Abstract
Throughout the eukaryotic lineage, small RNA silencing pathways protect the genome against the deleterious influence of selfish genetic elements such as transposons. In animals an elaborate small RNA pathway centered on PIWI proteins and their interacting piRNAs silences transposons within the germline, which transmits the genome to future generations. In contrast to other small RNA silencing pathways, we entirely lack a mechanistic understanding of this genome defense system. However, genetic and molecular work over the last ten years has uncovered a fascinating framework of this pathway that is conserved from sponges to mammals. This review discusses our current understanding of the piRNA pathway in Drosophila with an emphasis on origin and biogenesis of piRNAs.
Eukaryotic genomes harbor a variety of selfish genetic elements, stretches of DNA that gain a transmission advantage relative to the rest of the genome, while not increasing the organism’s fitness1. The best-understood and most widespread selfish elements are mobile elements called transposons2. The success of these “genome parasites” rests on their ability to multiply within the genome by transposition to new sites. This ultimately affects host fitness due to insertional mutagenesis and ectopic chromosomal recombination. Throughout the eukaryotic lineage, the threat posed by transposons is met by host defense systems that selectively silence them. Though early genetic studies have illustrated the existence of such defense-systems3,4, their molecular nature remained mysterious for a long time. This changed abruptly when the concept of small RNA pathways, which govern RNA mediated silencing phenomena was discovered5–8. Over the last ten years it has become increasingly evident that small RNA silencing pathways protect the genomes of plants, fungi and animals against transposons and other selfish elements9,10.
In this review, we discuss a small RNA silencing pathway that is selectively active in animal gonads where it safeguards the genome of reproductive cells against transposons. This so-called piRNA pathway centers on PIWI family proteins and their bound PIWI interacting RNAs (piRNAs). Our article focuses on the piRNA pathway in the Drosophila ovary, where a long history of genetic research combined with recent small RNA centered studies has revealed the conceptual framework of this genome surveillance system that is conserved from sponges to mammals.
We first outline the common logic of small RNA silencing pathways. We then describe the architecture of the Drosophila ovary as it allows to conceptually separate two distinct but related piRNA pathway modules. Separate discussions on these two modules constitute the major part of the review.
Concepts of small RNA silencing pathways
Common to all small RNA pathways is a silencing machine called the RNA induced silencing complex (RISC). Its central components are an Argonaute family protein and a bound small RNA11. Via complementary base pairing the small RNA guides RISC to cellular target RNAs, which typically results in target silencing. The remarkable elegance of small RNA pathways is based on their inherent simplicity. In one or the other form, an mRNA is a key intermediate of all gene expression programs. Thus, loading an Argonaute protein with a small RNA complementary to the target gene allows the inhibition of essentially every cellular process12,13.
Argonaute proteins have diversified during evolution and in most animals three small RNA pathways can be distinguished. These are the ubiquitous microRNA and small interfering (siRNA) pathways and the generally germline specific piRNA pathway (Box 1).
BOX1. Small RNA pathways.
Genetic studies have identified several concepts of small RNA mediated regulation years before RNA interference and related small RNA pathways were described28,85,86. Nevertheless, the milestones in the field were the discoveries of dsRNA as trigger for RNA interference and the identification of small RNAs and Argonaute proteins as the key components of all small RNA pathways5–8. For the first time, scientists had a molecular entry point into a world of novel and diverse biology. What followed was an explosion in our knowledge about mechanisms and functions of a still growing number of small RNA pathways.
Evolution has shaped a diverse array of pathways from the common principle of target repression via small complementary RNA guides. This is best illustrated by the radiation of Argonaute proteins, the universal binding partners for small RNAs, which in many cases are able to cleave (slice) the target upon successful recognition87,88.
In most animals, two classes of Argonaute proteins, the AGO subfamily and the PIWI subfamily can be distinguished (a third subfamily, the so-called WAGO proteins, has been identified only in nematodes89). AGO proteins are expressed ubiquitously and are loaded with microRNAs and endogenous siRNAs in response to specific dsRNA triggers. While microRNAs guide the regulation of endogenous gene expression programs90, siRNAs are mostly involved in the suppression of foreign gene expression13, be it from viruses of from selfish genetic elements. In flies, the siRNA pathway is much more elaborate than in mammals, presumably as insects lack the sophisticated adaptive immune system.
Most animals possess two or three PIWI family proteins that are typically expressed in gonads. Though flies and mice express both three PIWI proteins, pair wise orthologies cannot be determined. In fact, the three Drosophila PIWI clade proteins, Piwi, Aubergine and AGO3 are more related to each other than to the mouse PIWI members MILI, MIWI and MIWI2. This might suggest that PIWI proteins radiated within lineages from a single ancestral protein. Mutations in PIWI family proteins lead to sterility and severe defects in gametogenesis in all animals examined so far10,14.
A key advantage of small RNA pathways in the defense against foreign genetic elements is that the target sequence can also act as the trigger for small RNA biogenesis. Small RNAs are thereby inevitably coupled to their target, even if target sequences evolve rapidly. In most animals, the siRNA and piRNA pathways implement this principle. Within the siRNA pathway, intra- or inter-molecular double stranded RNA (dsRNA) originating from transposons or viral transcripts triggers siRNA production via Dicer. Loaded into RISC, these siRNAs guide target silencing in trans, and provide a protective layer in somatic cells12,13 (Box 2). The real battle, however, takes place in the germline, where transposons are particularly active due to their predominantly vertical transmission strategy. Here, the piRNA pathway silences selfish elements to ensure genetic stability across generations10,14. piRNA biology differs considerably from that of other small RNA pathways and almost nothing is known about piRNA biogenesis and their mode of action.
BOX2. Transposon silencing by endogenous siRNAs.
In flies, deep sequencing of small RNAs bound to AGO2 from somatic and germline tissues identified a large fraction of endogenous siRNAs (endo-siRNAs) with sequences corresponding to transposons and other genomic repeats33,34,36,91. Presumably, dsRNA from sense and antisense transcripts triggers their production. In ovaries, endo-siRNA profiles therefore overlap with those of piRNAs, yet they lack a similar antisense bias. Significant de-repression of several transposons at the RNA level has been observed in ovaries and flies mutant for AGO2, Dicer-2 or Loquacious, the three key factors for the endo-siRNA pathway. Thus, in gonads piRNAs and siRNAs collaborate in transposon silencing, though the extent of repression (at least for several elements) appears to be much higher for the piRNA pathway26,32. The Penelope element of Drosophila virilis, however, is largely controlled via endo-siRNAs with only very few piRNAs targeting this element being identified92. In non-gonadal tissues, the endo-siRNA pathway appears to be the only line of defense against transposons, though additional repression at the chromatin level cannot be excluded.
