ABSTRACT
We previously identified and characterized an intramolecular trans-sialidase (IT-sialidase) in the gut symbiont Ruminococcus gnavus ATCC 29149, which is associated to the ability of the strain to grow on mucins. In this work we have obtained and analyzed the draft genome sequence of another R. gnavus mucin-degrader, ATCC 35913, isolated from a healthy individual. Transcriptomics analyses of both ATCC 29149 and ATCC 35913 strains confirmed that the strategy utilized by R. gnavus for mucin-degradation is focused on the utilization of terminal mucin glycans. R. gnavus ATCC 35913 also encodes a predicted IT-sialidase and harbors a Nan cluster dedicated to sialic acid utilization. We showed that the Nan cluster was upregulated when the strains were grown in presence of mucin. In addition we demonstrated that both R. gnavus strains were able to grow on 2,7-anyhydro-Neu5Ac, the IT-sialidase transglycosylation product, as a sole carbon source. Taken together these data further support the hypothesis that IT-sialidase expressing gut microbes, provide commensal bacteria such as R. gnavus with a nutritional competitive advantage, by accessing and transforming a source of nutrient to their own benefit.
KEYWORDS: gut bacteria, glycoside hydrolase, intestinal mucin, intramolecular trans-sialidase, mucin glycans, Ruminococcus gnavus, sialic acid
Introduction
The gastrointestinal (GI) tract is inhabited by a diverse microbial community (microbiota) that influences host health through a number of mechanisms, including the production of metabolites, protection against pathogens, and interactions with the host immune system and physiology. The microbiota composition varies longitudinally along the GI tract but also transversally from the mucosa to the lumen.1 In the large intestine, the mucus shows a bi-layer organization, with the outer layer providing a habitat to the bacteria whereas the inner layer keeps them away from the epithelium surface.2 Mucins facilitate microbial tropism through the presentation of glycans which may impact colonization.3 The terminal mucin O-glycans have been proposed to serve as metabolic substrates, providing a nutritional advantage to bacteria which have adapted to the GI mucosal environment.4,5 Given the diversity and complexity of intestinal mucin glycan structures, strategies for deconstructing these molecules rely on the cooperative action of a number of proteases, sulfatases, and glycosidases encoded by the genome of mucin-degrading bacteria. So far, only a limited number of bacterial species/strains from the Bacteroidetes, Firmicutes, Actinobacteria, and Verrucomicrobia phyla have been studied for their ability to consume mucins and it is becoming apparent that the strategy used by these mucin-degraders is strain and species-dependent.5 Based on these studies, a number of glycoside hydrolases (GH) families as classified in the CAZy database (www.cazy.org)6 have been involved in mucin-degradation.5 These include neuraminidases/sialidases (GH33), fucosidases (GH29 and GH95), exo- and endo-β-N-acetylglucosaminidases (GH84 and GH85), β-galactosidases (GH2, GH20 and GH42), α-N-acetylglucosaminidases (GH89), endo-β1–4-galactosidases (GH98) and α-N-acetylgalactosaminidases (GH101, GH129) (www.cazy.org). Most bacteria appear to have incomplete packages of GHs for utilizing host mucins and so degradation of mucin in the human gut is likely to be carried out by a consortium of bacteria.
Ruminococcus gnavus is a prominent member of the gut microbial community,7,8 belonging to the Firmicutes division, Clostridia class and XIVa cluster, Lachnospiraceae family.9 In our previous work, we showed that the ability of R. gnavus to utilize mucins was strain-dependent and associated with sialic acid metabolism although R. gnavus is not able to grow on free sialic acid (Neu5Ac).10 We further showed that the mucin-degrader R. gnavus ATCC 29149 strain produces an intramolecular trans-sialidase (IT-sialidase) that cleaves off terminal α2–3-linked sialic acid from glycoproteins, releasing 2,7-anhydro-Neu5Ac instead of sialic acid,11 suggesting a novel mechanism of mucosal adaptation. Here we further explored the mucin-degradation strategy of R. gnavus by expanding the work to another mucin-degrading strain, R. gnavus ATCC 35913, for which we obtained the genome sequence, and carried out comparative sequence and transcriptomics analyses and confirmed the ability of the strains to grow on 2,7-anhydro-Neu5Ac, supporting the role of IT-sialidase in the mucin-degrading strategy utilized by R. gnavus.
Materials and methods
Materials
D-glucose (Glc), N-acetylneuraminic acid (Neu5Ac), 2′-(4-Methylumbelliferyl)-α-D-N-acetylneuraminic acid (4MU-Neu5Ac) and type III pig gastric mucin (PGM) were purchased from Sigma-Aldrich (St Louis, MO).10 Purified pig gastric mucin (pPGM) was obtained as previously described.12 6′-sialyllactose (6′SL) was kindly provided by Glycom A/S (Lyngby, Denmark). 3′-sialyllactose (3′SL) was purchased from Carbosynth Limited (Campton, UK).
Bacterial strains and growth conditions
R. gnavus strains were routinely grown in an anaerobic cabinet (Don Whitley, Shipley, UK) in BHI-YH as previously described.10 Growth on single carbon sources utilized anaerobic basal YCFA medium13 supplemented with 27.7 mM of specific mono- or oligosaccharides as indicated or 1% (wt/vol) pPGM. The growth assays were performed in 96-well plates with 200 μL of medium/well with a single point OD measurement at 595 nm after 44 hours. When the bacteria were grown with Glc or pPGM as sole carbon source with sampling for RNA extraction, the culture was performed in 14 mL tubes. Growth was determined spectrophotometrically by monitoring changes in optical density at 600 nm compared to the same medium without bacterium (Δ OD600 nm) hourly for 9 hours.
