Abstract
The multisubunit eukaryotic translation initiation factor eIF3 is thought to assist in the recruitment of ribosomes to mRNA. The expression of eIF3 subunits is frequently disrupted in human cancers, but the specific roles of individual subunits in mRNA translation and cancer remain elusive. Using global transcriptomic, proteomic, and metabolomic profiling, we found a striking failure of Schizosaccharomyces pombe cells lacking eIF3e and eIF3d to synthesize components of the mitochondrial electron transport chain, leading to a defect in respiration, endogenous oxidative stress, and premature aging. Energy balance was maintained, however, by a switch to glycolysis with increased glucose uptake, upregulation of glycolytic enzymes, and strict dependence on a fermentable carbon source. This metabolic regulatory function appears conserved in human cells where eIF3e binds metabolic mRNAs and promotes their translation. Thus, via its eIF3d-eIF3e module, eIF3 orchestrates an mRNA-specific translational mechanism controlling energy metabolism that may be disrupted in cancer.
Graphical abstract
Introduction
Protein synthesis through mRNA translation is the dominant determinant of cellular protein levels (Schwanhausser et al., 2011). Translation initiation is considered a rate-limiting step in protein synthesis that is governed by the availability and activity of eukaryotic translation initiation factors, eIFs (Sonenberg and Hinnebusch, 2009). eIF3 is the most complex translation initiation factor (Hinnebusch, 2006), comprising 13 subunits in mammals (Damoc et al., 2007; Querol-Audi et al., 2013) and 11 subunits in the fission yeast S. pombe (Sha et al., 2009; Zhou et al., 2005). eIF3 appears to encircle the 40S ribosome to serve as a scaffold orchestrating the recruitment of other eIFs involved in mRNA binding, scanning, and AUG recognition (Erzberger et al., 2014; des Georges et al., 2015; Querol-Audi et al., 2013). For certain mRNAs, the eIF3-dependent initiation mechanism involves direct interactions with RNA stem-loop structures or methylated adenosines within the 5’-UTR (Lee et al., 2015; Meyer et al., 2015).
Upregulation of eIF3 subunits is frequently observed in human cancers (Hershey, 2015). Overexpression of some subunits can drive de novo holo-complex formation and modest increases in protein synthesis along with cell transformation (Zhang et al., 2006), although the specific mechanisms leading to transformation remain unknown. A recent study suggested that eIF3 promotes the synthesis of proteins related to cell proliferation and demonstrated that eIF3-mediated synthesis of c-JUN promotes cell migration (Lee et al., 2015).
Not all eIF3 subunits are essential, however, suggesting that some subunits have regulatory functions such as mediating the translation of subsets of mRNAs under specific conditions (Choudhuri et al., 2013; Grzmil et al., 2010; Kim et al., 2007; Zhou et al., 2005). For example, eIF3e was first identified as a gene disrupted by integration of Mouse Mammary Tumor Virus during breast tumorigenesis (Asano et al., 1997; Marchetti et al., 1995), and eIF3e is downregulated in several human cancers (Buttitta et al., 2005; Hershey, 2015; Marchetti et al., 2001; Suo et al., 2015). Downregulation of eIF3e induces epithelial-mesenchymal transition in breast epithelial cells (Gillis and Lewis, 2012) and transdifferentiation of human mesynchemal stem cells into carcinoma-associated fibroblasts (Suo et al., 2015), but the molecular mechanisms underlying the apparent tumor suppressor function of eIF3e remain unknown.
As in breast cancer, eif3e is dispensable in fungi, including fission yeast (Smith et al., 2013; Zhou et al., 2005). Cells deleted for the gene encoding eIF3e (aka. Yin6p or Int6p) or its binding partner eIF3d (Moe1p) show a ~25% reduction in global protein synthesis and growth, and are hypersensitive to stress conditions (Bandyopadhyay et al., 2000, 2002). While these observations suggested potential mRNA-selective functions, the specific impact of eIF3e on protein synthesis and its role in breast tumorigenesis have remained elusive. We show here that eIF3e and eIF3d form a specificity module for the efficient synthesis of components of the mitochondrial electron transport chain (ETC) and that lack of eIF3d and eIF3e leads to a metabolic switch from respiration to glycolysis similar to what is frequently observed in cancer cells undergoing the Warburg effect. The data implicate the eIF3d-eIF3e module of eIF3 in a translational circuit to uphold metabolic balance that may be disrupted in human cancer.
Results
Cells deleted for eIF3 subunits e and d are deficient in initiation of translation
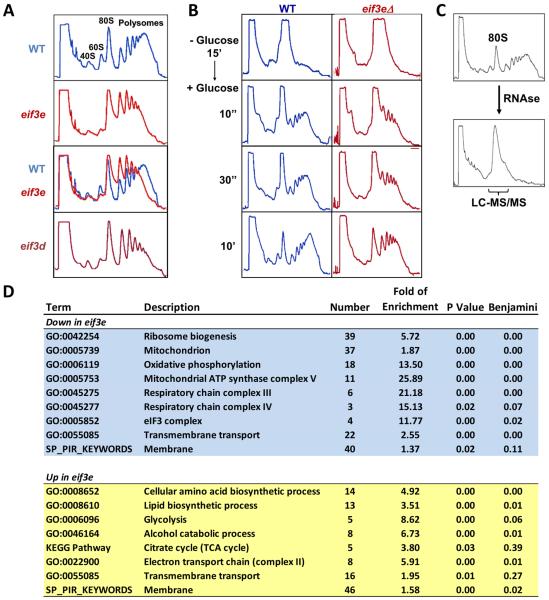
Using sucrose density gradient centrifugation, we observed that eif3e deleted cells have a disturbed polysome profile with accumulation of light polysomes (<5) at the expense of heavy polysomes (>5, Fig. 1A). A strain deleted for eif3d, encoding a direct interactions partner of eIF3e (Yen and Chang, 2000), has the same growth and stress phenotype (Bandyopadhyay et al., 2002) and showed a polysome profile identical to eif3e deleted cells (Fig. 1A). The abnormal profile suggested that the mutants are defective in recruiting ribosomes to mRNAs. To determine the efficiency of translation initiation, polysome run-off was induced by removing glucose from the culture media for 15 minutes, followed by glucose re-addition to induce initiation of translation. Whereas wild-type cells efficiently re-established polysomes containing more than 5 ribosomes within 10 seconds of glucose re-addition, the bulk of polysomes formed in eif3e mutant cells within the same period contained fewer than 4 ribosomes (Fig. 1B). This defect, which was even more pronounced at 30 seconds after glucose re-addition indicated a reduced efficiency in recruiting ribosomes to mRNAs.
Figure 1. Effect of deletion of eif3e and eif3d on protein synthesis.
A) Sucrose density gradient profile of the indicated wild-type (blue) and eif3 mutant (red) strains.
B) Polysome run-off was induced by glucose withdrawal for 15 minutes, and translation initiation was assesses by sucrose density gradient centrifugation upon re-addition of glucose for the indicated times. Glucose-induced protein synthesis was stopped at the indicated time points by the addition of 100 ug/ml cycloheximide.
C) Strategy for 80S proteomics. Cell lysate was digested with RNAse 1 and separated by sucrose density gradient centrifugation. Fractions containing 80S ribosomes were analyzed by liquid chromatography and tandem mass spectrometry (LC-MS/MS).
D) Gene ontology terms enriched in the list of proteins that were significantly up- (yellow) or down-regulated (blue) in 80S complexes isolated from eif3e mutant cells. Data are from a total of five repeat experiments.
Proteomic analysis of ribosome-associated proteins points to a role of eIF3e in the synthesis of mitochondrial ETC components
To gain further insight into the apparent initiation defect of eif3e mutant cells, we sought to assess the state of translating ribosomes. Cell lysate from wild-type and eif3e deleted cells was digested with RNAse, and 80S ribosomes and their associated proteins and nascent polypeptides were isolated and analyzed by liquid chromatography and tandem mass spectrometry (“80S proteomics”, Fig. 1C). In five independent experiments, 1622 proteins were quantified of which 261 showed a statistically significant difference (p ≤ 0.05) between the wild-type and eif3e mutant cells (Supplementary Data File 1).