In mammals, endo-siRNAs have so far been only identified in oocytes, where they have an important role in transposon control93,94. Also studies in C. elegans indicate a much more pronounced role of endo-siRNAs in the defense against transposons 95–97. Here, however the nemotode specific WAGO clade of Argonaute proteins makes direct comparisons to the above described siRNA pathway increasingly difficult.
In flies and mice, some endo-siRNAs are also generated from piRNA clusters34,35. This might simply reflect the ability of piRNA cluster transcripts to form low levels of dsRNA with complementary transposon transcripts. A significant mechanistic connection between the two pathways seems unlikely as piRNA pathway mutants show little impact on siRNA populations and siRNA pathway mutants do not affect piRNA pools31,32,34.
The Drosophila ovary: Evidence for distinct piRNA modules
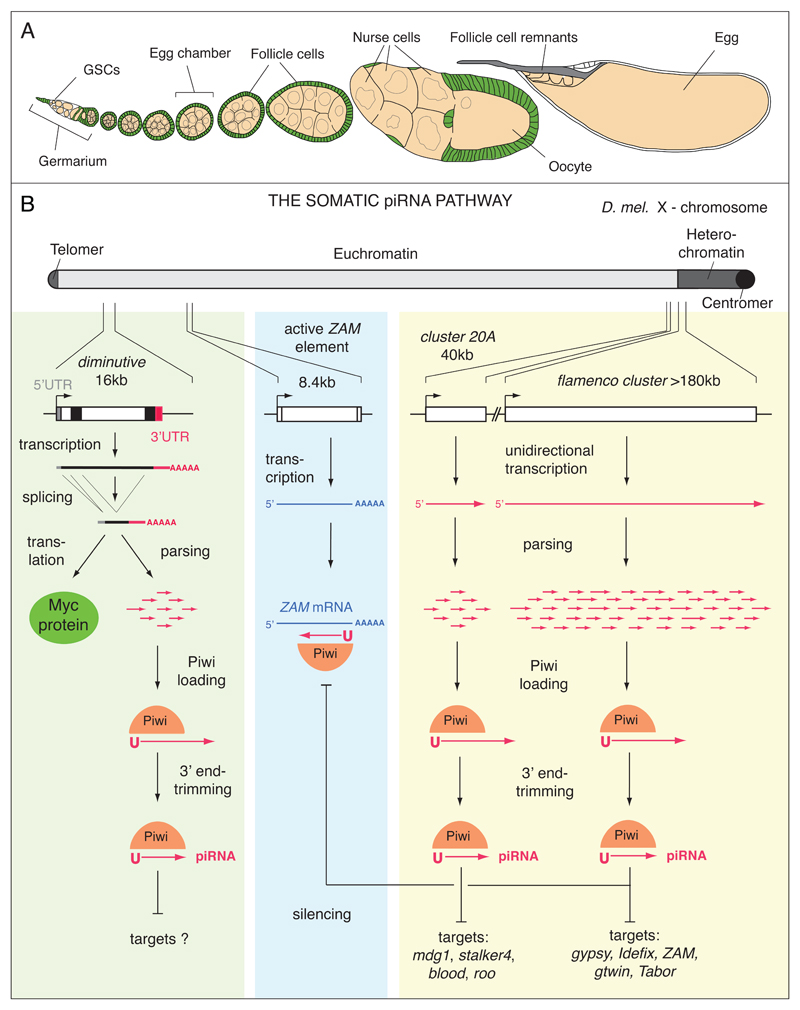
The Drosophila ovary consists of germline cells and somatic support cells (mostly follicle cells) that have key roles in maintaining and protecting the germline cells (Fig. 1A). Within the germline, nurse cells and oocyte share a common syncytial cytoplasm. Intricate connections also exist between germline and somatic cells, e.g. via exchange of developmental signals or nutrient flow into the germline. This exposes the oocyte genome to an internal and external threat: On the one hand, several transposons are highly active in germline cells and exploit cellular machineries to maximize access to the oocyte genome15. On the other hand, several retro-elements from the gypsy family form viral particles in somatic support cells that invade the germline, presumably via cellular transport vesicles 16–19. In both cell types, the piRNA pathway is the major line of defense against transposons. How silencing is achieved, however, differs significantly. In fact, while germline cells express three PIWI family members (Piwi, Aubergine (Aub) and AGO3), somatic support cells express exclusively Piwi20–22. In the following, we first describe the linear piRNA module that acts in ovarian somatic support cells. We then build on these concepts to introduce the more complex scenario in the germline, where a piRNA amplification module based on PIWI proteins is active.
Figure 1. A primary piRNA pathway is active in somatic cells of the Drosophila ovary.
A) The Drosophila oocyte is in direct contact with germline-derived cells (beige) and is surrounded by cells of somatic origin (green). This cartoon depicts an ovariole, the functional unit of the ovary (reproduced with kind permission from A. Spradling 110). Development proceeds from left (germarium) to right (mature egg). In the germarium, Germline Stem Cells (GSCs) divide asymmetrically into GSCs and differentiating cystoblasts. Four mitotic cystoblast cell divisions produce 15 nurse cells and an oocyte that remain connected by cytoplasmic bridges. Each of these germline cell clusters is surrounded by an epithelium of somatic follicle cells (green) to form an egg chamber that continuously grows until the oocyte matures into an egg. Follicle cells finally undergo apoptosis after depositing the eggshell. The deposited egg therefore lacks somatic cells. B) Shown is a schematic representation of the somatic piRNA pathway (primary piRNA module). For illustrative purposes, piRNA source and target loci from the X-chromosome (drawn at the top) are displayed. Colored boxes summarize primary piRNA biogenesis from piRNA clusters (yellow) and from 3’UTRs of protein coding genes (green). The blue box shows expression and silencing of ZAM, a prototypical LTR-retrotransposon, active in follicle cells. (yellow box) The piRNA clusters 20A and flamenco are located at the boundary between euchromatin and heterochromatin. Both contain almost exclusively transposon fragments oriented antisense to the unidirectional promoter. piRNA cluster transcripts (red) therefore give rise to antisense piRNAs. Unknown mechanisms parse piRNA cluster transcripts into shorter fragments that might enter Piwi. At this step, Piwi could preferentially select precursors with a 5’ Uridine (1U). Subsequently, the 3’ tail of Piwi bound RNAs is trimmed and 2’OH-methylated to generate mature piRNAs. (blue box) The sequence of mature piRNAs defines their target: Displayed is an active copy of the ZAM LTR-retrotransposon and its sense transcript (blue box). The green box summarizes piRNA biogenesis from genes (here diminuitive). The spliced dm transcript with 5’UTR, coding sequence and 3’UTR is shown. Mature dm mRNAs are either translated into Myc or act as piRNA precursors. piRNAs are preferentially processed from 3’UTR sequences, presumably by a similar mechanism as for piRNA cluster transcripts. Genic piRNAs are in sense orientation to the host gene and their targets (if any) remain to be identified.