Genomic DNA extraction from R. gnavus ATCC 35913 and sequencing
For the isolation of R. gnavus ATCC 35913-chromosomal DNA, cells from a 50 mL-overnight culture were harvested by centrifugation (10 000 g, 5 min, 4°C). The cell pellet was washed with 5 mL of TES buffer (10 mM Tris, 1 mM EDTA, 0.1 M NaCl, pH 8), resuspended in 5 mL of TES buffer supplemented with lysozyme (20 mg.mL−1) and incubated for 15 min at 37°C. Then, complete lysis was achieved by addition of 1 mL of 20% sodium dodecyl sulfate (SDS) and incubation for 10 min at 50°C. The mixture was then extracted by three consecutive treatments: first, with 5 mL of phenol pH 7.9 then with 5 mL of phenol-chloroform-isoamyl alcohol (25:24:1) and finally with 5 mL of chloroform-isoamyl alcohol (24:1). After precipitation with cold absolute ethanol, the genomic DNA was resuspended in 2 mL of TE buffer (10 mM Tris, 1 mM EDTA, pH 8). Traces of RNA were removed by a treatment with RNAse ONE (Promega, Madison, WI) used as recommended by the manufacturer. The DNA was again precipitated with 0.3 M sodium acetate (pH 5.2) and 70% ice-cold ethanol. Finally, it was dissolved in 1.5 mL of TE. Quality and quantity were assessed using NanoDrop 1000 UV-Vis Spectrophotometer and by electrophoresis on 0.7% agarose gel. The DNA was sequenced on the Roche 454 GS Flx platform using 500 ng of input DNA. The sequencing library was produced by the rapid library and emulsion PCR method and run on a single 454 picotiter plate at The Genome Analysis Center (TGAC). The 454 reads were assembled with Newbler (Roche) and annotated at first using Rast (NMPDR)14 with specific additions to the annotation from the authors, including some manual curation. The EMBL accession numbers of the annotated contigs are FCFA01000001-FCFA01000241.
Comparative CAZome analysis
The translated protein sequences of R. gnavus E1, ATCC 29149 and ATCC 35913 were compared to the full length sequences derived from the Carbohydrate-Active enZymes (CAZy) database (www.cazy.org)15 using BLAST.16 The sequences that had an e-value >0.1 were assigned to glycoside hydrolases (GH), glycoside transferases (GT), polysaccharide lyases (PL), carbohydrate esterases (CE), and carbohydrate binding module (CBM) families using a parallel procedure involving a BLAST search against partial sequences corresponding to individual GH, GT, PL, CE and CBM modules and a HMMer search17 using hidden Markov models built for each CAZy module family.15 The counts for each CAZy family of each strain were then compared and the putative function of the proteins of interest was evaluated by alignment with the sequences of biochemically characterized enzymes.14
Total RNA extraction from R. gnavus strains
Total RNA was extracted from 10 mL of mid- to late exponential phase cultures of ATCC 29149 and ATCC 35913 in YCFA supplemented with one carbon source (Glc or pPGM). Four biological replicates were performed for each carbon source. The RNA was stabilized prior to extraction by adding 1/5 vol of phenol (pH 4.3) : ethanol (1:9) mixture to 1 vol of culture then incubating 30 min on ice and finally pelleting the cells for 5 min at 10,000 g at 4°C. Cell pellets were stored at −80°C before extraction. Extraction was performed using a traditional method using phenol and chloroform. Genomic DNA contamination was removed by DNAse treatment using TURBO DNA-free kit (Life Technologies Ltd, Paisley, UK) according to supplier's recommendations. The purity, quantity and integrity of the extracted RNA were assessed before and after DNase treatment, with NanoDrop 1000 UV-Vis Spectrophotometer (Thermo Fischer Scientific, Wilmington, DE) and with Agilent RNA 600 Nano kit on Agilent 2100 Bioanalyzer (Agilent Technologies, Stockport, UK).
RNA sequencing
Transcriptome libraries were constructed using the Illumina TruSeq RNA sample preparation kit with modifications. The rRNA was depleted using Ribo-Zero™ rRNA Removal Kit for Gram-Positive Bacteria (Epicentre Illumina). The rRNA removal was confirmed with a Pico chip run on Bioanalyzer 2100 (Agilent) and the quantity measured with the Qubit RNA kit and Qubit fluorometer (Invitrogen). The resulting ribosomal depleted RNA was then fragmented for 8 min at 94°C using the Elute, Fragment, Prime buffer from Illumina TruSeq RNA kit. These conditions give final libraries of around 400 bp. The samples were then processed following the standard TruSeq RNA protocol.
The 4 Illumina libraries were normalized and equimolar pooled to 15 nM using elution buffer (Qiagen). The library pool was then diluted to 2 nM with NaOH and 5 μL transferred into 995 μL HT1 (Illumina) to give a final concentration of 10 pM. A portion (120 μL) of the diluted library pool was then transferred into a 200 μL strip tube, spiked with 1% PhiX Control v3 and placed on ice before loading onto the Illumina cBot. The flow cell was clustered using TruSeq Single End Cluster Generation Kit v3, following the Illumina SE_amplification_Linearization_Blocking_PrimerHyb_v8 recipe. Following the clustering procedure, the flow cell was loaded onto the Illumina HiSeq2000 instrument following the manufacturer's instructions. The sequencing chemistry used was TruSeq SBS Kit v3-HS using HiSeq Control Software 1.4.8 and RTA 1.12.4.2. The library pool was run in a single lane for 100 cycles. Reads in bcl format were demultiplexed based on the 6 bp Illumina index by CASAVA 1.8, allowing for a one base-pair mismatch per library, and converted to FASTQ format by bcl2fastq.
