Significance
A number of lipid mediators are known to contribute to inflammatory resolution. Fatty acid metabolites of cytochrome P450 (CYP) enzymes are found in abundance; however, their roles in inflammatory resolution are not known. Targeted lipidomics revealed that CYP450-epoxy-oxylipins were present during acute inflammation and inflammatory resolution. Using mice lacking soluble epoxide hydrolase, the major metabolizing pathway for CYP450-derived fatty acid mediators, and CYP450 epoxygenase inhibition specifically during resolution, we show that CYP450-derived lipids dramatically limit the accumulation of inflammatory monocytes during resolution. Moreover, all cells of the monocyte lineage examined showed a dramatic alteration in their proresolution phenotype following epoxygenase inhibition. These findings demonstrate that the CYP450-epoxy-oxylipins pathway has a critical role in monocyte lineage recruitment and resolution activity during inflammatory resolution.
Keywords: oxylipins, resolution, monocyte, phagocytosis, epoxygenase
Abstract
Resolution of inflammation has emerged as an active process in immunobiology, with cells of the mononuclear phagocyte system being critical in mediating efferocytosis and wound debridement and bridging the gap between innate and adaptive immunity. Here we investigated the roles of cytochrome P450 (CYP)-derived epoxy-oxylipins in a well-characterized model of sterile resolving peritonitis in the mouse. Epoxy-oxylipins were produced in a biphasic manner during the peaks of acute (4 h) and resolution phases (24–48 h) of the response. The epoxygenase inhibitor SKF525A (epoxI) given at 24 h selectively inhibited arachidonic acid- and linoleic acid-derived CYP450-epoxy-oxlipins and resulted in a dramatic influx in monocytes. The epoxI-recruited monocytes were strongly GR1+, Ly6chi, CCR2hi, CCL2hi, and CX3CR1lo. In addition, expression of F4/80 and the recruitment of T cells, B cells, and dendritic cells were suppressed. sEH (Ephx2)−/− mice, which have elevated epoxy-oxylipins, demonstrated opposing effects to epoxI-treated mice: reduced Ly6chi monocytes and elevated F4/80hi macrophages and B, T, and dendritic cells. Ly6chi and Ly6clo monocytes, resident macrophages, and recruited dendritic cells all showed a dramatic change in their resolution signature following in vivo epoxI treatment. Markers of macrophage differentiation CD11b, MerTK, and CD103 were reduced, and monocyte-derived macrophages and resident macrophages ex vivo showed greatly impaired phagocytosis of zymosan and efferocytosis of apoptotic thymocytes following epoxI treatment. These findings demonstrate that epoxy-oxylipins have a critical role in monocyte lineage recruitment and activity to promote inflammatory resolution and represent a previously unidentified internal regulatory system governing the establishment of adaptive immunity.
Monocytes and monocyte-derived macrophages play a critical role in chronic inflammation, in part via the production and release of lipid mediators (1). One such lipid precursor, arachidonic acid, is metabolized into families of biologically active mediators by the cyclooxygenase, lipoxygenase, and cytochrome P450 (CYP) pathways (2, 3). CYPs metabolize arachidonic acid by: (i) an epoxygenase activity that catalyzes the conversion of arachidonic acid to epoxyeicosatrienoic acids (EETs); (ii) a lipoxygenase-like activity that metabolizes arachidonic acid to midchain hydroxyeicosatetraenoic acids (HETEs); and (iii) ω- and ω-1-hydroxylase activity, which produces ω-terminal HETEs (3). In addition to arachidonic acid, CYPs with epoxygenase activity can also metabolize alternative polyunsaturated fatty acids such as linoleic acid and docosahexaenoic acid into a series of products including epoxyoctadacamonoenoic acids (EpOMEs) and 19,20-epoxydocosapentaenoic acid (EpDPE), respectively, whose functions remain poorly understood (3–5).
The main polyunsaturated fatty acid-metabolizing CYPs belong to the CYP2 family, in particular the CYP2J and CYP2C subfamilies (3, 4, 6, 7). Moreover, these CYP-lipid–metabolizing enzymes are the primary sources of eicosanoids in small blood vessels, the kidney, liver, lung, intestines, heart, and pancreas (3, 7). In most organs, EETs and related epoxygenase products are metabolically unstable and are rapidly metabolized. The major pathway that regulates EET metabolism is that catalyzed by epoxide hydrolases (8), which convert EETs to less biologically active dihydroxyeicosatrienoic acids (DHETs) (9). EpOMEs similarly get converted into dihydroxyoctadecenoic acids (DiHOMEs), whereas 19,20-EpDPE gets converted into 19,20-dihydroxydocosapentaenoic acid (DiHDPA). Elevating the levels of endogenous CYP products by disrupting (knockout) or inhibiting soluble epoxide hydrolase (sEH) reduces neointima formation (10), atherosclerosis, abdominal aortic aneurysm, dyslipidemia (11), hypertension (12), and diabetes (13) in different mouse models, all of which to some extent one could argue have a degree of nonresolving inflammation.
Over the last 15 y there has been a vast increase in our knowledge of fatty acid mediators that regulate inflammatory processes, particularly newly identified mediators such as the resolvins that mediate the resolution of inflammation (14–16). However, unlike cyclooxygenase and lipoxygenase products, the roles of CYP450 pathways in chronic inflammation remain unclear. The arachidonic acid products of the CYP epoxygenases, the EETs, can regulate vascular tone, smooth muscle cell mitogenesis, platelet aggregation, steroidogenesis, and endothelial and vascular smooth muscle cell activation (4, 5, 7, 17–19). We recently published that in human monocytes and macrophages, epoxygenases and some of their arachidonic acid products were antiinflammatory through their ability to activate the peroxisome proliferator-activated receptor (PPAR), in particular PPARα (20, 21). Overexpression of epoxygenase enzymes CYP2J2 and CYP2C8 or genetic disruption of sEH (sEH−/−) inhibits LPS-induced pulmonary inflammation (22, 23), and sEH−/− mice or treatment with sEH inhibitors is highly effective against inflammatory and neuropathic pain (24–27).
Monocytes are heterogeneous in mice and in humans (28). In mice, monocyte subsets can be divided based on the expression of Ly6c, Gr1, CC-chemokine receptor 2 (CCR2), and CX3C-chemokine receptor 1 (CX3CR1). Ly6chi monocytes are Gr1+, CCR2+, and CX3CR1lo, whereas Ly6clo monocytes are Gr1−, CCR2−, and CX3CR1hi (29, 30). Lipid mediators that regulate the recruitment and phenotype of monocytes are poorly understood.
Herein, using a sterile model of inflammatory resolution dependent on monocyte recruitment, we found that CYP-epoxygenase products not only accumulate in a temporal manner with monocyte recruitment but also limit proinflammatory monocyte recruitment and promote a proresolution phenotype in cells of the monocyte lineage.
Results
CYP450-Derived Lipid Metabolites Form Part of the Coordinated Eicosanoid Fatty Acid Metabolite Response in Acute Inflammation and Resolution.
