Significance
In analyzing complex networks, we are commonly interested in quantifying the influence that the network nodes exert on each other and in decoding the behavior of the network. We present the nonlinear multidimensional hidden state (NMHS) model, which addresses both of these unmet challenges by simultaneously decoding activity from parallel data streams and calculating the interaction strength among them. In NMHS models, each node in a network acts as a stochastic process that can influence the progression of other nodes in the network. We show that our procedure matches or outperforms state-of-the-art techniques in a multitude of scenarios, notably in systems with nonlinear interactions.
Keywords: causal analysis, functional connectivity, decoding, hidden Markov models, machine learning
Abstract
A universal need in understanding complex networks is the identification of individual information channels and their mutual interactions under different conditions. In neuroscience, our premier example, networks made up of billions of nodes dynamically interact to bring about thought and action. Granger causality is a powerful tool for identifying linear interactions, but handling nonlinear interactions remains an unmet challenge. We present a nonlinear multidimensional hidden state (NMHS) approach that achieves interaction strength analysis and decoding of networks with nonlinear interactions by including latent state variables for each node in the network. We compare NMHS to Granger causality in analyzing neural circuit recordings and simulations, improvised music, and sociodemographic data. We conclude that NMHS significantly extends the scope of analyses of multidimensional, nonlinear networks, notably in coping with the complexity of the brain.
In analyzing complex networks, there is a crucial need for tools to analyze interaction strength among nodes of the networks, extending from genetics to economics (1), demographics (2), and ecology (3). This need is newly pressing in neuroscience (4 –6) (Fig. 1A ). State-of-the-art recording techniques now allow simultaneous measurement of the activity of hundreds of neurons (7, 8). Interpreting these recorded data requires the identification of groups of neurons that strongly interact with each other, known as neural microcircuits, as well as the ability to link microcircuit activity to behavior (7, 9). Currently, Granger causality (GC) (1) and cross-correlation (7) are leading methods for interaction strength analysis (5, 6, 10). Neither method can effectively analyze the nonlinear interactions that are common in neural circuits and systems in other fields (1, 3 –5).
Fig. 1.
Description of the NMHS model. (A) Schematic diagram of a neuronal microcircuit with an extraneous neuron. An ideal interaction detection algorithm would identify and characterize the interactions among all neurons in the microcircuit, and identify the extraneous neuron as unconnected. (B) Each state describes a probability distribution of observed outputs. (C) The transitions of a process depend on the state of its neighbors, such that process B will undergo transitions (light blue or light red) corresponding to the current state of process A (dark blue or dark red). (D) At a given time, t, the output of a process depends solely on the state of that process, and the transitions of that process depend on both its current state and the states of its neighbors. (E) Flowchart of the NMHS method. We search for a model that optimizes the likelihood of the data. We conclude that there is an interaction between A and B if the likelihood of our optimized NMHS model is greater than the likelihood of an unconnected model. By analyzing the transition and emission matrices, we can characterize the strength of the interactions and decode the behavior of A and B. (F) When the state of one process affects the transition behavior of another process, the two processes are interacting. The more the transitions of the process are affected, the stronger the interaction.
Network decoding is an important need in many fields as well, and again, there is need for improved tools. Hidden Markov model (HMM)-based approaches are a leading method in the decoding field (11 –15), but these can only decode a single element at a time. Linear dynamical systems (LDSs) are another approach based on latent variables and are an effective way to model and characterize the behavior of populations of neurons (16). However, LDS models are designed for analyzing data from large populations of similar neurons: they are less appropriate for analyzing circuits made up of heterogeneous neurons. Moreover, LDS cannot use known interactions between populations of neurons to improve decoding and prediction for both populations.
A further motivating point is that most analytic methods now used are either strictly tools for interaction strength analysis or tools for decoding (5, 7, 17). There is a significant advantage to combining the two processes, because identifying nodes that strongly interact with each other enables robust network decoding (13). Here, we introduce the nonlinear multidimensional hidden state (NMHS) approach, which achieves both interaction strength analysis and decoding in networks with nonlinear interactions.
Results
The NMHS model is a generalization of the HMM (11) to systems encompassing two or more processes. For the purpose of analysis, a process may be any physical object or mathematical construct that produces a stream of data. Like the HMM, we model the activity of each process using hidden states. Each state describes a distribution of observed behaviors that account for the recorded activity (Fig. 1B ). The model tracks the evolution of states over time. At each time-step, the next state of a process is determined stochastically. Our model diverges from a classical HMM in that the transition probability of a process can be affected by other processes. The state transitions of each process are governed by a multidimensional transition matrix (Fig. 1C and Fig. S1 A–C ) that encodes the probability of transitioning to each possible state. Importantly, the probability of state transitions depends on both the current state of the process and the states of other processes in the network (Fig. 1D and Fig. S1D ).
Fig. S1.
Application of the model to different interaction scenarios. (A) A two-state NMHS model in which Neuron A affects Neuron B, but Neuron B does not affect Neuron A, i.e., a “unidirectional interaction” model. Note that when Neuron B changes state, Neuron A’s behavior is unaffected (TA,I = TA,II), but when Neuron A changes state, the behavior of Neuron B will change (TB,1 ≠ TB,2). (B) An NMHS model with no interactions. (C) A bidirectional model in which both neurons affect the behavior of the other. (D) Graphical representation of how the NMHS model evolves over time. Arrows represent causal interactions. State transitions can be affected by all neurons, but emissions are only affected by the current state of a neuron. (E) To determine the optimal threshold multiplier for selecting a interacting vs. noninteracting model, we compute a receiver operating characteristic curve using a simulation that is similar to the dataset of interest by plotting the detection rate against the false positive rate for a range of possible multipliers. (F) For each of six different simulated common-cause networks with varying interaction strengths, we train five different models. With an appropriate choice of threshold, we see that the weakly interacting networks are better explained by a noninteracting model, but strongly interacting networks are better explained by a model with a high total interaction strength. (G) Multiple models may sometimes explain the data equally well, as shown here with a common cause model. In this case, decoding and interaction strength estimation cannot proceed, but total interaction detection is not affected. (H) Detailed view of the five trained models from the common-cause network in G, with simulated interaction strength of 0.6. We see that all five models, although are different, have similar total interaction strengths.
To detect interactions, we construct an NMHS model that incorporates each process in a network or putative microcircuit, and we fit the model parameters to optimize . We then fit parameters to an HMM for each modeled process (Fig. 1E ). If the NMHS model explains the data better than the ensemble of HMMs, we consider the processes in the network to be interacting with one another.
NMHS can estimate the directional interaction strength from one process to another (Fig. 1F ). Given an NMHS model fitted to the data, the strength of a directional interaction is quantified by the degree to which state transitions of one process are found to be statistically dependent upon the state of the other process. Given two processes A and B, a change in A’s state may cause a change in B’s transition matrix: the interaction strength from A to B is the mean change in B’s transition matrix when A changes its state.
Network activity can be decoded by using the observations and the acquired model to infer the hidden state of each process at each point in time. Decoding provides insight into the behavior of the system by relating each process’s hidden state to recorded behavior. Such decoding is particularly valuable for applications in biological systems, given that a wide range of evidence has shown that hidden states underlie observed activity (12, 13, 18 –20).
We designed NMHS for analyzing complex nonlinear interactions in neural circuits that cannot be analyzed by current methods. NMHS models neurons as independent entities that may change their behavior based on the state of the network. Neurons are assigned their own emissions and states, but state transitions depend on the states of other neurons in the network. This interdependence matches the duality of neuronal interactions in the nervous system, where neurons act as individuals but modulate each other’s behavior (9, 21 –23). We demonstrate the power of NMHS by analyzing electrophysiological recording data, simulated models, simulated neural circuits, and static data.
First, we applied NMHS to analyze neural microcircuits in three regions of the rat brain: the prefrontal cortex (PFC), the dorsomedial striatum (DMS), and the pars compacta of the dopamine-containing substantia nigra (SNpc) (Fig. 2). Microcircuits were identified experimentally by applying antidromic and orthodromic microstimulation during microelectrode recordings (Fig. 2 A and B ). We categorized putative two- and three-neuron microcircuits as bidirectional or unidirectional based on responses to the microstimulation (24). Recordings of the identified circuits were made as rats performed a decision-making task (Fig. 2C ). We applied NMHS, GC (25), and cross-correlation analyses to the recording data. We used two control datasets. In one, each neuron’s recorded activity was selected from a different session, and all recordings were aligned to task events. This procedure yielded sets of recordings that were synchronized to task structure, but that necessarily represented activity without interaction. In the second control dataset, time bins were randomly permuted (shuffled). For microcircuits consisting of two neurons, NMHS correctly detected all anatomically identified microcircuits and found no spurious interaction in control data. GC and cross-correlation performed comparably (Fig. 2D and Fig. S2D ). In microcircuits of three neurons, NMHS again successfully identified all microcircuits. GC and cross-correlation spuriously identified a large number of interactions in the control group of neurons from different recording sessions, suggesting that they are not suitable for detecting interactions in spike-train data from circuits of three neurons (Fig. 2E and Fig. S2E ).
