Abstract
Many proteins contain intrinsically disordered regions (IDRs) lacking stable secondary and ordered tertiary structure. IDRs are often implicated in macromolecular interactions, and may undergo structural transitions upon binding to interaction partners. However, as binding partners of many protein IDRs are unknown, these structural transitions are difficult to verify and often are poorly understood. In this study we describe a method to identify IDRs that are likely to undergo helical transitions upon binding. This method combines bioinformatics analyses followed by circular dichroism spectroscopy to monitor 2,2,2-trifluoroethanol (TFE)-induced changes in secondary structure content of these IDRs. Our results demonstrate that there is no significant change in the helicity of IDRs that are not predicted to fold upon binding. IDRs that are predicted to fold fall into two groups: one group does not become helical in the presence of TFE and includes examples of IDRs that form β-strands upon binding, while the other group becomes more helical and includes examples that are known to fold into helices upon binding. Therefore, we propose that bioinformatics analyses combined with experimental evaluation using TFE may provide a general method to identify IDRs that undergo binding-induced disorder-to-helix transitions.
Keywords: intrinsically disordered region; protein-protein interaction; Anchor region; circular dichroism; disorder-to-helix transition; 2,2,2-trifluoroethanol
Graphical abstract
1. Introduction
The canonical view that protein structure dictates function requires a more nuanced understanding in light of the growing number of proteins recognized to bear intrinsically disordered regions (IDRs). IDRs lack the ability to form stable secondary structures such as α-helices and β-strands, and well-packed tertiary structures [1, 2]. IDRs tend to have sequences of lower complexity and an increased number of polar or charged residues rather than hydrophobic residues likely to pack in the interior of a protein [1, 3]. IDRs also tend to have low sequence conservation [4, 5]. They appear to be especially prevalent in eukaryotes, where at least 35-50% of proteins are predicted to bear IDRs [5, 6]. This prevalence implies that IDRs must confer some important evolutionary advantage in advanced organisms.
IDRs are increasingly recognized to mediate many functions essential for life. They appear to regulate protein lifetime by providing targets for proteolysis [7, 8]. Perhaps their most common function is to mediate macromolecular interactions, including protein-protein, protein-DNA, and protein-RNA interactions [2]. The low sequence conservation and conformational flexibility inherent in IDRs may provide IDRs with the evolutionary flexibility required to facilitate multiple protein-protein interactions [2, 5]. Consequently, IDRs are common among interaction hub proteins (regulatory and signaling proteins that bind a variety of diverse partners) [9]. IDRs often also serve as sites of phosphorylation, and in turn, phosphorylation appears to modulate interactions with other macromolecules [10-12]. Therefore, it is important to understand the structure-function relationship of IDRs and IDR-mediated binding.
Often, binding to appropriate partners appears to stabilize secondary structure elements within IDRs [1, 13], although it has recently been shown that up to one third of all residues are disordered in protein complexes [14]. Regions that have experimentally been shown to undergo disorder-to-order transitions have been called Molecular Recognition Features (MoRFs) [15, 16]. MoRFs have been further classified as α-MoRFs, β-MoRFs or σ-MoRFs, depending on whether they fold into α-helices, β-strands or stable coils, respectively. Understanding the structures of these regions in both unbound and bound states is important for understanding the mechanism of biological function of these regions, and ultimately for rational design of therapeutics targeting these interactions [13]. However, binding partners remain unknown for most IDRs, complicating experimental elucidation of structural transitions within IDRs. We hypothesized that it would be possible to identify α-MoRFs by bioinformatics analyses using programs such as ANCHOR [17] and DisoRDPbind [18] combined with experimental evaluation of chemically-induced structural transitions.
Therefore, in this study our goal was to develop a method to identify α-MoRFs in the absence of known binding partners. We first performed a bioinformatics analysis using the programs IsUnstruct [19], PONDR-FIT [20] and IUPred [21] to predict IDRs, then used the programs ANCHOR [17] and DisoRDPbind [18] to identify IDRs that are likely to be stabilized in ordered structures upon binding. JPred4 [22] was used to predict potential secondary structure. We used circular dichroism (CD) spectroscopy to monitor change in secondary structure content of diverse IDRs induced by 2,2,2-trifluoroethanol (TFE), which is known to stabilize α-helical structure in disordered amino acid sequences that have some native tendency to form helices [23-27]. Our results demonstrate that IDRs that are not predicted by ANCHOR to fold upon binding do not become helical in the presence of TFE. IDRs that are predicted to fold fall into two groups. One group becomes more helical upon addition of TFE, and includes all the known α-MoRFs that were included in our study. The other group does not become helical and includes examples of β-MoRFs. Therefore, we propose that predictions using programs such as ANCHOR combined with experimental evaluation using TFE may provide a general method to identify IDRs that undergo a binding-induced disorder-to-helix transition.
2. Materials and methods
2.1. Production of IDRs
Peptides corresponding to the selected IDRs (Table 1) were chemically synthesized, HPLC purified to >90% purity, and purity confirmed by electrospray mass spectrometry (RS Synthesis or EZBioLabs). For each peptide, a 1 mM stock solution in 10 mM phosphate pH 7.4, 100 mM ammonium sulfate was prepared.
Table 1.
IDR sequences used in this study
| Protein | Accession # |
Peptide Sequence |
|---|---|---|
| TRAP220 | Q925J9 | 637GNTKNHPMLMNLLKDNPA654 |
| BECN1 | Q14457 | 105DGGTMENLSRRLKVTGDLFDIMSGQT130 |
| GRIM | Q24570 | 1MAIAYFIPDQAQLLA15 |
| p53 | P04637 | 367SHLKSKKGQSTSRHKKLMFKTE388 |
| GOPC | Q9HD26 | 248RHKTVIRACRGRNDLKRPMQAPPGHDQDSL277 |
| ATG3 | Q9NT62 | 129ENKDNIRLQDCSALCEEEEDEDEGEAADME158 |
| ATG9A | Q7Z3C6 | 810 AEDGQSASRHPEPVPEEGSEDELPPQVHKV 839 |
| BECN1 | Q14457 | 76DGVSRRFIPPARMMSTESANSFTLIGEASD105 |
| AMBRA1 | Q9C0C7 | 527TQQAQEMLNNNIESE541 |
| GOPC | Q9H D26 | 255ACRGRNDLKRPMQAPPGHDQDSLKK279 |
| BECN1 | Q14457 | 50AQAKPGETQEEETNSGEEPFIETPRQDGV78 |
| ATG16L1 | Q676U5 | 59EKHDVPNRHEISPGHDGTWNDNQLQE84 |
| ATG2A | Q7Z 3C6 | 1244HPPPRPPSPTEIAGQKLSESPASLPSCPPV1273 |
Accession # corresponds to the full-length amino acid sequence downloaded from NCBI RefSeq protein database (http://www.ncbi.nlm.nih.gov/refseq).
