating double stranded RNA (11). miRNAs often bind to the 3'UTR region of the target mRNA, which directs the inhibition of its translation or degradation (12). For example, the product of Lin-4, controlling genes in Caenorhabditis elegans (C. elegans), is a 22 nucleotides RNA that is produced by a 60 nucleotides hairpin precursor, and inhibits translation of Lin14 through interaction with the 3'UTR of this mRNA (10). Distribution of miRNA regions in the human genome is in single or cluster form. Some of these regions, at least half of them, are presented in certain transcription units, such as introns and exons (13). miRNA biogenesis takes place in the nucleus and cytoplasm, while the primary miRNAs, transcribed and polyadenylated by RNA polymerase II, are several kilo-bases (Kbs) (5). Stem-loop structure of this transcript is recognized by a 650 kDa enzyme complex that is presented in the nucleus (14). This complex contains class 2 of the RNase III enzymes, called Drosha, which is specialized to cut a double-stranded RNA, as well as a RNA binding protein named DGCR8/Pasha (15). In the cytoplasm, another RNase enzyme (called Dicer) activity leads to generation of the mature miRNAs. Functionally, Dicer cleaves the terminal loop of pri-miRNA and produces double stranded 19-22 nucleotide miRNAs (16). Usually only one strand of the mature miRNAs, known as the guide strand, enters into the micro-ribonucleoprotein complex and creates a micro-RNA-induced silencing complex (miRISC), where the sequence of this strand determine binding region at the target mRNA (17,18). Since only one of the double strands has the ability to play the guidance role for directing the RISC to the 3'UTR region of the target mRNA, the second strand is deleted. RISC binding miRNAs pair to the 3'UTR region of the target mRNA homologous and control gene expression by inhibiting the cleavage or translation of mRNA targets (19,20). About one-third of the human genome is considered as potential regulatory targets by the several hundred miRNAs encoded in the genome. Such regulation happens post-transcriptionally and comprises the interaction of miRNA with the mRNA target site (Fig.1).
Fig.1.
microRNA and inhibiting gene expression.
Mammalian target of rapamycin signaling pathway
The mammalian target of rapamycin (mTOR) signaling pathway integrates inputs from both intracellular and extracellular signals to regulate different cellular processes including proliferation, growth, survival, motility, autophagy, protein synthesis and metabolism. mTOR is a downstream effector of the PI3K/ AKT pathway and consists of two biochemically distinct complexes, including mTORC1 and mTORC2. mTORC1 promotes anabolism, such as cell cycle progression, and inhibits catabolism by blocking autophagy. Signaling of this complex contributes to tumorigenesis through its major downstream targets and key regulators, namely 4E-BPs. It has been demonstrated that mTORC2 regulates cell survival, proliferation and metabolism. Furthermore, mTORC2 is responsible for phosphorylation and activating AKT, which may drive tumorigenesis (21,22). Recent studies have revealed different roles for mTOR in modulating lifespan, considering two processes that mTOR regulates, including protein synthesis and autophagy (23). Another study reported that mTOR is increased in association with BMAL1 deficiency, a transcription factor and core component of an internal time-keeping system called circadian clock. This event eventually contributes to premature aging and reduced lifespan (24). Wide-ranging researches have indicated that miRNAs-based regulation of the mTOR pathway plays a key role in cancer progression, and this pathway is a promising target by miRNAs for novel anticancer therapies (21). Jin et al. (25) in a study on the animal model identified a panel of 63 miRNAs during dermal wound healing, including miRNA-99 family (miRNA-99a, miRNA-99b, and miRNA-100). They demonstrated that miRNA-99 family members regulate AKT/mTOR signaling by targeting several genes such as IGF1R. Grundmann et al. (26) screened miRNAs involved in adaptive blood vessel growth following arterial occlusion. They showed that inhibition of miRNA-100 could be a novel approach for the modulation of mTOR-dependent processes, such as blood vessel growth. A growing body of evidences suggests that miRNAs may play a crucial role in cancer therapy and diagnosis, which mostly performed through the mTOR signaling pathway (Table 1).
Table 1.
