Abstract
Nearly all genes presently mapped to chicken chromosome 16 (GGA 16) have either a demonstrated role in immune responses or are considered to serve in immunity by reason of sequence homology with immune system genes defined in other species. The genes are best described in regional units. Among these, the best known is the polymorphic major histocompatibility complex-B (MHC-B) region containing genes for classical peptide antigen presentation. Nearby MHC-B is a small region containing two CD1 genes, which encode molecules known to bind lipid antigens and which will likely be found in chickens to present lipids to specialized T cells, as occurs with CD1 molecules in other species. Another region is the MHC-Y region, separated from MHC-B by an intervening region of tandem repeats. Like MHC-B, MHC-Y is polymorphic. It contains specialized class I and class II genes and c-type lectin-like genes. Yet another region, separated from MHC-Y by the single nucleolar organizing region (NOR) in the chicken genome, contains olfactory receptor genes and scavenger receptor genes, which are also thought to contribute to immunity. The structure, distribution, linkages and patterns of polymorphism in these regions, suggest GGA 16 evolves as a microchromosome devoted to immune defense. Many GGA 16 genes are polymorphic and polygenic. At the moment most disease associations are at the haplotype level. Roles of individual MHC genes in disease resistance are documented in only a very few instances. Provided suitable experimental stocks persist, the availability of increasingly detailed maps of GGA 16 genes combined with new means for detecting genetic variability will lead to investigations defining the contributions of individual loci and more applications for immunogenetics in breeding healthy poultry.
Keywords: Chicken, GGA 16, MHC, gene map, genetics of resistance to infectious disease
INTRODUCTION
The major histocompatibility complex (MHC) is a gene region possessed by all higher vertebrate species. Many of the genes within the MHC contribute to immunity. MHC encoded class I and class II molecules are central in defining the specificity of adaptive immune responses. MHC class I and class II molecules provide a context in which pathogens are recognized. Presentation by MHC molecules is essential for developing vaccinal immunity. MHC class I and class II molecules are typically highly polymorphic and polygenic. Other MHC genes, some of which are also polymorphic and polygenic, contribute to immunity in additional ways. The chicken MHC is especially interesting because polymorphic MHC class I and class II genes are localized into two regions (MHC-B and MHC-Y) on the same chromosome. The MHC-B and MHC-Y haplotypes assort independently as the result of an intervening region that supports highly frequent recombination. The chicken MHC is of interest since particular MHC-B haplotypes and, in a few instances, alleles at particular MHC-B loci are known to exert major genetic effects in the incidence of infectious diseases, such as Marek's disease (MD). Genetic lines of chickens selected for different MHC-B haplotypes have provided abundant evidence for the major contribution of the MHC to disease resistance. A detailed gene map exists for MHC-B in the red Jungle fowl reference genome in which two class I genes, two class IIβ genes and associated genes are located (Shiina et al., 2007). Additional sequence data is now available for the large family of BG genes (Salomonsen et al., 2014) and two nearby MHC class I-like genes, the CD1 genes (Maruoka et al., 2005; Miller et al., 2005; Salomonsen et al., 2014; Salomonsen et al., 2005; Shiina et al., 2007). While not yet fully sequenced, additional sections containing MHC-Y genes provide evidence for a number of specialized MHC class I, MHC class II, c-type lectin-like, and linked olfactory receptors (OR), and scavenger receptor (SRCR) genes (Miller et al., 2014; Rogers et al., 2003). A nomenclature guide provides the naming conventions for MHC-B and MHC-Y haplotypes and genes (Miller et al., 2004).
Not all investigators agree on where to draw the boundaries between genes within the major histocompatibility complex and those in adjacent regions. This is not surprising since the region is organized in diverse ways in different species. In addition, the original name – major histocompatibility complex – reflects an experimental outcome in a pre-genomic era. Now, with genomics, it is possible to have an overview that reveals far more about the genes within the region. In this review an inclusive definition has been adopted to encompass the different types of genes commonly found in the MHC genomic region. Most of these contribute in some way to immunity. The majority of the genes mapped to GGA 16 either have a demonstrated role typical of the MHC complex or are considered likely to have a role in immunity. The genes mapped in chicken to GGA 16 support consideration of the MHC as a region where a variety of co-functioning and co-evolving genes are concentrated together.
DISTRIBUTION OF GENES ON CHICKEN CHROMOSOME 16 (GGA 16)
GGA16 is one of the larger microchromosomes in the chicken karyotype. Genes have been mapped to the q-arm with no genes as yet assign to the p-arm (Figure 1) (Delany et al., 2009; Miller et al., 2014). In the current map, the first genes distal to the centromere on GGA 16 are multiple copies of OR and cysteine-rich domain SRCR genes. These were identified as likely mapping to GGA 16 by trisomy mapping(TM)/comparative genomic hybridization (aCGH) and then located by FISH (Miller et al., 2014). Distal to the OR and SRCR genes is the single NOR (nucleolar organizing region) in the chicken genome. This is the chromosomal region around which the nucleolus forms. The NOR occupies a large portion (5–7 Mb) of the q-arm and contains repetitive sets (∼150 copies) of 18S, 5.8S, and 28S ribosomal RNA genes. Located just distal to the NOR is the MHC-Y region.
Figure 1.
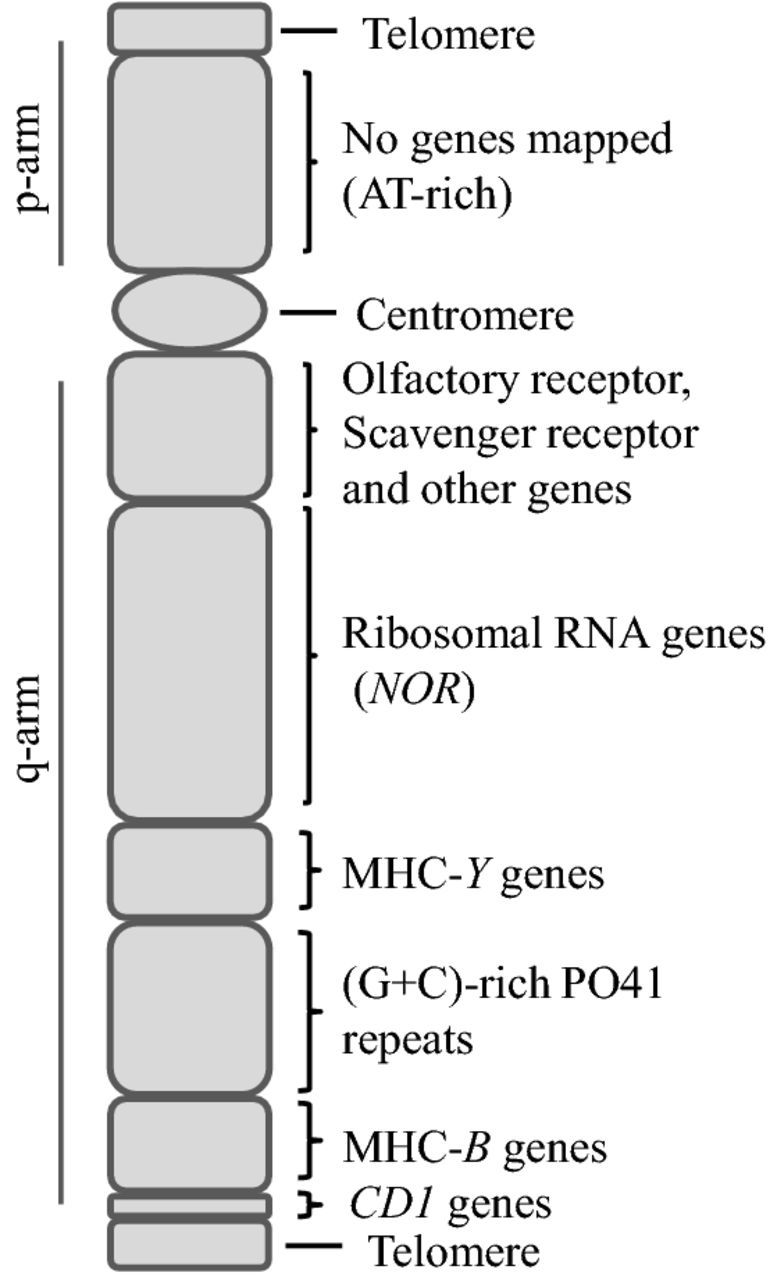
MHC-B and MHC-Y are located on the q-arm of chicken chromosome 16 (GGA 16) distal to the NOR and are separated by PO41 repeats. MHC-Y displays linkage with the olfactory receptor and scavenger receptor genes. MHC-B haplotypes assort independently from MHC-Y haplotypes. Two CD1 class I genes are located distal to MHC-B. Adapted from Miller et al. 2014.
Distal to MHC-Y is a long array of (G+C)-rich pattern of 41 nucleotides (PO41) tandem repeats (Solinhac et al., 2010). The chiasma observed in GGA 16 lampbrush chromosomes occur in the PO41 region suggesting that these tandem repeats are the source of the independent assortment of MHC-Y haplotypes from MHC-B haplotypes (Solinhac et al., 2010). The MHC-B region distal to the PO41 repeats has been studied extensively for its role in antigen presentation and disease resistance. Distal to MHC-B are additional genes including two members of the ancient CD1 gene family encoding specialized antigen presenting molecules.
Additional genes mapping to GGA 16 but not yet located at high resolution include BLA, encoding a class IIα chain (Kaufman et al., 1995a; Salomonsen et al., 2003); G9 (BAT8) encoding a methyl transferase mapping to the complement gene region in other species (Spike et al., 1996) and a fB gene, a gene for complement factor B (Kaufman et al., 1999). An MHC class II restriction fragment polymorphism has provided evidence of an additional class II gene that is not located (Juul-Madsen et al., 2000b). Additional candidates identified in the TM/CGH study as candidates for mapping to GGA 16 include members of the immunoglobulin gene superfamily that may also be contributing to immunity (Miller et al., 2014). Further sequencing and cytogenetic mapping studies are needed to confirm these assignments and to locate these genes.
OLFACTORY AND SCAVENGER RECEPTOR GENES
Eleven chrUn_random RefSeq sequences containing OR and SRCR genes were assigned to GGA 16 by TM/aCGH and mapped to the proximal region of the GGA 16 q-arm by FISH (Miller et al., 2014). Segregation of SRCR gene restriction fragments in analyses of fully pedigreed families has provided evidence that the region is polymorphic and that OR/SRCR genes segregate in linkage with MHC-Y region genes (Miller et al., 2014). Multiple types of OR genes are present in the OR/SRCR RefSeq sequences including member of the OR14J1 family. Interestingly, OR14J1 genes are also found in the extended region of the human MHC (Ehlers et al., 2000) so with this finding in the chicken genome there is evidence for a long-term association of OR14J1 genes with the MHC.
Nine SRCR genes were contained in the sequences assigned to GGA 16 in the TM/CGH study (Miller et al., 2014). The genes are easy to identify because they contain multiple sequences for the highly conserved and ancient scavenger receptor cysteine-rich domain (Herzig et al., 2010; Miller et al., 2014). In sequence alignments, the chicken SRCR genes are most similar to WC1/CD163 genes. Recent data suggest that cattle WC1 molecules are pattern recognition co-receptors that guide responses of γδ T cells to bacteria (Hsu et al., 2015; Telfer and Baldwin, 2015). The scavenger receptor CD163, a “jack-of-all-trades” molecule with restricted expression on the surfaces of monocytes and macrophages, is apparently multifunctional, mediating endocytosis of a variety of ligands (including hemoglobin, bacteria, and viruses), serving as an adhesion receptor (on erythroblasts) and as an immunomodulator operating under immunosuppressive conditions (Van Gorp et al., 2010). The chicken SRCR genes are candidates for serving in immunity and fit well within the inclusive definition of the MHC used in this review.
MHC-Y REGION GENES
The MHC-Y region, originally named Rfp-Y, was discovered when chicken MHC-B class I and class II gene probes revealed DNA restriction fragments from a fully pedigreed family that were inconsistent with the assigned MHC-B haplotypes (Briles et al., 1993). Soon after this discovery, two MHC class I, two MHC class II genes and a c-type lectin-like gene originally assigned to MHC-B (Guillemot and Auffray, 1989) were reassigned to MHC-Y and localized to GGA 16 (Miller et al., 1994; Miller et al., 1996). Assembly of the full sequence of the MHC-Y region is particularly challenging, but work is progressing (Miller, unpublished data). The published sequence data provides evidence for two c-type lectin-like genes, a full class I gene, a truncated class IIβ gene and CR1 repeat within the region (Rogers et al., 2003).
MHC-Y haplotypes are assigned on the basis of restriction fragment polymorphisms. Initially, these haplotypes were identified with MHC-B probes that cross-hybridize with MHC-Y class I and class II gene sequences (Briles et al., 1993; Miller et al., 1994; Miller et al., 1996; Pharr et al., 1997). MHC-Y specific probes include probe 18.1, from an intergenic region in MHC-Y (Guillemot and Auffray, 1989; Juul-Madsen et al., 1997) and 163/164f, a probe from a MHC-Y-specific region of the YF1*7.1 gene (Afanassieff et al., 2001).
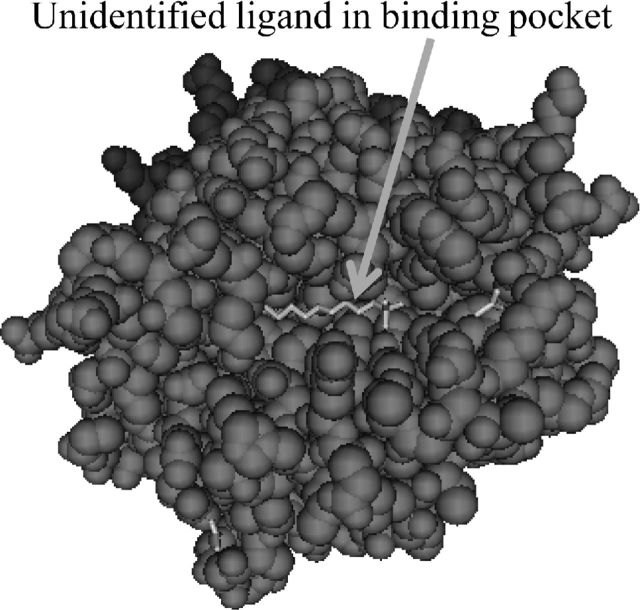
Early data demonstrated that MHC-Y antigens serve as weak alloantigens (Pharr et al., 1996). Similar results were reported in a study of three additional MHC-Y haplotypes (Thoraval et al., 2003). Other investigators (Afanassieff et al., 2001; Hunt et al., 2006) working with highly inbred and closed lines have shown that at least one MHC-Y class I genes is polymorphic and dynamically expressed. At least one MHC-Y class I (YF) molecule is alloimmunogenic (Hunt et al., 2006). Initial evidence of MHC-Y class I antigen binding groove specialization (Afanassieff et al., 2001; Afanassieff et al. 2000) was confirmed by structural studies (Figure 2) of YF1*7.1 (Hee et al., 2010; Hee et al., 2009) showing non-peptidic ligands logged within its hydrophobic binding groove. The structure of YF1*7.1 defined a new MHC class I paradigm; a molecule with the structure of a classical MHC class I molecule, but binding “non-classical” non-peptide ligands. A more recent structural study has revealed human MR1 (MHC class I (MHC-I)-like related molecule) as a second member of this group (Kjer-Nielsen et al., 2012). MR1 has a classical structure, but presents B vitamin metabolites of microbial origin. These findings suggest that MHC-Y class I molecules may contribute in guiding immune responses of a specialized nature.