rRNA depletion levels were assessed following read mapping. To carry this out, rRNA genes were initially manually curated on the ATCC 29149 genome. To compare the transcript expression levels across all samples, the RNA-seq reads were mapped onto the R. gnavus ATCC 29149 genome with the open source tool Bowtie v0.12.918 using default parameters. Differentially expressed genes were identified using the DESeq2 R package19 with raw counts normalized to the effective library size. Results were also assessed using DeSeq R package,20 edgeR21 and simple RPKM normalization methods to check the DESeq2 analysis. A QC check was performed using a sample correlation matrix and plotting it as a tree. The significance of differential expression was determined by the Benjamini–Hochberg22 corrected p-values of the Wald test for the negative binomial between two conditions. The threshold for significance was set to p < 0.05 and a Log2 (fold change) >1. Error bars were taken as the standard error of the Log2 fold change.
Synthesis and analysis of 2,7-anhydro-Neu5Ac
The synthesis of 2,7-anhydro-Neu5Ac was carried out using a modified method.23 Briefly, 4-Methylumbelliferyl (MU)-Neu5Ac was incubated with recombinant IT-sialidase11 in ammonium formate buffer (100 mM) at 37°C, pH 6.5 overnight. The reaction was terminated by the addition of an equal volume of ethanol and the precipitate removed by centrifugation (4000 g for 20 min). The supernatant was evaporated to dryness, dissolved in chloroform/methanol and subjected to Folch partitioning. Fractions containing 2,7-anhydro-Neu5Ac were identified by thin-layer chromatography (TLC) using silica gel 60 F254 (Merck) for the stationary phase and butanol/ acetic acid/ water at a ratio 2:1:1 for the mobile phase and vizualization by incubating the TLC into a potassium permanganate solution followed by heating. The purified product was analyzed by electrospray ionization mass spectrometry (ESI-MS) and H1 NMR. MS spectra were acquired on Expression CMS (Advion) with ESI ionization using direct injection operated in a negative ion mode. Advion Data Express (version 2.2.29.2) software package was used to evaluate the MS data. The 1H NMR spectrum was recorded in D2O at 400 MHz on a Bruker Avance spectrometer (Bruker BioSpin GmbH, Rheinstetten, Germany) running Topspin 3.2 software, and was fitted with a cryoprobe. 1H NMR spectrum was acquired with 16 scans, a spectral width of 8,223.6 Hz, an acquisition time of 3.98 s and a relaxation delay of 4.0 s. The spectrum was transformed with a 0.3-Hz line broadening, manually phased, baseline corrected and referenced by setting the water signal to 4.7 p.p.m.
Results and discussion
R. gnavus ATCC 35913 grows on mucin and 3′-sialyllactose but not on sialic acid or 6′-sialyllactose
Previous work showed that the ability of R. gnavus to utilize mucins is strain-dependent with ATCC 29149 but not E1 being able to grow on mucins.10 Here we tested the ability of R. gnavus ATCC 35913 to grow on mucin using purified porcine gastric mucin (pPGM) as source of carbon. R. gnavus ATCC 35913, has been isolated from fecal sample of a healthy human adult.24 Spectrophotometric measurements were made every hour for up to 9 hours (Fig. 1). The growth curves were analyzed using the in-house-developed DMFit program. For both strains the lag phase was just below 2 hours and the maximum ΔOD at 600 nm was identical. As previously reported for R. gnavus ATCC 29149,10 R. gnavus ATCC 35913 grew on Glc and on 3′-sialyllactose (Neu5Acα2–3Galβ1–4Glc, 3′SL) but was unable to grow in presence of sialic acid (Neu5Ac) or 6′-sialyllactose (Neu5Acα2–6Galβ1–4Glc, 6′SL) as sole carbon source (Table 1). These data suggest that the strategy used by the mucin-degrader R. gnavus ATCC 35913 is similar to that previously reported for ATCC 291495.5,10
Figure 1.
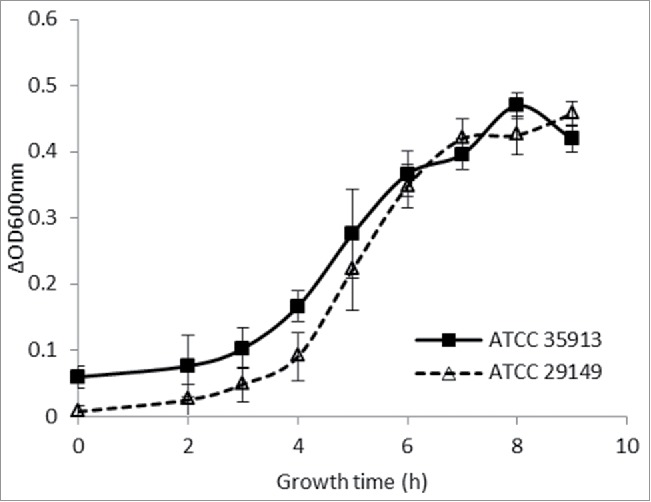
Growth curves of R. gnavus ATCC 29149 and ATCC 35913 on pPGM. The growth curves represent the average growth, measured at OD600nm, of at least 3 biological replicates.
Table 1.