2.2. Prediction of disorder and likelihood of folding upon binding
The amino acid sequences of the full-length proteins containing the thirteen selected IDRs were downloaded from the NCBI RefSeq protein database (http://www.ncbi.nlm.nih.gov/refseq) (Table 1). These sequences were analyzed with IsUnstruct (http://bioinfo.protres.ru/IsUnstruct/) [19], PONDR-FIT (http://www.disprot.org/metapredictor.php) [20], and IUPred (http://iupred.enzim.hu) [21] to predict whether the selected peptide sequences were intrinsically disordered (Table 2). The sequences were also analyzed with ANCHOR (http://anchor.enzim.hu) [17] and DisoRDPbind (http://biomine-ws.ece.ualberta.ca/DisoRDPbind/index.php) [18] to predict whether the selected IDRs were likely to undergo binding-associated structure stabilization (Table 2). Finally, these sequences were analyzed with Jpred4 (http://www.compbio.dundee.ac.uk/jpred/) [22] to predict secondary structure.
Table 2.
Predictions of disorder and binding regions
| Protein name (residues) |
Residues predicted to be intrinsically disordered |
Predicted binding region of IDR |
Predicted secondary structure (Jpred4) |
|||
|---|---|---|---|---|---|---|
| IsUnstruct | PONDR -FIT |
IUPRED | Anchor | DisoRDPbind (predicted binding partner) |
||
| Known MoRFs; contain Anchor regions | ||||||
| TRAP220 (637-654) |
637-654 | 637-654 | 637-654 | 637-654 | None | α: 643-648 |
| BECN1 (105-130) |
105-114 | 105-109 | 105, 108-112, 124, 128 |
117-127 | None | α: 111-124 |
| GRIM (1-15) |
1-3, 9-15 | 1-15 | None | 1-15 | 1-15 (RNA) 4-15 (protein) |
α: 10-15 |
| p53 (367-388) |
367-388 | None | 367-388 | 367-388 | 360-383 (DNA) |
None |
| IDRs with unidentified binding partners; contain Anchor regions | ||||||
| GOPC (248-277) |
258-277 | 248-277 | 255-277 | 248-257 | 269, 272-277 (DNA) |
β: 251-254 |
| ATG3 (129-158) |
137-158 | 129-158 | 139-158 | 136-143 | 139-158 (protein) | None |
| ATG9A (810-839) |
810-839 | 810-839 | 810-839 | 831-839 | 819, 821, 833-839 (protein) |
None |
| BECN1 (76-105) |
76-105 | 76-105 | 76-105 | 79-103 | 60-84 (protein) | None |
| AMBRA1 (527-541) |
527-541 | 527-541 | 527-541 | 530-538 | 527-541 (protein) | α: 527-535 |
| IDRs with unidentified binding partners; lack Anchor regions | ||||||
| GOPC (255-279) |
258-279 | 255-279 | 255-279 | None | 269, 272-279 (DNA) |
None |
| BECN1 (50-78) |
50-78 | 50-78 | 50-78 | None | 59-78 (protein) | None |
| ATG16L1 (59-84) |
59-84 | None | 59-84 | None | None | α: 65-66, 80-84 |
| ATG2A (1244- 1273) |
1244-1273 | 1244- 1273 |
1244- 1273 |
None | 1257-1260 (protein) |
None |
2.3. Circular Dichroism spectroscopy
Peptides were diluted to 25-100 μM in 10 mM phosphate pH 7.4, 100 mM ammonium sulfate and either 0%, 25%, or 40% v/v TFE. After a 1 hour incubation on ice, continuous scanning CD spectra were recorded from 190 to 240 nm at 4 °C in a 300 μL quartz cell (0.1 cm path length) on a Jasco J-815 spectropolarimeter equipped with a Peltier thermoelectric temperature control. Secondary structure content was estimated by analyzing the CD data using three analysis programs, SELCON3, CDSSTR, and CONTIN, from the CDpro suite within the Jasco software [28-30]. For each single peptide set (i.e. 0%, 25% and 40% TFE), a consistent reference protein database was used for SELCON3, CDSSTR, and CONTIN estimations. For each spectrum, the average secondary structure was calculated by averaging estimated secondary structure content obtained from the three CD data analysis programs (Tables 3-5).
Table 3.
Secondary structure content in consensus IDRs lacking Anchor regions
| Protein (residues in peptide) |
BECN1 (50-78) |
GOPC (255-279) |
ATG2A (1244-1273) |
ATG16L1 (59-84) |
|
|---|---|---|---|---|---|
| Average estimated secondary structure content (%) | |||||
| Helix (%) |
No TFE | 1 ± 2 | 6 ± 2 | 5 ± 2 | 4 ± 2 |
| 25% TFE | 7 ± 6 | 12 ± 8 | 18 ± 1 | 17 ± 1 | |
| 40% TFE | 1 ± 1 | 6 ± 3 | 16 ± 1 | 3 ± 2 | |
| Strand (%) |
No TFE | 16 ± 10 | 32 ± 1 | 19 ± 5 | 16 ± 6 |
| 25% TFE | 14 ± 11 | 28 ± 4 | 24 ± 2 | 17 ± 2 | |
| 40% TFE | 8 ± 3 | 35 ± 4 | 25 ± 1 | 35 ± 2 | |
| Coil (%) |
No TFE | 81 ± 8 | 61 ± 1 | 76 ± 2 | 79 ± 8 |
| 25% TFE | 80 ± 17 | 59 ± 4 | 58 ± 2 | 65 ± 3 | |
| 40% TFE | 89 ± 3 | 58 ± 1 | 58 ± 2 | 60 ± 6 | |
Table 5.
Secondary structure content in IDRs with Anchor regions with unidentified binding partners
| Protein (residues in peptide) |
ATG3 (129-158) |
ATG9A (810-839) |
BECN1 (76-105) |
AMBRA1 (527-541) |
GOPC (248-277) |
|
|---|---|---|---|---|---|---|
| Average estimated secondary structure content (%) | ||||||
| Helix (%) |
No TFE | 3 ± 5 | 3 ± 2 | 1 ± 1 | 2 ± 1 | 3 ± 1 |
| 25% TFE | 4 ± 1 | 5 ± 4 | 20 ± 3 | 37 ± 1 | 4 ± 1 | |
| 40% TFE | −3 ± 6 | 4 ± 0 | 33 ± 3 | 50 ± 4 | 20 ± 1 | |
| Strand (%) |
No TFE | 27 ± 8 | 20 ± 4 | 4 ± 2 | 4 ± 4 | 19 ± 4 |
| 25% TFE | 26 ± 3 | 28 ± 2 | 8 ± 6 | 10 ± 5 | 17 ± 6 | |
| 40% TFE | 22 ± 9 | 18 ± 5 | 7 ± 4 | 8 ± 5 | 20 ± 4 | |
| Coil (%) |
No TFE | 63 ± 15 | 76 ± 2 | 95 ± 4 | 95 ± 5 | 79 ± 4 |
| 25% TFE | 67 ± 5 | 59 ± 15 | 53 ± 5 | 53 ± 5 | 78 ± 4 | |
| 40% TFE | 76 ± 10 | 78 ± 5 | 43 ± 8 | 43 ± 9 | 60 ± 4 | |
3. Results
3.1. Selection of IDRs for analysis
We selected thirteen diverse IDRs for examination (Table 1), focusing on IDRs or fragments of IDRs that were no longer than 30 residues, to ensure that we did not include more than one Anchor region within the IDR being examined. IDRs that have experimentally been shown to be either α-MoRFs or β-MoRFs with known binding partners were identified by a literature search, and served as our control set (Table 2). The known α-MoRFs selected included TRAP220 residues 637-654 (PDB 1XDK) and BECN1 residues 105-130 (PDB 3DVU) [5, 31, 32]. A β-MoRF comprising GRIM residues 1-15 (PDB 1SE0, 1JD5) was used as a negative control for the analysis [33, 34]. We also included p53 residues 367-388, which can form either a α-helix (PDB 1DT7) or a β-strand (PDB 1MA3) depending on the binding partner [35, 36].