Fig. 2.
Verification of interactivity and decoding on neuronal microcircuits. (A) We first identified putative microcircuits: sets of two or three neurons such that the neurons are responders to stimulation of counterpart regions. (B) An example of a responding neuron, recorded in the DMS. In the raster plot (Left) and histogram (Right), spikes of the DMS neuron are aligned to stimulation of the PFC at time 0. (C) We recorded the activity of these putative microcircuits of neurons as a rat performed a decision-making task ( Supporting Information ). (D) We evaluated the ability of NMHS (Left) and GC (Right) to detect interactions in putative microcircuits. Microcircuits are categorized as bidirectional responders (both neurons respond to stimulus), unidirectional responders (one neuron responds to stimulus), unconnected (two recordings taken from different sessions, aligned to task events), or shuffled (recordings from connected circuits with observations shuffled over time). Cross-correlation analysis is shown in Fig. S2D . (E) The same evaluation, on microcircuits of three neurons (see also Fig. S2E ). (F) NMHS is also a valuable tool for decoding neuronal microcircuits. In an illustration with task recordings, NMHS and HMM find neuron hidden states with high (red), middle (yellow), or low (blue) activity in the SNpc (Left), DMS (Center), and PFC (Right) during the task. In hidden state plots, trials (horizontally stacked) are aligned to trial start, and black squares indicate the turn onset.
Fig. S2.
Preprocessing and decoding of neuronal data. (A) Bin size optimization for the LFP analysis shown in C. NMHS interactivity is calculated for many bin sizes, and the bin size that produces maximum interactivity is identified. (B) Dependence of false positive rate and detection rate on bin size. (C) An example of preprocessing LFP recordings for analysis by NMHS. In this example, recordings were binned into 59-ms intervals (vertical lines), and mean activity in each bin (green dots) was categorized by quartile (black horizontal lines). (D) Cross-correlation for the dataset plotted in Fig. 2D . (E) Cross-correlation for the dataset plotted in Fig. 2E . (F) Application of NMHS for the LPF analysis shown in A and B. In the hidden state plots, states (red: higher activity state, blue: lower activity state) of the motor cortex (Upper) and anterior caudate nucleus (Lower) are aligned to state transitions in the putamen (time 0). Hidden states in the motor cortex preceded the hidden states in the putamen by 50 ms, and there was no observed relation between the anterior caudate nucleus and putamen.
We then compared the decoding capabilities of HMMs (11) and NMHS. HMMs have been shown to be a powerful tool for decoding neural states (12, 13). Recordings were made from neurons in rodent microcircuits in the PFC, DMS, and SNpc as the rats ran in a T-maze to receive one of two chocolate milk rewards at the end-arms (Fig. 2C ). The activity of these three regions in decision-making tasks has been documented (24). SNpc neurons fire in anticipation of a reward (26). DMS neurons are active in the decision-making period. PFC neurons are activated after the turn and reach maximal activity when the rat consumes the reward (24). Recordings for each behavioral trial were divided into intervals based on behavioral events: the opening of the start gate, the turn, and the consumption of the reward. We selected circuits consisting of three neurons that we verified to be anatomically connected using microstimulation.
We used NMHS decoding to analyze the spike activity of the selected neural circuits. In the triplet shown in Fig. 2F , the SNpc neuron responded immediately to the opening of the start gate, whereas the DMS neuron was in an active state from gate opening to turn and the PFC neuron was active in relation to reward delivery. The decoded states of this triplet of neurons match the well-established relationship among the SNpc, DMS, and PFC in decision-making (24, 26). The hidden states decoded by the NMHS model matched the behavioral states of rats far better than those decoded by the HMM (Fig. 2F ).
We further applied NMHS decoding to local field potential (LFP) activity recorded in the putamen, caudate nucleus and motor cortex of monkeys performing reaching tasks (27). NMHS decoding demonstrated region-specific LFP synchronization between the putamen and motor cortex, which are known to be anatomically connected (Fig. S2F ). This match demonstrates NMHS’s ability to decode continuous neuronal data.
As further validation of NMHS, we applied the algorithm to simulated models with known interaction strengths. We compared the simulated and estimated models using the NMHS interaction strength for sequences of data from 28 two-process models with varying interaction structures and strengths. NMHS successfully estimated the strength of almost all pairwise interactions in two-process models (Fig. 3 A–B and Table S1). Additionally, NMHS successfully found the total interactivity (calculated as the maximum interaction strength of all pairwise interactions within the model) in a three-node model (Fig. S3A ).
Fig. 3.
Validation on simulated microcircuits. (A–D) Simulation data were generated from two-node, two-state, three-emission NMHS matrix models (A). Interactivity estimated using NMHS (B), GC (C), and cross-correlation (D) was plotted against the simulated interactivity. (Blue points represent how much node 2 is affected by node 1; red vice versa.) (E–H) A Hodgkin–Huxley model simulates a triplet of neurons with two excitatory synapses (E). The strength of each interaction in the triplet is estimated using NMHS (F), GC (G), and cross-correlation (H) and plotted against the simulated synaptic strength (gsyn). The remaining interactivity directions are shown in Fig. S3E . (I–L) Interactivity of a similar Hodgkin–Huxley model with one excitatory and one inhibitory synapse (I), estimated with NMHS (J), GC (K), and cross-correlation (L) (see also Fig. S3F ).
Table S1.
Number of models created for each connectivity structure
| Connectivity strength 2→1 | Connectivity strength 1→2 | ||
| Strong | Moderate | Weak | |
| Strong | 3 | 3 | 3 |
| Moderate | 3 | 3 | 3 |
| Weak | 3 | 3 | 3 |
Fig. S3.

Additional comparisons of performance of NHMS, GC, and cross-correlation on simulated microcircuits. (A) Estimated interactivity plotted as a function of simulated interactivity in a three-node matrix model calculated with NHMS (Left), GC (Center), and cross-correlation (Right). Simulated interactivity was varied by adding noise of various magnitudes to a baseline model described by transition and emission matrices. (B) Estimated interactivity as a function of simulated synaptic strength in a two-neuron model with an excitatory synapse. Data were generated using a Hodgkin–Huxley model where an upstream neuron synapses onto a downstream neuron. We varied the g syn parameter, which corresponds to simulated synaptic strength. The downstream neuron does not synapse onto the upstream neuron. Line colors represent the direction of estimated interactivity. (C) Estimated interactivity in a two-neuron model with an inhibitory synapse. (D) Estimated interactivity in a two-neuron model with an excitatory synapse that provides an oscillatory input. (E) Estimated interactivity of the remaining four potential interactions of the three-neuron model with two excitatory inputs shown in Fig. 3 E–H (only one interaction for cross-correlation). All four interactions have low interactivity values, as expected, because there is no simulated interaction between them. (F) Estimated interactivity of the remaining potential interactions of the model shown in Fig. 3 I–L .
To validate that our method successfully identifies interactions in neural microcircuits, we simulated five motifs of neuronal networks using the Hodgkin–Huxley model (28 –30) (Table S2). We found a strong correlation between simulated synaptic strength and estimated interaction strength in nonlinear scenarios (Fig. 3 E, F, I, and J and Fig. S3 B–F ). As we expected (4, 5, 10), given that GC and cross-correlation are designed for linear interactions (1, 3), neither GC nor cross-correlation detected interactions with comparable accuracy (Fig. 3 C, D, G, H, K, and L and Fig. S3). Additionally, to demonstrate NMHS performance in a situation when there are unobserved nodes in the network, resulting in incomplete information, we simulated two scenarios: one with a varying number of unobserved neurons (Fig. S4A ) and another with a single unobserved neuron, varying the synaptic strength (Fig. S4B ). As expected, with an increasing amount of unobserved information, the estimated interaction strength of the observed nodes decreases.
Table S2.
Parameters of Hodgkin–Huxley circuit models
| Model | No. of synapses | Presynaptic target duration, s | Presynaptic target frequency, Hz | Total simulation duration, s | |||
| Excitatory | Inhibitory | Low activity state | High activity state | Low activity state | High activity state | ||
| Model 1 (Fig. 3E and Fig. S3E ) | 2 | 0 | 3–10 | 2 | 7 | 50 | 50 |
| Model 2 (Fig. 3I and Fig. S3F ) | 1 | 1 | 1 | 1 | 7 | 50 | 50 |
| Model 3 (Fig. S3B ) | 1 | 0 | 3–10 | 2 | 7 | 50 | 50 |
| Model 4 (Fig. S3C ) | 0 | 1 | 3–10 | 2 | 0 | 50 | 10 |
| Model 5 (Fig. S3D ) | 1 | 0 | N/A | N/A | N/A | N/A | 25 |
| Model 6 (Fig. S4A ) | 1–12 | 0 | 1 | 1 | 0 | 50 | 50 |
| Model 7 (Fig. S4B ) | 2 | 0 | 3–10 | 2 | 7 | 50 | 50 |
N/A, not applicable.
