Significance
Transcriptional enhancers are elements within the genome that control when and where genes are expressed. Although they were identified more than 3 decades ago, how DNA sequence encodes enhancer function remains unclear. Here we show that enhancer syntax (the order, orientation, and spacing of transcription factor binding sites) is important for tissue-specific gene expression. Surprisingly, enhancers with low-affinity binding sites can mediate robust tissue specific patterns of gene expression when they are organized with optimal syntax. Such enhancers may be a vastly underappreciated feature of the regulatory genome.
Keywords: enhancer, gene regulation, transcription, enhancer grammar, regulatory principles
Abstract
Transcriptional enhancers are short segments of DNA that switch genes on and off in response to a variety of intrinsic and extrinsic signals. Despite the discovery of the first enhancer more than 30 y ago, the relationship between primary DNA sequence and enhancer activity remains obscure. In particular, the importance of “syntax” (the order, orientation, and spacing of binding sites) is unclear. A high-throughput screen identified synthetic notochord enhancers that are activated by the combination of ZicL and ETS transcription factors in Ciona embryos. Manipulation of these enhancers elucidated a “regulatory code” of sequence and syntax features for notochord-specific expression. This code enabled in silico discovery of bona fide notochord enhancers, including those containing low-affinity binding sites that would be excluded by standard motif identification methods. One of the newly identified enhancers maps upstream of the known enhancer that regulates Brachyury (Ci-Bra), a key determinant of notochord specification. This newly identified Ci-Bra shadow enhancer contains binding sites with very low affinity, but optimal syntax, and therefore mediates surprisingly strong expression in the notochord. Weak binding sites are compensated by optimal syntax, whereas enhancers containing high-affinity binding affinities possess suboptimal syntax. We suggest this balance has obscured the importance of regulatory syntax, as noncanonical binding motifs are typically disregarded by enhancer detection methods. As a result, enhancers with low binding affinities but optimal syntax may be a vastly underappreciated feature of the regulatory genome.
Previous studies have highlighted the importance of sequence constraints within developmental enhancers for tissue-specific patterns of gene expression in both Drosophila and Ciona embryos (1–6). For example, the 69-bp orthodenticle homeobox (Otx)-a enhancer mediates restricted expression in the Ciona neural plate in response to pleiotropic fibroblast growth factor (FGF) signaling (7–9). Specificity depends on a series of low-affinity binding sites for the transcription factors ETS (FGF signaling) and GATA (ectoderm determinant) (2). Modification of these sites to improve their binding affinities resulted in augmented levels of gene expression in the neural plate, as well as ectopic expression in additional tissues that respond to FGF signaling (2).
These observations prompted the suggestion that the evolution of developmental enhancers depends on the selection of submaximal binding sites. This “suboptimization” might also apply to the organization of enhancers, as changing the spacing of neighboring GATA and ETS binding sites resulted in a significant increase in enhancer activity (2). Modified Otx-a enhancers containing both optimal binding sites and optimal spacing of neighboring sites mediated intense expression in a variety of tissues responding to FGF, including the neural plate, notochord, anterior endoderm, and hindbrain (2). We define optimal binding sites as those with the highest affinity and optimal spacing with respect to the levels of expression. Here we sought to determine the relationship between binding affinities and syntax (e.g., spacing) in enhancer function.
A high-throughput screen of Otx-a enhancer variants (2) led to the identification of a synthetic enhancer that mediates robust expression in the notochord. Manipulation of this enhancer identified a fortuitous ZicL binding site as critical for activation in the notochord. This observation strengthens earlier evidence that notochord specification depends on the combinatorial activities of FGF/ETS signaling and the localized ZicL determinant, similar to the interplay of FGF/ETS and GATA in the activation of gene expression in the neural plate (10–12).
Manipulation of synthetic and native enhancers identified a notochord “regulatory code” that includes sequence and syntax constraints. Surprisingly, these features can compensate for each other to mediate tissue-specific expression. This code permitted the in silico identification of new notochord enhancers, including a Brachyury (Ci-Bra) shadow enhancer. This newly identified enhancer contains low-affinity ETS binding sites (relative affinity, ≤0.3), but optimal syntax (i.e., spacing and orientation). In contrast, another enhancer that was identified, which regulates the homeobox gene Mnx, contains higher-affinity binding motifs but suboptimal spacing between ETS and ZicL binding sites. We propose that a trade-off in binding affinity and syntax encodes tissue specificity. Enhancers with high-affinity sites possess suboptimal syntax, whereas those containing low-affinity sites use optimal syntax. This compensation might obscure the importance of regulatory syntax, as noncanonical binding motifs are typically disregarded by computational methods used for enhancer detection. We therefore suggest that enhancers such as the Ci-Bra shadow enhancer containing low-affinity binding sites and optimal syntax are a systematically underappreciated feature of the regulatory genome.