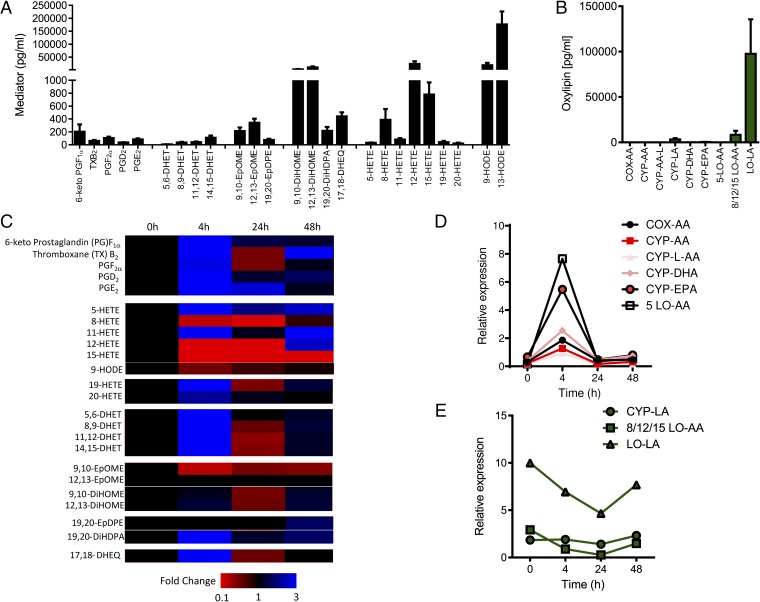
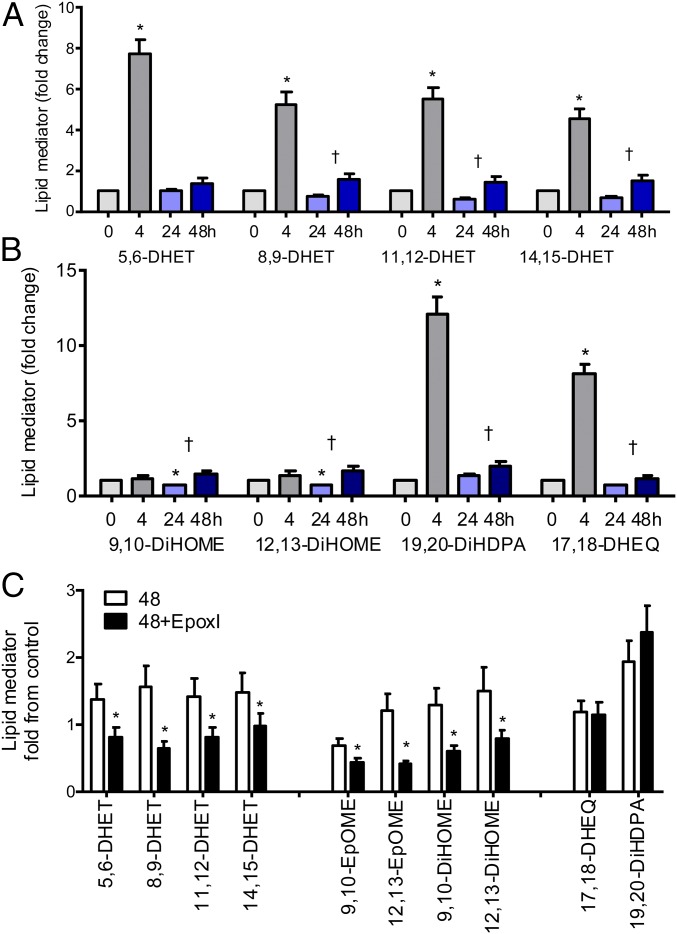
The naive mouse peritoneal cavity contains high levels of linoleic acid [9- and 13-hydroxy-octadecadienoic acid (HODE)] and arachidonic acid (8-, 12-, and 15-HETE)-derived lipoxygenase products as well as 9,10- and 12,13-DiHOME, which are epoxide diols of the linoleic acid epoxygenase products 9,10-EpOME and 12,13-EpOME (Fig. 1). Cyclooxygenase (COX), CYP450, 5-lipoxygenase (LO), and eicosapentaenoic acid (EPA)/docosahexaenoic acid (DHA)-derived mediators, including prostanoids, leukotrienes, EETs, and their metabolites, were found at much lower levels (Fig. 1). The liquid chromatography/tandem mass spectrometry (LC/MS/MS) assay we used was primarily designed to measure CYP450 and related products. We do not currently measure some of the other newly characterized lipid species known to regulate inflammatory resolution such as the resolvins, but clearly enzymatic pathways and lipid precursors for these pathways are all also present. During the acute inflammatory response elicited by zymosan (4 h), there was the expected burst of prostanoids, increases in 5-, 11-, 19-, and 20-HETE, and also increases in the CYP450 epoxygenase-derived oxylipins 5,6-, 8,9-, 11,12-, and 14,15-DHET and 17,18-dihydroxy-EEQ (DHEQ) (Fig. 1 C and D and SI Appendix, Fig. S1). The DHETs are the epoxide hydrolase diols of the P450 metabolites and could readily be detected, whereas parental EETs were not seen. The 15/12-lipoxygenase products initially found at high levels in the naive cavity dropped dramatically during acute inflammation (9- and 13-HODE and 8-, 12-, and 15-HETE), with only 9-HODE and 8-HETE returning to baseline levels over the 48-h time course (Fig. 1 C and E and SI Appendix). After acute inflammation, which is driven primarily by polymorphonuclear cells (PMNs), resolution begins and is associated with an accumulation of proresolution monocytes peaking 24–48 h after the initial inflammatory insult. After a drop at 24 h, this influx in monocytes was associated with a second phase of epoxy-oxylipin production of arachidonic acid (DHETs), linoleic acid (DiHOMEs), EPA (17,18-DHEQ), and DHA (19,20-DiHDPA) epoxygenase products (Fig. 2 A and B). Moreover, this phase was associated with the biosynthesis of new lipid products, as the addition of a selective epoxygenase inhibitor (epoxI; SKF525A; 30 mg/kg, i.p.) to zymosan-treated mice from 24 h onward led to a selective and specific inhibition of arachidonic acid and linoleic acid epoxy-oxylipins (Fig. 2C). EpoxI had no significant effect on COX, LO, or CYP450-lipoxygenase–like products (SI Appendix, Table S1), indicating the epoxI used is highly selective for the CYP-epoxygenase pathway.
Fig. 1.
LC/MS/MS analysis of oxylipin-generating pathways in the mouse peritoneal cavity during acute inflammation and resolution initiated by zymosan A. (A) Absolute levels of oxylipins determined in naive peritoneal cavity lavage fluid of male C57BL/6 mice. (B) Sum of oxylipin-generated COX, CYP450, CYP450-lipoxygenase–like (CYP-L), or LO pathways using either arachidonic acid (AA), linoleic acid (LA), DHA, or EPA as a substrate in the naive peritoneal cavity. (C) Heatmap showing fold changes in oxylipin formation following zymosan A treatment (1 mg, i.p.) from 0 to 4 h (peak of acute inflammation), 24 h, and 48 h (resolution). (D and E) Fold-normalized square root of oxylipins produced by COX-AA (prostanoids), CYP-AA (DHETs), CYP-L-AA (19- and 20-HETE), CYP-DHA (19,20-EpDPE and 19,20-DiHDPA), CYP-EPA (17,18-DHEQ), and 5-LO-AA (5-HETE) pathways (D) and CYP-LA (EpOMEs and DiHOMEs), 8/12/15-LO (8-, 12-, and 15-HETE), and LO-LA (HODEs) pathways (E) in response to zymosan A over 48 h. Pathways in D and E represent two distinct responses to challenge with zymosan A. Data represent the mean ± SEM from n = 4–8 mice per group.
Fig. 2.
Epoxygenase products form in a biphasic manner mirroring acute inflammation and inflammatory resolution: selective inhibition of CYP-AA and CYP-LA products by epoxI. (A and B) Mean fold change in (A) CYP-AA products: DHETs and (B) CYP-LA (DiHOME), CYP-DHA (DiHDPA), and CYP-EPA (DHEQ) products, 0–48 h. Each mediator was normalized to its paired levels found in the naive cavity in each experiment; the levels found in the naive cavity were given an arbitrary value of 1. *P < 0.05 by one-sample t test from 0 h. †P < 0.05 by unpaired t test between the values observed at 24 and 48 h. (C) Mean fold-change in epoxygenase product formation in the presence or absence of epoxI (30 mg/kg). EpoxI was given at 24 and 36 h. Epoxy-oxylipins were measured at 48 h and are represented as fold change from the respective mean levels observed in the naive cavity at 0 h. *P < 0.05 by unpaired t test. Data represents mean ± SEM.
We screened the mouse Cyp2 family to investigate which enzymes were present in the resolution-phase inflammatory cell populations, and found Cyp2c44, Cyp2j5, Cyp2j6, Cyp2j9, and Cyp2j13 epoxygenases along with Cyp2u1 (HETE and DHA metabolism) and Cyp2s1 (xenobiotic and retinoic acid metabolism) (Table 1 and SI Appendix, Fig. S2).
Table 1.
Expression of mouse Cyp2s in the resolving peritoneal cavity of mice
| Cyp | Liver | Exudate |
| Cyp2j5 | + | +/− |
| Cyp2j6 | + | ++ |
| Cyp2j9 | +/− | + |
| Cyp2j13 | Not tested | +/− |
| Cyp2c29 | + | − |
| Cyp2c37 | ND | ND |
| Cyp2c38 | + | − |
| Cyp2c39 | − | − |
| Cyp2c40 | ND | ND |
| Cyp2c44 | +++ | ++ |
| Cyp2c50 | ++ | − |
| Cyp2c54 | ++ | − |
| Cyp2c55 | + | − |
| Cyp2a1 | +++ | − |
| Cyp2u1 | ND | + |
| Cyp2s1 | ND | + |
Mice were treated with zymosan (1 mg, i.p.) for 24 h. The presence of mouse CYP2 mRNAs was determined by RT-PCR, using mouse liver as a positive control. The data summarize the findings from n = 6–8 mice. The number of + signs indicates the relative expression level; +/− indicates that not all samples tested showed positive expression. ND, not detected.
Epoxygenase Inhibition During Resolution Induces a Profound Accumulation of Ly6chi Monocytes.