Linkage between miRNA and mTOR signaling pathway
| Authors | Cell line(s) | Type of disease | Type of miRNA | Target* | Finding/Suggestion for miRNA |
|---|---|---|---|---|---|
| Zheng et al. (27) | SGC-7901 cell | Gastric cancer | miRNA-18a | Vacuolar protein sorting- associated protein 13D | Possible therapeutic strategy against malignancy |
| Wan et al. (28) | CNE and HeLa cells | Hypoxia- induced autophagy | miRNA-155 | Kinesin-like protein KIF1B Nuclear factor 1 A-type | A key regulator of autophagy via dysregulation of mTOR pathway |
| Shen and Houghton (29) | CB17SC SCID mice | Childhood sarcoma | miRNA-18a | Vacuolar protein sorting- associated protein 13D | Oncogenic growth signals may promote tumorigenesis by dampening the ATM checkpoint |
| miRNA-421 | Calmodulin-binding transcription activator 1 Arginine-glutamic acid dipeptide repeats protein | ||||
| Zhong et al. (30) | Colorectal carcinoma cell | Colorectal carcinoma | miRNA-30a | FUS-interacting serine- arginine-rich protein 1 | A potential therapeutic target to block CRC metastasis |
| miRNA-30b | Kinesin-like protein KIF1B MARCKS-related protein | ||||
| Wang et al. (31) | Mouse adult pancreatic islets | Diabetes | miRNA-7 | Kinesin-like protein KIF1B FKBP12-rapamycin complex-associated protein | As a therapeutic target for diabetes |
| Li et al. (32) | MG63 cells | Osteosarcoma | miRNA-223 | AR DNA-binding protein 43 Msx2-interacting protein FUS-interacting serine- arginine-rich protein 1 | Could be used in anticancer therapies in osteosarcoma |
| Cui et al. (33) | Renal cell Carcinoma cell | Renal cell carcinoma | miRNA-99a | Uncharacterized protein C1orf34 plasticity related gene 1 | May offer an attractive new target for diagnostic and therapeutic interventions |
| Iwaya et al. (34) | HT29 and CaR-1 cell | Colorectal carcinoma | miRNA-144 | PR domain zinc finger protein 2 Msx2-interacting protein | A meaningful prognostic marker |
| Gebeshuber and Martinez (35) | Breast cancer cell | Breast cancer | miRNA-100 | TAR DNA-binding protein 43 Uncharacterized protein C1 or f34 | A potential target for therapeutic approaches |
*; Predicted by target scan and mTOR; Mammalian target of rapamycin.
miRNAs link with cellular senescence, ageing and cancer
Well understanding of the cancer molecular mechanisms pathogenesis and active targeted therapies are necessary to improve patient treatment outcomes. miRNAs act as key components in cancer progression and as the potential therapeutic agents or targets. Numerous studies have suggested that miRNAs inhibit tumor proliferation and promote cellular senescence or ageing, but its function has yet to be elucidated. Other studies reported that miRNAs repress global translation, cell proliferation and initiates premature senescence (Table 2).
Table 2.
miRNA linking cellular senescence, ageing and cancer
| Authors | Type of disease | Type of miRNA | Target* | Finding/Suggestion for miRNA |
|---|---|---|---|---|
| Liu et al. (36) | Ovarian carcinoma | miRNA-506 | Calmodulin-binding transcription activator 1 Msx2-interacting protein | Inhibits proliferation while promotes senescence |
| Mazan-Mamczarz et al. (37) | Diffuse large B cell lymphoma | miRNA-520c-3p | Kinesin-like protein KIF1B Nuclear factor 1 A-type | A key regulator of autophagy via dysregulation of MTOR pathway. |
| Shen and Houghton (29) | Childhood sarcoma | miRNA-18a | Vacuolar protein sorting-associated protein 13D | Oncogenic growth signals may promote tumorigenesis by dampening the ATM checkpoint |
| miRNA-421 | Calmodulin-binding transcription activator 1 Arginine-glutamic acid dipeptide repeats protein | |||
| Zhong et al. (30) | Colorectal carcinoma | miRNA-30a | FUS-interacting serine- arginine-rich protein 1 | A potential therapeutic target to block CRC metastasis |
| miRNA-30b | Kinesin-like protein KIF1B MARCKS-related protein | |||
| Wang et al. (31) | Diabetes | miRNA-7 | Kinesin-like protein KIF1B FKBP12- rapamycin complex- associated protein | As a therapeutic target for diabetes |
| Li et al. (32) | Osteosarcoma | miRNA-223 | AR DNA-binding protein 43 Msx2-interacting protein FUS-interacting serine- arginine-rich protein 1 | Could be used in anticancer therapies in osteosarcoma |
| Cui et al. (33) | Renal cell carcinoma | miRNA-99a | Uncharacterized protein C1 or f34 plasticity related gene 1 | May offer an attractive new target for diagnostic and therapeutic intervention |
| Iwaya et al. (34) | Colorectal carcinoma | miRNA-144 | PR domain zinc finger protein 2 Msx2-interacting protein | A meaningful prognostic marker |
| Gebeshuber and Martinez (35) | Breast cancer | miRNA-100 | TAR DNA-binding protein 43 Uncharacterized protein C1 or f34 | A potential target for therapeutic approaches |
*; Predicted by target scan and mTOR; Mammalian target of rapamycin.
