Abstract
Heat stress is an important factor limiting plant growth, development, and productivity; thus, plants have evolved special adaptive mechanisms to cope with high-temperature stress. Non-coding RNAs (ncRNAs) are a class of regulatory RNAs that play an important role in many biological processes. Recently developed advanced technologies, such as genome-wide transcriptomic analysis, have revealed that abundant ncRNAs are expressed under heat stress. Although this area of research is still in its infancy, an increasing number of several classes of regulatory ncRNA (i.e., miRNA, siRNA, and lncRNA) related to heat stress responses have been reported. In this mini-review, we discuss our current understanding of the role of ncRNAs in heat stress responses in plants, especially miRNAs, siRNAs, and their targets. For example, the miR398-CSD/CCS-HSF, miR396-WRKY6, miR159-GAMYB, and TAS1-HTT-HSF pathways regulate plant heat tolerance. We highlight the hormone/development-related miRNAs involved in heat stress, and discuss the regulatory networks of miRNA-targets. We also note that DNA methylation and alternative splicing could affect miRNA expression under heat stress, and some lncRNAs could respond to heat stress. Finally, we briefly discuss future prospects concerning the ncRNA-related mechanisms of heat stress responses in plants.
Keywords: ncRNA, heat stress, miRNA, siRNA, lncRNA
Introduction
Abiotic stresses, such as heat, drought, salinity, and low temperature, seriously impact the growth and productivity of plants. Consequently, as sessile organisms, plants have evolved various sophisticated mechanisms to cope with multiple abiotic stresses. In particular, given the increasing evidence of climate change, the heat stress response mechanism in plants has received increasing interest. Heat stress (high temperature) hinders cellular homeostasis and can lead to leaf etiolation, severe retardation in growth and development, increased risk of disease, and even death (Bita and Gerats, 2013; Liu et al., 2014). Most previous research focused on the regulatory mechanisms linking heat response stress to genes or transcription factors, such as the heat stress transcription factors (HSFs) regulating the expression of heat-responsive genes, and further affecting the accumulation of heat shock proteins (HSPs) in plant thermotolerance (Wang et al., 2004; Huang and Xu, 2008). Recently, however, more emerging non-coding RNAs (ncRNAs) have been found to play important roles in heat responses, the regulatory mechanisms of which were revealed in plants.
The ncRNAs constitute a class of RNA which does not encode a protein and includes microRNAs (miRNAs), small interfering RNAs (siRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). These various types of ncRNA are involved in the transcriptional and post-transcriptional regulation of gene expression, and the modulation of RNA stability and translation (Hirayama and Shinozaki, 2010; de Lima et al., 2012; Khraiwesh et al., 2012). In recent years, a rapidly increasing number of ncRNAs have been reported to function in heat stress responses in plants (Table 1). In this mini-review, we summarize the plant ncRNAs involved in heat responses, focusing on miRNAs, siRNAs, and lncRNAs.
Table 1.
Non-coding RNAs responsive to heat stress in diverse plant species.
| NcRNA | Species | Target | References |
|---|---|---|---|
| miR156 | Ath ↑ Tae ↑ Bra(h,g) ↑ Osa ↓ | SPL | Xin et al., 2010; Yu et al., 2012; Sailaja et al., 2014; Stief et al., 2014 |
| miR159 | Tae ↑ Pvi ↑ | MYB | Xin et al., 2010; Wang et al., 2012; Hivrale et al., 2016 |
| miR160 | Ath ↑ Hvu(a) ↑ Tae ↑ Pvi ↑ Han ↑ Agr ↑ Pto (a–c) ↓ | ARF | Xin et al., 2010; Chen et al., 2012; May et al., 2013; Kruszka et al., 2014; Li M. Y. et al., 2014; Khaksefidi et al., 2015; Hivrale et al., 2016 |
| miR164 | Ath ↑ Agr ↑ Pvi ↑ | NAC | May et al., 2013; Li M. Y. et al., 2014; Hivrale et al., 2016 |
| miR166 | Ath ↑ Hvu ↑ Tae ↑ Pvi ↑ | HD-ZIPIII | Xin et al., 2010; May et al., 2013; Kruszka et al., 2014; Hivrale et al., 2016 |
| miR167 | Hvu(h) ↑ Pto(c–d) ↑ Tae ↑ Pvi ↑ Han ↑ Os ↓ | ARF | Xin et al., 2010; Chen et al., 2012; Kruszka et al., 2014; Sailaja et al., 2014; Khaksefidi et al., 2015; Hivrale et al., 2016 |
| miR168 | Tae ↑ Agr ↑ Pvi ↑ Osa ↓ Pto(a–b) ↓ | AGO1 | Xin et al., 2010; Chen et al., 2012; Li M. Y. et al., 2014; Sailaja et al., 2014; Hivrale et al., 2016 |