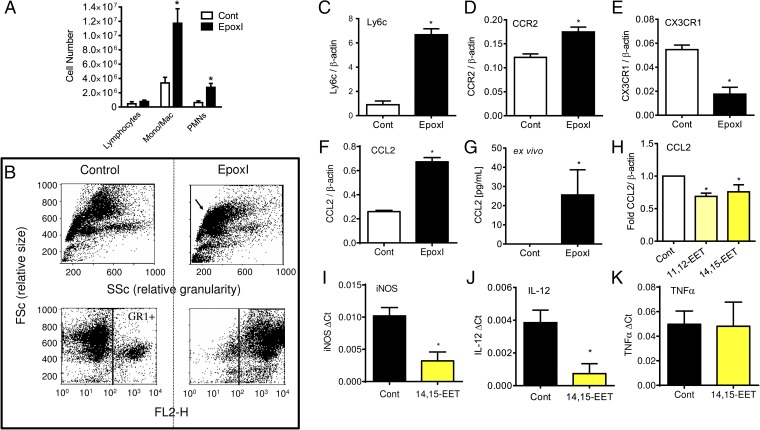
Epoxygenase inhibition was accompanied by a threefold increase in monocyte numbers during the 24- to 48-h period of inhibition and a small but significant increase in PMNs (Fig. 3A). These epoxI-recruited monocytes were strongly GR1+ (Ly6c and Ly6g antigens; Fig. 3B), and Ly6chi expression was confirmed by RT-PCR (Fig. 3C). Moreover, these recruited cells were CCR2hi (Fig. 3D), CX3CR1lo (Fig. 3E), and CCL2hi (Fig. 3F) at the mRNA level. CCL2 peptide was found to be highly elevated in the peritoneal cavity of epoxI-treated mice (vehicle 8 ± 8 pg/mL; epoxI 266 ± 190 pg/mL CCL2; n = 3). Substantial levels of CCL2 were produced exclusively from cells elicited from epoxI-treated mice ex vivo (Fig. 3G), suggesting they could be a major source of this elevated CCL2. EETs are too short-lived to be given as a bolus dose in vivo, and therefore to test whether EETs were antiinflammatory in mouse monocytes, we added back authentic 14,15-EET (1 μM; 7 h) to zymosan-elicited cells ex vivo. 14,15-EET reduced Ccl2 (Fig. 3H), inducible nitric oxide synthase (iNOS) (Fig. 3I), and IL-12 (Fig. 3J) mRNA expression in these cells ex vivo. Addition of 14,15-EET was not universally antiinflammatory in these elicited cells, as TNFα mRNA expression remained unaltered (Fig. 3H). These results are consistent with our previous findings, which reported an antiinflammatory role of epoxygenases in classically activated human monocytes and macrophages in vitro (21).
Fig. 3.
Epoxygenase inhibition during resolution causes Ly6chi monocyte recruitment and Ccl2 generation. (A) Change in lymphocytes, monocytes/macrophages, and PMNs in the peritoneal cavity of mice 48 h postzymosan treatment. At 24 and 36 h, mice were given either sterile PBS or epoxI (30 mg/kg, i.p.). Total cell numbers were counted on a hemocytometer and the proportion of each cell type was determined by FACS. Data represent the mean ± SEM from n = 6 mice per group; *P < 0.05 by unpaired t test between vehicle (Cont; PBS) and epoxI-treated mice. (B) Example of FACS analysis of total cell (Upper) and GR+ populations (Lower) of cells elicited by zymosan alone (control; Left) or in the presence of epoxI (Right) in terms of size (FSc) and granularity (SSc). (C–F) Relative expression of Ly6c (C), Ccr2 (D), CX3CR1 (E), and CCL2 (F) mRNA compared with β-actin in elicited cells from zymosan + control or epoxI-treated mice taken at 48 h. (G) CCL2 generation in cells from zymosan + control or epoxI-treated mice. Cells were elicited at 36 h and left for a further 8 h ex vivo and CCL2 was measured by ELISA. (H–K) CCL2 (H), iNOS (I), IL-12 (J), and TNFα (K) mRNA expression in zymosan-elicited cells at 36 h treated with 14,15-EET or 11,12-EET. Cells were elicited at 36 h and treated for a further 7 h with EETs. Data represent the mean ± SEM from n = 3–4 mice per group; *P < 0.05 by unpaired t test between vehicle (Cont; PBS) and epoxI-treated mice.
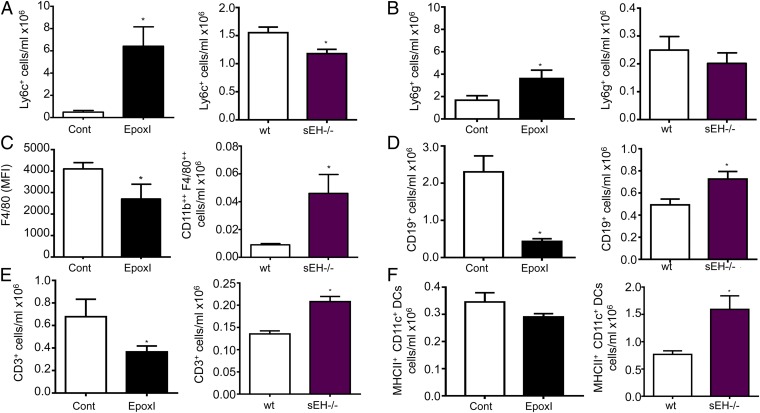
sEH−/− Mice Have an Opposing Phenotype to Epoxygenase-Inhibited Mice During Resolution.
To confirm the role of epoxy-oxylipins during resolution, we compared the findings of epoxI-treated mice with sEH−/− mice. sEH is considered the main pathway for epoxy-oxylipin metabolism and inactivation. Compared with epoxI-treated mice, which have reduced epoxy-oxylipins, sEH−/− mice have elevated oxylipins (31). In zymosan-treated mice, Ly6chi monocytes peaked at 48 h with epoxI treatment (SI Appendix, Fig. S3), whereas at 48 h in sEH−/− mice, Ly6chi monocytes were reduced compared with wild-type controls (Fig. 4A). There was a small increase in Ly6g+ cells at 48 h with epoxI, whereas in sEH−/− mice, Ly6g+ monocytes were slightly reduced compared with wild-type controls (Fig. 4B). With epoxI treatment, although there was no change in numbers of macrophage (F4/80)-positive cells (5.4 ± 0.5 compared with 5.9 ± 2.2 cells per milliliter, ×106), there was a reduction in the levels of F4/80 expression on the epoxI-elicited macrophages (Fig. 4C). In corollary, the opposite was found in sEH−/− mice: CD11b++ F480++ cells were greatly elevated compared with wild-type controls (Fig. 4C). CD19+ B cells (Fig. 4D) and CD3+ T cells (Fig. 4E) were reduced by epoxI and elevated in sEH−/− mice. There was also a similar trend to reduction of MHCII+ CD11c+ dendritic cells by epoxI, with a significant elevation of these MHCII+ CD11c+ dendritic cells in sEH−/− mice. Therefore, in all of the major indices of inflammatory cell accumulation tested, epoxy-oxylipin inhibition with epoxI showed opposing actions to epoxy-oxylipin elevation with the use of global sEH−/−.
Fig. 4.
Endogenous oxylipins regulate Ly6chi monocyte, F4/80 macrophage, and CD19 and CD3 cell populations during resolution. Changes in (A) Ly6chi cell populations, (B) Ly6g+ cell populations, (C) F4/80 expression (mean fluorescence intensity; MFI) (Left) and F4/80++ cell populations (Right), and (D) CD19+, (E) CD3+, and (F) MHCII+ CD11c+ dendritic cell populations in control and epoxI-treated mice (solid black bars; Left) and in wild-type (wt) and sEH−/− mice (Right). The proportion of cells in each group was determined by FACS and related back to cell numbers. Data represent the mean ± SEM from n = 4–5 mice per experimental group; *P < 0.05 by two-way ANOVA or unpaired t test.
Cells of the Monocyte Lineage Have a Disrupted Resolution Phenotype Following Epoxygenase Inhibition.
