ABSTRACT
We previously reported quantitation of gut microbiota in a panel of 89 different inbred strains of mice, and we now examine the question of sex differences in microbiota composition. When the total population of 689 mice was examined together, several taxa exhibited significant differences in abundance between sexes but a larger number of differences were observed at the single strain level, suggesting that sex differences can be obscured by host genetics and environmental factors. We also examined a subset of mice on chow and high fat diets and observed sex-by-diet interactions. We further investigated the sex differences using gonadectomized and hormone treated mice from 3 different inbred strains. Principal coordinate analysis with unweighted UniFrac distances revealed very clear effects of gonadectomy and hormone replacement on microbiota composition in all 3 strains. Moreover, bile acid analyses showed gender-specific differences as well as effects of gonodectomy, providing one possible mechanism mediating sex differences in microbiota composition.
KEYWORDS: genetics, gut microbiota interactions, hormones, inbred strains, sex-by-diet interactions, sex differences
Introduction
The individual variation in host-associated gut microbiota community structure is shaped by both environmental and host genetics factors.1.3 Diet, in particular, is one of the strongest factors affecting inter-individual and temporal variations in gut microbiota composition.4-7 While sex differences have a clear impact on physiology and behavior,8 it has proven difficult to demonstrate sex differences in gut microbial composition. While some studies suggested that gender has no or very limited effect on gut microbiota,9-11 other studies have provided at least suggestive evidence for differences in microbiota composition between sexes.12-16 Also, two recent studies demonstrated that the commensal microbial community can affect sex hormone levels.17,18
In previous studies using a panel of over 100 diverse inbred strains of mice we showed that diet5 and genetics3 have clear effects on gut microbiota composition. We now report on analysis of the role of sex differences and sex hormones in gut microbiota composition. We demonstrate that in a controlled environment male and female mice show significant differences in gut microbiota composition, although genetic differences obscured sex differences in examination of the entire population. Furthermore, using gonadectomy and hormone replacement, we were able to detect gut microbiota differences mediated by sex hormones and also significant differences in bile acid profiles between sexes as a result of gonadectomy. Finally, we examine the role of sex differences in the response to a high fat diet and identified sex-by-diet interactions.
Gender specific microbiota composition
To test whether sex differences exist in microbiota composition we compared 341 female and 348 male mice from 89 matched strains (Table S1). Mice from different strains and genders were housed in separate cages and we found that microbiotas were more similar overall within strains than between strains (p < 0.001 PERMANOVA of Bray-Curtis distances) (Fig. S1A). When considering each strain independently, clear differences in microbiota composition and diversity were observed between sexes (Fig. 1A). This was also supported by Procrustes analysis comparing the relative orientation of matched strains between genders (Monte-Carlo P value < 0.0001), where sex-specific differences were most apparent in C57BL/6J and C3H/HeJ strains (Fig. 1B). Both the magnitude and direction of change for multiple bacterial genera were different between strains, suggesting that the impact of gender may depend on the host genotype. However, no clear patterns differentiating samples from males and females were observed when the entire population was examined together using Bray-Curtis dissimilarity (Fig. 1C). It appears that the genetic variation between strains obscures sex differences when the entire population is examined together. We used permutational multivariate analysis of covariance to test whether a matrix of major PCoA axes are dependent on sex, strain or a sex-by-strain interactions. We observed that both sex and strain have significant effects on the microbiota (p < 0.001, PERMANOVA of Bray-Curtis distances).
Figure 1.