| miR169 | Tae ↑ Ath ↑ Pto ↓ | NF-Y | Xin et al., 2010; Chen et al., 2012; Guan et al., 2013 |
| miR171 | Ath ↑ Pto ↓ Pvi ↓ Ptc ↓ | SCL | Lu et al., 2008; Chen et al., 2012; Mahale et al., 2013; Hivrale et al., 2016 |
| miR172 | Tae ↓ Ath ↓ Han ↓ | AP2 | Xin et al., 2010; May et al., 2013; Khaksefidi et al., 2015 |
| miR319 | Pvi ↑ Tae ↓ | TCP | Kumar et al., 2014; Hivrale et al., 2016 |
| miR390 | Pvi ↑ | ARF | Hivrale et al., 2016 |
| miR393 | Tae ↑ Pvi ↑ Ath ↓ | TIR1/ AFB | Xin et al., 2010; Guan et al., 2013; Hivrale et al., 2016 |
| miR394 | Pto (a,b) ↓ Agr ↑ | F-box | Chen et al., 2012; Li M. Y. et al., 2014 |
| miR395 | Tae ↑ Agr ↑ Pvi ↑ Pto (a–j) ↓ | APS/AST | Chen et al., 2012; Kumar et al., 2014; Li M. Y. et al., 2014; Hivrale et al., 2016 |
| miR396 | Han ↑ Pvi ↑ | GRF, bHLH, WRKY | Giacomelli et al., 2012; Hivrale et al., 2016 |
| miR397 | Ath(a) ↓ Osa(b) ↑ | Laccases | Jeong et al., 2011; Mahale et al., 2013 |
| miR398 | Ath ↑ Han ↑ Tae ↑ Bra(a–b) ↓ Osa ↓ Pvi ↓ Pto(a-b) ↓ | CSD, CCS, COX5 | Xin et al., 2010; Chen et al., 2012; Yu et al., 2012; Guan et al., 2013; Lu et al., 2013; Sailaja et al., 2014; Khaksefidi et al., 2015; Hivrale et al., 2016 |
| miR400 | Ath ↓ | PPR | Yan et al., 2012; Li S. X. et al., 2014 |
| miR408 | Agr ↑ Sja(b) ↓ Pto ↓ Pvi ↓ | Plastocyanin | Chen et al., 2012; Li M. Y. et al., 2014; Liu et al., 2014; Hivrale et al., 2016 |
| miR529 | Pvi ↑ | SBP-box | Hivrale et al., 2016 |
| miR827 | Tae ↑ Pvi ↑ | SPX-MFS protein | Lin et al., 2010; Xin et al., 2010; Hivrale et al., 2016 |
| miR5175 | Hvu ↑ | ACC-like oxidase | Kruszka et al., 2014 |
| miR399 | Tae ↑ Bra ↓ | PHO2 | Xin et al., 2010; Yu et al., 2012 |
| SiRNA 002061_0636_3054.1 | Tae ↓ | Yao et al., 2010 | |
| SiRNA 005047_0654_1904.1 | Tae ↓ | Yao et al., 2010 | |
| SiRNA 080621_1340_98.1 | Tae ↓ | Yao et al., 2010 | |
| TAS1-siRNAs | Ath ↑ | HTT1, HTT2 | Li S. X. et al., 2014 |
| lnRNA5 | Tae ↑ | Xin et al., 2011 | |
| lnRNA27 | Tae ↑ | Xin et al., 2011 |
Ath, Arabidopsis thaliana; Hvu, Hordeum vulgare; Ptc, Populus trichocarpa; Osa, Oryza sativa; Tae, Triticum aestivum; Pvi, Panicum virgatum; Han, Helianthus annuus; Agr, Apium graveolens; Pto, Populus tomentosa; Sja, Saccharina japonica; Bra, Brassica rapa; NF-Y, nuclear transcription factor Y; SBP, squamosa promoter binding; APS/AST, ATP sulfurylase/affinity sulfate transporter; TIR1/AFB, transport inhibitor response 1/auxin-related F-box; bHLH, basic-helix-loop-helix; GRF, growth hormone releasing factor; PHO2, phosphate 2; ↑, upregulated; ↓, downregulated.
miRNAs
Plant miRNAs, a class of small (20–24 nucleotide) ncRNAs, negatively regulate gene expression by either mRNA degradation or translation inhibition (Rogers and Chen, 2013). Accumulating evidence has shown that miRNAs play essential roles in plant responses to heat stress (Table 1).
miRNA398
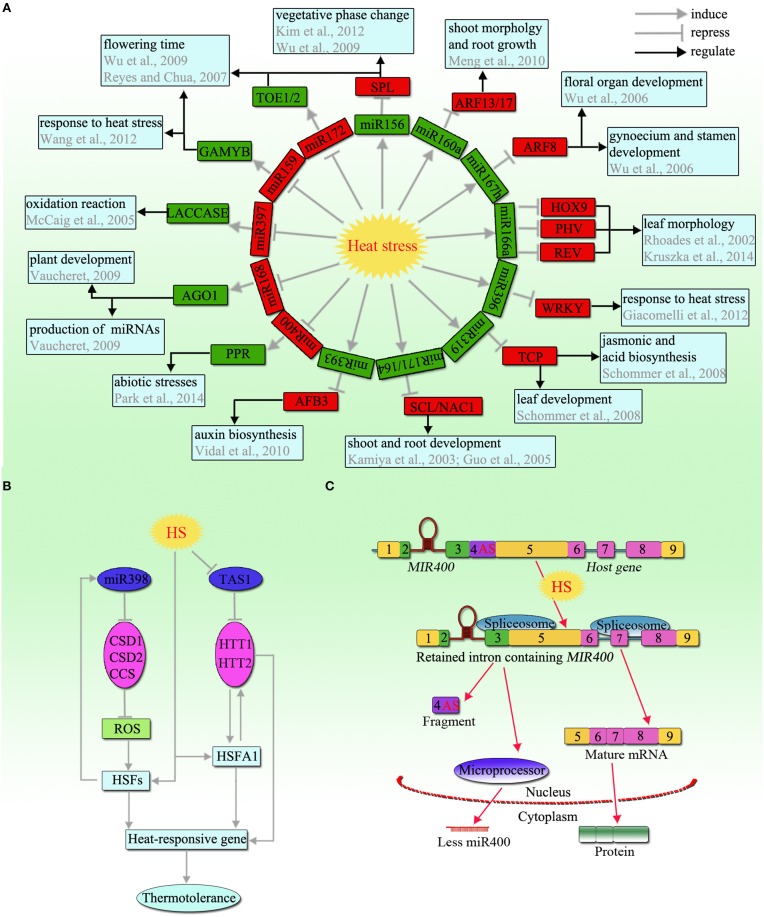
miR398 is a specific well-studied example of an miRNA involved in responses to diverse abiotic stresses, particularly heat stress. In Arabidopsis, miR398 has four target genes, namely, CSD1 and CSD2 (closely related copper/zinc superoxide dismutases), Cox5b-1 (a subunit of the mitochondrial cytochrome c oxidase), and CCS1 (a copper chaperone for SOD; Sunkar and Zhu, 2004; Zhu et al., 2011), which are highly conserved in land plants. Among these, the CSDs are important scavengers of reactive oxygen species (ROS), and CSD/CCS negatively regulates the accumulation of ROS (Mittler, 2002; Sunkar et al., 2006), which are also associated with HSF and HSP synthesis (Guan et al., 2013; Lu et al., 2013). miR398 was shown to be rapidly induced in response to heat stress, accompanied by the downregulation of its target genes (CSD1, CSD2, and CCS; Guan et al., 2013; Figure 1B). Transgenic plants expressing miR398-resistant versions of CSD1, CSD2, or CCS showed hypersensitivity to heat stress, while the csd1, csd2, and ccs mutants were more tolerant to heat stress, with increased HSF and HSP levels (Guan et al., 2013; Lu et al., 2013). In addition, miR398 and its target CSDs were also found in the heat stress responses of Brassica rapa and Populus tomentosa (Kotak et al., 2007; Yu et al., 2012), indicating that the miR398-CSD/CCS pathway is widely involved in the heat stress response in plants.