A linear piRNA module in somatic gonadal cells
piRNA biology in the ovary-soma exhibits an overall simple architecture (Fig. 1B). All somatic support cells express Piwi, the only nuclear PIWI family protein in flies23,24. Piwi binds a spectrum of predominantly transposon derived piRNAs20,25–27 and silences transposon expression by an unknown mechanism. The only other factor with an understood function is the X-chromosomal flamenco locus that serves as a major source for piRNAs20,28–31. To a large extent, deep-sequencing of piRNA populations provided our current insight into somatic piRNA biogenesis and their target spectrum and biological roles.
Somatic piRNAs and their origin
The purest datasets of somatic piRNAs were obtained from an ovarian somatic sheet cell line (OSS cells)21,22. We will use this dataset to illustrate key features of somatic piRNAs. We note that populations of Piwi bound piRNAs from entire ovaries and the population of piRNAs that is selectively found in ovaries but not in eggs (during final stages of oocyte development, germline cells dump their content into the growing oocyte and somatic support cells undergo apoptosis) strongly suggest that OSS cells accurately reflect the in vivo situation31,32.
Somatic Piwi bound piRNAs are ~22-30nt long and ~75% carry a 5’ Uridine, a pattern found for several Argonaute family proteins20,25,27. Over 60% of somatic piRNAs map the genome multiple times. This comes as no surprise given that 70% of them map to annotated transposons or transposon fragments, a strong enrichment compared to the average transposon content of ~10% in the assembled genome (Box 3). The remaining 30% of piRNAs map to non-annotated regions and protein coding genes. Given the observed piRNA composition, the cell evidently selects specific RNAs for piRNA processing. Significant insight into this and therefore piRNA biogenesis in general has been extracted from piRNAs that map to transposons and gene exons:
BOX3. Transposable elements in Drosophila.
The 180 Mb Drosophila genome harbors over 100 transposon families and members of all major classes (LINE and LTR type retro-elements as well as DNA-type elements) are represented98,99. Release 5 of the assembled genome contains 117 Mb of euchromatic and ~24 Mb of heterochromatic sequence. Assembly of the heterochromatic portion was an important prerequisite for identifying piRNA clusters100,101. Annotated transposon sequences make up 7% of euchromatin and ~75% of heterochromatin. Most heterochromatic insertions, however, are sequence fragments and around two thirds of the 560 full length and thus presumably active elements are located in euchromatin. The genomes of other Drosophilid species contain similar transposon-families. Both, transposon load and their individual sequences are, however, typically species specific102. Consequently, piRNA clusters differ in their content and serve as species-specific repositories of transposons that are or have been active in a population31.
Transposon derived piRNAs
In the soma, over 90% of transposon annotated piRNAs are antisense to active transposon transcripts21, a clear contrast to the siRNA pathway, where sense and antisense small RNA populations are equally abundant26,33–36. If mapped across transposon transcripts, piRNAs typically cover the entire sequence31,32. No obvious patterns indicate preferential processing from certain regions, an indication that RNA structure does not trigger piRNA biogenesis. However, in some cases, piRNA profiles exhibit pronounced boundaries and certain transposon regions do not give rise to piRNAs. Insight into the genomic origin of piRNAs offered a coherent explanation for both, the antisense bias of piRNAs and the irregularities of piRNA profiles across certain elements20,31.
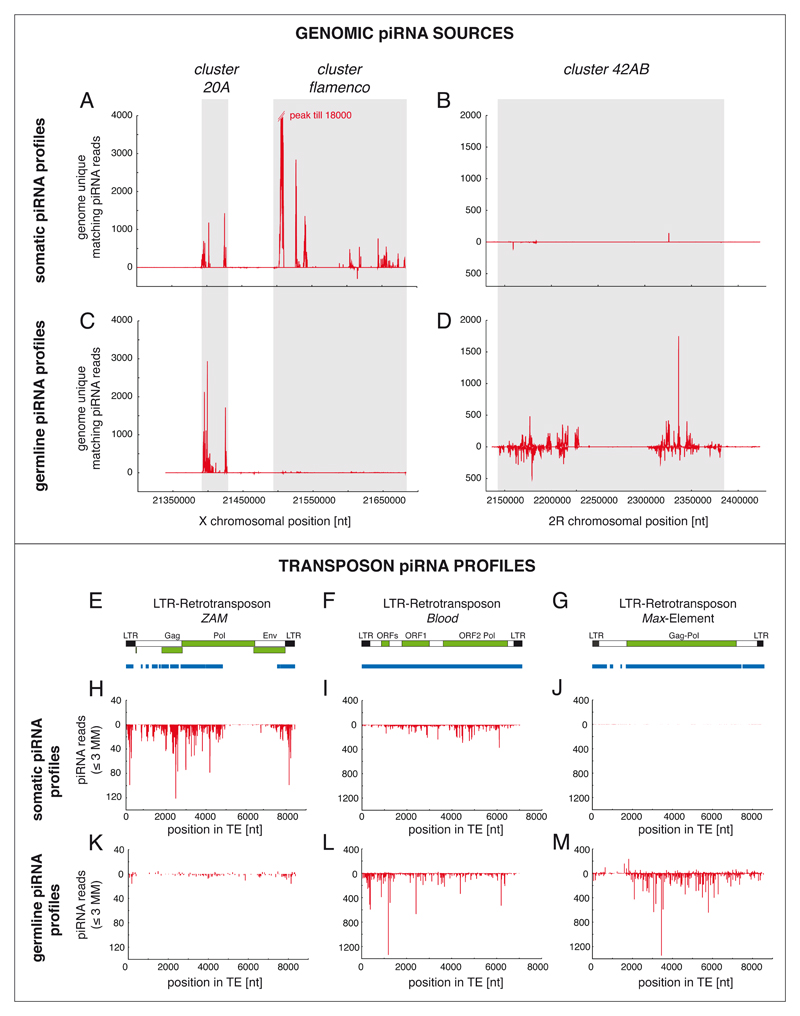
About 15% of somatic transposon-derived piRNAs map uniquely to the genome and only these allowed the confident identification of genomic piRNA origins20. This led to two conclusions: First, piRNAs appear to originate predominantly from broken transposon copies or their sequence fragments rather than from active copies. And secondly, piRNA generating transposon sequences are densely packed in a few genomic loci. These so-called piRNA clusters span dozens to hundreds of kilobases in length. They demarcate the regions with the highest density of broken, mutated and therefore immobile transposon fragments in the entire genome20,37. piRNA clusters are a conserved hallmark of piRNA pathways, though their repeat content varies widely38–41. In somatic ovarian cells, two piRNA clusters dominate and both are located on the X-chromosome roughly at the euchromatin/heterochromatin boundary. The larger one is the genetically identified flamenco locus, the smaller one is referred to as cluster 20A according to its cytology20,21,28–31. From both clusters, piRNAs are derived only from one genomic strand, arguing for uni-directional transcription. Moreover, ~90% of the transposon fragments in flamenco and 100% of those in cluster 20A are oriented antisense to the transcription direction. This immediately explains the massive antisense bias of transposon-derived piRNAs.