Results
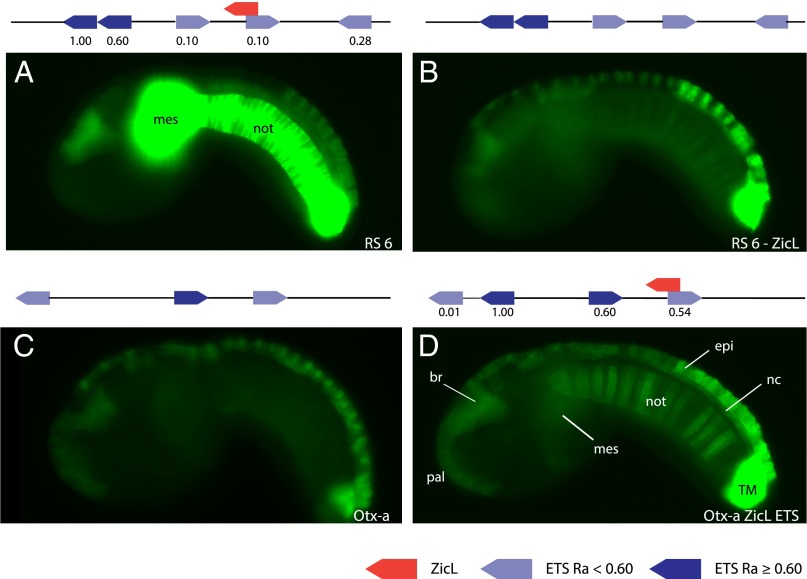
We previously identified more than 20,000 Otx-a enhancer variants that mediated significant expression in electroporated Ciona embryos (2). One of these variants, Random Synthetic Otx-a 6 (RS 6), was found to direct intense expression of a GFP reporter gene in the notochord (Fig. 1A and SI Appendix, Fig. S1). The enhancer also drives expression in the endogenous Otx-a pattern, the neural plate, but this activity is overshadowed by the intense expression in the notochord (SI Appendix, Fig. S1B). RS 6 possesses two distinctive features compared with the wild-type Otx-a enhancer (Fig. 1 A and C and SI Appendix, Fig. S1): an additional ETS motif with flanking sequences that mediate optimal binding affinity (CCGGAAGT; bold indicates flanking of core binding site that mediates highest affinity), and a ZicL binding site overlapping the proximal native ETS site.
Fig. 1.
ETS and ZicL mediate notochord expression. (A) Embryo electroporated with RS 6; GFP expression can be seen strongly (≥10% saturated pixels at 500 ms exposure time) in the notochord (not) and mesenchyme (mes), and moderately in the anterior brain and dorsal epidermis. Expression in this embryo is saturated, as all images in this figure are taken at the same exposure time. Please see SI Appendix, Fig. S1, for images of this embryo at other exposure times. (B) Embryo electroporated with RS 6 −ZicL, where the ZicL site has been mutated. GFP expression is only weakly (<10% saturated pixels at 800 ms) in the notochord. (C) Embryo electroporated with Otx-a; GFP expression can be seen in the anterior brain, palps, dorsal nerve cord, dorsal epidermis, and two tail muscle cells. No expression is seen in the notochord. (D) Embryo electroporated with Otx-a ZicL ETS, where the sequence was modified to add a ZicL and ETS site similar to RS 6. Moderate GFP expression is now seen in the notochord as well as locations of endogenous Otx-a expression anterior brain (br), palps (pal), dorsal nerve cord (nc), dorsal epidermis (epi), and two tail muscle cells (TM). A schematic of the sequence electroporated is shown above each image. Dark-blue arrows refer to ETS binding sites with a relative binding affinity (Ra) ≥0.60, which is classified as a high binding affinity; light-blue arrows refer to ETS binding sites with a binding affinity <0.60; and red arrows refer to a ZicL binding site. All images were taken at the same exposure time. For counting of expression, see SI Appendix, Fig. S1.
In the ascidian Halocynthia roretzi, Brachyury is directly activated by ETS and ZicL (10). In the ascidian Ciona intestinalis, ZicL is known to be important for activation of Brachyury (Ci-Bra) in the presumptive notochord (11, 12), whereas FGF signaling is essential for gene activity in the notochord (13). We therefore reasoned that ETS (FGF) and ZicL binding sites could be directly responsible for RS 6 activity in the notochord. Mutations that abolish the de novo optimal ETS site while retaining the two native ETS sites reduce expression of the GFP reporter gene in the notochord (SI Appendix, Fig. S1). Disruption of the novel ZicL binding site resulted in an even more dramatic reduction of expression in the notochord (Fig. 1B and SI Appendix, Fig. S1). Finally, mutations in both the de novo ETS and ZicL binding sites completely abolish expression in the notochord (SI Appendix, Fig. S1).
These observations suggest that the ZicL site, along with FGF/ETS signaling, is essential for the notochord-specific activity of the RS 6 enhancer. This regulatory logic is very similar to that seen for the Otx-a enhancer, which mediates expression in the neural plate in response to FGF/ETS signaling and a different localized tissue determinant (GATA rather than ZicL) (8). It is a logic used pervasively for combinatorial control of gene expression (14–16).