Figure 1.
An overview of non-coding RNAs in response to heat stress in plants. (A) miRNA-target network module involved in the heat stress response. The network is based on the changes in expression profiles of miRNAs and their targets in plants under heat stress. Black arrows represent regulatory effects (position or negative regulation); Green boxes: upregulated; red boxes: downregulated. (B) Schematic model of miR398 and TAS1, which play an important role in thermotolerance. (C) Schematic model of the heat-induced AS that led to a decrease in miR400 expression (modified from Yan et al., 2012). Under heat stress, an alternative splicing (AS) event occurs in the miR400-containing intron and generates a new host gene. In addition, a fragment containing the original branch site is excised, which induces the rest of the unrecognized intron including the miR400 hairpin to be retained in the host gene. The primary miR400 transcripts without splicing out are hardly processed into mature miR400 by Microprocessor. The number 4 represents the AS intron region. Abbreviations: SPL, squamosa promoter binding protein-like; ARF, auxin response factor; HOX, homeobox leucine zipper protein; PHV, phavoluta; REV, revoluta; TCP, teosinte branched/cycloidea protein; SCL, scarecrow-like; NAC, nascent polypeptide-associated complex; AFB, auxin receptor F-box proteins; PPR, pentatricopeptide repeat; AGO, argonaute; GAMYB, gibberellic acid MYB; TOE, target of eat; CSD, copper/zinc superoxide dismutase; CCS, copper chaperone for superoxide dismutase; TAS1, trans-acting siRNA precursor 1; HTT, heat-induced tas1 target; ROS, reactive oxygen species; HSF, heat stress transcription factor; HS, heat stress.
miR156 and miR172
miR156 and miR172 with their SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) and APETALA2 (AP2) targets control vegetative phase change and flowering (Wu et al., 2009). Under heat response conditions, miR156 was highly induced, and SPL was downregulated, which further induced FLOWERINGLOCUS T (FT) and FRUITFULL (FUL) expression in Arabidopsis (Kim et al., 2012; Stief et al., 2014). Similarly, miR156h and miR156g were particularly upregulated, and BracSPL2 was sharply downregulated under heat stress in B. rapa (Yu et al., 2012). Interestingly, miR156 isoforms were important for heat stress memory in Arabidopsis. miR156 can promote the sustained expression of heat stress-responsive genes through SPL genes, especially SPL2 and SPL11, and is critical only after heat stress (Cui et al., 2014; Stief et al., 2014). Owing to the conservation of miR156 and its target genes, it was proposed that the function of miR156 in heat stress memory may also be conserved in plants (Stief et al., 2014).
miR172 targets AP2-like genes, such as TARGET OF EAT1 (TOE1), TOE2, and SCHLAFMUTZE (SMZ). In contrast to miR156, miR172 is downregulated by heat stress in plants (Figure 1A), such as in Arabidopsis, wheat, and Helianthus annuus (Table 1), while TOE2 is upregulated (Li S. X. et al., 2014). Similarly, an elevated temperature also decreases miR172 expression and upregulates its target TOE1 (May et al., 2013). The findings indicate that a high temperature can alter the expression of all components in the miR156-SPL-miR172-AP2 pathway in a complex manner.
Phytohormone-related miRNAs
A certain type of miRNA is associated with hormone responses to heat stress. Auxin signaling-related miR160 targets the AUXIN RESPONSE FACTOR17 (ARF17) and ARF13 genes, which are involved in root, shoot, and flower development (Meng et al., 2010). Under heat stress, miR160 was found to be upregulated while its target ARF was downregulated in Hordeum vulgare and H. annuus (Kruszka et al., 2014; Khaksefidi et al., 2015; Figure 1A). However, opposing patterns of miR160 regulation were reported in wheat, in which miR160 was downregulated while its other target HSP70 was upregulated in response to heat stress (Kumar et al., 2014). Another auxin signaling-related miR167 that targets ARF8, which regulates floral organ and gynoecium and stamen development, was strongly decreased, while miR167h was increased significantly in response to heat stress (Wu et al., 2006; Kruszka et al., 2014; Figure 1A). In addition, many other miRNAs involved in the auxin signaling pathway, including miR390 and miR393, participate in the heat stress response (Vidal et al., 2010; Xin et al., 2010; Guan et al., 2013; Hivrale et al., 2016).
miR159 negatively regulates the gibberellic acid MYB (GAMYB) genes, which are important in seed germination and flower development (Reyes and Chua, 2007). In wheat, miR159 was downregulated with the upregulation of TaGAMYB after heat stress (Xin et al., 2010; Wang et al., 2012; Figure 1A). Rice mutants overexpressing TamiR159 mutants or Arabidopsis myb33myb65 double mutants (TaGAMYB1 homologous genes) were heat-sensitive, indicating that the overexpression of miR159 led to GAMYB downregulation to decrease plant heat tolerance. In addition, miR319 [targets the teosinte branched/cycloidea proteins (TCPs) regulating jasmonic acid biosynthesis] was found to be upregulated and its targets TCP2, TCP3, and TCP24 were downregulated under heat stress (Schommer et al., 2008; Li S. X. et al., 2014; Hivrale et al., 2016; Figure 1A). These findings indicate that many hormones related to miRNAs function in response to heat stress through miRNA-target gene networks.