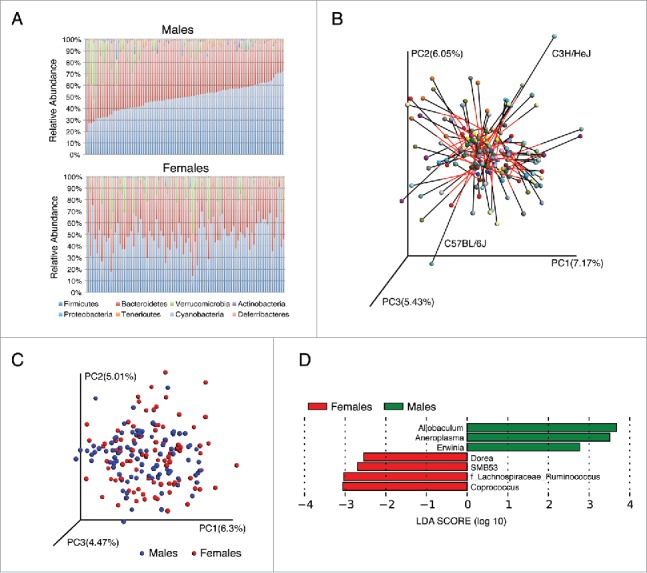
Sex differences in gut microbiota composition in 89 different inbred strains of mice. (A) Columns represent the average relative abundance of microbial phyla within 89 matched strains of male (n = 348) and female (n = 341) mice arranged in the same order. (B) Procrustes plot comparing β diversity between females and males of the same strains. The Bray-Curtis distances between the genders vary across the strains (M2 = 0.89, p < 0.001), highlighting the differences in microbial composition between sexes. (C) Bray-Curtis dissimilarity metric plotted in PCoA space comparing the gender microbial communities from different genders (89 matched strains). Each circle representing a different strain colored according to the gender. The first 3 principal components (PC1, PC2 and PC3) are shown, with the amount of variation explained are reported for each axes. Both sex and strain effects account for PC1 and PC3 variations (p < 0.0001, F test) and PC2 variation is explained only by strain effect (p < 0.001, F test). (D) Linear discriminant analysis (LDA) coupled with effect size measurements identified the most differentially abundant genus level taxa between female and male mice from 89 matched strains.
In order to further identify microbial taxa that account for the greatest differences between genders we performed Liner Discriminate Analysis (LDA) coupled with effect size measurements (LEfSe) and a multivariate linear model, which controlled for the effect of strain, as described in Methods. In the total cohort, the phylum Actinobacteria (0.81 ± 0.19 males; 0.18 ± 0.1 females) and Tenericutes (1.08 ± 0.22 males; 0.35 ± 0.12 females) were more abundant in male than female mice (p < 0.01) (Figs. 1A, S1B). At a finer taxonomic classification we detected multiple taxa with significantly altered relative abundance between male and female mice (Table S2). Genera with higher abundance in males as compared to female mice included Allobaculum, Anaeroplasma, and Erwinia, whereas SMB53 from family Colstridiaceae and 3 members of family Lachnospiraceae (Dorea, Coprococcus and Ruminococcus) were more abundant in female mice (Figs. 1D, S1C).
We also performed the analyses with strains that had 6 or more mice in both genders (total 15 strains, Table S1). When we analyzed all 15 strains together the same differences in composition as observed in the entire cohort were seen with the exception of the taxa, Oscillospira in females and an unclassified Bacteroidales in males (Fig. S2A). In addition, strain specific analysis revealed 7 strains out of 15 that showed additional significant sexually dimorphic taxa abundances (Table 1, Fig. S2B). To test if these differences were driven by host gender or reflected cage effects, we analyzed strains that had been sampled from multiple cages (total 5 strains). This analysis confirmed that all the observed differences between genders were consistently found in all cages (Fig. S2C).
Table 1.
The effect of sex on gut microbiota composition in HMDP strains.
| Relative abundance (%, Mean values ± SEM) |
||||
|---|---|---|---|---|
| Strain | Taxonomy | Females | Males | p-value (adjusted)* |
| BXD45/RwwJ | Allobaculum | 1.33 (±0.65) | 11.12 (±1.70) | 0.0244 |
| Clostridiaceae | 2.86 (±1.13) | 15.65 (±1.52) | 0.0025 | |
| Rikenellaceae | 0.014 (±0.002) | 0.002 (±0.001) | 0.0300 | |
| S24-7 | 44.16 (±2.88) | 29.02 (±2.09) | 0.0406 | |
| SMB53 | 1.44 (±0.10) | 0.37 (±0.09) | 0.0022 | |
| BXD85/Rww | Akkermansia | 0.02 (±0.01) | 37.8 (±1.11) | 0.0029 |
| Roseburia | 0.09 (±0.03) | 0.52 (±0.05) | 0.0178 | |
| Allobaculum | 0.003 (±0.002) | 7.00 (±0.97) | 0.0004 | |
| C57BL/6 | Coprococcus | 1.53 (±0.07) | 0.69 (±0.05) | 0.0270 |
| Bacteroides | 8.02 (±1.65) | 0.01 (±0.01) | 0.0040 | |
| C3H/He | Akkermansia | 0.01 (±0.004) | 23.14 (±2.40) | 0.0046 |
| Coprobacillus | 0.11 (±0.03) | 0.64 (±0.08) | 0.0252 | |
| Ruminococcus | 0.16 (±0.03) | 0.46 (±0.05) | 0.0340 | |
| Sutterella | 0 | 8.94 (±1.75) | 0.0302 | |
| BXD55/RwwJ | Lachnospiraceae | 2.77 (±0.23) | 1.51 (±0.22) | 0.0490 |
| AXB19a/PgnJ | Turicibacter | 0.19 (±0.04) | 0.01 (±0.008) | 0.0450 |
| BXD79/RwwJ | Lachnospiraceae | 1.59 (±0.20) | 6.09 (±0.93) | 0.0013 |
| Dorea | 0.25 (±0.08) | 0.01 (±0.004) | 0.0149 | |
| Coprococcus | 2.87 (±0.09) | 0.52 (±0.78) | 0.0167 | |
| Roseburia | 2.76 (±0.81) | 0.32 (±0.14) | 0.0187 | |
Note.
