Significance
The colonial tunicate, Botryllus schlosseri, undergoes natural self–nonself recognition that results in formation of a chimera. Following fusion, one chimeric partner is often eliminated in a process of allogeneic resorption, allowing for study of the induction and loss of tolerance. We provide evidence that elimination of one partner in a chimera is an immune cell-based rejection and that the principal cell type to mediate partner elimination is a cytotoxic morula cell (MC). Proinflammatory, blastogenic “takeover” programs render MCs cytolytic and, in collaboration with activated phagocytes, eradicate chimeric partners. These findings identify a conserved strategy for histocompatible elimination through the integrated function of both phagocytic programs and licensing of cytotoxic cells. The coordination of both dimensions of innate immune recognition elicits efficient elimination of chimeric partners.
Keywords: apoptosis, histocompatibility, innate immunity, inflammation, macrophages
Abstract
In a primitive chordate model of natural chimerism, one chimeric partner is often eliminated in a process of allogeneic resorption. Here, we identify the cellular framework underlying loss of tolerance to one partner within a natural Botryllus schlosseri chimera. We show that the principal cell type mediating chimeric partner elimination is a cytotoxic morula cell (MC). Proinflammatory, developmental cell death programs render MCs cytotoxic and, in collaboration with activated phagocytes, eliminate chimeric partners during the “takeover” phase of blastogenic development. Among these genes, the proinflammatory cytokine IL-17 enhances cytotoxicity in allorecognition assays. Cellular transfer of FACS-purified MCs from allogeneic donors into recipients shows that the resorption response can be adoptively acquired. Transfer of 1 × 105 allogeneic MCs eliminated 33 of 78 (42%) recipient primary buds and 20 of 76 (20.5%) adult parental adult organisms (zooids) by 14 d whereas transfer of allogeneic cell populations lacking MCs had only minimal effects on recipient colonies. Furthermore, reactivity of transferred cells coincided with the onset of developmental-regulated cell death programs and disproportionately affected developing tissues within a chimera. Among chimeric partner “losers,” severe developmental defects were observed in asexually propagating tissues, reflecting a pathologic switch in gene expression in developmental programs. These studies provide evidence that elimination of one partner in a chimera is an immune cell-based rejection that operates within histocompatible pairs and that maximal allogeneic responses involve the coordination of both phagocytic programs and the “arming” of cytotoxic cells.
The colonial marine species, Botryllus schlosseri, offers a unique platform to study mechanisms underlying loss of tolerance in a natural model system. Colonies undergo a genetically controlled histocompatibility reaction that can result in vascular fusion of distinct genotypes, creating a chimera (1, 2). Natural history studies among fused colonies show that partners rarely exist as stable chimeras (3–7). One chimeric partner is often eliminated in a process of allogeneic resorption, suggesting a break in immunological nonreactivity. Moreover, the eliminated partner may still persist, and even parasitize the nonresorbed partner, but at the level of the cell lineage (8, 9). Insight has been gained into fusion-partner resorption from observational studies showing physical similarities with the developmental period known as “takeover,” or blastogenic stage D (4, 10). B. schlosseri colonies are composed of clonogenic individuals, termed “zooids,” that undergo weekly cycles of death and regeneration, culminating in a massive wave of programmed cell death and removal, or takeover (11). These studies support the involvement of activated phagocytes in the elimination of tissues of the “losing” partner.
Here, we study the progression by which fused colonies eliminate chimeric partners and show that partner elimination seems to be an integrated function involving activation of phagocytic programs and licensing of cytotoxic cells. Neither process alone is sufficient to induce efficient chimeric partner elimination. Using prospective isolation of defined cells, we demonstrate that the resorption response can be adoptively transferred from allogeneic donors and that the principle cell type mediating this effect is a cytotoxic morula cell (MC). MCs have been implicated in rejection reactions among incompatible allogeneic colonies, leading to the formation of necrotic lesions called “points of rejection” (PORs) (12). Furthermore, we show developmental-regulated programmed cell death pathways, initiated during the takeover phase of colony life, “prime” MCs, thereby enhancing their capacity for alloreactivity. Using transcriptome data, we probed the role of specific takeover pathways to identify a proinflammatory cytokine-like factor, an IL-17 family member, that renders cells cytotoxic in allorecognition assays, suggesting that licensing signals active at the time of takeover contribute to alloreactivity. Therefore, a greater understanding by which phagocytic and cytotoxic recognition programs coordinate to eliminate allogeneic cells may uncover new strategies to promote or eliminate immunological nonreactivity.
Results
To begin to study the progression by which fused colonies eliminate chimeric partners, we generated 42 fused-juvenile chimeras from 177 F1 individuals, progeny of WT colonies collected in Monterey, CA. Chimeras were observed throughout the study period and scored for the degree of resorption of one partner, as follows: 0, none; I, mild; II, moderate; III, severe; IV, complete (Fig. 1B and Fig. S1). Consistent with previous observations, stable-mixed long-term chimerism occurs rarely. The vast majority of F1 chimeras exhibited morphologic evidence for allogeneic resorption (Tables S1 and S2), with 88% of colonies having progressed to a resorption score of III by 30 d. Furthermore, the onset of allogeneic resorption in 29 of 42 (69%) chimeras occurred during blastogenic stage D, corresponding to a developmental period of intensive phagocytosis of apoptotic bodies (Tables S1 and S2).
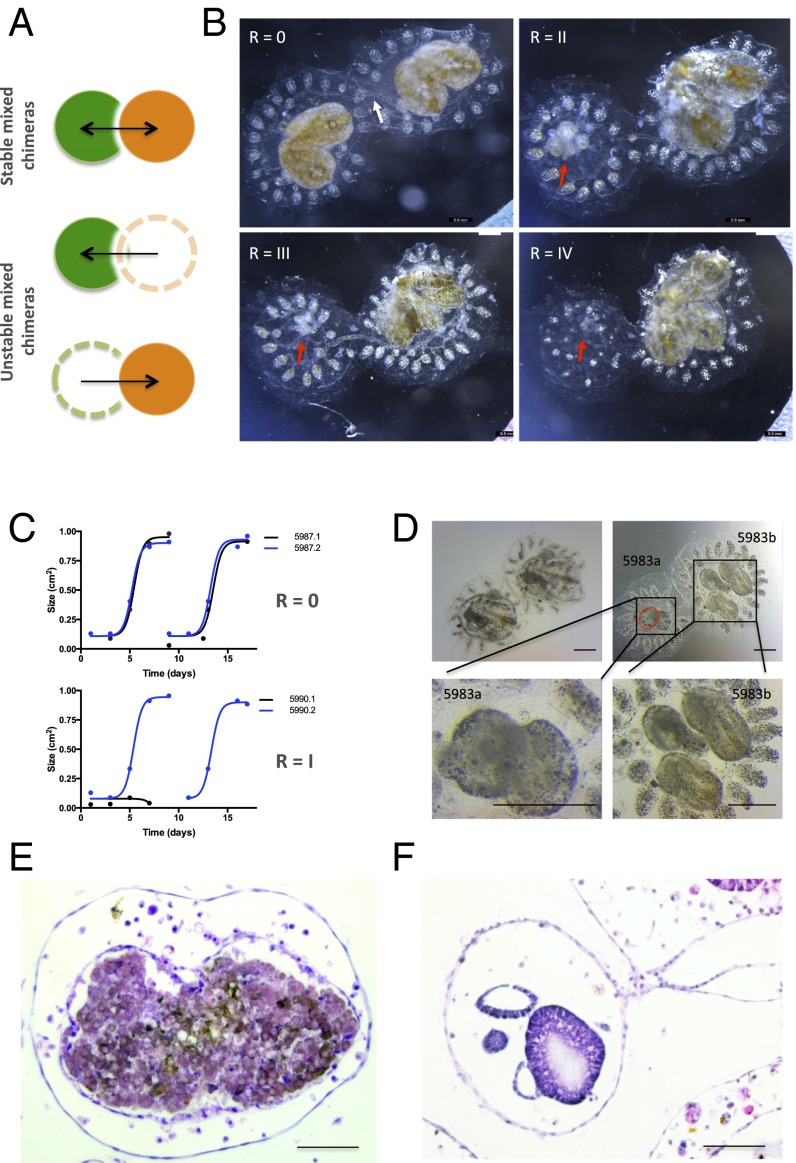
Fig. 1.