Gene Ontology (GO) enrichment analysis (Huang et al., 2008) revealed that the group of 130 proteins that were downregulated in the 80S fraction of eif3e deleted cells fell into three main categories: ribosome biogenesis, transmembrane transport, and oxidative phosphorylation, in particular subunits of mitochondrial respiration complexes III, IV, and V (Fig. 1D, Fig. S1A). The eIF3 complex (GO:5852) was also enriched owing to minor depletion of eIF3b and h and the virtually complete absence of eIF3d from 80S particles isolated from eif3e deleted cells (Fig. S1B). This finding is consistent with our previous demonstration that S. pombe eIF3 complexes missing subunit e are also devoid of eIF3d (Sha et al., 2009) and suggests that eIF3d and eIF3e are recruited into the complex as a module. At the same time, these data complement our previous evidence that a stable eIF3 complex can exist in the absence of eIF3d and eIF3e (Sha et al., 2009; Zhou et al., 2005).
The list of 131 proteins whose abundance was significantly increased in 80S complexes isolated from eif3e deleted cells was enriched for amino acid and lipid biosynthesis, transmembrane proteins, and glycolysis with its two downstream metabolic pathways alcohol fermentation and tricarboxylic acid (TCA) cycle. Only minor differences were found in ribosomal proteins (Fig. S1C), other initiation factors (Fig. S1D), and ribosome-associated chaperones (HSP70s, CCT complex) suggesting that overall 80S structure is intact in the absence of eif3e.
Cells deleted for eif3e are defective in synthesizing ETC proteins
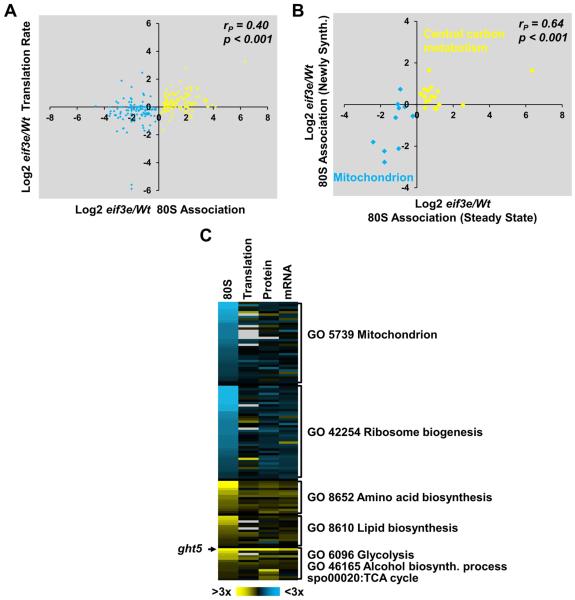
To assess whether the differences in the 80S-associated proteome observed between wild-type and eif3e mutant cells reflected differences in the synthesis of nascent proteins, we acquired a series of datasets on global protein dynamics employing stable isotope labeling techniques coupled with mass spectrometry. Differences in translation rates as determined in a pulsed SILAC experiment (Selbach et al., 2008) correlated significantly (r = 0.40; p < 0.001) with the differences observed in the 80S-associated proteome (Fig. 2A). In addition, out of the translation rates determined for 2597 proteins, 228 were significantly reduced in eif3e mutant cells, whereas 187 were increased. The lists of proteins with significantly altered rates of synthesis enriched GO terms that overlapped with those obtained for proteins with altered 80S association shown in Fig. 1D, indicating that the synthesis of components of the mitochondrial respiration chain is downregulated in eif3e deleted cells, whereas amino acid biosynthesis and glucose metabolism are upregulated (Fig. S2A, Supplementary Data File 1). Identical GO terms were also enriched in the set of 694 proteins whose steady-state levels significantly differed in eif3e mutant cells as determined by quantitative mass spectrometry (Fig. S2B, Supplementary Data File 1).
Figure 2. Effect of deletion of eif3e on the synthesis of metabolism-related proteins.
A) Correlation between changes in the association of a protein with 80S complexes and its translation rate as determined by pulsed SILAC in the absence of eif3e. Translation rates are averages of two replicates. Pearson coefficient and p value of an ANOVA regression analysis are indicated.
B) 80S complexes were isolated from cells pulse-labeled with 15N-NH4Cl to profile changes in the 80S association of newly synthesized metabolism-related proteins in eif3e mutant cells relative to wild-type (WT) cells. The resulting eif3e/WT log2 ratios were plotted on the vertical axis. Data are averages of two independent experiments. Corresponding steady-state 80S association ratios were plotted on the horizontal axis. Pearson coefficient and p value of an ANOVA regression analysis are indicated.
C) Intensity map of changes (log2) in 80S association, translation (pulsed SILAC), and steady-state protein and mRNA levels eif3e deleted cells /WT ratios. Proteins are sorted according to gene ontology terms. Missing values are in light grey.
We next directly measured changes in the synthesis rates of 80S-associated proteins in wild-type and eif3e mutant cells by labeling nascent proteins through growing cells in media containing heavy nitrogen (15N), followed by purification of 80S complexes and mass spectrometry. Observing changes in 14N/15N ratios of the identified peptides allowed us to assess the rate of synthesis of 232 proteins associated with 80S complexes (Supplementary Data File 1) of which 27 represented metabolism-related proteins that we had found altered in their 80S association in eif3e mutant cells. Plotting protein synthesis rates versus steady state levels of these 80S-associated proteins revealed a highly significant correlation (r = 0.64; p < 0.001, Fig. 2B), again indicating that mitochondrial proteins with reduced 80S ribosome association in eif3e mutant cells correspond to those with a reduced rate of synthesis. Conversely, increased association of glycolysis and TCA cycle proteins appears to reflect increased synthesis.
Although there was an overall positive correlation between a protein’s association with 80S complexes and its corresponding mRNA level (r = 0.45, p < 0.001, Fig. 2C), only 19 of the 261 proteins (= 7.3%) differentially associated with 80S ribosomes in wild-type versus eif3e mutant cells showed a statistically significant change at the mRNA level. In addition, decreased 80S association of 35 mitochondrial proteins was not correlated with their mRNA levels (Fig. S2D,E). The transcriptomic differences between wild-type and eif3e mutant cells (402 mRNAs, Supplementary Data File 1) did not enrich any of the metabolism related GO terms found in the proteomic datasets (80S proteome, pSILAC, Steady-State dataset Fig. S2C). Instead, there was marginally significant enrichment of the terms transmembrane transport and cellular stress response (Fig. S2C). In addition, thiamine biosynthesis was enriched in the set of 218 mRNAs that were upregulated in eif3e mutant cells (Fig. S2C). Comparison with our parallel proteomic datasets revealed that the corresponding biosynthetic enzymes as well as the thiamine transporter Thi9p were also upregulated at the protein level (Supplementary Data File 1), suggesting that eif3e mutant cells have an increased demand for thiamine, a vitamin essential for glucose oxidation at multiple levels. In summary, the integrated analysis of 80S proteomics, pSILAC, steady-state protein and mRNA datasets strongly suggested that the highly selective changes in the levels of metabolism-related proteins observed in the absence of the eIF3e are primarily driven by changes at the level of translating specific classes of transcripts.
Metabolomic analysis indicates a glycolytic switch in eIF3d and eIF3e deficient cells
To assess phenotypic consequences of the translational changes observed in eif3e mutants, we considered the possibility that the abnormal polysome profile may result from a defect in ribosome biogenesis. However, eif3e mutants did not display typical phenotypic features of ribosome biogenesis mutants such as flocculation (not shown) and “ribosome halfmers” (Li et al., 2013; Meyer et al., 2007) in the sucrose density gradient profile (Fig. S3A). Cells deleted for eif3e did, however, display a 10 - 20% reduction in total RNA levels, the bulk of which consists of ribosomal RNA (Fig. S3B). Thus, while ribosome biogenesis may be slightly reduced in eif3e mutant cells, these data suggest that this is a consequence rather than the cause of the reduced growth rate of the mutant.
