Abstract
Detection of intracellular DNA triggers activation of the STING-dependent interferon-stimulatory DNA (ISD) pathway, which is essential for antiviral responses. Multiple DNA sensors have been proposed to activate this pathway, including AIM2-like receptors (ALRs). Whether the ALRs are essential for activation of this pathway remains unknown. To rigorously explore the function of ALRs, we generated mice lacking all 13 ALR genes. We found that ALRs are dispensable for the type I interferon (IFN) response to transfected DNA ligands, DNA virus infection, and lentivirus infection. We also found that ALRs do not contribute to autoimmune disease in the Trex1−/− mouse model of Aicardi-Goutières Syndrome. Finally, CRISPR-mediated disruption of the human AIM2-like receptor IFI16 in primary fibroblasts revealed that IFI16 is not essential for the IFN response to human cytomegalovirus infection. Our findings indicate that ALRs are dispensable for the ISD response and suggest that alternative functions for these receptors should be explored.
Introduction
Innate immune sensing of nucleic acids is key for the induction of antiviral immune responses. Detection of intracellular DNA activates two pathways: the caspase-1 inflammasome and the interferon (IFN)-stimulatory DNA (ISD) pathway. Inflammasome activation is initiated following DNA sensing by AIM2, resulting in secretion of mature interleukin (IL)-1β and IL-18 and pyroptosis (Burckstummer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009). Evaluation of Aim2−/− mice has confirmed that AIM2 is essential for inflammasome activation following infection with multiple pathogens (Jones et al., 2010; Rathinam et al., 2010). Detection of intracellular DNA also promotes the IFN response via the IFN-stimulatory DNA (ISD) pathway, now known as the cGAS-STING pathway. This pathway detects DNA in a sequence-independent manner and signals via the adaptor stimulator of interferon genes (STING) (Ishikawa and Barber, 2008; Stetson and Medzhitov, 2006).
In 2013, the nucleotidyltransferase cyclic GMP-AMP synthase (cGAS) was identified as the key DNA sensor that activates the STING pathway (Sun et al., 2013; Wu et al., 2013). Multiple studies with cGas−/− mice have demonstrated that cGAS is essential for the IFN response to a variety of pathogens, including DNA viruses, retroviruses, and intracellular bacteria (Collins et al., 2015; Gao et al., 2013; Li et al., 2013b; Stavrou et al., 2015; Storek et al., 2015; Watson et al., 2015).
Several members of the AIM2-like receptor (ALR) gene family have also been proposed to activate the ISD pathway. ALRs are characterized by a Pyrin signaling domain and a DNA-binding HIN domain. The number of ALR genes varies widely among mammals, from a single ALR in cows, to four in humans (AIM2, IFI16, PYHIN1, and MNDA) and 13 in mice (Brunette et al., 2012; Cridland et al., 2012). Phylogenetic analysis indicates that, with the exception of AIM2, there is a complete lack of orthology among mammalian ALRs (Brunette et al., 2012; Cridland et al., 2012). Although AIM2 itself is well-defined as the key DNA sensor that activates the inflammasome, the function of the other ALRs is less clear, although several have been proposed to activate the ISD pathway.
Multiple observations support a role for ALRs in activation of the ISD pathway. First, several ALRs have been shown to bind viral DNA and interact with STING in co-immunoprecipitation experiments (Brunette et al., 2012; Stavrou et al., 2015; Unterholzner et al., 2010). Second, viral antagonists of the IFI16-mediated IFN response have been identified, including the human cytomegalovirus (HCMV) tegument protein pUL83 and the herpes simplex virus (HSV-1) E3 ubiquitin ligase ICP0 (Li et al., 2013a; Orzalli et al., 2012). Finally, knockdown of individual ALRs, including mouse Ifi203, mouse Ifi204, and human IFI16, has been shown to dampen the IFN response to infection with multiple pathogens, including HSV-1, HCMV, human immunodeficiency virus (HIV), murine leukemia virus (MLV), Francisella tularensis, and Mycobacterium tuberculosis (Jakobsen et al., 2013; Li et al., 2013a; Manzanillo et al., 2012; Orzalli et al., 2012; Stavrou et al., 2015; Storek et al., 2015). However, whether ALRs are essential for activation of the ISD pathway has not been tested rigorously, in part because of the potential for functional redundancy among the closely related ALRs.
To define the function of the AIM2-like receptors, we generated ALR−/− mice that lack all 13 mouse ALR genes. We found that the mouse ALRs are dispensable for the IFN response to transfected DNA ligands, DNA virus infection, and lentivirus infection. Moreover, the ALRs did not contribute to autoimmune disease in the Trex1-deficient mouse model of Aicardi-Goutières Syndrome. Finally, we used CRISPR to disrupt the human AIM2-like receptor IFI16 in primary human fibroblasts and found that IFI16 was dispensable for the IFN response to HCMV infection. Thus, our data reveal that ALRs are dispensable for activation of the ISD pathway and demonstrate that cGAS is the primary DNA sensor that drives the IFN response to DNA.
Results
Generation of ALR-deficient mice
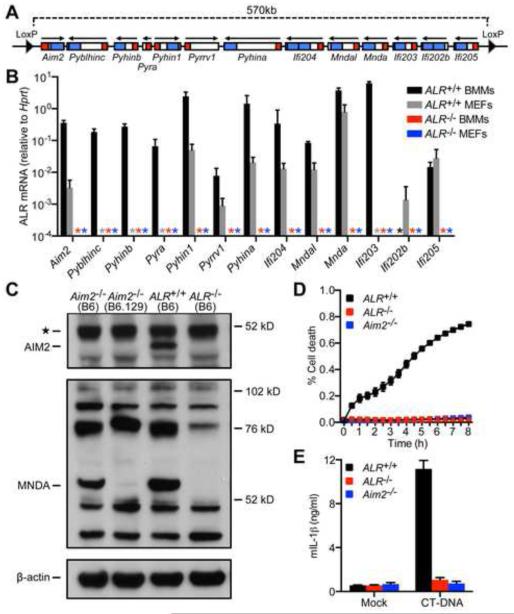
The mouse ALR locus consists of 13 consecutive genes spanning 570 kilobases on chromosome 1 (Figure 1A) (Brunette et al., 2012; Cridland et al., 2012). To generate ALR-deficient mice, we introduced LoxP sites flanking the ALR locus in C57BL/6NTac ES cells, derived mice containing the floxed locus, and then crossed these mice to Mox2-cre mice (Tallquist and Soriano, 2000) to delete this entire genomic interval (Figure 1A and Supplemental Figure 1). ALR−/− mice were viable and physically indistinguishable from their littermates (data not shown). Successful excision of the entire ALR locus was confirmed by PCR with primers flanking the ALR locus (Supplemental Figure 1B). Expression of all 13 ALR genes was undetectable by RT-PCR in macrophages and embryonic fibroblasts cultured from ALR−/− mice (Figure 1B) and AIM2 protein was undetectable in ALR−/− macrophages by Western blot (Figure 1C). Moreover, similar to Aim2−/− cells (Jones et al., 2010; Rathinam et al., 2010), ALR−/− macrophages, which lack AIM2 as well as the 12 additional ALRs, were protected from pyroptotic cell death and failed to secrete IL-1β in response to transfected calf-thymus DNA (CT-DNA, Figure 1D,E). Thus, we successfully generated ALR−/− mice.
Figure 1. Generation of ALR-deficient mice.