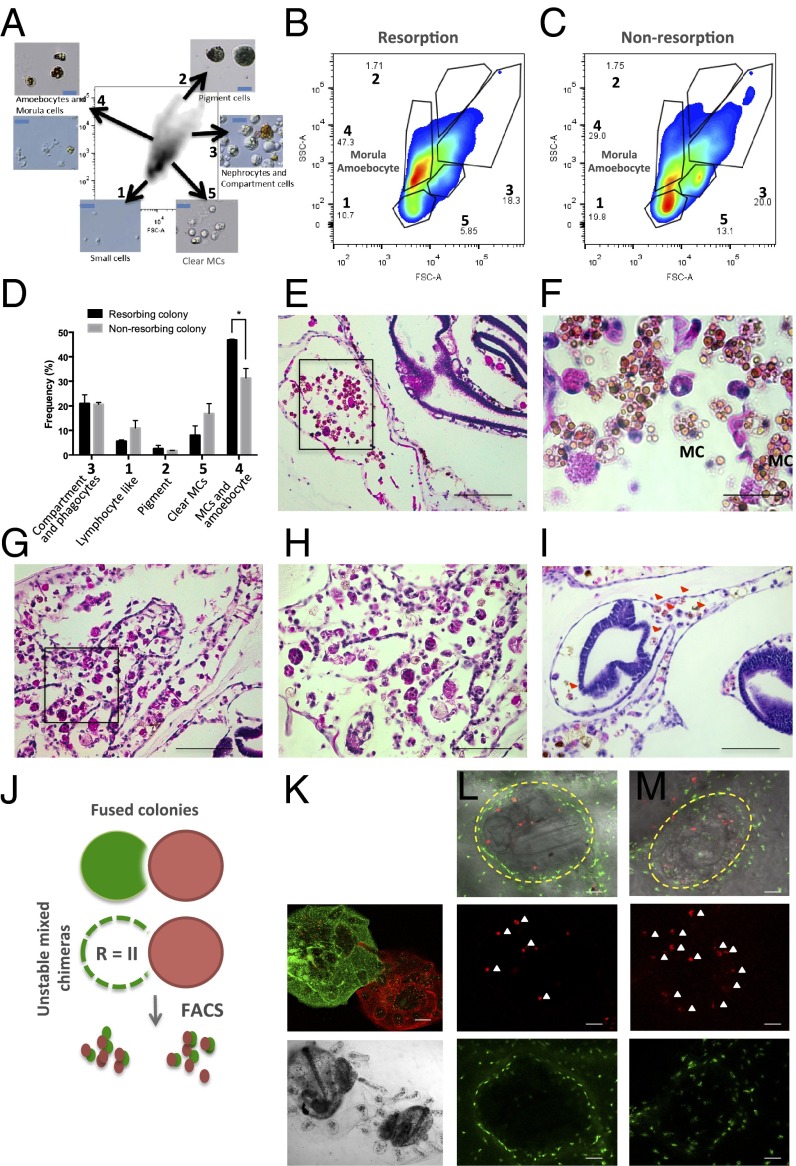
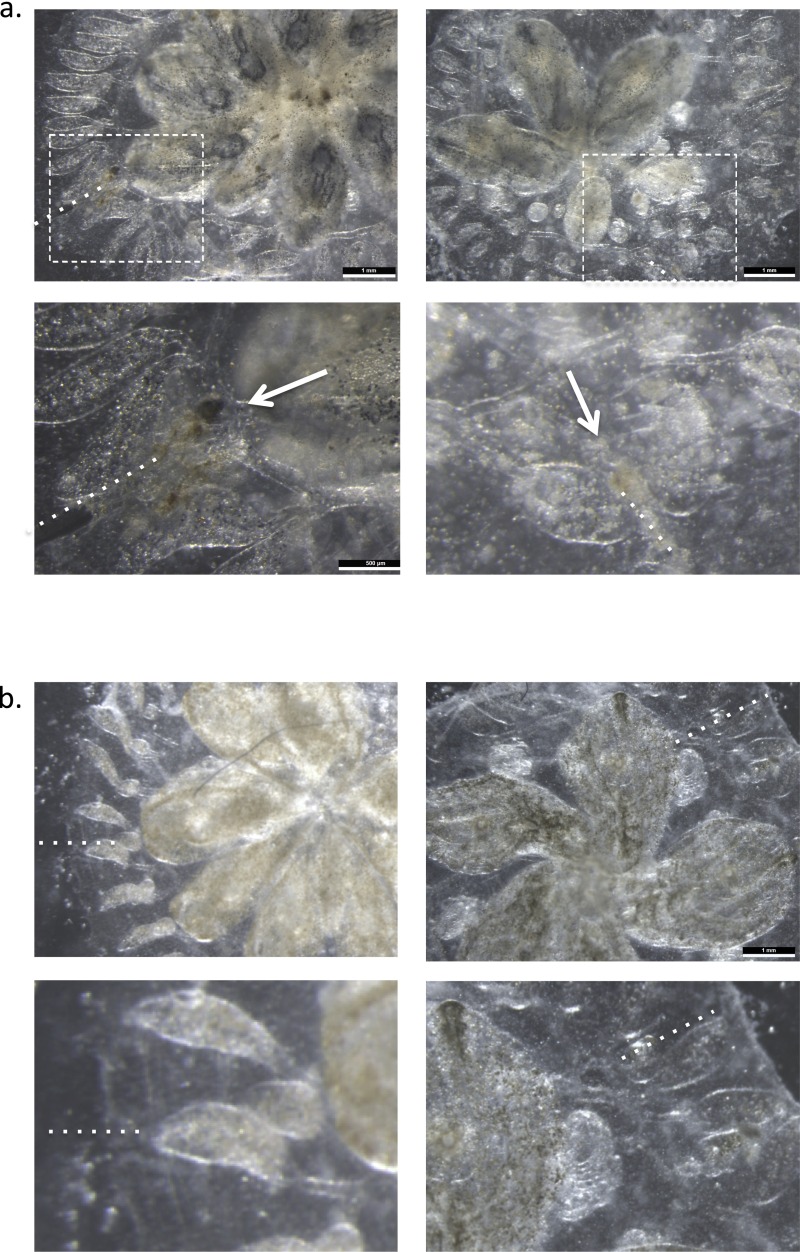
Effect of allogeneic resorption on asexual propagation. (A) Depiction of controlled fusion reactions among compatible, yet genetically distinct, fused partners that result in resorption of one partner (depicted with dotted lines). (B) Time lapse imaging of a juvenile fused chimera (white arrow depicts blood vessel anastomosis between two colonies) undergoing resorption. Images were acquired at 24-h intervals starting 5 d after fusion. Moderate tissue destruction (red arrow) was first observed 6 d after fusion (Top Right) and progressed to complete stage R = IV resorption within 72 h (Bottom Right) during the takeover period (stage D) of blastogenesis (nonresorbing colony, Bottom Left). (C) Growth curves (cm2) of primary buds in a stable chimera (depicted as blue and black lines) exhibit synchronized cycles of expansion and regression (Top). Unstable chimeras exhibit growth arrest in asexually propagating tissues (Bottom). (D) High power magnification of abnormal zooids and buds (Top Right, red dotted lines) in allogeneic resorbing colonies compared with nonresorbing colonies (Top Left). (Scale bars: Top, 500 μM; Bottom, 100 μM.) (E) Hematoxylin/eosin (H&E)-stained tissue section of a resorbing 1° bud showing effacement of developing structures with associated destructive changes and infiltration of cytotoxic and phagocytic cell populations compared with nonresorbing buds of the same developmental stage as in F. (Scale bars: Left, 200 μM; Right, 100 μM.)
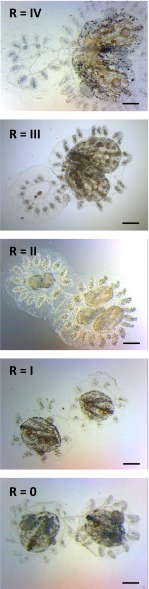
Fig. S1.
Resorption classification. Representative images of fused pairs undergoing allogeneic resorption and graded according to severity. Resorption status was graded on a 0 to IV scale: (Bottom) Stage 0 (no resorption observed); (Second From Bottom) stage I (mild resorption = zooid contraction); (Middle) stage II (moderate resorption = zooid contraction, 1° and 2° bud developmental arrest, chimeric blastogeneic asynchrony, residual cardiac activity); (Second From Top) stage III (severe resorption = only remnants visible, no cardiac activity); and (Top) stage IV (complete resorption = one partner has been fully eliminated; only supporting tissue (ampullae, blood vessels, and tunic) remain). (Scale bars: 2 mm.)
Table S1.
Progression to allogeneic resorption in juvenile B. schlosseri chimeras
| Chimera ID | Resorption | Nonresorption | Time to resorption, d postfusion | Stage at time of resorption (A–D) | ||
| RI | RIII | Stage = R1 | Stage > RIII | |||
| 5780.11 | Y | N | 4 | 6 | D | D |
| 5780.12 | Y | N | 6 | 11 | C | D |
| 5780.13 | Y | N | 10 | 12 | C | D |
| 5780.15 | Y | N | 5 | 10 | D | D |
| 5768.18 | Y | N | 6 | 13 | C | A |
| 5764.19 | Y | N | 4 | 11 | D | D |
| 5764.23 | Y | N | 7 | 17 | D | D |
| 5764.24 | Y | N | 7 | 10 | C | D |
| 5644.25 | Y | N | 13 | 17 | D | D |
| 5644.26 | N | Y | 0 | 0 | — | — |
| 5644.27 | Y | N | 3 | 5 | C | D |
| 5776.28 | Y | N | 7 | 17 | D | D |
| 5776.29 | N | Y | 0 | 0 | — | — |
| 5767.30 | Y | N | 5 | 28 | D | D |
| 5767.31 | Y | N | 2 | 6 | D | D |
| 5767.32 | Y | N | 2 | 4 | D | A |
| 5805.42 | Y | N | 12 | 18 | D | D |
| 5805.43 | Y | N | 18 | 25 | D | D |
| 5805.45 | Y | N | 8 | 13 | D | D |
| 5805.46 | Y | N | 8 | 13 | D | D |
| 5780.20 | Y | N | 4 | 10 | C | D |
| 5780.21 | Y | N | 4 | 9 | D | D |
| 5775.60 | Y | N | 9 | 15 | D | D |
| 5775.70 | N | Y | 0 | 0 | — | — |
| 5984.20 | Y | N | 16 | 23 | D | D |
| 5984.90 | Y | N | 9 | 14 | D | D |
| 5984.15 | Y | N | 14 | 18 | C | D |
| 5984.24 | Y | N | 18 | 22 | B | D |
| 5984.34 | Y | N | 12 | 16 | C | D |
| 5983.60 | N | Y | 0 | 0 | — | — |
| 5805.49 | Y | N | 18 | 25 | D | D |
| 5805.50 | Y | N | 10 | 12 | C | D |
| 5805.52 | Y | N | 6 | 13 | C | A |
| 5805.53 | Y | N | 12 | 16 | D | D |
Results from controlled allorecognition assays among outbred juvenile, oozooid B. schlosseri chimeras. Chimeras were observed throughout the study period for allogeneic resorption. Information includes the resorption status, time to progression to resorption after fusion, and developmental stage at time of resorption. Y, yes the phenomena was observed; N, no observed phenomena; —, indicate no observed resorption, therefore, no time to resorption or stage of resorption.
Table S2.