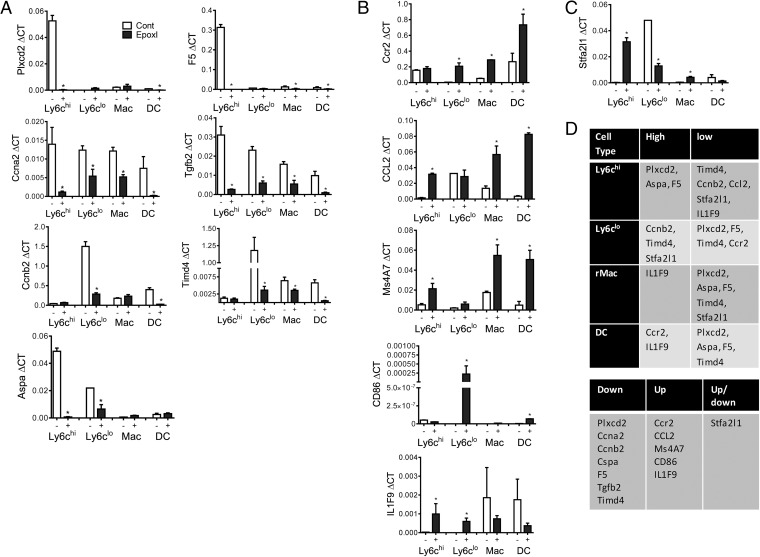
Using a recently identified novel panel of quantitative (q)RT-PCR of resolution monocyte markers (32), we examined the total inflammatory cell population for a changes in the resolution phenotype. The total epoxI-elicited cell population was Timd4lo, Tgfb2lo, and Plxdc2lo and IL1f9hi, CD86hi, and Ms4a7hi compared with cells from animals treated with vehicle/zymosan alone (SI Appendix, Fig. S4). At indicated time points there was also a significant difference in Ccnb2 (increased with epoxI) and Aspa and Stfa2l1 (decreased with epoxI) (SI Appendix, Fig. S4). Because epoxy-oxylipins appear to have the most dramatic effect on monocyte lineage cell recruitment, we examined these targets in FACS-sorted Ly6chi and Ly6clo monocyte populations, resident macrophages, and recruited dendritic cells (DCs) (Fig. 5). Normal-resolution Ly6chi monocytes were Plxcd2hi, Aspahi, and F5hi; Ly6clo monocytes were Ccnb2hi, Timd4hi, and Stfa2l1hi; resident macrophages were IL1F9hi; and dendritic cells were Ccr2hi and IL1F9hi (Fig. 5). Treatment with epoxI caused a significant down-regulation of Plxcd2 in Ly6chi monocytes and DCs; Ccna2 in Ly6chi and Ly6clo monocytes, resident macrophages, and DCs; Ccnb2 in Ly6clo monocytes and DCs; Aspa in Ly6chi and Ly6clo monocytes; F5 in Ly6chi, resident macrophages, and DCs; Tgfb2 in Ly6chi and Ly6clo monocytes, resident macrophages, and DCs; and Timd4 in Ly6clo monocytes, resident macrophages, and DCs (Fig. 5A). Treatment with epoxI caused an up-regulation of Ccr2 in Ly6clo monocytes, resident macrophages, and DCs; Ccl2 in Ly6chi monocytes, resident macrophages, and DCs; Ms4A7 in Ly6chi monocytes, resident macrophages, and DCs; CD86 in Ly6clo monocytes and DCs; and IL1F9 in Ly6chi and Ly6clo monocytes (Fig. 5B). However, Stfa2l1 was up-regulated in Ly6chi and resident macrophages but inhibited in Ly6clo monocytes. These findings collectively point toward a more inflammatory phenotype accompanying epoxygenase inhibition, with a clear induction in either or both Ccl2 and Ccr2 in all of the cell types and a decrease in resolution monocyte markers of apoptosis and repair (Timd4 and Tgfb2, respectively).
Fig. 5.
Epoxygenase inhibition alters the resolution phenotype of recruited cells of the monocyte lineage. Inflammation was initiated by zymosan (1 mg, i.p.), and mice were treated with vehicle control (PBS) or epoxI (30 mg/kg, i.p.) at 24 and 36 h. Cells were collected and pooled from n = 10 mice and Ly6chi, Ly6clo monocytes, resident macrophages, and recruited dendritic cells were sorted on a FACSAria as detailed in Materials and Methods. A qRT-PCR resolution monocyte panel (n = 3–6) was then used to examine the phenotype of each cell type. (A–C) In the presence of epoxI, (A) Plxcd2, Ccna2, Ccnb2, Aspa, F5, Tgfb2, and Timd4 were found to be down-regulated in the cell types; (B) Ccr2, CCL2, Ms4A7, CD86, and IL1F9 were up-regulated in the cell types; and (C) Stfa2l1 was up- and down-regulated in a cell type-specific manner. *P < 0.05 by unpaired t test. (D, Upper) Summary of the relative basal levels of each transcript in each cell type. (D, Lower) Summary of the effect of epoxI on the different transcripts. Data represents mean ± SEM.
Epoxygenase Inhibition Disrupts Resolution-Phase Macrophage Differentiation and Leads to Impaired Phagocytosis and Efferocytosis.
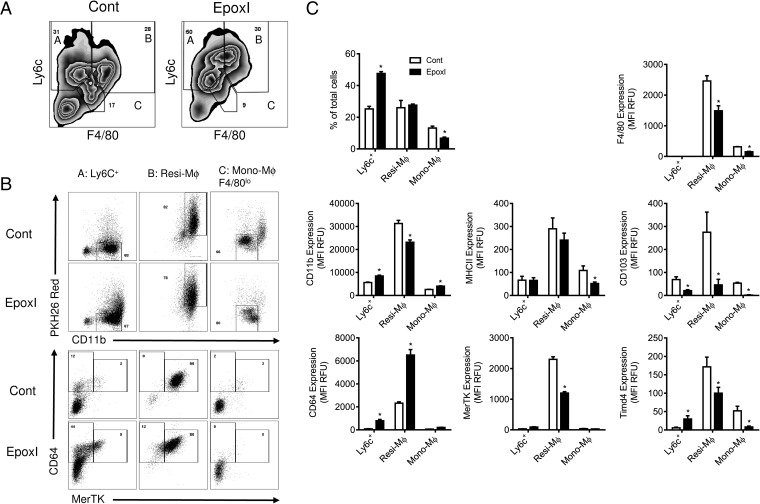
To examine functional changes in the monocyte lineage, we examined Ly6c+ monocytes, resident macrophages, and monocyte-derived macrophage populations. As previously shown, Ly6c+ monocytes were elevated, monocyte-derived macrophages were reduced, and resident macrophage levels remained unchanged (Fig. 6). Resident macrophages on a cell-per-cell level showed reduced expression of differentiation markers and phagocytosis receptors F4/80, CD11b, MerTk, Timd4, and CD103 but an increase in CD64 (Fig. 6C). A similar reduction in F4/80, MerTK, Timd4, and CD103 was also observed in monocyte-derived macrophages (Fig. 6C). In contrast, Ly6c+ monocytes demonstrated a small but significant increase in markers of differentiation CD11b, Timd4, and CD64, which was again accompanied by a decrease in CD103 (Fig. 6C).
Fig. 6.
Epoxygenase inhibition regulates monocyte and macrophage differentiation. Inflammation was initiated by zymosan (1 mg, i.p.), and mice were treated with vehicle control (PBS) or epoxI (30 mg/kg, i.p.) at 24 and 36 h. Cells were collected at 48 h and pooled from n = 9–18 mice. Ly6c+ monocytes, resident macrophages, and monocyte-derived macrophages were sorted on a LSR Fortessa as detailed in Materials and Methods. (A) Representative zebra plots of the cell populations from control and epoxI-treated mice expressing Ly6c and F4/80, with labeled populations representing A: Ly6c+ monocytes; B: resident macrophages (Resi-Mϕ); and C: monocyte-derived macrophages (Mono-Mϕ). (B) Representative dot plots of the Ly6c+ monocytes, resident macrophages, and monocyte-derived macrophages from control and epoxI-treated mice expressing PKH26 and CD11b (Upper) and CD64 and MerTK (Lower). (C) Changes in cell numbers, and expression (MFI in relative fluorescence units; RFUs) for F4/80, CD11b, MHCII, CD103, CD64, MerTK, and Timd4 in the Ly6c+, Resi-Mϕ, and Mono-Mϕ cell populations. Data are mean ± SEM from n = 5 mice per group; *P < 0.05 by unpaired t test.
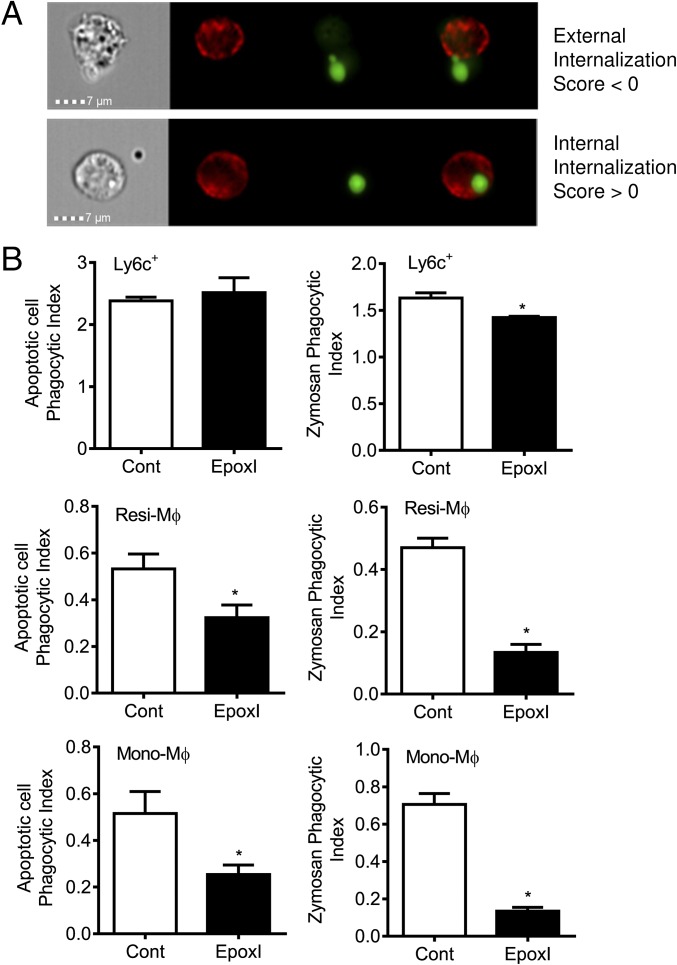
Using the ImageStream system, we were easily able to identify phagocytosis from particles and bodies sticking to cells (Fig. 7A). Ly6c+ monocytes had a small but significant decrease in the ability to phagocytose zymosan, but not apoptotic cells (Fig. 7B). In contrast, resident macrophages and monocyte-derived macrophages (Fig. 7B) both showed greatly reduced ability to phagocytose FITC-labeled zymosan A and efferocytose apoptotic cells ex vivo.