(A) Schematic of the LoxP-flanked mouse ALR locus for Cre-mediated deletion of all 13 mouse ALR genes. Pyrin and HIN domains are indicated in red and blue, respectively. (B) Quantification by RT-PCR of mouse ALR gene transcripts in bone marrow-derived macrophages (BMMs) or embryonic fibroblasts (MEFs) from ALR+/+ and ALR−/− mice (* = not detected). (C) AIM2 and MNDA protein expression evaluated in IFNβ-primed BMMs cultured from mice of the indicated genotype. Note that the band at 76kD (indicated by **) was not consistently observed in multiple experiments and does not match the expected size of IFI204. (D) Cell death of BMMs from ALR+/+, ALR−/−, or Aim2−/− mice transfected with calf-thymus DNA (CT-DNA) quantified using an IncuCyte imaging system. (E) Quantification of IL-1β secretion by BMMs from ALR+/+, ALR−/−, or Aim2−/− mice transfected with CT-DNA. Error bars represent mean ± SD. Data are representative of one experiment (B,C) or two experiments (D,E). See also Figure S1 and S2.
ALR expression is altered in Aim2−/− (B6.129) cells due to SNPs in the 129-derived ALR locus
We also confirmed loss of MNDA protein in ALR−/− macrophages using a polyclonal anti-MNDA/IFI204 antibody raised against a 51 amino acid peptide that shares 100% homology with IFI204 and MNDA (Figure 1C). We failed to detect a protein band consistent with the predicted size (~69kD) of IFI204 in macrophages using this antibody (Figure 1C). Unexpectedly, MNDA protein was not detected in Aim2−/− (B6.129) macrophages (Figure 1C). Moreover, expression levels of Mnda mRNAs and several additional ALRs were reduced, while expression of Ifi202b was increased in Aim2−/− (B6.129) macrophages compared to control cells (Supplemental Figure 2A). These Aim2−/− mice, which were used in the majority of our experiments, have been fully backcrossed to C57BL/6 (B6), but were generated by targeting Aim2 in 129 ES cells (Rathinam et al., 2010). Therefore, the genes flanking Aim2 on chromosome 1, including the ALR locus, are 129-derived (Vanden Berghe et al., 2015). To determine whether ALR gene expression was altered in Aim2−/− (B6.129) macrophages due to polymorphisms present in the 129 ALR locus, we compared ALR gene expression in macrophages cultured from Aim2−/− (B6) mice generated in B6 ES cells (Jones et al., 2010) to macrophages cultured from wildtype (WT) B6, WT 129, and Aim2−/− (B6.129) mice. MNDA protein was detected in WT B6 and Aim2−/− (B6) macrophages, but not WT 129 or Aim2−/− (B6.129) macrophages (Figure 1C and Supplemental Figure 2C). Moreover, with the exception of Aim2 itself, ALR gene expression was similar in WT B6 and Aim2−/− (B6) macrophages (Supplemental Figure 2A) and WT 129 and Aim2−/− (B6.129) macrophages (Supplemental Figure 2B). Thus, in contrast to a report suggesting that AIM2 regulates expression of Ifi202 (Panchanathan et al., 2010), ALR gene expression is altered in Aim2−/− (B6.129) macrophages due to polymorphisms present in the 129-derived ALR locus. Unless otherwise indicated, Aim2−/− (B6.129) mice were used throughout the remainder of this study.
ALRs are dispensable for the ISD response
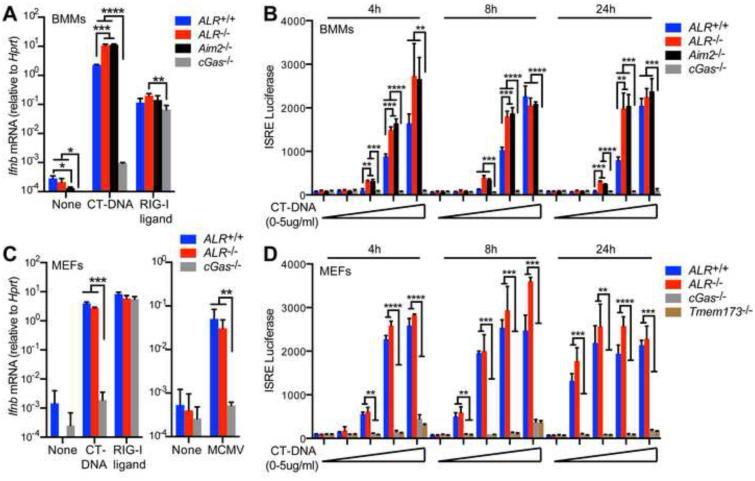
To determine whether the ALRs contribute to the ISD pathway, we analyzed the IFN response in cells cultured from ALR−/− mice compared to cells from ALR+/+, Aim2−/− or cGas−/− mice. Consistent with previous studies, the IFN response to transfected dsDNA (calf thymus DNA, CT-DNA) was enhanced in AIM2-deficient macrophages and reduced in cGAS-deficient macrophages (Figure 2A,B) (Corrales et al., 2016; Gray et al., 2015; Jones et al., 2010; Li et al., 2013b; Rathinam et al., 2010). The IFN response to transfected CT-DNA in ALR−/− macrophages was intact and similar to Aim2−/− macrophages (Figure 2A,B). Moreover, the IFN response to transfected CT-DNA in mouse embryonic fibroblasts (MEFs) was cGAS-dependent and completely ALR-independent (Figure 2C,D). This demonstrates that, in contrast to cGAS, the ALRs are not required in macrophages or fibroblasts for the IFN response to transfected DNA ligands.
Figure 2. ALRs are dispensable for the IFN response to transfected DNA.
(A) Quantification of Ifnb mRNA induction in BMMs from ALR+/+,ALR−/−, Aim2−/−, or cGas−/− mice transfected with CT-DNA or RIG-I ligand for 4h. (B) Quantification of type I IFN production using an IFN bioassay with supernatants from BMMs (A) of the indicated genotype transfected with 0-5μg CT-DNA (0μg, 0.01μg, 0.1μg, 1μg, or 5μg) for 4, 8, or 24h. (C) Quantification of Ifnb mRNA induction in mouse embryonic fibroblasts (MEFs) from ALR+/+,ALR−/−, or cGas−/− mice transfected with nucleic acid ligands as in (A) for 4h (left panel) or infected with MCMV (MOI=3) for 5h (right panel). (D) Quantification of type I IFN production using an IFN bioassay with supernatants from MEFs of the indicated genotype transfected with CT-DNA as in (B). Error bars represent mean ± SD. Data are representative of three experiments (A), two experiments (B,C), or one experiment (D). Statistical analysis was performed using an unpaired t-test corrected for multiple comparisons using the Holm-Sidak method. *p<0.05. **p < 0.01, ***p<0.001, ****p < 0.0001.
We also asked whether the ALRs were required for the IFN response to infection with the DNA virus mouse cytomegalovirus (MCMV). The IFN response to MCMV infection was intact and similar in ALR+/+ and ALR−/− fibroblasts, but was reduced in cGas−/− fibroblasts (Figure 2C). These data reveal that the ALRs are dispensable for activation of the ISD pathway.
ALRs are dispensable for the IFN response to retrovirus infection
Upon infection, retroviral RNA genomes are reverse-transcribed into DNA prior to integration into host chromosomes. Innate sensing of viral DNA is normally inhibited by the 3’ exonuclease Trex1, which degrades retroviral DNA in the cytosol. However, Trex1−/− cells mount a robust, STING-dependent IFN response to lentivirus infection (Yan et al., 2010). Multiple receptors have been proposed to sense reverse-transcribed retroviral DNA, including cGAS and the ALRs IFI203 (mouse) and IFI16 (human; (Gao et al., 2013; Jakobsen et al., 2013; Stavrou et al., 2015).