Summary of all observed fused colonies
| Resorption, % | 88.57 |
| Nonresorption, % | 11.43 |
| Mean time to first presentation of R = I, d | 7.62 |
| Mean time to progression to R = III, d | 12.62 |
| Stage D at R = I, % | 63.64 |
| Stage C at R = I, % | 33.33 |
| Stage B at R = I, % | 3.03 |
| Stage A at R = I, % | 0.00 |
Using time-lapse imaging, we observed a pattern of colony regression that began with chimeric partners breaking developmental synchrony with each other, and progressed to simultaneous loss of parental zooids and buds. Hallmark features observed in B. schlosseri naive and long-term stable mixed chimeras are synchronized cycles of death and regeneration, termed blastogenesis (Fig. 1C, Top and Fig. S2) (13, 14). Among chimeric partner “losers,” however, severe developmental defects were observed in asexually propagating tissues (Fig. 1C, Bottom and Fig. S3). Nascent buds emerging from the body walls of parental zooids exhibit overall poor growth that, on close inspection, revealed a wide variety of developmental defects. In the most severe forms, developing structures had been completely effaced by inflammatory infiltrate and showed extensive apoptotic changes (Fig. 1 D and E). Histologic and FACS analysis experiments revealed infiltrating cytotoxic MCs intermixed with amoebocyte cells (phagocytes) derived from “winning” partners at sites of colony resorption (Fig. 2 A–M). We labeled colonies with distinct lipophilic membrane fluorescent stains (CellTracker Green/Red) to follow the migration patterns of cells (Fig. 2 J–K). The concentration of MCs seemed to correlate with the severity of resorption, ranging from variable to scant MCs in mild forms, to heavy infiltration in the most severe cases. MCs have been implicated in rejection reactions among incompatible allogeneic colonies in a number of urochordate species (12, 15, 16). For comparison, we examined tissues matched for the developmental stage of takeover, where parental zooids contract and undergo rapid engulfment and elimination. Histologic examination of regressing parental zooids showed invasion by numerous phagocytic populations containing engulfed cellular corpses, but a near absence of cytotoxic MCs (Fig. 2 G and H). Interestingly, it has been suggested that there are two morphologies of MCs, distinguished by the appearance of their cytotoxic vacuoles. In the first, vacuoles appear clear. Upon phenoloxidase activation, such as in the setting of an allorecognition response, a melanization reaction occurs, giving rise to yellowish-green vacuoles with increased pigmentation (17–20). By FACS, we can differentiate these two populations, and, in accordance with the above in the resorbing colony, there is a significant increase in MCs bearing pigmented vacuoles (population 4 in Fig. 2 A–D). These findings suggest that the major effector cells to mediate allogeneic resorption reside within the MC compartment.
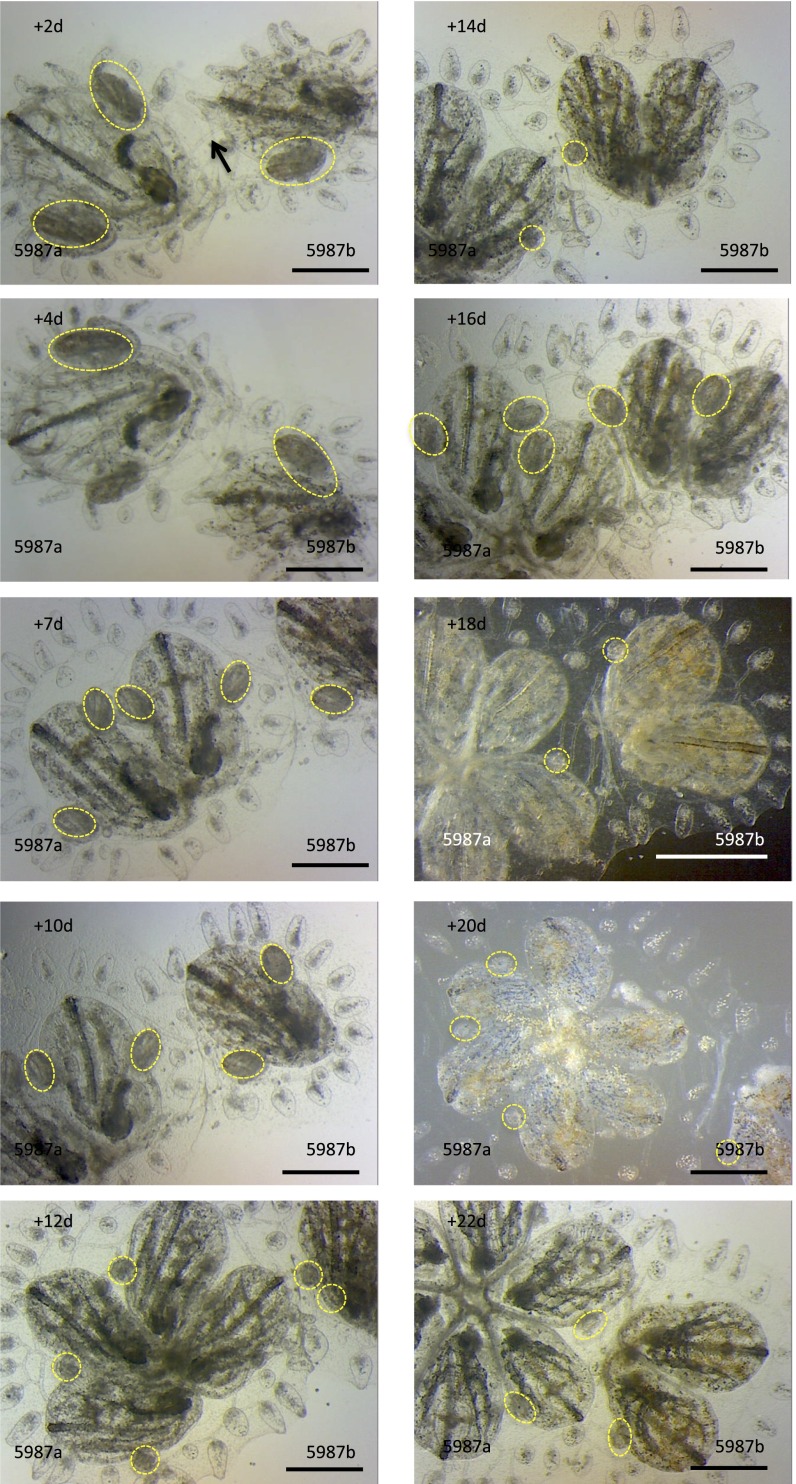
Fig. S2.
Stable, mixed chimeras exhibit synchronized cycles of death and regeneration. Histocompatible, but genetically distinct, juvenile colonies were brought together in controlled allorecognition reactions (arrow indicates anastomotic blood vessel) and imaged in the postfusion period at 48-h intervals. Colonies grow through weekly cycles of development termed blastogenesis. New buds emerge (yellow, dotted outlines) from the body wall of each parental zooid. Chimeric partners exhibit developmental synchrony with each other. (Scale bars: 2 mm.)
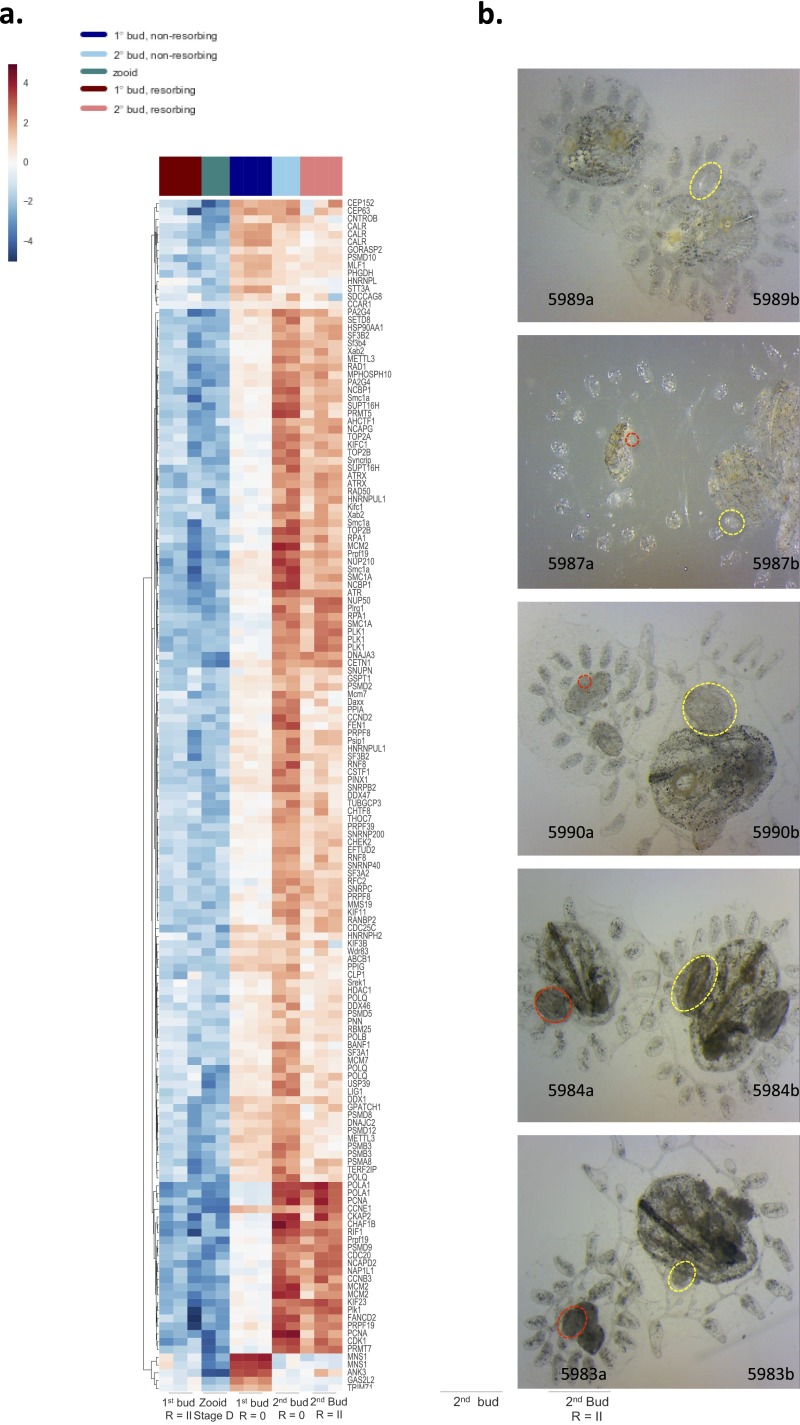
Fig. S3.
Effects of allogeneic resorption on developmental signaling. (A) Heat map and hierarchical clustering analysis among GO term developmental gene sets (GO terms: chromosome organization, cell cycle, RNA splicing, and DNA replication). Each gene is represented by a single row in the heat map; the color scale ranges from −5 to 5 cpm (blue, low; red, high). Aberrant developmental gene expression profiles are observed across both allogeneic resorbing 1° and 2° buds. (B) Representative images of fused pairs undergoing allogeneic resorption (red circles indicate abnormal developing primary and secondary buds; yellow circles indicate normal buds).
Fig. 2.