Fig. S4.
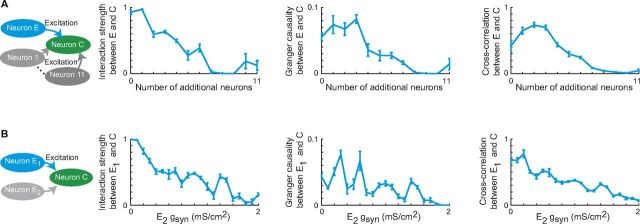
Detecting interaction strength in simulated circuits of Hodgkin–Huxley neurons. (A) Simulation of a multineuron network using a Hodgkin–Huxley model. We measure the interactivity from Neuron E to Neuron C (with a fixed simulated synaptic strength of g syn = 0.4), while increasing the number of unmeasured neurons in the network (Neurons 1–11, each with a synaptic strength of g syn = 0.2). We see that NMHS, GC, and cross-correlation all demonstrate a decreasing value with an increasing number of unmeasured neurons, because the activity of Neuron C is influenced more strongly by the unmeasured activity. (B) A similar simulation of a three-neuron network, keeping the synaptic strength of the measured pair (Neuron E1 and Neuron C) constant (g syn = 0.5), while increasing the synaptic strength between the unmeasured pair (Neuron E2 and Neuron C).
To expand our validation effort, we applied the NMHS algorithm to other networks of interacting processes. We chose another application in which independent agents interact and change behavior based on each other’s activity. We used recordings of coimprovising guitarists (Fig. 4A and Fig. S5A ) and applied NMHS to analyze guitar voices in a single band. Voices from separate bands were used as a control (Fig. 4B ). The algorithm determined significantly greater interaction strength between voices in a single band than between unrelated voices (Fig. 4B and Fig. S5 C and D ). The NMHS interactivity algorithm successfully identified the fact that all of the guitarists in a band listen to each other to varying degrees, which GC failed to detect (Fig. 4C ). NMHS’s decoding ability also provided insight into the guitarists’ behavior; by estimating the hidden states of the guitar recordings, we were able to evaluate the improvisation strategy (mimicking or contrasting) of the guitarists (Fig. S5E ). Furthermore, to validate NMHS performance in scenarios with incomplete information, we orchestrated recordings of three to six guitarists, analyzing the activity of only three musicians in the group. We found that NMHS performed equally well when up to half of the nodes were unobserved (Fig. 4D ).
Fig. 4.
Application to musical improvisation recordings and static data. (A) We collected recordings, in separate channels, of three guitarists improvising classic rock music. (B) NMHS determined interaction between three voices of a single band (coimprovised), three voices from separate bands (separately improvised), and randomly permuted recordings of three voices from a single band (shuffled). (C) NMHS attributed different interactivity from improvisation leaders to followers, from followers to leaders, and between followers. (D) Interaction strength detected by NMHS in the analysis of the activity of only three guitarists out of a larger group of three to six musicians. (E) In an application on a static network, NMHS analyzed sociodemographic data. In this static model, individuals in strata (boxes) are assigned states (blue and red ovals) that describe their wealth and education. (F) NMHS found interactivity between sociodemographic strata. (G) A probabilistic XOR model, where the output is produced by applying an XOR rule to a bin in the input streams with a set probability P. If the XOR rule is not applied, the output value is selected randomly. (H) NMHS performance in detecting interactivity strength between the input streams as a function of increasing P. (I) Cross-correlation performance in detecting the same interaction.
Fig. S5.
Preprocessing, interactivity, and decoding of musical improvisation recordings. (A) Pitch classifier used for extracting notes from guitar recordings shown in Fig. 4A . (B) Dependence of false positive rate and detection rate on bin size. (C) Cross-correlation of the dataset plotted in Fig. 4B . (D) NMHS was tested by comparing total interactivity in mixed groups with two voices (one band and one voice from a separate band) and in groups of two and three voices from a three-guitarist band. (E) NMHS assigns hidden states (red, higher activity state; blue, lower activity state) to the guitar recordings, which correlated with the pitches played by the improvisers. Recordings of the follower voices were aligned to state changes of the leader (time 0). The hidden state plot indicates that a band is synchronized with a short time delay and that we can identify the strategy of the followers (mimicking or contrasting) based on their hidden states.
Slightly modified, the NMHS algorithm also becomes a valuable tool for analysis of static data, such as those found in genomics, proteomics, and demographics (2). With static data, the algorithm measures the degree of interaction between observed variables. Because our algorithm looks at the dependence of the current state of the node on the previous state of itself as well as the neighbor, we modified it to instead consider the dependence of the current state of the node on the current state of the neighbor (but still the previous state of itself) (Fig. 4E ). Applied to three sociodemographic datasets, NMHS determined interaction between income and education level (Fig. 4F ) that aligns closely with research evidence (2). Additionally, we simulated another model (Fig. 4G ), where the output is produced by applying an exclusive or (XOR) rule to only a fraction of the sample bins. The sample bins where the XOR rule is not applied were random. We demonstrate that NMHS successfully detects strong total interactivity when the XOR rule is applied to a large fraction of sample bins (Fig. 4 H and I ).
Discussion
We show here that NMHS compares very favorably to GC, cross-correlation, and conventional HMM solutions in multiple analysis and decoding challenges. NMHS demonstrates great promise for analyzing neural microcircuits in particular. We emphasize that NMHS carries the important advantage of being a tool for both analysis and decoding of complex networks.
The NMHS model that we introduce here is closely related to the hierarchical HMM (HHMM) (14) and the factorial HMM (FHMM) (15). All three are variants of the HMM that impose constraints on the structure of the HMM to improve its performance at a specific task. HHMMs are designed for analyzing and decoding a stream of data that exhibit long-term structure, such as language: the model accomplishes this analysis by organizing the states of the model into a tree in which state transitions between the leaves of a node are common, but in which transitions up and down the levels of the tree are comparatively rare. By contrast, FHMMs are designed for analyzing processes that may have multiple independent hidden states. The FHMM is similar to the NMHS model, except that where the NMHS has a multidimensional transition matrix, the FHMM has a multidimensional emission matrix: the likelihood of an emission is conditioned on the value of each hidden state in the FHMM ( SI Materials and Methods , Comparison with Other Models ). In contrast to the FHMM and HHMM, NMHS is designed to analyze multiple streams of data, each representing a process that interacts with another. This design allows interaction strength to be inferred from the parameters of the model, and concurrently using information from multiple interacting processes makes it possible to decode network activity much more reliably than with other methods.
There are a number of limitations to consider when applying NMHS. First, a dynamic system analyzed must be describable in terms of states that make transitions over time. These states must be correlated across time; the recent past of a process should be an indicator of current activity.
The NMHS model is most suitable for analyzing interactions involving a small number of processes: the number of parameters in the model increases exponentially with the number of processes included in the model ( SI Materials and Methods , Scalability of the NMHS Model ), and thus estimating model parameters rapidly becomes computationally intractable as the number of processes increases. There are two potential approaches for increasing the maximum number of processes that can be modeled at once. First, we have successfully trained two- and three-process NMHS models using simulated annealing, an approach that can be applied to models with several processes. Second, approximation techniques such as variational inference can be used, as proposed by Ghahramani and Jordan (15). To analyze interactions within a collection of many processes, it is generally neither necessary nor desirable to use a single very large NMHS model. Larger NMHS models are only required for detecting complex interactions that cannot be characterized as a combination of multiple independent interactions involving fewer processes. For extremely large networks, we can group the network into smaller subnetworks within which nonlinear interactions are evident, and analyze each subnetwork using either the two- or three-process NMHS approach presented here, or the extended NMHS approach described above. By comparison with NMHS, cross-correlation is only defined for two signals and is not applicable to analyzing interactions involving more than two processes. GC can be scaled to larger interactions (31) but can only detect linear interactions. We have shown here that using GC analysis of three-neuron recordings leads to a high rate of false positive (Fig. 2E ).
Finally, when fitting parameters to the NMHS model, there may be multiple models that equally well describe the data (Fig. S1 F–H ). To determine whether this is the case, one must run parameter estimation with several different instantiations. If different models are found that account for the data equally well, interaction strength estimation and decoding cannot proceed. However, we have found that total interactivity detection is robust to this issue (Fig. S1 F and H ). Although the numerical value may be slightly higher or lower than that of the true value, it is unlikely that a noninteracting model can account for the data from an interacting pair or triplet of nodes as well as an interacting model can (Fig. S1 F and H ).