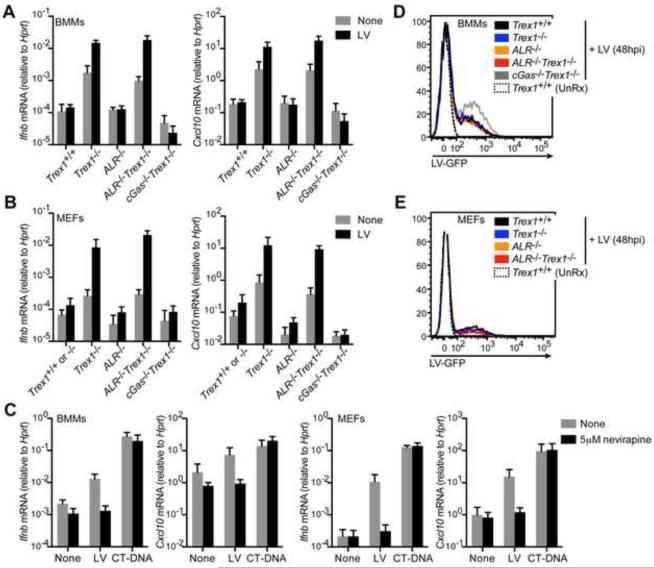
To explore whether ALRs are required for the IFN response to lentivirus infection, we infected cells with a VSV-G pseudotyped, self-inactivating lentivirus in which GFP expression was driven by an internal MND promoter. We infected BMMs and MEFs at an MOI of 1 and quantified induction of Ifnb and the IFN-stimulated gene Cxcl10 mRNA 20 hours after infection. Consistent with previous data (Yan et al., 2010), we observed robust induction of Ifnb and Cxcl10 mRNA in Trex1−/− cells, but not Trex1+/+ cells, in response to lentivirus infection (Figure 3A,B). This IFN response was dependent on reverse transcription, as pretreatment with the reverse transcription inhibitor nevirapine completely abrogated the IFN response to lentivirus but not CT-DNA (Figure 3C). We observed a similar induction of Ifnb and Cxcl10 mRNA in ALR+/+Trex1−/− and ALR−/−Trex1−/− cells at 6, 12, 20, and 24 hours after infection, indicating that the ALRs are dispensable for this IFN response (Figure 3A,B and Supplemental Figure 3). On the other hand, we observed no significant IFN response to lentivirus infection in cGas−/−Trex1−/− cells at any timepoint examined (Figure 3A,B and Supplemental Figure 3). This suggests that the IFN response to lentivirus infection is cGAS-dependent, but ALR-independent.
Figure 3. ALRs are dispensable for the IFN response to lentivirus infection.
(A,B) Quantification of Ifnb and Cxcl10 mRNA induction by RT-PCR in BMMs (A) and MEFs (B) from Trex1+/+, Trex1−/−, ALR−/−, ALR−/−Trex1−/−, and cGas−/−Trex1−/− mice uninfected (“None”) or infected with a VSVg-pseudotyped, self-inactivating lentivirus (“LV”, MOI=1) for 20h. (C) Quantification of Ifnb and Cxcl10 mRNA induction by RT-PCR in BMMs (left panels) and MEFs (right panels) from Trex1−/− cells treated with the reverse-transcriptase inhibitor nevirapine and infected with lentivirus (MOI=1) or transfected with 5μg CT-DNA for 20h. (D,E) Flow cytometric quantification of the frequency of infected (GFP+) BMMs (D) and MEFs (E) from mice of the indicated genotype 48h after infection with lentivirus (MOI=1). Error bars represent mean ± SD. All data are representative of two independent experiments. See also Figure S3.
We also explored whether ALRs influenced lentivirus infectivity by measuring the fraction of infected, GFP-positive macrophages and MEFs 48 hours after infection. Although Trex1−/− macrophages mount a robust IFN response to infection, surprisingly, we observed no significant reduction in the fraction of infected, GFP-positive Trex1−/− macrophages compared to Trex1+/+ cells (Figure 3D). The fraction of infected ALR−/−Trex1−/− macrophages was also similar to Trex1−/− cells, while on the other hand, we observed an increase in the fraction of infected cGas−/−Trex1−/− macrophages. This indicates that cGAS is a restriction factor for lentivirus infection in macrophages. The fraction of infected, GFP-positive MEFs was low and varied between two independent experiments, although there was a trend towards a reduction in infectivity in Trex1−/− cells compared to Trex1+/+ cells, consistent with previous data (Yan et al., 2010) (Figure 3E).
ALRs do not contribute to autoimmune disease in the Trex1-deficient mouse model of Aicardi Goutières Syndrome
Polymorphisms in the human TREX1 gene are associated with systemic lupus erythematosus (SLE), and loss of function mutations in TREX1 cause the rare autoimmune disease Aicardi-Goutières syndrome (AGS), which is characterized by elevated type I IFNs (Crow et al., 2015; Crow et al., 2006; Lebon et al., 1988; Lee-Kirsch et al., 2007; Rice et al., 2007). AGS can be modeled using Trex1−/− mice, which develop a lethal autoimmune disease characterized by elevated production of type I IFNs, autoantibodies, and multiorgan inflammation including inflammatory myocarditis (Morita et al., 2004; Stetson et al., 2008). Autoimmune disease is dependent on activation of the ISD pathway, as Trex1−/− mice that lack key components of this pathway, including cGAS, STING, or IRF3, are completely protected from premature death and autoimmune tissue destruction (Gall et al., 2012; Gao et al., 2015; Gray et al., 2015; Stetson et al., 2008).
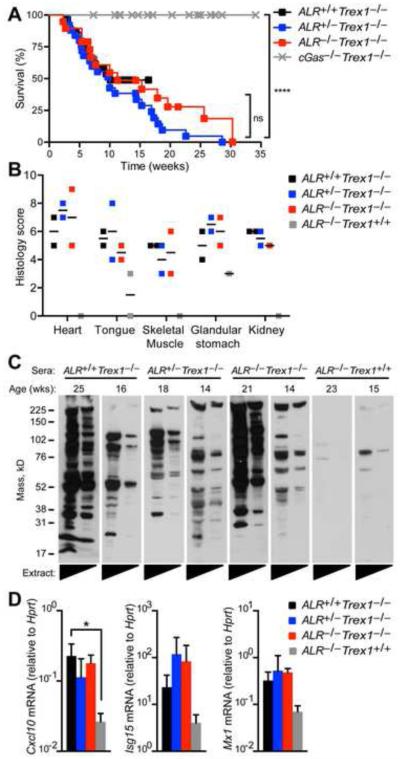
To determine whether ALRs contribute to disease in Trex1−/− mice, we intercrossed ALR−/− and Trex1−/− mice. ALR+/+Trex1−/−, ALR+/−Trex1−/−, and ALR−/−Trex1−/− mice succumbed to lethal disease with similar kinetics (Figure 4A), suggesting that the ALRs are not required to drive disease in Trex1−/− mice. Moreover, ALR−/−Trex1−/− mice developed inflammation in multiple tissues, including the heart, similar to ALR+/+Trex1−/− mice (Figure 4B). We detected autoantibodies against heart tissue antigens in serum from both ALR−/−Trex1−/− and ALR+/+Trex1−/− mice (Figure 4C). Finally, expression of IFN-stimulated genes was elevated in peripheral blood isolated from ALR−/−Trex1−/− and ALR+/+Trex1−/− mice compared to ALR−/− Trex1+/+ mice (Figure 4D). Thus, in contrast to cGAS and STING, which are absolutely required to drive disease, ALRs are dispensable for disease in Trex1-deficient mice.
