ABSTRACT
Using a murine Salmonella model of colitis, we recently reported that mice receiving a community of defined gut microbiota (MET-1) lost less weight, had reduced systemic inflammation and splenic S. typhimurium infection, and decreased neutrophil infiltration in the cecum, compared to vehicle controls. In addition, animals receiving MET-1 exhibited preserved tight junction protein expression (Zonula occludens-1, claudin-1), suggesting important effects on barrier function. In this addendum, we describe additional in vitro experiments examining effects of MET-1, as well as in vivo experiments demonstrating that MET-1 is protective in a DSS model of colitis after administration of antibiotics. Placed in the context of our findings and those of others, we discuss differences in our findings between the Salmonella colitis and DSS colitis models, provide speculation as to which bacteria may be important in the protective effects of MET-1, and discuss potential implications for other GI diseases such as IBD.
KEYWORDS: DSS, epithelial barrier, microbial ecosystem therapeutic, microbiota, salmonella
Introduction
The number of bacteria present in the gut eclipses the total number of eukaryotic cells in the human body by many fold, and the vast majority of these live in the nutrient rich environment of the intestinal lumen where they can thrive as a community. Commensal bacteria are beneficial to gastrointestinal health, thus the manipulation of the gut microbiota to increase commensals or optimize microbial communities has been an area of active interest with regards to disease management.1 An extension of this concept has been the use of fecal microbial therapy (FMT) to treat infectious colitis from pathogens such as Clostridium difficile.2,3 FMT entails transplanting healthy donor fecal material containing commensal bacteria into the colon of a patient to reconstitute a healthy microbial community, and has proven very successful for the treatment of recurrent C. difficile infection (CDI).4,5 However, FMT has a poorly defined and non-reproducible composition and carries risks of unintended infectious organisms.
Work in our laboratories has attempted to create a defined Microbial Ecosystem Therapeutic (MET)6,7 with known organisms, in order to make a safe, reproducible therapy that simulates the beneficial effects of FMT. The mixture, designed to replace a dysfunctional, damaged ecosystem with a healthy ecosystem composed of ‘native’ intestinal bacteria, is known as “Repoopulate” or MET-1. The composition of MET-1 is shown in Table 1. It consists of 33 strains of commensal bacteria isolated from the stool of a healthy human donor, as previously described.8 Strains were selected based on antimicrobial resistance profiles, as well as on their ability to grow as a robust community in vitro in a continuous culture chemostat.9 Additional information on the functional and metabolic capabilities (or “metabolome”) of MET-1 has been described elsewhere.10 MET-1 has been used successfully in 2 patients to cure recurrent C. difficile infection that was refractory to standard antibiotic therapies.8
Table 1.
Composition of MET-1.
| Strain Name | % match | Strain Name | % match |
|---|---|---|---|
| Acidaminococcus intestinia | 99% | Eubacterium rectale - 3 | 99% |
| Bacteroides ovatus | 99% | Eubacterium rectale - 4 | 99% |
| Bifidobacterium adolescentis - 1 | 99% | Eubacterium ventriosum | 99% |
| Bifidobacterium adolescentis - 2 | 99% | Fecalibacterium prausnitzii | 99% |
| Bifidobacterium longum - 1 | 99% | Lachnospira pectinoschiza | 95% |
| Bifidobacterium longum - 2 | 99% | Lactobacillus casei | 99% |
| Blautia stercoris | 99% | Lactobacillus paracasei | 99% |
| Clostridium cocleatum | 92% | Parabacteroides distasonis | 99% |
| Collinsella aerofaciens | 99% | Enterobacter aerogenesc | 99% |
| Dorea longicatena - 1 | 99% | Roseburia faecisd | 99% |
| Dorea longicatena - 2 | 99% | Roseburia intestinalis | 99% |
| Escherichia coli | 100% | Ruminococcus obeum | 99% |
| Butyricicoccus pullicaecorumb | 95% | Blautia lutie | 95% |
| Eubacterium eligens | 99% | Ruminococcus torques - 1 | 99% |
| Eubacterium limosum | 97% | Ruminococcus torques - 2 | 99% |
| Eubacterum rectale - 1 | 99% | Streptococcus mitisf | 99% |
| Eubacterium rectale - 2 | 99% |
Note. Updated strain identification of MET-1 mixture, as determined by full-length 16S sequencing. Using BLAST, sequences were matched against Greengenes database (July 2015).
Formerly identified as Acidaminococcus intestinalis,
Formerly identified as Eubacterium desmolans,
Formerly identified as Raoultella ornithinolytica,
Formerly identified as Roseburia faecalis,
Formerly identified as Ruminococcus obeum,
Formerly identified as Streptococcus parasanguinis.
In addition to protecting against C. difficile, there is increasing evidence that the gut microbiota play an important role in protecting the host against the deleterious effects of other intestinal pathogens, including Salmonella.11 Certain shifts in the gut microbiota (sometimes referred to as dysbiosis) contribute to this phenomenon. In animals, for example, treatment with a single dose of streptomycin will result in loss of colonization resistance, rendering mice more susceptible to developing Salmonella infection.12 The effect is similar regardless of whether streptomycin-sensitive or streptomycin-resistant strains of Salmonella are used, indicating that the antimicrobial susceptibility of the pathogen itself is of little importance.13 This observation has also been reported in humans, where exposure to antibiotics used to treat other infections increases the risk of developing Salmonella infection, with either Salmonella-resistant or Salmonella-sensitive strains.14,15
We have demonstrated that MET-1 is protective against Salmonella enterica serovar Typhimurium (S. typhimurium) in a murine antibiotic model of Salmonella colitis.16 C57BL/6 mice, pretreated with oral streptomycin prior to receiving MET-1 or vehicle control, were then gavaged with S. typhimurium. The S. typhimurium-infected mice receiving MET-1 lost less weight, had reduced inflammatory cytokine levels in serum, and decreased neutrophil infiltration in the cecum. MET-1 also preserved tight junction (TJ) protein expression (Zonula Occludens-1, claudin-1) in the cecum, and reduced metastatic spread of S. typhimurium infection to the spleen compared to vehicle controls. MET-1 did not appreciably kill Salmonella in vivo or decrease the intestinal burden of Salmonella in the intestine, suggesting that MET-1 confers protection through other mechanisms.
