Abstract
There is growing evidence that sperm DNA methylation is important in maintaining proper sperm health and function. Previous studies have associated sperm DNA methylation levels with sperm quality and function, however, little is known regarding the intra- and inter-individual variability in sperm methylation levels. This study characterizes this variation. Sperm epigenetic differences between successive semen samples from 12 patients were examined to identify the intra- and inter-individual differences globally across the genome, and in specifically defined genomic regions using the Illumina Infinium HumanMethylation450 BeadChips. Methylation analysis identified a bimodal distribution in the methylation levels that were non-uniformly distributed across the different genomic regions. The methylation levels were highly correlated in both the intra- and inter-individual comparisons. The intra-individual methylation levels were more highly correlated than the inter-individual comparison both globally and across the defined genomic regions, demonstrating that sperm DNA methylation levels are relatively stable between semen sample collections.
Introduction
Semen analysis is routinely performed in male reproductive clinics as a way to assess sperm quantity and quality. Mature sperm arise from an extremely complex and intricate biological process that requires the careful coordination of thousands of genes (Matzuk & Lamb 2002). The large intra- and inter-individual variability in the various parameters measured during a semen analysis is well-documented (Overstreet 1994), where even in healthy fertile men the total sperm count can range from 40 million to hundreds of millions (World Health Organization 2010). Although studies have begun to understand the molecular basis for these large variations, most of these have focused on the impact of environmental and genetic factors with very few investigating epigenetic causes.
The epigenome and higher-order chromatin structure are tightly integrated and impact normal cell physiology and function (Berger 2007, Feinberg 2007, Kouzarides 2007, Wen et al. 2012). Within mammalian sperm, aberrant DNA methylation is associated with decreases in sperm quality and function (Hammoud et al. 2013, Pacheco et al. 2011). Mammalian sperm cells are unique with regards to their chromatin structure, which may be heavily influenced by epigenetic marks present in the cell. Sperm cells undergo a histone-to-protamine transition during development that highly condenses chromatin to an extent 6–20 fold greater than nucleosome-bound chromatin, which contributes to the lack of transcriptional activity (Balhorn 2007, Balhorn et al. 1988, Dadoune 1995, Hecht 1990, Oliva & Dixon 1991, Ward & Coffey 1991).
The addition of a methyl group to cytosine residues may not only impart functional consequences on the sperm itself, but is also believed to be pivotal for both fertilization and early embryo viability (Anway et al. 2005, Bourc’his & Bestor 2004, Carrell & Hammoud 2010, Dada et al. 2012, Jenkins & Carrell 2012, Li et al. 2012, Okano et al. 1999, Romero et al. 2011, Yaman & Grandjean 2006). This is supported by evidence in human sperm demonstrating that histones are retained in the regulatory regions of genes associated with early embryonic development (Brykczynska et al. 2010, Hammoud et al. 2009), mediated by the presence of hypomethylated CpGs (Erkek et al. 2013).
The concept of epigenetic difference has been used to characterize inter- and intra-group DNA methylation levels (Flanagan et al. 2006), but few studies have examined it in the context of sperm DNA methylation. Furthermore, these studies have largely focused on comparing the variability between low and high quality sperm fractions from an individual (Jenkins & Carrell 2012, Krausz et al. 2012, Navarro-Costa et al. 2010). One recent study investigated the difference in the sperm methylation profiles from 2 different semen samples collected from the same individuals (Jenkins et al. 2014); however, this study was interested in the effects of age on sperm DNA methylation and compared the methylation profiles from the same individuals at a young and older age with a span of 9–19 years in between samples. No studies to date have addressed the issue of true intra-individual variability in the sperm DNA methylation landscape. Our study addresses this question by assessing the intra- and inter-individual sperm epigenetic differences using 2 semen samples collected less than 1.5 years apart.
Materials and Methods
Ethics Statement
The Committee on the Protection of Human Subjects: Rhode Island Hospital Institutional Review Board (Committee #403908) approved the study and written informed consent was obtained from all participants. Clinical investigation was conducted according to the principles expressed in the Declaration of Helsinki.
Individual Population, Semen Analysis, and Sperm Isolation
As part of an ongoing study of methylation patterns in sperm from men, men presenting to the Division of Urology at Rhode Island Hospital for diagnostic semen analyses as part of a fertility evaluation were consented for methylation analysis of their sperm. To examine intra-individual variation in these men, those that had at least two semen samples in the Division were included in this study. Samples from patients that did not yield enough DNA for analysis were excluded. In total, 19 patients had at least two semen samples. Seven patients were excluded because insufficient DNA was isolated to allow analysis. The remaining 12 patients had sufficient amounts of isolated DNA from both collections to perform DNA methylation analysis. Semen analysis parameters for all individuals at both collections were assessed and are summarized in Table 1, along with BMI values calculated from self-reported weights and heights. Following semen analysis, samples were washed with modified sperm washing medium (Irvine Scientific, Santa Ana, CA). Sperm cells were enriched by treating the samples with somatic cell lysis buffer (SCLB; 0.1% SDS, 0.5% Triton X-100) (Goodrich et al. 2007), washed with phosphate buffered saline and examined under a microscope to confirm the purification of sperm cells. Sperm cells were then pelleted and snap-frozen and stored at −80°C until further use.
Table 1.
Summary of individual population and semen parameter measurements.
| Individual ID |
Ageb | BMIa,c | Time between collections (days) |
Semen Parameter Values a |
||||
|---|---|---|---|---|---|---|---|---|
| Semen volume (mL) |
Sperm density (×106/mL) |
Total sperm count (×106) | Sperm motility (%) |
Round cell density (×106/mL) |
||||
| 1 | 35 | 32.1 | 307 | 4.5, 3.5 (−1.0) | 31, 47 (16) | 139.5, 164.5 (25.0) | 63, 56 (−7) | 1.0, 0.8 (−0.2) |
| 2 | 36 | 32.5, 33.2 | 21 | 2.0, 2.0 (0.0) | 70, 21 (−49) | 140.0, 42.0 (−98.0) | 57, 44 (−13) | 1.2, 0.7 (−0.5) |
| 3 | 41 | 27.0 | 7 | 3.5, 3.0 (−0.5) | 28, 62 (34) | 98.0, 186.0 (88.0) | 50, 57 (7) | 0.1, 0.1 (0.0) |
| 4 | 51 | 28.0 | 27 | 3.0, 2.0 (−1.0) | 82, 55 (−27) | 246.0, 110.0 (−136.0) | 47, 41 (−6) | 0.5, 0.8 (0.3) |
| 5 | 33 | 38.7 | 489 | 2.7, 2.5 (−0.2) | 61, 97 (36) | 164.7, 242.5 (77.8) | 68, 66 (−2) | 0.2, 0.0 (−0.2) |
| 6 | 29 | 26.5 | 27 | 5.0, 5.0 (0.0) | 39, 43 (4) | 195.0, 215.0 (20.0) | 66, 60 (−6) | 0.1, 0.0 (−0.1) |
| 7 | 32 | 28.2, 27.3 | 222 | 5.0, 4.3 (−0.7) | 92, 38 (−54) | 460.0, 163.4 (−296.6) | 53, 60 (7) | 0.7, 0.6 (−0.1) |
| 8 | 29 | 21.3 | 236 | 0.7, 1.8 (1.1) | 175, 87 (−88) | 122.5, 156.6 (34.1) | 49, 58 (9) | 1.1, 0.4 (−0.7) |
| 9 | 28 | 23.1, 22.5 | 80 | 2.3, 3.5 (1.2) | 56, 104 (48) | 128.8, 364.0 (235.2) | 71, 66 (−5) | 0.7, 0.6 (−0.1) |
| 10 | 36 | 24.4 | 7 | 2.1, 1.0 (−1.1) | 171, 311 (140) | 359.1, 311.0 (−48.1) | 51, 51 (0) | 0.1, 0.2 (0.1) |
| 11 | 29 | 23.7 | 329 | 4.0, 3.7 (−0.3) | 70, 54 (23) | 124.0, 199.8 (75.8) | 59, 67 (8) | 0.3, 0.0 (−0.3) |
| 12 | 30 | 25.6 | 7 | 4.3, 3.4 (−0.9) | 31, 77 (7) | 301.0, 261.8 (−39.2) | 51, 69 (−2) | 0.4, 0.4 (0.0) |
value of 1st sample, value of 2nd sample (change between 2nd and 1st value)
age at the time of 1st collection
BMI values calculated from self-reported weights and heights; single values are reported if BMI did not change
DNA Isolation, Bisulfite Modification, and Illumina Infinium HumanMethylation450 BeadChip Array
DNA was isolated from the collected sperm cells using a modified guanidine thiocyanate DNA extraction method (Griffin 2013). Isolated sperm DNA quality and quantity were assessed using a NanoDrop 1000 (Thermo Scientific, Wilmington, DE), and subsequently sent to Yale’s Center for Genome Analysis (Yale School of Medicine, West Haven, CT) for bisulfite conversion and genome-wide DNA methylation assessment using the Illumina Infinium HumanMethylation450 BeadChip (Illumina, San Diego, CA) according to the manufacturer’s protocol.
