Significance
The Runt-related transcription factors (RUNX) are critical regulators of development. Mutation or dysregulation of RUNX genes have been associated with diverse cancer types. Phosphorylation of the strictly conserved threonine 173 (T173) residue within the Runt domain of RUNX3 disrupts DNA binding activity, which facilitates the recruitment of RUNX proteins to mitotic structures. Moreover, RUNX3 deficiency is associated with delayed mitotic entry. The tight conservation of T173 and its flanking residues from unicellular organism to human, and the fact that mitosis is indispensable to all metazoans, strongly indicate that T173 phosphorylation is fundamental to the role of RUNX in these divergent organisms. Cancer-associated mutation T173I further corroborates the critical role of RUNX phosphorylation in mitosis.
Keywords: RUNX, phosphorylation, mitosis, centrosome, Aurora kinase
Abstract
The Runt-related transcription factors (RUNX) are master regulators of development and major players in tumorigenesis. Interestingly, unlike most transcription factors, RUNX proteins are detected on the mitotic chromatin and apparatus, suggesting that they are functionally active in mitosis. Here, we identify key sites of RUNX phosphorylation in mitosis. We show that the phosphorylation of threonine 173 (T173) residue within the Runt domain of RUNX3 disrupts RUNX DNA binding activity during mitotic entry to facilitate the recruitment of RUNX proteins to mitotic structures. Moreover, knockdown of RUNX3 delays mitotic entry. RUNX3 phosphorylation is therefore a regulatory mechanism for mitotic entry. Cancer-associated mutations of RUNX3 T173 and its equivalent in RUNX1 further corroborate the role of RUNX phosphorylation in regulating proper mitotic progression and genomic integrity.
Cell division is a highly ordered process comprising multiple steps that lead to dramatic changes to the cell architecture. Events such as cell rounding, chromatin condensation, spindle assembly, nuclear envelope disassembly, and cytokinesis involve numerous proteins, whose activities are tightly coordinated by mitotic kinases and proteasome-mediated degradation (1). Given that most transcription has stopped (2), the roles of transcription factors during mitosis are ill explored.
Runt-related transcription factors (RUNX) are master regulators of cell-fate decisions (3). Mutation or dysregulation of RUNX genes have been associated with diverse cancer types (3). Unlike many transcription factors (e.g., Ets-1 and Oct-1) (4), RUNX proteins are retained at the condensed mitotic chromatin, where they maintain epigenetic memory and ensure proper transmission of gene expression patterns to progeny cells; RUNX2 binds to the promoters of various cell cycle- and cell fate-related genes and regulates histone modifications during mitosis (5); RUNX2 and RUNX3 bind to regulatory regions of rRNA genes and are associated with their repression (6, 7). RUNX1 positively regulates the transcription of various spindle assembly checkpoint genes, such as BUB1 and NEK6 (8). These findings suggest that RUNX proteins are important for the accurate transmission of genetic information during mitosis and that defects in RUNX genes might contribute to aneuploidy and loss of cell identity.
Aside from binding to the chromatin, RUNX proteins also associate with microtubules (9, 10). RUNX3 molecules are detected at key mitotic structures such as the centrosome, mitotic spindle, and midbody (11). Likewise, RUNX-binding partner CBFβ was found at the midbody and implicated in cytokinesis (12). The reason why RUNX proteins are present at non-DNA sites (i.e., mitotic apparatus) during mitosis is unknown. An intriguing observation is the hyperphosphorylation of RUNX proteins during mitosis (13, 14). RUNX2 is phosphorylated by mitotic kinase CDK1-cyclin B1 (14, 15) and dephosphorylated at mitotic exit by the PP1/PP2A phosphatase (14). CDK1-mediated phosphorylation of RUNX2 enhanced DNA binding activity, suggesting a role for RUNX2 in G2/M progression (15). In addition, CDK1/2 phosphorylates RUNX1, promoting its degradation by CDC20-associated anaphase-promoting complex during the late stages of mitosis (13, 16). However, despite these findings, the significance of RUNX hyperphosphorylation in mitosis remains unclear.
Results
RUNX Proteins Are Hyperphosphorylated at Mitosis.
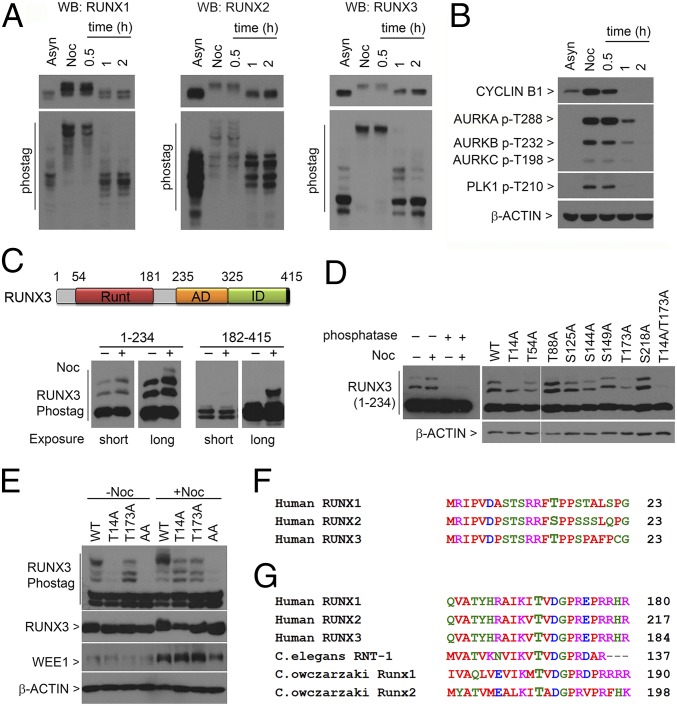
The localization of RUNX proteins at mitotic structures suggests direct involvement of RUNX proteins in mitosis. Because mitosis is regulated by phosphorylation events, we investigated RUNX phosphorylation using Phos-tag gel electrophoresis. In asynchronously (Asyn) growing cells, different migration patterns of phosphorylated RUNX1, -2, and -3 suggest unique phosphorylation signatures for each RUNX protein (Fig. 1A, lane Asyn). Cells were then arrested with nocodazole (Noc), released from nocodazole block, and analyzed for RUNX phosphorylation. During prometaphase, which was associated with high levels of cyclin B1 and activated mitotic kinases (Fig. 1B, lane Noc), we detected marked reduction of RUNX mobility (Fig. 1A, lane Noc). The retardation of RUNX mobility indicated multiple phosphorylation sites in RUNX during mitosis. The removal of nocodazole resulted in rapid dephosphorylation of RUNX proteins (Fig. 1A, lane 1 h) as cells progressed to telophase (Fig. S1).
Fig. 1.
T14 and T173 are important for RUNX3 hyperphosphorylation in mitosis. (A) U2OS cells were synchronized with nocodazole (Noc), collected by mitotic shake-off, replated in media without nocodazole, and harvested at the indicated times. Lysates were electrophoresed in conventional SDS/PAGE (Top) and Phos-tag gels (Bottom) for immunoblotting with the indicated antibodies. Asyn, asynchronous. (B) Lysates from A were immunoblotted with the indicated antibodies. (C, Top) Schematic of human RUNX3. Runt domain, DNA-binding domain; AD, activation domain; ID, inhibitory domain. (Bottom) Flag-tagged truncation constructs of RUNX3 spanning amino acids 1–234 and 182–415 were transfected into COS7 cells. After 28 h, cells were cultured with (+) or without (−) nocodazole. Nocodazole-treated cells were harvested by mitotic shake-off. Lysates were immunoblotted with anti-Flag antibodies. Short and long exposure times were indicated. (D) Phos-tag electrophoresis of lysates containing Flag-RUNX3 (amino acids 1–234). (Left) Lysates treated with lambda-protein phosphatase. (Right) Point mutants of Flag-RUNX3 (amino acids 1–234) were transfected into COS7 cells and treated with nocodazole. Immunoblots were probed with anti-Flag antibodies. WT, wild type. (E) T14 and T173 mutations were introduced into full-length Flag-tagged RUNX3 and transfected into COS7 cells. Transfectants were treated as in C and immunoblotted with the indicated antibodies. (F and G) Alignments of the regions flanking T14 and T173 in human RUNX3 with human RUNX1, RUNX2, C. elegans RNT-1, and C. owczarzaki RUNX1 and RUNX2. Protein sequence alignments were done with ClustalW (www.ebi.ac.uk/Tools/msa/clustalw2).