miRNAs, ageing and epigenetics
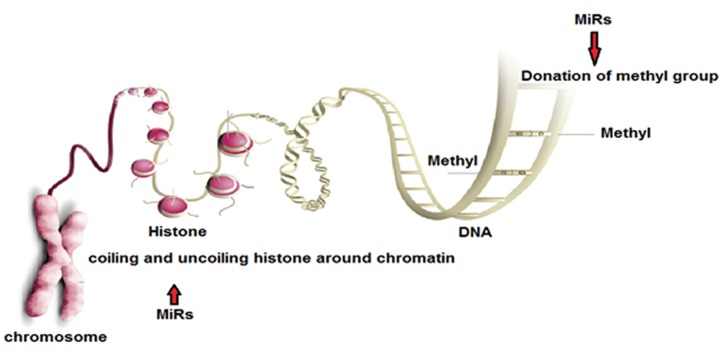
Ageing is a potent predictor of survival rate in cancers, while the biological mechanisms for the variation in clinical outcome are mostly unidentified. Determining genes and pathways, which are responsible for age-related survival changes, could facilitate the chance of novel therapeutic establishments. Bozdag et al. (38) have integrated various molecular and genetic methods to determine age-specific signatures at the genetic and epigenetic levels in glioblastoma multiforme. Ageing of higher organisms are regulated by the epigenetic variation over time. Some epigenetic changes do not follow any determined roles, suggesting that might be the outcome of epigenetics error accumulations. Thus, when this process takes place in adult stem cells, it could play an important role in ageing, through some unknown molecular mechanisms (39). Many researches have discussed various mechanism that miRNA could affect DNA methylation as an epigenetic change contributing to ageing and cancer (Table 3). Two main epigenetics components are DNA methylation, methyl marks add to a certain bases of a gene, and histone modification, combination of various molecules attached to the tails of histone proteins. Functionally, miRNAs could regulate gene expressions through two prominent mechanisms, including donation of the methyl group (40) and chromatin coiling/uncoiling (Fig.2) (41). Wakabayashi et al. (42) revealed that there is likely a cross-talk between miRNAs and epigenetic regulators, modulating neurogenesis in the adult mammalian brain.
Table 3.
miRNA affects DNA methylation as an epigenetic change contributing to ageing and cancer
| Authors | Type of disease | Type of miRNA | Target* | Finding/Suggestion for miRNA |
|---|---|---|---|---|
| Ng et al. (43) | Acute promyelocytic leukaemia | miRNA-34a | Kelch-like protein 17 (Actinfilin) Calmodulin-binding transcription activator 1 | Methylation of miRNA-34b/c may contribute to APL leukaemogenesis |
| miRNA-34b | Kelch-like protein 17 (Actinfilin) Tumor protein p73 (p53-related protein) | |||
| miRNA-34c | Kelch-like protein 17 (Actinfilin) PR domain zinc finger protein 16 | |||
| Xie et al. (44) | Hepatocellular carcinoma cancer | miRNA-34a | Kelch-like protein 17 (Actinfilin) Calmodulin-binding transcription activator 1 | DNA methylation might be involved in the inactivation of miRNA-34b in HCC |
| miRNA-34b | Kelch-like protein 17 (Actinfilin) Tumor protein p73 (p53-related protein) | |||
| miRNA-34c | Kelch-like protein 17 (Actinfilin) PR domain zinc finger protein 16 | |||
| Ko et al. (3) | Acute myeloid leukemia | miRNA-let-7a | Basement membrane-specific heparan sulfate proteoglycan core protein precursor | let-7a-3 methylation is a positive prognosticator for AML patients |
| Huang et al. (45) | Endometrial cancers | miRNA-203 | Msx2-interacting protein Macoilin AT-rich interactive domain-containing protein 1A | miRNA-203 methylation Level might represent a marker for the patients with endometrioid cancers |
| Verhoef et al.(46) | Cervical intraepithelial neoplasia grade 2 (CIN2) | miRNA-124a-2 | Arginine-glutamic acid dipeptide repeats protein retinoblastoma-associated factor 600 | DNA methylation of miRNA-124-2 on HPV-test-positive self-samples is non-inferior to cytology triage in the detection of CIN2 |
| Li et al. (47) | Non-small cell lung cancer cells (NSCLC) | miRNA-503 | Protein kinase C zeta type Vacuolar protein sorting-associated protein 13D | Epigenetic silencing of microRNA-503 regulates FANCA** expression in non-small lung cancer cell |
| Ben Gacem et al. (48) | Breast cancer | miRNA-124a-1 miRNA-124a-2 miRNA-124a-3 | Arginine-glutamic acid dipeptide repeats protein retinoblastoma-associated factor 600 | DNA methylation of miRNA-124a-1, miRNA-124a-2 and miR-124a-3 in breast cancer play a role in tumor growth and aggressiveness |