Imprinted Genes
A list of 187 imprinted genes in the human genome was compiled based on information from three sources: (1) experimentally determined imprinted genes listed in two databases (http://www.geneimprint.com and http://igc.otago.ac.nz/home.html) (n=62); (2) imprinted genes identified using the ChIP-SNP method (n=27) (Maynard et al. 2008); and (3) protein-coding genes from the 156 putatively imprinted sequences that correspond to known genes listed by NCBI (n = 106) (Luedi et al. 2007).
Statistical Analysis
The Illumina Infinium HumanMethylation450 BeadChip data were imported into R (R Core Team) and analyzed using different statistical packages available through Bioconductor (Gentleman et al. 2004). The probes were quality filtered with detection p-values < 0.05, then the data were normalized using functional normalization (Fortin et al. 2014) for batch correction between Infinium arrays, followed by BMIQ (Teschendorff et al. 2013) to normalize between the two probe types. Each of these methods was applied using the wateRmelon package (Pidsley et al. 2013). Probe-level data were filtered using a detection p-value > 0.05, and SNP-containing and cross-reacting probes were removed, resulting in 396,861 probes that passed the quality control measures.
Residual round cells in the semen samples can potentially impact the interpretation of sperm DNA methylation. To confirm the depletion of round cells from the purified sperm cells, Illumina HumanMethylation450 Beadchip data from flow-sorted neutrophils (granulocytes), lymphocytes (CD8+ and CD4+ T cells, CD56+ natural killer cells and CD19+ B cells) and CD14+ monocytes (Jaffe, Reinius et al. 2012) were compared against the methylation results from the collected semen samples. The mean methylation (β) values from 600 cell type-specific CpGs from the sorted cell populations were hierarchically clustered with the corresponding β-values from each collected semen samples.
The direct methylation (β) values and the logit-transformed (M) values were used for the comparison of methylation levels both within and between individuals. The contrast matrix (Supplementary Figure 1) schematically illustrates all of the permutations used for the intra- and inter-individual comparisons; 12 intra-individual and 264 inter-individual comparisons. The absolute epigenetic differences for the intra- and inter-individual comparisons were first calculated on a per-probe basis (|Δβ|- and |ΔM|-values), averaged across probes (sample mean), and then averaged across all comparisons (grand mean) and reported with the between-sample DNA methylation variance (MSB). For the intra-individual comparisons, the epigenetic difference (Δβ- and ΔM-values) introduced over time was also determined by calculating the sample and grand mean, and the between-sample variance between collected samples. Additionally, the Pearson correlation value was calculated for each pairwise intra- and inter-individual comparison shown in Supplementary Figure 1. Statistical analysis of the epigenetic differences, correlations, Wilcoxon rank sums and the Student’s t-tests were calculated in R. Differential methylation analysis was also performed in R using the limma package (Ritchie et al. 2015) for Bioconductor (Gentleman et al. 2004). Pathway analysis of annotated CpGs was performed using DAVID (Huang da et al. 2009).
Results
Regional Sperm DNA Methylation Profiles
The sperm DNA methylation profiles were assessed from 12 patients, each of whom provided two semen samples. The semen samples were collected from the patients presenting for fertility evaluations. Time between samples ranged from 7 to 489 days between successive sample collections with a mean and median value of 147 and 54 days respectively (Table 1). At the time of the first semen sample collection, patients ranged in age from 28 to 51 years (mean = 34, SD = 7) with a BMI of 27.6 (SD ± 4.9), and the average BMI values at the time of the second collection was 27.5 (SD ± 5.0). Global methylation levels at CpGs were assessed using Illumina Infinium HumanMethylation450 BeadChips. Probe-level data were filtered using a detection p-value > 0.05, and SNP-containing and cross-reacting probes were removed, resulting in 396,861 probes that passed the quality control measures.
It is important to recognize that the presence of any somatic cell (i.e. round cells) contamination in the purified sperm cell population may impact the measured DNA methylation levels. The number of round cells in each of the collected semen samples (Table 1) are relatively low with respect to the total sperm cell counts, and coupled with the SCLB treatment minimized the contaminating cell population. Any residual round cells in the enriched cell population would have a minimal effect on the measured DNA methylation values. This expectation was confirmed by hierarchical clustering of the sample β-values with the mean methylation (β) value of 600 leukocyte discriminating CpGs (Jaffe, Reinius et al. 2012). The clustering pattern revealed a distinctive sperm DNA methylation profile that clustered separately from the leukocyte cell populations (Supplementary Figure 2), documenting the absence of contaminating leukocytes in the purified sperm cell samples.
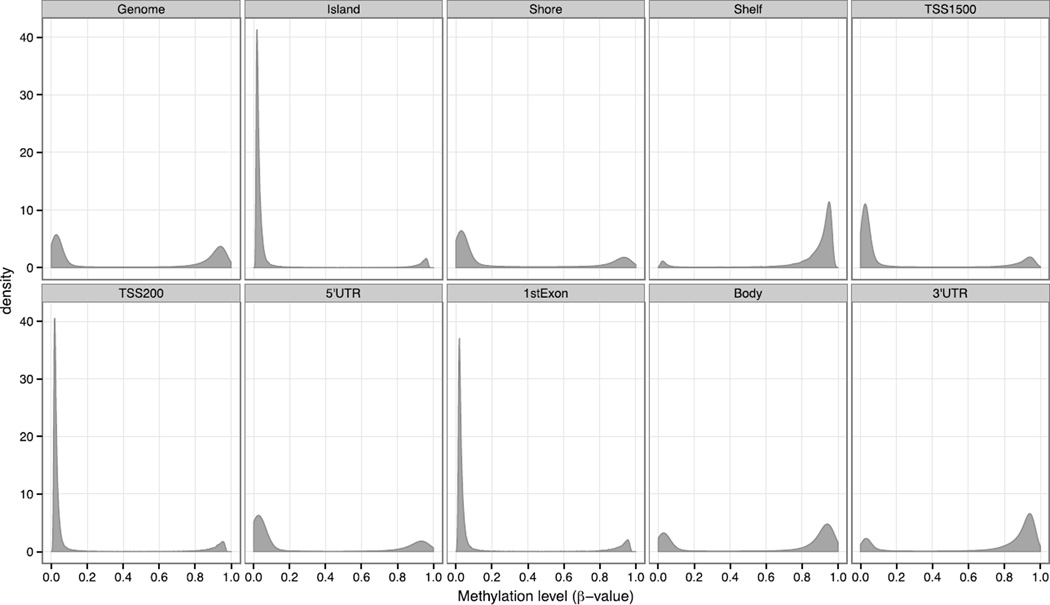
The β-values for a given site (CpG location in the genome) can range from 0 for unmethylated to 1 for fully methylated across all copies in the sample. The β-value distribution of each probe was examined globally throughout the genome, and across defined genomic regions: island, shore, shelf, within 1500 bp and 200 bp upstream of a transcriptional start site (TSS; TSS1500 and TSS200), first exon, body and the 5′ and 3′ untranslated regions (UTRs). At the level of the genome, sperm β-values of the 24 collected semen samples exhibited a distinctive bimodal profile where 49.5% of the measured CpGs were unmethylated (β-value ≤ 0.2), 42.0% were methylated (β-value ≥ 0.8), and 9% demonstrated an intermediate level of methylation (0.2 < β-value < 0.8; Figure 1 and Table 2). Although genome-wide analysis found a near equal distribution in the unmethylated and methylated probes, examination of the different genomic location categories identified regions where the probes were skewed to either unmethylated or methylated states (Figure 1). Probes in the CpG islands, TSS200 and 1st exons possessed a highly skewed β-value distribution, where greater than 80% of the CpGs were unmethylated. It is important to recognize that alternative TSSs and different gene splicing events can result in a probe being categorized into several gene feature categories, and skew the interpretations of the methylation profiles in the different genomic regions.