Fig. S1.
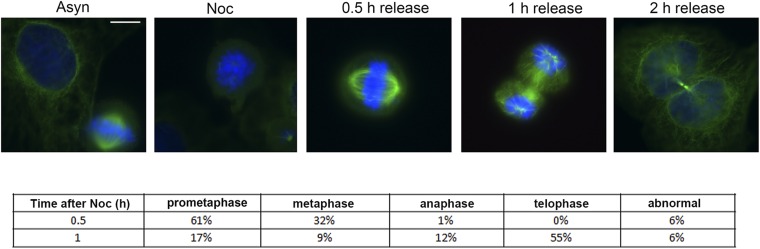
Immunofluorescence staining of U2OS cells after release from nocodazole treatment. Cells from Fig. 1 were analyzed by immunofluorescence microscopy with α-tubulin antibody (green). DNA was stained with DAPI (blue). Asyn, asynchronous cells; Noc, nocodazole treatment. (Scale bar, 10 µm.)
T173 and T14 Are Critical for Mitotic-Specific Hyperphosphorylation of RUNX3.
Because RUNX3 is believed to be the evolutionary founder of the human RUNX family (17), we focus on RUNX3 phosphorylation. To identify phosphorylation sites of RUNX3 in mitosis, we transiently expressed truncated forms of RUNX3 in COS7 cells and examined their phosphorylation state in the presence of nocodazole. In asynchronously growing cells, both the N and C terminus of RUNX3 were phosphorylated, as indicated by slower migrating forms (Fig. 1C). In mitotic cells, there was an increase in the phosphorylation of the N terminus of RUNX3 (amino acids 1–234) (Fig. 1C, lane 4 from Left). Phosphatase treatment led to the collapse of the two prominent slower migrating bands, whereas the remaining rapidly migrating band indicated completely dephosphorylated RUNX3 (Fig. 1D). Thus, there are multiple phosphorylation sites in the N terminus of RUNX3, where differences in mobility reflected the degree of phosphorylation. For untreated cells, the C terminus (amino acids 182–415) separated into two main bands, suggesting that ∼50% of the C terminus construct was phosphorylated (Fig. 1C, lanes 5 and 6). Nocodazole treatment resulted in an additional upward shift in mobility, indicating mitosis-specific phosphorylation of the C terminus domain (Fig. 1C, lanes 7 and 8). This result is consistent with studies that showed that the C termini of RUNX1 and RUNX2 harbor mitotic kinase CDK1 phosphorylation sites (13–16).
To identify phosphorylation sites in the RUNX3 N terminus, we introduced individual point mutations to abolish putative phosphorylation sites in the truncated RUNX3 protein (amino acids 1–234). Two mutants, threonine 14 alanine (T14A) and threonine 173 alanine (T173A), exhibited reduced phosphorylation during nocodazole treatment—instead of the two slower migrating bands, there was only one of intermediate mobility, aside from the unphosphorylated RUNX3 (Fig. 1D, lanes T14A and T173A). The double mutant T14A/T173A showed severe dephosphorylation (Fig. 1D, lane T14A/T173A), indicating that residues T14 and T173 are major phosphorylation sites in early mitosis. To eliminate any artifact arising from truncation of the RUNX3 protein, these point mutations, either individually or in combination, were introduced into the full length RUNX3 cDNA (Fig. 1E). In exponentially growing cells, the intensities of the two fastest migrating bands are reminiscent of the migration patterns of the RUNX3 C terminus truncation (compare Fig. 1C and Fig. 1E, −Noc lanes), indicating that ∼50% of the RUNX3 protein is phosphorylated at the C terminus domain (Fig. 1E). Moreover, the multiple bands in the wild type and T173A mutant reflected multiple phosphorylation sites at the N terminus in untreated cells. By contrast, phosphorylation of the T14A mutant was notably diminished (Fig. 1E, second lane from left). Prior phosphorylation at T14 is therefore important for subsequent phosphorylation events at the N terminus, even in nonmitotic cells. Upon nocodazole treatment, wild-type RUNX3 was hyperphosphorylated, relative to T14A and T173A mutants (Fig. 1E, compare lanes WT, T13A, and T173A). The double mutant T14A/T173A (AA) showed a significant loss of phosphorylation. This finding highlights the importance of both phospho-T173 and phospho-T14 for RUNX3 hyperphosphorylation during mitosis. The T14 residue is conserved in RUNX1 but present as serine in RUNX2 (Fig. 1F). Both were previously shown to be phosphorylated (18–20). T173 phosphorylation has not been reported. The T173 residue is conserved in all RUNX family members, including Runx1 and 2 of the unicellular holozoan Capsaspora owczarzaki and RNT-1 of the simple metazoan Caenorhabditis elegans (Fig. 1G).
Aurora Kinases Are Involved in RUNX Phosphorylation.
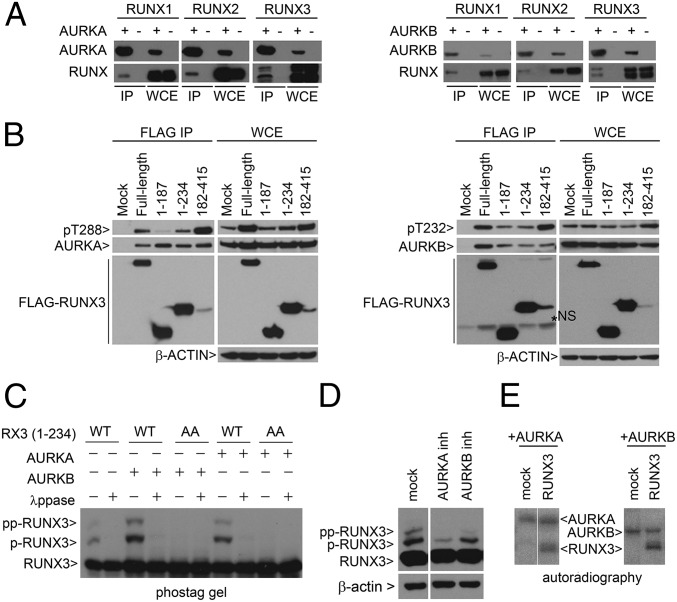
Computational prediction based on the phosphorylation site database (PHOSIDA) algorithm (21) suggests that T173 is a putative Aurora kinase phosphorylation site. Immunoprecipitation of cells cotransfected with RUNX and Aurora kinases showed that all RUNX proteins interacted with Aurora kinases (Fig. 2A). Whereas both the N and the C terminus of RUNX3 interacted with Aurora kinases, the C terminus bound activated Aurora kinases with higher affinity (Fig. 2B). Therefore, the C terminus of RUNX3 is either (i) a possible docking site for active Aurora kinases or (ii) promotes activation of Aurora kinases. Notably, coexpression of RUNX3 with Aurora A or B kinase led to a marked increase in RUNX3 phosphorylation (Fig. 2C). Adding small molecule inhibitors of Aurora A or B kinases during nocodazole treatment resulted in decreased phosphorylation of RUNX3 (amino acids 1–234) (Fig. 2D). Finally, in vitro kinase assays using recombinant proteins showed that Aurora A and B directly phosphorylated RUNX3 (Fig. 2E).
Fig. 2.