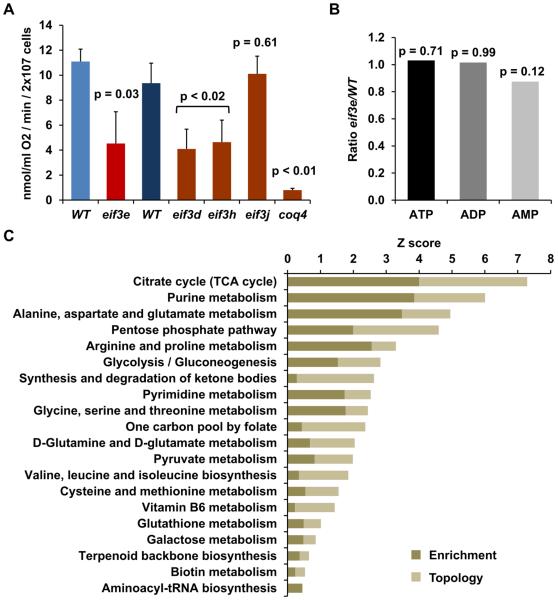
With respect to the large energy requirements of protein synthesis in exponentially growing cells – up to 30 % of the cellular ATP utilization (Rolfe and Brown, 1997) - we considered the possibility that failure to maintain the synthesis of mitochondrial respiration chain components might lead to a deficiency in ATP thereby causing inefficient protein synthesis. Exponentially growing S. pombe cells are known to derive ~50 – 65% of their energy from mitochondrial respiration (Zuin et al., 2008). Consistent with the proteomic data, cells deleted for eif3e maintained under exponential growth conditions showed a ~60% reduction in mitochondrial oxygen consumption rate (Fig. 3A). Similar reductions were observed with strains deficient in two other non-essential subunits, eIF3d and eIF3h (Tif38). Deletion of eIF3j (Hcr1), a sub-stoichiometric subunit thought to have modulatory function (Wagner et al., 2014), had no effect on respiration (Fig. 3A).
Figure 3. Metabolic re-programming in eif3 deficient cells.
A) Oxygen consumption rate was determined in the indicated wild-type (WT) and eif3 mutant strains. Two different WT strains were used that are syngeneic with the eif3 deleted strains to their right. Error bars represent standard deviations of 3 (WT, eif3e, eif3d, eif3h, eif3j) or 4 (WT, coq4) independent measurements. P values correspond to the results of a t-test with two-tailed distribution and unequal variance.
B) eif3e/WT ratios of the indicated metabolites as determined by LC-MS/MS in triplicates and statistical analysis in MetaboAnalyst. P values are results of a t-test.
C) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched in the sets of 90 metabolites and 521 proteins that significantly differed between wild-type and eif3e mutant cells. Plotted are the Z scores for pathway enrichment and pathway topology analysis derived from the MetaboAnalyst 3.0 tool suite (Xia et al., 2015). Since this type of integrated metabolite and protein expression analysis is only available for mammalian data, the putative human orthologues of the significantly changed S. pombe proteins were used for the analysis.
To determine whether reduced respiration coincides with reduced ATP levels, we generated a comprehensive metabolomic profile of wild-type and eif3e deficient cells by LC-MS. In triplicate experiments, ATP, ADP, and AMP levels did not significantly vary between wild-type and eif3e deleted cells (Fig. 3B), indicating that the translational phenotype of eif3e mutant cells is not due to ATP depletion. We then used the MetaboAnalyst tool suite (Xia et al., 2015), to identify a set of 90 metabolites (out of over 8000 LC-MS peaks) whose levels were significantly changed in eif3e mutant cells (Supplementary Data File 2). Pathway analysis of this metabolite set integrated with the list of significantly altered proteins identified by quantitative proteomics (Supplementary Data File 1), enriched numerous highly significant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Fig. 3C). The analysis revealed glucose utilization (i.e. glycolysis, pyruvate metabolism, TCA cycle), amino acid metabolism, and nucleotide metabolism as significantly upregulated KEGG pathways. Likewise, oxidative stress defense functions (glutathione metabolism and pentose phosphate pathway) were upregulated in eif3e deleted cells, a finding that is consistent with the well-established stress phenotype of eif3e mutant cells (Bandyopadhyay et al., 2000; Udagawa et al., 2008) as well as with our transcriptomic profile which indicated upregulation of oxidative stress response genes in the mutant (Fig. S2C). Consistent with a role of eIF3 in promoting the synthesis of the respiration chain, disruption of the ETC has previously been shown to lead to endogenous oxidative stress in S. pombe (Zuin et al., 2008). Biosynthesis pathways of several vitamin coenzymes essential for glucose, amino acid, and lipid metabolism was also found upregulated in cells lacking eif3e (i.e. biotin, pyridoxal 5'-phosphate, thiamine). The apparent high demand for thiamine of eif3e mutant cells is consistent with the upregulation of the thiamine transporter Thi9p noted above.
Cells deleted for eif3e and eif3d are dependent on glucose
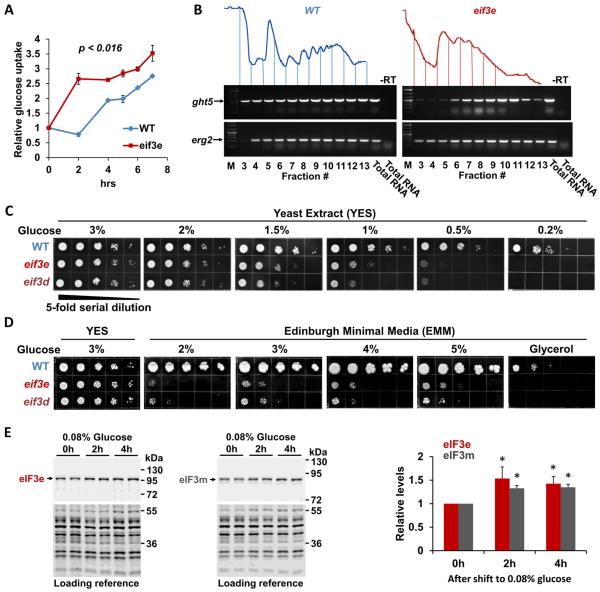
The combined proteomic and metabolomic data suggested that S. pombe cells lacking eIF3e function undergo a metabolic switch from respiration (which is downregulated) to glycolysis (which is upregulated) in order to maintain energy balance. To test this hypothesis, we determined glucose utilization by measuring the depletion of glucose from the media. Glucose uptake was significantly increased in cells lacking eif3e (Fig. 4A). This finding is consistent with the pronounced induction of the high affinity glucose transporter Ght5p (Saitoh et al., 2014) (Fig. 2C), which was more than 60-fold higher in the 80S fraction isolated from eif3e mutant cells (Supplementary Data file 1). Using RT-PCR, we assessed the presence of ght5 mRNA across the fractions of a sucrose density gradient. While in wild-type cells, a significant portion of the transcript was present in monosomal fractions, ght5 mRNA was quantitatively sequestered in polysomes of eif3e deleted cells, indicating strongly increased translation of ght5 mRNA in the mutant (Fig. 4B).
Figure 4. Effect of eIF3 deficiency on glucose uptake.
A) Glucose uptake was measured by determining the depletion of glucose from the media during the growth of wild-type (WT) and eif3e deleted cells using a YSI 2950 metabolite analyzer. Glucose concentration in the media was normalized to cell growth (i.e. OD600) to account for the slower growth rate of eif3e deleted cells. Data are averages of three biological replicates with error bars indicating standard deviations. Statistical significance was assessed with a t-test assuming two-tailed distribution and unequal variance.
B) RNA was purified from sucrose density gradient fractions prepared from WT and eif3e mutant cells and employed in RT-PCR reactions with primers specifically amplifying the mRNA encoding the hexose transporter ght5 and erg2 as a reference. Only fractions containing mRNA (3 – 12) were used for RT-PCR. Total RNA was used as reference. Reactions excluding reverse transcriptase are shown to prove that the band shown is derived from RNA not DNA.
C) Growth of eif3e and eif3d deleted cells under limiting glucose conditions. 5-fold serial dilutions of the indicated strains were plated onto rich yeast extract (YES) media containing decreasing glucose concentrations and growth was scored after 2 – 4 days.
D) Growth of eif3e and eif3d deleted cells on minimal media (EMM) containing increasing concentrations of glucose or on glycerol as the sole carbon source.