Figure 1.
Bimodal distribution of sperm DNA methylation levels (β-values) from 24 collected semen samples (12 patients, each with 2 semen samples).
Table 2.
Summary of the averaged sperm methylation levels (β-values) across collected semen samples.
| Probe methylation level | Genomic Regiona | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome | Island | Shore | Shelf | TSS1500 | TSS200 | 5′UTR | 1st Exon | Body | 3′UTR | ||
|
Global CpGs |
% unmethylatedb | 49.5 | 89.6 | 65.6 | 6.1 | 73.4 | 85.0 | 68.6 | 83.2 | 32.6 | 18.9 |
| % methylatedc | 42.0 | 8.2 | 23.2 | 81.3 | 19.0 | 11.9 | 25.4 | 13.3 | 58.3 | 69.1 | |
| % intermediate | 8.5 | 2.1 | 11.2 | 12.6 | 7.7 | 3.1 | 6.0 | 3.5 | 9.0 | 12.0 | |
|
Imprinted CpGs |
% unmethylatedb | 50.5 | 85.6 | 58.5 | 1.8 | 80.5 | 87.8 | 58.7 | 87.2 | 28.2 | 45.0 |
| % methylatedc | 41.4 | 11.4 | 30.2 | 87.8 | 12.6 | 8.9 | 34.8 | 8.5 | 62.6 | 45.3 | |
| % intermediate | 8.0 | 3.0 | 11.3 | 10.4 | 6.8 | 3.3 | 6.5 | 4.3 | 9.2 | 9.7 | |
probe genomic location are provided in the annotation file supplied by Illumina
β-value < 0.2
β-value > 0.8
Intra-Individual Sperm DNA Methylation Differences
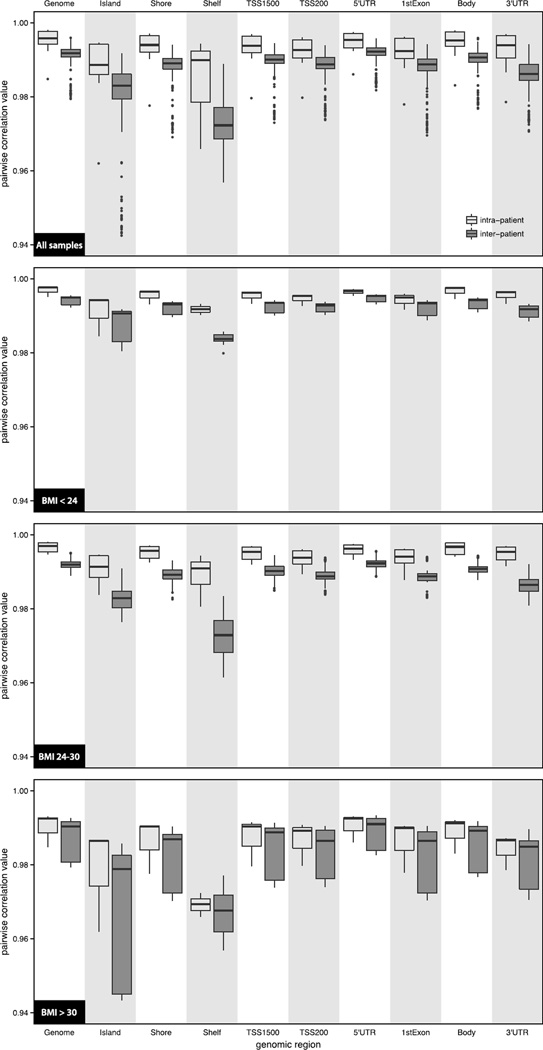
Pairwise comparisons of the intra-individual epigenetic differences were assessed using both β- and M-values, where the M-values are the logit transformation of the β-values. The intra-individual analysis investigated the sperm DNA methylation changes between the second and first collected semen samples from the same individual (DNA methylation levelsample 2 – DNA methylation levelsample 1), for the 12 patients in the study. The genome-wide distribution of the ΔM-values were normally distributed with a mean value of −0.0462 (MSB = 0.0049), implying that there was a slight decrease in the overall sperm DNA methylation levels in the second semen samples collected from the patients. Closer examination of the ΔM-values across the different regions of the genome, found that the methylation changes were dependent on the CpG locations (Table 3). For example, CpGs located in the shelves and 3′UTRs had a mean ΔM-value of −0.0201 (MSB = 0.0094) and −0.0365 (MSB = 0.0042), respectively, while CpGs in the islands had a mean ΔM-value of −0.0575 (MSB = 0.0215). Although the shift in DNA methylation varied by CpG location, the overall magnitude of the difference (|ΔM-value|) in each region were relatively equal (|ΔM-value| ≅ 0.3; Table 3). At the global level, the intra-patient Pearson correlation coefficient was 0.9950 (SD = 0.0038) illustrating a very high concordance in DNA methylation levels between donated semen samples. This is also observed throughout the different interrogated genomic regions (Figure 2 and Supplementary Table 1).
Table 3.
Intra- and inter-patient mean sperm epigenetic differences (ΔM-values)
| Genomic Regiona | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome | Island | Shore | Shelf | TSS1500 | TSS200 | 5′UTR | 1st Exon | Body | 3′UTR | |||
| Global CpGs | Intra | ΔM-value b | −0.0462 (0.0049) |
−0.0575 (0.0215) |
−0.0710 (0.0153) |
−0.0201 (0.0094) |
−0.0592 (0.0140) |
−0.0427 (0.0169) |
−0.0426 (0.0088) |
−0.0465 (0.0147) |
−0.0424 (0.0046) |
−0.0365 (0.0042) |
| |ΔM-value|b | 0.3065 (0.0099) |
0.3029 (0.0082) |
0.3241 (0.0130) |
0.2946 (0.0096) |
0.3098 (0.0094) |
0.2856 (0.0064) |
0.2967 (0.0074) |
0.2974 (0.0072) |
0.3079 (0.0106) |
0.3049 (0.0104) |
||
| Inter | |ΔM-value|b | 0.4038 (0.0062) |
0.3779 (0.0097) |
0.4346 (0.0102) |
0.3816 (0.0035) |
0.3974 (0.0080) |
0.3528 (0.0069) |
0.3755 (0.0061) |
0.3727 (0.0077) |
0.4056 (0.0056) |
0.4023 (0.0050) |
|
| Imprinted CpGs | Intra | ΔM-value b | −0.0616 (0.0093) |
−0.0810 (0.0260) |
−0.0785 (0.0203) |
−0.0220 (0.0190) |
−0.0811 (0.0264) |
−0.0613 (0.0204) |
−0.0696 (0.0120) |
−0.0815 (0.0303) |
−0.0498 (0.0085) |
−0.0780 (0.0139) |
| |ΔM-value|b | 0.3228 (0.0137) |
0.3257 (0.0139) |
0.3445 (0.0201) |
0.2968 (0.0117) |
0.3286 (0.0161) |
0.3081 (0.0103) |
0.3237 (0.0144) |
0.3326 (0.0147) |
0.3230 (0.0141) |
0.3448 (0.0210) |
||
| Inter | |ΔM-value|b | 0.4264 (0.0100) |
0.4142 (0.0165) |
0.4718 (0.0163) |
0.3873 (0.0046) |
0.4255 (0.0165) |
0.3788 (0.0102) |
0.4061 (0.0110) |
0.4214 (0.0164) |
0.4352 (0.0080) |
0.4641 (0.0224) |
|
probe genomic location are provided in the annotation file supplied by Illumina
values are reported as the mean of the mean patient methylation difference and the mean square between
Figure 2.
Intra- and inter-patient Pearson correlation analysis of the sperm DNA methylation levels. M-values from the intra- (light gray) and inter-patient (dark gray) comparisons are shown in the boxplots with the outliers identified as circles, for each of the defined genomic regions and by BMI levels. The intra- and inter-individual correlation values and associated p-values indicating the level of significance between them are summarized in Supplementary Table 1.