IDRs that have not been experimentally examined were selected from our prior bioinformatics analyses wherein we identified consensus IDRs, defined as regions of at least 25 consecutive residues predicted to be disordered by at least three of four disorder prediction programs [5]. The experimentally unverified IDRs selected included GOPC residues 248-277, ATG3 residues 129-158, ATG9A residues 810-839, BECN1 residues 76-105, AMBRA1 residues 527-541, GOPC residues 255-279, BECN1 residues 50-78, ATG16L1 residues 59-84, and ATG2A residues 1244-1273 (Table 1).
We assessed disorder in all selected sequences by analyzing the full-length protein sequences using the programs IsUnstruct [19], PONDR-FIT [20] and IUPred [21]. IsUnstruct estimates the energy of each residue in either the ordered or disordered state to predict whether each residue will be ordered or disordered based upon amino acid sequence [19]. Our IsUnstruct analyses (Table 2) indicate that all selected peptides are completely disordered except short stretches comprising GRIM residues 4-8, BECN1 residues 115-130, ATG3 residues 129-136, and GOPC residues 255-258.
PONDR-FIT predicts per-residue disorder based on a combination of multiple features such as the ratio of net charge and hydrophobicity, amino acid frequency in disordered regions, and sequence complexity; with accuracy of prediction increasing with the length of the predicted disordered region [20]. Our PONDR-FIT analyses (Table 2) indicate that all peptides we selected are completely disordered except for p53 residues 367-388, ATG16L1 residues 59-84, and a stretch comprising BECN1 residues 110-130.
IUPred, a program which is also incorporated into the PONDR-FIT suite, calculates a disorder probability for each residue by using the amino acid composition to estimate formation of energetically favorable interactions, thereby identifying residues that lack such interactions and do not contribute to a stable folded structure [37]. The long-disorder algorithm, a context-independent prediction that takes 100 flanking residues into consideration [21], predicts all selected peptides to be completely disordered, except for GRIM residues 1-15 and shorter stretches comprising BECN1 residues 113-123, GOPC residues 248-254, and ATG3 residues 129-138 (Table 2).
Thus, expect for stretches of BECN1 residues 105-130, all of the protein regions included in this study are predicted to be disordered by either IsUnstruct, PONDR-FIT, or IUPred; and most of them are predicted to be disordered by at least two programs. However, as BECN1 residues 105-130 have previously been experimentally shown to be disordered even within larger protein fragments [5], we included it in our study.
3.2. Prediction of IDRs likely to fold upon binding
Next we used the programs ANCHOR [17], DisoRDPbind [18] and JPred4 [22] to analyze full-length protein sequences to predict the ability of the selected IDRs to form folded structures (Table 2). The ANCHOR program analyzes amino acid sequence to predict short regions (that we term Anchor regions) within or flanking IDRs that likely stabilize the IDR in an ordered state upon binding. Of the selected IDRs, Anchor regions are predicted (Table 2) in all the known α-MoRFs and β-MoRFs: TRAP220 residues 637-654, BECN1 residues 105-130, GRIM residues 1-15 and p53 residues 367-388; as well as in five experimentally untested IDRs: GOPC residues 248-277, ATG3 residues 129-158, ATG9A residues 810-839, BECN1 residues 76-105 and AMBRA1 residues 527-541 (Table 2). Lastly, four experimentally untested IDRs were predicted to not contain Anchor regions: GOPC residues 255-279, BECN1 residues 50-78, ATG16L1 residues 59-84, and ATG2A residues 1244-1273. Although GOPC residues 255-279 contain three Anchor residues (255-257) (Table 2), this is insufficient to be considered an Anchor region, which must contain at least five residues [17]. Therefore, the IDR comprising GOPC residues 255-279 was included in the experimental set lacking Anchor regions.
DisoRDPbind predicts RNA, DNA, and protein binding regions within IDRs based on amino acid content, sequence complexity, predicted structure and disorder, and sequence alignments [18]. Of the known MoRFs that we selected, all are predicted to contain Anchor regions; however, only GRIM is predicted by DisoRDPbind to contain a protein-binding site (Table 2). Among the five experimentally unverified IDRs bearing Anchor regions (GOPC residues 248-277, ATG3 residues 129-158, ATG9A residues 810-839, BECN1 residues 76-105, and AMBRA1 residues 527-541), only GOPC residues 248-277 are not predicted by DisoRDPbind to contain a protein-binding region. Conversely however, two of the IDRs lacking Anchor regions, BECN1 residues 50-78 and ATG2A residues 1244-1273, are predicted by DisoRDPbind to contain protein-binding sites. Thus, the binding regions predicted by ANCHOR and DisoRDPbind are not consistent.
Full-length protein sequences were also analyzed by JPred4 to predict secondary structure [22] (Table 2). A minimal helix is defined by the formation of two consecutive hydrogen bonds, therefore; a minimal α-helix requires at least six residues in a helical conformation [38, 39]. A β-strand cannot exist unless stabilized by hydrogen bonds to another strand. Stable β-structure is defined by four consecutive hydrogen bonds in either a parallel or anti-parallel bonding pattern, thereby involving at least six residues (three residues in each of two strands) [38]. JPred4 analysis (Table 2) indicates some secondary structure in seven of the IDRs, however; only five of these predicted secondary structures were of sufficient length for the formation of stable α-helices or β-strands.
Three of these IDRs, BECN1 residues 105-130, TRAP220 residues 637-654 and GRIM residues 1-15 are known MoRFs, with known binding partners, and were predicted by JPred4 to contain at least six contiguous α-helical residues (Table 2). While TRAP220 residues 637-654 and BECN1 residues 105-130 are α-MoRFs, GRIM residues 1-15 form a β-MoRF, with residues 3-6 forming a β-strand that is hydrogen-bonded to one end of a β-sheet from its binding partner DIAP1 (Drosophila melanogaster inhibitor of apoptosis protein) [33, 34]. Only the prediction for TRAP220 (Table 2) is entirely consistent with available experimental data, as TRAP220 residues 643-648 have been shown to be become helical upon binding [31]. While Jpred4 only predicts that BECN1 residues 111-124 are helical (Table 2), BECN1 residues 108-126 have been shown to become helical upon binding [5, 32, 40], thus; JPred4 does not predict the complete extent of the helical structure adopted by this IDR upon binding. Finally, for p53, residues 383-384 have been shown to form a β-strand with two residues of the Sir2 enzyme [35], and residues 378-384 have been shown to become helical upon binding Ca2+-bound S100B (ββ) [36], but neither region is predicted by JPred4 to be structured (Table 2). Thus the JPred predictions do not appear to completely accurately predict the secondary structure induced upon binding to known partners.