Figure 2.
Structure of YF1*7.1 showing surfactant bound within the ligand binding cleft. From NCBI PBD ID: 3P73.
As a note of explanation, YF1*7.1 is named following the recommended chicken MHC nomenclature (Miller et al., 2004). Following standard conventions gene names are italicized. Briefly, “Y” designates the gene region. In chickens “F” is used to identify a chicken class I gene. The “1” is to distinguish the first F locus in the haplotype from additional class I loci in the haplotype, e.g., the YF7 haplotype has two class I loci, YF1 and YF2. An asterisk is used to separate locus from haplotype. “7.1” is the haplotype found in Reaseheath Line C and its derivatives. Because MHC-Y haplotypes are only partially defined, decimal designations (“7.1”, “7.2”, and so on) are used to distinguish apparently identical haplotypes originating from different genetic stock.
In early analyses of chicken MHC genes, six BLβ genes were described in the Line CB genome (Zoorob et al., 1990; Zoorob et al., 1993). Following MHC-Y identification and mapping, three of these genes, originally named BLβIII, BLβIV, and BLβV, were reassigned to MHC-Y and renamed YLB1, YLB2 and YLB3 (Miller et al., 2004; Miller et al., 1994; Miller et al., 1996). These three MHC-Y loci share sequence similarities and more limited polymorphism that set them apart from the MHC-B class IIβ loci (Zoorob et al., 1993).
The c-type lectin-like genes mapping to MHC-Y were identified in the first chicken MHC map as part of MHC-B (Bernot et al., 1994). Bernot and colleagues identified a locus (17.5) on cosmid clone cβ17 as a lectin-like gene for which a cDNA clone (17.5.3) was isolated. Southern blots with 17.5.3 provided evidence for a family of polymorphic genes The cβ17 clone was later assigned to MHC-Y (Miller et al., 1994). Two additional C-type lectin-like genes are currently mapped to MHC-Y (Rogers et al., 2003). Now identified as Ylec genes, these genes are similar to the mammalian genes encoding C-type lectin-like receptors that guide mammalian natural killer cell responses (Iizuka et al., 2003).
THE MHC-B REGION GENES
Serological and genetic data reported by Briles in 1950 provided the first evidence for the presence of MHC-B as an alloantigen system displayed on the surfaces of red blood cells (Briles et al., 1950). Gilmour's independent discovery nine years later confirmed the presence of this highly polymorphic system (Gilmour, 1959). Schierman and Nordskog (Schierman and Nordskog, 1961) linked the B alloantigen system to the chicken major histocompatibility complex by demonstrating that skin grafts were rejected more rapidly if donor and recipient were not matched for the B system compared with other blood alloantigens. Later, recombinant MHC-B haplotypes enabled Pink and colleagues (Pink et al., 1977) to define three classes of MHC-B antigens each with a different tissue distribution indicating the presence of at least three MHC loci. Further work showed that immune cell cooperation was restricted by MHC-B antigens (Maccubbin and Schierman, 1986; Vainio et al., 1984; Weinstock et al., 1989).
The first gene map for the B12 haplotype encompassed multiple class I and class II genes, additional loci and a portion of the NOR in four cosmid clone clusters (Guillemot et al., 1989). Following the discovery of MHC-Y (Briles et al., 1993; Miller et al., 1988), three of the four clusters, including the one containing rDNA genes, were reassigned to MHC-Y leaving MHC-B represented by a single cluster of genes (Miller et al., 1994). When sequenced, this cosmid revealed 19 genes within a 92 kb region including two class Iα genes, two class IIβ genes and two c-type lectin-like receptor genes (Kaufman et al., 1999). Subsequently, by sequencing B12 BAC clones, six TRIM genes and a guanine nucleotide-binding gene were located nearby (Ruby et al., 2005; Zoorob et al., 1996).
The Current MHC-B Gene Map
The foundation for the current Red Junglefowl (RJF) MHC-B region gene map (Figure 3) is the full sequence determinations made for three overlapping RJF BAC clones encompassing 242 kb (Shiina et al., 2007). The sequence revealed 46 genes within 242 kb. This extended the original MHC-B map from made from the B12 cosmid clone, adding one additional (pseudo) gene within the original map and fully organizing 21 additional genes upstream and five additional genes downstream of the B12 92 kb map. The recent publication of the B12 BG haplotype (Salomonsen et al., 2014) has made a significant addition to the MHC-B map. Twelve BG genes, identified in the B12 haplotype sequence, were found distributed in two tandem repeats upstream of the TRIM and Blec sub-region. These data were used to tentatively order the BG haplotype in the RJF whole genome shotgun (WGS) project. At least 15 RJF BG genes (apparently lacking the insert of dissimilar genes that disrupts the continuity of BG genes in B12 BG haplotype) are thought to be located upstream of BG3 (noted by a cluster of boxes at the top of Figure 3). The number of BG genes apparently differs among MHC-B haplotypes providing evidence for gene copy number variation within the chicken MHC.
Figure 3.
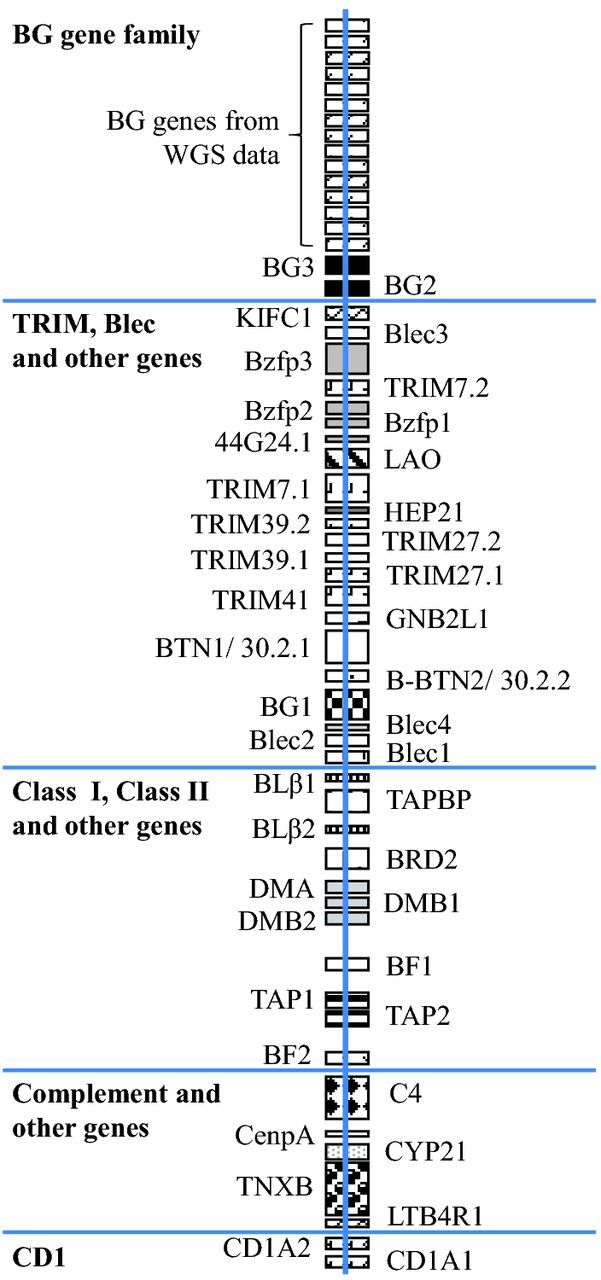
Gene Map for the red Jungle fowl MHC-B haplotype. Adapted from Shiina et al. 2007 and Salomonsen et al. 2014.
BG Gene Family Sub-Region
Almost all the genes within the BG family are clustered in tandem repeats in which indels, as well as simpler polymorphisms, distinguish haplotypes (Salomonsen et al., 2014). Sequence comparisons indicate that most BG genes are hybrid genes suggesting that recombination contributes to diversity among haplotypes. Only two genes fall outside the repeat cluster. One, BG0 is located away from the MHC on chromosome GGA 2. The other, BG1 is positioned more than 115 kb away from the cluster near the MHC class I and class II loci (Kaufman et al., 1999; Salomonsen et al., 2014; Shiina et al., 2007). All BG genes are apparently polymorphic. How they contribute to immunity remains to be revealed. Presently only BG1 provides a link with immune responses (Goto et al., 2009). Some BG proteins may participate in interactions with the cytoskeleton in the epithelial brush border (Bikle et al., 1996).
All BG molecules identified so far contain three structural elements. These are a single extracellular immunoglobulin-like domain of the V-region type bearing an “extra” cysteine for intermolecular disulfide linkage, a Type 1 transmembrane domain and a cytoplasmic tail composed of small domains (usually seven amino acids) predicted to form α-helical structures that pair into coiled coils (Kaufman et al., 1990; Miller et al., 1991). Diverse BG family members encoded by different loci and in distinct haplotypes maintain the three structural features but vary conspicuously in amino acid sequence and in the length of cytoplasmic tail even among isoforms encoded by the same locus (Hosomichi et al., 2008).
Specificity is apparent in the expression pattern of individual BG family members (Miller et al., 1991; Miller et al., 1990; Salomonsen et al., 2014; Salomonsen et al., 1991). Some members are especially well expressed on erythrocytes, where they are recognized as alloantigens that allow the B system haplotypes to be distinguished serologically. BG proteins are also especially well-expressed on epithelial cells, particularly those in the duodenum, caecum, and liver (Miller et al., 1990). They are also express on many types of cells important in immunity (Salomonsen et al., 1991). RT-PCR provides evidence for expression on T cells, B cells, thrombocytes, dendritic cells and other tissues (Salomonsen et al., 2014). Cell and tissue expression appears precise with pattern of expression correlating with two different types of promoters and 5′-untranslated regions (Salomonsen et al., 2014).
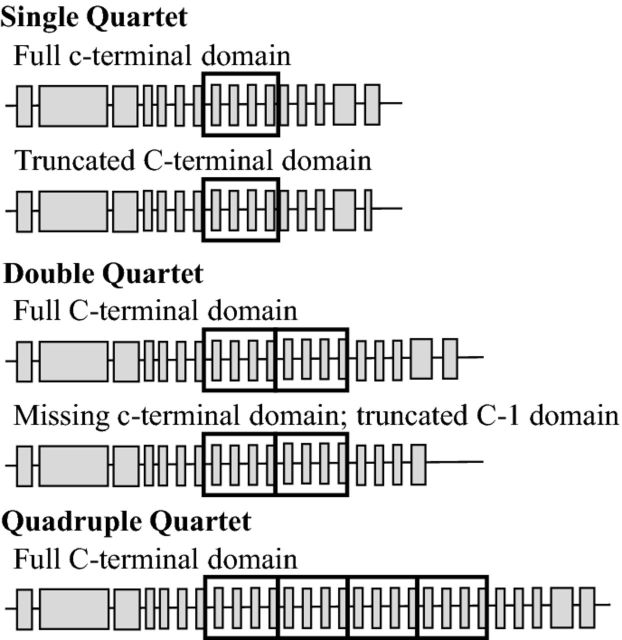
BG1, the isolated member of the family near the class I and class II genes, is one MHC-B locus for which a role in disease resistance has been defined (see Marek's disease section below). BG1 isoforms encoded within different standard MHC-B haplotypes display three types of variability. In addition to sequence polymorphisms, alleles differ structurally in the number of the exons encoding the BG cytoplasmic heptad repeats. BG1 alleles variously carry one, two and four copies of a quartet of exons and their associated introns (Figure 4). The significance of the variation in the coiled-coil region is not known. BG1 alleles also vary in the number and size of exons encoding the c-terminal domain, which is apparently present only in BG1 molecules (Figure 4). As a result an immunoreceptor tyrosine-based inhibition (ITIM) motif is present in some, but not all BG1 isoforms (Goto et al., 2009; Hosomichi et al., 2008).
Figure 4.
BG1 alleles have been observed to differ in the total number of exons. This occurs through duplication/deletions of a set (quartet) of four exons. Further variation occurs at the 3′-end where exons vary in size and number so that the immunoreceptor tyrosine-based inhibition motif is present in some isoforms but absent in others. Adapted from Hosomichi et al. 2008.
TRIM/Blec Sub-Region
There are 25 genes within the TRIM/Blec sub-region in addition to BG1, already described above. The nature of these genes is summarized in Table 1. Fourteen of the 25 are assessed as “expressed genes” because their sequences match cDNA and EST sequences (Shiina et al., 2007). Seven additional genes show structural integrity, have mRNA sequence accession numbers), but are not well characterized as to ORF. Nearly all the genes in this region are thought to contribute in some way in immune defense based on homology to counterparts in other species. Their functions are thought to be in innate immunity with the TRIM genes providing defense against viral infections. Among the Blec genes, Blec2 is considered likely to be a natural killer cell receptor perhaps working in concert with ligands encoded in the nearly MHC-B class I genes. Interestingly, the Blec2 locus, also named B-NK, is closely related to mammalian lectin-like natural killer cell receptors (Kaufman et al., 1999; Rogers et al., 2005; Rogers and Kaufman, 2008). Blec2 displays a relatively high level of diversity, bears a functional inhibitory motif and is well expressed in lymphoid tissues (Rogers et al., 2005; Rogers and Kaufman, 2008). In a comparison of sequences among 14 MHC-B haplotypes, Blec2 has a nonsynonymous (dN)/synonymous (dS) substitution ratio of 7.0698 for SNPs within amino acid coding sequences (Hosomichi et al., 2008). This is significantly different from the expectation of sequence diversity occurring by chance (dN/dS = 1), suggesting Blec2 is under positive selection. In contrast, Blec1 is less polymorphic, homologous to activation-induced lymphocyte receptors, found to be well expressed only in cecal tonsils, and is quickly up-regulated following cell activation (Rogers et al., 2005; Rogers and Kaufman, 2008).
Table 1.
