Summary
Through a clinical deep sequencing protocol, Wu and colleagues have identified multiple FGFR fusion proteins in diverse cancers. Pharmacologic inhibition of FGFR suppressed the growth of FGFR fusion positive tumor models, suggesting that these FGFR fusions are oncogenic drivers and highlighting the utility of streamlined clinical sequencing efforts to identify novel, actionable driver oncoproteins in human tumors.
The promise of “precision cancer medicine” is to translate an in depth understanding of the genomic alterations present in the tumor of an individual patient into a personalized and effective therapy (1). Many of the recent efforts in precision cancer medicine have been catalyzed by the development and use of deep sequencing technology to characterize tumor genomes and transcriptomes. However, some of the greatest clinical successes achieved through precision medicine began well before the advent of deep sequencing. Indeed, for decades chronic myeloid leukemia (CML) and acute promyelocytic leukemia (APL) have been classified as unique clinical entities due to the presence of cytogenetically evident translocations, BCR-ABL and PML-RARA respectively, creating tumor-specific fusion proteins that drive disease. The identification of the chimeric proteins fueled the effective clinical use of drugs that specifically target each fusion protein, transforming CML and APL from universally lethal to curable or chronic diseases for most patients. The current report by Wu and colleagues stands on the shoulders of these clinical successes by identifying novel, actionable fusion proteins in several tumor types.
Recent efforts suggest that we are only scratching the surface when it comes to the identification of chimeric fusion proteins in human cancer, and that cryptic translocations can give rise to putative oncogenic drivers that are prime therapeutic targets (2). Even in the era of deep sequencing, identifying chimeric oncoproteins remains challenging due to the need to bridge fusion boundaries with paired-end RNA sequencing (RNAseq). Yet the therapeutic potential of such discoveries is great, particularly if the identified fusion protein is a target for which therapies are already under development, as was the case with crizotinib and the EML4-ALK fusion in lung adenocarcinoma (3). How, then, can we identify systematically these rare genetic events that could arise within a heterogeneous tumor? Furthermore, how do we delineate passenger and driver alterations among the co-occurring genetic and transcriptional alterations that are identified in an individual tumor?
By using a systematic, patient-centered deep sequencing-based protocol at the University of Michigan (MI-ONCOSEQ), Wu and colleagues have identified recurrent fusions involving FGFR family members and a variety of binding partners (4). This protocol, similar to other efforts to make deep sequencing a clinical reality for all patients, prioritizes the collection of high quality tumor samples along with germline controls at diagnosis for patients with advanced disease and allows for integrative sequencing across tumor histologies. The rationale for such efforts is that knowledge of genomic and transcriptional alterations that could be addressed using available targeted therapies will direct physicians to select potentially life prolonging or even curative treatment for patients who have no such options currently. In addition to sequencing the entire cancer exome, the Michigan protocol includes isolation of RNA with paired-end RNASeq as a means of comprehensively interrogating the cancer transcriptome. One benefit of this is the ability to identify fusion transcripts, which these authors describe in four index patients. While each patient presented in this study had a unique combination of coding single nucleotide variants and copy number alterations in addition to an FGFR fusion, the strength of the parallel approach applied by Wu and colleagues allowed them to perceive the forest from the trees, and hypothesize that commonalities in FGFR fusion proteins were driving cholangiocarcinomas, breast cancers, and prostate cancers alike.
After identifying these index cases, the authors queried other databases that carried the depth of transcriptome sequencing needed to identify fusion genes. By examining an institutional cohort as well as The Cancer Genome Atlas (TCGA), they identified 24 total patient samples and cell lines with fusion proteins involving FGFR1, FGFR2, and FGFR3, all of which retained an intact kinase domain. These FGFR fusions join previously identified driver FGFR fusions in glioblastoma multiforme (5), bladder cancer (6), and myeloproliferative neoplasms (7). Again highlighting the importance of recognizing patterns in the genetic diversity of cancer, Wu and colleagues theorized a commonality in FGFR 5′ fusion partners, and demonstrated the enhanced dimerization of three FGFR fusions compared to wild type FGFR2 or FGFR3 alone. A proposed mechanism for the effects of these chimeric proteins, then, is enhanced dimerization with ligand-independent activation of the FGFR kinase domain. Functionally, the authors demonstrated enhanced growth of 293T cells and immortalized human mammary epithelial cells upon overexpression of different FGFR2- and FGFR3-fusion proteins.