Development-related miRNAs
Heat stress significantly affects plant development, such as root and leaf development, seed germination, and photosynthesis. miR164 targets nascent polypeptide-associated complex (NAC) transcription factors to regulate shoot and root development (Guo et al., 2005). In Arabidopsis, miR164 was induced and NAC1 was suppressed after high-temperature treatment (May et al., 2013; Li S. X. et al., 2014; Figure 1A). Particularly in wheat, miR164 could also directly target the heat shock protein HSP17, which showed upregulation under heat stress (Kumar et al., 2014). miR166 targets homeodomain-leucine zipper (HD-Zip) transcription factors regulating auxiliary meristem initiation and leaf morphology (Rhoades et al., 2002). In H. vulgare, miR166a was found to be upregulated while its targets PHV (PHAVOLUTA), REV (REVOLUTA), and HOX9 (homeobox leucine zipper protein HOX9-like) were downregulated in response to heat (Kruszka et al., 2014; Figure 1A).
miR171 plays an important role in the expression of SCARECROW-LIKE6- III (SCL6-III) and SCL6-IV (GRAS family genes), which are known to be involved in many developmental processes, such as the radial patterning of both roots and shoots (Kamiya et al., 2003). In response to heat stress, miR171 was upregulated and further suppressed the expression of GRAS genes in Arabidopsis (Barku et al., 2013). However, different results were obtained in Populus, namely, pto-miR171 and ptc-miR171 were downregulated (Lu et al., 2008; Chen et al., 2012). ARGONAUTE1 (AGO1) is one of the targets for miR168 and plays an important role in the production of miRNAs and in plant development (Vaucheret, 2009). miR168 was downregulated in response to heat, which would lead to a high level of AGO1 (Chen et al., 2012; Figure 1A), suggesting that the miRNA-mediated regulation system is active under heat stress. Additionally, some other development-related miRNAs, including miR396 and its target HaWRKY6, and miR397 and its target LACCASE, were found to be associated with heat stress (McCaig et al., 2005; Giacomelli et al., 2012; Figure 1A), implying that miRNAs and their respective targets function in a complex regulatory network developed to cope with heat stress, and are involved in plant thermotolerance mechanisms.
miRNA*
Mature miRNAs are excised as miRNA/miRNA* duplexes from a precursor that resembles a hairpin structure. miRNA* is a complementary strand of mature functional miRNA, and its expression level is very low compared with that of its miRNA counterpart. Previous investigations revealed the role of miRNA* in plant stress responses. For example, miR393* and miR399* were upregulated in Arabidopsis by a bacterial pathogen and phosphate deprivation stress, respectively (Navarro et al., 2006; Zhang et al., 2011). Recent studies indicated that miRNA* is also involved in the response to heat stress. For example, under heat stress, the expression of miRNA and miRNA* variants of miR156h-2 was upregulated. In contrast, the miRNA* variants of miR167a and miR400 were downregulated. In addition, the expression of both miR1885b.3 and miR1885b.3* was severely suppressed by heat stress (Yu et al., 2012). A similar result was found for miR169* and miR169 in switchgrass (Panicum virgatum) (Hivrale et al., 2016). Importantly, miR169* has been predicted to target bacterioferritin comigratory protein1 (BCP1) transcripts in Medicago truncatula (Devers et al., 2011), indicating that miRNA* may regulate complementary mRNA targets (Zhang et al., 2011; Manavella et al., 2013). These findings show that miRNA* is involved in heat response mechanisms in plants.
Regulation of miRNA expression by alternative splicing
Alternative splicing (AS) is common in plants and contributes to both transcriptomic and proteomic diversity (Syed et al., 2012). Previous studies showed that many genes undergoing AS are involved in the regulation of plant responses to stress (Qin et al., 2007; Matsukura et al., 2010; Guerra et al., 2015). For example, the expression of DEHYDRATION-RESPONSIVE ELEMENT BINDING 2B (DREB2B) was shown to be regulated by AS in response to heat stress in Zea mays (Qin et al., 2007). In addition, heat stress-related AS was shown to regulate the expression level of miRNAs. For example, under heat stress, intronic miR400 was cotranscribed with its host gene and downregulated, while the expression level of miR400 primary transcripts was increased (Yan et al., 2012). This is interesting because an AS event was induced by heat stress, occurred in the intron where MIR400 was located, and the heat stress-induced AS event inhibited mature miR400 expression (Figure 1C). Moreover, under heat stress, Arabidopsis seeds overexpressing miR400 had a lower germination rate (Yan et al., 2012). However, the miR400 target pentatricopeptide repeat (PPR), which is involved in plant development and abiotic stress, was upregulated by heat stress in Arabidopsis (Li S. X. et al., 2014; Park et al., 2014). In addition, some intronic miRNAs, including miR162a, miR788, miR838, miR844, miR848, miR853, and miR862, have potential AS isoforms, implying that through AS events, these intronic miRNAs may respond to stress in plants (Yan et al., 2012).
DNA methylation-related miRNAs
DNA methylation is an important epigenetic modification, and plays a key role in the regulation of plant growth and development; it also has crucial functions in regulating gene expression in response to abiotic stress in plants (Rakei et al., 2015). For example, the expression of methylated CycD3-1 and Nt-EXPA5 was found to be altered during heat stress in tobacco (Centomani et al., 2015). In addition, under heat stress, DNA methylation might affect the expression of miRNAs and their targets (Ci et al., 2015). In Populus simonii, miR393a, miR156i, miR167h, miR396e, and miR396g genes were methylated at CNG sites in heat-treated plants, while they were methylated at CG sites in cold-treated ones. Under heat stress, miR390c with mCG increased, while the expression of its target gene, ISOCITRATE DEHYDROGENASE (IDH), which participates in peroxisome biogenesis, was suppressed. Similarly, the expression of Ptc-miR156i and j with mCNG modification was increased, but that of their target genes, PHOSPHOLIPID/GLYCEROL ACYLTRANSFERASE FAMILY PROTEINS (LPCAT1 and LPCAT2), which are involved in ether lipid metabolism or glycerophospholipid metabolism, were suppressed under heat stress. In addition, miR396e/g were also induced, the targets of which are ACYL-COA OXIDASES (ACOX1 and ACOX3), which function in alpha-linolenic acid metabolism and fatty acid degradation and were downregulated under heat stress (Biswas and Mano, 2015; Ci et al., 2015). From these results, under heat stress, DNA methylation might regulate miRNA expression, further affecting the expression level of their targets, likely through the gene-silencing function of miRNAs (Ci et al., 2015; Song et al., 2015).