The flamenco cluster appears to be only expressed/processed in somatic ovarian cells31. Interestingly, most transposon fragments in flamenco belong to the gypsy-family of retrotransposons, precisely those that invade the germline via the somatic niche16–19,31.
These observations have led to a model in which the somatic piRNA pathway stores sequence information of transposons in specialized genomic regions. Their uni-directional transcripts are parsed into piRNAs, which -after loading into Piwi- allow the silencing of complementary transposons in trans30,31. In this scenario, insertions of gypsy-type elements antisense to the direction of cluster transcription were positively selected during evolution. Strong support for this model stems from an analysis of ZAM fragments located within flamenco: Only sequence regions of ZAM that are found within flamenco give rise to abundant piRNAs31 (Fig.3E). piRNA production in the soma should therefore be independent of the expression of active elements. Indeed, levels of gypsy-derived somatic piRNAs are not influenced by the presence of active gypsy elements in the genome42.
Figure 3. piRNA profiles along clusters and transposons are tissue specific.
This figure illustrates the pronounced differences in piRNA pools found in somatic and germline cells of the Drosophila ovary. Basis for these differences are tissue specific transcription of piRNA clusters and presence of the ping-pong cycle in germline cells only. Somatic graphs are based on the OSS cell data from ref.43 and germline graphs are based on early embryo libraries from ref.66. To enable comparison of these populations, profiles were always normalized to 1 millionn sequenced repeat-derived 23-30nt small RNAs. Panels A-D indicate that flamenco is a soma-specific piRNA cluster while cluster 42AB is germline specific (cluster coordinates are shaded in light grey). Cluster 20A is processed into piRNAs in both cell-types. Also apparent is the unidirectional transcription of flamenco and cluster 20A while cluster 42AB is transcribed in both orientations. In each panel, only genome-unique piRNAs were used and a 200 nt sliding window with step size of 20 nt was applied. Sense and antisense piRNAs are displayed as upwards and downwards peaks, respectively (E-G) Shown are schematics of the LTR retrotransposon ZAM, Blood and Max-Element. Blue bars display the respective transposon fragments found in piRNA clusters (antisense ZAM fragments within flamenco, a complete antisense Blood element in cluster20A and Max fragments in cluster 42AB). Transposon cartoons and cluster fragments are length matched to the piRNA profiles shown below. Panels H-M show profiles of somatic and germline piRNAs mapping to ZAM, Blood and Max. For each graph, piRNAs mapping with up to three mismatches to the indicated element were pooled. ZAM is a proto-typical element expressed and silenced in somatic cells, while Max is apparently only silenced (and presumably transcribed) in germline cells. Blood silencing is active in both cell-types. The ZAM fragments present in the flamenco piRNA cluster (blue) are in striking agreement with the observed piRNA profiles, suggesting that they are the major source of ZAM piRNAs. Similarly, piRNA profiles for Blood and Max are consistent with their respective fragments in piRNA clusters 20A and 42AB. Ping-pong signatures are significant only for Blood and Max in the germline samples (not shown).
Besides flamenco and cluster 20A several other, often smaller piRNA clusters have been cataloged based on OSS piRNAs21. These are, however, not yet assembled into chromosomal contigs. We note that flamenco ends in a genomic gap of unknown size and that some of these cluster fragments therefore likely correspond to pieces of flamenco.
piRNAs from exons
Based on the suspicious localization of piRNA clusters at the euchromatin/heterochromatin boundary, one might suggest that certain chromatin marks allow flagging cluster transcripts for piRNA biogenesis. This model was challenged with the surprising discovery that transcripts from several hundred genes are substrates for piRNA biogenesis and are the source of nearly 10% of somatic piRNAs22,43. About 95% of genic piRNAs are in sense orientation to the host transcript and typically originate from exons indicating that mature mRNAs are processing substrates.
Only a subset of cellular mRNAs gives rise to piRNAs and there is no apparent correlation between transcript abundance and piRNA levels43. Since exonic sense piRNAs will typically have no fully complementary targets within the cell, their function is unknown. It has been suggested that some exonic piRNAs target cellular transcripts via incomplete pairing22. The predicted target sites, however, are located in the intron of the target gene. It remains to be shown, whether this allows significant target regulation and how tolerant target recognition is towards incomplete complementarity between small RNA and target. Alternatively, the cell modulates expression of the piRNA host gene directly, as some of the mRNA transcripts are consumed during piRNA biogenesis. Interestingly, the gene giving rise to most piRNAs encodes the transcription factor Traffic jam, which is required for Piwi expression in somatic support cells, indicative of a classic negative feedback loop22,43.
Primary piRNA biogenesis
The linear biogenesis of piRNAs from precursor transcripts into PIWI proteins has been termed “primary piRNA biogenesis”20. In somatic support cells of the gonad piRNAs seem to be exclusively generated via primary processing. Little is known about this process at the mechanistic level. The above-mentioned features of piRNAs strongly suggest that single stranded transcripts (originating from piRNA clusters and genes) are substrates for the processing machinery. A P-element insertion at the beginning of flamenco abrogates piRNA production over the entire 180kb cluster, strong evidence for a long, single stranded transcript20,30,31. Moreover, piRNA biogenesis is independent of Dicer26, the key enzyme in the miRNA and siRNA pathways where dsRNAs serve as trigger molecules. piRNA profiles across exons or clusters do not correlate with any obvious RNA secondary structure elements. Nevertheless, pronounced peaks of genome-unique piRNAs across clusters are apparent (Fig.3A,D). Differences in sequence content between analyzed piRNA clusters and reference genome and the fact that only some cluster regions can generate “genome-unique” piRNAs are the likely basis for these irregularities. It therefore appears as if precursor transcripts are randomly processed into piRNAs.