Fig. 7.
Epoxygenase inhibition reduces the phagocytic activity of monocyte-derived and resident macrophages. Inflammation was initiated by zymosan (1 mg, i.p.), and mice were treated with vehicle control (PBS) or epoxI (30 mg/kg, i.p.) at 24 and 36 h. Cells were collected at 48 h and pooled from n = 9–18 mice. Ly6c+ monocytes, resident macrophages, and monocyte-derived macrophages were sorted on a FACSAria as detailed in Materials and Methods. Sorted cells were then tested for their ability to phagocytose CFSE-labeled apoptotic cells (thymocytes) or FITC-labeled zymosan BioParticles over 30 min. Cells were gated using F4/80 or Ly6c on an ImageStreamX Mark II. (A) ImageStream analysis differentiates cells that phagocytose apoptotic cells or zymosan (internalization score >0) from those where particles or bodies just stick to the cell (internalization score 0). (B) Ex vivo phagocytosis of apoptotic cells (Left) and zymosan (Right) by Ly6c+, Resi-Mϕ, and Mono-Mϕ cell populations from control and epoxI-treated mice. Data are mean ± SEM from n = 3–6 pooled samples; *P < 0.05 by unpaired t test.
Discussion
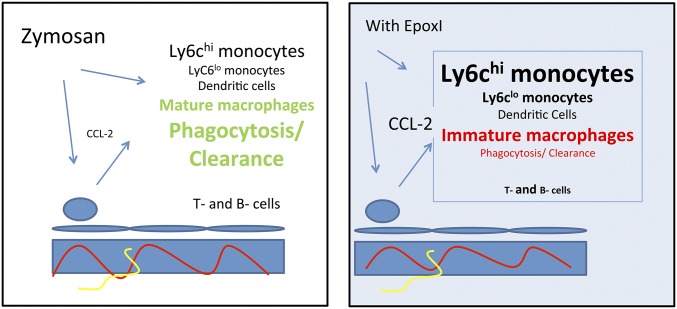
Our findings show that epoxy-oxylipin–generating pathways constitute an important control point limiting the accumulation of proinflammatory Ly6chi monocytes and the proinflammatory and clearance activity of cells of the monocyte lineage during inflammatory resolution (Fig. 8). Similar to cyclooxygenases and lipoxygenases, the epoxygenase pathways of arachidonic acid metabolism (SI Appendix, Fig. S5) are activated during both acute inflammation and resolution. There has been great recent interest in the therapeutic potential of sEH inhibitors as novel antiinflammatories. sEH inhibitors or genetic disruption of sEH in mice reduces inflammation in models of endotoxin-induced pulmonary inflammation (22), ischemia-reperfusion injury (33, 34), subarachnoid hemorrhage (35), and the murine ovalbumin model of asthma (36), and in more chronic models including atherogenic diet-induced fatty liver disease and adipose tissue (37) and atherosclerosis (38, 39). In contrast, the expression and roles of epoxy-oxylipins during inflammatory resolution have not been investigated.
Fig. 8.
Role of epoxy-oxylipins in the resolution phenotype. The resolution of zymosan-initiated inflammation involves monocytes, dendritic cells, and T- and B-cell recruitment and the differentiation of monocytes into resolution-type macrophages. In this process (as revealed by epoxI treatment and sEH−/− mice), epoxy-oxylipins, most likely EETs, limit Ccl2 and Ccr2 expression, Ly6chi monocyte accumulation, and T- and B-cell recruitment and encourage the formation of mature phagocytic resolution macrophages. Ly6chi monocytes, Ly6clo monocytes, dendritic cells, and monocyte-derived and resident macrophages are all activated in the presence of epoxI.
Human monocytes in vitro express CYP2J and CYP2C enzymes, and in particular CYP2J2 can be induced by TLR4/LPS activation (21). Rodents have expanded CYP2J and CYP2C subfamilies compared with humans, and we found the epoxygenase CYP2C44 (40) along with CYP2J6 and CYP2J9 to be present in all inflammatory exudates tested, although these findings do not rule out contributions from other local stromal or vascular cells. To test the role of epoxygenases, we used the epoxI SKF525A, which we routinely use in our in vitro assays (18–21, 41) and found to have an IC50 for human CYP2J2 of 1 μM (20). SKF525A is routinely used in vivo within the range of 5–50 mg/kg (42–45) and, although reported as a selective epoxygenase inhibitor, SKF525A had not been examined in the in vivo setting followed by lipidomic analysis to confirm this. We chose SKF525A as our epoxI because it is soluble in aqueous solutions (in our case PBS), and so could be tolerated far better over longer periods with limited vehicle effect. In our experiments only epoxygenase products and not prostanoids or lipoxygenase products were inhibited (by ∼50%) with 30 mg/kg of epoxI (SI Appendix, Table S1). epoxI was given at 24 h to look at resolution specifically, so the true levels of inhibition may be higher, as the kinetics of preformed metabolite removal in the cavity over the 24- to 48-h time period is not known.
The combination of these experiments using epoxI and sEH−/− mice leads to the conclusion that epoxy-oxylipins are produced during resolution by resident and incoming inflammatory cells and act in a paracrine/autocrine manner to limit Ly6chi monocyte accumulation and monocyte macrophage proinflammatory phenotype. Following epoxI treatment at the onset of resolution, the elicited cells were predominantly GR1+, Ly6chi, and CX3CR1lo and produced large amounts of the major Ly6chi chemokine CCL2. CCL2 was undetectable from cells elicited by zymosan alone. Interestingly, although limiting Ly6chi monocyte recruitment, epoxy-oxylipins promoted the differentiation of monocytes into F4/80++ macrophages. In vitro, we previously found that epoxygenases and 11,12-EET promote macrophage bacterial phagocytosis, which was associated with a reduction in CD11b (41). Similarly, here resident and monocyte-derived macrophages had reduced CD11b, along with markers of the mature macrophage phenotype and phagocytosis MerTK and Timd4, the latter being particularly important for efferocytosis (46). These markers correlated with both resident and monocyte-derived macrophages from epoxI-treated mice to have a dramatic reduction in the ability to phagocytose fresh zymosan and efferocytose apoptotic cells. LyC6+ monocytes had small but significantly elevated CD11b and CD64 expression but reduced CD103. There was, however, little change in the ability of monocytes to phagocytose material apart from a small reduction in the ability to phagocytose zymosan ex vivo. These findings suggest epoxy-oxylipins not only limit the recruitment of proinflammatory monocytes but also limit resident and monocyte-derived macrophages’ ability to clear inflammatory stimuli and apoptotic cells, further indicating a prominent role for epoxygenase pathways in promoting the resolution phenotype. Interestingly, CD19+ B cells and CD3+ T cells were also increased in sEH−/− mice (and reduced with epoxI treatment). Lymphocytes are dispensable for resolution in this model (47) but form an important part of a defense mechanism against reinfection (47). Although we have yet to investigate fully which lymphocyte subtypes are present, these findings nonetheless also provide the first evidence, to our knowledge, for a role for epoxy-oxylipins in acquired immune responses along with the roles in resolution reported here.
These findings all point to a proresolution role for epoxy-oxylipins in the proinflammatory monocyte lineage. CCL2 is known to be a central mediator of monocyte recruitment. EpoxI-treated animals produced large amounts of CCL2, as did the inflammatory cell population taken ex vivo. When we examined individual cell types, CCL2 was induced by epoxI in Ly6chi monocytes, resident macrophages, and DCs, whereas its receptor CCR2 (already expressed in Ly6chi monocytes) was also induced in Ly6clo monocytes, resident macrophages, and DCs. Using gene array analysis, we previously identified a panel of novel resolution monocyte markers (32) including the T-cell costimulator CD86, IL-1 family gene 9 (IL-1f9; IL-36), and CD20-like family member Ms4a7; these were all elevated after epoxI treatment, whereas the phosphatidylserine apoptotic cell receptors Timd4, Tgfb2, and Plxdc2 were all decreased. The roles of a number of these novel markers are still not known in resolution, but clearly epoxI-mediated changes in expression along with changes to functional clear antigens and apoptotic cells represent cells of a phenotype of highly dysregulated resolution compared with the typical resolution monocyte cell population.