| Wang et al. (49) | Chronic lymphocytic leukemia (CLL) | miRNA-9-3 | Nuclear inhibitor of protein phosphatase 1 Eyes absent homolog 3 Zinc finger MYM-type protein 6 | miRNA-9-3 is a tumor suppressor miRNA frequently methylated, and hence is silenced in CLL |
| Yamada et al. (50) | Acute lymphoblastic leukaemia | miRNA-128a | Integrator complex subunit 11 | Induction of miRNA128a expression by DNA demethylation is a novel mechanism of resistance to Fas-mediated apoptosis |
| Nadal et al. (51) | Lung adenocarcinoma | miRNA-34b | Kelch-like protein 17 (Actinfilin) Tumor protein p73 (p53-related protein) | Epigenetic inactivation of miRNA-34b/c by DNA methylation has independent prognostic value in patients with early-stage lung adenocarcinoma |
| miRNA-34c | Kelch-like protein 17 (Actinfilin) PR domain zinc finger protein 16 | |||
| Xing et al. (52) | Hepatocellular carcinoma | miRNA-122 | Arginine-glutamic acid dipeptide repeats protein Msx2-interacting protein | DNA Methylation of miRNA-122 might be involved in the development of hepatocellular carcinoma |
| Zhang et al. (53) | Nickel-induced cancer | miRNA-203 | Msx2-interacting protein Macoilin AT-rich interactive domain-containing protein 1A | DNA methylationassociated silencing of tumor suppressor miRNAs contributes to the development of Nickelinduced cancer |
| Zhang et al. (54) | Colorectal cancer (CRC) | miRNA-126 | D site-binding protein Protein LAP2 Uncharacterized protein C10orf6 | DNA methylation-induced silencing of miRNA-126 contributes to tumor invasion and angiogenesis in CRC |
*; Predicted by target scan and **; Fanconi anemia complementation group A protein.
Fig.2.
Two prominent mechanisms involved in regulation of gene expressions by miRs (miRNAs).
miRNA therapeutic applications
miRNA detection has opened a new window in our current perception of the gene expression regulation. Similar to protein-coding genes, several investigations have been performing to determine the expression level of these small RNAs in vitro or in vivo. Hence, miRNAs might undergo gain of function (GOF) or loss of function (LOF). This event could play an important role in various diseases-like protein-coding genes. Different mechanisms including genomic rearrangement, point mutation, and altering the pattern of promoter region methylation could be involved in regulation of miRNA expressions. Besides, this type of RNA plays an important role in expression and regulation of signaling pathways. It is necessary to evaluate the relationship between aberrant miRNA, like miRNA-128 and miRNA-30, expression levels and notch signaling in glioma and angiogenesis, respectively (55).
Several studies have shown that expression or inhibition of miRNAs can change the pattern of tumorigenesis or cancer progression (56-59). It has been demonstrated that expression of several miRNAs (e.g. miRNA-17, miRNA-155) might have oncogenic properties, while the others (e.g. miRNA-34, miRNA-16 and let-7) function as tumor suppressor (60, 61). Here, we suggest that oncogenic or inhibitory effect of miRNAs could raise a distinctive point to compare the normal cells with different types of cancer. Thus, analysis of miRNA expressions, as a molecular bio-marker, could help diagnose the patient’s disorder stage. For example, over-expression of miRNA-155 and down-regulation of let-7 indicated low survival chance in the patients with lung cancer (62, 63). Curiously, the expression pattern of some miRNAs is associated with different stages of tumorigenesis or metastasis, proposing their potential benefit to use as bio-markers (63). Generally, miRNAs can prevent cancer progression through inhibiting the other oncogenic miRNAs, by degrading mRNA through binding with miRNA, inducing tumor suppressor miRNAs or down-regulating the expression level of other miRNAs by regulating epigenetic factors, such as methylation of the gene promoter (64, 65). In contrast, anti-sense oligonucleotides paired with miRNAs can reduce the expression of these small RNAs (66).