Growth assay of the 3 strains of R. gnavus with several carbon sources.
| Carbon source |
||||||||
|---|---|---|---|---|---|---|---|---|
| Strain | none | Glc | 3′SL | 6′SL | MU-Neu5Ac | MU | Neu5Ac | 2,7-anhydro-Neu5Ac |
| E1 | − | + | − | − | − | − | − | − |
| ATCC29149 | − | + | + | − | + | − | − | + |
| ATCC35913 | − | + | + | − | + | − | − | + |
Note. +: growth; −: no growth; Glc was used as a positive control
Abbreviations: Glc, glucose; 3′SL, 3′-sialyllactose; 6′SL, 6′-sialyllactose; MU-Neu5Ac, 2′-(4-Methylumbelliferyl)-α-D-N-acetylneuraminic acid; MU, 4-Methylumbelliferol; Neu5Ac, N-acetylneuraminic acid; 2,7-anhydro-Neu5Ac, 2,7-anhydro-N-acetylneuraminic acid.
In silico and transcriptomics analysis confirm IT-sialidase as a major determinant for R. gnavus mucin-degrader strains
In order to gain insights into the molecular basis of R. gnavus ATCC 35913 mucin-degradation, we sequenced and analyzed the R. gnavus ATCC 35913 genome and compared its mucin-degrading potential to that of R. gnavus ATCC 29149 and E1 strains. The draft assembly possessed 241 contigs of average length 15510 bp and median length of 1792 bp. The longest contig was 222692 bp, while the shortest contig was 102 bp. The R. gnavus ATCC 35913 genome is 3.74 Mb long with a GC content of 43%, which is similar to the GC% of E1 and ATCC 29149 (42.44% and 42.9% respectively). Genomic analysis identified 99 full length genes and 12 fragments of genes encoding CAZymes (www.cazy.org), corresponding to approximately 2.87% of CDS dedicated to carbohydrate metabolism. R. gnavus ATCC 35913 genome contains the following number of CAZyme modules; 36 GTs, 4 CEs, 1 PL, 12 CBMs and 67 GHs. Nine of the CAZy genes encode two modules, almost always a CBM and a GH, although one of the fragments contains two GH2 domains. There are also 4 predicted proteins solely composed of a CBM (2 CBM32, 1 CBM50 and 1 CBM13) with no predicted associated catalytic module. Most of R. gnavus ATCC 35913 CAZome is represented by genes encoding GHs distributed into 26 GH families. A comparison of R. gnavus ATCC 35913 repertoire of GH families with that of R. gnavus ATCC 29149 and E1 strains is presented in Figure 2. Both mucin-degrading strains follow a similar GH representation with most GH families being common between the two strains apart from GH10 (xylanase), GH51 (endoglucanase, xylanase, L-arabinofuranosidase) and GH94 (phosphorylase) being specific of R. gnavus ATCC 35913 and GH113 (β-mannanase) being specific for R. gnavus ATCC 29149. Among the GH families which members have been implicated in mucin-degradation5 are GH2 β-galactosidases, GH29 and GH95 fucosidases and a GH33 sialidase. Interestingly, the genome of R. gnavus ATCC 35913 encodes a predicted GH33 sialidase sharing 100% identity with the enzyme which was characterized as an IT-sialidase in ATCC 29149, and is absent in the non-degrading strain E1. Similarly GH98 which encodes a predicted endo-β-galactosidase appears to be specific for the R. gnavus mucin-degrading strains. In contrast, all of the R. gnavus strains (including the E1 strain10) contain multiple GH29 and GH95 α-fucosidases, GH2 β-galactosidases and a high proportion of GH13 members, which is the major GH family acting on substrates containing α-glucoside linkages; a detailed analysis of their substrate specificity is warranted to assess their contribution to the carbohydrate preference of each strain.
Figure 2.
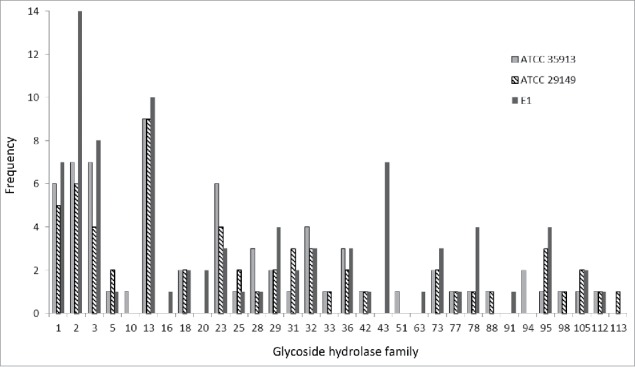
Comparison of the distribution of glycoside hydrolases (GHs) between R. gnavus strains. GHs are represented by light gray boxes for R. gnavus ATCC 35913, striped boxed for R. gnavus ATCC 29149 and dark gray boxes for R. gnavus E1.
To further assess the importance of these GHs in the ability of the mucin-degrader strains to grow on mucins, we analyzed the transcriptional activity of ATCC 29149 (Fig. 3A) and ATCC 35913 (Fig. 3B) by RNAseq analysis. The level of expression was compared to a reference dataset of the strains grown in minimal medium with Glc as the sole carbon source. These data confirmed the increased transcription of the GH33 sialidase gene (nanH) when ATCC 29149 was grown with mucin, previously observed by qPCR, and are in agreement with the implication of this extracellular enzyme in enabling R. gnavus ATCC 29149 to grow on mucin.10 Similarly the gene encoding GH33 sialidase from R. gnavus ATCC 35913 was highly upregulated in presence of mucins, the highest of all GHs (Fig. 3B). The GH98 endo-β1–4-galactosidase gene expression was also induced in presence of mucin for both strains. Other mucin-specific upregulated genes included some GH29 and GH95 fucosidases (Fig. 3A and B). It is also worth noting that other GH families, not implicated in mucin-degradation, showed gene expression induction including GH13 and GH1 (mainly exo-acting enzymes cleaving a β linkage) family members, which raises questions on the substrate specificity of these enzymes.
