Significance
The Drosophila coactivator cAMP-regulated transcription coactivator (Crtc) functions in neurons to promote energy balance. We found that Crtc and its binding partner cAMP response element binding protein (CREB) maintain metabolic homeostasis by stimulating the expression of the neuropeptide-Y–like neuropeptide short neuropeptide F (sNPF). Loss of sNPF disrupted gut epithelial integrity and increased immune response gene expression, leading to decreases in triglyceride and glycogen stores. Conversely, overexpression of Crtc or sNPF in neurons of wild-type flies attenuated the immune response and enhanced starvation resistance. By enhancing gut barrier integrity, the CREB/CRTC pathway maintains energy homeostasis in response to stress.
Keywords: CREB, CRTC, sNPF, cAMP, transcription
Abstract
The starvation-inducible coactivator cAMP response element binding protein (CREB)–cAMP-regulated transcription coactivator (Crtc) has been shown to promote starvation resistance in Drosophila by up-regulating CREB target gene expression in neurons, although the underlying mechanism is unclear. We found that Crtc and its binding partner CREB enhance energy homeostasis by stimulating the expression of short neuropeptide F (sNPF), an ortholog of mammalian neuropeptide Y, which we show here is a direct target of CREB and Crtc. Neuronal sNPF was found to promote energy homeostasis via gut enterocyte sNPF receptors, which appear to maintain gut epithelial integrity. Loss of Crtc–sNPF signaling disrupted epithelial tight junctions, allowing resident gut flora to promote chronic increases in antimicrobial peptide (AMP) gene expression that compromised energy balance. Growth on germ-free food reduced AMP gene expression and rescued starvation sensitivity in Crtc mutant flies. Overexpression of Crtc or sNPF in neurons of wild-type flies dampens the gut immune response and enhances starvation resistance. Our results reveal a previously unidentified tolerance defense strategy involving a brain–gut pathway that maintains homeostasis through its effects on epithelial integrity.
When faced with a pathogen, a host can use two defense strategies to survive infection: resistance and tolerance (1–3). Resistance mechanisms are a function of the immune system, which works to kill pathogens. Tolerance mechanisms reduce the negative impact of infection on host health without directly affecting pathogen levels. Although typically beneficial to the host, activation of the immune system comes at the expense of energy stores, and overactivation can lead to physiological damage in the form of metabolic disruption. Indeed, genetic mutations that increase resistance also affect tolerance (4).
Hosts have likely evolved tolerance mechanisms that maintain triglyceride stores, in part, by modulating resistance (1, 2). The tight junction protein big bang (Bbg) protects against bacterial infection, for example, by maintaining epithelial integrity in the gut; loss of Bbg expression triggers immune system activation and decreases survival. Indeed, persistent up-regulation of the immune response appears to decrease lifespan, in part, by interfering with insulin signaling and thereby reducing energy stores (5).
In Drosophila, bacterial infection stimulates the production of antimicrobial peptides (AMPs) in response to activation of the NF-κB–like pathways Imd and Toll (6). AMP expression is also up-regulated in hemocytes during fasting, albeit to a far lower level; it requires the forkhead box transcription factor FoxO (7, 8). Enhancing FOXO activity promotes longevity, in part, by increasing pathogen resistance (9, 10). However, dysregulated FOXO activation during pathogenic infections increases mortality, despite control of pathogen burden (11), due to the depletion of energy stores.
The Drosophila cAMP-regulated transcription coactivator (Crtc) is expressed primarily in neurons, where it protects against starvation and oxidative stress (12). Under basal conditions, Crtc, like its mammalian homologs, is phosphorylated by salt-inducible kinases and sequestered in the cytoplasm via interactions with 14-3-3 proteins (12, 13). In response to starvation or oxidative stress, Crtc undergoes dephosphorylation and nuclear translocation, where it associates with cAMP response element binding protein (CREB) over relevant promoters (14). Crtc mutant flies have reduced triglyceride and glycogen stores, and they are short-lived, although the underlying mechanism is unclear. Here, we reveal an unexpected role of the neuronal CREB/CRTC pathway in maintaining gut homeostasis by stimulating neuropeptide Y (NPY)-like signaling. Our results may provide insight into the treatment of stress-related gastrointestinal disorders.
Results
Crtc Suppresses Inflammation.
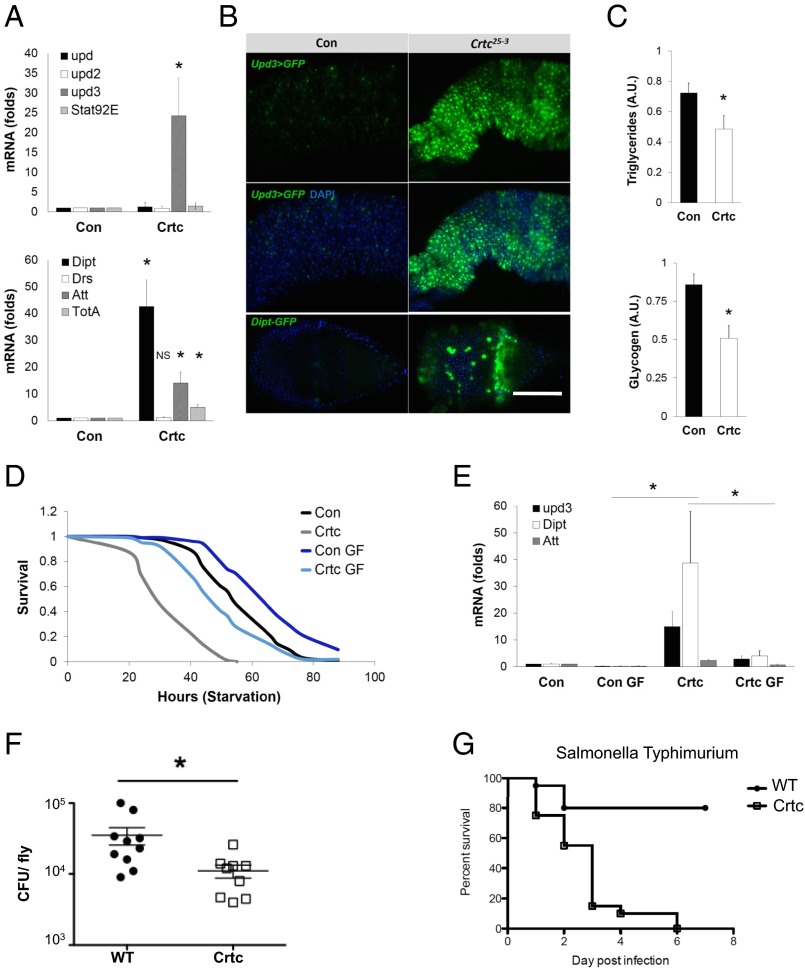
Although their food intake is comparable to controls, Crtc mutant flies have lower glycogen and lipid stores, and they are starvation-sensitive (12). In gene profiling studies, we noticed that the expression of unpaired 3 (upd3), which encodes a mammalian IL-6 homolog (15), was increased in Crtc mutant flies, whereas other members of the UPD/JAK/STAT pathway (upd, upd2, or Stat92) were not (Fig. 1A). Pointing to activation of the Imd pathway in Crtc flies, mRNA amounts of the stress peptide turandot A (TotA) and for the AMPs diptericin (Dipt) and attacin (Att) were also up-regulated.
Fig. 1.