siRNAs
Small interfering RNAs (siRNAs) are approximately 21–24 nucleotide endogenous RNAs derived from the DCL family that catalyze the processing of double-stranded RNA (dsRNA) precursors (Axtell, 2013). According to their biogenesis and function, they can be further classified as trans-acting siRNAs (ta-siRNAs), natural antisense transcript siRNAs (nat-siRNAs), or heterochromatic siRNAs (Sunkar and Zhu, 2007; Axtell, 2013).
Sunkar and Zhu (2004) demonstrated that siRNAs are involved in abiotic stress responses in plants. Subsequently, a nat-siRNA, derived from natural cis-antisense transcript pairs of SRO5 and P5CDH genes, was also found to regulate salt tolerance in Arabidopsis (Borsani et al., 2005). However, very little is known about the role of siRNAs in the heat stress response. In wheat seedlings, the expression levels of three siRNAs were downregulated by heat stress and upregulated by cold stress (Yao et al., 2010). ONSEN, a copia-type retrotransposon, was found to be activated in Arabidopsis seedlings under heat stress. In addition, the heat-induced accumulation of ONSEN was further significantly stimulated in mutants in which the biogenesis of siRNAs was impaired, indicating that siRNA-mediated regulation is responsible for the restriction of ONSEN transcript levels (Ito et al., 2011). Additionally, a high frequency of new ONSEN insertions was observed in the progeny of heat-stressed plants deficient in siRNAs (Matsunaga et al., 2012).
Trans-acting siRNAs (ta-siRNAs) are a specialized class of siRNAs that are generated by miRNA processing of a TAS gene transcript, the mode of action of which is very similar to that of miRNA (Axtell, 2013). The miR173-cleaved ta-siRNA (TAS1) targets HEAT-INDUCED TAS1 TARGET1 (HTT1) and HTT2 are involved in thermotolerance in Arabidopsis, and these targets were found to be highly induced by heat stress (Khraiwesh et al., 2012; Li S. X. et al., 2014; Figure 1B). The overexpression of HTT1 and HTT2 upregulated the accumulation of several Hsf genes to increase thermotolerance. Intriguingly, the HTT genes were also induced in mutants overexpressing HsfA1a under high temperatures. By contrast, the overexpression of TAS1a (TAS1 family) caused higher sensitivity to heat stress through the elevated accumulation of TAS1-siRNAs and reduced expression levels of the HTT genes, suggesting that the TAS1a gene negatively regulates HTT and reduces thermotolerance.
Nat-siRNA is another siRNA whose dsRNA precursor is formed by the hybridization of two independently transcribed RNAs (Axtell, 2013). In B. rapa, differential expression analysis revealed that nat-siRNAs derived from 12 cis-NATs were responsive to heat stress, most of which showed strand bias. In addition, most of the transcripts generating heat-responsive nat-siRNAs were upregulated under heat stress, while the transcripts from the opposite strands of the same loci were downregulated (Yu et al., 2013).
lncRNAs
lncRNAs in plants are more than 200 nt in length, distinguishing them from short ncRNAs (such as miRNA and siRNA). Based on their genomic locations, they are classified as antisense lncRNAs or intronic lncRNAs (Wierzbicki, 2012). Genome-wide scans have already revealed that lncRNAs are active in many plants (Zhang and Chen, 2013).
Despite limited reports on the mechanisms by which plant lncRNAs function, it was shown that they play vital roles in development and stress responses (Xin et al., 2011; Zhang and Chen, 2013). Several lncRNAs have been functionally characterized in plant stress-responsive pathways. For example, the lncRNAs COOLAIR (an antisense lncRNA) and COLDAIR (an intronic lncRNA) could be induced after vernalization to gradually suppress the expression of FLC (FLOWERING LOCUS C) (Heo and Sung, 2011). Under heat stress, lnc-173 was not induced, while its target gene SUCROSE SYNTHASE 4 was responsive to a high temperature (Di et al., 2014). In wheat, Xin et al. (2011) characterized 125 putative long non-protein-coding RNAs (npcRNAs) during powdery mildew infection and heat stress, four of which were miRNA precursors (TalnRNA5, TalnRNA8, TalnRNA19, and TahlnRNA27). Among them, TalnRNA27 and TalnRNA5 were upregulated under heat stress. Di et al. (2014) identified 245 poly(A)+ and 58 poly(A)− lncRNAs that are differentially expressed under stress responses in Arabidopsis, and differential expression is significantly depleted in heat stress. Furthermore, 15 heat-responsive lncRNAs were validated by qRT-PCR. In B. rapa, under heat stress, 34 specifically expressed lncRNAs were identified, 192 target genes were regulated by lncRNAs and most of them belonged to the heat respond genes (Song et al., 2016). In addition, in P. simonii, the expression level of PsiLncRNA00268512 was dynamic in response to heat stress (Song et al., 2015). Although some studies on the role of lncRNAs in plants have been performed, comprehensive surveys of lncRNA responses to heat stress are still lacking.
Conclusions and perspectives
Plant ncRNAs play important roles in heat responses via ncRNA-target pathways comprising the heat stress response networks of plants. With the development of sequencing technologies and genome-scale approaches, ncRNAs and their targets responsive to heat stress are being extensively studied in organisms from model plant species to agricultural crops and non-agricultural species. Given that the majority of research in this field has involved identifying ncRNAs from different plant species, the pursuit of several worthwhile lines of study, such as functional analyses of specific ncRNAs, quantification of the effects of ncRNAs on their targets, ncRNA spatiotemporal-specific expression patterns, and even emerging circRNAs, should provide great insight into the complex ncRNA-mediated regulatory networks controlling plant heat response and tolerance.