RUNX proteins interact with Aurora kinases A and B. (A) HEK293T cells were cotransfected with various combinations of RUNX1, RUNX2, RUNX3, Flag-AURKA, and GFP-AURKB expression vectors. Immunoprecipitation (IP) was performed using anti-Flag M2 beads to enrich for AURKA (Left) and GFP affinity beads to enrich for AURKB (Right), respectively. RUNX proteins were detected with anti–pan-RUNX antibody. WCE, whole cell extract. (B) COS7 cells were transfected with Flag-RUNX3 truncation constructs and GST-AURKA or AURKB expression constructs. Flag-RUNX3 constructs were precipitated by M2 beads. Bound AURKA (Left) or AURKB (Right) proteins were detected by immunoblot. (C) WT RUNX3 (amino acids 1–234) or double mutant AA (T14A/T173A) were transfected into COS7 with AURKA or AURKB expression plasmids. λppase, lambda phosphatase. Flag antibody was used to detect RUNX3 proteins. RUNX3, p-RUNX3, and pp-RUNX3 refer to the different degrees of phosphorylation. Absence of p refers to unphosphorylated RUNX3. (D) COS7 cells were transfected with Flag-RUNX3 (amino acids 1–234). Cells were treated with nocodazole, followed by inhibitors (inh) of AURKA (MLN8237, 5 µM) and AURKB (ZM4473439, 2 µM) kinase inhibitors for 4 h, and harvested by mitotic shake-off. (E) In vitro kinase reaction of recombinant RUNX3 with AURKA or AURKB in the presence of [γ32P]ATP. Proteins were resolved by SDS/PAGE and visualized by autoradiography.
Phosphorylated T173 RUNX Is Located at Mitotic Structures.
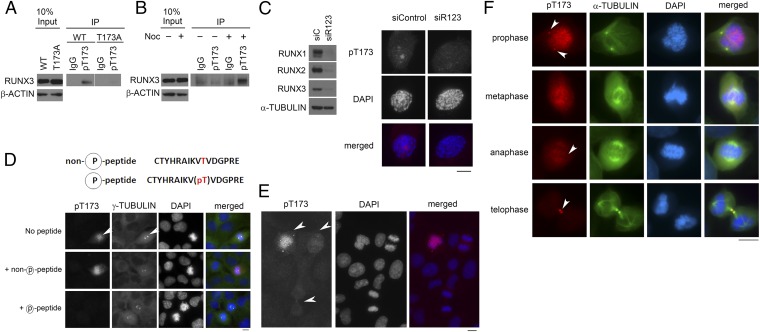
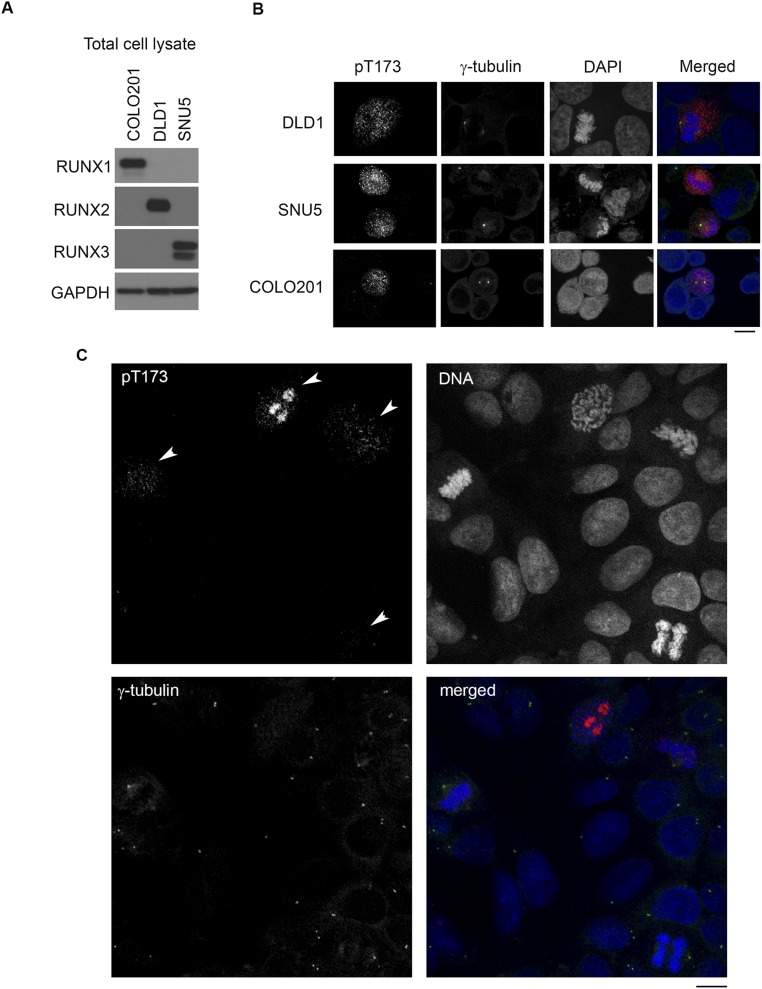
To ascertain if T173 is phosphorylated in vivo, we generated an antibody that specifically recognizes phosphorylated T173. Phospho-T173 antibody immunoprecipitated wild-type RUNX3 (amino acids 1–234), but not the nonphosphorylatable T173A mutant (Fig. 3A). Moreover, the phospho-T173 antibody binds preferentially to full-length RUNX3 from mitotic cells (Fig. 3B). The antibody was unfortunately not suitable for immunoblot. Nevertheless, these results indicate that T173 is preferentially phosphorylated in vivo during mitosis. We determined the cellular distribution of T173-phosphorylated RUNX by performing immunostaining with phospho-T173 antibody. Because the sequence of the synthetic phosphopeptide used as antigen is highly homologous for all RUNX proteins with only one amino acid difference in RUNX1 (Fig. 1G), the antibody staining likely reflects the subcellular localization of all RUNX proteins. siRNA-mediated knockdown of RUNX1, RUNX2, and RUNX3 (Fig. 3C), as well as competition with excess phospho-T173 peptide (Fig. 3D), significantly reduced staining intensity, thus validating the antibody for immunostaining. Cell lines COLO201, DLD1, and SNU5, with predominant RUNX1, RUNX2, or RUNX3 expression, respectively (Fig. S2A), were also prominently stained by the phospho-T173 antibody during mitosis, compared with neighboring interphase cells, thus indicating that the antibody detects all three RUNX proteins specifically in mitosis (Fig. S2B). Cells at early mitosis were strongly stained by phospho-T173 antibody (Fig. 3E for COS7; Fig. S2C for DLD1 cells). Staining intensity was strongest at regions not stained with DAPI (Fig. 3C), suggesting that T173 phosphorylation might inhibit RUNX association with chromatin. Moreover, we detected phospho-T173 staining at the centrosome (indicated by γ-tubulin in Fig. 3D) and midbody (indicated by α-tubulin in Fig. 3F). Phospho-T173 staining was detected at the centrosomes early in mitosis; as the cells progress to telophase, the phospho-T173 staining diminished and was mainly detected at the midbody (Fig. 3F). The elevated levels of phospho-T173 at the centrosome, midzone, and midbody are reminiscent of the respective locations of Aurora A and B (22).
Fig. 3.
Dynamic distribution of T173-phosphorylated RUNX3 in mitotic cells. (A) COS7 cells were transfected with Flag-tagged RUNX3 (amino acids 1–234) or T173A mutant and treated with nocodazole. Immunoprecipitation was done with phospho-T173 (pT173) antibodies. (B) Full-length Flag-tagged RUNX3 was immunoprecipitated from cells cultured with or without nocodazole. Normal rabbit IgG was used as control. (C, Left) Immunoblot of U2OS cells simultaneously transfected with RUNX1, RUNX2, and RUNX3 siRNA for 72 h (indicated as siR123). siControl, control siRNA. (Right) Fluorescence microscopy of cells treated with siRNA targeting RUNX1, RUNX2, and RUNX3. Cells are stained with pT173 antibody (red) and DAPI (blue). Cells at prophase are shown. (D) Competition with peptides to validate pT173 antibody. Cells were costained with pT173 (red), α-tubulin antibody (green), and DAPI (blue) to visualize DNA. Arrows indicate centrosomes. (E) Asynchronous COS7 cells were costained with pT173 antibody and DAPI. White arrows indicate cells at various stages of mitosis. (F) U2OS cells were stained with pT173 (red) and α-tubulin (green); α-tubulin staining was adjusted by Min/Max function of the Axiovision program to eliminate background staining. (Scale bar, 10 µm.)