Increased gut inflammation in Crtc mutant Drosophila. (A) qPCR analysis of upd family (Top) and AMP (Bottom) gene expression in 7- to 10-d-old control (Con) (w; Crtc25-3/+) and mutant (w; Crtc25-3) whole flies. (B) Upd3-GAL4;UAS-GFP, Dipt-GFP reporters in control and mutant flies. Top, GFP channel only. Middle and Bottom show GFP and DAPI. (Scale bar, 50 μm.) Genotypes are as follows: Con (w; Upd3-GAL4 UAS-GFP;Crtc25-3/+), Crtc25-3 (w; Upd3-GAL4 UAS-GFP;Crtc25-3), Con (w; Dipt-GFP;;Crtc25-3/+), and Crtc25-3 (w; Upd3-GAL4 UAS-GFP;;Crtc25-3). (C) Quantitation of triglyceride and glycogen amounts in control and Crtc25-3 mutant flies. A.U., arbitrary units. (D) Survival of flies subjected to starvation in controls (w; Crtc25-3/+) and mutants (w; Crtc25-3) (log-rank, P < 0.01). Effect of growth on germ-free (GF) food in control and Crtc25-3 mutant flies is shown. (E) AMP mRNA amounts in wild-type and Crtc mutant flies maintained on normal or germ-free food. Effect of Salmonella typhimurium injection on pathogen levels (F) at 48 h postinfection (cfu) and survival (G) of wild-type and Crtc mutant flies. For survival, *P < 0.0001; for the CFUs, *P = 0.0101 (data are mean ± SD).
Using promoter-driven GFP reporter transgenes to identify tissues in which Upd3 and Dipt transcriptional activities were elevated, we found that Upd3-GFP+ and Dipt-GFP+ cells were localized primarily in the midgut and proventriculus of Crtc mutant, but not control, flies, respectively (Fig. 1B), whereas other tissues were unremarkable.
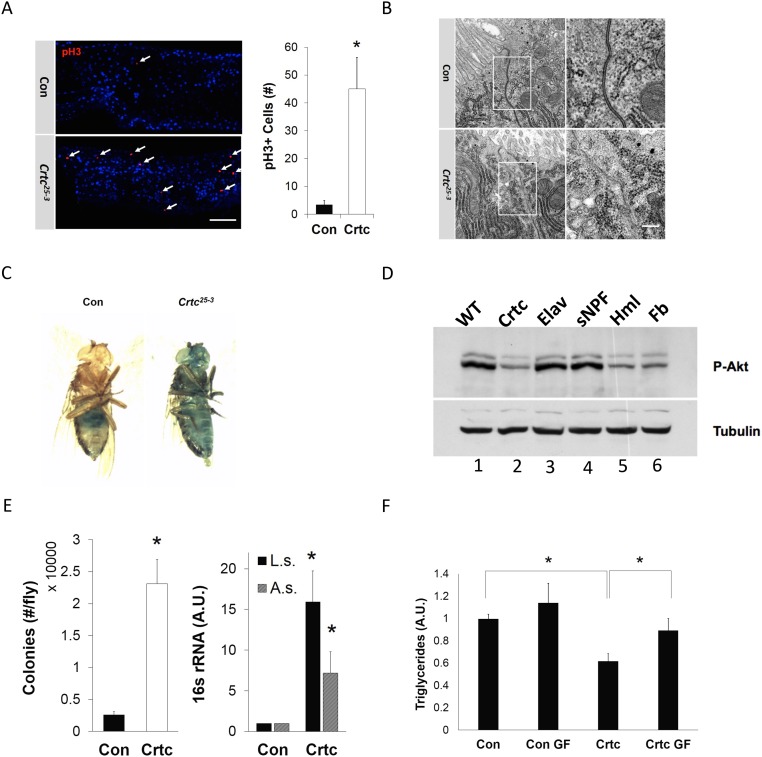
The induction of Upd3 in gut enterocytes often signals increases in stress due to bacterial infection or oxidative damage (16). Indeed, Upd3 has been shown to promote enterocyte turnover by increasing compensatory intestinal stem cell (ISC) proliferation. Using phospho-Ser10-histone 3 immunostaining as a marker for ISC activity, we found that numbers of mitoses in the midgut were increased more than 10-fold in Crtc mutants relative to controls (Fig. S1A). Collectively, these results indicate that Crtc suppresses Upd3 and Imd pathways in the gut.
Fig. S1.
Neuronal Crtc promotes gut homeostasis. (A) Mitotic cells [phospho-Ser10-histone 3 (PH3)+, red] in midguts of control (w; Crtc25-3/+) and mutant (w; Crtc25-3) flies. (Right) Total number of PH3+ cells from individual midgut [control (Con), n = 15; Crtc, n = 17]. (Scale bar, 50 μm.) (B) Transmission electron microscopy. Micrographs of midguts from control (Top) and Crtc mutant (Bottom) flies. The relative morphology of epithelial cell–cell junctions (boxed areas) is shown. (Magnification: 120,000×.) (Scale bar, 100 nm.) (C) Smurf assay of 7- to 10-d-old flies; representative flies showing relative gut permeability in control (zero of 20 Smurf+) and Crtc mutant (four of 20 Smurf+) flies (n = 20 per group). (D) Immunoblot of phospho-AKT amounts in control (WT) and Crtc mutant flies (lanes 1 and 2). Lanes 3–6 show phospho-AKT amounts in Crtc mutant flies following Crtc reexpression from neuronal (Elav, sNPF), hemocyte (Hml), or fat body (Fb) drivers. (E, Left) Numbers of bacterial colonies recovered from control (Crtc25-3/+) and mutant (Crtc25-3) larvae. (E, Right) 16S rRNA gene amounts for Acetobacter sp. (A.s.) and Lactobacillus sp. (L.s.) using DNA prepared from control (w; Crtc25-3/+) and mutant (w; Crtc25-3) intestines. (F) Triglyceride amounts in control and Crtc mutant flies maintained on normal or germ-free (GF) food. A.U., arbitrary units. *P < 0.05 (data are mean ± SD).
Epithelial integrity appears critical for innate immune homeostasis (17). Intestinal barrier dysfunction promotes increases in AMP gene expression that interfere with insulin signaling and reduce energy stores (5). Having seen that Upd3 is up-regulated in Crtc mutant flies, we wondered whether gut permeability is correspondingly affected. In transmission electron microscopy studies, electron densities at apical cell–cell junctions were reduced in Crtc mutant midgut relative to controls (Fig. S1B). Intestinal permeability was also increased in dye-based (Smurf) assays (Fig. S1C). As a result, amounts of active, phosphorylated AKT were decreased in Crtc flies, and these Crtc flies had lower amounts of triglyceride and glycogen relative to controls (Fig. 1C and Fig. S1D).
In many organisms, commensal bacteria promote nutrient absorption and contribute to immune homeostasis (18). The intestinal epithelium provides an important physical barrier that permits growth of these commensals without triggering overactivation of the immune system. Based on the loss of barrier integrity in Crtc mutant flies, we examined whether gut microbiota ecology was affected. In line with this notion, populations of the commensal bacteria Acetobacter sp. and Lactobacillus sp. were increased in Crtc mutants (Fig. S1E). We considered that the up-regulation of immune response genes in Crtc mutants may reflect these increases in resident bacteria. In that event, elimination of these microbes would rescue starvation sensitivity. To test this idea, we cultured Crtc mutant flies either under germ-free conditions or on normal medium. Germ-free growth not only reduced immune gene expression in Crtc mutant flies but also increased triglyceride stores and enhanced starvation resistance (Fig. 1 D and E and Fig. S1F).
