Abstract
De-identification is a shared task of the 2014 i2b2/UTHealth challenge. The purpose of this task is to remove protected health information (PHI) from medical records. In this paper, we propose a novel de-identifier, WI-deId, based on conditional random fields (CRFs). A preprocessing module, which tokenizes the medical records using regular expressions and an off-the-shelf tokenizer, is introduced, and three groups of features are extracted to train the de-identifier model. The experiment shows that our system is effective in the de-identification of medical records, achieving a micro-F1 of 0.9232 at the i2b2 strict entity evaluation level.
Keywords: Protected health information, De-identification, Medical records, Conditional random fields
1. Introduction
Clinical documents contain much valuable information that is significantly attractive to researchers. However, a large amount of protected health information (PHI) exists in clinical documents, and may reveal identities or other personal private information of the patients. To avoid exposing private information, PHI must be removed before researchers do research on these clinical documents. Hence, identifying PHI from clinical documents is essential.
De-identification is the process of identifying and removing PHI from medical records. However, manually de-identifying medical records is time consuming and has significant costs. In 2006, de-identification was first introduced into the Informatics for Integrating Biology and the Bedside (i2b2) project [1]. Automatically removing PHI from discharge summaries was challenging. In this task, Wellner et al.’s method [2] obtained the highest score, using an existing toolkit that was based on conditional random fields (CRFs). Methods for de-identification generally fall into three categories: rule-based methods, machine-learning methods and a combination of the two. Meystre et al. [3] selected 18 automated de-identification systems for different types of clinical documents for detailed analysis. The authors concluded that the rule-based methods perform better with PHI that is rare in clinical documents, whereas machine-learning methods are good at identifying PHI that is not in the dictionaries. For example, “Johnson and Johnson” is a PHI term of ORGANIZATION and appears less frequently in clinical documents, but rule-based methods can identify it as ORGANIZATION rather than NAME; machine-learning methods can strongly associate the word “Johnson” in “Johnson was admitted to hospital yesterday” with PATIENT, although “Johnson” is not in the dictionary of PATIENT. Rule-based methods need experts to construct rules or patterns manually to identify PHI entities. For example, Neamatullah et al. [4] proposed a pattern-matching de-identification system for nursing progress notes, which were obviously less structured than discharge summaries were, using dictionary look-ups, regular expressions and heuristics. In nursing progress notes, technical terminologies, non-standard abbreviations, ungrammatical statements, misspellings, incorrect punctuation characters, and capitalization errors occur frequently. The evaluation showed that their method experienced a high recall rate. Compared with rule-based methods, machine-learning methods such as conditional random fields, support vector machines (SVM) and so on are chosen by many more researchers for de-identification. Zuccon et al. [5] developed a tool to de-identify free-text health records based on conditional random fields. Linguistic features, lexical features, and features extracted by pattern matching were applied to train the classifier. Uzuner et al. [6] used support vector machines and local context to de-identify medical discharge summaries. They created an effective de-identifier and found that local context could improve the effect of de-identification. Boström and Dalianis [7] utilized an active learning method to train a random forest de-identifier on Swedish health records. Szarvas et al. [8] proposed an iterative learning method to de-identify discharge summaries using decision trees with local features and dictionaries. De-identification became a track in i2b2 shared tasks again in 2014. In the de-identification task, many of the teams employed methods using machine learning and rules [9].
Compared with the de-identification task in 2006, medical records and PHI categories in the 2014 i2b2 shared tasks have changed significantly. First, the content in the medical records is not tokenized, and many irregular terms exist in which one token may contain two words. For example, the token “Since6/03/04” actually means “Since 6/03/04”. Furthermore, many abbreviations exist in medical records, which may decrease the precision of the tokenizer and part of speech (POS) tagger. Moreover, the number of PHI categories in i2b2 2014 de-identification is far greater than in 2006. Consequently, de-identification of medical records in i2b2 2014 has become a new challenge for researchers.
In this paper, a new de-identifier of medical records, WI-deId, is proposed based on an implementation of conditional random fields. There has been a significant focus on preprocessing and feature generation from medical records. In the following sections, the details of the preprocessing procedure in WI-deId will be described, and features used in the classifier will be listed. Moreover, the experiment results and future directions of WI-deId will be discussed.
2. Methods
In this study, we utilized the medical records from task 1 of 2014 i2b2/UTHealth shared tasks. The dataset consists of 1,304 medical records annotated for de-identification; 790 of these are used for training, and the remaining 514 are used for testing. The PHI entities have been grouped into 7 main categories and 25 sub-categories, and these medical records have been annotated according to annotation guidelines formulated by specialists [10].