Fig. S2.
pT173 antibody recognizes RUNX1, RUNX2, and RUNX3 during mitosis. (A) Immunoblot comparing RUNX1, RUNX2, and RUNX3 expression in COLO201, DLD1, and SNU5 cells. A total of 30 µg of total cell lysate was loaded per lane. (B) Immunofluorescence microscopy with pT173 antibody. Cells were imaged by confocal microscopy (Olympus FluoView FV1000). Red, pT173; green, γ-tubulin; blue, DNA. (C) Asynchronous DLD1 cells were stained as in B. Arrows indicate cells at different stages of mitosis. (Scale bars, 10 µm.)
Mutation of T173 Results in Impairment of DNA Binding and Transcription Regulation.
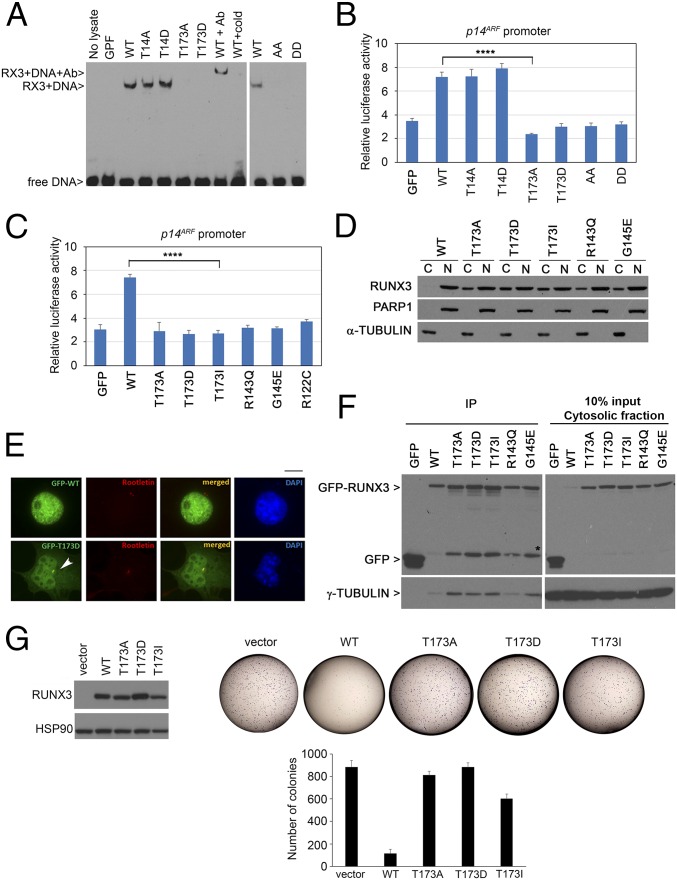
We assessed the effects of T14 and T173 mutations on RUNX3 DNA binding ability. In electrophoretic mobility assay, the T14A and T14D mutations did not affect DNA binding. By contrast, T173A and T173D mutants were defective in DNA binding (Fig. 4A). Interestingly, the T173I mutation was found in RUNX3 in the colon carcinoma cell line LS411, whereas its equivalent mutation in RUNX1 (T196I) was detected in chronic myelomonocytic leukemia (cancer.sanger.ac.uk/cosmic) (23–25); the equivalent residue in RUNX2 was found to be mutated, also to isoleucine, in the disease cleidocranial dysplasia (CCD) (26). This finding indicates that the T173 residue is important for the function of the Runt domain. Crystallography studies showed the T173 residue at the Runt–DNA interface, where it contacts the phosphate backbone of the DNA helix through polar interactions (27, 28). Replacing threonine with a negatively charged (i.e., mimicking phosphorylation) or a neutral amino acid led to defective DNA binding. We next evaluated the transactivation potential of the mutants. Consistent with their DNA binding properties, T14 mutants showed comparable abilities to induce a RUNX responsive promoter (p14ARF) fused to firefly luciferase reporter (Fig. 4B). Mutation at T173 resulted in impairment of transcriptional activation of the p14ARF promoter, similar to other cancer-derived mutations of the Runt domain (Fig. 4C).
Fig. 4.
T173 mutants are impaired in DNA binding and transactivation properties. (A) DNA binding abilities of GFP-RUNX3 and phosphomutants. Nuclear extracts of the transfectants were analyzed by electrophoretic mobility assay using a DNA probe with the RUNX binding site. RUNX3 antibody (Ab) and nonbiotinylated probe (cold) were used for supershift reaction and competition assay, respectively. WT, wild type; GFP, empty GFP vector control; AA, T14A/T173A; DD, T14D/T173D mutants. (B) HEK293T cells were transfected with GFP-RUNX3 constructs, p14ARF promoter fused to firefly luciferase, and β-galactosidase normalization control. Mean values ± SDs are obtained from triplicate experiments. Comparison of WT and T173A, ****P = 0.00003 (Student’s t test). (C) Luciferase reporter assay of Runt domain mutants with the p14ARF promoter. Comparison of WT and T173I, ****P = 0.0001. (D) COS7 cells transfected with GFP-RUNX3 constructs were subjected to biochemical fractionation to isolate cytosolic fraction (C) and nuclear fraction (N). Immunoblot was done with GFP, PARP, and α-tubulin antibodies to detect RUNX3 and validate purity of the nuclear and cytosolic fractions, respectively. (E) GFP-RUNX3 (WT) and GFP-T173D were transfected into COS7 cells for fluorescence microscopy. Rootletin antibody indicated centrosome (red) while DAPI counterstained for DNA (blue), respectively. White arrow indicates centrosome. (Scale bar, 10 µm.) (F, Left,) Immunoprecipitation (IP) of cytosolic fraction in D using GFP beads. (Right) Cytosolic fraction. RUNX3 and bound γ-tubulin were detected by immunoblot with GFP and γ-tubulin antibodies. * indicates GFP-linked degradation products. (G, Left) Expression of pRetro-X-flag-RUNX3 constructs in HeLa Tet-On cells (induced with doxycycline) as detected by immunoblot with anti-flag antibodies. (Right) Colony formation assay of the stable cells at Left. Vector, empty vector control. Representative photographs are shown. The graph shows mean colony numbers ± SDs from three independent experiments.
The strong conservation of T173 and its flanking residues suggest that T173 phosphorylation altered the role of the Runt domain. Subcellular fractionation studies showed increased accumulation of T173 mutants in the cytosolic fraction, whereas wild-type RUNX3 was predominantly nuclear (Fig. 4D). The R143Q and G145E mutations, which affect DNA binding (29), also showed cytoplasmic accumulation, indicating that the cytoplasmic localization of the T173 mutants could be due to their inability to bind DNA. We observed pronounced localization of T173D mutants at the centrosome, relative to the wild type (Fig. 4E). Moreover, immunoprecipitation of cytoplasmic RUNX3 revealed increased binding of the T173 mutants to the centrosomal marker γ-tubulin, compared with the wild type (Fig. 4F). Together, our data indicate that T173 phosphorylation is a molecular switch that dissociates RUNX3 from DNA, inhibits its transcriptional regulatory role, and promotes relocalization to the cytoplasm and centrosome during early mitosis.
RUNX3 suppresses tumor formation through DNA binding and transactivation of growth inhibitory genes (30). To assess the tumorigenic potential of the T173 mutants, stable wild-type RUNX3 and T173 mutant-expressing cells (Fig. 4G, Left) were subjected to colony formation assay. Wild-type RUNX3 inhibited colony formation, in contrast to the T173 mutants (Fig. 4G, Right). DNA binding and transcription activation are therefore critical for RUNX3-mediated tumor suppression, whereas mutation at T173 might promote cancer.
RUNX3 Is Involved in Regulating Mitotic Entry.