E) Effect of glucose restriction on the levels of eIF3e and eIF3m proteins. Strains harboring alleles of eif3e and eif3m modified with protein A epitope tags (Zhou et al., 2005) were shifted from media containing 3% glucose to medium containing 0.08% glucose for the indicated times. Cell lysates were loaded in duplicates and blotted with anti-protein A antibodies to detect eIF3e and eIF3m, respectively. Bands obtained with anti-PSTAIR antibodies were used as loading reference (bottom panels). Blots from biological replicates were quantified with the Licor Image Studio Lite package and results corrected for the loading reference were plotted. Statistical significance was assessed with a t-test assuming two-tailed distribution and unequal variance. Asterisks indicate p < 0.05.
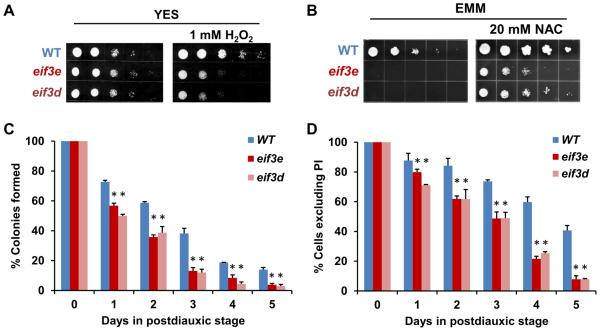
To test whether increased glucose uptake is relevant to cell physiology, we determined growth under glucose limiting conditions. Although yeast extract growth media (YES) typically contains 3% glucose, wild-type cells can maintain growth at glucose concentrations as low as 0.2% (Takeda et al., 2015). However, the growth of eif3e and eif3d mutant cells was inhibited at glucose concentrations below 1.5% (Fig. 4C). At 0.2%, both eif3 mutants were inviable (Fig. 4C). In minimal media (EMM), which contains 2% glucose and in which cells have an increased reliance on respiration (Zuin et al., 2008), eif3 mutant cells grew very poorly (Fig. 4D). Growth improved, however, 5 – 25-fold upon raising the glucose concentration to 5% (Fig. 4D). Cells deficient in eIF3e and eIF3d were entirely unable to grow in media containing non-fermentable glycerol as the carbon source (Fig. 4D), confirming that they are severely impaired in respiration.
Since eIF3 appears to promote the synthesis of ETC proteins, eIF3 subunits might be induced under conditions of increased respiration. Mitochondrial respiration is upregulated by shifting S. pombe cells to 0.08% glucose, a response that coincides with increased levels of the complex I subunit Cox2p (Takeda et al., 2015), which we found to require eIF3e for efficient synthesis (Supplementary Data File 1). Under the same glucose limiting conditions, both eIF3 subunits we tested, eIF3e and eIF3m, were upregulated between 30 – 40% after 4 hours (Fig. 4E), suggesting that eIF3 contributes to the upregulation of respiration upon glucose restriction.
Lack of eIF3e and eIF3d causes endogenous oxidative stress and premature aging
Consistent with the described stress sensitivity of eif3e and eif3d mutant cells (Bandyopadhyay et al., 2000; Udagawa et al., 2008), we found that these cells are sensitive to oxidative stress administered by the presence of 1 mM hydrogen peroxide in the media (Fig. 5A). To test whether poor growth on EMM is due to endogenous oxidative stress from which respiration deficient cells are known to suffer (Zuin et al., 2008), we supplemented the media with the anti-oxidant N-acetyl cysteine (NAC). NAC was able to substantially rescue the growth of eif3e and eif3d mutant cells on EMM, leading to a minimum 25-fold improvement (Fig. 5B). These phenotypic data combined with the mRNA data indicating induction of a stress response signature strongly suggest that the defect in mitochondrial respiration observed in eIF3 deficient cells leads to leakage of electrons from the ETC and subsequent oxidative stress.
Fig. 5. Stress sensitivity and premature aging of eif3e and eif3d deleted cells.
A) Sensitivity of eif3 deleted cells to H2O2 was assessed by spotting 5-fold serial dilutions on plates containing 1 mM H2O2.
B) Effect of the anti-oxidant N-acetyl cysteine on the growth of eif3 deleted strains in minimal media (EMM with 2% glucose).
C) Viability of wild-type (WT) and eif3 deleted strains in stationary phase as determined by colony formation. Data represent averages of biological replicates, and error bars indicate standard deviations. Statistical significance of the difference between WT and eif3 deleted cells at each time point was assessed with a t-test assuming two-tailed distribution and unequal variance. The asterisks indicate p < 0.05.
D) Viability of wild-type (WT) and eif3 deleted strains in stationary phase as determined by exclusion of propidium iodide. Data represent averages of biological replicates, and error bars indicate standard deviations. Statistical significance of the difference between WT and eif3 deleted cells at each time point was assessed with a t-test assuming two-tailed distribution and unequal variance. The asterisks indicate p < 0.05.
Since S. pombe cells with impaired mitochondrial function are known to display defects during postdiauxic stationary phase (Zuin et al., 2008), we determined the survival of eif3e and eif3d deleted cells in stationary phase which is a read-out of chronological aging (Roux et al., 2006). Studies in Saccharomyces cerevisiae established a “respiratory threshold” below which chronological longevity was drastically curtailed (Ocampo et al., 2012). Conversely, enhanced synthesis of ETC components prolongs lifespan in Drosophila under conditions of dietary restriction (Zid et al., 2009) . Accordingly, we found that after 5 days in the postdiauxic phase, respiration deficient S. pombe cells deleted for eif3e or eif3d had a 4- to 5-fold reduction in viability as determined by the ability to form colonies (Fig. 5C). Likewise, using exclusion of propidium iodide as a marker of cell integrity, cells deleted for eif3e or eif3d showed a ~5-fold reduction in survival (Fig. 5D). In summary, these data indicate an important metabolic homeostatic function of the eIF3d-eIF3e module in cellular physiology and aging.
eIF3e promotes the synthesis of electron transport proteins in human breast cells
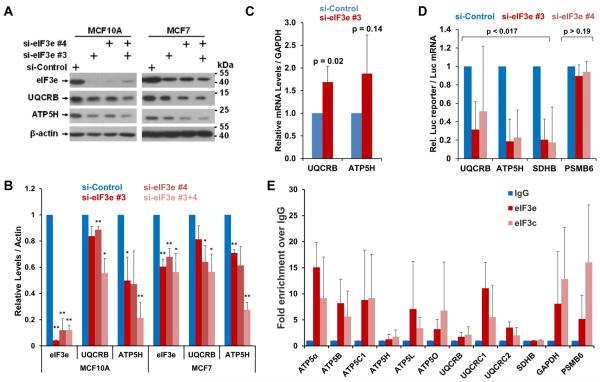
Since S. pombe Int6p and human eIF3e are functional orthologues (Crane et al., 2000), we asked whether human cells also rely on eIF3e for maintaining ETC integrity. Knockdown of eIF3e with two distinct siRNAs in MCF7 breast cancer cells and in non-tumorigenic MCF10A cells led to a marked downregulation of UQCRB and ATP5H (Fig. 6A,B), two complex III and V subunits whose S. pombe orthologues, ubiquinol-cytochrome-c reductase complex subunit 6 (SPCC737.02c) and F0-ATPase subunit D (SPBC29A10.13) require eIF3e for efficient synthesis (Supplementary Data File 1). The complex II subunit SDHB was also downregulated in eIF3e depleted MCF10A cells, whereas the complex I subunit NDUFB8 seemed unaffected (Fig. S4A,B).
Fig. 6. Effect of knockdown of eIF3e on the synthesis of ETC proteins in mammalian cells.
A) eIF3e was knocked down in non-tumorigenic MCF10A and tumorigenic MCF7 cells, followed by determination of the levels of the indicated ETC proteins by immunoblotting.
B) Blots in A) derived from biological replicates were quantified with the Licor Image Studio Lite package and results corrected for the loading reference were plotted in a bar graph (bottom panel). Statistical significance was assessed with a t-test assuming two-tailed distribution and unequal variance. Two asterisks indicate p < 0.05, one asterisk indicates p < 0.1. Error bars represent standard deviations.
C) eIF3e was knocked down in MCF7 cells, followed by determination of the levels of the indicated mRNA by quantitative RT-PCR relative to GAPDH mRNA as a reference. Averaged data from four biological replicates were plotted in a bar graph. Error bars indicate standard deviations. Statistical significance was assessed with a t-test assuming two-tailed distribution and unequal variance. P values for differences to the si-Control values are indicated. The corresponding mRNA data for MCF10A cells is shown in Fig. S4C.