S. typhimurium and C. difficile are 2 different pathogens with quite differing mechanisms of pathogenicity. S. typhimurium relies heavily on host cell invasion and C. difficile uses primarily a toxin-mediated mechanism. Our discovery that MET-1 can protect against both of these organisms and both of these mechanisms of pathogenesis offers insight into core functions of healthy microbial communities. We will discuss our work using defined microbiota in different animal models of colitis, and describe additional data implicating enhancement of barrier function as an important mechanism of action. We are still exploring multiple possible mechanisms of action for MET-1, but in this addendum we will focus specifically on effects on barrier function and microbial composition. We will also discuss potential implications for other GI diseases such as IBD.
Not just sitting on the fence: Gut microbiota protect by enhancing gut barrier integrity
One common unifying theme of colitis is inflammation leading to loss of barrier function. Intestinal barrier function is a key aspect of innate immunity, designed to physically limit bacteria to the intestinal lumen. Tight junctions between enterocytes at the apical aspect of the epithelium are comprised of transmembrane proteins that confer size and charge selectivity between the gut lumen and mucosa. In other words, tight junctions seal the space between adjacent epithelial cells. In addition to direct epithelial cell invasion, enteric bacteria pathogens can hijack host cell machinery by altering the structure and function of the TJ barrier. These effects may result from direct modification of TJ proteins, such as occludin, claudin and Zonula Occludens-1 (ZO-1), or by alteration of the perijunctional actomyosin ring.17 S. typhimurium specifically increases access to the basolateral compartment through disruption of the TJ complex and TJ protein localization, resulting in increased paracellular permeability and membrane “leakiness.”18
Our Salmonella colitis study showed that compared to saline pretreatment control, pretreatment with MET-1 preserved localization of TJ proteins measured (ZO-1, claudin-1), after Salmonella infection. We additionally found decreased spread of Salmonella to distant organ sites such as the spleen in infected MET-1 animals compared to infected vehicle controls, confirming preservation of barrier function by MET-1. Interestingly, our findings showed that MET-1 had minimal effect on Salmonella viability, and no effect on direct Salmonella invasion of intestinal epithelial cells.16 It thus appears that the mechanism by which MET-1 attenuates systemic infection is by inhibiting the ability of S. typhimurium to gain access to the systemic circulation through the paracellular pathway; MET-1 preserves the integrity of the TJ complex.
To examine this in more detail, we used an in vitro model of altered barrier function in which T84 human intestinal epithelial cell monolayers were grown on transwell semi-permeable supports and co-cultured with either S. tm/MET-1 or S. tm/vehicle control in the presence of C. difficile toxin (which rapidly destroys the integrity of the actin cytoskeleton); transepithelial electrical resistance (TER) was used to measure barrier integrity (Fig. 1A). In mice, intestinal inflammation and neutrophil infiltration contribute to epithelial barrier function disruption;19 however for the in vitro experiment, C. difficile toxin was used as a tool to disrupt the epithelial cell barrier. Whereas monolayers treated with toxin alone or Salmonella + toxin had a decrease in TER, in the MET-1 + Salmonella + toxin samples the TER decreased only marginally, indicating preserved barrier function with MET-1 compared to saline control (Fig. 1B). In the toxin-treated group receiving MET-1, fewer Salmonella bacteria were recovered from the basolateral compartment of the transwell compared to the vehicle control group (Fig. 1C). This data further supports the proposed mechanism that MET-1 enhances barrier function to decrease Salmonella translocation via a paracellular route.16
Figure 1.
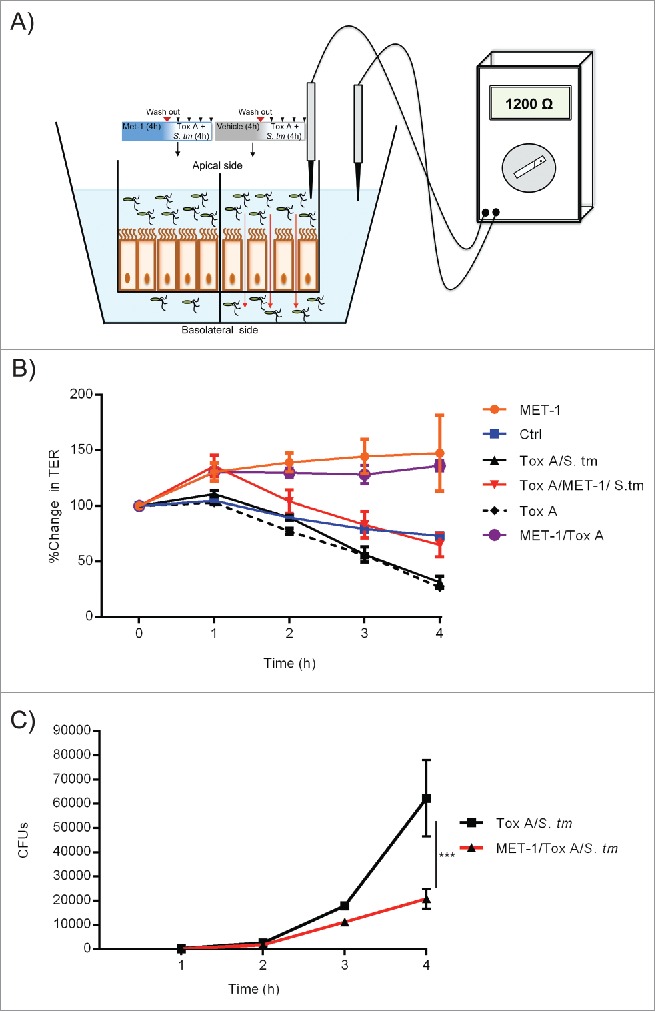
MET-1 enhances barrier function and decreases Salmonella translocation across epithelial cell monolayers via the paracellular pathway. (A) Schematic drawing of experimental design of T84 intestinal epithelial cell monolayers grown to confluence on transwell permeable supports and then treated with either MET-1 (left) or saline vehicle control (right) for 4 hours, followed by addition of S. typhimurium and C. difficile Tox A, to rapidly induce a breach in the barrier function of the monolayer and “open up” the paracellular pathway. (B) Measurement of transepithelial electrical resistance (TER), an indicator of barrier function, in T84 monolayers treated with various combinations of C. difficile toxin, Salmonella, and MET-1, showing preserved barrier function in MET-1 groups. (C) Quantification of the viable number of S. typhimurium (Colony Forming Units, or CFUs) on the basolateral side of the monolayer was determined by sampling the basolateral compartment every 60 minutes for 4 hours, and plating serial dilutions onto MacConkey plates.