TRIM/Blec sub-region genes within the MHC-B region. The genes are classified as: expressed if it is transcribed to mRNA and also has a reliable open reading frame and/or a known protein product; candidate if it is transcribed to mRNA (these have mRNA sequence accession numbers), but for which an ORF is uncertain or unknown, and pseudogene if partial gene sequences are present. Adapted from Shiina et al. 2007.
| Gene type | Gene copies | Names | Status | Conserved Domains/motifs | Immune Defense | Known or imputed function |
|---|---|---|---|---|---|---|
| Tripartite motif | 7 | TRIM7.1, TRIM7.2, TRIM27.1, TRIM27.2, TRIM 39.1, TRIM39.2, TRIM41 | Expressed/ Candidate | RING finger, B-box, coiled coil motifs, and PRY-SPRY (B30.2) | Likely | Possibly contributing resistance to pathogens |
| C-type lectin-like | 4 | Blec1, Blec2, Blec3, Blec4 (ps) | Expressed/ Candidate/ Pseudogene | C-type lectin-like genes similar to those guiding NK cell responses | Likely | Likely candidates for affecting early immune responses |
| Zinc finger protein | 3 | Bzfp1, Bzfp2, Bzfp3 | Expressed/Candidate | Zinc finger protein | Likely interaction modules binding nucleic acids, DNA, RNA, proteins, or other small molecules, | |
| transfer RNA | 4 | tRNA-Lys.1, tRNA-Lys.2, tRNA-Val, tRNA-Leu | tRNA | RNA cloverleaf structure | An RNA adaptor molecule physically linking the sequence of nucleic acids and the amino acid sequence of proteins | |
| B30.2 | 2 | B30.1and B30.2; B30.2-1 B30.2-2 | Expressed/Candidate | PRY-SPRY (B30.2) | Likely | Likely helping to regulate innate and adaptive immunity |
| Kinesin | 1 | KIFC1 (Kinesin Family Member C1) | Expressed | C-terminal motor, super-helical stalk, N-terminal tail | Candidate for sliding/cross-linking microtubules | |
| 44G24 | 1 | 44G24 | Candidate | Possibly DAXX | Function unknown. DAXX domain is reported to be associated with apoptosis | |
| L-amino acid oxidase | 1 | LAO | Expressed | LAAO | Perhaps | A flavoenzyme which catalyzes oxidative deamination of L-amino acids and could contribute in immunity by inducing apoptosis |
| HEP21 | 1 | HEP21 | Expressed | uPAR/CD59/Ly-6/ snake neurotoxin superfamily | Likely | Predominantly expressed in oviduct |
| Guanine nucleotide binding | 1 | GNB2L | Expressed | G protein | Perhaps | A G proteins transduces signals from surface receptors; may help control lymphocyte activation and proliferation |
| BG | 1 | BG1 | Expressed | IgV-like, transmembrane coiled-coil, ITIM motif | Likely | Likely cell activation/inhibition |
MHC Class I and Class II Gene Sub-Region
This core of the chicken MHC-B region contains genes involved in classical peptide antigen presentation. Within this sub-region are two MHC class I genes, two class IIβ genes for antigen presentation and genes for processing and loading antigen. Excellent early work greatly advanced the understanding of the chicken MHC-B class I and class II genes (Behar et al., 1988; Bourlet et al., 1988; Frangoulis et al., 1999; Guillemot et al., 1988; Guillemot et al., 1986; Kroemer et al., 1990; Zoorob et al., 1990; Zoorob et al., 1993). These studies provided both the basis for the first reported genomic sequence (B12) (Kaufman et al., 1999) as well as a bridge that sped the recognition of the MHC-Y gene region's complexity (Miller et al., 1994; Miller et al., 1996).
Both MHC class I genes, BF1 and BF2 encode classical MHC class I molecules (Kaufman et al., 1999; Koch et al., 2007; Sherman et al., 2008). Although fundamentally similar, the two loci are apparently functionally specialized. BF2 serves in antigen presentation whereas BF1 has apparently primarily another function. Several studies provide evidence that less BF1 mRNA is typically present in cells compared to BF2 mRNA (Livant et al., 2001; O'Neill et al., 2009; Shaw et al., 2007; Wallny et al., 2006). While all studies report that BF1 is less well expressed, the degree to which lower expression occurs may depend upon the tissue examined. Some ratios reported are in the range of ∼1:1.5 to 1:6 (Juul-Madsen et al., 2000a; O'Neill et al., 2009), while others are greater (Juul-Madsen et al., 2000a; Shaw et al., 2007). The significance of these differences in the level of expression is unknown. Promoter sequences differ between BF1 and BF2. Various deletions, insertions and rearrangements result in sequence divergence that likely affects the efficiency of expression and in some instances, the production of pseudogenes (Shaw et al., 2007). In addition, among the BF1 sequences themselves, two strongly separated lineages with differences in Enhancer A and promoter region sequences have been identified (O'Neill et al., 2009). These are interpreted as evidence that BF1 genes are under selection for variation.
BF1 encoded molecules may be ligands in NK cell interactions. A careful study of BF1 allele sequences (Ewald and Livant, 2004; Livant et al., 2004) revealed a conserved locus-specific motif that resembles the motif on human MHC HLA-C molecules recognized in natural killer cell interactions. This motif is conserved among BF1 alleles. In contrast, the corresponding region in BF2 alleles is highly variable. Ewald and Livant (2004) hypothesize that BF1 molecules serve primarily in innate immunity. In favor of this hypothesis is suppression of natural cell killing of target cells displaying BF1 on their cell surface compared to controls (Zhang et al., 2012b).
Considerable progress was made in defining the structure of the BF2 gene and demonstrating its function prior to the realization that this was the single gene in chickens for classical peptide antigen presentation (Fulton et al., 1995; Hunt et al., 1994; Kaufman et al., 1992; Kroemer et al., 1990; Pharr et al., 1994; Thacker et al., 1995). More recently, crystal structures and sequences of eluted peptides provide insights into the specificity of peptide binding in BF2 (Chappell et al., 2015; Koch et al., 2007; Sherman et al., 2008; Wallny et al., 2006). Because BF2 is the only classical class I isoform for presenting peptide antigens in chickens, the chicken provides a useful experimental model for investigating the specificity of antigen processing, binding and presentation and subsequent immune responses. A good example for this is the evidence that BF2 is the source of protective immunity against the Rous sarcoma virus challenge (defined as tumor regression over a time course in which an adaptive immune response likely occurs) (Hofmann et al., 2003). Immunization with a single v-src peptide predicted to bind in the BF2*12 binding groove was sufficient to induce a protective immune response. Findings with a vaccine for infectious bursal disease provide similar evidence between the effectiveness of a molecularly defined vaccine and BF2 binding preference (Butter et al., 2013). Further worked has defined CD8+ T-cell epitopes for avian influenza (Hou et al., 2012; Reemers et al., 2012). Alternative forms of transcripts may contribute further specificity to BF2 (and BF1) function (Dalgaard et al., 2003; Dalgaard et al., 2005).
In general, the same molecular interactions that occur in the peptide binding within other classical MHC class I molecules occur in the binding of peptides in the BF2 binding groove. Peptides are tethered through interactions between atoms in the peptide backbone at the amino and carboxyl termini and highly conserved residues at the margins of the ligand-binding groove. These features are confirmed in structural determinations for several BF2 isoforms (Chappell et al., 2015; Koch et al., 2007; Zhang et al., 2012a). As with other classical MHC class I molecules that bind peptide, the exact peptides bound are determined through preferential interactions with residues lining binding pockets arrayed along the length of the BF2 binding groove. Typically numerous different peptides are bound and presented in MHC class I molecules at the surfaces of cells. Although many of these are self peptides that elicit no response because of prior deletion of reactive T cells during development, these various peptides represent the bulk of peptides that elute from the MHC class I binding groove in tests to determine the peptide binding preferences of individual MHC class I molecules.
Peptide binding preferences are now available for BF2 isoforms. Motifs have been derived from sequencing of pooled peptides and from mass spectrometry (ms) identifications of individual peptides (Chappell et al., 2015; Sherman et al., 2008; Wallny et al., 2006). With the ends of the peptide bound to the margins of the groove through the main chain as is typical for MHC class I peptide binding and longer peptides assumed to be arching out of the groove (a structural study confirms this is correct (Chappell et al., 2015)) the binding preferences B2*2101 (based primarily on hydrophobic residues) were readily evident among 34 eluted endogenous peptides that varied in length from 8–12 residues. Motif scanning (http://www.hiv.lanl.gov/content/immunology/motif_scan/motif_scan) revealed that virtually all proteins produced by avian viral pathogens (other pathogens not examined) contain at least one BF2*2101 motif. Sherman and colleagues proposed that BF2*2101 is common in chicken populations because of its general utility (Sherman et al., 2008). It will be interesting to learn whether the general utility of antigen presentation in BF2*2101 is related to the constrained evolution of MDV pathogenicity observed in MHC-B21 chickens (Hunt and Dunn, 2013).
The 30 individual endogenous peptides eluted from BF2*1301 defined a quite different binding preference (Sherman et al., 2008). Most of the eluted peptides contained eight or nine amino acids, suggesting that these bind essentially in a fully extended confirmation, recently confirmed by crystallography of the BF2*0401 isoform, which is identical to BF2*1301 in binding groove sequence (Chappell et al., 2015). This motif is based mostly on acidic residues. When the BF2*1301 motif was evaluated by motif scanning, only a few proteins from a very limited array of viruses contained the BF2*1301 motif suggesting a selective advantage for this allele only in infections with a subset of pathogens (Sherman et al., 2008). The description of BF2*2101 and BF2*1301/BF2*0401 as “generalist” and “specialist”, respectively, in a recent structural study (Chappell et al., 2015) supports the initial observations of BF2 specialization by Sherman and colleagues and extends the characterization to levels of expression at the cell surface.
Also within the MHC class I and class II gene sub-region are two TAP (transporters associated with antigen processing) genes, which encode ATP-binding-cassette transporter molecules that deliver peptides from cell cytoplasm into the endoplasmic reticulum (ER) where they are loaded of MHC class I molecules. Interestingly, a MDV MHC class I immune evasion gene was recently shown to target TAP (Hearn et al., 2015). Also present is TAPBP encoding TAP-associated glycoprotein (also known as TAP-binding protein or tapasin), a protein that mediates interaction of MHC class I molecules with the TAP molecules and facilitates peptide antigen loading. Kaufman has postulated that TAP and TAPBP alleles have co-evolved with the adjacent BF2 alleles to optimize BF2 antigen presentation (Kaufman et al., 1999; Kaufman et al., 1995b; van Hateren et al., 2013; Walker et al., 2011; Walker et al., 2005).
MHC-B haplotypes have two class IIβ genes that can be described as major and minor forms (Jacob et al., 2000; Pharr et al., 1998; Pharr et al., 1993). These genes lie on either side of the tapasin gene. The major gene, always a member of the BLβII family, is always located between tapasin and RING3. The weakly expressed minor gene, located between Blec1 and tapasin, belongs to either the BLβII or the BLβVI family (Jacob et al., 2000; Zoorob et al., 1993). A third MHC-B class IIβ gene appears in some haplotypes, but has not been located (Jacob et al., 2000).
Molecules that assist in MHC class II loading, the DM molecules are class II molecules that remain within intracellular vesicles and have a central role in the loading of MHC-B class II molecules with peptide. The DM, TAP, and TAPBP genes are retained over evolutionary time, presumably co-evolving in their roles in peptide antigen presentation with the nearby MHC class I and class II genes. The remaining gene within this region is BRD2 (RING3), a chromatin binding protein that apparently has no direct role in immunity.
Complement Gene Sub-Region
This sub-region, often referred to as the class III region of the MHC, contains five genes in chickens. Four conserved genes, C4 (complement factor C4 with a role in inflammation), CenpA (centromere protein), CYP21 (steroid 21-hydroxylase) and TNXB (tenascin XB), are present in a highly conserved order in this sub-region. The fifth gene is LTB4R1 and likely encoding a leukotriene receptor with a role in inflammation. It is not located within or near the MHC in mammals, but has been located within the MHC of at least two avian species (Chen et al., 2015; Shiina et al., 2007).
The CD1 Gene Sub-Region
In contrast to mammals the CD1 genes in chickens are located on the same chromosome as the MHC. The two chicken CD1 genes, CD1-1 and CD1-2, are located a short distance away from the two classical MHC-B class I genes (Maruoka et al., 2005; Miller et al., 2005; Salomonsen et al., 2005). CD1 molecules are well studied in mammals and are known to contribute in immune responses against bacteria. Human CD1 molecules present lipids and glycolipids to specialized subsets of T cells that bear T cell receptors with remarkable specificity for the structural details of potential antigens including the capacity to distinguish self and non-self lipid antigens.
Not nearly as much is known about the two chicken CD1 molecules, although both structures have been resolved. The structures confirmed their relatedness to other members of this ancient gene family and revealed distinctive binding pockets not present in other CD1 molecules. The highly unique CD1-2 binding pocket is considered primordial because the binding groove is very small, actually more of a pore, and considered likely able to only accommodate fatty acids, or single alkyl chain lipids with up to 16 carbons (Zajonc et al., 2008). The CD1-1 gene encodes a molecule with a larger binding region of dual-pocket, dual-cleft structural design (Dvir et al., 2010). This molecule is more similar to mammalian CD1 in its capacity to bind large dual-chain lipids.
MHC POLYMORPHISM AND RESPONSES TO INFECTION
MHC-Y in Responses to Infection
The contribution of MHC-Y polymorphism in differential responses to infection is not well understood. Relatively few disease challenge trials have been done looking at how MHC-Y polymorphism might affect the incidence of disease. To date, most experiments that have been done have asked about whether MHC-Y contributes in responses against MD. No major influences have been found. In a Marek's virus challenge trial using progeny from fully pedigreed families in which three MHC-Y and two MHC-B haplotypes were segregating, birds homozygous for one MHC-Y haplotype were at a 2.3 times greater risk for developing tumors compared with all other MHC-Y genotypes combined (P < 0.02). In other experiments with progeny from two sets of paired lines displaying genetic differences in resistance to MD, no statistical significant differences noted between the MHC-Y haplotypes. However, these studies noted that birds with some MHC-Y genotypes tended to have greater longevity within the time course of the trial, lower viremia, and fewer tumors than others (Bacon et al., 1996; Pharr et al., 1996; Vallejo et al., 1997). A fourth trial found no association (Lakshmanan and Lamont, 1998).
The influence of MHC-Y haplotype on the regression of Rous sarcoma virus-induced tumors has also been examined. Since some MHC-B region haplotypes strongly influence RSV tumor growth (see section below), tests were designed in which the MHC-B influence was modest. In one test with B2B5 chickens, the MHC-Y genotype had a significant effect on the pattern of tumor growth, tumor fate and mortality rates (LePage et al., 2000). In another test carried out with birds homozygous for B19 that were selected over 18 generations for their capacity to regress or progress RSV tumors, MHC-Y haplotype was again found to have a significant, but here a more moderate effect on tumor growth (Pinard-van der Laan et al., 2004; Praharaj et al., 2004).
With the additional genome-wide assays that can be used in disease challenge trials, more data will be available defining the relationship between MHC-Y genes and immunity in chickens. It could be that antigen presentation by the MHC-Y class I genes has a primarily role in another arm of immune defense against non-viral pathogens. MHC-Y class I genes have been found to be up-regulated in several gene profiling studies examining responses to inoculation with bacteria and sheep red blood cells (Connell et al., 2012; Geng et al., 2015; Wu et al., 2015).