Semen samples collected from the same patients were collected over a period ranging from 7 to 489 days, which allowed for a longitudinal analysis of sperm DNA methylation changes over time. For this analysis, the patients were divided into two groups as determined by the length of time between semen sample donations; 75 days was chosen as an arbitrary divider because it divided the patient population into two equal groups. Coincidentally, this threshold cutoff period is approximately equal in duration to a spermatogenesis cycle and ensures that isolated sperm from samples donated >75 days apart arose from distinct germ cell population. The magnitude of the epigenetic differences (|ΔM-values|) across the defined genomic regions were approximately equal in patients who donated samples within and beyond 75 days of one another, illustrating that the magnitude of the DNA methylation differences were generally not location dependent. However, global sperm DNA methylation levels generally decreased over time as represented by the negative ΔM-values, and the decrease was greater in patients who donated semen samples > 75 days apart (Table 4). Although these trends were observed at the global level, analysis at the probe level using a linear model failed to identify any CpGs that were differentially methylated with respect to the samples being collected greater than or less than 75 days apart.
Table 4.
The average epigenetic difference (M-values) observed from semen samples collected either within or beyond 75 days from the same patient.
| Genomic Regiona | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome | Island | Shore | Shelf | TSS1500 | TSS200 | 5′UTR | 1st Exon | Body | 3′UTR | |||
| Global CpGs | < 75 days |
ΔM-valueb | −0.0276 (0.0061) |
0.0062 (0.0179) |
−0.0231 (0.0122) |
−0.0595 (0.0119) |
−0.0093 (0.0125) |
0.0050 (0.0168) |
−0.0117 (0.0097) |
−0.0038 (0.0147) |
−0.0437 (0.0074) |
−0.0473 (0.0057) |
| |ΔM-value|b | 0.3038 (0.0063) |
0.3005 (0.0045) |
0.3120 (0.0062) |
0.3092 (0.0112) |
0.3051 (0.0051) |
0.2899 (0.0040) |
0.2998 (0.0049) |
0.3005 (0.0044) |
0.3087 (0.0082) |
0.3074 (0.0089) |
||
| > 75 days |
ΔM-valueb | −0.0648 (0.0021) |
−0.1212 (0.0135) |
−0.1189 (0.0113) |
0.0193 (0.0022) |
−0.1092 (0.0082) |
−0.0903 (0.0097) |
−0.0736 (0.0044) |
−0.0891 (0.0087) |
−0.0412 (0.0010) |
−0.0258 (0.0017) |
|
| |ΔM-value|b | 0.3092 (0.0119) |
0.3052 (0.0106) |
0.3361 (0.0173) |
0.2800 (0.0060) |
0.3146 (0.0120) |
0.2813 (0.0077) |
0.2935 (0.0087) |
0.2942 (0.0087) |
0.3070 (0.0112) |
0.3025 (0.0102) |
||
| Imprinted CpGs | < 75 days |
ΔM-valueb | −0.0338 (0.0081) |
−0.0095 (0.0157) |
−0.0221 (0.0111) |
−0.0689 (0.0278) |
−0.0014 (0.0139) |
0.0023 (0.0189) |
−0.0253 (0.0083) |
−0.0075 (0.0208) |
−0.0574 (0.0125) |
−0.0306 (0.0027) |
| |ΔM-value|b | 0.3134 (0.0074) |
0.3110 (0.0054) |
0.3218 (0.0080) |
0.3128 (0.0123) |
0.3105 (0.0058) |
0.3022 (0.0041) |
0.3121 (0.0069) |
0.3164 (0.0061) |
0.3204 (0.0102) |
0.3197 (0.0099) |
||
| > 75 days |
ΔM-valueb | −0.0893 (0.0073) |
−0.1525 (0.0218) |
−0.1350 (0.0197) |
0.0250 (0.0026) |
−0.1609 (0.0218) |
−0.1250 (0.0104) |
−0.1139 (0.0098) |
−0.1555 (0.0237) |
−0.0421 (0.0028) |
−0.1254 (0.0183) |
|
| |ΔM-value|b | 0.3323 (0.0175) |
0.3404 (0.0196) |
0.3673 (0.0277) |
0.2809 (0.0086) |
0.3468 (0.0231) |
0.3139 (0.0147) |
0.3353 (0.0192) |
0.3488 (0.0204) |
0.3256 (0.0155) |
0.3699 (0.0273) |
||
probe genomic location are provided in the annotation file supplied by Illumina
values are reported as the mean of the mean patient methylation difference and the mean square between
Analysis of CpGs Associated with Imprinted Genes
The previous analysis was inclusive of all probes, including those that are associated with imprinted genes. The analysis was repeated focusing solely on the probes that are annotated as imprinted genes to determine if the methylation levels of imprinted CpGs are regulated differently. The bimodal distribution of sperm DNA methylation were very similar to the results from the global analysis with one notable exception; imprinted CpGs in the 3′UTR possessed nearly identical proportions of methylated and unmethylated levels, whereas at the global level, the 3′UTR CpGs were predominantly methylated (Table 2). Interestingly when comparing the regional epigenetic differences (ΔM-values) between all CpGs and only imprinted genes (Table 3), a consistent decrease in ΔM-values was observed in the imprinted CpGs of the intra-individual comparisons. The greatest decreases in ΔM-values were observed in the 3′UTR and 1st exon, with ΔM-values decreasing by 0.0415 and 0.035 respectively. The absolute epigenetic difference (|ΔM-values|) of imprinted CpGs was comparable to that of all the CpGs in both the intra- and inter-individual comparisons.
The effect of time on the epigenetic difference of imprinted CpGs in the intra-individual comparison was also investigated (Table 4). The |ΔM-values| from semen samples from patients who donated semen samples greater than and less than 75 days apart were similar for both all and only imprinted CpGs. However, when looking at the ΔM-values, which take into consideration the effect of time between samplings, subtle differences in the overall ΔM-values in each of the genomic regions are observed. For example, samples collected less than 75 days apart generally possessed greater sperm DNA methylation in the first sample compared to the second, especially within the CpG islands, 5′UTR, gene body and 3′UTR. Samples collected greater than 75 days apart were also generally more methylated in the first sample, and the regions with the largest difference between all and only imprinted CpGs were the TSS1500, 1st exon and 3′UTR.
Comparison of Intra- and Inter-Individual Sperm DNA Methylation Differences
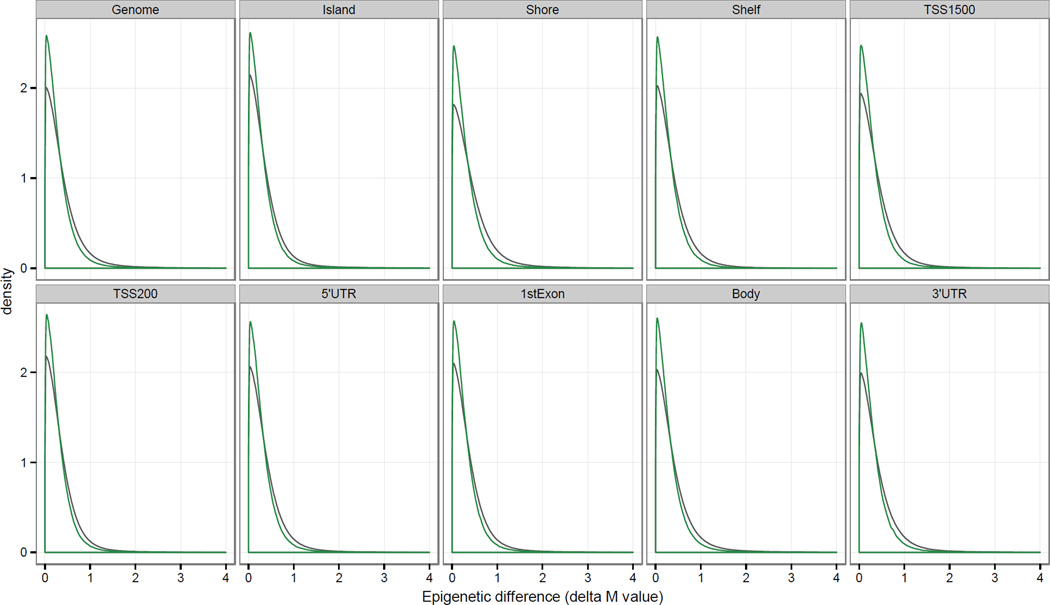
In addition to examining the intra-patient epigenetic differences, the difference in sperm DNA methylation between individuals was also investigated for all possible inter-patient semen samples (n = 264). Unlike the intra-patient comparison of DNA methylation differences where the time between sample collections provides biological insight, the inter-patient comparison analysis focused on the absolute epigenetic difference (|ΔM-values|; Table 3). Similar to the intra-patient comparison of |ΔM-values|, the values were approximately equal among the different genomic regions implying that alterations in sperm DNA methylation are not location dependent. Furthermore the |ΔM-values| from both the intra- and inter-patient comparisons were nearly equal demonstrating that the epigenetic differences are consistent between individuals. Correlation analysis of the |ΔM-values| was strongly correlated both within and between patients across the different genomic regions (r > 0.97), where the intra-patient correlation values were greater than their inter-patient counterpart by an average of 0.0053 (Figure 2 and Supplementary Table 1). Although the differences were small, they were statistically significant across each of the defined genomic regions (p < 0.05; Supplementary Table 1). The difference in the intra- and inter-patient |ΔM-values| is also visualized through their respective kernel density distribution (Figure 3) and Euclidean distance profiles (Supplementary Figure 3). The intra-individual profile peaks were sharper and narrower than the corresponding inter-individual peaks, illustrating the inter-patient DNA methylation variability due to the underlying genetic and/or environmental differences between individuals.