Materials and Methods
All animal procedures were approved by the Committee on Animal Care at the Massachusetts Institute of Technology.
Model.
The NMHS model is an extension of the HMM (5, 11) that is capable of learning the behavior of multiple interacting processes simultaneously. The NMHS model is related to a number of other HMM-based models that have been proposed for solving other problems (14, 15). When modeling a single process, NMHS is identical to a HMM. Like HMMs, a NMHS model can be trained through unsupervised learning—it requires only a sufficiently long stream of data for each process to be modeled. NMHS assumes that the processes being modeled are stateful: that is, the behavior at a time t can be predicted using the behavior at time t − 1. States are represented as distributions of possible behaviors: an emission matrix, E , describes the likelihood of observing a given emission e given the model is currently in state s: . The model automatically learns the best states to account for the data. Transitions between states are treated as stochastic events: the probability of a process making a transition to a new state is based on its current state and may be affected by other processes’ states. If i and j are the states of two interacting processes X , Y at time t, then the probability of process X making a transition to state k at t + 1 is given by . When modeling multiple processes, each process has its own transition and emission matrix.
To account for the idea that the transition probabilities of a process can be affected by other processes, we add dimensions to the transition matrix. Consider the case of modeling the interactions between two processes: the additional dimension in the transition matrix can be thought of as containing a “repertoire of behaviors”: each slice of the matrix along this dimension is an individual transition matrix that corresponds to one state of the neighboring process. Each of these transition matrices can describe a completely different behavior for the process. To illustrate this point, consider Fig. S1A . We can think of two processes: A with states 1 and 2 and B with states I and II. If process A exerts a strong influence on the behavior of process B, then layers 1 and 2 in B’s transition matrix will be very different, as is the case in Fig. S1A . Conversely, if B does not much influence A’s behavior, then layers I and II in A’s transition matrix will be quite similar to each other. Adding process C to our system also adds an additional dimension: now the behavior of each process is dependent on the states of both other processes. For each added process, an additional dimension and corresponding sets of layers are added. For example, the two processes in Fig. S1A are modeled with two states: the total size of the T matrix therefore is 2 × 2 × 2 = 8 elements. Were a third process added to the system, the number of elements in each process’s T-matrix would be 2 × 2 × 2 × 2 = 16.
The NMHS model can be expressed as a single HMM whose states and emissions are drawn from the product space of all of the process’s states and the product space of all of the process’s emissions. Compared with this gestalt HMM, the NMHS model enforces that a process’s emissions are independent from other process’s states and a process’s transitions are independent of other process’s transitions. The NMHS model is also more interpretable and requires fewer parameters ( SI Materials and Methods , Comparison with Other Models ). It is easy to understand directly from the model that a given layer of a transition matrix corresponds to the transition behavior, given the state of the neighbor. Furthermore, each process has its own emission matrix that describes its behavior independently from other processes.
Goal Function and Interactivity Detection.
We evaluate the fit of a model by calculating the likelihood function of the observations given the model parameters. For a two-process NMHS model with observation sequences and , transition matrices and , and emission matrices and , where , we have
and Bayes theorem gives us
For different parameters under the same model, we assume is the same, and we thus can compare the posterior probabilities of different parameterizations to determine which is better. We treat a set of one-process HMMs, one for each process being modeled, as the null hypothesis. For the two-process case, this gives us two HMMs with parameters N and forward probabilities f 1 (t) and f 2 (t). We then have .
Because != , we cannot directly compare and . However, if we make the assumption that is the same for all models, then we can estimate a scaling factor , and then compare to determine how good a fit the model is versus the null hypothesis.
To estimate , we train a number of interacting and noninteracting models on data known to represent an interaction, and take the mean difference between the interacting and noninteracting likelihoods. In practice, we have found that works well in most instances. We have also achieved good results by using the Bayesian information criteria (BIC) (32), which has the advantage that it does not require estimation of an additional parameter.
An alternative method of choosing an appropriate is by plotting a receiver operating characteristic curve (Fig. S1E ). Using a simulated model that is of similar size and structure to the dataset of interest, we select the that results in the highest probability of detection for a given probability of false positive. This method will work whether the goal function is the log-likelihood of the model, the BIC, or another appropriate goal function. An important note, however, is that the threshold value depends on the model itself so it must be calculated for each new model. For example, the optimal when comparing the BIC values of one- and three-node models was (Fig. S1F ).
Confidence in the Model.
In some instances, there can be multiple models that all account for the data well and meet the criterion. There are two possible issues that can lead to this situation. The first is insufficient data: with a small dataset, the NMHS model can be over-fit to the observations, and several distinct models might be able to account for the data. The second possible issue is that distinct models sometimes produce equivalent behavior and thus are in practice indistinguishable. An example is the “common cause” scenario, in which process A affects both process B and process C: a model wherein process A affects process B which in turn affects process C may produce equivalent behavior. In either instance, when multiple interacting models are found (Fig. S1 G and H ), it generally is not possible to quantify the interaction strength, or to decode the behavior of the processes. However, the total interactivity, calculated as the maximum of all of the pairwise interaction strengths in the model, is relatively robust to this issue. This phenomenon occurs because given a dataset that comes from a network with a high total interaction strength (e.g., > 0.5 in Fig. S1 F–H ), it is unlikely for three independent one-node models to explain it better than a single three-node model, even if that three-node model does not find the specific pairwise interactions correctly. Conversely, it is unlikely for a three-node model to explain a dataset that comes from three independent data streams better than three one-node models. Thus, with an appropriate estimate for , the models with low total interaction are better explained by three one-node models and are therefore labeled as noninteracting; the high total interaction models are better explained by a single three-node model and therefore are labeled as interacting (Fig. S1G ).
Determining the Multiple-Explanation Models.
To determine whether the multiple-explanation issue is present for a given dataset, we train several different models on that dataset, initializing the training algorithm with a different set of parameters each time. We perform enough initializations to sample the search space uniformly and densely. If a global-optimum approach is used to fit the model parameters, such as genetic algorithms or simulated annealing, it is necessary to keep track of all of the local optima encountered in the search and to compare the models that have log-likelihood above a predefined threshold. If we find that all of the trained models are similar to each other, it is likely that there is only one model that explains the data.
Quantifying Interaction Strength.
Once NMHS establishes that and that only one model explains the data well, we can quantify the interaction strength by analyzing the transition matrices. We calculate an interactivity index that signifies how strongly one process affects another.
Given processes A and B with N states for process A , and M states for process B , we calculate the interaction strength from B to A (i.e., how strongly A is affected by B ) as follows. Let A have the transition matrix . We first calculate the “specific interaction index” for each state z in A , given each pair of states x,y in B :
Given that T satisfies the Markov property, these values are bounded by the interval [0,1]. To measure pairwise interaction strength from B to A , we take the mean of all specific interaction indices:
This value is on the interval [0,1], where 0 corresponds to no interaction ( A and B are entirely independent of each other), and 1 corresponds to maximal interaction. In the NMHS Toolbox, this calculation can be performed for all pairs of nodes simultaneously in two- or three-process models using the interactivity() function. To calculate total interactivity, simply take the maximum interaction strength among all pairs of nodes.
SI Materials and Methods
Preprocessing Data for NMHS Model.
There are multiple ways to preprocess the raw data for the NMHS training and decoding algorithms, but in all instances, we need to determine three interrelated parameters: the bin size when binning the raw data, the number of states in the model, and the number of emissions in the model.
Finding the optimal bin size is critical for decoding and interactivity estimation (Fig. S2A ). If the bin size is too large, the critical information about state transitions will be lost within a single bin. On the other hand, if the bin size is too small, the critical information might be distributed throughout several preceding time points. In our model, activity at one point in time depends on the previous time point and nothing else. To determine the optimal bin size, we run our algorithm using several different bin sizes to find the one that returns the highest interactivity. In general, optimal bin size is a function of the process that we are measuring. It is possible, however, that a certain bin size can result in an undesirable rate of false positives. To verify that the chosen bin size is optimal, we simply run several simulations using models that are similar to the dataset of interest and plot the detection rate and false positive rate for several different bin sizes (Fig. S2B ). For example, in LFP recordings shown in Fig. S2C , the optimal bin size was 59 ms. For spike train recordings, it was 100 ms. For the musical improvisation recordings, it was 3 s.
The number of states depends on the number of emissions. Clearly, it must be smaller than the number of emissions. To find the optimal number of states, we run the training algorithm using several different possibilities for the number of states. We then compare the different models to find the best one using the BIC (32). Note that we cannot use log-likelihood to compare models with different sizes as we do when comparing models with different initializations ( Model ) due to overfitting. A model will always fit data more closely when given more parameters (11, 32). BIC, however, accounts for the number of parameters in the model. For example, for the LFP recordings, as well as for the musical improvisation data, we found that two states were optimal (Fig. S2F and Fig. S5E ). For spike trains, three states were optimal (Fig. 2F ).