Recently, the major rat monocyte epoxygenase Cyp2j4 was knocked out using a zinc finger nuclease-mediated gene-targeting approach (48). The resultant bone marrow-derived macrophages showed a highly profibrotic enriched transcriptome and activity and an induction of PPARγ (48). Although not an identical phenotype to those observed in our study, this adds to the literature showing that lipid-metabolizing CYPs have a critical role to play in the inflammatory phenotype of cells in the monocyte lineage.
EETs are too unstable to give as a bolus dose in vivo. Therefore, we tested the direct antiinflammatory effects of monocyte epoxygenases ex vivo. 14,15-EET, the most abundant epoxygenase-derived EET produced in the peritoneal cavity, inhibited inflammatory cell activation of iNOS and IL-12 ex vivo from zymosan-treated mice. Interestingly, the linoleic acid epoxygenase products 9,10-EpOME and 12,13-EpOME are highly expressed in the naive cavity. The levels of 9,10-EpOME drop rapidly during acute inflammation, whereas 12,13-EPOME changes very little. Our results therefore indicate a change in substrate availability or utilization by epoxygenases from linoleic acid in homeostasis to arachidonic acid during inflammation. Linoleic acid epoxygenase products are generally considered proinflammatory and cytotoxic (49–51), whereas EETs are antiinflammatory. How this concept fits with the high levels of EpOMEs and DiHOMEs in the naive peritoneal cavity is intriguing, and points to novel as yet undiscovered homeostatic roles for these mediators in the peritoneal cavity. In contrast to EETs, which inhibited Ccl2 in monocytes ex vivo, 9,10-EpOME induced CCL2 (SI Appendix, Fig. S6), suggesting that substrate utilization may be an additional layer of control over the effects of the epoxygenase pathway. High EpOME levels have been found in acute respiratory distress syndrome and in patients with extensive burns (52); however, the roles of EpOMEs in inflammation and homeostasis remain relatively poorly understood, although it is becoming clearer that many of the cytotoxic effects attributed to them are in fact due to their sEH metabolite DiHOMEs (51, 53–55). Clearly more work is needed to elucidate the function of these linoleic acid epoxygenase products in homeostasis as well as pathophysiology. At this stage, we cannot rule out potential effects of DHA and EPA CYP metabolites but, because we saw no significant change in their production with epoxI, we have here focused on arachidonic acid and linoleic acid epoxy-oxylipins.
The levels of 12/15-lipoxygenase, 9,10- and 12,13-HODE, and 12- and 15-HETE were high in the peritoneal cavity, consistent with the report that high levels of resident macrophages contain 12/15-lipoxygenase in mice, where it regulates immune function. Our findings are also consistent with this report in that 12/15-lipoxygenase products drop during acute inflammation (in our experiments only 9-HODE returned to basal levels by 48 h), and support the findings that 12/15-lipoxygenase has an important homeostatic immunoregulatory role in this model (56).
In conclusion, we have characterized the epoxygenase pathways during a model of inflammatory resolution. Inhibition of epoxygenases during this resolution phase established a proinflammatory environment that allows the recruitment of proinflammatory Ly6c monocytes and macrophages with a highly dysregulated resolution/clearance phenotype, whereas mice where epoxygenase products are elevated have an enhanced resolution phenotype. An efficient and active epoxygenase pathway therefore joins similar proresolution lipid pathways such as the resolvins in the requirement for effective inflammatory resolution. Drugs that elevate epoxygenase products or epoxygenase product mimetics therefore represent novel resolution targets for chronic inflammatory disorders.
Materials and Methods
Materials.
EETs were purchased from Cayman Chemical (Cambridge Bioscience). SKF525A was purchased from Biomol (Affiniti Research Products). Cytokines and the MCP-1 ELISA were obtained from R&D Systems. The TNFα ELISA was from eBioscience. Unless otherwise stated, all other reagents were purchased from Sigma-Aldrich.
Animal Models.
All experiments were completed with adult male mice on a pure C57BL/6 background. Control C57BL6/J mice were bred under standard conditions and maintained in a 12-h light/dark cycle at 22 °C and given food and tap water ad libitum in accordance with UK Home Office regulations. Mice with targeted disruption of sEH were rederived and backcrossed onto a C57BL/6 genetic background for more than 10 generations, as previously described (31, 52, 57). sEH−/− mice have significantly higher circulating epoxide:diol ratios compared with wild-type littermates, consistent with functional sEH disruption (57, 58). Peritonitis was induced by intraperitoneal injection of 1 mg type A zymosan in 0.5 mL PBS after 30 s of sonication on ice. Inflammatory cells were retrieved at the time points described in Results by injecting 2 mL sterile cell dissociation buffer (Life Technologies). Cells were counted by hemocytometer, and exudates were stored at −80 °C until further analysis. In some experiments, the epoxI SKF525A (30 mg/kg) or sterile PBS (0.5 mL vehicle) was given intraperitoneally 24 h after zymosan injection and at 12-h time points thereafter up until 96 h. These studies received institutional review board approval for the use of mice from the UK Home Office.
Lipid Analysis.
Eicosanoids and other fatty acid metabolites were extracted from inflammatory exudates by solid-phase extraction and eluted in ethyl acetate, essentially as described (22); see SI Appendix, Supplemental Methods for extended methods. Lipids were separated by reverse-phase HPLC on 2-μm, 150-mm, 5-μm Luna C18 columns (Phenomenex) and quantified using an MDS Sciex API 3000 triple quadrupole mass spectrometer (Applied Biosystems) with negative-mode electrospray ionization and multiple-reaction monitoring, as described (12). The relative response ratios of each analyte were used to calculate concentrations while correcting for surrogate losses via quantification relative to internal standards. The sensitivity of analytes ranged from 0.25 to 25 pg per sample.
FACS Analysis and Cell Sorting.
Flow cytometry and cell sorting were done on LSRII/LSRFortessa and FACSAria (BD Biosciences), respectively. Cells were incubated with Fc-blocker (AbD Serotec) and fluorescently labeled antibodies. Data were analyzed with FlowJo 7.0.1 software (Tree Star) using fluorescence minus one controls as the reference for setting gates. Antibodies were obtained from BD Biosciences unless stated [F4/80 (eBioscience), CD11b, CD11c, Ly6c, Ly6g, Gr1, CD3 (BioLegend), CD19, CD4, CD8, CD62l, CD44, MerTK, CD64, CD103, Timd4, and MHCII (eBioscience)] (59). To identify resident macrophages (59), PKH26-PCLred (0.35 mL, 500 nM; Sigma) was injected intraperitoneally 2 h before injection of zymosan. In cell-sorting experiments, monocytes and macrophages were sorted from a population of CD19− and CD3− cells as either Ly6c+F4/80+ or Ly6c−F4/80+; see SI Appendix, Fig. S7 for the gating strategy. For identification of Ly6g+ neutrophils and Ly6c+ monocytes, a combination of Gr1 and anti-Ly6c or anti-Ly6g was also used. Resident macrophages were characterized as PKHred++, DCs were characterized as PKHred− MHCII+ and CD11c+, Ly6chi monocytes were characterized as LyC6hi PKHred+ MHCII−, and Ly6clo monocytes were characterized as Ly6clo PKHred−, as previously described (59).
Cell Culture ex Vivo.
The peritoneal lavage was treated with ACK lysis buffer (Thermo Fisher Scientific) to remove erythrocytes. After being washed, peritoneal cells were suspended in DMEM supplemented with 10% (vol/vol) FBS and 50 μg/mL penicillin/streptomycin. The cells (2 × 106) were seeded on a 12-well plate and left to adhere for 45 min in a humidified CO2 incubator. Nonadherent cells were removed by three washes with DMEM. Remaining adherent cells (∼1 × 106) were incubated in 0.5 mL DMEM in the presence or absence of 1 μM 14,15-EET or 11,12-EET or vehicle (0.3% EtOH). After 6 h, cell-free supernatants were removed and cells were lysed using TRIzol (Invitrogen) for subsequent RNA extraction.
RT-PCR.