Discussion
Currently, there are several types of synthetically made miRNA. Antagomir is an example of this type of artificially made miRNAs. These RNA molecules are designed to inhibit miRNAs. The precise mechanism that anatgomir could inhibit miRNAs is not clear yet, although this mechanism might possibly be performed where these molecules could irreversibly bind to miRNAs. miRNA-based therapeutics could be applied through two approaches; in the first approach, miRNA antagonist applications (e.g. antagomir, anti-miRNA and LNA) contribute through GOF. In the second strategy, using inhibitory miRNAs (e.g. tumor inhibitors) could lead to LOF, compensating lack of natural intracellular miRNAs function. This strategy is similar to transferring protein-coding genes into cells during gene therapy, with even less limitations due to the small size of transferred DNA. Thus, it can easily be transferred into the cells using chemicals without any vector, like inhibitory RNA delivery. In addition, the nature of miRNA function is the other benefit which is mostly influenced by multiple oncogenic paths. Delivery of tumor suppressor miRNAs is mainly done by viral vectors. Another inhibitory transmission approaches, direct miRNAs to the target organ using plasmids, transposons and cationicliposome, as monoclonal antibodies embedded on their surface, epigenetic modifying drugs such as DNA methyltransferase inhibitors (including 5-aza-2'deoxycytidine), histonedeacetylase inhibitors (including 4-phenylbutyric acid) increase the expression of miRNA by reducing DNA methylation and increasing histoneacetylation level, as well as inhibiting cell proliferation through reversing the tumor suppressor effect of miRNA (67). Thus far, several miRNA inhibitors have been introduced to preclinical studies in animal models, one of the most prominent of which is let-7 (68,70). The expression of this miRNA inhibitor is reduced in some cancers, leading to inhibitory effects on the RAS protein family. Furthermore, reduction or loss of activity of this miRNA inhibitor leads to increase in the expression of these proto-oncogenes (71).
These miRNAs also affect other targets such as MYC, cyclin D and HMG2A, indicating the importance of such miRNAs in controlling several pathways related to cancer (72). miRNA-34a, as a target of P53, is another small RNA that prevents the growth of cancer cells by controlling the cell cycle (73). In addition to these direct applications of miRNA in cancer therapy, adjuvant administrations have been discovered for these RNAs. For example, it has been shown that transferring and expressing miRNA-302 in breast cancer cells enhances the sensitivity of these cells to radiotherapy (74). However, the important point, regarding miRNAs replacement therapy, is the risk of cellular toxicity. As demonstrated, miRNAs are required to be proceeded by the RISC. Transferring high amounts of miRNAs to the cells can, in contrast, decrease or omit the other natural miRNAs processing by this complex, which could negatively affect the cell survival. Recently in the field of cancer and ageing, scientists claimed that uric acid can be used to regulate ROS, preventing cancer and ageing. These findings highlight the role of miRNA-based inhibiting the SLC2A9 antioxidant pathway in cancer as a novel approach to kill malignant cells while a patient is fighting with the cancer (75).
Conclusion
The aforementioned evidences illustrate the complexity and the dynamic nature of miRNAbased linkage of ageing and cancer at genomics and epigenetics levels that might be crucial for the understanding of the age-related cancer mechanisms and ageing, in general.
Acknowledgments
The research has not received any financial support. The authors have no conflict of interest.
References
- 1.Dillin A, Gottschling DE, Nyström T. The good and the bad of being connected: the integrons of aging. Curr Opin Cell Biol. 2014;26:107–112. doi: 10.1016/j.ceb.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohile SG, Magnuson A. Comprehensive geriatric assessment in oncology. Interdiscip Top Gerontol. 2013;38:85–103. doi: 10.1159/000343608. [DOI] [PubMed] [Google Scholar]
- 3.Ko YC, Fang WH, Lin TC, Hou HA, Chen CY, Tien HF, et al. MicroRNA let-7a-3 gene methylation is associated with karyotyping, CEBPA promoter methylation, and survival in acute myeloid leukemia. Leuk Res. 2014;38(5):625–631. doi: 10.1016/j.leukres.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Repetto L, Venturino A, Fratino L, Serraino D, Troisi G, Gianni W, et al. Geriatric oncology: a clinical approach to the older patient with cancer. Eur J Cancer. 2003;39(7):870–880. doi: 10.1016/s0959-8049(03)00062-5. [DOI] [PubMed] [Google Scholar]
