Summary
Accurately detecting circulating endothelial cells (CECs) is important since their enumeration has been proposed as a biomarker to measure injury to the vascular endothelium. However, there is no single methodology for determining CECs in blood, making comparison across studies difficult. Many methods for detecting CECs rely on characteristic cell surface markers and cell viability indicators, but lack secondary validation. Here, a CEC population in healthy adult human subjects was identified by flow cytometry as CD45−, CD34dim that is comparable to a previously described CD45−, CD31bright population. In addition, nuclear staining with 7-aminoactinomycin D (7-AAD) was employed as a standard technique to exclude dead cells. Unexpectedly, the CD45−, CD34dim, 7-AAD− CECs lacked surface detectable CD146, a commonly used marker of CECs. Furthermore, light microscopy revealed this cell population to be composed primarily of large cells without a clearly defined nucleus. Nevertheless, immunostains still demonstrated the presence of the lectin Ulex europaeus and van Willebrand factor. Ultramicro analytical immunochemistry assays for the endothelial cell proteins CD31, CD34, CD62E, CD105, CD141, CD144 and vWF indicated these cells possess an endothelial phenotype. However, only a small amount of RNA, which was mostly degraded, could be isolated from these cells. Thus the majority of CECs in healthy individuals as defined by CD45−, CD34dim, and 7-AAD− have shed their CD146 surface marker and are senescent cells without an identifiable nucleus and lacking RNA of sufficient quantity and quality for transcriptomal analysis. This study highlights the importance of secondary validation of CEC identification.
Keywords: Vascular cell markers, gene expression, flow cytometry, viability, circulating endothelial cells
Introduction
The existence of circulating endothelial cells (CECs) was described over 30 years ago (1–3). Although CECs are an extremely rare population in healthy blood, accurately detecting and validating them by immunophenotyping may be important as a marker of vascular injury (4–6) in a variety of disease states including cancer, sickle cell anaemia, diabetes, and myocardial infarction (7–11). In addition, isolation and genomic assessment of CECs by microarray analysis has been proposed as a way to assess vascular function in patients with systemic sclerosis (12). More recently, levels of CECs have been used to monitor the effectiveness of chemotherapeutic agents (13–15).
Many different methods for the isolation of CECs from human blood have been reported (16). Isolation and identification of CECs has been complicated by the rarity of the population and the varying phenotypes, such as anuclear carcasses (1, 2), fragments (17) or multi-nucleated conglomerates (8). A variety of strategies have been described to capture CECs including gradient separation based on cell density (18), immunobeads directed at cell surface markers such as CD146 (19) and flow cytometry using multiple markers to negatively select leukocytes and positively identify endothelial cells (20). Many studies of CECs have focused on the detection of these cells using combinations of criteria including size, cellular complexity, cell surface markers and the presence of DNA (17, 21). Although some studies use viability markers such as 7-aminoactinomycin D (7-AAD), it is not clear if any of the CECs are functionally intact cells. To date, there is no consensus for a definition of a CEC or a technical method by which they may be reliably assessed (22). Existing methods of separation based on differential centrifugation and Ficoll gradient may not isolate the entire population of CECs present in the blood. Immunomagnetic bead capture of cells expressing CD146, a tight junction glycoprotein, may be compromised by the shedding of this surface feature from detached endothelial cells (23) and the presence of CD146 on a subset of circulating lymphocytes (24). Uniformly missing from published studies is the detection and validation of endothelial-specific proteins by methods other than immunostaining. In addition, isolated CECs have not been definitively shown to contain an intact nucleus and transcriptome, hallmarks of viable, fully functional cells.
To further clarify the aforementioned issues, lysed whole blood from healthy adult volunteers was used to collect cells by flow cytometry and high speed cell sorting. The sort gate was based on the absence of the surface marker CD45 to exclude leukocytes and on the presence of the glycoprotein cell adhesion factor CD34 to include endothelial cells. CD34 is found only on endothelial and progenitor cells. Endothelial cells are CD34dim while progenitor cells are CD34bright (25). Prior studies used a comparable population that also included CD31bright (26). However, the use of CD31bright could also capture large platelets (27). Earlier studies could not demonstrate any CD146 expression on the CD45−, CD31bright and CD34dim population (28). However, since CD146 has been used frequently as a marker for CEC in the published literature, the cell preparations were also tested for the presence of CD146. Finally, the novel technique of ultramicro analytical immunochemistry was used to provide evidence supporting the endothelial phenotype of the cells by interrogating the isolated cells for multiple endothelial markers. RNA analysis was performed to determine the feasibility of analysing expression profiles. The primary objectives of this study were to verify a rigorous and reliable method for CEC identification and isolation in healthy adults and to clarify the viability of these cells by assessing the integrity of their nucleus and RNA.
Materials and methods
Materials
For details, please refer to the Suppl. data (available online at www.thrombosis-online.com).
Study participants and blood sampling
The study protocol (03-H-0015) was approved by the institutional review board of the NHLBI at the NIH. Informed consent was obtained from all study participants. For the pilot study (n=4; mean age 45, range 41 to 49 years) each healthy donor had one 12 ml blood draw with the first 6 ml discarded to avoid contaminating the sample with endothelial cells from the venipuncture. For CD 146+ T cells, human umbilical vein endothelial cells (HUVEC) and putative CEC sorting each healthy donor (n=4; mean age 43, range 28 to 49 years) had three 50 ml blood draws with the first 10 ml discarded.
lmmunostaining of whole blood
A pilot study was done on four healthy donors. One hundred µl of ethylenediaminetetraacetic acid (EDTA) anticoagulated whole blood was blocked with normal mouse IgG at 4°C for 10 minutes (min) and stained with the following antibodies: CD31 FITC, CD146 PE, CD45 V450 and CD34 APC according to manufacturer’s instructions. Cells were lysed with BD FACS lyse™ and washed once. Data was acquired with Diva software on a FACS Aria™ (BD Biosciences, Plscataway, NJ, USA). A target of 50,000 mononuclear cell events was saved on each sample.
Blood samples for sorting putative CECs, CD14G+ T cells and HUVECs
Forty ml of blood from each of the study participants was placed into Vacutainer® tubes containing EDTA for processing of lysed whole blood for sorting putative CECs. An additional two sets of 40 ml of blood from each of the study participants was placed into Vacutainer® tubes containing sodium citrate. Both sets were used to isolate mononuclear cells. One of the mononuclear cell preparations was used to isolate CD146+ T cells from each donor. To the other set, HUVEC were added for subsequent retrieval by fluorescence- activated cell sorting (FACS). Blood was processed immediately after collection.
Mononuclear cell preparations
Mononuclear cells were isolated using a two-step approach. Citrate anticoagulated blood was first mixed with equal volumes of 3% dextran (M.W 200,000–300,000) and the erythrocytes were allowed to sediment for 40 min at a 30° angle. The red cell free supernatant was then layered over an equal volume of Histopaque-1077 and centrifuged at 1200 RPM at 15°C for 30 min with no brake.
Endothelial cell culture
HUVECs were cultured in EBMr medium supplemented with the EGM-2 bullet kit in Nunc™ tissue culture flasks according to manufacturer’s instructions and used at passages 3 or 4. For sorting HUVECs, trypsinised cultured HUVECs were added to mononuclear cell preparations.