D) Reporter constructs fusing the 5’-UTRs of the indicated ETC encoding mRNAs were fused to luciferase and transfected into MCF10A cells upon knockdown of eIF3e. Luciferase activity was normalized to luciferase mRNA measured by q-PCR. The graph shows averages of 3 independent experiments (Fig. S5B). Error bars indicate standard deviations. Statistical significance was assessed with a t-test assuming two-tailed distribution and unequal variance. P values for differences to the si-Control values are indicated. eIF3e knockdown efficiency is documented in Fig. S5C.
E) eIF3 complexes were immunopurified from MCF7 cells using antibodies against eIF3e and eIF3c as indicated. eIF3-associated mRNA was extracted and quantified by q-PCR. The graph shows fold change in the enrichment relative to the IgG control. Data represent averages of at least 3 independent experiments, and error bars indicate standard deviations.
Since the levels of mRNAs encoding ETC proteins were not decreased after eIF3e knockdown (Fig. 6C, S4C), we tested whether eIF3e regulates the synthesis of ETC proteins using reporter gene assays. Plasmids driving the expression of luciferase mRNA fused to the 5’-UTRs of ATP5H, UQCRB, and SDHB (Fig. S5A) were transfected into MCF10A cells 24 hours after knockdown of eIF3e. In three independent experiments, each with multiple technical replicates (Fig. S5B), the activity of 5’-UTR-luciferase fusions was strongly downregulated upon knockdown of eIF3e relative to control knockdown cells (Fig. 6 D). In contrast, luciferase reporter activity driven by the 5’UTR of PSMB6 was not affected by knockdown of eIF3e (Fig. 6D).
Since eIF3 was recently shown to regulate translation through direct binding of mRNA (Lee et al., 2015; Meyer et al., 2015), we prepared eIF3e and eIF3c immunoprecipitates and analyzed the associated mRNA by qPCR for the presence of select mRNAs encoding ETC components. Despite some variability, this assay, performed in triplicates in two different cell lines, MCF7 and HeLa, strongly suggested that eIF3 subunits associated with mRNAs encoding ETC subunits (Fig. 6E and S5D). eIF3 also bound the mRNA encoding the glycolytic enzyme GAPDH whose S. pombe orthologues are upregulated in the absence of eIF3e (Supplementary Data File 1). In summary, these data indicate that the function of eIF3e in maintaining the synthesis of mitochondrial ETC components may be conserved both in yeast and mammalian cells.
However, we could not detect a specific interaction of eIF3 with SDHB mRNA (Fig. 6E), even though SDHB protein and reporter activity were decreased in eIF3e knockdown cells (Fig. 6A, D, S5A). Conversely, whereas we detected robust interaction with mRNA encoding the proteasome subunit PSMB6 (Fig. 6E, S5D), a finding consistent with recent PAR-CLIP data (Meyer et al., 2015), its 5’-UTR did not mediate detectable eIF3e dependence in the reporter assay (Fig. 6D). It is therefore likely that mechanisms in addition to mRNA binding mediate the effect of eIF3 on translation.
Discussion
Control of energy metabolism at the level of mRNA translation
Compared to transcriptional pathways (Scarpulla et al., 2012), little is known about the control of mitochondrial biogenesis at the level of protein synthesis. Recent studies implicated the mTORC1/4E-BP/eIF4E axis in augmenting the synthesis of ETC subunits as well as mitochondrial ribosomal proteins (Gandin et al., 2016; Morita et al., 2013; Truitt et al., 2015). The findings presented here identified the eIF3 holo-complex as another critical promoter of the synthesis of ETC components and mitochondrial ribosomal proteins. Lack of this function as apparent in cell deleted for eif3d and eif3e causes respiratory deficiency and oxidative stress, a compensatory upregulation of glycolysis, strong dependence on glucose, and reduced lifespan (Fig. 7). These biochemical and cell biological observations are bolstered by negative genetic interactions of deletion of eif3e with the deletion of mitochondrial proteins as well as suppression of the eif3e mutant phenotype by overexpression of glycolytic and TCA cycle enzymes (Supplementary Data File 3). In response to glucose limitation, cells upregulate eIF3e and eIF3m, and probably the entire eIF3 holo-complex, in an apparent attempt to maximize the synthesis of respiration chain components. eIF3 function may therefore be a central node in the plasticity of cellular energy metabolism.
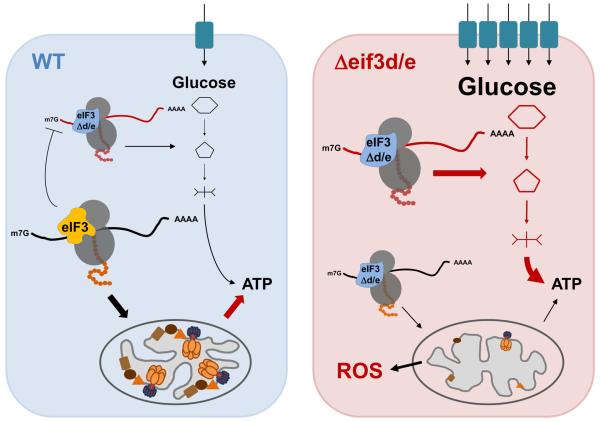
Fig. 7. Model for the role of eIF3 complexes in the translational control of energy metabolism.
An eIF3 complex containing all subunits including eIF3d and eIF3e directs the synthesis of mitochondrial ETC components and other mitochondrial proteins leading to efficient respiration and ATP production (left panel). In cells missing eif3e and eif3d, the synthesis of ETC components is diminished, leading to decreased respiration (right panel). As a result of inefficient electron transport, mitochondrial ROS production increases, resulting in endogenous oxidative stress and reduced life span. In an apparent attempt to balance the mitochondrial deficit, a retrograde mitochondria-to-nucleus signaling loop in eIF3d/e depleted cells leads to the induction of a transcriptional program upregulating the mRNAs encoding the high affinity glucose transporter Ght5p as well as glycolytic enzymes. These mRNAs appear preferentially translated by the eIF3 complex lacking subunits “d” and “e” (eIF3Δd/e), since their ribosomal occupancy is greatly increased in eif3e deleted cells. This suggests in inhibitory activity of the eIF3 holo-complex on the translation of mRNAs encoding glucose utilization components in wild-type cells whose biochemical mechanism - which our data suggest involves direct mRNA binding - remains subject to further investigations.
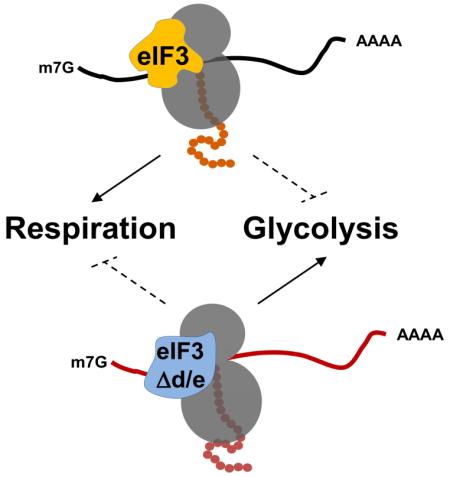
Transcript-selective control by eIF3: eIF3d-eIF3e as a specificity module
Our results show that cells deficient in eIF3e and eIF3d are not globally defective in protein synthesis as they are translating the compensatory response mRNAs encoding glycolytic enzymes and the glucose transporter Ght5p with greater efficiency than wild-type cells. Our findings thus strengthen our previous finding that S. pombe harbors two distinct eIF3 complexes distinguished by the presence or absence of the eIF3d-eIF3e dimer (Sha et al., 2009; Zhou et al., 2005). Consistent with this model, our quantitative mass spectrometry data show that a quasi stoichometric eIF3 complex containing all subunits except eIF3d is stably associated with 80S complexes in cells deleted for eif3e (Fig. S1B). The stability of this alternative complex, referred to as eIF3Δd/e (Fig. 7), likely accounts for the viability of cells lacking eif3d or eif3e, whereas most other eIF3 subunits are essential. Together with the exact overlap of their mutant phenotypes, their direct physical interaction, and their mutual dependency for proper protein levels and recruitment into the eIF3 holo-complex (Sha et al., 2009; Yen and Chang, 2000), it is likely that eIF3d and eIF3e together serve as specificity module to drive the synthesis of mitochondrial proteins.