Author contributions
JZ and BJ wrote the manuscript. QH, GC, LW, and BJ reviewed and updated the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was financially supported by the National Key Research and Development Program (2016YFD0600105) and the National Natural Science Foundation of China (No. 30870436).
References
- Axtell M. J. (2013). Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64, 137–159. 10.1146/annurev-arplant-050312-120043 [DOI] [PubMed] [Google Scholar]
- Barku M. M., Bashasab F., Sudipta G., Krishnaraj P. U. (2013). LNA mediated in situ hybridization of miR171 and miR397a in leaf and ambient root tissues revealed expressional homogeneity in response to shoot heat shock in Arabidopsis thaliana. J. Plant Biochem. Biotechnol. 23, 93–103. 10.1007/s13562-013-0191-0 [DOI] [Google Scholar]
- Biswas M. S., Mano J. I. (2015). Lipid peroxide-derived short-chain carbonyls mediate H2O2-induced and NaCl-induced programmed cell death in plants. Plant Physiol. 168, 885–898. 10.1104/pp.115.256834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bita C. E., Gerats T. (2013). Plant tolerance to high temperature in a changing environment: scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 4:273. 10.3389/fpls.2013.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O., Zhu J., Verslues P. E., Sunkar R., Zhu J. K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123, 1279–1291. 10.1016/j.cell.2005.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centomani I., Sgobba A., D'Addabbo P., Dipierro N., Paradiso A., De Gara L., et al. (2015). Involvement of DNA methylation in the control of cell growth during heat stress in tobacco BY-2 cells. Protoplasma 252, 1–9. 10.1007/s00709-015-0772-y [DOI] [PubMed] [Google Scholar]
- Chen L., Ren Y. Y., Zhang V. L., Xu V. L., Sun F. S., Zhang Z. Y., et al. (2012). Genome-wide identification and expression analysis of heat-responsive and novel microRNAs in Populus tomentosa. Gene 504, 160–165. 10.1016/j.gene.2012.05.034 [DOI] [PubMed] [Google Scholar]
- Ci D., Song Y. P., Tian M., Zhang D. Q. (2015). Methylation of miRNA genes in the response to temperature stress in Populus simonii. Front. Plant Sci. 6:921. 10.3389/fpls.2015.00921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L. G., Shan J. X., Shi M., Gao J. P., Lin H. X. (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80, 1108–1117. 10.1111/tpj.12712 [DOI] [PubMed] [Google Scholar]
- de Lima J. C., Loss-Morais G., Margis R. (2012). microRNAs play critical roles during plant development and in response to abiotic stresses. Genet. Mol. Biol. 35(Suppl. 1), 1069–1077. 10.1590/s1415-47572012000600023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devers E. A., Branscheid A., May P., Krajinski F. (2011). Stars and symbiosis: microRNA-and microRNA-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol. 156, 1990–2010. 10.1104/pp.111.172627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di C., Yuan J. P., Wu Y., Li J. R., Lin H. X., Hu L., et al. (2014). Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 80, 848–861. 10.1111/tpj.12679 [DOI] [PubMed] [Google Scholar]
- Giacomelli J. I., Weigel D., Chan R. L., Manavella P. A. (2012). Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol. 195, 766–773. 10.1111/j.1469-8137.2012.04259.x [DOI] [PubMed] [Google Scholar]
- Guan Q. M., Lu X. Y., Zeng H. T., Zhang Y. Y., Zhu J. H. (2013). Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 74, 840–851. 10.1111/tpj.12169 [DOI] [PubMed] [Google Scholar]
- Guerra D., Crosatti C., Khoshro H. H., Mastrangelo A. M., Mica E., Mazzucotelli E. (2015). Post-transcriptional and post-translational regulations of drought and heat response in plants: a spider's web of mechanisms. Front. Plant Sci. 6:57. 10.3389/fpls.2015.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H. S., Xie Q., Fei J. F., Chua N. H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 17, 1376–1386. 10.1105/tpc.105.030841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J. B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79. 10.1126/science.1197349 [DOI] [PubMed] [Google Scholar]
- Hirayama T., Shinozaki K. (2010). Research on plant abiotic stress responses in the postgenome era: past, present and future. Plant J. 61, 1041–1052. 10.1111/j.1365-313X.2010.04124.x [DOI] [PubMed] [Google Scholar]
- Hivrale V., Yun Z., Puli C. O. R., Jagadeeswaran G., Gowdu K., Kakani G., et al. (2016). Characterization of drought- and heat-responsive microRNAs in switchgrass. Int. J. Plant Sci. 242, 214–223. 10.1016/j.plantsci.2015.07.018 [DOI] [PubMed] [Google Scholar]
- Huang B., Xu C. P. (2008). Identification and characterization of proteins associated with plant tolerance to heat stress. J. Integr. Plant Biol. 50, 1230–1237. 10.1111/j.1744-7909.2008.00735.x [DOI] [PubMed] [Google Scholar]
- Ito H., Gaubert H., Bucher E., Mirouze M., Vaillant I., Paszkowski J. (2011). An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119. 10.1038/nature09861 [DOI] [PubMed] [Google Scholar]
- Jeong D. H., Park S., Zhai J. Z., Ranjan Gurazada S. G., Paoli E. D., Meyers B. C., et al. (2011). Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23, 4185–4207. 10.1105/tpc.111.089045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N., Itoh J., Morikami A., Nagato Y., Matsuoka M. (2003). The SCARECROW gene's role in asymmetric cell divisions in rice plants. Plant J. 36, 45–54. 10.1046/j.1365-313X.2003.01856.x [DOI] [PubMed] [Google Scholar]