Figure 3.
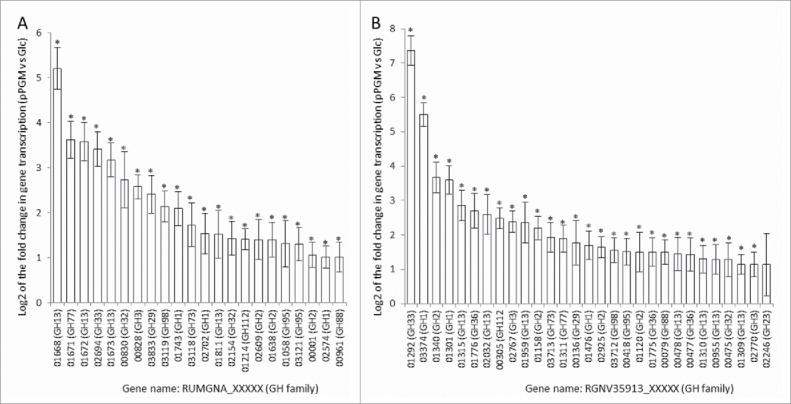
Relative level of transcription of GH genes in R. gnavus ATCC 29149 (A) and ATCC 35913 (B). The transcriptomic analysis has been performed by RNASeq from R. gnavus grown in presence of pPGM and compared to Glc as sole carbon source. The relative level of transcription was expressed as the Log2 of the fold change in gene transcription and the figures showed averages of 4 biological replicates for the GH genes that exhibited increased transcription (Log2 fold change > 1). Data were analyzed by DESeq2.19 The significance of differential expression was determined by the Benjamini-Hochberg21 corrected p-values of the Wald test of the negative binomial test per each set of two conditions. The transcription level was considered significantly increased when p < 0.05 and a Log2 (fold change) >1 and significant results were labeled with *. Error bars were plotted as the standard error of the Log2 fold change.
R. gnavus ATCC 35913 encodes a functional sialic acid utilization operon
R. gnavus ATCC 29149 encodes an 11.7-kb Nan operon containing genes encoding putative proteins involved in the utilization of sialic acid whereas this cluster is absent in the non-mucin degrading strain R. gnavus E1.10 Bioinformatics analysis of the R. gnavus ATCC 35913 genome suggests the presence of an 11.7- kb Nan cluster sharing 99.9% identity with the one present in ATCC 29149 and constituted of 11 open reading frames (ORFs) (Fig. 4A). The first gene of the cluster (RGNV35913_01299) is predicted to code for a GDSL-like protein belonging to the SGNH-hydrolase family, a diverse family of esterases and lipases, which comprises for example mucin-desulfating sulfatases (N-acetylglucosamine-6-sulfatases). The second gene (RGNV35913_01298) encodes a putative sugar isomerase belonging to the YhcH/YjgK/YiaL family. The following one (RGNV35913_01297) encodes a protein with homology with transcriptional regulators of the AraC family. The following 3 genes code for a predicted solute-binding protein (RGNV35913_01296) and two putative permeases (RGNV35913_01295 and RGNV35913_01294), components of a sugar ABC transporter; RGNV35913_01294 has specific homology with putative sialic acid transporters of the SAT2 family. The following gene has homology with an oxidoreductase from the Gfo/Idh/MocA family. The sialidase gene nanH (RGNV35913_01292) predicted to encode the GH33 enzyme comes next. Then nanE (RGNV35913_01291), which encodes a predicted N-acetylmannosamine-6P (ManNAc-6-P) epimerase converting ManNAc-6-P into N-acetylglucosamine-6-P (GlcNAc-6-P), followed by nanA (RGNV35913_01290) encoding a putative Neu5Ac lyase involved in the breaking down of Neu5Ac into ManNAc and pyruvate. nanK (RGNV35913_01289) is the last gene of the cluster, coding for a predicted ManNAc kinase, involved in the phosphorylation of ManNAc into ManNAc-6-P.
Figure 4.