How might the eIF3d-eIF3e module regulate specific mRNAs? Using RIP-CHIP, we have previously reported that eIF3 subunits interact with specific sets of mRNAs (Zhou et al., 2005). Re-examination of these datasets prompted by the 80S proteomics data presented here revealed a significant enrichment of the Gene Ontology term “Mitochondrial Part” (GO:0044429) within the list of the 100 mRNAs associating most abundantly with eIF3e (Supplementary Data File 4). Indeed, 12 of the top 100 mRNAs encode mitochondrial proteins, including mitochondrial ribosomal proteins as well as complex I, II, IV, and V subunits (Supplementary Data File 4). In the present work, we found that the interaction of eIF3 with mRNAs encoding ETC components is conserved in human cells. Likewise, recent PAR-CLIP data (Lee et al., 2015; Meyer et al., 2015) confirmed direct binding of eIF3 subunits to unique sets of mRNAs, which we found to be highly enriched in the Gene Ontology terms “Mitochondrion Organization” (GO:0007005, FDR = 10−33) and “Respiratory Electron Transport Chain (GO:0022904, FDR = 10−20.2) (Supplementary Data File 5). In conjunction with our demonstration that knockdown of eIF3e in human breast cells leads to downregulation of several ETC proteins, eIF3 likely promotes the synthesis of mitochondrial ETC components through direct interaction with their encoding mRNAs.
The efficient synthesis of Ght5p and glycolytic enzymes in eif3e deleted cells ostensibly relies on the alternative eIF3Δd/e complex, which we and others have shown to account for ~75% of the total eIF3 population in S. pombe (Ray et al., 2008; Sha et al., 2009; Zhou et al., 2005). Remarkably, the translational efficiency of ght5 mRNA is even higher in the absence of eIF3e than in its presence as it is quantitatively sequestered into polysomes in eif3e deleted cells (Fig. 4B). This suggests that holo-eIF3, which contains subunits d and e, actively represses ght5 mRNA translation. Our previous RIP-CHIP data (Zhou et al., 2005) revealed that eIF3e interacts with mRNAs for which we now show either decreased (e.g. ETC components) or increased (e.g. glucose transporters) translation in the absence of eIF3e (Supplementary Data File 4). Significantly, Lee et al. recently reported that eIF3 binding to two distinct mRNAs can either promote or inhibit their translation (Lee et al., 2015). Regardless, through dual stimulatory and inhibitory activities which remain to be defined molecularly, transcript-specific functions of eIF3 subunits appear to fine tune energy metabolism at the level of synthesis of high and low affinity glucose transporters.
eIF3 and cancer metabolism
As with other eIFs, upregulation of eIF3 subunits is frequently observed in human cancers (Hershey, 2015). Overexpression of individual subunits can drive de novo holo-complex formation and modest increases in global protein synthesis along with cell transformation (Zhang et al., 2006). With the re-emerging focus on the essential role of the mitochondrial ETC in cell proliferation, transformation, cancer stem cell maintenance, metastasis, and drug resistance (Birsoy et al., 2015; De Luca et al., 2015; LeBleu et al., 2014; Martinez-Outschoorn et al., 2014; Sullivan et al., 2015; Tan et al., 2015; Truitt et al., 2015; Wolf, 2014), it is conceivable that the ETC promoting function of eIF3 discovered in this report underlies, at least in part, its tumor promoting activity. In this model, eIF3 would be limiting for mitochondrially produced building blocks which fuel cancer cell anabolism and thus represent an intriguing cancer drug target.
Unlike most eIF3 subunits which are upregulated in cancer, eIF3e mRNA has been found downregulated in bulk samples of human breast and lung cancers (Buttitta et al., 2005; Marchetti et al., 2001). A recent study clarified that eIF3e protein downregulation in breast cancer occurs not in the epithelial carcinoma cells but in the stromal cancer-associated fibroblasts (CAFs) (Suo et al., 2015). In fact, knockdown of eIF3e is sufficient to confer CAF-like properties to normal human mammary fibroblasts (Suo et al., 2015). In situ studies of human breast cancer samples have revealed that CAFs have low OXPHOS activity but are highly glycolytic, whereas the epithelial cancer cells show the opposite metabolic profile, a phenomenon referred to as “Reverse Warburg” effect (Martinez-Outschoorn et al., 2014; Whitaker-Menezes et al., 2011). In this scenario, CAF catabolism and glycolysis will provide lactate and pyruvate to fuel the oxidative metabolism known to drive the tumor cells (Birsoy et al., 2015; De Luca et al., 2015; Martinez-Outschoorn et al., 2014; Sullivan et al., 2015; Tan et al., 2015; Wolf, 2014). Thus, based on our discovery of eIF3 as a promoter of ETC synthesis, the downregulation of eIF3e as well as all other eIF3 subunits ((Finak et al., 2008) and data not shown) specifically in CAFs may underlie metabolic synergy in the breast cancer microenvironment. If so, eIF3 inhibition as a therapeutic strategy may need to be approached with caution, at least in breast cancers showing the Reverse Warburg profile.
Experimental Procedures
Yeast strains
Strains deleted for eif3d and eif3e were described before (Yen and Chang, 2000). The sks2 gene was replaced with the NAT gene conferring resistance to nourseothricin. S. pombe strains were maintained in standard yeast extract (YES) or Edinburgh Minimal Media (EMM) unless otherwise noted.
Sucrose density gradient profiling
Cell lysates prepared in polysome lysis buffer (20 mM Tris-HCl, pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM DTT, 100 μg/ml cycloheximide) were separated on 10-50% (w/v) sucrose gradients. For immunoblotting, fractions were mixed with SDS sample buffer. For RNA isolation, collected fractions were precipitated in 1 volume of isopropanol at −80ºC, followed by purification via the RNeasy Kit (Quiagen).
Preparation of 80S complexes for LC-MS/MS analysis
Wild-type and eif3e deleted cells in exponential growth phase were lysed in a buffer containing 20 mM Tris pH 8, 140 mM KCl, 5 mM MgCl2, 100 μg/ml cycloheximide, 1% Triton X-100, 200 μg/ml heparin, and 1 mM PMSF and digested with RNAse1. The lysates were loaded onto 25% sucrose cushions and ribosomal pellets were analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS) (Brill et al., 2009).
For analysis of newly synthesized components, wild-type and eif3e deleted cells were switched from YES to EMM growth media containing 14N-NHCl4 as the nitrogen source for 4 h, followed by purification of 80S complexes and LC-MS/MS analysis. Statistically significant differences in the rate of synthesis of 80S-associated proteins between wild-type and eif3e mutant cells were derived from calculating 15N-WT and 15N-eif3e protein ratios from 15N-labeled peptides using IP2-Integrated Proteomics Pipeline software.
Quantitative proteomic profiles
Total lysate of wild-type and eif3e deleted cells were digested with trypsin, and peptides were separated by 2D-LC and analyzed on an Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific) operated in positive data-dependent acquisition mode. All mass spectra were analyzed with MaxQuant software version 1.5.0.25 (Cox and Mann, 2008). MS/MS spectra were searched against the Pombase protein sequence database ASM294 (version 2.23) using settings described in the Supplemental Methods.
For pulsed SILAC, wild-type and eif3e deleted cells were grown to exponential phase in YES medium, washed and switched to EMM containing 30 mg/L isotopically labeled 13C6 15N2 lysine (K8). Cell lysate was prepared after 2 hours and analyzed by 2DLC-MS/MS as described above. MaxQuant was used for quantifying K8-labeled peptides. Statistically significant differences in the rate of synthesis of individual proteins between wild-type and eif3e mutant cells were derived from calculating K8-WT and K8-eif3e protein ratios from K8-labeled peptides.
RNA seq analysis
Total RNA was extracted by phase separation using RNA-Bee™ (Amsbio) and chloroform, and samples were treated with RiboMinus Eukaryotic kit (Invitrogen) to deplete rRNA. The rRNA-depleted sample was subjected to enzymatic fragmentation and used for cDNA library construction. The cDNA was PCR amplified and sequenced on the SOLiD 4 platform. Data analysis followed similar steps as for ribosome profiling. Differentially expressed genes were called using the edgeR package. The RNA-seq data were deposited into the Gene Expression Omnibus repository (accession number GSE80349).