- Khaksefidi R. E., Mirlohi S., Khalaji F., Fakhari Z., Shiran B., Fallahi H., et al. (2015). Differential expression of seven conserved microRNAs in response to abiotic stress and their regulatory network in Helianthus annuus. Front. Plant Sci. 6:741. 10.3389/fpls.2015.00741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B., Zhu J. K., Zhu J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. BBA Gene Regul. Mech. 1819, 137–148. 10.1016/j.bbagrm.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. J., Lee J. H., Kim W., Jung H. S., Huijser P., Ahn J. H., et al. (2012). The miR156-SPL3 module regulates ambient temperature-responsive flowering via FT in Arabidopsis thaliana. Plant Physiol. 159, 461–478. 10.1104/pp.111.192369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Larkindale J., Lee U., Koskull-Doring P. V., Vierling E., Scharf K. D. (2007). Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 10, 310–316. 10.1016/j.pbi.2007.04.011 [DOI] [PubMed] [Google Scholar]
- Kruszka K., Pacak A., Swidabarteczka A., Nuc P., Alaba S., Wroblewska Z., et al. (2014). Transcriptionally and post-transcriptionally regulated microRNAs in heat stress response in barley. J. Exp. Bot. 65, 6123–6135. 10.1093/jxb/eru353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R. R., Pathak H., Sharma S. K., Kale Y. K., Nirjal M. K., Singh G. P. (2014). Novel and conserved heat-responsive microRNAs in wheat (Triticum aestivum L.). Funct. Integr. Genomics 15, 1–26. 10.1007/s10142-014-0421-0 [DOI] [PubMed] [Google Scholar]
- Li M. Y., Wang F., Xu Z. S., Jiang Q., Ma J., Tan G. F., et al. (2014). High throughput sequencing of two celery varieties small RNAs identifies microRNAs involved in temperature stress response. BMC Genomics 15:242. 10.1186/1471-2164-15-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. X., Liu J. X., Liu Z. Y., Li X. R., Wu F. J., He Y. K. (2014). Heat-induced tas1 target1 mediates thermotolerance via heat stress transcription factor A1a–directed pathways in Arabidopsis. Plant Cell 26, 1764–1780. 10.1105/tpc.114.124883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. I., Santi C., Jobet E., Lacut E., El K. N., Karlowski W. M., et al. (2010). Complex regulation of two target genes encoding spx-mfs proteins by rice miR827 in response to phosphate starvation. Plant Cell Physiol. 51, 2119–2131. 10.1093/pcp/pcq170 [DOI] [PubMed] [Google Scholar]
- Liu F. L., Wang W. J., Sun X. T., Liang Z. R., Wang F. J. (2014). RNA-Seq revealed complex response to heat stress on transcriptomic level in Saccharina japonica (Laminariales, Phaeophyta). J. Appl. Phycol. 26, 1585–1596. 10.1007/s10811-013-0188-z [DOI] [Google Scholar]
- Lu S. F., Sun Y. H., Chiang V. L. (2008). Stress-responsive microRNAs in Populus. Plant J. 55, 131–151. 10.1111/j.1365-313X.2008.03497.x [DOI] [PubMed] [Google Scholar]
- Lu X., Guan Q., Zhu J. (2013). Downregulation of CSD2 by a heat-inducible miR398 is required for thermotolerance in Arabidopsis. Plant Signal. Behav. 8:e24952. 10.4161/psb.24952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahale B. M., Fakrudin B., Ghosh S., Krishnaraj P. U. (2013). LNA mediated in situ hybridization of miR171 and miR397a in leaf and ambient root tissues revealed expressional homogeneity in response to shoot heat shock in Arabidopsis thaliana. J. Plant Biochem. Biotechnol. 23, 93–103. 10.1007/s13562-013-0191-0 [DOI] [Google Scholar]
- Manavella P. A., Daniel K., Ignacio R. S., Burbano H. A., Claude B., Detlef W. (2013). Tissue-specific silencing of Arabidopsis SU(VAR) 3-9 HOMOLOG8 by miR171a. Plant Physiol. 161, 805–812. 10.1104/pp.112.207068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura S., Mizoi J., Yoshida T., Todaka D., Ito Y., Maruyama K., et al. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics 283, 185–196. 10.1007/s00438-009-0506-y [DOI] [PubMed] [Google Scholar]
- Matsunaga W., Kobayashi A., Kato A., Ito H. (2012). The effects of heat induction and the siRNA biogenesis pathway on the transgenerational transposition of ONSEN, a copia-like retrotransposon in Arabidopsis thaliana. Plant Cell Physiol. 53, 824–833. 10.1093/pcp/pcr179 [DOI] [PubMed] [Google Scholar]
- May P., Liao W., Wu Y., Shuai B., McCombie W. R., Zhang M. Q., et al. (2013). The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nat. Commun. 4, 405–415. 10.1038/ncomms3145 [DOI] [PubMed] [Google Scholar]
- McCaig B. C., Meagher R. B., Dean J. F. (2005). Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta 221, 619–636. 10.1007/s00425-004-1472-6 [DOI] [PubMed] [Google Scholar]
- Meng Y., Chen D., Ma X., Mao C., Cao J., Wu P., et al. (2010). Mechanisms of microRNA-mediated auxin signaling inferred from the rice mutant osaxr. Plant Signal. Behav. 5, 252–254. 10.4161/psb.5.3.10549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. 10.1016/S1360-1385(02)02312-9 [DOI] [PubMed] [Google Scholar]
- Navarro L., Dunoyer P., Jay F., Arnold B., Dharmasiri N., Estelle M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. 10.1126/science.1126088 [DOI] [PubMed] [Google Scholar]
- Park Y. J., Lee H. J., Kwak K. J., Lee K., Hong S. W., Kang H. (2014). MicroRNA400-guided cleavage of pentatricopeptide repeat protein mRNAs renders Arabidopsis thaliana more susceptible to pathogenic bacteria and fungi. Plant Cell Physiol. 55, 1660–1668. 10.1093/pcp/pcu096 [DOI] [PubMed] [Google Scholar]
- Qin F., Kakimoto M., Sakuma Y., Maruyama K., Osakabe Y., Tran L. S., et al. (2007). Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 50, 54–69. 10.1111/j.1365-313X.2007.03034.x [DOI] [PubMed] [Google Scholar]
- Rakei A., Maali-Amiri R., Zeinali H., Ranjbar M. (2015). DNA methylation and physio-biochemical analysis of chickpea in response to cold stress. Protoplasma 253, 1–16. 10.1007/s00709-015-0788-3 [DOI] [PubMed] [Google Scholar]