To determine whether the activation of the Imd pathway in Crtc mutant flies enhances their resistance to gram-negative infection, we challenged them with the intracellular pathogen Salmonella. In keeping with the associated increases in AMP expression, Crtc mutant flies had reduced bacterial counts relative to control flies (Fig. 1F). Despite their ability to control the infection, however, Salmonella-infected Crtc mutants died more rapidly than infected wild-type flies, demonstrating that there were tradeoffs between the heightened resistance response and tolerance defenses (Fig. 1G). These data suggest that an overexuberant response to infection compromised survival of Crtc mutant flies relative to controls.
Creb Phenocopies the Metabolic Effects of Crtc.
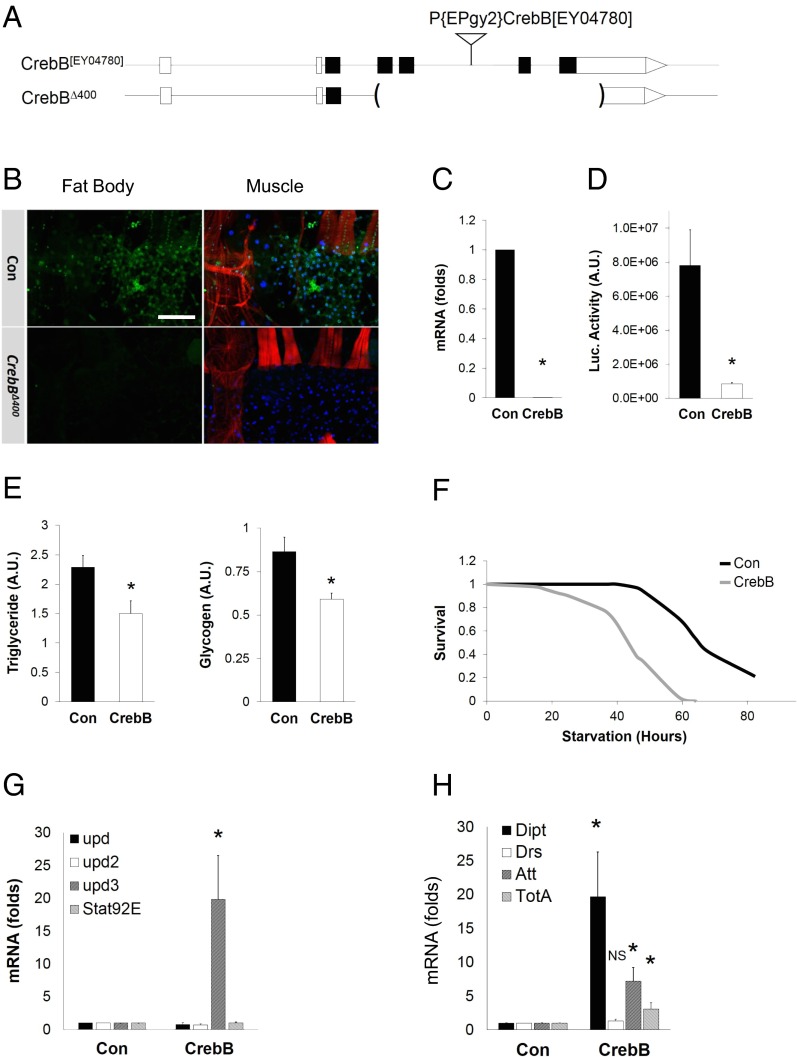
CRTCs have been found to mediate induction of cellular genes by binding to CREB over relevant promoters (14). Drosophila has a single CREB homolog, CrebB, which appears to function in a variety of settings, including learning and memory, circadian rhythmicity, and sleep (19, 20). To date, there exists a single mutant allele of CrebB, CrebBS162, which expresses truncated CrebB polypeptides of unknown activity (20). To obtain well-characterized CrebB mutations and to circumvent the lethality associated with the CrebBS162 chromosome, we generated a series of CrebB deletions by P-element–mediated imprecise excision. One of these deletions, CrebBΔ400, is a fully viable null allele that removes both the activation and leucine-zipper DNA-binding domains (Fig. 2A). CrebB mRNA and protein amounts are not detectable in CrebBΔ400 mutants (Fig. 2 B and C). Consistent with the absence of CrebB, CREB reporter gene activity is nearly eliminated in CrebBΔ400 flies (Fig. 2D).
Fig. 2.
Creb phenocopies effects of Crtc on energy balance and inflammation. (A) CrebB[EY04780] and CrebBΔ400 genomic loci. The insertion P{EPgy2}CrebB[EY04780] was used to delete activation and DNA-binding domain sequences in the CrebB gene. (B) Immunofluorescence analysis of CrebB expression (green) in control and CrebB mutant (w, CrebBΔ400) muscle (red, phalloidin) and fat body. (Scale bar, 50 μm.) (C) qPCR analysis of CrebB mRNA amounts in CrebB mutants (w, CrebBΔ400). (D) CRE-luciferase activity in controls (w, CrebBΔ400/+) and Creb mutants (w, CrebBΔ400). (E) Relative triglyceride and glycogen stores in control (w CrebBΔ400/+) and CrebB mutant (w CrebBΔ400) flies. (F) Survival of control and CrebB mutant flies on starvation medium (n = 100; log-rank, P < 0.01). (G and H) qPCR analysis of mRNA amounts for upd3 and AMPs. NS, not significant. *P < 0.05 [Student’s t test (C–E, G, and H) or log-rank test (F)]. Data (C, D, G, and H) are from three independent experiments (data are mean ± SD).
We examined metabolic effects of CrebB. Similar to Crtc mutants, CrebBΔ400 flies had reduced glycogen and lipid stores (Fig. 2E), and they were sensitive to starvation (Fig. 2F). Moreover, proinflammatory gene expression was up-regulated in CrebBΔ400 mutant flies compared with controls (Fig. 2 G and H).
Short Neuropeptide F Is a Direct Target of CrebB and Crtc.
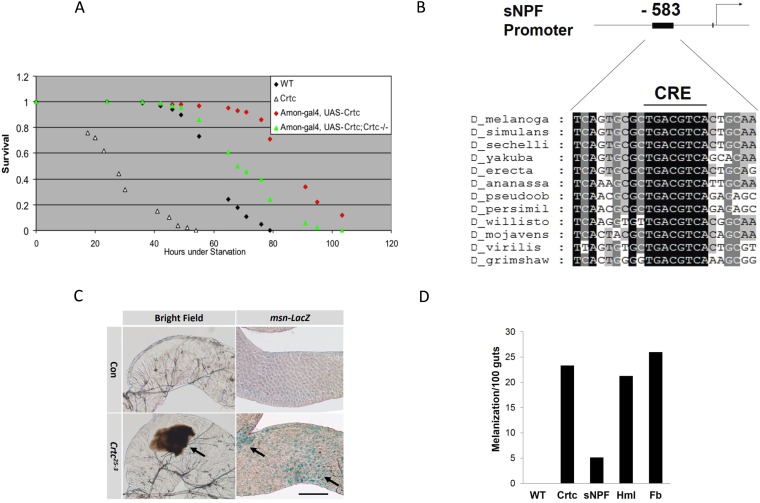
Having seen that panneuronal expression of fly Crtc fully rescues starvation sensitivity in Crtc mutants (12), we tested whether specific neuronal types could mediate effects of Crtc in this setting. Using the GAL4/upstream activation sequence (UAS) system to express Crtc in discrete neuronal subsets, we found that supplying Crtc function to the large class of peptidergic neurons (21) was sufficient to rescue starvation sensitivity in Crtc mutant flies (Fig. S2A). Of the roughly 60 neuropeptide-encoding genes in the Drosophila genome, we noted several whose expression appeared to be Crtc-dependent. In particular, the promoter for short neuropeptide F (sNPF) contains a cAMP response element (CRE) that is fully conserved across all Drosophila species (Fig. S2B). A member of the NPY family of neuropeptides, sNPF has been shown to function in energy-driven behavior (22).