MHC-B in Responses to Infection and Vaccination
Early findings that the major MHC-B haplotypes influence the MD incidence had a significant impact on research focused on the chicken MHC. Investigations have concentrated both on the mechanisms underlying this major MHC-B haplotype influence on MD and on learning whether MHC-B genetics affect immune responses to other diseases. The literature is now extensive. MHC-B has been examined for a role in resistance to a wide variety of diseases caused by viral, bacterial and parasitic pathogens.
Investigations into disease responses have been carried out in various ways. Some compare responses in inbred congenic and sometimes MHC-B recombinant lines. Others examine populations in which MHC-B types are segregating either in selected lines or, occasionally in natural or semi-natural populations. Here we attempt to provide a very brief overview of investigations into MD as well as diseases caused by Rous sarcoma virus (RSV), v-src DNA, other viruses, several bacterial infections, coccidiosis, and an infestation with avian ectoparasites. More information is available for the role of MHC-B in MD and RSV than in other diseases.
Marek's Disease (MD) Virus Infections and Vaccinations
The first association with MHC-B reported appeared in an abstract (Hansen et al., 1967). The outcome of a MD virus (JM Strain) challenge trial of fully pedigreed backcross families in which two MHC-B (B21 and B19) haplotypes were segregating clearly showed the major role of the MHC-B in resistance to Marek's disease (Briles et al., 1977). This study showed an eightfold difference in MD tumor incidence between MHC-B haplotype and genetic resistance to MD. Another study published the same year provided evidence for a role of MHC haplotype in active restriction of transplanted MD tumor growth (Longenecker et al., 1977). Further work helped to rank the contributions of additional MHC-B haplotypes in congenic lines and to reveal how MDV virulence affects MHC-B encoded resistance (Abplanalp et al., 1984). There is further evidence for the contribution of additional MHC-B haplotypes in MD (Bacon et al., 1983; Bacon et al., 1981). MHC-B haplotype also influences the efficacy of vaccination against MDV (Bacon and Witter, 1992, 1993, 1994a; Bacon and Witter, 1994b; Bacon and Witter, 1994c, 1995). Additional studies show that non-MHC-B genes also contribute in responses again MDV (Bacon et al., 2000).
Advances in identifying the individual genes within MHC-B contributing to the MHC-B influence on MD were made through the success of Elwood and Ruth Briles in identifying recombinant haplotypes (Briles and Briles, 1980). In early work, MHC-B linked resistance to MD was assigned to “a gene or genes, within or closely linked to the B-F region of the B complex” with one of several recombinant haplotypes isolated for this purpose (Briles et al., 1983). It is important to note here that the responsible gene was not identified. When genomic sequence for MHC-B became available it was possible to localize the crossover breakpoint just upstream of the TRIM7.2 gene (see location of TRIM7.2 in Figure 3) leaving all genes downstream from the breakpoint as candidates for contributing in MHC-B–linked resistance to MD (Shiina et al., 2007).
Further advances were made with two additional MHC-B recombinant haplotypes, BR2 and BR4, identified as “identical” by the serological markers then available (Briles and Briles, 1980; Plachy et al., 1984). Realizing that chromosomal recombination breakpoints rarely occur at precisely the same location, a small study tested these two recombinants in developing congenic lines (003.R2 and 003.R4) at the fourth backcross to determine if they had identical phenotypes with respect to MD resistance (Schat et al., 1994). The evidence found indicated that 003.R2 and 003.R4 chickens differed in resistance to MD. In a follow-up study, 003.R2 and 003.R4 lines were tested again with larger numbers of animals after 10 backcross generations. Highly significant differences in MD mortality and total MD incidence were found between the 003.R2 and 003.R4 lines (Goto et al., 2009). The 003.R2 birds exhibited 10% mortality during trial and 19% MD overall, while the 003.R4 birds showed 39% mortality during trial and 47% MD incidence. This study mapped the crossover breakpoints in the BR2 and BR4 haplotypes to different positions within the BG1 locus showing that the BG1*R4 and BG1*R2 alleles were completely identical in coding region sequence, but differed in the 3′-untranslated region (3′-UTR). In the BR4 3′-UTR a 225 bp insert of retroviral origin was present. The 3′-UTR difference had little influence on transcription but produced a consistent difference in assays for translation. The consequence of this modification is not yet fully understood, but it may place BG1*R4 transcripts under post-transcriptional control that results in altered BG1 activity. Given the presence of the ITIM motif (associated with attenuation of lymphoid cell activation) in both isoforms it could be that the difference in the MD incidence is related to BG1 signaling differences as the result of repression of BG1 translation encoded by the BR4 haplotype. This study provides strong evidence that at least part of the long observed genetic influence of MHC-B in MD can be attributed to BG1 and reveals another way in which the MHC-B region contributes to disease resistance in addition to specific antigen presentation.
Inoculation with Rous Sarcoma Virus (RSV) and v-src
Tumors induced by Rous sarcoma virus (RSV) are mediated by the oncogene v-src (Brugge and Erikson, 1977). A critical MHC-B influence on the fate of Rous sarcomas was first reported in two concurrent publications (Collins et al., 1977; Schierman et al., 1977). Inbred line crosses G-B1 (B13B13) and G-B2 (B6B6) were tested for their response against Rous sarcomas. Backcross progeny from the [G-B1 x G-B2] x G-B1 mating showed differential tumor growth with 94% tumor regression in the heterozygous B6B13 birds compared with 4% tumor regression in birds homozygous for B13 (Schierman et al., 1977). A simultaneous study with the F2 generation of inbred lines 61 (B2B2) x 151 (B5B5) showed differences in tumor regression among individuals in a challenge experiment. Tumor regression scores were 76% for B2B2, 35% for B2B5, and 0% for B5B5. In addition, metastasis, tumor spread to distant sites, was higher in genotype B5B5 than in B2B2 (Collins et al., 1977). A third study with two additional MHC-B haplotypes (B12 in Line CB and B4 in Line CC) in Prague, also confirmed a major MHC-B role in the regression of tumors induced by v-src (Plachy and Benda, 1981). Further studies with MHC-B recombinant haplotypes have been conducted to localize the gene or genes within MHC-B that determine Rous sarcoma tumor regression. Studies comparing the recombinant haplotype BG4-BF12 (in Line CB.R1) mapped resistance to the portion of MHC-B identified by BF alloantigens (Plachy and Benda, 1981). The opposite outcome, tumor progression, was found with two additional recombinant haplotypes, CC.R1 and CC.R2, in which the BF sub-region originated from B4 and the BG sub-region from B12 (BG12-BF4) (Plachy et al., 1984). Further studies with congenic recombinant lines produced on inbred Line UCD 003 background again mapped the MHC-B influence in RSV tumors to the sub-region identified by BF alloantigens. Line 003.R2 (BG23-BF2) and Line 003.R6 (BG23-BF21) had lower tumor scores compared with four other recombinants (White et al., 1994).
A hint for additional complexity of the MHC-B region role in disease resistance was seen in RSV tumor regression trials with MHC-B BR2 and BR4 recombinant haplotypes. Lines 003.R2 and 003.R4, as occurred with MD, have been reported to respond differently to RSV tumors (Schulten et al., 2009), suggesting a contribution from BG1. Other genes also influence the outcome of RSV infections. Tumor growth differed among MHC identical lines with dissimilar background genomes, which indicated a role for non-MHC genes. Furthermore, RSV tumor metastases differed according to non-MHC background as well as MHC-B genes (Taylor, 2004).
A direct response to v-src protein was supported in further studies. Injection of RSV v-src DNA produced tumors whose growth and outcome depended on chicken line (Halpern et al., 1991). Those tumors had no viral replication or viral structural sequence. MHC control of v-src-induced sarcomas was demonstrated in congenic lines 6.6-2 (B2B2), a regressing line and 6.15-5 (B5B5), a progressing line (Taylor et al., 1992), consistent with earlier findings with RSV (Collins et al., 1977). For v-src tumor metastases, the B2B2 line (6.6-2) had lower metastases than the B5B5 line (6.15-5) (Taylor et al., 1994). Lines 6.6-2 also had greater immunity to a second v-src challenge. The response was very likely directed against some antigen determined by v-src (Taylor et al., 1994; Taylor et al., 1992); perhaps the v-src C-tail antigen (Hofmann et al., 2003). Similar results were shown in the Prague congenic lines as v-src tumors regressed in Lines CB (B12B12), and CB.R1 (BG4-BF12) regressed v-src tumors compared with Line CC (B4B4) in which tumors progressed (Svoboda et al., 1992). Line CB had higher immunity against a second challenge of v-src or RSV following primary v-src injection compared with Line CC (Plachy et al., 1994). Clearly MHC-B makes a significant contribution to the immune responses underlying the regression of RSV induced tumors. BF2 molecules may contribute specificity in the presentation of v-src protein (Hofmann et al., 2003). Perhaps BG1 molecules affect cell responsiveness to initial infection.
Susceptibility to avian leukosis virus (ALV)
ALV infections are also affected by the MHC-B haplotype. In the F3 or F4 generation of inbred line crosses (63 × 151) producing B2B2, B2B5 and B5B5 progeny, B2 haplotype was associated with lymphoid leukosis resistance as well as resistance to MD and the ability to regress RSV tumors. Haplotype B5 was found generally associated with virus susceptibility indicating a general deficiency in anti-viral disease response (Bacon et al., 1981). In a second study including B2, B5 from another source (15I4) and B15, B5 and B15 were found to be highly and essentially equally susceptible to tumors induced by MDV, RSV, and ALV (Bacon et al., 1983). This finding further highlights the resistance conferred by B2 haplotype.
The MHC-B region was examined in an Australorp population for an influence in avian leukosis virus (ALV) group-specific (gs) antigen shedding related to virus infection. Haplotype frequencies for three MHC-B haplotypes were B8a = 66.7%, B9a = 15.6, and B21 = 17.8% in a base population. The data supported differential susceptibility to horizontal virus transmission based on MHC-B complex type but no MHC-B effect on vertical transmission (Yoo and Sheldon, 1992).
Responses to Newcastle Disease (ND) Virus
The role of MHC type has also been investigated in studies of immune responses to Newcastle disease virus (NDV), an avian paramyxovirus that causes respiratory disease that varies from extremely pathogenic to mild and unnoticed depending on strain. When tested with a live attenuated ND vaccine lines birds bearing different MHC types were found to vary in specific T cell responses (Norup et al., 2011). Significantly greater proliferation of CD4+ T cells was observed in birds of B13 haplotype compared to B130 haplotype in NDV antigen-specific recall responses (evidence of difference in adaptive immunity). MHC haplotypes also contribute in the antibody titers produced by immunization with NDV (Dunnington et al., 1992). A lower anti-NDV antibody titer was observed in B21B21 chickens compared to B13B13 and B13B21 birds in the white leghorn populations selected for high and low antibody responses to sheep red blood cell antigens. This observation provides additional evidence that MHC haplotype differences affect adaptive immune responses.
Responses to infectious bursal disease (IBD) Virus
IBD is highly contagious in young chickens. IBD virus targets are lymphoid tissues, especially the bursa. In an investigation of antibody responses to a live attenuated commercial IBDV vaccine among three different lines in which four different MHC-B haplotypes were segregating, birds with one haplotype (BW1) were found to have significantly higher serum antibody titers than birds with the three other haplotypes (B19, B21 and B131) (Juul-Madsen et al., 2002). BW1 was also associated with significant lower bursa lesion scores. In another study by the same research group with killed IBDV vaccine, MHC also affected antibody response (measured by ELISA) in additional lines (Juul-Madsen et al., 2006). Birds with some MHC haplotypes were low responders (B19, B12 and BW3) and others high responders (B201, B4, and BR5). Further work in this study highlights how various factors, such as various regions within the MHC-B region, age, and background genes, influence specific antibody responses. These studies provide another example of how adaptive immune responses are affected by the MHC genotype.
Infection with infectious bronchitis (IB) Virus
IBV, a coronavirus, causes a highly contagious, acute respiratory disease in chickens. The MHC basis for an earlier observation of a dominant autosomal resistance gene for IB (Bumstead et al., 1989) only recently became clear in a retrospective study of vaccination of young birds from MHC congenic strains (Bacon et al., 2004). These findings were supported by an additional study with more MHC-B haplotypes that also ranks the haplotypes and demonstrates dominance of the resistance gene (Banat et al., 2013). The contribution of MHC-B to IB is particularly intriguing because the rapid progression of disease following infection. Dramatic differences between MHC-B haplotypes in the innate immune response of macrophage maturation has been proposed as contributing to the IBV infection differences observed among some MHC-B types (Dawes et al., 2014).
MHC-B and Fowl Cholera
Pasteurella multocida infections cause fowl cholera in chickens. There is relatively little information available on the influence of host genetics in fowl cholera. One study using a three-marker assay for MHC-B (serological MHC-B type, humoral immune response to a amino acid polymer composed of glutamic acid, alanine and tyrosine, and RSV tumor responses) showed survival following inoculation with P. multocida was linked with MHC-B type in the Iowa State S1 line of white leghorn chickens (Lamont et al., 1987). In another study comparing indigenous and commercial breeds of chickens in Vietnam, the indigenous breed appeared more susceptible to fowl cholera than the commercial breed, with the susceptibility apparently linked to a select set of MHC haplotypes defined by LEI0258 (Schou et al., 2010).
Influence of MHC-B in Staphylococcal Infections
Two substantial studies provide evidence for MHC-B haplotype influencing the outcome of infections with Staphylococcus aureus. Comparisons with 10 UC Davis inbred congenic leghorn lines showed significant differences in mortality among chicks challenged at 3 d of age with a moderately pathogenic isolate and a highly pathogenic isolated of S. aureus (Cotter et al., 1992). There was lower mortality in the Line 331 (B2/B2) chicks compared to chicks in Line 336 (BQBQ), Line 335 (B19B19) and Line 330 (B21/B21). No significant difference was found among the lines in similar trials with birds at six wk of age. Another group studying bacterial skeletal disease in a genetically pure line of broiler breeder chickens observed highly significant association of two MHC-B homozygous genotypes (BA4BA4 and BA12BA12) with lameness in which Staphylococcus spp. was isolated from the skeletal lesions (Joiner et al., 2005).