P value were adjusted using Benjamini-Hochberg FDR.
Sex-by-diet interactions in microbiota composition
In a follow-up study, we examined male and female mice of strains C57BL/6J, C3H/HeJ, and DBA/2J on chow vs. high fat, high sucrose (HF/HS) diets. Previously, these strains were shown to exhibit very different responses to the HF/HS challenge in terms of fat gain and insulin resistance.5,19 Figure 2A shows Principal coordinate analysis (PCoA) of strains C57BL/6J, C3H/HeJ, and DBA/2J on a chow diet or following feeding of a HF/HS diet for 8 weeks. When considering each strain independently then all 3 strains showed a clear segregation by sexes as well as by diet (p value < 0.001, PERMANOVA of Bray-Curtis distances) (Fig. 2A). Furthermore, these diet-microbiota associations tended to be sex-dependent, as seen in Figures 2B and S3A. Similar sex-specific diet–microbiota correlations have been recently reported in natural fish populations and humans.13 The most significant sex-by-diet interactions were observed in strain DBA/2J. In addition to strain specific shifts, some taxa showed similar shifts for all 3 strains in both genders. For instance, compared to mice on a chow diet, the HF/HS fed mice had greater abundance of genera classified to the family Erysioplerotrichaceae (Fig. S3B).
Figure 2.
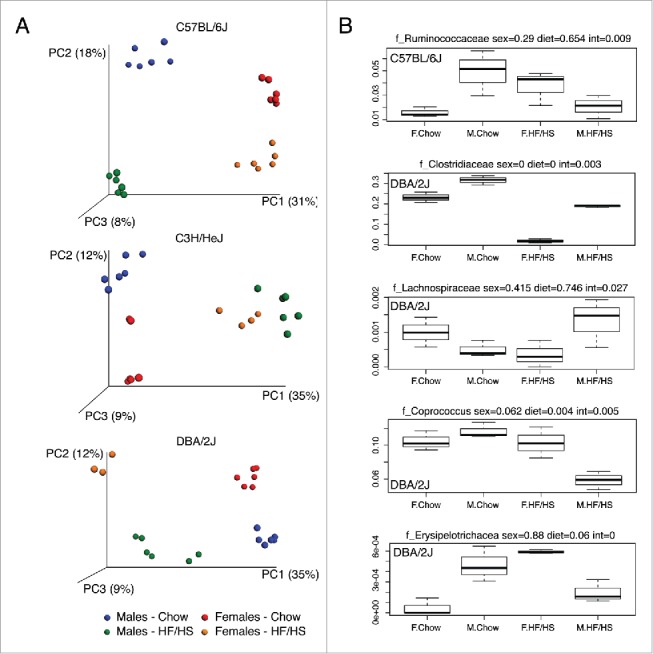
Principal coordinate analysis (PCoA) plot of unweighted UniFrac distances between male and female mice. C57BL/6J, C3H/HeJ, and DBA/2J strains fed a normal chow or high fat diets were studied as described in the text. PC1, PC2, and PC3 values for each mouse sample are plotted with the percent variation explained by each PC is shown in parentheses. (B) Examples of sex-by-diet interactions with 5 taxa in female and male from C57BL/6J and DBA/2J mice fed with chow or high fat diets. P values from MANOVA analysis are shown for sex, diet and sex-by-diet interactions (int).