Finally, the number of emissions is dependent on the bin size, which directly dictates the smallest and largest values within a bin. It is, however, possible to further reduce the number of emissions to reduce the number of parameters in the model for the purpose of computational speed or to compensate for insufficient data. If computational power or data size is not a limiting factor, we can use a method similar to one used to find the optimal number of states by comparing the BIC values for models with different numbers of emissions. For example, in the LFP recordings, neuronal spike train recordings and musical improvisation, we found, after binning, that it was optimal to separate the data into quartiles [i.e., the lowest 25% of the binned values are assigned to quartile 1, the next 25% are assigned to quartile 2, etc. (Fig. S2C )]. On the other hand, for the sociodemographic data, we used five and seven emissions, as dictated by the data without any reduction (Fig. 4E ).
Parameter Estimation with Modified Baum–Welch Algorithm.
The two-process HMM has two sequences of states. Let X t be the state of process 1 at time t, and let Y t be the state of process 2 at time t. The state transitions of the model are then governed by and . Let be the sequence of observations from process 1, of length τ, and let be the sequence observed from process 2, also of length τ. The emission probabilities are governed by and .
We train the two-process HMM using a modified Baum–Welch algorithm. Each iteration of Baum–Welch algorithm takes a model as input and attempts to yield an incrementally better model as output. Refinement continues until either a set number of iterations have occurred or the model’s likelihood stops improving. Because parameter estimation proceeds incrementally, it is susceptible to local minima. It is therefore important to run the Baum–Welch algorithm several times with different initializations to find a good minimum.
An explanation of the two-process Baum–Welch algorithm is given here. The three-process algorithm proceeds similarly.
We define the forward probabilities, f :
The forward probability at time t for states i, j corresponds to the probability that process 1 will be in state i and process 2 will be in state j at time t, given the current and previous observations. Note that : the probability of the observed data, given the current model parameters.
We define the backward probabilities, b :
The backward probabilities are a mirror image to the forward probabilities: describes the probability that processes 1 and 2 will be in states i and j given the observations that occur after t.
We also define a likelihood coefficient: , which describes the likelihood of processes 1 and 2 being in states i and j given at time t, given all observations in the dataset. The forward-backward algorithm for calculating b , f , and is implemented in the NMHS Toolbox for two- and three-process models in hmmtools/forward_backward2d() and hmmtools/forward_backward3d().
With these, we calculate the probability that processes 1 and 2 transition from states k and l to states i and j:
Because T and F are independent, we must calculate two intermediate values:
We use these to estimate a new T and R :
These matrices must be normalized to satisfy the Markov property: .
To estimate new emissions matrices, we use
Again, the new matrices are normalized to satisfy the Markov property. In the NMHS Toolbox, training is implemented for two-process and three-process models in the functions hmmtools/hmmtrain2d() and hmmtools/hmmtrain3d().
Decoding Hidden States.
If the NMHS algorithm succeeds in finding a unique model that best accounts for the data, that model can be used to decode the hidden states of the modeled processes based on observed behavior. Given the forward and backward probabilities:
We can then estimate the likelihood of each state over time:
This operation gives us a sequence for each process of the estimated hidden state at each time-step. In the case where the NMHS model explains the data better than an HMM and has thus found an interaction, NMHS decoding will have more explanatory power than the HMM decoding. When NMHS has found an interaction, the algorithm can use data from interacting processes to aid in decoding the process of interest. In the NMHS Toolbox, decoding is implemented for two- and three-process models in the functions hmmtools/hmmdecode2d() and hmmtools/hmmdecode3d().
One advantage of this decoding technique is that all of the processes being decoded have independent emission matrices, and thus the model can accommodate processes with widely differing outputs. No normalization is needed in this instance, because all observations are categorized into an emission. Additionally, as more data are accumulated, the NMHS model can be refined, leading to improved decoding results. Note, however, that when decoding the activity of noninteracting processes, NMHS is equivalent to HMM decoding, and thus no improvement in decoding performance will be observed. Fig. 2F shows the comparison of NMHS decoding and HMM decoding on electrophysiological data from rats. Compared with HMM states, the hidden states decoded by the NHMS model are a much better match to the observed behavioral states of rats.
Verification with Neuronal Spike Data Recorded in Rats.
To validate our algorithm, we recorded neuronal spike activity simultaneously in the PFC, DMS, and SNpc of rats performing a decision-making task. To identify the interactions for pairs and triplets of neurons, we performed electrical microstimulation in the brain regions of interest and used the evoked responses in the other regions to identify the putative microcircuits. We then preprocessed the recorded data from the decision-making task, estimated interactivity using NMHS and GC (25) (Fig. 2 D and E ) and decoded the hidden states (Fig. 2F ).
Rat decision-making T-maze task.
To illustrate the successful use of NMHS as a tool for interactivity estimation and decoding, we trained rats (male Long–Evans, 420–600 g) on a decision-making task in a T-maze consisting of a running track and two end-arms where chocolate milk rewards were placed (Fig. 2C ) (24). In each of 40 trials given daily, rats were offered a choice between pure chocolate milk and diluted chocolate milk. To start each trial, a click sound was played, indicating to the rat that the trial was beginning. The maze door then opened, allowing the rat to run down the running track and to make a decision between the two end-arms and corresponding reinforcement offers. Rats were then allowed to lick for the reward of their choosing. The intertrial interval was random, between 40 and 90 s. During each trial, neuronal activity was recorded with a Cheetah Data Acquisition System (Neuralynx). Details of behavioral training, device implantation, and recording procedures are described in ref. 24.
Putative identification of anatomically connected neurons.
We putatively identified neurons in the PFC, DMS, and SNpc that were anatomically connected by recording responses to electrical stimulations (Fig. 2A ). In each of the three brain regions, a bundle of tetrodes (n = 8, tungsten, impedance 100–400 kΩ) was implanted. In the center of each tetrode bundle was a tungsten bipolar stimulation electrode, constructed using two wires of a tetrode as an anode and other two wires as a cathode. Details of the devices and implantation are provided elsewhere (24).
The PFC, DMS, and SNpc were stimulated sequentially, via the central electrodes (0.5-ms square pulse, 15 μA, 2-s interval between pulses; 100 pulses) using ISO-Flex and Master 9 systems (AMPI). Simultaneous recordings were taken in the two responding brain regions: the recorded neuronal activity was sorted into putative single units using a custom-developed program (33), and the activity of each unit was aligned to the 100 stimulation pulses to determine whether the unit responded to the stimulation (Fig. 2 A and B ). Units that generated at least 10 spikes in response to the 100 stimulation pulses were considered as candidates for responders.
The candidates were then tested to isolate the neurons with statistically significant stimulation responses. A neuron in the responding brain region was considered a stimulation responder if its mean firing rate after stimulation was at least 4 SDs greater than it was during the baseline period (1-s interval preceding the stimulation pulse). The poststimulation windows were defined as the intervals 3–10 ms after the pulse.
After identifying neurons that responded to microstimulation, we defined two types of putative microcircuits. A pair of neurons that responded to microstimulation of both counterpart brain regions was labeled as bidirectionally connected. Pairs of neurons with only one neuron exhibiting a response to microstimulation were labeled as “unidirectional.” We used two sets of control data. In the first, labeled “shuffled,” we used recordings from putative microcircuits and permuted the order of the data in time. In the second control dataset, labeled “unconnected,” we sought to create data in which the task structure and synchronicity of events was preserved, but in which each recording was necessarily anatomically unconnected to each other. To construct the unconnected dataset, we aligned recordings based on the trial start (click) and the trial end (lick). We excluded trials with click-to-lick periods greater than 5 s, because they were too long to align well. The click-to-lick period of the remaining trials was 2.65 ± 0.69 s. Within each peri-trial period, we calculated the firing rate in each of 600 time bins. For each trial, we adjusted the bin size such that the starting click occurred between bins 300 and 301 and the first lick occurred between bins 360 and 361.
Verification with Primate LFP Recordings.
We applied NMHS to LFP activity recorded from the motor cortex and anterior caudate nucleus in macaque monkeys performing a reaching task, in which the monkeys controlled a cursor with a joystick, performing center-out-center movements, according to visual cues. Details of the recordings and tasks are described in ref. 27. We found the bin size using simulations ( Preprocessing Data for NMHS Model and Fig. S2 A and B ). We then quantized the analog LFP data into four levels (Fig. S2C ) and applied the NMHS algorithm to the resulting sequence of observations to decode the hidden states (Fig. S2F ).
Verification Using Simulated Data.
To validate our model, we conducted a series of tests on simulated data. We used two methods of generation: first, a known set of NMHS model matrices to generate pairs or triplets of state and observation sequences; and second, a Hodgkin–Huxley model to generate neuronal spike trains from pairs or triplets of simulated neuronal microcircuits.