Figure 3.
Intra- and inter-patient distribution of pairwise sperm epigenetic differences (|ΔM-values|) across genomic regions. Normalized and logit-transformed methylation (M) values were used to plot the genome-wide density distribution of the intra- (green) and inter-patient (gray) |ΔM-values|. Intra-patient epigenetic differences were calculated using the 2 collected samples from the same individuals (n=12), while the inter-patient comparison measured the difference between all samples and all patients (n=264).
The intra-patient Pearson correlation analysis found a near-perfect intra-individual correlation between the sperm DNA methylation levels from the 2 samples collected from each individual (r > 0.98). However, the true signal-to-noise ratio can potentially be muted if only a small fraction of the 396,262 probes on the beadchip exhibit high signal variability. Significant differences in the absolute value of the epigenetic difference (|Δβ-value|) were identified via a Wilcoxon test followed by a Benjamini-Hochberg multiple correction (q < 0.05). This identified 21,739 probes with significantly less β-value variability in semen samples from the same donor than in samples from different donors. Further restricting this set to CpGs with a difference in their mean inter- and intra- |Δβ-value| > 0.2 (|Δβ-valueCpG|inter - |Δβ-valueCpG|intra > 0.2) narrowed the list to 305 CpGs corresponding to 198 unique RefSeq identifiers and 130 distinct Entrez Gene IDs (Supplementary Table 2 summarizes the list of 305 CpGs with their q-values and available gene information). This group of CpGs possesses a higher degree of inter-patient variability with a significant difference in the methylation level between the inter- and intra-patient comparison. The associated 130 genes were used for pathway analysis to identify enriched Gene Ontology (GO) terms and KEGG pathways, and the results are summarized in Table 5. Functional analysis found GO and KEGG terms that were significant, or nearing significance (p < 0.05), but the multiple-corrected p-values failed to identify any significant terms.
Table 5.
Functional analysis of gene annotated CpGs with |Δβ-valueCpG|inter - |Δβ-valueCpG|intra > 0.2
| Category | Term | p-value | Benjamini-corrected p−value |
Gene Count |
|---|---|---|---|---|
| GOTERM_MF_FAT | GO:0003779 actin binding | 0.0263 | 0.9977 | 6 |
| GOTERM_MF_FAT | GO:0051015 actin filament binding | 0.0305 | 0.9704 | 3 |
| GOTERM_MF_FAT | GO:0005516 calmodulin binding | 0.0356 | 0.9358 | 4 |
| GOTERM_BP_FAT | GO:0000902 cell morphogenesis | 0.0389 | 1.0000 | 6 |
| KEGG_PATHWAY | hsa04010:MAPK signaling pathway | 0.0393 | 0.8809 | 5 |
| GOTERM_MF_FAT | GO:0005095 GTPase inhibitor activity | 0.0504 | 0.9470 | 2 |
| GOTERM_MF_FAT | GO:0005083 small GTPase regulator activity | 0.0529 | 0.9152 | 5 |
| GOTERM_MF_FAT | GO:0030695 GTPase regulator activity | 0.0572 | 0.8923 | 6 |
| GOTERM_BP_FAT | GO:0032989 cellular component morphogenesis | 0.0572 | 1.0000 | 6 |
| KEGG_PATHWAY | hsa04144:Endocytosis | 0.0598 | 0.8048 | 4 |
| GOTERM_MF_FAT | GO:0060589 nucleoside-triphosphatase regulator activity | 0.0617 | 0.8732 | 6 |
| KEGG_PATHWAY | hsa04612:Antigen processing and presentation | 0.0620 | 0.6771 | 3 |
| GOTERM_MF_FAT | GO:0005524 ATP binding | 0.0735 | 0.8854 | 13 |
| GOTERM_MF_FAT | GO:0032559 adenyl ribonucleotide binding | 0.0796 | 0.8767 | 13 |
| GOTERM_BP_FAT | GO:0030010 establishment of cell polarity | 0.1000 | 1.0000 | 2 |
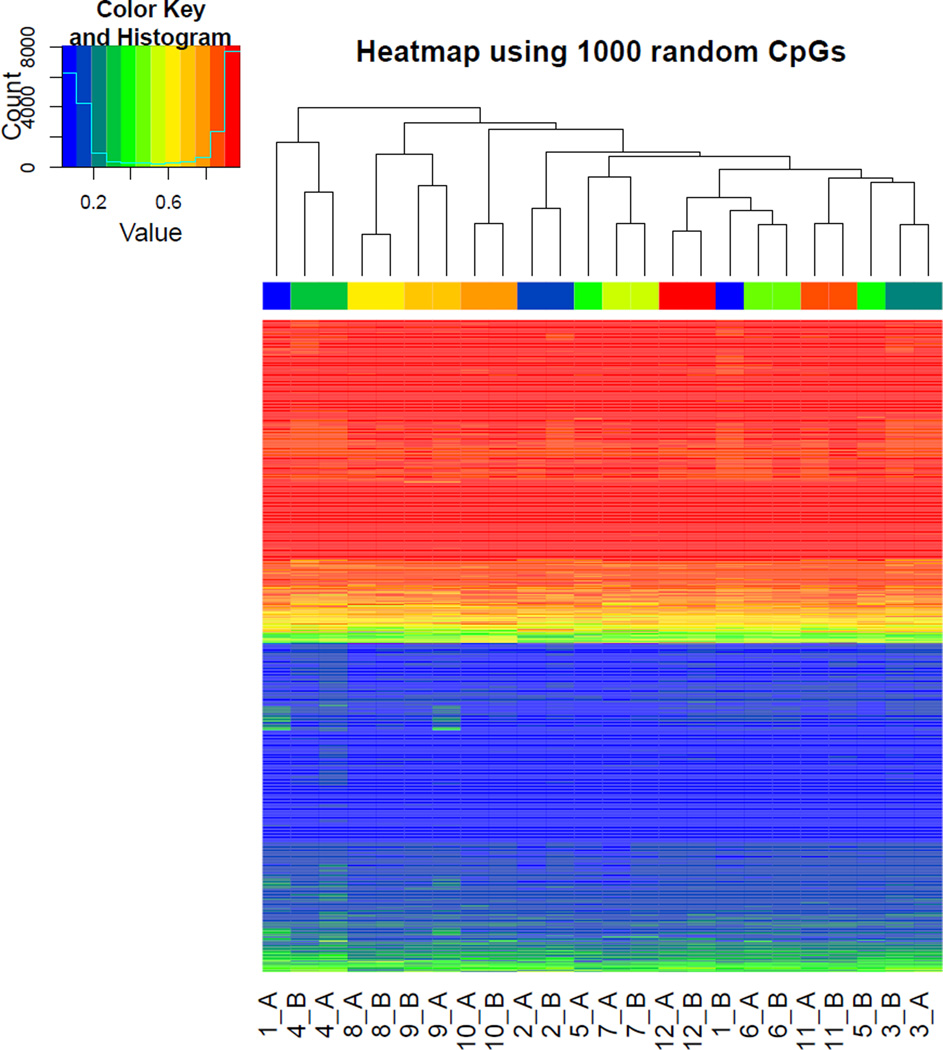
Hierarchical clustering of the β-values for 1,000 random CpGs found that the intra-individual sperm methylation profiles tended to cluster together (Figure 4). However, there were 2 notable exceptions; the two samples from patients #1 and #5 both clustered separately. Interestingly, these two patients were among the patients with the longest elapsed period between semen sample donations; 307 and 489 days respectively. It should be noted that 329 days passed in between sample donations from patient #11, yet his methylation profiles clustered together.