Profiling the cellular composition of allogeneic resorbing colonies. (A) B. schlosseri cell populations were analyzed and sorted using FACS according to their intrinsic size (FSC) and granularity (SSC) properties on log scale. Using this approach, we identified five populations of cells: 1, small lymphocyte-like cells; 2, pigment cells; 3, large phagocytes, nephrocytes, and compartment cells; 4, dark vacuolated morula cells (MCs) and amoebocytes; 5, clear MCs. In captions are representative images of sorted populations. (Blue scale bars: 20 µm.) (B and C) Representative FACS plots from a nonresorbing and resorbing partner. An increase in frequency of cells with intermediate/high side scatter (SSC), low forward scatter (FSC) was observed in resorbing partners (population 4). (D) Cellular composition of allogeneic resorbing and nonresorbing colonies; as above, significant increase of MCs and amoebocytes was observed in resorption (ANOVA, P < 0.05). (E and F) H&E-stained tissues show pronounced infiltration of MCs and prominent phagocytic infiltration, indicating severe allogeneic resorption. (Scale bars: Left, 200 μM; Right, 100 μM.) (G and H) H&E-stained tissues at the time of takeover show heavy infiltration of macrophages, but limited numbers of MCs. (I) Developing structures are infiltrated by allogeneic partner cells (red arrows). (J) Schematic of live cell fluorescent imaging of labeled colonies to identify source of infiltrating cells. (K–M) Partner cells traffic into developing buds (outlined in yellow). Allogeneic red cells (white arrows) infiltrate green-labeled colonies.
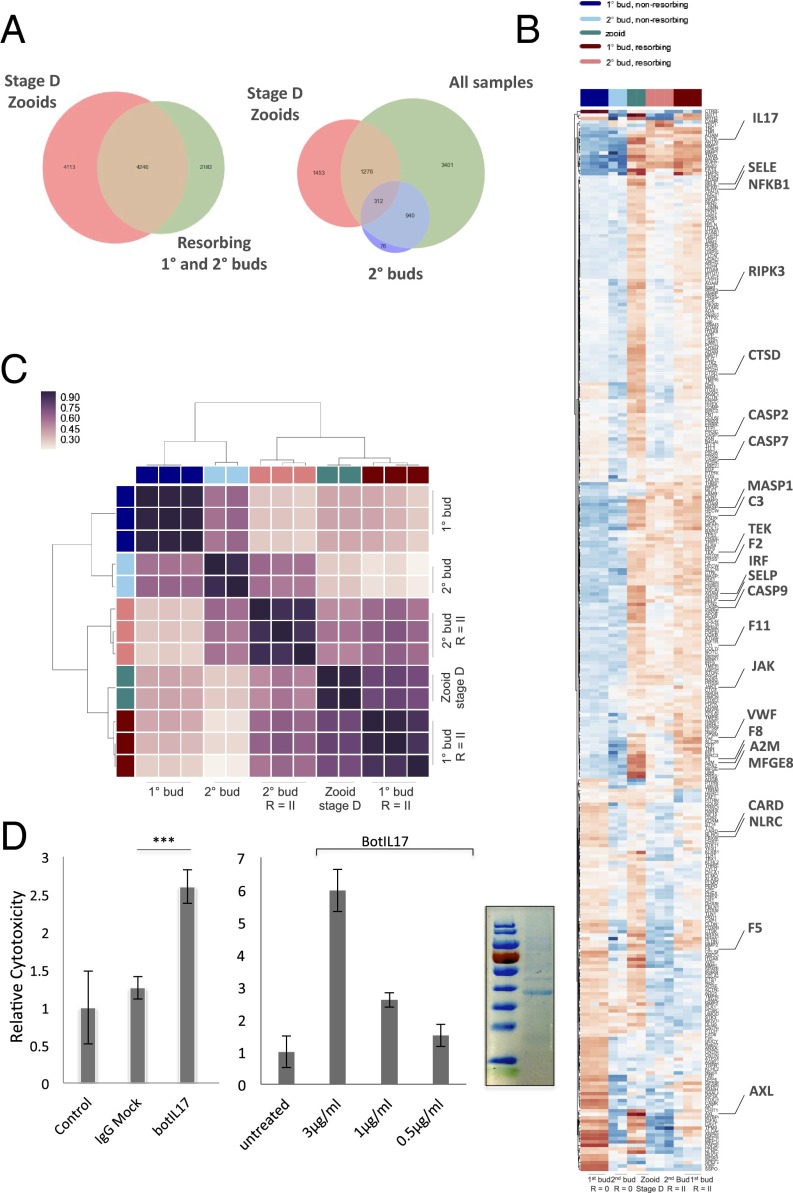
Next, we developed a cell transplantation model to test whether the resorption response could be adoptively acquired from allogeneic donors. MCs were prospectively purified using FACS from single cell suspensions of allogeneic colonies and delivered by ampullar microinjection into recipients (Fig. 3A and Fig. S4). Enrichment of MCs was carried out according to size [forward scatter (FSC)] and granularity [side scatter (SSC)] and included MCs with both clear and pigmented vacuoles, and as a control small lymphocyte-like cells. Because we hypothesized that developmental cell death pathways might “prime” cytotoxic cells before transfer, we collected cells from recipient colonies at the time of takeover: blastogenic stage D. Indeed, global RNA-seq expression profiles from adult zooids isolated during the takeover phase showed up-regulation of genes encoding proinflammatory and prophagocytic factors (Fig. 4 A and B and Figs. S5 and S6). In total, the transcript levels of 8,066 genes changed at least fourfold [false discovery rate (FDR) < 0.05] during takeover (Dataset S1). Of particular relevance, we observed significant up-regulation in key regulatory genes involved in host defense, such as the interleukin family member IL17 (60-fold increase), as well as genes encoding vascular adhesion molecules that promote leukocyte homing (SELP, SELE, Tie1), complement (C3, MASP1, MASP2), cell death (CASP2, CASP7, CASP9), lysosomal proteinases (CTSB, CTSF), coagulation components (F2, F8, KLK3, KLKB1), and TNF-associated proteins (TRAF3, TRAF4). These transcriptome profiles provide insights into how appropriately selected endogenous signals may be used to render cytotoxic cells at the time of adoptive transfer. In support of this finding, we used transcriptome data to probe the role of the proinflammatory cytokine IL-17 to test whether the cytotoxicity observed after transfer of MCs reflects an in vivo requirement for a proliferative stimulus. IL-17 is a key regulatory cytokine secreted by innate immune cell types and has been shown to facilitate clearance of extracellular bacteria and fungus (21). Its dysregulation results in excessive inflammation, leading to tissue damage, chronic inflammation (22), autoimmunity (23), and chronic graft-versus-host disease in higher vertebrates (24). In a number of invertebrate species, IL-17 genes have been identified (21, 25). Using an in vitro FACS-based cytotoxicity assay from whole cell suspensions of allogeneic colonies, recombinant Botrylllus IL-17 significantly up-regulated cellular cytotoxicity and had a dose-dependent enhancement in cytotoxicity (Fig. 5D). Thus, developmental cell death signaling pathways, such as IL17, augment the cytolytic activity of allogeneic effector cells.
Fig. 3.
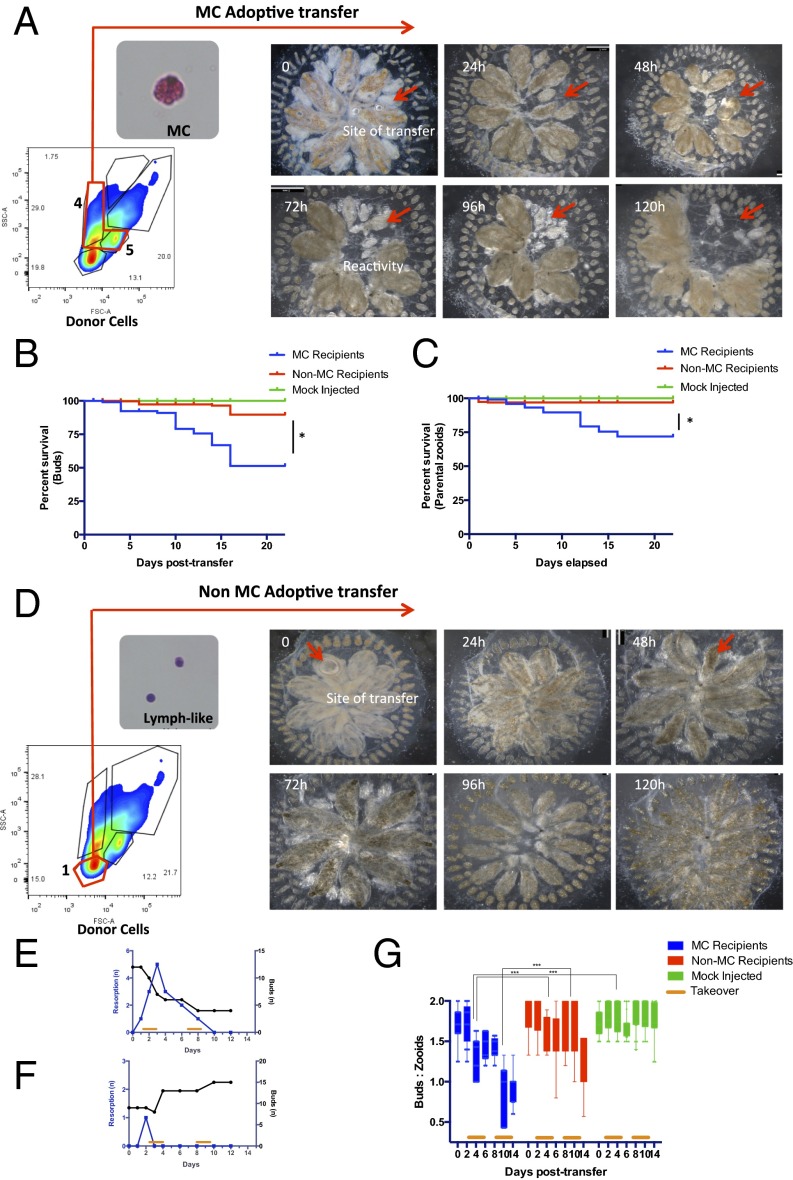
Cellular transfer of FACS-purified allogeneic MCs shows that the resorption response can be adoptively acquired. (A) Allogeneic MCs were prospectively isolated by FACS (populations 4 and 5), transferred via ampullar injection into recipients, and followed for features of allogeneic resorption. (Top) Time lapse imaging over 24-h intervals of a recipient colony after receiving 1 × 105 allogeneic MCs (red arrow indicates site of cellular transfer, followed by parental zooid and bud elimination). (B and C) Kaplan–Meir survival curves among recipient colonies show that transfer of allogeneic MCs eliminates recipient primary buds and zooids compared with non-MC and mock injected groups (P < 0.0001 by Log-rank test). (D) Transfer of 1 × 105 allogeneic control MC-less cells (small lymphocyte-like cell; population 1) had only minimal effects on recipient colonies. (E and F) Timing of bud and parental zooid loss in representative MC (E) and non-MC (F) recipients. (G) Box and whisker plots among groups showing the ratio of buds to parental zooids over time. Transfer of allogeneic MCs eliminates recipient primary buds, resulting in a decline in the bud-to-zooid ratio from 1.7 (±0.03) to 0.9 (±0.06) (P = 0.0001).