Figure 4. AIM2-like receptors are not required to drive autoimmune disease in the Trex1-deficient mouse model of Aicardi Goutières Syndrome.
(A) Survival curves of ALR+/+Trex1−/− (n=29), ALR+/−Trex1−/− (n=25), ALR−/−Trex1−/− (n=19), and cGas−/−Trex1−/− (n=29, monitored during a similar time period in our colony and published elsewhere (Gray et al., 2015)). All mice were on a pure C57BL/6 background. Statistical analysis was performed with a Log-rank (Mantel-Cox) test. (B) Histological scores of inflammation in the indicated tissues from ALR+/+Trex1−/−, ALR+/−Trex1−/−, ALR−/−Trex1−/−, and ALR−/−Trex1+/+ mice. All histological analysis was performed in a blinded manner. (C) Autoantibodies against heart antigens evaluated by blotting Rag2−/−Trex1−/− mouse heart extracts (neat and 1:5 diluted) with sera from mice of the indicated genotype. (D) Quantification by RT-PCR of IFN-stimulated gene transcripts in total peripheral blood cells from mice of the indicated genotype. Error bars represent mean ± SD. Data are representative of one experiment with 2-5 mice of each genotype.
Human IFI16 is dispensable for the IFN response to HCMV infection
Human have four ALRs (AIM2, IFI16, PYHIN1, and MNDA) that, with the exception of AIM2, lack mouse orthologues (Brunette et al., 2012; Cridland et al., 2012). In 2010, IFI16 was proposed to function as a DNA sensor for the ISD response (Unterholzner et al., 2010) and subsequent studies using siRNA approaches have suggested that IFI16 is required for the IFN response to herpes simplex virus (HSV-1), human cytomegalovirus (HCMV), and human immunodeficiency virus (HIV) infection (Jakobsen et al., 2013; Li et al., 2013a; Orzalli et al., 2012). However, the role of IFI16 in the ISD pathway has not yet been confirmed genetically in IFI16 knockout human cells. In contrast, studies with cGAS-deficient cells have shown that cGAS is essential for the IFN response to HSV-1 and HIV infection (Gao et al., 2013; Lahaye et al., 2013; Orzalli et al., 2015).
Infection with the DNA virus HCMV activates a STING-dependent IFN response (Li et al., 2013a). The DNA sensors involved in innate detection of HCMV are not well defined, although one recent study suggested that IFI16 is required for the IFN response to HCMV (Li et al., 2013a). The HCMV tegument protein pUL83 has been proposed to antagonize the IFN response (Abate et al., 2004; Browne and Shenk, 2003) by binding to IFI16, preventing activation of STING-TBK1-IRF3 signaling (Li et al., 2013a). However, whether UL83 antagonizes the IFN response to HCMV infection has been debated (Taylor and Bresnahan, 2006). Moreover, the contribution of cGAS to innate sensing of HCMV has not yet been explored.
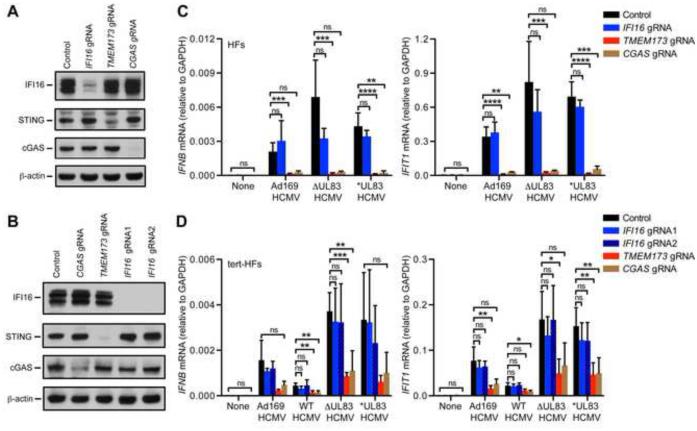
We used a lentiCRISPR approach to disrupt CGAS, IFI16, and TMEM173 (STING) in human fibroblasts (HFs) and confirmed protein loss by Western blot analysis (Figure 5A,B). In cGAS-deficient HFs, IFI16 protein levels were unchanged, in contrast to a recent study suggesting that cGAS promotes IFI16 protein stability (Orzalli et al., 2015). We infected STING-, cGAS-, or IFI16-deficient HFs with wild type HCMV (Ad169 or the parental strain AdCreGFP, referred to as WT HCMV) or two UL83-deficient viral strains: (1) ΔUL83 HCMV, in which the entire UL83 open reading frame has been replaced with a kanamycin resistance cassette, or (2) UL83stop (*UL83) HCMV, which contains stop codon mutations in UL83 (rather than deletion of the entire ORF (Taylor and Bresnahan, 2006)). We measured induction of IFNB and the IFN-stimulated gene IFIT1 6 hours post infection. We confirmed our observations in multiple experiments with three independently generated LentiCRISPR cell lines generated using primary fibroblasts (HFs) or tert-immortalized fibroblasts (tert-HFs) (Supplemental Figure 4). We observed a trend towards an increase in the IFN response to infection with ΔUL83 and UL83stop HCMV compared to Ad169 and WT HCMV (Figure 5C,D), although this varied among individual experiments (Supplemental Figure 4). The IFN response to ΔUL83 and UL83stop HCMV was impaired in cGAS and STING-deficient cells, but was intact in IFI16-deficient cells (Figure 5C,D) indicating that cGAS, rather than IFI16, is the key DNA sensor required for the IFN response to HCMV infection.
Figure 5. IFI16 is not required for the IFN response to HCMV infection.
(A,B) Primary human fibroblasts (HFs, A) or telomerase-immortalized HFs (tert-HFs, B) were transduced with LentiCRISPR lentivirus encoding Cas9 and the indicated guide RNAs (gRNA) and selected for three days in puromycin. Cells were harvested for evaluation of IFI16, STING, and cGAS protein expression by Western blot at least 12 days after transduction. (C,D) Quantification of IFNB and IFIT1 mRNA induction by RT-PCR in targeted HFs (C) and tert-HFs (D) described in panels (A) and (B), infected with the indicated HCMV viral strains (MOI = 3) for 6h. Error bars represent mean ± SD. Data are representative of five independent experiments (with at least two experiments with each gRNA). Statistical analysis was performed comparing control cells to cGAS, STING, and IFI16-targeted cells using an unpaired t-test corrected for multiple comparisons using the Holm-Sidak method. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S4.