Next, we considered whether claudin-2 would be affected by MET-1 pretreatment. Claudin-2 has been recently described as a “leaky” intestinal TJ protein which plays an important role in water and solute regulation and is upregulated by S. typhimurium infection.20 MET-1/S. tm mice were observed to generally produce more formed stool than VC/S. tm mice. As shown in Figure 2A, claudin-2 expression in MET-1/S. tm mice was confined primarily to the bottom of the crypts and more closely resembled uninfected controls (VC mice and MET-1 mice). In contrast (and as has been previously reported),20 in VC/S. tm mice the expression of claudin-2 was more diffusely spread up the length of the crypts. This site specificity appears important as the overall amount of claudin-2 in the VC/S. tm mice and MET-1/S. tm mice was unchanged (Fig. 2B). Taken together, these data indicate that even during Salmonella infection MET-1 maintains the localization of claudin-2 at the bottom of the crypts, where it is normally expressed under conditions of intestinal homeostasis.
Figure 2.
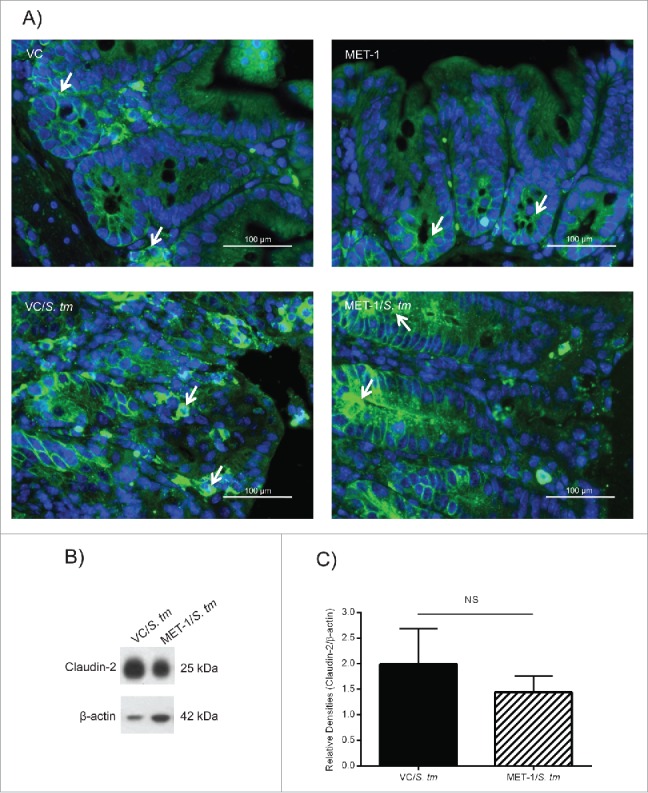
MET-1 pretreatment prevented the disruption of claudin-2 localization in the cecum caused by S. typhimurium infection. (A) Claudin-2 immunofluorescence staining (green) in ceca fixed in 10% formalin (400x magnification). Nucleic acid was stained using DAPI (blue). White arrows in picture show the green staining of claudin-2. Uninfected mice were pretreated with vehicle control (VC) or MET-1. Mice infected with S. typhimurium (S. tm) were also pretreated with either vehicle control (VC/S. tm) or MET-1 (MET-1/S. tm). VC/S. tm mice had increased claudin-2 epithelial cell immunofluorescence at the crypt base as well as in the mid and upper crypt zones and on the surface (compared to uninfected controls). This alteration in claudin-2 expression was attenuated in MET-1/S. tm mice. Scale bar represents 100 μm. (B) Expression of claudin-2 was measured by Western blot analysis of total cecal lysates (sample blot shown). Although panel (A) showed differences in localization, densitometric values calculated for the ratio of claudin-2 to β-actin showed no statistically significant difference between total expression of claudin-2 in MET-1/S. tm compared to VC/S. tm mice. Data were analyzed using a 1-way ANOVA with Bonferroni correction. N = 6 for each group. NS = not significant.
To further examine the effects of MET-1 on barrier function we utilized another murine model of colitis that disrupts barrier function, but does not involve infection with a specific pathogen. Dextran Sulfate Sodium (DSS) is a sulfated dextran molecule that can be dissolved in the drinking water of mice, leading to the development of colitis.21 DSS-induced colitis is one of the most commonly used mouse models of IBD and is thought to model human ulcerative colitis, although the mechanism of the disease is not well understood.22 It is thought that DSS can chemically compromise barrier function and allow for infiltration of microbes and luminal antigens to drive inflammation. The inability to maintain an intact barrier thus contributes to excessive translocation and sampling of intestinal microbes, resulting in a loss of homeostasis and resultant inflammation.
For this model, mice were pretreated 24 hours before MET-1 gavage with a combination of oral vancomycin and streptomycin. Both of these antibiotics are not absorbed into the circulation when taken by mouth, remaining in the gut lumen. Therefore, this combination of antibiotics has broad-spectrum effects on the gut microbiota, effectively creating a niche for other microbes to colonize. Following oral antibiotic treatment MET-1 was administered in a pretreatment regimen as described previously,16 and then mice were given 3% DSS. The hypothesized rationale for the efficacy of MET-1 pretreatment following oral antibiotics in the DSS model was that a decrease in indigenous microbes in the mouse colon would allow for stronger colonization of MET-1 following oral gavage, and that this microbial composition would be less conducive to the development of colitis following DSS administration. As predicted, MET-1 treated mice were less sick than control DSS mice (Fig. 3). Over the course of DSS exposure, mice that received MET-1 following antibiotics lost significantly less weight and displayed less histologic injury than controls. Effects on the immune response were further analyzed by measuring serum cytokine secretion levels. A decrease was noted in the average serum concentration for several inflammatory cytokines, including IL-1, IL-6, MCP-1, and IL-12 in antibiotic MET-1 DSS mice versus antibiotic saline DSS mice (Fig. 3B). The greatest cytokine decrease with DSS treatment occurred in chemokines important for acute phase inflammation, and myeloperoxidase (MPO) immunohistochemical staining also revealed reduced neutrophil infiltration into the colon of mice receiving MET-1 (data not shown). Both of these findings are similar to what was observed in the Salmonella colitis mouse model with MET-1.16
Figure 3.