3.3. All the selected IDRs lack α-helical structure in solution
CD spectra reflect the average signal from the entire molecular population over the time course of the measurement. Therefore, it is impossible to distinguish between the average secondary structure maintained in the entire population of the sample, versus more extensive secondary structure maintained in a smaller sub-population of the sample with a corresponding decrease of that secondary structure in the remainder of the population. Although the flexible structure of IDRs enables rapid (<millisecond) conformational exchange amongst multiple conformations including fully disordered and transient, partially-folded conformers in solution [41, 42], there is no reason to expect that a sub-population within a chemically identical set of molecules in a chemically homogeneous environment would be more ordered on average than the rest of the population. Since the molecules in solution are identical in terms of conformational behavior, we apply the first interpretation, which requires a definition of minimal length to define a stable secondary structure, rather than the second interpretation which requires a definition of minimal percentage of the population that should adopt a given secondary structure to be considered a physiologically relevant structural state. Further, the first interpretation has been commonly used in classical literature, and definitions of minimal length to define secondary structure are well established [38]. Lastly, we note that either interpretation would not ultimately impact the overall objective of this study, i.e. to propose a method that enables the identification of IDRs that are α-MoRFs.
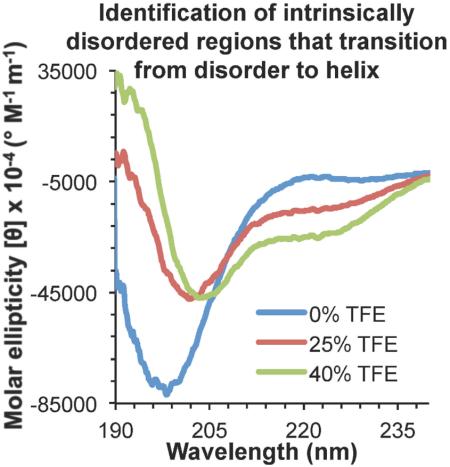
Secondary structure content estimated by analysis of CD spectra for each of the thirteen IDRs showed that none contained more than two residues in a helical conformation (Figures 1-3, Table 3-5), which is insufficient for the formation of a stable α-helix requiring a minimum of six residues. Formation of a minimal beta-structure requires six residues [38], and seven of the thirteen IDRs (Table 3-5) appear to have between six to eight residues in extended or β-conformation which may be sufficient for forming short stable β-hairpins (Table 3-5). Thus, while none of the selected IDRs had stable helical structure, several may have short β-strands. However, the marginal secondary structure content suggests that all the peptides tested are IDRs.
Figure 1. CD spectra of IDRs lacking Anchor regions.
Different spectra correspond to 0% TFE (blue), 25% TFE (red), or 40% TFE (green).
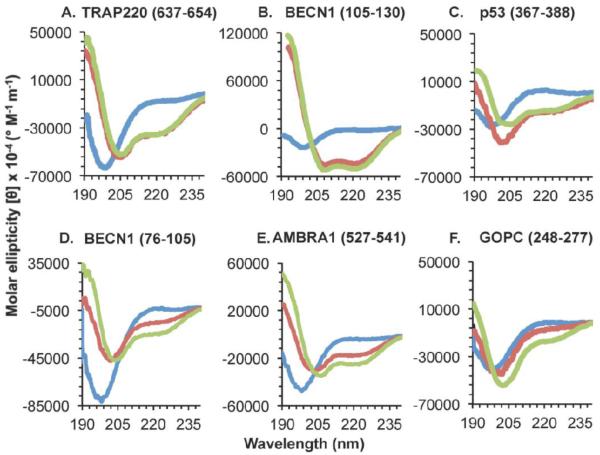
Figure 3. CD spectra of IDRs with Anchor regions that undergo a TFE-induced helical transition.
Different spectra correspond to 0% TFE (blue), 25% TFE (red), or 40% TFE (green). (A-C) Known α-MoRFs. (D-F) IDRs with experimentally unverified structural transitions.
3.4. IDRs lacking Anchor regions fail to undergo disorder-to-helix transitions
The secondary structure estimated from the CD spectra indicates that addition of TFE causes varying fluctuations in β-content of the different IDRs lacking Anchor regions (Table 3). In BECN1 residues 50-78, addition of 40% TFE, results in a decrease in the number of residues in β-conformation from five to two, clearly insufficient for the formation stable β-strands. However, GOPC residues 255-279 and ATG2A residues 1244-1273 show marginal increases of residues in β-content at 25% and 40% TFE, that would enable them to form short stable β-sheets. Finally, β-content in ATG16L (residues 59-84) remains constant at four residues upon addition of 25% TFE, but in 40% TFE, the number of residues in β-conformation increases to nine residues, which is sufficient for formation of stable β-strands.
Strikingly, the secondary structure content estimated from the CD spectra indicates that addition of TFE does not induce stable helical structure in the four IDRs lacking Anchor regions (Figure 1, Table 3). Helical content peaks in the presence of 25% TFE and is maintained or slightly decreased in 40% TFE; however, neither condition results in the six-residue minimum required for a stable helix. Thus, the changes in coil content estimated from the CD spectra of the four IDRs lacking Anchor regions appear to simply reflect fluctuations in helix and strand content (Table 3).
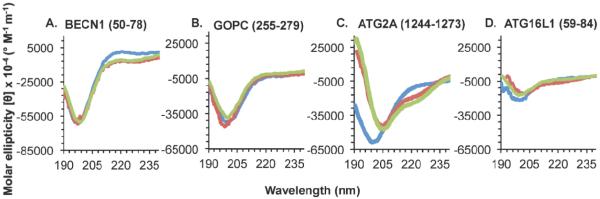
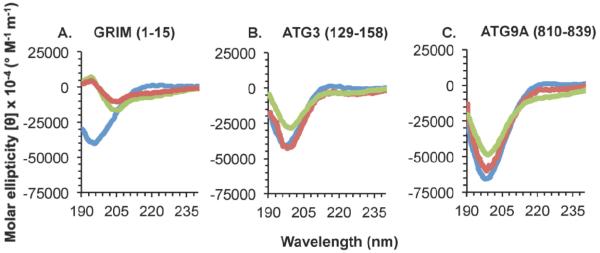
3.5. IDRs that bear Anchor regions, yet do not undergo TFE-induced helical transition, may be β- or σ-MoRFs
GRIM residues 1-15, which include a known β-MoRF and bear an Anchor region, do not become helical in the presence of TFE (Figure 2A, Table 4). The helical content, none at 0% and 25% TFE, and one residue at 40% TFE, is insufficient to stabilize a minimal helix, which requires at least six residues. However, while the number of residues in β-conformations increases marginally in 25% and 40% TFE, the three residues estimated to be in extended conformation in the presence of TFE, are also insufficient to form a stable β-strand, which requires six residues. These results demonstrate that TFE does not induce helicity in β-MoRFs, nor does it appear to support formation of stable β-structure.
Figure 2. CD spectra of IDRs with Anchor regions that do not undergo a TFE-induced helical transition.
Different spectra correspond to 0% TFE (blue), 25% TFE (red), or 40% TFE (green). (A) A known β-MoRF. (B-C) IDRs with experimentally unverified structural transitions.
Table 4.