Enhanced IFN response to transfected DNA in ALR- and AIM2-deficient cells
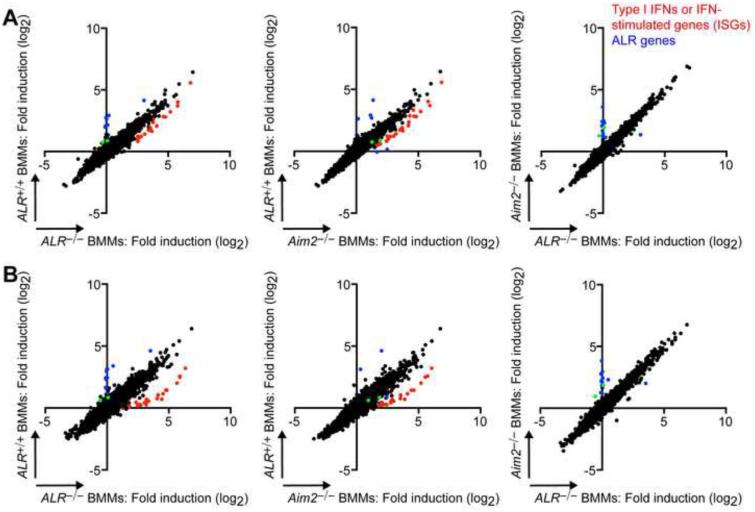
Our analysis of the ALR−/− cells indicated that ALRs were dispensable for the ISD response; therefore, we wondered whether the ALRs activated another innate signaling pathway upon intracellular DNA detection. To explore this possibility, we performed cDNA microarray analysis comparing ALR+/+, ALR−/−, and Aim2−/− BMMs and MEFs, treated for four or eight hours with CT-DNA. Analysis of gene expression changes in BMMs revealed enhanced induction of type I IFNs and IFN-stimulated genes (ISGs) in ALR−/− and Aim2−/− macrophages compared to ALR+/+ macrophages following CT-DNA transfection (Figure 6 and Supplementary Table 2). This is consistent with prior studies that observed an enhanced IFN response to transfected DNA in AIM2-deficient cells, likely due to lack of pyroptosis (Corrales et al., 2016; Jones et al., 2010; Rathinam et al., 2010). However, comparison of ALR−/− and Aim2−/− macrophages revealed only a small number of differentially expressed genes, the majority of which fell into two major groups: (1) ALR genes for which, as expected, the signal was very low in ALR−/− macrophages; (2) genes linked to Aim2 for which expression changes are likely related to their probable 129 origin in Aim2−/− (B6.129) mice (Supplemental Table 2). Of the few remaining differentially expressed genes, we chose to analyze two genes (Nrap and Rab3d, indicated by green dots in Figure 6) and failed to observe any significant differences in expression by RT-PCR (Supplemental Figure 5). Analysis of gene expression changes in MEFs revealed a slight increase in the induction of type I IFNs in ALR−/− and Aim2−/− fibroblasts compared to ALR+/+ fibroblasts in response to CT-DNA transfection (Supplemental Figure 6A,B and Supplemental Table 3) similar to the enhanced response that we observed in ALR−/− and Aim2−/− macrophages. However, we did not consistently detect an elevated IFN response to transfected CT-DNA in ALR−/− and Aim2−/− fibroblasts compared to ALR+/+ cells in other independent experiments (Figure 2C and Supplemental Figure 6C). Thus, AIM2 negatively regulates the IFN response to intracellular DNA in macrophages but not MEFs, suggesting cell type specificity in the interactions between AIM2 and the cGAS-STING pathway.
Figure 6. ALR-deficient and AIM2-deficient macrophages have an enhanced IFN response to transfected DNA.
Microarray analysis of gene expression changes in ALR+/+,ALR−/−, and Aim2−/− BMMs transfected with 5μg calf-thymus DNA (CT-DNA) for 4h (A) or 8h (B) plotted as fold induction relative to mock-treated cells (treated with lipofectamine alone). Selected type I IFN, cytokines, and IFN-stimulated genes are highlighted in red, and ALR genes are highlighted in blue. Nrap and Rab3d are indicated in green. Data represent one experiment. See also Figures S5 and S6.
We identified a number of differentially expressed genes in Aim2−/− MEFs compared to ALR−/− MEFs, including the ALR genes, as well as genes whose expression was altered in Aim2−/− MEFs, but similar in ALR+/+ and ALR−/− MEFs. Expression of nearly all genes in this latter category was not significantly changed by CT-DNA stimulation (Supplemental Table 3), suggesting that these differences were likely due to the distinct origins of the Aim2−/− fibroblasts, which were prepared from embryos harvested independently from the ALR+/+ and ALR−/− fibroblasts, rather than a unique ALR-dependent transcriptional response to DNA stimulation. Altogether, our microarray data demonstrate that AIM2 negatively regulates the ISD response in macrophages; however, we failed to detect a transcriptional response to CT-DNA transfection dependent on the other 12 mouse ALRs.
Unbiased exploration of ALR ligands and functions
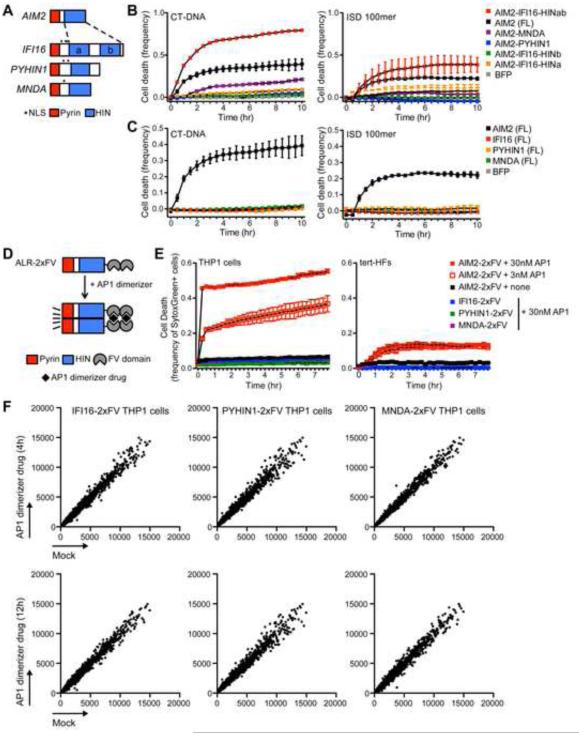
Given that all mouse ALRs and human IFI16 are dispensable for the IFN response to intracellular DNA, we considered two important questions. First, is dsDNA the relevant ligand for all ALRs? Second, can we bypass the ligand specificities of individual ALRs to explore their potential functions in activating pyroptosis or gene transcription? Both in vitro binding studies and crystal structure analyses have shown that the AIM2 and IFI16 HIN domains bind dsDNA in a sequence-independent manner (Hornung et al., 2009; Jin et al., 2012; Unterholzner et al., 2010), yet whether PYHIN1 and MNDA respond to dsDNA has not been tested. To explore whether the HIN domains of each human ALR were capable of being activated by dsDNA, we took advantage of the well-defined signaling function of the AIM2 Pyrin domain, which binds the adaptor ASC, leading to caspase-1 activation and pyroptotic cell death. We generated chimeric ALRs in which the AIM2 Pyrin domain was fused to the HIN domains of IFI16, PYHIN1, or MNDA (Figure 7A). We then reconstituted AIM2-deficient THP1 cells with each chimeric ALR protein (Supplemental Figure 7A) and measured cell death following transfection of dsDNA (CT-DNA or ISD 100-mer oligonucleotides (Stetson and Medzhitov, 2006)) as a readout of DNA-dependent activity of each HIN domain. Chimeric ALRs containing both IFI16 HIN domains, but not either HIN domain alone, induced robust pyroptosis in response to dsDNA, consistent with previous in vitro binding studies demonstrating that both HIN domains are required for maximal DNA binding activity (Figure 7B; (Unterholzner et al., 2010)). In contrast, we failed to reproducibly detect significant cell death following DNA stimulation in cells expressing chimeric ALRs containing the PYHIN1 or MNDA HIN domains (Figure 7B). As a control, we reconstituted AIM2-deficient THP1 cells with each native ALR. We observed DNA-induced pyroptosis in cells expressing AIM2, but not IFI16, PYHIN1, or MNDA (Figure 7C). This suggests that although the AIM2 and IFI16 HIN domains are clearly capable of binding dsDNA, the PYHIN1 or MNDA HIN domains either do not bind to dsDNA or are not sufficiently activated by dsDNA to be detected by this assay.