It is entirely unclear, how the cell selects cluster transcripts and those from a subset of genes for piRNA biogenesis. Are these transcripts marked in any special way for biogenesis? An experimental entry point into this question might well reside in the pool of genic piRNAs. In some cases the level of genic piRNAs per kilobase is approaching that of flamenco-derived piRNAs, indicating that genic piRNAs are not merely noisy by-products of cellular RNA metabolism. Somehow, these transcripts are special and it will be important to decipher the underlying molecular reason, be it sequence motifs or features like RNA half-life or translation efficiency.
The precise subcellular location for piRNA biogenesis is unknown. Piwi is enriched in the nucleus. Nevertheless, accumulating evidence suggests that primary piRNA biogenesis occurs in the cytoplasm. First, an overwhelming proportion of genic piRNAs originates from the 3’UTR with the first piRNAs mapping shortly downstream of the stop codon22,43. This indicates that ribosomal association precedes piRNA processing. Secondly, an N-terminally truncated Piwi protein that cannot localize to the nucleus is loaded efficiently with piRNAs22.
Though variable in length, piRNAs with the same core sequence typically share the same 5’ end and differ in their 3’ ends. The first base shows a strong bias for Uridine. Preferences for 5’ nucleotides are common among Argonaute proteins44–46 and a recent study lends structural support for the ability of Argonaute proteins to read out the identity of the bound RNA’s first base47. In a random processing model, it seems likely that Piwi selectively stabilizes pre-piRNAs starting with a 5’ Uridine. This might well explain observed local irregularities in piRNA profiles. In a second step, 3’ trimming of the pre-piRNA would generate the heterogeneous 3’ end. The footprint of Piwi on the pre-piRNA would determine piRNA length, explaining why piRNA populations bound to different PIWI family proteins differ in their length20. According to this, piRNA precursors are loaded as single stranded RNAs into Piwi. This is in contrast to siRNAs and miRNAs, which are loaded as small RNA duplexes into Argonaute proteins, after which one strand is removed48–52. A recent study from S. pombe, however, indicates that the proposed piRNA biogenesis model is not that exotic in the end: Though fission yeast Argonaute is primarily loaded with Dicer products (small RNA duplexes), it appears that initially it is loaded with so-called primal RNAs53. Strikingly, primal RNAs are preferentially derived from 3’UTR regions of cellular transcripts and centromeric repeats in a Dicer independent manner and appear to be trimmed at their 3’ end, potentially by the exosome53. Only upon target interaction and cleavage, an RNA dependent RNA Polymerase (RdRP) converts the target transcript into dsRNA, providing the substrate for Dicer processing into the much more abundant siRNAs. The resemblance of primal RNAs to primary piRNAs is provoking, though no RdRP dependent amplification seems to participate in the piRNA pathway.
The proteins involved in primary piRNA biogenesis are unknown with the exception of Zucchini; a predicted nuclease with a phospho-lipase D domain22,31,54. At which step Zucchini acts during biogenesis is, however, unclear. Though multiple other proteins have been identified as essential piRNA pathway members, genetic studies indicate that they are selectively involved in the more complex germline piRNA biology31,32,55. Without a doubt, several as yet to be identified factors must participate in biogenesis, loading and function of primary piRNAs and the availability of the OSS cell line56 promises rapid progress towards their identification and characterization.
The germline piRNA pathway and piRNA ping-pong
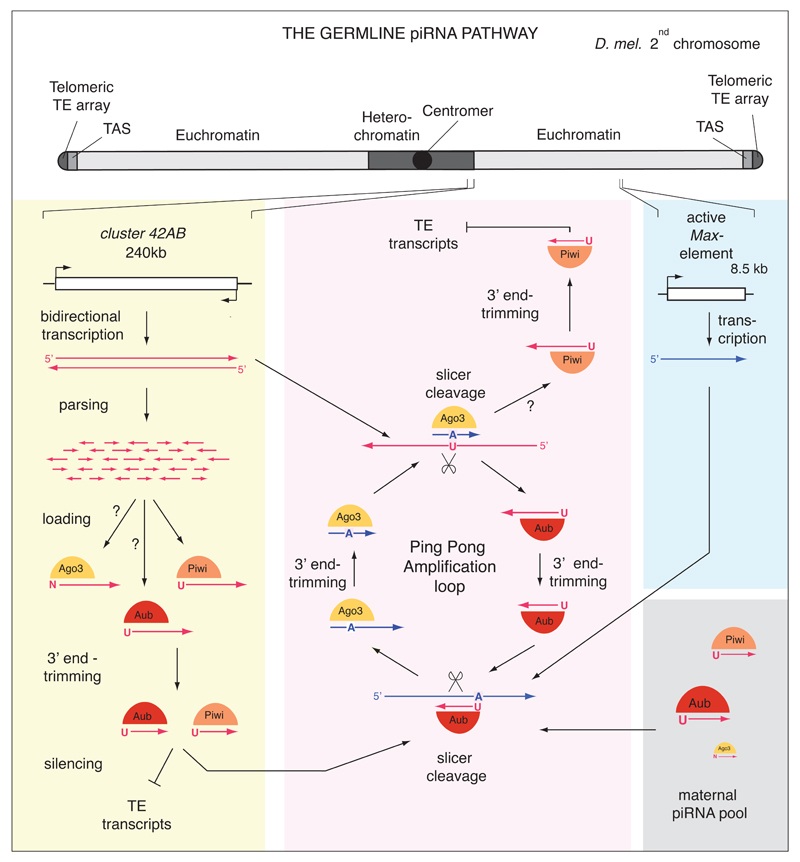
Considerable evidence indicates that the linear primary piRNA pathway feeding into Piwi is also active in germline cells31,32. piRNA biology in the germline is, however, much more complex: Ovarian germline cells express besides Piwi also Aubergine and AGO3, two related PIWI family proteins20,32,57,58. Sequence analysis of piRNAs selectively bound to Piwi, Aub and AGO3 revealed the existence of a sophisticated piRNA amplification loop that acts in parallel or on top of the above described primary piRNA pathway (Fig. 2). The central players in this so-called ping-pong cycle are Aub and AGO3, which localize to the cytoplasm of germline cells with an accumulation around the nucleus20,58. In the prevailing model, Aub is guided via an antisense piRNA to a sense transcript from an active transposon. Subsequent slicer cleavage of the target transcript triggers production of a novel sense piRNA, which is loaded into AGO3. The AGO3-piRNA complex in turn cleaves complementary piRNA cluster transcripts. This prompts biogenesis of a novel antisense and Aub-bound piRNA, whose sequence is identical to the initiator piRNA. As Aub and AGO3 presumably act catalytically58, the ping-pong cycle amplifies silencing competent piRNAs with the loop acting efficiently only in the presence of a target transcript (active transposon message). Indeed, ping-pong piRNAs are the most abundant cellular piRNAs20. A key conceptual difference to the primary piRNA pathway is that piRNA biogenesis in the germline depends in part on target expression. Elegant genetic experiments have hinted at this even before piRNAs were discovered59,60. Signatures of the ping-pong cycle have been found in sponges, planaria, moths, fish, frogs and mammals40,61–65 (Box 4). It is therefore one of the hallmarks of the piRNA pathway. Target dependent amplification of a small RNA response is common among small RNA pathways in fungi, plants and nematodes9. Here, however, cleavage of the target triggers dsRNA synthesis by RdRP enzymes, generating Dicer substrates. Most animals lack RdRP enzymes and the piRNA pathway utilizes instead sense and antisense RNAs from different transcripts and couples them via reciprocal Slicer cleavage.