Fig. S2.
Gut melanization in Crtc flies. (A) Effect of CRTC overexpression in peptidergic neurons on survival in flies subjected to starvation. Genotypes are: WT (w; Crtc25-3/+); Crtc (w; Crtc25-3); Amon-gal4, UAS-Crtc (w; amon-GAL4/+; Crtc25-3 UAS-Crtc/+); and Amon-gal4, UAS-Crtc, Crtc−/−(w; amon-GAL4/+; Crtc25-3 UAS-Crtc/Crtc25-3). (B) Relative conservation of a CRE in the sNPF gene promoter from various Drosophila species. (C) Light microscopic analysis showing melanization and msn-lacZ reporter activity in control and Crtc midgut. (Scale bar, 50 μm.) (D) Effect of Crtc reexpression in sNPF neurons (sNPF), Hml, or Fb on melanization in Crtc mutant flies (n = 25).
In transient transfection assays, overexpression of Crtc increased the activity of a wild-type, but not a CRE-mutant, sNPF reporter in S2 cells exposed to cAMP agonist (Fig. 3A). We also performed ChIP studies to determine if Crtc directly regulates sNPF transcription. Using the GAL4/UAS system to target expression of epitope-tagged Crtc to sNPF neurons, we observed robust recruitment of Crtc to the sNPF promoter relative to controls (Fig. 3B). Consistent with the role of CrebB and Crtc in stimulating sNPF gene expression, protein amounts for sNPF, detected by immunohistochemical staining, were substantially reduced in adult CrebB and Crtc mutant brains (Fig. 3C).
Fig. 3.
CREB/Crtc pathway promotes gut homeostasis by stimulating neuronal sNPF expression. (A) Relative activity of wild-type (sNPFP) and CRE mutant (sNPFP∆CRE) sNPF reporters in S2 cells. The effect of exposure to forskolin (FSK) and Crtc overexpression is shown. (B) ChIP assay showing recruitment of GFP-tagged Crtc to the sNPF promoter in sNPF neurons. Genotypes are GAL4 (sNPF-GAL4/+), Crtc (UAS-GFP::Crtc/+), and GAL4 > Crtc (sNPF-GAL4/UAS-GFP::Crtc). (C) Immunohistochemical analysis of sNPF staining in control (w; Crtc25-3/+) and Crtc mutant (w; Crtc25-3) flies, or in control (w CrebBΔ400/+) and CrebB mutant (w CrebBΔ400) flies. (Scale bar, 50 μm.) (D) Effect of Crtc or sNPF reexpression in sNPF neurons on starvation sensitivity of Crtc mutant flies. Genotypes are Crtc/+; GAL4/+ (w; Crtc25-3 sNPF-GAL4/+), Crtc; sNPF-Gal4 (w; Crtc25-3 sNPF-GAL4/Crtc25-3), Crtc; sNPF > Crtc (w; Crtc25-3 UAS-Crtc/Crtc25-3 sNPF-GAL4), and Crtc; sNPF > sNPF (w; Crtc25-3 UAS-sNPF/Crtc25-3 sNPF-GAL4) (log-rank, P < 0.01). (E and F) Relative mRNA amounts for inflammatory genes in control and Crtc mutant flies following Crtc reexpression in sNPF neurons (sNPF-GAL4; Rescue) (E) vs. hemocyte (Hml) or fat body (Fb) driver (F). Effect of Crtc overexpression (O/E) in control flies is also indicated. *P < 0.05. Genotypes are Con (w; Crtc25-3 UAS-Crtc/+), Crtc (w; Crtc25-3 UAS-Crtc/Crtc25-3), O/E (w; Crtc25-3 UAS-Crtc/sNPF-GAL4), Rescue (w;; Crtc25-3 UAS-Crtc/Crtc25-3 sNPF-GAL4), Hml (w; Crtc25-3 UAS-Crtc/Crtc25-3 hmlΔ-GAL4), and Fb (w; Fb-GAL4/+; Crtc25-3/UAS-Crtc, Crtc25-3). (G) Effect of sNPF reexpression from sNPF, Hml, or Fb drivers on triglyceride levels in Crtc mutants or control flies. *P < 0.05 [Student’s t test (B, and E–G) or log-rank test (D)]. Data are from three independent experiments (for B and E–G, data are SEM ± SD). (H) Effect of Crtc overexpression in sNPF neurons on starvation sensitivity in control and sNPF mutant flies. Genotypes are sNPFGAL4/+ (yw; sNPFNP6301/+), sNPFGAL4 (yw; sNPFNP6301), sNPFGAL4/+; UAS-Crtc (yw; sNPFNP6301/+; UAS-Crtc/+), and sNPFGAL4; UAS-Crtc (yw; sNPFNP6301; UAS-Crtc/+) (log-rank, P < 0.01).
We used the GAL4/UAS system in Crtc mutant flies to evaluate whether this coactivator is required specifically within sNPF neurons to regulate inflammation and starvation sensitivity. Supporting this idea, expression of Crtc under control of a sNPF-GAL4 driver reduced AMP gene expression and restored phospho-AKT amounts, thereby increasing triglyceride stores (Fig. 3 D–G and Fig. S1D); however, Crtc expression in other classes of neurons or in other tissues did not. Additionally, melanization, a characteristic feature of the Drosophila immune response, was evident in the posterior midgut and hindgut of Crtc mutants (Fig. S2C); reexpression of Crtc in sNPF neurons, but not in other tissues, rescued the melanization phenotype (Fig. S2D). Indeed, overexpression of Crtc in sNPF neurons of wild-type flies further enhanced their starvation resistance (Fig. 3H). Collectively, these results indicate that Crtc functions in some or all of the sNPF neurons to regulate starvation sensitivity and gut immunity.
sNPF Inhibits Inflammation.
To determine whether sNPF itself mediates the effects of Crtc on inflammation and energy metabolism, we evaluated starvation resistance and gut inflammation in sNPF mutants using two alleles: a P-element insertion (sNPFNP6301) and a sNPF partial deletion (sNPF∆119; Fig. S3A). Both sNPF mutant alleles act as strong loss-of-function alleles and show similar phenotypes. sNPF peptide is greatly reduced in either of the sNPF mutants by immunohistochemical analysis (Fig. S3B).
Fig. S3.