Metabolomic profiling by LC-MS
Metabolomic analysis was performed as described previously (Pluskal et al., 2010). Briefly, wild-type and eif3e deleted cells were quenched in methanol and disrupted using a Multi-Beads Shocker (Yasui Kikai, Osaka, Japan). Proteins were removed by centrifugal filtration and samples were concentrated by vacuum evaporation. Finally, each sample was re-suspended in 40 μl of 50% acetonitrile and 1 μl was used for liquid chromatography mass spectrometry (LC-MS) analysis on a Paradigm MS4 HPLC system (Michrom Bioresources, Auburn, USA) coupled to an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, USA). The data were analysed using MetaboAnalyst (http://www.metaboanalyst.ca/). Pathway enrichment analysis was done by hypergeometric test, topology analysis was based on degree centrality, and a combined gene-metabolite mode was used.
Studies in mammalian cells
eIF3e was knocked down in MCF7 and MCF10A cells using siRNA transfection. The levels of eIF3e and ETC proteins were analyzed by immunoblotting as described in the Supplemental Methods section. mRNA levels were determined by q-PCR. For reporter gene assays, constructs fusing the 5’-UTRs of ETC encoding mRNAs were fused to luciferase and transfected into MCF10A cells upon knockdown of eIF3e. Luciferase activity was normalized to luciferase mRNA measured by q-PCR. eIF3-mRNA interactions were tested by immunoprecipitating eIF3 complexes with eIF3e and eIF3c antibodies, followed by mRNA extraction and detection by q-PCR.
Statistical analysis
Statistical analyses of replicate datasets was performed with Microsoft Excel. Typically, data were averaged, standard deviations calculated, and statistical significance was assessed using the T.Test function assuming two-tailed distribution and unequal variance. The statistical analysis of proteomic and transcriptomic datasets is detailed in the Supplementary Methods section.
Supplementary Material
Highlights.
eIF3d and eIF3e promote synthesis of the mitochondrial ETC and respiration
eIF3 deficiency leads to glycolytic switch, oxidative stress, and reduced lifespan
eIF3e promotes the synthesis of ETC proteins through 5’-UTR-mediated mechanism
eIF3 subunits interact with mRNAs encoding ETC proteins
eTOC Blurb
eIF3 is frequently dysregulated in cancer. Shah et al. show that lack of eIF3d and eIF3e results in impaired synthesis of mitochondrial OXPHOS proteins, respiratory deficiency, glycolytic switch, oxidative stress, and reduced lifespan. Thus, the eIF3d-eIF3e module mediates mRNA-specific translational control of energy metabolism that may be disrupted in cancer.
Highlights.
Cells deleted for eif3e are defective in synthesizing mitochondrial ETC proteins
Metabolomic analysis indicates a glycolytic switch in eIF3e deficient cells
Lack of eIF3e and eIF3d causes glucose dependence, oxidative stress, and aging
eIF3e promotes the expression of ETC proteins in human breast cells
Acknowledgements
We thank Eric Chang for yeast strains, eIF3d antibodies, and discussions. Francois Bachand is thanked for Rps2p antibodies. This work was supported by grant GM105802 to D.A.W. Parts of this work were funded by P30 grants CA030199 and GM085764. D.A.W. is a scholar of the Foreign 1000 Talent Program funded by the Government of the People’s Republic of China.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
M.S. performed RT-PCR experiments from sucrose density gradient fractions, stress sensitivity and aging experiments, and all luciferase reporter assays. D.S. performed the eIF3 knockdown experiments in human cells and the RNA-IP experiments. J.S.S. did polysome profiling, prepared 80S complexes for mass spectrometry, RNA samples for RNA-seq, and performed glucose sensitivity experiments. T.P. and M.Y. performed the metabolomic studies. S.B. and E. H. measured respiration in various eIF3 deleted strains. K.M. and A.R.C performed all LC-MS/MS experiments. F.Q. performed the bioinformatics analysis of RNA-seq data. D.A.W. conceived the project, performed the pSILAC experiment, determined the effect of glucose withdrawal on eIF3 levels, performed the analysis of global proteomic, transcriptomic, and metabolomic datasets, prepared the figures, and wrote the manuscript.
Reference
- Asano K, Merrick WC, Hershey JWB. The Translation Initiation Factor eIF3-p48 Subunit Is Encoded by int-6, a Site of Frequent Integration by the Mouse Mammary Tumor Virus Genome. J. Biol. Chem. 1997;272:23477–23480. doi: 10.1074/jbc.272.38.23477. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Matsumoto T, Maitra U. Fission Yeast Int6 Is Not Essential for Global Translation Initiation, but Deletion of int6 + Causes Hypersensitivity to Caffeine and Affects Spore Formation. Mol. Biol. Cell. 2000;11:4005–4018. doi: 10.1091/mbc.11.11.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A, Lakshmanan V, Matsumoto T, Chang EC, Maitra U. Moe1 and spInt6, the Fission Yeast Homologues of Mammalian Translation Initiation Factor 3 Subunits p66 (eIF3d) and p48 (eIF3e), Respectively, Are Required for Stable Association of eIF3 Subunits. J. Biol. Chem. 2002;277:2360–2367. doi: 10.1074/jbc.M107790200. [DOI] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill LM, Motamedchaboki K, Wu S, Wolf DA. Comprehensive proteomic analysis of Schizosaccharomyces pombe by two-dimensional HPLC-tandem mass spectrometry. Methods. 2009;48:311–319. doi: 10.1016/j.ymeth.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttitta F, Martella C, Barassi F, Felicioni L, Salvatore S, Rosini S, D’Antuono T, Chella A, Mucilli F, Sacco R, et al. Int6 Expression Can Predict Survival in Early-Stage Non–Small Cell Lung Cancer Patients. Clin Cancer Res. 2005;11:3198–3204. doi: 10.1158/1078-0432.CCR-04-2308. [DOI] [PubMed] [Google Scholar]
- Choudhuri A, Maitra U, Evans T. Translation initiation factor eIF3h targets specific transcripts to polysomes during embryogenesis. PNAS. 2013;110:9818–9823. doi: 10.1073/pnas.1302934110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Crane R, Craig R, Murray R, Dunand-Sauthier I, Humphrey T, Norbury C. A Fission Yeast Homolog of Int-6, the Mammalian Oncoprotein and eIF3 Subunit, Induces Drug Resistance when Overexpressed. Mol. Biol. Cell. 2000;11:3993–4003. doi: 10.1091/mbc.11.11.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoc E, Fraser CS, Zhou M, Videler H, Mayeur GL, Hershey JWB, Doudna JA, Robinson CV, Leary JA. Structural Characterization of the Human Eukaryotic Initiation Factor 3 Protein Complex by Mass Spectrometry. Mol Cell Proteomics. 2007;6:1135–1146. doi: 10.1074/mcp.M600399-MCP200. [DOI] [PubMed] [Google Scholar]
- De Luca A, Fiorillo M, Peiris-Pagès M, Ozsvari B, Smith DL, Sanchez-Alvarez R, Martinez-Outschoorn UE, Cappello AR, Pezzi V, Lisanti MP, et al. Mitochondrial biogenesis is required for the anchorage-independent survival and propagation of stem-like cancer cells. Oncotarget. 2015;6:14777–14795. doi: 10.18632/oncotarget.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Stengel F, Pellarin R, Zhang S, Schaefer T, Aylett CHS, Cimermančič P, Boehringer D, Sali A, Aebersold R, et al. Molecular Architecture of the 40S·eIF1·eIF3 Translation Initiation Complex. Cell. 2014;158:1123–1135. doi: 10.1016/j.cell.2014.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- Gandin V, Masvidal L, Hulea L, Gravel S-P, Cargnello M, McLaughlan S, Cai Y, Balanathan P, Morita M, Rajakumar A, et al. nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 2016;25:636–648. doi: 10.1101/gr.197566.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Georges A, Dhote V, Kuhn L, Hellen CUT, Pestova TV, Frank J, Hashem Y. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015;525:491–495. doi: 10.1038/nature14891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis LD, Lewis SM. Decreased eIF3e/Int6 expression causes epithelial-to-mesenchymal transition in breast epithelial cells. Oncogene. 2012;32:3598–3605. doi: 10.1038/onc.2012.371. [DOI] [PubMed] [Google Scholar]