- 5.Salmanidis M, Pillman K, Goodall G, Bracken C. Direct transcriptional regulation by nuclear microRNAs. Int J Biochem Cell Biol. 2014;54:304–311. doi: 10.1016/j.biocel.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Chitnis NS, Pytel D, Bobrovnikova-Marjon E, Pant D, Zheng H, Maas NL, et al. miR-211 is a prosurvival microRNA that regulates chop expression in a PERK-dependent manner. Mol Cell. 2012;48(3):353–364. doi: 10.1016/j.molcel.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saki N, Abroun S, Soleimani M, Hajizamani S, Shahjahani M, Kast RE, et al. Involvement of microRNA in T-cell differentiation and malignancy. Int J Hematol Oncol Stem Cell Res. 2015;9(1):33–49. [PMC free article] [PubMed] [Google Scholar]
- 8.Wang F, Sun GP, Zou YF, Hao JQ, Zhong F, Ren WJ. MicroRNAs as promising biomarkers for gastric cancer. Cancer Biomark. 2012;11(6):259–267. doi: 10.3233/CBM-2012-00284. [DOI] [PubMed] [Google Scholar]
- 9.Yin Y, Li J, Chen S, Zhou T, Si J. MicroRNAs as diagnostic biomarkers in gastric cancer. Int J Mol Sci. 2012;13(10):12544–12555. doi: 10.3390/ijms131012544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev. 2003;67(4):657–685. doi: 10.1128/MMBR.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22(3):165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 15.MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr Opin Struct Biol. 2007;17(1):138–145. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Lim AK, Tao L, Kai T. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol. 2009;186(3):333–342. doi: 10.1083/jcb.200904063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol. 2008;23(10):578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14 Spec No 1:R121–132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- 20.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 21.Alqurashi N, Hashimi SM, Wei MQ. Chemical inhibitors and microRNAs (miRNA) targeting the mammalian target of rapamycin (mTOR) pathway: potential for novel anticancer therapeutics. Int J Mol Sci. 2013;14(2):3874–3900. doi: 10.3390/ijms14023874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pópulo H, Lopes JM, Soares P. The mTOR signalling pathway in human cancer. Int J Mol Sci. 2012;13(2):1886–1918. doi: 10.3390/ijms13021886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hands SL, Proud CG, Wyttenbach A. mTOR’s role in ageing: protein synthesis or autophagy? Aging (Albany NY) 2009;1(7):586–597. doi: 10.18632/aging.100070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khapre RV, Kondratova AA, Patel S, Dubrovsky Y, Wrobel M, Antoch MP, et al. BMAL1-dependent regulation of the mTOR signaling pathway delays aging. Aging (Albany NY) 2014;6(1):48–57. doi: 10.18632/aging.100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin Y, Tymen SD, Chen D, Fang ZJ, Zhao Y, Dragas D, et al. MicroRNA-99 family targets AKT/mTOR signaling pathway in dermal wound healing. PLoS One. 2013;8(5):e64434–e64434. doi: 10.1371/journal.pone.0064434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grundmann S, Hans FP, Kinniry S, Heinke J, Helbing T, Bluhm F, et al. MicroRNA-100 regulates neovascularization by suppression of mammalian target of rapamycin in endothelial and vascular smooth muscle cells. Circulation. 2011;123(9):999–1009. doi: 10.1161/CIRCULATIONAHA.110.000323. [DOI] [PubMed] [Google Scholar]
- 27.Zheng Y, Li S, Ding Y, Wang Q, Luo H, Shi Q, et al. The role of miR-18a in gastric cancer angiogenesis. Hepatogastroenterology. 2013;60(127):1809–1813. [PubMed] [Google Scholar]
- 28.Wan G, Xie W, Liu Z, Xu W, Lao Y, Huang N, et al. Hypoxia-induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy. 2014;10(1):70–79. doi: 10.4161/auto.26534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen C, Houghton PJ. The mTOR pathway negatively controls ATM by up-regulating miRNAs. Proc Natl Acad Sci USA. 2013;110(29):11869–11874. doi: 10.1073/pnas.1220898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong M, Bian Z, Wu Z. miR-30a suppresses cell migration and invasion through downregulation of PIK3CD in colorectal carcinoma. Cell Physiol Biochem. 2013;31(23):209–218. doi: 10.1159/000343362. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Liu J, Liu C, Naji A, Stoffers DA. MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic β-cells. Diabetes. 2013;62(3):887–895. doi: 10.2337/db12-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li G, Cai M, Fu D, Chen K, Sun M, Cai Z, et al. Heat shock protein 90B1 plays an oncogenic role and is a target of microRNA-223 in human osteosarcoma. Cell Physiol Biochem. 2012;30(6):1481–1490. doi: 10.1159/000343336. [DOI] [PubMed] [Google Scholar]