Figure 7. Unbiased exploration of ALR ligands and functions.
(A) Schematic of human ALR genes with Pyrin domains indicated in red and HIN domains indicated in blue. Chimeric ALRs were constructed in which the AIM2 Pyrin domain was fused to the HIN domains of IFI16, PYHIN1, or MNDA. (B,C) Cell death of AIM2-deficient THP1 cells reconstituted with lentiviral constructs expressing chimeric ALRs (B) or wild type ALRs (C) transfected CT-DNA or ISD 100-mer oligonucleotides (Stetson and Medzhitov, 2006) quantified using an IncuCyte imaging system. Error bars represent mean ± SD. Data are representative of three independent experiments. (D) Schematic of dimerizable ALR constructs in which two FV domains were fused to the C-terminus of each human ALR gene. (E) Cell death of THP1 cells (left panel) or tert-HFs (right panel) reconstituted with lentiviral constructs expressing ALR-2xFV dimerizable proteins treated with 3nM or 30nM of the AP1 dimerizer drug. Error bars represent mean ± SD. Data are representative of two (THP1 cells) or one (tert-HFs) independent experiment. (F) Microarray analysis of gene expression changes in ALR-2xFV-expressing THP1 cells 4h or 12h after treatment with 30nM AP1 (plotted as raw expression in vehicle-treated (Mock, X-axis) relative to AP1-treated cells (Y-axis). Data are representative of one experiment. See also Figure S7.
Since human PHYIN1 and MNDA HIN domains were unresponsive to dsDNA in the AIM2 chimera assay, we explored the consequences of ligand-independent oligomerization of human ALRs as an alternative approach to interrogate their potential functions. To do this, we generated fusion proteins in which we appended two Fv domains to the C-terminus of each full-length human ALR (Figure 7D). These Fv domains are specifically activated by a small molecule drug called AP1, leading to their oligomerization (Clackson et al., 1998). We transduced THP-1 cells and tert-HFs with these constructs and verified their expression by Western blot (Supplemental Figure 7B). We found that AP1 oligomerization of AIM2-2xFv resulted in rapid, dose-dependent induction of pyroptosis, confirming ligand-independent activation (Figure 7E). However, oligomerization of IFI16, PYHIN1, or MNDA in THP1 cells or IFI16 in tert-HFs resulted in no detectable cell death (Figure 7E).
To explore whether oligomerization of human ALRs resulted in any change in gene expression, we performed microarray analysis on THP-1 cells stably transduced with IFI16-2xFv, PYHIN1-2xFv, or MNDA-2xFv, comparing mRNA expression levels of resting cells to those in cells treated for 4 or 12 hours with AP1. We found no evidence for inducible gene expression in response to ALR oligomerization (Figure 7F). Together, these data suggest that, at least in the context of AP1-induced oligomerization, IFI16, PYHIN1, and MNDA activate neither the inflammasome nor any strong transcriptional responses in THP-1 cells.
Discussion
Here, we characterized mice that lack all 13 ALRs, as well as human cells lacking IFI16. We found that mouse ALRs are not required for the IFN response to transfected DNA, MCMV infection, and lentivirus infection and do not contribute to autoimmune disease in the Trex1−/− mouse model of Aicardi-Goutières Syndrome. We also demonstrated that IFI16 is dispensable for the IFN response to HCMV infection. Together, this suggests that, in contrast to cGAS, the ALRs are not essential for activation of the ISD pathway. Whether they contribute in a more subtle way, or whether they are essential for IFN responses to ligands and pathogens that we did not examine here remain important unanswered questions.
If the ALRs are not essential for the ISD response, what then might be their functions? One possibility is that the ALRs regulate the function of each other. Indeed, the mouse ALR IFI202 has been shown to bind to the AIM2 HIN domain and inhibit AIM2 oligomerization and caspase-1 inflammasome activation (Roberts et al., 2009; Yin et al., 2013). Another possibility is that, like AIM2 itself, the other ALRs may participate in inflammasome activation. IFI16 has been implicated in activating the inflammasome in CD4 T cells during abortive HIV infection (Monroe et al., 2014) and in endothelial cells infected with Kaposi sarcoma-associated herpesvirus (KSHV) (Kerur et al., 2011). However, the mechanism by which IFI16 might activate the caspase-1 inflammasome is unclear given that, unlike AIM2, the IFI16 Pyrin domain does not associate with the ASC adapter (Hornung et al., 2009). Moreover, we found that oligomerization of IFI16 was insufficient to induce pyroptosis in THP1 cells or tert-HFs. Nonetheless, it remains a possibility that IFI16 and/or other ALRs may activate the inflammasome in specific cell types and/or in response to certain pathogens.
Although not essential for the ISD response, the ALRs might contribute to the IFN response in a more subtle way, for example by enhancing the response at later time points. Consistent with this possibility, we observed robust induction of the IFN-stimulated genes IFIT1 and ISG15 in PMA-treated THP1 cells and HFs stably expressing HA-tagged IFI16 compared to cells expressing BFP (data not shown). Induction of ISGs was only observed in PMA-treated cells, indicating that overexpression of IFI16 alone is not sufficient for this response and another co-factor must be involved. We are currently exploring whether IFI16 induces ISG expression in a STING-dependent manner, or whether this occurs via another mechanism.
ALRs have been suggested to contribute to autoimmune disease. Gene intervals on chromosome 1 that span the ALR genes have been linked to SLE in humans (1q23 locus) and to autoimmunity in the lupus-prone mouse strains NZB (Nba2 locus) and BSXB (Bxs3 locus) (Haywood et al., 2004; Moser et al., 1998; Rozzo et al., 2001). However, the ALR locus is tightly linked to immune genes that are also proposed to modify lupus risk, including Fcγ receptor and SLAM family member genes. Thus, whether ALRs contribute to disease or are simply linked to other lupus susceptibility genes is not clear. Consistent with the latter possibility, more recent studies have found that the mouse ALR locus does not contribute to disease, while intervals of the Nba2 locus containing Fcγ receptor and SLAM family member genes each contribute to lupus-like symptoms (Jorgensen et al., 2010). Moreover, analysis of the ALR locus in two SLE cohorts failed to find evidence of SNP association (Fernando et al., 2011). Here, we found that ALRs are not required to drive autoimmune disease in Trex1-deficient mice. Our data also revealed that AIM2-dependent inflammasome activation does not contribute to disease in this mouse model. Whether ALRs contribute to autoimmune disease in other mouse models remains an open question, and our ALR-deficient mice provide a valuable tool address this question.
Although it is clear that the ALRs are not essential for the ISD response, there are still compelling reasons to think this family of genes has an important role in antiviral immunity. Most ALRs are IFN-stimulated genes, consistent with an antiviral function (Brunette et al., 2012). Evolutionary analysis has revealed extensive diversity within the ALR gene family, including multiple species-specific expansions, which may reflect evolutionary pressures from DNA viruses (Brunette et al., 2012; Cridland et al., 2012). Indeed, consistent with genetic conflict, a recent phylogenetic study provided evidence that both AIM2 and IFI16 have evolved under positive selection in primates (Cagliani et al., 2014). The ALR-deficient mice described here provide a valuable tool for future studies exploring the function of this family of receptors.