We modified the wild-type Otx-a enhancer to validate this model for notochord-restricted expression (Fig. 1 C and D and SI Appendix, Fig. S1). A ZicL site was created with four nucleotide substitutions straddling the 5′ portion of the proximal ETS site, as seen in the synthetic RS 6 enhancer. The modified WT enhancer mediates strong expression in the endogenous neural plate pattern, as well as the notochord (SI Appendix, Fig. S1). Notochord expression was further augmented by substituting five additional nucleotides to create an additional optimal ETS motif (Fig. 1D and SI Appendix, Fig. S1). The stronger expression mediated by RS 6 is probably a result of additional ETS sites not present in the modified Otx-a enhancer, as removal of these additional sites diminishes expression (SI Appendix, Fig. S2).
Because ZicL and two ETS sites appear to be sufficient for notochord expression, we surveyed the remainder of the RS Otx-a library for additional enhancer variants containing this combination of sites. There are 15 such variants, including RS 6 (Dataset S1 and SI Appendix, Fig. S1), but only two (RS 51 and RS 55) were found to mediate expression in the notochord. RS 51 and RS 55 have very little sequence similarity beyond conserved ETS and ZicL binding sites. It is therefore unlikely that disruption of cryptic repression elements contained in the wild-type Otx-a backbone is responsible for activation in the notochord. We sought to determine why so few of the synthetic Otx-a enhancer variants mediate expression in the notochord, despite sharing similar ETS and ZicL binding sites.
RS 6 and the inactive variant RS 52 share 40/69 bases and almost identical regulatory syntax (SI Appendix, Fig. S4A). However, only RS 6 activates expression in the notochord. This highlights the importance of identifying the functional features of the enhancer, rather than relying on sequence conservation to predict function. Manipulations of RS 52 to improve the affinity of the distal-most ETS site resulted in weak notochord expression (SI Appendix, Fig. S4). Altering the dinucleotide sequences flanking the remaining core binding sites to better match those contained in RS 6 led to intense notochord expression (SI Appendix, Fig. S4 C and D). Thus, as seen for Otx-a, the flanking dinucleotide sequences, and hence the affinities of ETS binding sites, are essential for enhancer activity (2). Relative binding affinities were calculated using median signal intensities of the universal protein binding microarray for mouse ETS-1 proteins from the UniProbe database (17) (thebrain.bwh.harvard.edu/uniprobe/index.php; see Datasets S1–S3 for all enhancer variants that were tested in this study).
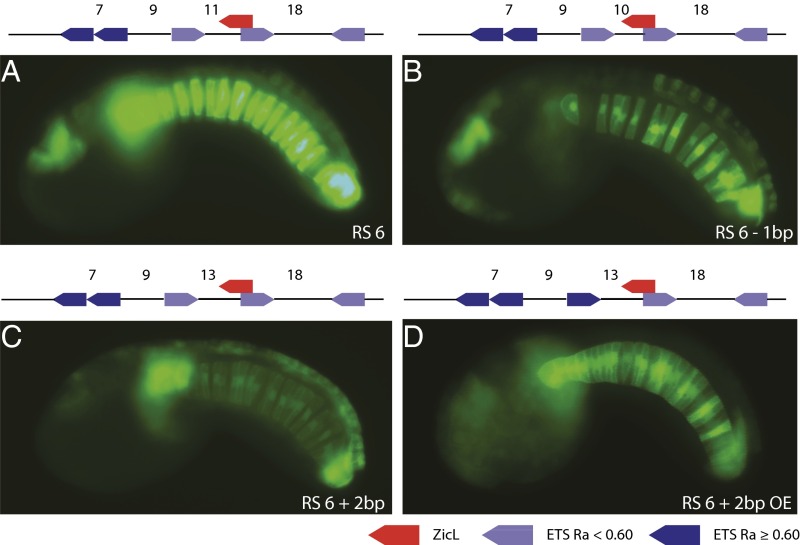
The fact that so few synthetic variants mediate notochord expression, despite having reasonable ETS and ZicL binding affinities, suggests a role for regulatory syntax. Having identified the sequence constraints for notochord expression, we examined two aspects of regulatory syntax: the orientation and spacing of linked sites. We previously showed that the spacing between neighboring GATA and ETS sites influenced the levels of Otx-a activity (2). We therefore examined the spacing of neighboring ZicL and ETS sites in the synthetic notochord enhancers. Distances are measured as the number of nucleotides from the core of one binding site to the core of the next. The spacing between linked 5′ ETS and 3′ ZicL sites in RS 6 is 11 bp, and deletion of a single nucleotide or addition of 2 bp led to a significant decrease in the levels of notochord expression (Fig. 2 B and C and SI Appendix, Fig. S5).
Fig. 2.