- 33.Cui L, Zhou H, Zhao H, Zhou Y, Xu R, Xu X, et al. MicroRNA-99a induces G1-phase cell cycle arrest and suppresses tumorigenicity in renal cell carcinoma. BMC Cancer. 2012;12:546–546. doi: 10.1186/1471-2407-12-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwaya T, Yokobori T, Nishida N, Kogo R, Sudo T, Tanaka F, et al. Downregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis. 2012;33(12):2391–2397. doi: 10.1093/carcin/bgs288. [DOI] [PubMed] [Google Scholar]
- 35.Gebeshuber CA, Martinez J. miR-100 suppresses IGF2 and inhibits breast tumorigenesis by interfering with proliferation and survival signaling. Oncogene. 2013;32(27):3306–3310. doi: 10.1038/onc.2012.372. [DOI] [PubMed] [Google Scholar]
- 36.Liu G, Sun Y, Ji P, Li X, Cogdell D, Yang D, et al. MiR506 suppresses proliferation and induces senescence by directly targeting the CDK4/6-FOXM1 axis in ovarian cancer. J Pathol. 2014;233(3):308–318. doi: 10.1002/path.4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazan-Mamczarz K, Zhao XF, Dai B, Steinhardt JJ, Peroutka RJ, Berk KL, et al. Down-regulation of eIF4GII by miR-520c-3p represses diffuse large B cell lymphoma development. PLoS Genet. 2014;10(1):e1004105–e1004105. doi: 10.1371/journal.pgen.1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bozdag S, Li A, Riddick G, Kotliarov Y, Baysan M, Iwamoto FM, et al. Age-specific signatures of glioblastoma at the genomic, genetic, and epigenetic levels. PLoS One. 2013;8(4):e62982–e62982. doi: 10.1371/journal.pone.0062982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huidobro C, Fernandez AF, Fraga MF. Aging epigenetics: causes and consequences. Mol Aspects Med. 2013;34(4):765–781. doi: 10.1016/j.mam.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 40.Hu W, Wang T, Xu J, Li H. MicroRNA mediates DNA methylation of target genes. Biochem Biophys Res Commun. 2014;444(4):676–681. doi: 10.1016/j.bbrc.2014.01.171. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Z, Zhang MQ. Histone modification profiles are predictive for tissue/cell-type specific expression of both protein-coding and microRNA genes. BMC Bioinformatics. 2011;12:155–155. doi: 10.1186/1471-2105-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wakabayashi T, Hidaka R, Fujimaki S, Asashima M, Kuwabara T. MicroRNAs and epigenetics in adult neurogenesis. Adv Genet. 2014;86:27–44. doi: 10.1016/B978-0-12-800222-3.00002-4. [DOI] [PubMed] [Google Scholar]
- 43.Ng HY, Wan TS, So CC, Chim CS. Epigenetic inactivation of DAPK1, p14ARF, mir-34a and -34b/c in acute promyelocytic leukaemia. J Clin Pathol. 2014;67(7):626–631. doi: 10.1136/jclinpath-2014-202276. [DOI] [PubMed] [Google Scholar]
- 44.Xie K, Liu J, Chen J, Dong J, Ma H, Liu Y, et al. Methylation-associated silencing of microRNA-34b in hepatocellular carcinoma cancer. Gene. 2014;543(1):101–107. doi: 10.1016/j.gene.2014.03.059. [DOI] [PubMed] [Google Scholar]
- 45.Huang YW, Kuo CT, Chen JH, Goodfellow PJ, Huang TH, Rader JS, et al. Hypermethylation of miR-203 in endometrial carcinomas. Gynecol Oncol. 2014;133(2):340–345. doi: 10.1016/j.ygyno.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verhoef VM, Bosgraaf RP, van Kemenade FJ, Rozendaal L, Heideman DA, Hesselink AT, et al. Triage by methylation-marker testing versus cytology in women who test HPV-positive on self-collected cervicovaginal specimens (PROHTECT-3): a randomised controlled non-inferiority trial. Lancet Oncol. 2014;15(3):315–322. doi: 10.1016/S1470-2045(14)70019-1. [DOI] [PubMed] [Google Scholar]
- 47.Li N, Zhang F, Li S, Zhou S. Epigenetic silencing of MicroRNA-503 regulates FANCA expression in non-small cell lung cancer cell. Biochem Biophys Res Commun. 2014;444(4):611–616. doi: 10.1016/j.bbrc.2014.01.103. [DOI] [PubMed] [Google Scholar]
- 48.Ben Gacem R, Ben Abdelkrim O, Ziadi S, Ben Dhiab M, Trimeche M. Methylation of miR-124a-1, miR-124a-2, and miR-124a-3 genes correlates with aggressive and advanced breast cancer disease. Tumour Biol. 2014;35(5):4047–4056. doi: 10.1007/s13277-013-1530-4. [DOI] [PubMed] [Google Scholar]
- 49.Wang LQ, Kwong YL, Kho CS, Wong KF, Wong KY, Ferracin M, et al. Epigenetic inactivation of miR-9 family microRNAs in chronic lymphocytic leukemia--implications on constitutive activation of NFκB pathway. Mol Cancer. 2013;12:173–173. doi: 10.1186/1476-4598-12-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamada N, Noguchi S, Kumazaki M, Shinohara H, Miki K, Naoe T, et al. Epigenetic regulation of microRNA-128a expression contributes to the apoptosis-resistance of human T-cell leukaemia jurkat cells by modulating expression of fas-associated protein with death domain (FADD) Biochim Biophys Acta. 2014;1843(3):590–602. doi: 10.1016/j.bbamcr.