- Grzmil M, Rzymski T, Milani M, Harris AL, Capper RG, Saunders NJ, Salhan A, Ragoussis J, Norbury CJ. An oncogenic role of eIF3e/INT6 in human breast cancer. Oncogene. 2010;29:4080–4089. doi: 10.1038/onc.2010.152. [DOI] [PubMed] [Google Scholar]
- Hershey JWB. The role of eIF3 and its individual subunits in cancer. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2015;1849:792–800. doi: 10.1016/j.bbagrm.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kim BH, Cai X, Vaughn JN, Von Arnim AG. On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol. 2007;8:R60. doi: 10.1186/gb-2007-8-4-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16:992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ASY, Kranzusch PJ, Cate JHD. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015;522:111–114. doi: 10.1038/nature14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li X, Sun L, Chen F, Liu Z, Gu Y, Gong X, Liu Z, Wei H, Huang Y, et al. Reduction of Ribosome Level Triggers Flocculation of Fission Yeast Cells. Eukaryotic Cell. 2013;12:450–459. doi: 10.1128/EC.00321-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith GH, Callahan R. Int-6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumor virus in mammary preneoplasia. J. Virol. 1995;69:1932–1938. doi: 10.1128/jvi.69.3.1932-1938.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Pellegrini S, Bertacca G, Callahan R. Reduced expression of INT-6/eIF3-p48 in human tumors. Int. J. Oncol. 2001;18:175–179. doi: 10.3892/ijo.18.1.175. [DOI] [PubMed] [Google Scholar]
- Martinez-Outschoorn U, Sotgia F, Lisanti MP. Tumor Microenvironment and Metabolic Synergy in Breast Cancers: Critical Importance of Mitochondrial Fuels and Function. Seminars in Oncology. 2014;41:195–216. doi: 10.1053/j.seminoncol.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Meyer AE, Hung N-J, Yang P, Johnson AW, Craig EA. The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. PNAS. 2007;104:1558–1563. doi: 10.1073/pnas.0610704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian S-B, Jaffrey SR. 5′ UTR m6A Promotes Cap-Independent Translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Gravel S-P, Chénard V, Sikström K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, et al. mTORC1 Controls Mitochondrial Activity and Biogenesis through 4E-BP-Dependent Translational Regulation. Cell Metabolism. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Ocampo A, Liu J, Schroeder EA, Shadel GS, Barrientos A. Mitochondrial Respiratory Thresholds Regulate Yeast Chronological Life Span and its Extension by Caloric Restriction. Cell Metabolism. 2012;16:55–67. doi: 10.1016/j.cmet.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluskal T, Nakamura T, Villar-Briones A, Yanagida M. Metabolic profiling of the fission yeast S. pombe: quantification of compounds under different temperatures and genetic perturbation. Mol. BioSyst. 2010;6:182. doi: 10.1039/b908784b. [DOI] [PubMed] [Google Scholar]
- Querol-Audi J, Sun C, Vogan JM, Smith MD, Gu Y, Cate JHD, Nogales E. Architecture of Human Translation Initiation Factor 3. Structure. 2013;21:920–928. doi: 10.1016/j.str.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe DF, Brown GC. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiological Reviews. 1997;77:731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- Roux AE, Quissac A, Chartrand P, Ferbeyre G, Rokeach LA. Regulation of chronological aging in Schizosaccharomyces pombe by the protein kinases Pka1 and Sck2. Aging Cell. 2006;5:345–357. doi: 10.1111/j.1474-9726.2006.00225.x. [DOI] [PubMed] [Google Scholar]
- Saitoh S, Mori A, Uehara L, Masuda F, Soejima S, Yanagida M. Mechanisms of expression and translocation of major fission yeast glucose transporters regulated by CaMKK/phosphatases, nuclear shuttling and TOR. Mol. Biol. Cell. 2014;26:373–386. doi: 10.1091/mbc.E14-11-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends in Endocrinology & Metabolism. 2012;23:459–466. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sha Z, Brill LM, Cabrera R, Kleifeld O, Scheliga JS, Glickman MH, Chang EC, Wolf DA. The eIF3 Interactome Reveals the Translasome, a Supercomplex Linking Protein Synthesis and Degradation Machineries. Molecular Cell. 2009;36:141–152. doi: 10.1016/j.molcel.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Gu Y, Querol-Audí J, Vogan JM, Nitido A, Cate JHD. Human-Like Eukaryotic Translation Initiation Factor 3 from Neurospora crassa. PLoS ONE. 2013;8:e78715. doi: 10.1371/journal.pone.0078715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo J, Medina D, Herrera S, Zheng Z-Y, Jin L, Chamness GC, Contreras A, Gutierrez C, Hilsenbeck S, Umar A, et al. Int6 reduction activates stromal fibroblasts to enhance transforming activity in breast epithelial cells. Cell & Bioscience. 2015;5:10. doi: 10.1186/s13578-015-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Starzynski C, Mori A, Yanagida M. The critical glucose concentration for respiration-independent proliferation of fission yeast, Schizosaccharomyces pombe. Mitochondrion. 2015;22:91–95. doi: 10.1016/j.mito.2015.04.003. [DOI] [PubMed] [Google Scholar]
- Tan AS, Baty JW, Dong L-F, Bezawork-Geleta A, Endaya B, Goodwin J, Bajzikova M, Kovarova J, Peterka M, Yan B, et al. Mitochondrial Genome Acquisition Restores Respiratory Function and Tumorigenic Potential of Cancer Cells without Mitochondrial DNA. Cell Metabolism. 2015;21:81–94. doi: 10.1016/j.cmet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Truitt ML, Conn CS, Shi Z, Pang X, Tokuyasu T, Coady AM, Seo Y, Barna M, Ruggero D. Differential Requirements for eIF4E Dose in Normal Development and Cancer. Cell. 2015;162:59–71. doi: 10.1016/j.cell.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T, Nemoto N, Wilkinson CRM, Narashimhan J, Jiang L, Watt S, Zook A, Jones N, Wek RC, Bahler J, et al. Int6/eIF3e Promotes General Translation and Atf1 Abundance to Modulate Sty1 MAPK-dependent Stress Response in Fission Yeast. Journal of Biological Chemistry. 2008;283:22063–22075. doi: 10.1074/jbc.M710017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Herrmannová A, Malík R, Peclinovská L, Valášek LS. Functional and Biochemical Characterization of Human Eukaryotic Translation Initiation Factor 3 in Living Cells. Mol. Cell. Biol. 2014;34:3041–3052. doi: 10.1128/MCB.00663-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker-Menezes D, Martinez-Outschoorn UE, Flomenberg N, Birbe R, Witkiewicz AK, Howell A, Pavlides S, Tsirigos A, Ertel A, Pestell RG, et al. Hyperactivation of oxidative mitochondrial metabolism in epithelial cancer cells in situ. Cell Cycle. 2011;10:4047–4064. doi: 10.4161/cc.10.23.18151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DA. Is Reliance on Mitochondrial Respiration a “Chink in the Armor” of Therapy-Resistant Cancer? Cancer Cell. 2014;26:788–795. doi: 10.1016/j.ccell.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucl. Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H-CS, Chang EC. Yin6, a fission yeast Int6 homolog, complexes with Moe1 and plays a role in chromosome segregation. PNAS. 2000;97:14370–14375. doi: 10.1073/pnas.97.26.14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Pan X, Hershey JWB. Individual Overexpression of Five Subunits of Human Translation Initiation Factor eIF3 Promotes Malignant Transformation of Immortal Fibroblast Cells. Journal of Biological Chemistry. 2006;282:5790–5800. doi: 10.1074/jbc.M606284200. [DOI] [PubMed] [Google Scholar]
- Zhou C, Arslan F, Wee S, Krishnan S, Ivanov AR, Oliva A, Leatherwood J, Wolf DA. PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol. 2005;3:14. doi: 10.1186/1741-7007-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP Extends Lifespan upon Dietary Restriction by Enhancing Mitochondrial Activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin A, Gabrielli N, Calvo IA, García-Santamarina S, Hoe K-L, Kim DU, Park H-O, Hayles J, Ayté J, Hidalgo E. Mitochondrial Dysfunction Increases Oxidative Stress and Decreases Chronological Life Span in Fission Yeast. PLoS ONE. 2008;3:e2842. doi: 10.1371/journal.pone.0002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.