Cells analyzed by qRT-PCR were lysed and RNA was isolated using TRIzol. The panel of resolution markers Timd4 (T-cell Ig and mucin domain-containing 4), Tgfb2, Plxdc2 (plexin domain-containing protein 2), IL1f9 (interleukin 1 family member 9), CD86, Ms4a7 (membrane-spanning four domains, subfamily A, member 7), Ccna2 (cyclin A2), Ccnb2 (cyclin B2), F5 (coagulation factor V), Aspa (aspartoacylase), and Stfa2l1 (stefin A2-like 1) were measured by qRT-PCR as previously described (32). Resolution monocytes were previously found to be enriched for cell-cycle/proliferation genes as well as for Timd4 and Tgfb2, key systems in the termination of leukocyte trafficking and clearance of inflammatory cells. Ly6c, CX3CR1, Ccl2, Ccr2, Cyp2j5, Cyp2j6, Cyp2j9, Cyp2j13, Cyp2c29, Cyp2c38, Cyp2c39, Cyp2c44, Cyp2c50, Cyp2c54, Cyp2c55, Cyp2a1, Cyp2u1, Cyp2s1, and β-actin were measured by RT-PCR. Primers are detailed in SI Appendix, Supplemental Methods.
Efferocytosis and Phagocytosis Assays.
Apoptotic cells (thymocytes) were produced from the thymuses of three naive control C57/BL mice killed 24 h presort. Harvested thymuses were passed through a 70-μm mesh and then lysed with ACK buffer for 3 min. Cells were washed twice with RPMI 1640 with 100 U/mL penicillin/streptomycin. To induce apoptosis, thymocytes were resuspended in media at 2 × 106 cells per mL and exposed to UV radiation (∼300 nm) for 20 min followed by incubation for 16–24 h at 37 °C with 5% CO2 using a Syngene UV transilluminator. Cells were then washed with PBS and labeled with 2 μM carboxyfluorescein succinimidyl ester (CFSE) according to the manufacturer’s protocol (Life Technologies; CellTrace CFSE).
Sorted cell populations, 2 × 105 cells per well, were plated in 24-well plates and resuspended in X-VIVO 15 media (Lonza) containing 10% FBS, 2 mM l-glutamine, and 1× pen-step. Cells were then challenged with apoptotic thymocytes (5:1 monocyte/macrophage) or FITC-labeled zymosan A BioParticles (Thermo Fisher Scientific) (10 particles:1 monocyte/macrophage) for 30 min at 37 °C and 5% CO2. Phagocytosis was stopped by placing the plates on ice. Cells were detached using 10 mM EDTA containing 4 mg/mL lignocaine for 20 min. Cells were stained for either F4/80 or Ly6c (allophycocyanin) for 20 min (see FACS Analysis and Cell Sorting, above) before being washed twice with PBS containing 2 mM EDTA. Cells were then fixed using 4% paraformaldehyde in PBS and analyzed on an ImageStreamX Mark II (Merck Millipore) (SI Appendix, Fig. S8).
Supplementary Material
Acknowledgments
We thank the Mass Spectrometry Core at the NIEHS for their technical assistance and Dr. Mark Paul Clark, Imperial College London, for his help with data analysis. This work was funded by grants from the British Heart Foundation (PG/08/070/25464, PG/11/39/28890) and the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences, Grant Z01 ES025034 (to D.C.Z.). D.W.G. holds a Wellcome Trust Senior Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521453113/-/DCSupplemental.
References
- 1.Mantovani A, Sica A, Locati M. Macrophage polarization comes of age. Immunity. 2005;23(4):344–346. doi: 10.1016/j.immuni.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Bishop-Bailey D, Wray J. Peroxisome proliferator-activated receptors: A critical review on endogenous pathways for ligand generation. Prostaglandins Other Lipid Mediat. 2003;71(1–2):1–22. doi: 10.1016/s0090-6980(03)00003-0. [DOI] [PubMed] [Google Scholar]
- 3.Zeldin DC. Epoxygenase pathways of arachidonic acid metabolism. J Biol Chem. 2001;276(39):36059–36062. doi: 10.1074/jbc.R100030200. [DOI] [PubMed] [Google Scholar]
- 4.Capdevila JH, Falck JR, Harris RC. Cytochrome P450 and arachidonic acid bioactivation. Molecular and functional properties of the arachidonate monooxygenase. J Lipid Res. 2000;41(2):163–181. [PubMed] [Google Scholar]
- 5.Konkel A, Schunck WH. Role of cytochrome P450 enzymes in the bioactivation of polyunsaturated fatty acids. Biochim Biophys Acta. 2011;1814(1):210–222. doi: 10.1016/j.bbapap.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Campbell WB, Fleming I. Epoxyeicosatrienoic acids and endothelium-dependent responses. Pflugers Arch. 2010;459(6):881–895. doi: 10.1007/s00424-010-0804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spiecker M, Liao JK. Vascular protective effects of cytochrome p450 epoxygenase-derived eicosanoids. Arch Biochem Biophys. 2005;433(2):413–420. doi: 10.1016/j.abb.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Zeldin DC, et al. Regio- and enantiofacial selectivity of epoxyeicosatrienoic acid hydration by cytosolic epoxide hydrolase. J Biol Chem. 1993;268(9):6402–6407. [PubMed] [Google Scholar]
- 9.Zeldin DC, et al. Metabolism of epoxyeicosatrienoic acids by cytosolic epoxide hydrolase: Substrate structural determinants of asymmetric catalysis. Arch Biochem Biophys. 1995;316(1):443–451. doi: 10.1006/abbi.1995.1059. [DOI] [PubMed] [Google Scholar]
- 10.Revermann M, et al. Soluble epoxide hydrolase deficiency attenuates neointima formation in the femoral cuff model of hyperlipidemic mice. Arterioscler Thromb Vasc Biol. 2010;30(5):909–914. doi: 10.1161/ATVBAHA.110.204099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang LN, et al. Inhibition of soluble epoxide hydrolase attenuated atherosclerosis, abdominal aortic aneurysm formation, and dyslipidemia. Arterioscler Thromb Vasc Biol. 2009;29(9):1265–1270. doi: 10.1161/ATVBAHA.109.186064. [DOI] [PubMed] [Google Scholar]
- 12.Lee CR, et al. Endothelial expression of human cytochrome P450 epoxygenases lowers blood pressure and attenuates hypertension-induced renal injury in mice. FASEB J. 2010;24(10):3770–3781. doi: 10.1096/fj.10-160119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo P, et al. Inhibition or deletion of soluble epoxide hydrolase prevents hyperglycemia, promotes insulin secretion, and reduces islet apoptosis. J Pharmacol Exp Ther. 2010;334(2):430–438. doi: 10.1124/jpet.110.167544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serhan CN, Petasis NA. Resolvins and protectins in inflammation resolution. Chem Rev. 2011;111(10):5922–5943. doi: 10.1021/cr100396c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serhan CN. The resolution of inflammation: The devil in the flask and in the details. FASEB J. 2011;25(5):1441–1448. doi: 10.1096/fj.11-0502ufm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stables MJ, Gilroy DW. Old and new generation lipid mediators in acute inflammation and resolution. Prog Lipid Res. 2011;50(1):35–51. doi: 10.1016/j.plipres.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Bishop-Bailey D, Thomson S, Askari A, Faulkner A, Wheeler-Jones C. Lipid-metabolizing CYPs in the regulation and dysregulation of metabolism. Annu Rev Nutr. 2014;34:261–279. doi: 10.1146/annurev-nutr-071813-105747. [DOI] [PubMed] [Google Scholar]
- 18.Thomson S, et al. Intimal smooth muscle cells are a source but not a sensor of anti-inflammatory CYP450 derived oxylipins. Biochem Biophys Res Commun. 2015;463(4):774–780. doi: 10.1016/j.bbrc.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Askari AA, et al. Basal and inducible anti-inflammatory epoxygenase activity in endothelial cells. Biochem Biophys Res Commun. 2014;446(2):633–637. doi: 10.1016/j.bbrc.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wray JA, et al. The epoxygenases CYP2J2 activates the nuclear receptor PPARalpha in vitro and in vivo. PLoS One. 2009;4(10):e7421. doi: 10.1371/journal.pone.0007421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bystrom J, et al. Endogenous epoxygenases are modulators of monocyte/macrophage activity. PLoS One. 2011;6(10):e26591. doi: 10.1371/journal.pone.0026591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng Y, et al. Endothelial CYP epoxygenase overexpression and soluble epoxide hydrolase disruption attenuate acute vascular inflammatory responses in mice. FASEB J. 2011;25(2):703–713. doi: 10.1096/fj.10-171488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oni-Orisan A, et al. Dual modulation of cyclooxygenase and CYP epoxygenase metabolism and acute vascular inflammation in mice. Prostaglandins Other Lipid Mediat. 2013;104–105:67–73. doi: 10.1016/j.prostaglandins.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inceoglu B, et al. Inhibition of soluble epoxide hydrolase reduces LPS-induced thermal hyperalgesia and mechanical allodynia in a rat model of inflammatory pain. Life Sci. 2006;79(24):2311–2319. doi: 10.1016/j.lfs.2006.