Sample processing for cell sorting
Mononuclear cells with added HUVECs as well as mononuclear cells alone were blocked with normal mouse IgG for 10 min at 4°C. These preparations were stained according to the manufacturer’s instructions using the following antibodies: CD45 Pacific Blue, CD3 FITC, CD14 PE, CD19 APC-Cy7 and CD146 Alexa 647. Negative fluorescence was set using unstained cells.
For CEC sorts, 40 ml of EDTA anticoagulated whole blood was processed according to previously described methods (26). Details are available in the online supplement. Samples were blocked with normal mouse IgG for 10 min at 4°C and stained with CD45 V450, CD34 APC, CD146 PE. Tubes were incubated in the dark at 4°C for 25 min followed by 5 min at room temperature. RBCs were lysed with Ammonium-Chloride-Potassium lysis buffer. Samples were checked at 3 min. If lysis was not complete samples were incubated for an additional 2 min. Samples were centrifuged for 5 min at 250 g with the brake on. Tubes were washed once with Hank’s balanced salt solution without calcium or magnesium (HBSS−) containing 1 mM EDTA. Cell pellets were resuspended in HBSS (−) with 2% (v/v) heat inactivated foetal calf serum (FCS) and 2 mM EDTA.
FACS cell sorting
Cell suspensions were filtered with 50 mm filters. 7-AAD was added prior to sorting. Cell sorting was performed using the FACS Aria™ with Diva software at 4°C using a 100 micron nozzle at 20 psi. CD146+ T-cells were sorted from mononuclear cell preparations using the following criteria: CD45+, CD3+, CD14−, CD19−, CD146+. HUVECs also sorted from mononuclear cell preparations were CD45−, CD3−, CD14−, CD19−, CD146+. The population of putative CECs were sorted from lysed whole blood using the following criteria: CD45−, CD34dim and then classified by the presence or absence of CD146 by applying a sequential gating strategy.
Cells for ultramicro analytical immunochemistry were collected in 25 µ1 phosphate-buffered saline (PBS) and frozen on dry ice immediately after collection. Cells for Cytospin™ slides were collected in 100 µ1 HBSS (−) with 20% heat inactivated FCS. Cells for RNA isolation and analysis were collected in RLT™ buffer with β-mercaptoethanol, taking care not to exceed a 1:10 dilution of RLT™ buffer in any sample to preserve the integrity of the RNA in the captured cells.
Sorted HUVECs were studied in parallel with putative CECs by light and immunofluorescent microscopy and by RNA analysis. Sorted CD146+ T cells, sorted HUVECs and putative CECs also underwent membrane protein analysis by ultramicro analytical immunochemistry.
Bright field and immunofluorescent microscopy
Cytospin™ slides were prepared and were either stained with the Diff-Quick™ staining kit for light microscopic analysis or fixed with formaldehyde and immunostained with TRlTC-labelled Ulex europaeus (UEA-1), FITC conjugated von Willebrand factor (vWF) and counterstained with 300 nM 4′,6-diamidino-2-phenylindole (DAPI). Staining protocols are available in the Suppl. data (available online at www.thrombosis-online.com).
Light and fluorescence microscopy were performed using an inverted Nikon Eclipse E-800 microscope with conventional filter packs (Nikon Instruments). The microscope was connected to a Nikon DXM1200F digital camera to document bright field and fluorescence microscopic images. Images were captured using Nikon ACT software.
Ultramicro analytical immunochemistry
FACS sorted samples of putative CECs (n=4), HUVECs (n=4) and CD146+ T cells (n=4) underwent prptein analysis by a single investigator (TP) blinded to sample group. Micro-recycling immunoaffinity chromatography was performed using a chip-based modification of a previously described technique (29). Details of the sample processing and analysis are available in the Suppl. data (available online at www.thrombosis-online.com).
RNA isolation and analysis
Total RNA purified from FACS sorted samples of putative CECs (n=4), HUVECs (n=4) and CD146+ T cells (n=4) was quantified using a Nanodrop ND-1000 and evaluated for RNA integrity using RNA Nano Chips. Assay details are available in the Suppl. data (available online at www.thrombosis-online.com).
PCR of FACS sorted cells
Two multiplex PCR assays were developed. One consisted of CD45, GAPDH and CD144 while the second was used to detect CD105, vWF and CD31. Primer-BLAST was used to select gene specific primers (30). Complete details for primer sequences, PCR conditions and quantitation can be found in the Suppl. data (available online at www.thrombosis-online.com).
Statistics
The flow data files were analysed with FCS Express4 (DeNovo Software). Data are shown as mean ± standard error of the mean (SEM). The R statistical program (31) was used for analysis. All tests were performed on data averaged from four separate donors. The results were analysed by performing two tailed paired t-tests with p < 0.05 indicating significant difference. Protein values that were below the level of detection (<0.2 fg/pg recovered protein) were assigned a value of 0.2 for the paired t-test calculations.
Results
CD45− populations of CD31 bright and CD34 dim cells
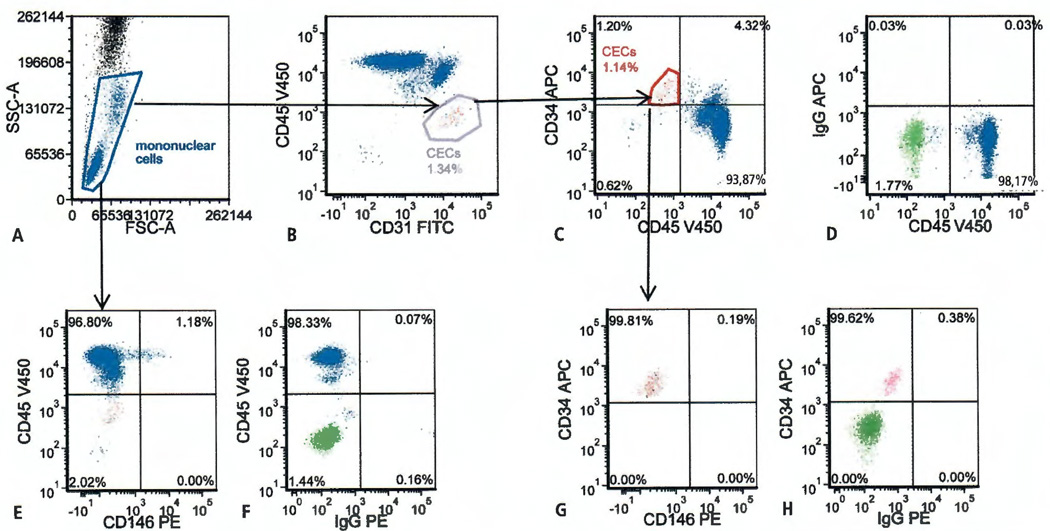
The initial data obtained from the FACS Aria™ pilot study confirmed a prior observation (26) that within the mononuclear cell gate (Figure 1A) there exists a unique population of CD45−, CD31bright cells (Figure 1B). Backgating this distinct CD45−, CD31bright population confirmed it identified a population that was also CD45−, CD34dim (Figure 1C). To determine the location of the CD45−, CD34dim population a fluorescence minus one control for APC with an overly of the unstained control was used (Figure 1D).
Figure 1. Four-colour flow cytometry evaluation of circulating endothelial cells (CECs) and CD146 expression.