Figure 2. In germline cells, the primary piRNA pathway and the Ping-Pong amplification loop are active.
Shown are representative examples of germline piRNA sources and targets originating from the 2nd chromosome (drawn at the top). Colored boxes show primary piRNA biogenesis from cluster 42AB (yellow), the adaptive module of the target dependent ping-pong amplification loop (red), expression and silencing of a typical active LTR-retrotransposon (Max-Element; blue) and the contribution of maternally inherited piRNAs (green). (yellow box) Cluster 42AB contains transposon fragments in both orientations and is bi-directionally transcribed. During primary piRNA biogenesis, cluster transcripts (red) presumably generate sense and antisense piRNAs. Unknown mechanisms parse the long piRNA precursor transcripts into shorter fragments that are loaded onto PIWI family proteins (Piwi, probably Aub and potentially AGO3). Piwi and Aub probably select RNA fragments with a 5’ Uridine (1U). Subsequently, the 3’ tail of pre-piRNAs are trimmed and 2’OH-methylated to generate mature piRNAs. piRISCs with antisense piRNAs are competent to silence sequence complementary transcripts of active transposons. Primary piRNA biogenesis in the germline is likely similar to the one in somatic cells. The blue box shows an active copy of the Max-Element (LTR-retrotransposon) and its transcribed sense transcript that is silenced by complementary piRISCs. The red box summarizes the ping-pong cycle. An Aub complexed piRNA (red) that is antisense to an active sense Max transcript (dark blue) guides slicing (scissors) of the transposon RNA, precisely 10nt downstream of its 5’ Uridine. The sliced Max transcript is predicted to be loaded onto AGO3 and typically has a profound bias for an Adenine at position 10 (10A). The AGO3 bound pre-piRNA is 3’ trimmed and 2’-OH methylated. This mature AGO3-piRNA complex in turn cleaves complementary cluster transcripts and triggers production of an Aub-loaded antisense piRNA, whose sequence is identical to the initiating piRNA. It is currently impossible to experimentally distinguish between Aub-piRNA complexes generated via primary piRNA biogenesis or via ping-pong. Weak ping-pong signatures exist between AGO3 and Piwi and could indicate that Piwi (besides primary biogenesis) also receives piRNAs via AGO3 mediated target slicing. (green box) At the end of oogenesis, mature Aub- and Piwi-piRNA complexes (to a lesser extent also AGO3) are efficiently loaded into the oocyte. Maternal Aub and to a lower extent also Piwi localize to the posterior pole of the mature egg, where the future germline will form. Maternal piRNAs might serve important roles in the starting phase of the pig-pong cycle.
BOX4. Commonalities and differences among animal piRNA pathways.
Based on primary sequence analyses, PIWI proteins are found throughout the animal kingdom. They are typically expressed in germline cells but in lower invertebrates also in cells responsible for regeneration (e.g. neoblasts in Planaria62,103). Primary piRNA biogenesis and signatures of the ping-pong amplification cycle are found in species ranging from sponges to mammals, indicating an ancient origin of the pathway’s key features61. A notable exception is C. elegans, where the two PIWI family proteins PRG1 and PRG2 are expressed in gonads yet bind 21U RNAs, a different class of small RNAs45,95–97. These appear to have distinct biogenesis features, do not exhibit ping-pong signatures and have a largely unclear function and target spectrum.
In mammals, three PIWI family proteins are expressed in testes and only one in ovaries104–107. Interestingly, the pathway seems largely dispensable for oogenesis, where an endo-siRNA pathway centered on AGO proteins cooperates in silencing transposable elements93,94. During mouse spermatogenesis, the three PIWI family proteins MIWI2, MILI and MIWI are expressed in different, yet overlapping temporal domains. MIWI2 and MILI are the key players in the genome defense pathway and their bound piRNAs show signatures of ping-pong and primary piRNA biogenesis108,109. MIWI2, the only nuclear mouse PIWI family protein is suggested to guide de novo DNA methylation at transposon loci, a process so far only reported for the mammalian piRNA pathway108,109. MIWI is expressed only after the pachytene stage of meiosis and binds primary piRNAs derived from a distinct set of piRNA clusters. Pachytene piRNAs are not enriched in transposon sequences, accumulate to very high levels and have an unclear function, but presumably distinct from transposon silencing38,39,41.
Germline piRNA clusters
RNAs in early embryos –prior to zygotic transcription- reflect by and large the pool of germline transcripts made during oogenesis31,66. piRNAs from young embryos are thus the best proxy for the germline specific piRNA pool. Germline piRNAs originate predominantly from several piRNA clusters but also from transcripts of active elements, in accordance with the ping-pong model. Evidence for this is best documented for the I-element (LINE family), where cluster resident fragments and active elements have sufficiently diverged at the sequence level to allow distinguishing them66.