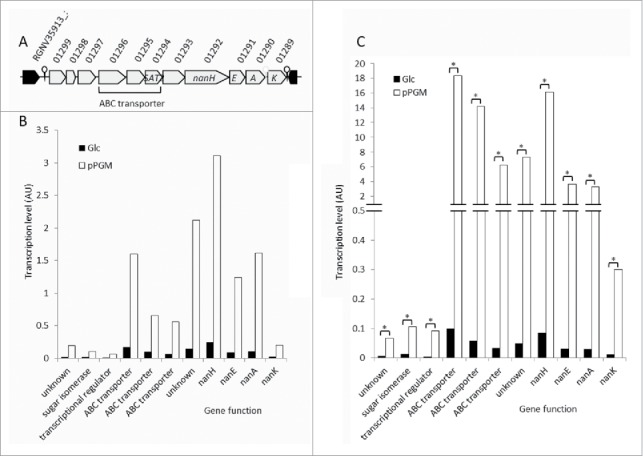
The Nan locus in R. gnavus ATCC 29149 and ATCC 35913. (A) Schematic representation of the nan genetic organization in ATCC 35913. RGNV35913_01299 encodes a putative GDSL-like protein. RGNV35913_01298encodes a putative sugar isomerase involved in sialic acid catabolism. RGNV35913_01297 encodes a protein with homology with transcriptional regulators of the AraC family. The following 3 genes code for a predicted solute-binding protein (RGNV35913_01296) and two putative permeases (RGNV35913_01295 and RGNV35913_01294), components of a sugar ABC transporter. The following gene has homology with oxidoreductases from the Gfo/Idh/MocA family. The sialidase gene nanH (RGNV35913_01292) predicted to encode the GH33 enzyme comes next. Then nanE (RGNV35913_01291), which encodes a predicted ManNAc-6-P epimerase is followed by nanA (RGNV35913_01290) encoding a putative Neu5Ac lyase. nanK (RGNV35913_01289) is the last gene of the cluster, coding for a predicted ManNAc kinase. The previously described R. gnavus ATCC 29149 nan cluster10 shares 99.9% identity with the one present in ATCC 35913. Level of transcription of nan genes in R. gnavus ATCC 29149 (B) or ATCC 35913 (C). R. gnavus was grown in basal YCFA medium supplemented with either glucose (Glc) or mucin (pPGM) as sole carbon source. Cells were collected during the exponential phase of growth; RNA was extracted from 4 biological replicates for each carbon sources. The level of transcription of each gene was determined by RNASeq. The transcription of each gene was compared when the bacterium grew with pPGM vs. Glc using the R package DESeqx; it was considered significantly increased when the transcript was present at least twice more frequently, with a padj value (p-value adjusted for multiple testing) <=0.05 (* padj<=0.05).
Previous data showed that the transcription of nanH was up-regulated when R. gnavus ATCC 29149 was grown in presence of mucin, as determined by microarrays and qPCR.10 Here we analyzed the transcriptional activity of this cluster by RNAseq analysis in both R. gnavus mucin-degrader strains grown on mucin as sole carbon source. We confirmed that all the genes involved in the release and transport of sialic acid - nanH and the 3 genes coding for an ABC transporter - as well as all the genes putatively involved in sialic acid metabolism inside the cell- were upregulated when R. gnavus ATCC 29149 and ATCC 35913 were grown with mucin (Fig. 4B and C). The average transcription of all the genes of the cluster (including nanA, nanE and nanK) was increased by at least 7 times when ATCC 29149 grew with pPGM as compared to Glc. Similarly, the change in transcription of the nan genes in ATCC 35913 is at least 8-fold and this increase was statistically significant for all 11 genes of the operon (Fig. 4C). These results indicate that these strains have adapted to scavenge sialic acid from host sialoglycans.
R. gnavus ATCC 29149 and ATCC 35913 grow on IT-sialidase transglycosylation product
In order to test the ability of R. gnavus ATCC 29149 and ATCC 35913 strains to utilize 2,7-anydro-Neu5Ac as a carbon source, we first performed the in vitro enzymatic synthesis of the transglycosylation product (not commercially available). The recombinant IT-sialidase from R. gnavus ATCC 29149 heterologously expressed in E. coli11 was used in presence of MU-Neu5Ac to produce 2,7-anhydro-Neu5Ac in amounts enabling screening the growth of R. gnavus strains in microtiter plates. The obtained product 2,7-anhydro-Neu5Ac was ∼80% pure according to ESI-MS and H1 NMR analysis (Fig. S1). The by-product of the reaction, identified as MU, did not interfere with the cell growth (data not shown). We showed that both R. gnavus ATCC 35913 and ATCC 29149 could grow on MU-Neu5Ac or 2,7-anhydro-Neu5Ac as sole carbon source, while the strains were unable to grow on Neu5Ac or MU. It is worth noting that E1 was unable to grow on any of these carbon sources (Table 1).
Taken together we propose a model whereby 2,7-anhydro-Neu5Ac is transported into the bacteria via the ABC transporter composed of a solute-binding protein (RUMGNA_02698 or RGNV35913_01296) and two putative permeases (RUMGNA_02697 and RUMGNA_02696, or RGNV35913_01295 and RGNV35913_01294) (Fig. 5A). It is not known whether this transporter is specific of the 2,7-anhydro-Neu5Ac or could also transport Neu5Ac. Once inside the cell, 2,7-anhydro-Neu5Ac could be hydrolyzed into Neu5Ac by the product of RUMGNA_02701 or RGNV35913_01299, which encodes a putative protein belonging to the GDSL-like family, a diverse family of esterases and lipases. This family contains SGNH-hydrolases, a sub-family of enzymes with multifunctional properties such as broad substrate specificity and regiospecificity, some of them have been shown to act as esterase on acetylated sugar e.g. NanS, an enzyme able to deacetylate the 9-O-acetyl Neu5Ac into Neu5Ac.25 RUMGNA_02701 or RGNV35913_01299 predicted protein possesses the four strictly conserved amino acids, Ser-Gly-Asn-His, involved in the catalytic function of these enzymes as well an Asp residue upstream of His, another feature of these enzymes.26 Neu5Ac would then be catabolized into GlcNAc-6-P by the successive action of NanA (Neu5Ac lyase), NanK (ManNAc kinase) and NanE (ManNAc-6-P epimerase) before entering the glycolytic pathway, in accordance with the predicted specificities of these enzymes.27 Alternatively it can be hypothesized that NanA shows specificity for 2,7-anhydro-Neu5Ac instead of Neu5Ac (Fig. 5B).
Figure 5.