A) Representative panel showing analysis gate used to exclude neutrophils, platelets, RBCs, and debris. B) Distinct CD31 bright population outside of the CD45 positive mononuclear cell population. C) CD31 bright identifies a distinct CD34 dim population. D) Overlay of the unstained control cell population (green) in the fluorescence minus one (FMO) APC control for CD34 APC vs CD45 V450. E) Mononuclear cell gate evaluated for CD146 staining shows a small percentage of CD45 positive, CD146 positive events. F) Overlay of the unstained control cell population (green) in the FMO PE control for CD146 PE vs CD45 V450. G) CEC gate evaluated for CD146 staining shows no CD146 staining. H) Overlay of the unstained control cell population (green) in the FMO PE control for CD146 PE vs CD34 APC.
Expression of CD146 on the surface of co45−, CD34dim cells
Since CD146 has been extensively used as a marker for CECs, the cell preparations were tested for the presence of CD146. In the pilot study CD146 expression (Figure 1E) was seen in 0.78 ± 0.13% (n=4) of the mononuclear cells based on the quadrant markers set by the FMO PE control for CD146 and CD45 V450 (Figure 1F). However, no CD146 expression was detected on the CD45−, CD34dim cells (Figure 1G), based on the quadrant markers set by the FMO PE control for CD146 and CD34 APC (Figure 1H).
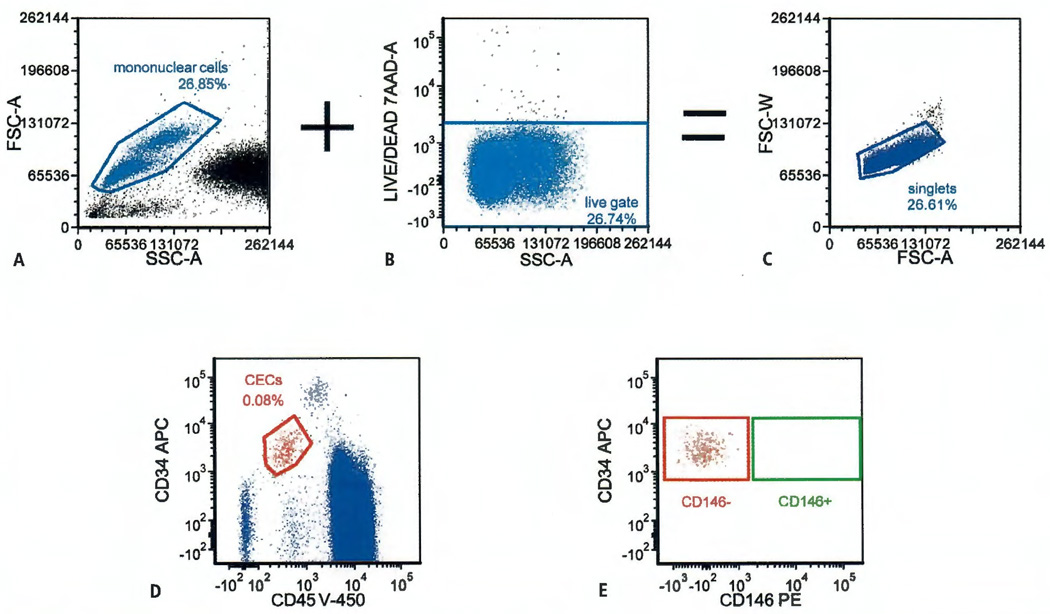
Based on these results, a modification of a published gating strategy (26) was devised to isolate putative CECs for further study (Figure 2). The mononuclear cell gate (Figure 2A) was combined with a 7-AAD negative (implies viability) gate (Figure 2B) to create a singlet gate (Figure 2C). The plot shown in Figure 2D was derived from this singlet gate. Using this strategy, the nuclei of nonviable cells should stain with 7-AAD and be eliminated from the collection. CD31 was not included in the sort tubes because CD45−, CD31dim cells were confirmed to be largely inclusive of the CD45−, CD34bright population. In addition, there was also the possibility that the CD31bright population, unlike the CD34dim population, might be contaminated with large platelets (32, 33). The CD45−, CD34dim, 7-AAD− population was gated and used for the sort plot (Figure 2E). Gates were set to separately collect the CD45−, CD34dim, 7-AAD−, CD146− population (red rectangle) and the CD45−, CD34dim, 7-AAD−, CD146+ population (green rectangle). For the CD45−, CD34dim, 7-AAD− population, the average number of CD146− events was 30,300 ± 6,600. However, using this gating strategy that included 7-AAD viability staining no CD146+ putative CEC events were available for collection. Since 7-AAD− events were initially presumed to be viable cells, validation studies were continued on the CD45−, CD34dim, CD146−, 7-AAD− cells (putative CECs). Subsequent histologic examination of these cells (see below) found that 7-AAD− events also captured cellular remnants since 7-AAD was unable to stain cells with disintegrating or absent nuclei.
Figure 2. Gating strategy for sorting circulating endothelial cells (CECs).
The mononuclear cell gate (A) and the viability gate (B) were combined to give a singlet gate (C). All subsequent plots were derived from the singlegate. D) Determination of the CD45 negative, CD34 dim CEC population used in the sort gates. E) Events inside the red rectangle were collected as CD146 negative CECs while those inside the green rectangle were collected as CD146 positive CECs.
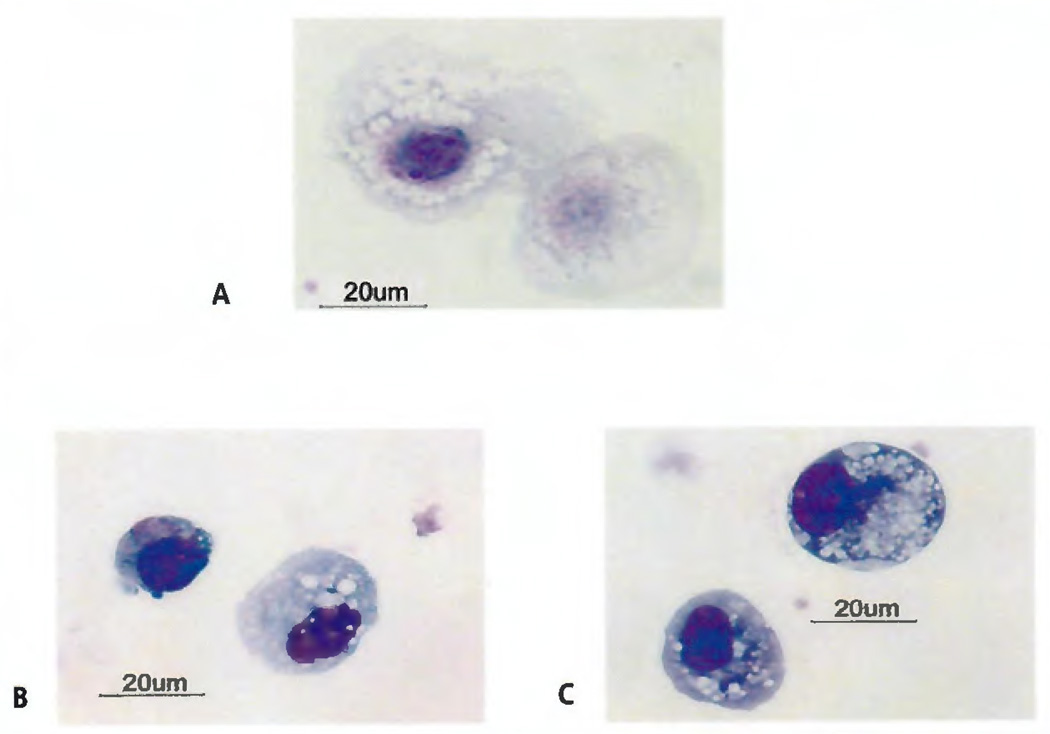
Light micrographs of FACS sorted CECs
Cytospins of sorted cells based on the CD45−, CD34dim, CD146−, 7-AAD− immunophenotype were prepared. While some of these putative CECs appeared to have a nucleus, the majority of these cells displayed various degrees of nuclear dissolution (Figure 3A). Intact, normal appearing nuclei were rare. In one of the four healthy volunteers, whose sorted cells were rigorously evaluated by light microscopy, CD45−, CD34dim, CD146−, 7-AAD− cells with any discernible nuclei were <20% of the total number evaluated. Considerable cytoplasmic vacuolisation was common. In contrast, Diff-Quick™ stains of Cytospin™ preparations of HUVECs made prior to their addition to a peripheral blood mononuclear cell preparation (Figure 3B) as well as after their addition and retrieval by FACS sorting (Figure 3C) revealed well-defined nuclear morphology with less cytoplasmic vacuolisation. Sorted HUVECs (Figure 3C) showed increased vacuolisation when compared to cells that were freshly harvested from culture (Figure 3B). However, the cytoplasm of both the freshly harvested (Figure 3B) and the sorted HUVECs (Figure 3C) appeared to be denser than the putative CECs (Figure 3A). Sorted HUVECs and putative CECs appeared to be similar in size.