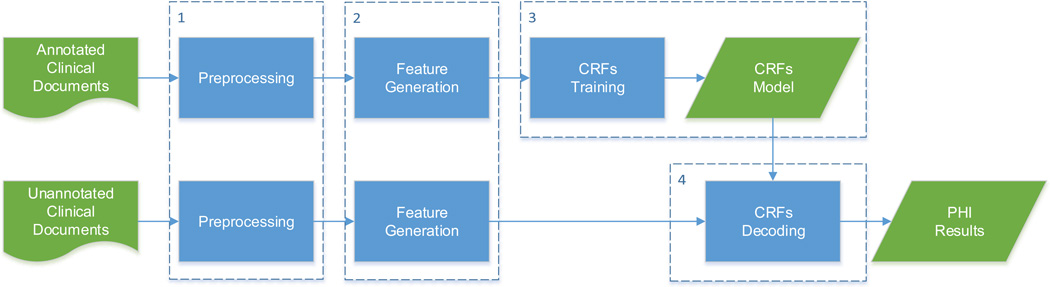
The flow diagram of the WI-deId system is shown in Figure 1. The WI-deId system consists of four main modules: 1) the preprocessing module, 2) the feature generation module, 3) the CRFs training module that trains a model based on the features generated from the second module, and 4) the CRFs decoding module that identifies the PHI entities from unseen data using the model trained in the third module. The details of the WI-deId system will be described in the following sections.
Figure 1.
Flow diagram of the WI-deId system
2.1 Preprocessing of Medical Records
Medical records in the dataset of 2014 i2b2 de-identification task are not tokenized, which makes automatically identifying PHI from the records very difficult. Therefore, focused on tokenization, the preprocessing module was sequentially performed in three steps. First, the sentence boundary detector and tokenizer modules in an existing open source toolkit, OpenNLP1, were utilized to split sentences and tokens in the medical records, respectively. Second, pattern-matching techniques were used to recognize some specific tokens such that when some tokens should be split into two parts, they are split based on the corresponding rules. All regular expressions used in the preprocessing module are listed in Table 1, and how to split the matched tokens is shown in the table. Finally, punctuation characters (“”,:;#*-=/<>[]{}) were segmented from other characters. For example, “2106-02-12” is split as “2016 - 02 - 12”.
Table 1.
Regular Expression Used for Tokenization
| Regular Expression | Whole Token of the Matched Part before Tokenization | after Tokenization |
|---|---|---|
| \d{2}/\d{2}/\d{4} | PEND01/26/2098 | PEND 01/26/2098 |
| \d{2}-year-old | 45-year-old | 45-year-old |
| [A-Za-z]+\d/\d{2} | CABG6/95 | CABG 6/95 |
| [a-z]\d{5} | x76221 | x 76221 |
| [A-Z]\d-\d{4} | X1-1335 | X 1-1335 |
| \d{3}[\-,]\d{3}[\-,]\d{4} | 47798497-045-1949 | 47798 497-045-1949 |
| [A-Z]{2}[a-z][a-z]+ | ALMarital | AL Marital |
| \d[A-Za-z][A-Za-z]+ | 34712RadiologyExam | 34712 RadiologyExam |
| [a-z][a-z]+[A-Z] | JaffreyMarital | Jaffrey Marital |
2.2 Feature Generation
Before model training, the WI-deId system must extract a large number of features from the medical records for training. The features, which are listed in Table 2, can be categorized into three groups: lexical features, orthographic features, and dictionary features.
Table 2.
Features Used in the CRFs Classifier
| Category | Feature | Feature Instantiations at "Valdez" in: Mr. Valdez describes undergoing |
|---|---|---|
| Lexical Features |
lowercase of the token | valdez |
| first four characters of the token | vald | |
| last four characters of the token | ldez | |
| POS tag of the token | NNP | |
| Orthographic Features |
shape of the token | Aaaaaa |
| length of the token | 6 | |
| whether the token contains a letter | 1 | |
| whether the token contains a capital letter | 1 | |
| whether the token begins with a capital letter | 1 | |
| whether all characters in the token are capital letters | 0 | |
| whether the token contains a digit | 0 | |
| whether all characters in the token are digits | 0 | |
| whether the token contains a punctuation character | 0 | |
| whether the token consists of letters and digits | 0 | |
| whether the token consists of digits and punctuation characters | 0 | |
| Dictionary Features |
whether the lowercase of the token is in the “city” dictionary | 1 |
| whether the lowercase of the token is in the “state” dictionary | 0 | |
| whether the lowercase of the token is in the “street” dictionary | 0 | |
| whether the lowercase of the token is in the “country” dictionary | 0 | |
| whether the lowercase of the token is in the “date” dictionary | 0 |
Lexical features contain the lowercase of the token and the first and last four characters of it [11]. These features can help the classifier “memorize” the categories of some tokens, which is necessary because some tokens often belong to one of the PHI categories, but some often belong to a non-PHI category. For instance, “April” is always recognized as a PHI term, and “at” is always identified as a non-PHI term. Moreover, the POS tag of a token is always very helpful in named entity recognition. WI-deId found the POS tag using the tool GENIA [12], which was trained on biomedical text, instead of using OpenNLP, whose POS tagger is trained on open domain corpus.