Regulatory compensation: suboptimal spacing can be balanced by improved binding affinities. (A) Embryo electroporated with RS 6; GFP expression can be seen strongly in the notochord and mesenchyme and moderately in the anterior brain and dorsal epidermis. (B) Embryo electroporated with RS 6 −1 bp, where 1 bp has been deleted between ETS and ZicL. GFP expression is weaker in the notochord compared with RS 6. (C) Embryo electroporated with RS 6 +2 bp, where 2 bp have been inserted between ETS and ZicL. GFP expression is weaker in the notochord compared with RS 6. (D) Embryo electroporated with RS 6 +2 bp optimized ETS (OE), where the ETS closest to the ZicL has been changed from CTGGAACT to CCGGAAGT, the optimal affinity ETS binding motif. GFP expression is stronger in the notochord than RS 6 +2 bp and equal to RS 6. The poorer spacing in RS 6 +2 bp has been compensated by the optimal ETS binding site. A schematic of the sequence electroporated is shown above each image. Dark-blue arrows refer to ETS binding sites with a binding affinity above 0.60, light-blue arrows refer to ETS binding sites with a binding affinity lower than 0.60, and red arrows refer to a ZicL binding site. All images were taken at the same exposure time.
As described previously, a modified version of the native Otx-a enhancer containing additional ZicL and ETS binding sites drives expression in the notochord, as well as the neural plate (Fig. 2). The most closely linked ETS and ZicL sites are separated by 11 bp, as seen in the synthetic RS 6 and RS 52 enhancers. The removal of 1 or 2 bp or the addition of 2 bp caused diminished notochord expression (SI Appendix, Fig. S5 and Dataset S2). Thus, for both RS 6 and the modified Otx-a enhancers, a distance of 11 bp between linked ETS and ZicL binding sites produces the highest levels of expression (summarized in Dataset S2).
Having identified sequence and spacing constraints for notochord expression, we next asked whether other features of enhancer syntax, such as the orientation of linked sites, are also important for enhancer activity. Toward this end, we reversed the orientation of the ETS site located closest to ZicL. Reversal of the site led to a complete loss of notochord expression, which suggests an additional tier of regulatory syntax, namely, orientation of linked sites.
The Otx-a enhancer contains a mixture of optimal and suboptimal features (2). For example, the proximal ETS and GATA sites possess low binding affinities but exhibit optimal spacing, whereas the distal sites have higher affinities but poor spacing (2). Such observations raise the possibility of a trade-off between binding affinities and regulatory syntax. To explore this issue, we improved the affinities of ETS binding sites within a modified RS 6 enhancer containing suboptimal spacing of 13 bp between neighboring sites rather than 11 bp. This significantly restored expression in the notochord (Fig. 2D), consistent with the idea of a trade-off between binding affinities and syntax.
The preceding analyses define a “regulatory code” for notochord-restricted expression: ZicL and two ETS sites, as well as constraints on spacing and orientation of sites. We applied this code to survey the complete Ciona genome and identified 69 putative enhancers (Dataset S3). None of these putative enhancers contains both optimal syntax and optimal affinity sites. To investigate the idea of trade-offs in affinity and syntax, we focused on two types of potential enhancers: those containing good binding sites but suboptimal syntax, and those containing low-affinity sites but optimal syntax. We analyzed one member of each class, a genomic DNA fragment from the Mnx 5′ flanking region (moderate binding affinities but poor syntax) and a putative Ci-Bra shadow enhancer (low-affinity sites but optimal syntax).
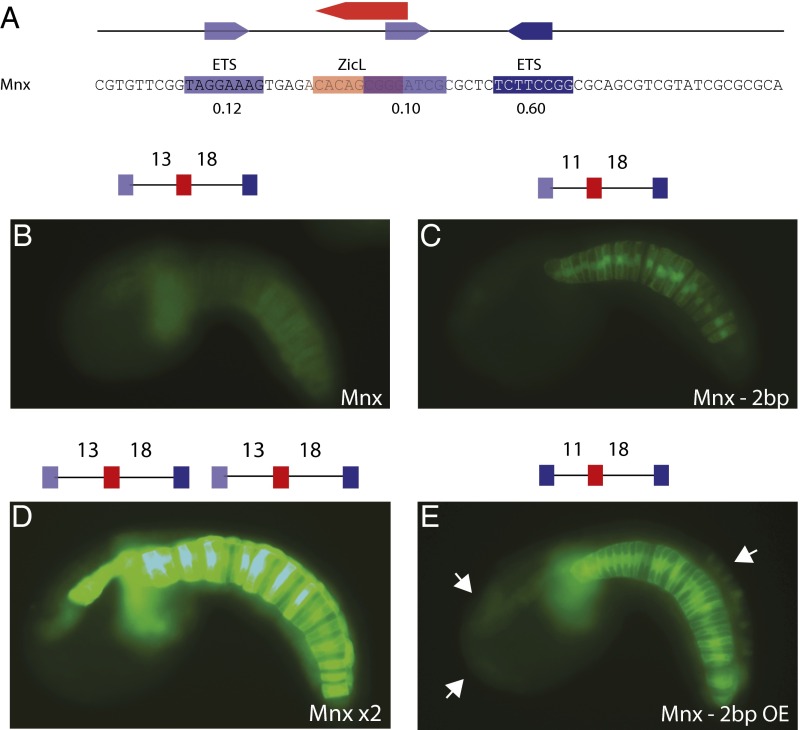
Mnx is expressed in both the floorplate and the notochord (18). A 70-bp sequence from the Mnx 5′ regulatory region, located 690 bp upstream of the transcription start site (18), was found to direct weak expression of a GFP reporter in the notochord of electroporated embryos (Fig. 3). This enhancer contains one low-affinity (relative affinity, ≤0.3) and one high-affinity (relative affinity, ≥0.6) ETS binding site, as well as suboptimal spacing (13 bp) between neighboring ZicL and ETS sites. Significantly stronger expression was obtained by simply reducing the spacing between the linked ETS and ZicL sites from 13 to 11 bp (Fig. 3C and SI Appendix, Fig. S6). Optimizing the binding affinity of the low ETS site (from a relative affinity of 0.25 to 1.00) in combination with optimal spacing resulted in even higher levels of notochord expression (Fig. 3E and SI Appendix, Fig. S6). However, as seen for the Otx-a enhancer, the combination of high-affinity binding sites and optimal spacing causes a loss in tissue specificity, with ectopic expression at additional sites of FGF signaling (7, 13, 19–22) (Fig. 3E, arrows, and SI Appendix, Fig. S6).