Experimental Procedures
Mice
Generation of ALR-deficient mice on a C57BL/6 background is described in the Supplemental Methods and Supplemental Figure 1. ALR−/− mice were intercrossed with Trex1−/− mice to generate ALR−/−Trex1−/− mice. Trex1−/−, Tmem173−/−, cGas−/− (Mb21d1−/−), and Rag2−/− mice have been described (Gall et al., 2012; Gray et al., 2015). 129P3/J mice (stock #000690) and Aim2−/− mice generated in 129 ES cells and backcrossed to C57BL/6J (stock #013144) (Rathinam et al., 2010) were purchased from Jackson Laboratories. Frozen bone marrow from Aim2−/− mice generated in C57BL/6 ES cells (Jones et al., 2010) was kindly provided by Denise Monack. All mouse studies were carried out with approval of the UW Institutional Animal Care and Use Committee.
Cell treatments and analysis
Primary mouse BMMs, MEFs, HFs, and THP1 cells were transfected with nucleic acid ligands using Lipofectamine 2000 (Life Technologies) as described (Brunette et al., 2012). For quantitative RT-PCR analysis, cells were harvested into RNA Stat-60 (Tel-Test, INC), and RNA was isolated, treated with DNase (Ambion), and reverse-transcribed into cDNA with EcoDry Premix (Clontech) or SuperScript III (Thermo Fisher). PCR was performed with EVA Green reagents (Bio-Rad Laboratories) on a Bio-Rad CFX96 Real-Time System. Type I IFN induction was measured using an IFN bioassay as described (Lau et al., 2015).
Lentivirus infections
VSV-G pseudotyped, self-inactivating lentivirus was prepared by transfecting 293T cells with 1.5μg pVSV-G, 3μg psPAX-2, and 6μg pRRL lentiviral vector in which GFP was cloned downstream of an MND promoter. Lentiviral supernatents were collected 48 hours later and concentrated by centrifugation at 8500xg overnight at 4°C. Viral stocks were titered on 293T cells by quantifying the %GFP+ cells by flow cytometry 48 hours after infection. BMMs or MEFs were seeded at 7.5×105 and 1×105 cells per well, respectively, in 12-well plates and infected the next day at a multiplicity of infection (MOI) of 1 for 6-24 hours as indicated. In some experiments, cells were pre-treated with 5μM nevirapine for 4-5 hours prior to infection with lentivirus in the presence of 5μM nevirapine for 20 hours.
HCMV and MCMV infections
HCMV strain Ad169, the GFP-expressing strain AdCREGFP (derived from Ad169), and the UL83-deficient strains ΔUL83 and UL83stop (derived from AdCREGFP and kindly provided by Wade Bresnahan (Taylor and Bresnahan, 2006)) were grown and titered in primary HFs. Loss of UL83 in ΔUL83 and UL83stop strains was confirmed by Western blot (data not shown). Cells were infected at a multiplicity of infection (MOI) of 3 PFU/cell by spinoculation at 1200rpm (290g) for one hour at room temperature. The inoculum was aspirated and replaced with fresh medium, and cells were incubated at 37°C for five additional hours.
MCMV strain K181 was grown and titered in NIH 3T3 cells. One day prior to infection, MEFs were seeded at 1×105 per well in a 12-well plate. Cells were infected at a MOI of 3 PFU/cell for 1h at 37°C. The inoculum was aspirated and replaced with fresh media, and cells were incubated at 37°C for four additional hours.
CRISPR/Cas9 gene targeting
For CRISPR/Cas9 gene targeting, we used a lentivirus in which a U6 promoter-driven guide RNA and MND promoter-driven Cas9-T2A-puromycin resistance cassette are constitutively expressed from a single, self-inactivating lentivirus. THP1 cells or HFs were transduced with lentivirus and selected with 5μg/ml puromycin (Thermo Fisher Scientific) for three days. Gene targeting was evaluated by restriction fragment length polymorphism (RFLP) using restriction sites that overlapped the CRISPR targeting sites, as well as western blot for endogenous protein. The guide RNA target sites are described in supplemental information.
Pathology
Tissues were fixed in 10% neutral buffered formalin and paraffin embedded. Tissue sections (5μm) were stained with hematoxylin and eosin and histological scores were evaluated in a blinded manner as previously described (Gall et al., 2012). Histology scores for the skeletal muscle refer to assessment of muscle on the head (scored separately from the tongue); histology scores for the kidney reflect maximum inflammation scores for the interstitium as well as the extent of pathology.
Detection of autoantibodies
Heart extracts were blotted with mouse sera as previously described (Stetson et al., 2008).
2xFV experiments
ALR-2xFV chimeric constructs were created by cloning full-length AIM2, IFI16, PYHIN1, and MNDA open reading frames upstream of a poly-glycine linker (GGGGR) followed by two FV domains (e.g. FKBP domains with the F36V mutation). These ALR-2xFV constructs were cloned into a pRRL lentiviral vector for SFFV promoter-driven ALR-2xFV-T2A-mCherry-T2A-blasticidin resistance expression. THP1 cells or tert-HFs were transduced with lentivirus and selected with 10μg/ml blasticidin (Thermo Fisher Scientific) for three days. The homodimerizer drug AP1 (Clontech, also called ‘B/B Homodimerizer’) was dissolved at 100μM in ethanol and diluted in culture media to a final concentration of 3nM or 30nM.
Statistical analysis
Statistical significance of difference between groups was assessed with unpaired t-test or Log-rank (Mantel-Cox) test as indicated in the figure legends. All analyses were performed with Graph Pad Prism 6 (GraphPad).
Supplementary Material
Acknowledgements
We are grateful to the University of Washington Histology and Imaging Core and B. Johnson for histology preparation and members of the Stetson lab for helpful discussions. Wade Bresnahan kindly provided ADCREGFP, ΔUL83, and UL83stop virus strains. Denise Galloway kindly provided telomerase-immortalized human foreskin fibroblasts. Denise Monack kindly provided bone marrow from Aim2−/− mice generated in C57BL/6 ES cells. D.B.S. is a scholar of the Rita Allen Foundation and a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease. This work was supported by grants from the NIH (AI113153 to D.B.S. and RO1AI027762 to A.G.). E.E.G. is supported by a Cancer Research Institute Irvington postdoctoral fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Microarray data were deposited in the NCBI GEO database (www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE80598 and GSE80766 (SuperSeries GSE80767).
Author Contributions
E.E.G. and D.B.S. designed experiments and wrote the manuscript. E.E.G., S.J.C., and D.W. performed experiments. J.M.S. performed histological analysis. S.J.C. and A.G. provided reagents and expertise in CMV biology.