MHC-B and Salmonella Species
Because Salmonella spp. are a major cause of foodborne illness in man, and poultry products are a major source, there is considerable interest in finding ways to reduce the incidence of Salmonella infections in flocks raised for food. Although most Salmonella serovars produce little disease in adult chickens, these bacteria often colonize the intestinal tracts of poultry. Immunity is considered a primary means of controlling colonization and hence there is an interest in immunogenetic links with infection. A study with inbred congenic lines showed that at as early as 3 d of age resistance to Salmonella is expressed (Cotter et al., 1998). Different MHC-B haplotypes contribute differently to resistance ((high in Line 342 (BCBC); lower in Line 253 (B18B18) and Line 254 (B15/B15). Using microsatellite markers for mapping, MHC class I was identified as a gene associated with Salmonella colonization (Lamont et al., 2002). Further work linked resistance to the MHC I and class II via a sequence polymorphism and SSCP (Liu et al., 2002), and to antibody response kinetics (Zhou and Lamont, 2003). The indigenous and commercial breeds of chickens in Vietnam also linked Salmonella-specific antibody responses to MHC-B haplotype (Schou et al., 2010). A QTL study of a (N × 61) × N backcross population identified MHC-B as making a small contribution to the Salmonella carrier-state (Tilquin et al., 2005). Differences in responses between birds with different MHC-B haplotypes may become less with increasing age (Barrow et al., 2004; Cotter et al., 1998).
MHC-B and Escherichia coli Infections
In a study comparing B21 and B13 haplotypes in birds 4 wk of age, MHC-B haplotype was implicated in the likelihood of an individual bird developing cellulitis, an inflammatory process induced by E. coli that causes tissue lesions (in this experiment induced by subcutaneous injection of E. coli). Interestingly B13 haplotype confers more resistance to cellulitis compared to B21 and two additional haplotypes BA9 and BA12 were intermediate in incidence (Macklin et al., 2002; Macklin et al., 2009). Among those birds which did developed cellulitis, there was no link between MHC-B haplotype and the severity of the lesions (Macklin et al., 2002; Macklin et al., 2009). In another study by another research group, the outcome of intratracheal inoculation of adult birds with a field strain of avian pathogenic E. coli mortality was found to be significantly lower in B21/B21 hens compared to B19/B21 hens (Cavero et al., 2009).
Coccidial Infections and MHC-B
Eimeria spp. (principally E. tenella, E. maxima, and E. acervulina) are obligate intracellular parasites that cause coccidiosis in birds. Infection produces intestinal destruction, blood loss, nutrient malabsorption, and mortality in the animals most severely affected. There is clearly a genetic influence with large differences in the coccidiosis resistance described between lines; see for example (Pinard-Van Der Laan et al., 1998; Uni et al., 1993). Additional studies have shown MHC-B genetic effects in coccidiosis in some genetically defined stocks (Caron et al., 1997; Clare et al., 1985; Kim et al., 2008); but not in others (Lillehoj et al., 1989; Ruff and Bacon, 1989). The outbreak of coccidiosis in a semi-natural red Jungle fowl population points to a single MHC-B haplotype conferring susceptibility, the contribution of which was masked by all of the six other haplotypes in heterozygous birds (Worley et al., 2010). The observation that multiple low-dose exposures to E. tenella stimulate immunity more effectively than a single higher-dose challenge suggests activation of adaptive immunity and that immunization occurs with time, implicating MHC-B in antigen presentation in the response. However, which individual genes contribute in responses to coccidial infections has not been determined.
MHC-B and the Incidence of Intestinal Ascarid Infections
With poultry increasingly being produced under conditions with free outdoor access the likelihood of diverse infections increases including infection with parasitic worms, such as Ascardia galli. As with other infectious disease in chickens, there is evidence for genetic factors influencing helminth infections (Schou et al., 2003; Schou et al., 2007). A statistically significant contribution from MHC-B in resistance to A. galli has been noted in indigenous and exotic chicken in Vietnam (Schou et al., 2007). The MHC-B region appears to affect infection intensity (Schou et al., 2010). A study comparing two inbred lines with different MHC-B haplotypes revealed significantly different levels of infection and antibody titers (Norup et al., 2013). Levels of infection and antibody titers were inversely related suggesting that humoral immunity may be involved in the disease response but that higher antibody titer does not result in clearing of infection.
MHC-B Haplotype Involvement in Resistance to Northern Fowl Mite (NFM) Colonization
Northern Fowl Mite (NFM; Ornithonyssus sylviarum) is the most common ectoparasitic arthropod in poultry production areas in the United States (Arends, 2003). MHC-B haplotypes appear to contribute differentially to the success NFM colonization. Trials that included UC Davis congenic lines homozygous for MHC-B haplotypes (UCD-254, B15; UCD-331, B2; UCD-253; B18 and UCD-330, B21) and a commercial layer line (Hy-Line W-36) in which three MHC-B types (B2, B15 and B21) segregate, revealed B21 birds to be significantly more resistant to mite infection compared with B15 birds (Owen et al., 2008). While it remains to be determined which MHC-B gene or genes contribute resistance to this ectoparasite, it has been suggested that this MHC-B encoded resistance is related to inflammatory response, with a greater degree of skin inflammation during an infestation conferring resistance by the accompanying swelling, thus limiting access of mites to blood sources in the skin (Owen et al., 2009).
Acknowledgments
The authors would like to honor the memory of Helle Risdahl Juul-Madsen, a major contributor in many aspects of chicken MHC research. We gratefully acknowledge the contributions of Ronald Goto, Beckman Research Institute, City of Hope, Duarte, CA and Lynda Caron, Department of Animal and Nutritional Sciences, University of New Hampshire, Durham, NH. We thank the National Science Foundation, NIH, the USDA NIFA Competitive Grants Program, and the New Hampshire and West Virginia Agricultural Experiment Stations (AES) for sustained support provided over many years. Our respective institutions facilitated our studies: City of Hope Beckman Research Institute (Miller), West Virginia University (Taylor) and the University of New Hampshire (Taylor), where the NH AES encouraged and supported maintenance of genetic research stocks until 2007. Scientific Article No. 3261 of the West Virginia Agricultural and Forestry Experiment Station, Morgantown.
REFERENCES
- Abplanalp H., Schat K. A., Calnek B. W. Resistance to Marek's disease of congenic lines differing in major histocompatibility haplotypes to 3 virus strains. In: Calnek B. W., Spencer J. L., editors. International Symposium on Marek's Disease. Ithaca, NY: American Association of Avian Pathologists, Inc.; 1984. pp. 347–358. [Google Scholar]
- Afanassieff M., Goto R. M., Ha J., Sherman M., Zhong L., Auffray C., Coudert F., Zoorob R., Miller M. M. At least one class I gene in restriction fragment pattern-Y (Rfp-Y), the second MHC gene cluster in the chicken, is transcribed, polymorphic and shows divergent specialization in antigen binding region. J. Immunol. 2001;166:3324–3333. doi: 10.4049/jimmunol.166.5.3324. [DOI] [PubMed] [Google Scholar]
- Afanassieff M., Goto R. M., Ha J., Zoorob R., Auffray C., Coudert F., Briles W. E., Miller M. M. Are chicken Rfp-Y class I genes classical or non-classical? In: Spencer J. L., editor. Major Histocompatibility Complex. Japan: Springer International Publishing AG; 2000. [Google Scholar]
- Arends J. J. External Parasites and Poultry Pests. In: Saif Y. M., editor. Diseases of Poultry. Iowa State Press; 2003. pp. 905–930. Editor-in-Chief ed. [Google Scholar]
- Bacon L., Vallejo R., Cheng H., Witter R. Failure of Rfp-Y genes to influence resistance to Marek's disease. In: Silva R. F., Cheng H. H., Coussens P. M., Lee L. F., Velicer L. F., editors. Current Research on Marek's Disease. American Association of Avian Pathologists, Kennett Square; 1996. pp. 69–74. [Google Scholar]
- Bacon L. D., Hunter D. B., Zhang H. M., Brand K., Etches R. Retrospective evidence that the MHC (B haplotype) of chickens influences genetic resistance to attenuated infectious bronchitis vaccine strains in chickens. Avian Pathol. 2004;33:605–609. doi: 10.1080/03079450400013147. [DOI] [PubMed] [Google Scholar]
- Bacon L. D., Hunt H. D., Cheng H. H. A review of the development of chicken lines to resolve genes determining resistance to diseases. Poult. Sci. 2000;79:1082–1093. doi: 10.1093/ps/79.8.1082. [DOI] [PubMed] [Google Scholar]
- Bacon L. D., Crittenden L. B., Witter R. L., Fadly A., Motta J. B5 and B15 associated with progressive Marek's disease, Rous sarcoma, and avian leukosis virus-induced tumors in inbred 15I4 chickens. Poult. Sci. 1983;62:573–578. doi: 10.3382/ps.0620573. [DOI] [PubMed] [Google Scholar]
- Bacon L. D., Witter R. L. Influence of turkey herpesvirus vaccination on the B-haplotype effect on Marek's disease resistance in 15.B-congenic chickens. Avian Dis. 1992;36:378–385. [PubMed] [Google Scholar]
- Bacon L. D., Witter R. L. Influence of B-haplotype on the relative efficacy of Marek's disease vaccines of different serotypes. Avian Dis. 1993;37:53–59. [PubMed] [Google Scholar]
- Bacon L. D., Witter R. L. B haplotype influence on the relative efficacy of Marek's disease vaccines in commercial chickens. Poult. Sci. 1994a;73:481–487. doi: 10.3382/ps.0730481. [DOI] [PubMed] [Google Scholar]
- Bacon L. D., Witter R. L. An influence of the MHC on efficacy of Marek's disease vaccines. Proc. 5th World Congress on Genetics Applied to Livestock Production. 1994b;20:261–264. [Google Scholar]
- Bacon L. D., Witter R. L. Serotype specificity of B-haplotype influence on the relative efficacy of Marek's disease vaccines. Avian Dis. 1994c;38:65–71. [PubMed] [Google Scholar]
- Bacon L. D., Witter R. L. Efficacy of Marek's disease vaccines in Mhc heterozygous chickens: Mhc congenic x inbred line F1 matings. J. Hered. 1995;86:269–273. doi: 10.1093/oxfordjournals.jhered.a111580. [DOI] [PubMed] [Google Scholar]
- Bacon L. D., Witter R. L., Crittenden L. B., Fadly A., Motta J. B-haplotype influence on Marek's disease, Rous sarcoma, and lymphoid leukosis virus-induced tumors in chickens. Poult. Sci. 1981;60:1132–1139. doi: 10.3382/ps.0601132. [DOI] [PubMed] [Google Scholar]
- Banat G. R., Tkalcic S., Dzielawa J. A., Jackwood M. W., Saggese M. D., Yates L., Kopulos R., Briles W. E., Collisson E. W. Association of the chicken MHC B haplotypes with resistance to avian coronavirus. Dev. Comp. Immunol. 2013;39:430–437. doi: 10.1016/j.dci.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow P. A., Bumstead N., Marston K., Lovell M. A., Wigley P. Faecal shedding and intestinal colonization of Salmonella enterica in in-bred chickens: the effect of host-genetic background. Epidemiol, Infect. 2004;132:117–126. doi: 10.1017/s0950268803001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar G., Bourlet Y., Frechin N., Guillemot F., Zoorob R., Auffray C. Molecular analysis of chicken immune response genes. Biochimie. 1988;70:909–917. doi: 10.1016/0300-9084(88)90232-5. [DOI] [PubMed] [Google Scholar]
- Bernot A., Zoorob R., Auffray C. Linkage of a new member of the lectin supergene family to chicken Mhc genes. Immunogenetics. 1994;39:221–229. doi: 10.1007/BF00188784. [DOI] [PubMed] [Google Scholar]
- Bikle D. D., Munson S., Komuves L. Zipper protein, a B-G protein with the ability to regulate actin/myosin 1 interactions in the intestinal brush border. J. Biol. Chem. 1996;271:9075–9083. doi: 10.1074/jbc.271.15.9075. [DOI] [PubMed] [Google Scholar]
- Bourlet Y., Behar G., Guillemot F., Frechin N., Billault A., Chausse A. M., Zoorob R., Auffray C. Isolation of chicken major histocompatibility complex class II (B-L) beta chain sequences: comparison with mammalian beta chains and expression in lymphoid organs. EMBO J. 1988;7:1031–1039. doi: 10.1002/j.1460-2075.1988.tb02910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briles W., Goto R., Auffray C., Miller M. A polymorphic system related to but genetically independent of the chicken major histocompatibility complex. Immunogenetics. 1993;37:408–414. doi: 10.1007/BF00222464. [DOI] [PubMed] [Google Scholar]
- Briles W., McGibbon W., Irwin M. On multiple alleles effecting cellular antigens in the chicken. Genetics. 1950;35:633–652. doi: 10.1093/genetics/35.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briles W. E., Briles R. W. A search for “duplicate” recombinants between B-F and B-G regions in the chicken B complex. Anim. Blood Groups Biochem. Genet. 1980;11:38–39. [Google Scholar]
- Briles W. E., Briles R. W., Taffs R. E., Stone H. A. Resistance to a malignant lymphoma in chickens is mapped to subregion of major histocompatibility (B) complex. Science. 1983;219:977–979. doi: 10.1126/science.6823560. [DOI] [PubMed] [Google Scholar]
- Briles W. E., Stone H. A., Cole R. K. Marek's disease: effects of B histocompatibility alloalleles in resistant and susceptible chicken lines. Science. 1977;195:193–195. doi: 10.1126/science.831269. [DOI] [PubMed] [Google Scholar]