Figure 4.
Hierarchical clustering of sperm DNA methylation levels. The β-values of 1,000 randomly selected CpGs were clustered vertically by CpG and horizontally by patient and sample (A = 1st sample, B = 2nd sample). The clustering patterns illustrate both the bimodal DNA methylation distributions shown in Figure 1, and how samples from the same patient generally clustered together.
Variation as a function of patient age, BMI, and measured sperm parameters
The variability in the sperm DNA methylation profiles were evaluated for their associations with various patient parameters, including age, BMI, time between semen samples, and semen characteristics. The epigenetic difference of each probe as calculated by |ΔM-values| was used to determine the global mean and standard deviation across the patients, and these values were used for the correlation analyses against patient age, BMI, time between semen collections, and changes in sperm volume, density, count, and motility. BMI values were well correlated with the |ΔM-values| across all probes measured with r = 0.66 (p = 0.019). Patients with a BMI value greater than 24 correlated significantly with the mean |ΔM-values| with r = 0.72 (p = 0.046). BMI values > 24 were included in this analysis because they approached the overweight classification as defined by the World Health Organization (BMI values ≥ 25). No significant correlation with age was observed in the patient population of this study. Neither the mean nor the standard deviation of the |ΔM-values| correlated with changes in sperm parameters or the time between semen sample collections.
The effects of BMI on the observed global and regional epigenetic differences were further evaluated by categorizing patients by their BMI values; BMI < 24, BMI between 24 and 30, and BMI > 30 (Table 6). Patients with BMI > 30 generally exhibited less DNA methylation in their second semen sample, while the absolute epigenetic difference (|ΔM-values|) were markedly increased compared to the other two BMI categories. The mean intra-individual |ΔM-values| were also considerably greater in the highest BMI category, and agree with the findings where BMI is correlated with |ΔM-values|.
Table 6.
Intra- and inter-patient mean sperm epigenetic differences (ΔM-values) by BMI classification.
| Genomic Regiona | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome | Island | Shore | Shelf | TSS1500 | TSS200 | 5′UTR | 1st Exon | Body | 3′UTR | |||
| BMI < 24 | Intra | ΔM-value b | −0.0320 (0.0011) |
−0.0406 (0.0049) |
−0.0472 (0.0055) |
−0.0098 (0.0009) |
−0.0434 (0.0031) |
−0.0331 (0.0020) |
−0.0307 (0.0012) |
−0.0308 (0.0023) |
−0.0224 (0.0004) |
−0.0298 (0.0003) |
| |ΔM-value|b | 0.2306 (0.0016) |
0.2286 (0.0020) |
0.2436 (0.0030) |
0.2235 (0.0004) |
0.2332 (0.0016) |
0.2131 (0.0005) |
0.2225 (0.0007) |
0.2237 (0.0009) |
0.2328 (0.0017) |
0.2310 (0.0013) |
||
| Inter | |ΔM-value|b | 0.3087 (0.0010) |
0.2672 (0.0014) |
0.3288 (0.0013) |
0.3003 (0.0005) |
0.2951 (0.0008) |
0.2515 (0.0004) |
0.2755 (0.0006) |
0.2673 (0.0007) |
0.3133 (0.0012) |
0.3079 (0.0011) |
|
| BMI 24 – 30 | Intra | ΔM-value b | −0.0189 (0.0082) |
−0.0095 (0.0276) |
−0.0137 (0.0172) |
−0.0285 (0.0105) |
−0.0127 (0.0196) |
−0.0056 (0.0267) |
−0.0165 (0.0152) |
−0.0185 (0.0224) |
−0.0221 (0.0070) |
−0.0260 (0.0055) |
| |ΔM-value|b | 0.2848 (0.0048) |
0.2970 (0.0050) |
0.2952 (0.0049) |
0.2730 (0.0053) |
0.2933 (0.0045) |
0.2845 (0.0040) |
0.2876 (0.0044) |
0.2943 (0.0048) |
0.2832 (0.0055) |
0.2785 (0.0053) |
||
| Inter | |ΔM-value|b | 0.4167 (0.0038) |
0.4009 (0.0083) |
0.4434 (0.0051) |
0.4014 (0.0023) |
0.4157 (0.0061) |
0.3850 (0.0080) |
0.4030 (0.0058) |
0.4039 (0.0078) |
0.4154 (0.0028) |
0.4133 (0.0026) |
|
| BMI > 30 | Intra | ΔM-value b | −0.1107 (0.0004) |
−0.1600 (0.0339) |
−0.1982 (0.0081) |
−0.0197 (0.0289) |
−0.1579 (0.0127) |
−0.1173 (0.0247) |
−0.1020 (0.0087) |
−0.1140 (0.0237) |
−0.1030 (0.0035) |
−0.0631 (0.0101) |
| |ΔM-value|b | 0.4439 (0.0016) |
0.4117 (0.0034) |
0.4794 (0.0061) |
0.4255 (0.0046) |
0.4395 (0.0025) |
0.3840 (0.0015) |
0.4107 (0.0005) |
0.4007 (0.0016) |
0.4490 (0.0020) |
0.4475 (0.0018) |
||
| Inter | |ΔM-value|b | 0.4729 (0.0088) |
0.4557 (0.0146) |
0.5273 (0.0177) |
0.4168 (0.0024) |
0.4655 (0.0119) |
0.3977 (0.0066) |
0.4233 (0.0060) |
0.4261 (0.0084) |
0.4745 (0.0081) |
0.4640 (0.0064) |
|
probe genomic location are provided in the annotation file supplied by Illumina
values are reported as the mean of the mean patient methylation difference and the mean square between
Additional correlation analyses were performed using intra-individual DNA methylation differences at the probe level. Using an absolute change in methylation (|Δβ-value|) ≥ 0.2 as threshold to identify a significant change in intra-patient DNA methylation identified 8 probes. These probes possessed the greatest change in sperm DNA methylation between the two collected semen samples, and their |ΔM-values| were used to identify correlations of intra-patient sperm DNA methylation with patients’ metadata information and changes in semen parameters. Two probes were significantly correlated with BMI; cg26006558 (r = 0.75; p = 0.005) and cg20386487 (r = 0.77; p = 0.034). However, only probe cg26006558 was significantly correlated with BMI values above 24 (r = 0.80; p = 0.018). Of the measured semen parameters, only changes in sperm concentration were found to significantly correlate with changes in sperm DNA methylation for two probes; cg14251216 (r = 0.66; p = 0.018) and cg20386487 (r = 0.72; p = 0.009). Detection of these probes suggest that altered methylation at these sites are associated with physiological changes and/or sperm quality. However, none of these probes possessed any gene annotation thereby limiting mechanistic insight into the regulatory nature of these CpGs.
Discussion
In this study, semen samples were collected twice from 12 different patients and used to determine the sperm DNA methylation variation across the genome. Globally, the CpGs possessed a clear bimodal distribution profile reflecting either an unmethylated or methylated state of nearly equal proportions. However, the methylation profiles were location-dependent and varied among the different genomic regions, consistent with observations seen across multiple tissue types (Lokk et al. 2014). For example, CpGs in CpG islands, in the 1st exon, and around the TSS were largely unmethylated and studies have demonstrated that methylation of these CpGs are linked to transcriptional silencing and gene inactivation (Brenet et al. 2011, Feng et al. 2010, Lister et al. 2009, Suzuki & Bird 2008, Zemach et al. 2010). These regions are known to be key epigenetic regulators of gene expression and their highly unmethylated state may reflect this role.