Tokens that are similar in shape may be classified into the same PHI category. We replaced uppercase letters, lowercase letters and digits in a token by “A”, “a” and “0”, respectively. Length of a token is a significant feature of tokens in some PHI categories. For example, the tokens in ZIP, AGE, and PHONE are mostly fixed length. Information about capital letters can help us identify PATIENT, DOCTOR and other PHI entities that mostly begin with a capital letter such as “Nick” and “Hayes”. Digit-related features can aid in identification of the PHI entities that contain digits such as ZIP, AGE, and DATE. Moreover, PHONE, DATE and some other PHI categories can benefit from punctuation features because there usually is a punctuation character in these PHI entities. Round brackets occur often in the entities of PHONE, such as “(784) 032-8966”, and “-” or “/” frequently appears in DATE.
In addition, we extracted PHI entities of CITY, STATE, STREET, COUNTRY, and DATE in the medical records for training and combined them with webpages2 of city, state, and country to generate the dictionaries. In these dictionaries, all of the elements are tokens, not phrases, and all of the tokens are lowercased. For example, “Los Angeles” is a PHI term of CITY, but “los” and “angeles” are separated elements in the dictionary of CITY. Dictionary features are generated by judging whether the lowercase of the token is in the dictionary. Note that the dictionary features cannot classify one token into its dictionary type directly; this function is different from post-processing. If one token belonging to COUNTRY does not appear in the training data, it will not be classified into COUNTRY in the testing data, although the token is one element of the COUNTRY dictionary. This behavior may weaken the effect of the dictionaries, but it also reduces problems caused when a token belongs to two or more dictionaries simultaneously.
In the WI-deId system, all of the above-mentioned features of the tokens within a ±2 context window of the current token are considered.
2.3 The CRFs Classifier
The proposed classifier was based on the conditional random fields algorithm which was widely used in named entity recognition. Given an observed sequence of tokens, x = x1x2 ⋯ xn, a CRF predicts a corresponding sequence of labels, y* = y1y2 ⋯ yn. y* maximizes the conditional probability P(y|x) for all y in the set of possible label sequences [13].
De-identification aims to automatically identify the boundaries of PHI entities and assign the PHI categories to them. The existing open source toolkit CRF++3 is utilized to classify the tokens in a sequence into the BIO scheme. “B” indicates a token is the beginning of a PHI entity, whereas “I” shows that a token is inside of a PHI entity, and “O” means that a token does not belong to any category of the PHI entities. In this classifier, the data files for training and testing using CRF++ are obtained in two steps. The first step is the preprocessing of medical records, which is described in Section 2.1. Then, the training and testing data files are created based on the features shown in Section 2.2. A CRFs model considering all of the categories can be learned after training on the training data file, and the tokens in the testing data file can be classified into one of these categories using the model.
3. Experiments
3.1 Evaluation
De-identification can be evaluated at either the entity level or token level [9]. Entity level evaluation contains two metrics: the "strict" metric measures whether the position and the type of a PHI result exactly matches the gold standard; the “relaxed” metric loosens the position constraint, allowing the end position of a PHI result to be off by two characters. The token level metric's constraint is less than the entity-level metrics. If a PHI entity in the gold standard is a phrase comprising several tokens and the system identifies these tokens as separate entities of this PHI's type, the system output is considered correct. For example, “Newton Hospital” is a single entity in the gold standard, but the system annotated “Newton” and “Hospital” as two separate entities. The token level metric treats this system annotation as correct when type attributes match. There are two sets of PHI categories: the i2b2 PHI categories, and the PHI categories that are defined by the Health Insurance Portability and Accountability Act (HIPAA). The i2b2 PHI categories contain some PHI types that are not included in HIPAA. The performance of the system is evaluated by calculating the precision, recall and F-measure of the metrics, which are composed of the entity level and token level metrics of the two category sets. The formulas of the precision, recall, and F-measure are shown in Eqs. (1)-(3), and they are evaluated at a micro level and at a macro level (micro means all of the PHI tags in the dataset are evaluated together; macro indicates that all of the PHI tags in each medical record are evaluated, then all the evaluations of the medical records in the dataset are averaged). The micro F-measure of i2b2 strict metric is chosen to rank in this task.
| (1) |
| (2) |
| (3) |
3.2 System Results
Using the evaluation metrics described above, the best official submission and two unofficial development runs are shown in Table 3. The best official run of the WI-deId system ranked fourth in the de-identification task of i2b2 2014, and the best unofficial development run (unofficial run2) would have ranked second. The details of the best unofficial system are described in Section 2. The following analysis in Sections 3.3 and 3.4 is also based on the best unofficial system.
Table 3.