Algorithm for generating data using an NMHS model.
For the first method for generating data, we used a known NMHS model—a set of transition and emission matrices (Fig. 3A and Fig. S3A ). We then used that model to generate either pairs or triplets of state sequences and corresponding observation sequences. Finally, from the observation sequences, we trained a new estimated model and compared the estimated model’s interactivity to the known model’s interactivity. The advantage of this method is that we use identical interactivity metrics to compare the known model to the estimated model, so we have a direct measure of error. Furthermore, because we have full control over the known model’s parameters, we can more freely explore the search space of the training algorithm.
To generate simulation data from ground truth model, we started with a set of transition and emission matrices that were either randomly generated (Fig. 3A ) or that were designed to have a desired interactivity structure (Fig. S3 A–C ). In the two-node case, where node 1 can take on N states with J emissions, and node 2 can take on M states with K emissions, transition matrix 1 is size N × N × M, and transition matrix 2 is size M × M × N. The emission matrices are N × J and M × K, respectively. We will only describe the two-node case here; the three-node case is analogous, simply with three transition matrices and three emission matrices, which results in an additional dimension in each transition matrix (the emission matrices remain two dimensional regardless of the number of nodes). For example, the first transition matrix in the three-node case will be size N × N × M × P, and the first emission matrix will be N × J.
In the two-node case, we generated two sequences of observations that simulate the measurable activity at each of the two nodes, as well as the two corresponding sequences of underlying hidden states that control the observed behavior. The algorithm is outlined as follows.
-
i)
Initialize two empty arrays obs1 and obs2 that will hold the observation sequences and two more, states1 and states2, which will hold the hidden-state sequences. All four arrays are of length L, where L is the desired number of time points in each sequence.
-
ii)
Assume that the two state sequences are initially both in state “1.” Given that this initial state is deterministic and always the same for all sequences, it will not be recorded in our sequences.
-
iii)
Once we have the state at time t, we look at the appropriate row in the transition matrices to determine probabilistically the next state (at time t + 1). This row will tell us the probability distribution of the state of the current node at time t + 1 given the state of the current node at time t and the state of the neighboring node at time t. We sample from this distribution to determine the upcoming state and append this value to the states1 array. The procedure is repeated for the states2 array.
-
iv)
Once we have the hidden states at time t + 1, look at the corresponding row in the emission matrices to probabilistically determine the value of the observation at that time. Row i in an emission matrix gives us the probability distribution of the current observation given that the current state is i. Use a random number generator to draw from that distribution and append the observation to the obs1 array. Repeat the same procedure for the obs2 array.
-
v)
Repeat steps iii and iv L times, looking only at the current state of both nodes to determine the next state of each node, and only the current state of each node to determine the current observation emitted from each node.
Data generated using an NMHS model.
By using this process, we created 27 different two-node models, where each node can take on one of two possible states and can produce one of three different emission values in a given state (Fig. 3A ). The transition matrices for each of the models were pseudorandomly generated to span the range of all plausible interactivity values (∼0–0.9), as well as to have all possible permutations of interactivity structure (Table S1).
To generate the transition matrices, we started with a seed matrix that represents either a maximally interacting node, where layer 1 is the identity matrix and layer 2 is the antidiagonal identity matrix (ones on the anti-diagonal, zeroes elsewhere), or a noninteracting node, where both layers are the identity matrix. (Note that in general an identity matrix and an anti-diagonal identity matrix do not always represent maximal interactivity.) We then took a weighted average of this matrix with a random matrix (values drawn from a uniform distribution). Finally, we normalized the matrix to satisfy the Markovian property. Corresponding emission matrices were generated analogously.
We used a similar procedure to test our algorithm on a three-node model (Fig. S3A ) with the interaction structure shown. We started with a set of matrices representing a model that exhibits maximal interactivity between the given pairs of nodes and added varying levels of noise to those matrices to vary the interaction strengths, while still preserving the interaction structure shown. Because of multiple models describing the data equally well, as discussed above, we could only reliably find the total interaction strength for data generated from this type of interactivity structure.
Generating data using a Hodgkin–Huxley model.
In the second method for generating data, we modeled neuronal microcircuits with two or three functionally connected neurons using a standard Hodgkin–Huxley formalism and a simple conductance-based synapse model. These simulations allow us to evaluate our algorithm’s performance in inferring the functional connectivity of biologically realistic neuronal circuits, while still knowing with certainty the underlying interactivity structure as well as having some measure of interaction strength.
The parameters of the model neurons are those used by Hodgkin and Huxley (30) to represent the squid giant axon. Both excitatory and inhibitory model synapses are described by the following equations, which have been used previously to approximate the behavior of AMPA- and GABAA-mediated synaptic currents (28, 29):
where represents the synaptic current, is the maximal synaptic conductance (which is the simulated synaptic strength parameter varied in the simulations), is the fraction of the receptors in the open state, and are the membrane potentials of the presynaptic and postsynaptic neurons shifted by −70 mV, is the synaptic reversal potential, and are the forward and backward rate constants, is the neurotransmitter concentration in the synaptic cleft, and is the maximal neurotransmitter concentration. For excitatory synapses, mV, whereas for inhibitory synapses, mV. For both excitatory and inhibitory synapses, mM−1ms−1, ms−1, and mM. The rate constants and are set relatively low to make the synapses slower and thus enhance the temporal summation of synaptic currents. Given that the resting membrane potential in Hodgkin and Huxley’s original model is 0 mV, when computing synaptic currents, the membrane potentials of the neurons are shifted by −70 mV to make them compatible with the voltage scale of the synaptic model. Because the inhibitory synaptic reversal potential is the same as the resting membrane potential of −70 mV, the model inhibitory synapses exhibit shunting inhibition. We solved the system of differential equations in MATLAB using the ode45 solver.
In our circuit models, one or more presynaptic neurons synapse on a postsynaptic neuron. Spiking activity in the presynaptic neurons is evoked by injecting a series of 5 µA/cm2, 5-ms current pulses. A single, isolated current pulse evokes a single action potential. The temporal pattern of these current pulses is stochastic and is based on two states: high-activity state and low-activity state. In each simulation, a nominal duration of each state is either specified or selected randomly from a uniform distribution in a range of possible durations. The selected represents the target mean duration of the state. At each time point of the simulation, the transitions between states are selected with a probability calculated based on this target . Similarly, in each state, a nominal current pulse frequency of each state is specified. The selected represents the target mean current pulse frequency. At each time point of the simulation, the probability of initiating a current pulse is calculated based on this target .
In Model 1 (Fig. 3E and Fig. S3E ), Neurons 1 and 2 (presynaptic) both make excitatory synapses on Neuron 3 (postsynaptic). As described above, spiking activity of Neurons 1 and 2 is driven by a series of current pulses whose temporal patterns are organized stochastically into high- and low-activity states with the durations and frequencies given in Table S1. Spiking activity of Neuron 3 is driven by the excitatory synaptic input from Neurons 1 and 2.
In Model 2 (Fig. 3I and Fig. S3F ), Neuron 1 makes an excitatory synapse on Neuron 3, whereas Neuron 2 makes an inhibitory synapse on Neuron 3. As in Model 1, spiking activity of the two presynaptic neurons is excited by a series of current pulses structured stochastically according to high- and low-activity states with the target durations and frequencies given in Table S1. Spiking activity of Neuron 3 is excited by synaptic input from Neuron 1 and inhibited by input from Neuron 2.
Model 3 (Fig. S3B ) is like Model 1 except that there is only one presynaptic neuron (and one excitatory synapse) rather than two.
In Model 4 (Fig. S3C ), a single presynaptic neuron makes an inhibitory synapse on the postsynaptic neuron. Spiking activity of the presynaptic neuron is evoked as in Models 1–3, with the durations and frequencies given in Table S2. Spiking activity of the postsynaptic neuron is driven by a series of current pulses generated in the same manner as for the high activity state described above. The target frequency of the postsynaptic neuron is 50 Hz for the duration of the simulation.
In Model 5 (Fig. S3D ), as in Model 3, a single presynaptic neuron makes an excitatory synapse on the postsynaptic neuron. Action potential oscillations are evoked in the presynaptic neuron by a constant input current of 10 µA/cm2. Phase-locked action potential oscillations are evoked in the postsynaptic neuron by excitatory synaptic drive from the presynaptic neuron. Our algorithm successfully detected that the neurons are functionally connected. However, it did not successfully determine the direction of the interaction. We argue that this is not a fundamental failure of the algorithm, but rather, is due to a lack of directional information contained in the spike data themselves. Because the neurons have phase-locked repetitive spikes, almost all of the spikes in one neuron both precede and follow a spike in the other with short latency.