Fig. 3.
Identification of the Mnx enhancer, using compensatory regulatory logic. (A) The 69-bp sequence of an Mnx enhancer located 690 bp upstream of the transcription start site for Mnx. Relative binding affinities are shown below sites. (B) Embryo electroporated with Mnx enhancer; GFP expression can be seen weakly in the notochord. (C) Embryo electroporated with Mnx −2 bp enhancer; GFP expression is stronger in the notochord compared with Mnx. (D) Embryo electroporated with Mnx ×2 enhancer; GFP expression can be strongly in the notochord and the floor plate and moderately in the mesenchyme. (E) Embryo electroporated with Mnx −2 bp with optimized ETS (OE) enhancer; GFP is strongly expressed in the notochord compared with Mnx −2 bp. White arrows point to ectopic expression in the palps, anterior brain, and dorsal epidermis. A schematic of the sequence electroporated is shown above each image. Dark-blue boxes refer to ETS binding sites with a binding affinity above 0.60, light-blue boxes refer to ETS binding sites with a binding affinity lower than 0.60, and red boxes refer to a ZicL binding site. All images were taken at the same exposure time to allow for direct comparison. For counting, see SI Appendix, Fig. S6.
We previously proposed that the trade-off between the levels and specificity of gene expression might be circumvented by the use of multiple copies of suboptimized enhancers (2). We further tested this idea by creating a GFP reporter gene containing two tandem copies of the native Mnx enhancer. This transgene mediates intense but specific expression in the endogenous location (Fig. 3D). We therefore suggest that additional Mnx “shadow” enhancers might be used to regulate the endogenous locus. Shadow enhancers are commonly used in vertebrates and Drosophila embryos (23–25), but have not been documented in Ciona (see following).
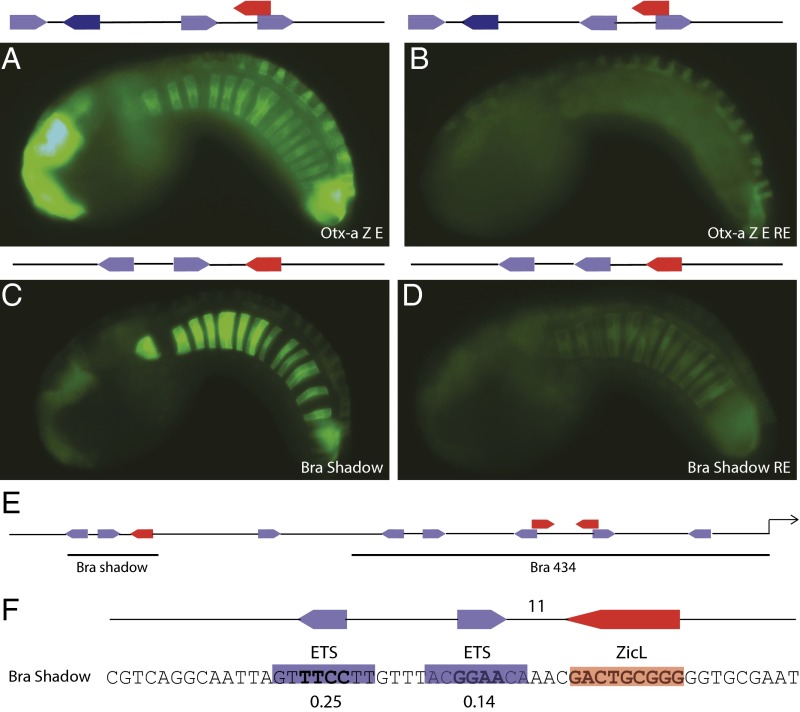
The other enhancer analyzed in our in silico survey of the Ciona genome represents a stringent test of the idea that regulatory syntax can compensate for low-affinity binding sites. A 70-bp sequence located 790 bp upstream of the Ci-Bra transcription start site and ∼300 bp upstream of the Ci-Bra proximal enhancer, contains two low-affinity ETS sites (relative affinity, ≤0.3), but optimal spacing and orientation of linked ZicL and ETS sites (Fig. 4). This sequence was found to mediate strong and localized expression in the notochord, and thereby represents the first shadow enhancer to be identified in Ciona (Fig. 4C). Standard computational methods would fail to identify this enhancer because of the poor quality of both ETS sites. It was recovered in our survey because of our knowledge of optimal syntax of linked ETS and ZicL sites. Indeed, disrupting this syntax by inversion of the proximal ETS site resulted in a substantial diminishment in enhancer activity (Fig. 4 and SI Appendix, Fig. S7). Similar results were obtained with the Mnx enhancer (SI Appendix, Fig. S8). We therefore suggest that the significance of regulatory syntax is underappreciated, as standard algorithms and chromatin immunoprecipitation methods select for enhancers containing high-affinity binding sites and are therefore likely to lack good syntax to maintain tissue-specific expression (Discussion).