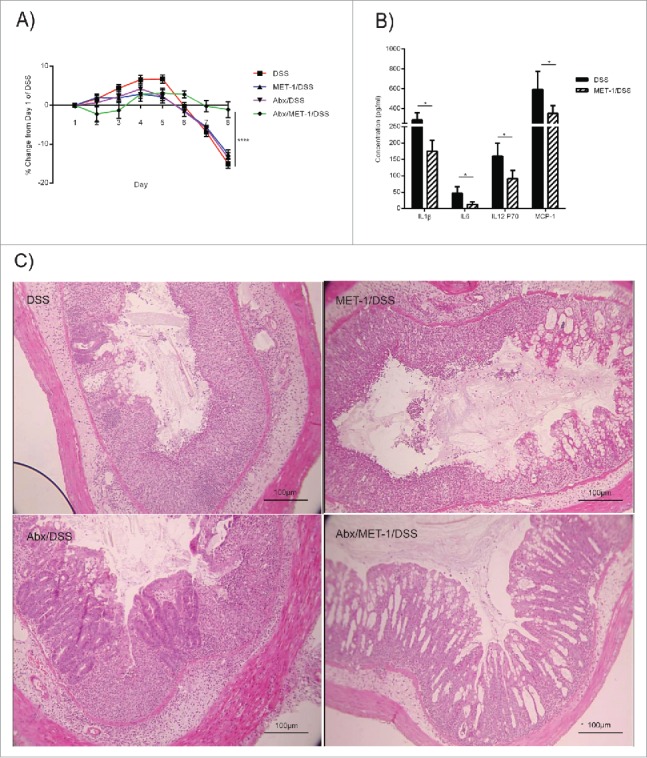
MET-1 treatment following oral antibiotics prevented DSS-induced weight loss and histological damage. (A) Female, 7 week old C57BL/6 mice received MET-1 treatment following oral antibiotics (0.1 mg Vancomycin, 20 mg Streptomycin, or saline as vehicle control), prior to acute exposure to 3% DSS. Data analyzed by 2 way ANOVA with Bonferroni correction, (* p < 0.001). (B) Serum cytokine levels of IL-1β, IL-6, IL-12p70, and MCP-1 were significantly reduced in DSS mice pretreated with antibiotic + MET-1 compared to DSS mice pretreated with antibiotic + vehicle control. Serum cytokine levels were measured using a Bio-Plex Pro mouse cytokine magnetic bead kit. Both groups received antibiotics prior to DSS. Data were analyzed by Student T-test (* p < 0.05). (C) Representative photo of H&E staining of colons. DSS treatment resulted in a loss of mucosal architecture, and submucosal edema. MET-1 pretreatment following oral antibiotics protected against histological damage. N = 4 for each treatment group (N = 5 for DSS group). Scale bar represents 100 μm.
In contrast, there was a relative preservation of mucin-2 in the colons of antibiotic MET-1 DSS mice compared to the other treatment groups, an effect not observed with the Salmonella colitis model (Fig. 4). Mucin-2 is the primary constituent of the secreted mucous layers in the colon and it has a significant impact on intestinal homeostasis, acting as a physical barrier atop the epithelium. Loss of function of mucin-2 results in the development of spontaneous colitis23 and mucin-2 is reduced in the DSS colitis model.24,25 The ability of the microbiota to regulate the composition of the intestinal mucous has been demonstrated in vivo.25,26 In vitro, the probiotic Lactobacillus plantarum strain 299v was shown to upregulate mucin-2 mRNA in HT-29 cells.27 Upregulation of mucin-2 by other probiotic Lactobacilli has also been reported.28 In contrast, bacteria from Deferribacteraceae, and Verrucomicrobiaceae families, including an increase in the mucin-degrading genera Akkermansia, and Mucispirillum, are increased in DSS-treated mice.29 The latter microbes are known inhabitants of the gut that possess mucolytic ability. Taken together, these findings suggest that specific microbes may contribute to modulating the intestinal mucous barrier.
Figure 4.
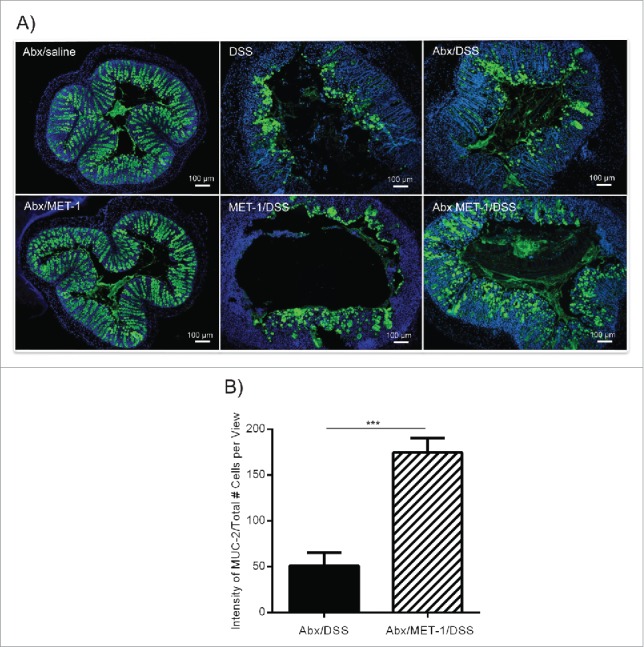
MET-1 pretreatment following oral antibiotics attenuates DSS-mediated loss of Mucin-2 in the colon. (A) Representative photos of distal colons from antibiotic saline and antibiotic MET-1 pretreated mice, saline and MET-1 pretreated DSS mice and antibiotic saline, antibiotic MET-1 pretreated DSS mice. Mucin-2 appears green, nuclei stained using DAPI (blue). Scale bar represents 100 μm. Mucin-2 quantification data in (B) shows a statistically significant difference between Abx/DSS treatment and Abx/MET-1/DSS treatment, with more Muc-2 in the MET-1 treatment group, N = 4 mice per group. Data were analyzed by ANOVA with Bonferroni correction (***p < 0.0005).