In Model 6 (Fig. S4A ), we simulate a network of 2–13 neurons using a Hodgkin–Huxley model. We start with a two-neuron model similar to the one in Fig. S3B , with Neuron E sending an excitatory input to Neuron C. We keep this primary synaptic connection from Neuron E to Neuron C with a g syn value of 0.4, because this is a moderate synaptic strength that could be affected by additional neurons in the network. Then, one by one, we add additional excitatory neurons with input projections to Neuron C, and monitor the resulting interaction strength from Neuron E to Neuron C. The additional neurons (numbered 1–11, with a g syn value of 0.2) affect the interaction strength between Neuron E and Neuron C inversely with the number of additional neurons. The additional neurons drive Neuron C, and thus the functional connection from Neuron E to Neuron C is decreased, as expected, because the final activity of Neuron C depends on the entire network, rather than just on Neuron E alone.
In Model 7 (Fig. S4B ), we simulate a three-neuron network using a Hodgkin–Huxley model similar to the one in Fig. 3E . We keep the “primary” excitatory synaptic strength from Neuron E1 at a constant value of g syn = 0.5. Once again, this is a moderate value that shows a strong initial interaction but can be affected by inputs from other neurons. We then change the value of the E2 synaptic strength from g syn = 0 up to g syn = 2. Similar to the multineuron network, we see that with increasing inputs from the nonprimary synapse, we have a decreasing interaction strength in the primary synapse, because the functional connection transfers from the original E1 synapse to the E2 synapse.
To preprocess the data before running the interactivity and decoding algorithm, we bin the spikes using 100-ms bins ( Preprocessing Data for NMHS Model ). We summarize the Hodgkin–Huxley model parameters in Table S2.
Verification Using Music Improvisation.
Three guitarists in a band were instructed to improvise on 12 notes of the A minor pentatonic scale, beginning at A2 and ending at C5. The guitarists were assigned either a leading or following role. The leader was instructed to indicate the rhythm, whereas the followers were instructed to follow the rhythm.
The musicians’ improvisations were recorded through separate channels (Behringer UM2 Audio Interface), with a sampling rate of 48 kHz. The notes were extracted from spectrograms of the recordings by finding the peak power frequency for each time sample and comparing it to the nearest frequency in the A minor pentatonic scale. The notes were then binned into time bins of 3 s, taking the average value of all of the nonzero values if more than one note was played in a time bin, or setting the binned value to zero if more than half of the notes in the bin were zero. The 13 binned values were then separated into quartiles ( Preprocessing Data for NMHS Model ) for a total of four possible emissions.
We then used the NMHS algorithm to estimate the interaction strength and to determine the hidden states for various orchestrations of the band: “connected” (all sequences are from the same recording session), “unconnected” (all three sequences are taken from different recording sessions), or “mixed” (two sequences are taken from one recording and one sequence from another). To verify that the three-node model accounts for the data better than one-node models, we compared the sum of one-node log-likelihoods of HMMs from individual recordings to the log-likelihood of the three-node NMHS model. If the log-likelihood of the three-node model was at least 3% or higher than the one-node model, we used the three-node model, and the one-node model otherwise (Fig. 4 B and C and Fig. S5D ).
Verification Using Social Demographic Data.
To demonstrate the wide range of applications of the NMHS algorithm, we used it to analyze static sociodemographic data and determine interactions between different variables in human populations. To analyze the data, we used sequences of values from four different variables—education, income, physical health, and year in which a house was built—where entry i in the sequence corresponds to the value from person i. Each variable represents a node in the network. As our algorithm looks at the dependence of the current state of the node on the previous state of itself as well as the neighbor, we slightly modified it to instead consider the dependence of the current state of the node on the current state of the neighbor (but still the previous state of itself) (Fig. 4E ). We then applied the two-node NMHS algorithm on every pair of variables and determined the interaction strength.
The target research population was the entire Israeli population aged 18 y or older. The research sample for all of the three presented databases was a representative sample of this population and the subpopulations composing it (Non-Ultra Orthodox Jews, Arabs, Ultra Orthodox Jews, Jewish immigrants from the former Soviet Union, Jewish immigrants from Ethiopia). The sample sizes of the first, second, and third database were, respectively, 1,415, 803, and 947 respondents.
A research institute specializing in sociological surveys collected the data. To minimize the nonresponse rate (and associated with it potential bias) and to improve the representativeness of the sample, up to three callbacks were made in the case of unanswered calls. The data for the first database were collected in May 2015, the second in May 2014, and the third in August to September 2014. The research variables are the following:
-
i)
Education: The level of education of the respondent was determined on the scale from 1 (basic education, did not finish high school) to 7 (doctoral degree or equivalent academic degree) for the first and second databases. For the third database, the scale for measuring education was from 1 (basic education or less) to 8 (master’s degree and above).
-
ii)
Income: The income of the respondent was measured according to five-point scale from 1 (much below the average) to 5 (much above the average).
-
iii)
Physical health: Self-assessment of physical health that appears in the first and second database was measured according to the five-point Likert scale (“For the statement ‘My physical health is good,’ please choose the number from 1 to 5 that most accurately reflects your opinion, when 1 indicates that you do not agree at all and 5 indicates that you very largely agree with the statement”).
-
iv)
A year in which the respondent’s house was built.
We found that the results of our analysis were in close accordance with those of previously conducted experimental studies of the data (2), namely, that there are significant reciprocal relationships between income and education (Fig. 4F ). This example shows that our algorithm can successfully be used to analyze static data with dependent dimensions.
Scalability of the NMHS Model.
The time-complexity of the forward-backward algorithm scales quadratically with the number of parameters in the transition matrix: time required to train the model thus scales exponentially with the number of processes to be modeled. Note, however, that this is an issue because NMHS searches through the entire space of possible interacting models. If S is the set of all elements in a circuit to be characterized, we can represent a set of interactions among the elements as a set of sets C, where each element, R, is a set of nodes in S that represents a causal relation between those nodes. In this case, the number of possible relations is equal to the size of the power set, 2S: the number of possible interactions grows exponentially with the size of the circuit. Higher dimensional NMHS models are not needed unless the circuit contains an interaction involving many processes that cannot be decomposed into simpler interactions. For instances in which interactions can be detected based on the activity of a small number of processes, NMHS is an efficient method of analyzing nonlinear interactions between a small number of elements. Training a three-process NMHS model with 10,000 observations on a 2.6-GHz Core i7 requires ∼5 min using unoptimized MATLAB code; training a two-process NMHS model under the same conditions requires ∼30 s. Optimized code and the use of high performance computing clusters may significantly increase the number of processes feasible to be analyzed. Additionally, we have successfully trained NMHS models using simulated annealing. Stochastic training algorithms, like simulated annealing, might further increase how many processes may be modeled at once.
Comparison with Other Models.
The NMHS model can be expresses as a single, much larger HMM whose transition and emission matrices fulfill certain constraints. In this composite HMM, the states are drawn from the product space of the states of each process in the NMHS model. This composite state represents the individual state of each process in the NMHS model. Given an NMHS model with N processes each with its own collection of states X i , we have:
Note that the transition matrix of each process can be expressed as a function of the model’s current composite state and the state of the process in the next time step. For the transition matrix of the n th process, we write this as . Given N transition matrices, we can construct the composite transition matrix, Ω, with:
where is the probability of transitioning from composite state C1 to composite state C2, and C[n] is the state of the n th process in the composite state. Given this construction, transitions for the state of one process must be conditionally independent from transitions of other processes. That is
Given the set of emissions of each process, Y, the construction of the composite emission, D, and the composite emission matrix, , follows similarly:
The emissions exhibit a similar independence:
The NMHS model has two advantages over the composite model. First, the independence properties of the emission and transition matrices are represented explicitly in the structure of the model rather than implicitly in its values: this makes the model much easier to interpret. Secondly, the NMHS model uses far fewer parameters. The size of the composite emission matrix, , is , whereas the size of the NMHS’s emissions matrices is . The size of the composite transition matrix, Ω, is , compared with NMHS’s .
Other variants of the HMM have been proposed for solving other tasks. The hierarchical HMM (14) was designed for modeling processes that exhibit long-term structure, such as language. The factorial HMM (15) was designed for scenarios in which multiple hidden states govern the behavior of a single process. Similar to the NMHS model, both of these models may be expressed as a larger composite HMM as well: they too are essentially placing constraints on the relationships between the values of the composite HMM. The factorial HMM model bears a great deal of similarity to the NMHS model. Where NMHS uses a multidimensional transition matrix, the factorial HMM uses a multidimensional emission matrix. The factorial HMM may also be trained with a modified Baum–Welch algorithm. Additionally, the factorial HMM exhibits the same scaling difficulties that the NMHS model does: the number of parameters (and therefore the asymptotic running time of the Baum–Welch algorithm) scales exponentially with the number of Markov chains in the model. Ghahramani and Jordan (15) propose two techniques for mitigating this issue: stochastic optimization and the use of variational methods to perform inference on an approximation of the full factorial model. We have performed tests to show that simulated annealing is an effective method of training the NMHS model with two and three processes. Variational methods might prove an effective and efficient way to estimate model parameters and have the appealing theoretical characteristic that each step of the technique is guaranteed to increase a lower bound on the model’s log-likelihood.