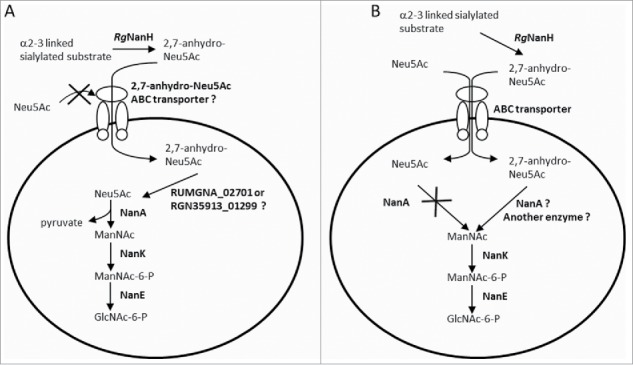
Proposed pathways for the catabolism of sialic acid in R. gnavus ATCC 29149 and ATCC 35913. RgNanH releases 2,7-anhydro-Neu5Ac from α2–3 linked sialylated substrates. (A) It can be hypothesized that 2,7-anhydro-Neu5Ac is transported inside the bacterium via a 2,7-anhydro-Neu5Ac-specific ABC transporter composed of a solute-binding protein (RUMGNA_02698 in ATCC 29149;RGNV35913_01296 in ATCC 35913) and two putative permeases (RUMGNA_02697 and RUMGNA_02696 in ATCC 29149; RGNV35913_01295 and RGNV35913_01294 in ATCC 35913) and then hydrolyzed into Neu5Ac, possibly by the action of RUMGNA_02701 or RGNV35913_01299, before being catabolized into GlcNAc-6-P following the traditional pathway by the successive action of NanA (Neu5Ac lyase), NanK (ManNAc kinase) and NanE (ManNAc-6-P epimerase). (B) Alternatively, both 2,7-anhydro-Neu5Ac and Neu5Ac could enter the cells via the ABC transporter but NanA would either be inactive or specific for 2,7-anhydro-Neu5Ac, explaining the absence of growth of the bacteria on sialic acid.
Gaining mechanistic insights into the strategies used by members of the gut microbiota to utilize mucin glycans is important to understand how the mucus-associated ecosystem is maintained. Due to their proximity to the host immune system, mucus-associated microorganisms may have a disproportionate impact on health.28 Several lines of evidence indicate that mucin-degrading bacteria such as R. gnavus may play an important role in dysbiosis by increasing total mucosa-associated bacteria in inflammatory bowel diseases.29 Furthermore R. gnavus is involved iso-bile acid (BA) biosynthesis,30,31 leading to the production of secondary BAs which play a role in modulating gut community composition.32 Identification of keystone and foundation taxa in the mucus-associated GI microbiota is important for novel diagnostic strategies and therapeutic modulation.33 Keystone species of the GI microbiota have been identified for bacteria that are instrumental to the degradation of dietary resistant starch.34 With regards to host mucin degradation, we believe that the presence of IT-sialidases will provide gut microbes such as R. gnavus with a competitive nutritional advantage, allowing the bacteria to thrive within mucosal environments by scavenging sialic acid from host mucus in a form, 2,7-anhydro-Neu5Ac, that can be used to their own benefit. Furthermore, once the terminal sugars and blood group antigens are removed and used to support growth of R. gnavus, the mucin core glycans are exposed to enzymatic degradation by other members of the gut microbiota, thus favoring metabolic cross-feeding within this mucosal niche. This dual strategy may fit the representation of R. gnavus as a keystone member of the mucus-associated gut microbiota.
Supplementary Material
Abbreviations
- CAZyme
carbohydrate-active enzyme
- CBM
carbohydrate binding module
- CDS
coding DNA sequence
- GH
glycoside hydrolase
- GI
gastrointestinal
- Glc
glucose
- IT-sialidase
intramolecular trans-sialidase
- MU
4-methylumbelliferyl
- Neu5Ac
N-acetylneuraminic acid
- ORF
open reading frame
- Ppgm
purified pig gastric mucin
- SL
sialyllactose
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors gratefully acknowledge the support of the Biotechnology and Biological Sciences Research Council (BBSRC). Sequencing was conducted at The Genome Analysis Center (TGAC). We would like to acknowledge Simon Anders and Michael Love for their help with the analysis of RNASeq dataThe genome sequence of R. gnavus ATCC35913 has been deposited in the European Nucleotide Archive (ENA-EMBL) with the study accession number, PRJEB12317; and annotated contigs are available with accession numbers FCFA01000001-FCFA01000241.
Funding
This research was partly funded by The Genome Analysis Center (TGAC) Capacity and Capability Challenge Program, the BBSRC Institute Strategic Program for The Gut Health and Food Safety (BB/J004529/1) and by the BB/L008602/1 BBSRC responsive mode grant.