Best Official Run and Two Unofficial Runs of the WI-deId System
| Best Official Run | Unofficial Run1 | Unofficial Run2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Micro P | Micro R | Micro F | Micro P | Micro R | Micro F | Micro P | Micro R | Micro F | |
| i2b2 Token | 0.9571 | 0.9051 | 0.9304 | 0.9664 | 0.9287 | 0.9471 | 0.9733 | 0.9304 | 0.9514 |
| i2b2 Strict | 0.9229 | 0.8505 | 0.8852 | 0.9468 | 0.8909 | 0.9180 | 0.9561 | 0.8925 | 0.9232 |
| i2b2 Relaxed | 0.9252 | 0.8526 | 0.8874 | 0.9485 | 0.8925 | 0.9196 | 0.9586 | 0.8949 | 0.9256 |
| HIPAA Token | 0.9708 | 0.9371 | 0.9536 | 0.9805 | 0.9541 | 0.9671 | 0.9844 | 0.9551 | 0.9695 |
| HIPAA Strict | 0.9414 | 0.8957 | 0.9180 | 0.9645 | 0.9305 | 0.9472 | 0.9695 | 0.9294 | 0.9490 |
| HIPAA Relaxed | 0.9444 | 0.8986 | 0.9209 | 0.9662 | 0.9322 | 0.9489 | 0.9724 | 0.9322 | 0.9519 |
Compared with the best unofficial system, the best official system has some differences in the preprocessing module and feature generation module. The best official system preprocessed the medical records without using pattern-matching techniques and generated only a part of the features listed in Section 2.2. The features generated in the best official system included current token, POS tag of current token, two previous tokens, POS tag of two previous tokens, two next tokens, POS tag of two next tokens, length of current token, whether the current token contains a digit, whether the current token contains a letter, whether the current token begins with a capital letter and whether the current token contains a punctuation character. Different from unofficial system2, unofficial system1 did not use the following features: the orthographic and dictionary features of the two previous tokens and the orthographic and dictionary features of the next two tokens.
As shown in Table 3, it is clear that the evaluation results of the HIPAA PHI categories are better than the results of the i2b2 PHI categories and that the token level outcomes exceed the corresponding entity level outcomes.
Table 4 lists the details of each PHI type in the best unofficial system output at the i2b2 strict entity evaluation level. In the seven main categories, NAME, DATE, AGE, CONTACT, and IDs received high F-measures of above 0.9, but PROFESSION had a quite low F-measure of 0.5929. Compared with the other main categories, the numbers of PHI terms of PROFESSION and CONTACT in the training dataset are obviously less, particularly PROFESSION. The lack of training instances is the fundamental reason for the poor performance in the PROFESSION category, and it also causes the low F-measures of some sub-categories such as ORGANIZATION, COUNTRY, LOCATION-OTHER, FAX, EMAIL and DEVICE. As is shown in Table 4, PROFESSION, HOSPITAL, STATE, COUNTRY, and LOCATION-OTHER achieved F-measures below 0.9; these categories do not belong to the HIPAA PHI categories. Consequently, the overall result of the i2b2 PHI categories is worse than that of the HIPAA PHI categories. In addition to the weak identification results for some PHI types, the WI-deId system also had good performance in several PHI types. For example, the F-measures of USERNAME, ZIP, DATE, and MEDICALRECORD exceeded 0.97.
Table 4.
Evaluations of Each PHI Type at the i2b2 Strict Entity Evaluation Level
| #Train | #Gold | #System | #Agree | P | R | F | ||
|---|---|---|---|---|---|---|---|---|
| NAME | PATIENT | 1316 | 879 | 761 | 729 | 0.9580 | 0.8294 | 0.8890 |
| DOCTOR | 2885 | 1912 | 1789 | 1676 | 0.9368 | 0.8766 | 0.9057 | |
| USERNAME | 264 | 92 | 89 | 88 | 0.9888 | 0.9565 | 0.9724 | |
| Overall | 4465 | 2883 | 2639 | 2552 | 0.9670 | 0.8852 | 0.9243 | |
| PROFESSION | 234 | 179 | 101 | 83 | 0.8218 | 0.4637 | 0.5929 | |
| LOCATION | HOSPITAL | 1437 | 875 | 737 | 673 | 0.9132 | 0.7691 | 0.8350 |
| ORGANIZATION | 124 | 82 | 22 | 21 | 0.9545 | 0.2561 | 0.4038 | |
| STREET | 216 | 136 | 125 | 120 | 0.9600 | 0.8824 | 0.9195 | |
| CITY | 394 | 260 | 257 | 218 | 0.8482 | 0.8385 | 0.8433 | |
| STATE | 314 | 190 | 171 | 160 | 0.9357 | 0.8421 | 0.8864 | |
| COUNTRY | 66 | 117 | 75 | 67 | 0.8933 | 0.5726 | 0.6979 | |
| ZIP | 212 | 140 | 133 | 133 | 1.0000 | 0.9500 | 0.9744 | |
| OTHER | 4 | 13 | 0 | 0 | 0.0000 | 0.0000 | 0.0000 | |
| Overall | 2767 | 1813 | 1520 | 1448 | 0.9526 | 0.7987 | 0.8689 | |
| AGE | 1233 | 764 | 696 | 667 | 0.9583 | 0.8730 | 0.9137 | |
| DATE | 7502 | 4980 | 4934 | 4831 | 0.9791 | 0.9701 | 0.9746 | |
| CONTACT | PHONE | 309 | 215 | 213 | 202 | 0.9484 | 0.9395 | 0.9439 |
| FAX | 8 | 2 | 0 | 0 | 0.0000 | 0.0000 | 0.0000 | |
| 4 | 1 | 0 | 0 | 0.0000 | 0.0000 | 0.0000 | ||
| Overall | 321 | 218 | 213 | 203 | 0.9531 | 0.9312 | 0.9420 | |
| IDs | MEDICALRECORD | 611 | 422 | 422 | 411 | 0.9739 | 0.9739 | 0.9739 |
| DEVICE | 7 | 8 | 0 | 0 | 0.0000 | 0.0000 | 0.0000 | |
| IDNUM | 261 | 195 | 175 | 151 | 0.8629 | 0.7744 | 0.8162 | |
| Overall | 879 | 625 | 597 | 568 | 0.9514 | 0.9088 | 0.9296 |
The “Overall” in each main category lists the performance of the corresponding main category. To calculate the “Overall” performance of the main category, all of the instances of the sub-categories in this main category are combined as instances of the main category. (The sub-categories whose count is zero in the gold standard dataset are not listed in this table.)