Sex hormone mediated shifts in microbiota
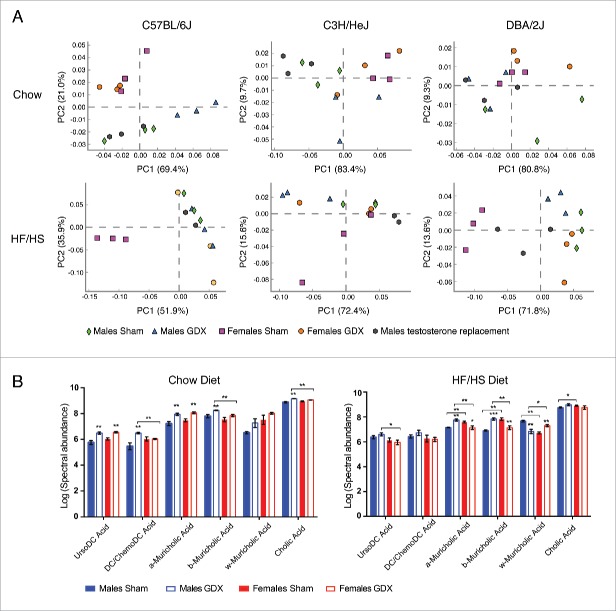
To determine whether differences between genders were mediated by sex hormones, we performed gonadectomy (GDX) with the same 3 strains (C57BL/6J, C3H/HeJ, and DBA/2J) and examined gut microbiota composition after 8 weeks. Principal component analysis revealed that all 3 strains of mice showed clear sex differences in gut microbiota composition on both chow and high fat diets (Fig. 3A). When analyzing all 3 mouse strains together in different treatment groups significant sex and strain effects were again detected on the microbiota (p value < 0.001, PERMANOVA of Bray-Curtis distacnes) (Fig. S4A). In male mice, the hormonal status affected the composition of microbiota more on chow diet, whereas in females this effect was more prevalent in response to the high fat diet (Fig. 3A). Administration of testosterone after gonadectomy prevented the significant gonadectomy-associated changes in gut microbiota composition in C57BL/6J and C3H/HeJ strains on both diets, but not in DBA/2J male mice (Fig. 3A). When analyzing all 3 strains together, then only the abundance of family Ruminococcacea was significantly different between GDX male (9.7% ± 1.2) compared to sham control male mice (5.7 % ± 0.09) on HF/HS diet (FDR <0.01). In females, a clear separation between sham control and GDX mice was detected on HF/HS diet with C57BL/6J and DBA/2J strains, where the abundance of Akkermansia was more abundant in sham controls than to GDX female mice (18.38% ± 0.05 vs. 0.02% ± 0.01, respectively; FDR < 0.01) (Figs. 3A and S4B). Again, differences between sham control and GDX mice in both genders were mostly strain-specific (Tables S3 and S4). Finally, we analyzed bile acid changes between C57BL/6J sham control and GDX mice in both genders. When fed a chow diet, the levels of several bile acid species in GDX mice were significantly higher compared to sham control mice in both genders (Fig. 3B, Table S5). Interestingly, significant gender specific differences in bile acid composition were observed in response to HF/HS diet, where several bile acids where significantly increased in GDX male mice compared to female GDX mice (Fig. 3B).
Figure 3.
The effect of gonadectomy in C57BL/6J, C3H/HeJ and DBA/2J mice. (A) Principal component analysis between male and female mice in normal chow or high fat/high-sucrose diet (B) from C57BL/6J, C3H/HeJ, and DBA/2J strains.(B) Bile acids measurements from gallbladder in male and female C57BL/6J mice on chow and HF/HS diet. Significance of differences was defined using unpaired T test with Holm-Sidak's correction for multiple hypothesis. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. DC, deoxycholic; GDX, gonadectomized.