To date, there have been numerous studies that have associated sperm DNA methylation levels with sperm quality and function (El Hajj et al. 2011, Hammoud et al. 2013, Jenkins et al. 2014, Krausz et al. 2012, Nanassy & Carrell 2011, Pacheco et al. 2011, Tunc & Tremellen 2009), but these studies have not generally examined intra-individual DNA methylation variability. However, one recent study investigated the aging effects on sperm DNA methylation profiles from semen samples longitudinally collected over 9+ years from 17 patients (Jenkins et al. 2014). In the current study, the change in sperm DNA methylation was examined from 12 patients over a shorter period time spanning weeks instead of years. The global change in sperm DNA methylation levels from two semen samples collected from the same patient found an overall reduction in the methylation state as represented by the decrease in the ΔM-value globally across the genome and within the defined genomic regions. Changes in methylation levels were most pronounced in CpG islands and shores, and least in the shelves and 3′UTRs (Table 3). However the magnitude of the changes (|ΔM-values|) were similar across all the regions. Although our study did not specifically examine the effects of age on DNA methylation, it found that patients who provided two semen samples >75 days apart generally exhibited a global decrease in DNA methylation over time compared to those whose samples were <75 days apart (Table 4). This differs from a study which found elevated global sperm DNA methylation levels in men over 45 years of age when compared to their younger counterparts (Jenkins et al. 2014). The conflicting results are difficult to interpret since our study only had one participant over the age of 45 years old.
The inherent genetic heterogeneity, and differences in lifestyle and environmental exposures between individuals is likely to influence sperm DNA methylation variability between individuals, and is illustrated by the shorter and broader methylation level density distribution and Euclidean distance profiles (Figure 3 and Supplementary Figure 3). A set of 305 CpGs (Supplementary Table 1) was identified that possessed higher methylation variability and greater methylation difference in the inter-patient comparisons with respect to the intra-patient comparisons. These CpGs may serve as indicators of an individual’s underlying genetics and/or environmental exposure, and have potential uses in the field of criminal forensics (Lee et al. 2015). Interestingly, the |ΔM-values| from the inter-patient comparison were similar to those of the intra-patient comparison. It should be noted that the interpretation of the inter-patient ΔM-value is ambiguous due to the fact that there is no time component when comparing semen samples collected from different individuals. However, the order of magnitude increase in the |ΔM-value| standard deviation reflects the increased inter-individual variability (Table 3). Despite the increase in sperm DNA methylation inter-patient variability, the Pearson correlation coefficients of the M-values between individuals were very strong (r > 0.973; Table 3). This suggests that the regulatory roles imparted by the methylation status of CpGs in sperm are highly conserved between individuals.
A growing number of studies are uncovering associations between sperm epigenetic changes with decreases in sperm quality and function (El Hajj et al. 2011, Hammoud et al. 2013, Jenkins et al. 2014, Krausz et al. 2012, Nanassy & Carrell 2011, Pacheco et al. 2011, Tunc & Tremellen 2009). In this study, the intra-individual epigenetic differences at the global level did not significantly correlate with changes in semen parameters, but at the individual probe level, 2 probes were significantly correlated with changes in sperm concentration: cg14251216 and cg20386487. Environmental exposures and conditions are also known to affect sperm quality and function, while also changing the sperm epigenetics. There is evidence linking obesity with decreases in sperm concentrations and total counts, as well as in the overall sperm health (Sermondade et al. 2013, Teerds et al. 2011). Studies in mice using high fat diets (Palmer et al. 2012) and prediabetic models (Wei et al. 2014) exhibited aberrant global sperm DNA methylation. The intra-individual comparisons in this study found a strong correlation between BMI and 2 measured probes (cg26006558 and cg20386487), which are consistent with the overall findings from the mice studies.
This study provides a detailed evaluation of the stability of sperm DNA methylation over time and between individuals. However, our small patient population size limits our ability to statistically identify the full spectrum of sperm CpGs that may associate with sperm quality and/or environmental exposures. Future studies to increase the patient population will aid in addressing this concern and help to identify CpGs whose methylation status are critical in maintaining sperm integrity.
Conclusions
In summary, this study provides a detailed comparison of the sperm DNA methylation profiles intra- and inter-individual variations. The intra-individual methylation levels generally decreased slightly over time across the defined genomic regions between samples but remained strongly correlated with one another. The methylation levels between patients were also strongly correlated, however, the correlation values were a fraction less than the intra-patient values. These results demonstrate that sperm DNA methylation levels are relatively stable between semen sample collections and between individuals. High fidelity of the methylation marks both in the intra- and inter-individual comparisons suggests that the methylation status of sperm CpGs have important regulatory roles in maintaining proper sperm function.
Supplementary Material
Supplementary Figure 1: Sample contrast matrix illustrating all of the intra- and inter-patient sample comparisons. The gray cells on the diagonal represent each of the samples where the numbers are the patient IDs, and the letters A and B are for the 1st and 2nd collected semen samples, respectively. The green cells represent the 12 intra-patient comparisons where the 1st and 2nd samples from each patient are compared (12 comparisons). The blue cells represent all the inter-patient comparisons performed (264 comparisons). The epigenetic differences were calculated for each comparison group (i.e. intra- and inter-) across the different genomic regions.
Supplementary Figure 2: Hierarchical clustering of the β-values for the top 600 most leukocyte discriminating probes (Jaffe, Reinius et al. 2012) with the corresponding β-values from the collected semen samples. The sperm DNA methylation signatures are distinct from those of the FACS-sorted peripheral blood cell populations; neutrophils (granulocytes [Gran]), lymphocytes (CD8+ [CD8T] and CD4+ [CD4T] T cells, CD56+ natural killer cells [NK] and CD19+ B cells [Bcell]) and CD14+ monocytes (Mono).
Supplementary Figure 3: Euclidian distance profiles of the intra- and inter-patient |ΔM-values| across the genomic regions.
Acknowledgments
This research is supported by the NIEHS Superfund Research Program P42 ES013660. This paper is subject to the NIH Public Access Policy.
Footnotes
Disclosures: K. Boekelheide is an occasional expert consultant for chemical and pharmaceutical companies and owns stock in Semma Therapeutics, an early stage biotechnology company developing a therapeutic for type 1 diabetes. These activities are unrelated to the current work but are mentioned in the spirit of full disclosure.