Fig. S4.
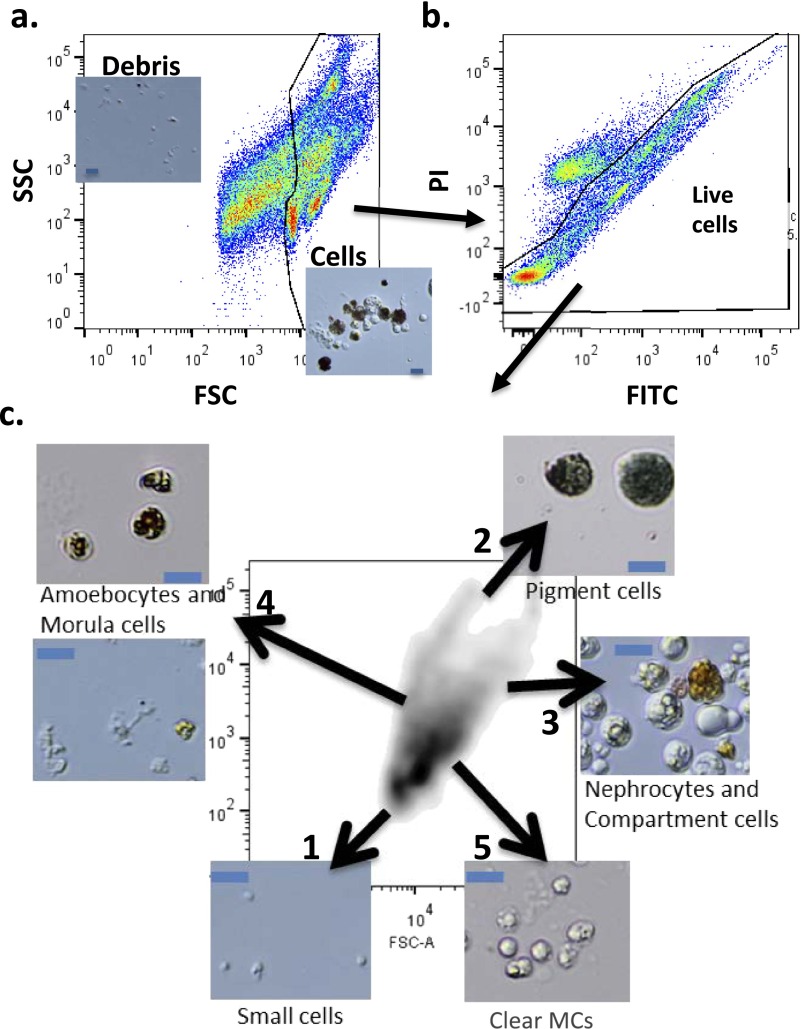
B. schlosseri cell populations analyzed by FACS. (A) B. schlosseri cell suspensions analyzed and sorted using FACS according to their intrinsic size (FSC) and granularity (SSC) properties on log scale. The gate separates cells from debris. (B) Gating on live cells using PI-negative gates. The fluorescent gates are in two dimensions due to B. schlosseri cells strong autofluorescence in every channel; the positive cells above the diagonal represent nonspecific fluorescence. (C) After excluding debris and dead cells, we used again SSC and FSC to characterize five main populations. Using this approach, we can identify five populations of cells, as follows: 1, small lymphocyte-like cells; 2, pigment cells; 3, large phagocytes, nephrocytes, and compartment cells; 4, dark vacuolated morula cells (MCs) and amoebocytes; 5, clear MCs. In captions are representative images of sorted populations based on the gating scheme. (Blue scale bars: 20 µm.) (C is enlarged Fig. 2A).
Fig. 4.
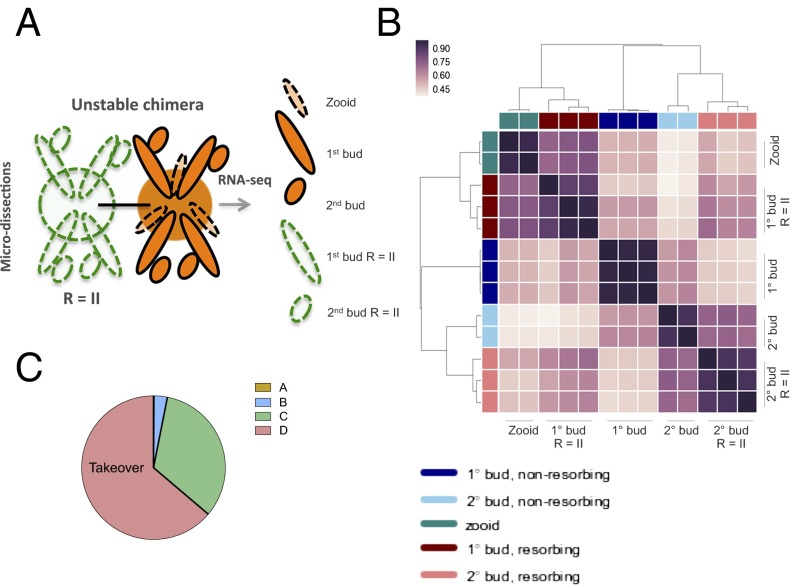
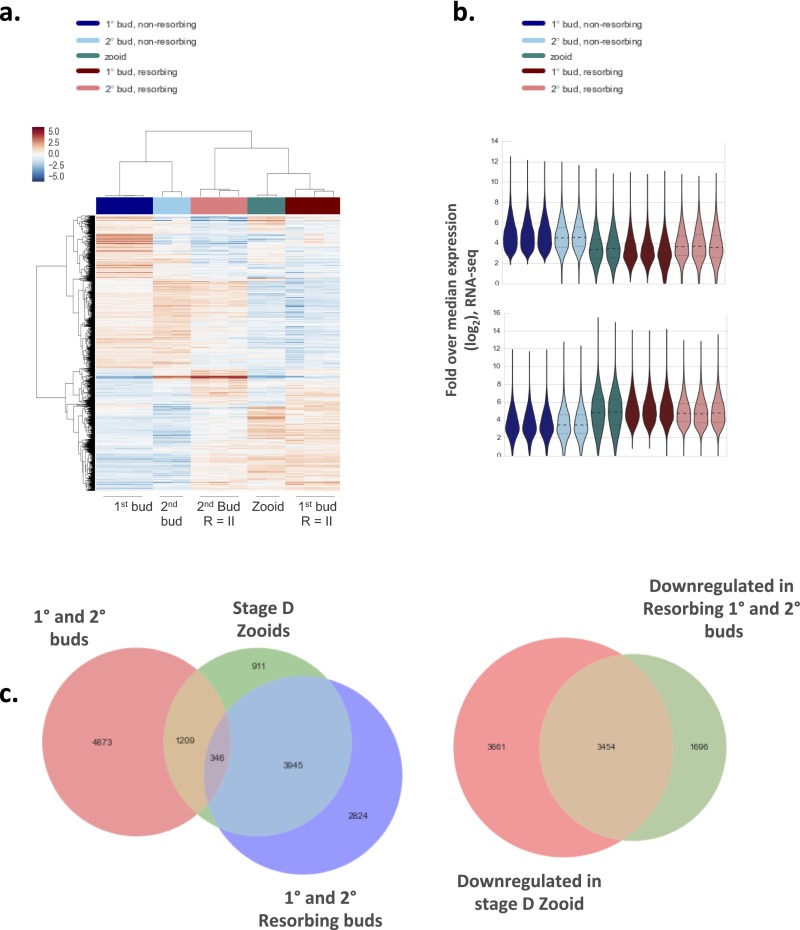
RNA-seq analysis of allogeneic resorbing colonies. (A) Schematic of RNA-seq experiments. Allogeneic, but compatible, defined lines were fused in colony allorecognition assays and then harvested for RNA-sequencing. Biologic replicates were obtained from stage R = II (resorption score = II) 1° (primary) and 2° (secondary) buds, corresponding to developmental stage D. (B) A hierarchical clustering matrix was generated using Pearson correlation coefficients from log2 counts per million across all genes. Widely divergent transcriptomes were observed across resorption status and developmental stage. The color scale indicates the degree of correlation (white, low correlation; purple, high correlation). (C) Pie chart showing blastogenic stage at time of chimeric resorption.
Fig. S5.
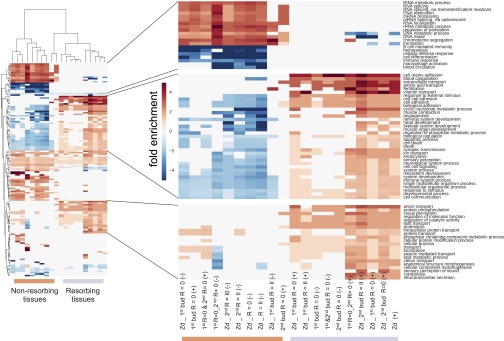
GO-category hierarchical clustering of differentially expressed genes across samples. GO category analysis of B. schlosseri microdissected tissue samples. Hierarchical clustering (using city block distance) of GO scores for categories that are statistically significant are displayed in a heat map. Each column represents the result for one comparison, with test group and direction of expression given as a column name. The values of entries are the multiplicative enrichment (reds) and depletion (blues) of genes falling within the given GO category. This analysis reveals two large clusters differentiated by GO term signatures: The left cluster (orange bar) has genes that are higher in buds than zooids and higher in the nonresorbing tissues; the right cluster (purple bar) has genes that are higher in zooids and resorbing tissues, mirroring development stage and resorption status. 1°, primary bud; 2°, secondary bud; Zd, Zooid stage D; R = 0, resorption score = 0; R = II, resorption score = II; (−), down-regulated; (+), up-regulated.