Fig. 4.
Optimal syntax compensates for poor affinities to encode tissue specific expression. (A) Embryo electroporated with Otx-a ZicL ETS (Otx-a Z E); GFP expression can be seen in the notochord as well as locations of endogenous Otx-a expression. (B) Embryo electroporated with Otx-a ZicL ETS reversed ETS (Otx-a Z E RE), where the sequence of the ETS closest to the ZicL was mutated to be the reverse complement. GFP expression in the notochord is lost. (C) Embryo electroporated with Bra shadow; GFP expression can be seen in the notochord. (D) Embryo electroporated with Bra shadow with reversed ETS (Bra shadow RE); GFP expression is diminished in the notochord. A schematic of the sequence electroporated is shown above each image. Dark-blue boxes refer to ETS binding sites with a binding affinity above 0.60, light-blue boxes refer to ETS binding sites with a binding affinity lower than 0.60, and red boxes refer to a ZicL binding site. All images were taken at the same exposure time. (E) The Bra shadow is 731 bp upstream of the Brachyury start site. (F) The Bra shadow employs the optimal syntax, the ETS and ZicL are 11 bp apart and are facing each other, but the relative affinity of the ETS binding site are poor at 0.25 and 0.14.
Discussion
The synthetic RS 6 enhancer was found to mediate strong expression in the notochord of Ciona embryos, in addition to its normal site of activity in the neural plate. Manipulation of this synthetic enhancer led to the demonstration that ETS and ZicL are the key mediators of notochord-restricted gene activity, providing clear evidence that ZicL works in concert with FGF signaling to specify the notochord during development.
Neural plate and notochord enhancers use a strikingly similar regulatory logic found pervasively in development (14–16). Namely, a restricted tissue determinant (GATA or ZicL) works in concert with a pleiotropic signaling system (e.g., FGF) to mediate localized expression in a specific tissue (8). Suboptimal binding causes diminished levels of gene expression compared with optimal binding affinities and spacing. However, this suboptimization appears to be important for tissue-specific patterns of gene expression in the neural plate and notochord. Enhancer optimization augments the levels of expression and causes ectopic activation in unwanted tissues (ref. 2; Fig. 3). Suboptimization is likely to increase the degeneracy of regulatory “codes” that relate enhancer sequences to gene activity. Such degeneracy has probably obscured past attempts to understand the regulatory logic of developmental enhancers, particularly the importance of syntax.
The further analysis of RS 6, as well as variants of the native Otx-a enhancer, raised the possibility of a trade-off between binding affinities and regulatory syntax. Enhancers containing high-affinity ETS binding sites possess suboptimal spacing between linked sites, whereas those containing lower-affinity sites possess optimal spacing. Enhancers containing both optimal ETS sites and optimal spacing exhibit strong expression in the notochord, as well as additional sites of FGF signaling such as the neural plate (summarized in SI Appendix, Fig. S9).
The Mnx enhancer contains one high-affinity ETS site and one low-affinity site, but suboptimal spacing of the nearest ZicL site (13 bp, rather than 11 bp). The removal of two nucleotides between these sites, to optimize the spacing, results in significantly stronger expression. It seems unlikely that this Mnx enhancer works alone to regulate expression in the notochord, and we suggest that additional “shadow” enhancers might help augment activity. Indeed, we identified the first shadow enhancer in Ciona by surveying the Ciona genome for optimal regulatory syntax, while relaxing the usual cutoff for minimal binding affinities (and thus deviations from “consensus” binding motifs). The Ci-Bra shadow enhancer mediates robust expression in the notochord despite containing two low-affinity ETS binding sites. The fact that the shadow enhancer mediates such robust expression attests to the importance of regulatory syntax. We suggest there is a continuum of enhancers with respect to the quality of binding sites and syntax (summarized in SI Appendix, Fig. S9). A typical enhancer might contain a mixture of both properties, as observed in the native Otx-a enhancer (2).