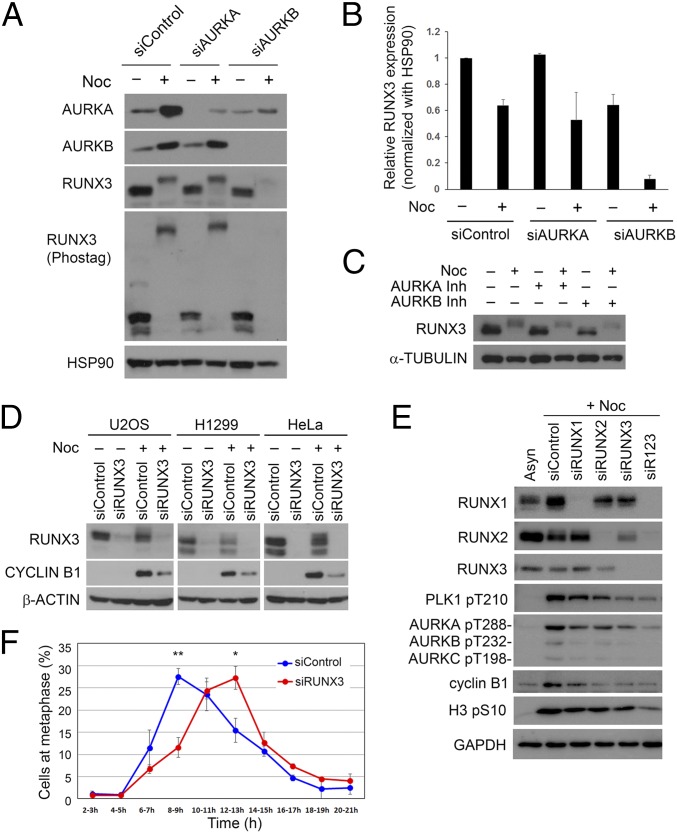
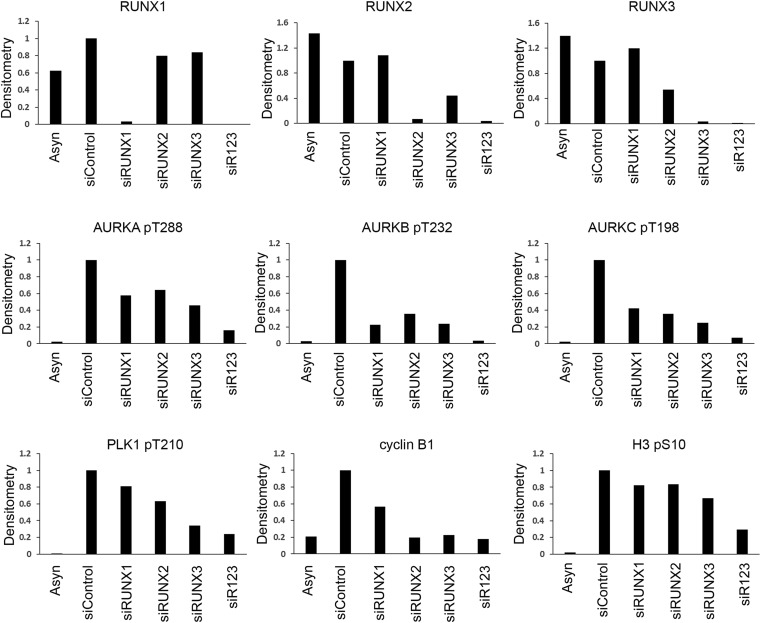
Because Aurora kinases promote RUNX3 phosphorylation, we investigated the effects of inhibiting Aurora kinases on RUNX3. Inhibition of Aurora B activity by siRNA-mediated depletion (Fig. 5 A and B) or Aurora B-specific inhibitor (Fig. 5C) resulted in down-regulation of endogenous RUNX3 protein levels, suggesting that Aurora B enhances RUNX3 protein stability. It is possible that Aurora B stabilizes RUNX3 to ensure proper mitotic progression. Indeed, RUNX3 depletion in three cell types, namely U2OS osteosarcoma, HeLa cervical, and NCI-H1299 lung cancer cells, was associated with reduced cyclin B1 levels during nocodazole treatment (Fig. 5D). We asked if abrogation of other RUNX family members affected cyclin B1 accumulation during nocodazole treatment. U2OS cells were depleted of RUNX proteins by siRNA and treated with nocodazole (Fig. 5E and Fig. S3). Compared with control siRNA-treated cells, individual depletion of RUNX1, RUNX2, or RUNX3 was associated with reduced accumulation of mitotic markers such as cyclin B1, phosphorylated Aurora, and PLK1 kinases. Notably, the effects of individual RUNX depletion were not uniform. Compared with RUNX1 knockdown, RUNX3 knockdown showed a more severe reduction in cyclin B1 and phospho-PLK1 (Fig. 5E and Fig. S3). RUNX2 depletion also revealed higher accumulation of cyclin B1, relative to RUNX1 depletion. Because RUNX3 knockdown also reduced RUNX2 expression (Fig. S3), we were unable to evaluate the relative contributions of individual RUNX proteins to mitosis. Importantly, depletion of all three RUNX proteins (siR123) resulted in a more pronounced reduction of mitotic markers such as phospho-Aurora kinases and phospho-Histone 3 (Fig. 5E and Fig. S3), suggesting functional redundancy for RUNX proteins in mitosis.
Fig. 5.
RUNX3 is involved in regulation of mitosis. (A) U2OS cells were treated with siRNA to deplete AURKA or AURKB proteins. Nocodazole-treated cells (Noc) were harvested by mitotic shake-off. RUNX3 and Aurora proteins were detected by immunoblot. (B) Densitometric analysis of RUNX3 expression after AURKA or AURKB knockdown as in A; three independent experiments were done. (C) U2OS cells were treated with nocodazole before addition of MG132 and Aurora kinase inhibitors (MLN8405 and ZM4473439) for 4 h. (D) Cells were treated with nontargeting siRNA (siControl) or RUNX3 siRNA (siRUNX3). Nocodazole was added for the last 16 h. Immunoblot was done with the indicated antibodies. (E) Immunoblot of U2OS cells treated with siRNA targeting RUNX1, RUNX2, and RUNX3. R123 indicates treatment with all three RUNX siRNA. (F) HeLa.Fucci cells were treated with RUNX3 siRNA and subjected to double thymidine block. Cells were monitored by time-lapse microscopy following release from block. Metaphase cells (visualized as mitotic rounding up) were scored. Numbers of cells counted for siControl and siRUNX3 are 447 and 492, respectively. The data represented mean ± SEM of three independent experiments. *P = 0.03, **P = 0.005 (Student’s t test).
Fig. S3.
Densitometric analysis of protein levels after siRNA knockdown of RUNX1, RUNX2, and RUNX3. The immunoblot in Fig. 5E was quantified for signal intensity with the ImageJ program. All graphs were plotted relative to GAPDH levels.