- Brugge J. S., Erikson R. L. Identification of a transformation-specific antigen induced by an avian sarcoma virus. Nature. 1977;269:346–348. doi: 10.1038/269346a0. [DOI] [PubMed] [Google Scholar]
- Bumstead N., Huggins M. B., Cook J. K. Genetic differences in susceptibility to a mixture of avian infectious bronchitis virus and Escherichia coli. Br. Poult. Sci. 1989;30:39–48. doi: 10.1080/00071668908417123. [DOI] [PubMed] [Google Scholar]
- Butter C., Staines K., van Hateren A., Davison T. F., Kaufman J. The peptide motif of the single dominantly expressed class I molecule of the chicken MHC can explain the response to a molecular defined vaccine of infectious bursal disease virus (IBDV) Immunogenetics. 2013;65:609–618. doi: 10.1007/s00251-013-0705-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron L. A., Abplanalp H., Taylor R. L., Jr Resistance, susceptibility, and immunity to Eimeria tenella in major histocompatibility (B) complex congenic lines. Poult. Sci. 1997;76:677–682. doi: 10.1093/ps/76.5.677. [DOI] [PubMed] [Google Scholar]
- Cavero D., Schmutz M., Philipp H. C., Preisinger R. Breeding to reduce susceptibility to Escherichia coli in layers. Poult. Sci. 2009;88:2063–2068. doi: 10.3382/ps.2009-00168. [DOI] [PubMed] [Google Scholar]
- Chappell P., Meziane el K., Harrison M., Magiera L., Hermann C., Mears L., Wrobel A. G., Durant C., Nielsen L. L., Buus S., Ternette N., Mwangi W., Butter C., Nair V., Ahyee T., Duggleby R., Madrigal A., Roversi P., Lea S. M., Kaufman J. Expression levels of MHC class I molecules are inversely correlated with promiscuity of peptide binding. eLife. 2015;4:e05345. doi: 10.7554/eLife.05345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. C., Lan H., Sun L., Deng Y. L., Tang K. Y., Wan Q. H. Genomic organization of the crested ibis MHC provides new insight into ancestral avian MHC structure. Sci. Rep. 2015;5:7963. doi: 10.1038/srep07963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare R. A., Strout R. G., Taylor R. L., Jr., Collins W. M., Briles W. E. Major histocompatibility (B) complex effects on acquired immunity to cecal coccidiosis. Immunogenetics. 1985;22:593–599. doi: 10.1007/BF00430307. [DOI] [PubMed] [Google Scholar]
- Collins W. M., Briles W. E., Zsigray R. M., Dunlop W. R., Corbett A. C., Clark K. K., Marks J. L., McGrail T. P. The B locus (MHC) in the chicken: Association with the fate of RSV-induced tumors. Immunogenetics. 1977;5:333–343. [Google Scholar]
- Connell S., Meade K. G., Allan B., Lloyd A. T., Kenny E., Cormican P., Morris D. W., Bradley D. G., O'Farrelly C. Avian resistance to Campylobacter jejuni colonization is associated with an intestinal immunogene expression signature identified by mRNA sequencing. PLoS ONE. 2012;7:e40409. doi: 10.1371/journal.pone.0040409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P. F., Taylor R. L., Abplanalp H. Differential resistance to Staphylococcus aureus challenge in major histocompatibility (B) complex congenic lines. Poult. Sci. 1992;71:1873–1878. doi: 10.3382/ps.0711873. [DOI] [PubMed] [Google Scholar]
- Cotter P. F., Taylor R. L., Jr., Abplanalp H. B-complex associated immunity to Salmonella enteritidis challenge in congenic chickens. Poult. Sci. 1998;77:1846–1851. doi: 10.1093/ps/77.12.1846. [DOI] [PubMed] [Google Scholar]
- Dalgaard T. S., Hojsgaard Skjodt S. K., Juul-Madsen H. R. Differences in chicken major histocompatibility complex (MHC) class Ialpha gene expression between Marek's disease-resistant and -susceptible MHC haplotypes. Scand. J. Immunol. 2003;57:135–143. doi: 10.1046/j.1365-3083.2003.01207.x. [DOI] [PubMed] [Google Scholar]
- Dalgaard T. S., Vitved L., Skjodt K., Thomsen B., Labouriau R., Jensen K. H., Juul-Madsen H. R. Molecular characterization of major histocompatibility complex class I (B-F) mRNA variants from chickens differing in resistance to Marek's disease. Scand. J. Immunol. 2005;62:259–270. doi: 10.1111/j.1365-3083.2005.01652.x. [DOI] [PubMed] [Google Scholar]
- Dawes M. E., Griggs L. M., Collisson E. W., Briles W. E., Drechsler Y. Dramatic differences in the response of macrophages from B2 and B19 MHC-defined haplotypes to interferon gamma and polyinosinic:polycytidylic acid stimulation. Poult. Sci. 2014;93:830–838. doi: 10.3382/ps.2013-03511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delany M., Robinson C., Goto R., Miller M. Architecture and organization of chicken microchromosome 16: order of the NOR, MHC-Y, and MHC-B subregions. J. Hered. 2009;100:507–514. doi: 10.1093/jhered/esp044. [DOI] [PubMed] [Google Scholar]
- Dunnington E. A., Larsen C. T., Gross W. B., Siegel P. B. Antibody responses to combinations of antigens in white Leghorn chickens of different background genomes and major histocompatibility complex genotypes. Poult. Sci. 1992;71:1801–1806. doi: 10.3382/ps.0711801. [DOI] [PubMed] [Google Scholar]
- Dvir H., Wang J., Ly N., Dascher C. C., Zajonc D. M. Structural basis for lipid-antigen recognition in avian immunity. J. Immunol. 2010;184:2504–2511. doi: 10.4049/jimmunol.0903509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers A., Beck S., Forbes S. A., Trowsdale J., Volz A., Younger R., Ziegler A. MHC-linked olfactory receptor loci exhibit polymorphism and contribute to extended HLA/OR-haplotypes. Genome Res. 2000;10:1968–1978. doi: 10.1101/gr.10.12.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald S. J., Livant E. J. Distinctive polymorphism of chicken B-FI (major histocompatibility complex class I) molecules. Poult. Sci. 2004;83:600–605. doi: 10.1093/ps/83.4.600. [DOI] [PubMed] [Google Scholar]
- Frangoulis B., Park I., Guillemot F., Severac V., Auffray C., Zoorob R. Identification of the Tapasin gene in the chicken major histocompatibility complex. Immunogenetics. 1999;49:328–337. doi: 10.1007/s002510050500. [DOI] [PubMed] [Google Scholar]
- Fulton J. E., Thacker E. L., Bacon L. D., Hunt H. D. Functional analysis of avian class I (BFIV) glycoproteins by epitope tagging and mutagenesis in vitro. Eur. J. Immunol. 1995;25:2069–2076. doi: 10.1002/eji.1830250740. [DOI] [PubMed] [Google Scholar]
- Geng T., Guan X., Smith E. J. Screening for genes involved in antibody response to sheep red blood cells in the chicken, Gallus gallus. Poult. Sci. 2015;94:2099–2107. doi: 10.3382/ps/pev224. [DOI] [PubMed] [Google Scholar]
- Gilmour D. G. Segregation of genes determining red cell antigens at high levels of inbreeding in chickens. Genetics. 1959;44:14–33. doi: 10.1093/genetics/44.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto R. M., Wang Y., Taylor R. L., Jr., Wakenell P. S., Hosomichi K., Shiina T., Blackmore C. S., Briles W. E., Miller M. M. BG1 has a major role in MHC-linked resistance to malignant lymphoma in the chicken. Proc. Natl. Acad. Sci. USA. 2009;106:16740–16745. doi: 10.1073/pnas.0906776106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F., Auffray C. A molecular map of the chicken B complex. Prog. Clin. Biol. Res. 1989;307:169–176. [PubMed] [Google Scholar]
- Guillemot F., Billault A., Pourquie O., Behar G., Chausse A., Zoorob R., Kreibich G., Auffray C. A molecular map of the chicken major histocompatibility complex: the class II beta genes are closely linked to the class I genes and the nucleolar organizer. EMBO J. 1988;7:2775–2785. doi: 10.1002/j.1460-2075.1988.tb03132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F., Kaufman J. F., Skjoedt K., Auffray C. The major histocompatibility complex in the chicken. Trends Genet. 1989;5:300–304. doi: 10.1016/0168-9525(89)90112-1. [DOI] [PubMed] [Google Scholar]
- Guillemot F., Turmel P., Charron D., Le Douarin N., Auffray C. Structure, biosynthesis, and polymorphism of chicken MHC class II (B-L) antigens and associated molecules. J. Immunol. 1986;137:1251–1257. [PubMed] [Google Scholar]
- Halpern M. S., England J. M., Coates L., Stoltzfus C. M., Mason W. S. Regression of v-src DNA-induced sarcomas is under host genetic control. Virology. 1991;180:857–860. doi: 10.1016/0042-6822(91)90107-m. [DOI] [PubMed] [Google Scholar]
- Hansen M. P., Van Zandy J. N., Law G. R. J. Differences in susceptibility to Marek's disease in chickens carrying two different B locus blood group alleles. Poult. Sci. 1967;46:1268. [Google Scholar]
- Hearn C., Preeyanon L., Hunt H. D., York I. A. An MHC class I immune evasion gene of Marek's disease virus. Virology. 2015;475:88–95. doi: 10.1016/j.virol.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hee C. S., Gao S., Loll B., Miller M. M., Uchanska-Ziegler B., Daumke O., Ziegler A. Structure of a classical MHC class I molecule that binds “non-classical” ligands. PLoS Biol. 2010;8:e1000557. doi: 10.1371/journal.pbio.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hee C. S., Gao S., Miller M. M., Goto R. M., Ziegler A., Daumke O., Uchanska-Ziegler B. Expression, purification and preliminary X-ray crystallographic analysis of the chicken MHC class I molecule YF1*7.1. Acta Crystallogr. 2009;F65:422–425. doi: 10.1107/S1744309109009026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig C. T., Waters R. W., Baldwin C. L., Telfer J. C. Evolution of the CD163 family and its relationship to the bovine gamma delta T cell co-receptor WC1. BMC Evol. Biol. 2010;10:181–199. doi: 10.1186/1471-2148-10-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A., Plachy J., Hunt L., Kaufman J., Hala K. v-src oncogene-specific carboxy-terminal peptide is immunoprotective against Rous sarcoma growth in chickens with MHC class I allele B-F12. Vaccine. 2003;21:4694–4699. doi: 10.1016/s0264-410x(03)00516-4. [DOI] [PubMed] [Google Scholar]
- Hosomichi K., Miller M. M., Goto R. M., Wang Y., Suzuki S., Kulski J. K., Nishibori M., Inoko H., Hanzawa K., Shiina T. Contribution of mutation, recombination, and gene conversion to chicken MHC-B haplotype diversity. J. Immunol. 2008;181:3393–3399. doi: 10.4049/jimmunol.181.5.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Guo Y., Wu C., Shen N., Jiang Y., Wang J. Prediction and identification of T cell epitopes in the H5N1 influenza virus nucleoprotein in chicken. PLoS ONE. 2012;7:e39344. doi: 10.1371/journal.pone.0039344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H., Chen C., Nenninger A., Holz L., Baldwin C. L., Telfer J. C. WC1 is a hybrid γδ TCR coreceptor and pattern recognition receptor for pathogenic bacteria. J. Immunol. 2015;194:2280–2288. doi: 10.4049/jimmunol.1402021. [DOI] [PubMed] [Google Scholar]
- Hunt H. D., Dunn J. R. The influence of host genetics on Marek's disease virus evolution. Avian Dis. 2013;57:474–482. doi: 10.1637/10315-080212-Reg.1. [DOI] [PubMed] [Google Scholar]
- Hunt H. D., Goto R. M., Foster D. N., Bacon L. D., Miller M. M. At least one YMHCI molecule in the chicken is alloimmunogenic and dynamically expressed on spleen cells during development. Immunogenetics. 2006;58:297–307. doi: 10.1007/s00251-005-0074-1. [DOI] [PubMed] [Google Scholar]
- Hunt H. D., Pharr G. T., Bacon L. D. Molecular analysis reveals MHC class I intra-locus recombination in the chicken. Immunogenetics. 1994;40:370–375. doi: 10.1007/BF01246678. [DOI] [PubMed] [Google Scholar]
- Iizuka K., Naidenko O. V., Plougastel B. F., Fremont D. H., Yokoyama W. M. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat. Immunol. 2003;4:801–807. doi: 10.1038/ni954. [DOI] [PubMed] [Google Scholar]
- Jacob J. P., Milne S., Beck S., Kaufman J. The major and a minor class II beta-chain (B-LB) gene flank the Tapasin gene in the B-F/B-L region of the chicken major histocompatibility complex. Immunogenetics. 2000;51:138–147. doi: 10.1007/s002510050022. [DOI] [PubMed] [Google Scholar]
- Joiner K. S., Hoerr F. J., van Santen E., Ewald S. J. The avian major histocompatibility complex influences bacterial skeletal disease in broiler breeder chickens. Vet. Pathol. 2005;42:275–281. doi: 10.1354/vp.42-3-275. [DOI] [PubMed] [Google Scholar]
- Juul-Madsen H. R., Dalgaard T. S., Afanassieff M. Molecular characterization of major and minor MHC class I and II genes in B21-like haplotypes in chickens. Anim. Genet. 2000a;31:252–261. doi: 10.1046/j.1365-2052.2000.00648.x. [DOI] [PubMed] [Google Scholar]
- Juul-Madsen H. R., Dalgaard T. S., Guldbrandtsen B., Salomonsen J. A polymorphic major histocompatibility complex class II-like locus maps outside of both the chicken B-system and Rfp-Y-system. Eur. J. Immunogenet. 2000b;27:63–71. doi: 10.1046/j.1365-2370.2000.00200.x. [DOI] [PubMed] [Google Scholar]
- Juul-Madsen H. R., Dalgaard T. S., Rontved C. M., Jensen K. H., Bumstead N. Immune response to a killed infectious bursal disease virus vaccine in inbred chicken lines with different major histocompatibility complex haplotypes. Poult. Sci. 2006;85:986–998. doi: 10.1093/ps/85.6.986. [DOI] [PubMed] [Google Scholar]
- Juul-Madsen H. R., Nielsen O. L., Krogh-Maibom T., Rontved C. M., Dalgaard T. S., Bumstead N., Jorgensen P. H. Major histocompatibility complex-linked immune response of young chickens vaccinated with an attenuated live infectious bursal disease virus vaccine followed by an infection. Poult. Sci. 2002;81:649–656. doi: 10.1093/ps/81.5.649. [DOI] [PubMed] [Google Scholar]
- Juul-Madsen H. R., Zoorob R., Auffray C., Skjodt K., Hedemand J. E. New chicken Rfp-Y haplotypes on the basis of MHC class II RFLP and MLC analyses. Immunogenetics. 1997;45:345–352. doi: 10.1007/s002510050215. [DOI] [PubMed] [Google Scholar]
- Kaufman J., Andersen R., Avila D., Engberg J., Lambris J., Salomonsen J., Welinder K., Skjodt K. Different features of the MHC class I heterodimer have evolved at different rates. Chicken B-F and beta 2-microglobulin sequences reveal invariant surface residues. J. Immunol. 1992;148:1532–1546. [PubMed] [Google Scholar]