References
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol. 2007;8:227. doi: 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhorn R, Reed S, Tanphaichitr N. Aberrant protamine 1/protamine 2 ratios in sperm of infertile human males. Experientia. 1988;44:52–55. doi: 10.1007/BF01960243. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, Scandura JM. DNA Methylation of the First Exon Is Tightly Linked to Transcriptional Silencing. PLoS ONE. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schübeler D, Stadler MB, Peters AH. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- Carrell DT, Hammoud SS. The human sperm epigenome and its potential role in embryonic development. Mol Hum Reprod. 2010;16:37–47. doi: 10.1093/molehr/gap090. [DOI] [PubMed] [Google Scholar]
- Dada R, Kumar M, Jesudasan R, Fernández JL, Gosálvez J, Agarwal A. Epigenetics and its role in male infertility. J Assist Reprod Genet. 2012;29:213–223. doi: 10.1007/s10815-012-9715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadoune JP. The nuclear status of human sperm cells. Micron. 1995;26:323–345. doi: 10.1016/0968-4328(95)00007-0. [DOI] [PubMed] [Google Scholar]
- El Hajj N, Zechner U, Schneider E, Tresch A, Gromoll J, Hahn T, Schorsch M, Haaf T. Methylation status of imprinted genes and repetitive elements in sperm DNA from infertile males. Sex Dev. 2011;5:60–69. doi: 10.1159/000323806. [DOI] [PubMed] [Google Scholar]
- Erkek S, Hisano M, Liang CY, Gill M, Murr R, Dieker J, Schübeler D, van der Vlag J, Stadler MB, Peters AH. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol. 2013;20:868–875. doi: 10.1038/nsmb.2599. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen SE. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JM, Popendikyte V, Pozdniakovaite N, Sobolev M, Assadzadeh A, Schumacher A, Zangeneh M, Lau L, Virtanen C, Wang SC, Petronis A. Intra- and interindividual epigenetic variation in human germ cells. Am J Hum Genet. 2006;79:67–84. doi: 10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin JP, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, Greenwood CM, Hansen KD. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15:503. doi: 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich R, Johnson G, Krawetz SA. The Preparation of Human Spermatozoal RNA for Clinical Analysis. Syst Biol Reprod Med. 2007;53:161–167. doi: 10.1080/01485010701216526. [DOI] [PubMed] [Google Scholar]
- Griffin J. Methods of sperm DNA extraction for genetic and epigenetic studies. Methods Mol Biol. 2013;927:379–384. doi: 10.1007/978-1-62703-038-0_32. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Cairns BR, Carrell DT. Analysis of gene-specific and genome-wide sperm DNA methylation. Methods Mol Biol. 2013;927:451–458. doi: 10.1007/978-1-62703-038-0_39. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht NB. Regulation of ‘haploid expressed genes’ in male germ cells. J Reprod Fertil. 1990;88:679–693. doi: 10.1530/jrf.0.0880679. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jaffe AE. FlowSorted.Blood.450k: Illumina HumanMethylation data on sorted blood cell populations. R package version 1.8.0 [Google Scholar]
- Jenkins TG, Aston KI, Pflueger C, Cairns BR, Carrell DT. Age-associated sperm DNA methylation alterations: possible implications in offspring disease susceptibility. PLoS Genet. 2014;10:e1004458. doi: 10.1371/journal.pgen.1004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TG, Carrell DT. The sperm epigenome and potential implications for the developing embryo. Reproduction. 2012;143:727–734. doi: 10.1530/REP-11-0450. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Krausz C, Sandoval J, Sayols S, Chianese C, Giachini C, Heyn H, Esteller M. Novel insights into DNA methylation features in spermatozoa: stability and peculiarities. PLoS ONE. 2012;7:e44479. doi: 10.1371/journal.pone.0044479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Jung SE, Oh YN, Choi A, Yang WI, Shin KJ. Epigenetic age signatures in the forensically relevant body fluid of semen: a preliminary study. Forensic Sci Int Genet. 2015;19:28–34. doi: 10.1016/j.fsigen.2015.05.014. [DOI] [PubMed] [Google Scholar]
- Li J, Harris RA, Cheung SW, Coarfa C, Jeong M, Goodell MA, White LD, Patel A, Kang SH, Shaw C, Chinault AC, Gambin T, Gambin A, Lupski JR, Milosavljevic A. Genomic hypomethylation in the human germline associates with selective structural mutability in the human genome. PLoS Genet. 2012;8:e1002692. doi: 10.1371/journal.pgen.1002692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokk K, Modhukur V, Rajashekar B, Märtens K, Mägi R, Kolde R, Koltšina M, Nilsson TK, Vilo J, Salumets A, Tõnisson N. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 2014;15:r54. doi: 10.1186/gb-2014-15-4-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–1730. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol. 2002;(4 Suppl):s41–S49. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- Maynard ND, Chen J, Stuart RK, Fan JB, Ren B. Genome-wide mapping of allele-specific protein-DNA interactions in human cells. Nat Methods. 2008;5:307–309. doi: 10.1038/nmeth.1194. [DOI] [PubMed] [Google Scholar]
- Nanassy L, Carrell DT. Analysis of the methylation pattern of six gene promoters in sperm of men with abnormal protamination. Asian J Androl. 2011;13:342–346. doi: 10.1038/aja.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Costa P, Nogueira P, Carvalho M, Leal F, Cordeiro I, Calhaz-Jorge C, Gonçalves J, Plancha CE. Incorrect DNA methylation of the DAZL promoter CpG island associates with defective human sperm. Hum Reprod. 2010;25:2647–2654. doi: 10.1093/humrep/deq200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Oliva R, Dixon GH. Vertebrate protamine genes and the histone-to-protamine replacement reaction. Prog Nucleic Acid Res Mol Biol. 1991;40:25–94. doi: 10.1016/s0079-6603(08)60839-9. [DOI] [PubMed] [Google Scholar]
- Overstreet JW. Clinical approach to male reproductive problems. Occup Med. 1994;9:387–404. [PubMed] [Google Scholar]
- Pacheco SE, Houseman EA, Christensen BC, Marsit CJ, Kelsey KT, Sigman M, Boekelheide K. Integrative DNA methylation and gene expression analyses identify DNA packaging and epigenetic regulatory genes associated with low motility sperm. PLoS ONE. 2011;6:e20280. doi: 10.1371/journal.pone.0020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer NO, Bakos HW, Fullston T, Lane M. Impact of obesity on male fertility, sperm function and molecular composition. Spermatogenesis. 2012;2:253–263. doi: 10.4161/spmg.21362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R, Y Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. URL https://www.R-project.org/ [Google Scholar]
- Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlen SE, Greco D, Soderhall C, Scheynius A, Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS ONE. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero Y, Meikar O, Papaioannou MD, Conne B, Grey C, Weier M, Pralong F, De Massy B, Kaessmann H, Vassalli JD, Kotaja N, Nef S. Dicer1 depletion in male germ cells leads to infertility due to cumulative meiotic and spermiogenic defects. PLoS ONE. 2011;6:e25241. doi: 10.1371/journal.pone.0025241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sermondade N, Faure C, Fezeu L, Shayeb AG, Bonde JP, Jensen TK, Van Wely M, Cao J, Martini AC, Eskandar M, Chavarro JE, Koloszar S, Twigt JM, Ramlau-Hansen CH, Borges E, Lotti F, Steegers-Theunissen RP, Zorn B, Polotsky AJ, La Vignera S, Eskenazi B, Tremellen K, Magnusdottir EV, Fejes I, Hercberg S, Levy R, Czernichow S. BMI in relation to sperm count: an updated systematic review and collaborative meta-analysis. Hum Reprod Update. 2013;19:221–231. doi: 10.1093/humupd/dms050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Teerds KJ, de Rooij DG, Keijer J. Functional relationship between obesity and male reproduction: from humans to animal models. Hum Reprod Update. 2011;17:667–683. doi: 10.1093/humupd/dmr017. [DOI] [PubMed] [Google Scholar]
- Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunc O, Tremellen K. Oxidative DNA damage impairs global sperm DNA methylation in infertile men. J Assist Reprod Genet. 2009;26:537–544. doi: 10.1007/s10815-009-9346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward WS, Coffey DS. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod. 1991;44:569–574. doi: 10.1095/biolreprod44.4.569. [DOI] [PubMed] [Google Scholar]
- Wei Y, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, Sun QY. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci USA. 2014;111:1873–1878. doi: 10.1073/pnas.1321195111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Wu H, Loh YH, Briem E, Daley GQ, Feinberg AP. Euchromatin islands in large heterochromatin domains are enriched for CTCF binding and differentially DNA-methylated regions. BMC Genomics. 2012;13:566. doi: 10.1186/1471-2164-13-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Department of Reproductive Health Research. 5th. Geneva: WHO Laboratory Manual for the Examination and Processing of Human Semen; 2010. [Google Scholar]
- Yaman R, Grandjean V. Timing of entry of meiosis depends on a mark generated by DNA methyltransferase 3a in testis. Mol Reprod Dev. 2006;73:390–397. doi: 10.1002/mrd.20430. [DOI] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Sample contrast matrix illustrating all of the intra- and inter-patient sample comparisons. The gray cells on the diagonal represent each of the samples where the numbers are the patient IDs, and the letters A and B are for the 1st and 2nd collected semen samples, respectively. The green cells represent the 12 intra-patient comparisons where the 1st and 2nd samples from each patient are compared (12 comparisons). The blue cells represent all the inter-patient comparisons performed (264 comparisons). The epigenetic differences were calculated for each comparison group (i.e. intra- and inter-) across the different genomic regions.
Supplementary Figure 2: Hierarchical clustering of the β-values for the top 600 most leukocyte discriminating probes (Jaffe, Reinius et al. 2012) with the corresponding β-values from the collected semen samples. The sperm DNA methylation signatures are distinct from those of the FACS-sorted peripheral blood cell populations; neutrophils (granulocytes [Gran]), lymphocytes (CD8+ [CD8T] and CD4+ [CD4T] T cells, CD56+ natural killer cells [NK] and CD19+ B cells [Bcell]) and CD14+ monocytes (Mono).
Supplementary Figure 3: Euclidian distance profiles of the intra- and inter-patient |ΔM-values| across the genomic regions.