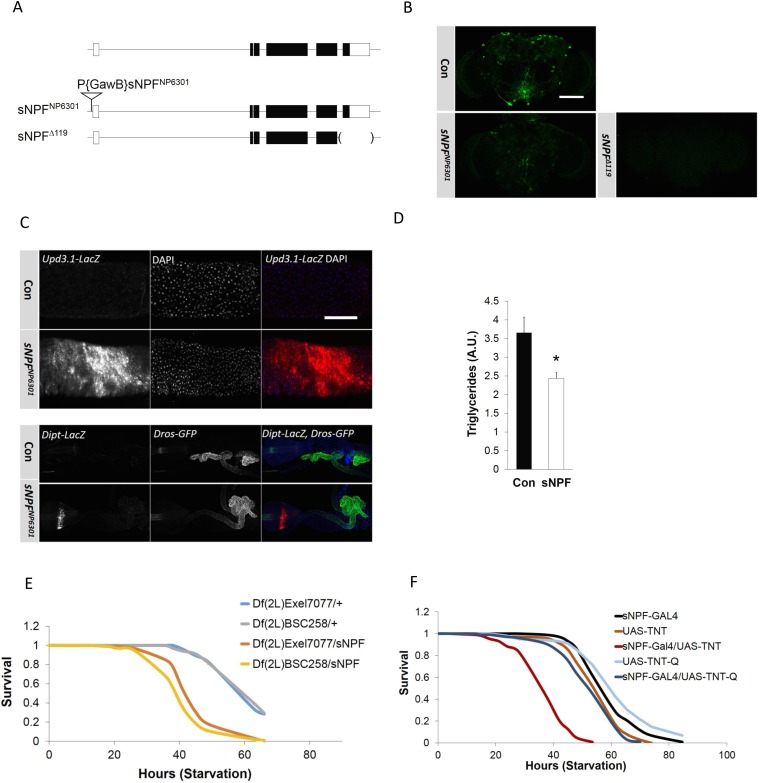
sNPF modulates starvation sensitivity and antimicrobial gene expression. (A) Diagram of wild-type, sNPFNP630, and sNPF∆119 mutant genomic loci. (B) Immunofluorescence analysis sNPF expression in control, sNPFNP630, and sNPF∆119 mutant brain. (Scale bar, 50 μm.) (C, Top) Upd3.1-LacZ reporter activity in control and sNPF mutants. Genotypes are as follows: Con (yw; sNPFNP6301/+; Upd3.1-LacZ/+) and sNPFNP6301 (yw; sNPFNP6301; Upd3.1-LacZ/+). (C, Bottom) Dipt-LacZ, Drs-GFP reporter activity detected using anti–β-Gal or anti-GFP antibody in control and sNPF mutants. Genotypes are as follows: Con (Dipt-LacZ, Drs-GFP; sNPFNP6301/+) and sNPFNP6301 (Dipt-LacZ, Drs-GFP/+; sNPFNP6301). (Scale bar, 50 μm.) (D) Triglyceride amounts in control and sNPF mutant flies. (E) P-element insertion in sNPFNP630 flies functions in trans to deletion mutant fly stocks. Starvation sensitivity in sNPFNP6301 flies crossed to flies with chromosomal deletions [Df(2L)Exel7077 or Df(2L)BSC258] that include the sNPF locus and scored for genetic complementation (n = 100). (F) Effect of wild-type (TNT) and inactive (TNT-Q) tetanus toxin expression in sNPF neurons on starvation sensitivity. TNT expression targeted to sNPF neurons with a GAL4-sNPF driver. *P < 0.05 [Student’s t test (D) or log-rank test (E and F)]. Data for D are from three independent experiments (mean ± SD).
mRNA amounts for proinflammatory genes (upd3, Dipt, Att, and TotA) were increased in sNPF mutants relative to controls (Fig. 4B). Similar to Crtc mutant flies, upd3-lacZ reporter activity was elevated in sNPF mutant midguts (Fig. S3C). Dipt-lacZ expression in the proventriculus of sNPF mutants was also substantially up-regulated, whereas Drs-GFP reporter activity was not (Fig. S3C).
Fig. 4.
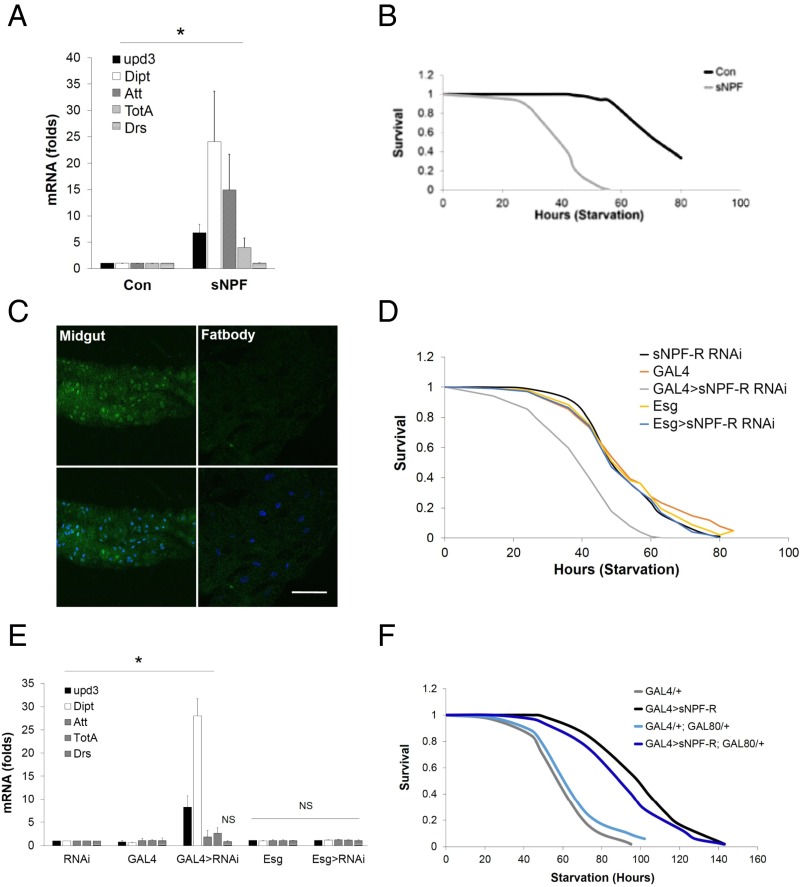
sNPF suppresses the gut immune response. (A) Starvation sensitivity of control and sNPF mutant (sNPFNP6301) flies (n = 100). (B) Relative mRNA amounts for proinflammatory genes in control and sNPFNP6301 mutant flies (n = 3). (C) Immunofluorescence analysis of UAS-GFP reporter expression in the midgut and fat body of sNPF-R promoter-GAL4 flies. (Scale bar, 50 μm.) Starvation sensitivity (D) and immune response gene expression (E) in flies depleted of sNPF-R with gut-specific GAL4 drivers are shown. Genotypes are as follows: sNPF-R RNAi (UAS-Dicer2, UAS-sNPF-R RNAi/+), GAL4 (Myo1A-Gal4/+), GAL4 > sNPF-R RNAi (Myo1A-GAL4/+; UAS-Dicer2, UAS-sNPF RNAi/+), escargot (Esg) (Esg-Gal4/+), and Esg > sNPF-R RNAi (Esg-GAL4/+; UAS-Dicer2, UAS-sNPF RNAi/+). (F) Effect of sNPF-R overexpression using a sNPF-R-GAL4 driver on starvation sensitivity. The effect of blocking sNPF-R expression in neuronal tissues using elav-GAL80 is indicated. Genotypes are GAL4/+ (sNPF-R-GAL4/+); GAL4 > sNPF-R (sNPF-R-GAL4/UAS-sNPF-R); GAL4/+, GAL80/+ (elav-GAL80; sNPF-R-GAL4/+); and GAL4 > sNPF-R; GAL80/+ (elav-GAL80; sNPF-R-GAL4/UAS-sNPF-R) (log-rank, P < 0.01). *P < 0.05 [Student’s t test (B and E) or log-rank test (A, D, and F)]. Data for B and E are from three independent experiments (data are mean ± SD).