Just like soma clusters, germline clusters are strongly enriched in transposon fragments and the most prominent ones map to euchromatin/heterochromatin boundaries20. As a much broader spectrum of transposable elements (LINE-, LTR- and DNA-type elements) is highly active in the germline, it comes as no surprise that germline piRNA clusters contain a more diverse collection of transposon fragments31. Figure 3 depicts the soma specific flamenco cluster, the germline specific cluster 42AB and the shared cluster 20A. Cluster 20A is the only germline piRNA cluster that resembles flamenco as it is uni-directionally transcribed and contains only antisense transposon fragments20,31. In the germline, it loads preferentially Piwi and to a lesser extent also Aub. All other germline clusters spawn piRNAs from both strands indicating bi-directional transcription and alleviating the pressure for transposons to integrate in a biased orientation as observed for flamenco31. piRNAs originating from these clusters are loaded into all three PIWI family proteins but absolute numbers cannot be derived as only a minority of piRNAs can unambiguously be mapped to clusters. Finally, germline piRNA clusters are also found at telomeres, where abundant piRNAs are derived from both, the telomeric arrays of HeT-A, TART and TAHRE elements as well as from subtelomeric satellite repeats20.
It is unclear whether piRNA cluster transcripts are essential for ping-pong or whether any sense and antisense transcripts could engage in it. Similarly unknown is how the cell prevents auto-amplification of piRNAs derived from bi-directionally transcribed clusters. Best evidence for this stems from the analysis of I-element piRNAs: I-element fragments within cluster 42AB give rise to high piRNA levels only in strains with active elements66. Perhaps, cluster transcripts are physically isolated in specific cellular domains and presented only to selected protein complexes to guarantee accurate progressing through the biogenesis cycle.
The connections between primary piRNA biogenesis and the ping-pong cycle are only poorly understood. The ping-pong signature is mostly confined to Aub/AGO3 and Aub/Aub pairs20,32. Ping-pong is almost entirely lost in aub mutants31. While Aub/Aub ping-pong prevails in AGO3 mutants, the resulting piRNA levels are severely diminished32. Piwi on the other hand is dispensable for the ping-pong cycle31. Nevertheless, a weak but significant ping-pong interaction occurs between Piwi and AGO320,32, suggesting that Piwi –while not providing input- could be a recipient in the cycle (Fig. 2). This might explain the antisense bias of Piwi bound piRNAs originating from germline clusters20 and might also explain the loss of nuclear Piwi in late stage ovarioles lacking AGO332. In such a model, primary piRNA biogenesis must also load Aub or AGO3. In fact, in Drosophila testes, germline cells express only Aub and AGO3, while Piwi is expressed in somatic support cells only67. It therefore remains to be shown, how primary piRNA biogenesis feeds into the ping-pong cycle.
The maternal piRNA pool
During oogenesis, the oocyte is connected to the 15 nurse cells via cytoplasmic bridges. Ultimately, nurse cells dump their cytoplasm into the maturing oocyte (Fig. 1A). Piwi and Aub localize to the oocyte’s posterior pole, where the primordial germ cells of the embryo will form57,66,68. Maternal piRNAs thus form a protective layer against transposons even before transcription initiates in future germ cells. Genetic experiments have suggested the existence of a maternal factor with essential roles in transposon silencing3,4. In this so-called hybrid dysgenesis phenomenon, crosses between naïve females and males carrying a novel transposon produce sterile offspring, while the reciprocal cross does not. Small RNA sequencing approaches have shown that inheritance of maternal piRNAs is required for an efficient ping-pong response in the F1 generation66. Three possibilities could explain this observation: (1) Maternal piRNAs are required to kick-start the ping-pong cycle. (2) Maternal piRNAs have an essential influence on the chromatin status of piRNA clusters and/or transposons. (3) The cellular Aub and AGO3 pools are limiting and in the absence of maternal piRNAs for a certain element, the low level of primary piRNAs is unable to compete with piRNAs abundantly inherited maternally.
In summary, the germline piRNA pathway is considerably more elaborate then the linear somatic piRNA pathway. Also here, we largely lack insight into the molecular and cellular details. Genetic studies have identified multiple proteins involved in the piRNA pathway23,31,32,54,55,57,69–78 and several are specifically required for the germline piRNA pathway31,32,55. Amongst those are several RNA helicases but also proteins with unknown functions. Recent studies have linked the piRNA pathway to Tudor biology79. Tudor domains bind symmetrically methylated Arginines in Aub, AGO3 and potentially Piwi80–82. The Drosophila genome encodes at least twenty Tudor-domain containing proteins and many of these are selectively expressed in the germline (unpublished observations). Given this and considering the complexity of piRNA biology, we expect the number of proteins with essential roles in this genome defense system to increase considerably.
Outlook
Research in the piRNA field is in the paradoxical situation that we understand a lot about conceptual frameworks but that we lack almost every mechanistic and molecular insight. A great deal of understanding other small RNA pathways has emerged from in vitro assays. No such attempt has been reported for the piRNA pathway, probably reflecting the complexity of this approach. Without a doubt though, in vitro systems coupled with genetics and structural approaches will be essential to understand the order and dynamics of the molecular events during piRNA biogenesis and silencing.
The second challenge will be to understand the nature of piRNA cluster transcripts and to decipher the protein-RNA network that forms the basis of the piRNA pathway. Here, we expect rapid progress by combinations of next generation sequencing approaches with technological advances in determining RNA-protein interactions83,84. All in all, these are exciting days for everybody working on this fascinating genome defense systems and if recent years are a measure, many surprises are yet to come.
Acknowledgements
This work was supported by a Marie Curie postdoctoral fellowship to K.A.S. by the European Union. An ERC Starting Grant from the European Union supports work in the Brennecke laboratory.