Fig. S6.
Profiling allogeneic resorption with RNA-seq. (A) Heat map and cluster analysis demonstrate clear differences among transcriptomes observed across developmental stage and resorption status. (B) Frequency distribution of expression values from RNA-seq bulk samples shown as a violin plot. Expression (vertical axis) is the log2-transformed fold change over median gene expression level across samples. The width of the violin indicates frequency at that expression level. The Top violin plot shows differentially expressed genes in nonresorbing tissues whereas the Bottom violin plot shows differentially expressed genes in resorbing tissues. (C) Venn diagrams among differentially expressed genes between samples. Takeover and allogeneic resorbing tissues share significant overlap in gene expression profiles.
Fig. 5.
Identification of licensing pathways from RNA-seq datasets. (A) Adult zooids were isolated during the takeover phase for RNA-sequencing and compared with R = II allogeneic resorbing tissues to identify shared pathways. Venn diagram plots for differentially expressed genes between nonresorbing 1° and 2° buds, zooids, and resorbing 1° and 2° buds. 1°, primary bud; 2°, secondary bud; R = II (resorption score = II). (B) Genes that were found to be differentially expressed in allogeneic resorbing tissues were intersected with significant GO terms (immune system process and cell adhesion) and displayed in a heat map. Each gene is represented by a single row in the heat map; the color scale ranges from −5 to 5 cpm (blue, low; red, high). Allogeneic and takeover tissues share expression profiles for a number of genes implicated in immunity (highlighted). (C) Hierarchical clustering matrix for differentially expressed genes among allogeneic resorbing tissue samples indicates clustering with stage D takeover pathways. The color scale indicates the degree of correlation (white, low correlation; purple, high correlation). (D) Bot IL17 significantly enhances cytolytic activity in allogeneic in vitro assays compared with untreated and human Fc control (ANOVA). (Left) Bot IL17, 1 µg/mL. (Right) Bot IL17 demonstrates a dose-dependent enhancement of lytic activity in allogeneic cytotoxicity assays. Assays were performed in 96-well plates overnight using differentially labeled cells from allogeneic colonies in 1:10 effector:target ratios, and analyzed by FACS. Error bars = SD. SDS/page analysis of purified recombinant IL17 (Far Right).
We next transplanted 1 × 105 allogeneic FACS-purified MCs, collected from donors at the time of takeover, and followed recipients for morphologic findings of resorption (Fig. 3A and Fig. S4). The cumulative data are presented in Fig. 3 and show that the resorption response could be adoptively acquired from allogeneic donors. Recipient colonies transplanted with purified MCs exhibited point-of-rejection morphology at ampullar injection sites and shared characteristic features of allogeneic resorption: Blood vessels, peripheral ampullae, and the surrounding gelatinous tunic remained intact whereas rapid elimination of parental zooids and nascent buds ensued at the site of cell transfer (Fig. 3A and Fig. S7). Morphologic findings of parental zooid and bud growth arrest and accompanying tissue injury were observed as early as day 2, and peaked at day 4 after cell transfer. By day 14 posttransfer, 33 of 78 (42%) recipient buds and 20 of 76 (20.5%) adult zooids had been eliminated, resulting in a decline in the bud-to-zooid ratio from 1.7 (±0.03) to 0.9 (±0.06) (P = 0.0001), a ratio insufficient to maintain colony size (Fig. 3 C and D). In contrast, transfer of allogeneic cell populations lacking MCs had only minimal effects on recipient colonies (Fig. 3 E–G). Thus, using prospective isolation of MCs, we show that the resorption response can be adoptively transferred from allogeneic donors and that the principle cell type mediating this effect is a cytotoxic MC.
Fig. S7.
Morphologic analysis of sites of ampullar injection. (A and B) Dissecting scope images of sites of ampullar injection of MCs (A) and non-MC populations (B) 8 h after cellular transplantation. Areas of necrosis and melanin-like pigmentation develop at sites of MC administration, but not in non-MC recipients (dotted lines mark direction of ampullar injection).
Last, similar to our observational studies, we observed the maximal effector response operative during the developmental period of takeover in adoptive cell transfer experiments. These data, we reasoned, reflect an in vivo requirement of transferred cells for a proliferative stimulus provided by recipient colonies. Therefore, we used an unbiased sequencing-based approach to identify additional pathways that may be contributing to the observed licensing effect on transferred cells (Fig. 4). We used global RNA-seq expression profiles to identify differentially expressed genes unique to each RNA-seq sample and then compared the extent of overlap between resorbing and takeover tissues (Fig. 4 A and B). Histological analysis of these tissues showed heavy infiltration of phagocytes and MCs, which are reflected in the transcriptome data (Fig. 2 D–I). We observed pronounced gene expression changes in takeover zooids (Fig. 4B), corresponding to major morphologic changes during this developmental period (Dataset S1). Of particular relevance, we observed significant up-regulation in genes related to autophagy, proteolysis, programmed cell death, and phagocytic clearance of cellular corpses (Dataset S1). These findings support prior observations showing intensive phagocytosis of apoptotic cells underlying colony-wide recycling between senescent zooids and developing buds during the takeover period (11, 26, 27).
We next compiled RNA-seq datasets from resorbing tissue microdissections and compared them with takeover expression profiles (Fig. 5 A and C). Tissue dissections were derived from defined mariculture lines. Using EdgeR, we showed significant overlap in transcriptomes between both processes. We identified 275 genes at the intersection of both datasets that increased by expression of at least 10-fold, and 55 are known to play a role in either acute inflammation or stress responses. These genes include evolutionarily conserved mediators of immune responses, such as the transcription factor NF-ΚB, a member of the Janus kinase (JAK) family of protein tyrosine kinases, two members of the TNF receptor-associated signal transduction family, TRAF3 and TRAF4, and a member of the IFN regulatory transcription factor (IRF) family (Fig. 5B and Dataset S1). We also identified genes encoding complement and coagulation components within this subset. Both processes initiate proteolytic cascades and have been observed downstream of pattern recognition proteins and damaged tissues in invertebrates (28). Members of these groups include genes encoding a primitive lectin-based complement activation system, such as complement components C3 and CFB, the C3b receptor CR1, and mannose binding lectin (MBL) (Ficolin 1) and MBL-associated serine proteases (MASP1 and MASP2) (29–31) (Fig. 5B and Dataset S1), as well as, thrombin (FII), factor VIII (FVIII), factor XI, plasminogen (PLG), and the kallikrein family proteins (KLK3, KLKB1) (Fig. 5B and Dataset S1). Lastly, we identified a network of genes involved in phagocytosis, shared by both datasets. These genes include the TAM receptor tyrosine kinase AXL and the phosphatidylserine binding bridge glycoprotein molecule MFG-E8, both of which are involved in the clearance of cellular corpses from tissues (Fig. 5C and Dataset S1) (25, 32). Taken together, these studies reveal multiple disparate immunogenic pathways induced during takeover, also used in the setting of allogeneic resorption, which may be contributing to the observed licensing effect on transferred cells.
Discussion
B. schlosseri chimeras offer a unique platform to unravel mechanisms that sustain immunological nonreactivity. Achievement of long-term persistence of allogeneic cells in hematopoietic cell transplantation (HCT) is an important therapeutic strategy for the induction of tolerance and as an approach to nonmyeloablative HCT in some malignancies, wherein the graft-vs.-host reaction by donor T cells includes tumor cell targets and also clears host hematopoietic cells from their niches (33). Our studies show that events that tip the chimeric balance toward partner elimination are linked to cyclical developmental cell death programs, whereby activation of proinflammatory factors has the effect of endowing cytotoxic MCs, the predominant cell type associated with tunicate rejection reactions among incompatible allogeneic participants in many urochordate species, with enhanced alloreactivity. These findings provide the strongest evidence to date that chimeric partner elimination seems to be an immune cell-based rejection that operates within histocompatible pairs, raising important questions regarding the role for protochordate histocompatibility loci in programmed cell death programs and allogeneic resorption. It will be of great interest in future studies to assess for allelic discordance among previously defined histocompatibility alleles among resorbing pairs. Based on our observations, one possibility is for mismatch of “minor” protochordate histocompatibility loci that promote alloreactivity within fused pairs.
The requirement of additional costimulatory signals to arm MCs follows our current understanding of the nature of innate immune effector cell recognition in higher vertebrates, such as natural killer (NK) cells. In this model, failure to receive licensing signals leaves NK cells in a hyporesponsive, “self-tolerant” state. Indeed, even in chimeric pairs, which undergo allogeneic resorption, we observe little morphologic evidence for alloreactivity before the onset of, or immediately after, the developmental period of takeover. The rapid transition to hyporesponsiveness suggests either repression of takeover processes or induction of counterregulatory systems. It is possible that inhibitory receptor and signaling pathways that dampen responsiveness to stimulatory signals are turned on during early events of blastogenesis. Elucidating these putative counterregulatory systems may yield insights into novel immune evasion strategies. Evidence for this model is the markedly shortened time period to resorption from adoptive transfer of licensed cells compared with the more gradual process of natural resorption.
Although our cell transplantation studies implicate primitive cytotoxic cells as the primary mediators of the allogeneic resorption response, it would be premature to conclude that complete elimination of a chimeric partner can be accomplished by direct cytolysis alone. During the death phase of colony life, where we observe maximal effector responses, circulating phagocytic cells engulf cell corpses within the dying organism. Histological analysis of resorbing tissues demonstrates a heavy infiltration of phagocytes, which is supported by transcriptome analysis, showing enrichment for a network of genes involved in phagocytosis. These findings suggest that elimination of chimeric partners involves the integrated function of both homeostatic cell turnover programs with cytolytic activation. Indeed, previous analyses carried out with isolated phagocytic cells revealed that engulfment was enhanced by incubation with cell lysate supernatants made from MCs, but not other cell types, suggesting that opsonins or other prophagocytic factors are derived from MCs and that phagocytosis requires cooperation from other activated cell types (34). These insights raise important questions about whether this strategy for immune elimination may apply to other settings, such as for the elimination of tumors or chronic infections, using compounds that potentiate phagocytosis in combination with agents that promote cytotoxicity.