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 51.Nadal E, Chen G, Gallegos M, Lin L, Ferrer-Torres D, Truini A, et al. Epigenetic inactivation of microRNA-34b/c predicts poor disease-free survival in early-stage lung adenocarcinoma. Clin Cancer Res. 2013;19(24):6842–6852. doi: 10.1158/1078-0432.CCR-13-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xing TJ, Xu HT, Yu WQ, Jiang DF. Methylation regulation of liver-specific microRNA-122 expression and its effects on the proliferation and apoptosis of hepatocellular carcinoma cells. Genet Mol Res. 2013;12(3):3588–3597. doi: 10.4238/2013.September.13.3. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, Zhou Y, Wu YJ, Li MJ, Wang RJ, Huang SQ, et al. Hyper-methylated miR-203 dysregulates ABL1 and contributes to the nickel-induced tumorigenesis. Toxicol Lett. 2013;223(1):42–51. doi: 10.1016/j.toxlet.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Wang X, Xu B, Wang B, Wang Z, Liang Y, et al. Epigenetic silencing of miR-126 contributes to tumor invasion and angiogenesis in colorectal cancer. Oncol Rep. 2013;30(4):1976–1984. doi: 10.3892/or.2013.2633. [DOI] [PubMed] [Google Scholar]
- 55.Azizidoost S, Bavarsad MS, Bavarsad MS, Shahrabi S, Jaseb K, Rahim F, et al. The role of notch signaling in bone marrow niche. Hematology. 2015;20(2):93–103. doi: 10.1179/1607845414Y.0000000167. [DOI] [PubMed] [Google Scholar]
- 56.Hong CC, Chen PS, Chiou J, Chiu CF, Yang CY, Hsiao M, et al. miR-326 maturation is crucial for VEGF-C-driven cortactin expression and esophageal cancer progression. Cancer Res. 2014;74(21):6280–6290. doi: 10.1158/0008-5472.CAN-14-0524. [DOI] [PubMed] [Google Scholar]
- 57.Luo J, Wu J, Li Z, Qin H, Wang B, Wong TS, et al. miR-375 Suppresses IGF1R expression and contributes to inhibition of cell progression in laryngeal squamous cell carcinoma. Biomed Res Int. 2014;2014:374598–374598. doi: 10.1155/2014/374598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang W, Ren F, Wu Q, Jiang D, Li H, Shi H. MicroRNA-497 suppresses angiogenesis by targeting vascular endothelial growth factor A through the PI3K/AKT and MAPK/ERK pathways in ovarian cancer. Oncol Rep. 2014;32(5):2127–2133. doi: 10.3892/or.2014.3439. [DOI] [PubMed] [Google Scholar]
- 59.Saki N, Abroun S, Soleimani M, Hajizamani S, Shahjahani M, Kast RE, et al. Involvement of microRNA in T-cell differentiation and malignancy. Int J Hematol Oncol Stem Cell Res. 2015;9(1):33–49. [PMC free article] [PubMed] [Google Scholar]
- 60.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 61.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67(16):7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 62.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 63.Alemar B, Gregorio C, Ashton-Prolla P. miRNAs as diagnostic and prognostic biomarkers in pancreatic ductal adenocarcinoma and its precursor lesions: a review. Biomark Insights. 2015;10:113–124. doi: 10.4137/BMI.S27679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6(6):590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 66.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25(46):6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 67.Lehmann U, Hasemeier B, Römermann D, Müller M, Länger F, Kreipe H. [Epigenetic inactivation of microRNA genes in mammary carcinoma] Verh Dtsch Ges Pathol. 2007;91:214–220. [PubMed] [Google Scholar]
- 68.Jin H, Liang Y, Wang X, Zhu J, Sun R, Chen P, et al. Association between a functional polymorphism rs712 within let-7-binding site and risk of papillary thyroid cancer. Med Oncol. 2014;31(10):221–221. doi: 10.1007/s12032-014-0221-3. [DOI] [PubMed] [Google Scholar]
- 69.Izzotti A. Molecular medicine and the development of cancer chemopreventive agents. Ann N Y Acad Sci. 2012;1259:26–32. doi: 10.1111/j.1749-6632.2012.06646.x. [DOI] [PubMed] [Google Scholar]
- 70.Ruzzo A, Graziano F, Vincenzi B, Canestrari E, Perrone G, Galluccio N, et al. High let-7a microRNA levels in KRASmutated colorectal carcinomas may rescue anti-EGFR therapy effects in patients with chemotherapy-refractory metastatic disease. Oncologist. 2012;17(6):823–829. doi: 10.1634/theoncologist.2012-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 72.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105(10):3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7(11):819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liang Z, Ahn J, Guo D, Votaw JR, Shim H. MicroRNA-302 replacement therapy sensitizes breast cancer cells to ionizing radiation. Pharm Res. 2013;30(4):1008–1016. doi: 10.1007/s11095-012-0936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Itahana Y, Han R, Barbier S, Lei Z, Rozen S, Itahana K. The uric acid transporter SLC2A9 is a direct target gene of the tumor suppressor p53 contributing to antioxidant defense. Oncogene. 2015;34(14):1799–1810. doi: 10.1038/onc.2014.119. [DOI] [PubMed] [Google Scholar]