These discoveries are important for the field of cancer biology because they increase the catalogue of driver oncoproteins, and timely given that several pharmacological inhibitors of FGFR family members are in clinical development. Based on the identification of activating mutations in FGFR3 in bladder and cervical cancer, FGFR4 in rhabdomyosarcoma, and FGFR2 in endometrial cancer, and amplification of FGFR1 in non-small cell lung cancer (NSCLC) (all reviewed in (8)), a number of trials are underway in advanced solid tumor patients testing kinase inhibitors with activity against FGFR proteins. This class of agents includes drugs with relative specificity for FGFR, such as PD173074 and the more recently developed AZD4547 (AstraZeneca) and BGJ398 (Novartis), as well as multi-kinase inhibitors such as pazopanib that block FGFR in addition to VEGFR, PDGFR, and other kinases.
For the purposes of their biochemical studies, Wu and colleagues tested the ability of PD173074 and pazopanib to slow the growth of the FGFR3-BAIAP2L1 positive bladder cancer line SW780 in comparison to cell lines carrying activating point mutations in FGFR3. Both drugs inhibited SW780 growth in a dose-dependent manner in tissue culture and murine explants. Intriguingly, neither agent had strong activity in cell lines carrying activating point mutations in FGFR3. Similarly, siRNA knockdown of FGFR3 had significantly more marked effects in fusion-gene positive cells compared to those with activating point mutations. The authors acknowledge that it is too early to say whether this is a general distinguishing feature between FGFR fusions and point mutations, but it is possible that the FGFR fusions identified by Wu and colleagues may be enriched for oncogenic drivers when compared to the landscape of all alterations in FGFR sequence and expression. The exciting finding of FGFR fusions in multiple cancer types, all with potential sensitivity to FGFR inhibitors, mirrors the broad therapeutic potential of ALK inhibitors in tumors driven by different ALK fusions, including NSCLC, anaplastic large cell lymphoma, and inflammatory myofibroblastic tumor (2).
These findings hold great promise for the treatment of patients with FGFR fusions, though there are several caveats to consider. First, it is important to remember that the cures seen in patients with CML or APL remain the exception despite the identification of many other targetable oncogenes. An open question is when and how FGFR fusion positive tumors could become resistant to FGFR inhibitor treatment. If we take crizotinib in the treatment of EML4-ALK positive NSCLC patients as an example, one notes a multitude of partial responses but few durable, complete responses to ALK-directed therapy (3). Substantial effort has been made to explain why oncoprotein-targeted therapy eventually fails in these patients, and initial evidence suggests a diversity of escape mechanisms (2). It is tempting to speculate that this diversity in fact represents a diversity in individual EML4-ALK cancers, such that the degree of oncogene dependence is not predetermined by the presence of the fusion gene but instead by the specific co-occurring genetic alterations in a given tumor cell. Along these lines, inhibition of FGFR4 in rhabdomyosarcoma has been shown to elicit either apoptosis or growth-arrest in alveolar and embryonal rhabdomyosarcoma histologies, respectively, despite the fact that identical FGFR mutations occur across both histologies (9, 10). The data presented by Wu and colleagues shed new light on the importance of considering not only the identify of the altered gene but also cellular and genetic variation in the tumor when implementing therapy targeting a putative oncoprotein driver and investigating how a tumor escapes from oncoprotein inhibition.
A second consideration is that the comprehensive and exciting studies outlined in this study unfortunately had a limited impact on the outcomes of the index patients described. Two patients died of progressive disease soon after enrollment, one before sequencing results were available; another patient had CNS disease, making him ineligible for FGFR-directed therapy. One cholangiocarcinoma patient who had no response to conventional chemotherapy has been enrolled on an FGFR inhibitor trial. These results should not, however, be a source of discouragement. The advancement of precision medicine will require early efforts to help pinpoint true driver alterations, followed by refinement to help us understand the contexts in which tailored therapy will be most effective when compared to conventional treatment options. The continued development of clinical sequencing pipelines such as that described by the authors as well as a knowledge network of functional annotation of putative driver alterations will help propel the field forward towards more rapid and powerful clinical implementation of genetically-informed treatments, that should lead to durable therapeutic responses and even cures in patients.