Discussion
Sexual dimorphism is a frequent characteristic for a variety of common disorders, including autoimmune, metabolic, cardiovascular and psychiatric diseases.20-22 A large body of data now indicates a clear contribution of gut microbiota to many of these disorders, but the mechanisms that mediate these associations are poorly understood, highlighting the need to better understand genetic and environmental factors affecting microbial composition. It is clear that the modern environment (including diet) has an enormous impact on shaping our microbial communities and these changes likely contribute to the increased risk of immune-related and other common disorders. However, it remains unclear how gender affects host-microbiota interactions and if it is connected to disease susceptibility. Thus, we aimed to examine sex differences in gut microbiota composition in a population of 89 common inbred strains. Since, at least in mice fed the same diet and raised in the same vivarium, genetic background is an important contributor to gut microbiota composition,3 we felt that it was optimal to examine sex differences across a variety of backgrounds. We detected a number of taxa that differed between male and female mice and showed that sex differences in gut microbiota composition depends in part on genetic background. Using gonadectomized and hormone treated mice, we were able to identify clear hormonal effects on gut microbiota composition. Finally, we showed that dietary effects on the composition and diversity of gut microbiota are dependent in part on sex-specific interactions.
Reports relating to gender effects on the composition of gut microbiota have been inconsistent. We suspected that the failure of some studies to find significant sex effects10,11 may have been due to noise introduced by factors such as diet, age, and host1,7,9 Indeed, when we examined the entire population of 89 inbred strains, differences in diversity and many differences in composition appeared to be obscured by genetic background effects on the microbiota. However, clear differences became prominent when mice with specific genetic backgrounds were examined. Our experiments also demonstrated, consistent with other recent studies, that sex is an important factor to consider when looking at interactions between gut microbiome and environmental factors such as diet.12,13
Among the factors that most likely mediate gender dependent interactions are sex hormones. Recent studies with pathogen-free nonobese diabetic (NOD) mice have shown a causal relationship between sex hormones, gut microbiota, and control of autoimmunity.17,18 Both studies demonstrated that colonization of germ free NOD mice with defined microbiota lead to changes in hormone levels and these hormone-microbiota interactions could trigger protective pathways for type 1 diabetes. Yurkovetskiy and coworkers,17 showed that after puberty gut microbiota differed in males and females, and that male castration eliminated this trend. Another study demonstrated that sexually dimorphic expression of mucosal immunity genes appears already in prepubescent mice.23 Here, using gonadectomy we also demonstrated that differences in gut microbiota composition between genders were clearly mediated at least in part by sex hormones. Furthermore, we showed that testosterone treatment after gonadectomy prevented the significant changes in gut microbiota composition that were seen in untreated males. Interestingly, the hormonal status of male mice clearly affected the composition of microbiota on chow and high fat diets, whereas in females this effect was more prevalent in response to the high-fat diet. Finally, we showed that hormonal changes strongly affect bile acid profiles and that significant gender-specific differences in bile acid profiles become more prominent in response to a high-fat high-sugar diet. It has been shown that the rate of bile acid synthesis and bile acid pool sizes tend to be higher in females than in males.24 Since bile acids have been shown to affect gut microbiota,25-27 our results provide one possible mechanism for sex differences in bile acid composition.
Taken together, these findings support the role of sex differences in shaping gut microbial communities. Sex hormones appear to be responsible in part, but the pathways involved are unknown. In addition, hormonal organizational (developmental) effects and non-gonadal (sex chromosome) effects may well play a role.28 Understanding how sex differences affect gut microbiota could ultimately lead to the identification of novel factors that influence disease susceptibility and improve diagnostic and clinical strategies.
Materials and methods
Sample collection
All mice were obtained from The Jackson Laboratory and were bred at UCLA for 2 or more generations for use in this study. Briefly, until 8 weeks of age mice were maintained on a chow diet (Ralson Purina Company) and then placed on a high-fat, high-sucrose diet (HF/HS) (Research Diets D12266B) for an additional 8 weeks.5 Samples were obtained from the cecum of 689 mice from 89 strains, with an average of 6 mice per strain (341 males and 348 females) (Table S1). Mice from different strains and genders were housed in separate cages, but in the same room throughout the study. Cecum and fecal samples were snap frozen with liquid nitrogen and stored at −80°C. The animal protocol for the study was approved by the Institutional Care and Use Committee (IACUC) at University of California, Los Angeles.