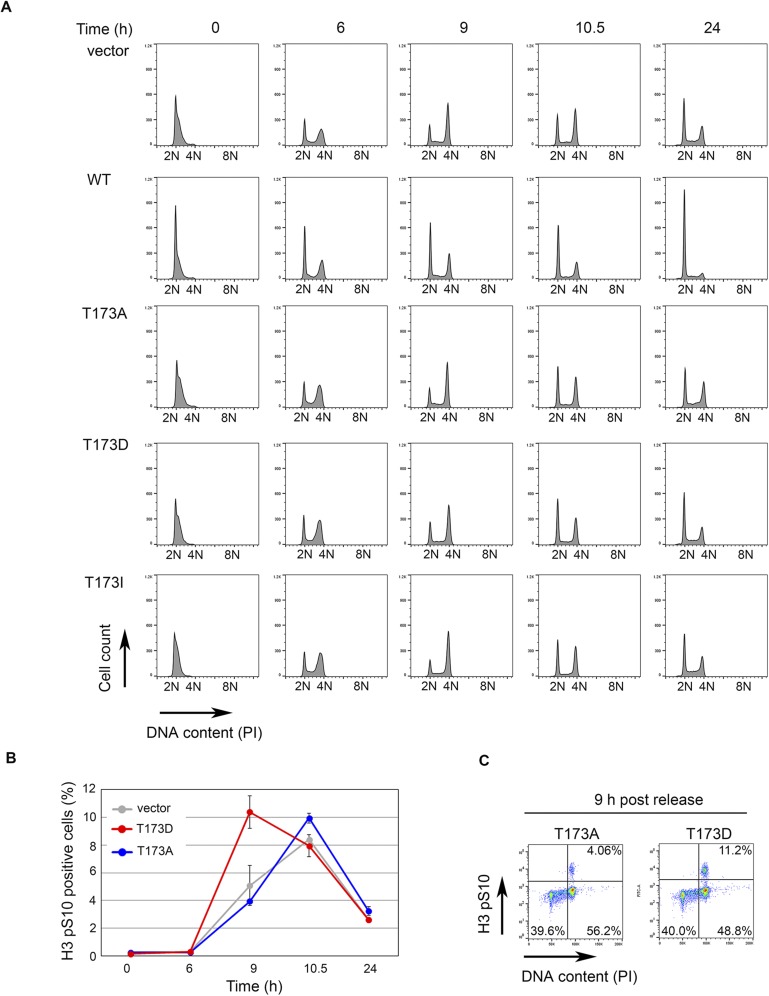
RUNX deficiency is either associated with (ii) failure to enter mitosis or (ii) premature mitotic exit. We asked whether RUNX3 deficiency affects mitotic entry by knocking down RUNX3 in HeLa fluorescent ubiquitination-based cell cycle indicator (Fucci) cells (Fig. 5F). Cells were synchronized at S phase by double thymidine block. After release from the block, time-lapse microscopy was used to follow the progression of cells from early S phase (i.e., cells with green fluorescence) to mitosis. RUNX3 depletion delayed the onset of mitosis, suggesting that RUNX3 promotes mitotic entry. To determine whether T173 phosphorylation contributes to mitotic entry, we subjected cells stably expressing wild-type RUNX3 and T173 mutants to thymidine block and monitored cell cycle progression following release from block (Fig. S4A). Induction of wild-type RUNX3 resulted in a drastic delay in cell cycle progression—a large population of cells remained at the G1 phase (Fig. S4A, WT). This finding is consistent with RUNX3’s transcriptional activity in suppressing cell growth. As expected, the T173 mutants did not delay cells in the G1 phase of the cell cycle and progressed to mitosis. Compared with the vector control and T173A mutant, the T173D mutant accelerated entry into mitosis, as shown by increased numbers of phospho-Histone 3 (Ser 10)-positive cells at 9 h postthymidine release (Fig. S4 B and C). This effect of T173D indicates a role for T173 phosphorylation in facilitating mitotic entry.
Fig. S4.
Flow cytometric analysis of cell cycle progression of pRetro-flag-RUNX3 stable HeLa Tet-On cell lines after release from thymidine block. (A) Histogram showing cell cycle profiles of the indicated p-RetroX-Flag-RUNX3 cell constructs at various time points after release from thymidine block. RUNX3 expression was induced with 0.5 µg/mL of doxycycline 16 h before release from block. Propidium iodide (PI) was used to quantify DNA content and the histogram of PI staining was plotted using FlowJo software. At time 0 h, cells were synchronized at G1/S phase with 2N DNA content. By 24 h, all cell lines except for WT RUNX3 have entered the next cell cycle. (B) Levels of mitotic cells of empty vector, T173A, and T173D cells in A were quantified by phospho-Histone 3 (Ser 10) (H3 pS10) staining. (C) Density plots of T173A and T173D at 9-h time point from B. Percentages of G2 and M subpopulations are indicated by elevated PI and H3 pS10 staining, respectively.
Discussion
Here, we uncover a key phosphorylation site that endows RUNX3 with an unexpected role in the regulation of mitosis. Phosphorylation of T173 inhibits the DNA-binding function of the Runt domain, which in turn facilitates the redistribution of RUNX proteins to mitotic structures such as the centrosome and midbody. Crystallography studies previously showed that T173 and its flanking residues constitute one of the key structural elements integral to the function of the Runt domain (27, 28). The disease derived-mutation T173I (26, 31) (cancer.sanger.ac.uk/cosmic) resulted in drastic destabilization of the Runt domain and impairment of DNA binding. Moreover, the ability of T173 mutants to form colonies on soft agar is indicative of a loss of growth suppressive function, suggesting that T173 phosphorylation must be tightly confined to mitosis. The frequent overexpression of Aurora kinases in cancer might nullify RUNX3 transcription activity during nonmitotic phases and promote tumor growth.
Earlier, we identified an acetylation site at lysine 171 (K171), which promotes interaction between RUNX3 and BRD2 to mediate transcription (32). The evolutionary conservation of the phospho-site T173 and acetyl-site K171, coupled with the strong preference of Aurora kinases for a positively charged residue at the −2 position (20), suggest cross-regulation between both types of modifications. One might expect acetylation/deacetylation of K171 to influence T173 phosphorylation during G2/M transition.
The localization of RUNX proteins at the chromatin, centrosome, and the midbody reflects multiple roles during mitosis. It is likely that only a subset of RUNX proteins is phosphorylated at T173 for recruitment to the mitotic structures, whereas another population of RUNX proteins (i.e., non-T173 phosphorylated) is retained at the mitotic chromatin for epigenetic regulation. The centrosome has been proposed to serve as a scaffold where regulatory proteins interact to determine cell cycle progression (33, 34). How RUNX3 interact with other centrosomal proteins is unclear. Nevertheless, the association of RUNX proteins with Aurora kinases indicates close relationships with regulators of mitotic progression and spindle assembly checkpoint.
The observation that Runx3+/− mice develop intestinal adenoma after long latencies of 16 mo after birth (35) is reminiscent of other mice models with mitotic checkpoint deficiencies—Mad2 heterozygous mice develop lung adenocarcinomas at 18–19 mo of age (36). Upon carcinogenic insult, Runx3+/− and Runx3−/− mice show accelerated tumor development compared with wild-type mice (35). Increased incidence of tumor formation in Runx3+/− mice may, in part, be due to the role of RUNX proteins in regulating mitotic fidelity or DNA repair (37).
In conclusion, our work highlights the strong influence of posttranslational modification on RUNX function. The tight conservation of T173 and K171 residues through divergent evolution indicates that all RUNX proteins possess non-DNA binding functions, which must be tightly regulated during cell cycle progression. More in-depth studies of RUNX posttranslational modification will enhance our understanding of RUNX’s pleiotropic activities in the cell cycle as well as cancer.
Materials and Methods
Cell Lines, Drug Treatment, Transfection, and siRNA Knockdown.
Cell lines were obtained from American Type Culture Collection (ATCC). For generation of permanent doxycycline (Dox)-inducible RUNX3 cell lines, RUNX3 and T173 mutants were cloned into pRetroX-Tight-puro-3× Flag (38). HeLa Tet-On cell line (Clontech) was infected with the constructs and selected with 0.5 µg/mL puromycin. Flag-RUNX3 expression was induced by doxycycline (0.5 µg/mL). DNA transfection was performed with TransIT-LT1 reagent (Mirus), whereas siRNA knockdown was performed with jetPRIME (Polyplus) using 35 nM siRNA (Dharmacon) for 72 h. All protocols were as recommended by the manufacturers. Nocodazole (Sigma) was used at 0.1 µg /mL for U2OS and 1 µg/mL for COS7. MLN8054 (0.5 µM), ZM447439 (2 µM), and MG132 (20 µM) were used. Double thymidine block was performed with 2 mM thymidine.
Phos-Tag Gel Electrophoresis and Immunoblot.
U2OS cells were synchronized by nocodazole for 18 h. Mitotic cells were collected by shake-off and replated in media without nocodazole. Cells were boiled in buffer containing 2% (wt/vol) SDS, 10% (vol/vol) glycerol, 100 mM NaCl, 10 mM Tris⋅HCl pH 7.5, 1 mM EDTA, 0.01 mM DTT, protease inhibitor (Roche), phosphatase inhibitor (Thermo Scientific), and 0.4 mM PMSF. Protein concentrations were determined by the BCA method (Pierce). Cell lysates (30 µg) were analyzed by conventional SDS/PAGE or with 20 µM Phos-tag reagent (Wako).
Detailed information regarding the antibodies, immunofluorescence, electrophoretic mobility shift assay, flow cytometry, immunoprecipitation, and other experimental procedures are provided in SI Materials and Methods.
SI Materials and Methods
Commercial Antibodies Used for Immunoblot.