Figure 3. Diff-Quick™ images of Cytospin™ cell preparations (100×).
A) Cells from CD45 negative, CD34 dim, CD146 negative sort. A size marker is shown with a platelet for comparison. B) Unsorted human umbilical vein endothelial cells freshly isolated from culture. C) Human umbilical vein endothelial cells spiked into mononuclear cell preparation and recovered by fluorescence-activated cell sorting.
Fluorescent staining for UEA-1, vWF and DAPI
To further evaluate the phenotype and morphologic integrity of the sorted CD45−, CD34dim, CD146−, 7-AAD− cells, Cytospin™ preparations were stained with anti-UEA-1, an antibody that specifically identifies UEA-1 bound to vascular endothelium (Figure 4A) and anti-vWF to label vWF normally found in the Weibel-Palade bodies of endothelial cells(Figure 4B). The nuclear stain DAPI was used to further assess for the presence or absence of a cell nucleus (Figure 4C). The UEA-1 stained cells (Figure 4A) displayed varying degrees of vWF staining (Figure 4B) suggesting endothelial phenotype. However, < 20% of cells evaluated showed nuclear DAPI staining (Figure 4C) suggesting most of the cells were anuclear. In addition at 100× (Figure 4D–E), most of the UEA-1 and vWF staining was concentrated in the centre of the cell. These observations could be consistent with senescent endothelial cells in various stages of disintegration. A typical Diff-Quick™ stain of these cells is shown for comparison (Figure 4F).
Figure 4. Fluorescent cell staining.
A) Ulex Europeus Agglutinin-1 (UEA-1) stain; B) Von Willebrand factor (vWF) stain; C) 4′,6-Diamidino-2-Phenylindole, Di lactate (DAPI) staining of a representative field at 20×; D) UEA at 100×; E) vWF stain at 100×; F) Light micrograph of a typical CD45 negative, CD34 dim, CD146 negative cell at 100×.
Validation of endothelial cell phenotype by ultramicro analytical immunochemistry
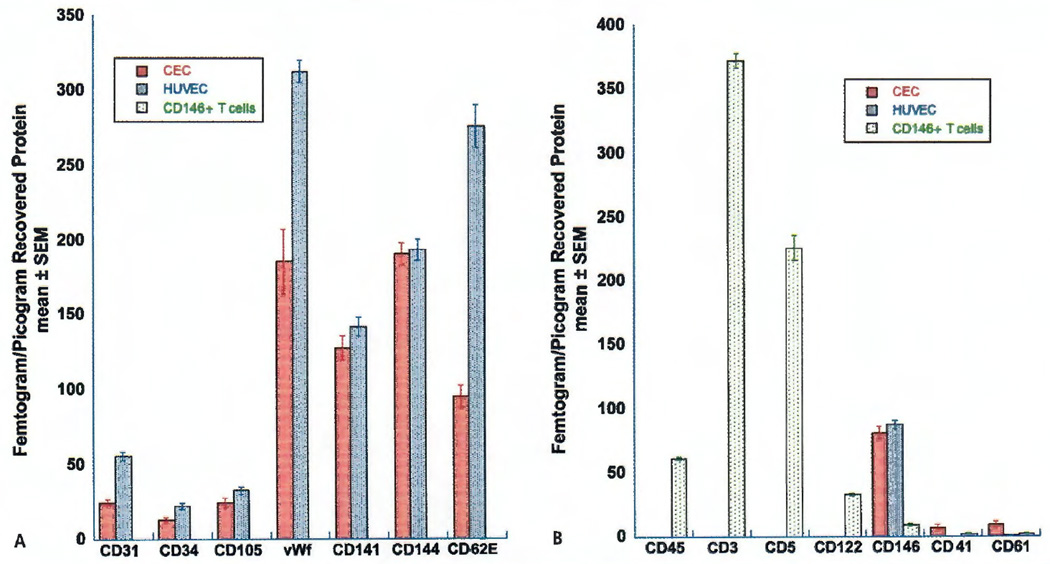
To validate the identity of the sorted CD45−, CD34dim, CD146−, 7-AAD− cells as endothelial cells, additional cells from each of the four CD45−, CD34dim, CD146−, 7-AAD− FACS sorts were evaluated by ultramicro analytical immunochemistry. This technique can detect proteins not only on the extracellular surface but also on the inner cytoplasmic surface of the cell membrane that could not be detected by surface staining and flow cytometry. CD146+ T cells and spiked HUVECs recovered from their four respective peripheral blood mononuclear cell preparations were similarly analysed and served as controls for T cell and endothelial cell markers, respectively.
The expression of seven endothelial cell markers on putative CECs, HUVEC, and CD146+ T lymphocytes was evaluated (Figure 5A). Putative CECs had protein levels of CD34, a marker found on endothelial cells and stem cells, that were similar to the CD34 levels found in HUVECs (13.03 ± 0.89 vs 22.30 ± 1.34 fg/pg recovered protein, p=0.05). The amount of CD141, an integral membrane protein expressed on the surface of endothelial cells, platelets, monocytes and neutrophils, was not statistically different in putative CECs compared to HUVECs (127.58 ± 5.17 vs 141.75 ± 4.45 fg/pg recovered protein, p>0.05). CD105 (endoglin) found on endothelial cells and stem cells, and CD144 (VE-cadherin) that is found only on endothelial cells, were also present in comparable amounts in putative CECs when compared to HUVECs (CD105: 24.15 ± 1.96 vs 32.68 ± 1.83 fg/pg recovered protein, p>0.05; CD144: 190.50 ± 5.34 vs 193.40 ± 4.98 fg/pg recovered protein, p>0.05). The following endothelial markers were also expressed in putative CECs but to a lesser extent than in HUVECs: CD31 (24.43 ± 1.63 vs 55.73 ± 1.92 fg/pg recovered protein, p<0.05), a marker expressed on endothelial cells and platelets; CD62E (94.93 ± 4.84 vs 276.5 ± 9.45 fg/pg recovered protein, p<0.05) a protein expressed after cytokine activation of endothelial cells; and vWF (185.3 ± 14.77 vs 313.08 ± 5.12 fg/pg recovered protein, p<0.05) found in the Weibel-Palade bodies of endothelial cells and a-granules of platelets. In the FACS sorted CD146+ T cell population all seven endothelial protein markers were below the limit of detection.