3.3 Comparison
Two groups of experiments were designed to explain the effectiveness of the preprocessing module and the feature sets used in the WI-deId system. In the first group, as a comparison system, the preprocessing module was removed from the WI-deId system. The second group was for analyzing the contribution of the three feature sets to the system performance by adding the feature sets in a greedy way.
The experimental result of the first group is shown in Table 5. Compared with WI-deId, the performance of the system without the preprocessing module dropped significantly. All of the F-measures of the comparison system were lower than WI-deId, particularly AGE, whose F-measure decreased more than 20 percent. This experiment shows the importance of the preprocessing module in the system.
Table 5.
Performance of the First Experiment at the i2b2 Strict Entity Evaluation Level
| F-measure | ||||||||
|---|---|---|---|---|---|---|---|---|
| NAME | PROFESSION | LOCATION | AGE | DATE | CONTACT | IDs | Total | |
| Comparison System | 0.8753 | 0.4314 | 0.8255 | 0.6999 | 0.9140 | 0.9296 | 0.9125 | 0.8638 |
| WI-deId | 0.9243 | 0.5929 | 0.8689 | 0.9137 | 0.9746 | 0.9420 | 0.9296 | 0.9232 |
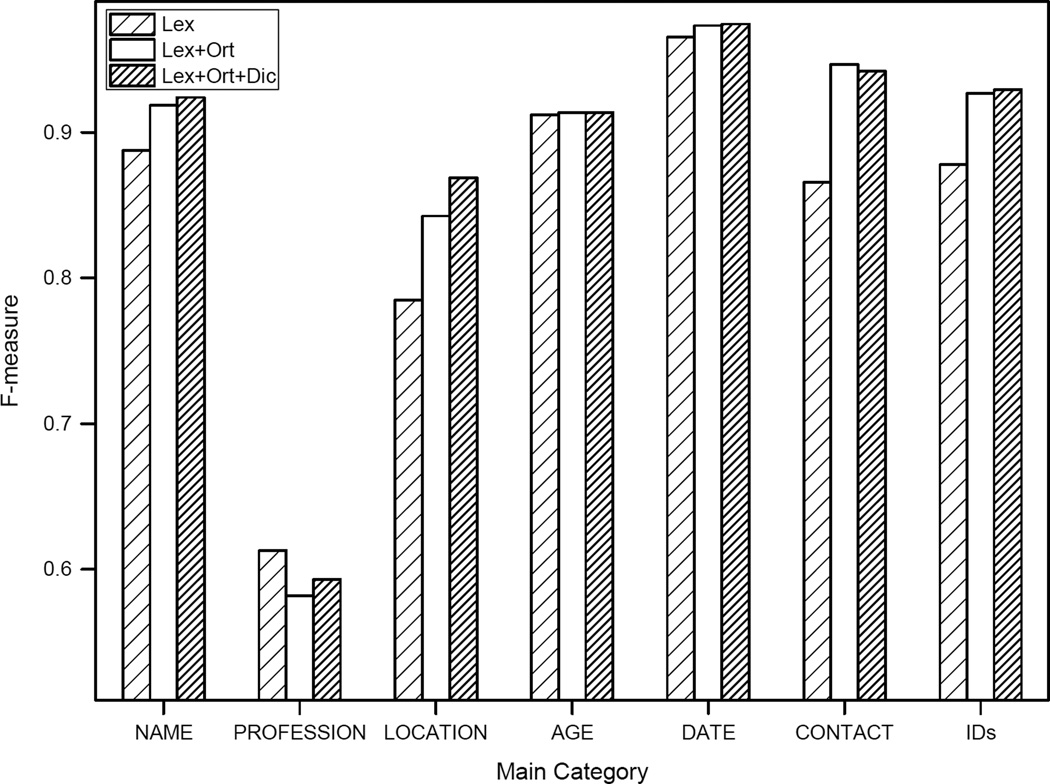
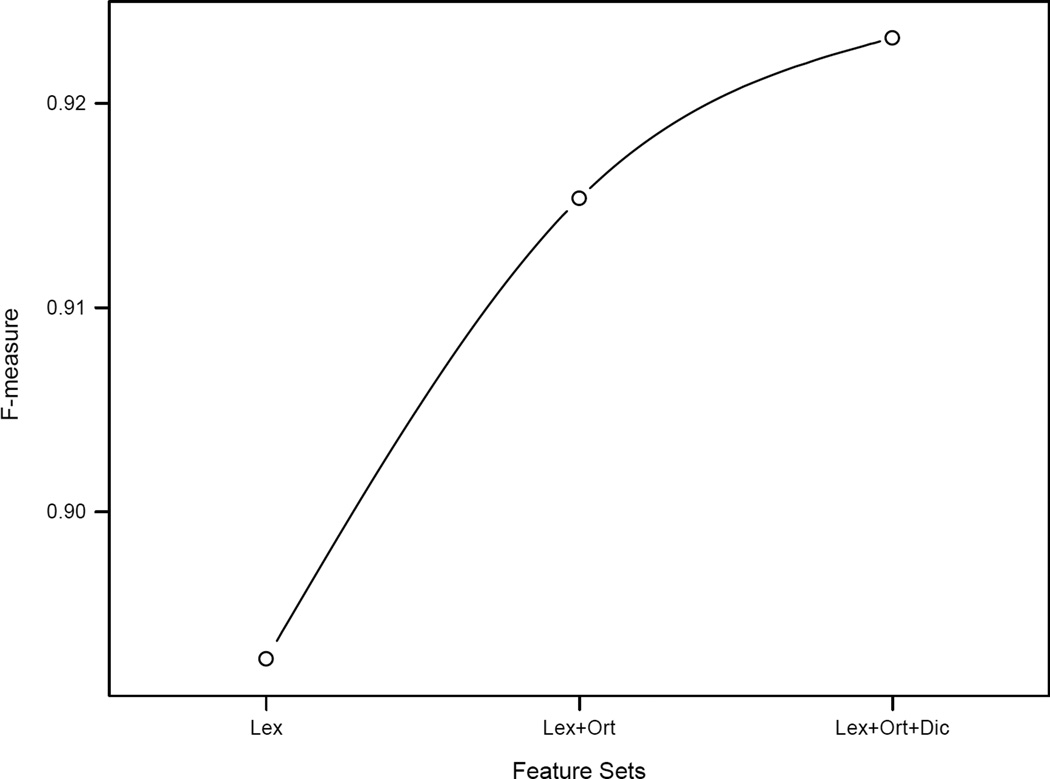
Figure 2 shows F-measures of the 7 main categories using three different feature sets (Lex, using lexical features; Lex+Ort, using lexical features and orthographic features; and Lex+Ort+Dic, using lexical features, orthographic features, and dictionary features). The major contribution of orthographic features lies in the performance of LOCATION, CONTACT, and IDs, which increased more than 5, 8, and 4 percent in the F-measure, respectively. Dictionary features focuses the contribution to the system result on LOCATION, which accords with the definition of those dictionaries. The curve shown in Figure 3 describes the improvement of the overall system performance by adding the feature sets.
Figure 2.
Performance of main categories using different feature sets at the i2b2 strict entity evaluation level
Figure 3.
Overall performance using different feature sets at the i2b2 strict entity evaluation level
3.4 Error Analysis
The errors in the WI-deId system were analyzed according to the error analysis method in [2]. In this method, errors were divided into four groups: type errors (entity is correct but type is wrong), extent errors (entity in system output has an additional or a missing part), missing errors (entity is in the gold standard but not in the system output) and spurious errors (entity is in the system output but not in the gold standard).
Table 6 lists the error distribution of the WI-deId system at the i2b2 strict entity evaluation level according to the four groups of errors. Missing errors constitute the highest proportion, 64.42%, causing the recall of the WI-deId system to be relatively lower than the precision. Furthermore, missing errors will expose the protected health information of the patients; thus, these are more serious than other errors.
Table 6.