Not all MET are created equal: Gut microbiota composition may be more important than we appreciate
During DSS there are compositional changes in the microbiota, including increases in a number of different taxa. DSS results in an increase in the Proteobacteria family Enterobacteriaceae,29 which is represented in low quantities in MET-1. Interestingly, this family of microbes was seen to be disproportionately high in 2 patients with CDI, but was almost completely eradicated 6 months following treatment with MET-1, suggesting that MET-1 inhibits growth of this family.8
Also in support of the role that microbes play in DSS-induced colitis is work demonstrating that certain probiotic organisms can curb disease progression. Oral pretreatment with a probiotic mixture containing 4 Lactobacillus spp. and four Bifidobacterium spp for one week prior to the induction of DSS-induced colitis was capable of significantly reducing weight loss and histological damage in mice. These changes were also correlated with restoration of certain microbes that were altered during colitis.30 The probiotic mixture VSL#3, comprised of 8 different strains belonging to the bacterial genera Bifidobacterium, Lactobacillus, and Streptococcus, was protective against DSS colitis by mechanisms involving limiting apoptosis in the colon and upregulation of tight junction proteins.31
The IL-10 knockout (Il-10−/−) mouse model is another commonly used animal model for IBD.32,33 Changes to the microbiota that occur with progression of intestinal inflammation have been reported in this model. Increases in Proteobacteria and particularly in E. coli, as well as decreases in Firmicutes, microbial richness and overall diversity occurring over time have been described.34 These changes have also been reported in UC and CD patients.35 As with the DSS model, however, it can be difficult to discern whether these alterations of the enteric microbiota are a cause or effect of colonic inflammation. In one Il-10−/− study, the investigators concluded that the observed changes in the microbiota were secondary to the host immune response, although they noted that the dysbiotic changes observed in the composition of the microbiota may serve to further perpetuate colonic inflammation by contributing to, and perhaps even amplifying, the host inflammatory response.34 Interestingly, FMT after ileocolic resection (ICR) in Il-10−/− mice seems to protect against ileal inflammation post-operatively, but increase colonic inflammation.36 The authors of this study concluded that increased inflammatory cytokines could act in concert with mucolytic pathobionts such as Klebsiella species, shown to be increased in the colonic samples, to culminate in the additive effect of increased bacterial translocation, a phenomenon previously reported to drive colitis and contribute to endotoxemia in this model.36,37
It is interesting to examine the composition of MET-1 in comparison to those taxa thought to be important in human IBD. The two major phyla in the healthy microbiota are consistently the Bacteroidetes and Firmicutes, highly represented in MET-1, whereas IBD is associated with an increase in Proteobacteria,35 present in very low abundance in MET-1. The family Lachnospiraceae, which are well represented in MET-1, also appear to be significantly reduced in IBD microbiota. Clinically, we have already successfully used MET-1 as treatment for 2 patients with refractory, chronic CDI.8 Treatment with MET-1 stimulated expansion of Lachnospiraceae in both patients that were treated for CDI, suggesting the MET-1 community created a positive environment for these beneficial bacteria.8 More specifically, the organism Faecalibacterium prausnitzii, which we consider to be an integral member of MET-1, has been shown to be decreased in IBD patients suffering relapse, while its abundance is thought to be an important factor in disease remission38 and is present to a high degree in MET-1.8 Conversely, taxa associated with the Proteobacteria family Enterobacteriaceae appear to be highly abundant in IBD39,40 but are represented by only 2 species in MET-1, and were significantly reduced in the 2 patients following MET-1 treatment.8 This is also true of Bacteroides and Desulfovibrio which are in high abundance in IBD patients41,42 but low and not present in MET-1, respectively.
The fact that there are a number of taxa increased in DSS-induced colitis in mice, as well as in IBD in humans, suggests that changes to these taxa may contribute to development of disease. Furthermore, it appears as though changes to the microbiota of mice in this model occur at the onset of disease, hinting at causation.43 This suggests that microbial transplant could be efficacious in preventing or correcting dysbiosis. Many of these taxa occur in low amounts or not at all in the MET-1 mixture and abundance of some of these taxa was drastically inhibited by MET-1 in humans, comprising only a very small proportion of the community following treatment. The opposite is also true; certain taxa which are known to be decreased in IBD patients are very abundant in MET-1 and flourish following transplant into a host. These parallels provide a compelling link between microbial contribution to the inflammatory disease state and the potential efficacy of MET-1 treatment.
In addition to MET-1, other groups have similarly demonstrated that engineered communities can be used to treat human disease. For example, Tvede and Rask-Madsen44 created a mixture of 10 strains of gut-derived bacteria grown in pure culture and then administered these, resuspended in saline, by enema to treat C. difficile infection. The mixture was comprised of Clostridia (C. innocuum, C. ramosum, C. bifermentans), Bacteroides (B. ovatus, B. vulgatus, B. thetaiotaomicron), Enterococcus faecalis (previously named Streptococcus faecalis), Ruminococcus productus (previously named Peptostreptococcus productus), and 2 strains of E. coli. Of the 6 recurrent CDI patients in the study, one received FMT, 4 received their defined mixture, and one patient who did not respond to FMT subsequently received the defined mixture, with good response. This study was performed in 1989 before the emergence of hypervirulent strains of C. difficile but it was nevertheless effective in achieving cure, demonstrating the feasibility of a defined gut microbiota approach. Using an animal model, an even more narrowly defined mixture of 6 strains of bacteria (Staphylococcus warneri, Enterococcus hirae, Lactobacillus reuteri), and 3 novel species (Anaerostipes sp. Bacteroidetes sp and Enterorhabdus sp.) was shown to be effective in protecting mice against C. difficile infection.45 Finally, as mentioned previously, defined probiotic mixtures such as VSL#3, which contains 8 different strains of bacteria, have shown some efficacy for other GI inflammatory conditions such as pouchitis.46,47
It is important to remember, however, that the compositional structure of a given microbiota tells only half the story – the abundance profiles of certain taxa may not always be correlated to their significance within a community48,49 and the mere presence or absence of certain species is likely less important than the net functional output of the ecosystem members. In addition, secreted products from the bacteria likely contribute significantly to their protective effects. Bacteria are not static beings, but instead respond rapidly to their environment and secrete a plethora of chemical compounds, some of which are used as signaling molecules both among microbes, and between microbes and host. Production of small molecules, such as short chain fatty acids, by the gut microbiota is heavily influenced by ecosystem structure and available nutrients, and also by the inflammatory environment of the gut itself.50 The same concept applies to organisms that can be found in a “dysbiotic” community. For example, E. faecalis secretes gelatinase, a metalloprotease which both degrades the epithelial junctional protein E-cadherin51 and acts on PAR-2 to increase intestinal permeability, an effect recapitulated by supernatants from ulcerative colitis patient fecal samples.52 Therefore, as we begin to unravel the mechanisms of protection by therapeutic ecosystems such as MET-1, a thorough understanding of the microbial ecosystem metabolome, and the influence of this on the host, is warranted. Such a study will be complex since the same microbes may respond differently to exposure to different ecosystems.53
Future directions
Dysbiosis may contribute to a variety of other gastrointestinal diseases and diseases in other organs. To better grasp the complexities of ecosystem function, future areas of study should focus on investigating effects of ecosystem members, on the bacterial metabolites released by these microbiota, and on the barrier function-enhancing capacity of defined microbiota, to determine which members of the community are critical for this important function. Further research in this area may lead to defined therapies that provide an optimally enhanced barrier function with the potential to protect against multiple other diseases, where barrier function disruption may play a critical role.