Figure 5. Protein analysis of fluorescence-activated sorted cells by ultramicro analytical immunochemistry.
A) Seven endothelial cell markers were assayed on membrane proteins isolated from circulating endothelial cells (CECs), human umbilical vein endothelial cells (HUVECs) and CD146 positive T cells. All seven endothelial cell markers were below the level of detection (<0.2 fg/pg recovered protein) on the CD146 positive T cells. B) Four leukocyte markers, two platelet markers and CD146 (expressed on both T cells and endothelial cells), were assayed on membrane proteins isolated from CE Cs, HUVECs and CD146 positive T cells. The leukocyte markers CD45, CD3, CD5 and CD122 were below the level of detection for the assay (<0.2 fg/pg recovered protein) in both CECs and HUVECs. vWF=Von Willebrand factor.
Only the CD146+ T cells were positive for the pan-leukocyte marker CD45; the T cell markers CD3 and CD5; and the T cell, B cell and monocyte marker CD122 (Figure 5B). The detection of CD41 and CD61 protein indicated a small amount of platelet contamination that could also be seen on the Diff-Quick™ stained cells. It is notable that while the analysed CD45−, CD34dim, CD146−, 7-AAD− cells were selected based on their lack of CD146 surface staining, the amount of CD146 protein determined by ultramicro analytical immunochemistry is similar for these cells and HUVECs (80.95 ± 3.22 vs 87.93 ± 2.21 fg/pg recovered protein, p>0.05) suggesting that CD146 was still present in the CD45−, CD34dim, CD146−, 7-AAD− cells but not accessible to flow cytometry as a surface feature. The amount of CD146 protein measured in the CD146+ T cell preparations is 10-fold lower, but its presence is consistent with prior observations of CD146 expression on T cells (24).
Additional markers besides those illustrated in Figure 5 were also evaluated. Membrane proteins isolated from CD45−, CD34dim, CD146−, 7-AAD− cells, HUVECs and CD146+ T cells were all below the limit of detection for the B cell markers CD19 and CD20; the B cell, neutrophil and NK cell marker IgG Fe receptor; the neutrophil, eosinophil and monocyte marker CD15; the monocyte marker CD14; and the plasma cell marker CD138. The level of the hematopoietic and endothelial progenitor cell marker CD133 was also below the level of detection in the CD45−, CD34dim, CD146−, 7-AAD− cell membrane preparations.
Evaluation of RNA from FACS sorted cells
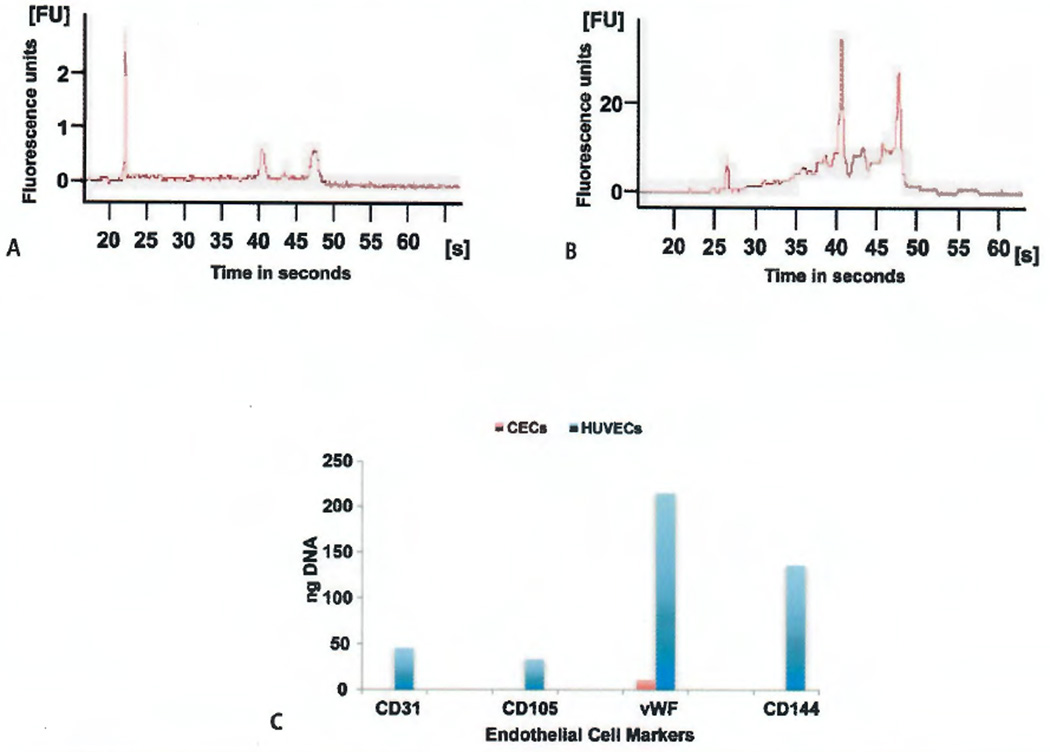
Calculation of the RNA Integrity Number (RIN) allows for the determination of the state of the entire RNA sample including the presence or absence of degradation products that is independent of sample concentration. A RIN number of 10 is indicative of RNA that is 100% intact while a RIN number of 2 or less indicates the RNA is totally degraded (34). Total RNA isolated from 30,000 FACS sorted events with the CEC phenotype CD45−, CD34dim, CD146−, 7-AAD− (Figure 6A) revealed only a small amount of RNA that was mostly degraded (RIN 1.95 ± 1.9). Total RNA from 20,000 sorted HUVEC events (Figure 6B) showed good preservation (RIN 6.75 ± 0.41).
Figure 6. Analysis of the RNA integrity and RT-PCR.
A) An electrophoretic trace of a representative sample of tRNA isolated from 30,000 circulating endothelial cells (CECs) collected by fluorescence-activated cell sorting. B) An electrophoretic trace of total RNA isolated from 20,000 human umbilical vein endothelial cells (HUVECs) recovered by fluorescence-activated cell sorting. C) Only the HUVECs were positive for all four endothelial cell mRNAs. A small amount of Von Willebrand factor (vWF) message was found in two of the CEC RNA preparations tested. For each sample, 10 µl from a 25 µl PCR reaction was evaluated.