16S rRNA gene sequencing and analysis
Microbial DNA was extracted and16S rRNA gene of the isolated DNA was sequenced using Illumina MiSeq platform as previously described.3 De-multiplexing 16S rRNA gene sequences, quality control and operational taxonomic unit (OTU) binning were performed using the open source pipeline Quantitative Insights Into Microbial Ecology (QIIME) version 1.7.0.29,30 The total number of sequencing reads was 15,010,979 (an average of 19,571 reads per sample) with an average length of 153 base pair reads. Sequences were binned into OTUs based on 97% identity using UCLUST31 against the Greengenes reference database.32 Each sample's sequences were rarefied to 17826 reads per strain to reduce the effect of sequencing depth. Microbial composition at each taxonomic level was defined using the summarize taxa function in QIIME. Beta-diversity (unweighted and weighted UniFrac metrics) and Bray-Curtis Dissimilarity were calculated using QIIME package. Comparison of group differences in microbiota within and between strains and gender (male and female) was performed using the adonis function for permutational multivariate analysis of variance (PERMANOVA) in R package vegan. Principal coordinates were transformed with Procrustes in order to facilitate direct comparison of trends in community structure between matched male and female strains (N = 89) using QIIME program. The criterion to select the best fit adopted by Procrustes analysis is the minimization of a residual sum of squares after matching (M2) which measures the remaining “lack of fit” of one configuration to the other. Then the M2 values might be used as a distance measure between any 2 PCoA comparisons where the smallest M2 indicates a small difference between 2 plots. The statistical significance of the goodness of the fit was measured by a Monte Carlo label permutation approach.
Gonadectomy studies
Male and female mice of strains C57BL/6J, C3H/HeJ and DBA/2J were gonadectomized under isoflurane anesthesia at 6 weeks of age. Scrotal regions of male mice were bilaterally incised, testes removed and the incisions closed with wound clips. Ovaries of female mice were removed through an incision just below the rib cage. The muscle layer was sutured and the incision closed with wound clips. In sham operated control mice, incisions were made and closed as described above. The gonads were briefly manipulated but remained intact. In mice that received hormonal replacement at time of surgery, a small incision was made at the base of the back of the neck. A 5 mg pellet of 5α-dihydrotestosterone 90 day release (Innovative Research of America, FL) was inserted and the incision closed. The animal protocol was approved by the Institutional Care and Use Committee (IACUC) at the University of California, Los Angeles. To examine the effect of diet, half of the mice were maintained on a chow diet and the other half placed on a high fat/high sucrose diet (Research Diets D12266B) at 8 weeks of age until 16 weeks of age as described above. There were 4 mice per group.
Gallbladder bile composition
Bile acids were measured in gallbladder of Sham control and gonodectomized C57BL/6J male and female mice on chow and HF/HS diets (on average 4 mice per group). Each mouse gallbladder bile sample was diluted with 1X PBS (Gibco#14190) to 1:20 and 1:1000 final dilution. The 1:20 dilution of bile sample was used for both cholesterol and phospholipid analysis, whereas the 1:1000 dilution was for total bile acid measurement. The assays were carried out on Olympus AU 680 and the reagents were from Backman Coulter # OSR 6116 (cholesterol), Wako #433-36201 (Phospholipids C), and BQ Kits # BQ042A-EALD (total bile acids). For individual bile acid analysis, all samples were protein precipitated with 3 volumes of cold methanol containing 0.1% formic acid and a stable label analog of deoxycholic acid, d4-deoxycholate, used as an internal standard. The samples were vortexed, spun down at 4K rpm for 10 min and the supernatant was collected and injected for LCMS analyses. All data was collected on a Thermo Q-Exactive mass spectrometer interfaced with Thermo Acella UHPLC pumps and Thermo PAL autosampler. The UHPLC column used was a Waters C18 BEH Acquity, 150 × 2.1, 1.7 u particles operated at 600 uL/min with starting conditions of 100% A (water, 0.1% formic), held for 0.5 min and ramped up to 80%A in 6 min, 40% A in 8 min and finally 100%B (98:2 acetonitrile:water, 0.1% formic acid) in 12 min with a hold for 3 min. LCMS data was collected in negative ion mode at 25K mass resolution and extracted ion chromatograms for all detected bile acids were generated with a 5 ppm mass accuracy.