Secondary structure content in known MoRFs
| Protein (residues in peptide) |
GRIM (1-15) |
TRAP220 (637-654) |
BECN1 (105-130) |
p53 (367-388) |
|
|---|---|---|---|---|---|
| Type of MoRF | β-MoRF | α-MoRF | α-MoRF | α- & β- MoRF | |
| Average estimated secondary structure content (%) | |||||
| Helix (%) |
No TFE | 0 ± 1 | 7 ± 3 | 2 ± 1 | 6 ± 4 |
| 25% TFE | 0 ± 4 | 62 ± 10 | 48 ± 2 | 34 ± 10 | |
| 40% TFE | 7 ± 7 | 50 ± 3 | 55 ± 2 | 31 ± 1 | |
| Strand (%) |
No TFE | 9 ± 8 | 29 ± 5 | 28 ± 7 | 31 ± 2 |
| 25% TFE | 19 ± 14 | 6 ± 3 | 12 ± 1 | 12 ± 4 | |
| 40% TFE | 22 ± 16 | 7 ± 6 | 8 ± 1 | 18 ± 2 | |
| Coil (%) |
No TFE | 89 ± 7 | 62 ± 6 | 65 ± 6 | 63 ± 2 |
| 25% TFE | 72 ± 7 | 32 ± 11 | 40 ± 2 | 54 ± 7 | |
| 40% TFE | 70 ± 20 | 43 ± 10 | 36 ± 2 | 50 ± 4 | |
Two of the Anchor region-containing IDRs for which binding partners have not yet been identified, ATG3 residues 129-158 and ATG9A residues 810-839, also do not undergo a disorder-to-helix transition in either 25% or 40 % TFE (Figure 2). These IDRs contain two or fewer helical residues (Table 5) even in 25% and 40% TFE, which is insufficient for the formation of a single stable helical turn. This failure to induce helicity indicates that ATG3 residues 129-158 and ATG9A residues 810-839 are not α-MoRFs. In contrast, in both these IDRs, there is sufficient β-content in TFE concentrations ranging from 0%-40% to permit the formation of β-strand structure; with the β-content increasing marginally at 25% TFE, then decreasing marginally at 40% TFE. Thus, TFE causes a marginal increase in the β-content, similar to that observed for the known β-MoRF, GRIM residues 1-15. However, as this increase is marginal, and given the limited number of known β-MoRFs available for testing, we cannot suggest that this method is a reliable way of identifying β-MoRFs.
3.6. IDRs that bear Anchor regions and undergo TFE-induced helical transition are α-MoRFs
TRAP220 residues 637-654, BECN1 residues 105-130, and p53 residues 367-388 are all known α-MoRFs with known binding partners. Consistent with this, addition of TFE induces helicity in each IDR (Figure 3 A-C). Although none of these IDRs have a helical content sufficient for formation of a stable helix in the absence of TFE, addition of TFE induces a significant disorder-to-helix transition in all three IDRs (Table 4). TFE-induced helical content peaks upon addition of 25% TFE, with 40% TFE inducing slight fluctuations. In TRAP220 residues 637-653, a maximum of eleven helical residues are stabilized at 25% and nine are maintained at 40% TFE. In BECN1 residues 105-130, 25% and 40% TFE induces the stabilization of thirteen and fourteen helical residues respectively. And finally, in p53 residues 367-388, 25% and 40% TFE stabilize eight and seven helical residues respectively. In each of these IDRs, the number of residues in β- and coil conformations decreases upon addition of TFE, suggesting that TFE destabilizes β-structure in these α-MoRFs.
All known α-MoRFs become helical in the presence of 25% and 40% TFE; however, the maximum number of helical residues induced is not always comparable to the number of helical residues actually observed in the corresponding crystal structures. Of the three known α-MoRFs, only in p53 does TFE induce a number of helical residues (Table 4) comparable to that observed in the crystal structure. Residues 378-384 (seven residues) of p53 become helical upon binding Ca2+-bound S100B(ββ) [36], which is equal to those stabilized by 40% TFE and just one less then the maximum observed (eight helical residues) in 25% TFE. However, in BECN1 residues 105-130, a maximum of fourteen residues are stabilized in helical conformations by 40% TFE, while upon binding to BCL2 homologs, BECN1 residues 108-126 (nineteen residues) form a helix [32, 40, 43], indicating that for this IDR the number of residues that undergo helical transformation may be underestimated by this method. Conversely, this method appears to slightly overestimate the number of TRAP220 residues that become helical upon binding. Only TRAP220 residues 643-648 (six residues) form a helix upon binding the retinoid receptor (RAR/RXR) ligand binding domain heterodimer [31]; however, eleven and nine helical residues are stabilized by 25% and 40% TFE respectively. Therefore, although this method stabilizes helical structure in α-MoRFs, the number of helical residues induced upon binding cannot be directly inferred from the secondary structure content estimations.
Amongst the Anchor-containing IDRs with unknown binding partners (Table 2), three IDRs: BECN1 residues 76-105, AMBRA1 residues 527-541, and GOPC residues 248-277, appear to undergo a TFE-induced disorder-to-helix transition. Helical content in BECN1 residues 76-105 increases from none in the absence of TFE, to six and ten, in 25% and 40% TFE respectively (Table 5, Figure 3D-F). In AMBRA1 residues 527-541, helical content increases from nothing in the absence of TFE to five and eight, in 25% and 40% TFE respectively. This increase occurs at the expense of residues in coil conformation, as both contain just one residue in β-conformation in the absence of TFE. Like the known α-MoRFs, the increase in helical content of BECN1 residues 76-105 and AMBRA1 residues 527-541 is observed in both 25% and 40% TFE. However, unlike the known α-MoRFs and BECN1 residues 76-105, for AMBRA1 residues 527-541, a stable helix comprising at least six residues is induced only in 40% TFE (Table 5); while in GOPC residues 248-277, 25% TFE is insufficient for inducing the disorder-to-helix transition, which occurs only upon addition of 40% TFE (Figure 3F, Table 5). Further, unlike the known α-MoRFs, despite containing β-content, the increase in helicity occurs only at the expense of residues in coil conformation, and residues in β-conformation do not appear to be destabilized by the addition of TFE (Table 4-5).
3.7. Effect of amino acid content on response to TFE
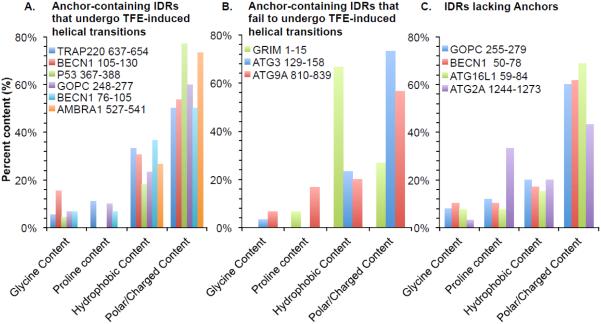
Amino acid composition of the various IDRs used in this study was analyzed (Figure 4). Relative to the average frequency of 37% hydrophobic residues and 51% polar or charged amino acids in proteins [44], all but one of the selected IDRs have a lower content of hydrophobic residues and a greater content of polar or charged residues, as is expected for typical intrinsically disordered regions (Figure 4). The known β-MoRF, GRIM residues 1-15, is an exception as 67% of this IDR is comprised of hydrophobic residues, which is higher than average; while only 30% of the residues are polar or charged, which is lower than average. All but two of the selected IDRs also have a lower proline content (Figure 4), compared to the average frequency of 5% found in proteins [44]. While the proline content of GRIM residues 1-15 is similar to the average; ATG2A residues 1244-1273 contain 33% prolines, which is substantially higher than the average proline content. Neither of these IDRs undergoes a transition to helix. Lastly, the glycine content of all the selected IDRs is approximately 7% (Figure 4), similar to that of average proteins [44]. However, while the amino acid composition of these peptides is consistent with these regions being IDRs, there does not appear to be a correlation between the amino acid composition of the IDRs and the TFE-induced helical transitions, beyond the absence of prolines in IDRs that do undergo a helical transition. This is consistent with findings that indicate that the accuracy of ANCHOR in predicting disordered binding regions is independent of amino acid content [17], and may also explain the ambiguity in the JPred predictions for these regions. Thus, beyond the lack of proline residues, the amino acid composition does not appear to be a direct predictor of potential transitions.