Since all of the HUVEC and CD45−, CD34dim, CD146−, 7-AAD− samples were positive for endothelial cell protein markers by ultramicro analytical immunochemistry (Figure 5A), the samples were tested by RT-PCR for the presence of endothelial cell specific RNA (Figure 6C). The housekeeping gene GAPDH was present in all of the HUVEC samples (HUVECs 174.4 ± 19.8 ng/10 µl PCR reaction) and most of the CD45−, CD34dim, CD146−, 7-AAD− samples (47.0 ± 12.7 ng/10 µl PCR reaction). However, only the RNA from sorted HUVECS showed CD31, CD105, and CD144 mRNA expression (Figure 6C). HUVEC samples showed a high content of mRNA for vWF while only two of the CD45−, CD34dim, CD146−, 7-AAD− samples had a small amount of detectable vWF mRNA (Figure 6C).
Discussion
Many of the current methodologies for CEC enumeration and isolation lack appropriate validation. This study modified and validated a previously described flow cytometry technique that identified CECs as CD45−, CD31bright and CD34dim population (26). Backgating analysis confirmed that the CD45−, CD31bright population co-identified with the CD45−, CD34dim population (26). The sort gate CD45−, CD34dim was used to decrease the possibility of including CD31bright large platelets (33). None of the CD45−, CD34dim 7-AAD− events showed CD146 surface expression. An earlier study was similarly unable to demonstrate any CD146 expression on CD45−, CD31bright, CD34dim cells (28). These results call into question prior studies comparing CEC counts in normal and disease states that relied exclusively on identification of CECs with CD146 (11, 18, 21).
Although none of the CD45−, CD34dim, 7-AAD− cells collected were positive for CD 146 surface expression, ultramicro analytical immunochemistry found the CD146 membrane bound protein content was similar in both HUVECs and sorted CECs. CD146 is an adhesion molecule that is normally found in the tight junctions between endothelial cells forming monolayers in the wall of the blood vessels (35). Studies have indicated that a large portion of CD 146 is intracellular and the ectodomain of this receptor can be shed from the surface of cells (23, 36, 37). In this study the absence of CD146 expression on the surface of cells by flow cytometry, but its presence by ultramicro analytical immunochemistry, which evaluates both the intra- and extracellular membrane surfaces, further supports the shedding of CD146 protein from the surface of endothelial cells that enter the circulation. Therefore, exclusive reliance on CD146 to identify CECs would capture only a small fraction of the actual population and could potentially be contaminated with substantial numbers of CD146+ lymphocytes (24). It is possible that the presence of CD146 surface expression may identify an even rarer population of newly shed endothelial cells that could contain intact nuclei.
Flow cytometry is a powerful tool for rapidly evaluating large numbers of events, but strategies used for identifying CECs have varied. The gating strategy used here is similar to prior studies (25, 26, 38) in that the initial gating strategy was based on CD45−, CD31+. Two of the other studies also showed a low level of CD34 expression (26, 38). Only one of these studies used viability staining and morphologic validation (38) and similarly found the majority of the events to be cellular remnants, but on the level of 1–5 µm microparticles. In contrast to these prior strategies the current study was able to isolate putative CECs without the use of KDR (25) or CD133 (25, 26, 38) or CD14 (38). Our study further simplifies their approaches by confirming there is a unique CD34 dim population distinct from the CD45+ mononuclear cells.
CEC studies by flow cytometry are complicated by the fact that many common cell markers used to identify endothelial cells are variably expressed by multiple cell populations (39, 40). One such population is the circulating progenitor cells which are described by the phenotype CD45dim, CD31+, CD34bright CD133+ (26). A subset of this population, the endothelial progenitor cells must be distinguished from CECs, but their phenotype by flow cytometry is also controversial (40). Their suggested phenotype CD45−/dim CD31+ CD34+, KDR+, CD133+ shares markers being used to identify CECs (25, 40). Thus, this study used ultramicro analytical immunochemistry, which analyses membrane bound proteins, to assess multiple markers including the level of the hematopoietic and endothelial progenitor cell marker CD133 which was found to be below the level of detection in the CD45−, CD34dim, CD146−, 7-AAD− population. In addition, the endothelial markers CD34, CD105, CD141, CD144 and CD146 were similarly expressed on HUVECs and the CD45−, CD34dim, CD146−, 7-AAD− population. The amount of vWF found in the CD45−, CD34dim, CD146−, 7-AAD− cells was almost half of the amount found in the HUVECs. These results are in agreement with an in vitro study linking vWF secretion and apoptosis in cultured HUVECs (41). CD 62E showed the lowest correlation between HUVECs and CD45−, CD34dim, CD146−, 7-AAD− cells. However, CD62E is transiently expressed in cytokine-activated endothelium, so a loss of this protein might be expected in shed senescent vascular endothelium. As expected, leukocyte and nonendothelial cell markers CD45, CD3, CD5, CD122, CD19, CD20, CD14, CD138 and IgGFc receptor were all below the limit of detection in the CD45−, CD34dim, CD146−, 7-AAD− population. Taken together, these results suggest these putative CECs (CD45−, CD34dim, CD146−, 7-AAD−) are consistent with an endothelial phenotype.
The morphology of CD45−, CD34dim, CD146− CECs was remarkably consistent in size with the HUVEC control. However, the pattern of UEA-1 and vWF staining of the CECs was not typical of viable endothelial cells. UEA-1 is a lectin stain that is routinely used to validate endothelial cells in the CD146 immunobead assay. It is normally found on the cell membrane. In endothelial cells, vWF factor is normally found in Weibel-Palade bodies in the cytoplasm. In the CECs characterised here, it appears that the distribution of UEA-1 lectin and vWF was concentrated in the nuclear area. These observations support the idea that most CECs in healthy individuals are senescent remnants or apoptotic cells which may result from program cell death triggered by the detachment of these anchorage-dependent cells from the surrounding extracellular matrix, a process known as anoikis (42).
It is also possible the observed degradation is an artefact of the technique. However, it is important to note the sorts were done using a 100 micron nozzle at 20 psi compared to a 70 micron nozzle at 70 psi that has been routinely used in FACS collections. The larger nozzle and lower pressure make mechanical cell damage less likely. In addition, the spiked HUVECs were retrieved intact after undergoing the same FACS sorting conditions. The large nozzle and low pressure may also have been instrumental in preserving cell morphology, thereby permitting for the first time the reproducible ability to obtain bright field images of CECs.
Many published studies employ a nuclear stain to ensure only nucleated cells are counted as CECs (17). Viability staining with 7-AAD requires leaky plasma membranes to detect dead cells by positive nuclear staining (43). DAPI staining of the CD45−, CD34dim, CD146−, 7-AAD− CEC population indicates that only a small fraction have intact nuclei. The majority are anucleated cells that will not be tagged by 7-AAD for exclusion. Electron micrographic studies of apoptotic HUVEC cells in culture showing dissolution of the nuclear compartment are consistent with this observation (44). Thus CEC identification algorithms using 7-AAD alone overestimate viability, since anuclear cells will be counted as viable.
RNA from the CD45−, CD34dim, CD146−, 7-AAD− CEC population was degraded, and RT-PCR analysis for the genes tested was unable to show any significant amount of endothelial cell specific mRNA expression. In contrast the RNA quality assessment assays and RT-PCR data from the sorted HUVEC cells showed that their RNA remained intact and expressed endothelial transcripts. These findings rule out degradation related to sample processing and flow sorting techniques. Based on these observations, we conclude that the majority of CECs in healthy adults contain mostly degraded RNA.