Lastly, one puzzling phenomenon in these studies is that, whereas fused partners rarely maintain stability at the level of the colony, the eliminated partner may still persist, but at the level of the cell lineage (8, 9). Those cells that escape allogeneic resorption may even parasitize the nonresorbed partner. Therefore, it is possible that immune editing of partners may play a role in the germ-line stem cell “competitions” wherein the gonads of a vascularly connected histocompatible kin are from the genome of the partner. This model leads to the prediction that immune responses iteratively select and or promote the generation of variants with increasing capacity to survive immune attack, similar to that which has been described in cancer clone evolution. Future studies will need to resolve whether winning chimeric partners can be rendered selectively resistant to histocompatible kin.
Experimental Procedures
Colony Development, Creation of Chimeras, and Tissue Dissections.
Mariculture procedures have been described previously (35). Briefly, B. schlosseri colonies were collected from the marina in Monterey, CA. Individual colonies were tied to 3- × 5-cm glass slides and placed 5 cm opposite another glass slide (called the settlement slide) in a slide rack. The slide rack was placed into an aquarium, and, within a few days, the tadpoles hatched, swam to the settlement slide, and metamorphosed into the adult body plan (oozoid). We transferred 177 F1 oozoids, progeny of WT colonies collected in Monterey, CA and placed them adjacent to each other, and then monitored them for fusion. Oozoids were allowed to adhere to slides for 5 min before transferring to the tanks. This resulted in controlled fusions (42 chimeras were formed in this manner), from a cohort of 177 colony allorecognition assays.
From this point on, colonies were monitored for formation of a chimera (fusion) (i.e., in the first 2 wk, an individual was united in a parabiotic union). Each individual and chimera was given a unique identifier. Rejection, fusion, and resorption were characterized visually. We recorded (i) age (days old), (ii) health (poor or good), and (iii) rejection, fusion, and resorption status according to the following scale: stage 0 (no resorption observed), stage I (mild resorption = zooid contraction), stage II (moderate resorption = zooid contraction, 1° and 2° bud developmental arrest, chimeric blastogeneic asynchrony, residual cardiac activity), stage III (severe resorption = only remnants visible, no cardiac activity), and stage IV [complete resorption = one partner has been fully eliminated; only supporting tissue (ampullae, blood vessels, and tunic) remain].
Tissue dissections were derived from stage R II resorbing-controlled fusion pairs between defined mariculture lines 944axByd196.6 × sc109e.92 and used for RNA-sequencing. Adult zooids and primary and secondary buds were isolated from the common tunic with insulin syringes and further sectioned with a blade along the frontal plane to facilitate isolation of specific tissues, flash frozen, and harvested for RNA. Three biologic replicates were obtained from (stage = R II) 1° and 2° buds, corresponding to developmental stage D.
RNA Extraction, Purification, and Transcriptome Sequencing.
Tissue dissections were stored at −80 °C, resuspended in TRIzol and liquid nitrogen to minimize RNA degradation, and then homogenized into a soft powder using vigorous mechanical disruption with a mortar and pestle. Homogenized-frozen tissue was allowed to thaw, and then RNA was isolated according to the manufacturer’s directions, with minor modifications. A linear polyacrylamide (LPA) carrier was added to enhance recovery of RNA, followed by DNase I treatment per the manufacturer’s instructions (Promega RQ1 DNase). Recovered RNA was then analyzed by an Agilent 2100 Bioanalyzer for quality analysis before library preparation. cDNA libraries were then prepared from high quality samples (RNA integrity number > 9) using Ovation RNA-seq v2 (Nugen); NEBnext DNA Master Mix for Illumina (New England Biolabs), and standard Illumina adapters and primers from IDT. Bar-coded library samples were then sequenced using RNA NextSEq. 500 (2 × 150 bp).
Cell Isolation and Injection.
Cell populations were isolated by FACS from allogeneic, outbred colonies at the time of takeover. Colonies were dissociated using a fine blade into cell suspensions and filtered through a 40-µm filter directly into staining media. Cell sorting was performed with a 100-µm nozzle size and sorted directly into staining media (3.3× PBS based; 75% of final volume) to minimize cellular stress. Enrichment of MCs was carried out according to size (FSC) and granularity (SSC) (populations 4 and 5). A control population of small lymphocyte-like cells (population 1) was isolated in parallel from allogeneic whole cell suspensions for comparison studies. Cells were resuspended to a concentration of 105 cells per microliter. We transferred 105 cells via ampullar injection using a manual microinjector and micromanipulator (Narishige) into recipient colonies.
SI Experimental Procedures
Colony-Labeling Studies and Microscopy.
Juvenile colonies were labeled with distinct fluorescent stains (CellTrace CFSE Green/CellTrace FarRed; Life Technologies) and brought together in controlled fusion reactions. To label colonies, stock solutions (CFSE dye, 5 mM; Far Red dye, 1 mM) were prepared in a dilution of 1 μL of dye to 1 mL of filtered sea water. Naive oozoids were then bathed in this solution for 60 min, allowed to recover and washed, and then used for colony allorecognition assays. Colonies were placed on µ-Dishes (µ-Dish, 50 mm, low uncoated; Ibidi) for observation. Images were obtained using a confocal fluorescent microscope (LSM700 Axio Observer.Z1; Zeiss).
FACS Analysis.
For FACS analysis, colonies were dissociated with a fine blade into cell suspensions and filtered through a 40-μm mesh using a sterile 1-mL syringe pump, and washed and collected in staining media: 3.3× PBS, 2% (vol/vol) FCS, and 10 mM Hepes buffer. After gating on propidium iodide negative cells (done on two-dimension plots due to natural fluorescence of Botryllus schlosseri cells), cells were analyzed using a size (FSC) and granularity (SSC) panel on log scale, and FACS sorted onto 18-well flat µ-Slides coated with poly-l-lysine (Ibidi). Five distinct populations were identified using this approach. Purified populations were analyzed according to morphology by light microscopy and defined according to previous cellular work characterizing B. schlosseri cell populations (16, 36). We identified five distinct morphologic populations based on FSC/SSC: 1, low FSC, low SSC, small lymphocyte-like cells; 2, mid FSC, high SSC, pigment cells; 3, high FSC, mid-high SSC, large phagocytes, nephrocytes, and compartment cells; 4, low FSC, high SSC, mix of amoebocytes and dark vacuolated MCs; 5, mid FSC, mid SSC, clear MCs (Fig. 3 A–C and Fig. S4). For hematoxylin/eosin (H&E) staining, paraffin-embedded samples were cut at 4 μm. After deparaffinizing the slides in xylenes, dehydrating in graded ethanols, and hydrating to water, they were stained with Harris hematoxylin for 5 min and 2% eosin Y for 1 min.
Protein Expression.
Full-length Botryllus IL-17 with a C-terminal hexa-His tag was cloned into a pAcGP67-A expression vector, and the protein was secreted by High Five insect cells grown in Insect Express media at 27 °C. The IL17 sequence was as follows: ATGCGCGAAAACGTCAGAGGTAACTTGAAGCGTTACAGGATCCCAATCACTCACGTAACCAAATCTGGATTGTCTGCCTTCGGTCAAGAGCTGGCTCCAAAGTGCCCTGTGTACAGTGCTTCAGAGACACGCGAAAGATCACTTAGCCCGTGGCGTTTAGTGTCCAATTTCGACCAACATCGCTATCCACAGTGGATTCCTGAGGCCAAGTGCCTTTGTAAGGGCTGTATTGATATGGCGACTGGAAAGGAGAACCGTCTGTTTGTTTCTACCGTGATCAAGGTCAAGGTCCGTTTCCTTCGTATGAACATGACTAGTTCCACTTCAACCTTTAGACAATGCACTGACGAGACCTGCTATCCATCCGATGAAGACGTATCGGTGGGATGCCATTGCGTTATCCCAGGAACGATTGTAGCCAAGAGATAA.
Supernatants containing Il-17 were mixed and concentrated before Ni-affinity purification, equilibrated in 1× PBS buffer, and concentrated to ∼1 mg/mL.
Cytotoxicity Assays.
For in vitro cytotoxicity assays, we developed a FACS-based cytotoxicity assay due to difficulty in radioactive labeling of slow growing cells (37, 38). Isolated B. schlosseri cells were labeled using carboxyfluorescein succinimidyl ester (CFSE) (5 μM; Life Technologies) and Far Red dye (1 μM; Life Technologies) for 30 min at 18 °C to distinguish effector and target cells and washed twice in staining media. Cells were incubated overnight in 96-well plates at 18 °C at a 10:1 effector:target (target cells 25,000 per well) ratio with or without recombinant Botryllus IL17 or control proteins diluted in 100 μL of staining media. After adding propidium iodide (PI) to wells for cell viability separation, wells were analyzed by FACS. Spontaneous lysis was measured as the percentage of PI-positive cells in gated target cells in wells without effector cells, and sample lysis was quantified as the percentage of PI-positive cells in gated target cells. Specific lysis was calculated as follows: Specific lysis % = 100 × [(sample lysis-spontaneous lysis)/(100-spontanius lysis)]. The gating on FACS analysis was on two-dimension plots due to the natural fluorescence of B. schlosseri cells. For recombinant protein cytotoxicity assays, different allogeneic colony combinations and mixtures were used to prevent histocompatibility bias.
Transcriptome and Gene Ontology Analysis.
A streamlined bioinformatics pipeline to align the reads and determine gene expression was written in Snakemake. The steps performed were as follows: low quality bases were trimmed from raw reads from the sequencer in the FASTQ format using Trimmomatic (version 0.32). Overlapping paired end reads were merged using FLASH (version 1.2.11) (39). Using bowtie2, biological vector sequences were removed by aligning to the UniVec core database. The filtered reads that remained were then aligned to the transcriptome of B. schlosseri (40) using BWA (“mem” algorithm). Aligned reads were sorted and indexed using samtools, PCR duplicates were removed using PICARD, and transcript level counts were determined.