Abbreviations and acronyms
- Abx
Antibiotics
- CDI
Clostridium difficile infection
- CFU
Colony Forming Unit
- DSS
Dextran Sulfate Sodium
- FMT
Fecal Microbial Therapy
- H&E
Hematoxylin and Eosin staining
- IBD
Inflammatory Bowel Disease
- ICR
Ileocolic resection
- LPS
Lipopolysaccharide
- LTA
Lipoteichoic Acid
- MET
Microbial Ecosystem Therapeutic
- MET-1
Microbial Ecosystem Therapeutic Mix (“Repoopulate”)
- S. tm
Salmonella typhimurium
- TLR
Toll-like receptor
- VC
Vehicle control
Disclosure of potential conflicts of interest
E.O.P and E.A-V. are co-founders of Nubiyota and have filed a patent for MET-1 through Parteq Innovations (Queen's University). The other authors have no conflict of interests to declare.
Funding
Funding for this work was supported in part by the Canada Foundation for Innovation and the Southeastern Ontario Academic Medical Organization (E.O.P). M.G.R. was supported by postdoctoral fellowship CONACYT-263618.
References
- [1].Backhed F, Fraser CM, Ringel Y, Sanders ME, Sartor RB, Sherman PM, Versalovic J, Young V, Finlay BB. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe 2012; 12:611-22; PMID:23159051; http://dx.doi.org/ 10.1016/j.chom.2012.10.012 [DOI] [PubMed] [Google Scholar]
- [2].Le Lay C, Fernandez B, Hammami R, Ouellette M, Fliss I. On Lactococcus lactis UL719 competitivity and nisin (Nisaplin((R))) capacity to inhibit Clostridium difficile in a model of human colon. Front Microbiol 2015; 6:1020; PMID:26441942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lee CH, Steiner T, Petrof EO, Smieja M, Roscoe D, Nematallah A, Weese JS, Collins S, Moayyedi P, Crowther M, et al.. Frozen vs Fresh Fecal Microbiota Transplantation and Clinical Resolution of Diarrhea in Patients With Recurrent Clostridium difficile Infection: A Randomized Clinical Trial. Jama 2016; 315:142-9; PMID:26757463; http://dx.doi.org/ 10.1001/jama.2015.18098 [DOI] [PubMed] [Google Scholar]
- [4].van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, et al.. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 2013; 368:407-15; PMID:23323867; http://dx.doi.org/ 10.1056/NEJMoa1205037 [DOI] [PubMed] [Google Scholar]
- [5].Youngster I, Russell GH, Pindar C, Ziv-Baran T, Sauk J, Hohmann EL. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. Jama 2014; 312:1772-8; PMID:25322359; http://dx.doi.org/ 10.1001/jama.2014.13875 [DOI] [PubMed] [Google Scholar]
- [6].Petrof EO, Claud EC, Gloor GB, Allen-Vercoe E. Microbial ecosystems therapeutics: a new paradigm in medicine? Benef Microbes 2013; 4:53-65; PMID:23257018; http://dx.doi.org/ 10.3920/BM2012.0039 [DOI] [PubMed] [Google Scholar]
- [7].Allen-Vercoe E, Reid G, Viner N, Gloor GB, Hota S, Kim P, Lee C, O'Doherty K, Vanner SJ, Weese JS, et al.. A Canadian Working Group report on fecal microbial therapy: microbial ecosystems therapeutics. Can J Gastroenterol 2012; 26:457-62; PMID:22803022; http://dx.doi.org/ 10.1155/2012/213828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC, Brown EM, Schroeter K, Allen-Vercoe E. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome 2013; 1:3; PMID:24467987; http://dx.doi.org/ 10.1186/2049-2618-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Donald M, Dower J, Bush R. Evaluation of a suicide prevention training program for mental health services staff. Community Ment Health J 2013; 49:86-94; PMID:22290304; http://dx.doi.org/ 10.1007/s10597-012-9489-y [DOI] [PubMed] [Google Scholar]
- [10].Yen S, McDonald JA, Schroeter K, Oliphant K, Sokolenko S, Blondeel EJ, Allen-Vercoe E, Aucoin MG. Metabolomic analysis of human fecal microbiota: a comparison of feces-derived communities and defined mixed communities. J Proteome Res 2015; 14:1472-82; PMID:25670064; http://dx.doi.org/ 10.1021/pr5011247 [DOI] [PubMed] [Google Scholar]
- [11].Antunes LC, McDonald JA, Schroeter K, Carlucci C, Ferreira RB, Wang M, Yurist-Doutsch S, Hira G, Jacobson K, Davies J, et al.. Antivirulence activity of the human gut metabolome. MBio 2014; 5:e01183-14; PMID:25073640; http://dx.doi.org/ 10.1128/mBio.01183-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stecher B, Hardt WD. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol 2011; 14:82-91; PMID:21036098; http://dx.doi.org/ 10.1016/j.mib.2010.10.003 [DOI] [PubMed] [Google Scholar]
- [13].Bohnhoff M, Miller CP. Enhanced susceptibility to Salmonella infection in streptomycin-treated mice. J Infect Dis 1962; 111:117-27; PMID:13968487; http://dx.doi.org/ 10.1093/infdis/111.2.117 [DOI] [PubMed] [Google Scholar]
- [14].Ryan CA, Nickels MK, Hargrett-Bean NT, Potter ME, Endo T, Mayer L, Langkop CW, Gibson C, McDonald RC, Kenney RT. Massive outbreak of antimicrobial-resistant salmonellosis traced to pasteurized milk. Jama 1987; 258:3269-74; PMID:3316720; http://dx.doi.org/ 10.1001/jama.1987.03400220069039 [DOI] [PubMed] [Google Scholar]
- [15].Pavia AT, Shipman LD, Wells JG, Puhr ND, Smith JD, McKinley TW, Tauxe RV. Epidemiologic evidence that prior antimicrobial exposure decreases resistance to infection by antimicrobial-sensitive Salmonella. J Infect Dis 1990; 161:255-60; PMID:2299207; http://dx.doi.org/ 10.1093/infdis/161.2.255 [DOI] [PubMed] [Google Scholar]