- Kaufman J., Bumstead N., Miller M., Riegert P., Salomonsen J. The chicken class II alpha gene is located outside the B complex. In: Davison T. F., Bumstead N., Kaiser P., editors. Advances in Avian Immunology Research. Abingdon, UK: Carfax Publishing Company; 1995a. pp. 119–127. [Google Scholar]
- Kaufman J., Salomonsen J., Skjodt K., Thorpe D. Size polymorphism of chicken major histocompatibility complex-encoded B-G molecules is due to length variation in the cytoplasmic heptad repeat region. Proc. Natl. Acad. Sci. USA. 1990;87:8277–8281. doi: 10.1073/pnas.87.21.8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman J., Milne S., Gobel T. W., Walker B. A., Jacob J. P., Auffray C., Zoorob R., Beck S. The chicken B locus is a minimal essential major histocompatibility complex. Nature. 1999;401:923–925. doi: 10.1038/44856. [DOI] [PubMed] [Google Scholar]
- Kaufman J., Volk H., Wallny H. J. A “minimal essential Mhc” and an “unrecognized Mhc”: two extremes in selection for polymorphism. Immunol. Rev. 1995b;143:63–88. doi: 10.1111/j.1600-065x.1995.tb00670.x. [DOI] [PubMed] [Google Scholar]
- Kim D. K., Lillehoj H. S., Hong Y. H., Park D. W., Lamont S. J., Han J. Y., Lillehoj E. P. Immune-related gene expression in two B-complex disparate genetically inbred Fayoumi chicken lines following Eimeria maxima infection. Poult. Sci. 2008;87:433–443. doi: 10.3382/ps.2007-00383. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L., Patel O., Corbett A. J., Le Nours J., Meehan B., Liu L., Bhati M., Chen Z., Kostenko L., Reantragoon R., Williamson N. A., Purcell A. W., Dudek N. L., McConville M. J., O'Hair R. A., Khairallah G. N., Godfrey D. I., Fairlie D. P., Rossjohn J., McCluskey J. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–723. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- Koch M., Camp S., Collen T., Avila D., Salomonsen J., Wallny H. J., van Hateren A., Hunt L., Jacob J. P., Johnston F., Marston D. A., Shaw I., Dunbar P. R., Cerundolo V., Jones E. Y., Kaufman J. Structures of an MHC class I molecule from B21 chickens illustrate promiscuous peptide binding. Immunity. 2007;27:885–899. doi: 10.1016/j.immuni.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Kroemer G., Zoorob R., Auffray C. Structure and expression of a chicken MHC class I gene. Immunogenetics. 1990;31:405–409. doi: 10.1007/BF02115020. [DOI] [PubMed] [Google Scholar]
- Lakshmanan N., Lamont S. J. Rfp-Y region polymorphism and Marek's disease resistance in multitrait immunocompetence-selected chicken lines. Poult. Sci. 1998;77:538–541. doi: 10.1093/ps/77.4.538. [DOI] [PubMed] [Google Scholar]
- Lamont S. J., Bolin C., Cheville N. Genetic resistance to fowl cholera is linked to the major histocompatibility complex. Immunogenetics. 1987;25:284–289. doi: 10.1007/BF00404420. [DOI] [PubMed] [Google Scholar]
- Lamont S. J., Kaiser M. G., Liu W. Candidate genes for resistance to Salmonella enteritidis colonization in chickens as detected in a novel genetic cross. Vet. Immunol. Immunopathol. 2002;87:423–428. doi: 10.1016/s0165-2427(02)00064-8. [DOI] [PubMed] [Google Scholar]
- LePage K. T., Miller M. M., Briles W. E., Taylor R. L., Jr Rfp-Y genotype affects the fate of Rous sarcomas in B2 B5 chickens. Immunogenetics. 2000;51:751–754. doi: 10.1007/s002510000180. [DOI] [PubMed] [Google Scholar]
- Lillehoj H. S., Ruff M. D., Bacon L. D., Lamont S. J., Jeffers T. K. Genetic control of immunity to Eimeria tenella. Interaction of MHC genes and non-MHC linked genes influences levels of disease susceptibility in chickens. Vet. Immunol. Immunopathol. 1989;20:135–148. doi: 10.1016/0165-2427(89)90094-9. [DOI] [PubMed] [Google Scholar]
- Liu W., Miller M. M., Lamont S. J. Association of MHC class I and class II gene polymorphisms with vaccine or challenge response to Salmonella enteritidis in young chicks. Immunogenetics. 2002;54:582–590. doi: 10.1007/s00251-002-0495-z. [DOI] [PubMed] [Google Scholar]
- Livant E. J., Brigati J. R., Ewald S. J. Diversity and locus specificity of chicken MHC B class I sequences. Anim. Genet. 2004;35:18–27. doi: 10.1111/j.1365-2052.2003.01078.x. [DOI] [PubMed] [Google Scholar]
- Livant E. J., Li L., Ewald S. J. Differential expression of class I MHC (B-F) loci in broiler breeder haplotypes. In: Schat K. A., editor. Current Progress on Avian Immunology Research. Ithaca, NY: American Association of Avian Pathologists; 2001. pp. 285–288. [Google Scholar]
- Longenecker B. M., Pazderka F., Gavora J. S., Spencer J. L., Stephens E. A., Witter R. L., Ruth R. F. Role of the major histocompatibility complex in resistance to Marek's disease: restriction of the growth of JMV-MD tumor cells in genetically resistant birds. Adv. Exp. Med. Biol. 1977;88:287–298. doi: 10.1007/978-1-4613-4169-7_27. [DOI] [PubMed] [Google Scholar]
- Maccubbin D. L., Schierman L. W. MHC-restricted cytotoxic response of chicken T cells: expression, augmentation, and clonal characterization. J. Immunol. 1986;136:12–16. [PubMed] [Google Scholar]
- Macklin K. S., Ewald S. J., Norton R. A. Major histocompatibility complex effect on cellulitis among different chicken lines. Avian Pathol. 2002;31:371–376. doi: 10.1080/03079450220141642. [DOI] [PubMed] [Google Scholar]
- Macklin K. S., Norton R. A., Ewald S. J. Effect of the major histocompatibility complex on the inhibition of induced cellulitis development in a broiler chicken model. Avian Dis. 2009;53:297–300. doi: 10.1637/8449-081808-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Maruoka T., Tanabe H., Chiba M., Kasahara M. Chicken CD1 genes are located in the MHC: CD1 and endothelial protein C receptor genes constitute a distinct subfamily of class-I-like genes that predates the emergence of mammals. Immunogenetics. 2005;57:590–600. doi: 10.1007/s00251-005-0016-y. [DOI] [PubMed] [Google Scholar]
- Miller M. M., Abplanalp H., Goto R. Genotyping chickens for the B-G subregion of the major histocompatibility complex using restriction fragment length polymorphisms. Immunogenetics. 1988;28:374–379. doi: 10.1007/BF00364237. [DOI] [PubMed] [Google Scholar]
- Miller M. M., Bacon L. D., Hala K., Hunt H. D., Ewald S. J., Kaufman J., Zoorob R., Briles W. E. 2004 Nomenclature for the chicken major histocompatibility (B and Y) complex. Immunogenetics. 2004;56:261–279. doi: 10.1007/s00251-004-0682-1. [DOI] [PubMed] [Google Scholar]
- Miller M. M., Goto R., Bernot A., Zoorob R., Auffray C., Bumstead N., Briles W. E. Two Mhc class I and two Mhc class II genes map to the chicken Rfp-Y system outside the B complex. Proc. Natl. Acad. Sci. USA. 1994;91:4397–4401. doi: 10.1073/pnas.91.10.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. M., Goto R., Young S., Chirivella J., Hawke D., Miyada C. G. Immunoglobulin variable-region-like domains of diverse sequence within the major histocompatibility complex of the chicken. Proc. Natl. Acad. Sci. USA. 1991;88:4377–4381. doi: 10.1073/pnas.88.10.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. M., Goto R., Young S., Liu J., Hardy J. Antigens similar to major histocompatibility complex B-G are expressed in the intestinal epithelium in the chicken. Immunogenetics. 1990;32:45–50. doi: 10.1007/BF01787328. [DOI] [PubMed] [Google Scholar]
- Miller M. M., Goto R. M., Taylor R. L., Jr., Zoorob R., Auffray C., Briles R. W., Briles W. E., Bloom S. E. Assignment of Rfp-Y to the chicken major histocompatibility complex/NOR microchromosome and evidence for high-frequency recombination associated with the nucleolar organizer region. Proc. Natl. Acad. Sci. USA. 1996;93:3958–3962. doi: 10.1073/pnas.93.9.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. M., Robinson C. M., Abernathy J., Goto R. M., Hamilton M. K., Zhou H., Delany M. E. Mapping genes to chicken microchromosome 16 and discovery of olfactory and scavenger receptor genes near the major histocompatibility complex. J. Hered. 2014;105:203–215. doi: 10.1093/jhered/est091. [DOI] [PubMed] [Google Scholar]
- Miller M. M., Wang C., Parisini E., Coletta R. D., Goto R. M., Lee S. Y., Barral D. C., Townes M., Roura-Mir C., Ford H. L., Brenner M. B., Dascher C. C. Characterization of two avian MHC-like genes reveals an ancient origin of the CD1 family. Proc. Natl. Acad. Sci. USA. 2005;102:8674–8679. doi: 10.1073/pnas.0500105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norup L. R., Dalgaard T. S., Pedersen A. R., Juul-Madsen H. R. Assessment of Newcastle disease-specific T cell proliferation in different inbred MHC chicken lines. Scand. J. Immunol. 2011;74:23–30. doi: 10.1111/j.1365-3083.2011.02534.x. [DOI] [PubMed] [Google Scholar]
- Norup L. R., Dalgaard T. S., Pleidrup J., Permin A., Schou T. W., Jungersen G., Fink D. R., Juul-Madsen H. R. Comparison of parasite-specific immunoglobulin levels in two chicken lines during sustained infection with Ascaridia galli. Vet. Parasitol. 2013;191:187–190. doi: 10.1016/j.vetpar.2012.07.031. [DOI] [PubMed] [Google Scholar]
- O'Neill A. M., Livant E. J., Ewald S. J. The chicken BF1 (classical MHC class I) gene shows evidence of selection for diversity in expression and in promoter and signal peptide regions. Immunogenetics. 2009;61:289–302. doi: 10.1007/s00251-008-0354-7. [DOI] [PubMed] [Google Scholar]
- Owen J. P., Delany M. E., Cardona C. J., Bickford A. A., Mullens B. A. Host inflammatory response governs fitness in an avian ectoparasite, the northern fowl mite (Ornithonyssus sylviarum) Int. J. Parasitol. 2009;39:789–799. doi: 10.1016/j.ijpara.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Owen J. P., Delany M. E., Mullens B. A. MHC haplotype involvement in avian resistance to an ectoparasite. Immunogenetics. 2008;60:621–631. doi: 10.1007/s00251-008-0314-2. [DOI] [PubMed] [Google Scholar]
- Pharr G. T., Bacon L. D., Dodgson J. B. A class I cDNA from SPAFAS line-11 chickens. Eur. J. Immunogenet. 1994;21:59–66. doi: 10.1111/j.1744-313x.1994.tb00176.x. [DOI] [PubMed] [Google Scholar]
- Pharr G. T., Dodgson J. B., Hunt H. D., Bacon L. D. Class II MHC cDNAs in 15I5 B-congenic chickens. Immunogenetics. 1998;47:350–354. doi: 10.1007/s002510050369. [DOI] [PubMed] [Google Scholar]
- Pharr G. T., Gwynn A. V., Bacon L. D. Histocompatibility antigen(s) linked to Rfp-Y (Mhc-like) genes in the chicken. Immunogenetics. 1996;45:52–58. doi: 10.1007/s002510050166. [DOI] [PubMed] [Google Scholar]
- Pharr G. T., Hunt H. D., Bacon L. D., Dodgson J. B. Identification of class II major histocompatibility complex polymorphisms predicted to be important in peptide antigen presentation. Poult. Sci. 1993;72:1312–1317. doi: 10.3382/ps.0721312. [DOI] [PubMed] [Google Scholar]
- Pharr G. T., Vallejo R. L., Bacon L. D. Identification of Rfp-Y (Mhc-like) haplotypes in chickens of Cornell lines N and P. J. Hered. 1997;88:504–512. doi: 10.1093/oxfordjournals.jhered.a023145. [DOI] [PubMed] [Google Scholar]
- Pinard-Van Der Laan M. H., Monvoisin J. L., Pery P., Hamet N., Thomas M. Comparison of outbred lines of chickens for resistance to experimental infection with coccidiosis (Eimeria tenella) Poult. Sci. 1998;77:185–191. doi: 10.1093/ps/77.2.185. [DOI] [PubMed] [Google Scholar]
- Pinard-van der Laan M. H., Soubieux D., Merat L., Bouret D., Luneau G., Dambrine G., Thoraval P. Genetic analysis of a divergent selection for resistance to Rous sarcomas in chickens. Genet. Sel. Evol. 2004;36:65–81. doi: 10.1186/1297-9686-36-1-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pink J. R. L., Droege W., Hala K., Miggiano V. C., Ziegler A. A three-locus model for the chicken major histocompatibility complex. Immunogenetics. 1977;5:203–216. [Google Scholar]
- Plachy J., Benda V. Location of the gene responsible for Rous sarcoma regression in the B-F region of the B complex (MHC) of the chicken. Folia Biol. (Praha). 1981;27:363–368. [PubMed] [Google Scholar]
- Plachy J., Hala K., Hejnar J., Geryk J., Svoboda J. src-specific immunity in inbred chickens bearing v-src DNA- and RSV-induced tumors. Immunogenetics. 1994;40:257–265. doi: 10.1007/BF00189970. [DOI] [PubMed] [Google Scholar]
- Plachy J., Jurajda V., Benda V. Resistance to Marek's disease is controlled by a gene within the B-F region of the chicken major histocompatibility complex in Rous sarcoma regressor or progressor inbred lines of chickens. Folia Biol. (Praha). 1984;30:251–258. [PubMed] [Google Scholar]
- Praharaj N., Beaumont C., Dambrine G., Soubieux D., Merat L., Bouret D., Luneau G., Alletru J. M., Pinard-Van der Laan M. H., Thoraval P., Mignon-Grasteau S. Genetic analysis of the growth curve of Rous sarcoma virus-induced tumors in chickens. Poult. Sci. 2004;83:1479–1488. doi: 10.1093/ps/83.9.1479. [DOI] [PubMed] [Google Scholar]
- Reemers S. S., van Haarlem D. A., Sijts A. J., Vervelde L., Jansen C. A. Identification of novel avian influenza virus derived CD8+ T-cell epitopes. PLoS ONE. 2012;7:e31953. doi: 10.1371/journal.pone.0031953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S., Shaw I., Ross N., Nair V., Rothwell L., Kaufman J., Kaiser P. Analysis of part of the chicken Rfp-Y region reveals two novel lectin genes, the first complete genomic sequence of a class I alpha-chain gene, a truncated class II beta-chain gene, and a large CR1 repeat. Immunogenetics. 2003;55:100–108. doi: 10.1007/s00251-003-0553-1. [DOI] [PubMed] [Google Scholar]
- Rogers S. L., Kaufman J. High allelic polymorphism, moderate sequence diversity and diversifying selection for B-NK but not B-lec, the pair of lectin-like receptor genes in the chicken MHC. Immunogenetics. 2008;60:461–475. doi: 10.1007/s00251-008-0307-1. [DOI] [PubMed] [Google Scholar]
- Rogers S. L., Gobel T. W., Viertlboeck B. C., Milne S., Beck S., Kaufman J. Characterization of the chicken C-type lectin-like receptors B-NK and B-lec suggests that the NK complex and the MHC share a common ancestral region. J. Immunol. 2005;174:3475–3483. doi: 10.4049/jimmunol.174.6.3475. [DOI] [PubMed] [Google Scholar]