Differential expression was performed using edgeR. In detail, the gene counts were compiled into a tabular format. This was loaded into R, and genes with at least five samples having at least five counts per million (cpm) were retained. The comparison between the resorbing and nonresorbing tissue was achieved using a paired design matrix. If “tissue” represents a primary or secondary bud and “status” whether the sample was resorbing or not, then the R command design <- model.matrix(∼tissue + status) achieves this classification. For all other comparisons, the makeContrasts function was used manually to select which sample populations to compare. In both cases, the default options for the generalized linear model (GLM) edgeR analysis were used. The P values computed were adjusted for multiple testing using the Benjamini–Hochberg method to produce a false discovery rate (FDR). Results were saved in tables (Dataset S1).
Genes that were differentially expressed between two populations of interest were selected that had homology to human genes in the Botryllus annotation. These genes were processed on the PANTHER website [Overrepresentation Test (release 20150430), PANTHER version 10.0 released 2015-05-15] to find GO-Slim biological processes that are highly associated with the gene sets. This task was done for subsets of genes both up- and down-regulated, and all results were aggregated into Dataset S1 and a GO clustermap.
Supplementary Material
Acknowledgments
We thank Aaron Newman, Norma F. Neff, Gary L. Mantalas, Seth Karten, and Theresa Storm for invaluable technical advice and laboratory support. This study was supported by NIH Grants 1R01AG037968 and R01GM100315 (to I.L.W., S.R.Q., and A.V.), the Virginia and D. K. Ludwig Fund for Cancer Research (I.L.W.), and in part by NIH Fellowship K12 HL087746 from the National Heart, Lung, and Blood Institute (to D.C.), 2T32AR050942-06A1 (to D.C.), a grant from the Siebel Stem Cell Institute (to I.L.W.), and by postdoctoral fellowships from the Human Frontiers Science Program Organization and Immunology Training Grant 5T32AI07290-28 of the US National Institutes of Health Molecular and Cellular Immunobiology Training Program, NIH NIAID (to B.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606276113/-/DCSupplemental.
References
- 1.Scofield VL, Schlumpberger JM, Weissman IL. Colony specificity in the colonial tunicate Botryllus and the origins of vertebrate immunity. Am Zool. 1982;22(4):783–794. [Google Scholar]
- 2.Voskoboynik A, et al. Identification of a colonial chordate histocompatibility gene. Science. 2013;341(6144):384–387. doi: 10.1126/science.1238036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirose E, Saito Y, Watanabe H. A new type of the manifestation of colony specificity in the compound ascidian, Botrylloides violaceus Oka. Biol Bull. 1988;175(2):240–245. [Google Scholar]
- 4.Rinkevich B, Weissman IL. Allogeneic resorption in colonial protochordates: Consequences of nonself recognition. Dev Comp Immunol. 1992;16(4):275–286. doi: 10.1016/0145-305x(92)90002-t. [DOI] [PubMed] [Google Scholar]
- 5.Rinkevich B, Weissman IL. A long-term study on fused subclones in the ascidian Botryllus schlosseri: The resorption phenomenon (Protochordata: Tunicata) J Zool (Lond) 1987;213(4):717–733. [Google Scholar]
- 6.Carpenter MA, et al. Growth and long-term somatic and germline chimerism following fusion of juvenile Botryllus schlosseri. Biol Bull. 2011;220(1):57–70. doi: 10.1086/BBLv220n1p57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinkevich B, Weissman IL. In: Invertebrate Historecognition. Grosberg RK, Hedgecock D, Nelson K, editors. Plenum; New York: 1988. pp. 93–109. [Google Scholar]
- 8.Stoner DS, Weissman IL. Somatic and germ cell parasitism in a colonial ascidian: Possible role for a highly polymorphic allorecognition system. Proc Natl Acad Sci USA. 1996;93(26):15254–15259. doi: 10.1073/pnas.93.26.15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rinkevich W. Chimeras in colonial invertebrates: A synergistic symbiosis or somatic and germ parasitism? Symbiosis. 1987;4:117–134. [Google Scholar]
- 10.Harp JA, Tsuchida CB, Weissman IL, Scofield VL. Autoreactive blood cells and programmed cell death in growth and development of protochordates. J Exp Zool. 1988;247(3):257–262. doi: 10.1002/jez.1402470309. [DOI] [PubMed] [Google Scholar]
- 11.Lauzon RJ, Ishizuka KJ, Weissman IL. A cyclical, developmentally-regulated death phenomenon in a colonial urochordate. Dev Dyn. 1992;194(1):71–83. doi: 10.1002/aja.1001940109. [DOI] [PubMed] [Google Scholar]
- 12.Rinkevich B, Tartakover S, Gershon H. Contribution of morula cells to allogeneic responses in the colonial urochordate Botryllus schlosseri. Mar Biol. 1998;131(2):227–236. [Google Scholar]
- 13.Berrill NJ. Regeneration and budding in tunicates. Biol Rev Camb Philos Soc. 1951;26(4):456–475. [Google Scholar]
- 14.Berrill NJ. The development of the bud in Botryllus. Biol Bull. 1941;80(2):169–184. [Google Scholar]
- 15.Ballarin L, Cima F, Sabbadin A. Morula cells and histocompatibility in the colonial ascidian Botryllus schlosseri. Zoolog Sci. 1995;12(6):757–764. [Google Scholar]
- 16.Ballarin L, Cima F. Cytochemical properties of Botryllus schlosseri haemocytes: Indications for morpho-functional characterisation. Eur J Histochem. 2005;49(3):255–264. [PubMed] [Google Scholar]
- 17.Ballarin L. Ascidian cytotoxic cells: State of the art and research perspectives. Inv Surv J. 2012;9:1–6. [Google Scholar]
- 18.Ballarin L, Cima F, Sabbadin A. Phenoloxidase and cytotoxicity in the compound ascidian Botryllus schlosseri. Dev Comp Immunol. 1998;22(5-6):479–492. doi: 10.1016/s0145-305x(98)00035-4. [DOI] [PubMed] [Google Scholar]
- 19.Oren M, et al. Marine invertebrates cross phyla comparisons reveal highly conserved immune machinery. Immunobiology. 2013;218(4):484–495. doi: 10.1016/j.imbio.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Oren M, Escande ML, Paz G, Fishelson Z, Rinkevich B. Urochordate histoincompatible interactions activate vertebrate-like coagulation system components. PLoS One. 2008;3(9):e3123. doi: 10.1371/journal.pone.0003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 22.Miossec P, Kolls JK. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov. 2012;11(10):763–776. doi: 10.1038/nrd3794. [DOI] [PubMed] [Google Scholar]
- 23.Zhu S, Qian Y. IL-17/IL-17 receptor system in autoimmune disease: Mechanisms and therapeutic potential. Clin Sci (Lond) 2012;122(11):487–511. doi: 10.1042/CS20110496. [DOI] [PubMed] [Google Scholar]
- 24.van der Waart AB, van der Velden WJFM, Blijlevens NM, Dolstra H. Targeting the IL17 pathway for the prevention of graft-versus-host disease. Biol Blood Marrow Transplant. 2014;20(6):752–759. doi: 10.1016/j.bbmt.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 25.Vizzini A, et al. Ciona intestinalis interleukin 17-like genes expression is upregulated by LPS challenge. Dev Comp Immunol. 2015;48(1):129–137. doi: 10.1016/j.dci.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Ballarin L, Burighel P, Cima F. A tale of death and life: Natural apoptosis in the colonial ascidian Botryllus schlosseri (Urochordata, Ascidiacea) Curr Pharm Des. 2008;14(2):138–147. doi: 10.2174/138161208783378798. [DOI] [PubMed] [Google Scholar]
- 27.Ballarin L, et al. Haemocytes and blastogenetic cycle in the colonial ascidian Botryllus schlosseri: A matter of life and death. Cell Tissue Res. 2008;331(2):555–564. doi: 10.1007/s00441-007-0513-4. [DOI] [PubMed] [Google Scholar]
- 28.Ariki S, et al. Factor C acts as a lipopolysaccharide-responsive C3 convertase in horseshoe crab complement activation. J Immunol. 2008;181(11):7994–8001. doi: 10.4049/jimmunol.181.11.7994. [DOI] [PubMed] [Google Scholar]
- 29.Fujita T, Endo Y, Nonaka M. Primitive complement system: Recognition and activation. Mol Immunol. 2004;41(2-3):103–111. doi: 10.1016/j.molimm.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 30.Endo Y, Takahashi M, Fujita T. Lectin complement system and pattern recognition. Immunobiology. 2006;211(4):283–293. doi: 10.1016/j.imbio.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Kuraya M, Ming Z, Liu X, Matsushita M, Fujita T. Specific binding of L-ficolin and H-ficolin to apoptotic cells leads to complement activation. Immunobiology. 2005;209(9):689–697. doi: 10.1016/j.imbio.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Hanayama R, et al. Identification of a factor that links apoptotic cells to phagocytes. Nature. 2002;417(6885):182–187. doi: 10.1038/417182a. [DOI] [PubMed] [Google Scholar]
- 33.Weissman IL, Shizuru JA. The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor-specific transplantation tolerance and treat autoimmune diseases. Blood. 2008;112(9):3543–3553. doi: 10.1182/blood-2008-08-078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith VJ, Peddie CM. Cell cooperation during host defense in the solitary tunicate Ciona intestinalis (L) Biol Bull. 1992;183(2):211–219. doi: 10.2307/1542208. [DOI] [PubMed] [Google Scholar]
- 35.Rinkevich B. Natural chimerism in colonial urochordates. J Exp Mar Biol Ecol. 2005;322(4):93–109. [Google Scholar]
- 36.Schlumpberger JM, Weissman IL, Scofield VL. Separation and labeling of specific subpopulations of Botryllus blood cells. J Exp Zool. 1984;229(3):401–411. doi: 10.1002/jez.1402290308. [DOI] [PubMed] [Google Scholar]
- 37.Lecoeur H, Février M, Garcia S, Rivière Y, Gougeon ML. A novel flow cytometric assay for quantitation and multiparametric characterization of cell-mediated cytotoxicity. J Immunol Methods. 2001;253(1-2):177–187. doi: 10.1016/s0022-1759(01)00359-3. [DOI] [PubMed] [Google Scholar]
- 38.Rosental B, et al. Proliferating cell nuclear antigen is a novel inhibitory ligand for the natural cytotoxicity receptor NKp44. J Immunol. 2011;187(11):5693–5702. doi: 10.4049/jimmunol.1102267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magoč T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voskoboynik A, et al. The genome sequence of the colonial chordate, Botryllus schlosseri. eLife. 2013;2:e00569. doi: 10.7554/eLife.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.