The results support the following identification scheme for CECs. CD45−, CD34dim should be used to enumerate the total number of CECs in circulation. CD31bright backgating may be used to confirm the CD45−, CD34dim CEC gate. Of this total, the vast majority·are CD146− cells and those with a positive nuclear stain are likely early apoptotic CECs, and the remaining cells lacking distinct nuclei are late apoptotic CECs. Our study also indicates that these CD146 extracellular surface negative CECs lack RNA suitable for transcript analysis by such techniques as microarray and quantitative RT-PCR. For the first time, additional validation is provided towards the identity of CD45−, CD31bright, CD34dim, CD146−, 7-AAD− circulating cell population utilising ultramicro analytical immunochemistry showing that these events have multiple detectable endothelial cell proteins (CD31 CD34, CD62E, CD105, vWF, CD141, CD144 and CD146). Overall our results provide insights into the fate of normally shed endothelium and serve as a basis for evaluating numerous studies using CECs as a biomarker in health and disease.
Supplementary Material
What is known about this topic?
Endothelial cells shed from the blood vessel walls comprise a very rare population in peripheral blood circulation.
Multiple poorly validated schemes exist for the identification of circulating endothelial cells (CECs) in human blood.
CECs have been proposed as a marker to assess vascular injury.
What does this paper add?
In a putative CD45−, CD34dim [CD31bright], 7-AAD− CEC population we were unable to detect CD146 surface expression, a marker that has been widely used to quantify CECs.
Ultramicro analytical immunochemistry was used to validate the endothelial phenotype of CD45−, (D34dim, CD146−, 7-AAD− cells in healthy adults.
FACS sorted cells isolated from whole blood with the phenotype CD45−, CD34dim, CD146−, 7-AAD− still express CD146 protein when examined as isolated membranes confirming the shedding of CD146 extracellular domain.
The paper demonstrates that the majority of 7-AAD− CECs in healthy adults are nonviable cells whose RNA is not suitable for transcript analysis illustrating the pitfall of relying on 7-AAD as a viability marker.
Acknowledgments
The authors would like to thank Kelly Byrne for her assistance in manuscript preparation and submission.
Financial support:
This work was funded by Critical Care Medicine Department, Clinical Center Intramural Research Program.
Footnotes
Conflicts of interest
None declared.
References
- 1.Bouvier CAGE, Cintron JR, et al. Circulating Endothelium as an Indicator of Vascular Injury. Thromb Diath Haemorrh. 1970;40(Suppl):163–168. [Google Scholar]
- 2.Hladovec JaR P. Circulating Endothelial Cells Isolated Together with Platelets and the Experimental Modification of their counts in Rats. Thrombosis Res. 1973;3:665–674. [Google Scholar]
- 3.Hladovec J, Prerovsky I, Stanek V, et al. Circulating endothelial cells in acute myocardial infarction and angina pectoris. Klin Wochenschr. 1978;56:1033–1036. doi: 10.1007/BF01476669. [DOI] [PubMed] [Google Scholar]
- 4.George F, Poncelet P, Laurent JC, et al. Cytofluorometric detection of human endothelial cells in whole blood using S-Endo 1 monoclonal antibody. J Immunol Methods. 1991;139:65–75. doi: 10.1016/0022-1759(91)90352-g. [DOI] [PubMed] [Google Scholar]
- 5.George FBC, Poncelet P, et al. Rapid isolation of human endothelial cells from whole blood using S-Endol monoclonal antibody coupled to immuno-magnetic beads: demonstration of endothelial injury after angioplasty. Thromb Haemost. 1992;67:147–153. [PubMed] [Google Scholar]
- 6.Janssens D, Michiels C, Guillaume G, et al. Increase in circulating endothelial cells in patients with primary chronic venous insufficiency: protective effect of Ginkor Fort in a randomized double-blind, placebo-controlled clinical trial. J Cardiovasc Pharmacol. 1999;33:7–11. doi: 10.1097/00005344-199901000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Bertolini F, Shaked Y, Mancuso P, et al. The multifaceted circulating endothelial cell in cancer: towards marker and target identification. Nat Rev Cancer. 2006;6:835–845. doi: 10.1038/nrc1971. [DOI] [PubMed] [Google Scholar]
- 8.Rowand JL, Martin G, Doyle GV, et al. Endothelial cells in peripheral blood of healthy subjects and patients with metastatic carcinomas. Cytometry A. 2007;71:105–113. doi: 10.1002/cyto.a.20364. [DOI] [PubMed] [Google Scholar]
- 9.Strijbos MH, Landburg PP, Nur E, et al. Circulating endothelial cells: a potential parameter of organ damage in sickle cell anemia? Blood Cells Mol Dis. 2009;43:63–67. doi: 10.1016/j.bcmd.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Lombardo MF, Iacopino P, Cuzzola M, et al. Type 2 diabetes mellitus impairs the maturation of endothelial progenitor cells and increases the number of circulating endothelial cells in peripheral blood. Cytometry A. 2012;81:856–864. doi: 10.1002/cyto.a.22109. [DOI] [PubMed] [Google Scholar]
- 11.Damani S, Bacconi A, Libiger O, et al. Characterization of circulating endothelial cells in acute myocardial infarction. Sci Transl Med. 2012;4:126ra33. doi: 10.1126/scitranslmed.3003451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tinazzi E, Dolcino M, Puccetti A, et al. Gene expression profiling in circulating endothelial cells from systemic sclerosis patients shows an altered control of apoptosis and angiogenesis that is modified by iloprost infusion. Arthritis Res Ther. 2010;12:R131. doi: 10.1186/ar3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossi E, Fassan M, Aieta M, et al. Dynamic changes of live/apoptotic circulating tumour cells as predictive marker of response to sunitinib in metastatic renal cancer. Br J Cancer. 2012;107:1286–1294. doi: 10.1038/bjc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manzoni M, Mariucci S, Delfanti S, et al. Circulating endothelial cells and their apoptotic fraction are mutually independent predictive biomarkers in Bevacizumab-based treatment for advanced colorectal cancer. J Cancer Res Clin Oncol. 2012;138:1187–1196. doi: 10.1007/s00432-012-1190-6. [DOI] [PubMed] [Google Scholar]
- 15.Jubb AM, Harris AL. Biomarkers to predict the clinical efficacy of bevacizumab in cancer. Lancet Oncol. 2010;11:1172–1183. doi: 10.1016/S1470-2045(10)70232-1. [DOI] [PubMed] [Google Scholar]
- 16.Erdbruegger U, Dhaygude A, Haubitz M, et al. Circulating endothelial cells: markers and mediators of vascular damage. Curr Stem Cell Res Ther. 2010;5:294–302. doi: 10.2174/157488810793351721. [DOI] [PubMed] [Google Scholar]
- 17.Mancuso P, Antoniotti P, Quarna J, et al. Validation of a standardized method for enumerating circulating endothelial cells and progenitors: flow cytometry and molecular and ultrastructural analyses. Clin Cancer Res. 2009;15:267–273. doi: 10.1158/1078-0432.CCR-08-0432. [DOI] [PubMed] [Google Scholar]