Error Distribution of the WI-deId System at the i2b2 Strict Entity Evaluation Level
| Error Number | Percentage | |
|---|---|---|
| Type Error | 159 | 12.04% |
| Extent Error | 202 | 15.29% |
| Missing Error | 851 | 64.42% |
| Spurious Error | 109 | 8.25% |
Table 7 shows the details of errors in groups by type errors, missing errors and spurious errors. The maximum number of type errors was produced by PATIENT and DOCTOR. Forty-four PATIENT entities were identified as DOCTOR, whereas 14 DOCTOR entities were marked as PATIENT. It is most difficult to distinguish between PATIENT and DOCTOR because they not only are similar in spelling but also belong to the same main category, NAME. Missing PHI entities account for 7.4% of the whole gold standard PHI entities (the number of missing PHI entities is 851, and the number of gold standard PHI entities is 11462). Moreover, PROFESSION, ORGANIZATION, LOCATION-OTHER, FAX, EMAIL and DEVICE have a missing percentage (calculated by dividing the missing number by the total number in each row in Table 7) of above 50%. COUNTRY also has a relatively high missing percentage. These are largely caused by the lack of training instances; for example, “Dutch” and “Finland” are PHI terms of COUNTRY in the gold standard but do not occur in training data as PHI. All of these characteristics lead to the low recall rate of the WI-deId system. Compared with missing errors, the proportions of spurious errors generated in the PHI categories are much lower. The highest occurrence rate of spurious errors, up to 9.6%, appears on PROFESSION.
Table 7.
Type Errors, Missing Errors, and Spurious Errors in the WI-deId System at the i2b2 Strict Entity Evaluation Level
| System Output |
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pa | Do | Us | Pr | Ho | Or | Str | Ci | Sta | Co | Zi | Ot | Ag | Da | Ph | Fa | Em | Me | De | Id | Missing | Total | |
| Pa | 729 | 44 | 1 | 2 | 1 | 82(9.5%) | 859 | |||||||||||||||
| Do | 14 | 1676 | 1 | 1 | 173(9.3%) | 1865 | ||||||||||||||||
| Us | 1 | 88 | 3(3.3%) | 92 | ||||||||||||||||||
| Pr | 83 | 88(51.5%) | 171 | |||||||||||||||||||
| Ho | 5 | 673 | 9 | 165(19.4%) | 852 | |||||||||||||||||
| Or | 1 | 1 | 9 | 21 | 6 | 39(50.6%) | 77 | |||||||||||||||
| Str | 120 | 7(5.5%) | 127 | |||||||||||||||||||
| Ci | 2 | 4 | 6 | 218 | 3 | 21(8.3%) | 254 | |||||||||||||||
| Sta | 2 | 10 | 160 | 14(7.5%) | 186 | |||||||||||||||||
| Co | 1 | 3 | 3 | 4 | 67 | 37(32.2%) | 115 | |||||||||||||||
| Zi | 133 | 6 | 1 | 0 | 140 | |||||||||||||||||
| Ot | 2 | 2 | 0 | 9(69.2%) | 13 | |||||||||||||||||
| Ag | 667 | 85(11.3%) | 752 | |||||||||||||||||||
| Da | 1 | 1 | 4831 | 1 | 88(1.8%) | 4922 | ||||||||||||||||
| Ph | 1 | 202 | 2 | 5(2.4%) | 210 | |||||||||||||||||
| Fa | 1 | 0 | 1(50.0%) | 2 | ||||||||||||||||||
| Em | 0 | 1(100%) | 1 | |||||||||||||||||||
| Me | 411 | 4 | 1(0.2%) | 416 | ||||||||||||||||||
| De | 8(100%) | 8 | ||||||||||||||||||||
| Id | 1 | 2 | 151 | 24(13.5%) | 178 | |||||||||||||||||
| Spurious | 1(0.1%) | 13(0.7%) | 1(1.1%) | 9(9.6%) | 19(2.7%) | 0 | 0 | 4(1.6%) | 2(1.2%) | 1(1.4%) | 0 | 0 | 16(2.3%) | 38(0.8%) | 2(0.9%) | 0 | 0 | 2(0.5%) | 0 | 1(0.6%) | 109 | |
| Total | 747 | 1748 | 89 | 94 | 712 | 21 | 120 | 255 | 166 | 72 | 133 | 0 | 683 | 4872 | 211 | 0 | 0 | 416 | 0 | 159 | 851 | |
The first two letters are used to represent all PHI types except for STREET and STATE, for which the first three letters are used. Type errors are bed, and missing errors and spurious errors are in i text. (The sub-categories whose count is zero in the gold standard dataset are not listed in this table.)
Table 8 shows the extent errors that occurred in system output. In this table, “Short” indicates a PHI term produced by a system that is a part of the corresponding PHI term in the gold standard. However, “Long” shows that a PHI term produced by the system exceeds the span of the corresponding PHI term in the gold standard, and “S&L” means that a PHI term produced by the system does not capture the entire text of the corresponding PHI term in the gold standard but spans more text. Almost one-third of the extent errors occurred on DATE, with 42 short errors and 20 long errors. For example, “5 August 2060” is a PHI term of DATE in the gold standard but the WI-deId system identified “August 2060” as DATE. This difference leads to a short error. Similarly, the system tagged “November 1 morning” as DATE, but only “November” is a PHI term in the gold standard. This difference generated a long error. Among the three groups of extent errors, “S&L” errors occurred the least, only 10 times, but “Short” errors accounted for more than half, up to 114 times. Similar to missing errors, “Short” errors will disclose a portion of protected health information, which is harmful to the patients.
Table 8.