In line with these increases in immune gene expression, sNPF mutant flies have lower triglycerides, and they are more sensitive to starvation compared with controls (Fig. 4A and Fig. S3 D and E). Consistent with a role for neuronal sNPF, blocking synaptic release in the sNPF neurons with tetanus toxin increased starvation sensitivity in wild-type flies (Fig. S3F).
Having seen that Crtc overexpression in sNPF neurons enhances starvation resistance in wild-type flies, we wondered whether these effects are indeed dependent on sNPF itself. By contrast with its salutary effects in a wild-type background, Crtc overexpression in sNPF mutant flies failed to promote starvation resistance (Fig. 3H). Taken together, these results demonstrate the importance of sNPF in mediating metabolic effects of Crtc.
Enterocyte sNPF Receptors Mediate Effects of Neuronal sNPF.
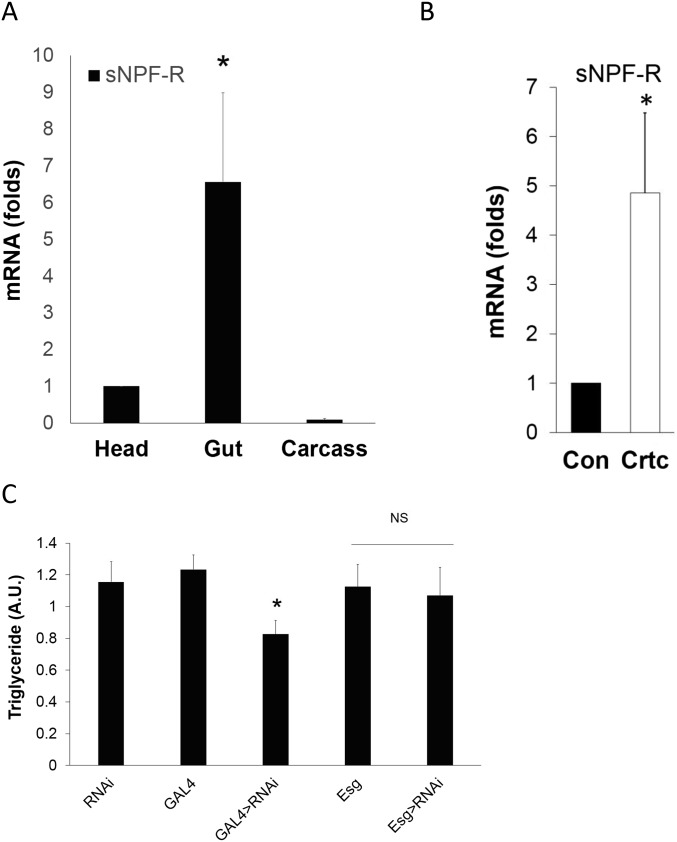
To determine the mechanism by which sNPF mediates effects of Crtc on starvation resistance, we evaluated sNPF receptor (sNPF-R) expression in tissues from adult flies. mRNA amounts for sNPF-R were detected in the head and at fivefold higher levels in the midgut (Fig. S4A). Similarly, UAS-GFP reporter expression was observed in enterocytes of transgenic flies expressing a sNPF-R promoter-GAL4 driver (Fig. 4C). Indeed, midgut sNPF-R expression was significantly increased in Crtc flies, likely reflecting the compensatory receptor up-regulation that often accompanies decreases in ligand binding (Fig. S4B).
Fig. S4.
Gut sNPF receptors mediate effects of sNPF on gut tolerance and starvation sensitivity. (A) Relative sNPF-R mRNA amounts in the fly head, gut, and carcass. (B) sNPF-R mRNA amounts in wild-type and Crtc25-3 flies. (C) Triglyceride content in control and mutant flies depleted of sNPF-R expression in the gut. Genotypes are sNPF-R RNAi (UAS-Dicer2, UAS-sNPF-R RNAi/+), GAL4 (Myo1A-Gal4/+), GAL4 > sNPF-R RNAi (Myo1A-GAL4/+; UAS-Dicer2, UAS-sNPF RNAi/+), escargot (Esg) (Esg-Gal4/+), and Esg > sNPF-R RNAi (Esg-GAL4/+; UAS-Dicer2, UAS-sNPF RNAi/+). NS, not significant. *P < 0.05, Student’s t test with three independent experimental repeats.
We tested the importance of sNPF-R in mediating effects of Crtc in the gut. Similar to disruption of sNPF, RNAi-mediated knockdown of sNPF-R with the enterocyte-specific GAL4 driver Myo1A-GAL4 (23) decreased fly starvation resistance compared with controls (Fig. 4D). By contrast, knockdown of sNPF-R in ISCs and enteroblasts (EBs) with an Esg-GAL4 driver (24) did not modulate starvation resistance. Indeed, knockdown of sNPF-R in enterocytes, but not ISCs or EBs, also up-regulated inflammatory peptide expression while decreasing energy stores (Fig. 4E and Fig. S4C). Conversely, overexpression of sNPF-R from its own promoter-driven GAL4 (sNPF-R-GAL4) increased starvation resistance in a wild-type background (Fig. 4F). sNPF-R-GAL4 drives expression in both the gut and nervous system. To determine in which tissue sNPF-R functions to increase starvation resistance, we blocked sNPF-R overexpression specifically in neuronal tissues by coexpression of the GAL80 repressor with elav-GAL80. The inclusion of elav-GAL80 did not attenuate sNPF-R effects on starvation resistance, indicating that the expression of SNPF-R specifically in the gut is required for metabolic effects (Fig. 4F). Collectively, these results demonstrate that enterocyte sNPF-R is required for salutary effects of Crtc on innate immunity and energy balance.
Discussion
Disruptions in energy balance are a component of the collateral damage associated with mounting an immune response. In addition to regulating the magnitude of an immune response, energy allocation must be properly regulated to minimize physiological damage during infection. We found that Drosophila sNPF, a mammalian NPY homolog, is regulated by CrebB/Crtc within the CNS, where it promotes energy balance by maintaining epithelial integrity and thereby attenuating overexuberant immune activation in the gut. The effects of sNPF were unexpected, given its role in food-seeking behavior (25). Indeed, food intake appears comparable between Crtc mutants and control flies (12).
The effects of sNPF are mediated by enterocyte sNPF-Rs, suggesting that the sNPF brain-gut signal is released by a subset of the sNPF+ neurons that directly innervate the gut (26). Although neuronal activity is known to contribute to energy homeostasis, our results suggest that the modulation of the gut immune system by CrebB/Crtc is a critical component in this process.
Epithelial tissues are typically colonized by both commensal and invasive microbes. sNPF appears to be actively expressed and released from the CNS in times of stress, providing nonautonomous control of gut immunity from the brain. Based on its widespread expression in the midgut, sNPF-R may provide ubiquitous attenuation of the innate immune response. Consistent with our observations in Drosophila, activation of the NPY receptor ortholog (NPR-1) in Caenorhabditis elegans also down-regulates inflammatory gene expression (27). Our studies extend these findings by showing how a neuronal fasting-inducible pathway modulates energy balance via its effects on the gut immune system.