Figure 4. Amino acid content of the selected IDRs.
Amino acid content of: (A) Anchor-containing IDRs that undergo TFE-induced helical transitions, (B) Anchor-containing IDRs that fail to undergo TFE-induced helical transitions, and (C) IDRs lacking Anchors.
4. Discussion and Conclusion
All but one of the sequences selected for this study were predicted to be entirely disordered by at least one, and often all three of the disorder predictors, IUPred, IsUnstruct, or PONDR-FIT, as none of these sequences contained contiguous non-disordered stretches that were long enough to form stable secondary structure. The only exception was BECN1 105-130, which has short regions that are not predicted to be disordered, but this sequence was included in our analysis as BECN1 residues 105-130 have previously been experimentally shown to be disordered even in the context of adjacent BECN1 domains [5]. Additionally, many of the IDRs selected for this study are preceded by long regions, which are also predicted to be disordered [5, 31, 33-36]. Lastly, our CD analysis verifies that, in solution, each of these IDRs is largely disordered.
We used ANCHOR and DisoRDPbind to predict regions that may fold upon binding, but found little agreement between predictions by these different programs. DisoRDPbind also did not accurately predict binding regions within known MoRFs. Interestingly, we find that JPred4 often predicts secondary structure of sufficient length to permit formation of stable secondary structure that overlap or lie within predicted Anchor regions, rather than predicting these regions as disordered. However the actual predictions of the type of secondary structure were not always accurate, nor did the lengths of the predicted folded structures always agree with experimental observations, perhaps due to the conformational flexibility of these regions.
Our results indicate that the combined approach described in this report, of performing sequence-based predictions using programs like ANCHOR followed by experimental assessment of structural transitions in the presence of TFE, is suitable for identifying IDRs that are α-MoRFs. Strikingly, we show that while TFE induces helicity in known α-MoRFs, it does not induce disorder-to-helix transitions in IDRs lacking Anchor regions, or in known β-MoRFs. This combined predictive and experimental approach did not produce any false positives or false negatives, i.e. we were able to use this approach to successfully identify all the α-MoRFs we tested, and no known β-MoRFs were misidentified as α-MoRFs. Thus, this approach appears to reliably identify α-MoRFs, and distinguish between these and β- or σ-MoRFs.
Although p53 residues 367-388 can be either a α- or β-MoRF depending on its binding partner [35, 36], it behaves as an α-MoRF in the presence of TFE where a helical transition is induced at the expense of β- and coil content. This is distinct from GRIM, the β-MoRF, in which β-content slightly increases while helical content is not increased by TFE. Therefore, this method can identify IDRs that are α-MoRFs even if they can adopt different conformations upon binding to different partners. Conversely, identification of an IDR as an α-MoRF does not preclude it from undergoing other disorder-to-order transitions upon binding to a different partner.
We have used the method proposed in this study to identify three new potential α-MoRFs: BECN1 residues 76-105, AMBRA1 residues 527-541, and GOPC residues 248-277; from amongst the IDRs for which binding partners have not yet been identified. Like the known α-MoRFs, the helical content of each of these IDRs increases in the presence of TFE. However, unlike the known α-MoRFs, in AMBRA1 residues 527-541, stable helical content is achieved only in 40% TFE; while in GOPC residues 248-277 an increase in helicity is seen only in 40% TFE. BECN1 residues 76-105 and AMBRA1 residues 527-541 lack significant β-content, hence their TFE-induced helical transition occurs at the expense of coil conformation. GOPC residues 248-277 contain β-structure; however, unlike the known α-MoRFs with β-content, the TFE-induced increase in helicity in GOPC residues 248-277 occurs at the expense of the coil-content, and the β-content is not affected by TFE. Therefore, although GOPC residues 248-277 may become helical upon binding to one partner, perhaps the lack of destabilization of β-structure, which is similar to that seen for β-MoRFs, suggests that it may adopt β-conformations upon binding to other partners.
We also identify two potential Anchor-containing IDRs that do not undergo helical transition in the presence of TFE. It is possible that these might be β- or σ-MoRFs. Based on the similarity in TFE-induced behavior to the known β-MoRFs, it is tempting to speculate that ATG3 residues 129-158 and ATG9A residues 810-839 are also β-MoRFs.
In this study we included two overlapping IDRs from GOPC: the first comprised of residues 248-277 and the second comprising residues 255-279. GOPC residues 248-257 comprise a ten-residue Anchor region, thus the first GOPC IDR includes the Anchor while the second contains only the last three residues of the Anchor. Strikingly, loss of the first seven Anchor residues abrogates the ability of the latter GOPC IDR to undergo the TFE-induced helical transition observed in GOPC residues 248-277. Identification of the Anchor region-containing GOPC residues 248-277 as a α-MoRF, demonstrates that the TFE-induced helical transition requires the presence of an Anchor region. This is reminiscent of an earlier study wherein residues within the Anchor region was demonstrated to be essential for nucleation of a binding-induced disorder-to-helix transition in the case of BECN1 residues 105-130 [5].
Thus, we propose that ANCHOR predictions combined with assessment of TFE-induced helicity serves to identify α-MoRFs. However, we cannot use this method to generally exclude the possibility that these regions may also form either σ- or β-MoRFs in the context of other binding partners. It would be interesting to eventually compare the predictions in this report to experimental structural transitions upon binding to different partners.
It is useful to have a convenient experimental diagnostic to identify IDRs that are α-MoRFs as that would help better identify disordered regions with a potential biological function in mediating macromolecular interactions. Further, a tool to identify α-MoRFs may help identify a potential binding partner for the IDR from a wider list of potential interaction partners for the full-length protein, by providing information about the IDR conformation and residues most likely to be involved in the interaction. This, in turn, would lead to a better mechanistic understanding of the function of proteins involved in IDR-mediated interactions. For example, within the BECN1 residues 76-105 shown by this study to be a α-MoRF, residue S90 has previously been shown to be phosphorylated by MK2 (members of the p38 mitogen-activated protein kinase signaling pathway PAPKAPK2), which binds helical substrates and is essential for the tumor suppressor function of BECN1 [45]. Identification of BECN1 residues 76-105 as a α-MoRF supports the hypothesis that MK2 binds directly to this region to phosphorylate and activate BECN1. This information may permit the analysis of MK2 structure to determine the binding determinants for interaction with, and phosphorylation of, BECN1.