Statistics
GraphPad Prism V6.0 (San Diego, California, USA) was used for analysis and graph preparation. For all graph data, the results are expressed as mean ± SD, and statistical analyses were performed using the 2-tailed non-parametric Mann-Whitney U test or Kruckal-Wallis test with Bebjamini-Hochberg FDR multiple comparison. Statistical significance of sample grouping for β-diversity analysis was performed using Permanova method (9999 permutations). Differences with a p value <0.05 were considered significant. A regression model in R was used to test the effect of sex and strain on microbiota variation using the first 3 PC scores (Bray-Curtis distances). To identify bacterial taxa whose sequences were differentially abundant between genders (males vs females) we used the STAMP33 and LEfSe (linear discriminant analysis (LDA) coupled with effect size measurements)34 programs. LEfSe is an algorithm for high-dimensional biomarker discovery that identifies genomic features (taxa) characterizing both statistical significance and biological relevance, allowing researchers to identify differentially abundant features that are also consistent with biologically meaningful categories (subclasses). We performed LEfSe analysis on the website http://huttenhower.sph.harvard.edu/galaxy. The differential features were identified on the genus level taxa, where gender was used as the class and strain as a subclass. LEfSe first performs the Kruskall-Wallis test (testing whether the values in different classes are differentially distributed), then a nonparametric Wilcoxon sum-rank test (testing whether all pairwise comparisons between subclasses within different classes significantly agreed with the class level trend) and then followed by LDA analysis to assess the effect size of each differentially abundant taxon.34 LEfSe analysis was performed under the following conditions: (1) the α value for the factorial Kruskal–Wallis test among classes is <0.01 and (2) the threshold on the logarithmic LDA score for discriminative features is >2.0. Multivariate association with linear model (MaAsLin) analysis was conducted to test for associations of microbial abundances with different genders while accounting for strain and cage effects.35 MaAsLin performs boosted, additive general linear models between metadata (the predictors) and microbial abundance (the responder). Multivariate analysis of variance (MANOVA) in R was used for analyzing sex, diet and sex-by-diet interactions. Significance of differences between bile acids in Sham and GDX mice where defined using unpaired T test with Holm-Sidak's correction for multiple hypothesis.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Hannah Qi, Zhiqiang Zhou, Judy Wu and Tieyan Han for expert assistance with mouse experiments.
Funding
This work was supported by National Institutes of Health (NIH) grants HL28481, HL30568 and DK094311 to A.J.L. and UCLA Iris Cantor Women's Health Center and UCLA CTSI to T.A.D. E.O. was supported by FP7-MC-IOF grant no 330381 and by the European Regional Development Fund (Project No. 2014–2020.4.01.15-0012). B.W.P. was supported by NIH training grant T32-HD07228.
Author contributions
A.J.L and T.A.D supervised the study. E.O, M.M and B.W.P oversaw collection of samples. E.O and M.M collected microbial data and performed the analyses. P.S and X.L performed bile acid analysis. E.O, M.M and A.J.L prepared the manuscript with comments from T.A.D.
References
- [1].Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science 2009; 326:1694-7; PMID:19892944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, et al.. Human genetics shape the gut microbiome. Cell 2014; 159:789-799; PMID:25417156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Org E, Parks BW, Joo JW, Emert B, Schwartzman W, Kang EY, Mehrabian M, Pan C, Knight R, Gunsalus R, et al.. Genetic and environmental control of host-gut microbiota interactions. Genome Res 2015; 25:1558-69; PMID:26260972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al.. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011; 334:105-8; PMID:21885731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau CD, Bennett BJ, et al.Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab 2013; 17:141-152; PMID:23312289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al.. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014; 505:559-63; PMID:24336217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Carmody RN, Gerber GK, Luevano JM Jr, Gatti DM, Somes L, Svenson KL, Turnbaugh PJ. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 2014; 17:72-84; PMID:25532804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].van Nas A, Guhathakurta D, Wang SS, Yehya N, Horvath S, Zhang B, Ingram-Drake L, Chaudhuri G, Schadt EE, Drake TA, et al.. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology 2009; 150:1235-49; PMID:18974276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kovacs A, Ben-Jacob N, Tayem H, Halperin E, Iraqi FA, Gophna U. Genotype is a stronger determinant than sex of the mouse gut microbiota. Microb Ecol 2011; 61:423-8; PMID:21181142 [DOI] [PubMed] [Google Scholar]