Following their activation, sNPF-Rs appear to promote energy balance by enhancing epithelial integrity. Although the mechanism underlying these effects is unclear, we note that disruption of the tight junction protein Bbg in flies also causes constitutive up-regulation of innate immunity genes (17). Future studies should reveal whether sNPF-R modulates the activity of Bbg or related proteins in enterocytes.
In mammals, inflammatory bowel diseases, such as ulcerative colitis, are often associated with profound weight loss, due, in part, to the chronic activation of the immune system. By reducing inflammatory gene expression and enhancing energy homeostasis, gut neuropeptides, such as NPY, may provide therapeutic benefit in this setting.
Methods
Fly Stocks.
All Drosophila melanogaster lines were maintained at 25 °C on standard food medium. GMR19H06-GAL4 (sNPF-Gal4) (28), GMR66A02-GAL4 (sNPF-R-GAL4) (28), UAS-CD8::GFP (29), Dipt-LacZ (30), Drs-GFP (31), r4-GAL4 (32), and Fb-gal4 (33) lines were obtained from the Bloomington Drosophila Stock Center. CRE-luc reporter flies (20) and Upd3-Gal4 (15), UAS-GFP, Dipt-GFP (34), and Crtc25-3 (12) lines have been described previously. The sNPFNP6301 line was purchased from the Drosophila Genetic Resource Center. The sNPF RNAi lines were purchased from the Vienna Drosophila Resource Center stock center.
Generation of Creb and sNPF Deletion Alleles.
CrebB deletion alleles were generated by mobilization of the P{EPgy2}insertion EY04780 with a ∆2-3 source of transposase (35). Potential deletions were screened by PCR. The CrebB∆400 deletion line has an ∼3.5-kb deletion that removes sequences encoding the CREB leucine zipper-DNA binding domain. The CrebB∆400 deletion break points were confirmed by sequencing.
sNPF deletion was generated using clustered regularly interspaced short palindromic repeats (CRISPRs) (36). Briefly, two pairs of chi-RNA targeting sequences were selected using CRISPR Resource (www.genome-engineering.org/crispr), sNPF-E2s, TACAGTTTCCCATCAACGCG, sNPF-E2as, CGCGTTGATGGGAAACTGTAC, sNPF-E5s, TGGCAATGACAATGGCAGCT, sNPF-E5as, and AGCTGCCATTGTCATTGCCAC. The sequences were cloned into pU6-BbsI-chiRNA. Targeting DNAs were used with phsp70-Cas9 to inject fly embryos. Positive deletions were selected by PCR and confirmed with sequencing.
Cultivation of Bacterial Flora.
Five third-instar larvae were collected, washed twice in PBS, and sterilized in 75% (vol/vol) ethanol for 2 min to eliminate bacteria on the larval surface. The larvae were then washed with sterile PBS and homogenized in 100 μL of sterile PBS. Ten microliters of the homogenate was diluted in sterile PBS and spread on LB plates. The plates were incubated at 25 °C for 3 d.
Salmonella Injections.
Salmonella typhimurium (a gift from D. Monack, Stanford University, Stanford, CA) was grown overnight in LB media shaking at 37 °C. Cultures were diluted, and 1,000 cfu was injected into male 5- to 7-d-old files. Flies were anesthetized with CO2 and injected with 50 nL of culture using a picospritzer and pulled glass needle. Flies were injected in the anterior abdomen on the ventrolateral surface. Flies were then placed in vials containing molasses medium and incubated at 29 °C, 65% humidity. The number of dead flies was counted daily. Using Prism (GraphPad) software, Kaplan–Meier survival curves were generated, and statistical analysis was done using log-rank analysis. Survival was tested at least three times and gave similar results for each trial. For cfu determination, infected flies were homogenized in PBS supplemented with 1% Triton X-100 and serially diluted. Dilutions were plated on LB agar plates and incubated overnight. The data were plotted using Prism software for three independent experiments. The P value was determined with a nonparametric two-tailed t test.
Cell Culture, Transfection, and Luciferase Assay.
Drosophila S2 cells were maintained as previously described (12), and 1.5-kb and 0.7-kb sNPF promoter fragments were cloned into a pGL3 vector. Transfections were performed as reported (12). After 2–3 d infection, cells were collected for protein or RNA analysis.
Starvation and Oxidative Stress.
Starvation and oxidative stress assays have been described (12).
Tissue Staining.
Primary and secondary antibodies were used as indicated: anti-LacZ (41-1a; 1:100) from the Developmental Studies Hybridoma Bank, rabbit anti-pH 3 (1:1,000; Cell Signaling), mouse anti-GFP (1:1,000; Life Technologies), Alexa Fluor 488- and 568-conjugated antibodies (1:1,000; Life Technologies), rabbit anticaudal (Paul McDonald, University of Texas at Austin, Austin), rabbit anti-sNPF (Jan Veenstra, Universite de Bordeaux, Bordeaux, France and Kweon Yu, Korea Research Institute of Bioscience and Biotechnology, Daejeon, South Korea), and mouse anti-CrebB (Jerry Yin, University of Wisconsin, Madison, WI). Tissues were counterstained with DAPI. Msn-lacZ activity was detected in transgenic flies by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining. Images from staining are representative of three independent experiments.
Quantitative PCR.
RNA was isolated from antennae of 10 female flies for each sample. The RNeasy kit (QIAGEN) was used to isolate RNA, and reverse transcription was performed using the Retroscript kit (Ambion) with random decamers. cDNA was subjected to quantitative PCR (qPCR) analysis using SYBR green detection on a Light Cycler 480 II thermocycler (Roche). All values are the average of four replicates, each measured in triplicate and normalized to rp49 mRNA.
qPCR Primers.
The following qPCR primers were used in this study:
upd-Forward (Fw) CTC CAC ACG CAC AAC TAC AAG
upd-Reverse (Re) GAT ACG CAA AGC TGG CCA CG
upd2-Forward (F) CGGAACATCACGATGAGCGAAT
upd2-Reverse (R) TCGGCAGGAACTTGTACTCG
upd3-Fw GTC TGA ATC TCA CTA GCA AAC
upd3-Re AGG TCC CAG TGC AAC TTG ATG
Stat92e-F CTGGGCATTCACAACAATCCAC
qStat92e-R GTATTGCGCGTAACGAACCG
Creb-Fw TCC CTG ATG CAA TTG GAT CC
Creb-Re GCC TCC CTG TTC TTC TGC AG
Crtc-Fw TAC TAT GAG GAG GCT GTT GC
Crtc-Re CTT CAA CTG TTA CTA CTG CTG
Lactobacillus sp. (L.s.)-Fw GAA AGA TGG CTT CGG CTA TCA
L.S.-Re CAA TAC CTG AAC AGT TAC TCT CA
Acetobacter sp. (A.s.)-Fw AGG GTC AAA GGC GCA AGT C
A.S.-Re TAC CGT CAT CAT CGT CCC CG
sNPF-Fw GGA AGC CAC AAC GAC TTC GC
sNPF-Re ATT TCG TAC TGC TGC TGG AG.
ChIP.