Information regarding binding-associated structural transitions in IDRs would not only provide a basis for understanding these interactions, but may also be useful for developing therapeutics targeting these proteins. For instance, a potential therapeutic was recently discovered that specifically binds p27, an IDR-containing protein that regulates the cell cycle via interaction with Cdk2/cyclinA. This molecule traps the disordered kinase binding domain of p27 in a conformation that prevents folding and binding to Cdk2/cyclin A [46].
An example of an IDR that was shown to be a α-MoRF and subsequently targeted for therapeutic development is the BECN1 BH3 domain (BECN1 105-130). An Anchor region within the BECN1 BH3 domain nucleates a helical transition upon binding BCL2 homologs, resulting in down-regulation of autophagy, a cellular homeostasis pathway important for innate immune defenses [5, 47]. An understanding of these structural transitions and the role of the Anchor region, enabled development of a peptide that selectively inhibits the interaction of BECN1 with the murine γ-herpesvirus68 BCL2, to prevent viral down-regulation of autophagy, while leaving cellular regulation unaffected [47]. However, unlike p27 and the BECN1 BH3 domain, binding partners for many Anchor regions remain unknown.
In a separate, parallel study, we used X-ray crystallography, CD spectroscopy, small angle X-ray scattering (SAXS), and electron paramagnetic resonance (EPR) to demonstrate that the BECN1 flexible helical domain (FHD) is partially disordered and partially helical (forming a 2.5 turn helix), when not in complex with other proteins, even in larger BECN1 fragments containing adjacent BECN1 domains [48]. Further, we found that although a binding partner for this region had not been identified, the FHD contains an Anchor region and undergoes TFE-induced helical transitions suggesting that it is a α-MoRF. Subsequently, in a 4.4 Å crystal structure (PDB 5DFZ) of the full-length yeast VPS34:VPS15:VPS30:VPS38 complex (VPS30 is the yeast homolog of BECN1), it was shown that VPS38, which binds primarily via the VPS30/BECN1 coiled-coil domain, also interacts in a coiled-coil manner with the BECN1 FHD (labeled CC1 in this structure) [49]. Therefore in this complex the FHD is completely helical, forming an eight-turn helix, verifying our prediction that the FHD is a α-MoRF. Thus, our method has successfully identified an IDR to be a α-MoRF; which was then confirmed by studies wherein a binding partner for this region was established, and the co-complex structure determined.
Identification of IDRs that are α-MoRFs by our method may enable development of therapeutics targeting interactions mediated by these IDRs by similar methods. Many of the IDR-containing proteins included in this study are implicated in diverse diseases. For instance, GOPC, which binds and modulates membrane proteins, including several oncogenes, is associated with glioblastoma and other cancers, as well as cystic fibrosis [50, 51]. Although a binding partner for the selected GOPC IDR remains to be identified, the GOPC α-MoRF identified here could potentially be targeted for therapeutic treatment by designing molecules that prevent helix formation, or bind very tightly to the helical conformation of the GOPC α-MoRF, preventing interactions that facilitate oncogenesis. Thus, this study provides a useful tool toward better understanding IDRs and their conformational transitions.
Highlights.
Many proteins contain intrinsically disordered regions (IDRs) that lack stable secondary or ordered tertiary structure.
IDRs are often implicated in macromolecular interactions, and may undergo structural transitions upon binding to interaction partners.
IDRs that undergo disorder-to-helix transitions can be identified by a combination of bioinformatics analyses and experimental stabilization of helical structure using 2,2,2-trifluoroethanol.
Acknowledgements
This work was supported by the following grants awarded to SCS: a NIH NINDS grant RO3 NS090939, a NSF grant MCB-1413525 and a North Dakota EPSCoR Track 1 grant 11A-1355466 Doctoral Dissertation Award for Y.M. (PI: SCS).
Abbreviations
- IDR
intrinsically disordered region
- TFE
2,2,2-trifluoroethanol
- MoRF
molecular recognition feature
- CD
circular dichroism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Wright P, Dyson H. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. Journal of Molecular Biology. 1999;293:321–31. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- [2].Oldfield C, Dunker A, Kornberg R. Intrinsically Disordered Proteins and Intrinsically Disordered Protein Regions. Annual Review of Biochemistry, Vol 83. 2014;83:553–84. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- [3].Ishida T, Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Research. 2007;35:W460–W4. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Brown CJ, Johnson AK, Dunker AK, Daughdrill GW. Evolution and disorder. Curr Opin Struct Biol. 2011;21:441–6. doi: 10.1016/j.sbi.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mei Y, Su M, Soni G, Salem S, Colbert C, Sinha S. Intrinsically disordered regions in autophagy proteins. PROTEINS: Structure, Function and Bioinformatics. 2014;82:565–78. doi: 10.1002/prot.24424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Orosz F, Ovadi J. Proteins without 3D structure: definition, detection and beyond. Bioinformatics. 2011;27:1449–54. doi: 10.1093/bioinformatics/btr175. [DOI] [PubMed] [Google Scholar]
- [7].Babu M, van der Lee R, de Groot N, Gsponer J. Intrinsically disordered proteins: regulation and disease. Current Opinion in Structural Biology. 2011;21:432–40. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- [8].van der Lee R, Lang B, Kruse K, Gsponer J, de Groot N, Huynen M, et al. Intrinsically Disordered Segments Affect Protein Half-Life in the Cell and during Evolution. Cell Reports. 2014;8:1832–44. doi: 10.1016/j.celrep.2014.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dunker A, Cortese M, Romero P, Iakoucheva L, Uversky V. Flexible nets - The roles of intrinsic disorder in protein interaction networks. Febs Journal. 2005;272:5129–48. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- [10].Wright P, Dyson H. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Bio. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Vuzman D, Levy Y. Intrinsically disordered regions as affinity tuners in protein-DNA interactions. Molecular Biosystems. 2012;8:47–57. doi: 10.1039/c1mb05273j. [DOI] [PubMed] [Google Scholar]
- [12].Huang K, Chadee A, Chen C, Zhang Y, Shyu A. Phosphorylation at intrinsically disordered regions of PAM2 motif-containing proteins modulates their interactions with PABPC1 and influences mRNA fate. Rna-a Publication of the Rna Society. 2013;19:295–305. doi: 10.1261/rna.037317.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tompa P. Unstructural biology coming of age. Curr Opin Struct Biol. 2011;21:419–25. doi: 10.1016/j.sbi.2011.03.012. [DOI] [PubMed] [Google Scholar]
- [14].Fong JH, Shoemaker BA, Garbuzynskiy SO, Lobanov MY, Galzitskaya OV, Panchenko AR. Intrinsic disorder in protein interactions: insights from a comprehensive structural analysis. PLoS Comput Biol. 2009;5:e1000316. doi: 10.1371/journal.pcbi.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mohan A, Oldfield C, Radivojac P, Vacic V, Cortese M, Dunker A, et al. Analysis of molecular recognition features (MoRFs) Journal Molecular Biology. 2006;362:1043–59. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- [16].Vacic V, Oldfield C, Mohan A, Radivojac P, Cortese M, Uversky V, et al. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res. 2007;6:2351–66. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Meszaros B, Simon I, Dosztanyi Z. Prediction of protein binding regions in disordered proteins. PLoS Computational Biology. 2009;5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Peng Z, Kurgan L. High-throughput prediction of RNA, DNA and protein binding regions mediated by intrinsic disorder. Nucleic Acids Research. 2015 doi: 10.1093/nar/gkv585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lobanov MY, Galzitskaya OV. The Ising model for prediction of disordered residues from protein sequence alone. Phys Biol. 2011;8:035004. doi: 10.1088/1478-3975/8/3/035004. [DOI] [PubMed] [Google Scholar]