- Reyes J. L., Chua N. H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49, 592–606. 10.1111/j.1365-313X.2006.02980.x [DOI] [PubMed] [Google Scholar]
- Rhoades M. W., Reinhart B. J., Lim L. P., Burge C. B., Bartel B., Bartel D. P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. 10.1016/S0092-8674(02)00863-2 [DOI] [PubMed] [Google Scholar]
- Rogers K., Chen X. (2013). Biogenesis, turnover and mode of action of plant microRNAs. Plant Cell 25, 2383–2399. 10.1105/tpc.113.113159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailaja B., Voleti S. R., Oleti D., Subrahmanyam N., Sarla V., Vishnuprasanth V. P., et al. (2014). Prediction and expression analysis of miRNAs associated with heat stress in Oryza sativa. Rice Sci 21, 3–12. 10.1016/S1672-6308(13)60164-X [DOI] [Google Scholar]
- Schommer C., Palatnik J. F., Aggarwal P., Chételat A., Cubas P., Farmer E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. 10.1371/journal.pbio.0060230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Liu G., Huang Z., Duan W., Tan H., Ying L., et al. (2016). Temperature expression patterns of genes and their coexpression with lncRNAs revealed by RNA-Seq in non-heading Chinese cabbage. BMC Genomics 17:297. 10.1186/s12864-016-2625-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Ci D., Tian M., Zhang D. Q. (2015). Stable methylation of a non-coding RNA gene regulates gene expression in response to abiotic stress in Populus simonii. J. Exp. Bot. 67, 1477–1492. 10.1093/jxb/erv543 [DOI] [PubMed] [Google Scholar]
- Stief A., Altmann S., Hoffmann K., Pant B. D., Scheible W., Bäurle I. (2014). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26, 1792–1807. 10.1105/tpc.114.123851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Kapoor A., Zhu J. K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18, 2051–2065. 10.1105/tpc.106.041673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Zhu J. K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019. 10.1105/tpc.104.022830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Zhu J. K. (2007). Micro RNAs and short-interfering RNAs in plants. J. Integr. Plant Biol. 49, 817–826. 10.1111/j.1744-7909.2007.00499.x [DOI] [Google Scholar]
- Syed N. H., Kalyna M., Marquez Y., Barta A., Brown J. W. S. (2012). Alternative splicing in plants–coming of age. Trends Plant Sci. 17, 616–623. 10.1016/j.tplants.2012.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. (2009). AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS ONE 4:e6442. 10.1371/journal.pone.0006442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal E. A., Araus V., Lu C., Parry G., Green P. J., Coruzzi G. M., et al. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 4477–4482. 10.1073/pnas.0909571107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. X., Vinocur B., Shoseyov O., Altman A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. 10.1016/j.tplants.2004.03.006 [DOI] [PubMed] [Google Scholar]
- Wang Y., Sun F., Cao H., Peng H. R., Ni Z. F., Sun Q. X., et al. (2012). TamiR159 directed wheat TaGAMYB cleavage and its involvement in anther development and heat response. PLoS ONE 7:e48445. 10.1371/journal.pone.0048445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A. T. (2012). The role of long non-coding RNA in transcriptional gene silencing. Curr. Opin. Plant Biol. 15, 517–522. 10.1016/j.pbi.2012.08.008 [DOI] [PubMed] [Google Scholar]
- Wu G., Park M. Y., Conway S. R., Wang J. W., Weigel D., Poethig R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. 10.1016/j.cell.2009.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. F., Tian Q., Reed J. W. (2006). Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218. 10.1242/dev.02602 [DOI] [PubMed] [Google Scholar]
- Xin M., Wang Y., Yao Y., Song N., Hu Z., Qin D., et al. (2011). Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC Plant Biol. 11:61. 10.1186/1471-2229-11-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M., Yu W., Yao Y., Xie C., Peng H., Ni Z., et al. (2010). Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 10:123. 10.1186/1471-2229-10-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K., Liu P., Wu C. A., Yang G. D., Xu R., Guo Q. H., et al. (2012). Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell 48, 521–531. 10.1016/j.molcel.2012.08.032 [DOI] [PubMed] [Google Scholar]
- Yao Y., Ni Z., Peng H., Sun F., Xin M., Sunkar R., et al. (2010). Non-coding small RNAs responsive to abiotic stress in wheat (Triticum aestivum L.). Funct. Integr. Genomics 10, 187–190. 10.1007/s10142-010-0163-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Wang H., Lu Y. Z., Ruiter M. D., Cariaso M., Prins M., et al. (2012). Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J. Exp. Bot. 63, 1025–1038. 10.1093/jxb/err337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Yang J., Li X., Liu X., Sun C., Wu F., et al. (2013). Global analysis of cis-natural antisense transcripts and their heat-responsive nat-siRNAs in Brassica rapa. BMC Plant Biol. 13:208. 10.1186/1471-2229-13-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. M., Zhao H. W., Gao S., Wang W. C., Surekha K. A., Huang H. D., et al. (2011). Arabidopsis argonaute 2 regulates innate immunity via miRNA393-mediated silencing of a golgi-localized SNARE gene, MEMB12. Mol. Cell 42, 356–366. 10.1016/j.molcel.2011.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. C., Chen Y. Q. (2013). Long noncoding RNAs: new regulators in plant development. Biochem. Biophys. Res. Commun. 436, 111–114. 10.1016/j.bbrc.2013.05.086 [DOI] [PubMed] [Google Scholar]
- Zhu C., Ding Y. F., Liu H. L. (2011). MiR398 and plant stress responses. Physiol. Plant. 143, 1–9. 10.1111/j.1399-3054.2011.01477.x [DOI] [PubMed] [Google Scholar]