- [16].Martz SL, McDonald JA, Sun J, Zhang YG, Gloor GB, Noordhof C, He SM, Gerbaba TK, Blennerhassett M, Hurlbut DJ, et al.. Administration of defined microbiota is protective in a murine Salmonella infection model. Sci Rep 2015; 5:16094; PMID:26531327; http://dx.doi.org/ 10.1038/srep16094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Berkes J, Viswanathan VK, Savkovic SD, Hecht G. Intestinal epithelial responses to enteric pathogens: effects on the tight junction barrier, ion transport, and inflammation. Gut 2003; 52:439-51; PMID:12584232; http://dx.doi.org/ 10.1136/gut.52.3.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kohler H, Sakaguchi T, Hurley BP, Kase BA, Reinecker HC, McCormick BA. Salmonella enterica serovar Typhimurium regulates intercellular junction proteins and facilitates transepithelial neutrophil and bacterial passage. Am J Physiol Gastrointest Liver Physiol 2007; 293:G178-87; PMID:17615177; http://dx.doi.org/ 10.1152/ajpgi.00535.2006 [DOI] [PubMed] [Google Scholar]
- [19].Fournier BM, Parkos CA. The role of neutrophils during intestinal inflammation. Mucosal Immunol 2012; 5:354-66; PMID:22491176; http://dx.doi.org/ 10.1038/mi.2012.24 [DOI] [PubMed] [Google Scholar]
- [20].Zhang YG, Wu S, Xia Y, Sun J. Salmonella infection upregulates the leaky protein claudin-2 in intestinal epithelial cells. PLoS One 2013; 8:e58606; PMID:23505542; http://dx.doi.org/ 10.1371/journal.pone.0058606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chassaing B, Aitken JD, Malleshappa M, Vijay-Kumar M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr Protoc Immunol 2014; 104:Unit 15 25; PMID:24510619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Vowinkel T, Kalogeris TJ, Mori M, Krieglstein CF, Granger DN. Impact of dextran sulfate sodium load on the severity of inflammation in experimental colitis. Dig Dis Sci 2004; 49:556-64; PMID:15185857; http://dx.doi.org/ 10.1023/B:DDAS.0000026298.72088.f7 [DOI] [PubMed] [Google Scholar]
- [23].Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, Büller HA, Dekker J, Van Seuningen I, Renes IB. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 2006; 131:117-29; PMID:16831596; http://dx.doi.org/ 10.1053/j.gastro.2006.04.020 [DOI] [PubMed] [Google Scholar]
- [24].Johansson ME, Gustafsson JK, Sjoberg KE, Petersson J, Holm L, Sjovall H, Hansson GC. Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PLoS One 2010; 5:e12238; PMID:20805871; http://dx.doi.org/ 10.1371/journal.pone.0012238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hudcovic T, Kolinska J, Klepetar J, Stepankova R, Rezanka T, Srutkova D, Schwarzer M, Erban V, Du Z, Wells JM, et al.. Protective effect of Clostridium tyrobutyricum in acute dextran sodium sulphate-induced colitis: differential regulation of tumour necrosis factor-alpha and interleukin-18 in BALB/c and severe combined immunodeficiency mice. Clin Exp Immunol 2012; 167:356-65; PMID:22236013; http://dx.doi.org/ 10.1111/j.1365-2249.2011.04498.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Petersson J, Schreiber O, Hansson GC, Gendler SJ, Velcich A, Lundberg JO, Roos S, Holm L, Phillipson M. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver Physiol 2011; 300:G327-33; PMID:21109593; http://dx.doi.org/ 10.1152/ajpgi.00422.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mack DR, Michail S, Wei S, McDougall L, Hollingsworth MA. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol 1999; 276:G941-50; PMID:10198338 [DOI] [PubMed] [Google Scholar]
- [28].Mattar AF, Teitelbaum DH, Drongowski RA, Yongyi F, Harmon CM, Coran AG. Probiotics up-regulate MUC-2 mucin gene expression in a Caco-2 cell-culture model. Pediatr Surg Int 2002; 18:586-90; PMID:12471471; http://dx.doi.org/ 10.1007/s00383-002-0855-7 [DOI] [PubMed] [Google Scholar]
- [29].Berry D, Schwab C, Milinovich G, Reichert J, Ben Mahfoudh K, Decker T, Engel M, Hai B, Hainzl E, Heider S, et al.. Phylotype-level 16S rRNA analysis reveals new bacterial indicators of health state in acute murine colitis. Isme J 2012; 6:2091-106; PMID:22572638; http://dx.doi.org/ 10.1038/ismej.2012.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nanda Kumar NS, Balamurugan R, Jayakanthan K, Pulimood A, Pugazhendhi S, Ramakrishna BS. Probiotic administration alters the gut flora and attenuates colitis in mice administered dextran sodium sulfate. J Gastroenterol Hepatol 2008; 23:1834-9; PMID:19120873; http://dx.doi.org/ 10.1111/j.1440-1746.2008.05723.x [DOI] [PubMed] [Google Scholar]
- [31].Mennigen R, Nolte K, Rijcken E, Utech M, Loeffler B, Senninger N, Bruewer M. Probiotic mixture VSL#3 protects the epithelial barrier by maintaining tight junction protein expression and preventing apoptosis in a murine model of colitis. Am J Physiol Gastrointest Liver Physiol 2009; 296:G1140-9; PMID:19221015; http://dx.doi.org/ 10.1152/ajpgi.90534.2008 [DOI] [PubMed] [Google Scholar]
- [32].Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 1993; 75:263-74; PMID:8402911; http://dx.doi.org/ 10.1016/0092-8674(93)80068-P [DOI] [PubMed] [Google Scholar]