The antibodies are as follows: anti-RUNX1 (39000; Activemotif), anti-RUNX1 (ab35962; Abcam), anti-RUNX2 (D1H7; Cell Signaling Technology), anti-RUNX3 (9647; Cell Signaling Technology), anti-RUNX3 (R3-5G4), anti–RUNX3-biotin (R3-5G4; eBioscience), anti–pan-RUNX (3D9; MBL International), anti-cyclin B1 (Cell Signaling Technology and Santa Cruz Biotechnology), anti–AURKA-pT288:AURKB-pT232:AURKC-T198 (D13A11; Cell Signaling Technology), anti–β-actin (Sigma-Aldrich), α-tubulin (Sigma-Aldrich), anti–γ-tubulin (GTU-88; Sigma-Aldrich), anti–p-Histone H3 serine 10 (D2C8; Cell Signaling Technology), anti-AURKA (1G4; Cell Technology), anti-AURKB (3094; Cell Signaling Technology), anti-PARP (C2-10; Santa Cruz Biotechnology), anti-Flag (F7425 rabbit polyclonal and M2 mouse monoclonal antibody; Sigma-Aldrich), anti-GAPDH (2118; Cell Signaling Technology), and anti-HSP90 (4877; Cell Signaling Technology).
Generation of Phospho-Specific Antibody.
Phospho-T173 antibody was generated by Biomatik: Phospho-T173 peptide CTYHRAIKV(pT)VDGPRE and its nonphosphorylated counterpart were synthesized for rabbit polyclonal antibody production; the antibodies were purified by phospho-peptide affinity column. Antibody specificity was verified by dot-blot and ELISA techniques (Biomatik).
Electrophoretic Mobility Shift Assay.
COS7 cells were transfected with 3 µg of the indicated pEGFP-RUNX3 constructs for 24 h. Reactions were done with a LightShift Chemiluminescent EMSA kit (Thermo Scientific). Nuclear extracts (1.6 µg) were preincubated with 2 µg poly(dI.dC) and recombinant CBFβ protein for 15 min before addition of 20 nM 3′-biotinylated double-stranded DNA probe with RUNX3 binding site (sense 5′-GTCTGGCCTGTGGTCTGGCT GT-3′; antisense 5′-ACAGCCAGACCACAGGCCA GAC-3′). RUNX consensus binding sequence is in bold. For competition or antibody addition, 50-fold excess of nonbiotinylated probe or 0.8 µg of anti-RUNX3 antibody (R3-5G4) was added, respectively. After incubation for 30 min, the samples were resolved by acrylamide gel electrophoresis, transferred to nylon membrane (Amersham), and UV cross-linked.
In Vitro Kinase Reaction.
Full-length RUNX3 was cloned into pET21d plasmid and expressed in bacteria as a histidine-tagged protein. The protein was purified with Nickel column (Qiagen) and eluted with imidazole as recommended by the manufacturer. Kinase reactions with recombinant AURKA and AURKB kinases (MBL International) were performed using 1 µL of kinase and 0.5 µL of [γ32P]ATP at 30 °C for 30 min.
Generation of Point Mutations of RUNX3.
All point mutations were generated by the QuikChange site-directed mutagenesis kit (Stratagene) and confirmed by DNA sequencing.
Immunofluorescence Staining.
COS7 or U2OS cells were transfected with pEGFP-RUNX3 or its mutant counterparts. At 28 h after transfection, cells were sequentially fixed with 4% (wt/vol) paraformaldehyde (10 min at 37 °C) and absolute methanol (10 min at −20 °C). Cells were permeabilized with 0.2% Triton X-100 and blocked with 5% (wt/vol) BSA before incubation with the indicated primary antibodies in 3% (wt/vol) BSA for 45 min at 37 °C. The following secondary antibodies were used: Alexa Fluor 488 anti-rabbit, Alexa Fluor 488 anti-mouse, Alexa Fluor 594 anti-rabbit, and Alexa Fluor 594 anti-mouse IgG (Molecular Probes). The coverslips were mounted with ProLong Gold antifade reagent with DAPI (Invitrogen). Images were acquired at 63× magnification using a Zeiss Observer Z1 inverted microscope and analyzed with the AxioVision 4.8 software. To verify the specificity of the phospho-T173 antibody by immunofluorescence, phospho-T173 antibodies were used at concentrations of 0.135 µg/mL for siRNA knockdown and 0.54 µg/mL for peptide competition experiments, respectively. Peptide competition was performed at 1 µg/mL.
Immunoprecipitation.
To verify the specificity of the phospho-T173 antibody by immunoprecipitation, COS7 cells were seeded on 15-cm dishes and transfected with the indicated plasmids (3 µg) for 48 h and treated with nocodazole for the last 16 h. Prometaphase-enriched cells were harvested by trypsinization. For phosphoantibody immunoprecipitation, 2 mg of cell lysate was incubated with 5 µg of antibody for 4 h. Bound proteins were analyzed by 10% (wt/vol) SDS/PAGE.
To ascertain binding of RUNX proteins with Aurora kinases, HEK293T cells were seeded on 10-cm dishes and transfected with Aurora kinase (7.5 µg) and EF-BOS RUNX (7.5 µg) expression vectors. After 48 h, cells were harvested and lysed for immunoprecipitation. For Flag immunoprecipitation or GFP immunoprecipitation, M2 Sepharose beads (Sigma-Aldrich) and GFP Trap beads (Chromotek) were used, respectively. Lysates (400 µg) were incubated 500 µL of binding buffer [25 mM Tris⋅HCl pH 8.0, 0.25% Nonidet P-40, 2.5% (vol/vol) glycerol, 0.5 mM EDTA, 150 mM NaCl, 1 mM DTT, phosphatase inhibitor (Thermo Scientific) and protease inhibitor mixture (Roche)] with the respective beads for 3 h at 4 °C. The GFP beads were washed copiously with binding buffer and eluted by boiling in Laemmli buffer. To elute Flag-tagged proteins from M2 beads, 3× Flag peptide (Sigma-Aldrich) was used. Bound proteins were analyzed by 10% (wt/vol) SDS/PAGE and immunoblot.
For immunoprecipitation of the cytosolic extract, COS7 cells were transfected with the GFP–RUNX3 constructs. After 24 h, cell fractionation was performed and 200 µg of cytosolic extract was incubated with GFP beads in binding buffer (conditions as described above).
Flow Cytometry.
Cells were harvested by trypsinization and fixed in 80% (vol/vol) ice cold ethanol. The cells were blocked with 0.5% BSA in PBS (Sigma) before incubation with phospho-Histone 3 (Ser 10) antibody (Alexa Fluor 488 conjugate) (9708, Cell Signaling Technology) for 1 h at room temperature (dilution of 1:65 in blocking buffer). After two washes with blocking buffer, the cells were resuspended in PBS containing 50 µg/mL propidium iodide, 20 µg/mL RNase A, and 0.1% Triton X-100 and incubated at room temperature for 30 min. Flow cytometry was conducted using a LSR II flow cytometer (BD Biosciences). The data were analyzed by FlowJo software.
Lambda Phosphatase Treatment.
COS7 cells were transfected with Flag-RUNX3 (amino acids 1–187) or Flag-RUNX3 (amino acids 182–415). After 24 h, the cells were lysed in buffer containing 50 mM Tris⋅HCL pH 8.0, 0.3 M NaCl, 5% (vol/vol) glycerol, 1 mM EDTA, 0.5% Nonidet P-40, 1 mM DTT, 0.4 mM PMSF, protease inhibitor (Roche) and phosphatase inhibitor (Thermo Scientific). Cell lysates (30 µg) were incubated with 20 units of lambda phosphatase (New England Biolabs) for 30 min at 30 °C and analyzed by 12% (wt/vol) SDS/PAGE (supplemented with Phos-tag).
Luciferase Reporter Assay.
HEK293T cells were seeded on a 24-well dish and cotransfected with 100 ng p14ARF promoter linked to firefly luciferase and 500 ng pEGFP-RUNX3 constructs and 50 ng of β-gal vector as internal control. After 48 h, luciferase activities were analyzed with Luciferase Reporter Assay (Promega). All reactions were performed in triplicate. The Beta-Glo assay (Promega) was used for the quantification of β-galactosidase to normalize luciferase readings.
Colony Formation Assay.