- [20].Xue B, Dunbrack R, Williams R, Dunker A, Uversky V. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dosztányi Z, Csizmok V, Tompa P, Simon In. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–4. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- [22].Drozdetskiy A, Cole C, Procter J, Barton G. JPred4: a protein secondary structure prediction server. Nucleic Acids Research. 2015;43:W389–W94. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dyson H, Wright P. Peptide Conformation and Protein-Folding. Current Opinion in Structural Biology. 1993;3:60–5. [Google Scholar]
- [24].Shiraki K, Nishkawa K, Goto Y. Trifluoroethanol-Induced Stabilization of the Alpha-Helical Structure of Beta-Lactoglobulin - Implication for Non-Hierarchical Protein-Folding. Journal of Molecular Biology. 1995;245:180–94. doi: 10.1006/jmbi.1994.0015. [DOI] [PubMed] [Google Scholar]
- [25].Buck M. Trifluoroethanol and colleagues: cosolvents come of age. Recent studies with peptides and proteins. Quarterly reviews of biophysics. 1998;31:297–355. doi: 10.1017/s003358359800345x. [DOI] [PubMed] [Google Scholar]
- [26].Roccatano D, Colombo G, Fioroni M, Mark AE. Mechanism by which 2,2,2-trifluoroethanol/water mixtures stabilize secondary-structure formation in peptides: A molecular dynamics study. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12179–84. doi: 10.1073/pnas.182199699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dunlap TB, Kirk JM, Pena EA, Yoder MS, Creamer TP. Thermodynamics of binding by calmodulin correlates with target peptide α-helical propensity. Proteins: Structure Function and Bioinformatics. 2013;81:607–12. doi: 10.1002/prot.24215. [DOI] [PubMed] [Google Scholar]
- [28].Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Analytical Biochemistry. 2000;287:252–60. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- [29].Sreerama N, Venyaminov S, Woody R. Estimation of protein secondary structure from circular dichroism spectra: Inclusion of denatured proteins with native proteins in the analysis. Analytical Biochemistry. 2000;287:243–51. doi: 10.1006/abio.2000.4879. [DOI] [PubMed] [Google Scholar]
- [30].Sreerama N, Venyaminov SY, Woody RW. Analysis of protein circular dichroism spectra based on the tertiary structure classification. Analytical biochemistry. 2001;299:271–4. doi: 10.1006/abio.2001.5420. [DOI] [PubMed] [Google Scholar]
- [31].Pogenberg V, Guichou J, Vivat-Hannah V, Kammerer S, Perez E, Germain P, et al. Characterization of the interaction between retinoic acid receptor/retinoid X receptor (RAR/RXR) heterodimers and transcriptional coactivators through structural and fluorescence anisotropy studies. Journal of Biological Chemistry. 2005;280:1625–33. doi: 10.1074/jbc.M409302200. [DOI] [PubMed] [Google Scholar]
- [32].Sinha S, Colbert CL, Becker N, Wei Y, Levine B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the γ-herpesvirus 68 Bcl-2 homolog M11. Autophagy. 2008;4:989–97. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Yan N, Wu J, Chai J, Li W, Shi Y. Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim. Nat Struct Mol Biol. 2004;11:420–8. doi: 10.1038/nsmb764. [DOI] [PubMed] [Google Scholar]
- [34].Wu J, Cocina A, Chai J, Hay B, Shi Y. Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides. Molecular Cell. 2001;8:95–104. doi: 10.1016/s1097-2765(01)00282-9. [DOI] [PubMed] [Google Scholar]
- [35].Avalos J, Celic I, Muhammad S, Cosgrove M, Boeke J, Wolberger C. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Molecular Cell. 2002;10:523–35. doi: 10.1016/s1097-2765(02)00628-7. [DOI] [PubMed] [Google Scholar]
- [36].Rustandi R, Baldisseri D, Weber D. Structure of the negative regulatory domain of p53 bound to S100B(beta beta) Nature Structural Biology. 2000;7:570–4. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- [37].Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. Journal of Molecular Biology. 2005;347:827–39. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- [38].Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- [39].Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins: Structure Function and Genetics. 1995;23:566–79. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- [40].Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3 only protein. The Journal of Biological Chemistry. 2007;282:13123–32. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- [41].Schulenburg C, Hilvert D. Protein conformational disorder and enzyme catalysis. Top Curr Chem. 2013;337:41–67. doi: 10.1007/128_2012_411. [DOI] [PubMed] [Google Scholar]
- [42].Zhang W, Ganguly D, Chen J. Residual structures, conformational fluctuations, and electrostatic interactions in the synergistic folding of two intrinsically disordered proteins. PLoS Comput Biol. 2012;8:e1002353. doi: 10.1371/journal.pcbi.1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ku B, Woo J-S, Liang C, Lee K-H, Hong H-S, Xiaofei E, et al. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral Bcl-2 of murine γ-Herpesvirus 68. PLoS Pathogens. 2008;4:e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].McCaldon P, Argos P. Oligopeptide biases in protein sequence and their use in predicting protein coding regions in nucleotide sequences. Proteins Strcture,Function and Genetics. 1988;4:99–122. doi: 10.1002/prot.340040204. [DOI] [PubMed] [Google Scholar]
- [45].Wei Y, An Z, Zou Z, Sumpter R, Su M, Zang X, et al. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. LID - 10.7554/eLife.05289 [doi] eLife. 2015;4 doi: 10.7554/eLife.05289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Iconaru L, Ban D, Bharatham K, Ramanathan A, Zhang W, Shelat A, et al. Discovery of Small Molecules that Inhibit the Disordered Protein, p27(Kip1) Scientific Reports. 2015;5 doi: 10.1038/srep15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Su M, Mei Y, Sanishvili R, Levine B, Colbert CL, Sinha S. Targeting γ-herpesvirus 68 Bcl-2–mediated down-regulation of autophagy. Journal of Biological Chemistry. 2014;289:8029–50. doi: 10.1074/jbc.M113.515361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mei Y, Ramanathan A, Glover K, Christopher , Stanley C, Sanishvili R, Chakravarthy S, et al. Conformational Flexibility Enables Function of a BECN1 Region Essential for Starvation-Mediated Autophagy. Biochemistry. 2016 doi: 10.1021/acs.biochem.5b01264. 10.1021/acs.biochem.5b01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rostislavleva K, Soler N, Ohashi Y, Zhang L, Pardon E, Burke JE, et al. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science. 2015;350:178–81. doi: 10.1126/science.aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Herrmann S, Ninkovic M, Kohl T, Pardo L. PIST (GOPC) modulates the oncogenic voltage-gated potassium channel K(v)10.1. Frontiers in Physiology. 2013;4 doi: 10.3389/fphys.2013.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cheng J, Moyer B, Milewski M, Loffing J, Ikeda M, Mickle J, et al. A golgi-associated PDZ domain protein modulates cystic fibrosis transmembrane regulator plasma membrane expression. Journal of Biological Chemistry. 2002;277:3520–9. doi: 10.1074/jbc.M110177200. [DOI] [PubMed] [Google Scholar]