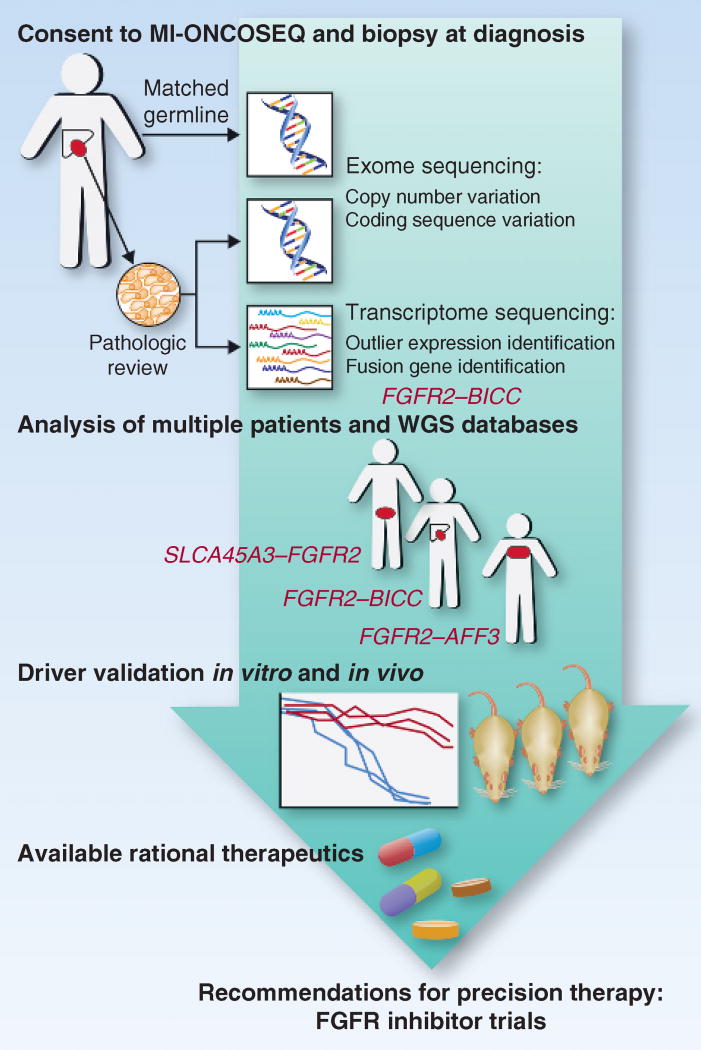
Figure 1.
A pipeline for translating clinical deep sequencing efforts into personalized recommendations for therapy. Patients with advanced cancers are enrolled onto the MI-ONCOSEQ protocol at diagnosis. Core biopsies are taken from accessible sites at presentation for extraction of tumor DNA and RNA after pathologic review of frozen sections confirms a high percentage of malignant cells. Matched normal DNA is obtained from a buccal swab or peripheral blood, allowing for whole-exome sequencing to identify copy number alterations or coding sequence variants. Paired-end sequencing of RNA (RNAseq) allows for identification of genes with abnormally high expression when compared with other patients or sites as well as fusion transcripts. The latter analysis uncovered an FGFR2-BICC fusion in the index patient described. FGFR fusions were also found in 3 other patients enrolled on this protocol, with further fusions identified after querying available RNAseq databases. Genetic and pharmacologic inhibition of FGFR in cell lines with FGFR fusions confirmed their central role in oncogenic transformation, allowing for a rational recommendation of FGFR-targeted therapy. An index patient was successfully enrolled on an early-phase trial of an FGFR inhibitor based on these efforts, showing the promise of the use of clinical deep-sequencing platforms to direct the mechanism-based management of patients with cancer. WGS, whole-genome sequencing.
Acknowledgments
The authors acknowledge funding support (to T.G.B) from the following sources: NIH Director’s New Innovator Award (DP2CA174497), National Cancer Institute (K08CA154787, R01CA169338), Howard Hughes Medical Institute (no number), Doris Duke Charitable Foundation (no number), American Lung Association (no number), National Lung Cancer Partnership (no number), Sidney Kimmel Foundation for Cancer Research (no number), Searle Scholars Program (no number). T.G.B. is a consultant and advisory board member of the Cancer Therapeutics Innovation Group. The other author has no disclosures to report.
References
- 1.Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 2.Shaw AT, Engelman JA. ALK in lung cancer: past, present, and future. J Clin Oncol. 2013;31:1105–11. doi: 10.1200/JCO.2012.44.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, et al. Identification of Targetable FGFR Gene Fusions in Diverse Cancers. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–5. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22:795–803. doi: 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao S, Nalabolu SR, Aster JC, Ma J, Abruzzo L, Jaffe ES, et al. FGFR1 is fused with a novel zinc-finger gene, ZNF198, in the t(8;13) leukaemia/lymphoma syndrome. Nat Genet. 1998;18:84–7. doi: 10.1038/ng0198-84. [DOI] [PubMed] [Google Scholar]
- 8.Dieci MV, Arnedos M, Andre F, Soria JC. Fibroblast growth factor receptor inhibitors as a cancer treatment: from a biologic rationale to medical perspectives. Cancer Discov. 2013;3:264–79. doi: 10.1158/2159-8290.CD-12-0362. [DOI] [PubMed] [Google Scholar]
- 9.Taylor JGt, Cheuk AT, Tsang PS, Chung JY, Song YK, Desai K, et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest. 2009;119:3395–407. doi: 10.1172/JCI39703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crose LE, Etheridge KT, Chen C, Belyea B, Talbot LJ, Bentley RC, et al. FGFR4 blockade exerts distinct antitumorigenic effects in human embryonal versus alveolar rhabdomyosarcoma. Clin Cancer Res. 2012;18:3780–90. doi: 10.1158/1078-0432.CCR-10-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]