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inceoglu B, et al. Soluble epoxide hydrolase and epoxyeicosatrienoic acids modulate two distinct analgesic pathways. Proc Natl Acad Sci USA. 2008;105(48):18901–18906. doi: 10.1073/pnas.0809765105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner K, et al. Comparative efficacy of 3 soluble epoxide hydrolase inhibitors in rat neuropathic and inflammatory pain models. Eur J Pharmacol. 2013;700(1–3):93–101. doi: 10.1016/j.ejphar.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kodani SD, Hammock BD. The 2014 Bernard B. Brodie Award Lecture—Epoxide hydrolases: Drug metabolism to therapeutics for chronic pain. Drug Metab Dispos. 2015;43(5):788–802. doi: 10.1124/dmd.115.063339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson JL, Newby AC. Macrophage heterogeneity in atherosclerotic plaques. Curr Opin Lipidol. 2009;20(5):370–378. doi: 10.1097/MOL.0b013e3283309848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swirski FK, et al. Ly-6Chi monocytes dominate hypercholesterolemia-associated monocytosis and give rise to macrophages in atheromata. J Clin Invest. 2007;117(1):195–205. doi: 10.1172/JCI29950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tacke F, et al. Monocyte subsets differentially employ CCR2, CCR5, and CX3CR1 to accumulate within atherosclerotic plaques. J Clin Invest. 2007;117(1):185–194. doi: 10.1172/JCI28549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinal CJ, et al. Targeted disruption of soluble epoxide hydrolase reveals a role in blood pressure regulation. J Biol Chem. 2000;275(51):40504–40510. doi: 10.1074/jbc.M008106200. [DOI] [PubMed] [Google Scholar]
- 32.Stables MJ, et al. Transcriptomic analyses of murine resolution-phase macrophages. Blood. 2011;118(26):e192–e208. doi: 10.1182/blood-2011-04-345330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Motoki A, et al. Soluble epoxide hydrolase inhibition and gene deletion are protective against myocardial ischemia-reperfusion injury in vivo. Am J Physiol Heart Circ Physiol. 2008;295(5):H2128–H2134. doi: 10.1152/ajpheart.00428.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JP, et al. Soluble epoxide hydrolase activity determines the severity of ischemia-reperfusion injury in kidney. PLoS One. 2012;7(5):e37075. doi: 10.1371/journal.pone.0037075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siler DA, et al. Soluble epoxide hydrolase in hydrocephalus, cerebral edema, and vascular inflammation after subarachnoid hemorrhage. Stroke. 2015;46(7):1916–1922. doi: 10.1161/STROKEAHA.114.008560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, et al. Soluble epoxide hydrolase inhibitor attenuates inflammation and airway hyperresponsiveness in mice. Am J Respir Cell Mol Biol. 2015;52(1):46–55. doi: 10.1165/rcmb.2013-0440OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.López-Vicario C, et al. Inhibition of soluble epoxide hydrolase modulates inflammation and autophagy in obese adipose tissue and liver: Role for omega-3 epoxides. Proc Natl Acad Sci USA. 2015;112(2):536–541. doi: 10.1073/pnas.1422590112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulu A, et al. Soluble epoxide hydrolase inhibitors reduce the development of atherosclerosis in apolipoprotein E-knockout mouse model. J Cardiovasc Pharmacol. 2008;52(4):314–323. doi: 10.1097/FJC.0b013e318185fa3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen L, et al. Inhibition of soluble epoxide hydrolase in mice promotes reverse cholesterol transport and regression of atherosclerosis. Atherosclerosis. 2015;239(2):557–565. doi: 10.1016/j.atherosclerosis.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeLozier TC, et al. CYP2C44, a new murine CYP2C that metabolizes arachidonic acid to unique stereospecific products. J Pharmacol Exp Ther. 2004;310(3):845–854. doi: 10.1124/jpet.104.067819. [DOI] [PubMed] [Google Scholar]
- 41.Bystrom J, et al. Inducible CYP2J2 and its product 11,12-EET promotes bacterial phagocytosis: A role for CYP2J2 deficiency in the pathogenesis of Crohn’s disease? PLoS One. 2013;8(9):e75107. doi: 10.1371/journal.pone.0075107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Somchit N, et al. Involvement of phenobarbital and SKF 525A in the hepatotoxicity of antifungal drugs itraconazole and fluconazole in rats. Drug Chem Toxicol. 2006;29(3):237–253. doi: 10.1080/01480540600651535. [DOI] [PubMed] [Google Scholar]
- 43.Ohhira S, Matsui H, Watanabe K. Effects of pretreatment with SKF-525A on triphenyltin metabolism and toxicity in mice. Toxicol Lett. 2000;117(3):145–150. doi: 10.1016/s0378-4274(00)00255-1. [DOI] [PubMed] [Google Scholar]
- 44.Ogata A, Yoneyama M, Sasaki M, Suzuki K, Imamichi T. Effects of pretreatment with SKF-525A or sodium phenobarbital on thiabendazole-induced teratogenicity in ICR mice. Food Chem Toxicol. 1987;25(2):119–124. doi: 10.1016/0278-6915(87)90144-x. [DOI] [PubMed] [Google Scholar]
- 45.Kozak W, et al. Inhibitors of alternative pathways of arachidonate metabolism differentially affect fever in mice. Am J Physiol. 1998;275(4 Pt 2):R1031–R1040. doi: 10.1152/ajpregu.1998.275.4.R1031. [DOI] [PubMed] [Google Scholar]
- 46.Thorp E, Tabas I. Mechanisms and consequences of efferocytosis in advanced atherosclerosis. J Leukoc Biol. 2009;86(5):1089–1095. doi: 10.1189/jlb.0209115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajakariar R, et al. Novel biphasic role for lymphocytes revealed during resolving inflammation. Blood. 2008;111(8):4184–4192. doi: 10.1182/blood-2007-08-108936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Behmoaras J, et al. Macrophage epoxygenase determines a profibrotic transcriptome signature. J Immunol. 2015;194(10):4705–4716. doi: 10.4049/jimmunol.1402979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viswanathan S, et al. Involvement of CYP 2C9 in mediating the proinflammatory effects of linoleic acid in vascular endothelial cells. J Am Coll Nutr. 2003;22(6):502–510. doi: 10.1080/07315724.2003.10719328. [DOI] [PubMed] [Google Scholar]
- 50.Edin ML, et al. Endothelial expression of human cytochrome P450 epoxygenase CYP2C8 increases susceptibility to ischemia-reperfusion injury in isolated mouse heart. FASEB J. 2011;25(10):3436–3447. doi: 10.1096/fj.11-188300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greene JF, Williamson KC, Newman JW, Morisseau C, Hammock BD. Metabolism of monoepoxides of methyl linoleate: Bioactivation and detoxification. Arch Biochem Biophys. 2000;376(2):420–432. doi: 10.1006/abbi.2000.1753. [DOI] [PubMed] [Google Scholar]
- 52.Hayakawa M, et al. Proposal of leukotoxin, 9,10-epoxy-12-octadecenoate, as a burn toxin. Biochem Int. 1990;21(3):573–579. [PubMed] [Google Scholar]
- 53.Hayakawa M, et al. Neutrophils biosynthesize leukotoxin, 9, 10-epoxy-12-octadecenoate. Biochem Biophys Res Commun. 1986;137(1):424–430. doi: 10.1016/0006-291x(86)91227-1. [DOI] [PubMed] [Google Scholar]
- 54.Ozawa T, et al. Biosynthesis of leukotoxin, 9,10-epoxy-12 octadecenoate, by leukocytes in lung lavages of rat after exposure to hyperoxia. Biochem Biophys Res Commun. 1986;134(3):1071–1078. doi: 10.1016/0006-291x(86)90360-8. [DOI] [PubMed] [Google Scholar]
- 55.Greene JF, Newman JW, Williamson KC, Hammock BD. Toxicity of epoxy fatty acids and related compounds to cells expressing human soluble epoxide hydrolase. Chem Res Toxicol. 2000;13(4):217–226. doi: 10.1021/tx990162c. [DOI] [PubMed] [Google Scholar]
- 56.Dioszeghy V, et al. 12/15-Lipoxygenase regulates the inflammatory response to bacterial products in vivo. J Immunol. 2008;181(9):6514–6524. doi: 10.4049/jimmunol.181.9.6514. [DOI] [PubMed] [Google Scholar]
- 57.Seubert JM, et al. Role of soluble epoxide hydrolase in postischemic recovery of heart contractile function. Circ Res. 2006;99(4):442–450. doi: 10.1161/01.RES.0000237390.92932.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panigrahy D, et al. Epoxyeicosanoids stimulate multiorgan metastasis and tumor dormancy escape in mice. J Clin Invest. 2012;122(1):178–191. doi: 10.1172/JCI58128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Newson J, et al. Resolution of acute inflammation bridges the gap between innate and adaptive immunity. Blood. 2014;124(11):1748–1764. doi: 10.1182/blood-2014-03-562710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.