HeLa Tet-On cells were infected with pRetro-Tight-puro 3× Flag RUNX3 constructs, selected with puromycin for 4 d and cultured in media containing 0.5 µg/mL of doxycycline and 0.25 µg/mL of puromycin for 2 wk. Colonies were stained with 0.01% crystal violet (wt/vol) and photographed. Colonies (at least 2 mm radius) were counted with the OpenCFU program (39).
Subcellular Fractionation.
Cell fractionation was performed as described (40). A total of 15 µg of cytoplasmic, nuclear, or total cell extract was analyzed by 10% (wt/vol) SDS/PAGE.
Acknowledgments
We thank Valerie Sin Wen Oh and Han Lei Goh for technical assistance. The p14ARF promoter was a kind gift from Scott Hiebert. This work was supported by the National Research Foundation Singapore, the Singapore Ministry of Education under its Research Centres of Excellence initiative, and the Singapore National Research Foundation under its Translational and Clinical Research Flagship Programme.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523157113/-/DCSupplemental.
References
- 1.Güttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10(3):178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- 2.Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22(6):197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- 3.Ito Y, Bae SC, Chuang LS. The RUNX family: Developmental regulators in cancer. Nat Rev Cancer. 2015;15(2):81–95. doi: 10.1038/nrc3877. [DOI] [PubMed] [Google Scholar]
- 4.Martínez-Balbás MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83(1):29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 5.Young DW, et al. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc Natl Acad Sci USA. 2007;104(9):3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young DW, et al. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007;445(7126):442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- 7.Pande S, et al. Subnuclear targeting of the Runx3 tumor suppressor and its epigenetic association with mitotic chromosomes. J Cell Physiol. 2009;218(3):473–479. doi: 10.1002/jcp.21630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Ami O, et al. Addiction of t(8;21) and inv(16) acute myeloid leukemia to native RUNX1. Cell Reports. 2013;4(6):1131–1143. doi: 10.1016/j.celrep.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 9.Pockwinse SM, et al. Microtubule-dependent nuclear-cytoplasmic shuttling of Runx2. J Cell Physiol. 2006;206(2):354–362. doi: 10.1002/jcp.20469. [DOI] [PubMed] [Google Scholar]
- 10.Stein GS, et al. Organization of transcriptional regulatory machinery in nuclear microenvironments: Implications for biological control and cancer. Adv Enzyme Regul. 2007;47:242–250. doi: 10.1016/j.advenzreg.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuang LS, et al. RUNX3 interactome reveals novel centrosomal targeting of RUNX family of transcription factors. Cell Cycle. 2012;11(10):1938–1947. doi: 10.4161/cc.20278. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Camacho C, van Wijnen AJ, Lian JB, Stein JL, Stein GS. Core binding factor β (CBFβ) is retained in the midbody during cytokinesis. J Cell Physiol. 2014;229(10):1466–1474. doi: 10.1002/jcp.24588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S, Zhang Y, Soosairajah J, Kraft AS. Regulation of RUNX1/AML1 during the G2/M transition. Leuk Res. 2007;31(6):839–851. doi: 10.1016/j.leukres.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Rajgopal A, et al. Mitotic control of RUNX2 phosphorylation by both CDK1/cyclin B kinase and PP1/PP2A phosphatase in osteoblastic cells. J Cell Biochem. 2007;100(6):1509–1517. doi: 10.1002/jcb.21137. [DOI] [PubMed] [Google Scholar]
- 15.Qiao M, et al. Cell cycle-dependent phosphorylation of the RUNX2 transcription factor by cdc2 regulates endothelial cell proliferation. J Biol Chem. 2006;281(11):7118–7128. doi: 10.1074/jbc.M508162200. [DOI] [PubMed] [Google Scholar]
- 16.Biggs JR, Peterson LF, Zhang Y, Kraft AS, Zhang DE. AML1/RUNX1 phosphorylation by cyclin-dependent kinases regulates the degradation of AML1/RUNX1 by the anaphase-promoting complex. Mol Cell Biol. 2006;26(20):7420–7429. doi: 10.1128/MCB.00597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bangsow C, et al. The RUNX3 gene--sequence, structure and regulated expression. Gene. 2001;279(2):221–232. doi: 10.1016/s0378-1119(01)00760-0. [DOI] [PubMed] [Google Scholar]
- 18.Wee HJ, Voon DC, Bae SC, Ito Y. PEBP2-beta/CBF-beta-dependent phosphorylation of RUNX1 and p300 by HIPK2: Implications for leukemogenesis. Blood. 2008;112(9):3777–3787. doi: 10.1182/blood-2008-01-134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiromizu T, et al. Identification of missing proteins in the neXtProt database and unregistered phosphopeptides in the PhosphoSitePlus database as part of the Chromosome-centric Human Proteome Project. J Proteome Res. 2013;12(6):2414–2421. doi: 10.1021/pr300825v. [DOI] [PubMed] [Google Scholar]
- 20.Kettenbach AN, et al. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal. 2011;4(179):rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gnad F, et al. PHOSIDA (phosphorylation site database): Management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8(11):R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marumoto T, Zhang D, Saya H. Aurora-A: A guardian of poles. Nat Rev Cancer. 2005;5(1):42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- 23.Mouradov D, et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74(12):3238–3247. doi: 10.1158/0008-5472.CAN-14-0013. [DOI] [PubMed] [Google Scholar]
- 24.Papaemmanuil E, et al. Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–3627, quiz 3699. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forbes SA, et al. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida T, et al. Functional analysis of RUNX2 mutations in Japanese patients with cleidocranial dysplasia demonstrates novel genotype-phenotype correlations. Am J Hum Genet. 2002;71(4):724–738. doi: 10.1086/342717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tahirov TH, et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell. 2001;104(5):755–767. doi: 10.1016/s0092-8674(01)00271-9. [DOI] [PubMed] [Google Scholar]
- 28.Bravo J, Li Z, Speck NA, Warren AJ. The leukemia-associated AML1 (Runx1)—CBF beta complex functions as a DNA-induced molecular clamp. Nat Struct Biol. 2001;8(4):371–378. doi: 10.1038/86264. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, et al. Energetic contribution of residues in the Runx1 Runt domain to DNA binding. J Biol Chem. 2003;278(35):33088–33096. doi: 10.1074/jbc.M303973200. [DOI] [PubMed] [Google Scholar]
- 30.Chi XZ, et al. RUNX3 suppresses gastric epithelial cell growth by inducing p21(WAF1/Cip1) expression in cooperation with transforming growth factor beta-activated SMAD. Mol Cell Biol. 2005;25(18):8097–8107. doi: 10.1128/MCB.25.18.8097-8107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matheny CJ, et al. Disease mutations in RUNX1 and RUNX2 create nonfunctional, dominant-negative, or hypomorphic alleles. EMBO J. 2007;26(4):1163–1175. doi: 10.1038/sj.emboj.7601568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee YS, et al. Runx3 inactivation is a crucial early event in the development of lung adenocarcinoma. Cancer Cell. 2013;24(5):603–616. doi: 10.1016/j.ccr.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15(6):303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 34.Gönczy P. Centrosomes and cancer: Revisiting a long-standing relationship. Nat Rev Cancer. 2015;15(11):639–652. doi: 10.1038/nrc3995. [DOI] [PubMed] [Google Scholar]
- 35.Ito K, et al. RUNX3 attenuates beta-catenin/T cell factors in intestinal tumorigenesis. Cancer Cell. 2008;14(3):226–237. doi: 10.1016/j.ccr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Michel LS, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409(6818):355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 37.Wang CQ, et al. Disruption of Runx1 and Runx3 leads to bone marrow failure and leukemia predisposition due to transcriptional and DNA repair defects. Cell Reports. 2014;8(3):767–782. doi: 10.1016/j.celrep.2014.06.046. [DOI] [PubMed] [Google Scholar]
- 38.Kitagawa M, Fung SY, Onishi N, Saya H, Lee SH. Targeting Aurora B to the equatorial cortex by MKlp2 is required for cytokinesis. PLoS One. 2013;8(6):e64826. doi: 10.1371/journal.pone.0064826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geissmann Q. OpenCFU, a new free and open-source software to count cell colonies and other circular objects. PLoS One. 2013;8(2):e54072. doi: 10.1371/journal.pone.0054072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schreiber E, Matthias P, Müller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17(15):6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]