References
- 1.Hurst GD, Werren JH. The role of selfish genetic elements in eukaryotic evolution. Nat Rev Genet. 2001;2:597–606. doi: 10.1038/35084545. [DOI] [PubMed] [Google Scholar]
- 2.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 3.Bucheton A, et al. The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: identification, cloning, and properties of the I factor. Cell. 1984;38:153–163. doi: 10.1016/0092-8674(84)90536-1. [DOI] [PubMed] [Google Scholar]
- 4.Rubin GM, et al. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell. 1982;29:987–994. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- 5.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 7.Zamore PD, et al. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 8.Hammond SM, et al. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 9.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 12.Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 15.Bucheton A. I transposable elements and I-R hybrid dysgenesis in Drosophila. Trends Genet. 1990;6:16–21. doi: 10.1016/0168-9525(90)90044-7. [DOI] [PubMed] [Google Scholar]
- 16.Chalvet F, et al. Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J. 1999;18:2659–2669. doi: 10.1093/emboj/18.9.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leblanc P, et al. Life cycle of an endogenous retrovirus, ZAM, in Drosophila melanogaster. J Virol. 2000;74:10658–10669. doi: 10.1128/jvi.74.22.10658-10669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelisson A, et al. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. Embo J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brasset E, et al. Viral particles of the endogenous retrovirus ZAM from Drosophila melanogaster use a pre-existing endosome/exosome pathway for transfer to the oocyte. Retrovirology. 2006;3:25. doi: 10.1186/1742-4690-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brennecke J, et al. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 21.Lau NC, et al. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009 doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito K, et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 23.Cox DN, et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox DN, et al. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 25.Saito K, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 27.Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 28.Prud'homme N, et al. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics. 1995;139:697–711. doi: 10.1093/genetics/139.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desset S, et al. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics. 2003;164:501–509. doi: 10.1093/genetics/164.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mevel-Ninio M, et al. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics. 2007;175:1615–1624. doi: 10.1534/genetics.106.068106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malone CD, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghildiyal M, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Czech B, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung WJ, et al. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawamura Y, et al. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 37.Bergman CM, et al. Recurrent insertion and duplication generate networks of transposable element sequences in the Drosophila melanogaster genome. Genome Biol. 2006;7:R112. doi: 10.1186/gb-2006-7-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lau NC, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 39.Aravin A, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 40.Aravin AA, et al. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 41.Girard A, et al. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 42.Pelisson A, et al. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robine N, et al. A broadly conserved pathway generates 3'UTR-directed primary piRNAs. Curr Biol. 2009;19:2066–2076. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mi S, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5' terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruby JG, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 46.Ghildiyal M, et al. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frank F, et al. Structural basis for 5'-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 48.Matranga C, et al. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 49.Miyoshi K, et al. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rand TA, et al. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 51.Leuschner PJ, et al. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maiti M, et al. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 2007;21:590–600. doi: 10.1101/gad.1497607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Halic M, Moazed D. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell. 2010;140:504–516. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pane A, et al. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klattenhoff C, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niki Y, et al. Establishment of stable cell lines of Drosophila germ-line stem cells. Proc Natl Acad Sci U S A. 2006;103:16325–16330. doi: 10.1073/pnas.0607435103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- 58.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 59.Jensen S, et al. Taming of transposable elements by homology-dependent gene silencing. Nat Genet. 1999;21:209–212. doi: 10.1038/5997. [DOI] [PubMed] [Google Scholar]
- 60.Jensen S, et al. Regulation of I-transposon activity in Drosophila: evidence for cosuppression of nonhomologous transgenes and possible role of ancestral I-related pericentromeric elements. Genetics. 2002;162:1197–1209. doi: 10.1093/genetics/162.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grimson A, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Friedlander MR, et al. High-resolution profiling and discovery of planarian small RNAs. Proc Natl Acad Sci U S A. 2009;106:11546–11551. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawaoka S, et al. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. Rna. 2009;15:1258–1264. doi: 10.1261/rna.1452209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Houwing S, et al. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 2008;27:2702–2711. doi: 10.1038/emboj.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lau NC, et al. Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO J. 2009;28:2945–2958. doi: 10.1038/emboj.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brennecke J, et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nishida KM, et al. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. Rna. 2007;13:1911–1922. doi: 10.1261/rna.744307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Megosh HB, et al. The Role of PIWI and the miRNA Machinery in Drosophila Germline Determination. Curr Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 69.Anne J, et al. Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development. 2007;134:137–146. doi: 10.1242/dev.02687. [DOI] [PubMed] [Google Scholar]
- 70.Boswell RE, Mahowald AP. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell. 1985;43:97–104. doi: 10.1016/0092-8674(85)90015-7. [DOI] [PubMed] [Google Scholar]
- 71.Chen Y, et al. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007;17:637–642. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cook HA, et al. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- 73.Gillespie DE, Berg CA. Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev. 1995;9:2495–2508. doi: 10.1101/gad.9.20.2495. [DOI] [PubMed] [Google Scholar]
- 74.Horwich MD, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 75.Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Patil VS, Kai T. Repression of Retroelements in Drosophila Germline via piRNA Pathway by the Tudor Domain Protein Tejas. Curr Biol. 2010 doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- 77.Saito K, et al. Pimet, the Drosophila homolog of HEN1, mediates 2'-O-methylation of Piwi- interacting RNAs at their 3' ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vagin VV, et al. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 2004;1:54–58. [PubMed] [Google Scholar]
- 79.Siomi MC, et al. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 24:636–646. doi: 10.1101/gad.1899210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kirino Y, et al. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol. 2009;11:652–658. doi: 10.1038/ncb1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nishida KM, et al. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 2009;28:3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kirino Y, et al. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA. 2010;16:70–78. doi: 10.1261/rna.1869710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chi SW, et al. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee RC, et al. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 86.Wightman B, et al. Posttranscriptional regulation of the heterochronic gene lin-14 by lin- 4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 87.Carmell MA, et al. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- 88.Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nat Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 89.Gu W, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 91.Okamura K, et al. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rozhkov NV, et al. Small RNA-based silencing strategies for transposons in the process of invading Drosophila species. RNA. 2010;16:1634–1645. doi: 10.1261/rna.2217810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 95.Das PP, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Batista PJ, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaminker JS, et al. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0084. RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Celniker SE, Rubin GM. The Drosophila melanogaster genome. Annu Rev Genomics Hum Genet. 2003;4:89–117. doi: 10.1146/annurev.genom.4.070802.110323. [DOI] [PubMed] [Google Scholar]
- 100.Smith CD, et al. The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science. 2007;316:1586–1591. doi: 10.1126/science.1139815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hoskins RA, et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 2007;316:1625–1628. doi: 10.1126/science.1139816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 103.Reddien PW, et al. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- 104.Kuramochi-Miyagawa S, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 105.Kuramochi-Miyagawa S, et al. Two mouse piwi-related genes: miwi and mili. Mech Dev. 2001;108:121–133. doi: 10.1016/s0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- 106.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 107.Carmell MA, et al. Miwi2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007 doi: 10.1016/j.devcel.2007.03.001. in press. [DOI] [PubMed] [Google Scholar]
- 108.Aravin AA, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kuramochi-Miyagawa S, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Frydman HM, Spradling AC. The receptor-like tyrosine phosphatase lar is required for epithelial planar polarity and for axis determination within drosophila ovarian follicles. Development. 2001;128:3209–3220. doi: 10.1242/dev.128.16.3209. [DOI] [PubMed] [Google Scholar]