NMHS Source Code
The source code, in its state at the time of publication, can be found in the SI Appendix . Please note that the source code is a prototype and is provided as assistance in implementing the algorithm described in the paper. It is not meant to be used out-of-the-box and may contain various bugs. The NMHS code base may continue to undergo development and bug fixes. For the latest code, please refer to the github repository at https://github.com/jfslocum/NMHS.
Copyright Notice
The Massachusetts Institute of Technology (MIT) desires to aid the academic and noncommercial research community while also raising awareness of our technology for potential commercial licensing and therefore agrees herein to grant a limited copyright license to the copyright-protected software for research and noncommercial purposes only, reserving and retaining all ownership rights and license in and to the intellectual property for all other uses.
Thus, subject to the terms herein, MIT grants a royalty-free, nontransferable, nonexclusive license under its copyrights to use, reproduce, display, perform, and modify the software solely for noncommercial research and/or academic testing purposes. To obtain any other license rights, including the right to use the software or filed patents for commercial purposes, you must contact the MIT Technology Licensing Office and enter into additional license agreements with MIT.
Supplementary Material
Acknowledgments
We thank Daniel Gibson and Yasuo Kubota for their help in many aspects of this work. This work was supported by National Institutes of Health Grant R01 MH060379, the Defense Advanced Research Project Agency and US Army Research Office Grant W911NF-10-1-0059, and the Saks Kavanaugh Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the github repository at https://github.com/jfslocum/NMHS.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606280113/-/DCSupplemental.
References
- 1.Granger CWJ. Investigating causal relations by econometric models and cross-spectral methods. Econometrica. 1969;37(3):424–438. [Google Scholar]
- 2.Putnam RD. Our Kids: The American Dream in Crisis. Simon and Schuster; New York: 2015. [Google Scholar]
- 3.Sugihara G, et al. Detecting causality in complex ecosystems. Science. 2012;338(6106):496–500. doi: 10.1126/science.1227079. [DOI] [PubMed] [Google Scholar]
- 4.Kispersky T, Gutierrez GJ, Marder E. Functional connectivity in a rhythmic inhibitory circuit using Granger causality. Neural Syst Circuits. 2011;1(1):9. doi: 10.1186/2042-1001-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oweiss KG. Statistical Signal Processing for Neuroscience and Neurotechnology. Academic; Cambridge, MA: 2010. [Google Scholar]
- 6.Schneidman E, Berry MJ, 2nd, Segev R, Bialek W. Weak pairwise correlations imply strongly correlated network states in a neural population. Nature. 2006;440(7087):1007–1012. doi: 10.1038/nature04701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown EN, Kass RE, Mitra PP. Multiple neural spike train data analysis: State-of-the-art and future challenges. Nat Neurosci. 2004;7(5):456–461. doi: 10.1038/nn1228. [DOI] [PubMed] [Google Scholar]
- 8.Stevenson IH, Kording KP. How advances in neural recording affect data analysis. Nat Neurosci. 2011;14(2):139–142. doi: 10.1038/nn.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneidman E, Bialek W, Berry MJ., 2nd Synergy, redundancy, and independence in population codes. J Neurosci. 2003;23(37):11539–11553. doi: 10.1523/JNEUROSCI.23-37-11539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cadotte AJ, DeMarse TB, He P, Ding M. Causal measures of structure and plasticity in simulated and living neural networks. PLoS One. 2008;3(10):e3355. doi: 10.1371/journal.pone.0003355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabiner LR. A tutorial on hidden Markov-models and selected applications in speech recognition. Proc IEEE. 1989;77(2):257–286. [Google Scholar]
- 12.Abeles M, et al. Compositionality in neural control: An interdisciplinary study of scribbling movements in primates. Front Comput Neurosci. 2013;7:103. doi: 10.3389/fncom.2013.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gat I, Tishby N, Abeles M. Hidden Markov modelling of simultaneously recorded cells in the associative cortex of behaving monkeys. Network-Comp Neural. 1997;8(3):297–322. [Google Scholar]
- 14.Fine S, Singer Y, Tishby N. The hierarchical hidden Markov model: Analysis and applications. Mach Learn. 1998;32(1):41–62. [Google Scholar]
- 15.Ghahramani Z, Jordan MI. Factorial hidden Markov models. Mach Learn. 1997;29(2-3):245–273. [Google Scholar]
- 16.Buesing L, Macke JH, Sahani M. Learning stable, regularised latent models of neural population dynamics. Network. 2012;23(1-2):24–47. doi: 10.3109/0954898X.2012.677095. [DOI] [PubMed] [Google Scholar]
- 17.Quian Quiroga R, Panzeri S. Extracting information from neuronal populations: Information theory and decoding approaches. Nat Rev Neurosci. 2009;10(3):173–185. doi: 10.1038/nrn2578. [DOI] [PubMed] [Google Scholar]
- 18.Burak Y, Rokni U, Meister M, Sompolinsky H. Bayesian model of dynamic image stabilization in the visual system. Proc Natl Acad Sci USA. 2010;107(45):19525–19530. doi: 10.1073/pnas.1006076107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinto DJ, Jones SR, Kaper TJ, Kopell N. Analysis of state-dependent transitions in frequency and long-distance coordination in a model oscillatory cortical circuit. J Comput Neurosci. 2003;15(2):283–298. doi: 10.1023/a:1025825102620. [DOI] [PubMed] [Google Scholar]
- 20.Gläscher J, Daw N, Dayan P, O’Doherty JP. States versus rewards: Dissociable neural prediction error signals underlying model-based and model-free reinforcement learning. Neuron. 2010;66(4):585–595. doi: 10.1016/j.neuron.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stern M, Sompolinsky H, Abbott LF. Dynamics of random neural networks with bistable units. Phys Rev E Stat Nonlin Soft Matter Phys. 2014;90(6):062710. doi: 10.1103/PhysRevE.90.062710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sompolinsky H. Computational neuroscience: Beyond the local circuit. Curr Opin Neurobiol. 2014;25:xiii–xviii. doi: 10.1016/j.conb.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Gutierrez GJ, Marder E. Modulation of a single neuron has state-dependent actions on circuit dynamics. eNeuro. 2014;1(1):ENEURO.0009-14.2014. doi: 10.1523/ENEURO.0009-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman A, et al. A corticostriatal path targeting striosomes controls decision-making under conflict. Cell. 2015;161(6):1320–1333. doi: 10.1016/j.cell.2015.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seth AK. A MATLAB toolbox for Granger causal connectivity analysis. J Neurosci Methods. 2010;186(2):262–273. doi: 10.1016/j.jneumeth.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 26.Schultz W. Predictive reward signal of dopamine neurons. J Neurophysiol. 1998;80(1):1–27. doi: 10.1152/jn.1998.80.1.1. [DOI] [PubMed] [Google Scholar]
- 27.Feingold J, Gibson DJ, DePasquale B, Graybiel AM. Bursts of beta oscillation differentiate postperformance activity in the striatum and motor cortex of monkeys performing movement tasks. Proc Natl Acad Sci USA. 2015;112(44):13687–13692. doi: 10.1073/pnas.1517629112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Destexhe A, Mainen ZF, Sejnowski TJ. Synthesis of models for excitable membranes, synaptic transmission and neuromodulation using a common kinetic formalism. J Comput Neurosci. 1994;1(3):195–230. doi: 10.1007/BF00961734. [DOI] [PubMed] [Google Scholar]
- 29.Destexhe A, Sejnowski TJ. Thalamocortical Assemblies: How Ion Channels, Single Neurons and Large-Scale Networks Organize Sleep Oscillations. Oxford Univ Press; New York: 2001. [Google Scholar]
- 30.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117(4):500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding M, Chen Y, Bressler SL. Granger causality: Basic theory and application to neuroscience. In: Schelter B, Winterhalder M, Timmer J, editors. Handbook of Time Series Analysis: Recent Theoretical Developments and Applications. Wiley; Wienheim, Germany: 2006. pp. 437–495. [Google Scholar]
- 32.Siddiqi SM, Gordon GJ, Moore AW. Fast state discovery for HMM model selection and learning. JMLR Workshop Conf Proc. 2007;2:492–499. [Google Scholar]
- 33.Friedman A, Keselman MD, Gibb LG, Graybiel AM. A multistage mathematical approach to automated clustering of high-dimensional noisy data. Proc Natl Acad Sci USA. 2015;112(14):4477–4482. doi: 10.1073/pnas.1503940112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.