We believe that current enhancer prediction methods are biased toward enhancers lacking syntax, as they emphasize high-affinity (consensus) binding motifs. Our findings that optimal syntax can compensate for low-affinity binding sites highlight the need for more sophisticated methods of enhancer analysis and the importance of regulatory syntax. We propose that enhancers with low-affinity sites but optimal syntax are a vastly underappreciated feature of the regulatory genome. Further high-throughput functional studies to map the grammatical constraints (sequence and syntax) of transcription factors will help identify and define such enhancers and their contributions to development.
Materials and Methods
Electroporation.
Adult C. intestinalis were obtained from M-Rep and maintained in artificial seawater (Crystal Sea Marine mix) at 18 °C, under constant illumination. Dechorionation, in vitro fertilization, and electroporation were carried out as described in ref. 26. For each electroporation, typically, eggs and sperm were collected from 20 adults, 70 μg DNA was resuspended in 100 μL buffer. Embryos were fixed at the appropriate developmental stage for 15 min in 4% (wt/vol) formaldehyde. The tissue was then cleared in a series of washes of 0.01% Triton-X in PBS. Samples were mounted in 50% (vol/vol) glycerol in PBS with 2% (wt/vol) DABCO compound for microscopy. Differential interference-contrast microscopy was used to obtain transmitted light micrographs with a Zeiss Axio Imager A2, using the ×20 EC Plan Neofluar objective. The same microscope was used to obtain GFP images. All constructs were electroporated at least twice in two completely separate experiments (biological replicates).
Counting Embryos.
For each experiment, once embryos had been mounted on slides, slide labels were covered with thick tape and randomly numbered by a laboratory member not involved in this project and randomized. In each experiment, all comparative constructs were present, along with a slide with WT Otx-a as a reference. The x-cite was turned on for 1hr before analysis to ensure the illumination intensity was constant. Fifty embryos were counted for each biological replicate. Weak, moderate, and strong expression definitions are based on exposure time required for 10% saturated pixels, calculated by the imaging software: strong, <500 ms; moderate, <800 ms; and weak, ≥800 ms.
Acquisition of Images.
For enhancers that were being compared, images were taken on the same day and from electroporations performed on the same day, using identical settings. For images, embryos were chosen that represented the average from counting data. Images are rotated and cropped, but have no other manipulations. At least four exposure times were taken for each construct: 100, 250, 500, and 1,000 ms. In each figure, the same exposure time for each image is shown to allow direct comparison. This occasionally leads to overexposed images being used for stronger constructs; for example, RS6. SI Appendix, Fig. S1 shows RS6 at differing exposure times.
Scoring Relative Affinities.
It is possible to determine relative binding affinities of the native sites by analyzing the frequency of selected sequences, using high-throughput binding datasets (27–29). Relative affinities were calculated using median signal intensities of the universal protein binding microarray data for mouse ETS-1 (17) proteins from the UniProbe database (thebrain.bwh.harvard.edu/uniprobe/index.php). The percentage of relative affinities represent the fold changes of median signal intensities of the native 8-mer motifs compared with the optimal 8-mer motifs for optimal ETS and GATA, respectively. Binding sites that match to the highest-affinity site are defined as optimal affinity sites with a relative affinity of 1.0. All sites with an affinity score of less than 1 are defined as suboptimal. Binding sites that have a relative affinity of ≥0.6 are described as high-affinity sites, a relative affinity of 0.3–0.6 is considered a moderate-affinity site, and those sites that have a relative affinity ≤0.3 are considered low-affinity sites. For simplicity, in figures, ETS sites are colored dark blue for high-affinity sites (≥0.6); all other sites (<0.6) are light blue.
Identification of Putative Notochord Enhancers.
We scanned the genome of C. intestinalis for a ZicL site and two ETS sites within a 200-bp region. We searched for sequences where the most proximal ETS points toward the ZicL, and these two binding sites are between 9 and 15 bp apart. We required one ETS to have a binding affinity of at least 0.10, and another ETS to have an affinity of at least 0.20. A total of 69 such regions were found in C. intestinalis. Of the two tested, both worked.
Supplementary Material
Acknowledgments
We thank members of the M.S.L. and D.S.R. laboratories for helpful discussions. This work was supported by a grant from the NIH (Grant NS076542).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 6330.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605085113/-/DCSupplemental.