- 18.Elshal M, Abdelaziz A, Abbas A, et al. Quantification of circulating endothelial cells in peripheral blood of systemic lupus erythematosus patients: a simple and reproducible method of assessing endothelial injury and repair. Nephrol Dial Transplant. 2009;24:1495–1499. doi: 10.1093/ndt/gfn650. [DOI] [PubMed] [Google Scholar]
- 19.Woywodt A, Blann AD, Kirsch T, et al. Isolation and enumeration of circulating endothelial cells by immunomagnetic isolation: proposal of a definition and a consensus protocol. J Thromb Haemost. 2006;4:671–677. doi: 10.1111/j.1538-7836.2006.01794.x. [DOI] [PubMed] [Google Scholar]
- 20.Mariucci S, Rovati B, Chatzileontiadou S, et al. A six-colour flow cytometric method for simultaneous detection of cell phenotype and apoptosis of circulating endothelial cells. Scand J Clin Lab Invest. 2009;69:433–438. doi: 10.1080/00365510802673175. [DOI] [PubMed] [Google Scholar]
- 21.Kraan J, Strijbos MH, Sieuwerts AM, et al. A new approach for rapid and reliable enumeration of circulating endothelial cells in patients. J Thromb Haemost. 2012;10:931–939. doi: 10.1111/j.1538-7836.2012.04681.x. [DOI] [PubMed] [Google Scholar]
- 22.Kraan J, Sleijfer S, Foekens JA, et al. Clinical value of circulating endothelial cell detection in oncology. Drug Discov Today. 2012;17:710–717. doi: 10.1016/j.drudis.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Bardin VM, Moal V, Anfosso F, Daniel L, Brunet P, Sampol J, Dignat George F. Soluble CD146, a novel endothelial marker, is increased in physiopathological settings linked to endothelial junctional alteration. Thromb Haemost. 2003;90:915–920. doi: 10.1160/TH02-11-0285. [DOI] [PubMed] [Google Scholar]
- 24.Elshal MF, Khan SS, Takahashi Y, et al. CD146 (Mel-CAM), an adhesion marker of endothelial cells, is a novel marker of lymphocyte subset activation in normal peripheral blood. Blood. 2005;106:2923–2924. doi: 10.1182/blood-2005-06-2307. [DOI] [PubMed] [Google Scholar]
- 25.Steurer M, Kern J, Zitt M, et al. Quantification of circulating endothelial and progenitor cells: comparison of quantitative PCR and four-channel flow cytometry. BMC Res Notes. 2008;1:71. doi: 10.1186/1756-0500-1-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duda DG, Cohen KS, Scadden DT, et al. A protocol for phenotypic detection and enumeration of circulating endothelial cells and circulating progenitor cells in human blood. Nat Protoc. 2007;2:805–810. doi: 10.1038/nprot.2007.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strijbos M, Kraan J, den Bakker M, et al. Cells meeting our immunophenotypic criteria of endothelial cells are large platelets 3. Cytometry B Clin Cytom. 2007;72:86–93. doi: 10.1002/cyto.b.20156. [DOI] [PubMed] [Google Scholar]
- 28.Duda DG, Cohen KS, di Tomaso E, et al. Differential CD146 Expression on Circulating Versus Tissue Endothelial Cells in Rectal Cancer Patients: Implications for Circulating Endothelial and Progenitor Cells As Biomarkers for Antiangiogenic Therapy. J Clin Oncol. 2006;24:1449–1453. doi: 10.1200/JCO.2005.04.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips TM. Multi-analyte analysis of biological fluids with a recycling immunoaffinity column array. J Biochem Biophys Methods. 2001;49:253–262. doi: 10.1016/s0165-022x(01)00202-0. [DOI] [PubMed] [Google Scholar]
- 30.Ye J, Coulouris G, Zaretskaya I, et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Team RDC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Accessed April 30, 2014]. Available at: http://www.R-project.org. [Google Scholar]
- 32.Newman PJ, Berndt MC, Gorski J, et al. PECAM-1 (CD31) cloning and relation to adhesion molecules of the immunoglobulin gene superfamily. Science. 1990;247:1219–1222. doi: 10.1126/science.1690453. [DOI] [PubMed] [Google Scholar]
- 33.Strijbos MH, Kraan J, den Bakker MA, et al. Cells meeting our immunophenotypic criteria of endothelial cells are large platelets. Cytometry B Clin Cytom. 2007;72:86–93. doi: 10.1002/cyto.b.20156. [DOI] [PubMed] [Google Scholar]
- 34.Mueller O, Lightfoot S, Schroder A. RNA Integrity Number (R1N) Standardization of RNA Quality Control. Tech Rep 5989-1165EN, Agilent Technologies, Application Note 2004. [Google Scholar]
- 35.Bardin N, Anfosso F, Masse JM, et al. Identification of CD146 as a component of the endothelial junction involved in the control of cell-cell cohesion. Blood. 2001;98:3677–3684. doi: 10.1182/blood.v98.13.3677. [DOI] [PubMed] [Google Scholar]
- 36.Boneberg EM, Illges H, Legler DF, et al. Soluble CD146 is generated by ectodomain shedding of membrane CD146 in a calcium-induced, matrix metalloprotease-dependent process. Microvasc Res. 2009;78:325–331. doi: 10.1016/j.mvr.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 37.Bardin N, Blot-Chabaud M, Despoix N, et al. CD146 and its Soluble Form Regulate Monocyte Transendothelial Migration. Arteriosder Thromb Vase Biol. 2009;29:746–753. doi: 10.1161/ATVBAHA.108.183251. [DOI] [PubMed] [Google Scholar]
- 38.Mund JA, Estes ML, Yoder MC, et al. Flow Cytometric Identification and Functional Characterization of Immature and Mature Circulating Endothelial Cells. Arterioscler Thromb Vase Biol. 2012;32:1045–1053. doi: 10.1161/ATVBAHA.111.244210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu H-K, Lee H-J, Choi H-N, et al. Characterization of CD45−/CD31+/CD105+ circulating cells in the peripheral blood of patients with gynecological malignancies. Clin Cancer Res. 2013;19:5340–5350. doi: 10.1158/1078-0432.CCR-12-3685. [DOI] [PubMed] [Google Scholar]
- 40.Fadini GP, Losordo D, Dimmeler S. Critical Reevaluation of Endothelial Progenitor Cell Phenotypes for Therapeutic and Diagnostic Use. Circ Res. 2012;110:624–637. doi: 10.1161/CIRCRESAHA.111.243386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antonova OA, Loktionova SA, Romanov YA, et al. Activation and damage of endothelial cells upon hypoxia/reoxygenation. Effect of extracellular pH. Biochemistry. 2009;74:605–612. doi: 10.1134/s0006297909060030. [DOI] [PubMed] [Google Scholar]
- 42.Aoudjit F, Vuori K. Matrix attachment regulates Fas-induced apoptosis in endothelial cells: a role for c-flip and implications for anoikis. J Cell Biol. 2001;152:633–643. doi: 10.1083/jcb.152.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmid I, Krall WJ, Uittenbogaart CH, et al. Dead cell discrimination with 7-amino-actinomycin D in combination with dual color immunofluorescence in single laser flow cytometry. Cytometry. 1992;13:204–208. doi: 10.1002/cyto.990130216. [DOI] [PubMed] [Google Scholar]
- 44.Xu W, Boadle R, Dear L, et al. Ultrastructural Changes in Endothelium during Apoptosis Indicate Low Microembolic Potential. J Vase Res. 2005;42:377–387. doi: 10.1159/000087213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.