- [10].Lay C, Rigottier-Gois L, Holmstrøm K, Rajilic M, Vaughan EE, de Vos WM, Collins MD, Thiel R, Namsolleck P, Blaut M, et al.. Colonic microbiota signatures across five northern European countries. Appl Environ Microbiol 2005; 71:4153-5; PMID:16000838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Huttenhower CH. C. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207-14; PMID:22699609; http://dx.doi.org/ 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shastri P, McCarville J, Kalmokoff M, Brooks SPJ, Green-Johnson JM. Sex differences in gut fermentation and immune parameters in rats fed an oligofructose-supplemented diet. Biol Sex Differ 2015; 6:13; PMID:26251695; http://dx.doi.org/ 10.1186/s13293-015-0031-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Org E, Parks B, Lusis AJ, Knight R, Caporaso JG, Svanbäck R. Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat Commun 2014; 5:4500; PMID:25072318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, Zhang Y, Shen J, Pang X, Zhang M, et al.. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci 2008; 105:2117-2122; PMID:18252821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al.. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464:59-65; PMID:20203603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, Cresci A, Silvi S, Orpianesi C, Verdenelli MC. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol 2006; 72:1027-33; PMID:16461645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. Gender bias in autoimmunity is influenced by microbiota. Immunity 2013; 39:400-12; PMID:23973225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013; 339:1084-8; PMID:23328391 [DOI] [PubMed] [Google Scholar]
- [19].Parks BW, Sallam T, Mehrabian M, Psychogios N, Hui ST, Norheim F, Castellani LW, Rau CD, Pan C, Phun J, et al.. Genetic architecture of insulin resistance in the mouse. Cell Metab 2015; 21:334-46; PMID:25651185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Whitacre CC. Sex differences in autoimmune disease. Nat Immunol 2001; 2:777-80; PMID:11526384; http://dx.doi.org/ 10.1038/ni0901-777 [DOI] [PubMed] [Google Scholar]
- [21].Danska JS. Sex Matters for Mechanism. Sci Transl Med 2014; 6:258fs40-258fs40; PMID:25320230; http://dx.doi.org/ 10.1126/scitranslmed.3009859 [DOI] [PubMed] [Google Scholar]
- [22].Arnold AP, Lusis AJ. Understanding the sexome: measuring and reporting sex differences in gene systems. Endocrinology 2012; 153:2551-5; PMID:22434084; http://dx.doi.org/ 10.1210/en.2011-2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Steegenga WT, Mischke M, Lute C, Boekschoten MV, Pruis MG, Lendvai A, Verkade HJ, Boekhorst J, Timmerman HM, Plösch T, et al.. Sexually dimorphic characteristics of the small intestine and colon of prepubescent C57BL/6 mice. Biol Sex Differ 2014; 5:11; PMID:25243059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Turley SD, Schwarz M, Spady DK, Dietschy JM. Gender-related differences in bile acid and sterol metabolism in outbred CD-1 mice fed low- and high-cholesterol diets. Hepatology 1998; 28:1088-94; PMID:9755247; http://dx.doi.org/ 10.1002/hep.510280425 [DOI] [PubMed] [Google Scholar]
- [25].Clarke SF, Murphy EF, Nilaweera K, Ross PR, Shanahan F, O'Toole PW, Cotter PD. The gut microbiota and its relationship to diet and obesity: new insights. Gut Microbes 2012; 3:186-202; PMID:22572830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Islam K, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011; 141:1773-81; PMID:21839040; http://dx.doi.org/ 10.1053/j.gastro.2011.07.046 [DOI] [PubMed] [Google Scholar]
- [27].Li T, Chiang JY. Bile acids as metabolic regulators. Curr Opin Gastroenterol 2015; 31:159-65; PMID:25584736; http://dx.doi.org/ 10.1097/MOG.0000000000000156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Link JC, Chen X, Arnold AP, Reue K. Metabolic impact of sex chromosomes. Adipocyte 2013; 2:74-9; PMID:23805402; http://dx.doi.org/ 10.4161/adip.23320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010; 7:335-6; PMID:20383131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 2013; 10:57-9; PMID:23202435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Edgar RC. Quality measures for protein alignment benchmarks. Nucleic Acids Res 2010; 38:2145-53; PMID:20047958; http://dx.doi.org/ 10.1093/nar/gkp1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012; 6:610-8; PMID:22134646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 2014; 30:3123-4; PMID:25061070; http://dx.doi.org/ 10.1093/bioinformatics/btu494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol 2011; 12:R60; PMID:21702898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, et al.. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012; 13:R79; PMID:23013615 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.