References
- 1.Crocker J, et al. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell. 2015;160(1-2):191–203. doi: 10.1016/j.cell.2014.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farley EK, et al. Suboptimization of developmental enhancers. Science. 2015;350(6258):325–328. doi: 10.1126/science.aac6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Small S, Kraut R, Hoey T, Warrior R, Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 1991;5(5):827–839. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- 4.Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72(5):741–752. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- 5.Swanson CI, Schwimmer DB, Barolo S. Rapid evolutionary rewiring of a structurally constrained eye enhancer. Curr Biol. 2011;21(14):1186–1196. doi: 10.1016/j.cub.2011.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramos AI, Barolo S. Low-affinity transcription factor binding sites shape morphogen responses and enhancer evolution. Philos Trans R Soc Lond B Biol Sci. 2013;368(1632):20130018. doi: 10.1098/rstb.2013.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertrand V, Hudson C, Caillol D, Popovici C, Lemaire P. Neural tissue in ascidian embryos is induced by FGF9/16/20, acting via a combination of maternal GATA and Ets transcription factors. Cell. 2003;115(5):615–627. doi: 10.1016/s0092-8674(03)00928-0. [DOI] [PubMed] [Google Scholar]
- 8.Rothbächer U, Bertrand V, Lamy C, Lemaire P. A combinatorial code of maternal GATA, Ets and beta-catenin-TCF transcription factors specifies and patterns the early ascidian ectoderm. Development. 2007;134(22):4023–4032. doi: 10.1242/dev.010850. [DOI] [PubMed] [Google Scholar]
- 9.Khoueiry P, et al. A cis-regulatory signature in ascidians and flies, independent of transcription factor binding sites. Curr Biol. 2010;20(9):792–802. doi: 10.1016/j.cub.2010.03.063. [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto J, Kumano G, Nishida H. Direct activation by Ets and Zic is required for initial expression of the Brachyury gene in the ascidian notochord. Dev Biol. 2007;306(2):870–882. doi: 10.1016/j.ydbio.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 11.Imai KS, Satou Y, Satoh N. Multiple functions of a Zic-like gene in the differentiation of notochord, central nervous system and muscle in Ciona savignyi embryos. Development. 2002;129(11):2723–2732. doi: 10.1242/dev.129.11.2723. [DOI] [PubMed] [Google Scholar]
- 12.Yagi K, Satou Y, Satoh N. A zinc finger transcription factor, ZicL, is a direct activator of Brachyury in the notochord specification of Ciona intestinalis. Development. 2004;131(6):1279–1288. doi: 10.1242/dev.01011. [DOI] [PubMed] [Google Scholar]
- 13.Yasuo H, Hudson C. FGF8/17/18 functions together with FGF9/16/20 during formation of the notochord in Ciona embryos. Dev Biol. 2007;302(1):92–103. doi: 10.1016/j.ydbio.2006.08.075. [DOI] [PubMed] [Google Scholar]
- 14.Brodu V, Mugat B, Roignant JY, Lepesant JA, Antoniewski C. Dual requirement for the EcR/USP nuclear receptor and the dGATAb factor in an ecdysone response in Drosophila melanogaster. Mol Cell Biol. 1999;19(8):5732–5742. doi: 10.1128/mcb.19.8.5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oosterveen T, et al. SoxB1-driven transcriptional network underlies neural-specific interpretation of morphogen signals. Proc Natl Acad Sci USA. 2013;110(18):7330–7335. doi: 10.1073/pnas.1220010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barolo S, Posakony JW. Three habits of highly effective signaling pathways: Principles of transcriptional control by developmental cell signaling. Genes Dev. 2002;16(10):1167–1181. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- 17.Wei GH, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29(13):2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imai KS, Levine M, Satoh N, Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312(5777):1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- 19.Shi W, Levine M. Ephrin signaling establishes asymmetric cell fates in an endomesoderm lineage of the Ciona embryo. Development. 2008;135(5):931–940. doi: 10.1242/dev.011940. [DOI] [PubMed] [Google Scholar]
- 20.Hudson C, Lemaire P. Induction of anterior neural fates in the ascidian Ciona intestinalis. Mech Dev. 2001;100(2):189–203. doi: 10.1016/s0925-4773(00)00528-1. [DOI] [PubMed] [Google Scholar]
- 21.Stolfi A, Wagner E, Taliaferro JM, Chou S, Levine M. Neural tube patterning by Ephrin, FGF and Notch signaling relays. Development. 2011;138(24):5429–5439. doi: 10.1242/dev.072108. [DOI] [PubMed] [Google Scholar]
- 22.Wagner E, Levine M. FGF signaling establishes the anterior border of the Ciona neural tube. Development. 2012;139(13):2351–2359. doi: 10.1242/dev.078485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321(5894):1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20(17):1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nolte C, Jinks T, Wang X, Martinez Pastor MT, Krumlauf R. Shadow enhancers flanking the HoxB cluster direct dynamic Hox expression in early heart and endoderm development. Dev Biol. 2013;383(1):158–173. doi: 10.1016/j.ydbio.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 26.Christiaen L, Wagner E, Shi W, Levine M. Electroporation of transgenic DNAs in the sea squirt Ciona. Cold Spring Harb Protoc. 2009;2009(12):pdbprot5345. doi: 10.1101/pdb.prot5345. [DOI] [PubMed] [Google Scholar]
- 27.Hume MA, Barrera LA, Gisselbrecht SS, Bulyk ML. UniPROBE, update 2015: New tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2015;43(Database issue):D117–D122. doi: 10.1093/nar/gku1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nitta KR, et al. March 17, 2015. Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. eLife 10.7554/eLife.04837.
- 29.Zhang H, Levine M, Ashe HL. Brinker is a sequence-specific transcriptional repressor in the Drosophila embryo. Genes Dev. 2001;15(3):261–266. doi: 10.1101/gad.861201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.