Chromatin was prepared and immunoprecipitated as described, with the following modifications. The heads of 0.5 g of flies were ground in liquid nitrogen with a mortar and pestle before cross-linking with formaldehyde. Chromatin was sheared by 15 10-s bursts of sonication at level 6 power with a Virtis Virsonic 100 Sonicator. IgG was used as a negative control, and monoclonal anti-GFP was used to pull down chromatin complexes. Precipitated DNA fragments were purified and resuspended in 50 μL of water for qPCR analysis.
Smurf Assay.
Seven- to 10-d-old flies were collected, starved for 2 h, and then transferred to normal food containing 10% blue food dye (FD&C blue dye no. 1). Flies were incubated in vials with blue dye at 29 °C. Flies with extended blue coloration not limited to the proboscis and crop were scored positive.
Statistical Analyses.
Statistical significance was determined using an unpaired two-tailed Student’s t test with unequal variance. Statistical significance for survival curves was determined using a log-rank test. Data are presented as mean ± SD. Two-tailed P values of ≤0.05 were considered significant. The target number of samples in each group was determined based on numbers reported in published studies. No statistical methods were used to predetermine sample size. Microsoft Excel was used for statistical analyses. The experiments were not randomized.
Acknowledgments
We thank Jan Veenstra and Kweon Yu for sNPF antisera, Paul McDonald for caudal antiserum, and Jerry Yin for CrebB antiserum. We also thank Huaqi Jiang, Jean-Luc Imler, the Bloomington Drosophila Stock Center, and the Vienna Drosophila RNAi Center for fly stocks. This work was supported by NIH Grant R37 DK083834 (to M.M.), NIH grant R01 DK077979 (to J.B.T.), and Leona M. and Harry B. Helmsley Charitable Trust Grant 2012-PG-MED002 (to M.M.). R.S. is supported by the Glenn Center for Aging Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606106113/-/DCSupplemental.
References
- 1.Ayres JS, Schneider DS. Tolerance of infections. Annu Rev Immunol. 2012;30:271–294. doi: 10.1146/annurev-immunol-020711-075030. [DOI] [PubMed] [Google Scholar]
- 2.Schneider DS, Ayres JS. Two ways to survive infection: What resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol. 2008;8(11):889–895. doi: 10.1038/nri2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. 2012;335(6071):936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Råberg L, Sim D, Read AF. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science. 2007;318(5851):812–814. doi: 10.1126/science.1148526. [DOI] [PubMed] [Google Scholar]
- 5.Rera M, Clark RI, Walker DW. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc Natl Acad Sci USA. 2012;109(52):21528–21533. doi: 10.1073/pnas.1215849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann JA. The immune response of Drosophila. Nature. 2003;426(6962):33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 7.Shim J, Mukherjee T, Banerjee U. Direct sensing of systemic and nutritional signals by haematopoietic progenitors in Drosophila. Nat Cell Biol. 2012;14(4):394–400. doi: 10.1038/ncb2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker T, et al. FOXO-dependent regulation of innate immune homeostasis. Nature. 2010;463(7279):369–373. doi: 10.1038/nature08698. [DOI] [PubMed] [Google Scholar]
- 9.Hwangbo DS, Gershman B, Tu M-P, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429(6991):562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 10.Giannakou ME, et al. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;305(5682):361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- 11.Dionne MS, Pham LN, Shirasu-Hiza M, Schneider DS. Akt and FOXO dysregulation contribute to infection-induced wasting in Drosophila. Curr Biol. 2006;16(20):1977–1985. doi: 10.1016/j.cub.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 12.Wang B, et al. The insulin-regulated CREB coactivator TORC promotes stress resistance in Drosophila. Cell Metab. 2008;7(5):434–444. doi: 10.1016/j.cmet.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi S, Kim W, Chung J. Drosophila salt-inducible kinase (SIK) regulates starvation resistance through cAMP-response element-binding protein (CREB)-regulated transcription coactivator (CRTC) J Biol Chem. 2011;286(4):2658–2664. doi: 10.1074/jbc.C110.119222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altarejos JY, Montminy M. CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12(3):141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agaisse H, Petersen U-M, Boutros M, Mathey-Prevot B, Perrimon N. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev Cell. 2003;5(3):441–450. doi: 10.1016/s1534-5807(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 16.Jiang H, et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137(7):1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonnay F, et al. big bang gene modulates gut immune tolerance in Drosophila. Proc Natl Acad Sci USA. 2013;110(8):2957–2962. doi: 10.1073/pnas.1221910110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuraishi T, Hori A, Kurata S. Host-microbe interactions in the gut of Drosophila melanogaster. Front Physiol. 2013;4:375. doi: 10.3389/fphys.2013.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin JCP, et al. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell. 1994;79(1):49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- 20.Belvin MP, Zhou H, Yin JCP. The Drosophila dCREB2 gene affects the circadian clock. Neuron. 1999;22(4):777–787. doi: 10.1016/s0896-6273(00)80736-9. [DOI] [PubMed] [Google Scholar]
- 21.Wegener C, Herbert H, Kahnt J, Bender M, Rhea JM. Deficiency of prohormone convertase dPC2 (AMONTILLADO) results in impaired production of bioactive neuropeptide hormones in Drosophila. J Neurochem. 2011;118(4):581–595. doi: 10.1111/j.1471-4159.2010.07130.x. [DOI] [PubMed] [Google Scholar]
- 22.Root CM, Ko KI, Jafari A, Wang JW. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell. 2011;145(1):133–144. doi: 10.1016/j.cell.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Apidianakis Y, Rahme LG. Drosophila melanogaster as a model for human intestinal infection and pathology. Dis Model Mech. 2011;4(1):21–30. doi: 10.1242/dmm.003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amcheslavsky A, Jiang J, Ip YT. Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell. 2009;4(1):49–61. doi: 10.1016/j.stem.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nässel DR, Enell LE, Santos JG, Wegener C, Johard HA. A large population of diverse neurons in the Drosophila central nervous system expresses short neuropeptide F, suggesting multiple distributed peptide functions. BMC Neurosci. 2008;9:90. doi: 10.1186/1471-2202-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veenstra JA, Agricola H-J, Sellami A. Regulatory peptides in fruit fly midgut. Cell Tissue Res. 2008;334(3):499–516. doi: 10.1007/s00441-008-0708-3. [DOI] [PubMed] [Google Scholar]
- 27.Styer KL, et al. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science. 2008;322(5900):460–464. doi: 10.1126/science.1163673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manning L, et al. A resource for manipulating gene expression and analyzing cis-regulatory modules in the Drosophila CNS. Cell Reports. 2012;2(4):1002–1013. doi: 10.1016/j.celrep.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22(3):451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 30.Reichhart JM, et al. Insect immunity: Developmental and inducible activity of the Drosophila diptericin promoter. EMBO J. 1992;11(4):1469–1477. doi: 10.1002/j.1460-2075.1992.tb05191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrandon D, et al. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 1998;17(5):1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167(1):311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grönke S, et al. Control of fat storage by a Drosophila PAT domain protein. Curr Biol. 2003;13(7):603–606. doi: 10.1016/s0960-9822(03)00175-1. [DOI] [PubMed] [Google Scholar]
- 34.Tzou P, et al. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity. 2000;13(5):737–748. doi: 10.1016/s1074-7613(00)00072-8. [DOI] [PubMed] [Google Scholar]
- 35.Robertson HM, et al. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118(3):461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gratz SJ, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194(4):1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]