References
- Abate DA, Watanabe S, Mocarski ES. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J Virol. 2004;78:10995–11006. doi: 10.1128/JVI.78.20.10995-11006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne EP, Shenk T. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc Natl Acad Sci U S A. 2003;100:11439–11444. doi: 10.1073/pnas.1534570100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunette RL, Young JM, Whitley DG, Brodsky IE, Malik HS, Stetson DB. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med. 2012;209:1969–1983. doi: 10.1084/jem.20121960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Cagliani R, Forni D, Biasin M, Comabella M, Guerini FR, Riva S, Pozzoli U, Agliardi C, Caputo D, Malhotra S, et al. Ancient and recent selective pressures shaped genetic diversity at AIM2-like nucleic acid sensors. Genome biology and evolution. 2014;6:830–845. doi: 10.1093/gbe/evu066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clackson T, Yang W, Rozamus LW, Hatada M, Amara JF, Rollins CT, Stevenson LF, Magari SR, Wood SA, Courage NL, et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc Natl Acad Sci U S A. 1998;95:10437–10442. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AC, Cai H, Li T, Franco LH, Li XD, Nair VR, Scharn CR, Stamm CE, Levine B, Chen ZJ, Shiloh MU. Cyclic GMP-AMP Synthase Is an Innate Immune DNA Sensor for Mycobacterium tuberculosis. Cell Host Microbe. 2015;17:820–828. doi: 10.1016/j.chom.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L, Woo SR, Williams JB, McWhirter SM, Dubensky TW, Jr., Gajewski TF. Antagonism of the STING Pathway via Activation of the AIM2 Inflammasome by Intracellular DNA. J Immunol. 2016 doi: 10.4049/jimmunol.1502538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland JA, Curley EZ, Wykes MN, Schroder K, Sweet MJ, Roberts TL, Ragan MA, Kassahn KS, Stacey KJ. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC evolutionary biology. 2012;12:140. doi: 10.1186/1471-2148-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GM, Gornall HL, Oojageer A, Anderson B, Pizzino A, Helman G, et al. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. American journal of medical genetics. Part A. 2015;167A:296–312. doi: 10.1002/ajmg.a.36887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, et al. Mutations in the gene encoding the 3'-5' DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando MM, de Smith AJ, Coin L, Morris DL, Froguel P, Mangion J, Blakemore AI, Graham RR, Behrens TW, Vyse TJ. Investigation of the HIN200 locus in UK SLE families identifies novel copy number variants. Annals of human genetics. 2011;75:383–397. doi: 10.1111/j.1469-1809.2011.00641.x. [DOI] [PubMed] [Google Scholar]
- Gall A, Treuting P, Elkon KB, Loo YM, Gale M, Jr., Barber GN, Stetson DB. Autoimmunity Initiates in Nonhematopoietic Cells and Progresses via Lymphocytes in an Interferon-Dependent Autoimmune Disease. Immunity. 2012;36:120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Li T, Li XD, Chen X, Li QZ, Wight-Carter M, Chen ZJ. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc Natl Acad Sci U S A. 2015;112:E5699–5705. doi: 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray EE, Treuting PM, Woodward JJ, Stetson DB. Cutting Edge: cGAS Is Required for Lethal Autoimmune Disease in the Trex1-Deficient Mouse Model of Aicardi-Goutieres Syndrome. J Immunol. 2015;195:1939–1943. doi: 10.4049/jimmunol.1500969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood ME, Rogers NJ, Rose SJ, Boyle J, McDermott A, Rankin JM, Thiruudaian V, Lewis MR, Fossati-Jimack L, Izui S, et al. Dissection of BXSB lupus phenotype using mice congenic for chromosome 1 demonstrates that separate intervals direct different aspects of disease. J Immunol. 2004;173:4277–4285. doi: 10.4049/jimmunol.173.7.4277. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen MR, Bak RO, Andersen A, Berg RK, Jensen SB, Tengchuan J, Laustsen A, Hansen K, Ostergaard L, Fitzgerald KA, et al. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci U S A. 2013;110:E4571–4580. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O'Rourke K, Chan S, Dong J, Qu Y, Roose-Girma M, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci U S A. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen TN, Alfaro J, Enriquez HL, Jiang C, Loo WM, Atencio S, Bupp MR, Mailloux CM, Metzger T, Flannery S, et al. Development of murine lupus involves the combined genetic contribution of the SLAM and FcgammaR intervals within the Nba2 autoimmune susceptibility locus. J Immunol. 2010;184:775–786. doi: 10.4049/jimmunol.0901322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye X, Satoh T, Gentili M, Cerboni S, Conrad C, Hurbain I, El Marjou A, Lacabaratz C, Lelievre JD, Manel N. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity. 2013;39:1132–1142. doi: 10.1016/j.immuni.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Lau L, Gray EE, Brunette RL, Stetson DB. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science. 2015;350:568–571. doi: 10.1126/science.aab3291. [DOI] [PubMed] [Google Scholar]
- Lebon P, Badoual J, Ponsot G, Goutieres F, Hemeury-Cukier F, Aicardi J. Intrathecal synthesis of interferon-alpha in infants with progressive familial encephalopathy. J Neurol Sci. 1988;84:201–208. doi: 10.1016/0022-510x(88)90125-6. [DOI] [PubMed] [Google Scholar]
- Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, de Silva U, Bailey SL, Witte T, Vyse TJ, et al. Mutations in the gene encoding the 3'-5' DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- Li T, Chen J, Cristea IM. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe. 2013a;14:591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013b;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, Greene WC. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE. Gene-targeted mice lacking the Trex1 (DNase III) 3'-->5' DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser KL, Neas BR, Salmon JE, Yu H, Gray-McGuire C, Asundi N, Bruner GR, Fox J, Kelly J, Henshall S, et al. Genome scan of human systemic lupus erythematosus: evidence for linkage on chromosome 1q in African-American pedigrees. Proc Natl Acad Sci U S A. 1998;95:14869–14874. doi: 10.1073/pnas.95.25.14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, Broekema NM, Diner BA, Hancks DC, Elde NC, Cristea IM, Knipe DM. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc Natl Acad Sci U S A. 2015;112:E1773–1781. doi: 10.1073/pnas.1424637112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A. 2012;109:E3008–3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchanathan R, Duan X, Shen H, Rathinam VA, Erickson LD, Fitzgerald KA, Choubey D. Aim2 deficiency stimulates the expression of IFN-inducible Ifi202, a lupus susceptibility murine gene within the Nba2 autoimmune susceptibility locus. J Immunol. 2010;185:7385–7393. doi: 10.4049/jimmunol.1002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J, Artuch R, Montalto SA, Bacino CA, Barroso B, et al. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. Am J Hum Genet. 2007;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Rozzo SJ, Allard JD, Choubey D, Vyse TJ, Izui S, Peltz G, Kotzin BL. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity. 2001;15:435–443. doi: 10.1016/s1074-7613(01)00196-0. [DOI] [PubMed] [Google Scholar]
- Stavrou S, Blouch K, Kotla S, Bass A, Ross SR. Nucleic Acid recognition orchestrates the anti-viral response to retroviruses. Cell Host Microbe. 2015;17:478–488. doi: 10.1016/j.chom.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Storek KM, Gertsvolf NA, Ohlson MB, Monack DM. cGAS and Ifi204 cooperate to produce type I IFNs in response to Francisella infection. J Immunol. 2015;194:3236–3245. doi: 10.4049/jimmunol.1402764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Taylor RT, Bresnahan WA. Human cytomegalovirus immediate-early 2 protein IE86 blocks virus-induced chemokine expression. J Virol. 2006;80:920–928. doi: 10.1128/JVI.80.2.920-928.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Berghe T, Hulpiau P, Martens L, Vandenbroucke RE, Van Wonterghem E, Perry SW, Bruggeman I, Divert T, Choi SM, Vuylsteke M, et al. Passenger Mutations Confound Interpretation of All Genetically Modified Congenic Mice. Immunity. 2015;43:200–209. doi: 10.1016/j.immuni.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J, Vance RE, Stallings CL, Virgin HW, Cox JS. The Cytosolic Sensor cGAS Detects Mycobacterium tuberculosis DNA to Induce Type I Interferons and Activate Autophagy. Cell Host Microbe. 2015;17:811–819. doi: 10.1016/j.chom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Sester DP, Tian Y, Hsiao YS, Lu A, Cridland JA, Sagulenko V, Thygesen SJ, Choubey D, Hornung V, et al. Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell reports. 2013;4:327–339. doi: 10.1016/j.celrep.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.