Extent Errors in the Output of the WI-deId System at the i2b2 Strict Entity Evaluation Level
| Pa | Do | Pr | Ho | Or | Str | Ci | Sta | Co | Ag | Da | Ph | Me | Id | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Short | 9 | 21 | 4 | 7 | 0 | 1 | 2 | 5 | 3 | 9 | 42 | 1 | 3 | 7 | 114 |
| Long | 5 | 18 | 3 | 15 | 1 | 0 | 0 | 0 | 0 | 4 | 20 | 1 | 3 | 8 | 78 |
| S&L | 0 | 2 | 0 | 3 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 10 |
| Total | 14 | 41 | 7 | 25 | 1 | 5 | 2 | 5 | 3 | 13 | 62 | 2 | 6 | 16 | 202 |
The first two letters are used to represent all PHI types except for STREET and STATE, for which the first three letters are used. (The sub-categories whose count is zero in the extent errors of system output are not listed in this table.)
Because tokenization is a key processing procedure in the WI-deId system, it also caused some errors in the system output. For example, "Sergio-Steven" is a PHI entity of PATIENT in the gold standard; it was tokenized into “Sergio - Steven” after preprocessing. Ultimately, the system identified “Sergio” as a PHI entity of PATIENT.
4. Conclusion
This work presents a system that is completely machine-learning-based; it uses neither a rule-based method nor a post-processing module. In this de-identification task, we achieved a micro F-measure of 0.8852 at the i2b2 strict entity evaluation level, which ranked fourth. After this task, the F-measure was promoted to 0.9232, which would have ranked second in the challenge rankings, and the content of this paper is the description of the improved system. The preprocessing of the corpus plays an extremely crucial role in the process of de-identification. The features chosen further enhance the performance of the WI-deId system. We did not use a sentence boundary detector or a tokenizer trained on clinical text; therefore, these domain-restricted tools can be used to update the preprocessing module in our system. Because the text in the medical record has a fixed width, a large proportion of the sentences are cut off, which destroys the integrity of the sentences and decreases the precision of the POS tagger. Future work should attempt to recover the integrity of these sentences.
Acknowledgments
The medical records used in this paper were provided by Partners HealthCare, and 2014 i2b2/UTHealth NLP Shared Tasks were supported by the following grants: NIH NLM (2U54LM008748, 5R13LM011411), NIH NIGMS (5R01GM102282). Thank you to the organizing committee of i2b2 and the annotators of the dataset. We are also thankful for the reviewers’ comments, which improved our paper significantly.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Uzuner Ö, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. Journal of the American Medical Informatics Association. 2007;14:550–563. doi: 10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wellner B, Huyck M, Mardis S, Aberdeen J, Morgan A, Peshkin L, et al. Rapidly retargetable approaches to de-identification in medical records. Journal of the American Medical Informatics Association. 2007;14:564–573. doi: 10.1197/jamia.M2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meystre SM, Friedlin FJ, South BR, Shen S, Samore MH. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC medical research methodology. 2010;10:70. doi: 10.1186/1471-2288-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neamatullah I, Douglass MM, Li-wei HL, Reisner A, Villarroel M, Long WJ, et al. Automated de-identification of free-text medical records. BMC medical informatics and decision making. 2008;8:32. doi: 10.1186/1472-6947-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuccon G, Kotzur D, Nguyen A, Bergheim A. De-identification of health records using Anonym: Effectiveness and robustness across datasets. Artificial intelligence in medicine. 2014;61:145–151. doi: 10.1016/j.artmed.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Uzuner Ö, Sibanda TC, Luo Y, Szolovits P. A de-identifier for medical discharge summaries. Artificial intelligence in medicine. 2008;42:13–35. doi: 10.1016/j.artmed.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boström H, Dalianis H. De-identifying health records by means of active learning. Recall (micro) 2012;97:97.0. [Google Scholar]
- 8.Szarvas G, Farkas R, Busa-Fekete R. State-of-the-art anonymization of medical records using an iterative machine learning framework. Journal of the American Medical Informatics Association. 2007;14:574–580. doi: 10.1197/j.jamia.M2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stubbs A, Kotfila C, Xu H, Uzuner Ö. Practical applications for NLP in Clinical Research: the 2014 i2b2/UTHealth shared tasks. Unpublished results. [Google Scholar]
- 10.Stubbs A, Uzuner Ö. De-identifying longitudinal medical records. Unpublished results. [Google Scholar]
- 11.Lv X, Guan Y, Deng B. Transfer learning based clinical concept extraction on data from multiple sources. Journal of biomedical informatics. 2014;52:55–64. doi: 10.1016/j.jbi.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Tsuruoka Y, Tateishi Y, Kim J-D, Ohta T, McNaught J, Ananiadou S, et al. Developing a robust part-of-speech tagger for biomedical text. Advances in informatics: Springer. 2005:382–392. [Google Scholar]
- 13.Lafferty J, McCallum A, Pereira FC. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. 2001 [Google Scholar]