References
- [1].Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016; 14:20-32; PMID:26499895; http://dx.doi.org/ 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Johansson ME, Ambort D, Pelaseyed T, Schütte A, Gustafsson JK, Ermund A, Subramani DB, Holmén-Larsson JM, Thomsson KA, Bergström JH, et al.. Composition and functional role of the mucus layers in the intestine. Cell Mol Life Sci 2011; 68:3635-41; PMID:21947475; http://dx.doi.org/ 10.1007/s00018-011-0822-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Juge N. Microbial adhesins to gastrointestinal mucus. Trends Microbiol 2012; 20:30-39; PMID:22088901; http://dx.doi.org/ 10.1016/j.tim.2011.10.001 [DOI] [PubMed] [Google Scholar]
- [4].Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. A refined palate: Bacterial consumption of host glycans in the gut. Glycobiol 2013; 23:1038-46; http://dx.doi.org/ 10.1093/glycob/cwt040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tailford LE, Crost EH, Kavanaugh D, Juge N. Mucin glycan foraging in the human gut microbiome. Front Genet 2015; 6:81; PMID:25852737; http://dx.doi.org/ 10.3389/fgene.2015.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 2014; 42:D490-5; PMID:24270786; http://dx.doi.org/ 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al.. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464:59-65; PMID:20203603; http://dx.doi.org/ 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kraal L, Abubucker S, Kota K, Fischbach MA, Mitreva M. The prevalence of species and strains in the human microbiome: a resource for experimental efforts. PLoS one 2014; 9:e97279; PMID:24827833; http://dx.doi.org/ 10.1371/journal.pone.0097279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ludwig W, Schleifer KH, Whitman WB. Revised road map to the phylum Firmicutes” In: Bergey's Manual of Systematic Bacteriology, 2nd ed., vol. 3 (The Firmicutes) De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB, eds., 2009; New York: Springer-Verlag; pp:1-13 [Google Scholar]
- [10].Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One 2013; 8:e76341; PMID:24204617; http://dx.doi.org/ 10.1371/journal.pone.0076341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tailford LE, Owen CD, Walshaw J, Crost EH, Hardy-Goddard J, Le Gall G, de Vos WM, Taylor GL, Juge N. Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation. Nat Commun 2015; 6:7624; PMID:26154892; http://dx.doi.org/ 10.1038/ncomms8624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gunning AP, Kirby AR, Fuell C, Pin C, Tailford LE, Juge N. Mining the “glycocode”-exploring the spatial distribution of glycans in gastrointestinal mucin using force spectroscopy. FASEB J 2013; 27:2342-54; PMID:23493619; http://dx.doi.org/ 10.1096/fj.12-221416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Duncan SH, Hold GL, Harmsen HJ, Stewart CS, Flint HJ. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol 2002; 52:2141-46; PMID:12508881 [DOI] [PubMed] [Google Scholar]
- [14].Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al.. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008; 9:75; PMID:18261238; http://dx.doi.org/ 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 2009; 37:D233-38; PMID:18838391; http://dx.doi.org/ 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-10; PMID:2231712; http://dx.doi.org/ 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- [17].Eddy SR. Profile hidden Markov models. Bioinformatics 1998; 14:755-63; PMID:9918945; http://dx.doi.org/ 10.1093/bioinformatics/14.9.755 [DOI] [PubMed] [Google Scholar]
- [18].Langmead B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics Chapter 2010; 11:Unit 11.17; http://dx.doi.org/ 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550; PMID:25516281; http://dx.doi.org/ 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010; 11: R106; PMID:20979621; http://dx.doi.org/ 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26:139-40; PMID:19910308; http://dx.doi.org/ 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Series B (Methodological) 1995; 57:289-300 [Google Scholar]
- [23].Teh LY, Chen LS. Isolation, purification and utilization of sialidase-L in the synthesis of 2,7-anhydro-N-acetylneuraminic acid and for the selective cleavage of sialylα2-3 D-galactose linkage. Patent US 5312747 A Filing date 8 Feb 1993. Publication date 17 May 1994
- [24].Hoskins LC, Agustines M, McKee WB, Boulding ET, Kriaris M, Niedermeyer G. Mucin degradation in human colon ecosystems. Isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J Clin Invest 1985; 75:944-53; PMID:3920248; http://dx.doi.org/ 10.1172/JCI111795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rangarajan ES, Ruane KM, Proteau A, Schrag JD, Valladares R, Gonzalez CF, Gilbert M, Yakunin AF, Cygler M. Structural and enzymatic characterization of NanS (YjhS), a 9-O-Acetyl N-acetylneuraminic acid esterase from Escherichia coli O157:H7. Protein Sci 2011; 20:1208-19; PMID:21557376; http://dx.doi.org/ 10.1002/pro.649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF. GDSL family of serine esterases/lipases. Prog Lipid Res 2004; 43:534-52; PMID:15522763; http://dx.doi.org/ 10.1016/j.plipres.2004.09.002 [DOI] [PubMed] [Google Scholar]
- [27].Ghosh S, Roseman S. The Sialic Acids. V. N-Acyl-D-Glucosamine 2-Epimerase. J Biol Chem 1965; 240:1531-36; PMID:14285488 [PubMed] [Google Scholar]
- [28].Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 2012; 10:323-35; PMID:22491358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 2010; 105:2420-8; PMID:20648002; http://dx.doi.org/ 10.1038/ajg.2010.281 [DOI] [PubMed] [Google Scholar]
- [30].Lee JY, Arai H, Nakamura Y, Fukiya S, Wada M, Yokota A. Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J Lipid Res 2013; 54:3062-9; PMID:23729502; http://dx.doi.org/ 10.1194/jlr.M039834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Devlin AS, Fischbach MA. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol. 2015; 11:685-90; PMID:26192599; http://dx.doi.org/ 10.1038/nchembio.1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Duboc H, Rajca S, Rainteau D, Benarous D, Maubert MA, Quervain E, Thomas G, Barbu V, Humbert L, Despras G, et al.. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013; 62:531-9; PMID:22993202; http://dx.doi.org/ 10.1136/gutjnl-2012-302578 [DOI] [PubMed] [Google Scholar]
- [33].Trosvik P, de Muinck EJ. Ecology of bacteria in the human gastrointestinal tract–identification of keystone and foundation taxa. Microbiome 2015; 3:44; PMID:26455879; http://dx.doi.org/ 10.1186/s40168-015-0107-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ze XL, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 2012; 6:1535-43; PMID:22343308; http://dx.doi.org/ 10.1038/ismej.2012.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