- Ruby T., Bed'Hom B., Wittzell H., Morin V., Oudin A., Zoorob R. Characterisation of a cluster of TRIM-B30.2 genes in the chicken MHC B locus. Immunogenetics. 2005;57:116–128. doi: 10.1007/s00251-005-0770-x. [DOI] [PubMed] [Google Scholar]
- Ruff M. D., Bacon L. D. Eimeria acervulina and Eimeria tenella in 15.B-congenic White Leghorns. Poult. Sci. 1989;68:380–385. doi: 10.3382/ps.0680380. [DOI] [PubMed] [Google Scholar]
- Salomonsen J., Chattaway J. A., Chan A. C., Parker A., Huguet S., Marston D. A., Rogers S. L., Wu Z., Smith A. L., Staines K., Butter C., Riegert P., Vainio O., Nielsen L., Kaspers B., Griffin D. K., Yang F., Zoorob R., Guillemot F., Auffray C., Beck S., Skjodt K., Kaufman J. Sequence of a complete chicken BG haplotype shows dynamic expansion and contraction of two gene lineages with particular expression patterns. PLoS Genet. 2014;10:e1004417. doi: 10.1371/journal.pgen.1004417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonsen J., Dunon D., Skjodt K., Thorpe D., Vainio O., Kaufman J. Chicken major histocompatibility complex-encoded B-G antigens are found on many cell types that are important for the immune system. Proc. Natl. Acad. Sci. USA. 1991;88:1359–1363. doi: 10.1073/pnas.88.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonsen J., Marston D., Avila D., Bumstead N., Johansson B., Juul-Madsen H., Olesen G. D., Riegert P., Skjodt K., Vainio O., Wiles M. V., Kaufman J. The properties of the single chicken MHC classical class II alpha chain (B-LA) gene indicate an ancient origin for the DR/E-like isotype of class II molecules. Immunogenetics. 2003;55:605–614. doi: 10.1007/s00251-003-0620-7. [DOI] [PubMed] [Google Scholar]
- Salomonsen J., Sorensen M. R., Marston D. A., Rogers S. L., Collen T., van Hateren A., Smith A. L., Beal R. K., Skjodt K., Kaufman J. Two CD1 genes map to the chicken MHC, indicating that CD1 genes are ancient and likely to have been present in the primordial MHC. Proc. Natl. Acad. Sci. USA. 2005;102:8668–8673. doi: 10.1073/pnas.0409213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schat K. A., Taylor R. L., Jr., Briles W. E. Resistance to Marek's disease in chickens with recombinant haplotypes to the major histocompatibility (B) complex. Poult. Sci. 1994;73:502–508. doi: 10.3382/ps.0730502. [DOI] [PubMed] [Google Scholar]
- Schierman L., Nordskog A. Relationship of blood type to histocompatibility in chickens. Science. 1961;134:1008–1009. doi: 10.1126/science.134.3484.1008. [DOI] [PubMed] [Google Scholar]
- Schierman L. W., Watanabe D. H., McBride R. A. Increased growth of Rous sarcomas in chickens pretreated with formalinized syngeneic tumor cells. Eur. J. Immunol. 1977;7:710–713. doi: 10.1002/eji.1830071012. [DOI] [PubMed] [Google Scholar]
- Schou T., Permin A., Roepstorff A., Sorensen P., Kjaer J. Comparative genetic resistance to Ascaridia galli infections of 4 different commercial layer-lines. Br. Poult. Sci. 2003;44:182–185. doi: 10.1080/0007166031000088335. [DOI] [PubMed] [Google Scholar]
- Schou T. W., Labouriau R., Permin A., Christensen J. P., Sorensen P., Cu H. P., Nguyen V. K., Juul-Madsen H. R. MHC haplotype and susceptibility to experimental infections (Salmonella enteritidis, Pasteurella multocida or Ascaridia galli) in a commercial and an indigenous chicken breed. Vet. Immunol. Immunopathol. 2010;135:52–63. doi: 10.1016/j.vetimm.2009.10.030. [DOI] [PubMed] [Google Scholar]
- Schou T. W., Permin A., Juul-Madsen H. R., Sorensen P., Labouriau R., Nguyen T. L., Fink M., Pham S. L. Gastrointestinal helminths in indigenous and exotic chickens in Vietnam: association of the intensity of infection with the Major Histocompatibility Complex. Parasitology. 2007;134:561–573. doi: 10.1017/S0031182006002046. [DOI] [PubMed] [Google Scholar]
- Schulten E. S., Briles W. E., Taylor R. L., Jr Rous sarcoma growth in lines congenic for major histocompatibility (B) complex recombinants. Poult. Sci. 2009;88:1601–1607. doi: 10.3382/ps.2009-00085. [DOI] [PubMed] [Google Scholar]
- Shaw I., Powell T. J., Marston D. A., Baker K., van Hateren A., Riegert P., Wiles M. V., Milne S., Beck S., Kaufman J. Different evolutionary histories of the two classical class I genes BF1 and BF2 illustrate drift and selection within the stable MHC haplotypes of chickens. J. Immunol. 2007;178:5744–5752. doi: 10.4049/jimmunol.178.9.5744. [DOI] [PubMed] [Google Scholar]
- Sherman M. A., Goto R. M., Moore R. E., Hunt H. D., Lee T. D., Miller M. M. Mass spectral data for 64 eluted peptides and structural modeling define peptide binding preferences for class I alleles in two chicken MHC-B haplotypes associated with opposite responses to Marek's disease. Immunogenetics. 2008;60:527–541. doi: 10.1007/s00251-008-0302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina T., Briles W. E., Goto R. M., Hosomichi K., Yanagiya K., Shimizu S., Inoko H., Miller M. M. Extended gene map reveals tripartite motif, C-type lectin, and Ig superfamily type genes within a subregion of the chicken MHC-B affecting infectious disease. J. Immunol. 2007;178:7162–7172. doi: 10.4049/jimmunol.178.11.7162. [DOI] [PubMed] [Google Scholar]
- Solinhac R., Leroux S., Galkina S., Chazara O., Feve K., Vignoles F., Morisson M., Derjusheva S., Bed'hom B., Vignal A., Fillon V., Pitel F. Integrative mapping analysis of chicken microchromosome 16 organization. BMC Genomics. 2010;11:616. doi: 10.1186/1471-2164-11-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spike C. A., Bumstead N., Crittenden L. B., Lamont S. J. RFLP mapping of expressed sequence tags in the chicken. J. Hered. 1996;87:6–9. doi: 10.1093/oxfordjournals.jhered.a022954. [DOI] [PubMed] [Google Scholar]
- Svoboda J., Plachy J., Hejnar J., Karakoz I., Guntaka R. V., Geryk J. Tumor induction by the LTR, v-src, LTR DNA in four B (MHC) congenic lines of chickens. Immunogenetics. 1992;35:309–315. doi: 10.1007/BF00189893. [DOI] [PubMed] [Google Scholar]
- Taylor R. L., Jr Major histocompatibility (B) complex control of responses against Rous sarcomas. Poult. Sci. 2004;83:638–649. doi: 10.1093/ps/83.4.638. [DOI] [PubMed] [Google Scholar]
- Taylor R. L., Jr., Ewert D. L., England J. M., Halpern M. S. Major histocompatibility (B) complex control of the growth pattern of v-src DNA-induced primary tumors. Virology. 1992;191:477–479. doi: 10.1016/0042-6822(92)90214-a. [DOI] [PubMed] [Google Scholar]
- Telfer J. C., Baldwin C. L. Bovine gamma delta T cells and the function of gamma delta T cell specific WC1 co-receptors. Cell. Immunol. 2015;296:76–86. doi: 10.1016/j.cellimm.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Taylor R. L., Jr., England J. M., Kopen G. C., Christou A. A., Halpern M. S. Major histocompatibility (B) complex control of the formation of v-src-induced metastases. Virology. 1994;205:569–573. doi: 10.1006/viro.1994.1681. [DOI] [PubMed] [Google Scholar]
- Thacker E. L., Fulton J. E., Hunt H. D. In vitro analysis of a primary, major histocompatibility complex (MHC)- restricted, cytotoxic T-lymphocyte response to avian leukosis virus (ALV), using target cells expressing MHC class I cDNA inserted into a recombinant ALV vector. J. Virol. 1995;69:6439–6444. doi: 10.1128/jvi.69.10.6439-6444.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoraval P., Afanassieff M., Bouret D., Luneau G., Esnault E., Goto R. M., Chausse A. M., Zoorob R., Soubieux D., Miller M. M., Dambrine G. Role of nonclassical class I genes of the chicken major histocompatibility complex Rfp-Y locus in transplantation immunity. Immunogenetics. 2003;55:647–651. doi: 10.1007/s00251-003-0618-1. [DOI] [PubMed] [Google Scholar]
- Tilquin P., Barrow P. A., Marly J., Pitel F., Plisson-Petit F., Velge P., Vignal A., Baret P. V., Bumstead N., Beaumont C. A genome scan for quantitative trait loci affecting the Salmonella carrier-state in the chicken. Genet. Sel. Evol. 2005;37:539–561. doi: 10.1186/1297-9686-37-6-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uni Z., Gutman M., Leitner G., Landesman E., Heller D., Cahaner A. Major histocompatibility complex class IV restriction fragment length polymorphism markers in replicated meat-type chicken lines divergently selected for high or low early immune response. Poult. Sci. 1993;72:1823–1831. doi: 10.3382/ps.0721823. [DOI] [PubMed] [Google Scholar]
- Vainio O., Koch C., Toivanen A. B-L antigens (class II) of the chicken major histocompatibility complex control T-B cell interaction. Immunogenetics. 1984;19:131–140. doi: 10.1007/BF00387856. [DOI] [PubMed] [Google Scholar]
- Vallejo R. L., Pharr G. T., Liu H. C., Cheng H. H., Witter R. L., Bacon L. D. Non-association between Rfp-Y major histocompatibility complex-like genes and susceptibility to Marek's disease virus-induced tumours in 6(3) x 7(2) F2 intercross chickens. Anim. Genet. 1997;28:331–337. doi: 10.1111/j.1365-2052.1997.00178.x. [DOI] [PubMed] [Google Scholar]
- Van Gorp H., Delputte P. L., Nauwynck H. J. Scavenger receptor CD163, a Jack-of-all-trades and potential target for cell-directed therapy. Mol. Immunol. 2010;47:1650–1660. doi: 10.1016/j.molimm.2010.02.008. [DOI] [PubMed] [Google Scholar]
- van Hateren A., Carter R., Bailey A., Kontouli N., Williams A. P., Kaufman J., Elliott T. A mechanistic basis for the co-evolution of chicken tapasin and major histocompatibility complex class I (MHC I) proteins. J. Biol. Chem. 2013;288:32797–32808. doi: 10.1074/jbc.M113.474031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. A., Hunt L. G., Sowa A. K., Skjodt K., Gobel T. W., Lehner P. J., Kaufman J. The dominantly expressed class I molecule of the chicken MHC is explained by coevolution with the polymorphic peptide transporter (TAP) genes. Proc. Natl. Acad. Sci. USA. 2011;108:8396–8401. doi: 10.1073/pnas.1019496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. A., van Hateren A., Milne S., Beck S., Kaufman J. Chicken TAP genes differ from their human orthologues in locus organisation, size, sequence features and polymorphism. Immunogenetics. 2005;57:232–247. doi: 10.1007/s00251-005-0786-2. [DOI] [PubMed] [Google Scholar]
- Wallny H. J., Avila D., Hunt L. G., Powell T. J., Riegert P., Salomonsen J., Skjodt K., Vainio O., Vilbois F., Wiles M. V., Kaufman J. Peptide motifs of the single dominantly expressed class I molecule explain the striking MHC-determined response to Rous sarcoma virus in chickens. Proc. Natl. Acad. Sci. USA. 2006;103:1434–1439. doi: 10.1073/pnas.0507386103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock D., Schat K. A., Calnek B. W. Cytotoxic T lymphocytes in reticuloendotheliosis virus-infected chickens. Eur. J. Immunol. 1989;19:267–272. doi: 10.1002/eji.1830190208. [DOI] [PubMed] [Google Scholar]
- White E. C., Briles W. E., Briles R. W., Taylor R. L., Jr Response of six major histocompatibility (B) complex recombinant haplotypes to Rous sarcomas. Poult. Sci. 1994;73:836–842. doi: 10.3382/ps.0730836. [DOI] [PubMed] [Google Scholar]
- Worley K., Collet J., Spurgin L. G., Cornwallis C., Pizzari T., Richardson D. S. MHC heterozygosity and survival in red junglefowl. Mol. Ecol. 2010;19:3064–3075. doi: 10.1111/j.1365-294X.2010.04724.x. [DOI] [PubMed] [Google Scholar]
- Wu G., Liu L., Qi Y., Sun Y., Yang N., Xu G., Zhou H., Li X. Splenic gene expression profiling in White Leghorn layer inoculated with the Salmonella enterica serovar Enteritidis. Anim. Genet. 2015;46:617–626. doi: 10.1111/age.12341. [DOI] [PubMed] [Google Scholar]
- Yoo B. H., Sheldon B. L. Association of the major histocompatibility complex with avian leukosis virus infection in chickens. Br. Poult. Sci. 1992;33:613–620. doi: 10.1080/00071669208417500. [DOI] [PubMed] [Google Scholar]
- Zajonc D. M., Striegl H., Dascher C. C., Wilson I. A. The crystal structure of avian CD1 reveals a smaller, more primordial antigen-binding pocket compared to mammalian CD1. Proc. Natl. Acad. Sci. USA. 2008;105:17925–17930. doi: 10.1073/pnas.0809814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Chen Y., Qi J., Gao F., Liu Y., Liu J., Zhou X., Kaufman J., Xia C., Gao G. F. Narrow groove and restricted anchors of MHC class I molecule BF2* 0401 plus peptide transporter restriction can explain disease susceptibility of B4 chickens. J. Immunol. 2012a;189:4478–4487. doi: 10.4049/jimmunol.1200885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Katselis G. S., Moore R. E., Lekpor K., Goto R. M., Hunt H. D., Lee T. D., Miller M. M. MHC class I target recognition, immunophenotypes and proteomic profiles of natural killer cells within the spleens of day-14 chick embryos. Dev. Comp. Immunol. 2012b;37:446–456. doi: 10.1016/j.dci.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Zhou H., Lamont S. J. Chicken MHC class I and II gene effects on antibody response kinetics in adult chickens. Immunogenetics. 2003;55:133–140. doi: 10.1007/s00251-003-0566-9. [DOI] [PubMed] [Google Scholar]
- Zoorob R., Behar G., Kroemer G., Auffray C. Organization of a functional chicken class II B gene. Immunogenetics. 1990;31:179–187. doi: 10.1007/BF00211553. [DOI] [PubMed] [Google Scholar]
- Zoorob R., Bernot A., Renoir D. M., Choukri F., Auffray C. Chicken major histocompatibility complex class II B genes: analysis of interallelic and interlocus sequence variance. Eur. J. Immunol. 1993;23:1139–1145. doi: 10.1002/eji.1830230524. [DOI] [PubMed] [Google Scholar]
- Zoorob R., Billault A., Severac V., Fillon V., Vignal A., Auffray C. XXV International Conference on Animal Genetics. Tours, France: International Society for Animal Genetics; 1996. Two chicken genomic libraries in the PAC and BAC cloning systems : Organization and characterization. [Google Scholar]