- [33].Scheinin T, Butler DM, Salway F, Scallon B, Feldmann M. Validation of the interleukin-10 knockout mouse model of colitis: antitumour necrosis factor-antibodies suppress the progression of colitis. Clin Exp Immunol 2003; 133:38-43; PMID:12823276; http://dx.doi.org/ 10.1046/j.1365-2249.2003.02193.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Maharshak N, Packey CD, Ellermann M, Manick S, Siddle JP, Huh EY, Plevy S, Sartor RB, Carroll IM. Altered enteric microbiota ecology in interleukin 10-deficient mice during development and progression of intestinal inflammation. Gut Microbes 2013; 4:316-24; PMID:23822920; http://dx.doi.org/ 10.4161/gmic.25486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 2007; 104:13780-5; PMID:17699621; http://dx.doi.org/ 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Perry T, Jovel J, Patterson J, Wong G, Fedorak RN, Thiesen A, Dicken B, Madsen KL. Fecal Microbial Transplant After Ileocolic Resection Reduces Ileitis but Restores Colitis in IL-10-/- Mice. Inflamm Bowel Dis 2015; 21:1479-90; PMID:26070001; http://dx.doi.org/ 10.1097/MIB.0000000000000383 [DOI] [PubMed] [Google Scholar]
- [37].Kennedy RJ, Hoper M, Deodhar K, Erwin PJ, Kirk SJ, Gardiner KR. Interleukin 10-deficient colitis: new similarities to human inflammatory bowel disease. Br J Surg 2000; 87:1346-51; PMID:11044159; http://dx.doi.org/ 10.1046/j.1365-2168.2000.01615.x [DOI] [PubMed] [Google Scholar]
- [38].Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al.. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 2008; 105:16731-6; PMID:18936492; http://dx.doi.org/ 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, Jian R, Doré J. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut 2003; 52:237-42; PMID:12524406; http://dx.doi.org/ 10.1136/gut.52.2.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Papa E, Docktor M, Smillie C, Weber S, Preheim SP, Gevers D, Giannoukos G, Ciulla D, Tabbaa D, Ingram J, et al.. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One 2012; 7:e39242; PMID:22768065; http://dx.doi.org/ 10.1371/journal.pone.0039242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol 2005; 43:3380-9; PMID:16000463; http://dx.doi.org/ 10.1128/JCM.43.7.3380-3389.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rowan F, Docherty NG, Murphy M, Murphy B, Calvin Coffey J, O'Connell PR. Desulfovibrio bacterial species are increased in ulcerative colitis. Dis Colon Rectum 2010; 53:1530-6; PMID:20940602; http://dx.doi.org/ 10.1007/DCR.0b013e3181f1e620 [DOI] [PubMed] [Google Scholar]
- [43].Nagalingam NA, Kao JY, Young VB. Microbial ecology of the murine gut associated with the development of dextran sodium sulfate-induced colitis. Inflamm Bowel Dis 2011; 17:917-26; PMID:21391286; http://dx.doi.org/ 10.1002/ibd.21462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tvede M, Rask-Madsen J. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet 1989; 1:1156-60; PMID:2566734; http://dx.doi.org/ 10.1016/S0140-6736(89)92749-9 [DOI] [PubMed] [Google Scholar]
- [45].Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, et al.. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog 2012; 8:e1002995; PMID:23133377; http://dx.doi.org/ 10.1371/journal.ppat.1002995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mimura T, Rizzello F, Helwig U, Poggioli G, Schreiber S, Talbot IC, Nicholls RJ, Gionchetti P, Campieri M, Kamm MA. Once daily high dose probiotic therapy (VSL#3) for maintaining remission in recurrent or refractory pouchitis. Gut 2004; 53:108-14; PMID:14684584; http://dx.doi.org/ 10.1136/gut.53.1.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Singh S, Stroud AM, Holubar SD, Sandborn WJ, Pardi DS. Treatment and prevention of pouchitis after ileal pouch-anal anastomosis for chronic ulcerative colitis. Cochrane Database Syst Rev 2015; 11:CD001176; PMID:26593456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. Isme J 2012; 6:1535-43; PMID:22343308; http://dx.doi.org/ 10.1038/ismej.2012.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ze X, Le Mougen F, Duncan SH, Louis P, Flint HJ. Some are more equal than others: the role of “keystone” species in the degradation of recalcitrant substrates. Gut Microbes 2013; 4:236-40; PMID:23549436; http://dx.doi.org/ 10.4161/gmic.23998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hollister EB, Gao C, Versalovic J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology 2014; 146:1449-58; PMID:24486050; http://dx.doi.org/ 10.1053/j.gastro.2014.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Steck N, Hoffmann M, Sava IG, Kim SC, Hahne H, Tonkonogy SL, Mair K, Krueger D, Pruteanu M, Shanahan F, et al.. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology 2011; 141:959-71; PMID:21699778; http://dx.doi.org/ 10.1053/j.gastro.2011.05.035 [DOI] [PubMed] [Google Scholar]
- [52].Maharshak N, Huh EY, Paiboonrungruang C, Shanahan M, Thurlow L, Herzog J, Djukic Z, Orlando R, Pawlinski R, Ellermann M, et al.. Enterococcus faecalis Gelatinase Mediates Intestinal Permeability via Protease-Activated Receptor 2. Infect Immun 2015; 83:2762-70; PMID:25916983; http://dx.doi.org/ 10.1128/IAI.00425-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Geirnaert A, Wang J, Tinck M, Steyaert A, Van den Abbeele P, Eeckhaut V, et al.. Interindividual differences in response to treatment with butyrate-producing Butyricicoccus pullicaecorum 25-3T studied in an in vitro gut model. FEMS Microbiol Ecol 2015; 91; PMID:25999470; http://dx.doi.org/ 10.1093/femsec/fiv054 [DOI] [PubMed] [Google Scholar]


